Abstract
Androgen receptor (AR) activity is required for prostate cancer development and progression. Thus, there is a major impetus to understand the regulation of AR action. We and others have previously shown that AR transactivation potential is dependent on the presence of an active SWI/SNF chromatin remodeling complex. However, the mechanisms underlying SWI/SNF regulation of the AR remained unsolved. We show here that the BAF57 subunit, an accessory component of the remodeling complex, is a critical regulator of AR function. We show that BAF57 is expressed in the luminal epithelia of the prostate and is required for AR-dependent transactivation in prostatic adenocarcinoma cells. Our data reveal that BAF57 can directly bind to the AR and is recruited to endogenous AR targets upon ligand activation. Loss of BAF57 or inhibition of BAF57 function severely compromised AR activity, as observed with both exogenous and endogenous AR targets. Rescue of BAF57 function restored AR activity, thus demonstrating a specific requirement of BAF57 for AR activity. This action of BAF57 proved to be dependent on SWI/SNF ATPase function. BAF57 has previously been implicated in nuclear receptor coactivator function, and we show that, although BAF57 facilitated coactivator activity, only a selected subset required BAF57 for coactivator function. Lastly, we demonstrate that both BAF57 and BRM are required for the proliferation of AR-dependent prostatic adenocarcinoma cells. In summary, these findings identify BAF57 as a critical modulator of the AR that is capable of altering AR activity, coactivator function, and AR-dependent proliferation.
The androgen receptor (AR) is a member of the nuclear receptor superfamily, whose activity is required for prostate development, growth, and survival (16, 26, 34). This dependence on AR function is exploited in prostate cancer therapy, wherein AR remains the major target for therapeutic intervention (1, 34). Given the importance of AR in treatment of this tumor type, considerable effort has been directed at revealing the mechanisms of AR action.
The AR is activated by ligand binding to induce transactivation potential. Testosterone, the most prevalent AR ligand in the sera, is converted to dihydrotestosterone (DHT) in the prostate through the action of 5-alpha reductase. DHT serves as a high-affinity ligand for the AR and stimulates a cascade of events that initiate AR activation (6, 16, 52). Initially, AR is released from heat shock proteins, undergoes homodimerization, and rapidly translocates to the nucleus, where it is targeted to androgen response elements (AREs) located within AR target gene promoters/enhancers. Once activated by ligand, AR recruits a host of cofactors that facilitate the formation of an active transcriptional complex (23, 59). In general, these cofactors act to release DNA from chromatin through either covalent modification of histones or remodeling of chromatin structure. Such modifications foster a state of open chromatin, allowing recruitment of basal transcription machinery and RNA polymerase II (27, 46, 62, 68).
Several classes of AR coactivators promote histone modifications. Members of the p160 coactivator family (including SRC1, SRC2/TIF2/GRIP1, and SRC3/RAC3/AIB1) serve as platforms for the recruitment of histone acetyltransferases (HATs), such as CBP/p300 or p/CAF. SRC1 and SRC3 also harbor intrinsic HAT activity, which likely contributes to their action as coactivators (12, 63, 67). Acetylation of core histones by the recruited HATs results in a displacement or loosening of DNA from the nucleosome, thus facilitating the recruitment of the subsequent cofactors and the basal transcription machinery. In addition, p/CAF has been shown to acetylate the AR itself, an event shown to be essential for AR activity (20). Thus, through their ability to modify both AR and core histones, the p160 coactivators play pivotal roles in AR activation. The importance of such coactivator regulation in prostate cancer is apparent, since aberrant expression of both SRC1 and TIF2 is associated with relapse after therapeutic intervention (21, 24). Many additional AR coactivators have been identified whose actions are less well described. For example, ARA70 and ARA55, in addition to enhancing AR activity, are able to increase the androgenic activity of anti-androgens such as hydroxyflutamide (22, 44). Recently, ARA70 expression was shown to be enhanced in the neoplastic prostate compared to benign tissue, thus further implicating a role for this coactivator in prostate cancer progression (29). Lastly, the E2 conjugating enzyme, Ubc9, has been shown to bind directly to and activate the AR, independent of its SUMO-1 conjugation function (51). The coordinate actions of these coactivators are thought to play pivotal roles in AR activation and likely influence tissue specific AR activity. The contribution of these coactivators to histone acetylation and cooperation with the p160 family has yet to be thoroughly examined.
Although the p160 coactivators initiate histone modifications through acetylation, it has been shown that several nuclear receptors also attract complexes that enzymatically regulate chromatin structure. Several classes of chromatin remodeling complexes have been described, including WINAC, NuRD, ISWI, and SWI/SNF (4, 39, 46, 70, 72). ARIP4 has been described as a SWI/SNF-like chromatin remodeling enzyme and was shown to modulate AR activity (57, 61). In general, chromatin remodeling complexes utilize the hydrolysis of ATP to disrupt histone-DNA association, thus altering chromatin structure to foster a state of either closed chromatin (resulting in gene repression) or open chromatin (resulting in enhanced gene transcription). Although the precise mechanisms of SWI/SNF action in vivo have yet to be elucidated, associated activities in vitro include alterations in nucleosome structure, nucleosome sliding, dinucleosome formation, and histone octamer transfer (17, 46). Such alterations in chromatin structure can be distinguished from histone modifications and are not dependent on histone acetylation (30). Moreover, SWI/SNF action has been shown to contribute to the dynamic activity of nuclear receptors, facilitating rapid mobilization of the receptors after SWI/SNF recruitment (18). The activity of several nuclear receptors, including the estrogen receptor (ER) and glucocorticoid receptor (GR), has been shown to be enhanced by the action of SWI/SNF (3, 10, 14, 18, 19, 31, 45, 50, 72, 74). SWI/SNF represents a class of large, 2-MDa chromatin remodeling complexes that contain a central ATPase (either BRM/SNF2α or BRG1/SNF2β) and a variable series of associated subunits termed BRG1-associated factors (BAFs). SWI/SNF exists as a multitude of biochemically diverse complexes, and although the function of each specialized complex remains unclear, it has been speculated that variances in combinatorial assembly dictate diverse biological outcomes. Supporting this view, it has been shown that BRG1 and BRM demonstrate specificity for protein-protein interaction and likely govern unique gene targets (35). Although some redundancies do exist, BRG1 is predominantly found in cell types undergoing proliferation or self-renewal, whereas BRM expression is increased after cellular differentiation. Interestingly, BRM was recently shown to be expressed in normal human prostatic epithelia, whereas BRG1 could not be detected in this tissue (53). We and others have shown that the AR requires SWI/SNF function for activity, and our studies highlighted a specific role for BRM complexes in AR regulation (30, 32, 43). However, the mechanism by which BRM specifically regulates AR activity remained undetermined.
It is speculated that the BAF subunits largely contribute to SWI/SNF specificity (47). Subunits commonly associated with the SWI/SNF complex include BAF155, BAF170, INI1/SNF5, BAF250, BAF60, and BAF57 (46, 71, 72). Several of these subunits have been shown to harbor critical functions. For example, the loss of the INI1/SNF5 subunit gives rise to rhabdoid tumors in humans and can promote tumorigenesis, thus identifying INI1/SNF5 as a tumor suppressor (69). Subsequent studies demonstrated that reintroduction of INI1/SNF5 into human tumor cell lines deficient in this subunit induced cell cycle arrest (5). Other subunits, including BAF250, BAF60a, and BAF57, have been shown to directly interact with the GR and to promote GR activation (28, 47). Lastly, the BAF57 subunit has been shown to interact with both the ER and the p160 coactivators. Although BAF57 function was dispensable for ER activation, BAF57 was essential for SRC1 to modulate the ER (3). Collectively, these observations implicate SWI/SNF subunits as critical regulators of nuclear receptor-SWI/SNF interactions.
BAF57 contains DNA-binding capability through its high-mobility-group (HMG) motif and a kinesin-like coiled-coil domain and is suggested to play an architectural role in the formation of higher-order protein complexes. Although these properties of BAF57 are dispensable for SWI/SNF chromatin remodeling activity (71), it is hypothesized that BAF57 may lend specificity in targeting to the SWI/SNF complex. This contention was supported by observations in transgenic mice, wherein mutations in the HMG domain of BAF57 inhibited specific SWI/SNF target gene regulation (9).
We show here that BAF57 is expressed in the AR-positive luminal epithelial layer of the prostate and that BAF57 strongly regulates AR function. We show that BAF57 directly binds to AR and is recruited to AR regulatory regions in a ligand-dependent fashion. Using both exogenous and endogenous prostate-specific AR targets, we show that AR requires BAF57 function for robust transcriptional response. We also demonstrate that this function of BAF57 requires SWI/SNF ATPase function, firmly linking BAF57 action to alterations in nucleosome structure. Similar to what was observed with ER, BAF57 also facilitated p160-mediated AR enhancement, and a similar profile was observed with Ubc9. Interestingly, ARA70 and ARA55 maintained activity in the absence of BAF57 but were augmented by BAF57 for AR activation. Thus, BAF57 exhibited coactivator specific modulation of AR activity. Given the observed necessity of BAF57 for AR target gene transcription and coactivator function, we examined the requirement of BAF57 and SWI/SNF for proliferation in AR-dependent prostate cancer cell growth. Strikingly, inhibition of either BAF57 or BRM function significantly attenuated cellular proliferation in this cell type. Collectively, these data establish a firm requirement of BAF57 for AR and coactivator function. Moreover, these data implicate BRM and BAF57 as putative therapeutic targets in prostate cancer therapy.
MATERIALS AND METHODS
Plasmids.
pSG5AR (wild-type AR), pSG5ARA70, pSG5ARA55, and pcDNA3-FLAGARA70N2 (22, 29, 73) expression plasmids were a gift from Chawnshang Chang (University of Rochester, Rochester, N.Y.). pARR2-Luc was generated as previously described (43). pCR3.1 h-SRC1A was obtained from Bert O'Malley (Baylor College of Medicine, Houston, Tex.) (58). The pCMV-TIF2 mammalian expression construct was a gift from Alvaro Puga (University of Cincinnati, Cincinnati, Ohio). pFLAG-Ubc9 was received from Jorma Palvimo (University of Helsinki, Helsinki, Finland) (51). tk-Renilla luciferase was purchased from Promega and pcDNA3.1 from Invitrogen. pEGFP H2B-GFP was a gift from G. Wahl (Salk Institute, La Jolla, Calif.). pGEX-KG GST is from Robert Brackenbury (University of Cincinnati) (25). pGEX 3X GST-BAF57 was provided by Gail Mandel (SUNY at Stony Brook, Stony Brook, N.Y.) and has been previously described (2). pCG-hBRM and pCG-dnBRM were provided by M. Yaniv (Institut Pasteur, Paris, France) (45). Dominant-negative AR expression plasmid (pSG5ARΔ46-408) was received from Ollie Janne (University of University of Helsinki) (49). pCMV-p16ink4a was previously described (40). pBabe-WTBAF57-FLAG and pBabe-BAF57ΔN-FLAG were constructed by PCR amplification of each gene from the pBJ5 vector by using primers that added BamHI (5′) and EcoRI/FLAG (3′). The PCR product was digested and inserted into the pBabe vector (which also encodes the puromycin resistance marker). The pHTP-BRMi construct uses a 64-mer double-stranded oligonucleotide to generate a shRNA targeting nucleotides 3388 to 3408 of the BRM mRNA. The fragment (or a scrambled control of the siRNA oligonucleotide sequence) was cloned into the pHTP-siRNA expression vector (kindly provided by William Reed, University of North Carolina at Chapel Hill) by using BglII and XhoI overhangs. The vector includes a puromycin resistance marker and places the expression oligonucleotide under the control of the human H1 promoter. pGreenLantern (Gibco) encodes the green fluorescent protein (GFP).
Immunohistochemistry.
