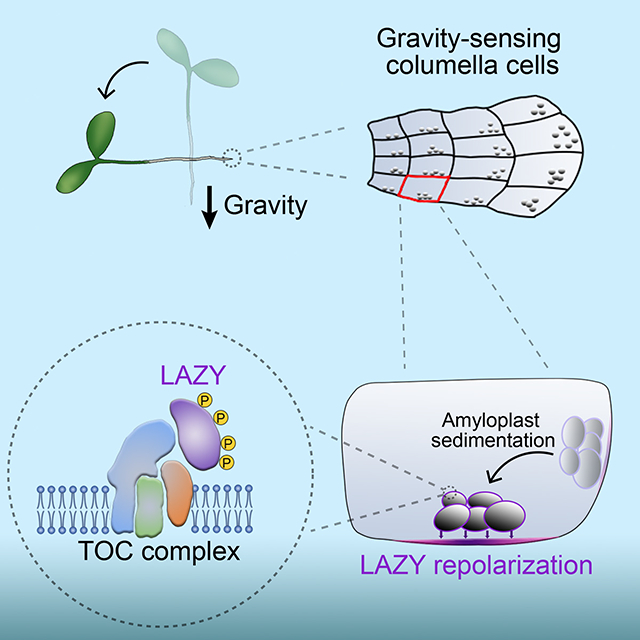
SUMMARY
Gravity controls directional growth of plants, and the classical starch-statolith hypothesis proposed more than a century ago postulates that amyloplast sedimentation in specialized cells initiates gravity sensing, but the molecular mechanism remains uncharacterized. The LAZY proteins are known as key regulators of gravitropism and lazy mutants show striking gravitropic defects. Here, we report that gravistimulation by reorientation triggers Mitogen-Activated Protein Kinase (MAPK) signaling-mediated phosphorylation of Arabidopsis LAZY proteins basally polarized in root columella cells. Phosphorylation of LAZY increases its interaction with several Translocons at the Outer envelope membrane of Chloroplasts (TOC) proteins on the surface of amyloplasts, facilitating enrichment of LAZY proteins on amyloplasts. Amyloplast sedimentation subsequently guides LAZY to relocate to the new lower side of the plasma membrane in columella cells, where LAZY induces asymmetrical auxin distribution and root differential growth. Together, this study provides a molecular interpretation for the starch-statolith hypothesis: the organelle movement-triggered molecular polarity formation.
Keywords: Gravitropism, Starch-statolith hypothesis, Amyloplast sedimentation, polarity, Phosphorylation, LAZY, TOC, MPK
In brief
Amyloplast sedimentation guides LAZY to relocate to the new lower side of the plasma membrane in columella cells to induce gravitropic growth in plants.
Graphical Abstract:
INTRODUCTION
Gravity is a key environmental factor acting on all organisms on earth. In plants, gravity usually directs roots to grow downwards (positive gravitropism) and aerial parts to grow upwards (negative gravitropism) 1,2, which may control agricultural traits such as drought tolerance3, nutrient absorption4, and so on. Gravitropism includes three key processes: gravity sensing, signal transduction, and differential growth response2,5. Since the direction and the magnitude of gravity are almost constant on the surface of the earth, gravitropism is regarded as a posture control, triggered by sensing the tilt of organs relative to the direction of gravity vector5.
The starch-statolith hypothesis and Cholodny-Went theory are the dogmas for explaining gravity sensing and response, respectively6. The starch-statolith hypothesis was proposed one hundred and twenty years ago, in which sedimentation of amyloplasts (starch-filled plastids) in the statocytes was considered to initiate gravity sensing7–9. The columella cells in roots and endodermal cells in shoots were demonstrated to be the statocytes for sensing gravity, and genetic disruption or laser ablation of these cells led to complete disruption of the gravitropism10–12. Gravity sensing is further divided into susception (physical response) and signal conversion from physical to physiological information5. The sedimentation of starchless plastids in phosphoglucomutase 1 (pgm1) mutants was impaired, resulting in a slower gravitropic response in both roots and shoots13,14. In contrast, disruption of actin accelerated amyloplast sedimentation, resulting in promoted gravitropism15. Generally, amyloplast sedimentation was considered to be the susception step of gravity sensing5, but the molecular roles of amyloplast sedimentation and the underlying mechanisms of signal conversion were unclear. The Cholodny-Went theory proposed that growth curvature is due to an unequal distribution of auxin between the two sides of the curving organ in plants16, and auxin efflux transporter PIN proteins were later proved to contribute to the redirection of auxin and subsequent differential growth in gravitropism17,18.
In the 1930s, an unusual corn mutant with a stalk growing towards the ground was described as “lazy” due to its prostrate phenotype (sleeping stature) 19,20. The rice lazy mutant with a similar phenotype was identified later21. Since then, the LAZY family genes were demonstrated to be critically important for the gravitropic responses of both shoots and roots in many plant species, including, rice3,21–23, maize20,24, Arabidopsis25–27, Medicago truncatula28, Prunus domestica26 and Lotus japonicus29. High-order lazy mutants in Arabidopsis showed more exaggerated phenotypes than single mutants, though the phenotypes reported by different groups varied28,30,31. Our previous study showed that light modulates gravitropism by controlling the expression level of LAZY432. A recent study showed that Arabidopsis LAZY4 (LZY3) proteins display polar distribution on the plasma membrane of the columella cells in lateral roots, and recruit RCC1-like domain (RLD) proteins from the cytoplasm to the plasma membrane to promote the re-localization of PIN3 and modulate auxin flow33. LAZY4 (AtDRO1) has also been reported to show nuclear localization, but its role there is unclear34.
Altered Response to Gravity 1 (ARG1) encodes a DnaJ-like protein and its mutation delayed gravitropism in both root and hypocotyl35. Mutating TOC34, TOC75, TOC120 or TOC132, components of the Translocons at the Outer envelope membrane of Chloroplasts (TOC) complexes, significantly enhanced the arg1 gravitropic defect, even though either toc132Q730Stop, toc75G658R, or arg1 single mutants showed weak to no gravitropic phenotypes36,37. The classical function of TOC proteins is to import proteins into the plastids38–41, but whether TOC proteins regulate gravitropism by importing unknown signal factors into amyloplasts is unclear37.
Mitogen-Activated Protein Kinase (MAPK) cascades are highly conserved in eukaryotic cells. In plants, the MAPK signaling modules regulate many aspects of growth and environmental responses42. The MKK4/MKK5-MPK3/MPK6 module regulates stomatal development and patterning43, the inflorescence architecture44, lateral root development45, and so on. The MKK7-MPK6 module positively regulates hypocotyl gravitropism46.
Here, we report that gravistimulation triggers the MKK5-MPK3 module to phosphorylate LAZY proteins, which may promote LAZY proteins to enrich on the surface of amyloplasts via interacting with the TOC proteins. Then, amyloplast sedimentation promotes the translocation of LAZY proteins to the new lower side of the plasma membrane in columella cells, in which relocation of LAZY proteins from the surface of amyloplasts to the adjacent plasma membrane is an important process. The repolarization of LAZY proteins leads to asymmetric auxin distribution and ultimately differential growth. Thus, our study reveals the molecular role of amyloplast sedimentation, providing insights into the molecular mechanism of the starch-statolith hypothesis.
RESULTS
LAZY2, 3, and 4 are requisite for the gravitropic responses in roots
Previous studies showed that simultaneously disrupting LAZY2 (NGR1), LAZY3 (NGR3) and LAZY4 (NGR2) in Arabidopsis via combined T-DNA insertions in the Col or Ws ecotypes resulted in various root gravitropism phenotypes, from negative gravitropism to no clear gravitropism, probably due to the complex ecotype backgrounds or growth conditions28,31. To further understand the functions of LAZY family members, we used a CRISPR/Cas9 system to generate Arabidopsis lazy2 lazy3 lazy4 (lazy234) triple mutants in the Col background. Three lines with different mutated alleles in the three LAZY genes were established for phenotypic analyses, designated lazy234-1, lazy234-2 and lazy234-3 (Figure S1A). With regards to gravitropism growth, either under white light or in the dark, primary roots of wild-type plants always grew towards the gravity vector, whereas those of lazy234 mutants grew randomly in all directions (Figures 1A and 1B), so did the lateral roots in lazy234 (Figures S1B and S1C). Our results are generally consistent with a previous study showing that the roots of the atlazy2,3,4 triple mutants generated by crossing T-DNA insertion lines were agravitropic under white light and displayed weakly negative gravitropism in the dark31. Although the genetic distinction and possible variation of growth conditions resulted in slight phenotypic differences, our and previous data all support that these three LAZYs are essential for positive gravitropism of roots. With regards to the amyloplast sedimentation after reorientation, similar behaviors were observed in wild-type and lazy234 mutants (Figures S1D and S1E), indicating that these LAZYs are dispensable for amyloplast sedimentation. As the asymmetric distribution of auxin promotes root gravitropism47, and DR5rev::GFP is widely used to indicate auxin distribution48, we mutated LAZY2, LAZY3, and LAZY4 in the DR5rev::GFP reporter line via CRISPR, resulting in losing gravitropic responses in roots (Figures S2A–S2D). After the gravistimulation via reorientation, DR5rev::GFP signals completely lost their differential distribution in DR5rev::GFP/lazy234 plants compared with wild-type (Figures 1C, 1D, S2E and S2F). Thus, these LAZY proteins are essential for gravity-triggered polar auxin distribution and bending responses in both primary and lateral roots.
Figure 1. Arabidopsis lacking LAZY2, 3 and 4 lose the gravitropic response and gravistimulation-induced auxin redistribution in roots.
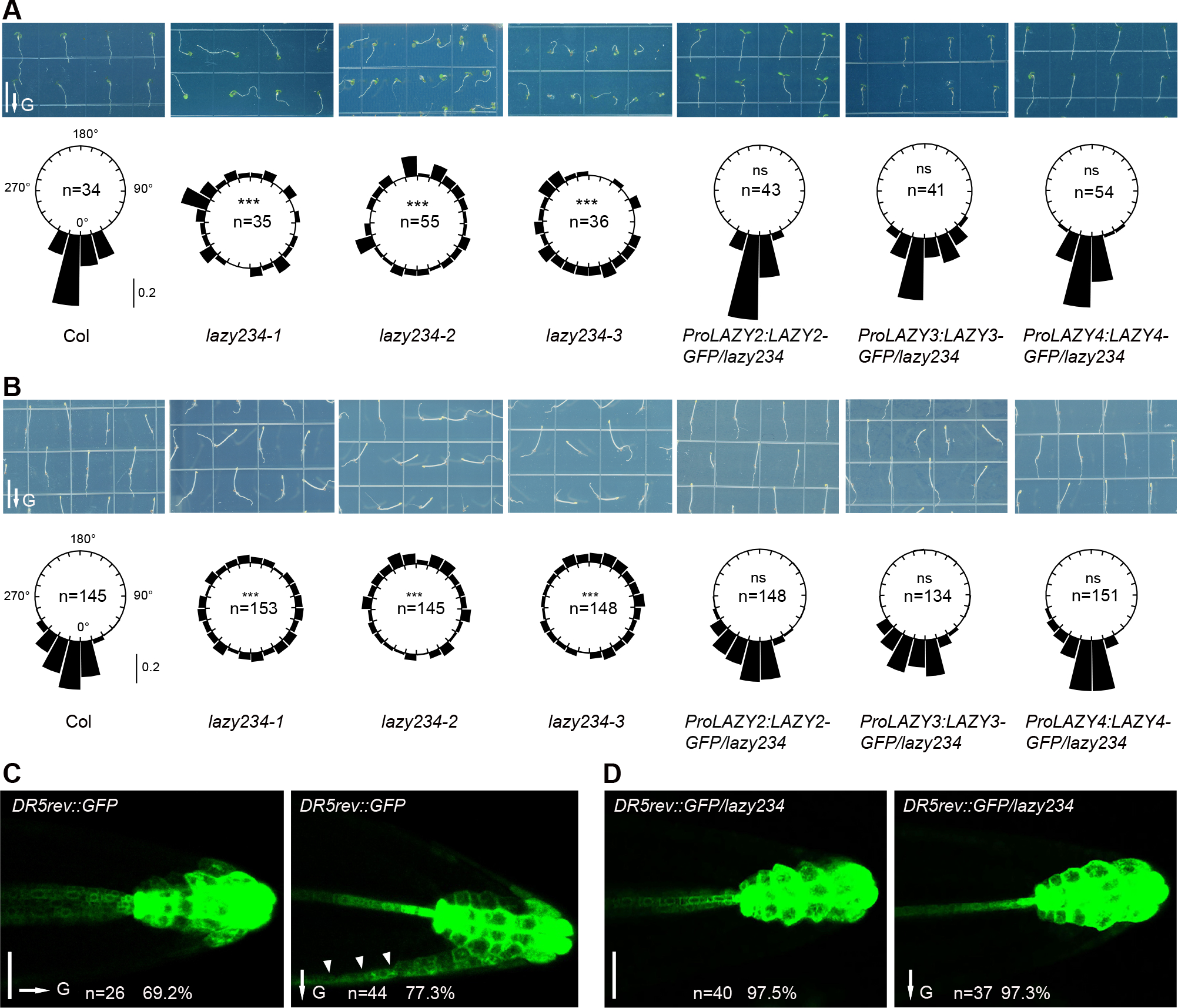
(A and B) Col, lazy234 triple mutants, and complementation lines were grown vertically under white light for 4 days (A) or in the dark for 3 days (B). Upper, representative seedlings. Scale bars, 1 cm. Lower, frequencies of root tip angles in each 15° division around a circle, and the bars indicate relative frequency. Root tip distribution was compared with Col, and significant differences were evaluated by the Kolmogorov-Smirnov test with Bonferroni correction (***, P < 0.001; ns, not significant, P > 0.05).
(C and D) Expression of DR5rev::GFP in wild-type and lazy234 primary roots before (left) and after gravistimulation via 90° reorientation (right) for around 4 hours. Random primary roots of DR5rev::GFP/ lazy234 were aligned to the gravity vector before the reorientation, therefore no gravity vector was marked in D, left. Scale bars, 50 μm. Arrowheads indicate the asymmetric accumulation of auxin on one side.
In A to D, arrows labeled “G” indicate the direction of gravity.
See also Figures S1, and S2.
LAZY2, 3 and 4 proteins are localized on the amyloplasts and plasma membrane in columella cells
To study protein subcellular localization, we generated endogenous promoter-driven LAZY-GFP transgenic lines in the lazy234 mutant background. Each of the transgenes similarly and efficiently rescued the gravitropic phenotypes of lazy234 primary roots (Figures 1A and 1B). In roots, the three LAZY proteins were predominantly expressed in columella cells, and LAZY4 was also expressed in the endodermis (Figures 2A–2C, and S3A), consistent with the previous results of promoter-driving GUS expression31. More strikingly, these LAZY proteins were not only localized on the cell periphery but also on the surface of the amyloplasts in the columella cells (Figures 2D–2F). Anti-GFP immunogold labelling confirmed the localization of LAZY proteins on the plasma membrane (Figure 2G). Since starch-statolith hypothesis proposed that amyloplast sedimentation initiates gravity sensing, the localization of LAZY proteins on the surface of amyloplasts suggested that they may play critical roles in an early stage of gravity sensing and signal transduction.
Figure 2. Arabidopsis LAZY2, 3 and 4 proteins are predominantly localized on the amyloplasts and plasma membrane of columella cells in roots.
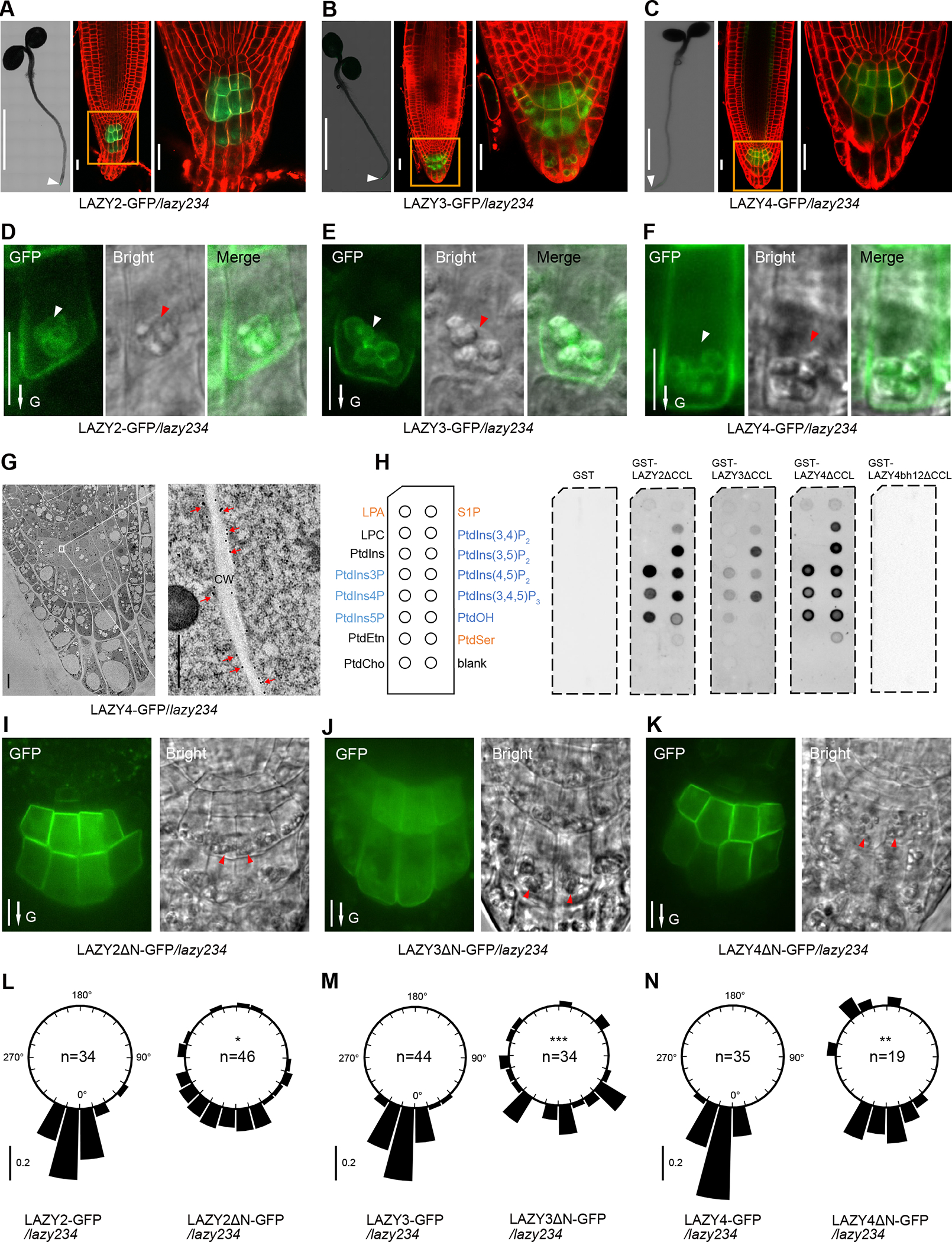
(A to C) Localization of GFP-tagged LAZY2, LAZY3, and LAZY4 proteins (green) at the root tip of seedlings grown in white light. For each figure panel, left, a confocal image (assembled) shows the predominant expression of LAZY-GFP at the root tip (white arrowhead); middle, a confocal image shows the subcellular localization of LAZY-GFP with an enlarged view (right). Cell outlines were stained with 5 μM FM4-64 for 2 minutes (red). Scale bars, 0.2 cm (left); 20 μm (middle and right). A, B and C are representative images of roots of 10, 8 and 13 seedlings, respectively.
