Abstract
Histone deacetylase 4 (HDAC4) negatively regulates skeletal myogenesis by associating with the myocyte enhancer factor 2 (MEF2) transcription factors. Our data indicate that the gene PC4 (interferon-related developmental regulator 1 [IFRD1], Tis7), which we have previously shown to be required for myoblast differentiation, is both induced by MyoD and potentiates the transcriptional activity of MyoD, thus revealing a positive regulatory loop between these molecules. Enhancement by PC4 of MyoD-dependent activation of muscle gene promoters occurs selectively through MEF2 binding sites. Furthermore, PC4 localizes in the nucleus of differentiating myoblasts, associates with MEF2C, and is able to counteract the HDAC4-mediated inhibition of MEF2C. This latter action can be explained by the observed ability of PC4 to dose dependently displace HDAC4 from MEF2C. Consistently, we have observed that (i) the region of PC4 that binds MEF2C is sufficient to counteract the inhibition by HDAC4; (ii) PC4, although able to bind HDAC4, does not inhibit the enzymatic activity of HDAC4; and (iii) PC4 overcomes the inhibition mediated by the amino-terminal domain of HDAC4, which associates with MEF2C but not with PC4. Together, our findings strongly suggest that PC4 acts as a coactivator of MyoD and MEF2C by removing the inhibitory effect of HDAC4, thus exerting a pivotal function during myogenesis.
Acquisition of myogenic identity, the initial step of myogenesis leading to the generation of skeletal myoblasts and the ensuing differentiation into multinucleated myotubes are controlled by a family of myogenic basic helix-loop-helix (bHLH) transcription factors, including MyoD, Myf5, myogenin, and MRF4/Myf-6/Herculin (7, 8, 14, 18, 53, 65, 83). When ectopically expressed in a number of cell types, the myogenic bHLH regulators are capable of initiating the skeletal muscle differentiation program (reviewed in reference 79). These transcription factors activate muscle gene transcription, forming heterodimers with ubiquitously expressed bHLH proteins, termed E proteins, and hence binding to the consensus E-box sequence (CANNTG) present in the promoters of many muscle-specific genes (4, 35). Targeted gene knockout and other experiments have revealed that each member of the myogenic bHLH family is expressed and plays a specific role at various stages of myogenesis (10, 36, 77). MyoD and Myf5 are essential for specifying and maintaining muscle cell identity (68), whereas myogenin is required for the differentiation of specified precursors (31, 60) and MRF4 contributes to the later maturation steps (62).
Skeletal muscle differentiation involves the concerted action of myogenic bHLH factors and of the myocyte enhancer factor 2 (MEF2) transcription factor family. This family consists of four proteins (MEF2A, -B, -C, and -D) that share two highly conserved amino-terminal sequence motifs (referred to as the MADS and MEF2 domains) responsible for DNA binding and dimerization, whereas the divergent carboxyl-terminal domains are important for gene activation and kinase responsiveness. MEF2 homo- or heterodimers bind an A/T-rich DNA sequence [C/TTA(A/T)4TAG/A] within the regulatory regions of several muscle-specific genes (reviewed in reference 3). Remarkably, ablation of MEF2C in mice causes a phenotype of embryonic lethality with cardiac malformation (39), whereas ablation of the single MEF2 gene in Drosophila melanogaster leads to an absence of differentiated somatic, cardiac, and visceral muscle, indicating that MEF2 is essential for muscle differentiation (5, 38).
There is evidence to indicate that MEF2 and myogenic bHLH proteins are engaged in reciprocal regulatory circuits that generate a positive-feedback loop between the two families of regulators (reviewed in references 42 and 58). MEF2 proteins, although unable to activate myogenesis by themselves, synergize with myogenic bHLH to regulate transcription through mechanisms involving their physical interaction and the transmission of a transcriptional activation signal (2, 56).
More recently, it has been shown that histone deacetylases (HDACs) play an important role in muscle differentiation. HDACs participate in the process of chromatin remodeling, by deacetylating histones and transcription factors, as corepressors in multiprotein complexes (for a review, see reference 81). Numerous data indicate that distinct HDAC families exert an inhibitory control on muscle-specific transcription. In fact, MyoD has been found to bind and be repressed by HDAC1, which belongs to class I of these enzymes (45, 63), whereas HDAC4, -5, and -7, members of class II, bind and repress MEF2, blocking the MyoD-induced muscle differentiation (16, 26, 37, 41, 54, 76). Class I HDACs appear to exert a negative control on the expression of late muscle genes (63), whereas class II HDACs inhibit the expression of both early and late muscle genes and their repression is removed by Ca-calmodulin kinase (CaMK), which triggers the nuclear export of HDAC4 and -5 (50, 89).
In this context the gene PC4, which has been previously shown by us to be required for muscle differentiation, plays its role. In fact, inhibition of PC4 expression in myoblasts, by antisense PC4 cDNA transfection or microinjection of anti-PC4 antibodies, prevents morphological and biochemical differentiation, impairing myogenin and myosin gene expression (27). Very recently, an important role for PC4 in muscle differentiation has been observed also in vivo. Mice lacking Tis7 (the murine homolog of PC4) display defective muscle regeneration, characterized by reduced differentiation potential of muscle satellite cells and decreased levels of MyoD and myogenin (73). PC4 was originally isolated as an immediate-early gene induced at the onset of the neuronal differentiation elicited by nerve growth factor in PC12 cells (72) or as a gene induced by tetradecanoyl phorbol acetate in NIH 3T3 cells (named Tis7) (74). The expression of PC4/Tis7, and of its human homolog (interferon-related developmental regulator 1 [IFRD1]) (9), is regulated during neuronal and muscle differentiation in cell lines and in vivo (27, 29, 33). PC4, as the gene is called here, is expressed in the mouse embryonic brain and skeletal muscle, attains an appreciable level in neural tissues at midgestation (embryonic day 10 [E10] to E12) and in back muscle at late gestation (E17), and presents maximal expression in adult skeletal muscle and heart (9, 33). Taken together, these observations strongly suggest a role of PC4 in terminal differentiation of skeletal muscle cells. Interestingly, recent reports have revealed a role for PC4 as a regulator of transcription involved in tissue regeneration after ischemic stroke and in loss of epithelial cell polarity (67, 75, 80).
We show here that PC4 can potentiate the transcriptional activity of MyoD and MEF2C and reverse the HDAC4-dependent inhibition of muscle gene transcription. Our data lead to the conclusion that PC4 elicits these two effects due to its ability to antagonize the association of HDAC4 with MEF2C. Thus, PC4 may play an important regulatory role in the control of myogenesis.
MATERIALS AND METHODS
Cell culture, cell lines, and transfection.
Clone 7 of the C2 line of mouse myoblasts (84) was obtained from M. Buckingham (Institut Pasteur, Paris, France) and was cultured in a humidified atmosphere of 5% CO2 at 37°C in growth medium (GM), i.e., Dulbecco modified Eagle medium supplemented with 20% fetal bovine serum (HyClone, Logan, Utah). Clone S4 of C2C12 cells overexpressing PC4 was obtained by stable transfection of myoblasts with the construct pBAP-neo-PC4, as previously described (27). Myoblasts were passaged before reaching cell-cell contact to avoid selection. C3H10T1/2 fibroblasts were also grown in GM. The C3H-ER-MyoD cell line (a C3H10T1/2 cell line stably expressing an estradiol-inducible MyoD protein) has been previously produced and characterized (11). To induce differentiation, myoblasts were exposed for 2 days (or as indicated) to differentiation medium (DM; Dulbecco modified Eagle medium containing 2% fetal bovine serum). Differentiation of C3H-ER-MyoD cells was induced in DM in the presence of 10−7 M estradiol.
Cell cultures were transfected by the liposome technique with Lipofectamine reagent (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions.
Genomic clone isolation, sequencing, and primer extension.
Approximately 500,000 plaques of a rat genomic library in an EMBL-3 vector were screened by filter hybridization, using rat PC4 full-length cDNA 32P labeled by random priming (19) as a probe, yielding eight clones with an average insert length of 20 kb. A further analysis of these genomic clones by digestion with restriction enzymes and Southern blotting, using the most 5′ region of PC4 cDNA (PstI-PstI fragment of 312 nucleotides [nt] excised from pCD-PC4 vector) (72) as a probe, indicated that four of them contained the region upstream to the 5′ untranslated region. The clone carrying the longest 5′ upstream sequence, named PC4G4, was cut into two fragments (5′XbaI-3′XbaI, 7.5 kb; 5′StuI-3′StuI, 3.6 kb), both hybridizing with 32P-labeled oligonucleotides complementary to the 5′ region of PC4 cDNA, which were subcloned in pBluescript (constructs named PC4G4/3.1 and PC4G4/12, respectively). The transcription initiation site was mapped by primer extension using a 25mer oligodeoxynucleotide complementary to nt 31 to 55 of PC4 cDNA (5′-GGCTGAGAGGCGAGTCTCCGGCTAA-3′). The [γ-32P]dATP-labeled oligonucleotide (by polynucleotide kinase) was annealed at 70°C with 20 μg of poly(A)+ RNA obtained from PC12 cells treated or not with nerve growth factor (NGF; 100 ng/ml) for 1 h, and extended at 37°C by using Moloney murine leukemia virus reverse transcriptase (100 U; Promega). The extended products were digested with RNase A and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 6% polyacrylamide gels with 7 M urea, in parallel with a dideoxy DNA sequencing reaction of clone PC4G4/3.1, performed by using the extension oligonucleotide as a primer (70).
Construction of the PC4 promoter reporter gene.
Clone PC4G4/12 (i.e., 5′StuI-3′StuI fragment of PC4G4; 3′StuI is located at nt −443) contained the PC4 sequence comprised between nt −778 and nt −438, relative to the transcription start site. Instead clone PC4G4/3.1 (5′XbaI-3′XbaI fragment of PC4G4; 5′XbaI is located at nt −560) started downstream to nt −560. PC4 (−778+160)-CAT was obtained by (i) cloning the fragment 5′HindIII-3′XbaI excised from PC4G4/12 (3′XbaI being located at nt −560) into the corresponding sites of the multiple cloning region of vector pSV0t2CAT (44), (ii) adding to the XbaI site of the obtained construct the insert 5′XbaI-3′SacI excised from clone PC4G4/3.1 (the 3′SacI site, located 35 nt before the ATG, was previously blunted and ligated to XbaI linkers), and (iii) removing from this construct the EMBL-3 lambda phage sequences (5′HindIII-3′SalI upstream fragment) and then blunting and religating the DNA ends. PC4 (−560+160)-CAT was obtained by subcloning the 5′XbaI-3′XbaI fragment excised from PC4 (−778+160)-CAT in vector pSV0t2CAT. PC4 (−133+160)-CAT was generated by subcloning the 5′BglII-3′XbaI fragment excised from PC4 (−560+160)-CAT and ligated to XbaI linkers in the XbaI site of vector pSV0t2CAT.
Plasmids, PC4 expression vectors, and mutants.
The following expression vectors were kindly provided as indicated. pEMC11s (also named pEMSV-MyoD) and its empty vector pEMSV Scribeα2 was provided by H. Weintraub (14). pcDNA1-MEF2C, pCDNA1-MEF2D and pVP16-MEF2C were provided by E. Olson (47, 57). pcDNA3.1-Myc-HDAC4 was provided by T. Kouzarides (54). Finally, activated SRα-CamKI was provided by A. Means (30). The following constructs and reporter plasmids were generously provided as follows. pGEM7z-MEF2A (human) was provided by S. Ferrari. Glutathione S-transferase (GST)-MyoD was provided by A. Lassar (34). pGEX-2T-MEF2C and pGEX-2T-MEF2C ΔMADS were provided by V. Sartorelli (71). pMyo84CAT, pMyo84(-E1)CAT, pMyo84(mutMEF2)CAT, pMyo84(mutMEF2/-E1)CAT were provided by E. Olson (17). pTK-MEF2x2CAT and the empty vector pBLCAT2 (86) were provided by S. Ferrari. 4RE-luciferase and 3x MEF2-luciferase were provided by E. Olson (41). Muscle creatine kinase luciferase (MCK-LUC) was provided by V. Sartorelli (64). Finally, pt184RTK-CAT was provided by H. Weintraub (78). pEB-myogenin was obtained by subcloning the open reading frame (ORF) in the vector pEB (46).
The expression vector pSCT was from B. Schäfer (23), who obtained it by adding an artificial polylinker to the vector pSCT GAL 556X (69). pSCT-PC4 was constructed by cloning in 5′BamHI-3′HindIII the corresponding fragment excised from vector pGEM3z-PC4b, containing the complete coding region of PC4 cDNA (nucleotides 129 to 1570, the ORF being from nucleotides 146 to 1495). pGEM3z-PC4b (29) was obtained by cloning into the SalI site of pGEM3z vector (Promega) the fragment 5′BanI-3′HindIII excised from pCD-PC4, containing the full-length PC4 ORF (subclone OB83R) (72), previously blunted and ligated to SalI linkers.
Hemagglutinin (HA)-tagged pSCT-PC4 was generated by cloning into pSCT the fragment 5′HindIII-3′BamHI, excised from pGEM3z-HA-PC4, containing the whole PC4 ORF in frame with an upstream 2× HA tag sequence (cloned in the 5′HindIII-3′SalI sites) preceded by a Kozak consensus sequence.
