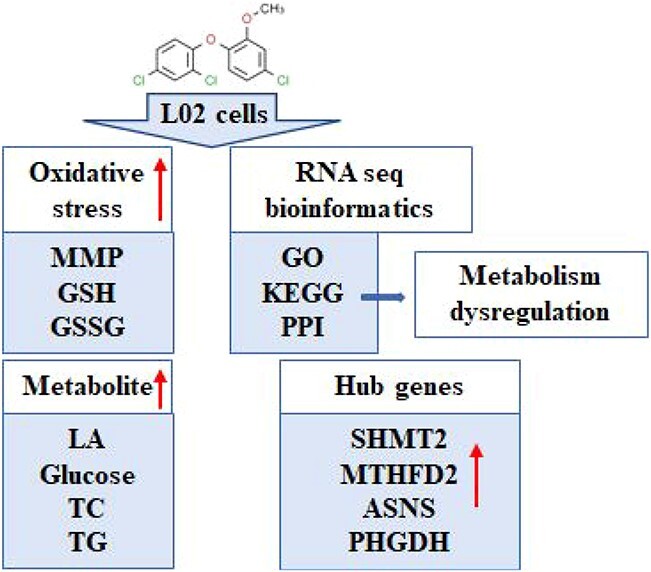
Abstract
Purpose: Methyl triclosan (MTCS) is one of the biomethylated by-products of triclosan (TCS). With the increasing use of TCS, the adverse effects of MTCS have attracted extensive attention in recent years. The purpose of this study was to investigate the cytotoxicity of MTCS and to explore the underlining mechanism using human hepatocyte L02 cells as in vitro model. Results: The cytotoxicity results revealed that MTCS could inhibit cell viability, disturb the ratio of reduced glutathione (GSH) and oxidized glutathione (GSSG), and reduce the mitochondrial membrane potential (MMP) in a dose-dependent manner. In addition, MTCS exposure significantly promoted the cellular metabolic process, including enhanced conversion of glucose to lactic acid, and elevated content of intracellular triglyceride (TG) and total cholesterol (TC). RNA-sequencing and bioinformatics analysis indicated disorder of glucose and lipid metabolism was significantly induced after MTCS exposure. Protein-protein interaction network analysis and node identification suggested that Serine hydroxy methyltransferase 2 (SHMT2), Methylenetetrahydrofolate dehydrogenase 2 (MTHFD2), Asparagine synthetase (ASNS) and Phosphoglycerate dehydrogenase (PHGDH) are potential molecular markers of metabolism imbalance induced by MTCS. Conclusion: These results demonstrated that oxidative stress and metabolism dysregulation might be involved in the cytotoxicity of MTCS in L02 cells.
Keywords: methyl triclosan, oxidative stress, RNA-sequencing, metabolism dysregulation
Graphical abstract
Introduction
Triclosan (TCS) is a world-widely applicated antibacterial material in toiletries, such as soap, hand sanitizer and toothpaste with an estimated annual use of more than 750 × 103 tons.1,2 Due to its extensive daily use and environmental deposition for decades, the concentration of TCS detected in aquatic ecosystems, wastewater and other related environments is increasing in recent years.3–8 TCS can enter the human body through skin contact with the surface of food packaging, posing a potential threat to health. So far, TCS had been detected in various human tissues including breast milk, blood serum, adipose and liver with concentrations correlating to the use of personal care products containing TCS.9–13 The urinary TCS levels in Chinese were even up to micrograms per liter, which has aroused great concern about the possible health risks and environmental hazards.7,14
Methyl triclosan (MTCS) is one of the biomethylated by-products of TCS,6,15,16 and its concentration in surface water is generally at ng/L levels.17,18 Unlike TCS, MTCS has no antibacterial properties due to substitution of hydroxyl by methyl. Meanwhile, the methyl in MTCS increases its resistance to photodegradation, leading to enhanced environmental durability of MTCS.19,20 Moreover, related research demonstrated that MTCS has greater bioaccumulation potentiality and biotransformation ability than TCS.13,21–23 Delorenzo et al. investigated the MTCS and TCS concentration in grass shrimp after exposure to 100 μg/L of TCS for two weeks. Only MTCS (but not TCS) was detected in adult grass shrimp in vivo, indicating that MTCS was more easily enriched in organism tissues than TCS.24 Research showed that MTCS with a bioaccumulation factor (BAF) of 1,200 was detected to be higher than that of TCS in snails collected from Pecan Creek, USA.25 Rudel et al. demonstrated that accumulation of MTCS was much higher than that of TCS in fish muscle tissue of snappers, which were collected from different sampling points in Germany.26 Macherius et al. found that after application of biosolids, the concentration of MTCS in the collected earthworms was significantly higher than that of TCS.21 It was reported that MTCS had obvious biological amplification effect in the food network with a nutrient amplification factor 3.85.27 Up to now, MTCS has been detected in different types of human body fluid samples, such as plasma samples (0.126–0.161 μg/L) and urine samples (0.211–0.254 μg/L).27–30 Considering the environment persistence and bioaccumulation capability of MTCS, the health risks and potential underlying mechanisms deserve deepen investigation.
As an endocrine disruptor, TCS is proved to influence various biological functions including neurodevelopment, immunity, reproduction, etc.31,32 The bio toxic effects of MTCS are weakly studied due to its lower toxicity because of methylation process. Recently, toxicology research reported that MTCS exposure could delay the larvae development of sea urchin (Paracentrotus lividus),31 disrupt the metabolomes of the zebrafish embryos involving synthesis of fatty acid, as well as energy and nitrogen metabolism,33,34 and induce toxic effects in hemocytes and gill cells of the European abalone Haliotis tuberculate.35 In human body, complex formation of MTCS with human serum albumin through hydrophobic forces is assumed to influence conformation of human serum albumin, and impair the endocrine function.36 These findings indicate the adverse effects of MTCS on the ecosystem. Our recent studies revealed that MTCS induced expression of cyclin A2 and cyclin-dependent kinase, resulting in cell cycle arrest at the S phase and mitochondrial apoptosis via oxidative stress in human hepatocellular carcinoma HepG2 cells.32 Under low concentration (0.5 and 1 μM) exposure, MTCS could induce cell proliferation and malignant transformation of human hepatocyte L02 cells.37 However, current available data is insufficient to clarify the potential toxicological mechanism of MTCS and to establish future safety standard.
