Abstract
In Saccharomyces cerevisiae, glucose depletion causes a profound alteration in metabolism, mediated in part by global transcriptional changes. Many of the transcription factors that regulate these changes act combinatorially. We have analyzed combinatorial regulation by Adr1 and Cat8, two transcription factors that act during glucose depletion, by combining genome-wide expression and genome-wide binding data. We identified 32 genes that are directly activated by Adr1, 28 genes that are directly activated by Cat8, and 14 genes that are directly regulated by both. Our analysis also uncovered promoters that Adr1 binds but does not regulate and promoters that are indirectly regulated by Cat8, stressing the advantage of combining global expression and global localization analysis to find directly regulated targets. At most of the coregulated promoters, the in vivo binding of one factor is independent of the other, but Adr1 is required for optimal Cat8 binding at two promoters with a poor match to the Cat8 binding consensus. In addition, Cat8 is required for Adr1 binding at promoters where Adr1 is not required for transcription. These data provide a comprehensive analysis of the direct, indirect, and combinatorial requirements for these two global transcription factors.
Cells respond to stresses such as variations in temperature, pH or osmotic or nutrient conditions by altering the expression of genes that allow the cell to respond to the new environment (10). These transcriptional changes are initiated by signals that converge on a limited number of transcription factors, which then act combinatorially to control many genes in multiple pathways. Microarrays can be used to identify these coordinated, combinatorial responses by detecting both the localization of key transcription factors and the corresponding changes in the transcriptome (1).
One example of a coordinated transcriptional response to environmental stress is the reprogramming of gene expression during glucose starvation. In Saccharomyces cerevisiae, glucose depletion causes a change of at least twofold in the RNA level of over 1,000 genes (12). Much of this global response is initiated through a signal transduction cascade that acts through the AMP-activated protein kinase Snf1 (8, 19). Among the targets of Snf1 are transcription factors that derepress genes required for alternative carbon source metabolism. Two of these factors are the Zn-cluster DNA-binding protein Cat8 and the Zn-finger DNA-binding protein Adr1 (41).
Snf1 activates Cat8 in part through phosphorylation of Mig1. This widely acting transcriptional repressor exits the nucleus when phosphorylated, relieving repression of dozens of genes, including CAT8 (13). Subsequent production of the Cat8 transcription factor and activation via Snf1-dependent phosphorylation (11, 36) up-regulates gluconeogenic and glyoxylate cycle genes as well as several transcription factors, such as SIP4 and HAP4 (6, 25).
SNF1 is also required for activation of many of the genes that are regulated by the Zn-finger transcriptional regulator Adr1 (49). Unlike Cat8, the precise mechanism by which Snf1 controls expression of Adr1-dependent genes is unknown. Adr1 from glucose-grown cells is able to bind DNA, as shown by electrophoretic mobility shift assays (44), but in glucose-grown cells in vivo it does not bind its cognate promoters (45, 50). Promoter binding in vivo requires Snf1, possibly for chromatin modification (32, 50). Chromatin modification apparently is required for Adr1 binding, because loss of histone deacetylation activity allows Adr1 to bind promoters in the presence of glucose (45). Additionally, nuclear extracts prepared from a strain lacking Snf1 are partially defective in preinitiation complex formation, particularly in the recruitment of mediator components (50). Thus, Snf1 may act at more than one level in regulating the activation of Adr1-dependent genes.
Adr1 and Cat8 influence the expression of a large number of genes required for metabolism of nonfermentable carbon sources, as shown by transcriptome analysis (23, 49). These data demonstrate the critical role of Adr1 and Cat8 in the response to nutrient stress, but they do not identify the genes that are regulated directly by Adr1 and Cat8. We are particularly interested in the genes that are coregulated directly by the two factors, so we combined the transcriptome data with global localization analysis to find genes that are both bound and regulated by Adr1 and Cat8. Naturally, this method misses genes that can be influenced by factors redundant to Adr1 or Cat8 and genes whose expression is weakly dependent on Adr1 or Cat8. Nonetheless, we can distinguish between regulation that is accompanied by factor binding from regulation that is indirect, for example through a regulatory network. Genes whose expression is dependent on ADR1 or CAT8 and whose promoters are bound by Adr1 or Cat8 are assumed to be direct regulatory targets. By these criteria, at least 14 genes are candidates for direct coregulation by both factors.
When two factors bind to different cis-acting elements at a common promoter, they may contribute to transcription in a cooperative and synergistic way or they may act independently, each influencing transcription in a strictly additive way (7, 30). If the factors interact, several mechanisms are possible. One factor may affect the binding of the other in a unilateral way, as the MADS box protein Mcm1 does with Ste12 (18). The factors may act bilaterally, with each influencing the binding of the other, as in the example of Ste12-Tec1 (2, 27, 33).
We investigated the interaction of Adr1 and Cat8 at coregulated promoters by using chromatin immunoprecipitation (ChIP) to detect in vivo binding of one factor in the absence of the other. We found that although Adr1 and Cat8 cooccupy coregulated promoters, they influence each others' binding at only a few. Where they affect each others' binding, they do so in a unilateral way. Our data suggest that Adr1 and Cat8 interactions are dependent on the relative position of their binding sites and how well their binding sites match the consensus sequence for the factors. We detected Adr1 binding at some genes that are minimally affected by Adr1, suggesting that binding may occur wherever a site is accessible.
MATERIALS AND METHODS
Yeast strains and growth of cultures.
Yeast strains were W303 based and are listed in Table 1. Deletions and epitope tags were introduced using published methods and plasmids (3, 22, 26). Culture methods, transformations, glucose repression in 5% glucose, and derepression in 0.05% glucose have been described elsewhere (49).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| Z1256 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 GAL+ [psi+] | 31 |
| Z1616 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 GAL+ [psi+] ADR1-myc9::TRP1 | 31 |
| NKY52b | MATaade2 can1-100 his3-11,15 leu2-13,112 trp1-1 ura3-1 CAT8-TAP-HA::KAN | This study |
| NKY53 and CTY-TY24 | MATaade2 can1-100 his3-11,15 leu2-13,112 trp1-1 ura3-1 CAT8-TAP-HA::KAN adr1Δ1::LEU2 | This study |
| CTY-TY16 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 Δcat8::KAN ADR1-HA::TRP1 | This study |
| CTY-TY18 | MATaade2-1 trp1-1 can1-100 leu2-2,112 his3-11,15 ura3 ADR1-myc::TRP1 CAT8-HA::KAN | This study |
| TYY203 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ADH2::YlpADH2/lacZ::TRP1 | 49 |
| TYY204 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 adr1Δ1::LEU2 ADH2::YlpADH2/lacZ::TRP1 | 49 |
| TYY458 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 Δcat8::KAN ADH2::YlpADH2/lacZ::TRP1 | 49 |
| TYY459 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 Δadr1::LEU2 Δcat8::KAN ADH2::YlpADH/lacZKAN | 49 |
| TYY495 | MATatrp1 leu2 ura3 his3 ade2 CAN1-100 adr1Δ1::LEU2 cat8Δ::KAN::TRP1 ADH2::YlpADH2/lacZ (trp1::HIS3) | This study |
| TYY497 | MATatrp1 leu2 ura3 his3 ade2 CAN1-100 adr1Δ1::LEU2 ADH2::YlpADH2/lacZ (trp1::HIS3) | This study |
ChIP, PCR, primers, and intergenic microarrays.
