Abstract
The Saccharomyces cerevisiae PAK (p21-activated kinase) family kinase Ste20 functions in several signal transduction pathways, including pheromone response, filamentous growth, and hyperosmotic resistance. The GTPase Cdc42 localizes and activates Ste20 by binding to an autoinhibitory motif within Ste20 called the CRIB domain. Another factor that functions with Ste20 and Cdc42 is the protein Bem1. Bem1 has two SH3 domains, but target ligands for these domains have not been described. Here we identify an evolutionarily conserved binding site for Bem1 between the CRIB and kinase domains of Ste20. Mutation of tandem proline-rich (PxxP) motifs in this region disrupts Bem1 binding, suggesting that it serves as a ligand for a Bem1 SH3 domain. These PxxP motif mutations affect signaling additively with CRIB domain mutations, indicating that Bem1 and Cdc42 make separable contributions to Ste20 function, which cooperate to promote optimal signaling. This PxxP region also binds another SH3 domain protein, Nbp2, but analysis of bem1Δ versus nbp2Δ strains shows that the signaling defects of PxxP mutants result from impaired binding to Bem1 rather than from impaired binding to Nbp2. Finally, the PxxP mutations also reduce signaling by constitutively active Ste20, suggesting that postactivation functions of PAKs can be promoted by SH3 domain proteins, possibly by colocalizing PAKs with their substrates. The overall results also illustrate how the final signaling function of a protein can be governed by combinatorial addition of multiple, independent protein-protein interaction modules.
Signal transduction pathways in eukaryotic cells use a variety of modular protein-protein interaction domains to provide functional linkages between individual signaling components (7, 51). Genetic exchange of sequences encoding these protein-protein interaction modules is thought to have facilitated the evolution of signaling proteins with new properties, by creating new combinations of protein contacts that in sum would govern the protein's function (51). An example of a ubiquitous interaction module is the SH3 domain (30, 45), which binds target proteins by recognizing proline-rich ligands that usually conform to the general consensus sequence PxxP (where P stands for proline and x stands for any residue). In the budding yeast Saccharomyces cerevisiae, genomic techniques have identified all proteins with SH3 domains, and peptide library screening methods have been used to define the optimum ligands for most of them (67). In theory, the existence of well-characterized signal transduction pathways combined with the ease of genetic manipulations makes the yeast system an ideal test bed for studying the role of SH3 domain-mediated interactions in signaling. Surprisingly, however, there are few instances in which an SH3-mediated interaction is known to participate in a well-defined signaling pathway in yeast. The best-characterized example is the hyperosmotic response (HOG) pathway, which requires interaction between an SH3 domain from the transmembrane sensor protein Sho1 and a proline-rich motif in the kinase/scaffold protein Pbs2 (40, 43, 59, 76). Another well-studied SH3 domain protein in yeast is Bem1, which functions in the control of polarized growth during both normal cell division and the mating response (4, 8, 9, 12, 24, 29) and which also has a role in facilitating signal transduction in response to mating pheromones (39, 47). Bem1 has no known enzymatic activity but has a PX (phox homology) domain that binds phosphoinositides, as well as several protein-protein interaction domains including two SH3 domains (SH3-1 and SH3-2) and a PB1 (phox and Bem1) domain. Its ability to interact with multiple proteins involved in polarized growth has led to the suggestion that Bem1 may serve as an organizing scaffold to help promote interactions between its binding partners (8, 9, 24, 29, 35). Despite considerable interest in the function of Bem1, no functionally significant target binding sites have been identified for either of its two SH3 domains (67). This study reports the identification of such a site in the protein kinase Ste20.
The budding yeast kinase Ste20 is a founding member of the p21-activated kinase (PAK) family of protein kinases (16). Originally identified as a component in the yeast mating pathway (33, 58), Ste20 also functions in signaling pathways that regulate filamentous growth and the osmotic stress response (37, 49, 57, 61). In addition, Ste20 is involved in the control of actin organization, polarized growth, and cell cycle transitions (15, 19, 23, 27, 28, 63, 73). In the mating pathway, Ste20 activates a mitogen-activated protein (MAP) kinase cascade in response to extracellular mating pheromones (reviewed in references 17, 20, and 25). Binding of these pheromones to G protein-coupled receptors triggers the release of Gβγ dimers, which bind Ste20 (36) and also recruit to the plasma membrane the kinase cascade scaffold protein Ste5 (22, 56, 69, 71). These Gβγ-Ste20 and Gβγ-Ste5 interactions result in the Ste5-associated kinase, Ste11 (a MAP kinase kinase kinase [MAPKKK]), being brought into close proximity with Ste20, which consequently phosphorylates and activates Ste11 (56, 68, 74). This activating event is then propagated through successive kinases in the mating pathway MAP kinase cascade.
Ste20 is normally enriched at the cell periphery, at regions undergoing polarized cell growth such as the tips of buds and mating projections (34, 53, 72). This polarized localization depends on interaction with the membrane-bound GTPase Cdc42 (1, 31, 34, 47, 53). Cdc42 itself is similarly localized and associates directly with the plasma membrane via posttranslational modification of its carboxyl terminus with a lipophilic prenyl moiety (77). In addition to localizing Ste20, Cdc42 activates Ste20 kinase activity, because the domain that binds Cdc42 (the CRIB domain) overlaps an autoinhibitory domain that regulates Ste20 kinase activity (31, 65). Point mutations in the Ste20 CRIB domain that precisely disrupt Cdc42 binding without disrupting the autoinhibitory function cause Ste20 to become refractory to activation by Cdc42 and cause a strong loss of signaling activity in vivo (1, 31). In contrast, mutations that disrupt the autoinhibitory function of the CRIB domain, or that remove the CRIB domain altogether, confer constitutive (Cdc42-independent) activity on Ste20 (31, 47). Thus, the localization and activity of Ste20 are normally intimately connected due to their dependence on Cdc42, though this association can be uncoupled by deleting the CRIB domain, which yields Ste20 that is active yet delocalized (1, 31, 34, 47, 53). Remarkably, although signaling by this CRIB deletion form of Ste20 is detectably reduced (e.g., in the mating response), it is not reduced as drastically as might be expected from its strong delocalization, given that signaling is thought to be initiated at the plasma membrane (31, 56, 69). One possible explanation for this relatively mild defect is that Ste20 might contain residual localization ability by binding to other polarized proteins.
Previous work (18, 35) has found that Ste20 can interact with Bem1, which shows a cortical localization similar to that of Cdc42 and Ste20 (2, 29). Interactions between PAKs and SH3 domain proteins are common in other systems (6, 11, 26, 38, 41). Some evidence suggests that Bem1 may serve a parallel or overlapping role with Cdc42, since overexpression of Bem1 can partially compensate for defects in Ste20 localization and signaling in certain cdc42 mutants (47). Nevertheless, the manner in which Bem1 affects pheromone signaling remains poorly defined, in part because Bem1 also binds other signaling proteins in the pheromone response pathway such as Ste5 and Far1 (35, 39). In this study, we investigate the binding between Ste20 and Bem1 and the contribution of this interaction to Ste20 function. We demonstrate that the regulatory (nonkinase) half of Ste20, which contains the Cdc42-binding domain, also contains a separate proline-rich motif that binds to Bem1. We then report on how mutations that perturb Bem1-Ste20 binding affect Ste20 function in several signaling pathways, both in the presence and in the absence of functional Cdc42-Ste20 interaction.
MATERIALS AND METHODS
Yeast strains.
Yeast strains are listed in Table 1. New strains were constructed as follows. PPY1249 is a segregant from a cross between the unpublished strain PPY650 (MATα ste20Δ) and YEL285 (MATa ADE2 ste20Δ285-582) (34). PPY1287 (ssk1Δ ste20Δ) was obtained via a cross between SO621 (MATa ssk1::HIS3 ste50::LEU2), a gift from S. O'Rourke, and the unpublished strain PPY1281 (MATα ste20-1::TRP1). PPY1341 and PPY1343 are segregants from a cross between strains PPY1329 and PPY1331, which were constructed as follows: PPY1329 contains a HIS2-marked FUS1-lacZ reporter integrated at the FUS1 locus, introduced by transformation with plasmid pFL-HIS2 (47) into strain MOSY0151 (MATα ste20::TRP1), a gift from D. Lew; PPY1331 (MATa bem1::ura3::LEU2) was derived by transformation of strain JMY1128 (bem1::URA3), a gift from D. Lew, with the marker swap construct pUC4-ura3::LEU2 (46). PPY1356 harbors an NBP2::myc13-kanR fusion, inserted at the genomic NBP2 locus by transformation of strain DLY1 with KpnI-digested pPP1446, which contains a 477-bp C-terminal fragment of NBP2 lacking the native stop codon in the BamHI site of the myc13-kanR plasmid pFA6a-13Myc-kanMX6 (70). PPY1415 and PPY1456 were derived from PPY1343 and PPY913, respectively, by replacing the complete NBP2 open reading frame with an nbp2::kanR fragment, obtained by PCR of the kanR cassette from pFA6a-kanMX6 (70) with primers whose 5′ ends contain 40 bp of homology to sequences immediately upstream and downstream of the NBP2 open reading frame. PPY1646 (ssk2Δ ssk22Δ ste20Δ) was created by deleting STE20 by use of plasmid pDH104 (74) in strain TM252 (40). PPY1691 was derived from the unpublished strain PPY860 (ste20Δ ste5Δ) by transformation with the hog1::hisG::URA3::hisG fragment from pBC99 (49), selection for uracil prototrophy, and then selection for 5-fluoroorotic acid (5-FOA) resistance.
TABLE 1.
Yeast strains used in this study
| Strain backgrounda | Strain name | Genotype or relevant genotype | Source |
|---|---|---|---|
| a | KBY211 | MATα ste20::ADE2 cla4::LEU2 + YCpTRP1-cla4-75ts | 28 |
| a | PPY866 | MATα FUS1::FUS1-lacZ::LEU2 ste20-1::TRP1 ste5::ADE2 ste4::ura3FOA | 56 |
| a | PPY913 | MATaFUS1::FUS1-lacZ::LEU2 ste20-3Δ::TRP1 | 31 |
| a | PPY1249 | MATaADE2 ste20-1::TRP1 | This study |
| a | PPY1287 | MATassk1::HIS3 ste20-1::TRP1 | This study |
| a | PPY1456 | MATaFUS1::FUS1-lacZ::LEU2 ste20-3Δ::TRP1 nbp2Δ::kanR | This study |
| a | PPY1691 | MATaFUS1::FUS1-lacZ::LEU2 ste20-1::TRP1 ste5::ADE2 hog1::hisG | This study |
| b | PPY1209 | MATahis3::hisG leu2::hisG trp1::hisG ura3-52 ste20-1::TRP1 | 31 |
| c | PPY760 | MATaade2 his3 leu2 trp1 LYS2::lexAop-HIS3 URA3::lexAop-lacZ far1::ADE2 | 10 |
| d | DLY1 | MATabar1 ade1 his2 leu2 trp1 ura3 | 8 |
| d | DLY4000 | MATabar1 ade1 his3 leu2 trp1 ura3 BEM1-myc12::HIS3 | 8 |
| d | PPY1341 | MATabar1 ade1 his2 leu2 trp1 ura3 FUS1::FUS1-lacZ::HIS2 ste20::TRP1 bem1::ura3::LEU2 | This study |
| d | PPY1343 | MATabar1 ade1 his2 leu2 trp1 ura3 FUS1::FUS1-lacZ::HIS2 ste20::TRP1 | This study |
| d | PPY1356 | MATabar1 ade1 his2 leu2 trp1 ura3 NBP2-myc13::kanR | This study |
| d | PPY1415 | MATabar1 ade1 his2 leu2 trp1 ura3 FUS1::FUS1-lacZ::HIS2 ste20::TRP1 npb2Δ::kanR | This study |
| e | PPY1646 | MATaleu2 trp1 ura3 ssk2::LEU2 ssk22::LEU2 ste20Δ-3::TRP1 | This study |
| e | PT2α | MATα hom3 ilv1 can1 | 56 |
Strain backgrounds: a, W303 (ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1); b, Σ1278b; c, S288C; d, 15Dau; e, other.