In this study, the cytotoxicity induced by MTCS was investigated with human immortalized hepatocytes cells (L02), which are widely used cell model for hepatotoxicity research with typical hepatocyte morphological features. Widespread transcriptomic changes were examined with RNA-sequencing (RNA-seq), and then bioinformatics analysis including biological functions, signaling pathways, and protein network construction were conducted. This study was designed to investigate the cytotoxic effects of MTCS in L02 cells, and to explore the potential molecular regulation mechanism. This study will provide experimental support to formulate risk assessment and control policies for MTCS.
Materials and methods
Reagents and chemical
The high-purity MTCS sample (CAS:4640-01-1) was obtained from Dr Ehrensorfer (Augsburg, Germany). The 200 mM storage solution of MTCS/dimethyl sulfoxide (DMSO) was kept at −20 °C away from light. The Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum (FBS) were purchased from Invitrogen (Paisley, UK). The methylthiazolyldiphenyl-tetrazolium bromide kit (MTT) and commercial kits for reduced glutathione (GSH) and oxidized glutathione disulfide (GSSG) were purchased from Beyotime (Shanghai, China). The lactic acid release (LA), glucose uptake, total cholesterol (TC) and triglyceride (TG) assay kits were purchased from Jian Cheng (Nanjing, China). A ReverTra Ace qPCR RT kit and SYBR Green Real-time PCR Master Mix were from Toyobo (Osaka, Japan). The DMSO and rhodamine 123 (Rh123) were from Sigma (Missouri, USA).
L02 cell culture and MTCS treatments
The L02 hepatocytes obtained from Professor Ping-Kun Zhou (Beijing Institute of Radiation Medicine, Beijing, China) were cultured with DMEM containing 10% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin in 5% CO2 incubator at 37 °C. L02 cells with 80% confluency were exposed to MTCS (0, 10, 20, and 40 μM) for 24 h. After MTCS treatment, cell viability, mitochondrial membrane potential (MMP), GSSG and GSH content, LA content, glucose uptake, TG and TC contents, and target gene mRNA levels were measured. The control group cells were treated with equal amounts of DMEM containing 0.1% DMSO (v/v). At least triplicate replicates were set for each concentration group, and experiments were repeated three times independently.
Cytotoxicity assay
Exponentially growing L02 cells were prepared and exposed to 10, 20, and 40 μM of MTCS for 24 h. MTT colorimetry assay was used to determine the cell viability, and the absorbance data of formazan at 490 nm was recorded using a microplate reader (Tecan, Mannedorf, Switzerland) to indirectly reflect the number of living cells. Fluorescent probe Rh123 (10 mM) is used to detect the MMP in living L02 cells after MTCS exposure. Fluorescence intensity of Rh123 was recorded using microplate reader (Tecan, Mannedorf, Switzerland) under excitation at 507 nm and emission at 529 nm.
Glutathione measurement
After MTCS exposure as described above, the supernatant of L02 cells culture medium was collected by centrifugation. The cell precipitate was lysated with ultrasonic crusher and centrifuged at 10,000g for 10 min at 4 °C. Contents of GSSG and GSH were measured using 5,5’-Dithiobis-(2-nitrobenzoic acid) colorimetric method according to the instruction of assay kit (Beyotime, Shanghai, China). The lysate supernatant was applicated to determine the total glutathione (GSH + GSSG) contents. Diluted GSH clearance auxiliary solution was used to measure the GSSG level in cell supernatant. The GSH levels were calculated based on the data of GSSG and total glutathione.
Metabolite detection
After MTCS treatment, supernatants of L02 cells were collected and used to measure LA content and glucose concentration under a spectrophotometer with optical density (OD) values at 530 nm and 505 nm, respectively, according to instruction of assay kits (Jian Cheng, Nanjing, China). In addition, the TC and TG concentrations were determined using cholesterol oxidase-peroxidase coupled (COD-PAP) and glycerol phosphate oxidase-peroxidase coupled (GPO-PAP) colorimetry with OD values at 510 nm (Jian Cheng, Nanjing, China).
Real time-quantitative polymerase chain reaction assay
Total RNA of L02 cells exposed to 0.1% DMSO or 40 μM MTCS for 12 h was extracted with TRIZOL reagents (Life technologies, California, USA). Reverse transcription and quantitative PCR reaction were then conducted with ReverTra Ace qPCR RT kit and SYBR Green Real-time PCR Master Mix (TOYOBO, Osaka, Japan), respectively, to measure target gene expression levels. The PCR amplification started at 95 °C for 60 s for initial denaturation, followed by 45 cycles of denaturation at 95 °C for 15 s, 63 °C for 15 s for annealing, and 72 °C for 45 s for extension. The target gene expression levels were calculated with the ΔΔCq method, and Gapdh was used as the internal reference for gene expression normalization. Sequences of target gene primers were presented in Table 1. The availability of RT-qPCR primers were validated through Primer-Blast on NCBI (National center for Biotechnology Information) website, and the validation results were displayed in Fig. S1 (Supplementary Materials 1).
Table 1.
Sequence of primer pairs for RT-qPCR.
| Genes | Primer | Primer sequence (5′-3′) |
|---|---|---|
| Shmt2 | Forward | TGGCAAGAGATACTACGGAGG |
| Reverse | GCAGGTCCAACCCCATGAT | |
| Mthfd2 | Forward | TACTCCATGGGGTGTGTGG |
| Reverse | TGGGCATTCCAAGGTTTT | |
| Phgdh | Forward | GCAGGATTGGGAGAGAGGTAG |
| Reverse | CAGCAAGCCTGTCGTGGAG | |
| Psat1 | Forward | ACAGGAGCTTGGTCAGCTAAG |
| Reverse | CATGCACCGTCTCATTTGCG | |
| Mmp9 | Forward | AGACCTGGGCAGATTCCAAAC |
| Reverse | CGGCAAGTCTTCCGAGTAGT | |
| E-cadherin | Forward | ATTTTTCCCTCGACACCCGAT |
| Reverse | TCCCAGGCGTAGACCAAGA | |
| Il-8 | Forward | TTTTGCCAAGGAGTGCTAAAGA |
| Reverse | AACCCTCTGCACCCAGTTTTC | |
| Il-6 | Forward | ACTCACCTCTTCAGAACGAATTG |
| Reverse | CCATCTTTGGAAGGTTCAGGTTG | |
|
c-myc
Gapdh |
Forward | GGCTCCTGGCAAAAGGTCA |
| Reverse Forward Reverse |
CTGCGTAGTTGTGCTGATGT GGAGCGAGATCCCTCCAAAAT GGCTGTTGTCATACTTCTCATGG |
RNA-seq
Total RNA of L02 cells exposed to 40 μM MTCS for 12 h was prepared using TRIZOL reagent (Life technologies, California, USA). Qualified RNA was used to construct library (QIAGEN, Heiden, Germany) after amplification, labeling and purification with RNeasy micro kit. Transcriptome sequencing was performed using the Illumina platform. Differential genes were screened according to the expression multiples and difference significance. Fold change of expression level greater than 1.5 compared to the control group was set as the screen criteria for differential expression genes (DEGs). Bioinformatic analysis was performed on the basis of Gene Ontology (GO) biological function, Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathway database, protein-protein international (PPI) network and Hubba genes.