PC4 deletion mutants (identified by amino acid residues unless otherwise indicated) were obtained through an intermediate step. The PC4 cDNA region was PCR amplified and cloned in 5′SalI-3′KpnI of pGEM3Z-HA (containing a 2× HA tag [see above]), and then the whole 5′HindIII-3′KpnI fragment (containing the HA-PC4 cDNA sequence) was excised and subcloned into the same sites of the pSCT polylinker. The following primers were used (SalI and KpnI sites are underlined in the forward and backward primer sequences, respectively): (i) HA-pSCT-PC4 1-295, corresponding to nt 149 to 1030 of PC4 cDNA (forward primer [5′-GCTGTCGACCCGAAGAACAAGAAGCGGAAC-3′] and backward primer [5′-AGGGTACCGGCCAATTCAAACAGAAGTGC-3′]), and (ii) HA-pSCT-PC4 290-449, corresponding to nt 1013 to 1495 of PC4 cDNA (forward primer [5′-GCTGTCGACCTTCTGTTTGAATTGGCCAGA-3′] and backward primer [5′-GTGGGTACCCTAGAAGAATTCTCCAACATC-3′]).
pGEM3z-PC4 1-118 and pGEM3z-PC4 1-295 (used to generate in vitro-translated proteins) were produced by excision of the fragments 5′XhoI-3′BamHI or 5′BalI-3′BamHI corresponding to amino acids (aa) 119 to 449 and aa 196 to 449, respectively, from the construct pGEM3z-PC4b; XhoI and BalI sites were blunted and ligated to BamHI linkers. pGEM3z-PC4 118-449 and pGEM3z-PC4 293-449 were obtained by subcloning the fragment 5′HindIII-3′BamHI amplified by PCR (nt 490 to 1570 and nt 1018 to 1570, respectively) into vector pGEM3z using as a template vector pGEM3z-PC4b, with the forward primers 5′-CAAAGCTTCCGCCAC CATGCTCGAGAGAAGAATGACT-3′ and 5′-CAAAGCTTCCGCCACCATGGAATT GGCCAGAGGAATG-3′ (the flanking 5′HindIII site is underlined), respectively, and with backward primer complementary to the T7 RNA polymerase promoter sequence present in the 3′ region of the pGEM3z-PC4b polylinker.
pcDNA3-HA-MEF2C was obtained by cloning the PCR-amplified MEF2C cDNA (using pcDNA1-MEF2C as a template) in frame in 5′SalI-3′XbaI of pGEM3Z-HA; the whole 5′HindIII-3′BamHI fragment containing the HA-MEF2C cDNA sequence was then subcloned into the same sites of pcDNA3 vector. Deletion mutants HDAC41-611 and HDAC4611-1084 were obtained by subcloning the fragment 5′EcoRII-3′XbaI, amplified by PCR using pcDNA3.1-Myc-HDAC4 as a template vector into vector pcDNA6-Myc (in frame to the Myc tag downstream).
All of the constructs described above were checked by sequence analysis.
mRNA analysis.
The extraction of total mRNA from C3H-ER-MyoD, C3H10T1/2, and C2C12 cell cultures and the following Northern analyses were performed as previously described (11).
Constructs for two-hybrid assay in C3H10T1/2 cells.
pMGAL4-PC4 was obtained by cloning the fragment 5′SalI-3′XbaI excised from pGEM 3Z-PC4ATG(−) (containing the PC4 ORF without ATG, preceded by a SalI site) into PM vector (Clontech), in frame with the GAL4 DNA-binding domain (DBD). pMGAL4-PC4 290-449 was constructed by cloning into 5′SalI-3′SalI sites of PM vector the PCR-amplified aa 290 to 449 region of PC4 in frame with the GAL4 DBD. The constructs were checked by sequence analysis.
Immunocytochemistry, confocal microscopy, and antibodies.
Endogenous PC4 protein was detected by immunofluorescence staining in C2C7 cell cultures grown in 35-mm dishes fixed for 10 min at room temperature in phosphate-buffered saline (PBS) containing 3.75% paraformaldehyde. Cultures were then washed three times in PBS, permeabilized by a 5-min incubation with 0.2% Triton X-100 in PBS, washed again three times in PBS, and incubated for 60 min at room temperature with the rabbit polyclonal A451 primary antibody (described in reference 29; diluted 1:75 in PBS). After three washes in PBS, cells were incubated 30 min with the secondary antibody, either fluorescein isothiocyanate (FITC)-conjugated (Jackson Immunoresearch) or TRITC (tetramethyl rhodamine isothiocyanate)-conjugated (Jackson Immunoresearch). Cells were finally washed in PBS and mounted with PBS-glycerol (3:1). Immunofluorescence was observed by using an Olympus BX51 microscope with a Diagnostic Instruments digital camera (model 1.3.0). To detect nuclei, cells were incubated at the end of the immunofluorescence staining procedure for 2 min in Hoechst 33258 dye diluted in PBS at 1 μg/ml (Sigma), washed twice in PBS, and mounted as described above. Detection of endogenous PC4 and HDAC4 by confocal microscopy was performed on C2C7 or C2C12 cells grown on polylysine-coated coverslips layered onto a 35-mm dish. The immunofluorescence protocol described above was followed (using A451 and the anti-HDAC4 goat polyclonal L-19 [Santa Cruz]), except that 0.1% bovine serum albumin in PBS was used in a 30-min preincubation with 1% goat serum, in the incubation with the primary antibody, and also in the following washes. A final 30-min incubation with RNase followed (0.1 mg/ml diluted in PBS). Nuclei were visualized by incubating the cells 4 min in propidium iodide (0.1 μg/ml in PBS). Coverslips were then mounted on slides. Omission of the primary antibody demonstrated minimal background staining. Fluorescently labeled preparations were observed by a confocal laser scanning microscope LEICA TCS 4D (Leica Microsystems) supplemented with an argon-krypton laser. The excitation and emission wavelengths used were 488 nm and 510 nm for FITC labeling and 568 and 590 nm, respectively, for TRITC labeling. The acquisitions were recorded by using pseudo-color representation.
Immunoprecipitations and immunoblots.
Myoblasts (either C2C7 or C2C12 clone S4 [27] as indicated) and NIH 3T3 and HEK293 cell cultures, grown in 90-mm dishes, transfected or naïve, were lysed by sonication in buffer containing 50 mM Tris-HCl (pH 7.4)-150 mM NaCl-1 mM EDTA-0.2% NP-40, with protease inhibitors, 1 mM Na3VO4, 10 mM β-glycerophosphate, 10 mM NaF, 5 mM ATP, and 5 mM MgCl2. Then, 1.5 mg of myoblast or NIH 3T3 lysate was immunoprecipitated with anti-PC4 coupled to CH-Sepharose 4B or with anti-HA agarose-conjugated (Santa Cruz), as indicated. Lysates of HEK293 cells used in the experiments of displacement were obtained in PBS containing 0.5% Triton X-100 and 1 mM EDTA; 1.5 mg of lysate was then immunoprecipitated with anti-HA monoclonal antibody (clone 12CA5; a gift from O. Segatto).
In lysates from transfected cultures, the Myc-HDAC4 and HA-MEF2C proteins, or also the HA-PC4 mutants, were revealed by Western blots with anti-Myc (clone 9E10; Santa Cruz) and anti-HA monoclonal antibodies. Endogenous HDAC4 was revealed by Western blots with anti-HDAC4 rabbit polyclonal (Cell Signaling Technology). PC4, MyoD, myogenin, and β-actin were revealed by Western blots with rabbit A451 antibody (29) and the mouse monoclonal antibodies 5.8A (15) (DakoCytomation), IF5D (82), and AC-15 (Sigma), respectively.
Reporter gene assays.
C3H10T1/2 cell cultures (35-mm dishes containing 105 cells seeded the day before transfection) were transfected with the indicated expression constructs by using the Lipofectamine reagent. Variations in the amounts of expression vectors were compensated by addition of the corresponding empty DNA plasmid vectors. Chloramphenicol acetyltransferase (CAT) assays were performed as described previously (24): cells were harvested in TNE (40 mM Tris-Cl [pH 7.5], 1 mM EDTA [pH 8], 150 mM NaCl) and lysed in 250 mM Tris-HCl (pH 7.8) and 1 mM dithiothreitol by three freeze-thaw cycles. CAT levels were measured in cell extract aliquots containing equal amounts of proteins (determined by the procedure described in reference 6) incubated with acetyl coenzyme A (0.4 mg/ml) and [14C]chloramphenicol (1.4 μCi/ml). Luciferase assays were performed by the Luciferase assay system (Promega) according to the manufacturer's instructions as previously described (28). The CAT and luciferase activity of each sample (Ai) was normalized for differences in transfection, measuring in each transfected cell extract the β-galactosidase (β-Gal) levels (Gi), as determined by a described procedure (66). The normalized activity of the reporter gene was thus equal to Ai × Gm/Gi, where Gm is the average value for each experiment. The fold activity was then obtained by dividing each normalized reporter activity value by the average number of reporter activity units of the corresponding control culture.
GST fusion proteins.
The construct pGEX-4T-PC4 was obtained by subcloning the coding region of PC4, amplified by PCR, in frame into 5′BamHI-3′SalI sites of the vector pGEX-4T3. The different GST fusion proteins (including GST-MEF2C and GST-MyoD) were purified through glutathione-Sepharose beads (Amersham-Pharmacia) and eluted as described by the manufacturer.
Pull-down assays were performed incubating 10 μl of GST proteins bound to glutathione-Sepharose resin beads with in vitro-programmed nuclease-treated rabbit reticulocyte lysates as described previously (28). For displacement assays of HDAC4-MEF2C complexes, equal amounts of lysates of transfected NIH 3T3 cells were incubated with either GST or increasing amounts of GST-PC4 overnight at 4°C. Afterward, lysates were immunoprecipitated with anti-HA agarose-conjugated antibody (Santa Cruz) for 2 h at 4°C. Bound proteins were collected by centrifugation and washed three times with lysis buffer. Immunoprecipitated proteins were analyzed by immunoblots with anti-Myc, anti-HA, and anti-GST antibodies.
Deacetylase assay.
The deacetylase activity was assayed by measuring the release of [3H]acetate from [3H]acetyl histones with an HDAC assay kit (Upstate Biotechnology). HEK293 cells transfected with Myc-HDAC4 were lysed by sonication in lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 0.2% NP-40, and 10% glycerol with inhibitors of proteases and phosphatases). Next, 2.4 mg of precleared cell lysates was immunoprecipitated by using 8 μg of anti-Myc antibody and protein G-Sepharose (Amersham) for 3 h at 4°C. Control samples were immunoprecipitated after preincubation with a synthetic peptide in 100-fold molar excess corresponding to the Myc epitope. Immunoprecipitated complexes were washed four times in lysis buffer (containing 0.5 M NaCl in the last two washes, as described in reference 22) and twice with HDAC buffer (10 mM Tris-HCl [pH 8.0], 10 mM NaCl, 10% glycerol). Equal aliquots of each purified protein sample were incubated with 75,000 cpm of [3H]acetylated H4 peptide in 100 μl of assay buffer for 5 to 7 h at 37°C with or without 200 nM trichostatin A (Sigma). Another protein aliquot was used in Western blot to determine the amounts of immunoprecipitated protein, to which the deacetylase activities were normalized. Free [3H]acetyl was measured with liquid scintillation counting.
RESULTS
MyoD induces PC4 gene expression during myogenesis.
We have previously shown that myoblasts in which the PC4 protein function has been inhibited, fail to express differentiation markers such as myogenin or myosin heavy chain, and do not undergo terminal differentiation (27).
To elucidate the role of PC4 during myogenesis, we sought in the first place to determine whether its expression is regulated by MyoD. To address this issue, we used a previously described cell line expressing a chimeric MyoD protein fused to the hormone-binding domain of the estrogen receptor (MyoD-ER), whose activity is inducible by treatment with estradiol. The C3H10T1/2 fibroblast cell line stably expressing such a conditional MyoD protein (C3H-MyoD-ER) differentiates efficiently in the presence of estradiol and retains the undifferentiated phenotype in hormone-free conditions (11).
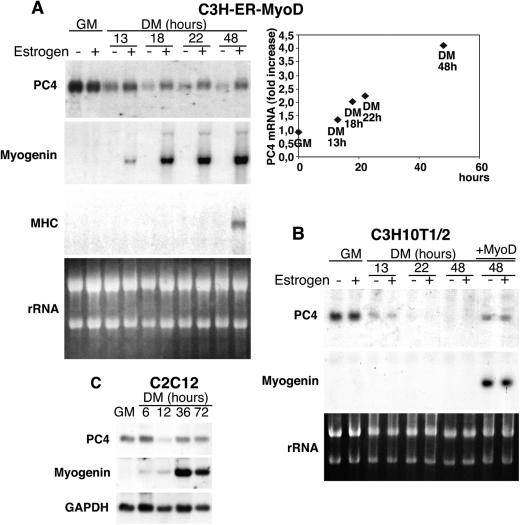
Figure 1A shows a Northern blot analysis of total RNA extracted from C3H-MyoD-ER cells cultured in GM or in differentiation medium (DM; with reduced serum concentration), either in the presence or in the absence of estradiol. PC4 mRNA was expressed in GM both in the presence and in the absence of hormone but decreased significantly in hormone-free DM, indicating that expression of PC4 is downregulated after serum removal. Conversely, the hormone-dependent induction of MyoD activity caused an increase of PC4 mRNA after 13 h in DM, concomitantly with the induction of myogenin expression. The PC4 mRNA levels increased up to fourfold during the following 48 h in DM (as indicated by the densitometric analysis of the blots shown in Fig. 1A), when late markers of differentiation (such as myosin heavy chain) were induced (Fig. 1A).
FIG. 1.