ChIP was performed as described previously (29, 49) with 45 min of dimethyl adipimidate and 15 min of formaldehyde cross-linking. Antihemagglutinin (F-7) or anti-myc (9E10) monoclonal antibodies (Santa Cruz Biotechnology) were used at 2 to 8 μg of antibody to 1 mg of whole-cell extract and precipitated with protein A-Sepharose CL-4B (Pharmacia). Sequential ChIP was performed as described by Geisberg and Struhl (20), using the wash and elution buffers of Kurdistani and Grunstein (29). Primers (IDT) for gene-specific PCRs are shown in Table 2. Products were separated on 2% agarose and visualized with ethidium bromide. The positive control was input (whole-cell extract), and the negative controls were either non-Adr1-, non-Cat8-regulated ACT1 or an untranscribed region of the chromosome VI-R telomere (TEL) (40).
TABLE 2.
Primers used in this study
| Primer | Sequence or reference |
|---|---|
| CTO ACT1A and B | 49 |
| CTO ADH2A and B | 49 |
| YIL057c | 49 |
| ACS1 D | 49 |
| ACT1-C and -D | 49 |
| CTA1-A and -B | 49 |
| CTO ACH1C | CGAGTTTGATCACTACAAGGAGGC |
| CTO ACH1D | CTACTTTGGTAGTTTGACGCTCCG |
| CTO ACS1E | AATGGCACGTGTATGTACGG |
| ADY2 F1 | GAGCACCCCGGGATTTGTTGTCCAAAACCTTG |
| ADY2 R1 | GCTCTCGAGGGGCTACACGTAAAAACCGTAGC |
| CTO ATO3A | TGGCTTGTGATTGCCATCTC |
| CTO ATO3B | TCACGGTCATCGTGAATAGC |
| CTO FBP1A | CCCATCAAACTGCATGGTCC |
| CTO FBP1B | TTGGCTCTTACGCCCTTAAC |
| HAP4 F (−985) | TTTTCTACTACAGGCCTCCGC |
| HAP4 R (−501) | AAATGGAGGAGGCAGAAGAA |
| CTO ICL1A | GGTTTTGCTACTCGTCATCC |
| CTO ICL1B | GGACTTTGGACTGACTTATGC |
| CTO JEN1C | GACTCGCAACGACTCCAATG |
| CTO JEN1D | TCGATCACTGACTTTCGCAC |
| CTO MDH2A | CATCTATATAGCGAAGTACG |
| CTO MDH2B | GGCAACACAAAATGCCACT |
| CTO MLS1A | CTCAGACGTAAAATTCGTGC |
| CTO MLS1B | CTCATGACAGAATCAAAACAC |
| CTO PUT4C | CATGTCTGCCAAATCTCCAG |
| CTO PUT4D | ACGTCATAGCTAGCGCAATG |
| SIP4 F (−388) | TTCTCTGTCAGAAAGGCGCAT |
| SIP4 R (−73) | TCCATGTCAAATGTCCCAAA |
| TEL55 | GCGTAACAAAGCCATAATGCCTCC (40) |
| TEL56 | CTCGTTAGGATCACGTTCGAATCC (40) |
The intergenic microarray analysis used ChIP DNA from three independent cultures that derepressed for 6 h and was cross-linked and immunoprecipitated as described above. Additional purification, labeling, and hybridization to microarrays of PCR products representing 6,361 yeast intergenic regions have been described elsewhere (31, 37). Enrichment in each ChIP preparation was determined by comparison to the whole-cell extract from which it was precipitated. Data for Adr1- or Cat8-bound DNA were averaged, and confidence estimates and promoter assignments were made as described previously (31). The complete data set can be downloaded from the supplemental material.
β-Gal assays and in-gel ADH activity assays.
β-Galactosidase (β-Gal) assays were performed on permeabilized cells as described in reference 21. Data are an average from at least three independent transformants. Alcohol dehydrogenase (ADH) activity was measured after electrophoresis of proteins in whole-cell extracts and visualization in situ as described previously (48).
Quantitative real-time PCR.
Quantitative real-time PCR was performed using a DNA Engine Opticon (MJ Research) and DyNAmo SYBR Green (Finnzymes) according to the manufacturers' directions. The concentrations of bound (ChIP) and input DNA were determined by comparison to genomic DNA standards.
RESULTS
Identification of direct targets of Adr1 and Cat8.
When a yeast culture is shifted from high to low glucose, several hundred genes require Adr1 or Cat8, directly or indirectly, for derepression as determined by DNA microarray analysis (49). To find genes that are directly regulated by these factors, promoters that are bound by Adr1 and Cat8 were identified by global localization, or ChIP-chip analysis (37). ChIP was used to isolate DNA bound by Adr1 or Cat8 after 6 h of derepression, a time point that corresponds to expression microarray data already available. Each ChIP preparation was hybridized to a microarray (chip) of yeast intergenic regions and compared to input DNA as described previously (31, 37). By this analysis, 137 promoters were bound by Adr1 at twofold or greater over background, at a 99.5% confidence level (P ≤ 0.005) (see data in the supplemental material). To find genes that are both bound and positively regulated by Adr1, the ChIP-chip data were compared with the list of 108 genes that are regulated twofold or greater by Adr1 in the transcriptome microarray (49). Twenty-nine genes were found in both data sets. Two others, ADY2 and PUT4, were included because they are Adr1 dependent for expression and binding of Adr1 had already been confirmed by gene-specific PCR of ChIP DNA. JEN1 was included because it is bound by Adr1, and the significance value for ADR1-dependent expression was just below the cutoff (49) (Table 3). A similar analysis for Cat8 (see data in the supplemental material) found 48 genes bound and 255 positively regulated twofold or greater. Twenty-seven appeared in both data sets. We included ADH2 because it is regulated by Cat8 and binding can be confirmed by gene-specific PCR of ChIP DNA (Table 4).
TABLE 3.