Data analysis
Count data were presented as mean ± standard error, and analyzed with one-way ANOVA followed by Bonferroni post hoc test using Excel 2010 software. The imaging data were processed using the ImageJ software. Difference with p-value less than 0.05 was considered statistic significant.
Results and discussion
Cytotoxicity induced by MTCS in L02 cells
In recent years, the increasing concentration of MTCS in aquatic environment and organisms has gradually attracted concern on its potential risks over ecological environment and human health. Macedo et al. found that both MTCS and TCS induced adverse effects on embryos and larvae of sea urchin in wastewater, and sea urchin larvae are more sensitive to MTCS.31 Gaume et al. revealed that exposure of 40 μM MTCS for 24 h had significant inhibitory effects on cell viability in abalone blood cells.35 In the present study, results of MTT test showed that MTCS exposure suppressed the cell viability of L02 cells in a dose-dependent manner. After treatment of 10, 20 and 40 μM of MTCS for 24 h, cell viability reduced to 92.6 ± 4.2%, 90.9 ± 2.3%, 85.2 ± 3.7% (p < 0.05), respectively, as compared with the control group (Fig. 1).
Fig. 1.
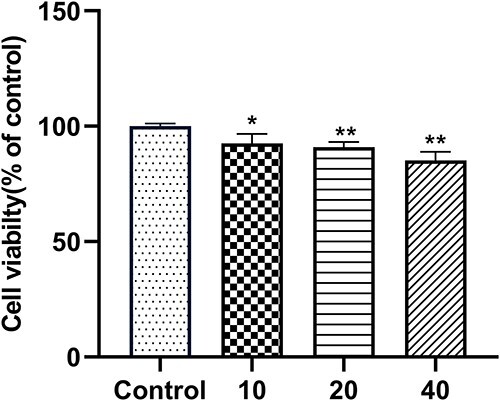
Cell viability of L02 cells after treatment of MTCS. L02 cells were treated with MTCS (10, 20 and 40 μM) for 24 h. Cell viability was detected with MTT assay. Control group were treated only with DMSO/DMEM (0.1%, v/v) only. * p < 0.05, ** p < 0.01, vs. control groups.
Oxidative stress is an important regulatory mechanism involved in most of exogenous compounds-induced cytotoxicity. Park et al. found that TCS caused dose- and time-dependent increase of reactive oxygen species (ROS) production and antioxidant superoxide dismutase (SOD) activity in the copepod T. japonicas.38 Zhong et al. reported that TCS exposure was closely correlated with the urinary levels of oxidative stress markers including 8-iso-prostaglandin-F2α and 8-hydroxy-deoxyguanosine.39 In addition, TCS exposure significantly induced the content of malondialdehyde (MDA) and reduced the glutathione system (GSH and GSSH) in the liver of adult zebrafish.40 Our previous studies have also proved that ROS production, contents of GSH and GSSG, and the expression of antioxidant enzyme HO-1 were significantly affected in response to MTCS exposure in L02 and HepG2 cells.37,41 Overproduction of ROS disrupts the mitochondrial membrane integrity and enhances the mitochondrial membrane permeability. The free diffusion of ions across the membrane could eventually result in loss of mitochondrial membrane potential (MMP). In this study, the MMP levels of L02 cells exposed to 10, 20, and 40 μM of MTCS reduced to 78.8 ± 1.5%, 74.3 ± 3.3%, and 63.8 ± 3.4% that of control groups (p < 0.05), respectively (Fig. 2A).
Fig. 2.
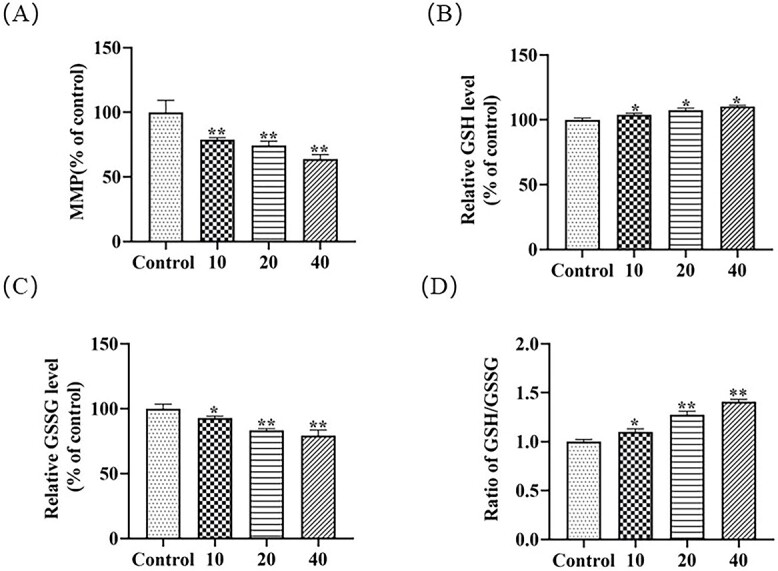
The oxidative stress response induced by MTCS exposure. L02 cells were exposed to MTCS (10, 20 and 40 μM) for 24 h. A): MMP measured by Rhodamine 123 fluorescent probe assay. B): Reduced glutathione (GSH) level and C): Oxidized glutathione (GSSG) level. D): The GSH/GSSG ratio. For control group, L02 cells were treated with DMSO/DMEM (0.1%, v/v) only. * p < 0.05, ** p < 0.01, vs. control group.
