Abstract
The development of Alzheimer’s disease (AD) drugs has recently witnessed substantial achievement. To further enhance the pool of drug candidates, it is crucial to explore non-traditional therapeutic avenues. In this study, we present the use of a photolabile curcumin-diazirine analogue, CRANAD-147, to induce changes in properties, structures (sequences), and neurotoxicity of amyloid beta (Aβ) species both in cells and in vivo. This manipulation was achieved through irradiation with LED light or molecularly generated light, dubbed as “molecular light”, emitted by the chemiluminescence probe ADLumin-4. Next, aided by molecular chemiluminescence imaging, we demonstrated that the combination of CRANAD-147/LED or CRANAD-147/ADLumin-4 (molecular light) could effectively slow down the accumulation of Aβs in transgenic 5xFAD mice in vivo. Leveraging the remarkable tissue penetration capacity of molecular light, phototherapy employing the synergistic effect of a photolabile Aβ ligand and molecular light emerges as a promising alternative to conventional AD treatment interventions.
Keywords: Alzheimer’s disease, Phototherapy, Chemiluminescence, Curcumin, Molecular light
Graphical Abstract
A photolabile curcumin analogue CRANAD-147 is reported. It could lead to changes in properties, structures (sequences) and neurotoxicity of amyloid beta (Aβ) species in vitro and in transgenic 5xFAD mice in vivo with molecularly generated light (dubbed as “molecular light”) from chemiluminescence probe ADLumin-4. It has great potential as an alternative approach for AD drug discovery.
Introduction
Photomedicine, a branch of medicine harnessing the therapeutic and diagnostic potential of light, has found wide application in various medical fields [1]. Traditionally, photomedicine has relied on light sources generated through physical methods, including incandescent light bulbs, lasers, and LEDs. However, an intriguing alternative light source originates at the molecular level, produced by chemiluminescent and bioluminescent compounds, as well as Cerenkov luminescent tracers, including luciferins, luminol, and 18F-FDG, respectively [2]. Here, we refer to this molecularly generated light as “molecular light”. Molecular light has been extensively used in preclinical animal studies for imaging purposes [2a, 3] and has been considered as “self-illuminated” molecules [4]. Our research has demonstrated that molecular light from Cerenkov luminescence of 18F-FDG could be used for in vivo uncaging reactions [2b]. Recently, several reports suggested that molecular light can sensitize photosensitizers to generate reactive oxygen species in the photodynamic therapy [2a]. However, the potential of molecular light in phototherapy for brain disorders remains largely unexplored.
One of the limitations of physically generated light in phototherapy is its limited tissue penetration and inability to target locations beyond superficial areas (such as skin and surfaces of internal cavities) [5]. In contrast, a promising alternative is the use of “molecular light”, which behaves like molecules and can be delivered to specific targets of interest, binding to biomacromolecules and enabling nearly limitless tissue penetration. While physically generated light requires a large quantity of photon flux to externally illuminate the target, but only a few photons can actually reach the intended site within the body [5a]. By contrast, the total photon flux from molecular light is considerably smaller, but can be delivered to the target of interest in close proximity, with distances as small as sub-nanometers, allowing for chemi- or bio-luminescence resonance energy transfer (CRET or BRET) if the energy from the molecular light matches with the excitation energy gap of its pair molecular. The proximity between target-of-interest and molecular light is approximately ~106-fold closer than that of physically produced light (nanometer vs. centimeter) (Scheme 1). In these regards, we speculate that molecular light may have an equal or even superior capacity to initiate photoreactions in vivo compared to physically generated light.
Scheme 1.
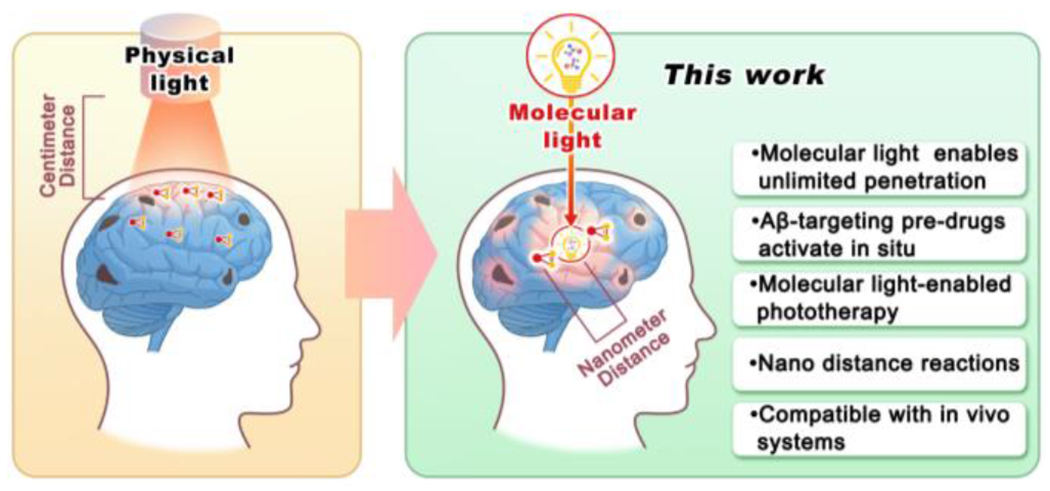
Illustration of phototherapy with physically produced light and near-field molecularly generated light (“molecular light”). Molecular light can be delivered as molecules to avoid tissue penetration limitations of light.
In this report, we focus on AD, a devastating neurodegenerative disorder with hallmarks of Aβ deposits. Aβ peptides are generated from amyloid peptide precursor (APP) by cleavage with β- and γ-secretases [6]. In the brains of AD patients, these soluble peptides gradually accumulate, leading to alternative tertiary conformations, self-assembly into amyloid protofilaments, and eventually the formation of amyloid plaques [7]. Current preclinical and clinical AD drugs primarily aim to the reduction of Aβ production, prevention of Aβ aggregation, and promotion of Aβ clearance [8]. Recent studies have explored strategies to reduce Aβ neurotoxicity by altering the amino acid structures within the Aβ sequence [9], particularly through catalytic photooxygenation of Aβ approaches [10]. However, these earlier studies have predominantly depended on the use of external physical light sources, resulting in limited light penetration of brains and posing a challenge for future clinical applications. In this report, we demonstrated that “molecular light” emitted by the chemiluminescence compound ADLumin-4 could effectively replace LED light in activating the photolabile curcumin-diazirine analogue CRANAD-147. This activation induces structural modifications to the amino acids within Aβ peptides and consequential attenuation of Aβ neurotoxicity. Furthermore, we propose the use of chemiluminescent ADLumin-4 as a deliverable light source for in vivo therapeutic treatment, which indeed significantly reduced Aβ burdens in 5xFAD mice when combined with CRANAD-147. Finally, we employ molecular imaging with our previously reported chemiluminescence probe ADLumin-1 [3a] to monitor the efficiency of phototherapy under LED light and ADLumin-4 (molecular light (Scheme 2A)) treatments. As a proof-of-concept, our study provides promising evidence for the potential application of “molecular light” in photodynamic therapy and the photochemistry of photolabile compounds both in cellular and in vivo settings, particularly in the attenuation of Aβ neurotoxicity.
Scheme 2.
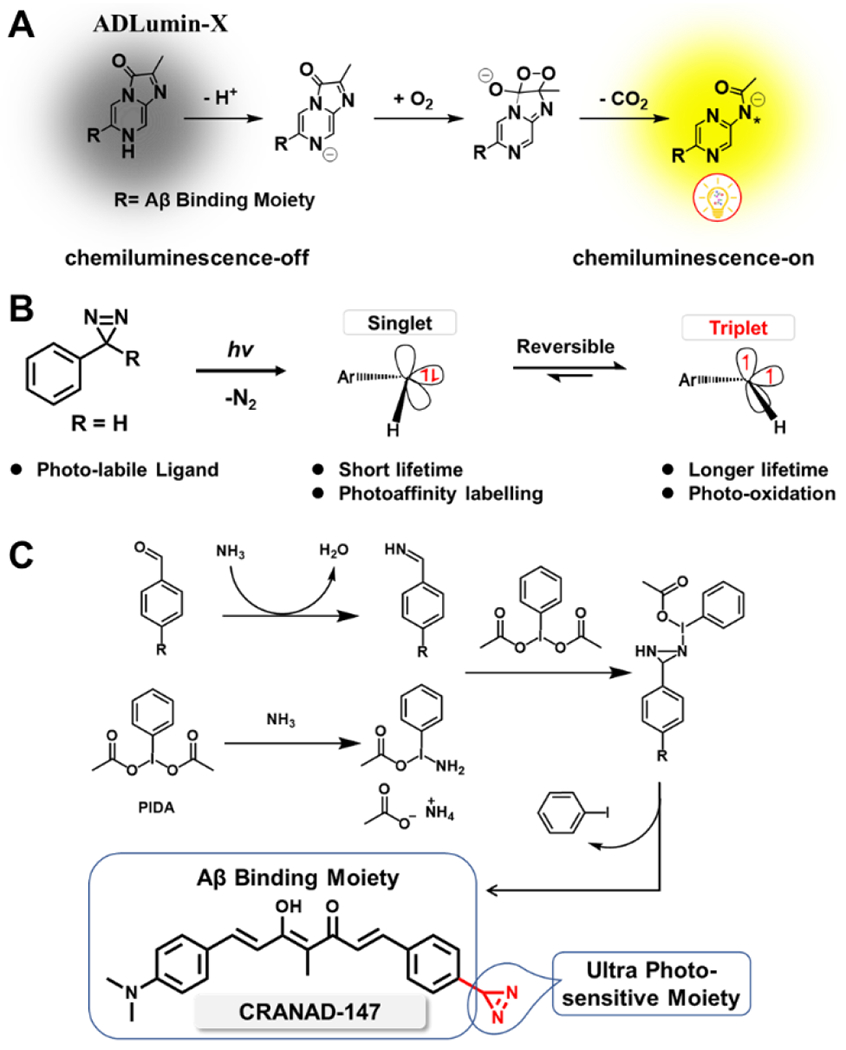
A) Proposed oxygen-dependent mechanism of ADLumin-4 for chemiluminescence generation.[3a, 25] B) Reaction mechanism and photoreaction of diazirines. C) The synthetic route for CRANAD-147, the designed photo-labile curcumin-diazirine analogue with two conjugated parts: the modified curcumin as the Aβ binding moiety and the aryl diazirine as the photosensitive moiety.