(D to F) Detailed localization of LAZY2/3/4-GFP protein (green) at the plasma membrane and around the amyloplast envelope in columella cells. White and red arrowheads point to the amyloplasts observed under the Confocal or DIC microscopy, respectively. Scale bars, 10 μm. D, E and F are representative images of roots of 15, 17 and 12 seedlings, respectively. For LAZY4-GFP in F, an image with a relatively strong appearance of amyloplast localization was selected for improved visualization of intracellular signals.
(G) Tracking LAZY4-GFP localization by immuno-gold labeling. The root tips of 4-day-old light-grown LAZY4-GFP/lazy234 seedlings were analyzed by immuno-electron microscopy using GFP antibody. Red arrows indicate the gold particles on the plasma membranes. CW, Cell Wall. Scale bars, 5 μm (left); 0.5 μm (right).
(H) Protein-lipid overlay assay with purified GST, GST-LAZY2ΔCCL, GST-LAZY3ΔCCL, GST-LAZY4ΔCCL and GST-LAZY4bh12ΔCCL using PBS buffer. LPA, lysophosphatidic acid; S1P, sphingosine-1-phosphate; LPC, lysophosphatidylcholine; PtdIns, phosphatidylinositol; PtdIns(x)P(y), mono/bis/tris phosphates, x is the phosphorylation position while y is the number of the phosphate groups; PtdEtn, phosphatidyl-ethanolamine; PtdCho, phosphatidylcholine; PtdOH, phosphatidic acid; PtdSer, phosphatidylserine. LAZY4bh12 means mutating K/R to A within two BH peaks (details in Figure S3C). ΔCCL means the deletion of the last 14 amino acids, which made these LAZY proteins express better in E.coli. Equal amounts of GST or GST-tagged proteins (2.5 μg) were used for each membrane. Anti-GST was used for the immunoblot.
(I to K) Localization of LAZY2ΔN-GFP, LAZY3ΔN-GFP, LAZY4ΔN-GFP proteins at the root tip of seedlings grown in white light. ΔN indicates the deletion of N terminus of LAZY proteins (details in methods). Red arrowheads indicate the amyloplasts observed under the DIC microscopy. Scale bars, 10 μm. I, J and K are representative images of roots of 16, 12 and 20 seedlings, respectively.
(L to N) Full length and N-terminal deletion LAZY2/3/4-GFP/lazy234 transgenic lines were grown vertically under white light for 4 days. Frequencies of root tip angles in each 15° division around a circle are shown, and the bar indicates relative frequency. Root tip distribution of truncated LAZY2/3/4-GFP/lazy234 transgenic lines was compared with full length lines, and significant differences were evaluated by Kolmogorov-Smirnov test with Bonferroni correction (***, P < 0.001; **, P < 0.01; *, P < 0.05).
In D to F, I to K, arrows labeled “G” indicate the direction of gravity.
See also Figure S3.
LAZY2, 3 and 4 proteins were localized on the membranes, but they have no transmembrane domain or lipid acylation modification sites that may contribute to membrane association. Previous studies proved that basic and hydrophobic (BH) regions within proteins may promote their association with membranes49,50. BH score analysis showed that each LAZY has potential membrane binding regions (Figure S3B). Protein-lipid overlay assays showed that these LAZY proteins could bind multiple membrane phospholipids, especially phosphatidylinositolphosphates (PIPs) (Figure 2H). Mutating amino acids K/R to A within the BH peaks of LAZY4 reduced the predicted BH score (Figures S3B and S3C), and disrupted its binding to the membrane phospholipids (Figure 2H). These results suggest that these LAZY proteins may associate with membrane via binding phospholipids.
By truncation analysis, we found that deletion of the N-terminal region (containing domain I) disrupted the amyloplast localization of LAZY2, LAZY3 and LAZY4 (Figures 2I–2K). Meanwhile, the deletion of the N-terminal region disrupted the functions of these LAZYs in complementing the gravitropic defects of lazy234 mutants (Figures 2L–2N). These data indicate that proper localization of LAZY proteins on the amyloplasts may be critical for their function in gravitropism.
Sedimentation of amyloplasts promotes the gravity-triggered redistribution of LAZY proteins
Since LAZY proteins were preferentially localized on the lower side of the plasma membrane in columella cells (Figures 2D–2F), we studied the redistribution of LAZY proteins under the regulation of gravity in both primary and lateral roots. Results showed that gravistimulation by re-orientating plants 90° could trigger re-polarization of all three LAZY proteins in columella cells of primary roots, with more proteins accumulated on the new lower side than the upper side (Figures 3A–3D, S3D and S3E). Consistently, in the columella cells of lateral roots, these LAZY proteins also displayed polarization and repolarization after re-orientating the seedlings 180° (Figures S3F and S3G).
Figure 3. Amyloplast sedimentation promotes the gravity-triggered redistribution of LAZY proteins to the lower side of columella cells.
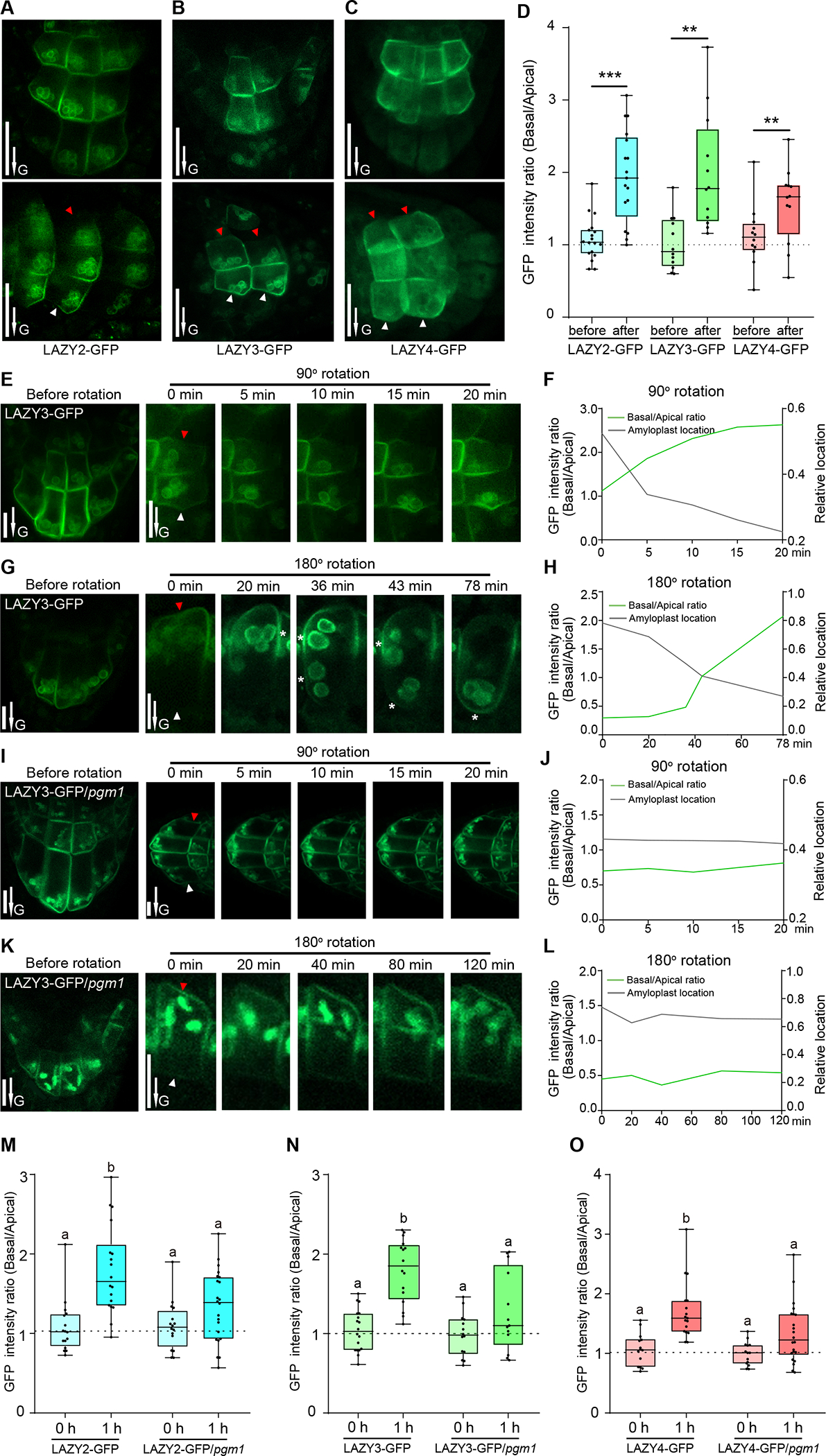
(A to D) LAZY proteins show polar distribution in columella cells under the control of gravity. (A to C) Seedlings were grown vertically on MS plates, and reoriented 90° and kept for 0.5 to 1 h of gravistimulation. Red and white arrowheads indicate the accumulation of LAZY-GFP proteins on the upper and lower sides of columella cells, respectively. Scale bars, 20 μm. (D) Statistical analysis of fluorescence intensity ratios of the two sides of columella cells (Basal/Apical) in panels A to C. Asterisks indicate Student’s t-test values (***, P < 0.001; **, P < 0.01; n = 18, 17, 12, 12, 12 and 12 respectively). The method for intensity ratio analysis is shown in Figures S3D and S3E.
(E to H) Gravity-triggered redistribution of LAZY proteins to the lower side of columella cells correlates with the sedimentation of amyloplasts. LAZY3-GFP seedlings were grown vertically and then reoriented 90° (E) or 180° (G), and fluorescence was collected at several time points. Red and white arrowheads indicate the upper and lower sides of columella cells, respectively. White stars in panel G indicate the positions of plasma membrane adjacent to amyloplasts, where the fluorescence is strong. Scale bars, 10 μm. The fluorescence intensity ratio (Basal/Apical) and average relative-locations of the amyloplasts (relative distances from the amyloplasts to the new bottoms of cells, cell heights were set as 1) are shown in F and H. E and G are representative images of 6 seedling roots for each rotation condition.
(I to L) Mutation of PGM1 delayed both amyloplast sedimentation and redistribution of LAZY3-GFP proteins triggered by gravistimulation. Data collection and analyses were the same as in E to H. I and K are representative images of 3 seedling roots for each rotation condition.
(M to O) Mutation of PGM1 delayed the redistribution of LAZY2, LAZY3 and LAZY4 proteins. The seedlings were grown vertically and then reoriented 90° and kept 1 h for gravistimulation. The statistical analyses of fluorescence intensity ratios of LAZY-GFP proteins on the two sides of columella cells (Basal/Apical) are shown. Significant differences in GFP intensity ratios were evaluated by one-way ANOVA with post-hoc Tukey’s HSD test (alpha=0.05; M, n = 16, 18, 16 and 22 respectively; N, n = 18, 16, 14 and 13 respectively; O, n = 14, 17, 14 and 22 respectively). Representative images are shown in Figure S4B.
In A, B, C, E, G, I and K, arrows labeled “G” indicate the direction of gravity. In E, G, I, and K, the images labeled as “Before rotation” were rotated from the images labeled as “0 min” after 90° or 180° rotation, to show the original orientation.
See also Figures S3 and S4.
We further studied whether the repolarization site of LAZY is related to where the amyloplasts sediment in the columella cells. Because LAZY3-GFP showed a strong amyloplast-localized fluorescence signal in our obtained transgenic lines, it was selected as the representative for detailed analysis. After the seedlings were rotated 90° or 180°, we found that redistributed LAZY3-GFP proteins to the new lower side of the columella cells highly correlated with where the amyloplasts sediment (Figures 3E–3H). Similar correlation was also observed for LAZY2-GFP and LAZY4-GFP (Figure S4A). Furthermore, to corroborate this correlation, we analyzed the distribution of LAZY proteins in the pgm1 mutant. Indeed, mutation of PGM1 clearly delayed the sedimentation of plastids after 90° or 180° rotation and we observed slower redistribution of LAZY proteins to the new lower side of the columella cells (Figures 3I–3L). Further statistical analysis clearly showed that gravistimulation-induced polar redistribution of LAZY2, 3, and 4 were all delayed in the pgm1 mutant (Figures 3M–3O, and S4B). These results suggest that the sedimentation of amyloplasts promote the redistribution of LAZY proteins after gravistimulation.
LAZY proteins translocate from the amyloplasts to the plasma membrane
In 2-D observation, multiple data showed that LAZY proteins were preferentially localized on the plasma membrane regions adjacent to amyloplasts (Figures 2D–2F, 3A–3C, 3E, 3G). To further confirm this conclusion, we rotated LAZY3-GFP seedlings 90° for gravistimulation, and collected the Z-series images when the amyloplasts were close to the new lower side of the columella cells. 3-D reconstruction of the fluorescence in columella cells showed that LAZY3-GFP proteins were only localized on the plasma membrane close to where amyloplasts were present (Figure 4A, Video S1). Further, we photo-bleached the fluorescence of LAZY3-GFP on the plasma membrane in columella cells, and found that the fluorescence in the lower side of the plasma membrane could be recovered rapidly in the columella cells with unbleached amyloplast fluorescence (LOI1, Figures 4B–4D, Video S2). In contrast, fluorescence recovery at the same position was much slower in the columella cells with bleached amyloplast fluorescence (LOI2, Figures 4B–4D, Video S2). These data indicate that LAZY proteins can be translocated from amyloplasts to the adjacent plasma membrane.
Figure 4. LAZY3 proteins translocate from the amyloplasts to the plasma membrane.
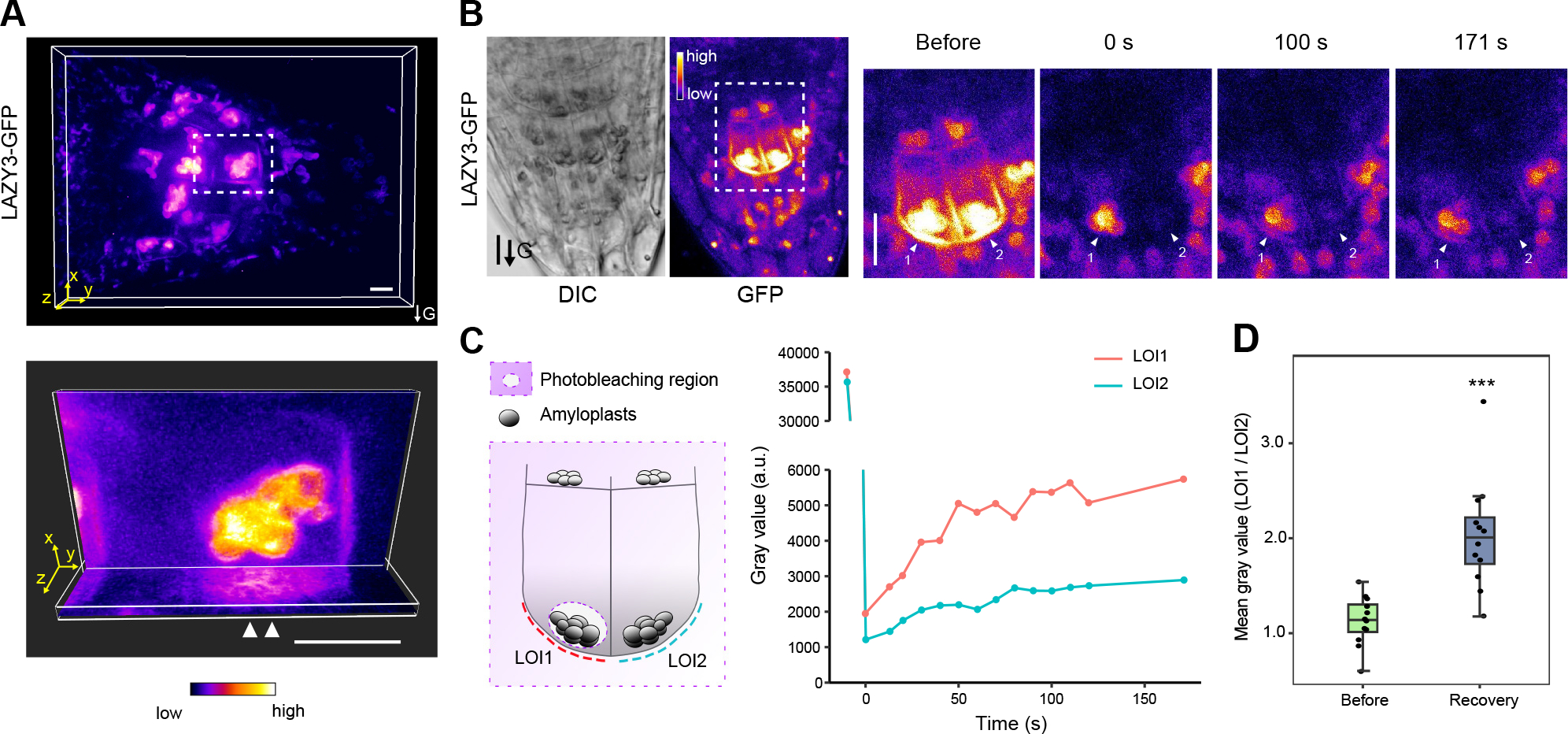
(A) 3-D reconstruction of the fluorescence of LAZY3-GFP in columella cells under gravistimulation. Seedlings of LAZY3-GFP/lazy234 were grown vertically under white light for 4 days, and then rotated 90° to the horizontal position for gravistimulation for around 0.5 h. Images were acquired using a spinning-disk confocal microscope with a z-series taken every 0.25 μm for a range of 10.5 μm. Upper, representative 3-D view of confocal image stacks of columella cells expressing LAZY3-GFP. Lower, ortho slicer view of XY plane and YZ plane of the cell in white dashed box (upper), extended section of 2.0 μm is shown for each slicer. White arrow heads indicate the accumulated signal on the lower side of plasma membrane adjacent to the amyloplasts. 3-D views were generated by Imaris. Scale bars, 10 μm. This is a representative image of 13 seedling roots.
