Abstract
The pocket protein family of tumor suppressors, and Rb specifically, have been implicated as controlling terminal differentiation in many tissues, including the heart. To establish the biological functions of Rb in the heart and overcome the early lethality caused by germ line deletion of Rb, we used a Cre/loxP system to create conditional, heart-specific Rb-deficient mice. Mice that are deficient in Rb exclusively in cardiac myocytes (CRbL/L) are born with the expected Mendelian distribution, and the adult mice displayed no change in heart size, myocyte cell cycle distribution, myocyte apoptosis, or mechanical function. Since both Rb and p130 are expressed in the adult myocardium, we created double-knockout mice (CRbL/L p130−/−) to determine it these proteins have a shared role in regulating cardiac myocyte cell cycle progression. Adult CRbL/L p130−/− mice demonstrated a threefold increase in the heart weight-to-body weight ratio and showed increased numbers of bromodeoxyuridine- and phosphorylated histone H3-positive nuclei, consistent with persistent myocyte cycling. Likewise, the combined deletion of Rb plus p130 up-regulated myocardial expression of Myc, E2F-1, and G1 cyclin-dependent kinase activities, synergistically. Thus, Rb and p130 have overlapping functional roles in vivo to suppress cell cycle activators, including Myc, and maintain quiescence in postnatal cardiac muscle.
Many tissues become quiescent with respect to proliferative growth as part of their terminal differentiation, with various degrees of reversibility. The heart is the first organ to form during embryogenesis and is especially susceptible to the consequences of cell loss (necrosis and apoptosis) in adult disorders. Although intriguing exceptions are seen in urodele amphibians and teleost fish (6, 41), mammalian cardiac myocytes characteristically proliferate during fetal life but exit the proliferative cell cycle soon after birth (49-51). The mechanisms underlying this permanent growth arrest are unclear; however, the retinoblastoma gene product (Rb) has been implicated in mediating not just quiescence with respect to growth but more specifically the irreversibility of cell cycle arrest associated with terminal differentiation in various lineages, including skeletal muscle (17), adipocytes (8), and macrophages (7), suggesting that this may be a general phenomenon. Among these lineages, Rb has perhaps been best studied in the differentiation of skeletal myocytes. MyoD and related members of the basic-helix-loop-helix family of myogenic transcription factors can induce both myogenic gene transcription and cell cycle exit (47). In contrast to skeletal myocytes that are deficient in p107 or p130, Rb−/− myocytes express early but not late markers of myogenic differentiation, fail to arrest in G1, and accumulate instead in the S and G2/M phases of the cell cycle (38, 39). Such data suggest that Rb, unlike the other pocket proteins, is required uniquely for normal myogenic cell cycle control and full differentiation.
Analogously, cardiac muscle differentiation and cell cycle exit have been suggested to display a similar dependence on pocket proteins, although the relationship of cycling to differentiation is not thought to be mutually exclusive as in skeletal muscle. The viral proteins simian virus 40 large T antigen and E1A can each promote G1 exit in cardiac myocytes, including mutants that specifically inhibit Rb family members (12, 19, 21, 22). Pocket proteins are the best-accepted substrate for phosphorylation by G1 cyclin-dependent kinases (Cdks). Hence, another strategy to assert their importance was the presumed functional inactivation of pocket proteins in vivo, with heart-specific transgenic mice that increase Cdk4 (52) or Cdk2 (28) activity. Both strains of mice displayed an increase in cardiac myocyte number and ongoing DNA synthesis in adult hearts. Like the studies in which viral proteins were used to inactivate Rb family members, the work with G1 Cdks suggests that one or more pocket proteins are necessary for terminal differentiation in cardiac muscle but does not discriminate among them. Developmental studies have shown that Rb is not expressed in the myocardium until late in gestation, concomitant with cell cycle exit by ventricular myocytes, supporting the premise that Rb may be the key to terminal differentiation in the myocardium as well (13, 53).
In mice, attempts to directly study the effects of Rb deficiency in the heart are complicated by the fact that Rb-null mice die at day 14 to 15 postcoitum from hematological and neurological deficits, including extensive apoptosis (9, 18, 25). Since Rb is not expressed in the developing myocardium until late gestation (13, 20), this makes the study of Rb's functional role in cardiac terminal differentiation much less straightforward. Recent developments in conditional mutagenesis allowing site-specific deletion of DNA with the Cre/loxP system permit the analysis of tissue-specific gene function in contexts that would otherwise result in lethality or require cumbersome chimeric models (42, 43). We report that heart-specific deletion of Rb results in mice with normal embryonic viability, life span, and heart size, with no overt defects in cardiac differentiation or function. Defects in cardiac cell cycle control and differentiation were uncovered only after disruption of the second pocket protein expressed in the adult myocardium, p130. Only combined deletion of Rb plus p130 led to the persistent expression of Myc, E2F-1, and G1 Cdk activities, all of which are normally down-regulated in the neonatal heart. Thus, although nonoverlapping functions of the two pocket proteins are seen in other settings (10, 59), the combined disruption of Rb by Cre-mediated recombination and of p130 by germ line deletion establishes unequivocally an essential role for these pocket proteins in controlling cardiac cell cycle exit and provides concrete in vivo evidence of their overlapping and redundant function in the heart.
MATERIALS AND METHODS
Animal studies.
Creation and characterization of mice carrying the conditional Rb allele have been previously described (35). These mice were created by inserting two loxP sites into the introns surrounding exon 19. Cre-mediated excision of the region flanked by the loxP sites results in a premature truncation of the Rb protein that is functionally equivalent to a null allele. The heart-restricted Cre mice (αMHC-Cre) have been previously described (1, 16). Recombination is induced exclusively in cardiac muscle and is nearly homogeneous in atrial and ventricular myocytes by mid-gestation, sparing other tissues and the nonmuscle components of the heart (16). These mice were backcrossed onto an FVB background and maintained homozygous for the αMHC-Cre transgene. Littermate controls were used throughout the study. M-mode echocardiography was performed as previously described (58). Animals were handled in accordance with institutional guidelines.
