Abstract
Progesterone receptor (PR), a member of the nuclear receptor superfamily, is a key regulator of several processes in reproductive function. We have studied the dynamics of the interaction of PR with a natural target promoter in living cells through the use of fluorescence recovery after photobleaching (FRAP) analysis and also have characterized the dynamics of the interaction of PR with the mouse mammary tumor virus (MMTV) promoter reconstituted into chromatin in vitro. In photobleaching experiments, PR in the presence of the agonist R5020 exhibits rapid exchange with the MMTV promoter in living cells. Two PR antagonists, RU486 and ZK98299, have opposite effects on receptor dynamics in vivo. In the presence of RU486, PR binds to the promoter and is exchanged more slowly than the agonist-activated receptor. In contrast, PR bound to ZK98299 is not localized to the promoter and exhibits higher mobility in the nucleoplasm than the agonist-bound receptor. Significantly, PR bound to R5020 or RU486 can recruit the SWI/SNF chromatin remodeling complex to the promoter, but PR activated with ZK98299 cannot. Furthermore, we found ligand-specific active displacement of PR from the MMTV promoter during chromatin remodeling in vitro and conclude that the interaction of PR with chromatin is highly dynamic both in vivo and in vitro. We propose that factor displacement during chromatin remodeling is an important component of receptor mobility and that ligand-specific interactions with remodeling complexes can strongly influence receptor nuclear dynamics and rates of exchange with chromatin in living cells.
Upon binding of ligands, steroid receptors, such as progesterone receptor (PR), glucocorticoid receptor (GR), and estrogen receptor (ER), recruit chromatin modifying or remodeling complexes, coregulators, and other transcription factors leading to the initiation of gene transcription (2, 10, 21). The steroid-regulated mouse mammary tumor virus (MMTV) promoter is a well-characterized model system with a well-defined, highly organized chromatin structure (3, 15, 16, 21, 37, 43). In the presence of an agonist, GR or PR binds to hormone response elements (HREs) located on nucleosomes (designated B/C) in the promoter and recruit the SWI/SNF chromatin remodeling complex (18). Chromatin remodeling by SWI/SNF in the presence of GR leads to the binding of secondary factors, including NF1 and Oct1, and eventually the initiation of transcription from the MMTV promoter (21).
The classical view of nuclear receptor function postulates the static binding of the liganded receptor to the promoter, which serves as a platform for the assembly of large transcriptional complexes (10, 29). Results obtained from recent advances in live-cell microscopy have led to the proposal of an alternative “hit-and-run” hypothesis (14, 30, 35, 36). According to this model, the receptor interacts transiently with the promoter, recruits other factors, and is itself dynamically displaced from HREs. Demonstration of the rapid exchange of green fluorescent protein (GFP)-tagged GR between chromatin and the nucleoplasmic compartment on a tandem array of MMTV promoters by fluorescence recovery after photobleaching (FRAP) analysis has provided evidence for the above model (30). In addition, the dissociation of GR from the promoter during chromatin remodeling has been demonstrated with in vitro-reconstituted MMTV chromatin (13, 14). Interestingly, although GR itself is displaced from the promoter, it participates in the binding of a secondary transcription factor, NF1 (14). Finally, rapid periodic binding and displacement of GR during chromatin remodeling in vitro have been demonstrated by a UV laser cross-linking assay (36), providing further support for the transient nature of the interaction of GR with the promoter. Rapid dynamic interactions of transcription cofactors, such as GRIP1 (1), SRC1 and CBP (41), and other transcription factors (32), have also been demonstrated in vivo. In contrast, the large subunit (RPB1) of RNA polymerase II manifests a much longer residence time (13 min), consistent with its function as a processive enzyme (1).
Among the nuclear receptors, only GR has been characterized for dynamic movement on a target promoter in living cells (30). Short residence times for ER in the nucleoplasm and for an ER-Lac repressor fusion on an artificially tethered array of lac operator elements have been reported (41). In contrast, residence times for ER on a time scale of 20 to 40 min have been described based on the results of chromatin immunoprecipitation assays (31, 38). Thus, it is not clear whether the transient interaction of receptors with target promoters in live cells is a general phenomenon of all nuclear receptors. Also, the mechanisms and factors influencing the observed short residence times of proteins are not well defined. We have therefore elected to characterize the behavior of PR on a natural target promoter both in living cells and during chromatin remodeling in vitro.
PR functions as a progesterone-activated transcription factor (26), and human PR exists as two isoforms, PRA and PRB. PRA differs from PRB by the absence of the N-terminal 164 amino acids (26). PRB is typically a stronger transcriptional activator than PRA, although the transcriptional activities of the two isoforms may vary depending on the cell type and promoter context (20). Two mechanistically different classes of antagonists have been described (25). However, there is considerable ambiguity regarding their mode of action (25). Type I antagonists, such as ZK98299 (onapristone), have been proposed to act by preventing the binding of PR to progesterone response elements (PREs) and to function as complete antagonists (25). However, other studies have suggested that ZK98299 stimulates PR binding to PREs and induces a receptor conformation distinct from that produced by RU486 or R5020. Type II antagonists, such as RU486 (mifepristone), can promote the binding of PR to PREs but often fail to induce gene activation (25). Unlike “pure” antagonists, RU486 and other mixed antagonists can either activate or repress gene transcription, depending on the cell type and promoter context (28).
We have investigated the dynamic interactions of PR with a natural target promoter in living cells and in vitro. Furthermore, we have examined the ability of PR to recruit a chromatin remodeling complex to the promoter when bound to different classes of antagonists and the influence of this interaction on PR dynamics and function in vivo. We also have investigated the interaction of PR with the MMTV template during receptor-dependent chromatin reorganization in vitro. We found that the receptor is lost from the template during chromatin remodeling; however, this displacement reaction was ligand specific. We propose (i) that the interaction of PRB with target promoters is highly dynamic both in vivo and in vitro, (ii) that chromatin remodeling is an important component of receptor mobility, and (iii) that the type of ligand associated with the receptor can have a dramatic impact on the interaction of the receptor with the chromatin remodeling apparatus.
MATERIALS AND METHODS
Cell culture and generation of an GFP-PRB stable cell line.
Cell line 5953 expressing enhanced GFP (EGFP)-human PRB (referred to in this study as GFP-PR) under the control of the Tet-Off inducible system (44) was obtained as a stable transfected derivative of a murine mammary adenocarcinoma cell line (3134). Cell line 3134 contains multiple copies of a bovine papillomavirus-MMTV long terminal repeat (LTR)-ras fusion. The GFP-human PRB chimeric construct was cloned into pTet-Splice (Life Technologies, Gaithersburg, Md.), and the resulting pTet-Splice-GFP-PRB construct was transfected along with a hygromycin B resistance plasmid, pTK-hygro, into a Tet-Off cell line (5858) made by transfecting pTet-Off (Clontech, Palo Alto, Calif.) into cell line 3134. Colonies were selected in media supplemented with 550 μg of hygromycin B (Invitrogen, Carlsbad, Calif.)/ml. Cell line 5953 was isolated by single-cell cloning of a strongly positive colony that showed both nuclear and cytoplasmic GFP fluorescence which became nuclear after the addition of hormone R5020. Cells were maintained in Dulbecco modified Eagle medium (DMEM) (Invitrogen, Grand Island, N.Y.) supplemented with 10% fetal bovine serum (Gemini, Woodland, Calif.), 0.1 mM nonessential amino acids, 2 mM l-glutamine, 1 mM sodium pyruvate, 1 mg of G418/ml, 550 μg of hygromycin B/ml, and 10 μg of tetracycline (FisherBiotech, Fair Lawn, N.J.)/ml at 37°C in 5% CO2 in a humidified incubator.
FRAP and time-lapse microscopy.
