Abstract
Bright (B-cell regulator of immunoglobulin heavy chain transcription) binding to immunoglobulin heavy chain loci after B-cell activation is associated with increased heavy chain transcription. Our earlier reports demonstrated that Bright coimmunoprecipitates with Bruton's tyrosine kinase (Btk) and that these proteins associate in a DNA-binding complex in primary B cells. B cells from immunodeficient mice with a mutation in Btk failed to produce stable Bright DNA-binding complexes. In order to determine if Btk is important for Bright function, a transcription activation assay was established and analyzed using real-time PCR technology. Cells lacking both Bright and Btk were transfected with Bright and/or Btk along with an immunoglobulin heavy chain reporter construct. Immunoglobulin gene transcription was enhanced when Bright and Btk were coexpressed. In contrast, neither Bright nor Btk alone led to activation of heavy chain transcription. Furthermore, Bright function required both Btk kinase activity and sequences within the pleckstrin homology domain of Btk. Bright was not appreciably phosphorylated by Btk; however, a third tyrosine-phosphorylated protein coprecipitated with Bright. Thus, the ability of Bright to enhance immunoglobulin transcription critically requires functional Btk.
Bright (B-cell regulator of immunoglobulin [Ig] heavy chain transcription) is a B-cell-restricted transcription factor that binds specific A-T-rich sequences. The protein consists of an acidic amino-terminal domain, a DNA-binding A-T-rich interaction domain, a putative transactivation domain, and a protein interaction domain (18). The carboxyl-terminal domain of Bright currently has no assigned function. Bright was originally identified in an antigen-specific B-cell line, BCg3R-1d, as a mobility-shifted complex induced after stimulation with interleukin-5 (IL-5) and antigen (54). Binding sites for Bright were originally identified 5′ of the basal promoter of the V1 S107 gene but also exist within the matrix association regions on either side of the intronic μ enhancer (53, 55). Bright binding to the 5′-flanking sequences of the V1 S107 variable heavy chain (VH) promoter correlated with two- to sixfold increases in μ heavy chain mRNA levels in response to IL-5 and antigen (54, 55). Deletion of Bright binding sites flanking the V1 promoter resulted in lack of antigen- and IL-5-stimulated μ heavy chain transcription (55). Bright expression is tightly regulated in normal murine lymphocytes, occurring in pre-B cells and late stages of B-cell differentiation (58). However, Bright is not present in detectable amounts in immature B cells, suggesting that it may not play a role in maintenance of Ig expression (58). On the other hand, Bright activity is induced in B cells activated in response to lipopolysaccharide (LPS), CD40 ligand stimulation, and anti-CD38 (55, 59). These data suggest that Bright enhances Ig heavy chain transcription above basal levels following B-cell activation.
Our earlier results revealed that Bruton's tyrosine kinase (Btk) associates with Bright to form a DNA-binding complex (59). Btk is a member of the Tec family of tyrosine kinases and plays an important role in intracellular signaling in activated B cells. B-cell receptor (BCR) cross-linking initiates activation of src, Btk, and syk family tyrosine kinases and subsequent phosphorylation of multiple protein substrates (1, 12, 41, 43). In normal B lymphocytes, stimulation of IL-5 (47) or antigen receptors (2, 3, 10, 46) leads to rapid translocation of Btk to the cell membrane, where it is activated by transphosphorylation via interactions with src and syk family kinases (25, 52, 61). BCR cross-linking also initiates calcium influx associated with Btk-dependent tyrosine phosphorylation of phospholipase Cγ (PLCγ) isoforms (12, 16, 21). It has been shown that in response to BCR engagement, Btk-dependent PLCγ2 activation mediates peak and sustained production of inositol-3-phosphate, which is required for sustained calcium signaling and transcriptional signals and which can lead to increased IgM secretion, cell maturation, proliferation, or cell death (17, 30, 39). B cells from X-linked immunodeficient (xid) mice and X-linked agammaglobulinemia patients exhibit reduced calcium responses following BCR cross-linking (12, 15). Thus, Btk plays a critical role in B-cell activation.
Btk is also required for normal B-lymphocyte development. Deficiencies in Btk result in X-linked agammaglobulinemia and blocks in B-cell development in patients (3, 17, 30, 39) and in xid mice. The Btk protein consists of an amino-terminal pleckstrin homology (PH) domain, a Tec homology (TH) domain, two Src homology domains (SH3 and SH2), and an SH1, or kinase domain, at the carboxyl end (reviewed in reference 13). In the mouse, a single amino acid change (R28C) in the PH domain causes the xid defect (33, 42). Although this mutation does not affect the kinase activity of the protein, xid mice exhibit blocks in B-cell development at the transitional 2 splenic immature B-cell stage (49). Multiple proteins interact with and are phosphorylated by Btk in vitro, including PLCγ2, WASp, Cbl, Stat5a, and BAP135 (12, 13, 21, 29, 40, 51). BAP135, originally identified as a Btk substrate in activated human B cells (63), was later discovered to be identical to a ubiquitously expressed transcription factor called TFII-I. The mechanism(s) by which Btk mutations result in B-cell deficiencies is not fully understood.
Our previous data supported a direct link between Bright and Btk. Although Bright and Btk were associated in wild-type murine B cells, we observed that xid splenic B cells with defective Btk were likewise deficient in Bright DNA-binding complex formation (59). Bright did not appear to be a Btk substrate in these experiments, although Btk and Bright were coprecipitated from wild-type B cells and antibody supershift assays indicated that Btk was a component of the Bright DNA-bound complex (59). Thus, the functional significance of the association between Bright and Btk is unclear. To determine if the interaction of Btk with Bright is functionally significant, we analyzed Bright transcriptional activity in the presence and absence of wild-type and mutated forms of Btk. Herein we present evidence that Btk is necessary for Bright function and that the Bright A-T-rich interaction domain and the Btk PHTH domains are important for that activity. In addition, a third tyrosine-phosphorylated protein coprecipitated with Bright. Furthermore, our data demonstrate that Bright complexes containing Btk bind to Ig heavy chain sequences within a B-cell line. These data demonstrate that Btk kinase activity is crucial for Bright function.
MATERIALS AND METHODS
Cell culture and transient transfections.
The adherent Chinese Hamster ovarian cell line (CHO; American Type Culture Collection, Manassas, Va.) was cultured in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, Calif.) with 10% heat-inactivated fetal calf serum (Atlanta Biologicals, Norcross, Ga.) and growth supplements as previously described (59). Transfections were performed using Fugene 6 transfection reagent as per the manufacturer's protocol (Roche, Indianapolis, Ind.). Briefly, 3 μg of each plasmid DNA and 30 μl of transfection reagent were incubated in serum-free Dulbecco's modified Eagle's medium for 15 min and then added to a 60% confluent T-75 flask.
Constructs.
The V1 reporter construct contained the S107 V1 heavy chain genomic sequence from bp −574 relative to the transcription start site to bp +146 and included the leader sequence, first intron, and 146 bases of additional coding sequence (55). It was subcloned into pGEM-4Z (Invitrogen) with a 1-kb XbaI fragment containing the Eμ enhancer. Constructs containing deletions of the V1 5′-flanking sequence were produced as described previously (55).
A full-length mouse Bright cDNA clone in pBKCMV (18) was used as a template to produce amino acid changes in the Bright coding sequence to produce a double point mutant (DPBr) with W299A and Y330A mutations that no longer binds DNA (34). Wild type and DPBr were tagged on the carboxyl terminus with the myc-His sequence by using the vector pcDNA4/TO/myc-His B (Invitrogen). Briefly, Bright was PCR amplified with oligonucleotides that added 5′ EcoRI and 3′ XbaI sites while deleting the endogenous stop codon and was ligated into these sites in the vector. Sequences were subcloned into the MiGR1 retroviral vector, which expresses green fluorescent protein (GFP) from an internal ribosomal entry site to allow visualization of transfected cells.
The Btk-GFP fusion construct was a gift from William Rodgers (Oklahoma Medical Research Foundation [OMRF]). The Btk wild-type, K430R, and xid genes were amplified with Pfu polymerase (Stratagene, La Jolla, Calif.) from vaccinia virus clones (12). The Btk PHTH deletion was generated by PCR mutagenesis, removing the first 211 codons of the wild-type gene. All Btk constructs were subcloned into pcDNA4/HisMaxC (Invitrogen) and pET15b (Novagen, Madison, Wis.) for eukaryotic and prokaryotic expression, respectively. All constructs were sequenced by the OMRF Sequencing Core Facility to confirm their identities prior to use.
Real-time quantitative RT-PCR.