Results and Discussions
Design and synthesis of CRANAD-147
Abundant evidence suggests that curcumin and its analogues exhibit a high binding affinity to Aβ species [11]. In our previous work, we have reported a series of curcumin-based imaging probes, known as CRANAD-Xs, targeting various Aβ species, including near-infrared fluorescence probes [12], two-photon probes [13], as well as reactive oxygen active probes [7e, 14]. Specifically, our previous studies showed evidence that curcumin analogues, namely CRANAD-3, -17, and -58, effectively bind to the Aβ13-20 (HHQKLVFF) fragment [12]. Similarly, Masuda et al. also demonstrated the interaction between curcumin and this Aβ fragment [15]. Based on these results, we propose using the curcumin scaffold as an anchor for targeting Aβ species. Notably, curcumin is known for its instability and limited bioavailability in vivo [16]; however, structural modification of curcumin, including replacing the reactive phenolic group, could significantly improve its stability in vitro and in vivo [16]. In our compound design, a modified curcumin structure was used as the anchor [12, 14b, 17]
Over the past decades, numerous photolabile reactions have been extensively explored in the fields of photochemistry and biological studies [18]. Various photolabile moieties have been widely employed for diverse applications [18a, 19]. Among these moieties, the diazirine group stands out due to exceptional light sensitivity and small size as a tag [20]. The first diazirine photoreaction was reported as early as 1973 [20–21], and since then, this photosensitive reaction has found applications in numerous biological studies [20]. In recent years, diazirine has emerged as an attractive photochemistry tool for various purposes, such as photo-crosslinking, bioconjugations, photo-labeling, target engagement identification, and protein-protein interactions [20]. Previously, the diazirine moiety was typically installed at the non-terminal position of an aliphatic chain, requiring short-wavelength light for activation and necessitating a prolonged irradiation [22]. The short-wavelength requirement and low sensitivity pose challenges in terms of biocompatibility. Considering the above facts, we propose to introduce the diazirine moiety into the long π-π conjugated curcumin system to extend its wavelength of the photoreaction and enhance its photosensitivity (extended π-π conjugation lowers the energy required for activation). In principle, diazirine derivatives have the potential as photosensitizers via the triplet state of the carbene [20, 23] (Scheme 2B). However, this feasibility has rarely been explored. In these regards, we designed CRANAD-147 by attaching the small diazirine moiety to the terminal of the curcumin scaffold (Scheme 2B and Fig. 1A).
Figure 1.
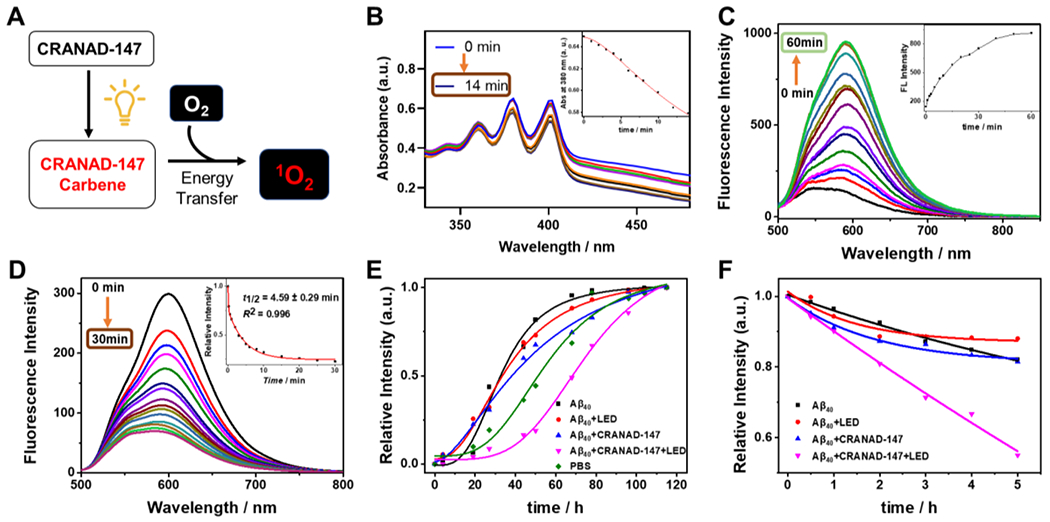
CRANAD-147 mediated generation of singlet oxygen (1O2), its binding with Aβ aggregates, and the changes of Aβ seeding capacity and degradation after LED irradiation. A) Schematic diagram of light-mediated 1O2 generation in the presence of CRANAD-147. B) The UV–Vis absorption spectra of ADPA for monitoring of 1O2 generated by CRANAD-147 (AD-147) under light irradiation. C) Fluorescence spectra of CRANAD-147 after incubation with Aβ aggregates with different time durations. D) The changes in fluorescence intensity of CRANAD-147 at different time points after LED irradiation. E) Relative fluorescence intensity of thioflavin T and time-course of Aβ aggregation in the seeding experiments, in which the Aβ seeds were pre-treated with different conditions. The fluorescence was measured with excitation and emission parameters for ThioT (λex/em= 435/495 nm). F) Time-course of the degradation of Aβs with proteinase K (based on ThioT fluorescence). The degradation was accelerated by the photoreaction of CRANAD-147 after LED irradiation.
Recently, Reboul’s group reported a series of terminal diazirines that can be used for para-hydrogen-induced hyperpolarization [24]. In their synthesis study, they found that phenyliodine (III) diacetate (PIDA) could catalyze imine oxidation to generate diazirine in the presence of NH3. Inspired by this methodology, we explored its application and successfully synthesized CRANAD-147 via a phenyl-imine intermediate (Scheme 2C). To the best of our knowledge, CRANAD-147 represents the first example of a diazirine positioned at the terminal position of a long conjugated aromatic system.
Photosensitivity studies of CRANAD-147
Firstly, we conducted photosensitivity studies to assess the activation efficiency of CRANAD-147 under visible light at 470 nm in PBS buffer solutions. We found that CRANAD-147’s half-life (t1/2) is 4.6 minutes upon 1-minute irradiation. High-performance liquid chromatography (HPLC) analysis revealed the emergence of a new peak with higher polarity upon irradiation (Fig S1). Mass spectrometry results indicated that the major peak at m/z=377.2 likely originated from methanol (solvent) attacking the carbene intermediate. Another apparent peak at m/z = 388.2 could be ascribed to the photoaffinity reaction with the solvent acetonitrile (Fig. S2). A product resulting from the water-carbene reaction was found as m/z=363.2. The presence of multiple products from the photoreaction aligns with the inherent reactivity of carbene intermediates.
To investigate the potential of CRANAD-147 as a photosensitizer, we monitored the generation of singlet oxygen (1O2) levels by measuring the absorbance of 9,10-anthracene dipropionic acid (ADPA), which acts as a trap for 1O2 [26]. Indeed, under the LED irradiation, the characteristic absorption peak of ADPA at about 380 nm gradually decreased with increasing irradiation time, indicating the presence of 1O2 generated by CRANAD-147 (Fig. 1B). These findings inspire us to further investigate the oxidization effect of CRANAD-147 towards Aβ aggregates.