Prior to live-cell imaging, the cells were transferred to Lab-Tek II chambers (Nalge Nunc International, Naperville, Ill.) at 40,000 cells per well. The cells were grown in medium without tetracycline for 2 days prior to the experiment. One day prior to imaging, the cells were grown in phenol red-free DMEM containing 5% charcoal-stripped serum. Cells were treated for 1 h at 37°C with ligand R5020 (30 nM), RU486 (100 nM), or ZK98299 (100 nM). FRAP analysis was carried out by using a Zeiss 510 laser scanning confocal microscope with a 100×/1.3-numerical-aperture oil immersion objective and a 40-mW argon laser. The stage temperature was maintained at 37°C with an ASI 400 Air Stream incubator (Nevtek). Five single imaging scans were acquired prior to bleaching with a bleach pulse of 160 ms by using 458-, 488-, and 514-nm laser lines at 100% laser power (laser output, 75%) without attenuation. Images of single z sections were collected at 0.5-s intervals by using a 488-nm laser line with laser power attenuated to 0.2%. Fluorescence intensities in the regions of interest were analyzed, and FRAP recovery curves were generated by using LSM software and Microsoft Excel as previously described (12). All of the quantitative data for FRAP recovery kinetics represent means ± standard deviations from at least 15 cells imaged in two independent experiments. For the time-lapse study of GFP-PR arrays, the cells were cultured as described above and treated with R5020 for 60 min at 37°C. After washes with phosphate-buffered saline (PBS), charcoal-stripped DMEM containing R5020, RU486, or ZK98299 was added to the cells. Images were collected immediately thereafter by using a Zeiss 510 laser scanning confocal microscope with a 100× objective at the desired time points.
RNA FISH and immunofluorescence analysis.
Cells were grown on 22-mm square coverslips deposited at the bottom of a six-well plate; culture conditions similar to those used for FRAP analysis were used for these experiments. Cells were subjected to immunofluorescence analysis and then to RNA fluorescence in situ hybridization (FISH) to detect MMTV transcripts. Following treatment with ligands as described above for 1 h at 37°C, cells were fixed with 4% paraformaldehyde for 20 min at room temperature. Coverslips were washed with PBS and permeabilized with 0.5% Triton X-100 and then incubated with primary antibody for 1 h at room temperature, followed by three washes with PBS. After incubation with secondary antibody for 1 h, coverslips were washed again with PBS and then processed for RNA FISH by fixing with 5% formaldehyde and rinsing with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). A digoxigenin-11-dUTP-labeled MMTV probe was prepared by using digoxigenin-nick translation mixture (Roche), denatured, and hybridized with coverslips overnight at 37°C in hybridization buffer (50 μg of tRNA, 4 μg each of Cot-1 DNA (Invitrogen, Carlsbad, Calif.) and salmon sperm DNA, formamide, 4× SSC). After hybridization, coverslips were washed with 2× SSC and 4× SSC, followed by incubation with anti-digoxigenin-rhodamine-conjugated secondary antibody (Roche) to detect the hybridized probe. GFP-PRB was detected by using mouse anti-GFP monoclonal antibody 3E6 (Molecular Probes), and BRG1 was detected by using antibody J1 (a gift from G. Crabtree and K. Zhao). The RNA FISH and immunofluorescence signals were quantified by using MetaMorph software (Universal Imaging, Downingtown, Pa.) after subtraction of the background nuclear fluorescence.
Reconstitution of MMTV chromatin.
An MMTV LTR PleI/NcoI fragment of 1.1 kb (positions 437 to 674) was immobilized on Dynal magnetic beads as described by Fletcher et al. (13). The immobilized fragment was reconstituted into chromatin by using Drosophila melanogaster late embryo extracts supplemented with mouse histone octamers as previously described (13). The reconstituted chromatin was washed as previously described (14) and finally stored in EX-N buffer (10 mM HEPES [pH 7.6], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA, 10% glycerol, 10 mM β-glycerolphosphate, 1 mM dithiothreitol [DTT], 0.05% NP-40, 1 mM aminoethylbenzenesulfonyl fluoride, 1 μg each of the proteasome inhibitors aprotinin, pepstatin, and leupeptin/ml) containing 2 mg of bovine serum albumin/ml.
Purification of PR and dominant-negative SWI/SNF (DN-SWI/SNF).
Polyhistidine-tagged human PR-B was expressed from baculovirus vector pBlueBacHis-2 (Invitrogen, San Diego, Calif.) in Sf9 insect cell cultures for 48 h at a multiplicity of infection of 1.0 as previously described (4). Ligands were bound to PR-B during expression in vivo by treating Sf9 insect cells during the last 24 h of infection with 200 nM agonist R5020, 200 nM RU486 (mifepristone; Sigma), or 500 nM ZK98299 (onapristone; Schering Pharma). PR-B bound to different ligands was purified by nickel affinity chromatography as previously described with minor modifications (4). Sf9 cell pellets from 500-ml cultures were lysed in lysis buffer (50 mM sodium phosphate buffer [pH 7.8], 5 mM imidazole, 10% glycerol, and 15 mM mercaptoethanol) containing 350 mM NaCl. The lysates were centrifuged at 100,000 × g for 30 min to generate soluble PR-B in the supernatant as a whole-cell extract. The whole-cell extract was passed over 2 ml of nickel affinity resin (nickel-nitrilotriacetic acid; QIAGEN), and the resin was washed extensively in lysis buffer with a high salt concentration (600 mM NaCl) followed by lysis buffer. Elution of bound PR from the nickel affinity resin was carried out with lysis buffer plus 250 mM imidazole by incubation for 5 min at 4°C. The eluted peak protein fractions detected at an optical density at 280 nm were pooled, and 1 μM ZnCl2, 1 mM DTT, and 1 mM EDTA were added prior to flash-freezing of aliquots at −80°C. Purified proteins were analyzed by silver-stained sodium dodecyl sulfate (SDS) gel electrophoresis and immunoblotting with PR-specific monoclonal antibody (1294) and determined to be of >90% purity. The concentration of purified PR was estimated by a combination of the Bradford (5a) protein assay and quantitative silver-stained SDS gel electrophoresis and was compared with known amounts of protein standards and optical densities at 280 nm.
Cell pellets of clone 5555 expressing FLAG-tagged dominant-negative BRG1 (DN-BRG1) were obtained from the National Cell Culture Center (Minneapolis, Minn.) and purified essentially as described previously for the purification of FLAG-tagged BRG1 from the FL-INI-11 cell line (14). Purified DN-SWI/SNF was probed with either an anti-FLAG monoclonal antibody (Upstate Biotechnology) or an anti-BRG1 antibody (sc-10768; Santa Cruz Biotechnology). DN-SWI/SNF was also probed with anti-BAF155 (sc-10756) and anti-Ini1 (sc-10768) antibodies obtained from Santa Cruz Biotechnology.
Chromatin and DNA pull-down assays.
Pull-down assays were performed essentially as described previously (14). In brief, pull-down assays were performed with 50 μl of pull-down buffer (20 mM HEPES [pH 7.3], 50 mM NaCl, 10% glycerol, 0.5 mM EDTA, 5 mM MgCl2, 0.5 mM EGTA, 10% glycerol, 10 mM β-glycerolphosphate, 1 mM DTT, 0.1% NP-40, 1 mM aminoethylbenzenesulfonyl fluoride, 1 μg each of proteasome inhibitors aprotinin, pepstatin, and leupeptin/ml) containing 2 mg of bovine serum albumin/ml and 10 μg of poly(dI-dC)/ml. A typical binding reaction was done with 50 ng of DNA or chromatin template, with or without purified PR (10 nM), with or without HeLa cell nuclear extracts (0.5 μg/μl), and with or without ATP (1 mM). Reaction mixtures were incubated at 30°C for 15 min. After two washes with pull-down buffer, the bound proteins were analyzed by SDS-polyacrylamide gel electrophoresis and Western blotting with the respective antibodies. In reactions with purified SWI/SNF or DN-SWI/SNF, 100 ng of purified proteins was used instead of HeLa cell nuclear extracts. His-tagged PR was detected by using anti-penta-His tag antibody (Qiagen), and BRG1 was detected by using antibody J1 or sc-10768.
RESULTS
Ligand-dependent binding of PRB to the MMTV promoter in vivo.