Transfected cells were identified by GFP expression 36 h posttransfection, and 200,000 GFP-positive cells (of >95% purity) were collected for RNA isolation using a MoFlo cell sorter (Cytomation, Inc., Fort Collins, Colo.) by the OMRF or University of Oklahoma Health Sciences Center Flow Cytometry cores. RNA was isolated using TRI reagent (Molecular Research Center, Inc., Cincinnati, Ohio), immediately treated with DNase I according to the manufacturer's protocol (Ambion, Austin, Tex.), extracted with phenol-chloroform (Clontech, Palo Alto, Calif.), and quantified by spectrophotometry. Reactions to generate cDNA included 300 ng of RNA, 1 mM deoxynucleoside triphosphate mix, 25 pM V1-specific primer or random primer (Integrated DNA Technologies, Coralville, Iowa), 40 U of the RNase inhibitor RNasin (Promega, Madison, Wis.), and 200 U of SuperScript II RNase H reverse transcriptase (RT; Invitrogen). Negative control reactions without RT were performed in parallel, and baseline CT values of 36 to 40 were routinely obtained.
Real-time quantitative RT-PCR was performed using the following specific TaqMan primers and probes to the V1 gene designed using PrimerExpress software (PE Applied Biosystems, Foster City, Calif.) and synthesized by Applied Biosystems and Integrated DNA Technologies: forward, 5′-TGTCCTGAGTTCCCCAATCC-3′; reverse, 5′-AAACCCAGTTTAACCACATCTTCAT-3′; probe, 5′-6-carboxyfluorescein-ACAATTCAGAAATCAGCACTCAGTCCTTGTCA-3′. Reactions were performed in 1× TaqMan Universal PCR master mix (Applied Biosystems) in triplicate in 96-well plates with a 250 nM concentration of each primer and 500 nM probe under the following conditions: 50°C for 2 min and 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 55°C for 1 min. A standard curve was generated by averaging CT values for triplicate reactions performed with 10-fold serial dilutions (from 20 to 10−4 ng) of plasmid containing the V1 gene. Three microliters of cDNA was used as template to quantify V1 transcription, standards were run with every experiment, and data were converted to nanograms. Data were analyzed using the ABI Prism 7700 SDS analysis software (Applied Biosystems).
Western blotting and immunoprecipitation.
Whole-cell extracts were prepared from transfected CHO cells 36 h posttransfection using hypotonic lysis as previously described (6). Briefly, cells were washed twice in phosphate-buffered saline (PBS), suspended in extraction buffer (20 mM HEPES [pH 7.9], 420 mM NaCl, 1 mM MgCl2, 0.2 mM EDTA, 20% glycerol) containing 500 μM dithiothreitol and protease inhibitors (500 μM phenylmethylsulfonyl fluoride, 20 μM leupeptin, 750 nM aprotinin, and 30 mM NaVO3), homogenized with pestles, and incubated on ice for 30 min. Lysates were collected after 15 min of 4°C centrifugation at 12,000 × g and dialyzed at room temperature for 2 h in storage buffer (20 mM HEPES [pH 7.9], 100 mM KCl, 0.2 mM EDTA, 20% glycerol) containing protease inhibitors. Protein concentrations were measured by modified Bradford assay (Bio-Rad, Hercules, Calif.), and Western blotting was performed as previously described (57). Blots were developed with goat anti-Btk (C-20; Santa Cruz Biotechnology, Santa Cruz, Calif.), rabbit anti-Bright (gift from P. W. Tucker, University of Texas, Austin), goat anti-BAP135/TFII-I peptide antibodies (prepared in this lab as previously described [63]), or mouse antiphosphotyrosine (4G10; Upstate USA, Inc., Charlottesville, Va.), and alkaline phosphatase-labeled rabbit anti-goat Ig, horseradish peroxidase-labeled anti-mouse IgG or alkaline phosphatase-labeled anti-rabbit Ig (Southern Biotechnology, Birmingham, Ala.) as appropriate.
Lysates were immunoprecipitated with anti-Btk (C-20), anti-myc (Invitrogen), or control antibodies (anti-Sp1 or goat Ig) by rocking 150 μg of cell lysate with antibodies in PBS containing protease inhibitors at 4°C for 2 h. Protein A/G Plus agarose beads (25 μl; 1:1 slurry; Santa Cruz) were added and incubated at 4°C for 12 h. Immunoprecipitates were washed five times with wash buffer (100 mM Tris-Cl, 500 mM NaCl, 0.1% Tween 20; pH 8) containing protease inhibitors, and proteins were eluted by addition of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis sample buffer and boiling for 5 min. Samples were run on SDS-8% polyacrylamide gels and transferred using standard protocols for Western blotting as described above.
Electrophoretic mobility shift assays (EMSAs).
Bright mobility shift assays were performed in 4% nondenaturing polyacrylamide gels as previously described (6). The prototypic Bright binding site (a 150-bp BamHI fragment called bf150) from the V1 S107 5′-flanking sequence (55) labeled with γ-32P was used as probe. Bright protein was produced in vitro with TNT-coupled rabbit reticulocyte lysates (Promega). Btk proteins were produced as recombinants in Escherichia coli and then purified from inclusion bodies (27). Protein samples processed in parallel from E. coli lacking the Btk plasmid served as a negative control.
Antibody-facilitated DNA precipitation.
BCg3R-1d cells stimulated for 20 h with 20 μg of LPS/ml to induce Bright activity (54) or negative control T-hybridoma cells, KD3B5.8 (gift of D. Farris, OMRF), were subjected to cross-linking and immunoprecipitation according to the method of Fernandez et al. (11). Briefly, cells were subjected to cross-linking for 10 min in 1% formaldehyde at 37°C, stopped with 0.125 M glycine for 5 min, and then washed twice in cold PBS containing protease inhibitors (500 μM phenylmethylsulfonyl fluoride, 750 nM aprotinin, and 20 μM leupeptin) before resuspension in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris; pH 8) with protease inhibitors. After incubation on ice for 10 min, the suspension was sonicated to reduce the DNA length to 200 to 1,000 bp and centrifuged at 4°C for 10 min, and the supernatant was used for immunoprecipitation. Sonicated extracts were precleared with protein A/G beads (Santa Cruz) plus 50 μg of salmon sperm DNA for 30 min at 4°C before incubation with either anti-Bright, anti-Btk, or control goat Ig overnight at 4°C. Immunocomplexes were precipitated with blocked protein A/G beads for 1 h. The beads were washed once in low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris, 150 mM NaCl; pH 8), once in high-salt buffer (containing 500 mM NaCl), once in LiCl buffer (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris; pH 8), and twice in Tris-EDTA buffer. Immunocomplexes were eluted with 0.1 M NaHCO3 and 0.2% SDS, and cross-linking was reversed by incubation at 65°C for 4 h. Ten micrograms of proteinase K (Invitrogen) and 2 μl of DNase-free RNase A (10 mg/ml; Roche) were added for 2 h at 50°C. The remaining immunoprecipitated DNA was phenol-chloroform purified and subjected to PCR using primers (forward, 5′-CTAGATCCACATGTATGATTT-3′; reverse, 5′-GTCTTTCAGACAATAGATTGG-3′) that amplify a 150-bp Bright binding region of the V1 gene (55). PCR conditions used were 95°C for 40 s, 60°C for 1 min, and 72°C for 2 min for 40 cycles, with a 10-min extension at 72°C.
RESULTS
Btk is critically required for Bright activation of an Ig reporter gene.
Previous studies demonstrated that both human and mouse Bright associate with Btk to produce DNA-binding complexes (35, 59). However, the functional relationship of the two proteins has not been shown. To determine whether Btk contributes to Bright's function as a transcription activator, Bright-induced transcription of an Ig promoter reporter construct was measured with and without Btk. The reporter construct contained two Bright binding sites previously shown to be necessary for upregulation of the V1 heavy chain gene in a B-cell line (55). In addition, the native mouse V1 Ig heavy chain promoter extended through the leader and first intron and into the following exon of the V1 coding sequence (6), allowing PCR amplification across an intron. V1 Ig mRNA expression was quantified by real-time PCR. A standard curve was prepared using V1 cDNA (Fig. 1A). Amplification produced a single band in ethidium bromide-stained gels, and sequencing confirmed its identity. Serial dilutions of plasmid DNA containing the V1 gene were amplified in the presence of the Taqman probe, and CT values for 10−3 to 10−1 ng of template DNA fell in the linear region of the curve.
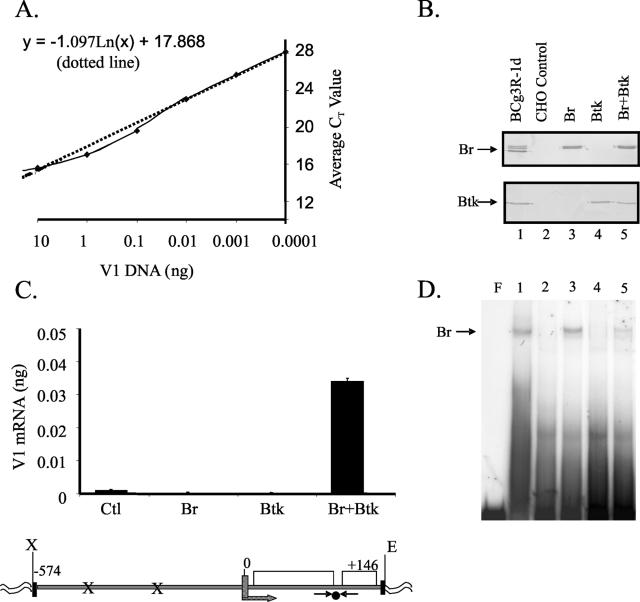
FIG. 1.