Induction of PC4 mRNA expression by MyoD. (A) C3H-ER-MyoD cells, expressing an estrogen-inducible MyoD protein, were cultured in GM or shifted to DM for increasing periods of time either in the presence or in the absence of estradiol (10−7 M) as indicated. Afterward, total RNA was extracted and analyzed by Northern blotting. Identical filters were probed for myogenin, myosin heavy chain, and PC4 mRNAs. Ethidium bromide staining of rRNA, on one of the filters, was photographed under UV light. The graph on the right shows densitometric quantification of the MyoD-mediated induction of PC4 mRNA expression, expressed as the ratio between the levels of PC4 mRNA measured in the presence or in the absence of estradiol. (B) Expression of PC4 mRNA was analyzed by Northern blot in parental C3H10T1/2 cells cultured in the same conditions as C3H-ER-MyoD cells. The mRNA from C3H10T1/2 cells infected with a MyoD-encoding retrovirus was also analyzed (infected cells were kept 48 h in differentiation conditions; lanes + MyoD). (C) Induction of PC4 mRNA expression during C2C12 myoblast differentiation. Cells were cultured in GM or shifted to DM for the times indicated. Total RNA was extracted and analyzed by Northern blotting, probing the same filter for PC4 and myogenin, as well as for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) to control for RNA integrity and quantity.
To verify that the estradiol treatment had no effect per se on the expression of PC4, the levels of PC4 mRNA were also measured in parental C3H10T1/2 fibroblasts under both growing and differentiation conditions with or without estrogen (Fig. 1B). PC4 mRNA was completely downregulated within 22 h after serum withdrawal, irrespective of the presence of hormone; however, a retrovirus-mediated ectopic expression of MyoD caused an upregulation of PC4 mRNA in differentiation conditions that was comparable to that elicited by the estrogen-regulated form of MyoD (Fig. 1B).
Finally, expression of PC4 mRNA was also analyzed in C2C12 myoblasts undergoing differentiation. In these cells, similarly to the fibroblasts, PC4 mRNA was expressed in proliferating conditions but decreased soon after serum withdrawal, to regain the initial expression levels at the onset of the differentiation process, in concomitance with the maximal induction of myogenin (Fig. 1C).
Together, these results indicate that MyoD induces PC4 mRNA expression from the early stages of terminal differentiation.
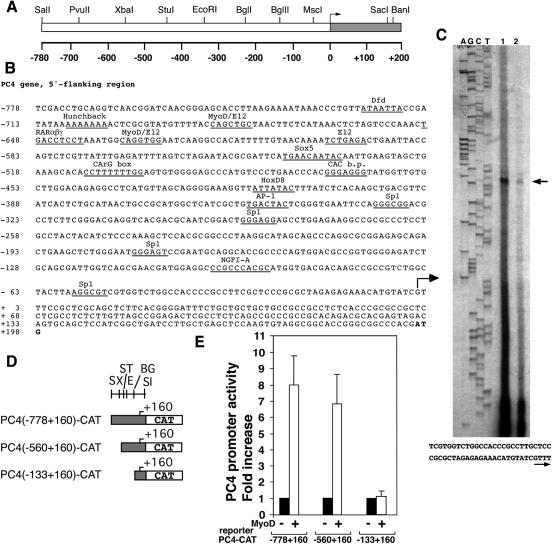
To verify whether the increase of PC4 mRNA levels elicited by MyoD was the consequence of upregulation of PC4 transcription, we sought to analyze the effect of MyoD on the activity of the PC4 gene promoter. We therefore isolated the 5′-flanking region of the rat PC4 gene, extending from nt −778 to nt +160 relative to the transcription initiation site (+1). This latter was determined by primer extension of poly(A)+ RNA isolated from rat PC12 cells induced for 2 h with NGF (given that PC4 is an NGF-inducible immediate-early gene; Fig. 2A to C). The same initiation site was mapped by using RNA from C2C7 myoblasts (data not shown). Inspection of the PC4 5′ flanking sequences (nt −778 to nt +160; Fig. 2B) did not show a TATA box but revealed the presence of a number of putative transcription factor consensus binding sites, including two E-boxes (nt −684 to −678; nt −634 to −628), the binding site of myogenic basic HLH factors (Fig. 2B) (4, 59).
FIG. 2.
Induction of PC4 promoter activity by MyoD. (A) Restriction map of the 5′ region of the rat PC4 gene. The transcribed region is represented in gray. (B) Nucleotide sequence of the 5′ region of the PC4 gene. Transcription factor consensus binding sequences are indicated. The transcription initiation site is indicated by the arrow. (C) Primer extension analysis of PC4 mRNA. RNA from rat PC12 cells either treated or untreated with NGF (lanes 1 and 2, respectively) was hybridized with the [γ-32P]dATP-labeled 31-55 PC4 oligonucleotide (see Materials and Methods). The arrow indicates the extension product of 105 nt. The left lanes show the DNA sequence of the genomic clone PC4G4/3.1, obtained with the same primer, to correlate extension product size and sequence. (D) Scheme of the constructs carrying different PC4 promoter sequences (gray box) fused to the CAT reporter gene. A restriction map is shown: S, SalI; X, XbaI; ST, StuI; E, EcoRI; BG, BglII; SI, SacI. (E) MyoD stimulates PC4 promoter activity. A total of 1.0 × 105 C3H10T1/2 cells, seeded onto 35-mm culture dishes the day before transfection, were transfected with each of the PC4 promoter reporters shown in panel D (1 μg) and with either the pEMSV-MyoD expression construct (1 μg) or the pEMVS Scribeα2 empty plasmid (1 μg). The cytomegalovirus (CMV)-β-Gal expression construct was cotransfected in each sample as an internal control. At 24 h after transfection cells were placed in DM for 48 h, and then lysates were collected and assayed for CAT and β-Gal activities. For each PC4 promoter construct the MyoD-dependent increase of CAT activity was calculated relative to the CAT activity in the absence of MyoD. Bars represent the average fold activity ± the standard error of the mean (SEM) of at least five independent experiments performed in duplicate. The CAT activities were measured as the percentage of the acetylated chloramphenicol/microgram of protein normalized to the β-Gal activity.
The 5′ region of the PC4 gene was cloned upstream to a CAT reporter gene in the vector pSV0t2CAT, obtaining the plasmid PC4(−778+160)-CAT. This plasmid was then transiently transfected into C3H10T1/2 fibroblasts with either a MyoD expression plasmid or its empty vector. The results shown in Fig. 2E indicate that indeed MyoD was able to induce the activity of the PC4 promoter construct PC4(−778 +160)-CAT up to sevenfold.
The most simple and testable mechanism by which MyoD could transactivate the PC4 promoter is by binding to the observed E-box consensus motifs. Thus, we generated a PC4 promoter 5′ deletion construct lacking the sequences that contain the E-boxes [PC4(−560+160)-CAT]. The activity of this PC4 promoter construct was still enhanced by MyoD (Fig. 2E), indicating that direct binding to the E-box motifs was not required for MyoD to mediate transcriptional induction of the PC4 promoter. In contrast, a 5′ deletion extending up to nt −133 caused a complete loss of promoter activation by MyoD (Fig. 2E). Thus, the sequences of the PC4 promoter targeted by MyoD appear to reside between nt −560 and nt −133. Further study will be required to identify them.
PC4 stimulates the MyoD-dependent activation of muscle-specific genes through MEF2.
The observations presented above, indicating that MyoD upregulates the transcription of the PC4 gene, together with our previous observations showing a requirement for PC4 during myogenesis (27), led us to question whether PC4 could act as a regulator of MyoD activity. To verify this hypothesis, we assessed the influence of PC4 on the MyoD-dependent activation of the myogenin gene promoter.
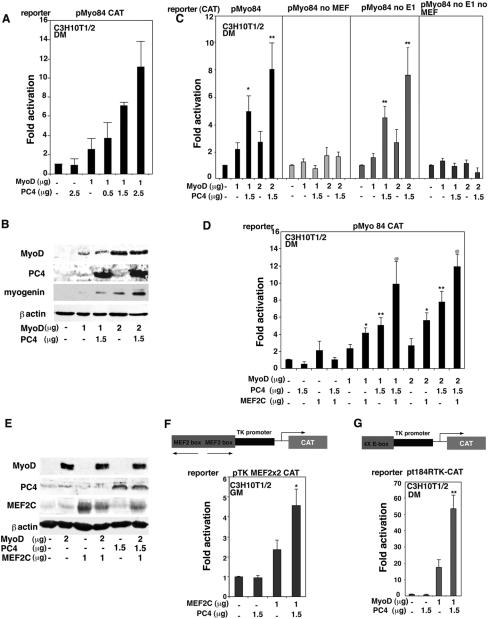
For our experiments, we used a reporter construct (pMyo84CAT) carrying 84 nucleotides of the myogenin promoter region, including an E-box and a MEF2 site (17, 25). These elements have been previously shown to mediate the responsiveness of the myogenin gene promoter to MyoD (17). Thus, the pMyo84CAT construct was transfected in C3H10T1/2 fibroblasts, together with MyoD and increasing amounts of PC4. We observed that PC4, in itself devoid of transcriptional activity, was able to potentiate the activation of the pMyo84 promoter by MyoD, dose dependently up to fourfold (Fig. 3A). We also tested whether PC4 could enhance the ability of MyoD to induce the endogenous myogenin gene. Cotransfection of PC4 with MyoD in C3H10T1/2 fibroblasts resulted in threefold higher levels of myogenin expression compared to those induced by MyoD alone (Fig. 3B).
FIG. 3.
PC4 enhances the transcriptional activity of MyoD through MEF2 binding sites. (A) PC4 synergizes with MyoD in stimulating the myogenin promoter activity. C3H10T1/2 cells were cotransfected with the pMyo84 reporter plasmid carrying the myogenin promoter (0.8 μg) and the indicated amounts of the pEMSV-MyoD and pSCT-PC4 expression vectors. The fold increase in CAT activity was calculated relative to the activity of the control sample (transfected with the empty vectors). In panel A, as well as in panels B, C, D, E, and G, cells were placed in DM 24 h after transfection and harvested 48 h later. (B) PC4 cooperates with MyoD to induce the endogenous myogenin gene. C3H10T1/2 fibroblasts were transiently transfected with MyoD alone or in conjunction with PC4; the levels of PC4, MyoD and endogenous myogenin were determined by Western blotting. (C) The synergism between PC4 and MyoD requires the MEF2 site of the myogenin promoter. C3H10T1/2 cells were transfected with the indicated amounts of pEMSV-MyoD, pSCT-PC4, and either the pMyo84-CAT reporter plasmid, the pMyo84(-E1) CAT, the pMyo84(mutMEF2) CAT (mutated in the E-box or MEF2 sites, respectively), or the pMyo84(mutMEF2/-E1)CAT (lacking both sites). Samples were analyzed as for panel A. ✽, P < 0.05; ✽✽, P < 0.02 (versus group with MyoD alone) (Student t test). (D) The synergism between MyoD and MEF2C is potentiated by PC4. C3H10T1/2 cells were transfected with the myogenin promoter construct pMyo84-CAT (0.8 μg) and with the indicated amounts of pEMSV-MyoD, pSCT-PC4, and pCDNA1-MEF2C. Samples were analyzed as for panel A. ✽, P < 0.05; ✽✽, P < 0.02 (versus group with MyoD alone); @, P < 0.03 (versus group transfected with pEMSV-MyoD and pCDNA1-MEF2C) (Student t test). (E) Western blot analysis of proteins expressed in C3H10T1/2 cells after a transfection experiment representative of those shown in panel D. (F) PC4 enhances the transcriptional activity of MEF2C. C3H10T1/2 cells were cotransfected with the indicated amounts of pSCT-PC4 or pCDNA1-MEF2C and with the reporter construct pTK-MEF2x2 CAT (carrying two MEF2C binding sites). Transfected cells were cultured in GM for 48 h before being harvested. Samples were analyzed as for panel A. ✽, P < 0.05 (versus MEF2C group). (G) PC4 enhances the E-box-mediated MyoD transcriptional activity. C3H10T1/2 were transfected with the indicated amounts of pEMSV-MyoD, pSCT-PC4, and the reporter construct pt184RTK-CAT (carrying four E-boxes). Samples were analyzed as for panel A. ✽✽, P < 0.02 (versus group with MyoD alone) (Student t test). In the assays in panels A, C, D, F, and G, CMV-β-Gal was cotransfected, and the empty DNA plasmid vectors were used in place of the corresponding expression vectors to keep DNA amounts constant. Luciferase activities were measured as units/microgram of protein normalized to the β-Gal activity. Bars represent the average fold activity ± the SEM of at least four independent experiments performed in duplicate.
Although MyoD can activate the myogenin promoter directly through the E-box, the MEF2 site also mediates activation by MyoD. In fact, the MEF2 site of the myogenin promoter has been shown to be required for myogenin transcription both in cultured cells and during embryonic development (17, 85), and a model has been proposed in which MEF2 factors, bound to DNA through the MEF2 site, can recruit MyoD, also to promoters devoid of E-boxes (56). We therefore sought to verify whether the potentiation of MyoD activity exerted by PC4 was mediated through the E-box or the MEF2 site. For this purpose we used either the reporter construct pMyo84(-E1)CAT, deleted in the E-box, or pMyo84(mutMEF2)CAT, mutated in the MEF2 site, or pMyo84(mutMEF2/-E1)CAT, lacking both of these sites. PC4 significantly potentiated the activation by MyoD of the pMyo84 promoter construct carrying only the MEF2 site but was without effect on the construct with the E-box only (Fig. 3C). This latter was stimulated by MyoD only weakly, in agreement with previous data (17) and with our observations indicating that optimal stimulation by MyoD occurs only in the presence of multiple E-boxes (see below, Fig. 3G). These data indicate that PC4 acts as a positive regulator of MyoD activity through MEF2.