Plasmids.
Plasmids used in this study are listed in Table 2. Signaling properties of Ste20 mutants were studied in two plasmid contexts, pRL116 and pPP1001. pRL116 (34) is a CEN URA3 plasmid expressing a GFP(S65T)-STE20 fusion gene from the native STE20 promoter. pPP1001 (31) is a CEN URA3 plasmid expressing the native, untagged STE20 gene from its own promoter. The degree to which these plasmids complement ste20Δ phenotypes has been discussed previously (31).
TABLE 2.
Plasmids used in this study
| Plasmid name | Description | Source |
|---|---|---|
| pRL116 | CEN URA3 GFP-STE20 | 34 |
| pPP964 | CEN URA3 GFP-ste20-S338A | 31 |
| pPP1010 | CEN URA3 GFP-ste20-Δ334-369 | 31 |
| pPP1211 | CEN URA3 GFP-ste20-P475G P477A | This study |
| pPP1212 | CEN URA3 GFP-ste20-S338A P475G P477A | This study |
| pPP1213 | CEN URA3 GFP-ste20-Δ334-369 P475G P477A | This study |
| pPP1001 | CEN URA3 STE20 | 31 |
| pPP1002 | CEN URA3 ste20-S338A | 31 |
| pPP1011 | CEN URA3 ste20-Δ334-369 | 31 |
| pPP1214 | CEN URA3 ste20-P475G P477A | This study |
| pPP1215 | CEN URA3 ste20-S338A P475G P477A | This study |
| pPP1216 | CEN URA3 ste20-Δ334-369 P475G P477A | This study |
| pBTM116 | 2μm TRP1 lexA DBD vector | 3 |
| pB20N2 | 2μm TRP1 lexA DBD-ste20 1-499 | 31 |
| pB20N | 2μm TRP1 lexA DBD-ste20 1-439 | This study |
| pB20BA | 2μm TRP1 lexA DBD-ste20 371-499 | This study |
| pB20BB | 2μm TRP1 lexA DBD-ste20 371-439 | This study |
| pB20BC | 2μm TRP1 lexA DBD-ste20 434-499 | This study |
| pPP1062 | 2μm TRP1 lexA DBD-ste20 1-499(Δ334-369) | 31 |
| pPP1180 | 2μm TRP1 lexA DBD-ste20 1-499(P475G P477A) | This study |
| pB20N2 point mutants | 2μm TRP1 lexA DBD-ste20 1-499(K469A, I471G,…S481A) | This study |
| pBTM-CDC24 | 2μm TRP1 lexA DBD-CDC24 | 10 |
| pDH37 | 2μm TRP1 lexA DBD-STE20 1-939 | 35 |
| pGAD424 | 2μm LEU2 GAL4 AD vector | 3 |
| pPP1027 | 2μm LEU2 GAL4 AD-CDC42-G12V C188S | 31 |
| pRL51 | 2μm LEU2 GAL4 AD-BEM1 157-551 | 35 |
| pRL5.2 | 2μm LEU2 GAL4 AD-BEM1 1-551 | 35 |
| pGADXP | 2μm LEU2 strongADH1pr GAL4 AD vector | 10 |
| pPP1355 | 2μm LEU2 strongADH1pr GAL4 AD-NBP2 | This study |
| pXP-BEM1 | 2μm LEU2 strong ADH1pr GAL4 AD-BEM1 | This study |
| pXP-BEM1-W108A | 2μm LEU2 strongADH1pr GAL4 AD-bem1(W108A) | This study |
| pXP-BEM1-W192A | 2μm LEU2 strongADH1pr GAL4 AD-bem1(W192A) | This study |
| pRD56 | CEN URA3 GAL1pr-GST vector | 55 |
| pRDSTE20ATG | CEN URA3 GAL1pr-GST-STE20 | 53, 55 |
| pRDSTE20RI | CEN URA3 GAL1pr-GST-ste20ΔN (Δ1-495) | 55 |
| pRD56-K649M | CEN URA3 GAL1pr-GST-ste20-K649M | 19 |
| pPP1254 | CEN URA3 GAL1pr-GST-ste20-P475G P477A | This study |
| pPP1327 | CEN URA3 GAL1pr-GST-ste20-L369G | This study |
| pRS316 | CEN URA3 | 64 |
| pH-GS5-CTM | CEN HIS3 GAL1pr-STE5-CTM | 56 |
| pH-SL2 | CEN HIS3 GAL1pr-His6-myc-STE5(P44L)-GST | 31 |
| pPP1202 | 2μm HIS3 BEM1 | This study |
Point mutations in STE20 were first generated in pRL116, by Pfu polymerase-mediated extension of complementary mutagenic oligonucleotides, according to the instructions in the QuikChange site-directed mutagenesis kit (Stratagene). To ensure the absence of spurious mutations, restriction fragments containing the desired mutations were transferred back into unmutagenized plasmids, and the transferred fragments were sequenced. The P475G P477A (PP-GA) mutation was transferred on a SalI-SgrAI fragment into the green fluorescent protein (GFP)-STE20 plasmids pRL116, pPP964, and pPP1010 to create pPP1211, pPP1212, and pPP1213, respectively, and into the non-GFP fusion constructs pPP1001, pPP1002, and pPP1011 to create pPP1214, pPP1215, and pPP1216, respectively. In addition, the P475G P477A mutation was introduced directly by site-directed mutagenesis of pB20N2 to create pPP1180. Point mutations are marked by the gain or loss of restriction sites: a loss of EcoRV for the S338A mutation and a gain of BlpI for the P475G P477A mutation.
The GAL1pr-GST-STE20 constructs encoding wild-type Ste20 (pRDSTE20ATG) and Ste20ΔN (pRDSTE20-RI), as well as the vector control (pRD56), have been described previously (19, 53, 55). Note that pRDSTE20ATG was also called CY409 in reference 53 and was mistakenly given the name of the empty vector, pRD56, in reference 19, as a consequence of which the GAL1pr-GST-ste20-K649M construct was designated pRD56-K649M (19). Variants pPP1254 (PP-GA) and pPP1327 (L369G) were derived from pRDSTE20ATG by transfer of the SalI-SgrAI fragment from pPP1211 or the BsrGI-SgrAI fragment from pPP1117.
The two-hybrid plasmids pB20N2 and pPP1062 have been described previously (31). Additional two-hybrid plasmids, pB20N, pB20BA, pB20BB, and pB20BC, were constructed by PCR amplification of the STE20 codons indicated in Table 2 (followed by a stop codon introduced by the downstream primer) and insertion into pBTM116 (3). Sequences in pB20N were inserted as an EcoRI-PstI fragment; those in pB20BA, pB20BB, pB20BC, and pPP1309 were inserted as BamHI-PstI fragments. In pB20N2, pB20BA, and pB20BC, a silent mutation from the downstream PCR primer disrupts the native EcoRI site near STE20 codon 499. The PP-GA mutation in pPP1180 was introduced by directed mutagenesis of pB20N2 as described above. Thirteen additional derivatives of pB20N2 containing point mutations at single codons from Lys469 to Ser481 (K469A through S481A) were made by first introducing a silent XhoI site (lowercase letters) overlapping STE20 codons 463 to 465 (TCc tcg agt) and then replacing the XhoI-PstI fragment with XhoI/PstI-cut PCR fragments in which mutations were introduced by the upstream (forward) primer.
The AD-Nbp2 plasmid pPP1355 was made by PCR amplification of full-length NBP2 and insertion into pGADXP as an EcoRI-SalI fragment. The activation domain (AD) fusions to Bem11-551 (pRL5.2) and Bem1157-551 (pRL51.1) have been described previously (35). Plasmid pXP-BEM1 was constructed by PCR amplification of full-length BEM1 and ligation into pGADXP as an EcoRI-BglII fragment. Coligation of two PCR fragments was used to generate pXP-BEM1-W108A (EcoRI-HindIII and HindIII-BglII) and pXP-BEM1-W192A (EcoRI-SalI and SalI-BglII); the W108A mutation introduces an MluI site, and the W192A mutation disrupts a BlpI site.
Plasmid pPP1202 (2μm HIS3 BEM1) is a HIS3-marked derivative of pPB321 (54), created by homologous recombination in S. cerevisiae using the EcoRI-digested marker swap construct pUC4-ura3::HIS3 (46).
Pheromone response, mating, and β-galactosidase assays.
Halo assays of growth arrest, patch mating assays, and FUS1-lacZ transcription assays in response to α-factor or galactose-inducible constructs were performed as described previously (31). For cross talk assays, transformants were grown overnight in selective glucose medium to mid-exponential phase; these cultures were split in two, diluted with an equal volume of growth medium with or without 2 M sorbitol (final concentration, 0 or 1 M), and then incubated for 3 h at 30°C before the β-galactosidase assay. Liquid β-galactosidase assays (31) and filter β-galactosidase assays (3) were performed as described previously.
Yeast cell lysates and coprecipitations.
For coprecipitation of Bem1-myc or Nbp2-myc with glutathione S-transferase (GST)-Ste20, transformants were first grown overnight at 30°C to an optical density at 660 nm of 0.6 to 1.0 in uracil-deficient medium (−Ura)-raffinose containing 0.1% glucose; then they were induced with galactose (2%) for 3 h. Equivalent numbers of cells (usually 1 × 108 to 4 × 108) were harvested, and clarified cell lysates were prepared by glass bead lysis as described elsewhere (31), except that additional Triton X-100 was not added after the glass bead lysis step. GST-Ste20 was precipitated from the clarified lysates by addition of 40 μl of a 50% slurry (in buffer B) of glutathione-Sepharose beads (Amersham Pharmacia Biotech), followed by mixing on a nutator for 1 h at 4°C. Beads were pelleted at 2,400 × g, washed four times with 200 μl of cold buffer B, resuspended in 25 μl of 1.25× sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer, boiled 2 min, and centrifuged 5 min. Eight microliters was run on duplicate sodium dodecyl sulfate-7% polyacrylamide gels, followed by transfer and immunodetection as described elsewhere (31). GST-Ste20 was detected by using a mouse anti-GST antibody at a 1:1,000 dilution (B-14; Santa Cruz Biotechnology). Bem1-myc and Nbp2-myc were detected by using a rabbit anti-myc antibody at 1:2,000 (a gift of the R. Vallee lab, Columbia University). Some Bem1-myc was always detectable in the precipitates, even from control lysates of cells expressing GST alone.