Bright activation of an Ig reporter gene requires Btk. (A) A standard curve for V1 DNA was generated by real-time PCR. Each point reflects the average of triplicate CT values from four experiments. The y intercept equation is shown. (B) Bright and Btk expression levels were assessed by Western blotting in CHO cells transfected with Bright (Br), Btk, and Bright plus Btk (lanes 3 to 5). Lane 1 contains extract from a control B-cell line (Bcg3R-1d), and lane 2 contains extract from the CHO cell line transfected with control vectors. (C) V1 expression was averaged from triplicate samples of three experiments with CHO cells expressing Btk, Bright, Bright plus Btk, and control vectors by using the standard curve in panel A. Values were calculated using the formula y = −1.097 ln(x) + 17.868 for each of the triplicate values, and standard errors of the means are shown. A diagram of the V1 reporter construct used is shown below. The primer (→←) and probe (•) locations are shown. Bright binding sites (x) are indicated. Exons are shown as boxes. (D) Extracts from panel B were assessed for Bright DNA-binding activity by EMSA with bf150. Lane numbers correspond to those shown in panel B. F indicates free probe (first lane), and Bright complexes are indicated with an arrow.
To determine whether Btk influences Bright activity, CHO cells that express neither Bright nor Btk were cotransfected with murine Bright and/or Btk expression vectors and the V1 reporter construct. Western blotting results showing protein expression of Bright and Btk in the transfected cells are illustrated in Fig. 1B. Control vectors containing the Bright coding sequence cloned in the reverse orientation, or the empty Btk vector, were used to maintain equal amounts of DNA in each transfection mixture. Both the Bright and control vectors coexpressed GFP from an internal ribosomal entry site. To obtain cell populations containing equivalent numbers of transfected cells, GFP+ cells were isolated from each transfected population by cell sorting after 48 h and mRNA was isolated for quantitative PCR analysis. Transfection efficiencies typically varied by less than 5%, and the purity of sorted cells was >95%. In each case, the mean fluorescence intensity of the sorted cells was equivalent. Reactions lacking RT were run in parallel to ensure that the data produced resulted from amplification of the V1 cDNA rather than from the transfected vector and gave baseline CT values. Figure 1C shows that cotransfection of Btk and Bright resulted in significantly increased V1 transcription in comparison with cells transfected with either Bright or Btk alone. Indeed, Bright and Btk singly transfected cells failed to show V1 transcription above control levels, while Bright and Btk increased transcription of the reporter construct from 3- to 12-fold. These findings show that Bright-dependent transcriptional activation in this cellular model is fully dependent upon coexpression of the Tec kinase, Btk.
The failure of Bright to transactivate V1 gene expression in the absence of Btk might result from the inability of Bright to bind DNA under those conditions. EMSA analyses of extracts from the transfectants described above were used to test the requirement of Btk for Bright DNA-binding activity (Fig. 1D). Bright complexes were not formed in extracts that did not contain Bright. However, Bright bound to DNA in the absence of cotransfected Btk (lane 3). Therefore, Bright DNA-binding activity in transfected CHO cells does not require Btk. However, Bright transcriptional activity critically requires Btk.
Bright DNA-binding activity is essential for transcription activation.
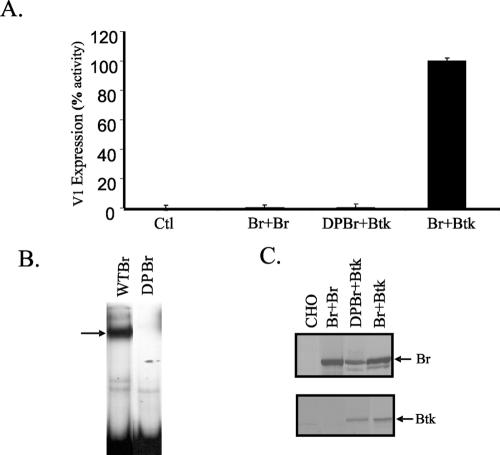
To confirm that the DNA-binding activity of Bright was required for V1 transcription, a mutant of Bright containing mutations W299A and Y330A, resulting in a protein that failed to bind DNA (DPBr) (34), was expressed with wild-type Btk in the CHO reporter system. Consistent with this model, V1 Ig transcripts were not generated in the presence of DPBr (Fig. 2). Mobility shift assays confirmed that DPBr did not bind DNA despite abundant protein expression (Fig. 2B and C). In addition, because Bright binds DNA as a dimer, we also asked if an increase in Bright expression levels might promote V1 transcription. Overexpression of Bright (up to twofold), however, failed to promote V1 transcription in the absence of coexpressed Btk (Fig. 2A). Therefore, Bright activity did not correlate with levels of Bright production in these cells. Rather, these data suggest that Bright DNA-binding activity is required for Bright function, but DNA-binding activity alone is not sufficient for transcriptional activity. Taken together, these findings suggested that Bright transcriptional activity is directly or indirectly regulated by Btk.
FIG. 2.
DNA-binding activity is necessary for Bright function as a transcription activator. (A) Average V1 expression was quantified in CHO cells transfected with DPBr and/or wild-type Bright plus Btk by real-time PCR using triplicate samples from three individual experiments. Br + Br, cells were transfected with twice the amount of Bright vector DNA as used with the Bright and Btk transfectants. Data are expressed as percent activity, with Bright plus Btk activity arbitrarily set at 100%. Standard error of the mean bars are shown. (B) EMSA showing Bright binding complex (arrow) present in extracts from wild-type Bright (WT Br) and DPBr cells cotransfected with Btk. (C) Western blots showing Bright and Btk expression levels in transfected CHO cells.
Bright activation of the V1 promoter is dependent on the 5′ Bright binding motif.
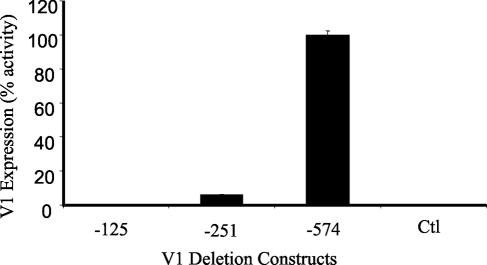
The full-length V1 promoter construct used in these studies (Fig. 1C) contains two Bright binding sites, one at approximately −550 and the other at −225 (55). Earlier analyses indicated that basal levels of V1 transcription in B lymphocytes required sequences from −125 to the transcription start site (55). These data were confirmed in V1 transgenic mice that expressed only 125 bases of the V1 promoter sequence (4). However, vectors containing the single Bright binding site at −225 resulted in a three- to sixfold enhancement of V1 transcription in B-cell lines, and similar results were obtained with a −574 construct containing two Bright binding motifs (55). These previous studies utilized mature B cells. Transcription of the V1 promoter has not previously been possible in nonlymphocytic cell lines (reference 18 and our unpublished data), probably due to a lack of Btk. To determine whether the transcriptional activation observed in the CHO transfectants required one or both of the Bright binding sites, we compared transcription levels of V1 in CHO cells transfected with Btk plus Bright by using either a full-length V1 construct with two Bright binding sites (−574) or truncated V1 constructs containing one (−251) or no (−125) Bright binding sites. The previous data presented in Fig. 1 and 2 were obtained with the −574 construct, where transcripts were quantified at 0.035 ng. This average value was therefore set as 100% activity (Fig. 3). The truncated vectors that contained no or a single Bright binding site produced 0.0075% (−125) and 3.5% (−251) activity, respectively, in comparison with the full-length construct (Fig. 3). Negative control values (0.001%) were obtained with the full-length vector in the absence of either Bright or Btk proteins as described previously. These results suggest that the presence of the −550 Bright binding site is essential for transcriptional activity in this system.
FIG. 3.
The distal Bright binding motif in the V1 promoter is necessary for Bright function. V1 deletion constructs containing zero (−125), one (−251), and two (−574) Bright binding sites were transfected with Bright plus Btk into CHO cells. V1 expression was measured using triplicate values from three real-time PCR experiments. The average value from the full-length (−574) construct was set at 100%, and the other values are presented as percent activity of that value. Standard error of the mean bars are shown. Control transfected cells contained the full-length vector (−574) with negative control Bright and Btk plasmids.