Animals were screened by Southern and PCR analyses as previously described (35). PCR analysis of Cre-mediated recombination in the RbLox mice was performed on genomic DNA extracted from the indicated organs (35). The primers flanking the LoxP sites were Rb18 (5′-GGCGTGTGCCATCAATG-3′) and Rb19 (5′-AACTCAAGGGAGACCTG-3′). These primers result in a 650-bp product for the wild-type allele and a 750-bp product for the LoxP-modified Rb allele. After Cre-mediated recombination, PCR yields a 185-bp product.
Protein and RNA analysis.
Western blot assays were performed on protein extracts from whole ventricles, in accordance with established protocols (32). Antibodies were obtained from Santa Cruz Biotechnology, Inc., unless otherwise noted. Immune complex assays for Cdk activity were performed as previously described (58). Total RNA was isolated from ventricles with an RNA STAT 60 kit (Tel-Test Inc.), and Northern blot assays were performed in accordance with established protocols (32). For Northern blotting, total RNA was isolated from ventricles with an RNA STAT 60 kit (Tel-Test Inc.). The oligonucleotide and cDNA probes used to determine heart-restricted gene expression have been reported previously (58). Ribonucleic protection assays (RPA) were done in accordance with the manufacturer's (Pharmingen) specifications.
Histologic analysis and BrdU injections.
Hearts were cut longitudinally and either frozen or fixed overnight in 4% paraformaldehyde buffered with phosphate-buffered saline and routinely processed. Hearts for cryostat sections were snap-frozen in OCT compound and stored at −80°C until used. If DNA synthesis was to be quantified, bromodeoxyuridine (BrdU) labeling was achieved by injecting 50 μg of BrdU per g of body weight intraperitoneally; the animals were euthanized, and their hearts were recovered 4 h later. To assess the prevalence of cell cycling, cryostat sections were probed with antibodies to BrdU (Roche) and serine-10-phosphorylated histone H3 (Upstate), in accordance with the manufacturer's recommendations. Secondary antibodies were purchased from Molecular Probes.
Isolation of cardiac myocytes and analysis.
Neonatal mouse cardiac myocytes were prepared as previously described (58). Cultured neonatal cells were serum starved for 48 h prior to virus infection. Adult mouse myocytes were isolated by retrograde perfusion (58). Myocyte dimensions were determined, and volumes were calculated by a computerized morphometric system (58). All myocytes were measured at the same magnification with the observer blinded to the genotype of the animals.
Myocyte-specific DNA quantification was performed by two-color flow cytometry with MF20 (Developmental Hybridoma Studies Bank), an antibody to muscle-specific myosin heavy chains, and propidium iodide to quantify DNA content (32).
ChIP.
Chromatin immunoprecipitation (ChIP) assays were done as previously described (57). Briefly, formaldehyde (Fisher Scientific) was added at a final concentration of 1% directly to neonatal rat ventricular myocytes (NRVM). After fixation, the NRVM were collected and resuspended in swelling buffer for 20 min and then Dounce homogenized. The nuclei were collected by microcentrifugation and then resuspended in sonication buffer and incubated on ice for 10 min. The samples were sonicated on ice and then microcentrifuged. The chromatin solution was precleared with Super A/G agarose (Santa Cruz) for 15 min at 4°C. Precleared chromatin was incubated with 1 μg of antibody or no antibody and rotated at 4°C for approximately 12 to 16 h. Subsequent immunoprecipitation, washing, and elution of immune complexes were done as previously described (57). Cross-links were reversed and RNA was removed with RNase A. The precipitated DNA was analyzed by PCR. PCRs were performed with 2 μl of immunoprecipitate or 2 μl of a 1:100 dilution of the total sample (positive control) under standard conditions.
Statistical analysis.
All data are presented as the mean ± the standard error of the mean. Results were compared by analysis of variance and Fisher's protected least-significant-difference tests, with a significance of P < 0.05.
RESULTS
Creation of heart-restricted Rb-deficient mice.
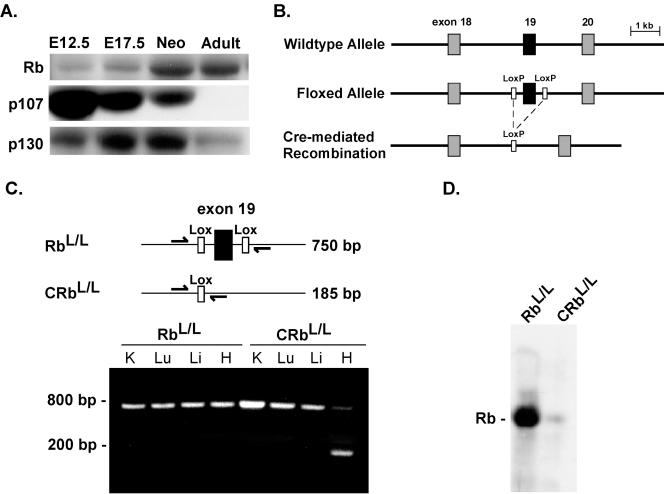
Preliminary studies were performed to determine the temporal program of pocket protein family member expression in embryonic, neonatal, and adult ventricular tissue. Total protein extracts from mouse myocardium tissue at various developmental stages were probed with antibodies for each protein, respectively. Rb is scant or undetectable in fetal mouse myocardium tissue at embryonic day 12.5 (E12.5), in agreement with prior surveys by in situ hybridization (20), but is up-regulated by the neonatal stage, and in adults, terminally differentiated cardiac tissue becomes the predominant pocket protein expressed (Fig. 1A). p107 expression has a pattern that is the reciprocal of that of Rb; i.e., it is highest in the embryonic heart and lowest in the adult heart. p130 expression peaks in the neonatal period and is subsequently down-regulated, and its level is low in the adult myocardium.
FIG. 1.