Filamentation assays.
Agar invasion assays were performed as described previously (13). Briefly, cells picked from freshly grown (2- to 4-day-old) colonies on −Ura plates were patched onto yeast extract-peptone-dextrose (YPD) plates, incubated for 30 h at 30°C, and then photographed before and after rinsing under a gentle stream of deionized water. To analyze filamentous morphology, cells were plated onto a low-glucose −Ura medium (14) and incubated at 30°C for 24 h prior to photomicroscopy.
Microscopy.
For GFP-Ste20 observations, transformants were grown at 30°C in −Ura-glucose or −Ura-His-glucose medium, and cells were examined without fixation by using a Nikon E600 epifluorescence microscope equipped with a 50× Plan oil immersion objective. Cell morphology was photographed by using a Nikon Labophot-2 microscope equipped with a 30× extra-long working distance objective and Hoffman modulation contrast optics.
RESULTS
Identification of a Bem1-binding site in the Ste20 regulatory domain.
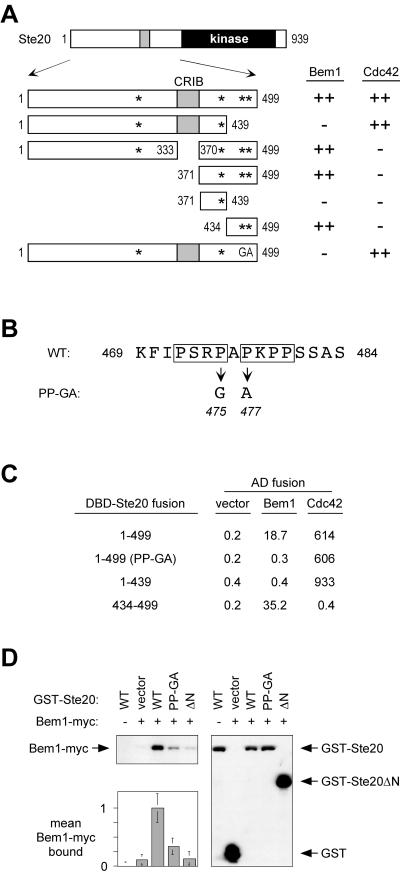
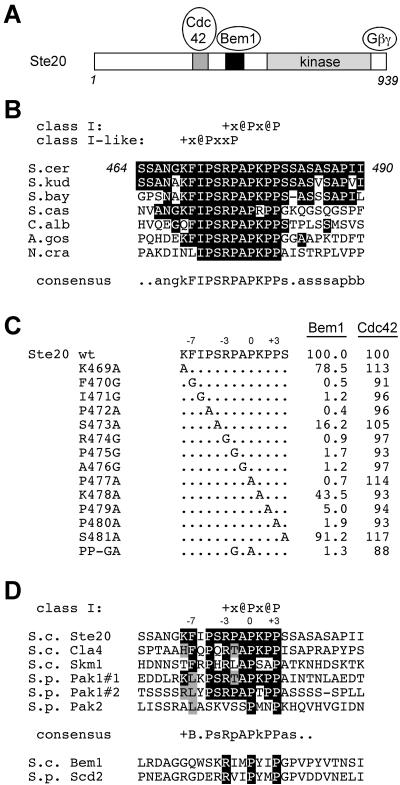
As described in the introduction, when active Ste20 is delocalized by removal of the Cdc42-binding (CRIB) domain, a measurable but mild signaling defect results. To explain why this defect was relatively small, we considered the possibility that interaction between Ste20 and Bem1 could provide some residual localization information. To determine whether Bem1 and Cdc42 could in fact contribute independent, additive effects on Ste20 function, we first mapped the binding site for Bem1 on Ste20. Using a two-hybrid assay and fusions to different fragments of Ste20, we found that residues 434 to 499, located between the CRIB and kinase domains of Ste20, were both necessary and sufficient for binding to Bem1 and were dispensable for binding to Cdc42 (Fig. 1A). Within this region lies a proline-rich segment (Fig. 1B) containing two adjacent matches to a core binding motif (PxxP) for SH3 domains (45), raising the possibility that these are recognized by a Bem1 SH3 domain. To test this notion, we made point mutations that simultaneously disrupt one proline in each motif (Pro475 to Gly and Pro477 to Ala) (Fig. 1B). Indeed, this double mutation (designated PP-GA) eliminated Bem1 binding by the two-hybrid assay as strongly as did deletion of residues 434 to 499 but had no effect on Cdc42 binding (Fig. 1A and C). By contrast, two other PxxP sites elsewhere in the Ste20 N terminus (Fig. 1A) were neither necessary nor sufficient for Bem1 binding (Fig. 1A and C).
FIG. 1.
A proline-rich domain between the CRIB and kinase domains of Ste20 contains a Bem1-binding site. (A) Fragments used to map a Bem1-binding motif within Ste20 residues 434 to 499 by two-hybrid analysis. Asterisks indicate PxxP motifs; GA represents mutations in two of these motifs, as shown in panel B. DNA binding domain fusions to Ste20 fragments (from top to bottom, pB20N2, pB20N, pPP1062, pB20BA, pB20BB, pB20BC, and pPP1180) were coexpressed in PPY760 with AD fusions to Bem1157-551 (pRL51.1), Cdc42G12V/C188S (pPP1027), or the vector (pGAD424). Interactions were scored as positive (++) or negative (−), in comparison to those for vector controls, by a filter β-galactosidase assay. Bem1 interaction results were similar when full-length Bem11-551 (pRL5.2) rather than Bem1157-551 was used, though in all cases the signal was stronger with the latter. (B) Sequence showing tandem PxxP motifs (boxed) in theminimal Bem1-binding region and mutations of Pro475 and Pro477 that constitute the PP-GA allele. WT, wild type. (C) Quantitative measurements of key two-hybrid interactions from panel A, using the same strain and plasmids (pRL51.1 for Bem1). Measurements are expressed as mean β-galactosidase units (n = 3 or 4; all standard deviations were within 40% of the mean). DBD, DNA binding domain. (D) Coprecipitation of GST-Ste20 and Bem1-myc is disrupted by the PP-GA mutation. Strains expressing myc12-tagged Bem1 (+) (DLY4000) or untagged Bem1 (−) (DLY1) were transformed with a vector (pRD56) or constructs expressing the indicated Ste20 mutants (WT, PP-GA, or ΔN) as galactose-inducible GST fusions. Following galactose induction, glutathione precipitates were analyzed by immunoblotting with anti-myc (top left) or anti-GST (right) antibodies. (Bottom left) Densitometric quantification of Bem1-myc signals from four separate experiments (mean ± standard deviation), with the mean density in the negative control (GST-Ste20WT in DLY1) assigned a value of zero and that in the positive control (GST-Ste20WT in DLY4000) assigned a value of 1.
To further substantiate these findings, we used a coprecipitation assay involving full-length GST-Ste20 and Bem1-myc fusion proteins. We found that the PP-GA mutation strongly decreased the amount of Bem1-myc that coprecipitated with GST-Ste20 (Fig. 1D). The residual interaction was eliminated by complete deletion of the 495 N-terminal residues of Ste20 (GST-Ste20ΔN). These coprecipitation results are consistent with those from the two-hybrid assays and indicate that the PP-GA mutation disrupts a predominant, though perhaps not exclusive, site of interaction between the full-length Ste20 and Bem1 proteins.
Additional evidence suggested that this tandem PxxP motif binds the second SH3 domain (SH3-2) in Bem1. First, the pattern of two-hybrid binding was the same regardless of whether the Bem1 constructs contained both SH3 domains (residues 1 to 551) or only the second SH3 domain (residues 157 to 551) (see Fig. 1A legend). Second, we made point mutations within full-length Bem1 in each SH3 domain (W108A in SH3-1 and W192A in SH3-2) at highly conserved tryptophans that are involved in binding proline-rich ligands (32). Only the mutation in the second SH3 domain (W192A) disrupted binding to Ste20; the mutation in the first SH3 domain (W108A) did not (Table 3). This was true when either of two Ste20 fragments (comprising residues 1 to 499 or 434 to 499) was used, while full-length Ste20 (residues 1 to 939) interacted negligibly with Bem1 by this assay, as observed previously (35). The W192A mutation also reduced the binding of Bem1 to Cdc24 (which served here as a control), but this was a much milder effect than the complete elimination of Ste20 binding. Together, therefore, these results show that the SH3-1 domain is dispensable, but an intact SH3-2 domain in Bem1 is required, for binding to the proline-rich motif in Ste20.
TABLE 3.
Effects of Bem1 SH3 domain mutations on interaction with Ste20
| DBD fusiona | Interactionb with the following AD-BEM1 (1-551) fusionc:
|
|||
|---|---|---|---|---|
| Vector | Wild type | W108A (SH3-1) | W192A (SH3-2) | |
| Ste20 1-939 | 0.1 ± 0.1 | 0.2 ± 0.1 | 0.2 ± 0.1 | 0.2 ± 0.1 |
| Ste20 1-499 | 0.1 ± 0.1 | 15.0 ± 6.6 | 21.3 ± 5.9 | 0.1 ± 0.1 |
| Ste20 434-499 | 0.1 ± 0.1 | 298.5 ± 69.2 | 430.1 ± 73.6 | 0.4 ± 0.6 |
| Cdc24 | 0.1 ± 0.1 | 143.6 ± 29.6 | 176.3 ± 31.4 | 47.2 ± 6.2 |
Plasmids pDH37, pB20N2, pB20BC, and pBTM-CDC24. DBD, DNA binding domain.
Expressed as mean β-galactosidase units ± standard deviation (n = 6) in the two-hybrid tester strain PPY760.
Plasmids pGADXP, pXP-BEM1, pXP-BEM1-W108A, and pXP-BEM1-W192A.
The Bem1-binding site collaborates with the CRIB domain for optimum signaling.