Bright function as a transcriptional activator requires Btk kinase activity and regions within the PHTH domains of Btk.
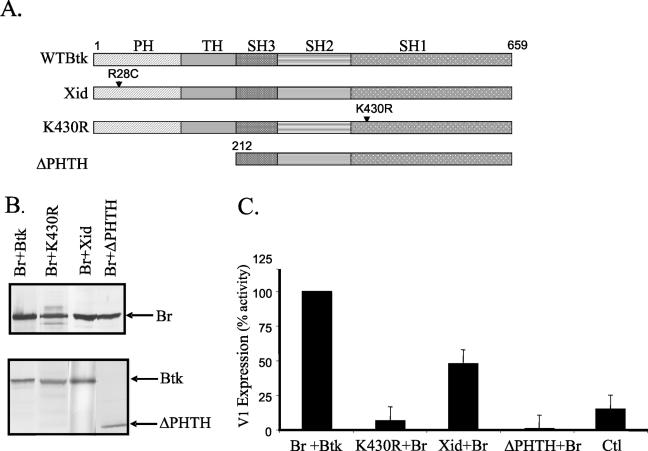
Mutated forms of Btk result in immunodeficiency disease in both mice and humans (14, 42, 50). To assess how inactive forms of Btk affect Bright activity, we investigated whether the K430R (kinase-inactive) (26), xid (R28C), and pleckstrin and tec homology domain deletion (ΔPHTH) mutants of Btk affected V1 transcription levels (Fig. 4A). CHO cells were cotransfected with Bright, the full-length V1 reporter construct, and with wild-type or mutant forms of Btk. Mutant and wild-type Btk protein expression in the transfectants was shown by Western blotting (Fig. 4B). Transfectants expressing kinase-inactive Btk and wild-type Bright exhibited <10% of the V1 transcription produced with wild-type Btk (Fig. 4C). Similarly, ΔPHTH Btk failed to enhance V1 transcription in this system. On the other hand, transfectants expressing the xid point mutation exhibited ∼50% of the V1 transcription observed with wild-type Btk. These data suggest that amino acid sequences within the PHTH domain are important for full activity. Moreover, the data indicate that Btk kinase activity is critically required for Bright transcription activation.
FIG. 4.
Functional Btk is required for Bright activity. (A) Schematic diagram depicting the pleckstrin (PH), tec (TH), and src (SH1 to -3) homology domains of wild-type Btk and the mutants used. R28C is the xid mutation; K430R renders Btk kinase inactive; ΔPHTH lacks the pleckstrin and tec homology domains. (B) Western blots showing expression of Bright (Br) and the Btk mutants in transfected CHO cell extracts. (C) V1 expression from the −574 full-length promoter construct was measured in CHO cells transfected with wild-type Bright and either wild-type or mutant Btk by real-time PCR as described in the previous figure legends. Each transfection was performed a minimum of three times, and data were calculated from triplicate samples in each experiment. Average values for each transfection are presented as percent activity of the values obtained with wild-type Btk plus Bright, which were set at 100%. Standard errors of the means are shown. Control transfected cells contained the empty Btk vector, the Bright reverse orientation vector, and the V1 reporter construct.
Btk is predominantly found in the cytoplasm of B cells, but we and others have observed Btk within the cell nucleus (32, 59). A possible explanation for the failure of the Btk mutants to support Bright function (Fig. 4) is that either the mutant Btk proteins, or Bright, fail to enter the nucleus in those cells. To test this possibility, the intracellular localization and distribution of Bright and Btk were examined by confocal microscopy. Immunostaining of transfected CHO cells with anti-Bright and anti-Btk revealed both cytoplasmic and nuclear localization of both wild-type proteins (data not shown). The K430R, xid, and ΔPHTH mutations in Btk did not grossly alter the intracellular localization of Bright. Therefore, there is no major effect upon the localization of Bright in this model system that would explain the inability of the mutant forms of Btk to facilitate Bright-induced transcription.
The PHTH domain of Btk is required for Btk-dependent enhancement of Bright DNA-binding activity.
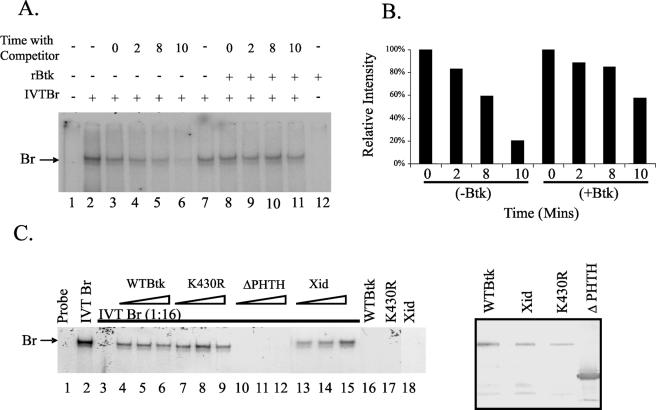
Our earlier studies showed that addition of recombinant wild-type Btk to suboptimal levels of Bright protein enhanced Bright DNA-binding activity in mobility shift assays (59). To determine if Btk acts by stabilizing Bright binding activity, we asked whether Bright binding affinity differed in the presence or absence of Btk. EMSAs were performed with in vitro-translated Bright and cold competitor DNA (Fig. 5A). In lanes 3 to 6 and 8 to 11, 100 M excess of unlabeled competitor DNA was added for 0, 2, 8, and 10 min prior to electrophoresis. In lanes 8 to 12, recombinant Btk was also added to the proteins. Without Btk, Bright binding was reduced by 80% after 10 min with the competitor DNA (Fig. 5B). In the presence of Btk, Bright binding activity was reduced by less than 50% after 10 min with competitor DNA. These data suggest that one function of Btk is to enhance Bright DNA-binding affinity.
FIG. 5.
Bright DNA-binding activity is facilitated by Btk, and this enhanced binding requires the PHTH domain of Btk. (A) EMSAs were performed using bf150 and in vitro-translated Bright (IVT Br) with or without the addition of exogenous recombinant wild-type Btk (rBtk). Unlabeled competitor DNA (100 M excess) was added to samples in some lanes for 0 to 10 min before electrophoresis was begun. The Bright complex is indicated with an arrow. (B) Densitometric quantification of the bands in lanes 3 to 6 and 8 to 11 from panel A shows stabilization of Bright DNA-binding activity in the presence of Btk. (C) EMSAs were performed using IVT Br or suboptimal levels (1:16 dilution, lanes 3 to 15) of IVT Br with increasing amounts (triangles) of rBtk. The arrow indicates the Bright complex. Probe alone is shown in the first lane. The last three lanes demonstrate the absence of binding activity produced by the maximum levels of each of the Btk proteins used in the absence of Bright. Expression of rBtk proteins is shown by the Western blot in the boxed panel at the right. Data are representative of three experiments.
The K430R, ΔPHTH, and xid forms of Btk failed to support V1 transcription activation by Bright. To investigate why these Btk mutants failed to support Bright function, we asked if they enhanced Bright DNA-binding activity similarly to wild-type Btk. At a dilution of 1:16, no Bright DNA-binding activity was evident in in vitro-translated extracts (Fig. 5C, lane 3). However, even low levels of recombinant wild-type Btk restored the DNA-binding activity of Bright (lanes 4 to 6). The K430R and xid mutants were also effective in restoring Bright DNA-binding activity. Wild-type, K430R, and xid Btk alone did not bind DNA without Bright (lanes 16 to 18). Relative protein levels of the Btk recombinant proteins used are shown in the right panel (Fig. 5C). On the other hand, Btk lacking the PHTH domains did not enhance Bright binding activity in this assay at any concentration of the mutant Btk protein (Fig. 5C, lanes 10 to 12). These data suggest that regions within the PHTH domains of Btk are required for facilitation of Bright DNA-binding activity by Btk.
Bright associates with each of the Btk mutants examined.