Heart-restricted deletion of Rb in vivo. (A) Western blot assays were performed on protein lysates extracted from ventricular tissue harvested at the indicated developmental time points. (B) Schematic diagram depicting the construction of the floxed Rb allele. (C) Results of PCR on genomic DNAs isolated from the indicated tissues in RbL/L and αMHC-CreRbL/L mice. (D) Western blot assay on anti-Rb immunoprecipitates from 750 μg of protein prepared from ventricular tissue.
To begin to investigate the role of Rb in cardiac muscle, mice with a floxed Rb allele (RbL/+) were mated with the αMHC-Cre transgenic line (αMHC-Cre+/+) and backcrossed until homozygous for Cre, thus creating mice that undergo deletion of the floxed allele and become deficient in Rb exclusively in cardiac myocytes (CRbL/+). Cre-mediated recombination of exon 19 of the gene for Rb would be predicted to delete critical sequences in Rb, resulting in an unstable, nonfunctional protein (Fig. 1B) (35). At 8 weeks of age, the loxP-tagged allele had undergone recombination in myocardium tissue, as demonstrated by the appearance of a 185-bp fragment representing the recombined allele; no recombination of Rb was seen in other tissues of heart-restricted Rb-null mice (CRbL/L) or in RbL/L mice in the absence of Cre (Fig. 1C). The residual unrecombined fragment likely represents a combination of Rb in myocytes where recombination has not occurred and the large number of nonmyocytes present in the adult heart. Heart-restricted recombination of the gene for Rb resulted in a greater than 90% reduction in Rb protein (Fig. 1D). CRbL/L mice were born with the expected Mendelian distribution, and no premature mortality was observed in mice followed through 1 year of age.
Rb-deficient cardiac myocytes exit the cell cycle appropriately and differentiate normally.
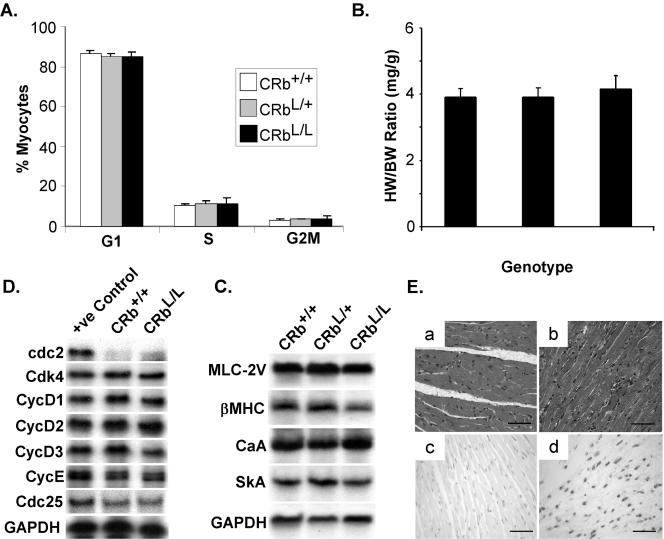
To determine if Rb-deficient cardiac myocytes display aberrant growth properties, we first examined freshly isolated neonatal cardiac myocytes from these mice. The DNA content of immunologically authenticated, MF20-positive cardiac myocytes was quantitated by two-color flow cytometry. As shown in Fig. 2A, Rb-null neonatal myocytes had exited the cell cycle appropriately and accumulated in G1, similar to control myocytes from Cre-homozygous mice that were wild type for Rb (CRb+/+). Adult heart-restricted Rb-deficient mice displayed no significant increase in the heart-to-body weight ratio at 16 weeks (3.8 ± 0.3 versus 4.1 ± 0.3 mg/g [no statistically significant difference]; Fig. 2B) and had no obvious defect in cardiac differentiation, as assessed by Northern blotting for representative genes encoding heart-restricted sarcomeric proteins (Fig. 2C). Likewise, RPA analysis of RNA isolated from ventricles of mice with the indicated genotypes for a panel of cell cycle-regulated genes, including known E2F targets (cdc2, CycE, and Cdc25), demonstrated no differences in expression levels between control (CRb+/+) and CRbL/L mice (Fig. 2D). Consistent with this, there were no significant differences in baseline myocyte cell cycling (see Fig. 3B). Histological analysis of CRbL/L myocardium tissue revealed normal appearing myocytes and no evidence of myocyte loss or fibrosis (Fig. 2E-b). As reported after tissue-restricted deletion of Rb in the central nervous system (CNS) (34), the hearts of heart-specific Rb-deficient mice did not demonstrate increased apoptosis (Fig. 2E-c).
FIG. 2.
Heart-restricted Rb-deficient mice are phenotypically normal. (A) Two-color flow cytometry was performed on neonatal cardiac myocytes isolated from animals of the indicated genotypes with fluorescein isothiocyanate-labeled MF20 to identify authentic cardiac myocytes and propidium iodide to quantitate DNA. The pooled results of three experiments are presented. (B) Heart weights (milligrams) normalized to body weight (grams) for 16-week-old mice with the indicated genotypes. (C) Total ventricular RNA from mice with the indicated genotypes was probed with the specified probes (myosin light chain 2V [MLC-2V], β-myosin heavy chain [βMHC], cardiac α-actin [CaA], skeletal α-actin [SkA], and glyceraldehyde-3-phosphate dehydrogenase [GAPDH]). (D) Representative RPA performed on total RNA prepared from the ventricles of wild-type or Rb-null mice. (E) H&E-stained myocardial sections from CRb+/+ (a) or CRbL/L (b) mice. Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling staining of CRbL/L myocardium tissue (c) or a positive control (d). Scale bars equal 50 μm.