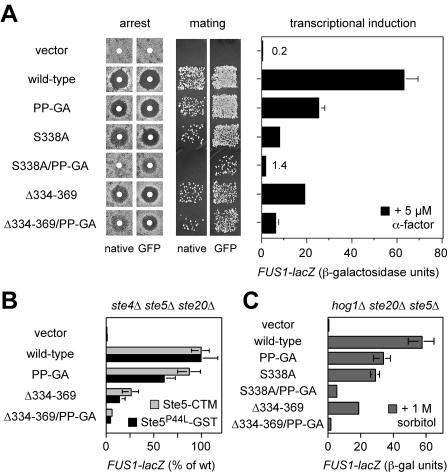
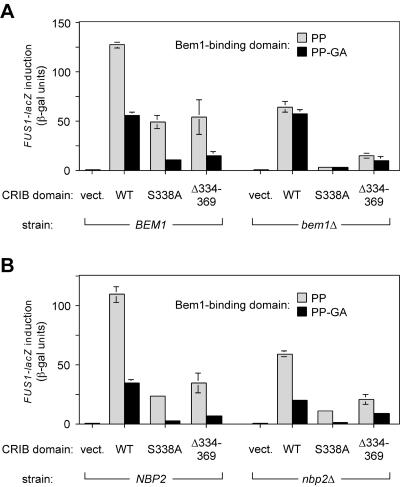
To study the effects of Bem1 binding on Ste20 signaling, the PP-GA mutation was introduced into vectors expressing full-length STE20 from its own promoter in either GFP-tagged or untagged (native) forms. Previous work showed that the GFP-tagged construct supplies a wild-type level of Ste20 function, whereas the untagged, native construct functions at a slightly reduced level (likely due to the lack of 3′ untranslated sequences [31]), which can mildly enhance phenotypic severity (31). When introduced into ste20Δ cells and tested for effects on pheromone response (Fig. 2A), the PP-GA mutant showed a mild defect in FUS1-lacZ induction (60% reduced from that of the wild type), though not strong enough to cause a clear phenotype in assays of growth arrest or mating (Fig. 2A). To address the relationship between Bem1 binding and Cdc42 binding, the PP-GA mutation was combined with two previously characterized mutations in the CRIB domain (31), which disrupt Cdc42 binding either mildly (S338A) or completely (Δ334-369). The partial effect of the PP-GA mutation became more pronounced when coupled with the mildest Cdc42-binding mutation, S338A, such that the combined mutant was strongly defective in FUS1-lacZ, arrest, and mating assays (Fig. 2A). The PP-GA mutation also reduced signaling when combined with the complete CRIB domain deletion, Δ334-369, suggesting that Bem1 binding can affect Ste20 signaling in the absence of Ste20-Cdc42 interaction. Altogether, these observations show that the Bem1-binding domain has a positive effect on Ste20 signaling, that Cdc42 and Bem1 make separable contributions to Ste20 function, and that these two interactions are not redundant but in fact collaborate to promote optimal signaling by Ste20.
FIG. 2.
Phenotypic consequences of point mutations (PP-GA) in the Bem1-binding site. In all panels, the vector is pRS316, and native STE20 and GFP-STE20 plasmids are as described in Table 2. (A) Mating pathway phenotypes of strains with the Ste20 PP-GA mutation, alone and in combination with mutations in the CRIB domain. Arrest (left) and mating (center) results are shown for both native and GFP-fused versions of each Ste20 mutant. Induction of the transcriptional reporter FUS1-lacZ by α-factor (5 μM; 2 h) is shown for the GFP fusions (right) as means ± standard deviations (n = 4). All assays used strain PPY913. (B) The effect of the PP-GA mutation on mating pathway signaling is independent of pheromone and Gβγ. Ste20-dependent signaling was activated in the absence of pheromone or Gβ (Ste4) by galactose-induced synthesis of Ste5-CTM (pH-GS5-CTM) or Ste5P44L-GST (pH-SL2); the host strain, PPY866 (ste4Δ ste5Δ ste20Δ), also harbored the indicated GFP-STE20 constructs. After a 3-h galactose treatment, FUS1-lacZ induction was measured (mean ± standard deviation; n = 9). To facilitate comparison between the two Ste5 reagents, results were expressed relative to those for wild-type (wt) Ste20, which yielded 134 (Ste5-CTM) and 88.3 (Ste5P44L-GST) mean raw units. (C) Hyperosmotic cross talk signaling shows that the effect of the PP-GA mutation is also independent of Ste5. Strain PPY1691 (hog1Δ ste20Δ ste5Δ) harboring the indicated GFP-STE20 constructs was treated with 1 M sorbitol for 3 h, after which FUS1-lacZ induction was measured (mean ± standard deviation; n = 3). Mean expression in cells not induced with sorbitol was <1 U (data not shown). Similar results were observed for hog1Δ ste20Δ and hog1Δ ste4Δ ste5Δ ste20Δ strains (data not shown).
Because Ste20Δ334-369 is a constitutively active kinase (31), the fact that its mating pathway signaling is reduced by the PP-GA mutation suggests that Bem1 can influence Ste20 at a postactivation step, possibly by promoting colocalization with substrates. To rule out the alternative possibility that the PP-GA mutation affects signaling by disrupting the binding of Ste20 to Gβγ or Ste5, each of which can interact with Ste20 (35, 36), we activated signaling in ways that bypass these factors. The requirement for pheromone and Gβγ was bypassed by expression of membrane-targeted Ste5 (Ste5-CTM [56]) or mutationally activated Ste5 (Ste5-P44L-GST [62]). Here, the PP-GA mutation mildly affected signaling by Ste20 with an intact CRIB domain and clearly decreased signaling by the Ste20Δ334-369 derivative, which lacks the CRIB domain (Fig. 2B). To bypass the requirement for Ste5 (as well as pheromone and Gβγ), we took advantage of Ste5-independent cross talk activation of the mating pathway, which occurs when hog1Δ cells are subjected to hypertonic stress (49). To ensure that Ste5 made no contribution, these experiments were performed in hog1Δ ste20Δ ste5Δ cells. As with pheromone signaling, cross talk signaling was moderately reduced by single mutations in either the CRIB or the Bem1-binding domain and was strongly reduced when mutations in the two domains were combined (Fig. 2C). In total, these results indicate that the PP-GA mutation affects the ability of Ste20 to activate downstream signaling rather than affecting its ability to be regulated by pheromone, Gβγ, Ste5, or Cdc42.
For several reasons, we do not believe that the PP-GA phenotypes are due to reduced Ste20 protein levels: (i) the mutation did not affect GST-Ste20 levels when expressed from a strong promoter (see Fig. 1D); (ii) we saw no consistent effect on levels of natively expressed GFP-Ste20 fusions, though these were only faintly detectable by immunoblotting even after immunoprecipitation with anti-GFP antibodies from large (50- to 100-ml) cultures (data not shown); and (iii) further experiments below show that the PP-GA mutation confers no defect in a specific genetic context (bem1Δ cells), making trivial explanations for the signaling phenotypes unlikely.
Role of the Bem1-binding domain in viability, polarization, and Ste20 localization.
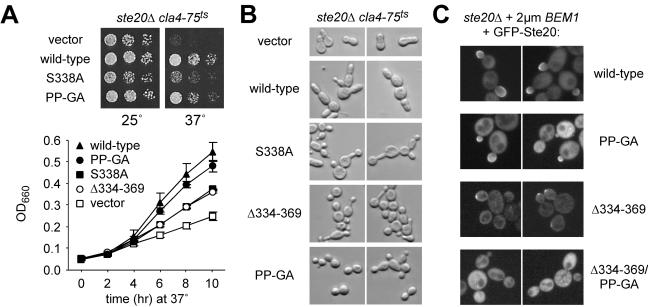
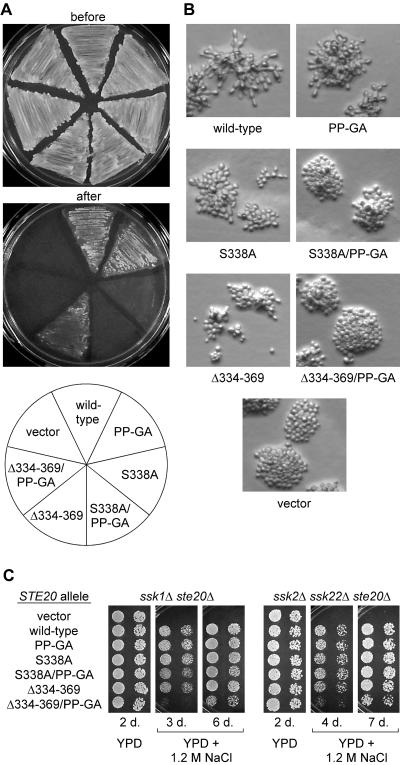
Ste20 is ordinarily not essential for viability, but ste20Δ cla4Δ cells are nonviable; therefore, Ste20 can perform an essential function that is redundant with that of its related PAK family kinase, Cla4, and that requires the Ste20 CRIB domain (31, 34, 53). This essential function of Ste20 appears to be less dependent on interaction with Bem1 than on interaction with Cdc42, as evidenced by the fact that the PP-GA mutant was better able to support growth upon Cla4 inactivation than was the S338A mutant (Fig. 3A, top). Nevertheless, quantitative measurement of growth rates (Fig. 3A, bottom) showed that the PP-GA mutant was slightly impaired, though not as strongly as the CRIB domain mutants (S338A and Δ334-369), which themselves retained partial function in this assay. The morphology of these cells was somewhat heterogeneous (Fig. 3B) but showed general trends suggesting a unique phenotype for the PP-GA mutant that is neither wild type nor null. The cla4 cells harboring the Ste20 PP-GA mutant had relatively normal bud necks, compared to those of the vector controls, but they tended to form rounder, less elongated buds than cells with wild-type Ste20, which showed elongated buds characteristic of cla4 STE20 cells (15). By comparison, the CRIB domain mutants showed more examples than the PP-GA mutant of both elongated buds and wide bud necks, plus additional morphologies (Fig. 3B) (data not shown), suggesting that the PP-GA morphology is not simply attributable to partial loss of function. While the PP-GA morphology resembles that of wild-type cells, it is not the phenotype of wild-type Ste20 in a cla4 background, and this suggests that the persistent apical bud growth of cla4 STE20 cells (15) depends on Ste20-Bem1 interaction, perhaps causing Ste20 to act apically rather than isotropically. This view is consistent with proposed roles for Bem1 in bud emergence, apical polarization, and symmetry breaking (8, 9, 29), as well as with the requirement for both Bem1 and Ste20 in the persistent apical growth of cdc34 mutants (24, 63).
FIG. 3.
