Abstract
The pluripotential cell-specific gene Nanog encodes a homeodomain-bearing transcription factor required for maintaining the undifferentiated state of stem cells. However, the molecular mechanisms that regulate Nanog gene expression are largely unknown. To address this important issue, we used luciferase assays to monitor the relative activities of deletion fragments from the 5′-flanking region of the gene. An adjacent pair of highly conserved Octamer- and Sox-binding sites was found to be essential for activating pluripotential state-specific gene expression. Furthermore, the 5′-end fragment encompassing the Octamer/Sox element was sufficient for inducing the proper expression of a green fluorescent protein reporter gene even in human embryonic stem (ES) cells. The potential of OCT4 and SOX2 to bind to this element was verified by electrophoretic mobility shift assays with extracts from F9 embryonal carcinoma cells and embryonic germ cells derived from embryonic day 12.5 embryos. However, in ES cell extracts, a complex of OCT4 with an undefined factor preferentially bound to the Octamer/Sox element. Thus, Nanog transcription may be regulated through an interaction between Oct4 and Sox2 or a novel pluripotential cell-specific Sox element-binding factor which is prominent in ES cells.
Mammalian pluripotential stem cells, which are defined by their ability to differentiate into a variety of specialized cellular lineages, are found in both preimplantation embryos and many adult tissues. They can also be isolated and maintained in vitro as embryonal carcinoma (EC) cells, embryonic stem (ES) cells, and embryonic germ (EG) cells (30).
The pluripotential state of cells is maintained under the regulation of some key genes whose expression is specific to pluripotential cells. The Oct4 gene, which is a member of the mammalian POU family of transcriptional factor genes, functions as a key regulator of the pluripotential state (16, 20). Sox2, known to act cooperatively at promoters with Oct4, activates transcription of the Fgf4, Utf1, Sox2, and Fbx15 genes (17, 33, 34, 42). Furthermore, the genes transcribed in the trophoectodermal lineage, Cdx-2 and Hand-1, are negatively regulated by Oct4 (20).
Another key molecule involved in the signaling pathway for maintaining the capacity for the self-renewal and pluripotency of mouse ES cells is leukemia inhibitory factor (LIF) (26, 38). LIF directs the activation of transcription factor STAT3 by phosphorylation through binding to the heterodimer of the LIF receptor and gp130 (6). Recently, it was also shown that the LIF signal is not sufficient to support the self-renewal of mouse ES cells under culture conditions in the absence of serum and feeder cells. An additional signal provided by bone morphogenetic proteins is required and induces the activation of the inhibitor of differentiation (Id) genes, which repress differentiation into the neuroectodermal lineage (41). In addition to the roles of these genes, it was demonstrated that Ezh2, a mammalian homologue of the Polycomb-group gene Enhancer of zeste in Drosophila (12), forms a complex with Eed (embryonic ectoderm development). This complex plays an important role in maintaining the pluripotency of ES cells and blastocyst inner cell mass cells through histone H3 lysine 27 methylation-based repression of specific homeotic genes (4, 7). Null mutation of the Oct4, Sox2, or Ezh2 gene results in early embryonic lethality (1, 16, 21); interestingly, however, loss of the Bmp4, Lif, Lif receptor, or Stat3 gene induces no obvious defect, at least in mouse preimplantation development (28, 31, 36, 39). It is known that LIF is dispensable for supporting the self-renewal and pluripotency of monkey and human ES cells (32).
NANOG is a newly identified homeodomain-bearing protein that may act as a transcription factor and that is transcribed specifically in pluripotential cells in mouse preimplantation embryos, ES cells, and EG cells (3, 15, 35) and monkey and human ES cells (8, 9). A critical requirement for Nanog in the maintenance of pluripotency has been suggested by the loss of pluripotency in Nanog-deficient ES cells and in Nanog-null embryos shortly after implantation (15). In addition, Nanog overexpression leads to the clonal expansion of ES cells by bypassing regulation by LIF-STAT3 signaling and maintenance of OCT4 levels (3). Thus, Nanog is an important regulator of pluripotency and self-renewal of ES cells and early embryonic cells. However, it remains largely unknown how the pluripotential cell-specific expression of Nanog is controlled and how the other stem cell-specific genes are implicated in Nanog expression.
To address the molecular mechanisms of pluripotential cell-specific expression, we investigated the regulatory elements that are involved in the control of Nanog transcription. We show that the undifferentiated state-specific expression of a green fluorescence protein (GFP) reporter gene in mouse ES cells can be induced by the addition of a 2.5-kb 5′-flanking region of Nanog, indicating that transcriptional cis regulatory elements exist in this region. Luciferase assays with deletion constructs of the 5′-flanking region revealed that the −332-bp fragment (−332 fragment) containing a pair of adjacent Octamer and Sox elements plays a crucial role in directing transcriptional up-regulation. Consistent with these results, we found that transcription was down-regulated by the introduction of sequence mutations in the Octamer and/or Sox elements. In nuclear extracts from F9 EC cells, specific binding of OCT4 to the Octamer element and of SOX2 to the Sox element was detected. Similar results were seen in EG cell extracts. In ES cell extracts, however, a complex of OCT4 and a novel pluripotential cell-specific Sox element-binding protein (PSBP) preferentially bound to the Octamer/Sox element. Nanog transcription is therefore regulated differently in ES, EG, and EC cells, and a novel factor (PSBP) may be involved in maintaining the ES cell-specific undifferentiated state.
MATERIALS AND METHODS
Cell culture and differentiation.
Mouse R1 ES, TMA-58G EG, and F9 EC cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Sigma) supplemented with 15% fetal bovine serum, 10−4 M 2-mercaptoethanol, and 1,000 U of recombinant LIF (Chemicon)/ml at 37°C in 5% CO2. Human ES cell line KhES-1 was cultured in DMEM with 20% knockout serum replacement (Invitrogen), 2 mM l-glutamine, nonessential amino acids, and 10−4 M 2-mercaptoethanol, according to 2001 guidelines for the derivation and utilization of human ES cells (Ministry of Education, Culture, Sports, Science and Technology of Japan). Mouse and human ES cells were maintained on mouse primary embryonic fibroblast feeder cells inactivated with mitomycin C. Embryoid bodies (EBs) were formed by suspension culturing for 5 days. Chemical differentiation induction was performed with 10−6 M all-trans-retinoic acid (Sigma). NIH 3T3 and COS-1 cells were cultured in DMEM containing 10% fetal bovine serum. Differentiation experiments were conducted with medium in the absence of LIF.
Transgenic cell lines.
The LR/Nanog-GFP transgene was constructed with a GFP-internal ribosome entry site (IRES)-puro-pA reporter cassette and with a 2.5-kb 5′-end genomic fragment and a 3.9-kb 3′-end genomic fragment of Nanog in vector pGEM-T Easy (Promega). After linearization, the transgene was electroporated into 107 R1 ES cells at 250 V and 500 μF with a Gene Pulser (Bio-Rad). Genomic DNA obtained from puromycin-resistant clones was screened by Southern blot hybridization analysis. A stable transformant ES cell line, −332-GFP TG, was obtained as a G418-resistant clone after cotransfection of vector p−332-pEGFP-N1 (Clontech) and vector pPgk-neo(TAKARA) into R1 ES cells.
Southern blot hybridization analysis.