The goal of this study was to understand the dynamics of PR in vitro and in vivo on a natural promoter in live cells. We chose PRB for the in vivo studies because PRB is typically a stronger activator of transcription than is PRA (20). GFP-PRA and GFP-PRB were previously shown to have transcriptional activities comparable to those of their non-GFP counterparts (27). A tetracycline-repressible stable murine cell line expressing GFP-PRB was generated from the 3134 cell line (see Materials and Methods). This cell line contains 200 copies of an MMTV Ras tandem array which enables the direct visualization GFP-PRB binding to the MMTV promoter array in live-cell microscopy, as previously described for GFP-GR (30). Earlier work showed that the hormone responses of promoters within this array are identical to that of a single-copy gene (16); therefore, this array provides a good model system for examining the dynamics of PR. Low-level background expression of GFP-PRB in this cell line was detected in the presence of tetracycline (Fig. 1A, lanes 1 and 3). Induction upon withdrawal of tetracycline from the medium was confirmed by Western blotting of cell extracts with anti-GFP (Fig. 1A, lanes 1 and 2) and anti-PR (Fig. 1A, lanes 3 and 4) antibodies. This stable cell line was used in all of the experiments described in this study.
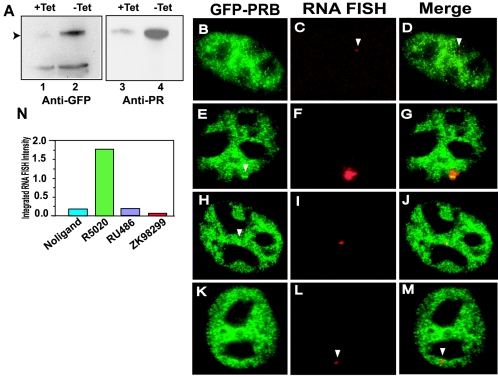
FIG. 1.
GFP-PRB colocalizes to the MMTV array with nascent MMTV transcripts, and antagonists inhibit transcription from the MMTV LTR. (A) Tetracycline-regulated expression of GFP-PRB. Total cell extracts prepared from cells grown in the presence (lanes 1 and 3) or the absence (lanes 2 and 4) of tetracycline were probed for the expression of GFP-PRB with either anti-GFP antibody (lanes 1 and 2) or anti-PR antibody (lanes 3 and 4) by Western blotting. (B to M) Colocalization of GFP-PRB to the MMTV promoter array by RNA FISH and immunofluorescence. GFP-PRB-expressing cells were subjected to RNA FISH analysis and immunofluorescence as described in Materials and Methods. Cells were treated with ethanol (B to D), R5020 (30 nM) (E to G), RU486 (100 nM) (H to J), or ZK98299 (100 nM) (K to M) for 1 h prior to fixation. Cells were fixed and processed for RNA FISH to detect MMTV transcripts and immunofluorescence to detect GFP-PR by using anti-GFP antibody. Anti-GFP antibody staining is shown in panels B, E, H, and K, and RNA FISH staining is shown in panels C, F, I, and L. Panels D, G, J, and M are overlays of panels B and C, panels E and F, panels H and I, and panels K and L, respectively. Colocalization of RNA FISH and GFP-PRB signals on the array can be observed for cells treated with R5020 (E to G) and RU486 (H to J). (N) Quantitation of MMTV RNA transcripts of GFP-PRB-expressing cells treated with different ligands. The histogram plot shows agonist-induced transcription and antagonist-induced inhibition of MMTV transcription. RNA FISH intensity of 30 cells from each ligand treatment described above was analyzed, and the total RNA FISH intensity is expressed in units measured in millions.
In the absence of a ligand, GFP-PRB was distributed in the nucleus (Fig. 1B) or in the cytoplasm (data not shown), as reported previously (27), and very low levels of basal MMTV transcription were detected by RNA FISH analysis (Fig. 1C). In cells treated with agonist R5020 for 60 min, a single bright fluorescent signal was detected (Fig. 1E) within the nucleus. Overlay of the MMTV RNA FISH signal with the immunostaining signal for GFP-PRB (Fig. 1G) confirmed targeted binding of GFP-PRB to the MMTV promoter in the presence of the agonist. Interestingly, when cells were treated with type II antagonist RU486, a similar bright fluorescent signal was observed and was colocalized with the MMTV RNA signal (Fig. 1H, I, and J). These data demonstrate that GFP-PRB can bind to the MMTV promoter in the presence of antagonist RU486. Significantly, about 3% of cells showed arrays in the presence of antagonist RU486 and 10% of cells did so in the presence of agonist R5020 (approximately 50 cells were analyzed for each treatment). In contrast, when cells were treated with type I antagonist ZK98299, targeted binding of GFP-PRB to the MMTV promoter was not observed (Fig. 1K and M), although low levels of the MMTV RNA FISH signal could still be detected (Fig. 1L). Quantitation of the MMTV RNA FISH signals obtained from cells treated with different ligands (Fig. 1N) showed that agonist R5020 induced about 10-fold activation, while antagonists RU486 and ZK98299 inhibited MMTV transcription (Fig. 1N). The MMTV promoter exhibited basal transcription even in the absence of a receptor, as indicated by the observation of RNA FISH signals in the absence of a ligand (Fig. 1C and N). These experiments demonstrated agonist-dependent binding of PRB to the MMTV promoter in living cells and showed that the two antagonists must differ in their mechanisms of action, as they differ dramatically in their ability to promote PRB binding to the promoter.
Effects of antagonists on the nuclear mobility of PRB in living cells.
Since antagonists RU486 and ZK98299 differentially affected PRB binding to the MMTV promoter, we examined the effects of these antagonists on the nuclear dynamics of PRB. FRAP analysis of GFP-PRB-expressing cells in the presence of agonist R5020 revealed a fast and complete recovery of GFP-PRB at the MMTV array after laser photobleaching (Fig. 2A to F). Recovery curves for 20 cells treated with R5020 were determined for PRB at the array (Fig. 2H) and in the nucleoplasm (Fig. 2I). Quantitative FRAP analysis of GFP-PRB at the MMTV array and in the nucleoplasm revealed similar recovery kinetics (Fig. 2H and I). The kinetics of fluorescence recovery of a GFP-tagged protein are a measure of its nuclear mobility, and the half-maximal recovery times (t1/2) for GFP-PRB at the MMTV array and in the nucleoplasm are each approximately 4 s (Fig. 2M). These data demonstrate that GFP-PRB in the presence of an agonist exhibits a fast and complete recovery in living cells, both at promoter targets and in the general nucleoplasmic compartment.
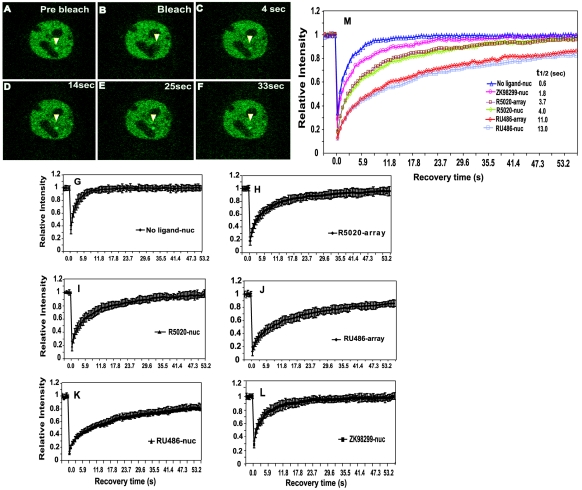
FIG. 2.
Effects of ligands on GFP-PRB exchange at the MMTV promoter array and in the nucleoplasm. (A to F) FRAP analysis of single z sections of GFP-PRB-expressing cells in the presence of agonist R5020. Cells were imaged before (A) and during (B) bleaching and during fluorescence recovery every 0.59 s. Images collected at 4 s (C), 14 s (D), 25 s (E), and 33 s (F) demonstrate the rapid recovery of the GFP-PRB signal at the MMTV array. (G to L) Recovery curves obtained from FRAP analysis of a GFP-PRB-expressing cell line in the nucleoplasm in the absence of a ligand (G); treated with agonist R5020 (30 nM) at the MMTV array and in the nucleoplasm, respectively (H and I); treated with RU486 (100 nM) at the MMTV array and in the nucleoplasm, respectively (J and K); and treated with ZK98299 (100 nM) in the nucleoplasm (L). For each condition, at least 20 cells from at least two independent experiments were analyzed. All ligand treatments were done for 60 min. (M) Overlay of FRAP recovery curves (G to L) for GFP-PRB under conditions of different ligand treatments at the MMTV array and in the nucleoplasm. The t1/2 for GFP-PRB in the presence of various ligands on the MMTV array and in the nucleoplasm are shown.