Prostates were resected from 9-week-old FVB/N mice. Individual lobes (ventral, dorsolateral, and anterior) were isolated, fixed by immersion in 10% neutral-buffered formalin, and subsequently embedded in paraffin. Blocks were sectioned (5 μm), mounted on slides, deparaffinized in xylene, and rehydrated in ethanol solutions. After antigen retrieval by boiling for 20 min in Antigen Unmasking Solution (Vector Laboratories), the sections were incubated for 15 min in peroxidase blocking reagent (Dako Cytomation) to quench endogenous peroxidase. The sections were then blocked for 20 min in diluted normal goat serum (Rabbit IgG Vectastain ABC Kit; Vector Laboratories), followed by incubation with anti-BAF57c rabbit polyclonal antibody (1:1,000 dilution in phosphate-buffered saline [PBS] solution) overnight at 4°C. After incubation for 30 min with anti-rabbit immunoglobulin G (IgG) biotinylated secondary antibody solution (Rabbit IgG Vectastain ABC Kit; Vector Laboratories), sections were incubated in ABC reagent for another 30 min. Antibodies were visualized with DAB Chromogen (Dako Cytomation), and nuclei were counterstained with hematoxylin.
Tissue culture and treatment.
BT459 cells were obtained from the American Type Culture Collection (ATCC) and cultured in RPMI supplemented with 10% heat-inactivated fetal bovine serum (FBS; Atlanta Biologicals, Norcross, Ga.), 2 mM l-glutamine, and 100 U of penicillin-streptomycin (Mediatech, Herndon, Va.)/ml. For reporter analyses, BT549 cells were cultured in phenol red-free RPMI medium and 10% charcoal dextran-treated FBS (CDT; Atlanta Biologicals). LNCaP cells received from the ATCC were cultured in improved minimal essential medium (IMEM) supplemented with 5% heat inactivated FBS, 2 mM l-glutamine, and 100 U of penicillin-streptomycin/ml. LNCaP reporter assays were performed in phenol red-free IMEM supplemented with 5% CDT. MCF7, CV1, and SW13 cells were obtained from the ATCC and maintained in Dulbecco modified Eagle medium supplemented with 10% FBS, 2 mM l-glutamine, and 100 U of penicillin-streptomycin/ml. Reporter analysis in these cell lines was carried out in phenol red-free medium containing 10% CDT.
LB1-luc cells were generated from the LNCaP cell line by cotransfection of the pARR2-Luc reporter construct (43) and the pBabe-PURO vector (which confers puromycin resistance). After selection in puromycin, individual colonies were isolated and screened for retention of ARR2 reporter activity. The resulting LB1-Luc cell line was tested by seeding 105 cells into a 12-well dish with IMEM medium containing 5% CDT. Cells were cultured in this condition of steroid deprivation for 3 days and were then stimulated with the indicated concentrations of DHT (Sigma). Cells were harvested and measured for luciferase activity corrected against total protein (as determined via the Bio-Rad-DC assay [Bio-Rad]).
Transfection and reporter analysis.
MCF7, CV1, and SW13 cells were seeded in Dulbecco modified Eagle medium containing 10% CDT. Cells were transfected the following day using the BES-calcium phosphate protocol as previously described (8). Cells were transfected with 0.5 μg of pARR2-Luc, 0.5 μg of pSG5AR, 0.25 μg of pTK-Renilla luciferase, 2.25 μg of pBabe-WTBAF57-FLAG, 1 μg of pCG-hBRM, or parental vector (to a total of 4.5 μg) as indicated. Posttransfection, cells were treated with 0.1% ethanol (EtOH) vehicle or 0.1 nM DHT for 24 h. We used 0.1 nM DHT since this is the optimal mitogenic dose for prostate cancer cells (0.1 to 1.0 nM DHT) (37). Cells were then harvested, lysed, and assayed for luciferase activity using the Dual Luciferase Assay System (Promega). For BT549 reporter analyses, cells were seeded in six-well dishes with RPMI containing 10% CDT. Cells were transfected the following day using the FuGENE6 (Roche Molecular Biochemicals) method as recommended by the manufacturer. Transfections consisted of 0.5 μg of pARR2-Luc reporter, 0.5 μg of pSG5AR, and 2.25 μg of pBabe-BAF57-FLAG (unless otherwise indicated), and/or 1 μg of coactivator (pCR3.1-SRC1, pCMV-TIF2, pFLAG-Ubc9, pSG5ARA70, or pSG5ARA55) or 1 μg of pCG-dnBRM or pSG5ARA70N2 as indicated. For each transfection, 0.25 μg of pTK-Renilla luciferase was used to normalize for transfection efficiency. Total plasmid DNA was brought to 4.5 μg with the parental (empty) vector. After transfection, fresh medium was added and cells were treated with either 0.1% EtOH or 0.1 nM DHT for 24 h. After stimulation, cells were harvested and lysed, and reporter analyses were carried out by using the Dual Luciferase Assay Reporter System (Promega). LNCaP reporter analysis was carried out in six-well dishes with IMEM containing 5% CDT. Cells were transfected by using the Lipofectin (Invitrogen) transfection reagent as recommended by the manufacturer. LNCaP cells were transfected with 1 μg of pARR2-Luc reporter and 2.25 μg of pBabe-BAF57ΔN-FLAG or parental vector (to 3.5 μg of total DNA). Next, 0.25 μg of pTK-Renilla luciferase was used to normalize for transfection efficiency. After transfection, cells were stimulated with 1 nM DHT. After 48 h, cells were harvested, and the Dual Luciferase Assay Reporter System was used to measure relative luciferase activity. Reporter assays were performed from at least in six independent transfections for each condition. Averages and standard deviations are shown. Statistical analysis was performed by using the Student t test, with a P value of <0.05 indicating statistical significance. Transfection method was chosen based on the optimal method for each cell line (determined empirically using pTK-Renilla luciferase). Where indicated, parallel transfections were performed in which the internal control plasmid was replaced by pGreenLantern, and cells were harvested to monitor relative AR expression levels via immunoblot.
Immunoblotting.
For protein analysis, cells were harvested by trypsinization (for mouse prostate, individual lobes were microdissected and ground), and lysis was carried out in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1.0% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris [pH 8.0]) supplemented with protease inhibitors and phenylmethylsulfonyl fluoride (PMSF). After brief sonication and clarification, equal protein was subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to an Immobilon-P membrane (Millipore). The membrane was immunoblotted with the indicated antibodies by standard techniques and visualized by chemiluminescence (Western Lightning; Perkin-Elmer Life Sciences). The antibodies used were generated against BAF57c (a gift from Weidong Wang), AR (N-20; Santa Cruz), GFP (B2; Santa Cruz), and CDK4 (H-22; Santa Cruz).
Immunoblotting to verify BRM knockdown via small interfering RNA (siRNA) was carried out by transfection with 7 μg of either pHTP-vector or pHTP-BRMi, along with cotransfection of 1 μg of pEGFP H2B-GFP into LNCaP cells. Posttransfection, cells were selected by using 0.625 μg of puromycin (Sigma)/ml. Cells were simultaneously harvested after >90% of the population was H2B-GFP positive (ca. 3 days). Pellets were rinsed in cold phosphate-buffered saline (PBS) and resuspended in buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol [DTT], 0.5 mM PMSF, and 0.5 mg of leupeptin and 1 mg of aprotinin/ml), followed by incubation on ice for 20 min. Next, 6.5 μl of 10% NP-40 was then added, and the pellets were vortexed and centrifuged for 5 min at 4°C. The supernatant was removed, and the pellet was resuspended in 50 μl of cold buffer C (20 mM HEPES [pH 7.9], 400 mM NaCl, 15 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF, and 0.5 mg of leupeptin and 1 mg of aprotinin/ml). Equal amounts of protein were subjected to SDS-7.5% PAGE and transferred to an Immobilon-P membrane (Millipore). Immunoblotting was performed with the indicated antibodies by standard techniques. Antibodies used were generated against BRM (courtesy of Moshe Yaniv) and actin (Sigma).
GST pull-down.
AR protein was produced and radiolabeled with [35S]methionine by using the TnT-Coupled Reticulocyte Lysate system (Promega) for in vitro transcription and translation according to the manufacturer's specifications and the NEG-772 Easytag Express protein labeling mix (Perkin-Elmer Life Sciences). Glutathione S-transferase (GST) or GST-BAF57 was transformed into BL21 bacteria and induced by using 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 to 5 h. After centrifugation, bacteria were washed one time in STE (150 mM NaCl, 25 mM Tris [pH 8.0], and 1 mM EDTA) and then resuspended in 3 ml of STE. Pellets were lysed upon the addition of 0.1 mg of lysozyme/ml and 1.5% Sarkosyl, sonicated, and clarified by centrifugation. The lysate was then incubated with 300 μl of glutathione-agarose beads at 4°C overnight. Beads were washed four times in PBS and then incubated with the in vitro-translated protein for 3 h at 4°C. Reactions were performed in 500 μl of NETN (20 mM Tris-HCl [pH 8.0], 1 mM EDTA, 100 mM NaCl, 0.5% NP-40) supplemented with PMSF and protease inhibitors and in the presence of either 0.1% EtOH or 10 nM DHT, as indicated. “Input” (3% of total) and “bound” fractions were separated by SDS-10% PAGE, stained with Coomassie blue, and subjected to fluorography by using Fluoro-Hance (Research Products International Corp., Mount Prospect, Ill.). Labeled protein was detected by autoradiography and quantified on a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Coimmunoprecipitation.
Immunoprecipitations were performed as previously described (28). LNCaP cells were harvested, washed in PBS, and lysed in immunoprecipitation buffer (IP buffer; 20 mM HEPES [pH 7.9], 250 mM NaCl, 10% [vol/vol] glycerol, 0.1% [vol/vol] Tween 20, 0.2 mM EDTA, 2 mM DTT) supplemented with protease inhibitors, 0.5 mM PMSF, and 100 ng of leupeptin and aprotinin per ml. After sonication and clarification, protein was quantified by using the Bio-Rad DC protein assay kit. A total of 1 mg of protein was used for each immunoprecipitation reaction. Lysate was precleared with 1% preimmune rabbit serum and 20 μl of protein-A Sepharose (Amersham) in 0.5 ml of IP buffer for 1 h at 4°C. Anti-AR, anti-BAF57, and preimmune serum (negative control) were incubated with the lysate overnight at 4°C. Then, 20 μl of protein-A Sepharose was added for a 1-h incubation. Beads were subjected to five washes in IP buffer, resuspended in 2× SDS-PAGE buffer, and boiled to denature proteins. Samples were then subjected to SDS-PAGE and immunoblot analysis with antibody generated against AR (AR-N20; Santa Cruz).
ChIP.
LNCaP or LB1-Luc cells were seeded into 15-cm dishes in phenol red-free IMEM supplemented with 5% CDT. After 4 days of incubation, cells were stimulated with either 0.1% EtOH or 10 nM DHT for 60 min. A higher than physiological dose was utilized for chromatin immunoprecipitation (ChIP) analysis (consistent with the literature) in order to enhance AR on target promoters (59). Cells were then treated with 1% formaldehyde diluted in PBS to allow for protein cross-linking to DNA, followed by quenching with 0.125 M glycine. Cells were then washed with ice-cold PBS, treated with trypsin, and pelleted. Pellets were washed in PBS containing 0.5 mM phenylmethylsulfonyl fluoride (PMSF) and 100 ng of leupeptin and aprotinin per ml. Cells were lysed in cell lysis buffer (5 mM PIPES [pH 8.0], 85 mM KCl, 0.5% NP-40, 0.5 mM PMSF, and 100 ng of leupeptin and aprotinin per ml) for 10 min. Nuclei were lysed in 400 μl of nucleus lysis buffer (1% sodium dodecyl sulfate [SDS], 50 mM Tris-Cl [pH 8.1], 10 mM EDTA, 0.5 mM PMSF, 100 ng of leupeptin and aprotinin per ml) for 10 min, and sonication was performed to shear DNA. Immunoprecipitations were carried out with 80 μl of lysate and antibodies to AR (N-20; Santa Cruz), acetylated histone H4 (catalog no. 06-866; Upstate Biotechnology), BAF57c (specific for C terminus), BAF57h (specific for HMG domain; a gift from Weidong Wang) (71), and a control IgG antibody (preimmune sera). Chromatin and antibody were incubated in RIPA buffer (150 mM NaCl, 1.0% NP-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris [pH 8.0]) for 1.5 h at 4°C, at which time protein-A Sepharose (Amersham) was added for an additional incubation. Beads were washed three times with super-RIPA (RIPA buffer plus 300 mM NaCl), once with RIPA buffer, and once with TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA), with 10-min rotations between each wash at room temperature. Next, 300 μl of ChIP extraction buffer (1% SDS, 0.1 M NaHCO3) was added, along with 20 μl of 5 M NaCl and RNase A. Cross-linking was reversed by incubation at 65°C (4 h up to overnight). DNA was purified by using the QIAquick PCR purification kit (Qiagen). PCR amplification was performed on recovered DNA products as well as input chromatin (10% of input for Fig. 6A and C; 1% of input in Fig. 6B) in the presence of [α-32P]dCTP, which allows accurate quantification of the PCR products over a broad range of DNA concentrations (60). To monitor recruitment to the prostate specific antigen (PSA) enhancer, PCR was performed by using G/H primers, as previously described (59). These primers are specific for a region containing an androgen responsive element (AREIII) contained within the enhancer region of the PSA gene. For PCR, the parameters were as follows: 94°C for 5 min; followed by 28 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s; and finally 72°C for 10 min. To monitor recruitment to the probasin promoter, the primers 5′-CATCTACCATTCCAGTTAAG (forward) and 5′-CCAGCGGTTCCATCC (reverse) were used. PCR parameters were 94°C for 5 min; followed by 40 cycles of 94°C for 45 s, 49°C for 45 s, and 72°C for 45 s. PCR products were separated on a 6% polyacrylamide gel and visualized with a PhosphorImager.