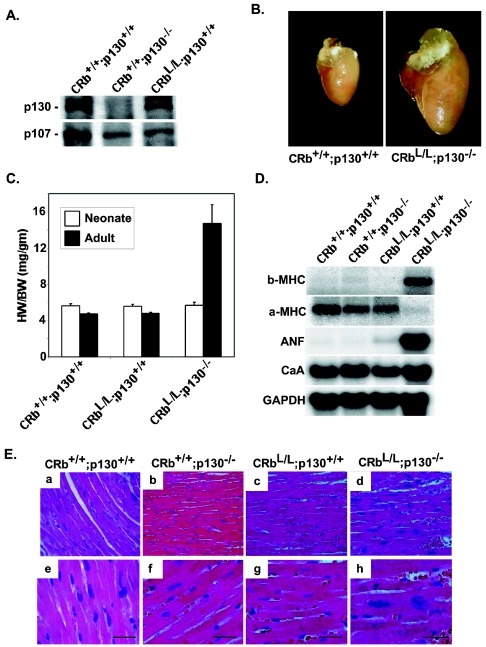
FIG. 3.
p130 functionally compensates for the loss of Rb in cardiac myocytes. (A) Ventricular lysates from animals with the indicated genotypes were probed by Western blotting. (B) Representative perfusion-fixed hearts are shown. (C) Quantitative results of heart weight-to-body weight ratios of neonatal (6 per group) or adult (12 per group) animals. *, P < 0.0001 for adult CRbL/L p130−/− mice versus CRb+/+ p130+/+ or CRbL/L p130+/+ mice. (D) Total ventricular RNA from mice with the indicated genotypes was probed with the specified probes. (C) H&E-stained ventricular tissue at low (a to d) and high (e to h) magnifications. Scale bars equal 25 μm.
To determine if loss of Rb affected left ventricular function, transthoracic echocardiograms were performed on six mice from each genotype at 16 weeks of age (Table 1). Compared with either control (CRb+/+) mice or the heart-specific heterozygotes (CRbL/+), CRbL/L mice demonstrated a similar posterior wall thickness and intraventricular septal thickness. Likewise, no significant differences were seen in functional parameters as measured by aortic ejection time, left ventricular fraction shortening, and circumferential shortening velocity.
TABLE 1.
Left ventricular function in 16-week-old Rb-null micea
| Parameter | CRb+/+ | CRbL/+ | CRbL/L |
|---|---|---|---|
| VST (mm) | 0.57 ± 0.04 | 0.56 ± 0.04 | 0.60 ± 0.05 |
| EDD (mm) | 3.82 ± 0.11 | 3.85 ± 0.16 | 3.75 ± 0.08 |
| PWT (mm) | 0.63 ± 0.03 | 0.61 ± 0.03 | 0.65 ± 0.02 |
| ESD (mm) | 2.77 ± 0.13 | 2.82 ± 0.10 | 2.79 ± 0.16 |
| Ao-ET (ms) | 53.7 ± 2.7 | 51.9 ± 2.5 | 51.8 ± 5.3 |
| HR (bpm) | 474 ± 9 | 505 ± 30 | 511 ± 41 |
| LV% FS | 27.5 ± 2.0 | 27.9 ± 2.6 | 25.8 ± 2.8 |
| Vcf (mm/s) | 5.11 ± 0.27 | 5.38 ± 0.63 | 5.03 ± 0.66 |
Echocardiographic analysis of the left ventricle: IVST, intraventricular septal thickness; EDD, end-diastolic diameter; PWT, posterior wall thickness; ESD, end-systolic diameter; Ao-ET, aortic ejection time; HR, heart rate; bpm, beats per minute; LV%FS, left ventricular fractional shortening; Vcf, circumferential shortening velocity. There were six mice per group. No significant differences were found between groups in any of the parameters.
p130 functionally compensates for the loss of Rb in cardiac muscle.
The lack of an obvious basal phenotype in CRbL/L mice contrasts with in vitro data suggesting that pocket proteins are necessary for normal cell cycle exit in cardiac myocytes (22). Although functional redundancy between Rb and p130 in vivo has not been described, one explanation for the lack of an overt phenotype in these mice is that p130 is required or functionally substitutes for the loss of Rb in adult cardiac myocytes. This is particularly important since p130 expression was increased in Rb-deficient myocardium tissue (Fig. 3A). Therefore, to establish to what extent the loss of Rb is complemented by the continued presence of p130, we engineered mice lacking both Rb and p130 in the myocardium. We bred our CRbL/L mice to conventional p130 germ line-null mice, which are viable and display no overt phenotype (11). The resultant CRbL/+ p130+/− mice were backcrossed again to CRbL/L mice, and the F2 mice (CRbL/L p130+/−) were used for subsequent breeding. In contrast to p130−/− mice in a BALB/cJ background (24), no phenotype was seen in mice deficient only in p130, and identical results were obtained when CRbL/+ p130−/− F2 mice were used for breeding. Although mice deficient in both proteins (CRbL/L p130−/−) are born with the expected Mendelian frequency, 36% of these mice developed signs of generalized edema and respiratory distress presumably related to congestive heart failure or died suddenly by 12 weeks of age, and only 48% of the CRbL/L p130−/− mice survived to 6 months of age. CRbL/L p130−/− mice display a 3.1-fold increase in the heart/body weight ratio at 8 weeks of age compared to CRbL/L littermates (14.7 ± 2.1 versus 4.7 ± 0.1 [P < 0.0001]; Fig. 3B and C). This enlargement was not apparent immediately after birth. There was no significant difference in neonatal heart size between genotypes, consistent with the notion that the primary role for these proteins is in the postnatal period (Fig. 3C). CRbL/L p130−/− mice surviving to 12 weeks of age demonstrated evidence of LV dilation and systolic dysfunction by echocardiogram (Table 2). While the hearts of CRbL/L p130−/− mice surviving to 6 months of age were also enlarged compared to control hearts (8.25 ± 0.65 versus 4.65 ± 0.45; P < 0.005), this increase was less dramatic than at 8 weeks, suggesting that the mice with the greatest cardiac enlargement were the most susceptible to cardiac dysfunction and death.
TABLE 2.