Interestingly, in the presence of antagonist RU486, GFP-PRB manifested a slower recovery, with t1/2 of 11 s at the MMTV array (Fig. 2J and M) and 13 s in the nucleoplasm (Fig. 2K and M). In contrast, GFP-PRB in the presence of complete antagonist ZK98299 showed a much faster recovery in the nucleoplasm (t1/2 of 1.8 s) (Fig. 2L and M) than did the receptor activated with R5020 or RU486 (Fig. 2G and M). In the absence of a hormone, GFP-PRB manifested very rapid mobility in the nucleoplasm (Fig. 2G) compared to cells treated with ligands (Fig. 2M). A comparison of the receptor kinetics revealed that a unliganded receptor was the fastest to recover, followed by receptors liganded to ZK98299, R5020, and RU486 (Fig. 2M). Complete fluorescence recovery of GFP-PRB was observed with all of the ligands, including RU486, when FRAP analysis was performed for longer durations (data not shown), suggesting the lack of an immobile fraction for the receptor. These results demonstrate a highly dynamic interaction of PR with chromatin and large ligand-specific differences in PRB dynamics in living cells.
Since the binding of GFP-PRB to the MMTV array was not observed in the presence of ZK98299, we could not directly characterize the effect of this antagonist on receptor dynamics at the promoter. To examine the effects of ZK98229, cells were initially treated with R5020 to allow GFP-PRB to bind to the promoter. Cells so prepared then were treated with ZK98299 and observed for potential replacement of PRB bound to R5020 on the array. Cells initially were treated with R5020 for 1 h, washed with PBS several times, and incubated with medium containing ZK98299, RU486, or R5020. Promoter arrays then were observed by time-lapse live-cell microscopy (Fig. 3A to H) at different time intervals. Binding of GFP-PRB to the MMTV arrays was observed initially between 0 and 30 min after replacement of the agonist with antagonist ZK98299 (Fig. 3A and B). However, the arrays diminished in both size and fluorescence signal intensity by 60 min (Fig. 3C) and eventually completely disappeared by 90 min (Fig. 3D), as confirmed by quantitation of the array signal intensities from 20 cells (data not shown). In contrast, when cells were pretreated with R5020 followed by a chase with RU486 (Fig. 3E and H) or with R5020 (data not shown), there was no significant change either in array size or fluorescence signal intensity. This finding was confirmed by quantitation of the array signal intensities from 20 different cells (data not shown). These results demonstrate that the loss of arrays in the presence of ZK98299 is a specific effect.
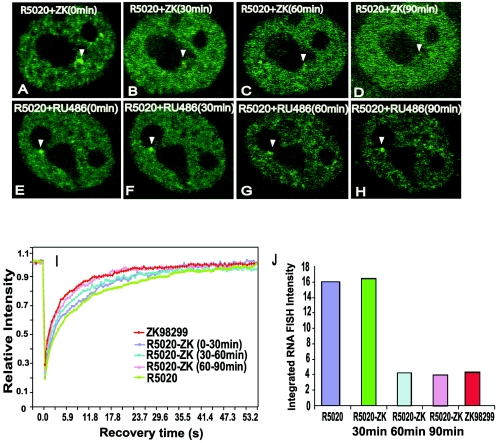
FIG. 3.
Chase of GFP-PRB bound to R5020 from MMTV promoter arrays by antagonist ZK98299. (A to H) Time-lapse microscopy images of GFP-PRB-expressing cells under conditions of different ligand treatments. (A to D) Cells were pretreated for 1 h with R5020, washed with PBS, and then treated with ZK98299 for 90 min. The images represent single z sections of images collected after 0 min (A), 30 min (B), 60 min (C), and 90 min (D) of chase with ZK98299. A loss of GFP-PRB bound to R5020 on arrays by chase with ZK98299 can be seen. (E to H) Pretreatment of GFP-PRB-expressing cells with R5020 for 1 h followed by chase with RU486 for 90 min as described for ZK98299. (I) FRAP recovery curves for GFP-PRB-expressing cells during chase of R5020 from MMTV arrays by antagonist ZK98299. Curves for cells treated with ZK98299 alone or with R5020 alone or for cells pretreated with R5020 (60 min) and then chased with ZK98299 for 30, 60, or 90 min are shown. At least 20 cells from two independent experiments were analyzed by FRAP for each ligand treatment. (J) Quantitation of RNA FISH signals from the MMTV promoter arrays over the time course of the chase of GFP-PRB bound to R5020 by ZK98299. RNA FISH was performed on cells treated under various conditions as described above; the histogram shows the signals for at least 25 analyzed cells. The total RNA FISH intensity values are represented in arbitrary units.
The rate of exchange of GFP-PRB with promoter arrays was determined by quantitative FRAP analysis during the period of R5020 replacement by ZK98299. Importantly, between 0 and 30 min (Fig. 3I), the receptor exhibited recovery rates similar to those of the R5020-bound receptor. In contrast, between 30 and 60 min (Fig. 3I) after the addition of ZK98299, the receptor exhibited faster recovery than the R5020-bound receptor while the arrays were becoming smaller and less intense (Fig. 3C). From 60 to 90 min, after which the arrays completely disappeared (Fig. 3D), the receptor showed more rapid recovery comparable to that of the ZK98299-bound receptor in the nucleoplasm (Fig. 3I).
RNA FISH analysis was performed under the treatment conditions described above to determine transcription levels. The quantitative analysis of RNA FISH signals (Fig. 3J) showed increased transcription rates during 0 to 30 min of chase with ZK98229, as with R5020 treatment alone. Significant decreases in transcription rates were observed at both 30 to 60 min and 60 to 90 min after the chase with ZK98229. These decreases in transcription rates were similar to that observed with ZK98299 alone (Fig. 3J).
These experiments demonstrated that antagonist ZK98299 treatment results in the dissociation of GFP-PRB from the MMTV array, accompanied by an increase in the rate of exchange of the receptor at the promoter and a decrease in the MMTV transcription level. Thus, the MMTV array-bound receptor cycles on and off the promoter and can exchange ligands.
Ligand-specific recruitment of the SWI/SNF complex by PR.
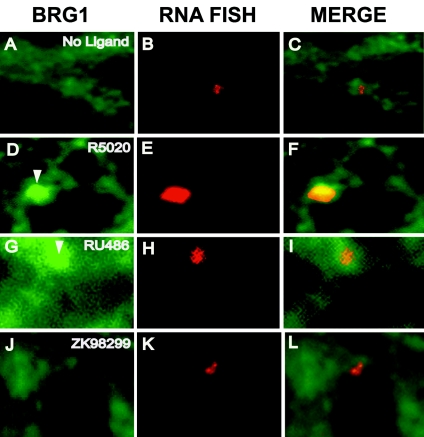
The SWI/SNF remodeling complex was previously implicated in nuclear receptor dynamics (14, 36). Although the hBRM complex (human homologue of the drosophila Bramha complex) clearly can stimulate the function of the androgen receptor (28a), present data suggest that the BRG1 subunit of the SWI/SNF complex is the primary participant with regard to PR action (34). We therefore examined the potential PR-dependent recruitment of this complex to the MMTV promoter. R5020-dependent recruitment of the BRG1 subunit of the SWI/SNF complex by PRB to the MMTV promoter array was detected by immunofluorescence (Fig. 4D to F). In contrast, non-ligand-bound (Fig. 4A to C) or ZK98299-bound (Fig. 4J to L) GFP-PRB showed no significant recruitment of BRG1 to the MMTV promoter array. Interestingly, the activation of PRB by partial antagonist RU486 led to the localization of BRG1 at the array (Fig. 4G to I), albeit less efficiently than R5020, since the BRG1 fluorescence signal was smaller in size and intensity, as confirmed by quantitation of the immunofluorescence signals from 20 different cells (data not shown). These experiments demonstrated the ligand-specific recruitment of BRG1 to the MMTV promoter array by PRB.