FIG. 6.
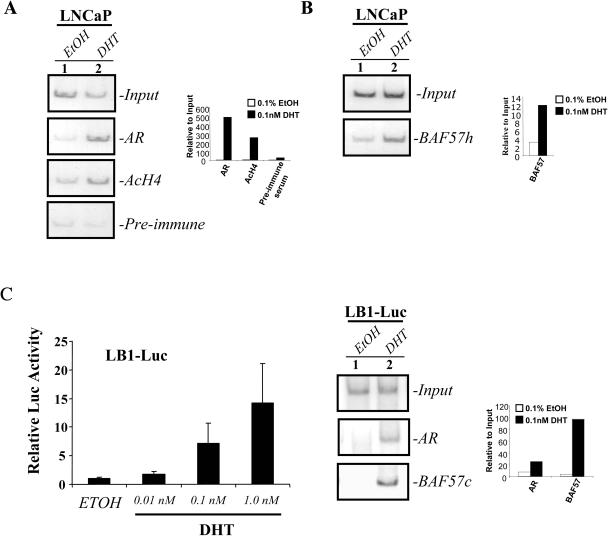
BAF57 is recruited to the AR regulatory regions in the presence of ligand. LNCaP cells (5 × 106) were cultured in steroid-free medium for 4 days and then stimulated with either 0.1% EtOH vehicle (lane 1) or 10 nM DHT (lane 2) for 60 min. After treatment the cells were formaldehyde cross-linked, and chromatin was recovered for use in ChIP assays. (A and B) Antibodies to acetylated histone H4 (AcH4), AR, BAF57h, and preimmune serum were used for ChIP analyses of the PSA enhancer region, as indicated. (C) A total of 105 LB1-Luc cells (with integrated pARR2-Luc) were serum starved for 3 days and then treated with the indicated concentrations of DHT for 24 h. Reporter analysis was performed and demonstrated activation of the AR in a dose-dependent manner (left panel). For the ChIP analysis (right panel), LB1-Luc cells (5 × 106) were cultured in steroid-free medium for 4 days and then stimulated with either 0.1% EtOH vehicle (lane 1) or 10 nM DHT (lane 2) for 60 min. After treatment, the cells were formaldehyde cross-linked, and chromatin was recovered for use in ChIP assays. Antibodies to AR and BAF57c were used for ChIP analyses of the ARR2 promoter region.
RT-PCR.
LNCaP cells were seeded in 6-cm dishes and transfected with the Lipofectin transfection reagent with 1 μg of pEGFP H2B-GFP to monitor for transfection efficiency, 1 μg of pBabe-PURO for rapid puromycin selection, and 4 μg of one of the following expression plasmids: pCMV-NEO BAM (vector control), pBabe-BAF57ΔN-FLAG, or dominant-negative AR (pSG5ARΔ46-408, positive control). After transfection, cells were allowed to recover for ca. 24 h. Transfected cells were selected by using 0.625 μg of puromycin (Sigma)/ml. Selection was carried out until >90% of cells were H2B-GFP positive (ca. 3 days). RNA was isolated by using the TRIzol Reagent (Gibco) as recommended by the manufacturer. To quantify expression, reverse transcription-PCR (RT-PCR) was performed with amplification of the cDNA performed in the presence of [32P]dCTP to quantify expression over a large linear range, as previously described (60). Primers were used for PSA (primer pair 5′-CTTGTAGCCTCTCGTGGCAG-3′ and 5′-GACCTTCATAGCATCCGTGAG-3′) and GAPDH (loading control; primer pair 5′-CCACCCCATGGCAAATTCCATGGCA-3′ and 5′-TCTAGACGGCAGGTCAGGTCCACC-3′). PCR conditions were as follows: 94°C for 2 min; followed by 23 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 30 s; and finally 72°C for 10 min. PCRs were separated by PAGE, and signal was quantified on a PhosphorImager. For comparison, maximal AR activity (PSA and GAPDH) in the presence of vector was set to 100. Representative data from two independent experiments are shown.
BrdU incorporation.
LNCaP cells were seeded onto poly-l-lysine coated coverslips, followed by transfection with Lipofectin, as described above. Cells were transfected with 0.5 μg of H2B-GFP, along with 4.5 μg of one of the following expression plasmids: pBabe-PURO, pBabe-BAF57ΔN-FLAG, dominant-negative AR (pSG5ARΔ46-408), pHTP vector, pHTP-scrambled, pHTP-BRMi, or pCMV-p16ink4a. Posttransfection (3 days for the pBabe-PURO/BAF57ΔN/dnAR experiment or 4 days for the pHTP-vector/pHTP-BRMi/pCMV-p16ink4a experiment), bromodeoxyuridine (BrdU; Amersham Biosciences) labeling reagent was added, followed by incubation for 16 h. Detection of BrdU by indirect immunofluorescence was performed as previously described (41). A minimum of 150 H2B-GFP transfected cells per sample were counted for BrdU incorporation. The results are plotted as the percent inhibition versus vector control, and the average of at least three replicates from two independent experiments is shown.
RESULTS
BAF57 is expressed in the prostatic epithelia.
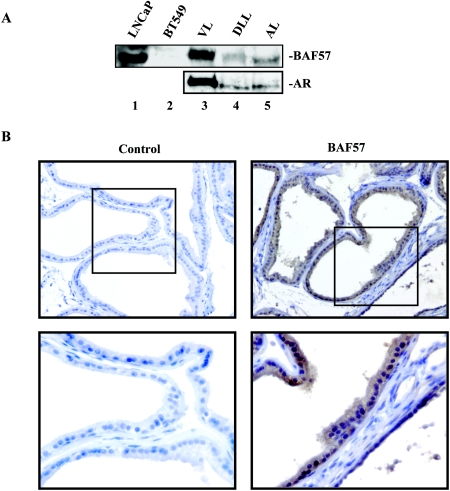
We and others have previously shown that SWI/SNF function is required for AR activity and that AR is preferentially assisted by the BRM-containing SWI/SNF complex (30, 32, 43). Correspondingly, BRM (but not BRG1) has been detected in normal human prostatic epithelia (53). However, the mechanism by which SWI/SNF is recruited to AR targets remained unsolved. Similar to the inability of the core ATPase, BRG1, to bind to the estrogen and glucocorticoid receptors (14, 28), we failed to observe in vitro interaction of BRM with recombinant AR (data not shown), thus indicating that AR does not bind directly to the core ATPase. It has been previously demonstrated that selected SWI/SNF subunits may modulate association with sequence-specific binding to transcription factors. For example, the GR has been shown to interact directly with BAF250, BAF60a, and BAF57 (28, 47). The BAF57 subunit was also shown to interact with both the ER and the p160 coactivators and was critical for p160 function in these assays (3). We demonstrate here that BAF57 is highly expressed in prostatic epithelia. As can be seen in Fig. 1A, BAF57 was detected in all lobes of the murine prostate (lanes 3 to 5). To ascertain the cellular origin of BAF57 expression in the prostate, immunohistochemistry was performed (Fig. 1B). As seen in the figure, BAF57 was predominantly expressed in the luminal epithelial layer and was enriched in the nuclei (right two panels [magnifications: top, ×20, bottom, ×40]) compared to the no primary antibody control (left two panels [magnifications: top, ×20; bottom, ×40]). These observations indicate that BAF57 is highly expressed in the AR-positive, AR-dependent prostatic epithelia. Given the requirement of SWI/SNF for AR function, a potential relationship between BAF57 and AR was investigated.
FIG. 1.
BAF57 is expressed in the luminal epithelia. Mouse prostates were resected and the individual lobes (ventral lobe [VL, lane 3], dorsolateral lobe [DLL, lane 4], and anterior lobe [AL, lane 5]) were isolated and prepared for either immunoblotting or immunohistochemistry as described in the text. (A) BAF57 and AR were detected by immunoblotting. LNCaP cell lysate (lane 1) and BT549 cell lysate (lane 2) were used as positive and negative controls, respectively. (B) Tissue from mouse prostate was sectioned and stained by standard immunohistochemistry techniques with no primary antibody control (left panels) or BAF57 (right panels). Antibody reactivity was visualized with diaminobenzidene chromogen, and nuclei were counterstained with hematoxylin. Magnifications, ×20 (top panels) and ×40 (bottom panels).
BAF57 binds directly to the AR and is associated with AR in prostate cancer cells.
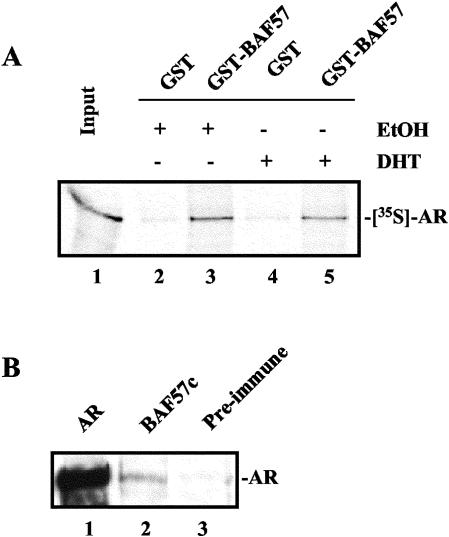
To examine the putative role of BAF57 in AR function, GST pull-down assays were used (Fig. 2A). Recombinant AR bound significantly to GST-BAF57 (compared to GST control), since in the absence of ligand ca. 60% of input was captured compared to only 7% with GST (compare lanes 2 and 3). Inclusion of ligand did not enhance AR binding (compare lanes 3 and 5). Thus, these data indicate that BAF57 can directly bind AR, independent of ligand. To verify these findings, coimmunoprecipitations were performed with AR-positive prostate cancer cells (LNCaP). Cell lysate was subjected to immunoprecipitation analysis with antibodies to AR, BAF57, and preimmune serum, as indicated. Immunoblotting with antibody to AR revealed an endogenous interaction between BAF57 and the AR (Fig. 2B). Combined, these data demonstrate that AR associates with BAF57 in prostate cancer cells.
FIG. 2.
BAF57 binds the AR independent of ligand. (A) Immobilized GST or GST-BAF57 was incubated with [35S]methionine-labeled AR in the presence of 0.1% EtOH vehicle (lanes 2 and 3) or 10 nM DHT (lanes 4 and 5) for 3 h at 4°C, washed, and denatured. Input and bound protein was subjected to SDS-PAGE, and 35S-labeled AR was detected by autoradiography. (B) Clarified protein lysates from LNCaP cells was utilized for coimmunoprecipitation with antibodies to AR (lane 1), BAF57c (specific to C terminus of BAF57) (71) (lane 2), or preimmune serum (lane 3). The resultant immunoprecipitates were subjected to SDS-PAGE, and the presence of AR was detected by immunoblotting.