As a ROS scavenger, the non-enzymatic antioxidant glutathione system play important roles in countering and eliminating cellular damage caused by excessive ROS. Research data demonstrated that GSH contents were positively correlated with increase of ROS production in response to copper in T. japonicus42 and UV-B exposure in Bombus koreanus.43 TCS induced the GSH contents in concentration-dependent and time-dependent manners in T. japonicas, indicating that GSH induction can be considered as cellular primary protection against ROS accumulation.38 In our study, compared with control groups, the GSH contents increased to 104.0 ± 1.1%, 107.4 ± 1.6%, and 110 ± 0.8%, respectively (Fig. 2B). Under oxidative stress state, GSH is induced to scavenge the excess of ROS and is oxidized to GSSG via glutathione peroxidase (GPx). On the other hand, GSSG can be conversed to GSH depending on the activity of glutathione reductase (GR). After exposure of MTCS (10, 20, and 40 μM) for 24 h, GSSG levels decreased to 92.6 ± 1.7%, 83.2 ± 1.3%, 79.2 ± 4.3% respectively in L02 cells (Fig. 2C). The ratio of GSH/GSSG also showed an increased trend (Fig. 2D). The fluctuation of GSH/GSSG contents and increase of the GSH/GSSG ratio in this study indicated that the recycling of GSH from GSSG by GR is beyond the conversion of GSH to GSSG, which may be due to the fact that MTCS is a comparatively weak oxidative stress inducer.
MTCS induced glucose and lipid metabolism in L02 cells
It has been demonstrated that TCS exposure increased the content of triglycerides and cholesterol, enhanced lipogenesis and lipid transport, and inhibited lipid oxidation in zebrafish.44 To investigate the impacts of MTCS on lipid metabolism in L02 cells, the levels of TC and TG after exposure were detected. Under normal physiological conditions, TC is an important component of cell membrane and the synthetic precursor of many hormones.45,46 TG are transesterification product of fatty acids and glycerol, participating in the transportation of fats.47 Compared with the control group, MTCS exposure (20 and 40 μM) increased the TC levels to 166.2 ± 40.0% and 181.2 ± 28.2%, and TG levels to 128.7 ± 11.3% and 143.5 ± 21.7%, respectively (Fig. 3A and B, p < 0.05). It has been demonstrated that TCS treatment increased the glycolysis process and promoted energy metabolism in HepG2 cells.40 Our previous study also found that TCS promoted the glycolysis process in HepG2 cells via activation of the PI3K pathway.48 In this study, 20 and 40 μM MTCS induced approximately 111.8 ± 6.4% and 126.9 ± 4.2% increase of glucose uptake as compared with the control group (Fig. 3C), respectively; and lactic acid production was elevated to 112.4 ± 1.2% and 114.3 ± 4.4% that of control (Fig. 3D). Glucose is an essential component and important carbon source during the energy metabolic processes.49 Lactic acid (LD) is a carboxylic acid containing hydroxyl participating in various biological processes such as energy metabolism, ischemic injury response, and cell proliferation.50 During the glycolysis process, cells commonly enhances the adenosine triphosphate production rate through increased glucose uptake, and produces a large amount of lactic acid to modulate the cell micro-environment. The increase of glucose uptake and lactate excretion indicated that MTCS may affect the cellular glucose metabolism process. The elevation of TC and TG content indicated that MTCS promoted lipid synthesis and caused fat accumulation in L02 cell. Our results suggested that MTCS might influence the metabolic homeostasis in L02 cells.
Fig. 3.
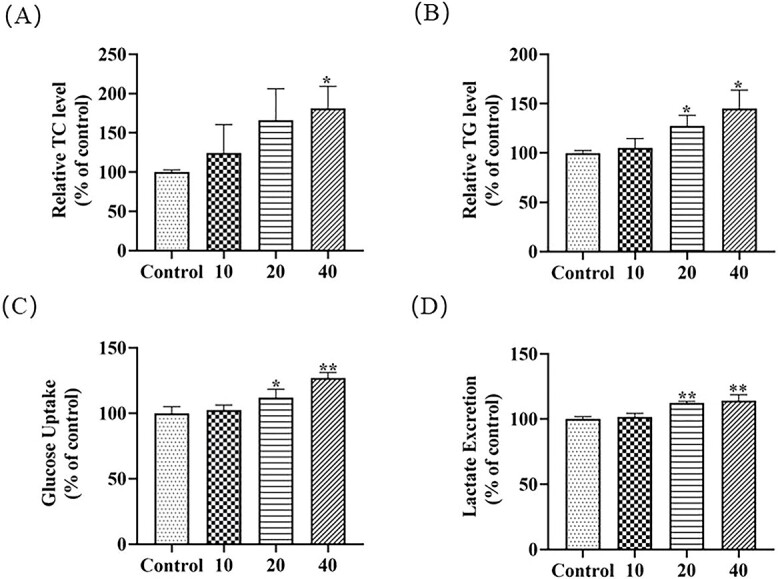
Effects of MTCS on lipid metabolism in L02 cells. L02 cells were treated with MTCS (10, 20 and 40 μM) for 24 h. A): Triglyceride (TG); B) Total cholesterol (TC); C): Glucose uptake and D): Lactic acid release were assayed with corresponding commercial detection kits (Jian Cheng, Nanjing, China). Control group cells were treated with DMSO/DMEM (0.1%, v/v) only. * p < 0.05, ** p < 0.01, vs. control group.
Transcriptomic widespread alterations induced by MTCS exposure
RNA-seq has become a paramount approach for transcriptome profiling analysis, which is widely used in medicine, pharmacology and environmental biology. To explore the molecular toxicological mechanisms and screen sensitive target biomarkers of MTCS, RNA-seq51 on total 20,024 spotted genes were conducted to evaluate the transcriptomic alterations induced by MTCS exposure in L02 cells. DEGs were screened using criteria as fold change >1.5 (up-regulated) or < 0.67 (down-regulated) and p value <0.05 compared with the control. There were totally 428 DEGs identified in this study, including 185 up-regulated and 243 down-regulated. The detailed information of DEGs is shown in the Supplementary Materials 2. Meanwhile, RT-qPCR was performed to validate the RNA-seq data as described in methods. As shown in Table S1 (Supplementary Materials 1), the results of RNA-seq were almost identical to that of RT-qPCR.