Left ventricular function in 12-week-old Rb- and p130- null micea
| CRbL/L p130+/+ | CRbL/L p130−/− | |
|---|---|---|
| VST (mm) | 0.71 ± 0.02 | 0.77 ± 0.03 |
| EDD (mm) | 3.91 ± 0.16 | 4.5 ± 0.18b |
| PWT (mm) | 0.76 ± 0.02 | 0.78 ± 0.02 |
| ESD (mm) | 2.73 ± 0.15 | 3.41 ± 0.19b |
| Ao-ET (ms) | 55.2 ± 1.5 | 57.5 ± 2.7 |
| HR (bpm) | 537 ± 26 | 444 ± 22b |
| LV% FS | 30.4 ± 1.7 | 24.3 ± 2.1b |
| Vcf (mm/s) | 5.54 ± 0.35 | 4.28 ± 0.39b |
Results are presented as the mean±the standard error of the mean. For definitions of abbreviations, see Table 1, footnote a. There were nine mice per group.
P < 0.05 by analysis of variance.
To determine if this increased growth was associated with altered cardiac gene expression, Northern blot assays were performed on RNA prepared from ventricles of mice with the indicated genotypes (Fig. 3D). As shown, CRbL/L p130−/− mice display increased expression of ANF and βMHC, heart-specific genes normally associated with the fetal ventricle but also with hypertrophy of the adult myocardium, and also showed a reciprocally decreased expression of αMHC, the normally predominant adult isoform. Histological examination of the hearts of these mice revealed large, hyperchromatic nuclei and increased myocyte diameters (Fig. 3E-d, h). Similar to the single-protein knockout (CRbL/L) mice, CRbL/L p130−/− mice displayed no evidence of myocardial apoptosis (by terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling staining) or fibrosis (data not shown).
Rb- and p130-null cardiac myocytes continue to cycle.
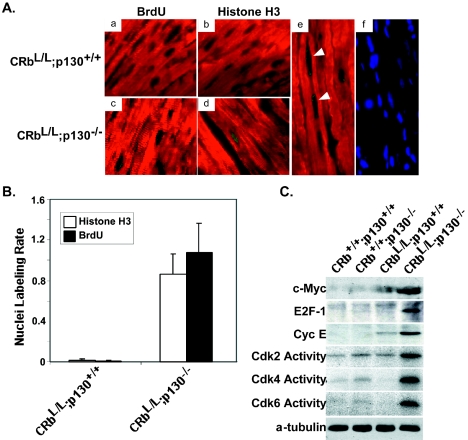
Neither the increase in heart size, nor the activation of “fetal” cardiac genes, nor the histological changes provoked by combined deletion of Rb and p130 in the heart would distinguish exclusively hypertrophic growth from an admixture of myocyte enlargement and persistent myocyte cycling. To establish whether or not the loss of Rb and p130 resulted in abnormal cell cycle regulation in cardiac myocytes, we determined the prevalence of BrdU incorporation (S phase) and Cdc2-dependent phosphorylation of histone H3 (serine-10; M phase). Ventricular myocardium tissue from CRbL/L p130−/− mice demonstrated a 135-fold increase in BrdU-positive myocyte nuclei (15,750 ± 1,150 versus 116 ± 60 per 106 myocytes; P < 0.0001) and a 48-fold increase in phosphorylated histone H3-positive nuclei (10,710 ± 2,517 versus 225 ± 161 per 106 myocytes; P < 0.001) compared to hearts lacking just Rb (CRbL/L; Fig. 4A and B). Interestingly, although hearts from neonatal CRbL/L p130−/− mice were similar in size to hearts with both Rb and p130 or hearts lacking Rb alone (Fig. 3C), myocardium tissue from these mice already demonstrated enhance cell cycling (2.01-fold ± 0.34-fold increase in phosphorylated histone H3-positive nuclei; P < 0.05).
FIG. 4.
CRbL/L p130−/− null myocytes display defects in cell cycle regulation in vivo. (A) Detection of BrdU incorporation (green; a versus c) or the M phase-specific marker phosphorylated histone H3 (green; b versus d) by immunofluorescence in cardiac myocytes identified with MF20 (red) from CRbL/L p130−/− mice versus CRbL/L p130+/− or CRbL/L p130+/+ mice. Nuclei were stained with 4′,6′-diamidino-2-phenylindole (blue). Multiple BrdU-positive staining nuclei are visible in a single CRbL/L p130−/− myocyte (e and f; white arrows). (B) Percentages of BrdU- or histone H3-positive nuclei from sham- and transaortic constriction-treated mice (five per group) were quantified. (C) Representative Cdk assays and Western blot assays of ventricular lysates from mice with the indicated genotypes.
A search for genes specifically up-regulated in CRbL/L p130−/− myocardium tissue with microarrays identified a number of E2F-regulated genes whose expression was increased. E2F target cyclin E was specifically up-regulated in Rb- and p130-deficient hearts (Fig. 4C). Consistent with the persistence of cyclin E in adult hearts, Cdk2 activity, along with that of the other G1 Cdks Cdk4 and Cdk6, was specifically increased in myocardium tissue lacking both pocket proteins (Fig. 4C). Myc and E2F-1 were also increased in adult ventricular muscle by the loss of Rb plus p130 (Fig. 4D). Myc and E2F are critical and interdependent transcription factors required for normal cell cycle progression in most cell types (27, 46). Both Myc and E2F-1 are E2F gene targets that are normally down-regulated when cardiac myocytes exit the cell cycle but can provoke cell cycle reentry in adult cardiac myocytes when ectopically expressed (2, 58).
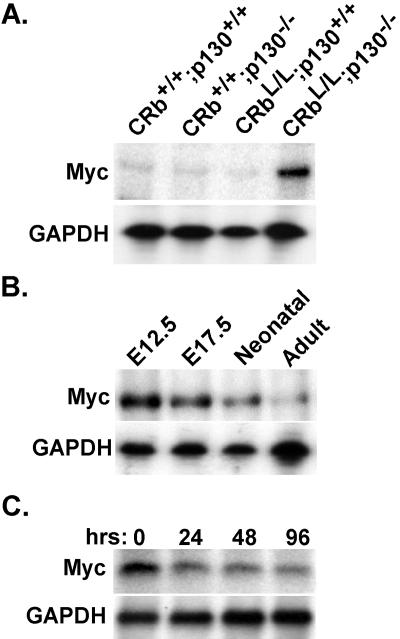
Expression of the gene for Myc is increased in myocardium tissue by the combined loss of Rb plus p130, but not by either mutation alone.