BAF57 is required for receptor activity.
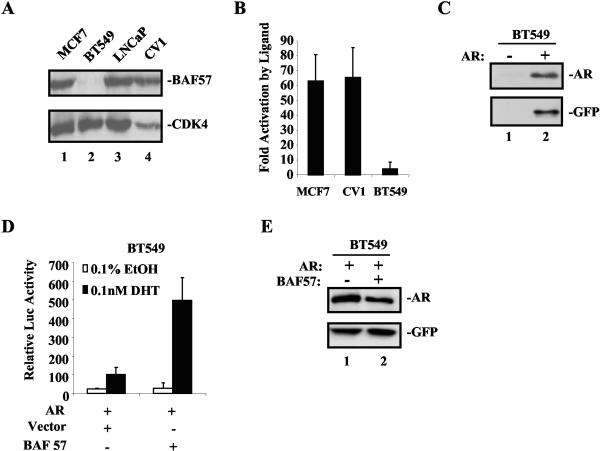
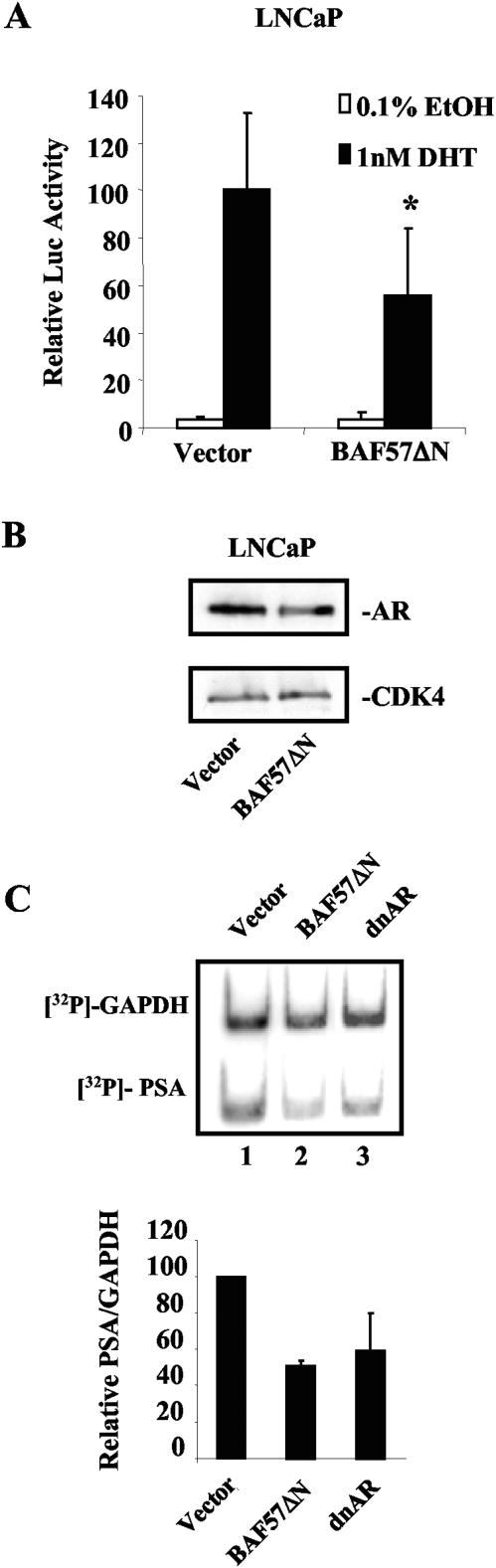
To assess the functional role of BAF57 on AR activity, a cell line deficient in BAF57 (BT549) was utilized. Although null for BAF57 (Fig. 3A), these cells do express other core SWI/SNF proteins (including both BRM and BRG1) (3, 13). In contrast, MCF7, LNCaP, and CV1 cells are competent for both BAF57 expression (Fig. 3A) and SWI/SNF function (43). To compare relative AR activity in these lines, reporter assays were used with a reporter for a well-described AR target gene, probasin (pARR2-Luc) (76). Transfections and reporter analyses were performed as indicated in the Materials and Methods section. As shown in Fig. 3B, MCF7 and CV1 cells demonstrated robust AR activity (63- and 65-fold inductions, respectively). LNCaP cells also exhibited strong activation of AR target genes (Fig. 4A) (11, 76). In contrast, in the BAF57-deficient BT549 cell line, AR activation was significantly impaired (only a fourfold increase versus the EtOH control) (Fig. 3B). This deficiency was not attributed to a lack of ectopic AR expression, as determined by immunoblot (Fig. 3C, compare lanes 1 and 2) (GFP expression denotes transfection). To establish the role of BAF57 in this aberrant response, reporter analyses were repeated in the presence of restored BAF57. Addition of BAF57 to BT549 cells restored ligand-dependent AR activation (Fig. 3D), indicating a functional role for BAF57 in regulating AR activity. The difference in pARR2-Luc activity was not attributed to altered AR expression levels, since parallel experiments with a GFP expression plasmid (pGreenLantern) as an internal control demonstrated equal AR protein levels in the presence or absence of BAF57 (Fig. 3E, compare lanes 1 and 2).
FIG. 3.
BAF57 is critical for AR activity. (A) MCF7 (lane 1), BT549 (lane 2), LNCaP (lane 3), and CV1 (lane 4) cells were harvested, lysed, and subjected to SDS-PAGE. Immunoblots were performed with antibodies to BAF57 and CDK4 (loading control). (B) MCF7, CV1, and BT549 cells (2 × 105) were transfected with 0.5 μg of pARR2-Luc reporter, 0.5 μg of pSG5AR, and 0.25 μg of pTK-Renilla luciferase (to normalize for transfection efficiency). Posttransfection, cells were stimulated with either 0.1% EtOH vehicle or 0.1 nM DHT for 24 h. Cells were harvested, lysed, and monitored for luciferase activity by utilizing the Dual Luciferase Assay Reporter System. AR activity is presented as the fold activation by ligand (DHT) versus EtOH. Averages and standard deviations are shown. (C) BT549 cells either untransfected (lane 1) or transfected as described above with pGreenLantern (indicating transfection) and pSG5AR (lane 2) were harvested 24 h posttransfection. Immunoblot analysis demonstrates AR expression in transfected BT549 cells. (D) To elucidate BAF57 function on AR reporter activity, BT549 cells (2 × 105) were transfected with 0.5 μg of pARR2-Luc reporter, 0.5 μg of pSG5AR, 0.25 μg of pTK-Renilla luciferase, and either 2.25 μg of parental vector or pBabe-WTBAF57. Posttransfection, cells were treated with either 0.1% EtOH vehicle or 0.1 nM DHT for 24 h. Cells were harvested and lysed, and the luciferase activity was measured by using the dual Luciferase Assay Reporter System. Ligand-induced AR activity in the presence of vector was set to 100, and the relative luciferase activity is shown. (E) BT549 cells (2 × 105) were transfected with 0.5 μg of pGreenLantern GFP (transfection control), 0.5 μg of SG5AR, and either 2.25 μg of vector (lane 1) or BAF57 expression plasmid (lane 2). Immunoblotting was performed for AR and GFP at 24 h posttransfection.
FIG.4.
Inhibition of BAF57 results in reduced endogenous AR activity. (A) LNCaP cells (2 × 105) were transfected with 1 μg of pARR2-Luc reporter, 0.25 μg of pTK-Renilla luciferase, and either 2.25 μg of pBabe-BAF57ΔN (dominant-negative BAF57) or parental vector (to 4.5 μg of total DNA). Cells were treated with either 0.1% EtOH vehicle or 1 nM DHT for 48 h (with restimulation after 24 h) and then harvested and lysed. The luciferase activity was measured as in Fig. 3D, and the AR activity in the presence of vector alone plus DHT was set to 100. The average relative luciferase activity and standard deviations are shown. ✽, P < 0.05. (B) LNCaP cells transfected with either vector (lane 1) or BAF57ΔN (lane 2) were harvested, and the expression of endogenous AR and CDK4 (loading control) was determined by immunoblotting. (C) LNCaP cells (4.5 × 105) were transfected with 1 μg of pBabe-PURO, 1 μg of H2B-GFP, and 4 μg of parental vector (lane 1), BAF57ΔN (lane 2), or dominant-negative AR (dnAR) (lane 3) expression plasmids. Posttransfection, cells were selected with puromycin until >90% of cells were H2B-GFP positive (ca. 3 days). Cells were then harvested, and RNA was isolated and subjected to RT-PCR. cDNAs were amplified in the presence of [32P]dCTP to accurately quantify the signal (upper panel). Product levels were quantified by using a PhosphorImager. Relative levels of PSA over GAPDH from two independent experiments, and standard deviations are shown (lower panel).
Inhibition of BAF57 results in reduction of endogenous AR activity.
To further elucidate the function of BAF57 on AR target gene transcription, BAF57 activity was inhibited in the AR-positive prostatic adenocarcinoma cell line, LNCaP. These cells are dependent on AR for proliferation and express BAF57 (Fig. 3A). A well-characterized dominant-negative allele of BAF57, BAF57ΔN, was utilized to suppress endogenous function. BAF57ΔN lacks the proline-rich region and the HMG domain (N terminus) and has been shown to block BAF57 activity and downregulate endogenous BAF57 expression (via immunoblot analysis) in the thymocytes of BAF57ΔN transgenic mice (9). Importantly, this allele has been shown to retain association with SWI/SNF. For these experiments, LNCaP cells were transfected in the absence of ligand with pARR2-Luc and either pBabe-BAF57ΔN or vector control to monitor the impact on endogenous AR activity (Fig. 4A). As expected, ligand induced robust activation of the pARR2-Luc target gene by endogenous AR in the presence of empty vector (28-fold over basal activity). In contrast, there was a significant reduction in endogenous AR activation of the reporter upon transfection of pBabe-BAF57ΔN (an ∼2-fold reduction in activity), thereby demonstrating the importance of BAF57 for endogenous AR function. Furthermore, endogenous AR levels remained constant upon transfection of vector control and pBabe-BAF57ΔN relative to control (CDK4) (Fig. 4B). Transient introduction of pBabe-BAF57ΔN into LNCaP cells demonstrated no effect on BAF57 expression levels (data not shown), thus indicating that in this system, BAF57-ΔN functions through its defined dominant-negative capabilities. Ectopic expression of wild-type BAF57 has been previously demonstrated to also act in a dominant-negative fashion in cell lines containing endogenous BAF57, presumably by titrating endogenous complexes (3). Combined, our observations indicate that inhibition of BAF57 activity reduces AR function.
To verify this hypothesis, the effect of BAF57ΔN on an endogenous AR target gene (PSA) was examined. For these experiments, LNCaP cells were transfected with pBabe-puro (encoding a puromycin resistance marker), pEGFP H2B-GFP (to monitor transfection and/or selection), and either vector control, pBabe-BAF57ΔN, or pSG5-ARΔ46-408 (dominant-negative AR) as a positive control. After rapid selection with puromycin, RNA was isolated, and RT-PCR amplification was performed in the presence of [32P]dCTP to examine PSA mRNA expression (Fig. 4C). As expected, dnAR significantly reduced PSA mRNA expression, relative to vector control (upper panel, compare lanes 1 and 3). Strikingly, BAF57-ΔN similarly attenuated PSA expression (upper panel, compare lanes 1 and 2). Quantification of these data indicate that BAF57-ΔN inhibits PSA expression similar to dominant-negative AR (lower panel). Together, these data confirm that disruption of BAF57 function inhibits AR activity on both exogenous and endogenous AR targets.
SWI/SNF ATPase activity is required for BAF57 function.