Binding of CRANAD-147 with Aβ aggregates and Aβ property changes after light irradiation
We initially investigated the binding affinity of CRANAD-147 towards Aβs. Upon binding to Aβ40 aggregates, CRANAD-147 displayed a modest 11.3-fold increase in fluorescence after 60 minutes of incubation. Meanwhile, the quantum yield increased from 0.003 to 0.026. The time course of fluorescence increase demonstrated that a binding equilibrium was reached within 60 minutes (Fig. 1C). We also performed fluorescence titration with Aβ aggregates and obtained a good binding with Kd value of 2.89 ± 0.43 μM (Fig. S3). This binding affinity is comparable to that of curcumin itself with Aβ aggregates [27]. These results suggested that the introduction of diazirine moiety is compatible with the curcumin scaffold’s binding to Aβs.
We reasoned that the diazirine group would be activated under light irradiation and produce highly reactive carbene intermediates that can react with the nearby Aβ peptides that are binding with the curcumin scaffold. To verify this speculation, we first recorded the fluorescence intensity change of CRANAD-147 in the presence of Aβ40 aggregates over time (Fig. 1D) after exposing the mixture under an LED array light (4 W/cm2, 470 nm) for 2 seconds at each time point. We observed significant intensity decreases upon light irradiation with a half-life of 4.59 minutes (for 2.5 μM CRANAD-147 with 500 nM Aβ40 aggregates). We also performed control experiments with thioflavin T (ThioT), which can bind Aβ aggregates but is not photolabile. Although we also observed fluorescence intensity decreases, its half-lifetimes (13.8 minutes) were much longer than that of CRANAD-147 (Fig. S4). These results suggested that CRANAD-147 is photolabile and undergoes conversion to new products.
To further investigate the reaction between CRANAD-147 and Aβs, we carried out fluorescence rescue experiments by adding fresh CRANAD-147 to the photo-reaction solution. Interestingly, we only observed less than 10% intensity recovery, suggesting that the properties of Aβ aggregates were irreversibly altered during the irradiation (Fig. S5). Consequently, we investigated whether the photo-reaction can lead to property changes of Aβ aggregates. Seeding experiments with CRANAD-147/Aβ/LED and CRANAD-147/Aβ/dark were conducted, revealing that the seeds from the light-irradiation group exhibited significantly reduced capacity to Aβ aggregation formation (half-time of aggregation: 33.6 vs. 68.2 hours) (Fig. 1E). In addition, proteinase K digesting experiments demonstrated that the irradiated Aβ/CRANAD-147 mixture underwent much faster degradation (Fig. 1F). Collectively, these findings suggest that this photoreaction could alter the properties of Aβ aggregates.
Given that the neurotoxicity of CRANAD-147 during its therapeutic action is a concern, we specifically investigated whether the photo-oxidation process was confined within Aβ aggregates, minimizing reactions outside the aggregates. To this end, we used catechol as a cocatalyst since it has been reported to increase photon-induced free radical reaction efficiency [28] while almost no binding capability to Aβ aggregates. Thus, if the reactive oxygen leaks from the Aβ aggregate, it will react with catechol and produce downstream oxidative side-products. As expected, the fluorescence of CRANAD-147 diminished after light irradiation, even in the presence of high catechol concentrations. More importantly, no acceleration of the fluorescence decay was observed, suggesting that catechol can’t interact with the reactive oxygen to catalyze the production of more radicals. Interestingly, we observed that catechol could lead to lower fluorescence intensities of CRANAD-147, which is probably due to the competitive effect of catechol absorption. Collectively, these results indicate no leakage of sensitized oxygen into the surroundings of Aβ aggregates (Fig. S6). This result strongly suggests that the photoreaction of Aβ peptides remains confined to the Aβ aggregates, with minimal impact on nearby proteins.
We further investigated the property changes of Aβs using SDS-PAGE gel electrophoresis. After gel electrophoresis, silver staining and western blotting were performed to image the gels (Fig. 2A/B and Fig. S7). From the images, several new bands that migrated slower than the control group could be unambiguously identified, suggesting that Aβs were crosslinked while being irradiated in the presence of CRANAD-147. According to the standard gel ladder, these bands could be ascribed to soluble monomers, dimers, trimers, and tetramer. Moreover, time-dependent irradiation demonstrated that the intensity of the bands corresponding to soluble oligomers significantly increased with higher light doses (Fig. 2C, Fig. S8). We reasoned that these Aβ oligomeric bands contained oxidized Aβs instead of native Aβs. Indeed, we observed that there was less precipitation from the tube under irradiation with CRANAD-147, compared to the tube without irradiation (Fig. 2D), suggesting the irradiation resulted in better solubility of Aβs, likely due to polarity increases caused by oxidation of amino acid residues. Consistent with this phenomenon, particle size analysis revealed smaller sizes in the irradiated samples (Fig. 2E). This modification could be the reason why the irradiated Aβs can be easily degraded by proteinase K (Fig. 1F).
Figure 2.
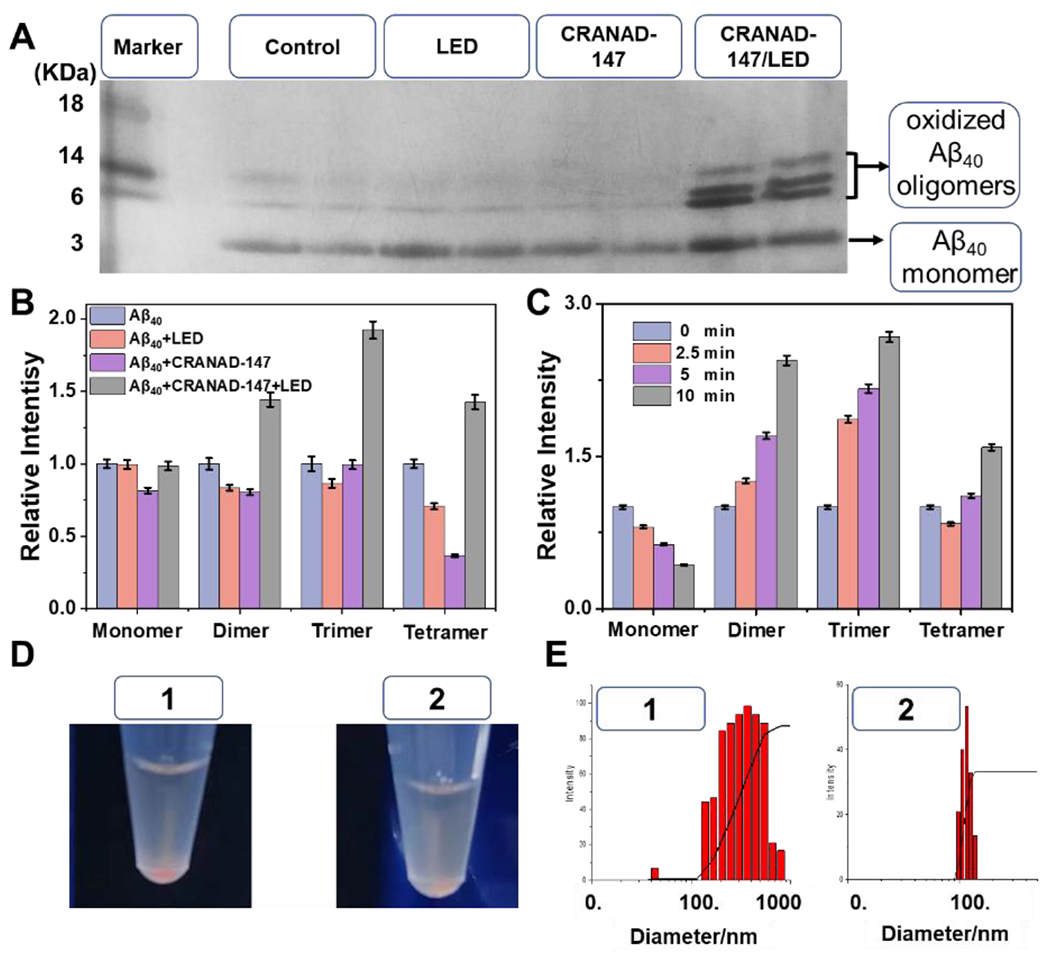
The property changes of Aβs after LED irradiation. A) SDS-Page gel of Aβs after different treatments. B) Relative intensity of each band from different groups by normalizing with the corresponding band in the control group (Aβ40 only) in (A). C) Relative intensity of monomeric and oligomeric Aβs after different irradiation times with CRANAD-147. The normalization is based on the control group (0 min). D) Photographs of mixtures of Aβs and CRANAD-147 with/without LED irradiation (left: more precipitates). E) Particle sizes of the mixtures CRANAD-147 after LED irradiation. In (D) and (E): No.1 without irradiation and No.2 with LED irradiation.