(B) Fluorescence of LAZY3-GFP on plasma membrane in columella cells in FRAP experiment. Left, representative images of roots expressing LAZY3-GFP; right, zoomed image of the region in white dashed box (left) at different time points showing fluorescence recovery. Arrows labeled with numbers indicate a symmetrical pair of columella cells with similar fluorescence before photobleaching. LAZY3-GFP was bleached by 488 nm laser in two labeled cells and adjacent cells except for the amyloplast region in cell #1. Bleached region is also diagrammed in (C). The fluorescence was collected immediately after photobleaching (0 s) and at indicated time after photobleaching. Scale bars, 10 μm. These are representative images of 12 seedling roots.
(C) Fluorescence recovery on plasma membrane of representative cells in (B). Left, diagram showing the bleached region. LOI, line of interest on which fluorescence signal intensity was measured. Right, the recovery rate of signal intensity of LOI1 and LOI2.
(D) Statistical analysis of signal intensity ratios of LOI1/LOI2 before bleaching and after recovery in different roots. Symmetric columella cells were selected for analysis as in (B). Intensity was measured on LOI1 and LOI2 after a recovery of 100 s (***, P < 0.001; n = 12).
Gravistimulation induces phosphorylation of LAZY4 via MKK5-MPK3
To study how gravistimulation mediates the translocation of LAZY proteins, we performed Immunoprecipitation-Mass Spectrometry (IP-MS) analysis using LAZY4-GFP as the representative due to its highest expression level (based on fluorescence) among the three LAZYs. LAZY4-GFP seedlings were grown vertically or treated with gravistimulation (5 min, 10 min, 0.5 h, 2 h and 24 h) for phospho-proteomic analysis. The mass spectrometric analysis identified 10 phosphorylation sites in total, most of which showed up-regulated phosphorylation levels after gravistimulation (Figure S5 and Table S1). Phosphorylated peptide “FLNCPSSLEVDR” containing phosphorylation sites S139/S140 could be detected stably in all the replicates, thus it was selected as the representative to show the details of mass spectrometry results. Phosphorylation levels of this peptide were clearly induced by gravistimulation but returned to the original status after 24 h (Figure 5A and Table S2). Furthermore, to identify possible kinases, we searched the IP-MS data of LAZY4-GFP and found the protein abundance of two kinases, MPK3 and MKK5, was increased dramatically after gravistimulation and decreased to the original level after 24h (Figure 5B and Table S3), a pattern highly similar to the phosphorylation dynamics of LAZY4 (Figure 5A). Since 5-min gravistimulation was already sufficient to induce a strikingly increased association of MPK3 to LAZY4, as well as increased phosphorylation level of LAZY4 (Figures 5A and 5B), the phosphorylation of LAZY4 may take place shortly (within 5 min) after gravistimulation. Additionally, the MPK3 proteins co-immunoprecipitated by LAZY4 are highly phosphorylated (Figure 5C and Table S4). Consistently, in vitro pull-down experiments also showed that the interactions of LAZY4 with MPK3 and MKK5 were detectable only after the incubation with both kinases (Figure 5D). In vitro phosphorylation assay confirmed that MKK5-activated MPK3 can phosphorylate LAZY4, and the phosphorylation sites matched well with the phosphorylation sites in plants (Figure 5E and Table S1). These data suggest that gravistimulation rapidly induces the phosphorylation of LAZY4 by MKK5 and MPK3.
Figure 5. Gravity-induced phosphorylation of LAZY4 by MKK5-MPK3.
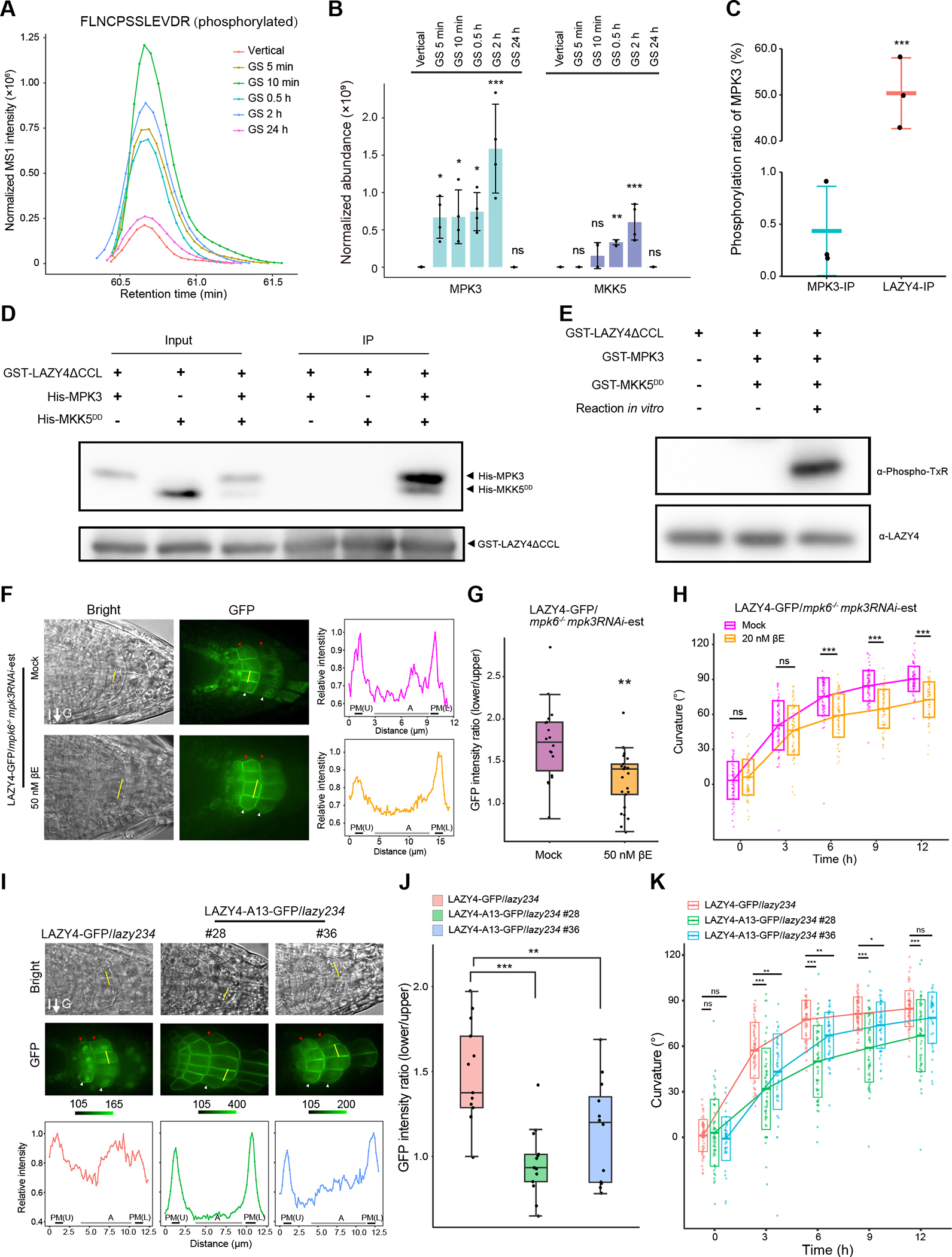
(A) Gravistimulation induces the phosphorylation of LAZY4. LAZY4-GFP seedlings were grown vertically or gravistimulated (90° reorientation) for 5 min, 10 min, 0.5 h, 2 h and 24 h. Then, LAZY4-GFP proteins were immunoprecipitated for Mass-Spectrometry (MS) analysis. The MS result was summarized in Figure S5 and Table S1, and the normalized abundance of the representative phosphopeptide “FLNCPSSLEVDR” (S139 and S140 sites) are shown. The result is a representative of four replicates.
(B) LAZY4 shows stronger interaction with MPK3 and MKK5 after gravistimulation in Arabidopsis. The original data is from the experiment in panel A, including four replicates. The barplot shows the comparison of MPK3 and MKK5 abundance co-immunoprecipitated by LAZY4-GFP (mean ± SD). MS1 abundance of all identified peptides of MPK3 and MKK5 in the MS results were summed and normalized using MS1 abundance of bait protein LAZY4-GFP. Protein abundance of MPK3 and MKK5 in different samples was compared with that in the vertical, and significant difference was evaluated by one-way ANOVA with post-hoc Dunnett’s t-test (***, P < 0.001; **, P < 0.01; *, P < 0.05; n = 4).
(C) LAZY4 prefers to interact with phosphorylated MPK3 in Arabidopsis. MPK3-GFP and LAZY4-GFP seedlings were grown vertically and then gravistimulated (90° reorientation) for 0.5 h. The phosphorylation ratio of the MPK3 peptides containing T196/Y198 sites was calculated using the MS data of MPK3-GFP IP samples (MPK3-IP) and LAZY4-GFP Co-IP samples (LAZY4-IP). Data are represented as mean ± SD. Significant differences were evaluated by Student’s t-test (***, P < 0.001; n = 3).
(D) LAZY4 interacts with MPK3 and MKK5 in vitro. Recombinant GST-LAZY4ΔCCL, His-MPK3 and His-MKK5DD proteins purified from E.coli were incubated at 4 °C overnight. For lane 3, His-MPK3 and His-MKK5DD was mixed and treated for 30 min at 37 °C with ATP, and then incubated with GST-LAZY4ΔCCL. Glutathione Sepharose 4B was used for immunoprecipitation. Pellet fractions were separated by SDS-PAGE, and visualized by anti-His and anti-GST immunoblots. GST-LAZY4ΔCCL was used since it is easier to express in E.coli than full length GST-LAZY4.
(E) MPK3 and MKK5 phosphorylate LAZY4 in vitro. The recombinant proteins were mixed and reacted as indicated. For in vitro reactions recombinant GST-MPK3 and GST- MKK5DD proteins were mixed and treated for 30 min at 37 °C with ATP, and then GST-LAZY4ΔCCL was added and treated for 60 min under the same conditions. The proteins were resolved by SDS-PAGE, and visualized by anti-Phospho-TxR (recognizing the phosphorylation of T18 in LAZY4) and anti-LAZY4 immunoblots. The LAZY4 proteins were also analyzed by Mass-Spectrometry, and the phosphorylation sites are summarized in Table S1.
(F and G) Mutation of MPK3 and MPK6 disrupts the localization of LAZY4-GFP onto the surface of amyloplasts and polar redistribution of LAZY4-GFP on the plasma membrane. (F) LAZY4-GFP/mpk6−/−mpk3RNAi-est seedlings were grown vertically on MS plates containing DMSO (Mock) or 50 nM β-estradiol (βE) to induce RNAi of MPK3 expression for 3 days. The seedlings were transferred to MS plates containing 10 μM CHX to inhibit protein synthesis, and reoriented 90° for 0.5 to 1-h gravistimulation. Left and middle, representative confocal images of LAZY4-GFP; Right, fluorescence intensity profile along the yellow line marked in the representative cells. Max intensity in each profile was set to 1.0, and relative intensity was calculated for plots. PM (U), upper side of the plasma membrane; PM (L), lower side of the plasma membrane; A, amyloplasts. Red and white arrowheads indicate the accumulation of LAZY4-GFP proteins on the upper and lower sides of columella cells, respectively. (G) Statistical analysis of fluorescence intensity ratios of the two sides of columella cells (Basal/Apical) in panel F. Asterisks indicate Student’s t-test values (**, P < 0.05; n = 18 and 24 respectively).
(H) Mutation of MPK3 and MPK6 delays the gravitropic responses of roots. LAZY4-GFP/mpk6−/−mpk3RNAi-est seedlings were grown vertically under white light on MS plates containing DMSO (Mock) or 20 nM βE to induce RNAi of MPK3 expression. Seedlings were grown vertically for 4 days and reoriented 90° for gravistimulation. The crossbars indicate mean ± SD. At each time point, significant difference of root tip angles between mock group and treated group was evaluated by Student’s t-test (***, P < 0.001; ns, not significant, P > 0.05; n = 72 and 65 respectively).
(I and J) Mutation of phosphorylation sites within LAZY4 disrupts the localization of LAZY4-GFP onto the surface of amyloplasts and polar redistribution of LAZY4-GFP on the plasma membrane. (I) Localization of Arabidopsis LAZY4-GFP and LAZY4-A13-GFP proteins in columella cells under gravistimulation for 0.5 to 1 h with CHX treatment. Upper and middle, representative confocal images of LAZY4-GFP and LAZY4-A13-GFP; Lower, fluorescence intensity profile along the yellow line marked in the representative cells. The method for profile analysis is the same as in panel F. Scale bars, 10 μm. Red and white arrowheads indicate the upper and lower sides of columella cells respectively. (J) Statistical analysis of fluorescence intensity ratios of the two sides of columella cells (Basal/Apical) in panel I. Fluorescence intensity ratios were compared with the first group, and significant difference was evaluated by one-way ANOVA with post-hoc Dunnett’s t-test (***, P < 0.001; **, P < 0.01; n = 13, 13 and 12 respectively).
(K) Mutation of phosphorylation sites disrupts the gravitropic responses of roots. LAZY4-GFP/lazy234 and LAZY4-A13-GFP/lazy234 seedlings were grown vertically for 4 days, and then rotated 90° to the horizontal position for gravistimulation. The crossbars indicate mean ± SD. At each time point, root tip angles were compared with the first group, and significant difference was evaluated by one-way ANOVA with post-hoc Dunnett’s t-test (***, P < 0.001; **, P < 0.01; ns, not significant, P > 0.05; n = 66, 55 and 48 respectively).
In F and I, arrows labeled “G” indicate the direction of gravity.
MPK3 and MPK6 are often functionally redundant in regulating developmental processes and the null allele of the double mutant is seedling lethal43. To evaluate the regulation of LAZY4 by MPK3 and MPK6 in plants, we reduced the expression of MPK3 via inducible RNAi in mpk6 using the mpk6−/−mpk3RNAi-est line45. We found that the double mutants disrupted the localization of LAZY4-GFP onto the surface of amyloplasts and polar redistribution of LAZY4-GFP on the plasma membrane under gravistimulation (Figures 5F and 5G), resulting in delayed gravitropic response (Figure 5H). Further, we mutated all 13 phosphorylation sites identified in plants and by in vitro kinase assays to be Ala (A) (Figure S5 and Table S1), which was named as LAZY4-A13, a phosphodead version of LAZY4. We transformed LAZY4-A13-GFP into the lazy234 mutant, and selected the lines with fluorescence intensity comparable to LAZY4-GFP for further analysis. Like the effects of the mutation of MPK3 and MPK6, these mutations on phosphorylation sites disrupted the localization of LAZY4-A13-GFP onto the surface of amyloplasts and polar redistribution of LAZY4-A13-GFP on the plasma membrane under gravistimulation (Figures 5I and 5J). In consistent, these mutations disrupted the functions of LAZY4 in complementing the gravitropic defects of lazy234 mutants (Figure 5K). Together, these data demonstrated that phosphorylation of LAZY4 by the MAPK cascade is important for its function in gravitropism.