To further investigate this point, we evaluated the functional interaction between PC4 and MEF2C, which is the MEF2 isoform with the highest ability to induce myogenic conversion when cotransfected with MyoD (56). In addition, MEF2C is specifically expressed in muscle and developing brain (43, 49, 61), where the expression of PC4 is high (9, 72). Therefore, we cotransfected an expression construct for MEF2C, together with MyoD, PC4, and the pMyo84 reporter. We observed that PC4 was able to further enhance the stimulation of pMyo84 promoter activity that resulted from the synergistic action of MyoD and MEF2C (Fig. 3D). MyoD and MEF2C protein levels in transfected cells were not influenced by PC4, as indicated by Western blot analysis (Fig. 3E).
To distinguish whether the target of PC4 was MyoD or MEF2C, we sought to analyze the effect of PC4 on the activity of each of these two factors independently. For this we used either an MEF2-responsive reporter construct, carrying two MEF2 sites (pTK-MEF2x2 CAT; Fig. 3F), or a MyoD-responsive reporter construct, carrying four tandemly repeated MyoD binding sites (pt184RTK-CAT; Fig. 3G). It was found that PC4 potentiated the transactivation mediated by either MEF2C or MyoD of the corresponding reporter construct (Fig. 3F and G). It has to be noted that, while in the experiments with the MyoD-responsive reporter the transfected MyoD probably induced the endogenous MEF2C (13, 48), in the experiments with the MEF2x2 reporter the transfected MEF2C did not elicit expression of MyoD (56). Therefore, the ability of PC4 to stimulate the MEF2C-dependent activation of pTK-MEF2x2 CAT reporter (Fig. 3F) suggests that PC4 can interact functionally with MEF2C independently of MyoD.
As a whole, the experiments shown in Fig. 3 indicate that PC4 can stimulate the transcriptional activation exerted either by MyoD or by MEF2C. The observed synergism between PC4 and MyoD can occur either through MEF2 binding sites, or through a sufficient number of E-boxes, which are possibly necessary to allow the formation of myogenic complexes competent for cooperation with MEF2 factors (56).
Translocation of PC4 to the nucleus during muscle differentiation.
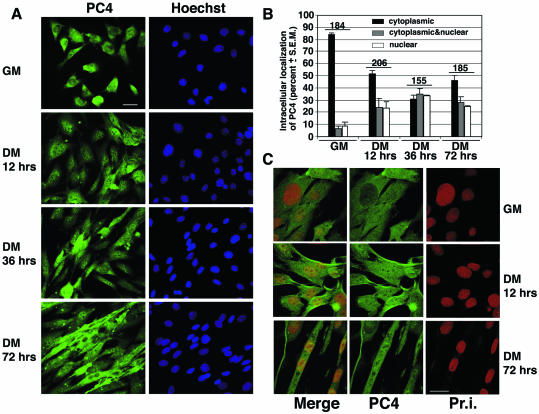
The ability of PC4 to modulate the transcriptional activity of MyoD and MEF2C raised the question as to whether PC4, which is prevalently cytoplasmic (27), can translocate to the nucleus during differentiation. To assess this point, we evaluated the intracellular distribution of the endogenous PC4 protein in differentiating C2C7 myoblasts by immunofluorescence (Fig. 4A to C). In undifferentiated C2C7 myoblasts, growing in GM, PC4 was found mainly in the cytoplasm, although in a low percentage of cells (<15%) it was localized in the nucleus or both in the nucleus and in the cytoplasm (Fig. 4A and B). The number of cells showing exclusively nuclear or nuclear and cytoplasmic staining increased gradually during the first 36 h of differentiation up to ca. 70%, whereas, during the same time period, the percentage of cells displaying cytoplasmic localization of PC4 decreased correspondingly. The cytoplasmic staining of PC4 increased again 72 h after the onset of differentiation in multinucleate myotubes, in which, however, PC4 remained detectable in a high percentage of nuclei (Fig. 4A and B). The localization of PC4 in the nucleus of terminally differentiated myocytes was confirmed by confocal microscopy, which also revealed a dot-like pattern of PC4 nuclear staining (Fig. 4C). These results suggest that the subcellular distribution of PC4 is dynamic and show that PC4 molecules enter the nucleus in significant numbers during muscle differentiation.
FIG. 4.
Nuclear localization of endogenous PC4, analyzed by immunofluorescence and confocal microscopy. (A) A total of 7 × 104 C2C7 cells were seeded in 35-mm dishes and shifted 24 h later to DM to initiate differentiation. At the indicated times after the shift to DM, cells were fixed, permeabilized, and stained for immunofluorescence detection. Endogenous PC4 was visualized by using the anti-PC4 rabbit polyclonal antibody A451 (29), followed by incubation with goat anti-rabbit FITC-conjugated antibody. Nuclei were detected by Hoechst 33258 dye (corresponding photomicrographs on the right). Bar, 30 μm. (B) Percentage of cells with endogenous PC4 staining that was cytoplasmic only (black bars), nuclear and cytoplasmic (gray bars), or nuclear only (white bars). Values are calculated for each category as the percentages of the total number of cells scored at each time point (within three fields for each experiment). Means ± the SEM are from three independent experiments. The total number of cells counted for each time point is indicated at the top of the corresponding bar. (C) Confocal microscopy of endogenous PC4. A total of 7 × 104 C2C7 cells were seeded onto circular coverslips placed in 35-mm dishes. Cultures were treated, and endogenous PC4 was detected as in panel A. Nuclei are visualized in red by propidium iodide (Pr.i.). The panels on the left (merge) show the overlay of the green and red staining (in orange-green), which indicates the presence of endogenous PC4 protein in the nucleus. Bar, 10 μm.
A deletion analysis carried out on the PC4 molecule indicated that the carboxyl-terminal region of PC4 (aa 290 to 445), isolated from the rest of the molecule, localizes to the nucleus (data not shown). Whether this region contains authentic nuclear localization signals has yet to be determined. Furthermore, we found that PC4 accumulates in the nucleus in the presence of leptomycin B, a specific inhibitor of the nuclear export receptor CRM1 (data not shown), indicating that PC4 is also subject to active nuclear export.
In vitro and in vivo interaction of PC4 with MEF2C.
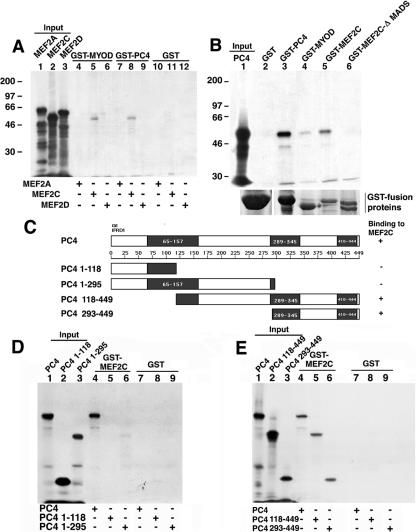
The experiments described above indicate that PC4 potentiates MyoD and MEF2C activity, which might imply its physical interaction with these molecules. This is also suggested by the observation that PC4 translocates to the nucleus during differentiation. Thus, we performed GST pull-down assays to analyze the ability of PC4 to bind MyoD and the MEF2 isoforms A, C, and D.
MEF2C, among the in vitro-translated isoforms of MEF2, was specifically able to associate with GST-PC4 and, similarly, in vitro-translated PC4 associated with GST-MEF2C (Fig. 5A, lane 8; Fig. 5B, lane 5). The efficiency of the binding of MEF2C to PC4 was comparable to that of MEF2C to MyoD (Fig. 5A). Indeed, MEF2C and bHLH factors (MyoD and myogenin) have been shown to physically interact and to synergistically regulate transcription (56). Furthermore, an MEF2C mutant deleted in the MADS domain (which is responsible, with the neighboring MEF2 domain, for DNA binding and dimerization) (57) was unable to bind PC4 (Fig. 5B lane 6). This pointed to the MADS domain as the requisite for the interaction of MEF2C with PC4.
FIG. 5.
PC4 interacts in vitro with MEF2C. (A) Binding of in vitro-translated 35S-labeled MEF2A, MEF2C, and MEF2D to GST-MyoD and GST-PC4. (B) Binding of in vitro-translated 35S-labeled PC4 to GST-PC4, GST-MyoD, GST-MEF2C, and GST-MEF2C-ΔMADS. The amount of GST fusion proteins bound to the glutathione-Sepharose resin beads used in the GST pull-down assay (10 μl), as detected by Coomassie blue staining, is shown below each lane. (C) Schematic representation of PC4 deletion mutants. The gray boxes identify the regions most conserved within the IFRD1 protein family (corresponding in the rat sequence to aa 65 to 157, aa 289 to 345, and aa 410 to 444) (9). (D and E) Binding to GST-MEF2C of PC4 mutants with deletions at the carboxyl terminus (D) or at the amino terminus (E). (A, B, D, and E) The indicated [35S]methionine-labeled products in vitro translated in rabbit reticulocyte lysates were incubated with the different GST-fused proteins bound to glutathione-Sepharose 4B. Bound proteins were eluted and analyzed by SDS-8% PAGE, followed by autoradiography. The labeled input products loaded were about 30% (A and B) or 10% (D to E) of the amount used in the pull-down incubations.
In addition, a weak interaction occurred between PC4 and GST-MyoD (Fig. 5B, lane 4). Also, PC4 unexpectedly turned out to be able to strongly homodimerize (Fig. 5B, lane 3).
To determine the region(s) of PC4 that mediate the binding to MEF2C, we constructed four PC4 deletion mutants, carrying different domains of the PC4 protein, defined by the presence of sequences conserved within the IFRD protein family (Fig. 5C) (9). The PC4 mutant proteins were translated in vitro and their binding to MEF2C was assessed by means of GST pull-down assays. The amino-terminal and central regions of PC4 (mutant proteins PC41-118 and PC41-295) showed null or very weak binding to GST-MEF2C, whereas both mutants PC4118-449 and PC4293-449 bound strongly, thus indicating that the carboxyl-terminal region of PC4 is required to bind MEF2C (Fig. 5D and E).
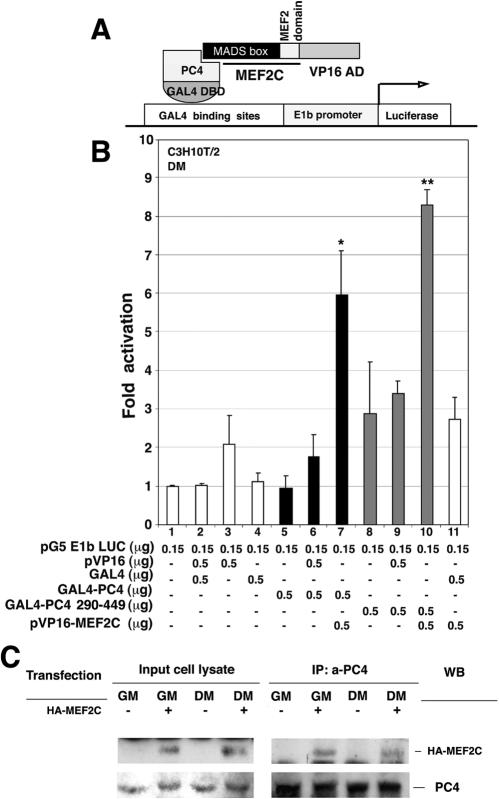
To define whether PC4 can interact with MEF2C also in vivo, we used a protein-protein-interaction assay system (20). PC4 was fused to the yeast GAL4 DBD to obtain the chimeric construct referred to as pMGAL4-PC4 (Fig. 6A). When introduced into C3H10T1/2 cells, pMGAL4-PC4 failed to activate the expression of the GAL4-dependent luciferase reporter gene pG5E1b-LUC (Fig. 6B, lane 5), indicating that PC4 does not harbor transcription activation domains. To test the interaction between PC4 and MEF2C in this system, we used a truncated MEF2C protein encoding the MADS and MEF2 domains (aa 1 to 117) of MEF2C, fused to the VP16 activation domain (pVP16-MEF2C). This region of MEF2C was found to contain the sequences required for binding PC4 in vitro (as shown above in Fig. 5B). When pMGAL4-PC4 was expressed with pVP16-MEF2C, the activity of the reporter gene was induced ∼6-fold (Fig. 6B, lane 7). Furthermore, the mutant pMGAL4-PC4 290-449, encoding the GAL4 DBD fused to the carboxyl-terminal region of PC4, was seen to be even more efficient in this assay. In fact, when coexpressed with VP16-MEF2C, it was able to induce transcription more than eightfold above the basal activity of the pG5E1b-LUC reporter (Fig. 6B, lane 10). These results indicate that PC4 and MEF2C can interact in mammalian cells and confirm that the MADS domain of MEF2C is the determinant of the interaction with PC4 and that the carboxyl-terminal region of PC4 contains the MEF2C binding domain.
FIG. 6.