Identification of structural change of amino acids of Aβ peptides
We first characterized the Aβ changes with LC-MS. We found several clusters of MS peaks. As shown in Fig. 3C, a cluster of m/z = 860.8, 870.0, 872.9, and 876.8 represents Aβ40 monomer with five charges (5+), oxidized Aβ40 monomer with one, two, and three oxygens with five charges (5+), respectively. Similarly, clusters of m/z= 1083.2, 1087.2, and 1091.2, m/z = 1443.9, 1449.4, and 1454.6 represent Aβ monomers and oxidized monomers with four (4+) and three charges (3+), respectively. All the results suggested that the monomeric bands in Fig. 2A contained plenty of oxidized monomers. Interestingly, we also found clusters of oligomerized Aβs and oxidized oligomers (Fig. 3D, tetramers, and octamers). We further used MALDI-MS to confirm the existence of irreversible oligomerized Aβs, evident by peaks of dimer (m/z=8653.7), trimer (m/z=12990.9), and larger oligomeric Aβs. Importantly, we noticed that m/z=17348.7, 26030.5, 30361.1, 34693.9 were not exact masses of the corresponding oligomeric native Aβs (Fig. 3E). Instead, for each Aβ unit, an 8-Dalton higher mass could be found when it is compared to the native Aβ40 peptide, indicating half of the Aβ units were oxidized (+16 Dalton) in these oligomers (Fig. 3E). As expected, we could not find such characteristic oxidized peaks and oligomerized peaks from the non-irradiated control groups (Fig. S9, S10, S11).
Figure 3.
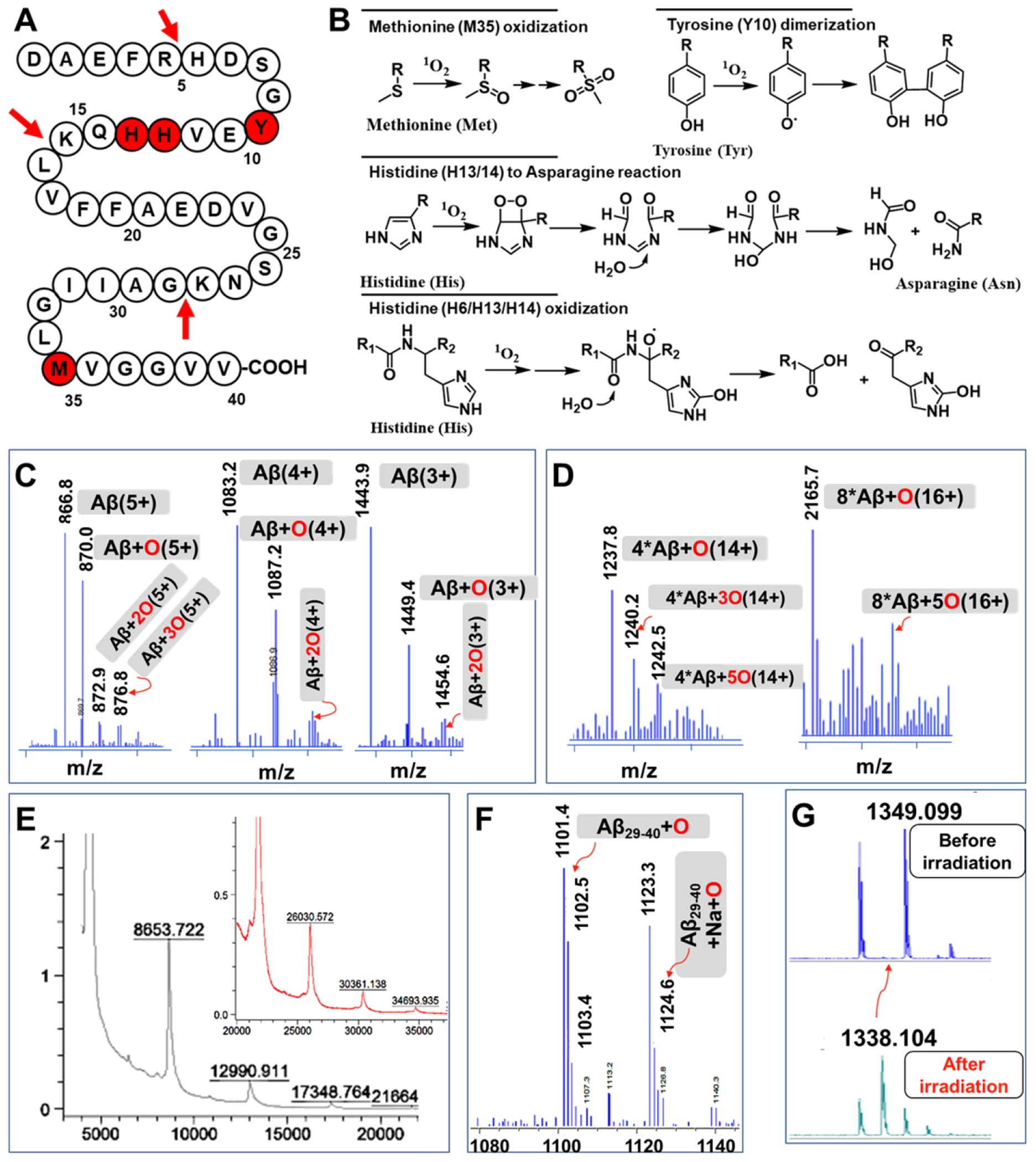
MS characterization of structural changes of Aβs after LED irradiation. A) Schematic diagram of the different modifications (red spheres) of Aβ40 during the light-mediated oxidation, and trypsin cleavage site (red arrows). B) The possible modifications of amino acids in the presence of 1O2. C) Mass spectra of oxidized monomeric Aβs with different charges. D) MS of oxidized oligomeric Aβs. E) MALDI-TOF MS of oligomeric Aβs and their oxidized products. F) MS of oxidized Aβ29-40. G) MALDI-MS of oxidized Aβ6-16 fragment.
To further investigate the photo-oxidized products, the sample mixtures were digested by trypsin and analyzed by LC-MS. It is well-documented that trypsin will selectively cleavage the C-terminal of Arginine (R) and lysine (K) (as shown in Fig. 3A) [29]. In the case of the Aβ40 peptide, the possible trypsinized fragments include Aβ1-5 (DAEFR), Aβ6-16 (HDSGYEVHHQK), Aβ17-28 (LVFFAEDVGSNK), and Aβ29-40 (GAIIGLMVGVV). From the LC-MS of the trypsinized mixture, as shown in Fig. S12, fragments Aβ1-5 and Aβ17-28 could be easily found since their m/z signatures are readily identifiable. M/z =637.2 and 319.2 (2+), and m/z =1325.4 and 663.3 (2+) were corresponding to Aβ1-5 and Aβ17-28, respectively. Aβ29-40 also could be found (m/z=1085.6 and 543.3) (Fig. S12); however, interestingly, several high molecular weight peaks could also be observed. After careful analysis, we assigned the peaks to Aβ29-40+O (m/z =1102.5, m/z =1123.4 plus sodium ion) (Fig. 3F/S12), suggesting that Aβ29-40 fragment was oxidized and this oxidation is in accordance with the above MALDI-MS results. Methione-35 is the likely amino acid to be oxidized since it is the most vulnerable residue for the oxidation [30]. It has been reported that oxidation of Met-35 to the sulfoxide could lead to less neurotoxicity of Aβs by altering the production of toxic Aβ oligomers [30c, 31]. Nonetheless, fragment Aβ6-16 was difficult to identify after irradiation, suggesting some structural changes occurred for this fragment. Likely, the Histidine-13 or -14 was oxidized and degraded [30, 32].
To further find evidence, we used high-resolution MALDI-MS to investigate the trypsinized mixture without liquid-chromatography separation. Consistent with the data from LC-MS, as shown in Fig. S12, the trypsinized fragments Aβ1-5, Aβ17-28, Aβ29-40, and Aβ29-40+O could be found. Notably, there was one new peak (m/z =1338.1, Fig. 3G/S13) could be easily observed for the irradiated sample. Given that histidine (H) could be photo-oxidized into Aspartic acid (D) or Asparagine (N) and tyrosine could also be photo-oxidized[32–33], we assigned m/z= 1338.1 to a modified Aβ6-16 fragment NDSGY(+O)EVNH(+2O)QK, in which two histidine (H6 and/or H13 or H14) were converted into Asparagine, and one histidine and tyrosine were oxidized. Taken together, both LC-MS and MALDI-MS data provided evidence that the structure of Aβ6-16 was changed. Collectively, our MS results suggested several structural modifications (covalent oligomerization, oxidation of amino acid residue) of Aβ peptides under the LED irradiation with CRANAD-147 (Fig. 3A/B). These structural modifications are similar to previously reported results from the Kanai group [34]. However, they didn’t identify which amino acids or fragments were modified.