Effects of Ste20 mutations in Bem1- versus Cdc42-binding domains on viability, polarization, and localization. (A) Ste20PP-GA shows a slight defect in performing the Cla4-redundant, essential function of Ste20. (Top) Strain KBY211 (ste20Δ cla4-75ts) was transformed with the indicated GFP-STE20 plasmids, and then fivefold serial dilutions were spotted onto −Ura plates and incubated for 3 days at 25 or 37°C. (Bottom) KBY211 cultures harboring the indicated GFP-STE20 plasmids were grown in −Ura liquid medium to logarithmic phase at 23°C and then were diluted back and shifted to 37°C. Culture densities were measured over time by optical density at 660 nm (OD660) as indicated. Results are means ± standard deviations from three trials; in each trial the same rank order of growth rate was observed. (B) Morphology of cells in panel A after 4 h at 37°C (similar results were observed after 3, 8, or 9 h). Note that the PP-GA mutant generally allows for normal bud neck constriction, compared to that of vector controls, but bud growth is less elongated than with wild-type Ste20, possibly implying a defect in apical polarization. Cells harboring CRIB domain mutants were heterogeneous but were on average more elongated than the PP-GA mutant and showed more examples of wide bud necks (especially the S338A mutant [data not shown]). (C) The Bem1-binding domain can contribute to localization of GFP-Ste20. Localization of the indicated GFP-Ste20 derivatives was examined in cells (PPY1249) overexpressing Bem1 from a high-copy-number plasmid (pPP1202). This method allowed occasional detection of the Ste20 Δ334-369 derivative at polarized growth sites (though rare in comparison to wild-type Ste20), whereas this phenomenon was never observed with the combined Ste20 Δ334-369 PP-GA derivative. The PP-GA mutation alone causes a mild decrease in the intensity of enrichment at bud tips rather than affecting the number of cells showing such enrichment.
Cortical localization of GFP-Ste20 was only mildly decreased by the PP-GA mutation (data not shown); this decrease was not as severe as that of even the mildest Cdc42-binding mutant, Ste20S338A (31), suggesting that Bem1 may assist in localizing Ste20 to the cell cortex (47) but makes a less critical contribution than does Cdc42. The fact that the CRIB domain mutations (S338A and Δ334-369) on their own cause strong delocalization (31, 34, 53) made it difficult to assess whether combination with the PP-GA mutation would confer a more severe defect in localization, as it does for signaling. To circumvent this difficulty, we examined GFP-Ste20 localization in cells overexpressing Bem1; Bem1 overexpression had been shown previously to restore GFP-Ste20 cortical localization in cdc42 mutant cells (47). Under these conditions, the Δ334-369 derivative of GFP-Ste20 was detected at sites of polarized growth in some cells, whereas the combined Δ334-369 PP-GA derivative was not (Fig. 3C). When the CRIB domain was left intact, the PP-GA mutant showed polarized localization similar to that of wild-type Ste20, but in general it was less intensely enriched at these locations. These results support the general notion that Bem1 can help localize Ste20, though Cdc42 is the predominant localizing factor in otherwise-normal cells.
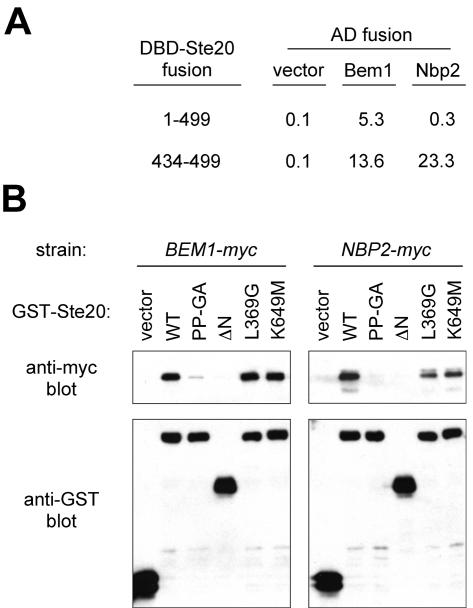
The Bem1-binding domain of Ste20 also binds the SH3 domain protein Nbp2.
Of the 24 SH3 domain proteins encoded in the S. cerevisiae genome, only Bem1 has previously been implicated in pheromone signaling, though others (e.g., Cdc25, Boi1, Boi2, Myo3, and Myo5) can serve as Ste20 substrates or binding partners or can affect Ste20-related functions (18, 52, 73). Therefore, it was conceivable that the phenotypic effects of the Ste20 PP-GA mutation reflect a defect in binding to an SH3 domain protein other than Bem1. A recent phage display screen (67) identified consensus ligands for 20 yeast SH3 domains (not including the Bem1 SH3-2 domain). The SH3 domain of Nbp2, a potential adaptor protein with roles in stress response pathways (42, 48), preferentially bound the consensus sequence PxRPaPxxP, which matches the Bem1-binding region in Ste20 (Fig. 1B). Hence, this Ste20 sequence might bind more than one SH3 domain. Indeed, we found that Nbp2 displayed a two-hybrid interaction with the isolated Bem1-binding fragment of Ste20 (residues 434 to 499), though it interacted negligibly with a larger Ste20 fragment (residues 1 to 499) that can bind Bem1 (Fig. 4A). Furthermore, Nbp2 coprecipitated with a GST fusion to full-length Ste20 in a manner that, like Bem1, required the Ste20 N terminus and was disrupted by the PP-GA mutation (Fig. 4B). In contrast, mutations causing kinase hyperactivity (L369G [31]) or inactivity (K649M [19, 34, 53, 55, 74]) did not disrupt Ste20 interaction with either protein (Fig. 4B), suggesting that the other mutations (PP-GA and ΔN) disrupt binding by perturbing the binding site rather than by indirect effects on Ste20 activity. Together, these results show that both Bem1 and Nbp2 can associate with Ste20 and that they have parallel binding requirements. Thus, without further analysis, conclusions regarding the role of the Bem1-binding site in Ste20 signaling become somewhat ambiguous given the intracellular presence of Nbp2 and multiple other SH3 domains that could have ligand-binding preferences overlapping those of Bem1.
FIG. 4.
The Bem1-binding domain of Ste20 also binds another SH3 domain protein, Nbp2. (A) Two-hybrid analysis showing interaction of Nbp2 with the isolated Bem1-binding fragment (Ste20 434-499) but not with a larger Bem1-interacting fragment (Ste20 1-499). DNA binding domain (DBD) fusions (plasmids pB20N2 and pB20BC) were coexpressed in PPY760 with vector (pGAD424) or AD fusions to Bem1157-551 (pRL51.1) or Nbp2 (pPP1355). Interactions were quantified in parallel by mean β-galactosidase units (n = 3; standard deviations were within 40% of the mean for all means greater than 1). (B) GST coprecipitation assays showing that Bem1 and Nbp2 have similar requirements for binding to full-length Ste20. Strains expressing Bem1-myc12 (DLY4000) (left) or Nbp2-myc13 (PPY1356) (right) were transformed with galactose-inducible constructs expressing GST alone(vector; pRD56) or the indicated GST-Ste20 fusion (pRDSTE20ATG [wild type {WT}], pPP1254 [PP-GA], pRDSTE20RI [ΔN], pPP1327 [L369G], or pRD56-K649M). Following galactose induction, glutathione precipitates were analyzed by immunoblotting with anti-myc (top) or anti-GST (bottom) antibodies. The Bem1-myc and Nbp2-myc experiments for which results are shown here were conducted simultaneously; similar results were observed in multiple independent trials. The relevant portions of the anti-myc blots from the Bem1 and Nbp2 experiments were juxtaposed solely to facilitate comparison and do not imply that Bem1-myc12 (82 kDa) and Nbp2-myc13 (48 kDa) have similar electrophoretic mobilities. Note that for both Bem1 and Nbp2, coprecipitation with GST-Ste20 is disrupted by the PP-GA and ΔN mutations but not by the L369G or K649M mutation.
Phenotypic effects of Ste20 PxxP motif mutations are attributable to disrupted Bem1 binding.
To address whether the pheromone response defects resulting from the Ste20 PP-GA mutation relate to Bem1, we compared the effects of this mutation in BEM1 versus bem1Δ cells (Fig. 5A). In support of the relevance of the PxxP motifs to Bem1 binding, deletion of BEM1 caused signaling defects that were similar in magnitude to those caused by the PP-GA mutation, regardless of the status of the CRIB domain. Most importantly, the PP-GA mutation conferred no defect on bem1Δ cells (Fig. 5A, right), indicating that the intact (PP) domain contributes its positive signaling role only when Bem1 is present. These results also show that the PP-GA phenotypes do not result from trivial defects such as misfolding or instability, as the mutation has no consequence when Bem1 is absent. Note that mutations in the CRIB domain (S338A and Δ334-369) reduced signaling both in the presence and in the absence of Bem1 (Fig. 5A), demonstrating that bem1Δ cells are not generally insensitive to perturbations in Ste20 function but instead are specifically insensitive to the PP-GA mutation. In contrast to the results for bem1Δ cells, the PP-GA mutation still caused a pheromone response defect in nbp2Δ cells, and did so in every CRIB domain context tested (Fig. 5B), indicating that the Bem1-binding domain can perform its signaling role irrespective of the presence or absence of Nbp2.
FIG. 5.
The phenotypic effect of Ste20 PxxP mutation depends on Bem1 rather than on Nbp2. The level of FUS1-lacZ induced by α-factor was assayed for GFP-Ste20 fusions that contained combinations of mutations in two domains—the indicated CRIB domain variant (wild type [WT], S338A, or Δ334-369) combined with a Bem1-binding domain that was either intact (PP) or mutant (PP-GA). Control transformants harbored the pRS316 vector (vect.). Deletion mutant strains were compared to congenic wild-type cells in the same strain background. Because both bem1Δ and nbp2Δ mutations impair growth with severities that depend on the strain background, the experiments for which results are shown here were performed in strain backgrounds where the mutants are the least growth impaired (15Dau for BEM1 versus bem1Δ; W303 for NBP2 versus nbp2Δ), but similar results were obtained for nbp2Δ mutants in the strain background (15Dau; PPY1343 versus PPY1415) where the deletion caused a stronger growth defect (data not shown). (A) Deletion of BEM1 mimics the effect of the PP-GA mutation, and in bem1Δ cells the PP-GA mutation confers no further pheromone response defect, indicating that the effect of the PP-GA mutation can be attributed to a disruption of Bem1-Ste20 interaction. Strains: PPY1343 (BEM1) and PPY1341 (bem1Δ). (B) In nbp2Δ cells, the PP-GA mutation still confers a signaling defect. Strains: PPY913 (NBP2) and PPY1456 (nbp2Δ).
Altogether, these results indicate that the signaling defect resulting from the PP-GA mutation can be attributed to a disruption in Ste20-Bem1 interaction and that this interaction contributes to signaling in a manner that is clearly separable from interactions of Ste20 with other binding partners such as Cdc42 and Nbp2. It is also clear that in the absence of Bem1, the ability of the CRIB domain to bind Cdc42 is still critical (Fig. 5A, right), because the S338A mutation caused an exceptionally strong defect in bem1Δ cells and this defect was suppressed somewhat by deletion of the entire CRIB domain, which (like Cdc42 binding) releases Ste20 from autoinhibition. This behavior suggests that binding and activation by Cdc42 still regulate Ste20 signaling even in the absence of Bem1 or the Bem1-binding domain, and therefore Bem1 is not required as a cofactor in the activation of Ste20 by Cdc42. Instead, interaction with Bem1 makes Ste20 signaling more efficient regardless of the status of Cdc42-Ste20 interaction.
Role of the Bem1-binding domain in other Ste20 signaling pathways.