In vivo interaction between PC4 and MEF2C. (A) Schematic representation of the mammalian two-hybrid assay, as applied to our molecular model. (B) Interaction of PC4 and PC4 290-449 with MEF2C as evaluated by the two-hybrid assay. C2C7 cells were transfected with pG5E1b-LUC, pMGAL4-PC4, pMGAL4-PC4 290-449, or pVP16-MEF2C or with the empty vectors as indicated. At 24 h after transfection the cells were placed in DM for 48 h before harvesting and luciferase assay measurements. ✽, P < 0.05; ✽✽, P < 0.02 (versus the GAL4 control group [lane 11] and also versus the corresponding pVP16 control group [i.e., lanes 6 or 9, respectively]) (Student t test). (C) Coimmunoprecipitation of PC4 and MEF2C. C2C12 cells (clone S4 constitutively overexpressing PC4 [27]) were transfected with HA-MEF2C or with the empty vector, kept in GM or in DM for 48 h after transfection, and then lysed and immunoprecipitated with the polyclonal A451 anti-PC4 antibody, covalently bound to Sepharose resin. The anti-HA or the anti-PC4 antibodies were used for Western blot analysis of the immunoprecipitated complexes (IP: a-PC4 panel) and of the input cell lysate (1/30 of the immunoprecipitated lysate).
As a complementary approach to test the in vivo interaction between PC4 and MEF2C, we performed a coimmunoprecipitation assay. C2C12 myoblasts constitutively overexpressing PC4 (clone S4) (27) were transfected with HA-MEF2C; afterward, the cells were either maintained in growing conditions (GM) or transferred to differentiation conditions (DM). The Western blot analysis of anti-PC4 immunoprecipitates indicated that MEF2C was able to associate with PC4, either in proliferating or in differentiating cells (Fig. 6C, panel IP: a-PC4).
Reversal by PC4 of the HDAC4-mediated inhibition of MEF2C and MyoD.
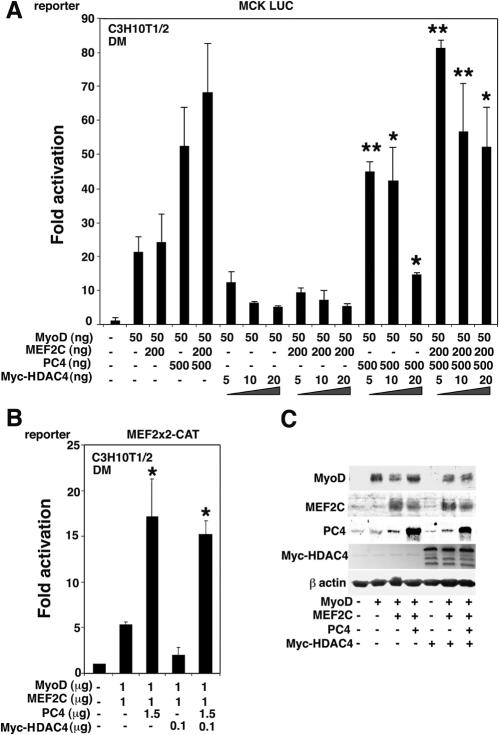
The data reported above showed that PC4 stimulates the transcriptional activity of MyoD/MEF2C and can physically interact with MEF2C. A large body of evidence has recently highlighted that the activity of MEF2 transcription factors is regulated by class II deacetylases during the process of muscle differentiation (16, 41, 54, 76). HDAC4 has been shown to repress MEF2C activity through association with the MADS domain (41, 76). Given that PC4 interacts with the same domain of MEF2C, one possibility was that PC4 could potentiate the activity of MEF2C by interfering with the inhibitory action of HDAC4. To ascertain this, we tested whether PC4 could overcome the HDAC4-mediated inhibition of the muscle creatine kinase (MCK) promoter activity. The MCK regulatory region is known to contain both MyoD and MEF2 binding sites (1). As expected, HDAC4 inhibited the stimulation of the MCK LUC reporter gene mediated by MyoD and MEF2C (Fig. 7A). However, cotransfection of PC4 was able to rescue the MyoD activity, reaching a complete reversal of the HDAC4-imposed inhibition when MEF2C was also present (Fig. 7A).
FIG. 7.
PC4 rescues the transcriptional activity of MEF2C and MyoD from the inhibition exerted by HDAC4. (A) The MCK LUC reporter was cotransfected in C3H10T1/2 cells with pEMSV-MyoD, pcDNA1-MEF2C, pcDNA-Myc-HDAC4, or PC4 in the indicated combinations. At 24 h after transfection, the cells were placed in DM and left for 48 h before luciferase assay determination. Luciferase activities were measured as units per microgram of protein normalized to β-Gal activity and then calculated as the fold activation relative to the activity of control samples (transfected with empty vectors). Bars represent the average fold activity ± the SEM of at least three independent experiments performed in duplicate. ✽, P < 0.05; ✽✽, P < 0.02 (versus the corresponding group without PC4) (Student t test). (B) The MEF2-responsive reporter was cotransfected in C3H10T1/2 cells with combinations of the expression vectors pEMSV-MyoD, pcDNA1-MEF2C, pcDNA-Myc-HDAC4, and pSCT-PC4, as indicated. Transfected cells were treated and analyzed as described in panel A. ✽, P < 0.05 (versus the corresponding group without PC4) (Student t test). (C) Western blot analysis showing the expression levels of the constructs transfected in the experiments depicted in panels A and B.
We then monitored the influence of PC4 on the HDAC4-mediated inhibition of the MEF2C-responsive pTK-MEF2x2 CAT reporter construct in conditions promoting differentiation. As shown in Fig. 7B, PC4 was indeed able to overcome the HDAC4-mediated repression of the MEF2C-dependent reporter. Western blot analysis of the protein extracts used in transcription assays indicated that PC4 did not alter the expression levels of the MyoD, MEF2C, and HDAC4 transfected constructs (Fig. 7C).
PC4 can dissociate HDAC4 from MEF2C.
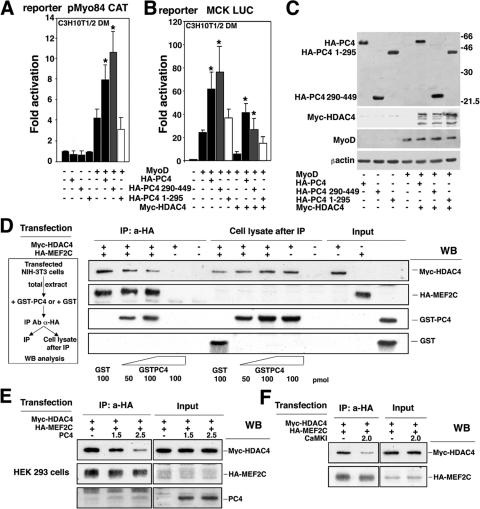
Given that PC4 was able to overcome the HDAC4-dependent inhibition of MEF2C activity, we sought to determine whether such an effect of PC4 was dependent on its ability to interact with MEF2C. To address this issue, we analyzed the ability of the amino- or the carboxyl-terminal domains of PC4 to synergize with MyoD and to counteract HDAC4 (as shown in Fig. 5D and E, the carboxyl-terminal region of PC4 binds MEF2C). Thus, we coexpressed MyoD and either PC4, PC41-295, or PC4290-449 in C3H10T1/2 cells with the reporter plasmid pMyo84 CAT or MCK LUC. The carboxyl-terminal region of PC4, but not the amino-terminal region, enhanced the MyoD-dependent activation of both pMyo84 CAT and MCK LUC reporters as efficiently as full-length PC4 (Fig. 8A and B). Similarly, only the carboxyl-terminal region of PC4 was significantly able to rescue the MCK promoter activity from HDAC4 inhibition (Fig. 8B). Western blot analysis of protein extracts from transfected cells indicated that PC4 full-length and the PC4 deletion mutants were expressed at similar levels (Fig. 8C). Thus, the ability of PC4 to synergize with MyoD and to counteract the inhibitory action of HDAC4 appears to correlate with the ability of PC4 to physically interact with MEF2C.
FIG. 8.
PC4 antagonizes HDAC4 and displaces it from MEF2C. (A and B) The carboxyl-terminal region of PC4 synergizes with MyoD and reverses the inhibition by HDAC4. (A) The pMyo84 CAT reporter was cotransfected in C3H10T1/2 cells with pEMSV-MyoD and with either HA-pSCT-PC4 or the deletion mutants HA-pSCT-PC4 290-449 and HA-pSCT-PC4 1-295, as indicated. (B) C3H10T1/2 cells were transfected with the MCK LUC reporter, with the expression constructs used for panel A, and with pcDNA-Myc-HDAC4, in the indicated combinations. In panels A and B, at 24 h after transfection cells were placed in DM for 48 h before luciferase assay determination. The luciferase activity was measured as units per microgram of protein normalized to the β-Gal activity; the fold activations were calculated relative to the control sample (transfected with empty vectors). The average fold activity ± the SEM is shown for at least three independent experiments performed in duplicate. ✽, P < 0.05 (versus the corresponding group without HA-pSCT-PC4 or without HA-pSCT-PC4 1-295 transfected) (Student t test). (C) Analysis by Western blotting of the expression levels of the constructs transfected, in one that is experiment representative of those shown in panels A and B. (D) PC4 displaces HDAC4 from MEF2C in an in vitro assay. Lysates of NIH 3T3 cells, cotransfected with Myc-HDAC4 and HA-MEF2C (or alternatively, with the empty vectors), were incubated with increasing amounts of purified GST-PC4 or GST protein and then immunoprecipitated with anti-HA antibody. The immunoprecipitated complexes (left panel, IP), as well as the cell lysates after immunoprecipitation (central panel) and the input cell lysates (1/30 of the total lysate), were analyzed by Western blotting with anti-Myc, anti-HA, or anti-GST antibodies. (E) PC4 displaces HDAC4 from MEF2C in an in vivo assay. Lysates of HEK293 cells, cotransfected with Myc-HDAC4 and HA-MEF2C (0.75 μg each) and with increasing amounts of PC4, were then immunoprecipitated with anti-HA antibody. The immunoprecipitated complexes (panel IP) and the input cell lysates (1/30 of the total lysate) were analyzed by Western blotting with the indicated antibodies. (F) Activated CaMKI dissociates HDAC4-MEF2C complexes. HEK293 cells were cotransfected with Myc-HDAC4 and HA-MEF2C (0.75 μg each) and with either CaMKI or its empty vector. Lysates of transfected cells were then immunoprecipitated with anti-HA. The immunoprecipitated complexes (panel IP) and the input cell lysates (1/30 of the total lysate) were analyzed by Western blotting with the indicated antibodies.
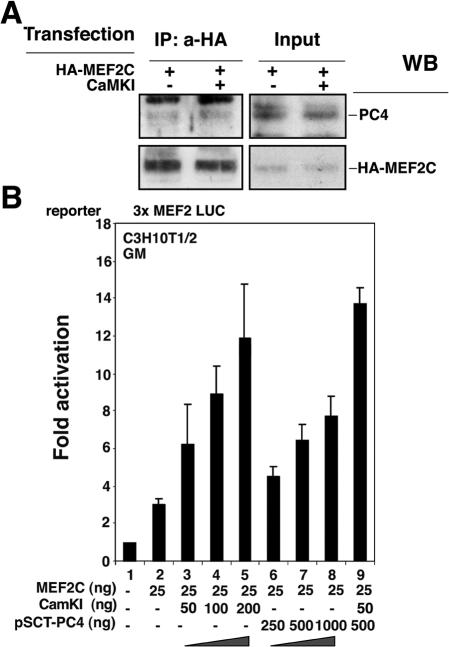
Because HDAC4 and PC4 bind to the same region of MEF2C (the MADS domain), we sought to determine whether PC4 could impair the association of HDAC4 with MEF2C. To test this hypothesis, we analyzed the formation of HDAC4-MEF2C complexes either in the presence or in the absence of recombinant PC4. Myc-HDAC4 and HA-MEF2C were expressed in NIH 3T3 mouse fibroblasts, and their interaction was analyzed by anti-HA immunoprecipitation, followed by Western blot detection of the immunoprecipitated products (Fig. 8D). When the extracts were preincubated with increasing amounts of purified GST-PC4 protein, immunoprecipitation of extracts with anti-HA led to the recovery of HDAC4 in amounts inversely proportional to the quantity of immunoprecipitated GST-PC4, as revealed by Western blotting with anti-Myc or anti-GST antibodies (Fig. 8D, see IP: a-HA). Consistently, in the supernatants after immunoprecipitation the amount of Myc-HDAC4 increased proportionally to the GST-PC4 added (Fig. 8D, see cell lysates after immunoprecipitation). Thus, the binding of PC4 and HDAC4 to MEF2C appear to be mutually exclusive.
As an alternative approach, we sought to evaluate the effects of transfection of PC4 on the MEF2C-HDAC4 complex in vivo. Therefore, we cotransfected HA-MEF2C and Myc-HDAC4 with increasing amounts of PC4. We found that the level of HDAC4 protein coimmunoprecipitating with MEF2C was lower in samples where PC4 was coexpressed, with a decrease proportional to the amount of transfected PC4 (Fig. 8E, panel IP: a-HA). These results confirm the hypothesis that PC4 can displace HDAC4 from MEF2C.
It has been recently shown that CaMK enhances the transcriptional activity of MEF2C by negatively regulating the interaction between HDAC4 and ME2C (40). Thus, we wanted to compare the effects of PC4 and CaMK. HDAC4 was almost completely displaced from MEF2C by an activated form of CaMK (Fig. 8F), similarly to what was observed with PC4 (Fig. 8E). Based on this observation, one would expect that the activated form of CaMK would favor the association of PC4 with MEF2. Indeed, we found that the ability of PC4 to bind to MEF2C was somewhat increased by cotransfection of activated CaMK (about twofold; Fig. 9A). Finally, we observed that, although both PC4 and activated CaMK stimulated the transcriptional activity of MEF2 to a comparable extent (Fig. 9B), the simultaneous transfection of submaximal concentrations of PC4 and activated CaMK resulted in an additive effect, suggesting cooperativity between the two molecules (Fig. 9B).