FIG. 4.
Ligand-dependent recruitment of BRG1 by PRB to the MMTV promoter array. Cells were subjected to RNA FISH analysis, and BRG1 was detected by immunofluorescence of cells that were treated with ethanol (A to C), R5020 (D to F), RU486 (G to I), or ZK98299 (J to L). (A, D, G, and J) Anti-BRG1 staining. (B, E, H, and K) RNA FISH signals. (C, F, I, and L) Overlay of BRG1 and RNA FISH signals. R5020- and RU486-dependent recruitment of BRG1 to the MMTV array by PR can be seen.
Dynamic behavior of PR on the MMTV promoter during in vitro chromatin remodeling.
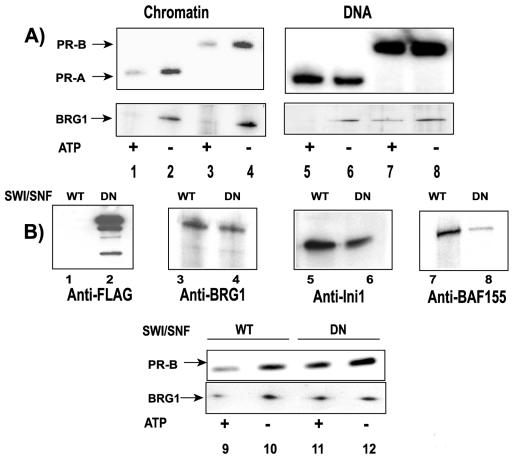
We previously described an in vitro-reconstituted chromatin remodeling system (14) that accurately recapitulates the in vivo MMTV chromatin transition. Using this system, we investigated the dynamic behavior of PR during the chromatin remodeling reaction. Purified His-tagged PRA and PRB bound to R5020 were incubated with biotinylated MMTV chromatin or MMTV naked DNA immobilized on streptavidin beads. Reactions were carried out in the presence of HeLa cell nuclear extracts, which provide nuclear activities required for efficient receptor binding. The assays were performed in the presence or in the absence of ATP for 15 min at 30°C. Template-bound proteins then were analyzed by Western blotting. Probing for PR with anti-His tag antibody (Fig. 5A, upper panels, lanes 1 to 8) demonstrated strong binding of R5020-activated PRA and PRB to both chromatin and DNA in the absence of ATP (Fig. 5A, lanes 2 and 4 and lanes 6 and 8, respectively). During incubation with ATP, which stimulates chromatin remodeling activities in nuclear extracts, displacement of PR from MMTV chromatin templates was observed (Fig. 5A, lanes 1 and 3). However, no significant displacement of PR from MMTV naked DNA templates in the presence of ATP was observed (Fig. 5A, lanes 5 and 7), indicating that the dissociation of PR from the promoter is chromatin specific. Protein fractions eluted from the beads were also probed for BRG1 (Fig. 5A, lower panel, lanes 1 to 8). In contrast to the chromatin-specific displacement observed for PR, BRG1 was lost from both MMTV chromatin templates (Fig. 5A, lower panel, lanes 1 and 3) and MMTV naked DNA templates (Fig. 5A, lower panel, lanes 5 and 7) in the presence of ATP. These results demonstrated the active displacement of PRA and PRB from MMTV chromatin during the chromatin remodeling reaction.
FIG. 5.
Displacement of PR from MMTV chromatin during chromatin remodeling in vitro. (A) Template pull-down assays performed with MMTV chromatin (lanes 1 to 4) and with MMTV DNA (lanes 5 to 8). Templates were incubated with purified PRA-R5020 (lanes 1, 2, 5, and 6) or purified PRB-R5020 (lanes 3, 4, 7, and 8) in the presence of HeLa cell nuclear extracts (lanes 1 to 8) and with ATP (lanes 1, 3, 5, and 7) or without ATP (lanes 2, 4, 6, and 8). The pull-down reaction mixtures were washed and probed with either anti-His tag antibody to detect PR (upper panels, lanes 1 to 8) or anti-BRG1 antibody (lower panels, lanes 1 to 8) to detect proteins that were bound to the promoter by Western blotting. Displacement of PR in the presence of ATP can be seen. (B) Lack of displacement of PRB-R5020 from MMTV chromatin in the presence of DN-SWI/SNF. Purified FLAG-tagged DN-BRG1 (lanes 2 and 4) or wild-type (WT) SWI/SNF (lanes 1 and 3) was analyzed with either anti-FLAG antibody (lanes 1 and 2) or anti-BRG1 antibody (lanes 3 and 4). Purified wild-type SWI/SNF (lanes 5 and 7) or purified DN-SWI/SNF (lanes 6 and 8) was probed with either anti-Ini1 antibody (lanes 5 and 6) or anti-BAF155 antibody (lanes 7 and 8) by Western blotting to detect the association of Ini1 and BAF155 with SWI/SNF. Pull-down reactions were performed with MMTV chromatin in the presence of purified wild-type SWI/SNF (lanes 9 and 10) or DN-SWI/SNF (lanes 11 and 12) and with PRB-R5020 in the presence (lanes 9 and 11) or in the absence (lanes 10 and 12) of ATP. Proteins associated with the template were analyzed for PR (upper panel, lanes 9 to 12) and BRG1 (lower panel, lanes 9 to 12) by Western blotting.
To demonstrate that the displacement of PR was directly mediated by the chromatin remodeling activity present in the HeLa cell nuclear extracts, template pull-down assays were performed with either purified wild-type SWI/SNF or FLAG-tagged DN-SWI/SNF. DN-BRG1, which does not hydrolyze ATP due to a mutation in its ATP binding site (8), was purified by affinity chromatography and could be detected by anti-FLAG and anti-BRG1 antibodies (Fig. 5B, lanes 2 and 4, respectively). Wild-type SWI/SNF (14) was used as a control (Fig. 5B, lanes 1 and 3) in these experiments. The purified DN-BRG1 complex was probed with antibodies against Ini1 and BAF155, two components of the multisubunit SWI/SNF complex, to confirm the association of these two endogenous proteins with DN-BRG1. DN-BRG1 could form a complex with Ini1 and BAF155 (Fig. 5B, lanes 6 and 8, respectively). The association of Ini1 and BAF155 with purified wild-type BRG1 (Fig. 5B, lanes 5 and 7, respectively) was used as a positive control for this experiment. Because both PRA and PRB could be displaced from chromatin in the presence of an antagonist, further experiments were carried out only with PRB.
Significant displacement of R5020-activated PRB from chromatin templates was observed when template pull-down assays were performed in the presence of purified wild-type SWI/SNF and ATP (Fig. 5B, upper panel, lanes 9 and 10), similar to the findings obtained with HeLa cell extracts (Fig. 5A). These data suggested that SWI/SNF present in the HeLa cell extracts mediated the dissociation of PR from the MMTV chromatin templates. In the presence of purified DN-SWI/SNF (Fig. 5B, upper panel, lanes 11 and 12), the binding of PRB-R5020 to MMTV chromatin was observed in the absence or in the presence of ATP (Fig. 5, lane 12). However, when DN-SWI/SNF was present, the ATP-dependent displacement of PRB-R5020 from chromatin templates was not observed (Fig. 5B, upper panel, lane 11). Also, although wild-type BRG1 (Fig. 5B, lower panel, lane 9) could be displaced from the templates, DN-BRG1 (Fig. 5B, lower panel, lane 11) was not lost from MMTV chromatin. It is important to note that DN-BRG1 is fully capable of binding to chromatin in the absence of ATP. These results implicated a role for active chromatin remodeling in thedissociation of PR from chromatin.
Ligand-specific dissociation of PR from the MMTV promoter during chromatin remodeling.