Genomic DNA was digested with restriction enzymes, electrophoresed through 1.0% agarose, and transferred to Hybond N+ nylon membranes (Amersham) by alkali blotting. Membranes were hybridized at 42°C overnight with either a 5′ probe (500 bp) or a 3′ probe (750 bp) labeled with 32P-dCTP by using a Megaprime DNA labeling system (Amersham). Membranes then were washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) at 65°C for 30 min and with 0.1× SSC-0.1% SDS at 65°C for 15 min.
Construction of reporter and expression vectors.
Deletion fragments of the mouse Nanog promoter were PCR amplified from the mouse genome with a common antisense primer that spans an XhoI restriction site (+50 bp from the transcriptional start site, 5′-CTACTCGAGCGCAGCCTTCCCACAGAAA-3′) and various sense primers into which an XhoI restriction site was introduced (−2,342 bp, 5′-CTACTCGAGTGGTGTAAACAGTGGGTCTG-3′; −332 bp, 5′-CTACTCGAGATCGCCAGGGTCTGGA-3′; and −153 bp, 5′-CTACTCGAGCCTGCAGGTGGGATTAACT-3′). The PCR products were digested with XhoI and ligated into the XhoI site of pGL3-Basic (Promega) or cytomegalovirus promoterless vector pEGFP-N1 (Clontech) (−m332 or −m153). PCR fragments of the human Nanog promoter were amplified from human ES cell DNA with a sense primer (−380 bp from the transcriptional start site) having the sequence 5′-GCTGGTTTCAAACTCCTGACTTC-3′ and an antisense primer (+24 bp) having the sequence 5′-TCCTGGAGTCTCTAGATT-3′ and ligated into vector pGEM-T Easy. NotI-NotI (−380 to +24 bp), NotI-PstI (−123 to +24 bp), and NotI-StyI (−101 to +24 bp) fragments were blunt ended and recloned into the SmaI site of cytomegalovirus promoterless vector pEGFP-N1. Oligonucleotide-directed mutations were introduced into the Octamer and/or Sox elements by PCR with primers having nucleotide replacements.
Oct4 and Sox2 open reading frames (ORFs) amplified by reverse transcription-PCR with primers EcoRI-Oct4-ORF-F (5′-CCGAATTCGGATGGCTGGACACCTGGCTTCAG-3′), BglII-Oct4-ORF-R (5′-AGAGATCTTTAACCCCAAAGCTCCAGGTTC-3′), EcoRI-Sox2-ORF-F (5′-CCGAATTCGGATGTATAACATGATGGAGACGG-3′), and BglII-Sox2-ORF-R (5′-AGAGATCTTCACATGTGCGACAGGGGCAGT-3′) were subcloned into vector pGEM-T Easy. EcoRI-BglII fragments of Oct4 and Sox2 were ligated into expression vectors pCMV-Myc and pCMV-HA (Promega), respectively.
For cotransfection reporter assays, three tandem repeats of the Octamer and Sox elements, which were produced by ligation of synthetic oligonucleotides (Nanog-O/S-F, 5′-GATCCTTACAGCTTCTTTTGCATTACAATGTCCATGGTGGA-3′; and Nanog-O/S-R, 5′-GATCTCCACCATGGACATTGTAATGCAAAAGAAGCTGTAAG-3′), were cloned into vector pTK-Luc (Clontech) to produce pTAL-Luc.
Transient expression assays were performed with Lipofectamine 2000 (Invitrogen). Samples were analyzed 2 days after transfection.
Luciferase reporter assays.
ES (5.0 × 105), F9 (5.0 × 105), and NIH 3T3 (2.5 × 105) cells were incubated in six-well tissue culture plates for 24 h. Each reporter construct (1.25 pmol) was cotransfected with vector phRL-TK (0.125 pmol) (Promega) as an internal control by using Lipofectamine 2000. Cell extracts were prepared 48 h after transfection, and luciferase activities were evaluated by using a dual-luciferase assay system (Promega). The luciferase activity of each construct was calculated relative to that of control vector pGL3-Basic. All transfection experiments were repeated in triplicate. For cotransfection reporter assays, construct 3×Oct/Sox-pTK-Luc (0.06 pmol) was cotransfected with vectors pCMV-Myc-Oct4 (0.6 pmol), pCMV-HA-Sox2 (0.6 pmol), and phRL-TK (0.006 pmol) into NIH 3T3 cells. The promoter activities are reported as means ± standard errors.
Western blot hybridization analysis.
Cell extracts (20 μg/lane) were separated through 12% polyacrylamide by SDS-polyacrylamide gel electrophoresis and transferred to Protran nitrocellulose membranes (Schleicher & Schuell). After blocking was done with 3% skim milk in phosphate-buffered saline for 1 h, the membranes were incubated with anti-NANOG (1:1,000 dilution) (9), anti-OCT4 (1:500) (Santa Cruz), anti-SOX2 (1:500) (Chemicon), anti-GFP (1:1,000) (Clontech), anti-Myc (1:1,000) (Covance), anti-hemagglutinin (HA) (1:1,000) (Covance), anti-histone H3 (1:3,000) (AbCam), or anti-β-actin (1:3,000) (AbCam) antibody overnight at 4°C. After washing was done with 0.1% Triton X-100 in phosphate-buffered saline, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody (1:3,000) (Amersham) for 90 min. Bands were detected with an ECL Western blotting detection kit (Amersham).
Electrophoretic mobility shift assays (EMSAs).
Expression vectors pCMV-Myc-Oct4 and pCMV-HA-Sox2 were transfected into COS-1 cells by using Lipofectoamine 2000. Whole-cell lysates were collected 30 h after transfection (11). Nuclear extracts of F9 EC, TMA-58G EG, and R1 ES cells were prepared as reported previously (22). Double-stranded synthetic oligonucleotide probes were labeled with 32P-dCTP. Whole-cell extracts (10 μg) and nuclear extracts (10 μg) were preincubated for 10 min on ice in the presence of 2 μg of poly(dG-dC) (Amersham) in 20 μl of reaction buffer (10 mM Tris-HCl [pH 7.5], 1 mM MgCl2, 0.5 mM dithiothreitol, 10% glycerol) and then incubated with 0.1 ng of radiolabeled probes for 30 min. Competition or supershift assays were performed by adding 1- to 300-fold excess cold competitors or 1 μg of rabbit polyclonal anti-OCT4, goat polyclonal anti-SOX2, or rabbit polyclonal anti-SOX2 antibody (each from Santa Cruz) prior to treatment with radiolabeled probes. Probe DNA-protein complexes were separated by electrophoresis at 150 V through 4% polyacrylamide in 0.25× Tris-borate-EDTA buffer at 4°C for 135 min and visualized by autoradiography.
Chromatin immunoprecipitation assays.
Chromatin immunoprecipitation was performed as described previously (10). Protein and DNA were cross-linked by incubation in 1% formaldehyde. The chromatin then was sonicated to an average DNA fragment length of 200 to 1,000 bp. Soluble chromatin reacted with or without 2 μg of rabbit polyclonal anti-OCT4 or goat polyclonal anti-SOX2 antibody (each from Santa Cruz) was purified and collected in elution buffer (0.1 M NaHCO3, 1% SDS). Cross-linking then was reversed with elution buffer containing RNase A (0.03 mg/ml) and NaCl (0.3 M) by incubation for 4 h at 65°C. Supernatant obtained without antibody was used as an input control. Following treatment with proteinase K for 1 h at 45°C, the DNA was purified and analyzed by PCR with the following primers: Nanog-O/S-ChIP-F, 5′-GTCTTTAGATCAGAGGATGCCCC-3′; Nanog-O/S-ChIP-R, 5′-CTACCCACCCCCTATTCTCCCA-3′; Fgf4-O/S-ChIP-F, 5′-AGACTTCTGAGCAACCTCCCGAA-3′; and Fgf4-O/S-ChIP-R, 5′-CAACTGTCTTCTCCCCAACACTCT-3′.