Replacement of LED light with molecular light for in vitro studies
In this report, we propose to use the molecularly generated light from chemiluminescence probes to replace LED light. Our previous report showed that ADLumin-1 is an excellent Aβ targeting chemiluminescence probe [3a]. It can transfer the energy from itself to the near-infrared fluorescence probe CRANAD-3 via CRET [3a]. Herein, given that the emission between ADLumin-1 and CRANAD-147 only have a 10-nm separation, suggesting that ADLumin-1 and CRANAD-147 are not an ideal CRET pair. In order to solve this problem, we synthesized ADLumin-4 (Fig. 4A, Scheme 2A), which has a shorter wavelength of emission. Compared with ADLumin-1, ADLumin-4 has only one double bond, and its emission is 50 nm shorter, and this emission has a much better spectral overlap match with the absorption spectrum of CRANAD-147. Indeed, the energy transfer efficiency was quite high between ADLumin-4 and CRANAD-147, evident by the low signal at 500 nm after CRET (Fig. 4B, red line). The luminescent emission spectrum of ADLumin-4 is shown in Fig. S14. We further investigated the CRET-induced photoreaction of CRANAD-147 in the presence of Aβs. To this end, CRANAD-147 (1.0 μM) was first incubated with Aβ aggregates for 60 min. After that, different concentrations of ADLumin-4 were added. Next, we used the SDS-PAGE gel electrophoresis to analyze the products. As shown in Fig. 4C/D, the oligomeric bands could be easily observed from the CRANAD-147/ADLumin-4 pair, which is consistent with the results from LED irradiation of Aβ/CRANAD-147. These results suggested that molecular light from ADLumin-4 could be used to replace LED light for in vitro studies. We also performed proteinase K hydrolysis with Aβ/CRANAD-147/ADLumin-4, and found that, similar to the LED irradiation, ADLumin-4 could accelerate the hydrolysis of the aggregated Aβs (Fig. 4E). Lastly, we performed LC-MS analysis for the mixture of CRANAD-147/ADLumin-4/Aβs, and found oxidized monomers (m/z=1449.6), tetramers (m/z=1237.9), octamers (m/z=2165.1) (Fig. 4F), which were similar to the results from CRANAD-147/LED/Aβs mixture (Fig. 3C/D).
Figure 4.
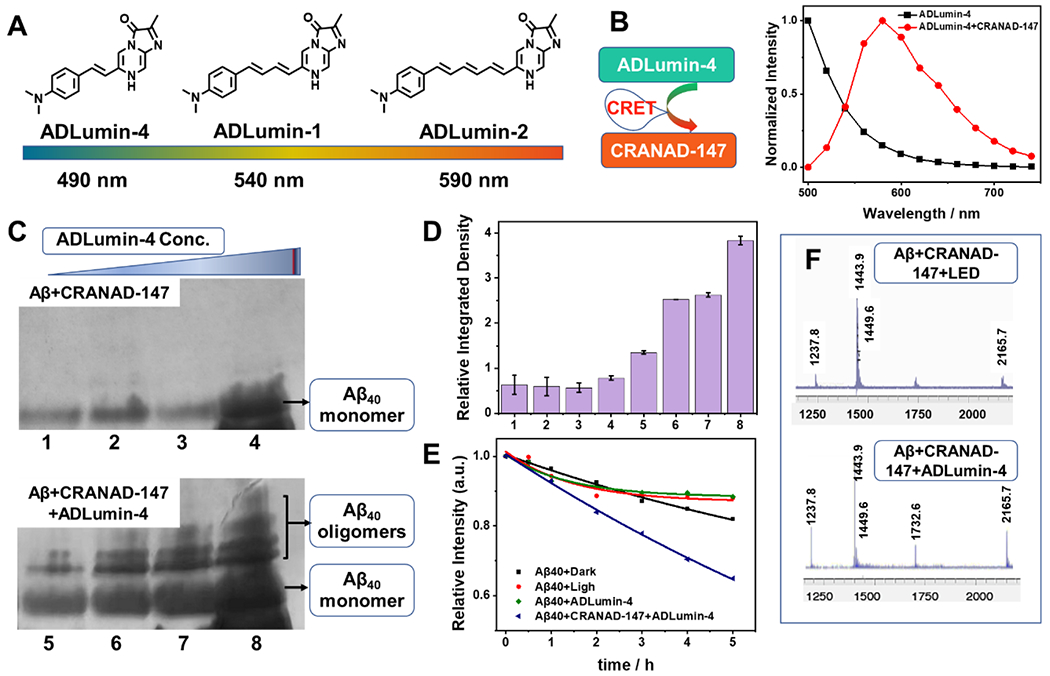
Replacement of LED irradiation with molecular light from ADLumin-4. A) Structures and maximum emissions of ADLumin-Xs (X= −1, −2, and −4). B) CRET between ADLumin-4 and CRANAD-147 suggested high transfer efficiency. C) SDS-Page of Aβ bands (upper panel: Aβ controls without CRANAD-147) with increased concentrations of ADLumin-4 (lower panel). D) Quantitative analysis of the oligomeric bands in (C). E) Time-course of proteinase K degradation of Aβs measured with ThioT fluorescence (Note: the control groups were the same as in Fig. 1F). F) MS of oxidized monomeric and oligomeric Aβs with LED (upper) and with molecular light from ADLumin-4 (bottom).
Lastly, to validate the 1O2 production from CRANAD-147 with molecular light (ADLumin-4), we used ADPA. As shown in Fig. S15, with the molecular light, the characteristic absorption peak of ADPA at around 380 nm decreased gradually as the irradiation time increased, indicating the generation of 1O2. Taken together, our data (SDS-Page gel, proteinase K, and LC-MS) suggested that molecular light from ADLumin-4 could initiate a photo-oxidation reaction in vitro in the presence of Aβs.
CRANAD-147 attenuated Aβ’s cell toxicity with LED irradiation and molecular light
We evaluated the relationship between Aβ property changes and its toxicity. First, we examined cell viability with different treatments of Aβ oligomers using an adenosine triphosphate (ATP) assay kit. ATP can be used to measure cell proliferation and cell cycle dynamics, and its concentration decreases if cell viability decreases [35]. As shown in Fig. 5A, we found that the viability result from SH-SY5Y cells, a neuroblastoma cell line, were apparently increased from the Aβ/CRANAD-147/LED group, compared to the group only with Aβ or Aβ/CRANAD-147. Similarly, after incubating with ADLumin-4 and CRANAD-147 together with Aβs, the cell viability exhibited a noticeable increase, compared to other groups. Next, the neurotoxicity of Aβs was evaluated using an adenylate kinase assay method. Adenylate kinase is an essential intracellular enzyme [36]. When a cell is damaged, the kinase leaks through the cell membrane into the medium. Therefore, measuring the medium concentrations of adenylate kinase can be used to report cell toxicity. As shown in Fig. 5B, the Aβs/dark group showed the highest toxicity towards the neuroblastoma cells, while the Aβs/CRANAD-147/LED group and Aβs/CRANAD-147/ADLumin-4 groups showed decreased toxicity.
Figure 5.
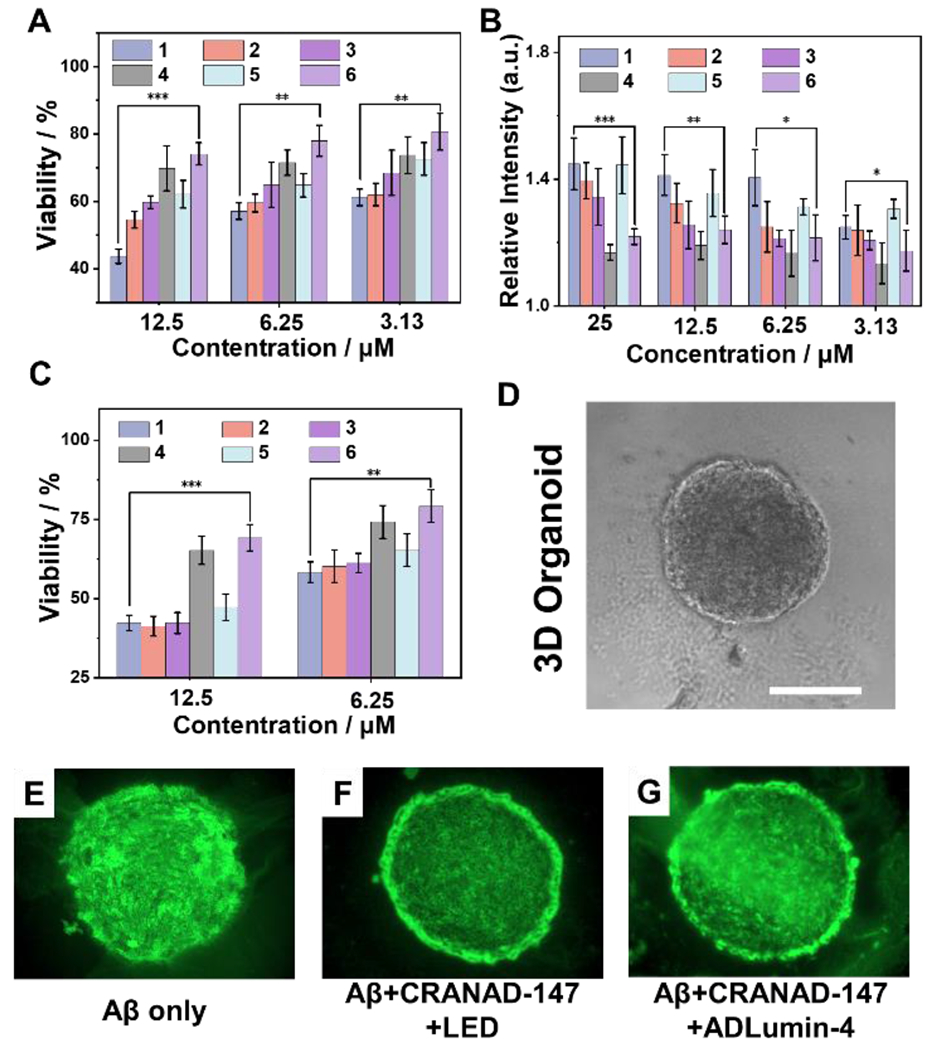
Cell viability studies with different treatments. A) Cell viability assays by ATP kit of SH-SY5Y cells with different Aβs. B) Aβ neurotoxicity reported via adenylate kinase assays with SH-SY5Y cells. C) Cell viability assessment with ATP method for 3D organoids from iPS cell cultures. Group 1: Aβ only; group 2: Aβ + LED; group 3: Aβ + CRANAD-147; group 4: Aβ + CRANAD-147 + LED; group 5: Aβ + ADLumin-4; group 6: Aβ + CRANAD-147 + ADLumin-4. *** P < 0.001, ** P < 0.01, * P < 0.05. D-G) Representative images of 3D organoid slices visualized by ApopTag(®) for apoptosis induced by Aβs. Scale bar: 500 μm.