Adr1-bound and positively regulated genes
| Genea | Function | Expression (ADR1/Δadr1) | Binding (ChIP/input)b |
|---|---|---|---|
| ALD4* | Aldehyde dehydrogenase | 5.5 | 22.9 |
| ATO3* | Ammonia transporter | 6.4 | 15.6 |
| ACS1* | Acetyl-CoA synthase | 9.0 | 11.7 |
| YIL057C* | Uncharacterized ORF | 16.0 | 10.9 |
| ADH2* | Alcohol dehydrogenase | 6.8 | 10.5 |
| JEN1* | Lactate transporter | (3.3) | 10.5 |
| CTA1* | Catalase | 16.0 | 9.7 |
| YIR016W | Uncharacterized ORF | 2.5 | 8.9 |
| CYB2 | Lactate dehydrogenase | 4.6 | 7.4 |
| GIP2* | Protein phosphatase interactor | 3.6 | 6.9 |
| FOX2 | Beta-oxidation enzyme | 16.0 | 6.1 |
| YKR075C | Uncharacterized ORF | 2.5 | 8.8 |
| PTR2 | Peptide transporter | 2.5 | 5.3 |
| SPO20 | tSNARE | 5.6 | 5.2 |
| CAR2 | Omithine aminotransferase | 3.6 | 5.2 |
| IME1 | Meiosis regulator | 4.5 | 4.8 |
| DIC1 | Dicarboxylic acid transporter | 6.3 | 4.7 |
| GUT2 | Glycerol 3-P dehydrogenase | 2.4 | 4.6 |
| SPG1 | Uncharacterized ORF | 15.0 | 4.4 |
| SUE1 | Uncharacterized ORF | 3.8 | 4.4 |
| YER121W | Uncharacterized ORF | 5.5 | 4.3 |
| CIT3* | Mitochondrial citrate synthase | 10.0 | 4.1 |
| POT1* | Acetyl-CoA C-acyltransferase | 14.0 | 4.1 |
| ALT2 | Uncharacterized ORF | 3.2 | 4.0 |
| YGL081W | Uncharacterized ORF | 2.3 | 3.8 |
| YPL201C | Uncharacterized ORF | 4.5 | 3.7 |
| CSM4 | Chromosome segregation protein | 3.0 | 3.7 |
| GUT1* | Glycerol kinase | 2.4 | 3.7 |
| ICL2* | Methylisocitrate lyase | 7.0 | 2.4 |
| LSC2 | Succinyl CoA ligase | 2.0 | 5.2 |
| PUT4* | Proline permease | 2.8 | (2.0) |
| ADY2* | Acetate transporter | 20.0 | (−1.0) |
*, confirmed by gene-specific PCR of Adr1-bound ChIP DNA.
Values in parentheses were below the significance cutoff.
TABLE 4.
Cat8-bound and positively regulated genes
| Genea | Function | Expression (CAT8/Δcat8) | Binding (ChIP/input)b |
|---|---|---|---|
| MDH2* | Malate dehydrogenase | 10.0 | 12.7 |
| REG2 | Protein phosphatase regulator | 5.0 | 9.8 |
| MLS1* | Malate synthase | 12.0 | 8.9 |
| ACS1* | Acetyl-CoA synthase | 3.5 | 8.2 |
| PCK1 | PEP carboxykinase | 3.6 | 7.7 |
| YGR067c | Uncharacterized ORF | 7.1 | 7.3 |
| ADY2* | Acetate transporter | 9.3 | 7.2 |
| SFC1 | Succinate-fumarate transporter | 6.2 | 6.8 |
| ACH1* | Acetyl-CoA hydrolase | 2.1 | 6.6 |
| BAT2 | Amino acid transaminase | 2.4 | 6.3 |
| ALD6 | Aldehyde dehydrogenase | 2.6 | 5.2 |
| FBP1* | Fructose 1,6-bisphosphatase | 7.4 | 5.0 |
| PRM4 | Uncharacterized ORF | 2.8 | 4.9 |
| LSR1 | snRNA | 2.4 | 4.8 |
| ICL1* | Isocitrate lyase | 20.0 | 4.2 |
| JEN1* | Lactate transporter | 2.1 | 3.6 |
| ATO3* | Ammonia transporter | 4.6 | 3.4 |
| PTR2 | Peptide transporter | 5.7 | 3.2 |
| ODC1 | Mitochondrial membrane transport | 2.6 | 3.2 |
| YAT2 | Carnitine acetyltransferase | 2.0 | 2.9 |
| IDP2 | Isocitrate dehydrogenase | 2.5 | 2.9 |
| GAP1 | Amino acid permease | 9.1 | 2.8 |
| CAT2 | Carnitine O-acetyltransferase | 3.5 | 2.8 |
| GDH2 | Glutamate dehydrogenase | 3.7 | 2.8 |
| CLB2 | Cyclin-dependent kinase regulator | 2.5 | 2.7 |
| PUT4* | Proline permease | 1.7 | 2.7 |
| DLD1 | Lactate dehydrogenase | 2.1 | 2.6 |
| ADH2* | Alcohol dehydrogenase | 2.7 | (1.9) |
*, confirmed by gene-specific PCR of Cat8-bound ChIP DNA.
Value in parentheses was below the significance cutoff.
In vivo Adr1 binding has already been established for many of the promoters identified by the ChIP-chip microarray as Adr1 bound (ADH2, CTA1, ACS1, GUT1, and POT1 [50], ADY2, ALD4, CIT3, GIP2, and ICL2 [49]). For Cat8, in vitro binding has been demonstrated by electrophoretic mobility shift assays to the CSRE (carbon source-responsive element) sequences from ACS1, ADH2, ICL2, ICL1, FBP1, MDH2, MLS1, and PCK1 (9, 28, 35, 36, 39, 47, 51). All of these genes are known to be regulated by Cat8 under derepressing conditions. To confirm in vivo binding of Cat8 to promoters identified by the ChIP-chip microarray analysis, we used gene-specific PCR on Cat8-bound ChIP DNA, testing a selection of promoters that included some known to be Cat8 bound in vitro and some not previously tested. Gene-specific PCR of ChIP DNA confirmed that Cat8 binds in vivo to the promoters of ACH1, ACS1, ADH2, ADY2, ATO3, FBP1, ICL1, JEN1, MDH2, MLS1, and PUT4 under derepressing conditions (Fig. 1; see also Fig. 5, below). Cat8 was not present at the ADH2 promoter under repressing conditions (data not shown), consistent with its very low expression in repressed cells (24).
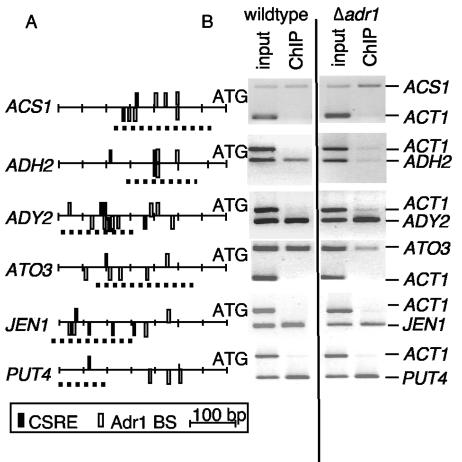
FIG. 1.
Cat8 binds in vivo to positively regulated promoters. (A) Promoter maps, generated as described in reference 49, showing CSREs and Adr1-binding sites for seven promoters. All promoter maps show 700 bp upstream of the ATG except for HAP4, which shows 800 to 100 bp upstream. Dotted lines indicate the region amplified by PCR in panel B. (B) PCR using primers to the indicated promoters was performed on Cat8-bound ChIP DNA from logarithmically growing wild-type cells transferred to 0.05% glucose for 6 h. See Fig. 5 for additional ChIP data on Cat8.