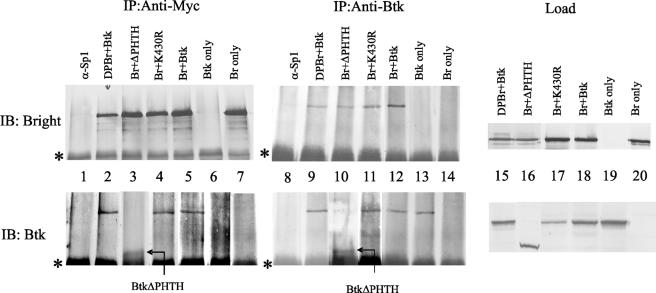
Our earlier studies showed that Btk and Bright interact either directly or indirectly. To determine if the Btk mutants failed to facilitate Bright-induced transcription of the V1 gene because they failed to bind Bright, coimmunoprecipitation experiments were performed. Extracts from the CHO cells transfected with wild-type Bright and wild-type or mutant Btk were immunoprecipitated with antibodies to the myc tag on the carboxyl end of Bright. Precipitated proteins were detected with anti-Bright and anti-Btk (Fig. 6). While the isotype control (anti-Sp1) did not precipitate Bright or Btk (lane 1), wild-type Bright coprecipitated Btk (lane 5). Anti-myc failed to precipitate Btk in the absence of Bright (lane 6). On the other hand, Btk coprecipitated with dominant negative Bright (lane 2), indicating that DPBr is still able to interact with Btk. This finding suggests that the absence of transcription activation observed with DPBr in Fig. 2 was due to the mutant Bright's inability to bind DNA rather than failure to associate properly with Btk. The K430R mutant (lane 4) also precipitated with Bright, consistent with its ability to increase Bright affinity for DNA, as shown in Fig. 5. Bright associated weakly with the ΔPHTH mutant (lane 3), although this Btk mutant did not increase Bright affinity for DNA. Likewise, in every case, immunoprecipitation of the Btk mutants with an antibody that recognized the SH1 domain also coprecipitated Bright (Fig. 6, lanes 9 to 12). These data show that Bright-Btk association is not disrupted by point mutations in the Bright DNA-binding domain or in the Btk and PH domains. Furthermore, the PHTH domain is not required for Bright/Btk complex formation. These data suggest that the increased affinity of Bright for DNA is facilitated by the PHTH domain of Btk but that additional regions of Btk, or associations with a third protein, account for its ability to associate with Bright.
FIG. 6.
Formation of Btk/Bright complexes is not dependent on Btk kinase activity or its PHTH domain, or on the DNA-binding activity of Bright. Whole-cell extracts from wild-type (Btk) or mutant (ΔPHTH, K430R) Btk and wild-type (Br)- or DPBr-transfected CHO cells were immunoprecipitated (IP) with antibodies reactive with the myc tag on Bright (lanes 1 to 7) or with anti-Btk (lanes 8 to 14). Proteins were then immunoblotted (IB) with anti-Bright (top panels) or anti-Btk (bottom panels) antibodies. An unrelated antibody, anti-Sp1 (lanes 1 and 8), was used as a control for both the Btk and anti-myc monoclonal antibodies. In lanes 6, 7, 13, and 14, either Btk or Bright was expressed singly. Lanes 15 to 20 (load) show levels of proteins prior to immunoprecipitation. The asterisk indicates the immunoprecipitated Ig heavy chain band.
A Btk substrate associates with Bright.
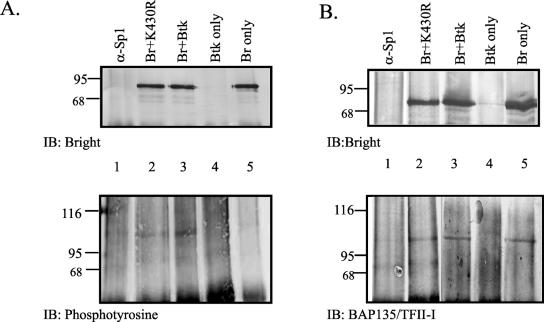
Although kinase-inactive Btk coprecipitated with Bright (Fig. 6), it was not effective in enhancing Ig transcription (Fig. 4). Our previous data suggested that Bright was not appreciably tyrosine phosphorylated (59). Therefore, we hypothesized that a third protein, and Btk substrate, associates with Bright and Btk. CHO cells were transfected with myc-tagged Bright and/or Btk proteins, immunoprecipitated with anti-myc, and immunoblotted for both Bright and phosphotyrosine (Fig. 7A). A 107-kDa band reactive with antiphosphotyrosine coprecipitated with Bright in lane 3. A similar band of weaker intensity was also evident in extracts that contained Bright without Btk (lane 5) and Bright with kinase-inactive Btk (lane 2). No phosphorylated band was observed at 70 kDa, the expected size of phosphorylated Bright, in extracts that contained Bright. These data suggest that a third tyrosine-phosphorylated protein of 107 kDa coprecipitates with Bright. Although the intensity of the phosphorylated band was always greater in the extracts containing wild-type Btk and Bright, tyrosine phosphorylation of this protein was not solely dependent upon Btk.
FIG. 7.
A third protein associates with Bright. Extracts from CHO cells transfected with myc-tagged Bright and/or Btk were immunoprecipitated with anti-myc antibody (lanes 2 to 5) or the isotype control, anti-Sp1 (lane 1), and immunoblotted (IB) with anti-Bright, antiphosphotyrosine (A), or anti-BAP135/TFII-I (B) antibodies. Positions of molecular weight markers are indicated. Data are representative of two separate experiments.
A previously identified substrate of Btk in activated B cells is the transcription factor BAP135/TFII-I (9, 63). To narrow down the identity of the phosphorylated protein in Fig. 7A, similar immunoprecipitation experiments were performed and blots were developed with anti-BAP135/TFII-I (Fig. 7B). In each case, Bright coprecipitated from CHO cells with a 107-kDa protein that reacted with the anti-BAP/135 antibodies. These results are consistent with the hypothesis that a third protein that is serologically related to BAP135/TFII-I is tyrosine phosphorylated by Btk and associates with Bright and Btk. Furthermore, these data suggest that Bright can associate with this protein in the absence of Btk.
Bright/Btk complexes interact with a heavy chain promoter in a B-cell line.
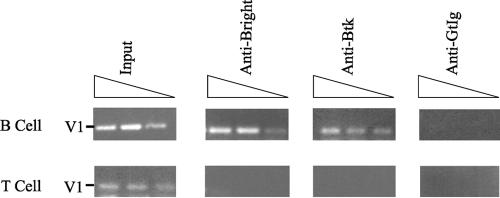
Although the data from the previous experiments strongly suggested that Btk is required to enhance transcription of the V1 heavy chain, these data were all derived from artificially transfected non-B cells. To determine if Bright and Btk interact with the V1 promoter in B cells, we took advantage of a B-cell line that expresses a V1 μ heavy chain gene, BCg3R-1d (54). Modified chromatin immunoprecipitation assays were conducted using anti-Bright, anti-Btk (C20), or control antibodies with LPS-stimulated BCg3R-1d cells that express Bright (55, 58) and a T-cell hybridoma (KD3B5.8) that does not express either Bright or Btk. PCR primers were designed to amplify a region containing the Bright binding site between −574 and −425 of the V1 promoter (55). Figure 8 shows amplification of the V1 Bright site using 10% of the input DNA obtained from either BCg3R-1d or KD3B5.8. Both anti-Bright and anti-Btk antibodies immunoprecipitated DNA from the B-cell line contained the V1 Bright site, but neither antibody precipitated that region of DNA from the T-cell line. In addition, control antibodies were unable to precipitate the V1 Bright binding site from either the B- or T-cell line. These data suggest that both Bright and Btk are present as a complex on the V1 promoter in the BCg3R1-d B-cell line. Furthermore, these data are consistent with experiments in transfected CHO cells that suggest that Bright and Btk association facilitates V1 transcription.
FIG. 8.
Bright/Btk complexes bind Bright sites within a B-cell line. Anti-Bright, anti-Btk, or control goat antibodies (gtIg) were used in modified chromatin immunoprecipitation experiments with lysates of the B-cell line BCg3R-1d and the T-cell hybridoma KD3B5.8. Immunoprecipitated DNA was PCR amplified at final dilutions of 1:100, 1:500, and 1:1,000 (represented by triangles) for the presence of a Bright binding site (V1). Ten percent of the DNA used for each immunoprecipitation was used as a positive control (input). Data are representative of three experiments.
DISCUSSION
Our previous studies have suggested that Btk and Bright can associate to form a DNA-binding complex (59). However, it was not clear whether this association had any functional significance. In the present study, we developed a novel model of Bright activity using a nonlymphoid cell line. Using this model, we clearly demonstrated that Bright-dependent transcription of an Ig promoter construct is fully dependent upon coexpressed, functional Btk. While Bright DNA binding was required for its transcriptional activity, this was not sufficient to enhance Ig transcription. Furthermore, kinase-inactive Btk did not facilitate Bright-activated transcription in this system, suggesting that Btk kinase activity is critical for Bright function. Consistent with these findings, a third tyrosine-phosphorylated protein was found to coprecipitate with Bright in CHO cells. This protein is serologically similar to the previously identified Btk substrate, BAP135/TFII-I. Moreover, modified chromatin immunoprecipitation experiments demonstrated that Bright/Btk complexes bound to Bright binding sites in the V1 promoter in a B-cell line, consistent with a role for Bright/Btk complexes within B cells. These data are the first to demonstrate Bright activity in a non-B-cell line and suggest that Btk provides additional B-cell-specific information necessary for Ig heavy chain expression.