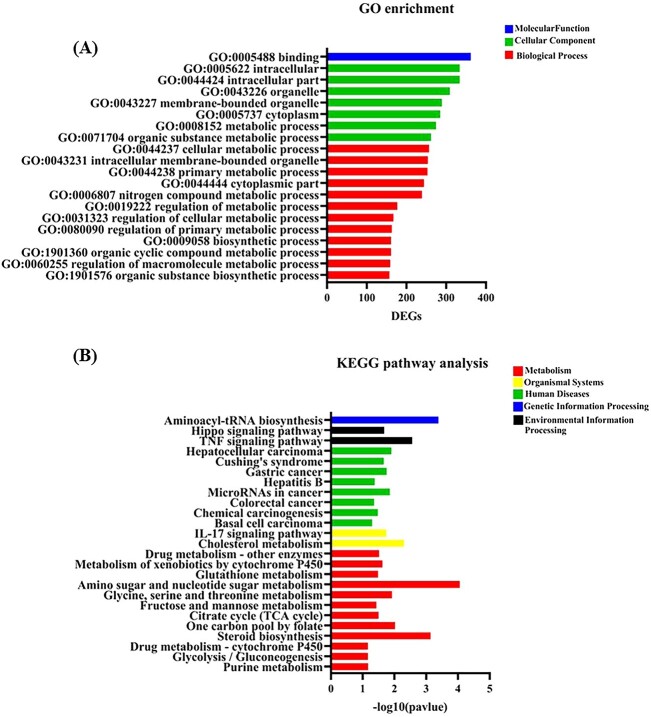
Annotation and enrichment analysis of DEGs were then conducted to analyze the biological function distribution of DEGs based on Gene ontology (GO) database.52 GO is an ontology database that describes the gene functions according to standard classification of Biological Process (BP), Cellular Component (CC), and Molecular Function (MF). Based on the screen criteria of p value less than 0.05, total 931 GO terms were enriched (displayed in Supplementary Materials 2). Then further filtering according to the number of enriched DEGs in each GO term were applied to select the top 20 GO terms. As shown in Fig. 4A, 10 of 12 BP category GO terms were involved in substance and energy metabolism process. Some of these GO terms enriched over 200 DEGs, such as GO:0006807 (nitrogen compound metabolic process), GO:0044237 (cellular metabolic process), GO:0044238 (primary metabolic process), GO:0071704 (organic substance metabolic process), and GO:0008152 (metabolic process). These results suggested a rapid response of L02 cells to MTCS exposure on metabolism balance.
Fig. 4.
The transcriptomic changes induced by MTCS in L02 cells. L02 cells were treated with MTCS (40 μM) for 12 h, and then RNA seq was conducted as described in methods. A): The top 20 enriched GO terms selected on the basis of enriched DEGs number. B): The significant enriched 26 signaling pathways.
KEGG is an online database resource for systematic analysis of gene function from the perspective of genes and molecular networks. Taking p < 0.05 as the filtering standard, 26 pathways were screened in KEGG pathways analysis (displayed in Supplementary Materials 2), which were mainly enriched in “Human Diseases” and “Metabolism” related pathways (Fig. 4B). Among the metabolic related pathways, the enrichment degree of Amino sugar and nucleoside sugar metabolism pathway (hsa00520) in carbohydrate metabolism is most prominent, followed by the steroid biosynthesis pathway (hsa00100) of lipid metabolism. Previously, Fu et al. have reported alterations of metabolic related genes associated with energy and lipid metabolism induced by TCS or/and MTCS exposure in zebrafish.33,34 Furthermore, Fu et al. also found that TCS or/and MTCS disrupted the carbon metabolism process in zebrafish embryos.44 These results demonstrated that metabolism dysregulation is one of the major effects of MTCS, which needs to be further investigated in future.
Key node identification and mechanism analysis
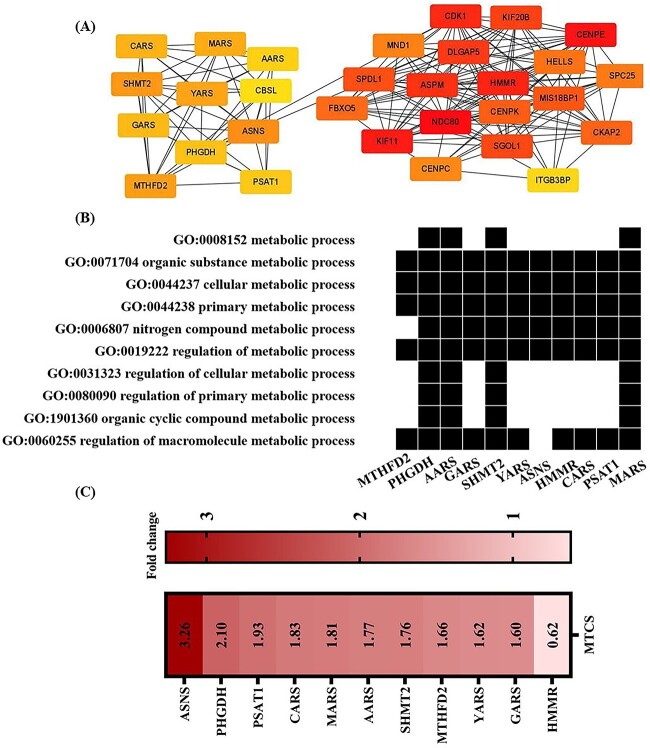
The interactions between proteins are dynamic and interrelated, combining with molecular pathways to constitute the protein-protein interaction (PPI) network. The construction of PPI network contributes to describe the correlations among related proteins, and ultimately elucidate meaningful molecular regulatory networks in organisms.53 To explore the underlying toxic molecular mechanism and identify the crucial biomarkers of MTCS effect, Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) biological database was applicated to acquire preliminary network data (combined score > 0.4), which was subsequently used to map the overall PPI network using Cytoscape (version 3.7.2).54 As shown in Fig. S2, there are total 323 nodes and 1,540 edges in the whole network (Supplementary Materials 1). In the network diagram, nodes refer to potential proteins, and the edges between nodes represent protein-protein interactions. Totally 428 DEGs were further filtered using the cytoHubba plug-in in Cytoscape,55 and the degree algorithm was used to identify hub genes with pivotal roles (Supplementary Materials 2).56 The top 30 hub genes were selected according to the highest connectivity, and interactions among these genes were displayed in Fig. 5A.
Fig. 5.
Key hubba gene identifying induced by MTCS in L02 cells. L02 cells were treated with MTCS (40 μM) for 12 h, and then RNA seq and PPI network construction was performed as described in methods. A): DEGs were filtered using the cytoHubba plug-in in Cytoscape, and the degree algorithm was used to identify hub genes. The top 30 hub genes according to degree algorithm were screened and the interaction between these genes was constructed. B): 11 hubba genes enriched in 10 metabolic related GO terms. C): The expression of 11 hubba genes.