RESULTS
Pluripotential state-specific expression of the LR/Nanog-GFP transgene.
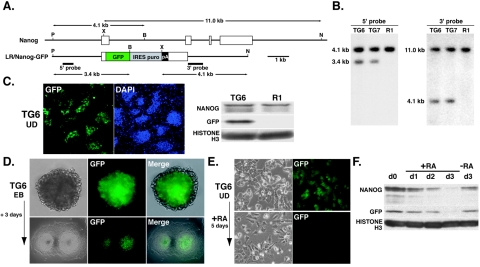
To explore the function of Nanog in regulating the stability of pluripotency and self-renewal of cells, we constructed a vector (LR/Nanog-GFP) with a 2.5-kb genomic fragment upstream of the 5′ end of the Nanog ORF and a 3.9-kb genomic fragment downstream of the 3′ end of the ORF (Fig. 1A). The full ORF of Nanog was replaced with a GFP-IRES-puro-pA reporter and selection cassette. By electroporation of the construct into R1 ES cells, 9 out of 11 GFP-positive clones examined were isolated as clones with random integration of the transgene(s). Southern blot hybridization analysis indicated that two clones (TG6 and TG7) carried intact 5′- and 3′-end genomic fragments. The 5′-end transgenic fragment was detected as a 3.4-kb band and the endogenous fragment was detected as a 4.1-kb band in PvuII-BglII-digested genomic DNA with a 5′-end-specific probe. The 3′-end transgenic fragment was detected as a 4.1-kb band and the endogenous fragment was detected as an 11.0-kb band in XbaI-NcoI-digested genomic DNA with a 3′-end-specific probe (Fig. 1B). The transgene-specific bands were less intense than the endogenous bands in both cases, indicating that both clones contained a single integrated copy of the transgene. All of the undifferentiated TG6 and TG7 colonies were positive for GFP, as verified by both fluorescence microscopy and Western blot hybridization analysis (Fig. 1C).
FIG. 1.
Undifferentiated state-specific expression of the LR/Nanog-GFP transgene. (A) Structure of the LR/Nanog-GFP transgene containing 5′- and 3′-flanking regions. P, PvuII; X, XbaI; B, BglII; N, NcoI. (B) Southern blot hybridization analysis of TG6 and TG7 transgenic cell lines and the parental R1 ES cell line. The transgene-specific 3.4-kb PvuII-BglII fragment and the 4.1-kb XbaI-NcoI fragment were detected with 5′ and 3′ probes, respectively. (C) Expression of GFP in undifferentiated (UD) TG6 ES cells. GFP expression was visualized by fluorescence microscopy and Western blot hybridization analysis with anti-GFP antibody. Histone H3 was used as a control. (D) Expression of GFP restricted to undifferentiated ES cells located in the middle of 5-day-old EBs and in the center of colonies 3 days after culturing of 5-day-old EBs. (E) Down-regulation of GFP expression by in vitro differentiation with retinoic acid (RA) treatment for 5 days. (F) Western blot hybridization analysis of GFP and endogenous NANOG during RA-induced cell differentiation. Histone H3 was used as a control.
To address whether the down-regulation of transgene expression would occur upon cell differentiation, as seen at the endogenous locus, GFP expression in 5-day-old EBs and in a 3-day-old culture of 5-day-old EBs without LIF or feeder cells was analyzed (Fig. 1D). GFP expression was not detectable in endodermal cells on the EB surface or in differentiated cells at the periphery of the adhesive colonies. To determine whether the down-regulation of GFP expression was linked to that of endogenous NANOG expression, ES clones were differentiated by chemical induction with retinoic acid (RA). After 5 days of continuous RA treatment, the cell phenotype changed significantly and GFP became undetectable (Fig. 1E). Western blot hybridization analysis revealed that decreasing GFP levels correlated well with decreasing NANOG levels after RA treatment (Fig. 1F). These data demonstrated that regulatory elements required for pluripotential state-specific regulation of Nanog gene expression are located on the transgene.
Octamer and Sox elements are required for Nanog expression.
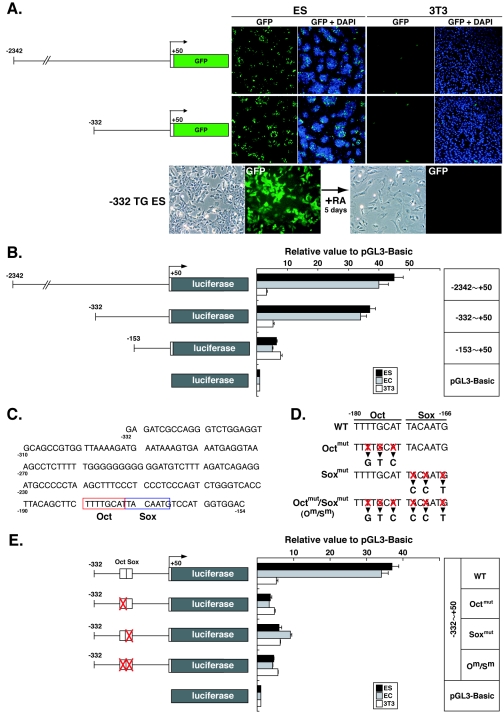
To further characterize the region required for Nanog expression, two additional GFP reporter transgenes (carrying a −2342 or −332 5′-end genomic fragment but no 3′-end fragment) were constructed and transfected into ES cells. In addition, the constructs were transfected into NIH 3T3 fibroblast cells, which do not express endogenous Nanog. A pCMV-GFP construct was used in parallel to control for transfection efficiency. With both experimental constructs, GFP was highly expressed in ES cells but not in fibroblasts (Fig. 2A), while the control construct was highly expressed in both cell types (data not shown). These results suggested that regulatory elements essential for pluripotential state-specific expression are located in the 382-bp region immediately upstream of Nanog. To confirm this notion, stably transformed −332-GFP TG ES cells were generated and differentiated in vitro through RA treatment for 5 days. GFP expression that was observed in undifferentiated −332-GFP TG ES cells was completely suppressed on differentiation.
FIG. 2.
Octamer and Sox elements are required for Nanog expression. (A) Transient expression of GFP transgenes with −2342 or −332 5′-end fragments in R1 ES and NIH 3T3 cells. Transcriptional down-regulation of GFP was detected in differentiated −332-GFP TG ES (−332 TG ES) cells by treatment with retinoic acid (RA). (B) Luciferase assays with deletion constructs in R1 ES, F9 EC, and NIH 3T3 cells. Luciferase activities are shown relative to those of pGL3-Basic. Bars represent the means ± standard errors of three independent experiments. (C) DNA sequence of the mouse 5′-flanking region between positions −332 and −154. Octamer (Oct) and Sox elements are outlined in red and blue, respectively. (D) Sequence mutations introduced into Octamer and/or Sox elements. (E) Luciferase assays with the −332 5′-end fragment with or without mutations in Octamer and/or Sox elements in R1 ES, F9 EC, and NIH 3T3 cells. Luciferase activities are shown relative to those of pGL3-Basic. Bars represent the means ± standard errors of three independent experiments.