Considering SH-SY5Y is a cancer cell line, it may not be a good model to mimic normal neuronal cells. In this regard, we used 3D organoids of induced pluripotent stem cell (iPSC) derived neurons to perform the above ATP cell viability studies (Fig. 5D). The results showed that CRANAD-147/LED and CRANAD-147/ADLumin-4 groups could significantly reduce Aβ’s toxicity (Fig. 5C). We further performed a TUNEL assay with the 3D organoid slices via imaging with ApopTag(®) kits. The results suggested that CRANAD-147/LED and CRANAD-147/ADLumin-4 groups had less apoptosis (Fig. 5E–G).
In vivo therapeutic studies with CRANAD-147
Based on the above promising results in vitro, we investigated whether photolabile CRANAD-147 has therapeutic effects under LED light and with molecular light. Firstly, we performed mimic studies in a biologically relevant environment. To this end, we used brain homogenates from wild-type mice to examine whether structural modifications of Aβ could be observed. We externally added Aβs to the brain homogenates, and the CRANAD-147/Aβ group was irradiated with 470 nm LED light for 5 min. As shown in (Fig. S16), after being treated with the CRANAD-147/LED group displayed an apparent band (band 4) that could not be found in all the other groups, suggesting that CRANAD-147 can selectively bind to Aβ to induce changes in its properties in biologically relevant conditions.
To validate whether CRANAD-147 and ADLumin-4 can bind to Aβ species in biologically relevant environments, we incubated the compounds with brain slices from a 5xFAD mouse. As expected, both compounds can label the plaques in the brain (Fig. 6A), suggesting that both compounds are specifically binding to Aβ deposits.
Figure 6.
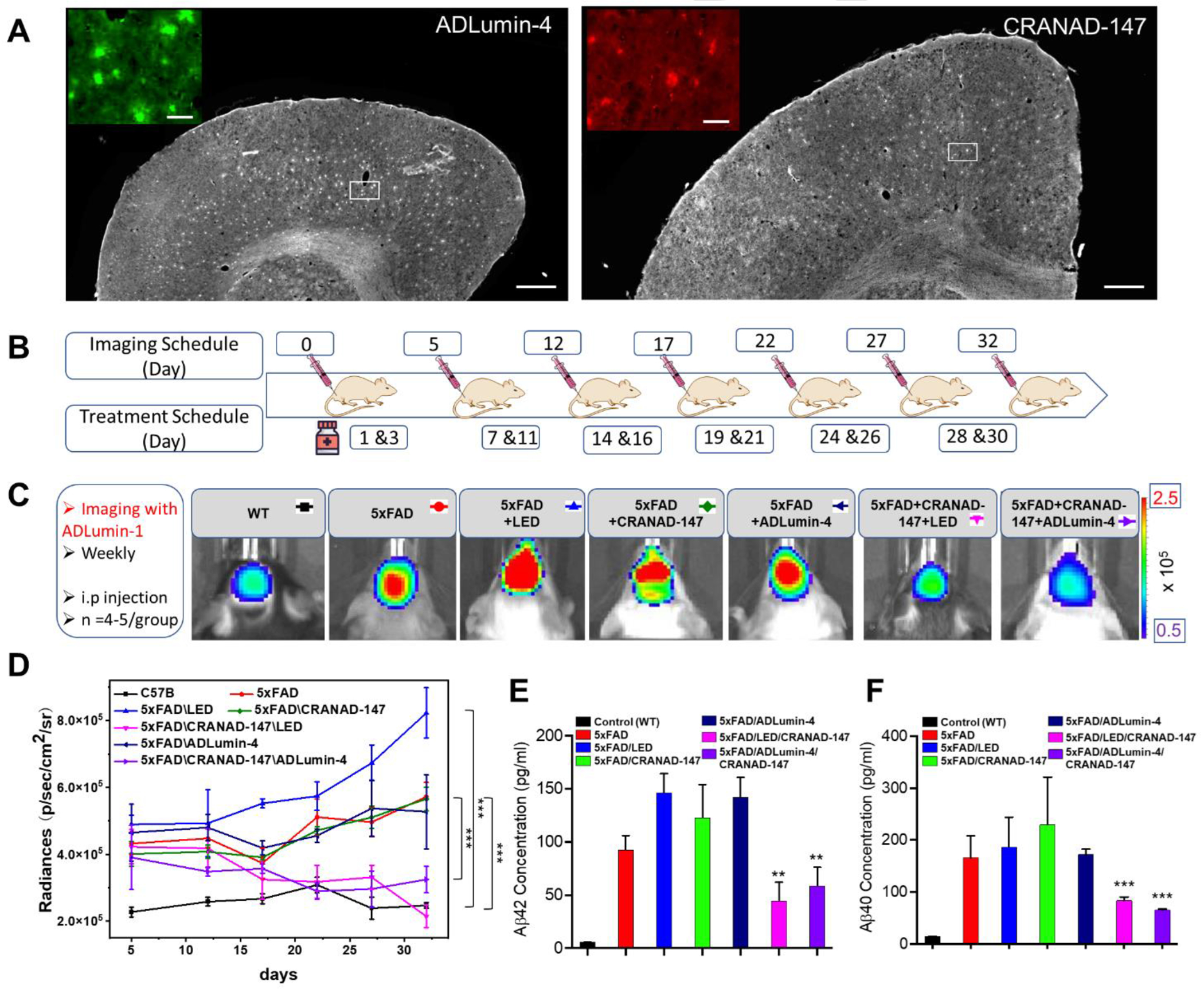
Histological staining, in vivo therapy and in vivo imaging. A) Histological staining of brain slides of a 5xFAD mouse (8-month-old) with ADLumin-4 (left) and CRANAD-147(right). Scale Bars: 500 μm. Insert: Zoomed-in image of the white box area. Scale Bars: 50 μm. B) Diagram of in vivo imaging and treatment schedule. C) Representative images of different groups at Day 32. D) Quantitative analysis of chemiluminescence intensity of images captured at 30 minutes after i.p injection of ADLumin-1. There are significant differences between controls and experimental groups with LED irradiation (pink) or molecular light from ADLumin-4 (purple). E) Immunoassay-measured concentrations of Aβ42, and F) Concentrations of Aβ40, depicting statistical significance analysis between 5xFAD groups and 5xFAD/CRANAD-147/ADLumin-4 groups. *** P < 0.001, ** P < 0.01
To investigate whether CRANAD-147/LED and CRANAD-147/ADLumin-4 combination can reduce Aβ burdens in vivo, our initial focus was on investigating the biostability of CRANAD-147 and ADLumin-4 in the phototherapy system. In this regard, we employed an in situ CRET effect to assess the simultaneous presence of both ADLumin-4 and CRANAD-147 in the mouse brains. As shown in Figure S17, the signals from the CRET pair group are apparently higher than that from the ADLumin-4-only group for over 2.5 hours. Given that CRANAD-147 itself can’t emit chemiluminescence, the increased signals from the CRET pair indicate the occurrence of CRETing and better tissue penetration with longer emissions from CRANAD-147. The results also suggest the considerable co-existence stability of the CRET pair and good stabilities of both ADLumin-4 and CRANAD-147 in vivo. Next, we treated 6 groups of transgenic 5xFAD mice under different treatments, as shown in Fig. 6B, for one month. The mice were administrated CRANAD-147 (2.5 mg/kg) every two days and irradiated with LED light (15 minutes) or treated with ADLumin-4 (10.0 mg/kg) (administrated together with CRANAD-147). We monitored the progress every five days with our previously reported chemiluminescence probe ADLumin-1 via i.p. injection at different treatment time points. In the meantime, we also used a group of wild-type (WT) mice, which do not have overexpressed “humanized” Aβ and thus have no changes in Aβ concentrations, as the control group to ensure our monitoring method is reliable.