Among the top 30 hub genes, there are many genes involved in metabolism process. The waterfall chart of GO terms and DEGs (Fig. 5B) showed that there were 11 hub genes mainly enriched in biological function terms related to metabolism process. The expression of 11 hub genes were displayed in Fig. 5C, which would be the potential molecular markers in the metabolic disturbance effect of MTCS. Serine hydroxy methyltransferase 2 (SHMT2) is mainly involved in maintenance of one-carbon metabolism, regulating the conversion of serine to glycine, and transfer of methyl group to tetrahydrofolate in the mitochondrial matrix.57 Methylenetetrahydrofolate dehydrogenase 2 (MTHFD2) is also an important mitochondrial enzyme during one-carbon metabolism, and dysregulation of MTHFD2 is frequently observed in many cancers.58 The serine synthesis pathway (SSP) is of great significance in cell metabolism, and phosphoglycerate dehydrogenase (PHGDH) is one of key regulators in SSP process.46 Asparagine synthetase (ASNS) is one metabolic enzymes crucial for glutamine homeostasis.59 ASNS can convert aspartate and glutamine to asparagine and glutamate in ATP-dependent metabolic reaction.60 Combining these above findings, SHMT2, MTHFD2, ASNS and PHGDH genes have the potential of molecular markers to indicate the metabolic imbalances induced by MTCS.
Conclusion
MTCS inhibited the cell viability and mitochondrial membrane potential in L02 cells through induction of oxidative stress. Furthermore, MTCS exposure increased the levels of glucose and lactic acid, triglyceride and cholesterol. The results of biological function and pathway analysis indicated the disorder of glucose and lipid metabolism induced by MTCS exposure. The gene interaction analysis and node identification indicated that SHMT2 MTHFD2, ASNS, PHGDH genes are potential molecular markers of metabolism imbalance induced by MTCS. These results demonstrated that metabolism dysregulation is one of the major effects of MTCS exposure in L02 cells.
Supplementary Material
Contributor Information
Jing An, Institute of Environmental Pollution and Health, School of Environmental and Chemical Engineering, Shanghai University, Nanchen Road 333, Shanghai 200444, PR China.
Yuting Yi, Institute of Environmental Pollution and Health, School of Environmental and Chemical Engineering, Shanghai University, Nanchen Road 333, Shanghai 200444, PR China.
Jingjing Jiang, Institute of Environmental Pollution and Health, School of Environmental and Chemical Engineering, Shanghai University, Nanchen Road 333, Shanghai 200444, PR China.
Weiwei Yao, Institute of Environmental Pollution and Health, School of Environmental and Chemical Engineering, Shanghai University, Nanchen Road 333, Shanghai 200444, PR China.
Guofa Ren, Institute of Environmental Pollution and Health, School of Environmental and Chemical Engineering, Shanghai University, Nanchen Road 333, Shanghai 200444, PR China.
Yu Shang, Institute of Environmental Pollution and Health, School of Environmental and Chemical Engineering, Shanghai University, Nanchen Road 333, Shanghai 200444, PR China.
Author contributions
Y.T.Y. and J.J.J. performed the experiments and data collection. W.W.Y. produced the figured and scripts. G.F.R. performed the statistical analysis. Y.S. contributed to reagents, materials, and analysis tools. J.A. (*) were responsible for the research activity planning and execution. All authors wrote the manuscript
Funding
This work was supported by National Natural Science Foundation of China (No: 41977366, 42077388); Guangdong Foundation for Program of Science and Technology Research (Grant No: 2020B1212060053); Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program (2017BT01Z134).
Conflict of interest statement. The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Availability of data and materials
All the data are contained in the manuscript.
References
- 1. Escarrone AL, Caldas SS, Primel EG, Martins SE, Nery LE. Uptake, tissue distribution and depuration of triclosan in the guppy Poecilia vivipara acclimated to freshwater. Sci Total Environ. 2016:560–561:218–224. [DOI] [PubMed] [Google Scholar]
- 2. Jurewicz J, Wielgomas B, Radwan M, Karwacka A, Klimowska A, Dziewirska E, Korczak K, Zajdel R, Radwan P, Hanke W. Triclosan exposure and ovarian reserve. Reprod Toxicol. 2019:89:168–172. [DOI] [PubMed] [Google Scholar]
- 3. Buth JM, Steen PO, Sueper C, Blumentritt D, Vikesland PJ, Arnold WA, McNeill K. Dioxin photoproducts of triclosan and its chlorinated derivatives in sediment cores. Environ Sci Technol. 2010:44(12):4545–4551. [DOI] [PubMed] [Google Scholar]
- 4. Dhillon GS, Kaur S, Pulicharla R, Brar SK, Cledón M, Verma M, Surampalli R. Triclosan: current status, occurrence, environmental risks and bioaccumulation potential. Int J Environ Res Public Health. 2015:12(5):5657–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haggard DE, Noyes PD, Waters KM, Tanguay RL. Phenotypically anchored transcriptome profiling of developmental exposure to the antimicrobial agent, triclosan, reveals hepatotoxicity in embryonic zebrafish. Toxicol Appl Pharmacol. 2016:308:32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Santos LHMLM, Freixa A, Insa S, Acuña V, Sanchís J, Farré M, Sabater S, Barceló D, Rodríguez-Mozaz S. Impact of fullerenes in the bioaccumulation and biotransformation of venlafaxine, diuron and triclosan in river biofilms. Environ Res. 2019:169:377–386. [DOI] [PubMed] [Google Scholar]
- 7. Wang X, Gao M, Gao J, Wang X, Ma M, Wang H. Extraction of triclosan and methyltriclosan in human fluids by in situ ionic liquid morphologic transformation. J Chromatogr B. 2018:1092:19–28. [DOI] [PubMed] [Google Scholar]
- 8. Zhao RS, Wang X, Sun J, Hu C, Wang XK. Determination of triclosan and triclocarban in environmental water samples with ionic liquid/ionic liquid dispersive liquid-liquid microextraction prior to HPLC-ESI-MS/MS. Microchim Acta. 2011:174(1–2):145–151. [Google Scholar]