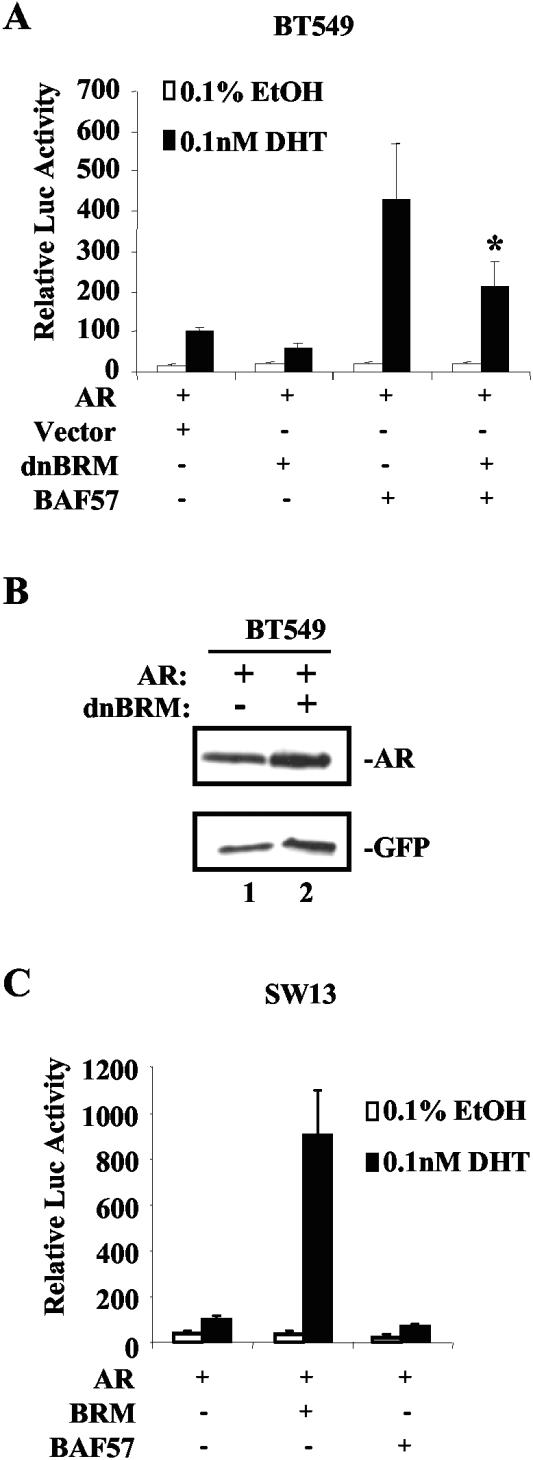
Although these data indicate a requirement of BAF57 for AR transactivation potential, the necessity of SWI/SNF ATPase activity for BAF57 function has not been demonstrated. This point is particularly salient since BAF57 can bind DNA independent of the SWI/SNF complex (71). Therefore, we initially tested the ability of BAF57 to promote AR function in coordination with the preferred core ATPase, BRM, in BT549 cells. Although null for BAF57, BT549 cells have been shown previously to express the BRM ATPase (13). Ectopic expression of dominant-negative BRM (pCG-dnBRM) was utilized to examine the impact of the core ATPase in the absence of BAF57. This allele of BRM has been previously described (45) and encodes an ATPase defective mutant protein which can still assemble into SWI/SNF but fails to remodel chromatin. Consistent with our previous observations (Fig. 3D), ligand induced a similar fold induction (i.e., eightfold) over basal activity (EtOH control) (Fig. 5A). Moreover, dnBRM failed to promote receptor activity and slightly inhibited residual AR activity. This may be indicative of the ability of SWI/SNF to regulate AR activity even in the absence of BAF57 (albeit inefficiently). As expected, BAF57 restored AR activation (Fig. 5A). However, BAF57-mediated AR activation was significantly reduced upon expression of dnBRM, thus indicating that BAF57 requires SWI/SNF ATPase function to efficiently promote AR activity. Parallel experiments with GFP expression as an internal control demonstrated that expression of dnBRM had no impact on AR protein levels (Fig. 5B). To verify the requirement of BAF57 for SWI/SNF function, SW13 cells were utilized. These cells are null for both BRG1 and BRM and thus lack SWI/SNF activity. However, these cells do express BAF57 (13). Consistent with our published findings (43), SW13 cells failed to support significant AR activity in reporter assays (Fig. 5C); however, expression of BRM restored AR function. Moreover, BAF57 failed to promote AR activity in this model system, thus demonstrating that BAF57 action is dependent on a functional SWI/SNF complex.
FIG.5.
BAF57 action is dependent upon SWI/SNF ATPase activity. (A) BT549 cells (2 × 105) were transfected with 0.5 μg of pARR2-Luc reporter, 0.25 μg of pTK-Renilla luciferase, 0.5 μg of pSG5AR, and 2.25 μg of pBabe-WTBAF57, 1 μg of dominant-negative BRM (dnBRM), or parental vector (to 4.5 μg of total DNA) as indicated. Cells were treated posttransfection with 0.1% EtOH or 0.1 nM DHT for 24 h. Cells were harvested and lysed, and the dual luciferase activity was measured. Ligand-activated AR activity was set to 100, and relative luciferase activity and standard deviations are shown. ✽, P < 0.05. (B) BT549 cells were transfected with 0.5 μg of SG5AR, 0.5 μg of pGreenLantern GFP, and 1 μg of vector (lane 1) or dnBRM (lane 2) expression plasmid. Cell lysates were obtained, and immunoblot analysis was performed for AR and GFP. (C) SW13 cells (2 × 105) were transfected with 0.5 μg of pARR2-Luc reporter, 0.25 μg of pTK-Renilla luciferase, 0.5 μg of pSG5AR, and 2.25 μg of pBabe-WTBAF57, 1 μg of BRM, and parental vector (to 4.5 μg of total DNA) as indicated. Posttransfection, cells were treated with 0.1% EtOH or 0.1 nM DHT for 24 h. Cells were harvested, lysed, and dual luciferase activity was measured. Ligand-activated AR activity was set to 100, and relative luciferase activity and standard deviations are shown.
BAF57 is recruited to AR target regulatory regions in the presence of ligand.
Having demonstrated the requirement of BAF57 for AR function, we examined the recruitment of BAF57 to paradigm AR targets in the prostate using ChIP. Isolated chromatin from LNCaP cells was immunoprecipitated with antibodies directed against either AR, BAF57 (either BAF57h [Fig. 6B], directed against the HMG domain, or BAF57c [Fig. 6C], directed against the C terminus), acetylated histone H4, or a preimmune serum. Recruitment of protein to the PSA regulatory region was determined by using primers specific to the PSA enhancer encompassing a well-defined ARE (11, 59). A similar result was observed at an ARE contained within the proximal PSA promoter (data not shown). PCR amplification was performed in the presence of [32P]dCTP and quantified by using autoradiography (Fig. 6A and B, right panels) as previously described (60). As shown (Fig. 6A), minimal AR or acetylated histone H4 could be detected at the PSA enhancer in the presence of EtOH vehicle. However, upon ligand stimulation, AR was recruited to the PSA enhancer, and histone acetylation was apparent (compare lanes 1 and 2). These data are consistent with previously published reports (59). As shown in Fig. 6B, BAF57 was detected at the PSA enhancer and was significantly enriched after ligand stimulation (compare lanes 1 and 2). To further confirm the recruitment of BAF57 to AR targets, ChIP analyses were repeated with an LNCaP derived cell line, LB1-Luc. LB1-Luc cells contain a stably integrated pARR2-Luc promoter as described in Materials and Methods. As can be seen in Fig. 6C, these cells exhibit robust AR activity in a dose-dependent manner upon the addition of ligand (left panel). ChIP analysis with isolated chromatin from the LB1-Luc cells revealed the recruitment of AR and BAF57 to the pARR2-Luc promoter upon stimulation with DHT (right two panels). Taken together, these data confirm the presence of BAF57 on AR regulatory regions in the presence of ligand.
BAF57 functions cooperatively with p160 HAT coactivators to activate transcription of AR target genes.
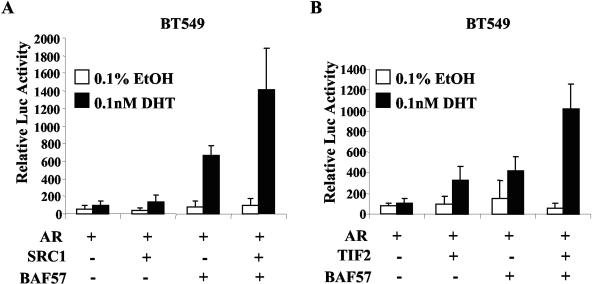
A role for BAF57 has been previously established in p160 HAT coactivator mediated function, using ER as a model (3). Interestingly, p160 coactivators have been shown to target histones H3 and H4 for acetylation (38, 42, 64). Since BAF57 recruitment was observed when histones were acetylated (Fig. 6), we analyzed the impact of BAF57 in coordination with p160 coactivators (Fig. 7) to enhance AR activity. For these experiments, reporter analyses were performed in BT549 cells as in Fig. 3. As shown, SRC1 expression failed to generate significant AR activity in the presence of ligand compared to vector control. As expected, BAF57 restored significant AR activity, a finding consistent with our previous results (Fig. 3D). However, the combination of SRC1 and BAF57 led to markedly enhanced AR activity (10.3-fold over SRC1 alone and 2.1-fold over BAF57 alone) (Fig. 7A). Interestingly, unlike SRC1 alone, TIF2 by itself appeared to significantly enhance AR activity similar to BAF57 alone (Fig. 7B). However, a similar profile to SRC1 was observed upon cotransfection of TIF2 and BAF57 (3.2-fold versus TIF2 alone and 2.5-fold versus BAF57 alone). These data indicate that BAF57 may cooperate with p160 coactivators to enhance AR activity.
FIG. 7.
BAF57 cooperates with p160 coactivators. BT549 cells (2 × 105) were transfected with 0.5 μg of pARR2-Luc reporter, 0.25 μg of pTK-Renilla luciferase, 0.5 μg of pSG5AR, and 2.25 μg of pBabe-WTBAF57, as well as 1 μg of coactivator (pRC3.1-SRC1 [A] or pCMV-TIF2 [B]) or parental vector (to 4.5 μg of total DNA) as indicated. Posttransfection, cells were treated with 0.1% EtOH or 0.1 nM DHT for 24 h. Cells were harvested and lysed, and the dual luciferase activity was measured. Ligand-activated AR activity was set to 100, and the average relative luciferase activity and standard deviations are shown.
BAF57 facilitates non-p160 AR coactivator function.
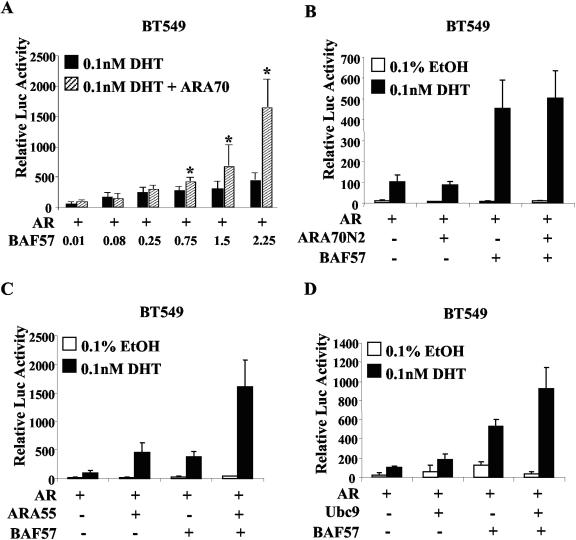
Upon establishing a role for BAF57 in p160 coactivator AR function, BAF57 influence on non-p160 coactivator function was examined. BT549 cells were utilized and transfected with the indicated effector plasmids as described above. Interestingly, ARA70 supported a dose-dependent response to BAF57 (Fig. 8A). A significant induction of AR activity was stimulated by addition of both ARA70 and BAF57 (versus BAF57 alone) at higher concentrations of BAF57, as indicated. A more than additive effect was observed at the highest doses (8.4-fold over ARA70 alone [2-fold over EtOH; data not shown] and 3.6-fold over BAF57 alone). In contrast, a mutant of ARA70 which retains the ability to interact with AR through the FXXLF AR interacting motif but lacks the ability to coactivate AR activity (ARA70N2) failed to cooperate with BAF57 or induce AR activity (29) (Fig. 8B). These observations indicate that a functional coactivator is necessary for enhanced AR activation by BAF57. As shown in Fig. 8C, BAF57 also facilitated ARA55 activity, stimulating AR activity 3.5-fold over ARA55 alone and 4.2-fold over BAF57 alone (Fig. 8C). Lastly, the impact of BAF57 on the E2 SUMO-conjugating enzyme Ubc9 was assessed. Ubc9 activates AR in a manner independent of its ability to conjugate SUMO-1 to AR through poorly defined mechanisms (51). Similar to the selected SRC1, Ubc9 alone failed to significantly activate AR target gene transcription, but an enhanced effect was observed in combination with BAF57 (5-fold over Ubc9 alone and 1.7-fold over BAF57 alone), indicating BAF57 is able to facilitate Ubc9 coactivator function (Fig. 8D). Together, these data indicate that the Ubc9 requirement for BAF57 is similar to that of the p160 coactivators. In contrast, whereas ARA70 and ARA55 do not require BAF57, they synergize with BAF57 for AR activation.