To evaluate the level of transcriptional activity, reporter gene expression was monitored by luciferase assays with deletion fragments of the 5′-flanking region in R1 ES, F9 EC, and NIH 3T3 cells (Fig. 2B). Consistent with the results of the GFP reporter assays in ES cells, the −332 fragment induced high-level luciferase expression in both ES and EC cells. While the slight decrease in activity seen with the −2342 and −332 fragments was statistically significant (P < 0.05), a much more notable decrease was observed with the −153 fragment, resulting in approximately 15% activity in comparison to that obtained with the −2342 fragment. This level of activity was similar in all cell lines tested, suggesting that this activity was nonpluripotential state-specific basal promoter activity. These results suggested that a key cis-acting pluripotency-specific element(s) lies within the region between positions −332 and −154.
Sequence analysis demonstrated the presence of conserved Octamer (TTTTGCAT) and Sox (TACAATG) elements between positions −166 and −180 (Fig. 2C). To examine the functions of these elements, triple point mutations were introduced by replacement of DNA residues in each element (Fig. 2D). Luciferase assays with −332 fragments carrying mutated elements clearly showed a dramatic reduction in luciferase activities in all cases, to levels similar to that found with the −153 fragment (Fig. 2E). Thus, factors that interact with the Octamer and Sox elements are likely to play a crucial role in regulating the expression of Nanog in a pluripotential state-specific manner.
Octamer and Sox elements regulate expression in human ES cells.
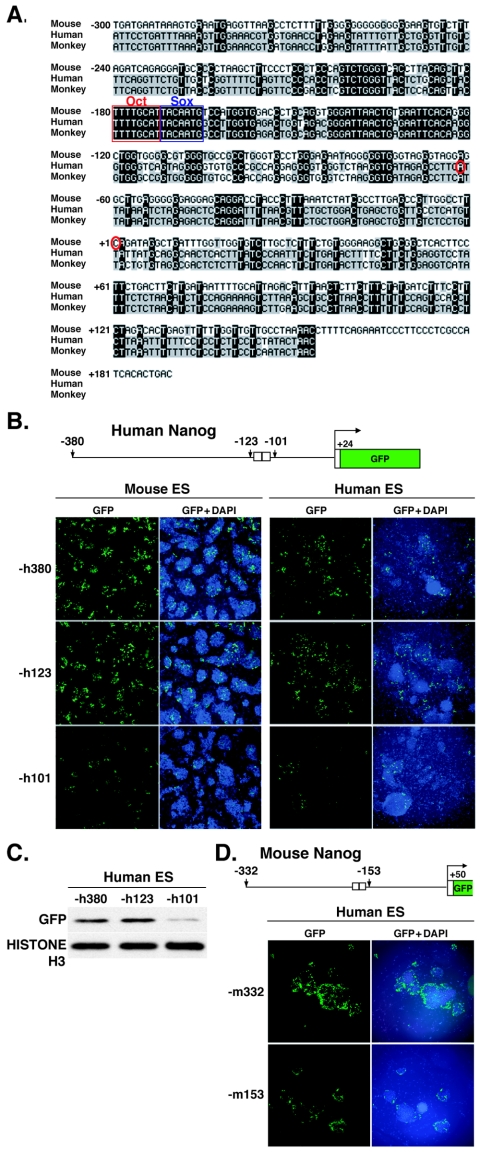
Nanog has also been identified in humans and monkeys, in which specific expression in the nuclei of undifferentiated ES cells has been shown by immunocytochemical analysis with anti-NANOG antibody (9). Comparative DNA sequence analysis of the region spanning from −300 bp to the translational start site revealed a high degree of conservation between humans and monkeys (93.2%) but not between humans and mice (54.1%) or between monkeys and mice (51.7%). However, the Octamer and Sox elements present in both humans and monkeys showed 100% identity to the mouse sequence (Fig. 3A).
FIG. 3.
Expression of the human Nanog reporter gene in mouse and human ES cells. (A) Comparative DNA sequence analysis of the Nanog 5′-flanking regions of mice, monkeys, and humans. Octamer (Oct) and Sox elements are outlined in red and blue, respectively. Red circles show putative transcriptional start sites. Identical nucleotides are highlighted in black. (B) Transient GFP expression under the regulation of the human Nanog promoter in mouse and human ES cells. The −h380 and −h123 5′-end fragments contain both Octamerand Sox elements, while the −h101 5′-end fragment does not. (C) Western blot hybridization analysis of GFP expression in human ES cells. Histone H3 was used as a control. (D) Transient GFP expression under the regulation of the mouse Nanog promoter in human ES cells. The −m332 5′-end fragment contains the Octamer and Sox elements, while the −m153 5′-end fragment does not.
To address whether the Octamer and Sox elements are required as cis regulatory elements for Nanog transcription in humans, human −h380, −h123, and −h101 fragments were linked to GFP and transfected into mouse and human ES cells (Fig. 3B). The −h380 and −h123 fragments produced relatively high-level expression in both mouse and human ES cells, while the −h101 fragment, in which the Octamer and Sox elements were missing, promoted very low-level expression. For human ES cells, these findings were confirmed by Western blot hybridization analysis (Fig. 3C). Similarly, the mouse −m332 fragment promoted high-level expression in human ES cells, while the −m153 fragment induced low-level expression (Fig. 3D). A similar considerable reduction in GFP expression was also seen with −m332 fragments carrying mutations in the Octamer and Sox elements (Fig. 2D) (data not shown). These results suggested that the Octamer and Sox elements play a similar role in regulating Nanog transcription in humans and mice.
OCT4 and SOX2 bind to the Octamer and Sox elements.
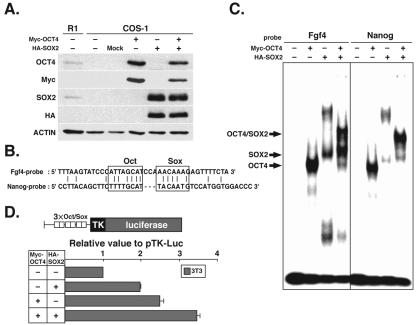
OCT4 and SOX2 are expressed in pluripotential cells and are well characterized as factors that bind to the Octamer and Sox elements, respectively. Indeed, it has been shown that they bind to these elements in regions proximal to the pluripotential cell-specific Fgf4, Utf1, Sox2, and Fbx15 genes, where they act synergistically to activate transcription (17, 33, 34, 42). We therefore investigated the abilities of OCT4 and SOX2 to bind to the Nanog Octamer and Sox elements by EMSAs. Exogenous Myc-tagged Oct4 (Myc-Oct4) and HA-tagged Sox2 (HA-Sox2) were transfected into COS-1 cells, in which endogenous OCT4 and SOX2 are repressed. Western blot hybridization analyses with antibodies against OCT4, SOX2, Myc, and HA showed that exogenous OCT4 and SOX2 were abundant in whole extracts from COS-1 cells carrying the transgene(s) (Fig. 4A).
FIG. 4.