Phosphorylation of LAZY4 promotes its interaction with TOC proteins on the surface of amyloplasts
Previous studies showed that mutating genes encoding TOC34, TOC75, TOC120 and TOC132, components of the Translocon of Outer Membrane of Chloroplasts (TOC) complexes, locating on the surface of both chloroplasts and amyloplasts, enhanced the gravitropic defects of arg136,37. Thus, these TOC proteins played positive roles in gravitropism, but the underlying mechanism was unclear. Since LAZY proteins also localize on the surface of amyloplasts (Figures 2 and 3), and the phosphorylation of LAZY4 was induced by gravistimulation (Figure 5), we examined the possible interactions between LAZY4 and TOC proteins and studied whether phosphorylation affected the interactions. Among the 13 phosphorylation sites, we assayed all of them by gradually mutating them to Ala (A) or Asp (D), i.e. LAZY4-A5/D5, the 5 Ser/Thr sites located in the phosphorylated peptide in domain I; LAZY4-A9/D9, the 9 sites located in domain I and III; and LAZY4-A13/D13, all 13 sites mutated (detailed in Table S1). Using the yeast two-hybrid assay, we found that LAZY4 and phosphodead LAZY4 (A5, A9 and A13) did not interact with any TOC proteins (Figures 6A, S6A, S6B and Table S1). In contrast, one phosphomimic LAZY4 variant (LAZY4-D9) interacted strongly with TOC34, TOC120 and TOC132, all of which are receptors on the outer membrane of plastids and usually used for importing non-photosynthetic preproteins (Figures 6A, S6A, S6B and Table S1). Previous study showed that a TOC132 variant without N-terminus that only retains the GTP-binding and membrane domains (TOC132GM) could restore the gravitropic response of arg1 toc132 to that of arg1 single mutant37. We found that all three phosphomimic LAZY4 variants (LAZY4-D5, LAZY4-D9, and LAZY4-D13) showed strong interaction with TOC132GM (Figures 6A, S6A, S6B, and Table S1). In contrast, the central pore component TOC75 did not interact with these phosphomimic LAZY4 proteins. Since LAZY4-D9 showed the strongest interaction with TOC proteins among the LAZY4 variants, it was selected to be transformed into the lazy234 mutant for examination of localization and results showed a high co-localization pattern with TOC132-RFP around the amyloplasts in the columella cells (Figures 6B and 6C). These data suggest that phosphorylation of LAZY4 proteins enhances their interaction with the TOC proteins facing the cytosol, which may promote the translocation of LAZY4 proteins onto the surface of amyloplasts.
Figure 6. Phosphorylation of LAZY4 promotes its interaction with TOC proteins, which control the redistribution of LAZY4 and the gravitropic response.
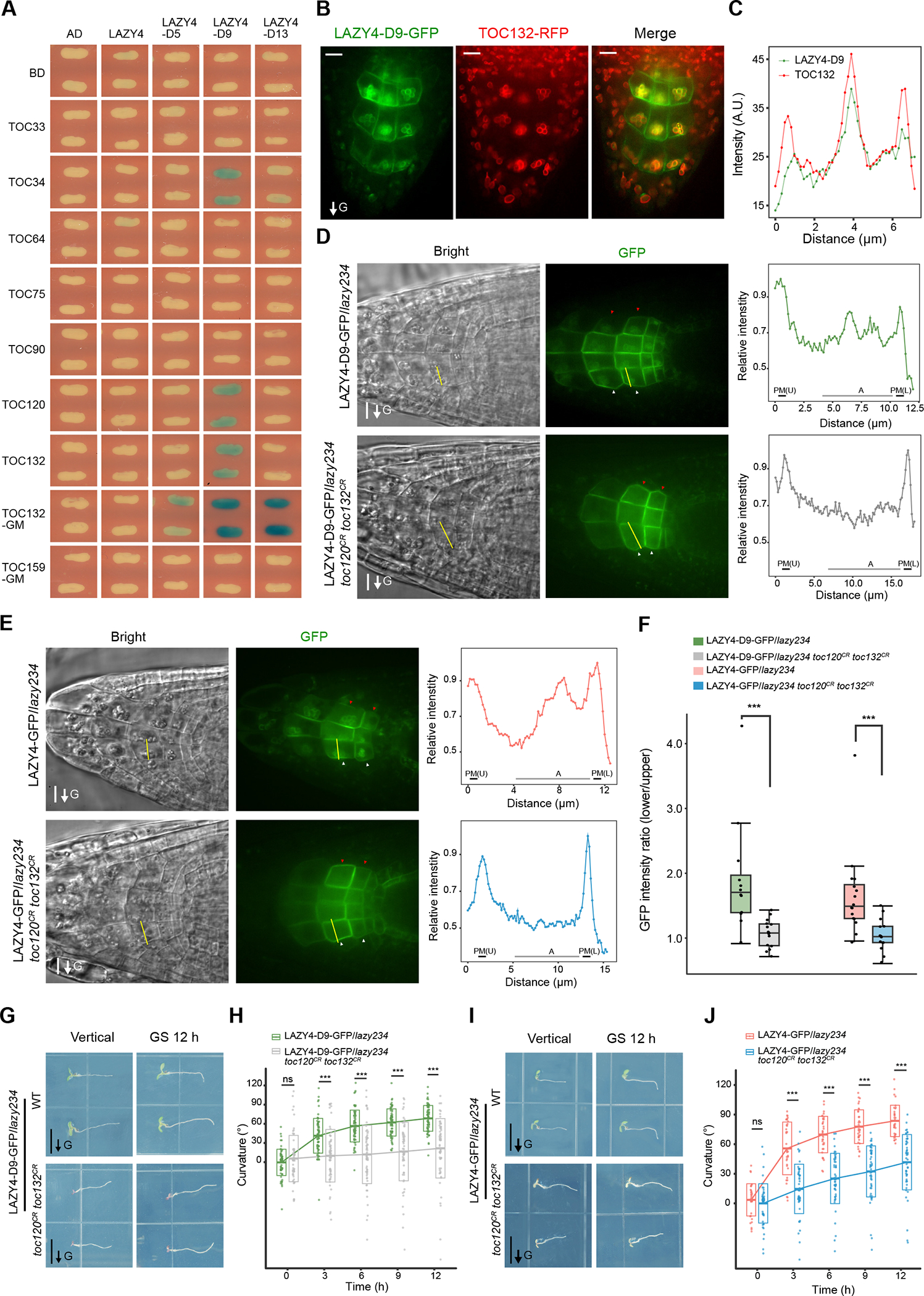
(A) Yeast two-hybrid assays to test interactions between LAZY4 variants and TOC proteins. The phosphorylation sites of LAZY4 were mutated to Asp (D) as indicated in Table S1. Blue indicates positive interactions. Reporter: LacZ; substrate, X-Gal. TOC132GM and TOC159GM indicate the GTP-binding and membrane domains of these TOC proteins. The yeasts were grown for 24 h, and results from 40-h growth with more LAZY4 variants are shown in Figure S6B.
(B and C) Co-localization of LAZY4-D9-GFP and TOC132-RFP in root columella cells. (B) TOC132-RFP was transformed into LAZY4-D9-GFP/lazy234, and the seedlings were grown vertically on MS plates for 4 days. Root tip fluorescence was collected. Scale bars, 10 μm. (C) Statistical analysis of fluorescence intensities of LAZY4-D9-GFP and TOC132-RFP in columella cells. Green and red lines show the respective fluorescence intensities of GFP and RFP along the yellow dotted line marked in panel B. a.u., arbitrary units. These are representative images of roots of 20 seedlings.
(D to F) Mutation of TOC120 and TOC132 disrupts the localization of LAZY4 onto the surface of amyloplasts and polar redistribution of LAZY4 on the plasma membrane. (D) LAZY4-D9-GFP/lazy234 and LAZY4-D9-GFP/lazy234 toc120CR toc132CR seedlings were grown vertically on MS plates for 4 days. The seedlings were transferred to other MS plates, and reoriented 90° for 0.5 to 1-h gravistimulation. Red and white arrowheads indicate the accumulation of LAZY-GFP proteins on the upper and lower sides of columella cells, respectively. Left and middle, representative confocal images of LAZY4-GFP; Right, fluorescence intensity profile along the yellow line marked in the representative cells. Max intensity in each profile was set to 1.0, and relative intensity was calculated for plots. Scale bars, 10 μm. PM (U), upper side of the plasma membrane; PM (L), lower side of the plasma membrane; A, amyloplasts. (E) LAZY4-GFP/lazy234 and LAZY4-GFP/lazy234 toc120CR toc132CR seedlings were grown vertically on MS plates for 4 days. The seedlings were transferred to MS plates with 10 μM CHX to inhibit protein synthesis during the 0.5 to 1-h gravistimulation. The following experiments and analysis were the same as in panel D. Scale bars, 10 μm. (F) Statistical analysis of fluorescence intensity ratios of the two sides of columella cells (Basal/Apical) in panel D and E. Asterisks indicate Student’s t-test values (***, P < 0.001; n = 13, 15, 16 and 14 respectively).
(G to J) Mutation of TOC120 and TOC132 disrupts the gravitropic responses of roots. Seedlings were grown vertically on MS plates for 4 days, and then rotated 90° to the horizontal position for gravistimulation. (G and I) Representative seedlings. Scale bars, 0.5 cm. (H and J) The crossbars indicate mean ± SD. At each time point, significant differences of root tip angles between two groups were evaluated by Student’s t-test (***, P < 0.001; ns, not significant, P > 0.05; H, n = 71 and 65 respectively; J, n = 35 and 53 respectively).
TOC proteins are essential for amyloplast localization and polar redistribution of LAZY4 proteins
To analyze the physiological significance of the interaction between phosphorylated LAZY4 and TOC proteins, we mutated TOC120 and TOC132 simultaneously with CRISPR/Cas9 in LAZY4-D9-GFP/lazy234 and LAZY4-GFP/lazy234 since these two TOC genes showed redundancy in chlorophyll synthesis (Figure S6A) 38,39. The strong association of LAZY4-D9 with the amyloplast was disrupted by mutations in TOC120 and TOC132 (Figures 6D and S6C), indicating that the localization of LAZY4 on amyloplasts is dependent on TOC120 and TOC132. Gravistimulation-induced polar redistribution of LAZY4-D9 was disrupted in this toc120CR toc132CR mutant background (Figures 6D and 6F). Consistently, mutation of TOC120 and TOC132 disrupted the localization of LAZY4-GFP proteins onto the amyloplasts and their polar redistribution on the plasma membrane (Figures 6E, 6F, and S6C). Both materials with toc120 and toc132 mutations showed strong gravitropic defects (Figures 6G–6J). Thus, these TOC proteins are the key anchor proteins on the surface of amyloplasts that bind and facilitate the redistribution of LAZY proteins that are required for asymmetric root growth in plant gravitropic responses.
DISCUSSION
Gravity sensing, signal transduction, and differential growth are three sequential processes in plant gravitropism2,5. The starch-statolith hypothesis is the most widely-accepted theory explaining gravity sensing and is considered a dogma in the field of plant gravitropism5–9,51, but the underlying molecular effects of amyloplast sedimentation are unknown. Here, we revealed that the molecular effect of amyloplast sedimentation is to promote the redistribution of LAZY proteins via the TOC proteins on the amyloplast surface. Based on this and previous studies, we suggest the following model for gravity sensing in plants (Figure 7): (i) During regularly vertical growth, amyloplasts settle to the bottom in root columella cells, and more LAZY proteins accumulate on the lower side of the plasma membrane. (ii) When plants tilt relative to the direction of gravity vector, gravistimulation induces the interaction between MKK5-MPK3 kinases and LAZY, resulting in increased phosphorylation of LAZY proteins. (iii) Phosphorylated LAZY proteins show stronger interactions with TOC34/120/132 proteins on the surface of amyloplasts, which may promote their translocation from the plasma membrane to the amyloplasts. (iv) Amyloplast sedimentation brings and guides the LAZY proteins toward the new lower side of columella cells. In this process, LAZY proteins may keep on trafficking between the amyloplasts and adjacent plasma membrane. When the amyloplasts approach the new bottom, LAZY proteins are translocated from the surface of amyloplasts to the new lower side of the plasma membrane, re-establishing their polarity to promote asymmetric distribution of auxin and bending growth. (v) When the roots resume vertical growth, the LAZY proteins are dephosphorylated by unknown mechanisms, returning to their original state. Although the underlying mechanism of gravistimulation-triggered interaction between MPK3 and LAZY4 still need further investigation, the phosphorylation status of LAZY4 and its interaction with MPK3 can be regarded as two molecular indicators of plant responses to gravistimulation (Figure 5), which must be very useful in future studies in this field. Polarized LAZYs may regulate the asymmetric distribution of auxin through RLD and PIN proteins to mediate differential growth33. Since LAZY orthologs are prevalent and have also been demonstrated to regulate gravitropism in many other plant species, including rice3,21–23, maize20,24, Medicago truncatula28, Prunus domestica26 and Lotus japonicus29, and TOC proteins are conserved in plant kingdom38–41, it is possible that the mechanism revealed in this study is also applicable to other plants.
Figure 7. Model for gravity sensing in plant roots.
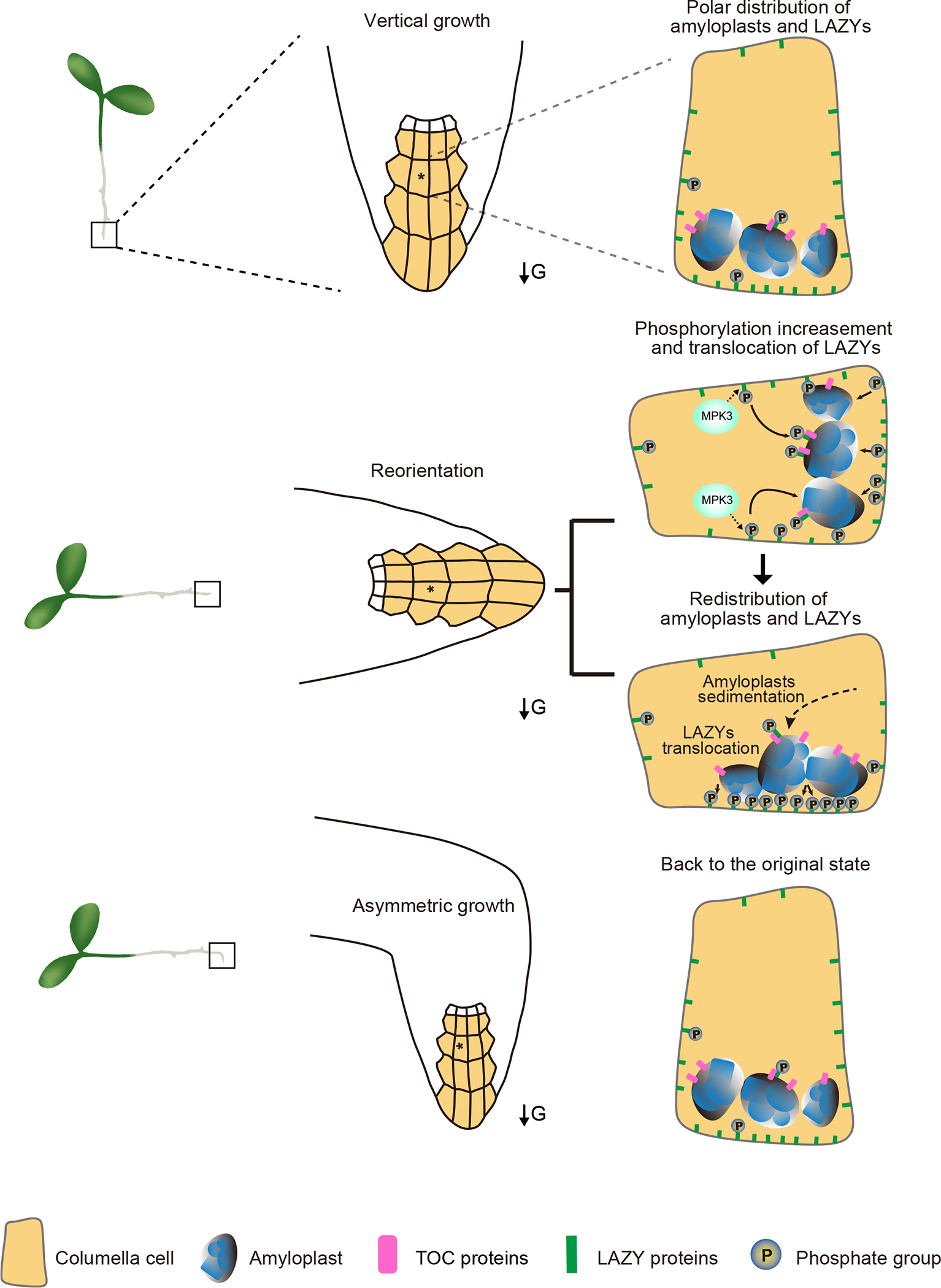
Under vertical growth, more LAZY proteins accumulate on the lower side of the plasma membrane in columella cells of roots. Gravistimulation via reorientation triggers the interactions between the MKK5-MPK3 kinase module and LAZY proteins, resulting in increased phosphorylation of LAZY proteins. Subsequently, phosphorylated LAZYs may translocate onto the surface of amyloplasts by directly interacting with TOC proteins. Amyloplast sedimentation guides the LAZY proteins to distribute onto the new lower side of the plasma membrane in columella cells, in which relocation of LAZY proteins from the surface of amyloplasts to the adjacent plasma membrane is an important process. The repolarization of LAZY proteins induces asymmetrical auxin distribution and differential growth. When the roots resume vertical growth, the phosphorylation status and distribution of LAZY proteins return to their original state. The square frames indicate the root tips. The stars indicate the representative columella cells in root tips. Arrows labeled “G” indicate the direction of gravity.
For the starch-statolith hypothesis, two models have been proposed to explain how the physical stimulus of amyloplast sedimentation is transformed into a biochemical signal responsible for the gravitropic curvature in plants52. One model postulates that sedimenting statoliths may activate mechano-sensitive ion channels via exerting a pressure on sensitive membranes or cytoskeletons within the statocyte cells53,54, but no such plant mechanosensitive ion channel has been identified. Another model proposes that the contact of statoliths with membrane-bound receptors rather than pressure or tension exerted by the weight of statoliths may achieve graviperception55, but the information about the receptor is unavailable. Our study proposes a model solely involving organelle movement that triggers the repolarization of the key gravitropism regulator LAZYs (Figure 7), suggesting that the molecular effect of amyloplast sedimentation probably doesn’t need mechano-sensitive ion channels or receptors as proposed in the previous models. Interestingly, although the classical function of TOC proteins is to import proteins into plastids38–41, it seems that relocation of LAZYs does not require them to be imported into amyloplasts (Figures 2, 3, 5 and 6). This unconventional working mechanism of TOCs makes repolarization of LAZYs simple and fast, resulting in a rapid gravitropic response. Since both movement of organelles and polarity formation are common phenomena, by revealing that the movement of a specific organelle directly triggers the protein re-polarity within cells, our study may also inspire other studies related to polarity.