FIG. 8.
BAF57 facilitates non-p160 AR coactivator function. (A) BT549 cells (2 × 105) were transfected with 0.5 μg of pARR2-Luc reporter, 0.25 μg of pTK-Renilla luciferase, 0.5 μg of pSG5AR, and 0.01 to 2.25 μg of pBabe-WTBAF57 in the absence (▪) or presence of 1 μg of pSG5ARA70 (▨). As described in Materials and Methods, all transfections were brought to a total 4.5 μg by using empty vector. Posttransfection, cells were treated with 0.1% EtOH or 0.1 nM DHT for 24 h. Cells were harvested and lysed, and the luciferase activity was measured. The ligand-activated AR activity was set to 100 (data not shown), and the average relative luciferase activity and standard deviations are shown (P < 0.05). (B to D) BT549 cells (2 × 105) were transfected with 0.5 μg of pARR2-Luc reporter, 0.25 μg of pTK-Renilla luciferase, 0.5 μg of pSG5AR, and 2.25 μg of pBabe-WTBAF57, vector, or the indicated coactivator (pSG5ARA55 and pFLAG-Ubc9) or mutant (pcDNA3-ARA70N2) expression plasmid (1 μg). Posttransfection, cells were treated with 0.1% EtOH or 0.1 nM DHT for 24 h. Cells were harvested and lysed, and luciferase activity was measured. The ligand-activated AR activity in the absence of BAF57 was set to 100, and the average relative luciferase activity and standard deviations are shown (P < 0.05).
Inhibition of either BRM or BAF57 attenuates AR-dependent LNCaP cell proliferation.
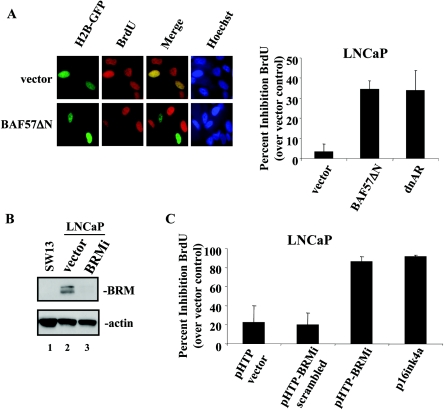
Our studies demonstrate a specific requirement of BAF57 for both ligand-induced and coactivator-enhanced AR activation. Since AR activity is essential for the proliferation of prostate cancer cells, we examined the necessity of both BRM and BAF57 in this process. Initially, BAF57 function was depleted via transfection of the dominant-negative expression plasmid (pBabe-BAF57ΔN) into LNCaP cells. This approach was utilized since attempts to target BAF57 using siRNA technology have not yet proven successful (data not shown). This experiment was performed by transfection of LNCaP cells with pEGFP-H2B-GFP (to mark transfected cells) and either parental vector, pBabe-BAF57ΔN, or pSG5ARΔ46-408 (dnAR). As expected, dnAR attenuated LNCaP cell proliferation (34% inhibition) (Fig. 9A, right panel). Representative data from two independent experiments are shown in Fig. 9A (left panel) and demonstrate that cells transfected with pBabe-BAF57ΔN were dramatically inhibited for BrdU incorporation. The percent inhibition of BrdU incorporation over vector control was measured (Fig. 9A, right panel). Interestingly, inhibition of BAF57 slows proliferation of LNCaP cells (35% inhibition) over vector control in a manner similar to the positive control, dnAR.
FIG. 9.
Inhibition of BAF57 or BRM attenuates AR-dependent proliferation. (A) LNCaP cells (105) were transfected with 0.5 μg of pEGFP-H2B-GFP and 4.5 μg of parental vector, pBabe-BAF57ΔN, or dominant-negative AR (pSG5AR-Δ46-408). At 2 days posttransfection, cells were labeled with BrdU for 16 h and fixed. The percentage of transfected cells (H2B-GFP positive) incorporating BrdU was determined (left panel, representative image). The effect of dnAR or BAF57ΔN is shown as the percent inhibition of BrdU incorporation over vector control (right panel). (B) LNCaP cells were transfected with 0.5 μg of H2B-GFP and 4.5 μg of either pHTP-vector (lane 2) or pHTP-BRMi (lane 3). After transfection, cells were selected by using puromycin. Once cells were >90% H2B-GFP positive, they were lysed and subjected to immunoblot analyses for BRM and actin (loading control). SW13 cells (lane 1) are shown as a positive control for BRM deficiency. (C) LNCaP cells were transfected with 0.5 μg of H2B-GFP and 4.5 μg of parental vector, pHTP-BRMi scrambled, pHTP-BRMi, or pCMV-p16ink4a. The percentage of transfected cells (H2B-GFP positive) incorporating BrdU was determined. The effect of the BRM attenuation is shown as the percent inhibition of BrdU incorporation over vector control.
To examine the role of the ATPase, BRM expression was targeted by using siRNA technology. The efficacy of the construct was demonstrated by immunoblotting, as seen in Fig. 9B, wherein LNCaP cells were transfected with pHTP parental vector or pHTP-BRMi (both of which confer puromycin resistance), as indicated, and immunoblotted to detect endogenous BRM. As shown, transfection of pHTP-BRMi resulted in significant knockdown of endogenous BRM in LNCaP cells compared to vector (compare lanes 2 and 3). SW13 cells were included as a negative control, and actin was detected to demonstrate equal loading. The effect of BRM ablation on LNCaP cellular proliferation was subsequently assessed by BrdU incorporation (Fig. 9C). Cells were transfected as indicated, pEGFP-H2B-GFP-positive (transfected) cells were screened for BrdU incorporation, and impact on proliferation was plotted as the percent inhibition of BrdU incorporation versus vector control (Fig. 9C). As expected, expression of p16ink4a significantly inhibited LNCaP cell growth (4.1-fold over vector and 4.7-fold over scrambled siRNA control). The proliferation of LNCaP cells transfected with expression plasmid for the BRM siRNA was significantly reduced compared to vector control (3.9-fold reduction) or scrambled BRM siRNA control (4.4-fold reduction). Thus, the inhibition of BRM severely retards LNCaP cellular proliferation. Together, these data clearly establish a requirement of both BAF57 and the BRM ATPase for proliferation of AR-dependent prostate cancer cells.
DISCUSSION
The SWI/SNF chromatin remodeling complex is involved in activating gene transcription of nuclear receptor targets. We have previously shown that AR requires SWI/SNF for activity, although the mechanisms underlying this requirement remained obscure. Here, we identify the BAF57 subunit as a major regulator of SWI/SNF effects on the AR in the prostate. We show that BAF57 is expressed in the AR-positive luminal epithelia of the prostate (Fig. 1), which are dependent on serum androgen for proliferation and survival. Subsequently, a ligand independent in vitro interaction was observed between BAF57 and AR, and AR was found to be in association with BAF57 in vivo (Fig. 2). Combined, these data indicated that BAF57 may be the mechanism by which SWI/SNF interfaces with the AR. Consistent with this hypothesis, cells deficient in BAF57 exhibited significant defects in AR activity, and rescue of BAF57 in such cells effectively restored AR activity (Fig. 3). These findings were confirmed in AR-dependent prostate cancer cells, wherein expression of the endogenous AR target, PSA, was attenuated to a level similar to that observed with dominant-negative AR (Fig. 4). Collectively, these observations define a requirement of BAF57 for AR transactivation potential. This function of BAF57 was shown to be dependent on SWI/SNF function, since either loss or inhibition of BRM ATPase activity significantly compromised BAF57 action (Fig. 5). Although the in vitro interaction between AR and BAF57 occurred independent of ligand, we show that BAF57 is recruited to endogenous AR targets in response to ligand activation, thus supporting the hypothesis that BAF57 regulates ligand-dependent AR activation (Fig. 6). The impact of BAF57 was not limited to AR alone, since BAF57 also influenced AR coactivator function. Active BAF57 was essential for AR potentiation exerted by either SRC1 or Ubc9 and, moreover, BAF57 cooperated with this selected subset of coactivators for AR activation. In contrast, although BAF57 was not essential for TIF2, ARA70, or ARA55 AR coactivator function, BAF57 facilitated the ability of these coactivators to induce AR activation (Fig. 7 and 8). Given the observed requirement of BAF57 for AR and coactivator function, we examined the requirement of BAF57 and SWI/SNF for proliferation. Strikingly, inhibition of either BAF57 or BRM function severely retarded proliferation in AR-dependent prostate cancer cells (Fig. 9). Thus, these data establish a critical role for BAF57-mediated SWI/SNF function in AR-dependent cells.
Although the SWI/SNF complexes are widely expressed, their function results in a multitude of diverse biological outcomes. For example, association of SWI/SNF with the GR is required for GR-mediated transcriptional activation (45, 74). In contrast, both core ATPase subunits (BRG1 and BRM) associate with the retinoblastoma tumor suppressor protein, RB, to regulate RB-dependent transcriptional repression of E2F target genes. This mechanism of repressive action is controversial in that it has been demonstrated to occur through direct interaction between the ATPase and RB (15, 54, 65, 66, 75), as well as through upregulation of the cdk inhibitor p21cip1 (36). This function of RB is essential for its antiproliferative function, and loss of SWI/SNF association renders all tested cells resistant to RB-mediated growth arrest (54, 66). Thus, SWI/SNF can confer either transcriptional activation or repression. Specificity for SWI/SNF action is purported to be governed by the biochemical diversity and plasticity of SWI/SNF subunit composition, wherein selected subunits likely regulate tissue or cell-type-specific protein-protein interactions.
Targeted deletion of the core ATPases in mouse model systems first revealed nonredundancies in SWI/SNF action. Deletion of BRG1 is an embryonic lethal event (7), whereas BRM−/− mice are viable and demonstrate increased body weight (55). Intriguingly, BRM-null mice exhibited slightly reduced testicular weight, indicating a putative defect in androgen-dependent development (55). A specificity for the ATPase has also been shown biochemically, wherein BRG1 was shown to preferentially regulate selected zinc finger proteins, whereas BRM interacted preferentially with members of the Notch pathway (35). Variations in cell type expression may partially underlie specificity of ATPase action, since BRG1 is primarily expressed in proliferating or self-renewing cells, and BRM protein levels are enriched in differentiated cell types. Differentiated prostatic epithelia and prostatic adenocarcinoma cells require AR for survival and, consistent with the role of BRM in such cell types, we have shown that the AR is preferentially activated by BRM-containing SWI/SNF complexes (43). Furthermore, in support of this observation, normal human prostate epithelia were recently shown to express the BRM ATPase; in contrast, BRG1 was not detected (53). Thus, the core ATPase lends some specificity to SWI/SNF modulation of the AR. However, no direct interaction between the AR and BRM was observed in vitro (data not shown), thus indicating that SWI/SNF action must be either indirect or through association with alternate SWI/SNF subunits.