Binding of exogenous OCT4 and SOX2 to Octamer and Sox elements in COS-1 cells. (A) Western blot hybridization analysis of exogenous Myc-tagged OCT4 and HA-tagged SOX2 expression with anti-OCT4, anti-SOX2, anti-Myc, and anti-HA antibodies. Actin was used as a control. (B) DNA sequences of Nanog and Fgf4 probes. Octamer (Oct) and Sox elements are outlined. (C) EMSA with Nanog and Fgf4 probes and COS-1 cells. Bands of the OCT4-DNA, SOX2-DNA, and OCT4/SOX2-DNA complexes are indicated. (D) Cotransfection reporter assays with Oct4 and Sox2 expression constructs in NIH 3T3 cells. Bars represent the means ± standard errors of three independent experiments.
DNA fragments containing the Octamer and Sox elements of Nanog or Fgf4 were synthesized and radiolabeled for use as specific EMSA probes (Fig. 4B). Following incubation of the Fgf4 probe with COS-1 cell extracts, Myc-OCT4, HA-SOX2, and Myc-OCT4/HA-SOX2 complexes were clearly detectable, as previously reported (42). Incubation of the Nanog probe with COS-1 cell extracts also resulted in clearly detectable binding of Myc-OCT4 and Myc-OCT4/HA-SOX2 complexes (Fig. 4C). Importantly, the affinity of binding of SOX2 alone to the Nanog Sox element was relatively weak, whereas SOX2 in combination with OCT4 resulted in the formation of a much stronger ternary protein-DNA complex. The specificity of OCT4 and/or SOX2 binding was confirmed by supershift analyses with anti-Myc and anti-HA antibodies (unpublished data).
To examine whether OCT4 and SOX2 activate Nanog expression by binding to their respective elements, reporter assays were performed with NIH 3T3 cells and with a pTK-Luc construct into which three tandem repeats of the Octamer/Sox element had been introduced (Fig. 4D). Cotransfection of this construct with Myc-Oct4 and HA-Sox2 led to an ∼3.5-fold increase in luciferase activity. Taken together, our data clearly show that the Nanog Octamer and Sox elements are able to recruit OCT4 and SOX2, respectively, leading to the up-regulation of Nanog gene expression.
Binding of OCT4 and SOX2 to Octamer and Sox elements in EC cell extracts.
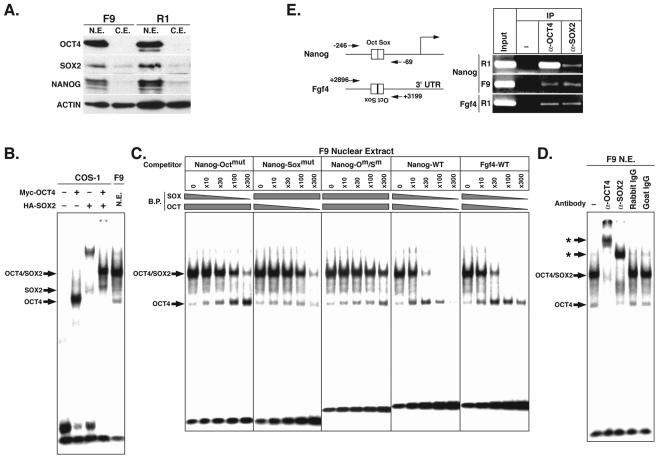
F9 cells are EC cells that have defective pluripotency and that were derived from embryonic day 6.0 (E6.0) embryos through carcinogenesis (27). As in R1 ES cells, OCT4, SOX2, and NANOG are expressed in the nuclei of F9 EC cells, as shown by Western blotting hybridization analyses (Fig. 5A). To examine the abilities of endogenous OCT4 and SOX2 to bind to the Nanog Octamer and Sox elements, nuclear extracts from F9 cells were prepared for EMSAs. Incubation with the radiolabeled Nanog probe resulted in a prominent band at the position of the OCT4/SOX2 complex, while an independent SOX2-specific band was barely detectable (Fig. 5B). The complex mobilities correlated well with those found when COS-1 cell extracts containing exogenous Myc-OCT4 and HA-SOX2 were used.
FIG. 5.
Binding of endogenous OCT4 and SOX2 to Octamer and Sox elements in F9 EC cells. (A) Western blot hybridization analysis of endogenous OCT4, SOX2, and NANOG in nuclear extracts (N.E.) and cytoplasmic extracts (C.E.) of F9 EC and R1 ES cells. (B) EMSA with the Nanog probe and nuclear extracts of COS-1 and F9 EC cells. Bands of the OCT4-DNA, SOX2-DNA, and OCT4/SOX2-DNA complexes are indicated. (C) Competition assays with unlabeled probes with or without mutations in Octamer and/or Sox elements. Relative amounts of binding proteins (B.P.) are indicated by grey bars. Bands of the OCT4-DNA and OCT4/SOX2-DNA complexes are indicated. Octmut (Om), Nanog probe with triple mutations in the Octamer element; Soxmut (Sm), Nanog probe with triple mutations in the Sox element. (D) Supershift assay with anti-OCT4 or anti-SOX2 antibody and F9 EC cell nuclear extracts. Bands of the OCT4-DNA and OCT4/SOX2-DNA complexes are indicated by arrows, and supershifted bands are indicated by asterisks. Rabbit IgG and goat IgG were used as controls. (E) Chromatin immunoprecipitation assay demonstrating the in vivo potential of OCT4 and SOX2 to bind to Nanog and Fgf4 Octamer and Sox elements, respectively.
To further investigate the factors binding to the Octamer and Sox elements, binding competition assays were performed with unlabeled Nanog probe or Nanog probe mutated in the Octamer element (Octmut), the Sox element (Soxmut), or both (Fig. 5C). Depletion of a factor(s) binding to the Sox element by the Octmut competitor probe resulted in a decrease in OCT4/SOX2 complex binding and an increase in OCT4 binding that correlated well with increasing competitor probe concentration. Depletion of a factor(s) binding to the Octamer element by the Soxmut competitor probe also caused a decrease in OCT4/SOX2 complex binding but no reciprocal increase in OCT4 binding. It should be noted that the binding of SOX2 alone was barely detectable due to a low affinity of binding to the Nanog Sox element (Fig. 4C). As expected, the Octmut/Soxmut competitor probe had little effect, while the unlabeled Nanog probe induced a decrease in the binding of OCT4 and the OCT4/SOX2 complex. The binding of OCT4 and the OCT4/SOX2 complex was also verified by competition assays with depletion by the unlabeled Fgf4 probe. These data demonstrated that endogenous OCT4 and OCT4/SOX2 complex are able to bind to the Nanog Octamer and Sox elements in F9 cells.
While we were unable to demonstrate specificity for SOX2 in these initial experiments, further analysis by supershift assays clearly demonstrated that both OCT4 and SOX2 are involved, since the inclusion of anti-OCT4 and anti-SOX2 antibodies resulted in reduced complex mobility (supershift), while the inclusion of rabbit and goat control IgGs had no effect (Fig. 5D). Thus, endogenous OCT4 and SOX2 form ternary protein-DNA complexes with the Nanog Octamer and Sox elements in F9 EC cell extracts.
We next examined the in vivo potential for the binding of OCT4 and SOX2 to the Nanog elements in ES and EC cells by chromatin immunoprecipitation assays with anti-OCT4 and anti-SOX2 antibodies (Fig. 5E). Exogenously expressed OCT4 and SOX2 bound to the Octamer and Sox elements, respectively, in both R1 ES and F9 EC cells, as seen for the Fgf4 Octamer and Sox elements.