In addition to pheromone response, Ste20 functions in the filamentous growth and HOG pathways (37, 49, 57, 61). In each case, as with pheromone response, Ste20 functions downstream of Cdc42 and stimulates a MAP kinase cascade by activation of the MAPKKK Ste11 (31, 46, 57, 68). We tested the Ste20 mutants for function in the filamentous growth pathway by an agar invasion test (Fig. 6A) and by examining the morphology of cells grown on low-glucose medium (Fig. 6B). Previous work indicated a role for Cdc42-Ste20 interaction, as agar invasion was strongly defective when Cdc42 binding alone was disrupted (by an S338A H345G mutation) but was largely restored when the autoinhibitory effect of the CRIB domain was also eliminated (31). Here, the PP-GA mutation had no evident effect on its own, but it eliminated the residual function of the Δ334-369 derivative, as revealed by comparison of the Δ334-369 and Δ334-369 PP-GA mutants (Fig. 6A). The S338A mutant was more defective than the Δ334-369 mutant, but a faint residual haze of adherence was reproducibly observed; this haze was eliminated in the S338A PP-GA combined mutant. A similar hierarchy of phenotypes was apparent when these cells were examined for filamentous morphology (Fig. 6B): (i) the PP-GA mutation alone caused at most a mild reduction in the extent of elongated, branched morphology; (ii) each of the two mutations in the CRIB domain, S338A and Δ334-369, caused a stronger reduction in elongation and branching but with some residual filamentous characteristics still apparent (e.g., slightly elongated cells, and uneven colony edges with cells occasionally protruding from the colony mass); (iii) these residual features were eliminated when the PP-GA mutation was combined with either of the CRIB domain mutations.
FIG. 6.
Effects of Ste20 PxxP mutations on filamentous growth and hyperosmotic resistance. (A) Agar invasion. Strain PPY1209 (Σ1278b ste20Δ) was transformed with the indicated GFP-Ste20 fusion constructs and assayed for invasive growth as described in Materials and Methods. Representative examples of cells remaining adherent after gentle rinsing with water, as well as before rinsing, are shown. Note that invasive growth conferred by the Δ334-369 allele is eliminated by the PP-GA mutation. (B) Filamentous colony morphology. The transformants used for panel A were grown on low-glucose medium for 24 h. Cells with wild-type Ste20 become elongated and form branched microcolonies under these conditions. Note that the disruption of these features becomes gradually more severe as mutations in the Cdc42-binding and Bem1-binding motifs are combined. (C) Hyperosmotic resistance. Strains PPY1287 (ssk1Δ ste20Δ) and PPY1646 (ssk2Δ ssk22Δ ste20Δ) were transformed with the indicated GFP-Ste20 fusion constructs. Fivefold serial dilutions were spotted onto YPD or YPD plus 1.2 M NaCl and incubated at 30°C for 2 to 7 days (d.) as indicated. Note that only the Δ334-369 PP-GA double mutant has a strong defect in osmoresistance and that even this mutant retains some function, as revealed at longer incubation times.
Function in the HOG pathway was assessed by growth on high-osmolarity media (Fig. 6C). To make growth dependent on the Ste20 branch of the HOG pathway, ssk1Δ or ssk2Δ ssk22Δ deletion mutations were used to inactivate a parallel, redundant branch (49, 57). None of the single Ste20 mutations caused an observable defect, but the Δ334-369 PP-GA combined mutation caused a strong reduction in growth on high-osmolarity media (Fig. 6C), with partial function evident upon extended incubation (6 or 7 days). Although a previous study reported a HOG signaling defect for the Δ334-369 allele (57), our tests did not uncover this defect in either of two strain backgrounds. The reasons for this difference are not clear (possibilities include differences in growth media and plasmid backbones) but may ultimately represent differences in detection—i.e., while the earlier study detected the partial defect in the Δ334-369 allele, our conditions may have been more permissive, allowing detection of the stronger Δ334-369 PP-GA defect. In this view, each result is informative, and it would be inaccurate to classify these alleles as simply functional or nonfunctional. Overall, however, the results in both the filamentation and HOG pathways were largely reminiscent of those in the mating pathway, in that single mutations in the Bem1-binding and Cdc42-binding domains conferred only partial defects, whereas double mutations combined to produce stronger phenotypes.
Testing the requirement for conserved residues in the Bem1-binding domain.
Previous attempts to determine the recognition sequence for the Bem1 SH3-2 domain were unsuccessful, due to poor expression in Escherichia coli (67, 76). To gain some insight into this issue, we examined evolutionary conservation of the Bem1-binding site in Ste20 and probed the conserved sequence with additional mutations. Comparison of this site among fungal orthologs of Ste20 (Fig. 7B) shows that the strongest conservation occurs within a block of 11 residues (FIPSRPAPKPP; residues 470 to 480 in Ste20), which includes both of the tandem PxxP motifs targeted by the PP-GA mutation. Functional PxxP motifs often exist within more-constrained 7-residue sites: class I (+x@Px@P) and class II (@Px@Px+) (where + and @ stand for positively charged and aliphatic residues, respectively) sites (45). Both tandem motifs in Ste20 have positively charged residues at the −3 position with respect to PxxP (i.e., KFIPSRP and RPAPKPP), characteristic of class I sites (45), though only the second motif is a perfect match with the class I consensus (Fig. 7B). Conservation of residues in both motifs could imply that they function redundantly or additively, or simply that one binding motif consists of more than the prototypical seven residues. We introduced point mutations across a 13-residue region (KFIPSRPAPKPPS) that includes the core 11-residue sequence plus 1 residue to either side (Fig. 7C). Consistent with the conservation, Bem1 binding was minimally affected by mutations at the periphery (K469A and S481A) but was strongly disrupted by mutation at most of the 11 core residues, including each single mutation present in the original PP-GA double mutant (P475G and P477A). Mutations in the core sequence with the least impact (S473A and K478A) affect analogous positions within each of the tandem class I-like motifs, and indeed SH3 domains rarely show preferences at this position (45, 67). Overall, these results show that the tandem motifs are not redundant, and the possibility that they function additively also seems unlikely, because removal of the positive charge at the −3 position was tolerated in the first motif (K469A in KFIPSRP) but not in the second motif (R474G in RPAPKPP). Nevertheless, the second motif is not sufficient; mutations at flanking residues (e.g., F470, I471, P472) were strongly disruptive. Therefore, while not exhaustively defining the optimal ligand, these observations suggest that the Bem1 SH3-2 domain recognizes sequence features spread over a broad region of 11 residues, including a 7-residue class I motif plus four N-terminally flanking residues. The involvement of these flanking residues is reminiscent of some other SH3 domains (60, 66, 67), and it supports the functional significance of their evolutionary conservation (see Discussion).
FIG. 7.
Evolutionary conservation of the Bem1 binding site in Ste20 orthologs and paralogs. Standard single-letter symbols for amino acid residues are used. In addition, the symbols +, @, b, and x stand for basic, aliphatic, hydrophobic, and any residues, respectively. (A) Schematic diagram indicating relative positions of defined domains in Ste20. (B) Alignment of sequences from Ste20 orthologs from fungi of increasing phylogenetic distance. Sequences are from S. cerevisiae (S.cer), Saccharomyces kudriavzevii (S.kud), Saccharomyces bayanus (S.bay), Saccharomyces castelli (S.cas), Candida albicans (C.alb), Ashbya gossypii (A.gos), and Neurospora crassa (N.cra). Each sequence shown lies between the CRIB and kinase domains. Identities to the Ste20 sequence are boxed black. (C) Bem1 binding is disrupted by mutation of conserved residues in the Ste20 motif. The indicated point mutations (from K469A to S481A) were incorporated into a DBD-Ste201-499 fusion construct (pB20N2), and interaction with Bem1157-551 (pRL51.1) or Cdc42 (pPP1027) was tested in the two-hybrid tester strain PPY760 by a quantitative β-galactosidase assay. For ease of comparison, signals were normalized to those of wild-type (wt) pB20N2 (taken as 100%), whose mean values were 15.0 (Bem1) and 271.5 (Cdc42) β-galactosidase units; standard deviations (n = 3)were within 15% of the mean in all cases and are omitted for clarity. Note that the mutations do not disrupt interaction with Cdc42 and thus are specific for Bem1. Also, similar Bem1-binding results were observed by using a full-length Bem1 (residues 1 to 551) construct (pXP-BEM1), and no appreciable interaction signals were observed by using a vector (pGAD424) control (data not shown). (D) Alignment of Ste20 paralogs and orthologs from S. cerevisiae (S.c.) and Schizosaccharomyces pombe (S.p.). A variable number of PxxP motifs (from 1 to 12) are present in each protein. Shown here are the only motifs that are perfect matches to class I (+x@Px@P) or class II (@Px@Px+) consensus sequences (45). Exceptions are as follows: Pak2 has no perfect matches, so its only PxxP motif is shown for comparison; Ste20 has one class I match and one class II match, but only the class I match is shown because it is the one identified in this study as a Bem1-binding site. Pak1 has two class I matches (#1 and #2), each of which is shown. Within the core motif, identities to Ste20 are boxed black, and conservative or common substitutions are boxed grey. The top six sequences are from PAK family kinases and thus represent candidate trans ligands for the SH3-2 domain of Bem1 or its ortholog (with the exception of the Pak2 sequence, which is a poor match and which lies within the CRIB domain). The bottom two sequences lie within the PX domains of Bem1 and its S. pombe ortholog, Scd2, and have been proposed to bind in cis to SH3-2 domains within the same protein (21). These sequences match the class I consensus but lack the additional conserved sequence features common to the PAK motifs.
DISCUSSION
In this study we identify and functionally characterize a proline-rich motif within Ste20, a PAK family kinase, that provides a binding site for the SH3 domain protein Bem1. Mutations in this motif (PP-GA) reduce pheromone-responsive signaling in a way that mimics a bem1Δ mutation, and the intact Bem1-binding domain enhances signaling only in cells that express Bem1, clearly demonstrating the physiological significance of the Bem1-Ste20 interaction. Bem1 and Cdc42 bind to distinct domains in Ste20, and combined disruption of Bem1-binding and Cdc42-binding domains impairs Ste20 signaling more strongly than either disruption alone, indicating that these interactions contribute in an additive fashion to optimal function. The additive effect of Bem1 and Cdc42 interactions and their physical independence offer what is perhaps the most broadly applicable aspect of our observations: they provide a concrete example of how changes in function can be conferred incrementally on a protein by the acquisition (e.g., during evolution) of new protein-protein interaction modules (51).