The ability of SWI/SNF subunits to direct the complex to sequence-specific transcriptional complexes and thus regulate discrete biological processes has strong precedent. For example, loss of the SNF5 subunit is associated with human rhabdoid tumorigenesis, and SNF5−/− mice develop lymphomas or rhabdoid tumors with complete penetrance (56). SNF5 tumor suppressor activity is thought to occur in part through the cell cycle inhibitor p16ink4a (5, 48). The BAF250 subunit, which contains intrinsic DNA-binding capabilities through an AT-rich DNA-binding domain, directs SWI/SNF activity to the GR (47). Thus, BAF250 may utilize both DNA and protein-binding functions in combination to confer specificity of action. In contrast, BAF60a likely utilizes two distinct protein-binding surfaces (the first for GR and BRG1 and the second for BAF155 and BAF170) to regulate GR specificity (28). BAF60a has also been shown to function as to discriminately activate a subset of Fos/Jun dimers, thus governing specificity of AP-1 action (33). Lastly, BAF57 harbors multiple activities that likely assist in the regulation of sequence-specific DNA-binding factors (2, 3, 71).
BAF57 is composed of an N-terminal HMG DNA-binding domain and a kinesin-like coiled-coil region. Given this structure and its ability to bind four-way junction DNA, BAF57 has been suggested as a potential targeting subunit of the SWI/SNF complex that regulates selected target genes (71). This hypothesis has been tested with the ΔHMG allele of BAF57, which does not disrupt SWI/SNF complex formation or SWI/SNF recruitment but rather lacks DNA-binding potential. Transgenic mice expressing this allele in T cells demonstrate a specific derepression of the CD4 locus (9). We report here that BAF57 interacts directly with the AR and activates AR function through SWI/SNF. Although AR can bind BAF57 independent of ligand (similar to what was reported for GR [28]), BAF57 was recruited to AR targets in a ligand-dependent manner. Since we have also observed that BAF57 resides predominantly in the nucleus (data not shown), interaction between AR and BAF57 is hypothesized to occur in this cellular compartment. Mechanisms regulating the AR-BAF57 interaction will be the subject of future studies. The ER has also been shown to interact with BAF57, but ER binding is enhanced by ligand, thus indicating a disparate mode of interaction. In support of this view, ER was shown to retain transactivation capacity in BAF57 null cells (3), whereas AR activity was markedly reduced by BAF57 loss or inhibition. The requirement of BAF57 for AR was apparent in both transient assays and upon analysis of PSA, an endogenous AR target gene. It is of interest that previous analyses of SWI/SNF effects on this AR target revealed that in transient assays with an ectopically expressed PSA regulatory region, the presence of the PSA enhancer relieved the requirement for SWI/SNF for AR function (43), thus indicating that DNA composition or architecture likely influences the necessity for SWI/SNF. Although such studies are useful for dissecting the mode of SWI/SNF action on discrete templates, the data shown herein establish a solid requirement for BAF57 and SWI/SNF action on prostate-specific AR targets in the context of chromatin.
Independent of nuclear receptor association, BAF57 has been shown to regulate coactivator function. Specifically, BAF57 has been shown to interact directly with the p160 coactivators and is required for the ability of these coactivators to modulate ER function (3). Similarly, we show that SRC1 maintains little AR coactivation function in the absence of BAF57 and cooperates with BAF57 to augment AR activity. In contrast to SRC1, TIF2 maintains activity in the absence of BAF57; however, the addition of BAF57 facilitates AR action similar to SRC1. Surprisingly, potentiation of AR by Ubc9 demonstrated a similar requirement, indicating that the BAF57 requirement extends beyond the p160 class of coactivators. Although the mechanism by which Ubc9 modulates AR remains elusive, these data put forth the hypothesis that Ubc9 may activate p160 function. In contrast, ARA70 and ARA55 potentiated AR gene transcription independent of BAF57 and actually synergized with BAF57 for AR activation. Thus, this class of coactivators maintain function independent of SWI/SNF. Collectively, these data indicate that, like the nuclear receptors, coactivators demonstrate discrete requirements for SWI/SNF function. Given the importance of selected coactivators in prostate cancer progression, knowledge of SWI/SNF requirement will be an important component of targeting coactivator function in cancer therapy.
Inhibition of AR activity is the main line of treatment for prostate cancer therapy, and recurrent, late-stage tumors are hallmarked by inappropriate restoration of AR activity (1, 34). We have shown that inhibition of either BRM or BAF57 significantly retards proliferation in prostate cancer cells. These observations again underscore the plasticity of the SWI/SNF requirement, since loss of SWI/SNF ATPase activity or selected subunits is often associated with growth advantage, such as is observed upon SNF5 loss or in response to RB upon loss of SWI/SNF ATPase activity (5, 54, 56, 66, 69). Together, these data implicate a specific response for BAF57 in AR-dependent growth and further identify the importance of the BAF subunits in SWI/SNF function.
In summary, these data reveal an essential role for BAF57 in regulating SWI/SNF modification of AR. Moreover, our studies demonstrate that BAF57 strongly influences the impact of coactivators on AR activation. These functions of BAF57 are critical, since inhibition of BAF57 or BRM function blocked proliferation of androgen-dependent prostate cancer cells. Together, these studies implicate BAF57 as a major regulator of AR and prostate cancer proliferation, thus identifying BAF57 as a valid target for potential therapeutic intervention.
Acknowledgments
We thank Weidong Wang for the generous gift of the BAF57 antibodies. We thank Erik Knudsen, Christin Petre, Chris Mayhew, Hasan Siddiqui, and Janet Hess-Wilson for critical reading of the manuscript. We thank Robert Matusik, Tom Case, Simon Hayward, and Janni Mirosevich for training in prostate tissue microdissection and immunohistochemistry.
This study was supported by DAMD 02-1-0037 (K.E.K.) and RO1CA099996 (K.E.K.). B.W., G.R., and E.H. are supported by grants CA91048 and CA102848. K.A.L. is supported by NIEHS training grant 2 T32 ES07250-16.
REFERENCES
- 1.Balk, S. P. 2002. Androgen receptor as a target in androgen-independent prostate cancer. Urology 60:132-139. [DOI] [PubMed] [Google Scholar]
- 2.Battaglioli, E., M. E. Andres, D. W. Rose, J. G. Chenoweth, M. G. Rosenfeld, M. E. Anderson, and G. Mandel. 2002. REST repression of neuronal genes requires components of the hSWI.SNF complex. J. Biol. Chem. 277:41038-41045. [DOI] [PubMed] [Google Scholar]
- 3.Belandia, B., R. L. Orford, H. C. Hurst, and M. G. Parker. 2002. Targeting of SWI/SNF chromatin remodeling complexes to estrogen-responsive genes. EMBO J. 21:4094-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belandia, B., and M. G. Parker. 2003. Nuclear receptors: a rendezvous for chromatin remodeling factors. Cell 114:277-280. [DOI] [PubMed] [Google Scholar]
- 5.Betz, B. L., M. W. Strobeck, D. N. Reisman, E. S. Knudsen, and B. E. Weissman. 2002. Re-expression of hSNF5/INI1/BAF47 in pediatric tumor cells leads to G1 arrest associated with induction of p16ink4a and activation of RB. Oncogene 21:5193-5203. [DOI] [PubMed] [Google Scholar]
- 6.Brinkmann, A. O., L. J. Blok, P. E. de Ruiter, P. Doesburg, K. Steketee, C. A. Berrevoets, and J. Trapman. 1999. Mechanisms of androgen receptor activation and function. J. Steroid Biochem. Mol. Biol. 69:307-313. [DOI] [PubMed] [Google Scholar]
- 7.Bultman, S., T. Gebuhr, D. Yee, C. La Mantia, J. Nicholson, A. Gilliam, F. Randazzo, D. Metzger, P. Chambon, G. Crabtree, and T. Magnuson. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6:1287-1295. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi, T. H., M. Wan, K. Zhao, I. Taniuchi, L. Chen, D. R. Littman, and G. R. Crabtree. 2002. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature 418:195-199. [DOI] [PubMed] [Google Scholar]
- 10.Chiba, H., M. Muramatsu, A. Nomoto, and H. Kato. 1994. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 22:1815-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleutjens, K. B., H. A. van der Korput, C. C. van Eekelen, H. C. van Rooij, P. W. Faber, and J. Trapman. 1997. An androgen response element in a far upstream enhancer region is essential for high, androgen-regulated activity of the prostate-specific antigen promoter. Mol. Endocrinol. 11:148-161. [DOI] [PubMed] [Google Scholar]
- 12.Collingwood, T. N., F. D. Urnov, and A. P. Wolffe. 1999. Nuclear receptors: coactivators, corepressors and chromatin remodeling in the control of transcription. J. Mol. Endocrinol. 23:255-275. [DOI] [PubMed] [Google Scholar]
- 13.Decristofaro, M. F., B. L. Betz, C. J. Rorie, D. N. Reisman, W. Wang, and B. E. Weissman. 2001. Characterization of SWI/SNF protein expression in human breast cancer cell lines and other malignancies. J. Cell Physiol. 186:136-145. [DOI] [PubMed] [Google Scholar]
- 14.DiRenzo, J., Y. Shang, M. Phelan, S. Sif, M. Myers, R. Kingston, and M. Brown. 2000. BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation. Mol. Cell. Biol. 20:7541-7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunaief, J. L., B. E. Strober, S. Guha, P. A. Khavari, K. Alin, J. Luban, M. Begemann, G. R. Crabtree, and S. P. Goff. 1994. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79:119-130. [DOI] [PubMed] [Google Scholar]
- 16.Feldman, B. J., and D. Feldman. 2001. The development of androgen-independent prostate cancer. Nat. Rev. Cancer 1:34-45. [DOI] [PubMed] [Google Scholar]
- 17.Flaus, A., and T. Owen-Hughes. 2001. Mechanisms for ATP-dependent chromatin remodeling. Curr. Opin. Genet. Dev. 11:148-154. [DOI] [PubMed] [Google Scholar]
- 18.Fletcher, T. M., N. Xiao, G. Mautino, C. T. Baumann, R. Wolford, B. S. Warren, and G. L. Hager. 2002. ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling. Mol. Cell. Biol. 22:3255-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fryer, C. J., and T. K. Archer. 1998. Chromatin remodeling by the glucocorticoid receptor requires the BRG1 complex. Nature 393:88-91. [DOI] [PubMed] [Google Scholar]
- 20.Fu, M., C. Wang, J. Wang, X. Zhang, T. Sakamaki, Y. G. Yeung, C. Chang, T. Hopp, S. A. Fuqua, E. Jaffray, R. T. Hay, J. J. Palvimo, O. A. Janne, and R. G. Pestell. 2002. Androgen receptor acetylation governs trans activation and MEKK1-induced apoptosis without affecting in vitro sumoylation and trans-repression function. Mol. Cell. Biol. 22:3373-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujimoto, N., A. Mizokami, S. Harada, and T. Matsumoto. 2001. Different expression of androgen receptor coactivators in human prostate. Urology 58:289-294. [DOI] [PubMed] [Google Scholar]
- 22.Fujimoto, N., S. Yeh, H. Y. Kang, S. Inui, H. C. Chang, A. Mizokami, and C. Chang. 1999. Cloning and characterization of androgen receptor coactivator, ARA55, in human prostate. J. Biol. Chem. 274:8316-8321. [DOI] [PubMed] [Google Scholar]
- 23.Gelmann, E. P. 2002. Molecular biology of the androgen receptor. J. Clin. Oncol. 20:3001-3015. [DOI] [PubMed] [Google Scholar]