A novel factor binds to Octamer and Sox elements in ES cell extracts.
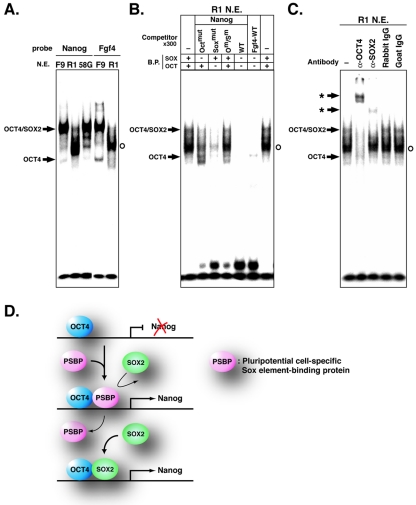
R1 ES cells, TMA-58G EG cells, and F9 EC cells share important properties, which include a robust capacity for self-renewal and expression of NANOG, OCT4, and SOX2 (Fig. 5A). However, full pluripotency is retained only by ES cells. Notably, R1 ES cells were isolated from normal inner cell mass cells of blastocyst-stage E3.5 embryos, while F9 EC cells were derived from E6.0 embryos through carcinogenesis and TMA-58G EG cells were derived from homing primordial germ cells in genital ridges of E12.5 embryos. Thus, some of the factors involved in maintaining pluripotency in ES cells may not be present in EC or EG cells. We therefore repeated the EMSA, binding competition, and supershift assays with nuclear extracts from R1 ES cells and TMA-58G EG cells. Strikingly, the major complex that formed when ES cell extracts were used was clearly distinct from the OCT4/SOX2 complex detected in EC cell extracts or in EG cell extracts (Fig. 6A). The same complex was detected in ES cell extracts with both the Nanog and the Fgf4 probes. A decrease in the band intensity of the ES cell-specific major complex in binding competition assays with the Octmut or Soxmut competitor probe demonstrated that the ES cell major complex required both the Octamer and the Sox elements for stable binding (Fig. 6B). This finding was confirmed by a lack of competition when the Octmut/Soxmut competitor probe was used. Supershift assays revealed that ES cell major complex mobility was reduced following incubation with anti-OCT4 antibody whereas, in contrast to the findings for F9 cell nuclear extracts (Fig. 5D), anti-SOX2 antibody had little effect on complex mobility (Fig. 6C). This finding was confirmed with a second anti-SOX2 antibody. These data showed that OCT4 is an essential component of the ES cell major complex but that SOX2 is not. Thus, another as-yet-undefined component (PSBP) preferentially associates with OCT4 to form an ES cell-specific complex on the Octamer and Sox elements in the Nanog promoter region.
FIG. 6.
Binding of endogenous OCT4 and PSBP to Octamer and Sox elements in R1 ES cells. Bands of the OCT4-DNA and OCT4/SOX2-DNA complexes are indicated by arrows, bands of the OCT4/PSBP-DNA complex are indicated by open circles, and supershifted bands are indicated by asterisks. (A) EMSA with Nanog and Fgf4 probes and nuclear extracts of F9 EC, TMA-58G EG, and R1 ES cells. (B) Competition assay with unlabeled probes with or without mutations in Octamer and/or Sox elements. (C) Supershift assay with anti-OCT4 or anti-SOX2 antibody and R1 ES cell nuclear extracts. Rabbit IgG and goat IgG were used as controls. (D) Schematic model for transcriptional regulation of Nanog in ES cells. In R1 ES cells but not in F9 EC cells, the OCT4/PSBP complex dominantly up-regulates Nanog transcription by binding to the Octamer/Sox element. PSBP binds to the Sox element with a greater affinity than SOX2.
As EMSAs carried out with the Fgf4 probe gave results similar to those obtained with the Nanog probe, it is evident that the ES cell-specific OCT4/PSBP complex is not specific for the Nanog sequence (Fig. 6A). This conclusion was confirmed by preincubation with anti-OCT4 antibody, which resulted in a loss of the OCT4/PSBP complex, whereas preincubation with anti-SOX2 antibody had no effect (unpublished data). Therefore, in ES cells, some genes that have Octamer and Sox elements as cis regulatory elements may be regulated in part by the synergistic action of OCT4 and PSBP in preference to OCT4 and SOX2.
DISCUSSION
The pluripotential state-specific gene Nanog is transcribed under the control of a regulatory region that lies within 332 bp upstream of the transcriptional start site. This region contains Octamer and Sox elements, which are highly conserved among the 5′-flanking regions of the mouse, monkey, and human Nanog genes. Indeed, the mouse and human Octamer and Sox elements were sufficient for up-regulating GFP reporter gene activity in both mouse and human ES cells. In nuclear extracts of F9 EC cells, OCT4 and SOX2, which are coexpressed in undifferentiated stem cells, were capable of binding to the Octamer and Sox elements, respectively. Interestingly, in nuclear extracts of ES cells, OCT4 dominantly bound to the Octamer element, while an undefined factor (PSBP) preferentially bound to the Sox element (Fig. 6D). Thus, Nanog expression may be dominantly controlled by an interaction between OCT4 and PSBP in ES cells.
It is evident that the Octamer and Sox elements are required for the up-regulation of mouse and human Nanog transcription and that factors that bind to these elements act to promote Nanog transcription through synergistic molecular interactions. Adjacent Octamer and Sox elements have been identified as cis regulatory elements in the Fgf4, Sox2, Utf1, and Fbx15 genes, which are expressed in EC and ES cells and during embryogenesis (17, 33, 34, 42). Sox2 is a member of the Sox (SRY-related HMG box) family that bears DNA-binding HMG domains and that is implicated in transcriptional regulation. The gene is expressed in pluripotential embryonic cells and neuronal cells (1). In pluripotential embryonic cells, expression is governed by at least two regulatory regions, the 5′-flanking region containing the CCAAT box and the 3′-flanking region containing the Octamer and Sox elements (34, 37). Fgf4 is a member of the fibroblast growth factor family that is expressed in blastocyst inner cell mass cells and in developing embryos, as well as in ES and EC cells (19). Both Octamer and Sox elements are located in the intragenic 3′ untranslated region, while a GT-box motif is located in the 3′-flanking region. Both regions are required for mediating optimal transcriptional Fgf4 activation (13, 14). Thus, the expression of some pluripotential embryonic cell-specific genes appears to require the action of Octamer and Sox elements in combination with other gene-specific cis regulatory elements. For Nanog, luciferase assays with ES cell extracts demonstrated that activity controlled by the −2342 5′-end genomic fragment was about 15% higher than that controlled by the −332 5′-end genomic fragment (Fig. 2B); this finding implied that an unidentified cis regulatory element(s) lying in the region from position −332 to position −2342 may function, in combination with the Octamer and Sox elements, in enhancing and determining the specificity of Nanog expression.