The Bem1-Ste20 interaction is pertinent to other cases where SH3 domain proteins are thought to promote localization and/or activation of PAKs (6, 11, 26, 38, 41). Our findings suggest that SH3-PAK interactions can contribute to signal transduction even after activation by the GTPase, as disruption of the Bem1-Ste20 interaction or deletion of BEM1 reduces signaling by the Cdc42-independent hyperactive mutant, Ste20Δ334-369 (Fig. 2 and 5A) (47). This Bem1 dependence also implies that although Ste20Δ334-369 appears to be largely delocalized (31, 34, 53), it may still signal predominantly at the cell periphery, because that is where Bem1 is enriched (2, 29). Therefore, the simplest explanation for the effects of Bem1-Ste20 binding is that Bem1 helps colocalize activated Ste20 with its substrates, for instance, by maintaining a pool of active Ste20 at the cell periphery. This can explain why Bem1 binding enhances pheromone signaling, which is initiated at the plasma membrane (56, 69), but what about other signaling, such as that by Ste5P44L-GST (Fig. 2B) or hyperosmotic cross talk (Fig. 2C)? In fact, in each of these alternative settings there is reason to suspect that signaling is still localized. For example, signaling by Ste5P44L-GST is reduced when Ste20 is delocalized by mutations that disrupt Cdc42 binding, irrespective of Ste20 activity levels (e.g., Fig. 2B) (31); hence, Ste5P44L-GST might still signal preferentially at the cell periphery, despite not being obviously enriched there (62). Also, hyperosmotic cross talk is initiated by the polarized transmembrane proteins Sho1 and Msb2 (49, 50), and hence the contribution of Ste20 may also occur primarily at these polarized sites.
Our mutational analysis of the Ste20 Bem1-binding site suggests that the Bem1 SH3-2 domain recognizes a rather broad motif, with important sequence features spread over 11 residues (Fig. 7C), including a 7-residue class I consensus SH3-binding sequence (45) plus N-terminally flanking sequence. This 11-residue motif is strongly conserved among Ste20 orthologs (Fig. 7B), suggesting that it confers a function that is under selective pressure. The disruption of Bem1 binding by mutations in Ste20 residues F470 and I471, at positions −7 and −6 (with respect to PxxP), is reminiscent of SH3 domains from Src family kinases, which show a preference for hydrophobic residues at these positions (60, 66). Hydrophobic residues are also present at the −7 position of PxxP motifs in other proteins that bind the Bem1 SH3-2 domain, such as Boi1, Boi2, and Mps1 (5, 44), though the functional significance of those interactions remains unclear. Whether other yeast SH3 domains show biases at these positions would have been missed in previous work due to the use of peptide libraries with only nine residues (67), but biases were found at positions −5 and −4 in several cases, and three SH3 domains showed preferences for tandem PxxP motifs similar to the Ste20 sequence (i.e., PxxPxPxxP). Altogether, these observations suggest that while SH3 domains can recognize short sequences such as the 7-residue class I or class II motifs (45), residues at the flanking 4 to 5 positions can also influence binding (60, 66).
Similarities to the Bem1-binding domain are also present in more-distant Ste20 orthologs and in the other two S. cerevisiae PAKs, Cla4 and Skm1 (Fig. 7D). Bem1 does bind Cla4 (8, 24), likely mediated by the Cla4 sequence FQPQRTAPKPP (residues 451 to 461) (Fig. 7D), which resembles the Ste20 motif both in sequence and in its positioning between the CRIB and kinase domains. The Schizosaccharomyces pombe ortholog of Ste20, Pak1/Shk1, contains two strong matches to the Bem1-binding motif (Fig. 7D): one (#1) upstream of the CRIB domain and another (#2) between the CRIB and kinase domains. Indeed, fragments that include either motif can bind Scd2, the S. pombe homolog of Bem1 (11). Each of these other sequences contains some of the flanking features present in Ste20 (Fig. 7D). Interestingly, Bem1 and Scd2 themselves each contain a class I motif within their PX domains, which are proposed to bind in cis to their own SH3-2 domains, possibly generating an autoinhibited conformation (21). While each sequence matches the class I motif consensus, they lack the additional conserved features of the broader Bem1-binding motif in Ste20 (Fig. 7D). This lack could be due to other functional constraints on the PX domain, but a more tantalizing possibility is that it reflects negative selection against an optimal binding motif, which if present as an intramolecular ligand (in cis) might bind so tightly as to always outcompete trans ligands.
Finally, our observations show that two different SH3 domains, the Nbp2 SH3 domain and the Bem1 SH3-2 domain, can bind the same target site in vivo. At present, there is little to suggest that Ste20 function is affected by Nbp2, which so far is implicated only in stress response pathways (42, 48), but this remains a possibility given that the homolog of Nbp2 in S. pombe, Skb5, can activate the kinase activity of the S. pombe Ste20 homolog, Pak1/Shk1 (75). Recent work on the SH3-mediated interaction between Sho1 and Pbs2 suggests that other yeast SH3 domains have been under negative selection to reduce binding to the Pbs2 ligand, ensuring highly specific binding of Pbs2 to only the Sho1 SH3 domain (76). Our findings suggest the converse possibility that single SH3 ligands may interact with multiple different SH3 domains in the same cell, which may also be implied by the multitude of SH3 binding partners for the yeast protein Las17 (67). It remains to be determined whether this serves a functional purpose in the case of Ste20 interactions with Bem1 versus Nbp2, or in other situations in which multiple SH3 domains share ligand preferences.
Acknowledgments
We are grateful to D. McCollum, A. Neiman, D. Lew, S. O'Rourke, H. Saito, R. Vallee, and M. Whiteway for plasmids, strains, and antibodies and to R. Lamson for additional technical assistance.
This work was supported by a grant from the NIH (GM57769) to P.M.P.
REFERENCES
- 1.Ash, J., C. Wu, R. Larocque, M. Jamal, W. Stevens, M. Osborne, D. Y. Thomas, and M. Whiteway. 2003. Genetic analysis of the interface between Cdc42p and the CRIB domain of Ste20p in Saccharomyces cerevisiae. Genetics 163:9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayscough, K. R., J. Stryker, N. Pokala, M. Sanders, P. Crews, and D. G. Drubin. 1997. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137:399-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel, P. L., and S. Fields. 1995. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 254:241-263. [DOI] [PubMed] [Google Scholar]
- 4.Bender, A., and J. R. Pringle. 1991. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:1295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender, L., H. S. Lo, H. Lee, V. Kokojan, V. Peterson, and A. Bender. 1996. Associations among PH and SH3 domain-containing proteins and Rho-type GTPases in yeast. J. Cell Biol. 133:879-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bokoch, G. M., Y. Wang, B. P. Bohl, M. A. Sells, L. A. Quilliam, and U. G. Knaus. 1996. Interaction of the Nck adapter protein with p21-activated kinase (PAK1). J. Biol. Chem. 271:25746-25749. [DOI] [PubMed] [Google Scholar]
- 7.Bork, P., J. Schultz, and C. P. Ponting. 1997. Cytoplasmic signalling domains: the next generation. Trends Biochem. Sci. 22:296-298. [DOI] [PubMed] [Google Scholar]
- 8.Bose, I., J. E. Irazoqui, J. J. Moskow, E. S. Bardes, T. R. Zyla, and D. J. Lew. 2001. Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J. Biol. Chem. 276:7176-7186. [DOI] [PubMed] [Google Scholar]
- 9.Butty, A. C., N. Perrinjaquet, A. Petit, M. Jaquenoud, J. E. Segall, K. Hofmann, C. Zwahlen, and M. Peter. 2002. A positive feedback loop stabilizes the guanine-nucleotide exchange factor Cdc24 at sites of polarization. EMBO J. 21:1565-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butty, A. C., P. M. Pryciak, L. S. Huang, I. Herskowitz, and M. Peter. 1998. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science 282:1511-1516. [DOI] [PubMed] [Google Scholar]
- 11.Chang, E., G. Bartholomeusz, R. Pimental, J. Chen, H. Lai, L. Wang, P. Yang, and S. Marcus. 1999. Direct binding and in vivo regulation of the fission yeast p21-activated kinase Shk1 by the SH3 domain protein Scd2. Mol. Cell. Biol. 19:8066-8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chenevert, J., K. Corrado, A. Bender, J. Pringle, and I. Herskowitz. 1992. A yeast gene (BEM1) necessary for cell polarization whose product contains two SH3 domains. Nature 356:77-79. [DOI] [PubMed] [Google Scholar]
- 13.Cook, J. G., L. Bardwell, and J. Thorner. 1997. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature 390:85-88. [DOI] [PubMed] [Google Scholar]
- 14.Cullen, P. J., and G. F. Sprague, Jr. 2000. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 97:13619-13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cvrckova, F., C. De Virgilio, E. Manser, J. R. Pringle, and K. Nasmyth. 1995. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 9:1817-1830. [DOI] [PubMed] [Google Scholar]
- 16.Dan, I., N. M. Watanabe, and A. Kusumi. 2001. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 11:220-230. [DOI] [PubMed] [Google Scholar]
- 17.Dohlman, H. G., and J. W. Thorner. 2001. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70:703-754. [DOI] [PubMed] [Google Scholar]
- 18.Drees, B. L., B. Sundin, E. Brazeau, J. P. Caviston, G. C. Chen, W. Guo, K. G. Kozminski, M. W. Lau, J. J. Moskow, A. Tong, L. R. Schenkman, A. McKenzie III, P. Brennwald, M. Longtine, E. Bi, C. Chan, P. Novick, C. Boone, J. R. Pringle, T. N. Davis, S. Fields, and D. G. Drubin. 2001. A protein interaction map for cell polarity development. J. Cell Biol. 154:549-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eby, J. J., S. P. Holly, F. van Drogen, A. V. Grishin, M. Peter, D. G. Drubin, and K. J. Blumer. 1998. Actin cytoskeleton organization regulated by the PAK family of protein kinases. Curr. Biol. 8:967-970. [DOI] [PubMed] [Google Scholar]
- 20.Elion, E. A. 2000. Pheromone response, mating and cell biology. Curr. Opin. Microbiol. 3:573-581. [DOI] [PubMed] [Google Scholar]
- 21.Endo, M., M. Shirouzu, and S. Yokoyama. 2003. The Cdc42 binding and scaffolding activities of the fission yeast adaptor protein Scd2. J. Biol. Chem. 278:843-852. [DOI] [PubMed] [Google Scholar]
- 22.Feng, Y., L. Y. Song, E. Kincaid, S. K. Mahanty, and E. A. Elion. 1998. Functional binding between Gβ and the LIM domain of Ste5 is required to activate the MEKK Ste11. Curr. Biol. 8:267-278. [DOI] [PubMed] [Google Scholar]
- 23.Goehring, A. S., D. A. Mitchell, A. H. Tong, M. E. Keniry, C. Boone, and G. F. Sprague, Jr. 2003. Synthetic lethal analysis implicates Ste20p, a p21-activated protein kinase, in polarisome activation. Mol. Biol. Cell 14:1501-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gulli, M., M. Jaquenoud, Y. Shimada, G. Niederhauser, P. Wiget, and M. Peter. 2000. Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol. Cell 6:1155-1167. [DOI] [PubMed] [Google Scholar]