Limitations of the study
Although our study provides critical insights into the molecular mechanism for the starch-statolith hypothesis, there are a few open issues. First, the underlying mechanism of gravistimulation-triggered interaction between MKK5-MPK3 and LAZY proteins needs further studies. What are the earliest signaling events after gravistimulation and whether MAPKKKs and/or other signaling molecules participate in this process remain the most exciting and challenging questions in the field. Second, we clearly showed that phosphorylation of LAZY4 increased its interaction with TOC proteins, and mutating TOCs disrupted the localization of LAZY4 onto amyloplasts, resulting in severe gravitropic defects. Mutating the 13 phosphorylation sites within LAZY4 leads to obvious gravitropic defects, but the defects are not as severe as lazy null mutant or toc120CR toc132CR mutants. It is possible that more phosphorylation sites than we identified are involved in the interactions between LAZY and TOC proteins in vivo. Alternatively, there may exist other uncharacterized mechanisms besides phosphorylation that also contribute to the interaction. Third, the detailed trafficking processes of LAZY proteins between amyloplasts and the plasma membrane need future investigation. For example, how LAZY proteins are unloaded from amyloplasts and how the MAPK-mediated phosphorylation events crosstalk with the RLD-mediated cellular processes to ensure the changes of LAZY polarization in gravitropism.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Haodong Chen (chenhaodong@tsinghua.edu.cn)
Materials availability
Constructs and plant seeds generated in this study will be available from the lead contact upon request.
Data and code availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD045213.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Plant materials and growth conditions
All wild-type plants used in this study were the Columbia ecotype (Col) of Arabidopsis thaliana. Several mutants and transgenic lines used in this study were described previously: pgm113,14, mpk6−/−mpk3RNAi-est line45, pMPK3-MPK3-GFP45 and DR5rev::GFP48,56. Generation of plant materials was described in the method details.
The seeds were surface-sterilized by soaking in 15% (v/v) NaClO for around 10 min and washing in sterile distilled water for at least three times. After surface-sterilization, the seeds were plated on MS medium (4.4 g/L MS, 1% (w/v) sucrose, 0.8–1% (w/v) agar, pH 5.8), and cold-treated at 4 °C for 2 to 3 days in the dark. The seedlings were grown under continuous white light (70 μmol m−2 s−1) or dark conditions as described in the figure legends. For growing in the dark, the plated seeds were illuminated under white light (70 μmol m−2 s−1) for 6 h to induce germination before dark incubation. Arabidopsis seedlings were grown at 22 °C in a growth chamber. Adult Arabidopsis plants were grown under a 16 h/8 h day/night photoperiod at 22 °C.
METHOD DETAILS
Generation of mutants and transgenic plants
For mutating Arabidopsis LAZY2, LAZY3, and LAZY4 simultaneously, we generated a modified Cas9 vector named as pEC-Cas9. First, rbcS-E9 terminator was cloned from pHEE401E57, and inserted between the BamH I and EcoR I sites of p35S-Cas9-SK58 to replace the Nos terminator, resulting in p35S-Cas9-rbcS-E9t. Second, p35S-Cas9-rbcS-E9t was further modified by deleting a Xho I site upstream of the double CaMV 35S promoter and adding a Nhe I site between Sal I and Hind III to generate p35S-Cas9-rbcS-E9t-M. Third, Ec1.2enEc1.1 promoter was cloned from pHEE401E57 and inserted between Nhe I and Xho I sites to generate pEc1.2enEc1.1p-Cas9-rbcS-E9t (abbr.: pEC-Cas9). Two sgRNAs were designed for each LAZY gene (LAZY2, LAZY3, and LAZY4), and were inserted into the Bbs I sites of the pAtU6-26-M59. The primers used for annealing are listed in Table S5. The expression cassettes of six sgRNAs were digested with Kpn I and Sal I and ligated into pEC-Cas9. Then, the Cas9 cassettes were subcloned into the Kpn I and EcoR I sites of pCAMBIA1300 (Hyg) and pJIM19 (Gent) vectors to generate pCAMBIA1300-LAZY2/3/4 sgRNA and pJIM19-LAZY2/3/4 sgRNA. These binary constructs were transformed into wild type and DR5rev::GFP to generate lazy2 lazy3 lazy4 (lazy234) and DR5rev::GFP/lazy234 mutant lines. The mutants were analyzed using the primers listed in Table S5, and the lines lazy234-1, lazy234-2, lazy234-3, and DR5rev::GFP/lazy234 were obtained.
To construct LAZY-GFP transgenic lines driven by the native promoter, we amplified the genomic DNAs of LAZY2, LAZY3 and LAZY4 including promoter regions by the primers listed in Table S5. Genomic fragments of LAZY2, LAZY3 and LAZY4 fused with GFP at the C-terminus were inserted into the SbfI/Xba I, SbfI/Xho I and SbfI/Spe I sites of the plant binary vector pJIM19 (Bar) to construct pJIM19-LAZY2-GFP, pJIM19-LAZY3-GFP and pJIM19-LAZY4-GFP. Then, these LAZY-GFP constructs were transformed into wild type (Col) and lazy234 respectively, and transgenic plants were selected with glufosinate ammonium (20 μg/mL). The obtained plants include ProLAZY2:LAZY2-GFP (abbr.: LAZY2-GFP/Col, only used in lateral roots fluorescence analysis in Figures S3F and S3G), ProLAZY2:LAZY2-GFP/lazy234 (abbr.: LAZY2-GFP/lazy234 or LAZY2-GFP), ProLAZY3:LAZY3-GFP/lazy234 (abbr.: LAZY3-GFP/lazy234 or LAZY3-GFP), and ProLAZY4:LAZY4-GFP/lazy234 (abbr.: LAZY4-GFP/lazy234 or LAZY4-GFP). These transgenic lines were crossed with pgm1 to generate LAZY2-GFP/lazy234 pgm1 (abbr.: LAZY2-GFP/pgm1), LAZY3-GFP/lazy234 pgm1 (abbr.: LAZY3-GFP/pgm1) and LAZY4-GFP/lazy234 pgm1 (abbr.: LAZY4-GFP/pgm1). LAZY4-GFP/lazy234 was crossed with mpk6−/−mpk3RNAi-est line to generate LAZY4-GFP/lazy23 mpk6−/−mpk3RNAi-est (abbr.: LAZY4-GFP/mpk6−/−mpk3RNAi-est).
To construct GFP-fused truncated LAZY driven by the LAZY native promoter, we amplified the promoter and truncated coding sequences of LAZY using the primers listed in Table S5. The LAZY2ΔN and LAZY4ΔN related fragments were inserted into pCAMBIA1300 (Hyg), and LAZY3ΔN related fragments were inserted into pJIM19 (Bar). Then, these constructs were transformed into lazy234 triple mutants. Transgenic plants were selected with hygromycin B (50 μg/mL) or glufosinate ammonium (20 μg/mL). The obtained plants include ProLAZY2:LAZY2ΔN-GFP/lazy234 (abbr.: LAZY2ΔN-GFP/lazy234), ProLAZY3:LAZY3ΔN-GFP/lazy234 (abbr.: LAZY3ΔN-GFP/lazy234), and ProLAZY4:LAZY4ΔN-GFP/lazy234 (abbr.: LAZY4ΔN-GFP/lazy234).
To construct ProLAZY4:LAZY4-D9-GFP/lazy234 and ProLAZY4:LAZY4-A13-GFP/lazy234 transgenic plants, we amplified the promoter and coding sequences of LAZY4 from wild type plants using the primers listed in Table S5. Point mutations were generated by overlap PCR and the mutation sites are listed in Table S1. Then, the fragments with GFP were inserted into pJIM19 (Bar), and transformed into lazy234 triple mutants. Transgenic plants were selected with glufosinate ammonium (20 μg/mL). The obtained plants include ProLAZY4:LAZY4-D9-GFP/lazy234 (abbr.: LAZY4-D9-GFP/lazy234) and ProLAZY4:LAZY4-A13-GFP/lazy234 (abbr.: LAZY4-A13-GFP/lazy234).
To construct LAZY4-GFP/lazy234 toc120CR toc132CR and LAZY4-D9-GFP/lazy234 toc120CR toc132CR, we designed two sgRNAs for each TOC (TOC120 and TOC132), and were inserted into the Bbs I sites of the pAtU6-26-M56 vector. The primers used for annealing are listed in Table S5. The expression cassettes of four sgRNAs were digested with Kpn I and Sal I and inserted into the Kpn I and EcoR I sites of pUBQ10:Cas9-P2A-GFP (Gent) 59 vector. This construct was transformed into LAZY4-GFP/lazy234 and LAZY4-D9-GFP/lazy234 transgenic lines. The mutants were analyzed using the primers listed in Table S5.
To construct TOC132-RFP/LAZY4-D9-GFP/lazy234 transgenic lines, we amplified the genomic DNAs of TOC132 including promoter regions and coding sequences by the primers listed in Table S5. Genomic fragments of TOC132 were inserted into the Sal I and BamH I sites of the plant binary vector pCAMBIA1300(Hyg)-mRFP. Then, the pCAMBIA1300(Hyg)-TOC132-mRFP construct was transformed into LAZY4-D9-GFP/lazy234 transgenic lines, and transgenic plants were selected with hygromycin B (50 μg/mL).
Gravitropic phenotype analyses
In experiments studying gravitropism of primary roots under regular growth, seedlings were grown 4 days on vertically-oriented MS plates under white light or darkness. In experiments studying the tropic responses of primary roots after gravistimulation, seedlings were grown vertically under white light for around 4 days, and then the plates were re-orientated 90°. In experiments studying growth orientations of lateral roots (LR), seedlings were grown vertically on MS plates under white light. Then, the primary roots were aligned to the gravity vector on the 7th day, and the plants continued to grow for several additional days. Root growth angles were measured by Image J software60. The distribution frequency of root growth angles was plotted using the R statistics program (http://www.rproject.org).
Microscopic observation and analyses
For fluorescence observation of Arabidopsis seedlings under vertical growth or gravistimulation, the fluorescent images were collected using the confocal laser scanning microscope (Zeiss LSM800) or spinning-disk confocal microscopy (Andor Dragonfly 200) with a vertical objective table. 488 nm laser was used to detect the fluorescence of LAZY-GFP (including mutated variants), and 561 nm laser was used to detect FM4-64 or TOC132-RFP. Differential interference contrast microscopy (DIC) was used to collect the images of the amyloplasts. Around four-day-old seedlings grown under white light were used for fluorescence observation unless specified otherwise.
Signal intensity was calculated by ImageJ60. Signal intensity ratios of LAZY-GFP proteins in columella cells after gravistimulation were calculated by comparing the fluorescence of the lower outer and upper outer lateral plasma membranes of central columella cells or lateral columella cells (the columella cells adjacent to central columella cells). The mean gray value of several regions of interest was calculated as the background, and subtracted from the signal intensity of the images acquired by spinning-disk confocal microscopy. Signal ratio for one root was calculated as an average of signal intensity ratios within this root61. The details of signal intensity ratio analysis are shown in Figures S3D and S3E.
Immuno-electron microscopy
The roots of LAZY4-GFP/lazy234 seedlings were fixed by high-pressure freezing (HPF, Leica EM HPM100) with 5% sucrose as a cryoprotectant. The freeze-substitution solution was 0.4% (w/v) uranyl acetate in acetone, and the freeze-substitution was carried out as follows. First, the samples were kept at −90 °C for 48 hours, and then the temperature was slowly raised to −50 °C in 20 hours. Then, the samples were rinsed in ethanol for three times in two hours. Second, the temperature was slowly raised to −35 °C and kept for 48 hours, and then the solution was changed to Lowicryl HM20 resin (Electron Microscopy Sciences, 14340) with gradually increasing concentration (50%, 75%, 100% v/v in ethanol). The samples in pure resin were raised to room temperature, and the resin was changed for another three times. Third, the resin was polymerized with UV at −35 °C for 48 h in a Leica AFS apparatus. Resin blocks with samples were cut into 80-nm ultrathin sections and mounted on nickel grids with a single slot, and blocked with PBS containing 0.5% BSA-c (Aurion, 900.022) and 0.05% Tween for 10 min. GFP antibody (1:10, Abcam, ab290) diluted in blocking solution was added on each cover sheet, and kept for 30 min at room temperature. Then, the sections were washed 5 times with PBS (2 min for each wash), and labeled with protein A Gold 10 nm (1:50; Dept. of Cell Biology, UMC Utrecht, the Netherlands) for 30 min at room temperature. After several washes with PBS, the sections were fixed in 1% (w/v) glutaraldehyde, washed with distilled water and counterstained with 2% uranyl acetate solution. The sections on grids were imaged at 80 kV in a JEOL JEM-1400 Flash (HC) TEM using a CMOS camera (XAROSA, EMSIS).
Protein-lipid overlay assay
Recombinant GST-tagged proteins were expressed and purified using E.coli Transetta (DE3). Strips with 15 different phospholipids on a nitrocellulose membrane were blocked in blocking buffer (3% w/v BSA in PBST) at 4 °C overnight. Then, 0.5 μg/mL GST-tagged proteins in blocking buffer were incubated with the membranes for 1 h at room temperature. The membranes were washed three times with PBST, and then incubated with anti-GST (1:2000 in blocking buffer) for 1 h at room temperature. Further, the membranes were washed three times with PBST, and then incubated with second antibody (1:8000 in blocking buffer) for 1 h at room temperature. Chemiluminescence were detected using GE Healthcare Amersham ECL kit.
Immunoprecipitation-Mass Spectrometry (IP-MS)
The following buffers were prepared for carrying out IP-MS on LAZY4-GFP and MPK3-GFP. Lysis buffer: 1x RIPA buffer (Abcam: ab156034), 1 mM PMSF, 1x protease inhibitor cocktail (Mei5bio: MF182-plus-01), and 1x phosphatase inhibitor cocktail (Mei5bio: MF183-01). Dilution buffer: 50 mM Tris-HCl (pH 7.5), 1 mM Beta-glycerophosphate, 1 mM Sodium orthovanadate, 0.5% (w/v) Sodium deoxycholate, 1mM EGTA, 1mM EDTA, and 150 mM Sodium chloride. Wash buffer: 100 mM Tris-HCl (pH 7.5), 10 mM EDTA, 10 mM EGTA, and 5 mM KCl. Four-day-old Arabidopsis seedlings (around 4 g) were collected and ground into powder with liquid nitrogen, and proteins were extracted using lysis buffer and diluted to around 1 mg/mL with dilution buffer. Immunoprecipitation was then performed with GFP-Trap beads (ChromoTek, gtma-20), using 50 μL GFP-Trap beads for around 6 mg proteins. The proteins were eluted from the beads with 50–60 μL 0.2 M glycine solution (pH 2.5) by gently shaking for 30–60 s, followed by magnetic or centrifuging separation. Then, the supernatant was transferred into another empty centrifuge tube and 10% (v/v) Tris-base (pH 10.4) was added into the tube.
The protein samples were analyzed on an Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Thermo). After dithiothreitol reduction and iodoacetamide alkylation, the proteins were digested with trypsin (Sequencing grade modified; Promega) overnight at 37 °C. The resulting tryptic peptides were desalted with C18 tips (Pierce #87784) and dried in a vacuum centrifuge concentrator. The peptides were resolved using 0.1% formic acid and loaded onto a trap column (Acclaim PepMap™ 100 75 μm × 2 cm nanoViper, C18, 3 μm, 100Å, Thermo) connecting to an analytical column (Acclaim PepMap™ RSLC 75 μm × 15 cm nanoViper, C18, 2 μm, 100Å, Thermo) on a nanoflow HPLC Easy-nLC 1200 system (Thermo Fisher Scientific), using a 75 min LC gradient at 280 nL/min. Buffer A is consisted of 0.1% (v/v) formic acid in H2O and Buffer B is consisted of 0.1% (v/v) formic acid in 80% acetonitrile. The gradient was set as follows: 4%–8% B in 5 min; 8%–20% B in 45 min; 20%–30% B in 10 min; 30%–90% B in 13 min; 90% B in 2 min. Proteomic analyses were performed on a Thermo Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) using a nano-electrospray ion source with electrospray voltages of 2.2 kV. Data-dependent acquisition was performed using Xcalibur software in profile spectrum data type. The MS1 full scan was set at a resolution of 120,000 @ m/z 200, AGC target 5e5 and maximum IT 90 ms by orbitrap mass analyzer (300–1500 m/z), followed by MS2 scans generated by HCD fragmentation at a resolution of 30,000 @ m/z 200, AGC target 5e4. The fixed first mass of MS2 spectrum was set as 110.0 m/z. Isolation window was set as 1.6 m/z. The normalized collision energy (NCE) was set as NCE 30%.
The MS spectrometry data were analyzed by software Proteome Discoverer or Maxquant. The phosphorylation sites of LAZY4 or MPK3 were identified based on MS2 results. MS1 area intensities of phosphorylated peptides were calculated for quantification, normalized by the total area intensity of phosphorylated and non-phosphorylated peptides. For analyzing the phosphorylation ratio of MPK3 Co-Immunoprecipitated by LAZY4-GFP, two replicates detecting the peptide containing the key phosphorylation sites T196/Y198 at GS 0.5 h from the time series experiments were used, and the third replicate was carried out additionally.
GST pull-down
Recombinant proteins were expressed and purified using E.coli Transetta (DE3). Recombinant His-tagged MPK3 (~10 μg) was activated by incubation with recombinant His-MKK5DD (~1 μg, mutated MKK5 with constitutive activity) in the presence of 50 μM ATP in 50 μL of reaction buffer (25 mM Tris, pH 7.5, 10 mM MgCl2 and 1mM DTT) at 37 °C for 30 min. These proteins were mixed with GST-LAZY4ΔCCL (~100 μg) at 4 °C overnight, and then incubated with Glutathione Sepharose 4B (GE Healthcare) for 3 h at 4 °C. Alternatively, these proteins were mixed with GST-LAZY4ΔCCL and incubated with Glutathione Sepharose 4B (GE Healthcare) at 4 °C overnight. Resins were washed three times with wash buffer (First, 50 mM Tris-HCl pH 7.5, 150 mM NaCl; Second, 50 mM Tris-HCl pH 7.5, 200 mM NaCl; Third, 50 mM Tris-HCl pH 7.5, 250 mM NaCl). Pellet proteins were analyzed by Western blot using anti-His and anti-GST antibodies.