Consistent with increased Myc protein levels in CRbL/L p130−/− hearts, Myc transcripts were persistently expressed in the adult myocardium in this genetic background (Fig. 5A). Deletion of either pocket protein, singly, did not influence Myc expression in the adult heart. Normally, Myc is highly expressed in fetal ventricles when the myocytes are proliferative (E12.5) but is down-regulated throughout gestation and is detected at only minute levels in the adult myocardium (Fig. 5B). This pattern is similar to that observed in rat ventricular tissue (48) and occurs in a temporal pattern consistent with the exit of cardiac myocytes from the cell cycle (51). To verify that this down-regulation of the Myc promoter was occurring in authentic cardiac myocytes, we analyzed Myc expression in purified NRVM, since they represent a well-characterized model of cardiomyocyte growth arrest and a developmental time point at which Myc is normally being down-regulated as cardiac myocytes exit the cell cycle. Although Myc transcripts are already expressed at low levels in neonatal cardiac myocytes compared with mid-gestation myocardium tissue, Myc mRNA is down-regulated even further after culture in serum-free medium (55) (Fig. 5C). Hence, this repression of myc gene expression in cardiac myocytes might provide an accessible and readily manipulated system for mechanistic studies, provided that reasonable fidelity to the in vivo phenotype were substantiated.
FIG. 5.
Myc transcripts are increased in CRbL/L p130−/− null myocardium tissue. (A) Expression of Myc was determined in the ventricles of mice with the indicated genotypes by RPA. (B) RPA for Myc in total ventricular RNA from the indicated developmental time points. (C) Total RNA from NRVM differentiated for the indicated times was probed for Myc expression. Equal loading was confirmed by probing for glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
E2F-4 and Rb/p130 complexes bind the E2F site in cardiac muscle.
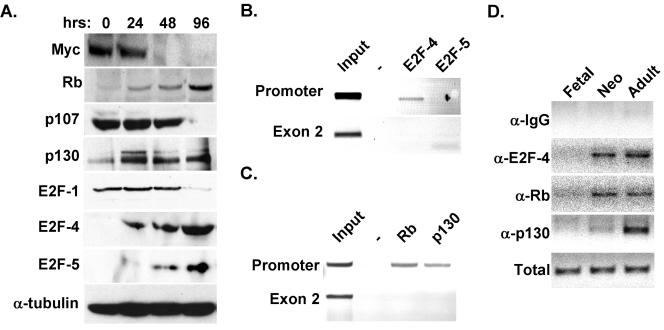
To address this point, clarify the expression of E2F family members in cultured cardiac myocytes, and ensure that they parallel those occurring upon cardiac maturation in vivo, extracts from purified cardiac myocytes were examined by Western blotting. Analogous to the reduction of Myc mRNA, Myc protein was down-regulated with culture (Fig. 6A). Likewise, E2F-1 was down-regulated with an increase in the duration of culture in serum-free medium, whereas E2F-4 and -5 were up-regulated (Fig. 6A), which was also seen in adult ventricular tissue (Fig. 7C). Neither E2F-2, -3, nor -6 was detected in the cardiac myocytes at any time point tested.
FIG. 6.
E2F-4 and p130/Rb bind the Myc E2F site in vivo. (A) Protein extracts from primary fetal rat cardiac myocytes, differentiated for the indicated times in serum-free medium, were probed with antibodies to the indicated proteins. Equal protein loading was confirmed with an antibody to α-tubulin. (B and C) ChIPs were performed on sonicated genomic DNA from formaldehyde-treated NRVM with the indicated antibodies. Antibodies to E2F-4 and -5 (B) or Rb and p130 (C) were used. PCR was done on the immunoprecipitated DNA with primers flanking either the E2F site within the promoter or an irrelevant site within exon 2 to demonstrate the specificity of the reaction. (D) ChIPs were performed on genomic DNA isolated from ventricular tissue at the indicated developmental time points. IgG, immunoglobulin G.
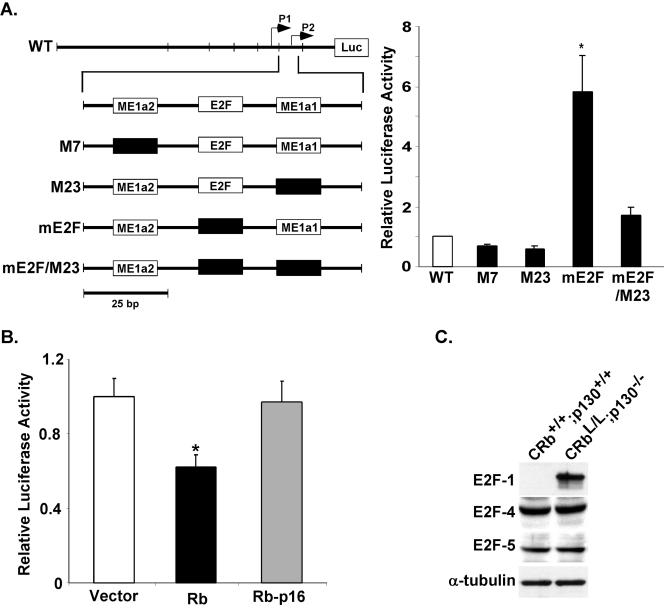
FIG. 7.
Mutation of Myc E2F derepresses Myc transcription specifically in cardiac myocytes. (A) Site-directed mutations were introduced into the ME1a1 and E2F sites of the full-length promoter as shown. The indicated constructs and a constitutively expressed cytomegalovirus lacZ control were transfected into cardiac myocytes. Luciferase activity normalized to LacZ was analyzed after 48 h in serum-free medium. Results are reported relative to the full-length promoter (wild type, WT). *, P < 0.001 for the mE2F construct versus the wild-type promoter. (B) Neonatal cardiac myocyte cells were transfected with the Myc promoter along with the empty vector, wild-type Rb (pCMV-Rb), or an Rb mutant defective for E2Fbinding (pCMV-Rb-p16). Results are reported as the mean ± the standard error of the mean, relative to vector-transfected myocytes. (C) Protein extracts prepared from ventricles from mice with the indicated genotypes were blotted with the indicated antibodies.