- 24.Gregory, C. W., B. He, R. T. Johnson, O. H. Ford, J. L. Mohler, F. S. French, and E. M. Wilson. 2001. A mechanism for androgen receptor-mediated prostate cancer recurrence after androgen deprivation therapy. Cancer Res. 61:4315-4319. [PubMed] [Google Scholar]
- 25.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 26.Hayward, S. W., M. A. Rosen, and G. R. Cunha. 1997. Stromal-epithelial interactions in the normal and neoplastic prostate. Br. J. Urol. 79(Suppl. 2):18-26. [DOI] [PubMed] [Google Scholar]
- 27.Heinlein, C. A., and C. Chang. 2002. Androgen receptor (AR) coregulators: an overview. Endocrinol. Rev. 23:175-200. [DOI] [PubMed] [Google Scholar]
- 28.Hsiao, P. W., C. J. Fryer, K. W. Trotter, W. Wang, and T. K. Archer. 2003. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol. Cell. Biol. 23:6210-6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu, Y. C., S. Yeh, S. D. Yeh, E. R. Sampson, J. Huang, P. Li, C. L. Hsu, H. J. Ting, H. K. Lin, L. Wang, E. Kim, J. Ni, and C. Chang. 2004. Functional domain and motif analyses of androgen receptor coregulator ARA70 and its differential expression in prostate cancer. J. Biol. Chem. 279:33438-33446. [DOI] [PubMed] [Google Scholar]
- 30.Huang, Z. Q., J. Li, L. M. Sachs, P. A. Cole, and J. Wong. 2003. A role for cofactor-cofactor and cofactor-histone interactions in targeting p300, SWI/SNF, and Mediator for transcription. EMBO J. 22:2146-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ichinose, H., J. M. Garnier, P. Chambon, and R. Losson. 1997. Ligand-dependent interaction between the estrogen receptor and the human homologues of SWI2/SNF2. Gene 188:95-100. [DOI] [PubMed] [Google Scholar]
- 32.Inoue, H., T. Furukawa, S. Giannakopoulos, S. Zhou, D. S. King, and N. Tanese. 2002. Largest subunits of the human SWI/SNF chromatin-remodeling complex promote transcriptional activation by steroid hormone receptors. J. Biol. Chem. 277:41674-41685. [DOI] [PubMed] [Google Scholar]
- 33.Ito, T., M. Yamauchi, M. Nishina, N. Yamamichi, T. Mizutani, M. Ui, M. Murakami, and H. Iba. 2001. Identification of SWI.SNF complex subunit BAF60a as a determinant of the transactivation potential of Fos/Jun dimers. J. Biol. Chem. 276:2852-2857. [DOI] [PubMed] [Google Scholar]
- 34.Jenster, G. 1999. The role of the androgen receptor in the development and progression of prostate cancer. Semin. Oncol. 26:407-421. [PubMed] [Google Scholar]
- 35.Kadam, S., and B. M. Emerson. 2003. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell 11:377-389. [DOI] [PubMed] [Google Scholar]
- 36.Kang, H., K. Cui, and K. Zhao. 2004. BRG1 controls the activity of the retinoblastoma protein via regulation of p21CIP1/WAF1/SDI. Mol. Cell. Biol. 24:1188-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, I. Y., J. H. Kim, D. J. Zelner, H. J. Ahn, J. A. Sensibar, and C. Lee. 1996. Transforming growth factor-β1 is a mediator of androgen-regulated growth arrest in an androgen-responsive prostatic cancer cell line, LNCaP. Endocrinology 137:991-999. [DOI] [PubMed] [Google Scholar]
- 38.Kinyamu, H. K., and T. K. Archer. 2004. Modifying chromatin to permit steroid hormone receptor-dependent transcription. Biochim. Biophys. Acta 1677:30-45. [DOI] [PubMed] [Google Scholar]
- 39.Kitagawa, H., R. Fujiki, K. Yoshimura, Y. Mezaki, Y. Uematsu, D. Matsui, S. Ogawa, K. Unno, M. Okubo, A. Tokita, T. Nakagawa, T. Ito, Y. Ishimi, H. Nagasawa, T. Matsumoto, J. Yanagisawa, and S. Kato. 2003. The chromatin-remodeling complex WINAC targets a nuclear receptor to promoters and is impaired in Williams syndrome. Cell 113:905-917. [DOI] [PubMed] [Google Scholar]
- 40.Knudsen, E. S., and J. Y. Wang. 1997. Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol. Cell. Biol. 17:5771-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knudsen, K. E., K. C. Arden, and W. K. Cavenee. 1998. Multiple G1 regulatory elements control the androgen-dependent proliferation of prostatic carcinoma cells. J. Biol. Chem. 273:20213-20222. [DOI] [PubMed] [Google Scholar]
- 42.Kuo, M. H., and C. D. Allis. 1998. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20:615-626. [DOI] [PubMed] [Google Scholar]
- 43.Marshall, T. W., K. A. Link, C. E. Petre-Draviam, and K. E. Knudsen. 2003. Differential requirement of SWI/SNF for androgen receptor activity. J. Biol. Chem. 278:30605-30613. [DOI] [PubMed] [Google Scholar]
- 44.Miyamoto, H., S. Yeh, G. Wilding, and C. Chang. 1998. Promotion of agonist activity of antiandrogens by the androgen receptor coactivator, ARA70, in human prostate cancer DU145 cells. Proc. Natl. Acad. Sci. USA 95:7379-7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muchardt, C., and M. Yaniv. 1993. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 12:4279-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 47.Nie, Z., Y. Xue, D. Yang, S. Zhou, B. J. Deroo, T. K. Archer, and W. Wang. 2000. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol. Cell. Biol. 20:8879-8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oruetxebarria, I., F. Venturini, T. Kekarainen, A. Houweling, L. M. Zuijderduijn, A. Mohd-Sarip, R. G. Vries, R. C. Hoeben, and C. P. Verrijzer. 2004. P16INK4a is required for hSNF5 chromatin remodeler-induced cellular senescence in malignant rhabdoid tumor cells. J. Biol. Chem. 279:3807-3816. [DOI] [PubMed] [Google Scholar]
- 49.Palvimo, J. J., P. J. Kallio, T. Ikonen, M. Mehto, and O. A. Janne. 1993. Dominant negative regulation of trans-activation by the rat androgen receptor: roles of the N-terminal domain and heterodimer formation. Mol. Endocrinol. 7:1399-1407. [DOI] [PubMed] [Google Scholar]
- 50.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 51.Poukka, H., P. Aarnisalo, U. Karvonen, J. J. Palvimo, and O. A. Janne. 1999. Ubc9 interacts with the androgen receptor and activates receptor-dependent transcription. J. Biol. Chem. 274:19441-19446. [DOI] [PubMed] [Google Scholar]
- 52.Quigley, C. A., A. De Bellis, K. B. Marschke, M. K. el-Awady, E. M. Wilson, and F. S. French. 1995. Androgen receptor defects: historical, clinical, and molecular perspectives. Endocrinol. Rev. 16:271-321. [DOI] [PubMed] [Google Scholar]
- 53.Reisman, D. N., J. Sciariotta, T. W. Bouldin, B. E. Weissman, and W. Funkhouser, Jr. The expression of the SWI/SNF ATPase subunits BRG1 and BRM in normal human tissues. Appl. Immunohistochem. Mol. Morphol., in press. [DOI] [PubMed]
- 54.Reisman, D. N., M. W. Strobeck, B. L. Betz, J. Sciariotta, W. Funkhouser, Jr., C. Murchardt, M. Yaniv, L. S. Sherman, E. S. Knudsen, and B. E. Weissman. 2002. Concomitant down-regulation of BRM and BRG1 in human tumor cell lines: differential effects on RB-mediated growth arrest versus CD44 expression. Oncogene 21:1196-1207. [DOI] [PubMed] [Google Scholar]
- 55.Reyes, J. C., J. Barra, C. Muchardt, A. Camus, C. Babinet, and M. Yaniv. 1998. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α). EMBO J. 17:6979-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roberts, C. W., M. M. Leroux, M. D. Fleming, and S. H. Orkin. 2002. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell 2:415-425. [DOI] [PubMed] [Google Scholar]
- 57.Rouleau, N., A. Domans'kyi, M. Reeben, A. M. Moilanen, K. Havas, Z. Kang, T. Owen-Hughes, J. J. Palvimo, and O. A. Janne. 2002. Novel ATPase of SNF2-like protein family interacts with androgen receptor and modulates androgen-dependent transcription. Mol. Biol. Cell 13:2106-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rowan, B. G., N. Garrison, N. L. Weigel, and B. W. O'Malley. 2000. 8-Bromo-cyclic AMP induces phosphorylation of two sites in SRC-1 that facilitate ligand-independent activation of the chicken progesterone receptor and are critical for functional cooperation between SRC-1 and CREB binding protein. Mol. Cell. Biol. 20:8720-8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shang, Y., M. Myers, and M. Brown. 2002. Formation of the androgen receptor transcription complex. Mol. Cell 9:601-610. [DOI] [PubMed] [Google Scholar]
- 60.Siddiqui, H., D. A. Solomon, R. W. Gunawardena, Y. Wang, and E. S. Knudsen. 2003. Histone deacetylation of RB-responsive promoters: requisite for specific gene repression but dispensable for cell cycle inhibition. Mol. Cell. Biol. 23:7719-7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sitz, J. H., M. Tigges, K. Baumgartel, L. G. Khaspekov, and B. Lutz. 2004. Dyrk1A potentiates steroid hormone-induced transcription via the chromatin remodeling factor Arip4. Mol. Cell. Biol. 24:5821-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sommer, S., and S. A. Fuqua. 2001. Estrogen receptor and breast cancer. Semin. Cancer Biol. 11:339-352. [DOI] [PubMed] [Google Scholar]
- 63.Spencer, T. E., G. Jenster, M. M. Burcin, C. D. Allis, J. Zhou, C. A. Mizzen, N. J. McKenna, S. A. Onate, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1997. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389:194-198. [DOI] [PubMed] [Google Scholar]
- 64.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strobeck, M. W., K. E. Knudsen, A. F. Fribourg, M. F. DeCristofaro, B. E. Weissman, A. N. Imbalzano, and E. S. Knudsen. 2000. BRG-1 is required for RB-mediated cell cycle arrest. Proc. Natl. Acad. Sci. USA 97:7748-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strobeck, M. W., D. N. Reisman, R. W. Gunawardena, B. L. Betz, S. P. Angus, K. E. Knudsen, T. F. Kowalik, B. E. Weissman, and E. S. Knudsen. 2002. Compensation of BRG-1 function by Brm: insight into the role of the core SWI-SNF subunits in retinoblastoma tumor suppressor signaling. J. Biol. Chem. 277:4782-4789. [DOI] [PubMed] [Google Scholar]
- 67.Torchia, J., C. Glass, and M. G. Rosenfeld. 1998. Co-activators and co-repressors in the integration of transcriptional responses. Curr. Opin. Cell Biol. 10:373-383. [DOI] [PubMed] [Google Scholar]
- 68.Urnov, F. D., and A. P. Wolffe. 2001. Chromatin remodeling and transcriptional activation: the cast (in order of appearance). Oncogene 20:2991-3006. [DOI] [PubMed] [Google Scholar]
- 69.Versteege, I., N. Sevenet, J. Lange, M. F. Rousseau-Merck, P. Ambros, R. Handgretinger, A. Aurias, and O. Delattre. 1998. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394:203-206. [DOI] [PubMed] [Google Scholar]
- 70.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang, W., T. Chi, Y. Xue, S. Zhou, A. Kuo, and G. R. Crabtree. 1998. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc. Natl. Acad. Sci. USA 95:492-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, W., J. Cote, Y. Xue, S. Zhou, P. A. Khavari, S. R. Biggar, C. Muchardt, G. V. Kalpana, S. P. Goff, M. Yaniv, J. L. Workman, and G. R. Crabtree. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15:5370-5382. [PMC free article] [PubMed] [Google Scholar]
- 73.Yeh, S., and C. Chang. 1996. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc. Natl. Acad. Sci. USA 93:5517-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoshinaga, S. K., C. L. Peterson, I. Herskowitz, and K. R. Yamamoto. 1992. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science 258:1598-1604. [DOI] [PubMed] [Google Scholar]
- 75.Zhang, H. S., A. A. Postigo, and D. C. Dean. 1999. Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4a, TGFβ, and contact inhibition. Cell 97:53-61. [DOI] [PubMed] [Google Scholar]
- 76.Zhang, J., T. Z. Thomas, S. Kasper, and R. J. Matusik. 2000. A small composite probasin promoter confers high levels of prostate-specific gene expression through regulation by androgens and glucocorticoids in vitro and in vivo. Endocrinology. 141:4698-4710. [DOI] [PubMed] [Google Scholar]