As shown in Fig. 6C/D, during the time course, there was no significant chemiluminescence intensity increase from the WT group, suggesting that our monitoring method is trustworthy. As expected, we observed a steady increase in intensity from the 5xFAD groups that were not treated with CRANAD-147. Similarly, we observed increasing trends of chemiluminescence signals from groups that were treated with CRAND-147 only (no LED irradiation) or treated with ADLumin-4 only (no CRANAD-147). By contrast, no significant increases in chemiluminescence signals were observed when the 5xFAD mice were treated with CRANAD-147/LED (pink line) or CRANAD-147/ADLumin-4 (purple line), indicating the combinations of CRANAD-147 with light that from LED or ADLumin-4 molecules are necessary to reduce Aβ burdens during treatments.
To validate the above imaging results, we sacrificed the mice at the end of the imaging session, and the brains were homogenized and extracted with TBST buffers. With the brain extracts, Aβ concentrations were measured via MSD (Meso Scale Discovery) immunoassays [37]. As expected, the groups of CRANAD-147/LED (pink) or CRANAD-147/ADLumin-4 (purple) showed significantly lower concentrations of Aβ42 and Aβ40 (Fig. 6E/F), which is consistent with the above in vivo imaging data.
Conclusion
In the report, we presented novel approaches for phototherapy of AD via attenuating Aβ’s neurotoxicity and reducing the accumulation of Aβs. Likely, the photoreaction with photolabile CRANAD-147 changed the properties of Aβ via the oxidation of vulnerable amino acids such as methionine and histidine. These oxidization processes can convert neurotoxic Aβ fibers to amorphous forms, which greatly increase solubility and accelerate its degradation by the proteasome. Remarkably, we demonstrated that molecularly generated light (molecular light) could have similar effects as LED light on the photoreaction of the photolabile CRANAD-147, suggesting that molecular light could have the potential to overcome the limitations of externally applied light. Lastly, we demonstrated that the combination of a photolabile compound and LED light or molecular light could reduce Aβ burdens during treatment. Taken together, our studies offer promising new approaches for AD therapy and open up a new avenue for in vivo photochemistry.
Supplementary Material
Acknowledgments
This work was supported by NIH R01AG055413, R21AG065826, and S10OD028609 awards (C.R.).
Footnotes
Supporting information for this article is given via a link at the end of the document.
Researcher Twitter usernames: @RanLabMGH
References
- [1].Moore KC, Photomed. Laser Surg 2013, 31, 563–564. [DOI] [PubMed] [Google Scholar]
- [2].a) Zhang Y, Hao Y, Chen S, Xu M, Front. Chem 2020, 8, 770. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ran C, Zhang Z, Hooker J, Moore A, Mol. Imaging Biol 2012, 14, 156–162. [DOI] [PubMed] [Google Scholar]
- [3].a) Yang J, Yin W, Van R, Yin K, Wang P, Zheng C, Zhu B, Ran K, Zhang C, Kumar M, Shao Y, Ran C, Nat. Commun 2020, 11, 4052. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yang X, Qin X, Ji H, Du L, Li M, Org. Biomol. Chem 2022, 20, 1360–1372 [DOI] [PubMed] [Google Scholar]; c) Liu S, Su Y, Lin MZ, Ronald JA, ACS Chem. Biol 2021, 16, 2707–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Zambito G, Chawda C, Mezzanotte L, Curr. Opin. Chem. Biol 2021, 63, 86–94 [DOI] [PubMed] [Google Scholar]; e) Syed AJ, Anderson JC, Chem. Soc. Rev 2021, 50, 5668–5705 [DOI] [PubMed] [Google Scholar]; f) Li S, Ruan Z, Zhang H, Xu H, Eur. J. Med. Chem 2021, 211, 113111. [DOI] [PubMed] [Google Scholar]; g) Love AC, Prescher JA, Cell Chem. Biol 2020, 27, 904–920 [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Su TA, Bruemmer KJ, Chang CJ, Curr. Opin. Biotechnol 2019, 60, 198–204 [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Zhang X, Kuo C, Moore A, Ran C, PLoS One 2013, 8, e62007. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Yevtodiyenko A, Bazhin A, Khodakivskyi P, Godinat A, Budin G, Maric T, Pietramaggiori G, Scherer SS, Kunchulia M, Eppeldauer G, Polyakov SV, Francis KP, Bryan JN, Goun EA, Nat. Commun 2021, 12, 2680. [DOI] [PMC free article] [PubMed] [Google Scholar]; k) Kagalwala HN, Gerberich J, Smith CJ, Mason RP, Lippert AR, Angew. Chem. Int. Ed 2022, 61, e202115704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].(a) Kim EH, Park S, Kim YK, Moon M, Park J, Lee KJ, Lee S, Kim YP, Sci. Adv 2020, 6, eaba3009. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Xu X, An H, Zhang D, Tao H, Dou Y, Li X, Huang J, Zhang J, Sci. Adv 2019, 5, eaat2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].a) Hong GS, Antaris AL, Dai HJ, Nat. Biomed. Eng 2017, 1, 0010 [Google Scholar]; b) Feng H, Zhao Q, Zhang B, Hu H, Liu M, Wu K, Li X, Zhang X, Zhang L, Liu Y, Angew. Chem. Int. Ed 2023, 62, e202215215. [DOI] [PubMed] [Google Scholar]
- [6].Selkoe DJ, Nature 1999, 399, A23–A31. [DOI] [PubMed] [Google Scholar]
- [7].a) Anfinsen CB, Science 1973, 181, 223–230 [DOI] [PubMed] [Google Scholar]; b) Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D, Nature 2005, 435, 773–778 [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Riek R, Eisenberg DS, Nature 2016, 539, 227–235 [DOI] [PubMed] [Google Scholar]; d) Miao J, Miao M, Jiang Y, Zhao M, Li Q, Zhang Y, An Y, Pu K, Miao Q, Angew. Chem. Int. Ed 2023, 62, e202216351. [DOI] [PubMed] [Google Scholar]; e) Wang P, Yu L, Gong J, Xiong J, Zi S, Xie H, Zhang F, Mao Z, Liu Z, Kim JS, Angew. Chem. Int. Ed 2022, 61, e202206894. [DOI] [PubMed] [Google Scholar]; f) Shin J, Verwilst P, Choi H, Kang S, Han J, Kim NH, Choi JG, Oh MS, Hwang JS, Kim D, Mook-Jung I, Kim JS, Angew. Chem. Int. Ed 2019, 58, 5648–5652. [DOI] [PubMed] [Google Scholar]
- [8].a) Tanzi RE, Moir RD, Wagner SL, Neuron 2004, 43, 605–608 [DOI] [PubMed] [Google Scholar]; b) Das B, Yan R, CNS Drugs 2019, 33, 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Cummings J, Lee G, Zhong K, Fonseca J, Taghva K, Alzheimer’s Dement. 2021, 7, e12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].a) Zimbone S, Giuffrida ML, Sabatino G, Di Natale G, Tosto R, Consoli GML, Milardi D, Pappalardo G, Sciacca MFM, ACS Chem. Neurosci 2023, 14, 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Du Z, Li M, Ren J, Qu X, Acc. Chem. Res 2021, 54, 2172–2184. [DOI] [PubMed] [Google Scholar]
- [10].a) Lee BI, Lee S, Suh YS, Lee JS, Kim AK, Kwon OY, Yu K, Park CB, Angew. Chem. Int. Ed 2015, 54, 11472–11476 [DOI] [PubMed] [Google Scholar]; b) Liu Z, Ma M, Yu D, Ren J, Qu X, Chem. Sci 2020, 11, 11003–11008 [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Yu D, Guan Y, Bai F, Du Z, Gao N, Ren J, Qu X, Chem. Eur. J 2019, 25, 3489–3495 [DOI] [PubMed] [Google Scholar]; d) Taniguchi A, Shimizu Y, Oisaki K, Sohma Y, Kanai M, Nat. Chem 2016, 8, 974–982 [DOI] [PubMed] [Google Scholar]; e) Li Y, Du Z, Liu X, Ma M, Yu D, Lu Y, Ren J, Qu X, Small 2019, 15, e1901116. [DOI] [PubMed] [Google Scholar]