Given its critical role in growth and differentiation more generally, the transcriptional regulation of Myc and its regulatory sequences have been well characterized (4, 15, 37, 54). The Myc promoter contains two transcription initiation sites, P1 and P2 (Fig. 6). However, the P2 initiation site mediates transcription in most normal cells, including cardiac muscle (37). A proximal minimal promoter containing three elements, termed ME1a2, E2F, and ME1a1, regulates transcription from this initiation site (37). Use of the ME1a2 or ME1a1 elements is tissue dependent, and these have been reported to bind a variety of cell type-specific and ubiquitous factors. The role of the E2F site is more controversial. It has been reported to potentiate (54), inhibit (15), or have no effect on (4) Myc transcription. To resolve whether the increased expression of Myc was a direct consequence of the loss of Rb and p130 or an indirect effect related to the cycling of the myocytes, we sought to examine the role of Rb and p130 in down-regulating Myc in normal ventricular myocytes. We performed ChIP assays to determine what E2F-pocket protein complexes, if any, were interacting with the Myc E2F site in cultured, differentiated cardiac myocytes. Two sets of primers were used, either flanking the E2F site within the Myc promoter or directed to an unrelated site within the second exon of the gene for Myc to demonstrate the specificity of the assay. Although both E2F-4 and -5 are present in differentiated cardiac muscle, only E2F-4 was bound to the E2F site in the endogenous Myc promoter (Fig. 6B). Interestingly, both of the pocket proteins in differentiated cardiac myocytes, Rb and p130, were recruited to the Myc E2F site (Fig. 6C). As expected, the negative control sequence from exon 2 was not detected after any of the four ChIP procedures. Next, to determine if E2F-4-p130/Rb complexes were specifically recruited to the endogenous Myc promoter at a developmental time consistent with down-regulation of the endogenous Myc-encoding gene, we performed ChIP assays on samples prepared from fetal (E17.5), neonatal (P2), and adult (3-month) ventricular muscle. E2F-4 was first recruited to the Myc E2F site in the neonatal heart and remained associated with it in the adult cardiac myocardium (Fig. 6D). Rb and p130 both associate with the endogenous Myc promoter in neonatal and adult ventricles, consistent with their proposed role in the repression of this promoter in quiescent cells.
Mutation of the E2F site derepresses Myc transcription selectively in cardiac myocytes.
If recruitment of Rb and p130 to the Myc E2F site in the neonatal period contributes to the developmental down-regulation of Myc, we would expect that mutation of this site would derepress transcription activity in differentiated myocytes. Therefore, we introduced previously characterized mutations into the full-length human Myc promoter (37). These mutations have been shown to disrupt the binding of their cognate partners. Mutations of ME1a2 (M7) and ME1a1 (M23) resulted in 33 and 40% reductions in the activity of the full-length promoter, respectively (Fig. 7A). In contrast, ablation of the E2F site resulted in a significant 5.8-fold increase in activity, compared to that achieved with the wild-type promoter (P < 0.001). This increase was abolished when the ME1a1 site was mutated in combination with the E2F site (mE2F/m23), consistent with the functional hierarchy previously reported for these two elements (3). This suggests that the E2F site normally functions to repress transcription of the Myc promoter in postmitotic cardiac myocytes.
To confirm that Rb negatively regulates Myc expression through this E2F site in cardiac myocytes, neonatal myocytes were transfected with the Myc promoter along with wild-type Rb or Rb-p16, an inactive mutant that is unable to bind E2F or suppress growth (14) (Fig. 7B). As shown, wild-type Rb suppressed the activity of the Myc promoter by 38% (P < 0.01). However, the Rb mutant unable to interact with E2Fs had no significant effect on Myc transcriptional activity. E2F-4, which is expressed in the adult p130+/+ myocardium (Fig. 7C), is also expressed in CRbL/L p130−/− hearts. These data support a model in which down-regulation of Myc postnatally is related to recruitment of pocket proteins and the persistent expression in cardiac myocytes lacking both Rb and p130 is related to the inability of E2F-4 to recruit inhibitory complexes to the Myc promoter.
DISCUSSION
Several lines of evidence have suggested that one or more pocket proteins are necessary for cardiac cell cycle control (22, 52). Although both Rb and p130 are expressed in the adult myocardium, we assumed that Rb was the critical molecule since it has been postulated to be crucial for terminal differentiation in skeletal muscle and p130 nullizygous mice are viable and appear phenotypically normal (11), at least in most genetic backgrounds (24). Rb−/− skeletal myocytes demonstrated marked baseline cell cycle abnormalities in vitro (38) and were able to reenter S phase in response to serum (47). In contrast, heart-restricted Rb-deficient mice develop normally and do not display cardiac cell cycle defects. The lack of an obvious phenotype in Rb-null myocardium tissue raised the question that if pocket proteins are necessary for normal cell cycle exit in cardiac myocytes, as previously suggested (22), to what extent does p130 functionally substitute for the loss of Rb. CRbL/L p130−/− mice created to address this question displayed defects in cardiac cell cycle regulation and differentiation. Previous studies have shown that p107 and p130 perform overlapping growth-regulatory and/or differentiation functions that are not fulfilled by Rb (11). Likewise, embryos deficient in both Rb and p107 (Rb−/− p107−/−) died at approximately 11.5 days of gestation, 2 days earlier than embryos homozygous for Rb alone (26), and Rb+/− p107−/− mice have pronounced growth retardation and an increased mortality rate during the first 3 weeks after birth. The Rb+/− p107−/− pups that survived to adulthood did not show any altered tumor predisposition compared with Rb+/− mice but developed multiple dysplastic lesions of the retina (26). These results provided the first in vivo evidence that pocket proteins have overlapping but also distinct tissue-specific functions.