FIG. 5.
Cat8 binding is affected by Δadr1 at some promoters. (A) Promoter maps for Adr1-bound and Cat8-bound and regulated promoters showing CSREs and Adr1-binding sites. Lines under the promoters indicate the region amplified by PCR in panel B. (B) PCR using primers to the indicated promoters was performed on Cat8-bound ChIP DNA from wild-type or isogenic Δadr1 cells after 6 h in 0.05% glucose.
We tested for Cat8 binding at the HAP4 and SIP4 promoters because of previous studies showing in vitro binding (6, 25). SIP4 encodes a homolog of Cat8 that participates in the activation of some CAT8-dependent genes, and HAP4 encodes a transcription factor that activates genes expressed abundantly in aerobically grown cells (41). Gene-specific PCR of Cat8-bound DNA did not detect significant in vivo binding to either promoter (Fig. 1).
Coregulated promoters are cooccupied by Adr1 and Cat8.
To determine which genes are regulated directly by both Adr1 and Cat8, we compared the data sets containing the direct targets of each factor and then made individual evaluations of candidate genes using gene-specific expression or binding data from the literature (4, 5, 9, 24, 28, 36, 39, 46, 47, 50, 51), from Fig. 1, and from our unpublished data. Fourteen genes were strong candidates for direct coregulation by Adr1 and Cat8 (Table 5). Four showed significant binding and expression in all microarrays (ACH1, ACS1, ATO3, and PTR2). Note that ACH1 does not appear in Table 4, which contains only positively regulated genes. Three candidates were not significant for binding in the global localization microarray but had been independently confirmed by gene-specific PCR of ChIP DNA (ADH2, ADY2, and PUT4 [49, 50]). Seven are included because they were strongly positive in three of the four assays and had measurements just below the significance cutoff in the fourth (ALD6, BAT2, CIT3, DIC1, GDH2, JEN1, and REG2).
TABLE 5.
Adr1- and Cat8-bound and -regulated genes
| Gene | Function | Expression ADR1/Δadr1 | Binding Adr1 (ChIP/input) | Expression CAT8/Δcat8 | Binding Cat8 (ChIP/input) |
|---|---|---|---|---|---|
| ACH1 | Acetyl-CoA hydrolase | −14.6 | 5.0 | 2.1 | 6.6a |
| ACS1 | Acetyl-CoA synthase | 9.0a,b | 11.7a | 3.5b | 8.2a |
| ADH2 | Alcohol dehydrogenase | 6.8b | 10.5a | 2.7b | (1.9)a |
| ADY2 | Acetate transporter | 20.0b | (−1.0)a | 9.3 | 7.2a |
| ALD6 | Aldehyde dehydrogenase | (−3.9)c | 3.7 | 2.6 | 5.2 |
| ATO3 | Ammonia transporter | 6.4 | 15.6a | 4.6 | 3.4a |
| BAT2 | Amino acid transaminase | (1.4) | 16.0 | 2.4 | 6.3 |
| CIT3 | Citrate synthase | 10.0 | 4.1a | (−1.4) | 3.4a |
| DIC1 | Dicarboxylic acid transporter | 6.3 | 4.7 | 1.8 | (2.4) |
| GDH2 | Glutamate dehydrogenase | (1.7) | 7.0 | 3.7 | 2.8 |
| JEN1 | Lactate transporter | (3.3) | 10.5a | 2.1b | 3.6a |
| PTR2 | Peptide transporter | 2.5 | 5.3 | 5.7 | 3.2 |
| PUT4 | Proline permease | 2.8 | (2.0)a | 1.7 | 2.7a |
| REG2 | Phosphatase regulator | (1.5) | 11.4 | 5.0 | 9.8 |
The list of coregulated candidate genes in Table 5 was generated from separate lists of genes regulated by Adr1 and genes regulated by Cat8. It suggests but does not demonstrate that Adr1 and Cat8 bind to the same promoters at the same time. Cooccupancy at a subset of coregulated promoters was tested using the sequential ChIP method (20), in which chromatin is immunoprecipitated for one bound factor, released, and then immunoprecipitated for the second factor. Adr1 and Cat8 were found together at the promoters of all coregulated genes that were tested (Fig. 2 and data not shown).
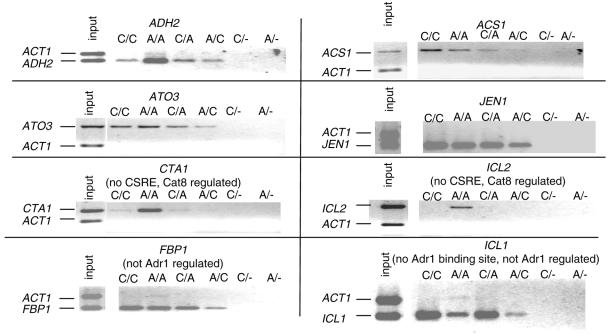
FIG. 2.
Adr1 and Cat8 cooccupy coregulated promoters. Sequential ChIP was performed on cells with myc-tagged Adr1 and hemagglutinin-tagged Cat8 and derepressed for 6 h. A/C, sample was precipitated for bound Adr1 and then bound Cat8; C/A, sample was precipitated for bound Cat8 and then bound Adr1. PCR using primers to the indicated promoters was performed on input (whole-cell extract) and ChIP samples. As positive controls, each factor was subjected to sequential precipitations with the same antibody (lanes C/C and A/A are Cat8 and Adr1 precipitations, respectively). As negative controls, each factor was subjected to a single precipitation followed by a mock sequential precipitation using protein A-Sepharose only (lanes C/- and A/-).
Other results shown in Fig. 2 demonstrate the importance of using both expression and binding data to confirm direct regulation by a transcription factor. Regulation without binding can be seen in Cat8's effect on ICL2 expression. This promoter showed a 2.1-fold Cat8 dependence for derepression, yet lacked a CSRE and Cat8 binding was negligible. Thus, we conclude that the effect of Cat8 on ICL2 expression is indirect. Conversely, binding without positive regulation was demonstrated by Adr1's cooccupation of the ICL1 and FBP1 promoters with Cat8, even though these genes are not Adr1 dependent for expression in low glucose and ICL1 lacks a match to the Adr1-binding site consensus. Adr1 binding was also detected at MDH2 and MLS1, which are not Adr1 dependent (see Fig. 6, below).
FIG. 6.
Adr1 binding is affected by Δcat8 at some promoters. (A) Promoter maps for three Adr1- and Cat8-coregulated promoters (ACS1, ADH2, and ATO3) and three promoters that are bound but not positively regulated by Adr1 (FBP1, MDH2, and MLS1). Lines under the promoters indicate the region amplified by PCR in panel B. (B) PCR using primers to the indicated promoters was performed on Adr1-bound ChIP DNA from wild-type or isogenic Δcat8 cells after 6 h in 0.05% glucose.
Adr1 and Cat8 can act independently or synergistically.