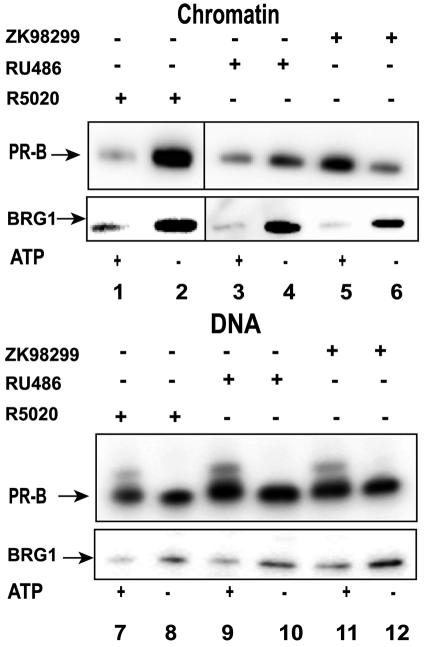
Because large ligand-specific differences were observed in the dynamics of PRB in vivo, we examined the effects of antagonists on the in vitro dynamics of PR. Template pull-down assays were performed with either MMTV chromatin (Fig. 6, upper panel, lanes 1 to 6) or DNA (Fig. 6, upper panel, lanes 7 to 12) templates in the presence of purified wild-type SWI/SNF (Fig. 6, lanes 1 to 12). PRB activated with R5020 (Fig. 6, lanes 1, 2, 7, and 8), RU486 (Fig. 6, lanes 3, 4, 9, and 10), or ZK98299 (Fig. 6, lanes 5, 6, 11, and 12) was incubated with DNA or chromatin templates in the presence or in the absence of ATP. In the absence of ATP, PR activated with R5020, RU486, or ZK98299 could interact with chromatin and DNA (Fig. 6, upper panel, lanes 2, 4, 6, 8, 10, and 12). However, ATP-dependent displacement of PR from chromatin templates was observed only in the presence of R5020 and RU486 (Fig. 6, upper panel, lanes 1 and 3). Interestingly, a lack of displacement or even increased binding of PR was detected in the presence of antagonist ZK98299 (Fig. 6, upper panel, lane 5). These findings indicated a ligand-dependent effect on the displacement of PR from chromatin during the process of remodeling. As expected, the displacement of PR from DNA templates was not observed (Fig. 6, upper panel, lanes 7, 9, and 11), irrespective of the ligand bound to PR. The displacement of BRG1 from both DNA and chromatin templates was observed with receptors activated with either an agonist or an antagonist (Fig. 6, lower panel, lanes 1 to 12). In addition, the ligand used to activate PR did not affect the amount of BRG1 bound to the chromatin templates. These results demonstrated that the dynamic interaction of PR with chromatin is strongly affected by the nature of the activating ligand and provided evidence that receptor displacement in vitro is mediated primarily through chromatin remodeling.
FIG. 6.
Ligand-specific displacement of PR in the presence of antagonists during chromatin remodeling in vitro. Pull-down assays were performed either with chromatin (lanes 1 to 6) or with DNA (lanes 7 to 12), in the presence of purified wild-type SWI/SNF (lanes 1 to 12), with (lanes 1, 3, 5, 7, 9, and 11) or without (lanes 2, 4, 6, 8, 10, and 12) ATP, and with PRB bound to agonist R5020 (lanes 1, 2, 7, and 8), antagonist RU486 (lanes 3, 4, 9, and 10), or antagonist ZK98299 (lanes 5, 6, 11, and 12). The bound proteins were probed by Western blotting to detect PR (upper panels, lanes 1 to 12) or BRG1 (lower panels, lanes 1 to 12). No displacement of PRB-ZK98299 in the presence of ATP was observed.
DISCUSSION
The findings presented here demonstrate a rapid and transient interaction of PR at a natural promoter in vivo and in vitro, suggesting that dynamic interactions with target promoters are a common feature of steroid receptors. We report large ligand-specific differences in the dynamics of PR and its ability to recruit chromatin remodelers. Based on these observations, we propose that receptors liganded to antagonists not only recruit different coregulators but also differ in their interactions with chromatin remodelers and that these interactions in turn have a major impact on receptor dynamics and function.
Ligand-specific interaction of PR with chromatin.
Agonist R5020 and antagonist RU486 can promote the binding of PRB to HREs in DNA, with RU486 being reported to have a higher-affinity interaction than R5020 in vitro (9, 11). Evidence for PR binding in the presence of R5020 and RU486 has been obtained mainly from gel shift experiments with naked DNA (9). Our experiments for the first time show the targeted binding of PRB on a natural target promoter in living cells in the presence of R5020 or RU486. Significantly, although RU486 inhibits MMTV transcription, it does not affect the ability of PRB to bind to the promoter. Contrary to previous reports showing that ZK98299 does not promote receptor binding to PREs (24, 42), we found that PRB-ZK98299 will bind to specific response elements on a natural target promoter in vitro. However, the affinity of the interaction of PR with the HRE in the presence of ZK98299 has been reported to be lower than that observed with R5020 or RU486 (7, 19). Although no significant recruitment of PRB-ZK98299 to the MMTV promoter in vivo was observed, PRB-ZK98299 showed slower recovery kinetics than non-ligand-bound PR in live cells. This finding indicates that PRB-ZK98299 must interact with chromatin and other nuclear components in vivo. Furthermore, significant binding of PRB in the presence of ZK98299 was observed in vitro in our template pull-down experiments with MMTV chromatin and naked DNA (Fig. 5 and 6). Thus, our experiments suggest that PRB-ZK98299 is capable of interacting with chromatin both in vivo and in vitro.
Role of chromatin remodeling in steroid receptor dynamics.
Chromatin remodeling complexes are involved in gene activation by several members of the nuclear receptor superfamily (5, 6, 18, 23, 33, 39, 45). The standard model of receptor action (based largely on chromatin immunoprecipitation data) holds that receptors remain stably bound to the template during recruitment of these complexes. However, we have shown (with template pull-down assays) that GR is surprisingly mobile on the template during remodeling in vitro (14). More recently, using an ultrafast laser cross-linking assay, we observed that GR manifests a highly transient and periodic interaction with the template (36).
The findings described here show that PR is also mobile on the template during chromatin remodeling. Our results obtained with DN-SWI/SNF (Fig. 5) provide direct evidence for the role of the SWI/SNF remodeling complex in the dissociation of PR from MMTV chromatin in vitro. Our in vivo results show that PR bound to different ligands can differentially mobilize BRG1 to the promoter. We propose a model wherein PR, when bound to ligands which can recruit BRG1 to the promoter, exhibits a longer residence time on the template. Conversely, in the presence of ligands which do not recruit BRG1, PR has a shorter residence time. The slower recovery kinetics and longer residence time for PR in the presence of R5020 or RU486 versus ZK98299 may be explained by the observed recruitment of BRG1 and the binding of PR to the promoter in vivo (Fig. 1 and 4). In addition, active displacement of PRB from MMTV chromatin in the presence of R5020 or RU486 was observed during chromatin remodeling in vitro (Fig. 6). Significantly, although RU486 inhibits MMTV transcription, it can still promote PR-mediated recruitment of BRG1 and target the receptor to the promoter, in accordance with our proposed model. Inhibition of MMTV transcription and a slower rate of recovery of PRB in the presence of RU486 versus R5020 likely are mediated by events downstream from chromatin remodeling, including interactions with coregulators and transcription factors. We also note that receptor-dependent chromatin remodeling at the MMTV promoter was detected in cells expressing PR in the presence of RU486 or R5020 (34). We suggest that the shorter residence time observed for PRB bound to ZK98299 results from the lack of recruitment of BRG1 in the presence of ZK98299 (Fig. 4). The ZK98299 receptor is not engaged in chromatin remodeling and other subsequent events. Therefore, the receptor does not dwell at the promoter during remodeling; thus, residence times are short.