FIG. 9.
Cooperativity between CaMK and PC4. (A) Activated CaMKI weakly increases the amount of PC4 associated with MEF2C. Clone S4 of C2C12 cells cultured in proliferating conditions were cotransfected with HA-MEF2C (6 μg) and with either activated CaMKI (SRα-CaMKI; 6 μg) or the empty vector. MEF2C was then immunoprecipitated by the anti-HA monoclonal antibody, and the immunoprecipitated complexes were analyzed by Western blotting by using the anti-PC4 A451 or anti-HA antibody, as indicated. (B) PC4 and CaMK cooperate in stimulating MEF2 transcriptional activity. C3H10T1/2 cells were cotransfected with the reporter construct 3x MEF2 LUC (carrying three MEF2 binding sites [40]; 100 ng) with pCDNA1-MEF2C and with increasing amounts of either pSCT-PC4 or activated CaMKI, as indicated. Doses of PC4 and CaMKI above 1000 ng and 200 ng, respectively, did not induce further stimulation, indicating that these were maximally effective doses, within the linear range of the dose-response curve. Cotransfection of pSCT-PC4 and activated CaMKI (lane 9) produced additive effects. Transfected cells were cultured in GM for 48 h before being harvested. Luciferase activities were measured as units per microgram of protein normalized to β-Gal activity and are expressed as the fold activation relative to the activity of the control sample (transfected with the empty vectors). Bars represent the average fold activity ± the SEM of three independent experiments performed in duplicate.
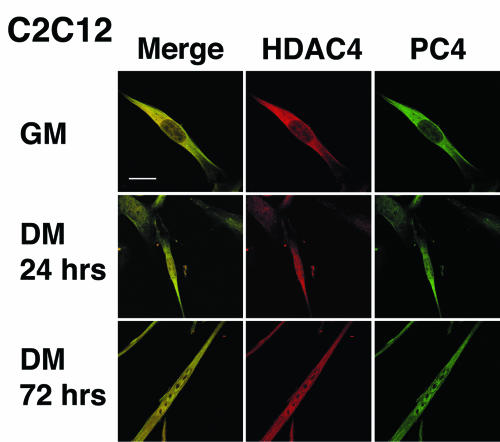
A functional requisite for the displacement of HDAC4 from MEF2C by PC4 is the colocalization of these molecules in the nucleus of differentiating myocytes. Since it is known that MEF2C is an essentially nuclear protein (49), whereas HDAC4 and PC4 translocate from the cytoplasm to the nucleus during differentiation (55, 89; the present study), we tested whether endogenous PC4 and HDAC4 indeed colocalize. As shown by confocal microscopy, PC4 and HDAC4 colocalized in the cytoplasm of proliferating C2C12 myoblasts. In differentiating myoblasts (after 24 h in DM) and, more evidently, in mature myotubes (72 h in DM), PC4 and HDAC4 progressively accumulated and colocalized in the nucleus of the majority of cells, although a significant amount of these proteins was still detectable in the cytoplasm (Fig. 10). This indicated that a functional interaction between PC4, MEF2C, and HDAC4 can take place in the nucleus during differentiation.
FIG. 10.
Colocalization of endogenous PC4 and HDAC4 in proliferating and differentiating myoblasts. Endogenous HDAC4 and PC4 were visualized by confocal microscopy in C2C12 cells by using anti-HDAC4 and anti-PC4 rabbit polyclonal antibodies, followed by incubation with goat anti-rabbit TRITC- and FITC-conjugated antibodies, respectively. In proliferating myoblasts HDAC4 is prevalently cytoplasmic, whereas in differentiating myotubes it is also nuclear, as shown in representative photomicrographs. The pattern of localization of HDAC4 overlaps with that of PC4, as indicated by the yellow pseudo-color produced by the overlay (merge) of the green and red staining. Bar, 10 μm.
Physical and functional interaction of PC4 and HDAC4.
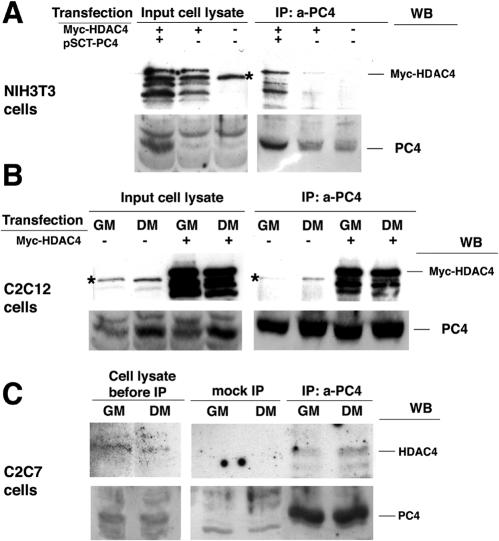
It has been recently reported that Tis7 (the mouse PC4 homolog) can interact with class I HDACs (75). To verify whether PC4 could also bind HDAC4, lysates of NIH 3T3 cells cotransfected with Myc-HDAC4 and PC4 were immunoprecipitated with the anti-PC4 (A451) antibody and subjected to Western blot analysis with anti-Myc antibody to reveal the HDAC4 protein. As shown in Fig. 11A, HDAC4 could be detected in association with the immunoprecipitated PC4 protein. To verify the association of PC4 with HDAC4 also in muscle cells, we immunoprecipitated PC4 from the C2C12 myoblast clone S4 (constitutively overexpressing PC4) transfected with Myc-HDAC4. As shown in Fig. 11B, PC4 associated with HDAC4, both in proliferation and in differentiation conditions (i.e., GM and DM). Moreover, the ability of endogenous PC4 and endogenous HDAC4 to coimmunoprecipitate in extracts from C2C7 cells indicated that their association occurs also in physiological conditions (Fig. 11C). A higher association of PC4 with HDAC4 was observed in differentiated myotubes.
FIG. 11.
PC4 associates with HDAC4. (A) NIH 3T3 cells were cotransfected with pcDNA-Myc-HDAC4 and pSCT-PC4 (or with the empty vector) as indicated, lysed 48 h after transfection, and immunoprecipitated with the anti-PC4 antibody A451 covalently bound to Sepharose resin. Immunoprecipitated complexes (IP: a-PC4 panel) and input cell lysates were analyzed by Western blotting with anti-Myc or anti-PC4 antibodies. Asterisks in panels A and B indicate a background protein (see reference 32). (B) C2C12 cells (clone S4 constitutively overexpressing PC4) were transfected with pcDNA-Myc-HDAC4 (or with the empty vector) as indicated, grown in GM or DM for 48 h after transfection, and then lysed and immunoprecipitated with the anti-PC4 antibody A451 covalently bound to Sepharose resin. The immunoprecipitated complexes (IP: a-PC4 panel), as well as the input cell lysates, were analyzed by Western blotting with anti-Myc or anti-PC4 antibodies. (C) C2C7 cells, cultured in GM or DM for 48 h, were lysed and immunoprecipitated with the anti-PC4 antibody covalently bound to Sepharose resin. Immunoprecipitated complexes (IP: a-PC4 panel) and the input cell lysates were analyzed by Western blotting with anti-HDAC4 or anti-PC4 antibodies. As a control (mock IP panel), the immunoprecipitates obtained by using Sepharose resin coupled to preimmune serum were analyzed by Western blotting. For each experiment shown in panels A to C, 1.5 mg of protein lysate was immunoprecipitated and fractionated by SDS-PAGE. Western blot analysis of input lysates was carried out on 1/30 of the lysates used for immunoprecipitation.
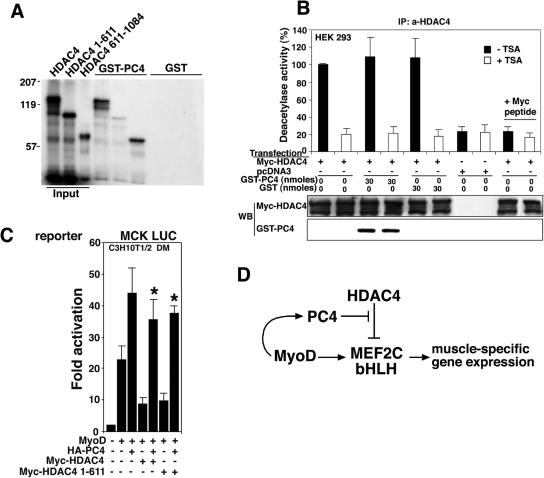
To better understand the modalities of the interaction between HDAC4 and PC4 observed in vivo, we sought to define whether they could associate in vitro and to identify the region of HDAC4 required to bind PC4. Thus, using GST pull-down assays, we tested the ability of GST-PC4 to bind in vitro-translated HDAC4 and its deletion mutants (HDAC41-611 and HDAC4611-1084). HDAC4 wild type and HDAC4611-1084 were both able to bind PC4 efficiently, whereas HDAC41-611 was not (Fig. 12A).
FIG. 12.
Physical and functional interaction between HDAC4 and PC4. (A) The carboxyl-terminal region of HDAC4 binds PC4. GST pull-down assays were performed by incubating equal amounts of in vitro-translated [35S]methionine-labeled HDAC4 proteins, full-length or truncated mutants, with equimolar amounts of either GST-PC4 or GST proteins bound to glutathione-Sepharose 4B resin. Bound proteins were eluted, separated by SDS-8% PAGE, and analyzed by using a phosphorimager. (B) PC4 does not influence the enzymatic activity of HDAC4. HEK293 cells, grown in 90-mm dishes, were transfected with 12 μg of either pcDNA-Myc-HDAC4 or the empty vector. The precleared lysates were incubated with GST or GST-PC4 and then immunoprecipitated with anti-Myc antibody. As a control, a synthetic peptide corresponding to the Myc epitope was also added in molar excess to samples before immunoprecipitation. Immunoprecipitated proteins, treated or not with trichostatin A (200 nM), were then incubated with preacetylated histones. Deacetylase activities are shown as percent values relative to the level of control samples (transfected with HDAC4 and without GST protein added) set to 100 (the absolute mean dpm values were about 1,400). The bars are the average ± the SEM of three independent experiments performed in duplicate, normalized to the amount of immunoprecipitated proteins by Western blotting (a representative blot is shown in the lower panel). (C) PC4 counteracts the inhibitory effect exerted by the HDAC4 amino-terminal region. The MCK LUC reporter was cotransfected in C3H10T1/2 cells with pEMSV-MyoD (50 ng) and the indicated combinations of HA-pSCT-PC4 (1 μg) and pcDNA-Myc-HDAC4 and pcDNA-Myc-HDAC4 1-611 (5 ng). At 24 h after transfection, cells were placed in DM for 48 h before luciferase assay determination. Luciferase activity was measured as units per microgram of protein normalized to the β-Gal activity. The average fold activity ± the SEM shown (relative to control samples transfected with empty vectors) was calculated from three independent experiments performed in duplicate. ✽, P < 0.05 (versus the corresponding group without PC4) (Student t test). (D) Working model for PC4 coactivation of MyoD/MEF2C. See the Discussion for more information.
These data indicate that PC4 binds the carboxyl-terminal region of HDAC4, which harbors the deacetylase domain. Therefore, we wanted to analyze whether PC4 could inhibit the enzymatic activity of HDAC4. For this purpose, we measured the deacetylase activity associated with HDAC4 complexes immunoprecipitated from HEK293 cells transfected with HDAC4, either in the presence or in the absence of GST-PC4 (Fig. 12B). The results in Fig. 12B show that PC4 coimmunoprecipitated with HDAC4 and yet did not influence the HDAC4-associated deacetylase activity.
These results, taken together, indicate that PC4 can bind the carboxyl-terminal region of HDAC4 without inhibiting its deacetylase activity and also that HDAC4 binds PC4 and MEF2C through separate domains. Thus, we sought to test whether PC4 could counteract the inhibition exerted by the amino-terminal region of HDAC4 (HDAC41-611), which is known to bind to and repress MEF2C as effectively as full-length HDAC4 (12). Therefore, we coexpressed the MCK LUC reporter, MyoD, and HDAC4 or HDAC41-611 in C3H10T1/2 fibroblasts, with or without PC4. As expected, HDAC41-611 was able to inhibit the stimulation by MyoD of the MCK promoter activity as efficiently as did full-length HDAC4, but its effect was completely reversed by PC4 (Fig. 12C). Thus, PC4 can counteract the repressor activity mediated by the amino-terminal region of HDAC4 which, notably, does not contain the PC4 binding site. This result clearly indicates that the ability of PC4 to overcome the HDAC4-dependent inhibition of MyoD activity does not rely on its ability to bind HDAC4.
DISCUSSION
In this study we have characterized the functional role of the PC4 gene in myoblast differentiation. Our results show that expression of PC4 mRNA is induced during the process of terminal differentiation, being upregulated in differentiating C2C12 myoblasts, as well as in a fibroblast cell line expressing a conditional MyoD protein. This MyoD-mediated induction of PC4 expression occurs at the transcriptional level, as indicated by the observation that MyoD stimulates the activity of the PC4 promoter.
As for the function of PC4, we found that the PC4 protein, which in itself is devoid of transcriptional activity, is able to significantly potentiate the MyoD-dependent transactivation of muscle gene promoters and that this effect of PC4 occurs selectively through MEF2 sites. Consistently, PC4 enhances the synergism of MyoD and MEF2C in activating muscle-specific promoters. A direct action of PC4 on MEF2C is suggested also by the ability of PC4 to increase the activation, elicited by MEF2C, of a synthetic reporter carrying only MEF2 sites. The observation that PC4 can override the repression imposed by HDAC4 on the muscle-inducing function of MyoD further supports the hypothesis that MEF2C is the target of PC4 action. Indeed, it has been well established that class II deacetylases suppress the myogenic function of MyoD by interacting with the MADS domain of MEF2 factors and repressing their transcriptional activity (16, 37, 41, 54, 76).