The promoters that are coregulated and cooccupied by Adr1 and Cat8 gave us an opportunity to investigate the interaction of these factors as a model for combinatorial transcriptional control. Two activators that regulate a single promoter may act independently, exerting strictly additive effects on transcription, or they may act synergistically. In the latter case, the presence of both activators increases transcription more than the additive effect of each factor alone.
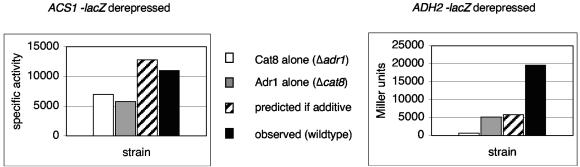
Kratzer and Schüller (28) showed that Adr1 and Cat8 coregulate ACS1, which encodes an acetyl coenzyme A synthetase, in an additive way (Fig. 3). Expression of ACS1 in the presence of only one factor was measured using a strain deleted for the other factor. Adding the levels of ACS1 expression in the presence of either factor alone approximated the level of expression in a wild-type strain with both Adr1 and Cat8.
FIG. 3.
Additive and synergistic interactions between Adr1 and Cat8. β-Gal activity was measured in derepressed strains bearing the indicated reporters on plasmids. The ADH2 plasmid is CEN, and the ACS1 plasmid is 2μm. Data for ACS1 from reference 28 are used with permission. White bars are the activity from Δadr1, gray bars are from Δcat8, striped bars are the expected activity if Cat8 and Adr1 activation were additive, and black bars are the actual activity measured in CAT8 ADR1 wild type.
On the other hand, Adr1 and Cat8 appear to act synergistically at ADH2 (17, 47), which encodes the glucose-repressible ADH required for ethanol catabolism. We confirmed these results by using an ADH2 promoter-lacZ reporter (Fig. 3).
Adr1 binding is sufficient to stimulate Cat8 activation of ADH2.
Synergism between Adr1 and Cat8 could involve cooperative DNA binding or cooperative activation, or both. If cooperative activation is involved, deleting the major activation domain of Adr1 would cause loss of synergism. If synergism involves only DNA binding, loss of the activation domain would not affect synergism. To distinguish these possibilities, we transformed a Δadr1 CAT8 strain with pJS21, a plasmid expressing the amino-terminal 172 amino acids of Adr1. This portion of Adr1 contains the entire DNA-binding domain and a nuclear targeting signal but lacks the major activation domain. As controls, the same strain was transformed with pJS20, which is a plasmid that expresses full-length Adr1, or with vector alone. Activation of ADH2 was tested with an in-gel assay for ADH activity (Fig. 4). The strain with Cat8 but no Adr1 (Δadr1) showed no ADHII after 6 h of derepression. When the plasmid expressing the DNA-binding domain of Adr1 (BD) was present, ADH2 expression was seen as a band of ADHII activity. Cat8 is the activator in this strain, because expressing the DNA-binding domain alone, in a Δcat8 strain, does not activate ADH2 expression.
FIG. 4.
Adr1 binding alone assists Cat8 activation of ADH2. In-gel assays for ADH activity are shown for strains that are Δadr1CAT8 (TYY497) and Δadr1 Δcat8 (TYY495) with one of the following plasmids: empty vector pRS314 (-) (42); pJS21, which expresses the first 172 amino acids of Adr1, containing the entire DNA-binding domain but lacking the major activation domain (BD) (43); or pJS20, which expresses wild-type full-length Adr1 (ADR1) (43). Activity levels in Miller units from an integrated β-Gal reporter (16) in the same strains are shown below. In-gel activity assays were done with extracts prepared after 6 h of derepression in 0.05% glucose; reporter assays were done with cells derepressed for 24 h. The upper ADHI band from constitutively expressed ADH 1 served as a loading control for the in-gel activity assay.
Reporter assays using an ADH2 promoter-lacZ construct showed similar results (Fig. 4, numbers below gel). The DNA-binding domain of Adr1 did not activate ADH2/lacZ in the absence of Cat8. However, a strain with Cat8 and the DNA-binding domain of Adr1 induced expression from the ADH2/lacZ reporter above the background level seen in the same strain with the vector alone.
These results indicate that DNA binding by Adr1 influences the ability of Cat8 to activate the ADH2 promoter. To find out whether Adr1 affects Cat8 binding or if it affects a later step in activation, we used gene-specific PCR against ChIP DNA to probe for binding of Cat8 in the absence of Adr1 at coregulated promoters.
Adr1 is important for Cat8 binding at the ADH2 promoter.
Adr1 and Cat8 might act synergistically at ADH2 but independently at ACS1 because they influence each other's binding at one promoter but not the other. To test this hypothesis, we examined the ability of each factor to bind cooccupied promoters in the absence of the other by using ChIP DNA from derepressed cells and gene-specific PCR. Cat8 bound to the independently regulated ACS1 promoter in the absence of Adr1. However, binding of Cat8 at the synergistically regulated ADH2 promoter was reduced in the Δadr1 strain (Fig. 5). Real-time quantitative PCR of Cat8-bound ChIP DNA showed that Cat8 binding to ADH2 decreased by 2.5-fold in the Δadr1 strain compared to the isogenic wild type. Student's t test of eight assays confirmed that the difference was significant, with a P value of 0.001. Cat8 protein levels, measured by Western blotting, were comparable in wild-type and Δadr1 strains (data not shown). Deleting ADR1 may have a severe effect on ADH2 expression because it eliminates both Adr1 and Cat8 binding at this promoter.
Binding of Adr1 and Cat8 is independent at most promoters.
Other promoters were tested for binding of Cat8 in the absence of Adr1 and Adr1 in the absence of Cat8. Cat8 binding to ACH1, ACS1, ADY2, FBP1, JEN1, and PUT4 was unaffected or only slightly affected by lack of Adr1, but binding at the ATO3 promoter was noticeably reduced. At the promoter of YIL057c, which has a CSRE but is not positively regulated by Cat8, weak binding in an Adr1 wild type was reduced in an Δadr1 mutant (Fig. 5 and data not shown).
Adr1 binding to ACS1, ADH2, ADY2, ATO3, CIT3, CTA1, ICL2, JEN1, PUT4, and YIL057c was unaffected by lack of Cat8 (Fig. 6 and data not shown). All are Adr1 dependent for derepression in low glucose. Four other promoters, FBP1, ICL1, MDH2, and MLS1, were tested because they are bound by Adr1 but are not Adr1 dependent for derepression. All four are bound and regulated by Cat8 (Table 4). In the absence of Cat8, Adr1 binding was reduced at ICL1, MDH2, and MLS1, where there is no apparent Adr1 consensus-binding sequence or the Adr1-binding site is buried within Cat8-binding sites. The decreased binding of Adr1 at these promoters cannot be due to reduced levels of Adr1 in the Δcat8 mutant, because binding at other promoters was unaffected. Absence of Cat8 had little or no effect on Adr1 binding at the FBP1 promoter, where the Adr1 binding site is several hundred base pairs from the CSRE (Fig. 6).