Although model systems such as ours allow evaluation of the importance of protein-protein interactions through the use of ectopically expressed mutant proteins, the data obtained do not always faithfully reflect physiologic associations and expression levels. Antibody supershift experiments suggested that Bright and Btk form DNA-binding complexes in protein extracts from both mouse and human B cells (35, 59). Cotransfection of Bright and Btk in COS7 cells also gave results similar to those observed with the CHO cells (data not shown). Furthermore, we have presented evidence here from modified chromatin immunoprecipitation analyses that Btk and Bright form a complex on the V1 promoter in the B-cell line BCg3R-1d (Fig. 8). Therefore, while we cannot rule out the possibility that some of the protein associations observed in our in vitro model system may partially reflect the abundant levels of the ectopically expressed proteins, or inappropriate intracellular localization of some of the proteins, multiple lines of evidence now support a role for functional Btk as a critical component of a Bright transcription complex.
Our previous work (59) and the data shown in Fig. 7 indicate that Bright is not appreciably phosphorylated on tyrosine residues and is, therefore, unlikely to act as a direct substrate for Btk. Our present data, however, clearly indicate a requirement for Btk kinase activity in Bright function. Together, these data suggest that an additional protein component(s), including a Btk substrate, is likely to be required for Bright-dependent transcriptional activity. TFII-I enhances transcription of promoters that lack TATA boxes and regulates promoter activity of the T-cell receptor β locus through interactions with an initiator element (9, 37, 60). Although some IgH gene promoters contain good consensus TATA boxes, the V1 heavy chain promoter used in this system is TATA-less (7).
Data from Fig. 7 showed that a 107-kDa tyrosine-phosphorylated protein coimmunoprecipitated with Bright in CHO cell extracts (Fig. 7). While this protein reacts serologically with antibodies against TFII-I, its precise identity is currently unknown. There are at least three gene families encoding TFII-I-related proteins, and individual members of these families are found in most cell types and can be expressed as multiple isoforms (19). Nothing is known regarding hamster TFII-I family proteins. Furthermore, cotransfection of kinase-inactive Btk with Bright did not entirely eliminate the ability of the 107-kDa protein to react with antiphosphotyrosine antibodies (Fig. 7). Therefore, we cannot conclude from these experiments that Btk phosphorylates the 107-kDa protein. TFII-I was shown previously to be phosphorylated by JAK2, ERK, and Src, as well as by Btk (8, 23, 24, 44). In addition, residual phosphorylation of TFII-I without Btk has been observed by others (44). Both JAK2 and Btk can phosphorylate tyrosine 248 of TFII-I (24, 44). Additional experiments will be required to determine if Bright/Btk complexes associate with TFII-I in B lymphocytes and if Bright activity requires tyrosine phosphorylation of the associated proteins. Nonetheless, results from our studies are consistent with data from other labs in which the Btk-associated protein BAM11 exhibited increased transcription activity of a reporter construct ectopically expressed with Btk (20, 22). In these studies, addition of TFII-I to the system further enhanced transcription activation (20). Similarly, our data are consistent with a model whereby Bright acts to tether Btk and a Btk substrate on the V1 Ig promoter such that transcription activation can occur.
Transcription of the V1 promoter construct required binding of Bright to the Bright binding motifs. DPBr, which was unable to bind DNA, was also ineffective at enhancing V1 transcription (Fig. 2A and B) even though it associated with Btk (Fig. 7). Deletion of the Bright binding motifs in the V1 promoter also inhibited V1 transcription (Fig. 3). It is not clear why the presence of a single Bright binding site (−250 in the V1 construct) did not activate V1 transcription. However, the Bright binding motifs at −250 and −550 are not identical in sequence, and only the −550 site has been shown to be a matrix association region (55, 56). Another possibility is that the intronic enhancer and its spacing relative to Bright are important for activity. Experiments to address the function of the enhancer in this response are under way. In any case, data presented in Fig. 5 suggest that Btk contributes to Bright DNA-binding activity by increasing its binding affinity. Together, these data suggest a link between Bright DNA-binding activity and the requirement for fully functional Btk.
The PH domain is a characteristic feature of the Tec family of tyrosine kinases. It is important for protein-protein interactions (28) and for binding to phosphatidylinositol-3,4,5-bisphosphate (45) and protein kinase C in mast cells and B cells (64). In addition, the PHTH domain has been shown to be important for interaction with PI5K, Vav, G proteins, F-actin, the tyrosine kinase FAK, phosphotyrosine phosphatase PTPD1, and the substrate for BCR downstream signaling 1 (BRDG1) (40, 45, 48, 62). Our data suggest that regions within the PH and TH domains are also important for Bright and Btk activity. When the PHTH deletion mutant of Btk was coexpressed with Bright, this mutant failed to upregulate V1 transcription. Furthermore, unlike the full-length forms of Btk, it failed to facilitate Bright DNA-binding activity in vitro. While these results could be explained by improper folding of the ΔPHTH proteins, immunoprecipitation experiments suggested that it associates weakly with Bright. However, our data now indicate that Bright interacts with a TFII-I-related protein in CHO cells in the absence of Btk (Fig. 7B). Therefore, we cannot exclude the possibility that Btk interacts indirectly with Bright through its association with this third protein. Additional studies will be required to further elucidate these interactions.
A single amino acid change in the PH domain causes the xid defect in mice (42, 50). The xid mutant coprecipitated with Bright (data not shown) and was capable of enhancing Bright DNA-binding activity when added to in vitro-translated Bright. Moreover, the xid mutant cooperated with Bright to induce V1 transcription, although it was only half as efficient as wild-type Btk. This finding is consistent with the fact that the xid mutant retains kinase activity. Failure to activate transcription as efficiently as wild-type Btk may be due to conformational changes in the PH domain that affect the affinity of its association with Bright or a third TFII-I-related protein component required for V1 transcription. Others have reported that BAP135/TFII-I constitutively associates with wild-type Btk and kinase-inactive Btk, but not xid Btk (38). Intriguingly, xid mice fail to produce B cells that use the V1 heavy chain in response to immunization with phosphorylcholine, although the V1 gene is used predominantly in this response by both female littermates and other wild-type mice (5). Our data suggest that the requirement for Btk for appropriate V1 expression may in part reflect its requirement for Bright-induced V1 promoter activity.
In the mouse, the xid mutation, or complete deficiency of Btk protein expression, causes blocks in B-cell development at the immature B-cell stage and results in impaired responses to type II T-independent antigens, deficiencies in isotype switching to IgG3, and low serum Ig production. In humans, mutations in Btk generally result in less than 1% normal serum Ig levels caused by failure of the majority of B cells to differentiate beyond the pro-B-cell stage (36). Thus, the murine disease is less severe than the human defect. It is interesting to speculate that the ability of xid Btk to partially activate Ig transcription of the mouse V1 promoter might contribute to the presence of the immature B cells in the xid mouse.
Although Bright enhances heavy chain transcription three- to sixfold after B-cell activation (55), it is not clear how this transcriptional upregulation contributes to B-cell differentiation. Indeed, the role that Bright plays in B-cell development remains incompletely defined. However, the fact that Bright function critically requires Btk kinase activity suggests that Bright could also be an important mediator of B-cell differentiation. Other studies demonstrated the importance of Btk kinase activity for normal B-cell development. In Btk-deficient mice where the kinase-inactive form of Btk was introduced as a transgene, the defective Btk rescued some of the defects associated with Btk deficiency but not others (31). Regulation of Ig λ L chain usage was kinase independent, and the modulation of pre-B-cell marker expression was only partially dependent on kinase activity (31). Bright-deficient mice will be required to determine if Bright, like Btk, is critically important for specific events in early or late B-cell development.
Acknowledgments
We thank Amy Windell for technical assistance and P. W. Tucker (Austin, Tex.), M. Coggeshall, and L. Thompson (OMRF) for critically reading the manuscript. We would also like to thank Kerry Humphrey for assistance preparing the manuscript.
This work was supported by National Institutes of Health grants AI044215 and AI45864 (C.F.W.) and T32 AI07633 (J.C.N.).