A key question raised by our findings is whether the two observed activities of PC4, i.e., the ability to potentiate the transcriptional activity of MyoD and to rescue MEF2C from the inhibition by HDAC4, rely on a common mechanistic basis.
HDAC4 contains several domains active in transcriptional repression that can represent possible targets for regulation by PC4. The deacetylase catalytic domain of HDAC4, canonical within the class II HDACs family, resides in the carboxyl-terminal region of the protein (22, 26). It has been shown that this domain of HDAC4 acts by recruiting a corepressor complex composed of N-CoR, SMRT, and HDAC3 (21). The interaction of HDAC4 with MEF2C occurs through a separate region localized within the amino-terminal of HDAC4 (41, 76), which also effectively inhibits the MEF2 transcriptional activity. In fact, the amino-terminal region of HDAC4 can interact with, and recruit to MEF2C, other transcriptional corepressors, such as HDAC1 (12), carboxyl-terminal-binding protein (87), and heterochromatin protein 1 (88).
We show that PC4 specifically binds MEF2C within the MADS domain (Fig. 5A and B), the same region through which MEF2C interacts with HDAC4, and that PC4 can dissociate HDAC4 from MEF2C in a concentration-dependent manner (Fig. 8D and E). Such displacement of HDAC4 from MEF2C occurs concomitantly with an increased binding of PC4 to MEF2C, which indicates that PC4 and HDAC4 bind to MEF2C in a mutually exclusive fashion.
The observed dissociation of HDAC4 from MEF2C induced by PC4 can plausibly account for the ability of PC4 to counteract the HDAC4-dependent repression of the MyoD/MEF2C transcriptional activity. This is also consistent with the observation that a carboxyl-terminal fragment of PC4, which binds efficiently the MADS domain of MEF2C (to which also HDAC4 associates), was able to reverse the inhibition by HDAC4, whereas the amino-terminal region of PC4, which does not bind MEF2C, was not (Fig. 8A and B). Notably, the carboxyl-terminal region of PC4 was also able to act cooperatively with MyoD in the activation of muscle reporter genes, displaying an efficiency even higher than that of PC4 wild type. This evidence, taken together, strongly suggest that the ability of PC4 to potentiate the MyoD-dependent transcription and to reverse the transcriptional inhibition exerted by HDAC4 rely on the ability of PC4 to antagonize the recruitment of HDAC4 onto MEF2C by associating with MEF2C.
We also show here that PC4 can interact in vivo and in vitro with HDAC4 and that this interaction occurs within the carboxyl-terminal region of HDAC4, which harbors the deacetylase domain. Nonetheless, PC4 does not affect the deacetylase activity of HDAC4. Furthermore, we found that PC4 can counteract the inhibition of MyoD activity exerted by the amino-terminal region of HDAC4, which binds MEF2C but lacks both the deacetylase domain and the PC4 binding site (Fig. 12A and C). This observation clearly indicates that PC4 can override the HDAC4-mediated inhibition of MEF2C without the requirement to bind HDAC4 and also further supports our idea that PC4 acts by inhibiting the interaction between HDAC4 and MEF2C.
Together, these results indicate that in differentiating myocytes PC4 behaves as coactivator of MyoD/MEF2C. However, it has recently been shown that in epithelial cells PC4, upregulated after c-jun activation, associates with Sin3 and with class I HDACs and acts as corepressor by HDAC dependently repressing gene expression (75). Based on these findings and on our data indicating an association of PC4 with HDAC4, it seems likely that PC4 can participate in repressor complexes containing both class I and class II HDACs. Whatever the function of PC4 in these repressor complexes, our data appear to exclude the concomitant presence of PC4 and HDAC4 in complexes with MEF2C. A corollary to this observation is that PC4 can be bound either to an MEF2C or to an HDAC4 molecule. The outcome would be that PC4, depending on the target to which is associated, may act as a coactivator or corepressor. In agreement with this model is the recent finding that the conditional overexpression of PC4 in a kidney (NRK) or in a neuronal (PC12) cell line induced the expression of about 300 genes in both cell lines, as defined by differential microarray analysis, whereas only a few genes were repressed (67).
It has been previously shown that HDAC4 shuttles between nuclear and cytoplasmic compartments during muscle differentiation (55, 89). In fact, endogenous HDAC4 is prevalently cytoplasmic in proliferating myoblasts, being subjected to active nuclear export, whereas it becomes progressively nuclear in differentiating myoblasts (55, 89) (Fig. 10). Likewise, we found that PC4, which shows cytoplasmic localization in myoblasts, moves to the nucleus in a high percentage of differentiating myocytes, colocalizing with HDAC4 and MEF2C (Fig. 4 and 10). These observations are consistent with the hypothesis that when MEF2C levels increase at the onset of differentiation (48) PC4, by translocating to the nucleus, inhibits the interaction of HDAC4 with MEF2C, thus releasing MEF2C from repression and also possibly rendering HDAC4 available for other targets or for nuclear export. In this model PC4 may function as a novel regulator of MEF2C activity additional to activated CaMK (Fig. 9), a kinase that has been shown to favor the dissociation of nuclear HDAC4 from MEF2C and its nuclear export by phosphorylating HDAC4 at the beginning of differentiation (40, 50, 51, 89).
Our previous data indicated an essential role of PC4 in myogenesis, since ablation of PC4 in myocytes caused a strong reduction of myogenin and myosin expression (27). The present study provides evidence that the physiological role of PC4 in myogenesis is to synergize with MyoD by antagonizing the inhibition of MEF2C by HDAC4. Notably, it has been observed that an overexpression of MyoD can overcome the inhibition by class II HDACs (12, 41). A molecular basis for this effect could lie in our finding showing that MyoD induces the expression of PC4, thus denoting the existence of a positive feedback loop between MyoD and PC4 (Fig. 12D). On the whole, PC4 appears to act as a positive cofactor, necessary for MyoD to attain a threshold activity, above which myogenesis can efficiently take place. The recent data obtained from Tis7-null mutant mice, showing defects in myogenesis, are completely consistent with this view, also considering that they focus on the requirement of PC4 in the process of muscle regeneration (73). Remarkably, the unique role for which MyoD is required in vivo, as indicated by knockout mice models, is to allow adult muscle regeneration rather than embryonic muscle development, for which other myogenic regulatory factors can compensate (52). Indeed, PC4 is highly expressed in vivo in adult skeletal muscle but only poorly during embryonic development (9), thus suggesting a prevalent role in postdevelopmental myogenesis.
Acknowledgments
We are grateful to Guriqbal Basi for the gift of the rat EMBL-3 genomic library, to Stefano Lorenzetti for the construction of pcDNA3-HA-MEF2C, to Giuseppina Corrente for experiments regarding PC4 pull-down performed when she was undergraduate student, to Fabio Cavaliere for the production of constructs carrying PC4, and to the numerous colleagues who kindly provided reagents. We are indebted to Livio Baron for excellent technical assistance. F.T. gratefully remembers the late Franco Tatò for the encouragement he provided while this study was in progress.
This research was supported by Telethon (project A46), by Progetto Finalizzato CNR Terapia Preclinica Molecolare in Oncologia, by European Community Grant QLG3-CT-2000-00072, and by the Donazione Maria Bianchi, which was made possible thanks to Roberto Salmoiraghi, mayor of Campione d'Italia.
REFERENCES
- 1.Amacher, S. L., J. N. Buskin, and S. D. Hauschka. 1993. Multiple regulatory elements contribute differentially to muscle creatine kinase enhancer activity in skeletal and cardiac muscle. Mol. Cell. Biol. 13:2753-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black, B. L., J. D. Molkentin, and E. N. Olson. 1998. Multiple roles for the MyoD basic region in transmission of transcriptional activation signals and interaction with MEF2. Mol. Cell. Biol. 18:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, B., and E. N. Olson. 1998. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell. Dev. Biol. 14:167-196. [DOI] [PubMed] [Google Scholar]
- 4.Blackwell, T. K., and H. Weintraub. 1990. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science 250:1104-1110. [DOI] [PubMed] [Google Scholar]
- 5.Bour, B. A., M. A. O'Brien, W. L. Lockwood, E. S. Goldstein, R. Bodmer, P. H. Taghert, S. M. Abmayr, and H. T. Nguyen. 1995. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 9:730-741. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Braun, T., E. Bober, B. Winter, N. Rosenthal, and H. H. Arnold. 1990. Myf-6, a new member of the human gene family of myogenic determination factors: evidence for a gene cluster on chromosome 12. EMBO J. 9:821-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun, T., G. Buschhausen-Denker, E. Bober, E. Tannich, and H. H. Arnold. 1989. A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversion in 10T1/2 fibroblasts. EMBO J. 8:701-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buanne, P., B. Incerti, D. Guardavaccaro, V. Avvantaggiato, A. Simeone, and F. Tirone. 1998. Cloning of the human interferon-related developmental regulator (IFRD1) gene coding for the PC4 protein, a member of a novel family of developmentally regulated genes. Genomics 51:233-242. [DOI] [PubMed] [Google Scholar]
- 10.Buckingham, M., L. Bajard, T. Chang, P. Daubas, J. Hadchouel, S. Meilhac, D. Montarras, D. Rocancourt, and F. Relaix. 2003. The formation of skeletal muscle: from somite to limb. J. Anat. 202:59-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cenciarelli, C., F. De Santa, P. L. Puri, E. Mattei, L. Ricci, F. Bucci, A. Felsani, and M. Caruso. 1999. Critical role played by cyclin D3 in the MyoD-mediated arrest of cell cycle during myoblast differentiation. Mol. Cell. Biol. 19:5203-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan, J. K. L., L. Sun, X.-J. Yang, G. Zhu, and Z. Wu. 2003. Functional characterization of an amino-terminal region of HDAC4 that possesses MEF2 binding and transcriptional repressive activity. J. Biol. Chem. 278:23515-23521. [DOI] [PubMed] [Google Scholar]
- 13.Cserjesi, P., and E. N. Olson. 1991. Myogenin induces muscle-specific enhancer binding factor MEF-2 independently of other muscle-specific gene products. Mol. Cell. Biol. 11:4854-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis, R. L., H. Weintraub, and A. B. Lassar. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51:987-1000. [DOI] [PubMed] [Google Scholar]
- 15.Dias, P., D. M. Parham, D. N. Shapiro, S. J. Tapscott, and P. J. Houghton. 1992. Monoclonal antibodies to the myogenic regulatory protein MyoD1: epitope mapping and diagnostic utility. Cancer Res. 52:6431-6439. [PubMed] [Google Scholar]
- 16.Dressel, U., P. J. Bailey, S. C. Wang, M. Downes, R. M. Evans, and G. E. Muscat. 2001. A dynamic role for HDAC7 in MEF2-mediated muscle differentiation. J. Biol. Chem. 276:17007-17013. [DOI] [PubMed] [Google Scholar]
- 17.Edmondson, D. G., T. C. Cheng, P. Cserjesi, T. Chakraborty, and E. N. Olson. 1992. Analysis of the myogenin promoter reveals an indirect pathway for positive autoregulation mediated by the muscle-specific enhancer factor MEF-2. Mol. Cell. Biol. 12:3665-3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edmondson, D. G., and E. N. Olson. 1989. A gene with homology to the myc similarity region of MyoD1 is expressed during myogenesis and is sufficient to activate the muscle differentiation program. Genes Dev. 3:628-640. [DOI] [PubMed] [Google Scholar]
- 19.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 20.Finkel, T., J. Duc, E. R. Fearon, C. V. Dang, and G. F. Tomaselli. 1993. Detection and modulation in vivo of helix-loop-helix protein-protein interactions. J. Biol. Chem. 268:5-8. [PubMed] [Google Scholar]
- 21.Fischle, W., F. Dequiedt, M. J. Hendzel, M. G. Guenther, M. A. Lazar, W. Voelter, and E. Verdin. 2002. Activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell 9:45-57. [DOI] [PubMed] [Google Scholar]
- 22.Fischle, W., S. Emiliani, M. J. Hendzel, T. Nagase, N. Nomura, W. Voelter, and E. Verdin. 1999. A new family of human histone deacetylases related to Saccharomyces cerevisiae HDA1p. J. Biol. Chem. 274:11713-11720. [DOI] [PubMed] [Google Scholar]
- 23.Genini, M., P. Schwalbe, F. A. Scholl, A. Remppis, M. G. Mattei, and B. W. Schäfer. 1997. Subtractive cloning and characterization of DRAL, a novel LIM-domain protein down-regulated in rhabdomyosarcoma. DNA Cell Biol. 16:433-442. [DOI] [PubMed] [Google Scholar]
- 24.Gorman, C. M., L. F. Mofat, and B. H. Howard. 1982. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol. Cell. Biol. 2:1044-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gossett, L. A., D. J. Kelvin, E. A. Sternberg, and E. N. Olson. 1989. A new myocyte-specific enhancer-binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Mol. Cell. Biol. 9:5022-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grozinger, C. M., C. A. Hassig, and S. L. Schreiber. 1999. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc. Natl. Acad. Sci. USA 96:4868-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guardavaccaro, D., M. T. Ciotti, B. W. Schäfer, A. Montagnoli, and F. Tirone. 1995. Inhibition of differentiation in myoblasts deprived of the interferon-related protein PC4. Cell Growth Differ. 6:159-169. [PubMed] [Google Scholar]