In vitro kinase assay
Recombinant proteins were expressed and purified using E.coli Transetta (DE3). Recombinant GST-tagged MPK3 (10 μg) was activated by incubation with recombinant GST-MKK5DD (1 μg) in the presence of 50 μM ATP in 50 μL of reaction buffer (25 mM Tris, pH 7.5, 10 mM MgCl2 and 1mM DTT) at 37 °C for 30 min. Then, GST-LAZY4ΔCCL (~100 μg) was added and reacted in the same reaction buffer with 10 mM ATP at 37 °C for 60 min. Equal volumes of 2x SDS loading buffer were added and boiled at 95 °C for 10 min to stop the reaction. Phosphorylation of LAZY4 proteins was analyzed by mass-spectrometry and Western blot.
Antibody generation
We generated a polyclonal LAZY4 antibody designated anti-LAZY4. The synthetic peptide KQEITHRPSISSASSHHPR derived from the Arabidopsis LAZY4 was injected into rabbits as antigen, using 400 μg for the first injection and 200 μg for the following three times. Polyclonal anti-LAZY4 antibodies were purified from rabbit serum using the synthetic peptide by affinity chromatography.
Yeast two hybrid assays
The yeast two-hybrid assays were performed as described previously62. The coding sequences of TOC33, TOC34, TOC64, TOC75 (TOC75-III), TOC90, TOC120, TOC132, TOC132-GM, TOC159-GM, LAZY4, LAZY4-D5/9/13 and LAZY4-A5/9/13 were amplified using the primers listed in Table S5, and the mutation sites are listed in Table S1. The TOC fragments were subcloned into the pLexA vector, and LAZY4, LAZY4- D5/9/13 and LAZY4- A5/9/13 were subcloned into the pB42AD vector. The LexA fusion constructs (bait) and the activation domain fusion constructs (prey) were co-transformed into yeast strain EGY48[p8op-lacZ] (CLONTECH). Clones containing both constructs were selected on medium lacking His, Trp and Ura, and transferred onto minimal SD/Gal/Raf/-His-Trp-Ura agar plates containing X-gal for testing β-Galactosidase activity63.
QUANTIFICATION AND STATISTICAL ANALYSIS
Root tip angles and fluorescence intensity were measured using ImageJ (https://imagej.net/software/fiji/). MS1 intensity was evaluated by Proteome Discoverer (Thermo Fisher Scientific, https://www.thermofisher.cn/cn/zh/home/industrial/mass-spectrometry/liquid-chromatography-mass-spectrometry-lc-ms/lc-ms-software/multi-omics-data-analysis/proteome-discoverer-software.html). Student’s t-test was performed in comparison of two independent groups. For multiple comparison, one-way ANOVA with post-hoc Tukey’s HSD test or Dunnett’s t-test was performed using IBM SPSS software (https://www.ibm.com/spss). The distribution of root tips was evaluated by Kolmogorov-Smirnov test with Bonferroni correction. The statistical details, including the statistical tests used and exact value of n for each measurement, were presented in figure legends and figures. The significant differences were indicated as following: ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant, P > 0.05.
Supplementary Material
Figure S1. Arabidopsis LAZY2, 3 and 4 are essential for lateral root gravitropism, while they have no effect on amyloplast sedimentation, related to Figure 1
(A) Gene structures and mutation sites of Arabidopsis LAZY2, LAZY3 and LAZY4 in the three alleles of lazy2 lazy3 lazy4 (lazy234) triple mutants. For each LAZY gene, two sgRNAs were designed to edit the genome. PAM sequences are labeled in red, and sgRNA sequences are underlined adjacent to the PAM in Col. The mutation details are shown for each allele, including deletion, insertion and inversion.
(B and C) Arabidopsis lateral roots lacking LAZY2, 3 and 4 were agravitropic. (B) Seedlings were grown vertically on MS plates under white light, and the primary roots were aligned to the gravity vector on day 7 and continued to grow until day 16. Scale bars, 1 cm. Arrows labeled “G” indicate the direction of gravity. (C) The growth angles of lateral root (LR) tips from the gravity vector were measured on day 16. Lateral root tip angles were compared with Col, and significant difference was evaluated by one-way ANOVA with post-hoc Dunnett’s t-test (***, P < 0.001; n = 30, 30, 30 and 43 respectively).
(D and E) Amyloplast sedimentation in Col and lazy234 is similar. (D) Four-day old seedlings were grown on MS plates under white light. Then, amyloplasts in the columella cells of wild-type and lazy234 seedlings after 90° reorientation were imaged using a microscope with a vertical stage. Bars, 5 μm. Arrowheads indicate the positions of amyloplasts. (E) The relative distances from the amyloplasts to the new bottom of columella cells after 90° reorientation were measured at several time points. The average relative-distances of several amyloplasts within a cell was used to indicate the relative position of the amyloplasts in the cell. The distance from the top to the bottom of the cells was set as 1. The Student’s t-test was performed to compare the distances of Col and lazy234 mutant at each time point. ns represents no significant difference between Col and lazy234 mutant (ns, not significant, P > 0.05; n = 7 and 11 respectively).
Figure S2. Both the primary and lateral roots of DR5rev::GFP/lazy234 lost gravitropic responses, and gravistimulation-induced asymmetrical distribution of auxin was disrupted in their lateral roots, related to Figure 1
(A) Gene structures and mutation sites of Arabidopsis LAZY2, LAZY3 and LAZY4 in the DR5rev::GFP/lazy234. For each LAZY gene, two sgRNAs were designed to edit the genome. PAM sequences are labeled in red, and sgRNA sequences are underlined adjacent to the PAM in Col. The mutation details are shown for each allele, including deletion, insertion and substitution.
(B) Growth orientation of DR5rev::GFP/lazy234 primary roots. Seedlings were grown vertically under white light for 4 days. Frequencies of root angles in each 30° division around a circle are shown (n = 53 and 57 respectively).
(C and D) Growth orientation of DR5rev::GFP/lazy234 lateral roots. Seedlings were grown vertically under white light, and the primary roots were aligned to the gravity vector on day 7. The growth angles of lateral root (LR) tips from the gravity vector were measured on day 13. Student’s t-test was used to assess the statistical significance by comparing DR5rev::GFP/lazy234 to DR5rev::GFP (***, P < 0.001; n = 38 and 40 respectively).
(E) Expression of DR5rev::GFP in the stage 2 lateral roots of wild type. Two categories of GFP fluorescence patterns were observed and the proportions of each category are indicated. Left, symmetric distribution; Right, higher accumulation on the lower side. Arrowheads indicate the asymmetric accumulation of auxin.
(F) Expression of DR5rev::GFP in the stage 2 lateral roots of lazy234. Three categories of GFP fluorescence patterns were observed and the proportions of each category are indicated. Left, symmetric distribution; Middle, higher accumulation on the upper side. Right, higher accumulation on the lower side. Arrowheads indicate the asymmetric accumulation of auxin.
In B, C, E and F, arrows labeled “G” indicate the direction of gravity. In B and C, scale bars, 1 cm; in E and F, scale bars, 50 μm.
Figure S3. The localization of LAZY2, 3 and 4 proteins in the columella cells of Arabidopsis primary and lateral roots, related to Figures 2 and 3
(A) The localization of Arabidopsis LAZY2, LAZY3 and LAZY4 proteins in root tips. Four-day old seedlings were grown on MS plates under white light, and then imaged by confocal microscope. The root tips were treated with 5 μM FM4-64 for 2 minutes, and the fluorescence of LAZY-GFP proteins and FM4-64 were collected from the root tips. Scale bars, 20 μm. The merged images are the same as those in the Figures 2A to 2C, whereas the images of separated channels are shown here for better view of the GFP signals. These are representative images of roots of 10, 8 and 13 seedlings, respectively.
(B) BH score predictions of LAZY2, LAZY3, LAZY4, and LAZY4 with mutated bh sites (LAZY4bh12). Predictions were made on https://hpcwebapps.cit.nih.gov/bhsearch/. Blue grid line refers to 0.6, which is a threshold for possible PtdIns binding.
(C) The schematic of LAZY4bh12 mutating K/R to A within two basic hydrophobic peaks.
(D and E) Measurement of LAZY-GFP fluorescence intensity ratio. Images showing two representative patterns of LAZY-GFP fluorescence in columella cells, with clear signals only in central columella (CC) cells (B), or in both of central columella (CC) cells and lateral columella (LC) cells (C). In each panel, the left is the original image, and the right image shows how the intensity ratio between basal and apical is calculated. Upper or lower side of the plasma membrane in columella cells were selected as region of interest (ROI) for measurement of LAZY-GFP fluorescence intensity. Signal ratio (basal/apical) for one root was calculated as an average of signal intensity ratios within this root.
(F and G) LAZY2, 3 and 4 proteins show polar distribution in the columella cells of Arabidopsis lateral roots under the regulation of gravity. (D) LAZY2, LAZY3 and LAZY4 proteins are mainly accumulated on the lower side of columella cells in the lateral roots (LRs) at the stage 2 under both regular growth (upper) or after 180° reorientation (lower). The primary roots of ProLAZY2:LAZY2-GFP (LAZY2-GFP/Col), ProLAZY3:LAZY3-GFP/lazy234 (LAZY3-GFP) and ProLAZY4:LAZY4-GFP/lazy234 (LAZY4-GFP) growing under white light conditions were aligned to the gravity vector on the 7th day and continued to grow until the 12th day. The fluorescence of LAZY-GFP was collected before (Upper in each panel), or after the seedlings were reoriented 180° and grown for another 6 h (Lower in each panel). Double-headed arrows show the apical-basal axis of the plant bodies. Red and white arrow heads indicate the accumulation of LAZY-GFP proteins on the apical and basal sides of columella cells respectively. For the whole seedlings, scale bars, 1 cm; for all the others, scale bars, 20 μm. Arrows labeled “G” indicate the direction of gravity. (E) The statistical analysis of LAZY-GFP fluorescence intensity ratios of the two sides of the lateral roots before and after 180° rotation. Asterisks indicate Student’s t-test values (***, P < 0.001; n = 12, 14, 12, 16, 12 and 13 respectively).
Figure S4. Amyloplast sedimentation promotes the redistribution of LAZY proteins, related to Figure 3
(A) Gravity-triggered redistribution of LAZY2 and LAZY4 proteins to the lower side of columella cells correlates with the sedimentation of amyloplasts. The seedlings of LAZY2-GFP (without CHX treatment) and LAZY4-GFP (with 10 μM CHX treatment to inhibit protein synthesis) were reoriented to a position with amyloplasts on the top of the columella cells, and kept for fluorescence collection at several time points. The area inside yellow box was zoomed to show details of fluorescence. White dash lines indicate the contour of root tips. Scale bars, 10 μm.
(B) Mutation of PGM1 delayed the redistribution of LAZY2-GFP, LAZY3-GFP and LAZY4-GFP proteins in root columella cells. Seedlings were grown vertically and then reoriented 90° and kept 1 h for gravistimulation (GS 1 h). Red and white arrowheads indicate the accumulation of LAZY-GFP proteins on the upper and lower sides of columella cells respectively. Scale bars, 20 μm. Statistical data are shown in Figures 3M to 3O.
In A and B, arrows labeled “G” indicate the direction of gravity.
Figure S5. Phosphorylation sites of LAZY4 protein in plants or in in vitro kinase assay, related to Figure 5
(A) This schematic summarizes the phosphorylation sites identified by MS in the IP-MS experiment in Figures 5A and 5B, and in the in vitro kinase assays in Figure 5E. The detailed information about the phosphorylation of these sites in each experiment is shown in Table S1.
(B) MS/MS spectra. X axis shows the m/z value of each fragment ion. Y axis shows the relative abundance (left) or ion intensity (right) of the corresponding daughter ions. Peptide sequences are displayed above the spectra.
Figure S6. Interactions between LAZY4 variants and TOC proteins, and mutation sites of TOCs, related to Figure 6
(A) Distinct TOC complexes in Arabidopsis. In Arabidopsis TOC complexes, TOC75 is a central pore component localized within the outer membranes of plastids, and other TOC proteins are receptors facing the cytosol. TOC33 and TOC159 are components of a class of TOC complexes that specifically import photosynthesis-related preproteins. TOC34, TOC120 and TOC132 are components of another class of TOC complexes that specifically import non-photosynthetic or house-keeping preproteins. TOC90 may be partially redundant with TOC159, and associates with TOC33 or TOC34. Abbreviations: TOC, Translocons at the Outer envelope membrane of Chloroplasts; OM, Outer Membrane; IMS, Intermembrane Space. The models of TOC complexes were drawn based on information from a previous review38.
(B) Yeast two-hybrid assays to test interactions between LAZY4 variants and TOC proteins. Interactions between LAZY4 variants and TOC proteins were examined by yeast growth on media with X-Gal but lacking Trp, His and Ura. The phosphorylation sites of LAZY4 were mutated to Asp (D) or Ala (A) as indicated in Table S1. Blue indicates positive interactions. Reporter: LacZ; substrate, X-Gal. These results were from the same experiment shown in Figure 6A, but with longer growth time (40 h) and more LAZY4 variants (mutated to A).
(C) Mutation sites of Arabidopsis TOC120 and TOC132 in the LAZY4-GFP/lazy234 toc120CR toc132CR (LAZY4-GFP/toc120CR toc132CR) and LAZY4-D9-GFP/lazy234 toc120CR toc132CR (LAZY4-D9-GFP/toc120CR toc132CR) seedlings. Two alleles of LAZY4-GFP toc120CR toc132CR showed similar phenotypes. For each TOC gene, two sgRNAs were designed to edit the genome. PAM sequences are labeled in red, and sgRNA sequences are underlined adjacent to the PAM in Col. The mutation details are shown for each allele, including deletion, insertion and substitution.
Table S2. MS1 intensity of phosphopeptide “FLNCPSSLEVDR”, related to Figure 5
Table S3. MPK3 and MKK5 Co-immunoprecipitated by LAZY4-GFP, related to Figure 5
Table S4. Phosphorylation of MPK3 in MPK3-GFP IP samples and LAZY4-GFP Co-IP samples, related to Figure 5
Table S5. Primers used in this study, related to STAR Methods
Video S1. 3-D view of LAZY3-GFP fluorescence in columella cells after gravistimulation, related to Figure 4
Growth condition, acquisition parameters and data-process details were described in Figure 4A. The 3-D construction view was rotated in the video for observation from different angles.