There is significant evidence that Nanog plays a key role in maintaining the pluripotency of ES cells and embryonic cells. Nanog-deficient ES and embryonic cells show a complete loss of pluripotency (9, 15), whereas Nanog overexpression results in the clonal expansion of ES cells via the bypassing of regulation by LIF-STAT3 signaling and maintenance of OCT4 levels (3). Therefore, it is important to understand how the expression of Nanog acts in harmony with the expression of other embryonic factors through molecular communications in the stem cell-specific regulatory network. Our data clearly show that the ternary protein-DNA complexes of OCT4/PBSP and Octamer and Sox elements efficiently formed even in the presence of SOX2. The data also suggest that this complex formation is essential for the activation of Nanog transcription in ES cells. During embryonic development, mouse Nanog RNA and protein expression can be detected from the morula stage to the epiblast stage of E7.5 embryos (8, 9, 15). The pattern of expression of Oct4 and Sox2 is temporally similar. Interestingly, in E7.5 embryos, Nanog expression is spatially enhanced in the caudal region (primitive streak region) of the epiblast (8, 9), whereas Sox2 expression is restricted to the presumptive neuroectoderm in the anterior and is excluded from the posterior (primitive streak region) (1). These data suggest that the OCT4/PSBP complex may activate Nanog expression in vivo. In primordial germ cells of E7.5 embryos, Oct4 and PGC7/Stella are expressed, whereas Nanog is repressed (9, 23, 40). However, Nanog is expressed in primordial germ cells at E11.5 and in EG cells derived from E12.5 embryos (3, 9). Our EMSA data (Fig. 6A) suggest that OCT4 and SOX2 may function in this up-regulation of Nanog transcription, although the profile of expression of SOX2 in germ cells is not fully understood. Thus, even though the control of Nanog expression is closely linked to the Oct4 regulatory network, Oct4 expression alone is insufficient for inducing Nanog expression. We suggest that the expression of Nanog is tightly regulated by competing coactivators (SOX2 and PSBP) in different cell types, which have different affinities for the Nanog Sox element sequence.
To understand the relationship between Nanog and Oct4, Nanog transcription in Oct4-deficient embryos was analyzed (3). mRNA in situ hybridization analysis demonstrated that Nanog expression was maintained in the blastocyst inner cell mass cells, suggesting that other pluripotential cell-specific factors may contribute to alternative transcriptional regulatory mechanisms. It has been shown that OCT1 and OCT6 are expressed in pluripotential embryonic cells (24, 29) and have the capacity to bind to Octamer elements in the Fgf4, Sox2, Utf1, and Rex-1 (Zfp-42) promoter regions (2, 5, 17, 34). Thus, instead of OCT4, OCT1 or OCT6 may participate to form the DNA-protein complex with SOX2 or PSBP on the Nanog Octamer and Sox elements. In fact, it has been reported that OCT6 but not OCT1 can form a complex with SOX2 on the Octamer and Sox elements in Sox2 (34). However, the affinity of OCT1 and OCT6 for binding to the Nanog Octamer element (TTTTGCAT) is low relative to that for binding to the consensus Octamer element (ATTAGCAT) (18). It remains to be explored whether other pluripotential genes contribute to the up-regulation of Nanog activity through interactions with the Octamer and Sox elements and whether Nanog expression in Oct4-null mutants is quantitatively equivalent to that in normal embryos.
ES cells promise to serve as an unlimited cell source of therapeutic materials for use in regenerative medicine. In clinical applications, it would be crucial to monitor the undifferentiated state of ES cells through numerous cell divisions and to selectively eliminate populations of spontaneously differentiated cells in cultures. Furthermore, following tissue-specific cell differentiation induction, a tool for the selective elimination of pluripotential ES cells is desirable to avoid contamination with a potential source for generating malignant tumors in vivo. For such purposes, the Nanog minimum promoter encompassing the Octamer and Sox elements may be a suitable tool for positive and negative selection of undifferentiated stem cells. Furthermore, genetically engineered human stem cells containing the herpes simplex virus thymidine kinase gene have been generated for selective elimination of undifferentiated ES cells with ganciclovir after in vitro and in vivo differentiation (25). In this context, the human Nanog promoter is an ideal candidate element for regulating the expression of a pluripotential cell-specific suicide gene. Further understanding of the mechanisms that regulate Nanog gene expression will also contribute to the field of stem cell engineering and its application to regenerative medicine.
Acknowledgments
We thank Andras Nagy for kindly providing the R1 ES cell line, Justin F. X. Ainscough for critical reading of the manuscript, and Emiko Moribe and Yoshimitsu Toyoda for DNA sequencing.
REFERENCES
- 1.Avilion, A. A., S. K. Nicolis, L. H. Pevny, L. Perez, N. Vivian, and R. Lovell-Badge. 2003. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17:126-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Shushan, E., J. R. Thompson, L. J. Gudas, and Y. Bergman. 1998. Rex-1, a gene encoding a transcription factor expressed in the early embryo, is regulated via Oct-3/4 and Oct-6 binding to an octamer site and a novel protein, Rox-1, binding to an adjacent site. Mol. Cell. Biol. 18:1866-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chambers, I., D. Colby, M. Robertson, J. Nichols, S. Lee, S. Tweedie, and A. Smith. 2003. Functional expression cloning of nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113:643-655. [DOI] [PubMed] [Google Scholar]
- 4.Czermin, B., R. Melfi, D. McCabe, V. Seitz, A. Imhof, and V. Pirrotta. 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111:185-196. [DOI] [PubMed] [Google Scholar]
- 5.Dailey, L., H. Yuan, and C. Basilico. 1994. Interaction between a novel F9-specific factor and octamer-binding proteins is required for cell-type-restricted activity of the fibroblast growth factor 4 enhancer. Mol. Cell. Biol. 14:7758-7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis, S., T. H. Aldrich, N. Stahl, L. Pan, T. Taga, T. Kishimoto, N. Y. Ip, and G. D. Yancopoulos. 1993. LIFR beta and gp130 as heterodimerizing signal transducers of the tripartite CNTF receptor. Science 260:1805-1808. [DOI] [PubMed] [Google Scholar]
- 7.Erhardt, S., I. H. Su, R. Schneider, S. Barton, A. J. Bannister, L. Perez-Burgos, T. Jenuwein, T. Kouzarides, A. Tarakhovsky, and M. A. Surani. 2003. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development 130:4235-4248. [DOI] [PubMed] [Google Scholar]
- 8.Hart, A. H., L. Hartley, M. Ibrahim, and L. Robb. 2004. Identification, cloning and expression analysis of the pluripotency promoting Nanog genes in mouse and human. Dev. Dyn. 230:187-198. [DOI] [PubMed] [Google Scholar]
- 9.Hatano, S., M. Tada, H. Kimura, S. Yamaguchi, T. Kono, T. Nakano, H. Suemori, N. Nakatsuji, and T. Tada. 2005. Pluripotential competence of cells associated with Nanog activity. Mech. Dev. 122:67-79. [DOI] [PubMed]