We found that at most promoters, Adr1 and Cat8 did not require each other for binding. However, at ADH2 and to some extent ATO3, Cat8 clearly requires Adr1. The well-defined Cat8-binding sequence, CSRE, lends itself to an analysis of Adr1 dependence and promoter context.
Relative position and binding site sequence may determine if binding is cooperative or independent.
We tested the hypothesis that Adr1 dependence of Cat8 binding correlates with promoter features like CSRE sequence or distance between the Adr1- and Cat8-binding sites. Adr1-binding sites and CSREs were identified in coregulated promoters by using the Cat8-binding sequence (23), the Adr1-binding site (49), and the RSA tools website (embnet.cifn.unam.mx/rsa-tools). Cat8-binding sequences were compared to the consensus determined in reference 38, revealing multiple mismatches in the CSREs at ADH2 and ATO3, where Adr1 affects Cat8 binding (Table 6). Some of these mismatches severely affect in vitro binding of Cat8 (38). ADH2 has an additional upstream CSRE with only two mismatches, but we do not know if it is bound by Cat8. If it is, the distance of nearly 200 bp from the Adr1-binding site may reduce the influence of Adr1. Promoters where Cat8 binding was independent of Adr1, like ACS1, ADY2, FBP1, ICL1, MDH2, and MLS1, have a perfect or nearly perfect match to the CSRE consensus sequence or, in the case of PUT4, have CSREs that are several hundred base pairs from the Adr1-binding site. Adr1 may stabilize Cat8 at promoters where Cat8 binding is weak, like ADH2, and may have little or no influence at promoters where binding is already strong.
TABLE 6.
Adr1 affects Cat8 binding at weak CSREs
| Gene | Cat8 binding to promoter in Δadr1 | CSRE sequences (from furthest upstream to closest to ATG)a |
|---|---|---|
| ACS1 | Yes | GCCGTTCGTCCG (1) |
| TCCATTTCGCCG (1) | ||
| ADH2 | Reduced | TCCGTCTCTCCGG (2) |
| GCCGGAACACCG (4) | ||
| GCCTTGTGGCCC (2) | ||
| ADY2 | Yes | TCCGGAGCTCCG (3) |
| ACCACTCAGCCG (1) | ||
| GCCGCCCAACCG (3) | ||
| ATO3 | Reduced | GCCGCACCGCCG (4) |
| FBP1 | Yes | TCCATCCGTCCG (1) |
| TCCGGGTGTCCG (2) | ||
| ICL1 | Yes | TCCATTCATCCG (0) |
| JEN1 | Yes | TCCACTAGACCG (1) |
| MLS1 | Yes | TCCATTGAGCCG (0) |
| TCCATTGGGCCG (0) | ||
| CCCGGCGAGCCG (2) | ||
| GCCGGCTCGCCGG (4) | ||
| MDH2 | Yes | TCCATTTGGCCG (0) |
| CCCTTTAATCCG (0) | ||
| CCCATTCGGCCG (0) | ||
| PUT4 | Yes | TCCGGCAGCCCG (3) |
| CCCGGGTACCCG (3) | ||
| CCCGGGCTGCCG (3) |
Values in parentheses are the number of mismatches to consensus (see reference 38). Sequence portions in italics are mismatches that allowed in vitro binding; portions in boldface are mismatches that severely reduced in vivo binding.
DISCUSSION
Transcriptome analysis of genome-wide expression in the presence and absence of a transcription factor is an excellent tool for detecting targets of the factor. Nonetheless, these data do not distinguish between directly and indirectly regulated genes. Global localization analysis (ChIP-chip), in which intergenic microarrays are probed with ChIP DNA, provides a wealth of data on the location of a transcription factor but does not reveal if binding influences transcription. Comparing data from transcriptome and global localization data sets, each produced under conditions in which the factor is known to have an impact on gene expression, identifies direct regulatory targets of a transcription factor. Although targets may be missed because of redundant factors, weak effects on transcription, or different rates of activation, we used this approach successfully to find genes coregulated by the carbon source-responsive activators Adr1 and Cat8 (Tables 3 to 5).
The data in these tables are probably conservative lists of the genes regulated directly by Adr1 and Cat8, since legitimate targets can be excluded at either the transcriptome or the ChIP-chip step. For example, in our transcriptome analysis, Cat8-dependent derepression of ADH2 was masked by background expression from genes encoding non-glucose-repressed ADH isozymes (49). In the global localization analysis, Adr1 binding at the ADY2 promoter, which can be detected by gene-specific PCR, was not above background, probably because the spot representing its chromosomal locus covered several intergenic regions (C. Tachibana and T. I. Lee, unpublished observations).
Our results stress the advantage of using both expression and binding data to confirm direct regulatory targets of a factor. We found that both Adr1 and Cat8 affect expression of more genes than they actually bind, suggesting that they regulate genes indirectly, through transcription factor networks (31). For example, activation of the transcription factor gene HAP4 is CAT8 dependent. Specific binding to its CSRE-like sequence was detected in CAT8 but not Δcat8 extracts (6). We cannot detect Cat8 binding in vivo to the HAP4 CSRE-like region, so the observed Cat8-dependent binding might be due to another Cat8-dependent factor, as suggested by Brons et al. (6). A strong candidate is Sip4, which binds to some CSREs and requires CAT8 for full expression (23, 25, 39). Our failure to detect Cat8 binding to the SIP4 promoter in vivo, despite evidence of binding in vitro (25), might reflect different sensitivities of the assays.
The global localization analysis showed binding at several promoters where transcriptome analysis did not indicate positive regulation (21 for Cat8 and 107 promoters for Adr1). Some of these may be false positives, and some may be bound yet not regulated, at least under the conditions that we tested. Precedent for the latter situation comes from chromosome-wide analysis of the mammalian transcription factor NF-κB binding, which showed that DNA-binding proteins may be present at a binding site, even when the nearest gene is not differentially regulated by the factor (34). Only in specific contexts, for example the presence of other transcription factors, does binding of a factor influence gene expression. Based on our data, direct binding alone is not an infallible predictor of gene regulation. In vivo binding of Adr1 can be detected at several promoters (FBP1, ICL1, MDH2, and MLS1) whose expression undergoes minor changes, if any, when ADR1 is deleted. The combination of statistically significant binding and expression provides the best evidence that a gene is regulated directly by a transcription factor.
Fourteen genes are strong candidates for direct regulation by both Adr1 and Cat8 (Table 5). Adr1 and Cat8 cooccupy the promoters of coregulated genes (Fig. 2) but do not show the same pattern of interaction at all of them. Their effect on transcription is independent at ACS1 but synergistic at ADH2. They bind independently at most coregulated promoters, but at others one does not bind fully without the other (Cat8 at ADH2 or ATO3; Adr1 at ICL1, MDH2, and MLS1). Promoter context may play a role in Adr1-Cat8 interactions, because Cat8 binding is affected by the absence of Adr1 at promoters where the Cat8-binding CSRE sequence is a weak match to the consensus (Fig. 5 and Table 6).