Video S2. Time-lapse record of LAZY3-GFP fluorescence recovery on plasma membrane in columella cells in FRAP experiment, related to Figure 4
Growth condition and bleaching parameters were described in Figures 4B and 4C. The fluorescence was acquired by the confocal laser scanning microscope (Zeiss LSM800) with a vertical objective table. Arrowheads labeled with numbers indicate a symmetric pair of columella cells with similar fluorescence before photobleaching.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-GST | CMC-TAG | Cat# AT0027 |
| Mouse monoclonal anti-GST | Sigma-Aldrich | Cat# G1160 |
| Mouse monoclonal anti-His | Sigma-Aldrich | Cat# H1029 |
| Rabbit polyclonal anti-TxR | Cell Signaling Technology | Cat# 2351S |
| Rabbit polyclonal anti-LAZY4 | This paper | N/A |
| Goat Anti-Rabbit Mouse IgG-HRP | Abmart | Cat# M21003 |
| Bacterial and Virus Strains | ||
| Trans5α Chemically Competent Cell | Transgen | Cat# CD201–01 |
| Transetta (DE3) Chemically Competent Cell | Transgen | Cat# CD801–02 |
| GV3101 | Beyotime | Cat# D0392 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Murashige and Skoog Basal Medium | Sigma-Aldrich | Cat# M5519 |
| Agar | Affymetrix USB | Cat# 10906 |
| BASTA | Sigma-Aldrich | Cat# 45520 |
| Hygromycin B | Thermo Fisher | Cat# 10687–010 |
| MES hydrate | Sigma-Aldrich | Cat# M8250 |
| MgCl2•6H2O | Sigma-Aldrich | Cat# M2670 |
| CaCl2 | Sigma-Aldrich | Cat# C5670 |
| D-Mannitol | Sigma-Aldrich | Cat# M4125 |
| Poly (ethylene glycol) | Sigma-Aldrich | Cat# 81240 |
| KCl | Sigma-Aldrich | Cat# P9541 |
| LB broth | AmericanBio | Cat# AB01198–01000 |
| Trizma Base | Sigma-Aldrich | Cat# T1503 |
| NaCl | Sigma-Aldrich | Cat# S3014 |
| Glucose | Sigma-Aldrich | Cat# C5767 |
| Kanamycin sulfate | Amresco | Cat# 0408 |
| Rifampicin | Sigma-Aldrich | Cat# R3501 |
| Ampicillin | Amresco | Cat# 0339 |
| Silwet L-77 | GE Healthcare | Cat# SL77080596 |
| SynaptoRed (FM4-64) | Millipore | Cat# C574799 |
| Dimethyl sulfoxide | Sigma-Aldrich | Cat# D8418 |
| DTT | Sigma-Aldrich | Cat# D0632 |
| Protease inhibitors | Mei5bio | Cat# MF182 |
| Phosphatase inhibitors | Mei5bio | Cat# MF183 |
| RIPA buffer | Abcam | Cat# Ab156034 |
| GFP-Trap beads | ChromoTek | Cat# gtma-20 |
| Minimal SD Base | Takara | Cat# 630411 |
| Minimal SD Base/Gal/Raf | Takara | Cat# 630420 |
| DO Supplement-Ura | Takara | Cat# 630416 |
| DO Supplement-His/-Trp/-Ura | Takara | Cat# 630424 |
| RNeasy Plant Mini kit | Qiagen | Cat# 74904 |
| DNeasy Plant Mini kit | Qiagen | Cat# 69104 |
| X-gal | Takara | Cat# 9031 |
| TRIS | VWR LifeScience | Cat# 0497 |
| ExBlue Protein Stain | ZOMANBIO | Cat# ZD305A |
| Nonfat Dry Milk | Cell Signaling Technology | Cat# 9999 |
| Clarity™ Western ECL Substrate | BIO-RAD | Cat# 170–5061 |
| GST-tag Protein Purification Kit | Beyotime | Cat# P2262 |
| His-tag Protein Purification Kit | Beyotime | Cat# P2226 |
| IPTG | Sigma-Aldrich | Cat# I6758 |
| SDS-PAGE Gel Preparation Kit | Coolaber | Cat# SK6010 |
| Tween-20 | Solarbio | Cat# T8220 |
| PMSF | Mei5Bio | Cat# MF105 |
| BSA | Sigma | Cat# B2064 |
| ATP | NEB | Cat# P0756S |
| Glycine | VWR LifeScience | Cat# 0167 |
| Deposited Data | ||
| Raw peptide data | This paper, see www.proteomexchange.org | PXD045213 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Arabidopsis lazy234–1 | This paper | N/A |
| Arabidopsis lazy234–2 | This paper | N/A |
| Arabidopsis lazy234–3 | This paper | N/A |
| Arabidopsis ProLAZY2:LAZY2-GFP | This paper | N/A |
| Arabidopsis ProLAZY2:LAZY2-GFP/lazy234 | This paper | N/A |
| Arabidopsis ProLAZY3:LAZY3-GFP/lazy234 | This paper | N/A |
| Arabidopsis ProLAZY4:LAZY4-GFP/lazy234 | This paper | N/A |
| Arabidopsis DR5rev::GFP | Ulmasov et al.48 | N/A |
| Arabidopsis DR5rev::GFP/lazy234 | This paper | N/A |
| Arabidopsis ProLAZY2:LAZY2ΔN-GFP/lazy234 | This paper | N/A |
| Arabidopsis ProLAZY3:LAZY3ΔN-GFP/lazy234 | This paper | N/A |
| Arabidopsis ProLAZY4:LAZY4ΔN-GFP/lazy234 | This paper | N/A |
| Arabidopsis ProLAZY2:LAZY2-GFP/lazy234 pgm1 | This paper | N/A |
| Arabidopsis ProLAZY3:LAZY3-GFP/lazy234 pgm1 | This paper | N/A |
| Arabidopsis ProLAZY4:LAZY4-GFP/lazy234 pgm1 | This paper | N/A |
| Arabidopsis pMPK3-MPK3-GFP | Huang et al.45 | N/A |
| Arabidopsis mpk6−/−mpk3RNAi-est | Huang et al.45 | N/A |
| Arabidopsis ProLAZY4:LAZY4-GFP/lazy23 mpk6−/− mpk3RNAi-est | This paper | N/A |
| Arabidopsis ProLAZY4:LAZY4-A 13-GFP /lazy234 | This paper | N/A |
| Arabidopsis ProLAZY4:LAZY4-D9-GFP /lazy234 | This paper | N/A |
| Arabidopsis ProTOC132:TOC132-RFP/ProLAZY4:LAZY4-D9-GFP/lazy234 | This paper | N/A |
| Arabidopsis ProLAZY4:LAZY4-D9-GFP/lazy234 toc120CR toc132CR | This paper | N/A |
| Arabidopsis ProLAZY4:LAZY4-GFP/lazy234 toc120CR toc132CR | This paper | N/A |
| Oligonucleotides | ||
| Primers are listed in table S5 | This paper | N/A |
| Recombinant DNA | ||
| pHEE401E | Wang et al.57 | N/A |
| p35S-Cas9-SK | Feng et al.58 | N/A |
| pAtU6-26-M | Wang et al.59 | N/A |
| pEC-Cas9 | This paper | N/A |
| pCAMBIA 1300-LAZY2/3/4 sgRNA | This paper | N/A |
| pJIM19-LAZY2/3/4 sgRNA | This paper | N/A |
| pJIM19-ProLAZY2:LAZY2-GFP | This paper | N/A |
| pJIM19-ProLAZY3:LAZY3-GFP | This paper | N/A |
| pJIM19-ProLAZY4:LAZY4-GFP | This paper | N/A |
| pCAMBIA1300-ProLAZY2-LAZY2ΔN-GFP | This paper | N/A |
| pJIM19-ProLAZY3:LAZY3ΔN-GFP | This paper | N/A |
| pCAMBIA1300-ProLAZY4:LAZY4ΔN-GFP | This paper | N/A |
| pJIM19-ProLAZY4:LAZY4-A13-GFP | This paper | N/A |
| pJIM19-ProLAZY4:LAZY4-D9-GFP | This paper | N/A |
| pCAMBIA 1300-TOC132-mRFP | This paper | N/A |
| pUBQ10:Cas9-P2A-GFP (Gent)-TOC132-TOC120 | This paper | N/A |
| pGEX4T-1-GST-LAZY2ΔCCL | This paper | N/A |
| pGEX4T-1-GST-LAZY3ΔCCL | This paper | N/A |
| pGEX4T-1-GST-LAZY4ΔCCL | This paper | N/A |
| pGEX4T-1-GST-LAZY4bh12ΔCCL | This paper | N/A |
| pGEX4T-1-GST-MKK5DD | This paper | N/A |
| pGEX4T-1-GST-MPK3 | This paper | N/A |
| pET28a-His-MKK5DD | This paper | N/A |
| pET28a-His-MPK3 | This paper | N/A |
| pB42-AD-LAZY4 | This paper | N/A |
| pB42-AD-LAZY4-A5 | This paper | N/A |
| pB42-AD-LAZY4-D5 | This paper | N/A |
| pB42-AD-LAZY4-A9 | This paper | N/A |
| pB42-AD-LAZY4-D9 | This paper | N/A |
| pB42-AD-LAZY4-A13 | This paper | N/A |
| pB42-AD-LAZY4-D13 | This paper | N/A |
| plexA-BD-TOC33 | This paper | N/A |
| plexA-BD-TOC34 | This paper | N/A |
| plexA-BD-TOC64 | This paper | N/A |
| plexA-BD-TOC75 | This paper | N/A |
| plexA-BD-TOC90 | This paper | N/A |
| plexA-BD-TOC120 | This paper | N/A |
| plexA-BD-TOC132 | This paper | N/A |
| plexA-BD-TOC132-GM | This paper | N/A |
| plexA-BD-TOC159-GM | This paper | N/A |
| Software and Algorithms | ||
| Image J | NIH, USA | http://rsb.info.nih.gov/ij |
| R (version 4.2.2) | R Core Team (2022). Vienna, Austria. | https://www.R-project.org/ |
| IBM SPSS statistics (version 24) | SPSS Inc., Chicago IL, USA | https://www.ibm.com/spss |
| Proteome Discoverer (versions 1.4, 2.2, 2.5) | Thermo Fisher | https://www.thermofisher.cn/cn/zh/home/industrial/mass-spectrometry/liquid-chromatography-mass-spectrometry-lc-ms/lc-ms-software/multi-omics-data-analysis/proteome-discoverer-software.html |
| MaxQuant (version 1.6.5.0) | Cox, J. and Mann, M (2008) | https://www.maxquant.org/ |
| Other | ||
| PVDF-Western Blotting Membranes | Roche Diagnostics | 03010040001 |
| PIP Strips | Echelon Biosciences | P-6001 |
Highlights.
Gravistimulation triggers MAPK-mediated phosphorylation of LAZY proteins
Phosphorylation increases LAZY’s interaction with TOCs on the surface of amyloplasts
Amyloplast sedimentation carries LAZY proteins to the new bottom of columella cells
LAZY translocates from amyloplasts to adjacent plasma membrane to form new polarity
ACKNOWLEDGMENTS
We thank Prof. Tongda Xu for mpk6−/−mpk3RNAi-est and pMPK3-MPK3-GFP seeds, Profs. Jian-Kang Zhu and Qijun Chen for CRISPR/Cas9 plasmids, Profs. Li-Jia Qu, Tongda Xu, William Terzaghi, Genji Qin, Li Yu, Guangshuo Ou, Yule Liu, Yijun Qi, Shanjin Huang, Daoxin Xie and Haiteng Deng for helpful comments, and Huifang Qin and Jingxi Sun for technical assistance. We thank the National Center for Protein Sciences and Core Facilities of Life Sciences at Peking University in Beijing, China, particularly Dr. Dong Liu and Dr. Qi Zhang for assistance with the mass spectrometry experiments, and Dr. Yiqun Liu, Dr. Yingchun Hu, and Miss. Pengyuan Dong for assistance with ImmunoEM. We thank the Protein Chemistry and Proteomics Facility at Tsinghua University particularly Dr. Yuling Chen for assistance with the mass spectrometry experiments. We thank Cryo-EM Facility of China National Center for Protein Sciences (Beijing) at Tsinghua University particularly Dr. Ying Li for assistance with HPF and immunolabeling. This study was supported by the National Natural Science Foundation of China (32022005, 31621001, 32170287), Tsinghua University Dushi Program (20231080035), and the Tsinghua-Peking Center for Life Sciences. J.D. was funded by grants from the National Institute of Health (GM1851907) and the National Science Foundation (2049642).
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Knight T (1806). On the direction of the radicle and germen during the vegetation of seeds. Philos. Trans. R. Soc. Lond. Ser. B 96, 99–108. 10.1098/rstl.1806.0006. [DOI] [Google Scholar]
- 2.Vandenbrink JP, and Kiss JZ (2019). Plant responses to gravity. Semin. Cell Dev. Biol. 92, 122–125. 10.1016/j.semcdb.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, Kitomi Y, Inukai Y, Ono K, Kanno N, et al. (2013). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–1102. 10.1038/ng.2725. [DOI] [PubMed] [Google Scholar]
- 4.Huang G, Liang W, Sturrock CJ, Pandey BK, Giri J, Mairhofer S, Wang D, Muller L, Tan H, York LM, et al. (2018). Rice actin binding protein RMD controls crown root angle in response to external phosphate. Nat Commun 9, 2346. 10.1038/s41467-018-04710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morita MT (2010). Directional gravity sensing in gravitropism. Annu. Rev. Plant Biol. 61, 705–720. 10.1146/annurev.arplant.043008.092042. [DOI] [PubMed] [Google Scholar]
- 6.Chin S, and Blancaflor EB (2022). Plant Gravitropism: From Mechanistic Insights into Plant Function on Earth to Plants Colonizing Other Worlds. Methods Mol Biol 2368, 1–41. 10.1007/978-1-0716-1677-2_1. [DOI] [PubMed] [Google Scholar]
- 7.Nemec B (1900). Uber die Art der Wahrnehmung des Schwerkraftreizes bei den Pflanzen. Ber. Deut. Bot. Ges. 18, 241–245. 10.1111/J.1438-8677.1900.TB04905.X. [DOI] [Google Scholar]
- 8.Darwin F (1903). The statolith theory of geotropism. Nature 67, 571–572. 10.1038/067571a0. [DOI] [Google Scholar]
- 9.Haberlandt G (1900). Ueber die Perception des geotropischen Reizes. Ber. Deut. Bot. Ges. 18, 261–272. 10.1111/j.1438-8677.1900.tb04908.x. [DOI] [Google Scholar]
- 10.Blancaflor EB, Fasano JM, and Gilroy S (1998). Mapping the functional roles of cap cells in the response of Arabidopsis primary roots to gravity. Plant Physiol. 116, 213–222. 10.1104/pp.116.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukaki H, Wysocka-Diller J, Kato T, Fujisawa H, Benfey PN, and Tasaka M (1998). Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J. 14, 425–430. 10.1046/j.1365-313x.1998.00137.x. [DOI] [PubMed] [Google Scholar]
- 12.Tsugeki R, and Fedoroff NV (1999). Genetic ablation of root cap cells in Arabidopsis. Proc Natl Acad Sci U S A 96, 12941–12946. 10.1073/pnas.96.22.12941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caspar T, and Pickard BG (1989). Gravitropism in a starchless mutant of Arabidopsis : Implications for the starch-statolith theory of gravity sensing. Planta 177, 185–197. 10.1007/BF00392807. [DOI] [PubMed] [Google Scholar]
- 14.Kiss JZ, Hertel R, and Sack FD (1989). Amyloplasts are necessary for full gravitropic sensitivity in roots of Arabidopsis thaliana. Planta 177, 198–206. 10.1007/BF00392808. [DOI] [PubMed] [Google Scholar]
- 15.Zheng Z, Zou J, Li H, Xue S, Wang Y, and Le J (2015). Microrheological insights into the dynamics of amyloplasts in root gravity-sensing cells. Mol Plant 8, 660–663. 10.1016/j.molp.2014.12.021. [DOI] [PubMed] [Google Scholar]
- 16.Went FW, and Thimann KV (1937). Phytohormones (New York: The macmillan company; ). 10.5962/bhl.title.5695. [DOI] [Google Scholar]
- 17.Friml J, Wisniewska J, Benkova E, Mendgen K, and Palme K (2002). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415, 806–809. 10.1038/415806a. [DOI] [PubMed] [Google Scholar]
- 18.Kleine-Vehn J, Ding Z, Jones AR, Tasaka M, Morita MT, and Friml J (2010). Gravity-induced PIN transcytosis for polarization of auxin fluxes in gravity-sensing root cells. Proc Natl Acad Sci U S A 107, 22344–22349. 10.1073/pnas.1013145107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkins MT, and Gerhardt F (1931). A gene influencing the composition of the culm in maize. Iowa Ag Exp Sta Research Bull 11, 121–151. https://dr.lib.iastate.edu/handle/20.500.12876/62445. [Google Scholar]
- 20.Van Overbeek J (1936). “Lazy,” an A-geotropic form of maize: “Gravitational indifference” rather than structural weakness accounts for prostrate growth-habit of this form. J Heredity 27, 93–96. 10.1093/oxfordjournals.jhered.a104191. [DOI] [Google Scholar]
- 21.Jones JW, and Adair CR (1938). A “lazy” mutation in rice. J Heredity 29, 315–318. 10.1093/oxfordjournals.jhered.a104527. [DOI] [Google Scholar]
- 22.Li P, Wang Y, Qian Q, Fu Z, Wang M, Zeng D, Li B, Wang X, and Li J (2007). LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 17, 402–410. 10.1038/cr.2007.38. [DOI] [PubMed] [Google Scholar]
- 23.Yoshihara T, and Iino M (2007). Identification of the gravitropism-related rice gene LAZY1 and elucidation of LAZY1-dependent and -independent gravity signaling pathways. Plant Cell Physiol. 48, 678–688. 10.1093/pcp/pcm042. [DOI] [PubMed] [Google Scholar]
- 24.Dong Z, Jiang C, Chen X, Zhang T, Ding L, Song W, Luo H, Lai J, Chen H, Liu R, et al. (2013). Maize LAZY1 mediates shoot gravitropism and inflorescence development through regulating auxin transport, auxin signaling, and light response. Plant Physiol. 163, 1306–1322. 10.1104/pp.113.227314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshihara T, Spalding EP, and Iino M (2013). AtLAZY1 is a signaling component required for gravitropism of the Arabidopsis thaliana inflorescence. Plant J. 74, 267–279. 10.1111/tpj.12118. [DOI] [PubMed] [Google Scholar]
- 26.Guseman JM, Webb K, Srinivasan C, and Dardick C (2017). DRO1 influences root system architecture in Arabidopsis and Prunus species. Plant J. 89, 1093–1105. 10.1111/tpj.13470. [DOI] [PubMed] [Google Scholar]
- 27.Che X, Splitt BL, Eckholm MT, Miller ND, and Spalding EP (2023). BRXL4-LAZY1 interaction at the plasma membrane controls Arabidopsis branch angle and gravitropism. Plant J. 113, 211–224. 10.1111/tpj.16055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge L, and Chen R (2016). Negative gravitropism in plant roots. Nat Plants 2, 16155. 10.1038/nplants.2016.155. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Xu S, Tian L, Liu L, Huang M, Xu X, Song G, Wu P, Sato S, Jiang H, and Wu G (2020). LAZY3 plays a pivotal role in positive root gravitropism in Lotus japonicus. J. Exp. Bot. 71, 168–177. 10.1093/jxb/erz429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taniguchi M, Furutani M, Nishimura T, Nakamura M, Fushita T, Iijima K, Baba K, Tanaka H, Toyota M, Tasaka M, and Morita MT (2017). The Arabidopsis LAZY1 Family Plays a Key Role in Gravity Signaling within Statocytes and in Branch Angle Control of Roots and Shoots. Plant Cell 29, 1984–1999. 10.1105/tpc.16.00575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshihara T, and Spalding EP (2017). LAZY Genes Mediate the Effects of Gravity on Auxin Gradients and Plant Architecture. Plant Physiol. 175, 959–969. 10.1104/pp.17.00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang P, Wen Q, Yu R, Han X, Deng XW, and Chen H (2020). Light modulates the gravitropic responses through organ-specific PIFs and HY5 regulation of LAZY4 expression in Arabidopsis. Proc Natl Acad Sci U S A 117, 18840–18848. 10.1073/pnas.2005871117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furutani M, Hirano Y, Nishimura T, Nakamura M, Taniguchi M, Suzuki K, Oshida R, Kondo C, Sun S, Kato K, et al. (2020). Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control. Nat Commun 11, 76. 10.1038/s41467-019-13729-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waite JM, Collum TD, and Dardick C (2020). AtDRO1 is nuclear localized in root tips under native conditions and impacts auxin localization. Plant Mol. Biol. 103, 197–210. 10.1007/s11103-020-00984-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sedbrook JC, Chen R, and Masson PH (1999). ARG1 (altered response to gravity) encodes a DnaJ-like protein that potentially interacts with the cytoskeleton. Proc Natl Acad Sci U S A 96, 1140–1145. 10.1073/pnas.96.3.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanga JP, Boonsirichai K, Sedbrook JC, Otegui MS, and Masson PH (2009). A role for the TOC complex in Arabidopsis root gravitropism. Plant Physiol. 149, 1896–1905. 10.1104/pp.109.135301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strohm AK, Barrett-Wilt GA, and Masson PH (2014). A functional TOC complex contributes to gravity signal transduction in Arabidopsis. Front Plant Sci 5, 148. 10.3389/fpls.2014.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andres C, Agne B, and Kessler F (2010). The TOC complex: preprotein gateway to the chloroplast. Biochim Biophys Acta 1803, 715–723. 10.1016/j.bbamcr.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Kubis S, Patel R, Combe J, Bedard J, Kovacheva S, Lilley K, Biehl A, Leister D, Rios G, Koncz C, and Jarvis P (2004). Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell 16, 2059–2077. 10.1105/tpc.104.023309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin Z, Wan L, Zhang Y, Li X, Cao Y, Liu H, Fan S, Cao D, Wang Z, Li X, et al. (2022). Structure of a TOC-TIC supercomplex spanning two chloroplast envelope membranes. Cell 185, 4788–4800 e4713. 10.1016/j.cell.2022.10.030. [DOI] [PubMed] [Google Scholar]