- 9. Allmyr M, Harden F, Toms LM, Mueller JF, McLachlan MS, Adolfsson-Erici M, Sandborgh-Englund G. The influence of age and gender on triclosan concentrations in Australian human blood serum. Sci Total Environ. 2008:393(1):162–167. [DOI] [PubMed] [Google Scholar]
- 10. Bai X, Zhang B, He Y, Hong D, Song S, Huang Y, Zhang T. Triclosan and triclocarbon in maternal-fetal serum, urine, and amniotic fluid samples and their implication for prenatal exposure. Environ Pollut. 2020:266(Pt 1):115117. [DOI] [PubMed] [Google Scholar]
- 11. Geens T, Neels H, Covaci A. Distribution of bisphenol-A, triclosan and n-nonylphenol in human adipose tissue, liver and brain. Chemosphere. 2012:87(7):796–802. [DOI] [PubMed] [Google Scholar]
- 12. Pannu MW, O'Connor GA, Toor GS. Toxicity and bioaccumulation of biosolids-borne triclosan in terrestrial organisms. Environ Toxicol Chem. 2012:31(3):646–653. [DOI] [PubMed] [Google Scholar]
- 13. Wang L, Asimakopoulos AG, Kannan K. Accumulation of 19 environmental phenolic and xenobiotic heterocyclic aromatic compounds in human adipose tissue. Environ Int. 2015:78:45–50. [DOI] [PubMed] [Google Scholar]
- 14. Yang D, Kong S, Wang F, Tse LA, Tang Z, Zhao Y, Li C, Li M, Li Z, Lu S. Urinary triclosan in South China adults and implications for human exposure. Environ Pollut. 2021:286:117561. [DOI] [PubMed] [Google Scholar]
- 15. Bester K. Triclosan in a sewage treatment process—balances and monitoring data. Water Res. 2003:37(16):3891–3896. [DOI] [PubMed] [Google Scholar]
- 16. Bester K. Fate of triclosan and triclosan-methyl in sewage treatment plants and surface waters. Arch Environ Contam Toxicol. 2005:49(1):9–17. [DOI] [PubMed] [Google Scholar]
- 17. Chen X, Nielsen JL, Furgal K, Liu Y, Lolas IB, Bester K. Biodegradation of triclosan and formation of methyl-triclosan in activated sludge under aerobic conditions. Chemosphere. 2011:84(4):452–456. [DOI] [PubMed] [Google Scholar]
- 18. Liang Y, Song H, Wu Y, Gao S, Zeng X, Yu Z. Occurrence and distribution of triclosan and its transformation products in Taihu Lake, China. Environ Sci Pollut Res Int. 2022:29(56):84787–84797. [DOI] [PubMed] [Google Scholar]
- 19. Clayborn AB, Toofan SN, Champlin FR. Influence of methylation on the antibacterial properties of triclosan in Pasteurella multocida and Pseudomonas aeruginosa variant strains. J Hosp Infect. 2011:77(2):129–133. [DOI] [PubMed] [Google Scholar]
- 20. Lindström A, Buerge IJ, Poiger T, Bergqvist PA, Müller MD, Buser HR. Occurrence and environmental behavior of the bactericide triclosan and its methyl derivative in surface waters and in wastewater. Environ Sci Technol. 2002:36(11):2322–2329. [DOI] [PubMed] [Google Scholar]
- 21. Macherius A, Lapen DR, Reemtsma T, Römbke J, Topp E, Coors A. Triclocarban, triclosan and its transformation product methyl triclosan in native earthworm species four years after a commercial-scale biosolids application. Sci Total Environ. 2014:472:235–238. [DOI] [PubMed] [Google Scholar]
- 22. Wang X, Ouyang F, Feng L, Wang X, Liu Z, Zhang J. Maternal urinary Triclosan concentration in relation to maternal and neonatal thyroid hormone levels: a prospective study. Environ Health Perspect. 2017:125(6):067017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen X, Liang J, Bao L, Gu X, Zha S, Chen X. Competitive and cooperative sorption between triclosan and methyl triclosan on microplastics and soil. Environ Res. 2022:212(Pt D):113548. [DOI] [PubMed] [Google Scholar]
- 24. Delorenzo ME, Keller JM, Arthur CD, Finnegan MC, Harper HE, Winder VL, Zdankiewicz DL. Toxicity of the antimicrobial compound triclosan and formation of the metabolite methyl-triclosan in estuarine systems. Environ Toxicol. 2008:23(2):224–232. [DOI] [PubMed] [Google Scholar]
- 25. Coogan MA, La Point TW. Snail bioaccumulation of triclocarban, triclosan, and methyltriclosan in a North Texas, USA, stream affected by wastewater treatment plant runoff. Environ Toxicol Chem. 2008:27(8):1788–1793. [DOI] [PubMed] [Google Scholar]
- 26. Rüdel H, Böhmer W, Müller M, Fliedner A, Ricking M, Teubner D, Schröter-Kermani C. Retrospective study of triclosan and methyl-triclosan residues in fish and suspended particulate matter: results from the German Environmental Specimen Bank. Chemosphere. 2013:91(11):1517–1524. [DOI] [PubMed] [Google Scholar]
- 27. Goodbred S, Rosen MR, Patiño R, Alvarez D, Echols K, King K, Umek J. Movement of synthetic organic compounds in the food web after the introduction of invasive quagga mussels (Dreissena bugensis) in Lake Mead, Nevada and Arizona, USA. Sci Total Environ. 2021:752:141845. [DOI] [PubMed] [Google Scholar]
- 28. Allmyr M, Adolfsson-Erici M, McLachlan MS, Sandborgh-Englund G. Triclosan in plasma and milk from Swedish nursing mothers and their exposure via personal care products. Sci Total Environ. 2006:372(1):87–93. [DOI] [PubMed] [Google Scholar]
- 29. Calafat AM, Ye X, Wong LY, Reidy JA, Needham LL. Urinary concentrations of triclosan in the U.S. population: 2003–2004. Environ Health Perspect. 2008:116(3):303–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Geens T, Neels H, Covaci A. Sensitive and selective method for the determination of bisphenol-A and triclosan in serum and urine as pentafluorobenzoate-derivatives using GC-ECNI/MS. J Chromatogr B Analyt Technol Biomed Life Sci. 2009:877(31):4042–4046. [DOI] [PubMed] [Google Scholar]
- 31. Macedo S, Torres T, Santos MM. Methyl-triclosan and triclosan impact embryonic development of Danio rerio and Paracentrotus lividus. Ecotoxicology. 2017:26(4):482–489. [DOI] [PubMed] [Google Scholar]
- 32. Wang L, Mao B, He H, Shang Y, Zhong Y, Yu Z, Yang Y, Li H, An J. Comparison of hepatotoxicity and mechanisms induced by triclosan (TCS) and methyl-triclosan (MTCS) in human liver hepatocellular HepG2 cells. Toxicol Res (Camb). 2019:8(1):38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fu J, Gong Z, Bae S. Assessment of the effect of methyl-triclosan and its mixture with triclosan on developing zebrafish (Danio rerio) embryos using mass spectrometry-based metabolomics. J Hazard Mater. 2019:368:186–196. [DOI] [PubMed] [Google Scholar]