At promoters where Adr1 and Cat8 affect each others' binding, one may be altering the chromatin structure in a way that increases the accessibility of binding sites for the other factor. For example, at ADH2, chromatin remodeling is ADR1 dependent (14, 15) and Adr1 binding is itself influenced by the acetylation state of the nucleosomes (45). One possibility for the dependence of Cat8 on Adr1 at some promoters (ADH2 and ATO3) is that Adr1-dependent chromatin remodeling is required for efficient Cat8 binding at these promoters. In this case, even though complete ADR1-dependent chromatin remodeling is dependent on an Adr1 that contains an activation domain, expression of an ADR1 allele containing the DNA-binding domain but lacking the major activation domain would suffice for a primitive state of remodeled chromatin (15). Since this same fragment of Adr1 suffices for synergism between Adr1 and Cat8 (Fig. 4), we speculate that DNA binding by Adr1 allows Cat8 to bind to a weak CSRE and promote a low level of transcription. Promoters at which Cat8 binding is independent of Adr1 may have a chromatin conformation that is already permissive for Cat8 binding. Conversely, Cat8-dependent chromatin remodeling at promoters like ICL1, MDH2, and MLS1 may be required to uncover binding sites before Adr1 can bind.
Alternatively, a subfraction of complexed Adr1 and Cat8 may be involved in regulating those genes where Cat8 binding is dependent on Adr1 or vice versa. We have detected a minor fraction of Adr1 that can be coimmunoprecipitated with Cat8 from DNase-treated cell extracts (unpublished data). This Adr1-Cat8 complex might contain the factors that are dependent on each other for binding. According to this model, promoters at which Cat8 is dependent on Adr1 for binding would be bound by this preformed complex. These two alternatives may be resolved by investigations currently in progress to determine the effects of histone modifications and the roles of chromatin remodeling complexes at Adr1 and Cat8 coregulated promoters.
Supplementary Material
Acknowledgments
This work was supported by research grants GM-26079 and DK-067276, National Institutes of Health, to E.T.Y.
We are grateful to H.-J. Schüller for permission to use previously published data, members of the E.T.Y. lab for helpful discussions, and R. Biddick, J. Infante, K. Dombek, and J. Hansen for critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Allocco, D. J., I. S. Kohane, and A. J. Butte. 2004. Quantifying the relationship between co-expression, co-regulation and gene function. BMC Bioinformatics 5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baur, M., R. K. Esch, and B. Errede. 1997. Cooperative binding interactions required for function of the Ty1 sterile responsive element. Mol. Cell. Biol. 17:4330-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumberg, H., T. A. Hartshorne, and E. T. Young. 1988. Regulation of expression and activity of the yeast transcription factor ADR1. Mol. Cell. Biol. 8:1868-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bojunga, N., and K. D. Entian. 1999. Cat8p, the activator of gluconeogenic genes in Saccharomyces cerevisiae, regulates carbon source-dependent expression of NADP-dependent cytosolic isocitrate dehydrogenase (Idp2p) and lactate permease (Jen1p). Mol. Gen. Genet. 262:869-875. [DOI] [PubMed] [Google Scholar]
- 5.Bojunga, N., P. Kotter, and K. D. Entian. 1998. The succinate/fumarate transporter Acr1p of Saccharomyces cerevisiae is part of the gluconeogenic pathway and its expression is regulated by Cat8p. Mol. Gen. Genet. 260:453-461. [DOI] [PubMed] [Google Scholar]
- 6.Brons, J. F., M. De Jong, M. Valens, L. A. Grivell, M. Bolotin-Fukuhara, and J. Blom. 2002. Dissection of the promoter of the HAP4 gene in S. cerevisiae unveils a complex regulatory framework of transcriptional regulation. Yeast 19:923-932. [DOI] [PubMed] [Google Scholar]
- 7.Carey, M., Y. S. Lin, M. R. Green, and M. Ptashne. 1990. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature 345:361-364. [DOI] [PubMed] [Google Scholar]
- 8.Carlson, M. 1999. Glucose repression in yeast. Curr. Opin. Microbiol. 2:202-207. [DOI] [PubMed] [Google Scholar]
- 9.Caspary, F., A. Hartig, and H. J. Schuller. 1997. Constitutive and carbon source-responsive promoter elements are involved in the regulated expression of the Saccharomyces cerevisiae malate synthase gene MLS1. Mol. Gen. Genet. 255:619-627. [DOI] [PubMed] [Google Scholar]
- 10.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12:323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charbon, G., K. D. Breunig, R. Wattiez, J. Vandenhaute, and I. Noel-Georis. 2004. Key role of Ser562/661 in Snf1-dependent regulation of Cat8p in Saccharomyces cerevisiae and Kluyveromyces lactis. Mol. Cell. Biol. 24:4083-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 13.De Vit, M. J., J. A. Waddle, and M. Johnston. 1997. Regulated nuclear translocation of the Mig1 glucose repressor. Mol. Biol. Cell 8:1603-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Mauro, E., S. G. Kendrew, and M. Caserta. 2000. Two distinct nucleosome alterations characterize chromatin remodeling at the Saccharomyces cerevisiae ADH2 promoter. J. Biol. Chem. 275:7612-7618. [DOI] [PubMed] [Google Scholar]
- 15.Di Mauro, E., L. Verdone, B. Chiappini, and M. Caserta. 2002. In vivo changes of nucleosome positioning in the pretranscription state. J. Biol. Chem. 277:7002-7009. [DOI] [PubMed] [Google Scholar]
- 16.Dombek, K. M., and E. T. Young. 1997. Cyclic AMP-dependent protein kinase inhibits ADH2 expression in part by decreasing expression of the transcription factor gene ADR1. Mol. Cell. Biol. 17:1450-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donoviel, M. S., N. Kacherovsky, and E. T. Young. 1995. Synergistic activation of ADH2 expression is sensitive to upstream activation sequence 2 (UAS2) orientation, copy number and UAS1-UAS2 helical phasing. Mol. Cell. Biol. 15:3442-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Errede, B., and G. Ammerer. 1989. STE12, a protein involved in cell-type-specific transcription and signal transduction in yeast, is part of protein-DNA complexes. Genes Dev. 3:1349-1361. [DOI] [PubMed] [Google Scholar]
- 19.Ferre, P., D. Azzout-Marniche, and F. Foufelle. 2003. AMP-activated protein kinase and hepatic genes involved in glucose metabolism. Biochem. Soc. Trans. 31:220-223. [DOI] [PubMed] [Google Scholar]
- 20.Geisberg, J. V., and K. Struhl. 2004. Cellular stress alters the transcriptional properties of promoter-bound Mot1-TBP complexes. Mol. Cell 14:479-489. [DOI] [PubMed] [Google Scholar]