REFERENCES
- 1.Afar, D. E. H., H. Park, B. W. Howell, D. J. Rawlings, J. Cooper, and O. N. Witte. 1996. Regulation of Btk by Src family tyrosine kinases. Mol. Cell. Biol. 16:3465-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki, Y., K. J. Isselbacher, B. J. Cherayil, and S. Pillai. 1994. Tyrosine phosphorylation of Blk and Fyn Src homology 2 domain-binding proteins occurs in response to antigen-receptor ligation in B cells and constitutively in pre-B cells. Proc. Natl. Acad. Sci. USA 91:4204-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki, Y., K. J. Isselbacher, and S. Pillai. 1994. Bruton tyrosine kinase is tyrosine phosphorylated and activated in pre-B lymphocytes and receptor-ligated B cells. Proc. Natl. Acad. Sci. USA 91:10606-10609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avitahl, N., and K. Calame. 1996. A 125 bp region of the Ig VH1 promoter is sufficient to confer lymphocyte-specific expression in transgenic mice. Int. Immunol. 8:1359-1366. [DOI] [PubMed] [Google Scholar]
- 5.Brown, M., M. Stenzel-Poore, and M. Rittenberg. 1985. Immunologic memory to phosphocholine VII. Lack of T15 V1 gene utilization in Xid anti-PC hybridomas. J. Immunol. 135:3558-3563. [PubMed] [Google Scholar]
- 6.Buchanan, K. L., S. E. Hodgetts, J. Byrnes, and C. F. Webb. 1995. Differential transcription efficiency of two Ig VH promoters in vitro. J. Immunol. 155:4270-4277. [PubMed] [Google Scholar]
- 7.Buchanan, K. L., E. A. Smith, S. Dou, L. M. Corcoran, and C. F. Webb. 1997. Family-specific differences in transcription efficiency of Ig heavy-chain promoters. J. Immunol. 159:1247-1254. [PubMed] [Google Scholar]
- 8.Cheriyath, V., Z. P. Desgranges, and A. L. Roy. 2002. c-Src-dependent transcriptional activation of TFII-I. J. Biol. Chem. 277:22798-22805. [DOI] [PubMed] [Google Scholar]
- 9.Cheriyath, V., C. D. Novina, and A. L. Roy. 1998. TFII-I regulates Vβ promoter activity through an initiator element. Mol. Cell. Biol. 18:4444-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Weers, M., G. S. Brouns, S. Hinshelwood, C. Kinnon, R. K. B. Schuurman, R. W. Hendriks, and J. Borst. 1994. B-cell antigen receptor stimulation activates the human Bruton's tyrosine kinase, which is deficient in X-linked agammaglobulinemia. J. Biol. Chem. 269:23857-23860. [PubMed] [Google Scholar]
- 11.Fernandez, L. A., M. Winkler, and R. Grosschedl. 2001. Matrix attachment region-dependent function of the immunoglobulin μ enhancer involves histone acetylation at a distance without changes in enhancer occupancy. Mol. Cell. Biol. 21:196-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fluckiger, A. C., Z. M. Li, R. M. Kato, M. I. Wahl, H. D. Ochs, R. Longnecker, J. P. Kinet, O. N. Witte, A. M. Scharenberg, and D. J. Rawlings. 1998. Btk/Tec kinases regulate sustained increases in intracellular Ca2+ following B-cell receptor activation. EMBO J. 17:1973-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fruman, D. A., A. B. Satterthwaite, and O. N. Witte. 2000. xid-like phenotypes: a B cell signalosome takes shape. Immunity 13:1-3. [DOI] [PubMed] [Google Scholar]
- 14.Genevier, H. C., and R. E. Callard. 1997. Impaired Ca2+ mobilization by X-linked agammaglobulinaemia (XLA) B cells in response to ligation of the B cell receptor (BCR). Clin. Exp. Immunol. 110:386-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genevier, H. C., S. Hinshelwood, H. B. Gaspar, K. P. Rigley, D. Brown, S. Saeland, F. Rousset, R. J. Levinsky, R. E. Callard, C. Kinnon, and R. C. Lovering. 1994. Expression of Bruton's tyrosine kinase protein within the B cell lineage. Eur. J. Immunol. 24:3100-3105. [DOI] [PubMed] [Google Scholar]
- 16.Guo, B. C., R. M. Kato, M. Garcia-Lloret, M. I. Wahl, and D. J. Rawlings. 2000. Engagement of the human pre-B cell receptor generates a lipid raft-dependent calcium signaling complex. Immunity 13:243-253. [DOI] [PubMed] [Google Scholar]
- 17.Healy, J. I., and C. C. Goodnow. 1998. Positive versus negative signaling by lymphocyte antigen receptors. Annu. Rev. Immunol. 16:645-670. [DOI] [PubMed] [Google Scholar]
- 18.Herrscher, R. F., M. H. Kaplan, D. L. Lelsz, C. Das, R. Scheuermann, and P. W. Tucker. 1995. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: a B cell-specific trans-activator that describes a new DNA-binding protein family. Genes Dev. 9:3067-3082. [DOI] [PubMed] [Google Scholar]
- 19.Hinsley, T. A., P. Cunliffe, H. J. Tipney, A. Brass, and M. Tassabehji. 2004. Comparison of TFII-I gene family members deleted in Williams-Beuren syndrome. Protein Sci. 13:2588-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirano, M., Y. Kikuchi, S. Nisitani, A. Yamaguchi, A. Satoh, T. Ito, H. Iba, and K. Takatsu. 2004. Bruton's tyrosine kinase (Btk) enhances transcriptional co-activation activity of BAM11, a Btk-associated molecule of a subunit of SWI/SNF complexes. Int. Immunol. 16:747-757. [DOI] [PubMed] [Google Scholar]
- 21.Humphries, L. A., C. Dangelmaier, K. Sommer, K. Kipp, R. M. Kato, N. Griffith, I. Bakman, C. W. Turk, J. L. Daniel, and D. J. Rawlings. 2004. Tec kinases mediate sustained calcium influx via site-specific tyrosine phosphorylation of the phospholipase Cγ Src homology 2-Src homology 3 linker. J. Biol. Chem. 279:37651-37661. [DOI] [PubMed] [Google Scholar]
- 22.Kikuchi, Y., M. Hirano, M. Seto, and K. Takatsu. 2000. Identification and characterization of a molecule, BAM11, that associates with the pleckstrin homology domain of mouse Btk. Int. Immunol. 12:1397-1408. [DOI] [PubMed] [Google Scholar]
- 23.Kim, D. W., and B. H. Cochran. 2000. Extracellular signal-regulated kinase binds to TFII-I and regulates its activation of the c-fos promoter. Mol. Cell. Biol. 20:1140-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, D. W., and B. H. Cochran. 2001. JAK2 activates TFII-I and regulates its interaction with extracellular signal-regulated kinase. Mol. Cell. Biol. 21:3387-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurosaki, T., and M. Kurosaki. 1997. Transphosphorylation of Bruton's tyrosine kinase on tyrosine 551 is critical for B cell antigen receptor function. J. Biol. Chem. 272:15595-15598.9188445 [Google Scholar]
- 26.Li, T. J., D. J. Rawlings, H. Park, R. M. Kato, O. N. Witte, and A. B. Satterthwaite. 1997. Constitutive membrane association potentiates activation of Bruton tyrosine kinase. Oncogene 15:1375-1383. [DOI] [PubMed] [Google Scholar]
- 27.Lin, X. L., Y. Z. Lin, and J. Tang. 1994. Relationships of human immunodeficiency virus protease with eukaryotic aspartic proteases. Methods Enzymol. 241:195-224. [DOI] [PubMed] [Google Scholar]
- 28.Lowry, W. E., J. Huang, M. Lei, D. Rawlings, and X.-Y. Huang. 2001. Role of the PHTH module in protein substrate recognition by Bruton's agammaglobulinemia tyrosine kinase. J. Biol. Chem. 276:45276-45281. [DOI] [PubMed] [Google Scholar]
- 29.Mahajan, S., A. Vassilev, N. Sun, Z. Ozer, C. Mao, and F. M. Uckun. 2001. Transcription factor STAT5A is a substrate of Bruton's tyrosine kinase in B cells. J. Biol. Chem. 276:31216-31228. [DOI] [PubMed] [Google Scholar]
- 30.Melamed, D., R. J. Benschop, J. C. Cambier, and D. Nemazee. 1998. Developmental regulation of B lymphocyte immune tolerance compartmentalizes clonal selection from receptor selection. Cell 92:173-182. [DOI] [PubMed] [Google Scholar]
- 31.Middendorp, S., G. M. Dingjan, A. Maas, K. Dahlenborg, and R. W. Hendriks. 2003. Function of Bruton's tyrosine kinase during B cell development is partially independent of its catalytic activity. J. Immunol. 171:5988-5996. [DOI] [PubMed] [Google Scholar]
- 32.Mohamed, A. J., L. Vargas, B. F. Nore, C. M. Backesjo, B. Christensson, and C. I. Smith. 2000. Nucleocytoplasmic shuttling of Bruton's tyrosine kinase. J. Biol. Chem. 275:40614-40619. [DOI] [PubMed] [Google Scholar]