- 10.Kimura, H., M. Tada, N. Nakatsuji, and T. Tada. 2004. Histone code modifications on pluripotential nuclei of reprogrammed somatic cells. Mol. Cell. Biol. 24:5710-5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kubota, H., S. Matsumoto, S. Yokota, H. Yanagi, and T. Yura. 1999. Transcriptional activation of mouse cytosolic chaperonin CCT subunit genes by heat shock factors HSF1 and HSF2. FEBS Lett. 461:125-129. [DOI] [PubMed] [Google Scholar]
- 12.Laible, G., A. Wolf, R. Dorn, G. Reuter, C. Nislow, A. Lebersorger, D. Popkin, L. Pillus, and T. Jenuwein. 1997. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 16:3219-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luster, T. A., L. R. Johnson, T. K. Nowling, K. A. Lamb, S. Philipsen, and A. Rizzino. 2000. Effects of three Sp1 motifs on the transcription of the FGF-4 gene. Mol. Reprod. Dev. 57:4-15. [DOI] [PubMed] [Google Scholar]
- 14.Luster, T. A., and A. Rizzino. 2003. Regulation of the FGF-4 gene by a complex distal enhancer that functions in part as an enhanceosome. Gene 323:163-172. [DOI] [PubMed] [Google Scholar]
- 15.Mitsui, K., Y. Tokuzawa, H. Itoh, K. Segawa, M. Murakami, K. Takahashi, M. Maruyama, M. Maeda, and S. Yamanaka. 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113:631-642. [DOI] [PubMed] [Google Scholar]
- 16.Nichols, J., B. Zevnik, K. Anastassiadis, H. Niwa, D. Klewe-Nebenius, I. Chambers, H. Scholer, and A. Smith. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95:379-391. [DOI] [PubMed] [Google Scholar]
- 17.Nishimoto, M., A. Fukushima, A. Okuda, and M. Muramatsu. 1999. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol. Cell. Biol. 19:5453-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishimoto, M., S. Miyagi, T. Katayanagi, M. Tomioka, M. Muramatsu, and A. Okuda. 2003. The embryonic Octamer factor 3/4 displays distinct DNA binding specificity from those of other Octamer factors. Biochem. Biophys. Res. Commun. 302:581-586. [DOI] [PubMed] [Google Scholar]
- 19.Niswander, L., and G. R. Martin. 1992. Fgf-4 expression during gastrulation, myogenesis, limb and tooth development in the mouse. Development 114:755-768. [DOI] [PubMed] [Google Scholar]
- 20.Niwa, H., J. Miyazaki, and A. G. Smith. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24:372-376. [DOI] [PubMed] [Google Scholar]
- 21.O'Carroll, D., S. Erhardt, M. Pagani, S. C. Barton, M. A. Surani, and T. Jenuwein. 2001. The Polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 21:4330-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadowski, H. B., and M. Z. Gilman. 1993. Cell-free activation of a DNA-binding protein by epidermal growth factor. Nature 362:79-83. [DOI] [PubMed] [Google Scholar]
- 23.Saitou, M., S. C. Barton, and M. A. Surani. 2002. A molecular programme for the specification of germ cell fate in mice. Nature 418:293-300. [DOI] [PubMed] [Google Scholar]
- 24.Scholer, H. R., A. K. Hatzopoulos, R. Balling, N. Suzuki, and P. Gruss. 1989. A family of octamer-specific proteins present during mouse embryogenesis: evidence for germline-specific expression of an Oct factor. EMBO J. 8:2543-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuldiner, M., J. Itskovitz-Eldor, and N. Benvenisty. 2003. Selective ablation of human embryonic stem cells expressing a “suicide” gene. Stem Cells 21:257-265. [DOI] [PubMed] [Google Scholar]
- 26.Smith, A. G., J. K. Heath, D. D. Donaldson, G. G. Wong, J. Moreau, M. Stahl, and D. Rogers. 1988. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336:688-690. [DOI] [PubMed] [Google Scholar]
- 27.Stewart, C. 1980. Aggregation between teratocarcinoma cells and preimplantation mouse embryos. J. Embryol. Exp. Morphol. 58:289-302. [PubMed] [Google Scholar]
- 28.Stewart, C. L., P. Kaspar, L. J. Brunet, H. Bhatt, I. Gadi, F. Kontgen, and S. J. Abbondanzo. 1992. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature 359:76-79. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki, N., H. Rohdewohld, T. Neuman, P. Gruss, and H. R. Scholer. 1990. Oct-6: a POU transcription factor expressed in embryonal stem cells and in the developing brain. EMBO J. 9:3723-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tada, T., and M. Tada. 2001. Toti-/pluripotential stem cells and epigenetic modifications. Cell Struct. Funct. 26:149-160. [DOI] [PubMed] [Google Scholar]
- 31.Takeda, K., K. Noguchi, W. Shi, T. Tanaka, M. Matsumoto, N. Yoshida, T. Kishimoto, and S. Akira. 1997. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. USA 94:3801-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomson, J. A., J. Itskovitz-Eldor, S. S. Shapiro, M. A. Waknitz, J. J. Swiergiel, V. S. Marshall, and J. M. Jones. 1998. Embryonic stem cell lines derived from human blastocysts. Science 282:1145-1147. [DOI] [PubMed] [Google Scholar]
- 33.Tokuzawa, Y., E. Kaiho, M. Maruyama, K. Takahashi, K. Mitsui, M. Maeda, H. Niwa, and S. Yamanaka. 2003. Fbx15 is a novel target of Oct3/4 but is dispensable for embryonic stem cell self-renewal and mouse development. Mol. Cell. Biol. 23:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomioka, M., M. Nishimoto, S. Miyagi, T. Katayanagi, N. Fukui, H. Niwa, M. Muramatsu, and A. Okuda. 2002. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res. 30:3202-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, S. H., M. S. Tsai, M. F. Chiang, and H. Li. 2003. A novel NK-type homeobox gene, ENK (early embryo specific NK), preferentially expressed in embryonic stem cells. Gene Expr. Patterns 3:99-103. [DOI] [PubMed] [Google Scholar]
- 36.Ware, C. B., M. C. Horowitz, B. R. Renshaw, J. S. Hunt, D. Liggitt, S. A. Koblar, B. C. Gliniak, H. J. McKenna, T. Papayannopoulou, B. Thoma, L. Cheng, P. J. Donovan, J. J. Peschon, P. F. Bartlett, C. R. Willis, B. D. Wright, M. K. Carpenter, B. L. Davison, and D. P. Gearing. 1995. Targeted disruption of the low-affinity leukemia inhibitory factor receptor gene causes placental, skeletal, neural and metabolic defects and results in perinatal death. Development 121:1283-1299. [DOI] [PubMed] [Google Scholar]
- 37.Wiebe, M. S., P. J. Wilder, D. Kelly, and A. Rizzino. 2000. Isolation, characterization, and differential expression of the murine Sox-2 promoter. Gene 246:383-393. [DOI] [PubMed] [Google Scholar]
- 38.Williams, R. L., D. J. Hilton, S. Pease, T. A. Willson, C. L. Stewart, D. P. Gearing, E. F. Wagner, D. Metcalf, N. A. Nicola, and N. M. Gough. 1988. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 336:684-687. [DOI] [PubMed] [Google Scholar]
- 39.Winnier, G., M. Blessing, P. A. Labosky, and B. L. Hogan. 1995. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 9:2105-2116. [DOI] [PubMed] [Google Scholar]
- 40.Yeom, Y. I., G. Fuhrmann, C. E. Ovitt, A. Brehm, K. Ohbo, M. Gross, K. Hubner, and H. R. Scholer. 1996. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development 122:881-894. [DOI] [PubMed] [Google Scholar]
- 41.Ying, Q. L., J. Nichols, I. Chambers, and A. Smith. 2003. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115:281-292. [DOI] [PubMed] [Google Scholar]
- 42.Yuan, H., N. Corbi, C. Basilico, and L. Dailey. 1995. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 9:2635-2645. [DOI] [PubMed] [Google Scholar]