- 34. Fu J, Tan YXR, Gong Z, Bae S. The toxic effect of triclosan and methyl-triclosan on biological pathways revealed by metabolomics and gene expression in zebrafish embryos. Ecotoxicol Environ Saf. 2020:189:110039. [DOI] [PubMed] [Google Scholar]
- 35. Gaume B, Bourgougnon N, Auzoux-Bordenave S, Roig B, Le Bot B, Bedoux G. In vitro effects of triclosan and methyl-triclosan on the marine gastropod Haliotis tuberculata. Comp Biochem Physiol C Toxicol Pharmacol. 2012:156(2):87–94. [DOI] [PubMed] [Google Scholar]
- 36. Lv W, Chen Y, Li D, Chen X, Leszczynski J. Methyl-triclosan binding to human serum albumin: multi-spectroscopic study and visualized molecular simulation. Chemosphere. 2013:93(6):1125–1130. [DOI] [PubMed] [Google Scholar]
- 37. An J, Yao W, Tang W, Jiang J, Shang Y. Hormesis effect of methyl Triclosan on cell proliferation and migration in human hepatocyte L02 cells. ACS Omega. 2021:6(29):18904–18913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Park JC, Han J, Lee MC, Seo JS, Lee JS. Effects of triclosan (TCS) on fecundity, the antioxidant system, and oxidative stress-mediated gene expression in the copepod Tigriopus japonicus. Aquat Toxicol. 2017:189:16–24. [DOI] [PubMed] [Google Scholar]
- 39. Zhong R, He H, Jin M, Lu Z, Deng Y, Liu C, Shen N, Li J, Wang H, Ying P, et al. Genome-wide gene-bisphenol A, F and triclosan interaction analyses on urinary oxidative stress markers. Sci Total Environ. 2022:807(Pt 1):150753. [DOI] [PubMed] [Google Scholar]
- 40. Gyimah E, Dong X, Qiu W, Zhang Z, Xu H. Sublethal concentrations of triclosan elicited oxidative stress, DNA damage, and histological alterations in the liver and brain of adult zebrafish. Environ Sci Pollut Res Int. 2020:27(14):17329–17338. [DOI] [PubMed] [Google Scholar]
- 41. Li X, An J, Li H, Qiu X, Wei Y, Shang Y. The methyl-triclosan induced caspase-dependent mitochondrial apoptosis in HepG2 cells mediated through oxidative stress. Ecotoxicol Environ Saf. 2019:182:109391. [DOI] [PubMed] [Google Scholar]
- 42. Rhee JS, Yu IT, Kim BM, Jeong CB, Lee KW, Kim MJ, Lee SJ, Park GS, Lee JS. Copper induces apoptotic cell death through reactive oxygen species-triggered oxidative stress in the intertidal copepod Tigriopus japonicus. Aquat Toxicol. 2013:132–133:182–189. [DOI] [PubMed] [Google Scholar]
- 43. Kim RO, Rhee JS, Won EJ, Lee KW, Kang CM, Lee YM, Lee JS. Ultraviolet B retards growth, induces oxidative stress, and modulates DNA repair-related gene and heat shock protein gene expression in the monogonont rotifer, Brachionus sp. Aquat Toxicol. 2011:101(3–4):529–539. [DOI] [PubMed] [Google Scholar]
- 44. Fu J, Gong Z, Bae S. Ecotoxicogenomic analysis of zebrafish embryos exposed to triclosan and mixture triclosan and methyl triclosan using suppression subtractive hybridization and next-generation sequencing. J Hazard Mater. 2021:414:125450. [DOI] [PubMed] [Google Scholar]
- 45. Michikawa M. Cholesterol paradox: is high total or low HDL cholesterol level a risk for Alzheimer's disease? J Neurosci Res. 2003:72(2):141–146. [DOI] [PubMed] [Google Scholar]
- 46. Yang J, Zhang X, Liu Z, Yuan Z, Song Y, Shao S, Zhou X, Yan H, Guan Q, Gao L, et al. High-cholesterol diet disrupts the levels of hormones derived from anterior pituitary basophilic cells. J Neuroendocrinol. 2016:28(3):12369. [DOI] [PubMed] [Google Scholar]
- 47. Pundir CS, Narang J. Determination of triglycerides with special emphasis on biosensors: a review. Int J Biol Macromol. 2013:61:379–389. [DOI] [PubMed] [Google Scholar]
- 48. An J, He H, Yao W, Shang Y, Jiang Y, Yu Z. PI3K/Akt/FoxO pathway mediates glycolytic metabolism in HepG2 cells exposed to triclosan (TCS). Environ Int. 2020:136:105428. [DOI] [PubMed] [Google Scholar]
- 49. Yamada K. Aberrant uptake of a fluorescent l-glucose analogue (flg) into tumor cells expressing malignant phenotypes. Biol Pharm Bull. 2018:41(10):1508–1516. [DOI] [PubMed] [Google Scholar]
- 50. Sun S, Li H, Chen J, Qian Q. Lactic acid: no longer an inert and end-product of glycolysis. Physiology (Bethesda). 2017:32(6):453–463. [DOI] [PubMed] [Google Scholar]
- 51. Kukurba KR, Montgomery SB. RNA sequencing and analysis. Cold Spring Harb Protoc. 2015:2015(11):951–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000:25(1):25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sufyan M, Ali Ashfaq U, Ahmad S, Noor F, Hamzah Saleem M, Farhan Aslam M, El-Serehy HA, Aslam S. Identifying key genes and screening therapeutic agents associated with diabetes mellitus and HCV-related hepatocellular carcinoma by bioinformatics analysis. Saudi J Biol Sci. 2021:28(10):5518–5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003:13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003:31(1):258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. CytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014:8(S4):S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Choi YJ, Lee G, Yun SH, Lee W, Yu J, Kim SK, Lee BH. The role of SHMT2 in modulating lipid metabolism in hepatocytes via glycine-mediated mTOR activation. Amino Acids. 2022:54(5):823–834. [DOI] [PubMed] [Google Scholar]
- 58. Shi Y, Xu Y, Yao J, Yan C, Su H, Zhang X, Chen E, Ying K. MTHFD2 promotes tumorigenesis and metastasis in lung adenocarcinoma by regulating AKT/GSK-3β/β-catenin signalling. J Cell Mol Med. 2021:25(14):7013–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang S, Ding K, Shen QJ, Zhao S, Liu JL. Filamentation of asparagine synthetase in Saccharomyces cerevisiae. PLoS Genet. 2018:14(10):e1007737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lomelino CL, Andring JT, McKenna R, Kilberg MS. Asparagine synthetase: function, structure, and role in disease. J Biol Chem. 2017:292(49):19952–19958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data are contained in the manuscript.