- 21.Guarente, L. 1983. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 101:181-191. [DOI] [PubMed] [Google Scholar]
- 22.Guldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haurie, V., M. Perrot, T. Mini, P. Jeno, F. Sagliocco, and H. Boucherie. 2001. The transcriptional activator Cat8p provides a major contribution to the reprogramming of carbon metabolism during the diauxic shift in Saccharomyces cerevisiae. J. Biol. Chem. 276:76-85. [DOI] [PubMed] [Google Scholar]
- 24.Hedges, D., M. Proft, and K. D. Entian. 1995. CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 15:1915-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiesinger, M., S. Roth, E. Meissner, and H. J. Schuller. 2001. Contribution of Cat8 and Sip4 to the transcriptional activation of yeast gluconeogenic genes by carbon source-responsive elements. Curr. Genet. 39:68-76. [DOI] [PubMed] [Google Scholar]
- 26.Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963-972. [DOI] [PubMed] [Google Scholar]
- 27.Kohler, T., S. Wesche, N. Taheri, G. H. Braus, and H. U. Mosch. 2002. Dual role of the Saccharomyces cerevisiae TEA/ATTS family transcription factor Tec1p in regulation of gene expression and cellular development. Eukaryot. Cell 1:673-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kratzer, S., and H. J. Schuller. 1997. Transcriptional control of the yeast acetyl-CoA synthetase gene, ACS1, by the positive regulators CAT8 and ADR1 and the pleiotropic repressor UME6. Mol. Microbiol. 26:631-641. [DOI] [PubMed] [Google Scholar]
- 29.Kurdistani, S. K., and M. Grunstein. 2003. In vivo protein-protein and protein-DNA crosslinking for genomewide binding microarray. Methods 31:90-95. [DOI] [PubMed] [Google Scholar]
- 30.Laybourn, P. J., and J. T. Kadonaga. 1992. Threshold phenomena and long-distance activation of transcription by RNA polymerase II. Science 257:1682-1685. [DOI] [PubMed] [Google Scholar]
- 31.Lee, T. I., N. J. Rinaldi, F. Robert, D. T. Odom, Z. Bar-Joseph, G. K. Gerber, N. M. Hannett, C. T. Harbison, C. M. Thompson, I. Simon, J. Zeitlinger, E. G. Jennings, H. L. Murray, D. B. Gordon, B. Ren, J. J. Wyrick, J. B. Tagne, T. L. Volkert, E. Fraenkel, D. K. Gifford, and R. A. Young. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799-804. [DOI] [PubMed] [Google Scholar]
- 32.Lo, W. S., L. Duggan, N. C. Emre, R. Belotserkovskya, W. S. Lane, R. Shiekhattar, and S. L. Berger. 2001. Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293:1142-1146. [DOI] [PubMed] [Google Scholar]
- 33.Madhani, H. D., and G. R. Fink. 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275:1314-1317. [DOI] [PubMed] [Google Scholar]
- 34.Martone, R., G. Euskirchen, P. Bertone, S. Hartman, T. E. Royce, N. M. Luscombe, J. L. Rinn, F. K. Nelson, P. Miller, M. Gerstein, S. Weissman, and M. Snyder. 2003. Distribution of NF-κB-binding sites across human chromosome 22. Proc. Natl. Acad. Sci. USA 100:12247-12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahner, A., M. Hiesinger, and H. J. Schuller. 1999. Deregulation of gluconeogenic structural genes by variants of the transcriptional activator Cat8p of the yeast Saccharomyces cerevisiae. Mol. Microbiol. 34:146-156. [DOI] [PubMed] [Google Scholar]
- 36.Randez-Gil, F., N. Bojunga, M. Proft, and K. D. Entian. 1997. Glucose derepression of gluconeogenic enzymes in Saccharomyces cerevisiae correlates with phosphorylation of the gene activator Cat8p. Mol. Cell. Biol. 17:2502-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings, I. Simon, J. Zeitlinger, J. Schreiber, N. Hannett, E. Kanin, T. L. Volkert, C. J. Wilson, S. P. Bell, and R. A. Young. 2000. Genome-wide location and function of DNA binding proteins. Science 290:2306-2309. [DOI] [PubMed] [Google Scholar]
- 38.Roth, S., J. Kumme, and H. J. Schuller. 2004. Transcriptional activators Cat8 and Sip4 discriminate between sequence variants of the carbon source-responsive promoter element in the yeast Saccharomyces cerevisiae. Curr. Genet. 45:121-128. [DOI] [PubMed] [Google Scholar]
- 39.Roth, S., and H. J. Schuller. 2001. Cat8 and Sip4 mediate regulated transcriptional activation of the yeast malate dehydrogenase gene MDH2 by three carbon source-responsive promoter elements. Yeast 18:151-162. [DOI] [PubMed] [Google Scholar]
- 40.Rundlett, S. E., A. A. Carmen, N. Suka, B. M. Turner, and M. Grunstein. 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392:831-835. [DOI] [PubMed] [Google Scholar]
- 41.Schuller, H. J. 2003. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 43:139-160. [DOI] [PubMed] [Google Scholar]
- 42.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sloan, J. S., K. M. Dombek, and E. T. Young. 1999. Post-translational regulation of Adr1 activity is mediated by its DNA binding domain. J. Biol. Chem. 274:37575-37582. [DOI] [PubMed] [Google Scholar]
- 44.Taylor, W. E., and E. T. Young. 1990. cAMP-dependent phosphorylation and inactivation of yeast transcription factor ADR1 does not affect DNA binding. Proc. Natl. Acad. Sci. USA 87:4098-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verdone, L., J. Wu, K. van Riper, N. Kacherovsky, M. Vogelauer, E. T. Young, M. Grunstein, E. Di Mauro, and M. Caserta. 2002. Hyperacetylation of chromatin at the ADH2 promoter allows Adr1 to bind in repressed conditions. EMBO J. 21:1101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vincent, O., and J. M. Gancedo. 1995. Analysis of positive elements sensitive to glucose in the promoter of the FBP1 gene from yeast. J. Biol. Chem. 270:12832-12838. [DOI] [PubMed] [Google Scholar]
- 47.Walther, K., and H. J. Schuller. 2001. Adr1 and Cat8 synergistically activate the glucose-regulated alcohol dehydrogenase gene ADH2 of the yeast Saccharomyces cerevisiae. Microbiology 147:2037-2044. [DOI] [PubMed] [Google Scholar]
- 48.Williamson, V. M., J. Bennetzen, E. T. Young, K. Nasmyth, and B. D. Hall. 1980. Isolation of the structural gene for alcohol dehydrogenase by genetic complementation in yeast. Nature 283:214-216. [DOI] [PubMed] [Google Scholar]
- 49.Young, E. T., K. M. Dombek, C. Tachibana, and T. Ideker. 2003. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J. Biol. Chem. 278:26146-26158. [DOI] [PubMed] [Google Scholar]
- 50.Young, E. T., N. Kacherovsky, and K. Van Riper. 2002. Snf1 protein kinase regulates Adr1 binding to chromatin but not transcription activation. J. Biol. Chem. 277:38095-38103. [DOI] [PubMed] [Google Scholar]
- 51.Zaragoza, O., O. Vincent, and J. M. Gancedo. 2001. Regulatory elements in the FBP1 promoter respond differently to glucose-dependent signals in Saccharomyces cerevisiae. Biochem. J. 359:193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.