Significantly, in cells treated with ZK98299, there is a loss of hypersensitivity at the nucleosome B/C transition, indicating a lack of chromatin remodeling (34). Although we did not detect PRB bound to ZK98299 at the promoter in live cells, PR-bound R5020 arrays that were chased with ZK98299 exhibited recovery curves faster than those seen with ZK98299 alone in the nucleoplasm (Fig. 3). The chase of R5020 arrays with ZK98299 could result from the loss of R5020-bound PRB from the array or from its replacement by PRB bound to ZK98299. Interestingly, FRAP recovery under conditions of R5020 withdrawal (data not shown) revealed mobility faster than that seen with R5020 and slower than that seen with ZK98299 alone. The intermediate mobility observed might be mediated through a mixture of receptors either liganded with PR-R5020 or unliganded. These results suggested that the chase of arrays is not just due to the release of R5020 but might be mediated by ZK98299. PR in the presence of ZK98299, apart from having recovery kinetics different from those seen with R5020 and RU486 in vivo, was not displaced from MMTV chromatin during remodeling in vitro (Fig. 6). Although in vivo PRB-ZK98299 is unable to recruit BRG1 to the promoter array (Fig. 4), we observed that BRG1 was present on MMTV chromatin templates in the presence of ZK9829 in vitro (Fig. 6). Furthermore, PRB bound to ZK98299 did not influence the displacement of BRG1 itself from chromatin in vitro (Fig. 6). We propose that PRB bound to ZK98299 is unable to interact productively and direct the remodeling activity of BRG1 already on a template. This defect causes the lack of displacement of PRB-ZK98299 from chromatin in vitro and provides a possible model for the lack of targeted binding of PRB-ZK98299 in vivo.
Alternatively, the absence of targeted binding of PRB to the MMTV promoter array in the presence of ZK98299 could be due to the inability of the receptor to bind to HREs. However, our in vitro data show significant binding of PR bound to ZK98299 to the promoter (Fig. 6). In addition, we cannot exclude the possibility that PRB bound to ZK98299 might have weak or low-affinity transient interactions with chromatin in vivo. In the presence of ZK98299, PR might not occupy all of the HREs at the MMTV promoter due to the absence of cooperative binding to multiple HREs. This lack of cooperative binding could result from the inability of PRB bound to ZK98299 to recruit SWI/SNF. In this context, rapid periodic binding and displacement of GR from the promoter during chromatin remodeling in vitro have been proposed to be highly cooperative processes (22).
Although we have focused in this report on the role of the BRG1 component of the SWI/SNF complex in PR dynamics, it is possible that other remodeling activities participate in the movement of PR and other nuclear receptors. In particular, hBRM has been shown to dramatically stimulate androgen receptor action at the probasin promoter (28a). It therefore seems likely that multiple remodeling complexes are involved in receptor mobility in living cells.
Steroid receptor dynamics on chromatin.
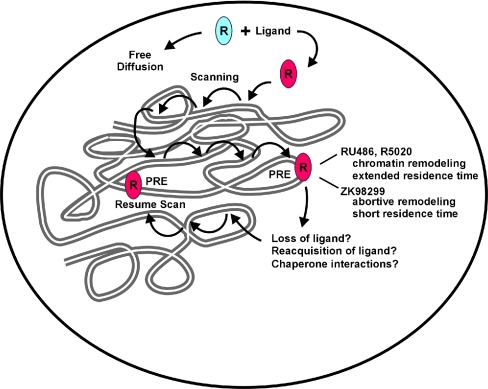
Based on our results, we propose a model in which several parameters affect and/or influence the observed rapid and transient binding of the receptors within the nucleus (Fig. 7). Energy-independent passive diffusion, the search process for target DNA binding sites, and intrinsic disassociation and reassociation of proteins with chromatin and other nuclear components could each influence the nuclear movement of proteins. We have focused in this report on the role of chromatin remodeling proteins in receptor mobility and the selective effects of specific ligands on these processes. It is clear, however, that other processes must be involved in overall nuclear movement. Molecular chaperones were recently shown to be localized to hormone-regulated promoters (17), and a direct ATP-dependent effect of chaperones on the mobility of GR and PR was recently demonstrated (40). A thorough exploration of each of these mechanisms will be necessary for a complete understanding of the dynamic movement of transcription factors in living cells.
FIG. 7.
Model depicting the various factors that influence the nuclear mobility of receptors and their effect on antagonist-mediated PR nuclear dynamics. Diffusion, genome-wide scanning of receptors for target sites, the affinity of ligands, and on-off rates of liganded receptors from promoters are important parameters for nuclear dynamics of receptors. Binding of the ligand-bound receptor at the promoter initiates chromatin remodeling events, leading to longer residence times and slower recovery of PR bound to R5020 or RU486. PR in the presence of ZK98299 does not recruit the SWI/SNF chromatin remodeler, leading to a shorter residence time and faster recovery curves for PR activated with this ligand. The displaced receptor may interact with chaperones, possibly aiding in hormone binding or refolding of the receptor, implying a role for chaperones in nuclear mobility.
Acknowledgments
We thank G. Crabtree (Stanford University, Stanford, Calif.) and K. Zhao (National Institutes of Health [NIH]) for the generous gift of anti-BRG1 antibody J1. We also thank Carl Wu (NIH) for the Drosophila embryos used for the preparation of chromatin assembly extracts. We acknowledge Anthony Imbalzano (University of Massachusetts Medical School) for the DN-BRG1 construct and the National Cell Culture Center for providing the cell pellets of FL-INI-11 (FLAG-tagged BRG1) and FLAG-tagged DN-BRG1. We thank Jim McNally, Tom Misteli, and Sagar Sengupta for critical reading of the manuscript and discussions. Live-cell microscopy was carried out at the Fluorescence Imaging Facility, Laboratory of Receptor Biology and Gene Expression, National Cancer Institute.
REFERENCES
- 1.Becker, M., C. T. Baumann, S. John, D. Walker, M. Vigneron, J. G. McNally, and G. L. Hager. 2002. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 3:1188-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belandia, B., and M. G. Parker. 2003. Nuclear receptors: a rendezvous for chromatin remodeling factors. Cell 114:277-280. [DOI] [PubMed] [Google Scholar]
- 3.Blomquist, P., Q. Li, and O. Wrange. 1996. The affinity of nuclear factor 1 for its DNA site is drastically reduced by nucleosome organization irrespective of its rotational or translational position. J. Biol. Chem. 271:153-159. [DOI] [PubMed] [Google Scholar]
- 4.Boonyaratanakornkit, V., V. Melvin, P. Prendergast, M. Altmann, L. Ronfani, M. E. Bianchi, L. Taraseviciene, S. K. Nordeen, E. A. Allegretto, and D. P. Edwards. 1998. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol. Cell. Biol. 18:4471-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourachot, B., M. Yaniv, and C. Muchardt. 1999. The activity of mammalian brm/SNF2α is dependent on a high-mobility-group protein I/Y-like DNA binding domain. Mol. Cell. Biol. 19:3931-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Debril, M. B., L. Gelman, E. Fayard, J. S. Annicotte, S. Rocchi, and J. Auwerx. 2004. Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. J. Biol. Chem. 279:16677-16686. [DOI] [PubMed] [Google Scholar]
- 7.Delabre, K., A. Guiochon-Mantel, and E. Milgrom. 1993. In vivo evidence against the existence of antiprogestins disrupting receptor binding to DNA. Proc. Natl. Acad. Sci. USA 90:4421-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la Serna, I. L., K. A. Carlson, D. A. Hill, C. J. Guidi, R. O. Stephenson, S. Sif, R. E. Kingston, and A. N. Imbalzano. 2000. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol. Cell. Biol. 20:2839-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeMarzo, A. M., S. A. O:nate, S. K. Nordeen, and D. P. Edwards. 1992. Effects of the steroid antagonist RU486 on dimerization of the human progesterone receptor. Biochemistry 31:10491-10501. [DOI] [PubMed] [Google Scholar]
- 10.Dilworth, F. J., and P. Chambon. 2001. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene 20:3047-3054. [DOI] [PubMed] [Google Scholar]