- 28.Guardavaccaro, D., G. Corrente, F. Covone, L. Micheli, I. D'Agnano, G. Starace, M. Caruso, and F. Tirone. 2000. Arrest of G1-S progression by the p53-inducible gene PC3 is Rb dependent and relies on the inhibition of cyclin D1 transcription. Mol. Cell. Biol. 20:1797-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guardavaccaro, D., A. Montagnoli, M. T. Ciotti, A. Gatti, L. Lotti, C. Di Lazzaro, M. R. Torrisi, and F. Tirone. 1994. Nerve growth factor regulates the subcellular localization of the nerve growth factor-inducible protein PC4 in PC12 cells. J. Neurosci. Res. 37:660-674. [DOI] [PubMed] [Google Scholar]
- 30.Haribabu, B., S. S. Hook, M. A. Selbert, E. G. Goldstein, E. D. Tomhave, A. M. Edelman, R. Snyderman, and A. R. Means. 1995. Human calcium-calmodulin dependent protein kinase I: cDNA cloning, domain structure and activation by phosphorylation at threonine-177 by calcium-calmodulin dependent protein kinase I kinase. EMBO J. 14:3679-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hasty, P., A. Bradley, J. H. Morris, D. G. Edmondson, J. M. Venuti, E. N. Olson, and W. H. Klein. 1993. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 364:501-506. [DOI] [PubMed] [Google Scholar]
- 32.Huang, E. Y., J. Zhang, E. A. Miska, M. G. Guenther, T. Kouzarides, and M. A. Lazar. 2000. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 14:45-54. [PMC free article] [PubMed] [Google Scholar]
- 33.Iacopetti, P., G. Barsacchi, F. Tirone, and F. Cremisi. 1996. Expression of the PC4 gene in the developing rat nervous system. Brain Res. 707:293-297. [DOI] [PubMed] [Google Scholar]
- 34.Lassar, A. B., J. N. Buskin, D. Lockshon, R. L. Davis, S. Apone, S. D. Hauschka, and H. Weintraub. 1989. MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer. Cell 58:823-831. [DOI] [PubMed] [Google Scholar]
- 35.Lassar, A. B., R. L. Davis, W. E. Wright, T. Kadesch, C. Murre, A. Voronova, D. Baltimore, and H. Weintraub. 1991. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell 66:305-315. [DOI] [PubMed] [Google Scholar]
- 36.Lassar, A., and A. Münsterberg. 1994. Wiring diagram: regulatory circuits and control of skeletal myogenesis. Curr. Opin. Cell Biol. 6:432-442. [DOI] [PubMed] [Google Scholar]
- 37.Lemercier, C., A. Verdel, B. Galloo, S. Curtet, M. P. Brocard, and S. Khochbin. 2000. mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J. Biol. Chem. 275:15594-15599. [DOI] [PubMed] [Google Scholar]
- 38.Lilly, B., B. Zhao, G. Ranganayakulu, B. M. Paterson, R. A. Schulz, and E. N. Olson. 1995. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science 267:688-693. [DOI] [PubMed] [Google Scholar]
- 39.Lin, Q., J. Schwarz, C. Bucana, and E. N. Olson. 1997. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 276:1404-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu, J., T. A. McKinsey, R. L. Nicol, and E. N. Olson. 2000. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc. Natl. Acad. Sci. USA 97:4070-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu, J., T. A. McKinsey, C. L. Zhang, and E. N. Olson. 2000. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell 6:233-244. [DOI] [PubMed] [Google Scholar]
- 42.Ludolph, D. C., and S. F. Konieczny. 1995. Transcription factor families: muscling in on the myogenic program. FASEB J. 9:1595-1604. [DOI] [PubMed] [Google Scholar]
- 43.Lyons, G. E., B. K. Micales, J. Schwarz, J. F. Martin, and E. N. Olson. 1995. Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J. Neurosci. 15:5727-5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Magenta, A., C. Cenciarelli, F. De Santa, P. Fuschi, F. Martelli, M. Caruso, and A. Felsani. 2003. MyoD stimulates RB promoter activity via the CREB/p300 nuclear transduction pathway. Mol. Cell. Biol. 23:2893-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mal, A., M. Sturniolo, R. L. Schiltz, M. K. Ghosh, and M. L. Harter. 2001. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 20:1739-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martelli, F., C. Cenciarelli, G. Santarelli, B. Polikar, A. Felsani, and M. Caruso. 1994. MyoD induces retinoblastoma gene expression during myogenic differentiation. Oncogene 9:3579-3590. [PubMed] [Google Scholar]
- 47.Martin, J. F., J. M. Miano, C. M. Hustad, N. G. Copeland, N. A. Jenkins, and E. N. Olson. 1994. A Mef2 gene that generates a muscle-specific isoform via alternative mRNA splicing. Mol. Cell. Biol. 14:1647-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin, J. F., J. J. Schwarz, and E. N. Olson. 1994. Myocyte enhancer factor (MEF) 2C: a tissue-restricted member of the MEF-2 family of transcription factors. Proc. Natl. Acad. Sci. USA 90:5282-5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McDermott, J. C., M. C. Cardoso, Y. T. Yu, V. Andres, D. Leifer, D. Krainc, S. A. Lipton, and B. Nadal-Ginard. 1993. hMEF2C gene encodes skeletal muscle- and brain-specific transcription factors. Mol. Cell. Biol. 13:2564-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKinsey, T. A., C. L. Zhang, J. Lu, and E. N. Olson. 2000. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408:106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2001. Control of muscle development by dueling HATs and HDACs. Curr. Opin. Genet. Dev. 11:497-504. [DOI] [PubMed] [Google Scholar]
- 52.Megeney, L. A., B. Kablar, K. Garrett, J. E. Anderson, and M. A. Rudnicki. 1996. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 10:1173-1183. [DOI] [PubMed] [Google Scholar]
- 53.Miner, J. H., and B. Wold. 1990. Herculin, a fourth member of the MyoD family of myogenic regulatory genes. Proc. Natl. Acad. Sci. USA 87:1089-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miska, E. A., C. Karlsson, E. Langley, S. J. Nielsen, J. Pines, and T. Kouzarides. 1999. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 18:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miska, E. A., E. Langley, D. Wolf, C. Karlsson, J. Pines, and T. Kouzarides. 2001. Differential localization of HDAC4 orchestrates muscle differentiation. Nucleic Acids Res. 29:3439-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Molkentin, J. D., B. L. Black, J. F. Martin, and E. N. Olson. 1995. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83:1125-1136. [DOI] [PubMed] [Google Scholar]
- 57.Molkentin, J. D., B. L. Black, J. F. Martin, and E. N. Olson. 1996. Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol. Cell. Biol. 16:2627-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Molkentin, J. D., and E. N. Olson. 1996. Defining the regulatory networks for muscle development. Curr. Opin. Genet. Dev. 6:445-453. [DOI] [PubMed] [Google Scholar]
- 59.Murre, C., P. S. McCaw, and D. Baltimore. 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56:777-783. [DOI] [PubMed] [Google Scholar]
- 60.Nabeshima, Y., K. Hanaoka, M. Hayasaka, E. Esumi, S. Li, I. Nonaka, and Y. Nabeshima. 1993. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature 364:532-535. [DOI] [PubMed] [Google Scholar]
- 61.Naya, F. J., C. Wu, J. A. Richardson, P. Overbeek, and E. N. Olson. 1999. Transcriptional activity of MEF2 during mouse embryogenesis monitored with a MEF2-dependent transgene. Development 126:2045-2052. [DOI] [PubMed] [Google Scholar]
- 62.Olson, E. N., H. H. Arnold, P. W. Rigby, and B. J. Wold. 1996. Know your neighbors: three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell 85:1-4. [DOI] [PubMed] [Google Scholar]
- 63.Puri, P. L., S. Iezzi, P. Stiegler, T. T. Chen, R. L. Schiltz, G. E. Muscat, A. Giordano, L. Kedes, J. Y. Wang, and V. Sartorelli. 2001. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol. Cell 8:885-897. [DOI] [PubMed] [Google Scholar]
- 64.Puri, P. L., V. Sartorelli, X. J. Yang, Y. Hamamori, V. V. Ogryzko, B. H. Howard, L. Kedes, J. Y. Wang, A. Graessmann, Y. Nakatani, and M. Levrero. 1997. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell 1:35-45. [DOI] [PubMed] [Google Scholar]
- 65.Rhodes, S. J., and S. F. Konieczny. 1989. Identification of MRF4: a new member of the muscle regulatory factor gene family. Genes Dev. 3:2050-2061. [DOI] [PubMed] [Google Scholar]
- 66.Rosenthal, N. 1987. Identification of regulatory elements of cloned genes with functional assays. Methods Enzymol. 152:704-720. [DOI] [PubMed] [Google Scholar]
- 67.Roth, A., R. Gill, and U. Certa. 2003. Temporal and spatial gene expression patterns after experimental stroke in a rat model and characterization of PC4, a potential regulator of transcription. Mol. Cell. Neurosci. 22:353-364. [DOI] [PubMed] [Google Scholar]
- 68.Rudnicki, M. A., P. N. Schnegelsberg, R. H. Stead, T. Braun, H. H. Arnold, and R. Jaenisch. 1993. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75:1351-1359. [DOI] [PubMed] [Google Scholar]
- 69.Rusconi, S., Y. Severne, O. Georgiev, I. Galli, and S. Wieland. 1990. A novel expression assay to study transcriptional activators. Gene 89:211-221. [DOI] [PubMed] [Google Scholar]
- 70.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sartorelli, V., J. Huang, Y. Hamamori, and L. Kedes. 1997. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol. Cell. Biol. 17:1010-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tirone, F., and E. M. Shooter. 1989. Early gene regulation by nerve growth factor: induction of an interferon related gene. Proc. Natl. Acad. Sci. USA 86:2088-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vadivelu, S. K., R. Kurzbauer, B. Dieplinger, M. Zweyer, R. Schafer, A. Wernig, I. Vietor, and L. A. Huber. 2004. Muscle regeneration and myogenic differentiation defects in mice lacking TIS7. Mol. Cell. Biol. 24:3514-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Varnum, B. C., R. W. Lim, and H. R. Herschman. 1989. Characterization of TIS 7, a gene induced in Swiss 3T3 cells by the tumor promoter tetradecanoyl phorbol acetate. Oncogene 4:1263-1265. [PubMed] [Google Scholar]
- 75.Vietor, I., S. K. Vadivelu, N. Wick, R. Hoffman, M. Cotten, C. Seiser, I. Fialka, W. Wunderlich, A. Haase, G. Korinkova, G. Brosch, and L. A. Huber. 2002. TIS7 interacts with the mammalian SIN3 histone deacetylase complex in epithelial cells. EMBO J. 21:4621-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang, A. H., N. R. Bertos, M. Vezmar, N. Pelletier, M. Crosato, H. H. Heng, J. Th'ng, J. Han, and X. J. Yang. 1999. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol. Cell. Biol. 19:7816-7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weintraub, H. 1993. The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell 75:1241-1244. [DOI] [PubMed] [Google Scholar]
- 78.Weintraub, H., R. Davis, D. Lockshon, and A. Lassar. 1990. MyoD binds cooperatively to two sites in a target enhancer sequence: occupancy of two sites is required for activation. Proc. Natl. Acad. Sci. USA 87:5623-5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weintraub, H., R. Davis, S. Tapscott, M. Thayer, M. Krause, R. Benezra, T. K. Blackwell, D. Turner, R. Rupp, S. Hollenberg, Y. Zhuang, and A. Lassar. 1991. The myoD gene family: nodal point during specification of the muscle cell lineage. Science 251:761-766. [DOI] [PubMed] [Google Scholar]
- 80.Wick, N., A. Schleiffer, L. A. Huber, and I. Vietor. 2004. Inhibitory effect of TIS7 on Sp1-C/EBPα transcription factor module activity. J. Mol. Biol. 336:589-595. [DOI] [PubMed] [Google Scholar]
- 81.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]
- 82.Wright, W. E., M. Binder, and W. Funk. 1991. Cyclic amplification and selection of targets (CASTing) for the myogenin consensus binding site. Mol. Cell. Biol. 11:4104-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wright, W. E., D. A. Sassoon, and V. K. Lin. 1989. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell 56:607-617. [DOI] [PubMed] [Google Scholar]
- 84.Yaffe, D., and O. Saxel. 1977. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 270:725-727. [DOI] [PubMed] [Google Scholar]
- 85.Yee, S. P., and P. W. Rigby. 1993. The regulation of myogenin gene expression during embryonic development of the mouse. Genes Dev. 7:1277-1289. [DOI] [PubMed] [Google Scholar]
- 86.Yu, Y. T., R. E. Breitbart, L. B. Smoot, Y. Lee, V. Mahdavi, and B. Nadal-Ginard. 1992. Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev. 6:1783-1798. [DOI] [PubMed] [Google Scholar]
- 87.Zhang, C. L., T. A. McKinsey, J. R. Lu, and E. N. Olson. 2001. Association of COOH-terminal-binding protein (CtBP) and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J. Biol. Chem. 276:35-39. [DOI] [PubMed] [Google Scholar]
- 88.Zhang, C. L., T. A. McKinsey, and E. N. Olson. 2002. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol. Cell. Biol. 22:7302-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao, X., A. Ito, C. D. Kane, T. S. Liao, T. A. Bolger, S. M. Lemrow, A. R. Means, and T. P. Yao. 2001. The modular nature of histone deacetylase HDAC4 confers phosphorylation-dependent intracellular trafficking. J. Biol. Chem. 276:35042-35048. [DOI] [PubMed] [Google Scholar]