- 25.Gustin, M. C., J. Albertyn, M. Alexander, and K. Davenport. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hing, H., J. Xiao, N. Harden, L. Lim, and S. L. Zipursky. 1999. Pak functions downstream of Dock to regulate photoreceptor axon guidance in Drosophila. Cell 97:853-863. [DOI] [PubMed] [Google Scholar]
- 27.Hofken, T., and E. Schiebel. 2002. A role for cell polarity proteins in mitotic exit. EMBO J. 21:4851-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holly, S. P., and K. J. Blumer. 1999. PAK-family kinases regulate cell and actin polarization throughout the cell cycle of Saccharomyces cerevisiae. J. Cell Biol. 147:845-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irazoqui, J. E., A. S. Gladfelter, and D. J. Lew. 2003. Scaffold-mediated symmetry breaking by Cdc42p. Nat. Cell Biol. 5:1062-1070. [DOI] [PubMed] [Google Scholar]
- 30.Kay, B. K., M. P. Williamson, and M. Sudol. 2000. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14:231-241. [PubMed] [Google Scholar]
- 31.Lamson, R. E., M. J. Winters, and P. M. Pryciak. 2002. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol. Cell. Biol. 22:2939-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larson, S. M., and A. R. Davidson. 2000. The identification of conserved interactions within the SH3 domain by alignment of sequences and structures. Protein Sci. 9:2170-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leberer, E., D. Dignard, D. Harcus, D. Y. Thomas, and M. Whiteway. 1992. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein βγ subunits to downstream signalling components. EMBO J. 11:4815-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leberer, E., C. Wu, T. Leeuw, A. Fourest-Lieuvin, J. E. Segall, and D. Y. Thomas. 1997. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 16:83-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leeuw, T., A. Fourest-Lieuvin, C. Wu, J. Chenevert, K. Clark, M. Whiteway, D. Y. Thomas, and E. Leberer. 1995. Pheromone response in yeast: association of Bem1p with proteins of the MAP kinase cascade and actin. Science 270:1210-1213. [DOI] [PubMed] [Google Scholar]
- 36.Leeuw, T., C. Wu, J. D. Schrag, M. Whiteway, D. Y. Thomas, and E. Leberer. 1998. Interaction of a G-protein β-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature 391:191-195. [DOI] [PubMed] [Google Scholar]
- 37.Liu, H., C. A. Styles, and G. R. Fink. 1993. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262:1741-1744. [DOI] [PubMed] [Google Scholar]
- 38.Lu, W., S. Katz, R. Gupta, and B. J. Mayer. 1997. Activation of Pak by membrane localization mediated by an SH3 domain from the adaptor protein Nck. Curr. Biol. 7:85-94. [DOI] [PubMed] [Google Scholar]
- 39.Lyons, D. M., S. K. Mahanty, K. Y. Choi, M. Manandhar, and E. A. Elion. 1996. The SH3-domain protein Bem1 coordinates mitogen-activated protein kinase cascade activation with cell cycle control in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4095-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maeda, T., M. Takekawa, and H. Saito. 1995. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269:554-558. [DOI] [PubMed] [Google Scholar]
- 41.Manser, E., T. H. Loo, C. G. Koh, Z. S. Zhao, X. Q. Chen, L. Tan, I. Tan, T. Leung, and L. Lim. 1998. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol. Cell 1:183-192. [DOI] [PubMed] [Google Scholar]
- 42.Mapes, J., and I. M. Ota. 2004. Nbp2 targets the Ptc1-type 2C Ser/Thr phosphatase to the HOG MAPK pathway. EMBO J. 23:302-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marles, J. A., S. Dahesh, J. Haynes, B. J. Andrews, and A. R. Davidson. 2004. Protein-protein interaction affinity plays a crucial role in controlling the sho1p-mediated signal transduction pathway in yeast. Mol. Cell 14:813-823. [DOI] [PubMed] [Google Scholar]
- 44.Matsui, Y., R. Matsui, R. Akada, and A. Toh-e. 1996. Yeast src homology region 3 domain-binding proteins involved in bud formation. J. Cell Biol. 133:865-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayer, B. J. 2001. SH3 domains: complexity in moderation. J. Cell Sci. 114:1253-1263. [DOI] [PubMed] [Google Scholar]
- 46.Mosch, H. U., R. L. Roberts, and G. R. Fink. 1996. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5352-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moskow, J. J., A. S. Gladfelter, R. E. Lamson, P. M. Pryciak, and D. J. Lew. 2000. Role of Cdc42p in pheromone-stimulated signal transduction in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:7559-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohkuni, K., A. Okuda, and A. Kikuchi. 2003. Yeast Nap1-binding protein Nbp2p is required for mitotic growth at high temperatures and for cell wall integrity. Genetics 165:517-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Rourke, S. M., and I. Herskowitz. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12:2874-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Rourke, S. M., and I. Herskowitz. 2002. A third osmosensing branch in Saccharomyces cerevisiae requires the Msb2 protein and functions in parallel with the Sho1 branch. Mol. Cell. Biol. 22:4739-4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pawson, T., and P. Nash. 2003. Assembly of cell regulatory systems through protein interaction domains. Science 300:445-452. [DOI] [PubMed] [Google Scholar]
- 52.Perlman, R., D. Yablonski, G. Simchen, and A. Levitzki. 1993. Cloning of the STE5 gene of Saccharomyces cerevisiae as a suppressor of the mating defect of cdc25 temperature-sensitive mutants. Proc. Natl. Acad. Sci. USA 90:5474-5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peter, M., A. M. Neiman, H. O. Park, M. van Lohuizen, and I. Herskowitz. 1996. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 15:7046-7059. [PMC free article] [PubMed] [Google Scholar]
- 54.Peterson, J., Y. Zheng, L. Bender, A. Myers, R. Cerione, and A. Bender. 1994. Interactions between the bud emergence proteins Bem1p and Bem2p and Rho-type GTPases in yeast. J. Cell Biol. 127:1395-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Polverino, A., J. Frost, P. Yang, M. Hutchison, A. M. Neiman, M. H. Cobb, and S. Marcus. 1995. Activation of mitogen-activated protein kinase cascades by p21-activated protein kinases in cell-free extracts of Xenopus oocytes. J. Biol. Chem. 270:26067-26070. [DOI] [PubMed] [Google Scholar]
- 56.Pryciak, P. M., and F. A. Huntress. 1998. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 12:2684-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raitt, D. C., F. Posas, and H. Saito. 2000. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 19:4623-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramer, S. W., and R. W. Davis. 1993. A dominant truncation allele identifies a gene, STE20, that encodes a putative protein kinase necessary for mating in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90:452-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reiser, V., S. M. Salah, and G. Ammerer. 2000. Polarized localization of yeast Pbs2 depends on osmostress, the membrane protein Sho1 and Cdc42. Nat. Cell Biol. 2:620-627. [DOI] [PubMed] [Google Scholar]
- 60.Rickles, R. J., M. C. Botfield, X. M. Zhou, P. A. Henry, J. S. Brugge, and M. J. Zoller. 1995. Phage display selection of ligand residues important for Src homology 3 domain binding specificity. Proc. Natl. Acad. Sci. USA 92:10909-10913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roberts, R. L., and G. R. Fink. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8:2974-2985. [DOI] [PubMed] [Google Scholar]
- 62.Sette, C., C. J. Inouye, S. L. Stroschein, P. J. Iaquinta, and J. Thorner. 2000. Mutational analysis suggests that activation of the yeast pheromone response mitogen-activated protein kinase pathway involves conformational changes in the Ste5 scaffold protein. Mol. Biol. Cell 11:4033-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sheu, Y. J., Y. Barral, and M. Snyder. 2000. Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:5235-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Simon, M. N., C. De Virgilio, B. Souza, J. R. Pringle, A. Abo, and S. I. Reed. 1995. Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature 376:702-705. [DOI] [PubMed] [Google Scholar]
- 66.Sparks, A. B., J. E. Rider, N. G. Hoffman, D. M. Fowlkes, L. A. Quillam, and B. K. Kay. 1996. Distinct ligand preferences of Src homology 3 domains from Src, Yes, Abl, Cortactin, p53bp2, PLCγ, Crk, and Grb2. Proc. Natl. Acad. Sci. USA 93:1540-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tong, A. H., B. Drees, G. Nardelli, G. D. Bader, B. Brannetti, L. Castagnoli, M. Evangelista, S. Ferracuti, B. Nelson, S. Paoluzi, M. Quondam, A. Zucconi, C. W. Hogue, S. Fields, C. Boone, and G. Cesareni. 2002. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295:321-324. [DOI] [PubMed] [Google Scholar]
- 68.van Drogen, F., S. M. O'Rourke, V. M. Stucke, M. Jaquenoud, A. M. Neiman, and M. Peter. 2000. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 10:630-639. [DOI] [PubMed] [Google Scholar]
- 69.van Drogen, F., V. M. Stucke, G. Jorritsma, and M. Peter. 2001. MAP kinase dynamics in response to pheromones in budding yeast. Nat. Cell Biol. 3:1051-1059. [DOI] [PubMed] [Google Scholar]
- 70.Wach, A., A. Brachat, C. Alberti-Segui, C. Rebischung, and P. Philippsen. 1997. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13:1065-1075. [DOI] [PubMed] [Google Scholar]
- 71.Whiteway, M. S., C. Wu, T. Leeuw, K. Clark, A. Fourest-Lieuvin, D. Y. Thomas, and E. Leberer. 1995. Association of the yeast pheromone response G protein βγ subunits with the MAP kinase scaffold Ste5p. Science 269:1572-1575. [DOI] [PubMed] [Google Scholar]
- 72.Wu, C., T. Leeuw, E. Leberer, D. Y. Thomas, and M. Whiteway. 1998. Cell cycle- and Cln2p-Cdc28p-dependent phosphorylation of the yeast Ste20p protein kinase. J. Biol. Chem. 273:28107-28115. [DOI] [PubMed] [Google Scholar]
- 73.Wu, C., V. Lytvyn, D. Y. Thomas, and E. Leberer. 1997. The phosphorylation site for Ste20p-like protein kinases is essential for the function of myosin-I in yeast. J. Biol. Chem. 272:30623-30626. [DOI] [PubMed] [Google Scholar]
- 74.Wu, C., M. Whiteway, D. Y. Thomas, and E. Leberer. 1995. Molecular characterization of Ste20p, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J. Biol. Chem. 270:15984-15992. [DOI] [PubMed] [Google Scholar]
- 75.Yang, P., R. Pimental, H. Lai, and S. Marcus. 1999. Direct activation of the fission yeast PAK Shk1 by the novel SH3 domain protein, Skb5. J. Biol. Chem. 274:36052-36057. [DOI] [PubMed] [Google Scholar]
- 76.Zarrinpar, A., S. H. Park, and W. A. Lim. 2003. Optimization of specificity in a cellular protein interaction network by negative selection. Nature 426:676-680. [DOI] [PubMed] [Google Scholar]
- 77.Ziman, M., D. Preuss, J. Mulholland, J. M. O'Brien, D. Botstein, and D. I. Johnson. 1993. Subcellular localization of Cdc42p, a Saccharomyces cerevisiae GTP-binding protein involved in the control of cell polarity. Mol. Biol. Cell 4:1307-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]