- 11.Edwards, D. P., M. Altmann, A. DeMarzo, Y. Zhang, N. L. Weigel, and C. A. Beck. 1995. Progesterone receptor and the mechanism of action of progesterone antagonists. J. Steroid Biochem. Mol. Biol. 53:449-458. [DOI] [PubMed] [Google Scholar]
- 12.Elbi, C. C., D. A. Walker, G. Romero, W. P. Sullivan, D. O. Toft, G. L. Hager, and D. B. DeFranco. 2004. Molecular chaperones function as steroid receptor nuclear mobility factors. Proc. Natl. Acad. Sci. USA 101:2876-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fletcher, T. M., B.-W. Ryu, C. T. Baumann, B. S. Warren, G. Fragoso, S. John, and G. L. Hager. 2000. Structure and dynamic properties of the glucocorticoid receptor-induced chromatin transition at the mouse mammary tumor virus promoter. Mol. Cell. Biol. 20:6466-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fletcher, T. M., N. Xiao, G. Mautino, C. T. Baumann, R. Wolford, B. S. Warren, and G. L. Hager. 2002. ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling. Mol. Cell. Biol. 22:3255-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fragoso, G., S. John, M. S. Roberts, and G. L. Hager. 1995. Nucleosome positioning on the MMTV LTR results from the frequency-biased occupancy of multiple frames. Genes Dev. 9:1933-1947. [DOI] [PubMed] [Google Scholar]
- 16.Fragoso, G., W. D. Pennie, S. John, and G. L. Hager. 1998. The position and length of the steroid-dependent hypersensitive region in the mouse mammary tumor virus long terminal repeat are invariant despite multiple nucleosome B frames. Mol. Cell. Biol. 18:3633-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freeman, B. C., and K. R. Yamamoto. 2002. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science 296:2232-2235. [DOI] [PubMed] [Google Scholar]
- 18.Fryer, C. J., and T. K. Archer. 1998. Chromatin remodeling by the glucocorticoid receptor requires the BRG1 complex. Nature 393:88-91. [DOI] [PubMed] [Google Scholar]
- 19.Gass, E. K., S. A. Leonhardt, S. K. Nordeen, and D. P. Edwards. 1998. The antagonists RU486 and ZK98299 stimulate progesterone receptor binding to deoxyribonucleic acid in vitro and in vivo, but have distinct effects on receptor conformation. Endocrinology 139:1905-1919. [DOI] [PubMed] [Google Scholar]
- 20.Giangrande, P. H., E. A. Kimbrel, D. P. Edwards, and D. P. McDonnell. 2000. The opposing transcriptional activities of the two isoforms of the human progesterone receptor are due to differential cofactor binding. Mol. Cell. Biol. 20:3102-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hager, G. L. 2001. Understanding nuclear receptor function: from DNA to chromatin to the interphase nucleus. Prog. Nucleic Acids Res. Mol. Biol. 66:279-305. [DOI] [PubMed] [Google Scholar]
- 22.Hager, G. L., A. K. Nagaich, T. A. Johnson, D. A. Walker, and S. John. 2004. Dynamics of nuclear receptor movement and transcription. Biochim. Biophys. Acta 1677:46-51. [DOI] [PubMed] [Google Scholar]
- 23.Ichinose, H., J. M. Garnier, P. Chambon, and R. Losson. 1997. Ligand-dependent interaction between the estrogen receptor and the human homologues of SWI2/SNF2. Gene 188:95-100. [DOI] [PubMed] [Google Scholar]
- 24.Klein-Hitpass, L., A. C. Cato, D. Henderson, and G. U. Ryffel. 1991. Two types of antiprogestins identified by their differential action in transcriptionally active extracts from T47D cells. Nucleic Acids Res. 19:1227-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leonhardt, S. A., and D. P. Edwards. 2002. Mechanism of action of progesterone antagonists. Exp. Biol. Med. 227:969-980. [DOI] [PubMed] [Google Scholar]
- 26.Li, X., and B. W. O'Malley. 2003. Unfolding the action of progesterone receptors. J. Biol. Chem. 278:39261-39264. [DOI] [PubMed] [Google Scholar]
- 27.Lim, C. S., C. T. Baumann, H. Htun, W. Xian, M. Irie, C. L. Smith, and G. L. Hager. 1999. Differential localization and activity of the A and B forms of the human progesterone receptor using green fluorescent protein chimeras. Mol. Endocrinol. 13:366-375. [DOI] [PubMed] [Google Scholar]
- 28.Liu, Z., D. Auboeuf, J. Wong, J. D. Chen, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 2002. Coactivator/corepressor ratios modulate PR-mediated transcription by the selective receptor modulator RU486. Proc. Natl. Acad. Sci. USA 99:7940-7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Marshall, T. W., K. A. Link, C. E. Petre-Draviam, and K. E. Knudsen. 2003. Differential requirement of SWI/SNF for androgen receptor activity. J. Biol. Chem. 278:30605-30613. [DOI] [PubMed] [Google Scholar]
- 29.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465-474. [DOI] [PubMed] [Google Scholar]
- 30.McNally, J. G., W. G. Mueller, D. Walker, R. G. Wolford, and G. L. Hager. 2000. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287:1262-1265. [DOI] [PubMed] [Google Scholar]
- 31.Metivier, R., G. Penot, M. R. Hubner, G. Reid, H. Brand, M. Kos, and F. Gannon. 2003. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751-763. [DOI] [PubMed] [Google Scholar]
- 32.Misteli, T. 2001. Protein dynamics: implications for nuclear architecture and gene expression. Science 291:843-847. [DOI] [PubMed] [Google Scholar]
- 33.Muchardt, C., and M. Yaniv. 1993. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 12:4279-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mymryk, J. S., and T. K. Archer. 1995. Dissection of progesterone receptor-mediated chromatin remodeling and transcriptional activation in vivo. Genes Dev. 9:1366-1376. [DOI] [PubMed] [Google Scholar]
- 35.Nagaich, A. K., G. V. Rayasam, E. D. Martinez, T. A. Johnson, C. Elbi, S. John, and G. L. Hager. 2004. Subnuclear trafficking and gene targeting by nuclear receptors, p. 213-220. In T. Tomoshige, E. Charmandari, and G. P. Chrousos (ed.), Glucocorticoid action: basic and clinical implications. New York Academy of Science, New York, N.Y. [DOI] [PubMed]
- 36.Nagaich, A. K., D. A. Walker, R. G. Wolford, and G. L. Hager. 2004. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol. Cell 14:163-174. [DOI] [PubMed] [Google Scholar]
- 37.Perlmann, T., and O. Wrange. 1988. Specific glucocorticoid receptor binding to DNA reconstituted in a nucleosome. EMBO J. 7:3073-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reid, G., M. R. Hubner, R. Metivier, H. Brand, S. Denger, D. Manu, J. Beaudouin, J. Ellenberg, and F. Gannon. 2003. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol. Cell 11:695-707. [DOI] [PubMed] [Google Scholar]
- 39.Salma, N., H. Xiao, E. Mueller, and A. N. Imbalzano. 2004. Temporal recruitment of transcription factors and SWI/SNF chromatin-remodeling enzymes during adipogenic induction of the peroxisome proliferator-activated receptor gamma nuclear hormone receptor. Mol. Cell. Biol. 24:4651-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stavreva, D. A., W. G. Mueller, G. L. Hager, C. L. Smith, and J. G. McNally. 2004. Rapid exchange of the glucocorticoid receptor at promoter sites is coupled to transcription and regulated by chaperones and proteasomes. Mol. Cell. Biol. 24:2682-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stenoien, D. L., A. C. Nye, M. G. Mancini, K. Patel, M. Dutertre, B. W. O'Malley, C. L. Smith, A. S. Belmont, and M. A. Mancini. 2001. Ligand-mediated assembly and real-time cellular dynamics of estrogen receptor alpha-coactivator complexes in living cells. Mol. Cell. Biol. 21:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Truss, M., J. Bartsch, and M. Beato. 1994. Antiprogestions prevent progesterone-receptor binding to hormone-responsive elements in-vivo. Proc. Natl. Acad. Sci. USA 91:11333-11337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venditti, P., L. Di Croce, M. Kauer, T. Blank, P. B. Becker, and M. Beato. 1998. Assembly of MMTV promoter minichromosomes with positioned nucleosomes precludes NF1 access but not restriction enzyme cleavage. Nucleic Acids Res. 26:3657-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker, D., H. Htun, and G. L. Hager. 1999. Using inducible vectors to study intracellular trafficking of GFP-tagged steroid/nuclear receptors in living cells. Methods 19:386-393. [DOI] [PubMed] [Google Scholar]
- 45.Yoshinaga, S. K., C. L. Peterson, I. Herskowitz, and K. R. Yamamoto. 1992. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science 258:1598-1604. [DOI] [PubMed] [Google Scholar]