- 41.Liu H, Li A, Rochaix JD, and Liu Z (2023). Architecture of chloroplast TOC-TIC translocon supercomplex. Nature. 10.1038/s41586-023-05744-y. [DOI] [PubMed] [Google Scholar]
- 42.Xu J, and Zhang S (2015). Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 20, 56–64. 10.1016/j.tplants.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 43.Wang H, Ngwenyama N, Liu Y, Walker JC, and Zhang S (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19, 63–73. 10.1105/tpc.106.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meng X, Wang H, He Y, Liu Y, Walker JC, Torii KU, and Zhang S (2012). A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24, 4948–4960. 10.1105/tpc.112.104695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang R, Zheng R, He J, Zhou Z, Wang J, Xiong Y, and Xu T (2019). Noncanonical auxin signaling regulates cell division pattern during lateral root development. Proc Natl Acad Sci U S A 116, 21285–21290. 10.1073/pnas.1910916116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jia W, Li B, Li S, Liang Y, Wu X, Ma M, Wang J, Gao J, Cai Y, Zhang Y, et al. (2016). Mitogen-Activated Protein Kinase Cascade MKK7-MPK6 Plays Important Roles in Plant Development and Regulates Shoot Branching by Phosphorylating PIN1 in Arabidopsis. PLoS Biol. 14, e1002550. 10.1371/journal.pbio.1002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ottenschlager I, Wolff P, Wolverton C, Bhalerao RP, Sandberg G, Ishikawa H, Evans M, and Palme K (2003). Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc Natl Acad Sci U S A 100, 2987–2991. 10.1073/pnas.0437936100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ulmasov T, Murfett J, Hagen G, and Guilfoyle TJ (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bailey MJ, and Prehoda KE (2015). Establishment of Par-Polarized Cortical Domains via Phosphoregulated Membrane Motifs. Dev. Cell 35, 199–210. 10.1016/j.devcel.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brzeska H, Guag J, Remmert K, Chacko S, and Korn ED (2010). An experimentally based computer search identifies unstructured membrane-binding sites in proteins: application to class I myosins, PAKS, and CARMIL. J. Biol. Chem. 285, 5738–5747. 10.1074/jbc.M109.066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su SH, Keith MA, and Masson PH (2020). Gravity Signaling in Flowering Plant Roots. Plants (Basel) 9. 10.3390/plants9101290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su SH, Gibbs NM, Jancewicz AL, and Masson PH (2017). Molecular Mechanisms of Root Gravitropism. Curr. Biol. 27, R964–R972. 10.1016/j.cub.2017.07.015. [DOI] [PubMed] [Google Scholar]
- 53.Leitz G, Kang BH, Schoenwaelder ME, and Staehelin LA (2009). Statolith sedimentation kinetics and force transduction to the cortical endoplasmic reticulum in gravity-sensing Arabidopsis columella cells. Plant Cell 21, 843–860. 10.1105/tpc.108.065052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perbal G, and Driss-Ecole D (2003). Mechanotransduction in gravisensing cells. Trends Plant Sci. 8, 498–504. 10.1016/j.tplants.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 55.Limbach C, Hauslage J, Schafer C, and Braun M (2005). How to activate a plant gravireceptor. Early mechanisms of gravity sensing studied in characean rhizoids during parabolic flights. Plant Physiol. 139, 1030–1040. 10.1104/pp.105.068106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zadnikova P, Petrasek J, Marhavy P, Raz V, Vandenbussche F, Ding Z, Schwarzerova K, Morita MT, Tasaka M, Hejatko J, et al. (2010). Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137, 607–617. 10.1242/dev.041277. [DOI] [PubMed] [Google Scholar]
- 57.Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, and Chen QJ (2015). Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol 16, 144. 10.1186/s13059-015-0715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, Cao F, Zhu S, Zhang F, Mao Y, and Zhu JK (2013). Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23, 1229–1232. 10.1038/cr.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang J, and Chen H (2020). A novel CRISPR/Cas9 system for efficiently generating Cas9-free multiplex mutants in Arabidopsis. aBIOTECH 1, 6–14. 10.1007/s42994-019-00011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grones P, Abas M, Hajny J, Jones A, Waidmann S, Kleine-Vehn J, and Friml J (2018). PID/WAG-mediated phosphorylation of the Arabidopsis PIN3 auxin transporter mediates polarity switches during gravitropism. Sci Rep 8, 10279. 10.1038/s41598-018-28188-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Serino G, Tsuge T, Kwok S, Matsui M, Wei N, and Deng XW (1999). Arabidopsis cop8 and fus4 mutations define the same gene that encodes subunit 4 of the COP9 signalosome. Plant Cell 11, 1967–1980. 10.1105/tpc.11.10.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dong J, Tang D, Gao Z, Yu R, Li K, He H, Terzaghi W, Deng XW, and Chen H (2014). Arabidopsis DE-ETIOLATED1 represses photomorphogenesis by positively regulating phytochrome-interacting factors in the dark. Plant Cell 26, 3630–3645. 10.1105/tpc.114.130666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Arabidopsis LAZY2, 3 and 4 are essential for lateral root gravitropism, while they have no effect on amyloplast sedimentation, related to Figure 1
(A) Gene structures and mutation sites of Arabidopsis LAZY2, LAZY3 and LAZY4 in the three alleles of lazy2 lazy3 lazy4 (lazy234) triple mutants. For each LAZY gene, two sgRNAs were designed to edit the genome. PAM sequences are labeled in red, and sgRNA sequences are underlined adjacent to the PAM in Col. The mutation details are shown for each allele, including deletion, insertion and inversion.
(B and C) Arabidopsis lateral roots lacking LAZY2, 3 and 4 were agravitropic. (B) Seedlings were grown vertically on MS plates under white light, and the primary roots were aligned to the gravity vector on day 7 and continued to grow until day 16. Scale bars, 1 cm. Arrows labeled “G” indicate the direction of gravity. (C) The growth angles of lateral root (LR) tips from the gravity vector were measured on day 16. Lateral root tip angles were compared with Col, and significant difference was evaluated by one-way ANOVA with post-hoc Dunnett’s t-test (***, P < 0.001; n = 30, 30, 30 and 43 respectively).
(D and E) Amyloplast sedimentation in Col and lazy234 is similar. (D) Four-day old seedlings were grown on MS plates under white light. Then, amyloplasts in the columella cells of wild-type and lazy234 seedlings after 90° reorientation were imaged using a microscope with a vertical stage. Bars, 5 μm. Arrowheads indicate the positions of amyloplasts. (E) The relative distances from the amyloplasts to the new bottom of columella cells after 90° reorientation were measured at several time points. The average relative-distances of several amyloplasts within a cell was used to indicate the relative position of the amyloplasts in the cell. The distance from the top to the bottom of the cells was set as 1. The Student’s t-test was performed to compare the distances of Col and lazy234 mutant at each time point. ns represents no significant difference between Col and lazy234 mutant (ns, not significant, P > 0.05; n = 7 and 11 respectively).
Figure S2. Both the primary and lateral roots of DR5rev::GFP/lazy234 lost gravitropic responses, and gravistimulation-induced asymmetrical distribution of auxin was disrupted in their lateral roots, related to Figure 1
(A) Gene structures and mutation sites of Arabidopsis LAZY2, LAZY3 and LAZY4 in the DR5rev::GFP/lazy234. For each LAZY gene, two sgRNAs were designed to edit the genome. PAM sequences are labeled in red, and sgRNA sequences are underlined adjacent to the PAM in Col. The mutation details are shown for each allele, including deletion, insertion and substitution.
(B) Growth orientation of DR5rev::GFP/lazy234 primary roots. Seedlings were grown vertically under white light for 4 days. Frequencies of root angles in each 30° division around a circle are shown (n = 53 and 57 respectively).
(C and D) Growth orientation of DR5rev::GFP/lazy234 lateral roots. Seedlings were grown vertically under white light, and the primary roots were aligned to the gravity vector on day 7. The growth angles of lateral root (LR) tips from the gravity vector were measured on day 13. Student’s t-test was used to assess the statistical significance by comparing DR5rev::GFP/lazy234 to DR5rev::GFP (***, P < 0.001; n = 38 and 40 respectively).
(E) Expression of DR5rev::GFP in the stage 2 lateral roots of wild type. Two categories of GFP fluorescence patterns were observed and the proportions of each category are indicated. Left, symmetric distribution; Right, higher accumulation on the lower side. Arrowheads indicate the asymmetric accumulation of auxin.
(F) Expression of DR5rev::GFP in the stage 2 lateral roots of lazy234. Three categories of GFP fluorescence patterns were observed and the proportions of each category are indicated. Left, symmetric distribution; Middle, higher accumulation on the upper side. Right, higher accumulation on the lower side. Arrowheads indicate the asymmetric accumulation of auxin.
In B, C, E and F, arrows labeled “G” indicate the direction of gravity. In B and C, scale bars, 1 cm; in E and F, scale bars, 50 μm.
Figure S3. The localization of LAZY2, 3 and 4 proteins in the columella cells of Arabidopsis primary and lateral roots, related to Figures 2 and 3
(A) The localization of Arabidopsis LAZY2, LAZY3 and LAZY4 proteins in root tips. Four-day old seedlings were grown on MS plates under white light, and then imaged by confocal microscope. The root tips were treated with 5 μM FM4-64 for 2 minutes, and the fluorescence of LAZY-GFP proteins and FM4-64 were collected from the root tips. Scale bars, 20 μm. The merged images are the same as those in the Figures 2A to 2C, whereas the images of separated channels are shown here for better view of the GFP signals. These are representative images of roots of 10, 8 and 13 seedlings, respectively.
(B) BH score predictions of LAZY2, LAZY3, LAZY4, and LAZY4 with mutated bh sites (LAZY4bh12). Predictions were made on https://hpcwebapps.cit.nih.gov/bhsearch/. Blue grid line refers to 0.6, which is a threshold for possible PtdIns binding.
(C) The schematic of LAZY4bh12 mutating K/R to A within two basic hydrophobic peaks.
(D and E) Measurement of LAZY-GFP fluorescence intensity ratio. Images showing two representative patterns of LAZY-GFP fluorescence in columella cells, with clear signals only in central columella (CC) cells (B), or in both of central columella (CC) cells and lateral columella (LC) cells (C). In each panel, the left is the original image, and the right image shows how the intensity ratio between basal and apical is calculated. Upper or lower side of the plasma membrane in columella cells were selected as region of interest (ROI) for measurement of LAZY-GFP fluorescence intensity. Signal ratio (basal/apical) for one root was calculated as an average of signal intensity ratios within this root.
(F and G) LAZY2, 3 and 4 proteins show polar distribution in the columella cells of Arabidopsis lateral roots under the regulation of gravity. (D) LAZY2, LAZY3 and LAZY4 proteins are mainly accumulated on the lower side of columella cells in the lateral roots (LRs) at the stage 2 under both regular growth (upper) or after 180° reorientation (lower). The primary roots of ProLAZY2:LAZY2-GFP (LAZY2-GFP/Col), ProLAZY3:LAZY3-GFP/lazy234 (LAZY3-GFP) and ProLAZY4:LAZY4-GFP/lazy234 (LAZY4-GFP) growing under white light conditions were aligned to the gravity vector on the 7th day and continued to grow until the 12th day. The fluorescence of LAZY-GFP was collected before (Upper in each panel), or after the seedlings were reoriented 180° and grown for another 6 h (Lower in each panel). Double-headed arrows show the apical-basal axis of the plant bodies. Red and white arrow heads indicate the accumulation of LAZY-GFP proteins on the apical and basal sides of columella cells respectively. For the whole seedlings, scale bars, 1 cm; for all the others, scale bars, 20 μm. Arrows labeled “G” indicate the direction of gravity. (E) The statistical analysis of LAZY-GFP fluorescence intensity ratios of the two sides of the lateral roots before and after 180° rotation. Asterisks indicate Student’s t-test values (***, P < 0.001; n = 12, 14, 12, 16, 12 and 13 respectively).
Figure S4. Amyloplast sedimentation promotes the redistribution of LAZY proteins, related to Figure 3
(A) Gravity-triggered redistribution of LAZY2 and LAZY4 proteins to the lower side of columella cells correlates with the sedimentation of amyloplasts. The seedlings of LAZY2-GFP (without CHX treatment) and LAZY4-GFP (with 10 μM CHX treatment to inhibit protein synthesis) were reoriented to a position with amyloplasts on the top of the columella cells, and kept for fluorescence collection at several time points. The area inside yellow box was zoomed to show details of fluorescence. White dash lines indicate the contour of root tips. Scale bars, 10 μm.
(B) Mutation of PGM1 delayed the redistribution of LAZY2-GFP, LAZY3-GFP and LAZY4-GFP proteins in root columella cells. Seedlings were grown vertically and then reoriented 90° and kept 1 h for gravistimulation (GS 1 h). Red and white arrowheads indicate the accumulation of LAZY-GFP proteins on the upper and lower sides of columella cells respectively. Scale bars, 20 μm. Statistical data are shown in Figures 3M to 3O.
In A and B, arrows labeled “G” indicate the direction of gravity.
Figure S5. Phosphorylation sites of LAZY4 protein in plants or in in vitro kinase assay, related to Figure 5
(A) This schematic summarizes the phosphorylation sites identified by MS in the IP-MS experiment in Figures 5A and 5B, and in the in vitro kinase assays in Figure 5E. The detailed information about the phosphorylation of these sites in each experiment is shown in Table S1.
(B) MS/MS spectra. X axis shows the m/z value of each fragment ion. Y axis shows the relative abundance (left) or ion intensity (right) of the corresponding daughter ions. Peptide sequences are displayed above the spectra.
Figure S6. Interactions between LAZY4 variants and TOC proteins, and mutation sites of TOCs, related to Figure 6
(A) Distinct TOC complexes in Arabidopsis. In Arabidopsis TOC complexes, TOC75 is a central pore component localized within the outer membranes of plastids, and other TOC proteins are receptors facing the cytosol. TOC33 and TOC159 are components of a class of TOC complexes that specifically import photosynthesis-related preproteins. TOC34, TOC120 and TOC132 are components of another class of TOC complexes that specifically import non-photosynthetic or house-keeping preproteins. TOC90 may be partially redundant with TOC159, and associates with TOC33 or TOC34. Abbreviations: TOC, Translocons at the Outer envelope membrane of Chloroplasts; OM, Outer Membrane; IMS, Intermembrane Space. The models of TOC complexes were drawn based on information from a previous review38.
(B) Yeast two-hybrid assays to test interactions between LAZY4 variants and TOC proteins. Interactions between LAZY4 variants and TOC proteins were examined by yeast growth on media with X-Gal but lacking Trp, His and Ura. The phosphorylation sites of LAZY4 were mutated to Asp (D) or Ala (A) as indicated in Table S1. Blue indicates positive interactions. Reporter: LacZ; substrate, X-Gal. These results were from the same experiment shown in Figure 6A, but with longer growth time (40 h) and more LAZY4 variants (mutated to A).
(C) Mutation sites of Arabidopsis TOC120 and TOC132 in the LAZY4-GFP/lazy234 toc120CR toc132CR (LAZY4-GFP/toc120CR toc132CR) and LAZY4-D9-GFP/lazy234 toc120CR toc132CR (LAZY4-D9-GFP/toc120CR toc132CR) seedlings. Two alleles of LAZY4-GFP toc120CR toc132CR showed similar phenotypes. For each TOC gene, two sgRNAs were designed to edit the genome. PAM sequences are labeled in red, and sgRNA sequences are underlined adjacent to the PAM in Col. The mutation details are shown for each allele, including deletion, insertion and substitution.
Table S2. MS1 intensity of phosphopeptide “FLNCPSSLEVDR”, related to Figure 5
Table S3. MPK3 and MKK5 Co-immunoprecipitated by LAZY4-GFP, related to Figure 5
Table S4. Phosphorylation of MPK3 in MPK3-GFP IP samples and LAZY4-GFP Co-IP samples, related to Figure 5
Table S5. Primers used in this study, related to STAR Methods
Video S1. 3-D view of LAZY3-GFP fluorescence in columella cells after gravistimulation, related to Figure 4
Growth condition, acquisition parameters and data-process details were described in Figure 4A. The 3-D construction view was rotated in the video for observation from different angles.
Video S2. Time-lapse record of LAZY3-GFP fluorescence recovery on plasma membrane in columella cells in FRAP experiment, related to Figure 4
Growth condition and bleaching parameters were described in Figures 4B and 4C. The fluorescence was acquired by the confocal laser scanning microscope (Zeiss LSM800) with a vertical objective table. Arrowheads labeled with numbers indicate a symmetric pair of columella cells with similar fluorescence before photobleaching.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD045213.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