- 33.Nisitani, S., A. B. Satterthwaite, K. Akashi, I. L. Weissman, O. N. Witte, and M. I. Wahl. 2000. Posttranscriptional regulation of Bruton's tyrosine kinase expression in antigen receptor-stimulated splenic B cells. Proc. Natl. Acad. Sci. USA 97:2737-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nixon, J. C., J. Rajaiya, and C. F. Webb. 2004. Characterization of dominant negative forms of the transcription factor, Bright. J. Biol. Chem. 279:52465-52472. [DOI] [PubMed] [Google Scholar]
- 35.Nixon, J. C., J. B. Rajaiya, N. Ayers, S. Evetts, and C. F. Webb. 2004. The transcription factor, Bright, is not expressed in all human B lymphocyte subpopulations. Cell. Immunol. 228:42-53. [DOI] [PubMed] [Google Scholar]
- 36.Nomura, K., H. Kanegane, H. Karasuyama, S. Tsukada, K. Agematsu, G. Murakami, S. Sakazume, M. Sako, R. Tanaka, Y. Kuniya, T. Komeno, S. Ishihara, K. Hayashi, T. Kishimoto, and T. Miyawaki. 2000. Genetic defect in human X-linked agammaglobulinemia impedes a maturational evolution of pro-B cells into a later stage of pre-B cells in the B-cell differentiation pathway. Blood 96:610-617. [PubMed] [Google Scholar]
- 37.Novina, C. D., V. Cheriyath, and A. L. Roy. 1998. Regulation of TFII-I activity by phosphorylation. J. Biol. Chem. 273:33443-33448. [DOI] [PubMed] [Google Scholar]
- 38.Novina, C. D., S. Kumar, U. Bajpai, V. Cheriyath, K. M. Zhang, S. Pillai, H. H. Wortis, and A. L. Roy. 1999. Regulation of nuclear localization and transcriptional activity of TFII-I by Bruton's tyrosine kinase. Mol. Cell. Biol. 19:5014-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nussenzweig, M. C. 1998. Immune receptor editing: revise and select. Cell 95:875-878. [DOI] [PubMed] [Google Scholar]
- 40.Qiu, Y., and H. J. Kung. 2000. Signaling network of the Btk family kinases. Oncogene 19:5651-5661. [DOI] [PubMed] [Google Scholar]
- 41.Rawlings, D. J. 1999. Bruton's tyrosine kinase controls a sustained calcium signal essential for B lineage development and function. Clin. Immunol. 91:243-253. [DOI] [PubMed] [Google Scholar]
- 42.Rawlings, D. J., D. C. Saffran, S. Tsukada, D. A. Largaespada, J. C. Grimaldi, L. Cohen, R. N. Mohr, J. F. Bazan, M. Howard, N. G. Copeland, N. A. Jenkins, and O. N. Witte. 1993. Mutation of the unique region of Bruton's tyrosine kinase in immunodeficient XID mice. Science 261:358-361. [DOI] [PubMed] [Google Scholar]
- 43.Rawlings, D. J., A. M. Scharenberg, H. Park, M. I. Wahl, S. Lin, R. M. Kato, A. C. Fluckiger, O. N. Witte, and J. P. Kinet. 1996. Activation of BTK by a phosphorylation mechanism initiated by SRC family kinases. Science 271:822-825. [DOI] [PubMed] [Google Scholar]
- 44.Sacristán, C., M. I. Tussié-Luna, S. M. Logan, and A. L. Roy. 2004. Mechanism of Bruton's tyrosine kinase-mediated recruitment and regulation of TFII-I. J. Biol. Chem. 279:7147-7158. [DOI] [PubMed] [Google Scholar]
- 45.Saito, K., K. F. Tolias, A. Saci, H. B. Koon, L. A. Humphries, A. Scharenberg, D. J. Rawlings, J. P. Kinet, and C. L. Carpenter. 2003. BTK regulates PtdIns-4,5-P2 synthesis: importance for calcium signaling and PI3K activity. Immunity 19:669-678. [DOI] [PubMed] [Google Scholar]
- 46.Saouaf, S. J., S. Mahajan, R. B. Rowley, S. A. Kut, J. Fargnoli, A. L. Burkhardt, S. Tsukada, O. N. Witte, and J. B. Bolen. 1994. Temporal differences in the activation of three classes of non-transmembrane protein tyrosine kinases following B-cell antigen receptor surface engagement. Proc. Natl. Acad. Sci. USA 91:9524-9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato, S., T. Katagiri, S. Takaki, Y. Kikuchi, Y. Hitoshi, S. Yonehara, S. Tsukada, D. Kitamura, T. Watanabe, O. Witte, and K. Takatsu. 1994. IL-5 receptor-mediated tyrosine phosphorylation of SH2/SH3-containing proteins and activation of Bruton's tyrosine and Janus 2 kinases. J. Exp. Med. 180:2101-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Satterthwaite, A. B., and O. N. Witte. 2000. The role of Bruton's tyrosine kinase in B-cell development and function: a genetic perspective. Immunol. Rev. 175:120-127. [PubMed] [Google Scholar]
- 49.Su, T. T., and D. J. Rawlings. 2002. Transitional B lymphocyte subsets operate as distinct checkpoints in murine splenic B cell development. J. Immunol. 168:2101-2110. [DOI] [PubMed] [Google Scholar]
- 50.Thomas, J. D., P. Sideras, C. I. E. Smith, I. Vorechovsky, V. Chapman, and W. E. Paul. 1993. Colocalization of X-linked agammaglobulinemia and X-linked immunodeficiency genes. Science 261:355-358. [DOI] [PubMed] [Google Scholar]
- 51.Tsukada, S., M. I. Simon, O. N. Witte, and A. Katz. 1994. Binding of beta gamma subunits of heterotrimeric G proteins to the PH domain of Bruton tyrosine kinase. Proc. Natl. Acad. Sci. USA 91:11256-11260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wahl, M. I., A. C. Fluckiger, R. M. Kato, H. Park, O. N. Witte, and D. J. Rawlings. 1997. Phosphorylation of two regulatory tyrosine residues in the activation of Bruton's tyrosine kinase via alternative receptors. Proc. Natl. Acad. Sci. USA 94:11526-11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, Z., A. Goldstein, R.-T. Zong, D. Lin, E. J. Neufeld, R. H. Scheuermann, and P. W. Tucker. 1999. Cux/CDP homeoprotein is a component of NF-μNR and represses the immunoglobulin heavy chain intronic enhancer by antagonizing the Bright transcription activator. Mol. Cell. Biol. 19:284-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Webb, C. F., C. Das, R. L. Coffman, and P. W. Tucker. 1989. Induction of immunoglobulin μ mRNA in a B cell transfectant stimulated with interleukin-5 and a T-dependent antigen. J. Immunol. 143:3934-3939. [PubMed] [Google Scholar]
- 55.Webb, C. F., C. Das, S. Eaton, K. Calame, and P. W. Tucker. 1991. Novel protein-DNA interactions associated with increased immunoglobulin transcription in response to antigen plus interleukin-5. Mol. Cell. Biol. 11:5197-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Webb, C. F., C. Das, K. L. Eneff, and P. W. Tucker. 1991. Identification of a matrix-associated region 5′ of an immunoglobulin heavy chain variable region gene. Mol. Cell. Biol. 11:5206-5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Webb, C. F., K. L. Eneff, and F. H. Drake. 1993. A topoisomerase II-like protein is part of an inducible DNA-binding protein complex that binds 5′ of an immunoglobulin promoter. Nucleic Acids Res. 21:4363-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Webb, C. F., E. A. Smith, K. L. Medina, K. L. Buchanan, G. Smithson, and S. Dou. 1998. Expression of Bright at two distinct stages of B lymphocyte development. J. Immunol. 160:4747-4754. [PubMed] [Google Scholar]
- 59.Webb, C. F., Y. Yamashita, N. Ayers, S. Evetts, Y. Paulin, M. E. Conley, and E. A. Smith. 2000. The transcription factor Bright associates with Bruton's tyrosine kinase, the defective protein in immunodeficiency disease. J. Immunol. 165:6956-6965. [DOI] [PubMed] [Google Scholar]
- 60.Wu, Y. X., and C. Patterson. 1999. The human KDR/flk-1 gene contains a functional initiator element that is bound and transactivated by TFII-I. J. Biol. Chem. 274:3207-3214. [DOI] [PubMed] [Google Scholar]
- 61.Yang, W., S. N. Malek, and S. Desiderio. 1995. An SH3-binding site conserved in Bruton's tyrosine kinase and related tyrosine kinases mediates specific protein interactions in vitro and in vivo. J. Biol. Chem. 270:20832-20840. [DOI] [PubMed] [Google Scholar]
- 62.Yang, W. C., Y. Collette, J. A. Nunes, and D. Olive. 2000. Tec kinases: a family with multiple roles in immunity. Immunity 12:373-382. [DOI] [PubMed] [Google Scholar]
- 63.Yang, W. Y., and S. Desiderio. 1997. BAP-135, a target for Bruton's tyrosine kinase in response to B cell receptor engagement. Proc. Natl. Acad. Sci. USA 94:604-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yao, L., Y. Kawakami, and T. Kawakami. 1994. The pleckstrin homology domain of Bruton tyrosine kinase interacts with protein kinase C. Proc. Natl. Acad. Sci. USA 91:9175-9179. [DOI] [PMC free article] [PubMed] [Google Scholar]