- [11].a) Yang F, Lim GP, Begum AN, Ubeda OJ, Simmons MR, Ambegaokar SS, Chen PP, Kayed R, Glabe CG, Frautschy SA, Cole GM, J. Biol. Chem 2005, 280, 5892–5901 [DOI] [PubMed] [Google Scholar]; b) Ryu EK, Choe YS, Lee KH, Choi Y, Kim BT, J. Med. Chem 2006, 49, 6111–6119 [DOI] [PubMed] [Google Scholar]; c) Thapa A, Jett SD, Chi EY, ACS Chem. Neurosci 2016, 7, 56–68. [DOI] [PubMed] [Google Scholar]
- [12].a) Ran C, Xu X, Raymond SB, Ferrara BJ, Neal K, Bacskai BJ, Medarova Z, Moore A, J. Am. Chem. Soc 2009, 131, 15257–15261 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang X, Tian Y, Li Z, Tian X, Sun H, Liu H, Moore A, Ran C, J. Am. Chem. Soc 2013, 135, 16397–16409 [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhang X, Tian Y, Zhang C, Tian X, Ross AW, Moir RD, Sun H, Tanzi RE, Moore A, Ran C, Proc. Natl. Acad. Sci. U. S. A 2015, 112, 9734–9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang X, Tian Y, Yuan P, Li Y, Yaseen MA, Grutzendler J, Moore A, Ran C, Chem. Commun 2014, 50, 11550–11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].a) Yang J, Yang J, Liang SH, Xu Y, Moore A, Ran C, Sci. Rep 2016, 6, 35613. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yang J, Zhang X, Yuan P, Yang J, Xu Y, Grutzendler J, Shao Y, Moore A, Ran C, Proc. Natl. Acad. Sci. U. S. A 2017, 114, 12384–12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Masuda Y, Fukuchi M, Yatagawa T, Tada M, Takeda K, Irie K, Akagi K, Monobe Y, Imazawa T, Takegoshi K, Bioorg. Med. Chem 2011, 19, 5967–5974. [DOI] [PubMed] [Google Scholar]
- [16].Nelson KM, Dahlin JL, Bisson J, Graham J, Pauli GF, Walters MA, J. Med. Chem 2017, 60, 1620–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].a) Li Y, Yang J, Liu H, Yang J, Du L, Feng H, Tian Y, Cao J, Ran C, Chem. Sci 2017, 8, 7710–7717 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mourtas S, Canovi M, Zona C, Aurilia D, Niarakis A, La Ferla B, Salmona M, Nicotra F, Gobbi M, Antimisiaris SG, Biomaterials 2011, 32, 1635–1645. [DOI] [PubMed] [Google Scholar]
- [18].a) Groppi J, Baroncini M, Venturi M, Silvi S, Credi A, Chem. Commun 2019, 55, 12595–12602 [DOI] [PubMed] [Google Scholar]; b) Barth CW, Shah VM, Wang LG, Masillati AM, Al-Fatease A, Husain Rizvi SZ, Antaris AL, Sorger J, Rao DA, Alani AWG, Gibbs SL, Biomaterials 2022, 284, 121490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].a) Hoffmann N, Chem. Rev 2008, 108, 1052–1103 [DOI] [PubMed] [Google Scholar]; b) Liu H, Luo H, Xue Q, Qin S, Qiu S, Liu S, Lin J, Li JP, Chen PR, J. Am. Chem. Soc 2022, 144, 5517–5526. [DOI] [PubMed] [Google Scholar]
- [20].a) Dubinsky L, Krom BP, Meijler MM, Bioorg. Med. Chem 2012, 20, 554–570 [DOI] [PubMed] [Google Scholar]; (b) Hill JR, Robertson AAB, J. Med. Chem 2018, 61, 6945–6963. [DOI] [PubMed] [Google Scholar]
- [21].Smith RA, Knowles JR, J. Am. Chem. Soc 1973, 95, 5072–5073. [DOI] [PubMed] [Google Scholar]
- [22].Das J, Chem. Rev 2011, 111, 4405–4417. [DOI] [PubMed] [Google Scholar]
- [23].Zhu Z, Bally T, Stracener LL, McMahon RJ, J. Am. Chem. Soc 1999, 121, 2863–2874. [Google Scholar]
- [24].Glachet T, Marzag H, Saraiva Rosa N, Colell JFP, Zhang G, Warren WS, Franck X, Theis T, Reboul V, J. Am. Chem. Soc 2019, 141, 13689–13696. [DOI] [PubMed] [Google Scholar]
- [25].Yeh AH, Norn C, Kipnis Y, Tischer D, Pellock SJ, Evans D, Ma P, Lee GR, Zhang JZ, Anishchenko I, Coventry B, Cao L, Dauparas J, Halabiya S, DeWitt M, Carter L, Houk KN, Baker D, Nature 2023, 614, 774–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ruiz-Gonzalez R, Cortajarena AL, Mejias SH, Agut M, Nonell S, Flors C, J. Am. Chem. Soc 2013, 135, 9564–9567. [DOI] [PubMed] [Google Scholar]
- [27].Yanagisawa D, Shirai N, Amatsubo T, Taguchi H, Hirao K, Urushitani M, Morikawa S, Inubushi T, Kato M, Kato F, Morino K, Kimura H, Nakano I, Yoshida C, Okada T, Sano M, Wada Y, Wada KN, Yamamoto A, Tooyama I, Biomaterials 2010, 31, 4179–4185. [DOI] [PubMed] [Google Scholar]
- [28].Josephson B, Fehl C, Isenegger PG, Nadal S, Wright TH, Poh AWJ, Bower BJ, Giltrap AM, Chen L, Batchelor-McAuley C, Roper G, Arisa O, Sap JBI, Kawamura A, Baldwin AJ, Mohammed S, Compton RG, Gouverneur V, Davis BG, Nature 2020, 585, 530–537. [DOI] [PubMed] [Google Scholar]
- [29].Olsen JV, Ong SE, Mann M, Mol. Cell. Proteomics 2004, 3, 608–614. [DOI] [PubMed] [Google Scholar]
- [30].a) Cheignon C, Tomas M, Bonnefont-Rousselot D, Faller P, Hureau C, Collin F, Redox Biol 2018, 14, 450–464 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Varadarajan S, Kanski J, Aksenova M, Lauderback C, Butterfield DA, J. Am. Chem. Soc 2001, 123, 5625–5631 [DOI] [PubMed] [Google Scholar]; c) Butterfield DA, Sultana R, J. Amino Acids 2011, 2011, 198430. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Friedemann M, Helk E, Tiiman A, Zovo K, Palumaa P, Tougu V, Biochem. Biophys. Rep 2015, 3, 94–99 [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Razzokov J, Yusupov M, Bogaerts A, Sci. Rep 2019, 9, 5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Misiti F, Clementi ME, Giardina B, Neurochem. Int 2010, 56, 597–602. [DOI] [PubMed] [Google Scholar]
- [32].Yang J, Zhang X, Zhu Y, Lenczowski E, Tian Y, Yang J, Zhang C, Hardt M, Qiao C, Tanzi RE, Moore A, Ye H, Ran C, Chem. Sci 2017, 8, 6155–6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Miyahara Y, Shintani K, Hayashihara-Kakuhou K, Zukawa T, Morita Y, Nakazawa T, Yoshida T, Ohkubo T, Uchiyama S, Sci. Rep 2020, 10, 6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].a) Nagashima N, Ozawa S, Furuta M, Oi M, Hori Y, Tomita T, Sohma Y, Kanai M, Sci. Adv 2021, 7, eabc9750. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Li C, Wang J, Liu L, Front. Chem 2020, 8, 509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Riss TL, Moravec RA, Niles AL, Duellman S, Benink HA, Worzella TJ, Minor L, in Assay Guidance Manual (Eds.: Markossian S, Grossman A, Brimacombe K, Arkin M, Auld D, Austin C, Baell J, Chung TDY, Coussens NP, Dahlin JL, Devanarayan V, Foley TL, Glicksman M, Gorshkov K, Haas JV, Hall MD, Hoare S, Inglese J, Iversen PW, Kales SC, Lal-Nag M, Li Z, McGee J, McManus O, Riss T, Saradjian P, Sittampalam GS, Tarselli M, Trask OJ Jr., Wang Y, Weidner JR, Wildey MJ, Wilson K, Xia M, Xu X), Eli Lilly & Company and the National Center for Advancing Translational Sciences, Bethesda (MD), 2004. [PubMed] [Google Scholar]
- [36].Dzeja P, Terzic A, Int. J. Mol. Sci 2009, 10, 1729–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].a) Zhang C, Browne A, Child D, Tanzi RE, J. Biol. Chem 2010, 285, 28472–28480 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wagner SL, Zhang C, Cheng S, Nguyen P, Zhang X, Rynearson KD, Wang R, Li Y, Sisodia SS, Mobley WC, Tanzi RE, Biochemistry 2014, 53, 702–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