Equivalent in vivo data for functional redundancy between Rb and p130 have not been reported until now, although hints of overlapping roles in cell cycle control exist. Rb−/− p130−/− fibroblasts, compared to comparable cell lines with one or more pocket protein family members deleted, demonstrated the most severe defects in G1 arrest after serum withdrawal (10). By contrast, however, somatic deletion of Rb in mouse skin elicits defects in melanocyte survival that were not exacerbated even after concurrent inactivation of p107 and p130 (60). Given the tissue-restricted, developmentally regulated expression patterns of E2Fs and Rb family members in vivo shown by us (Fig. 1A and 6A) and others (23, 29), the degree to which different Rb proteins can functionally compensate for one another may vary depending on the cellular context. However, results of gene expression profiling on RNA isolated from single- or double-null hearts (30) demonstrate that even in cardiac muscle, where Rb and p130 clearly have overlapping roles, they do have distinct gene targets and unique functions may exist for each family member particularly in the context of physiological stress.
We demonstrated a dramatic increase in cycling adult cardiac myocytes from CRbL/L p130−/− mice. Despite this 100-fold increase over control mice, only 1% of the total myocytes were identified as cycling by markers specific for the S and G2/M phases. However, this is likely an underestimate of the percentage of cycling myocytes since these assays do not reflect the total pool of cycling myocytes but only identify those specifically in the cell cycle phase being assayed. Likewise, since we clearly demonstrated the presence of cycling nuclei in adult highly organized cardiac myocytes (Fig. 4A), these results cannot be accounted for simply by an effect on the subpopulation of immature cardiac progenitors recently described (5, 40). It is also questionable whether the αMHC-Cre transgene would even be expressed in this subpopulation of immature cells; however, we are testing whether their cycling and proliferation are also increased by the lack of p130/Rb. In some, but not all, cases the increased proliferation appeared to lead to myocardial dysfunction and early lethality. The reason for the variability in the penetrance of this phenotype is speculative. Genetic modifiers of pocket protein function have been described previously (24), although these are unlikely to account for the results of our study. The strain-specific cardiac defects and embryonic lethality reported by LeCouter et al. in their p130-null mice in certain inbred mouse backgrounds was not seen with the p130 mice used in this study (11), even when they were placed in a similar genetic background (30). Additionally, the mice we used for the present study were crossed into the background of FVB, an unrelated strain. However, whether the variable penetrance of the lethal cardiac defect in the double-knockout mice represents a limitation of the Cre-Lox system secondary to variable recombination versus novel modifier genes needs to be determined.
In cardiomyocytes, cell cycle exit begins during late gestation and is essentially complete by 2 weeks postnatally (51). Investigations into the mechanisms that regulate this G1 arrest in cardiac myocytes are limited (31); however, since factors such as Myc (58) and E2F-1 (2) provoke G1 exit in adult cardiac myocytes in vivo when overexpressed, their down-regulation must be a critical step in this process. Myc expression was derepressed specifically in CRbL/L p130−/− myocytes, suggesting that either Rb or p130 is sufficient for its down-regulation. This is consistent with the ChIP results demonstrating that both Rb and p130 are recruited to the E2F site in a temporal pattern consistent with Myc's down-regulation in cardiac muscle. Although novel to cardiac muscle, an inhibitory effect of the E2F site in Myc has been reported in other differentiated cell types. A previous study performed with B cells indicated that the E2F site negatively regulates the Myc promoter as well (3). Similarly, transcriptional repression of the gene for E2F-1 is mediated through binding of E2F-4/Rb and E2F-4/p130 complexes (15). Thus, recruitment of Rb and p130 may be a mechanism to down-regulate these factors in myocardium tissue and perhaps differentiated tissue more generally. In contrast, although it has been reported that Myc can be induced by E2F, we believe that the persistent expression of E2F-1 does not contribute to transcription of Myc. Mutation of the Myc E2F site had no effect on Myc transcriptional activity in fetal cardiac myocytes, a developmental time point when E2F-1 is normally expressed. Likewise, we were never able to identify E2F-1 associated with the Myc promoter by ChIP (30). This model of the regulatory elements in the c-myc promoter is consistent with a previous report suggesting that binding of E2F results in transcriptional repression (3).
The absence of significant baseline cardiac apoptosis in our heart-restricted Rb-null and CRbL/L p130−/− mice was unexpected given the dramatic programmed cell death seen in the conventional Rb-null mice (33) and in vitro studies in which disrupting Rb function with E1A induced apoptosis in cardiac myocytes (22). However, although CNS apoptosis was a prominent feature of the conventional Rb knockouts, several recent studies involving CNS-specific excision of Rb in vivo (34, 35, 56) did not demonstrate significant apoptosis. One group proposed that the apoptosis seen in the Rb-null embryos was in part related to a concomitant defect in erythropoiesis and that hypoxia is a necessary cofactor in the death of CNS neurons in the developing Rb mutant embryo (34). Apoptosis plays a critical role in a number of cardiovascular diseases (44), and it will be interesting to determine if Rb plays a protective role in the heart to proapoptotic stimuli, such as ischemia-reperfusion. Because our present results can exclude neither chronic compensatory effects nor a critical requirement for Rb in establishment versus maintenance of the postmitotic phenotype, it will be intriguing in the future to test the interplay between p130 and Rb with respect to growth and apoptosis by means of heart-restricted drug-activated Cre, disrupting the gene for Rb acutely in the adult myocardium (36, 45).
In summary, we have created heart-restricted Rb-deficient mice that appear phenotypically normal at baseline. However, deleting the other pocket protein family member expressed in adult hearts, p130, revealed overlapping roles for Rb and p130 in mediating cardiac cell cycle exit and differentiation. The effect presumably lies in their ability to inhibit the transcription of Myc and other cell cycle-promoting factors in the adult myocardium. These data are consistent with the growing body of literature suggesting that pocket proteins are critical to the regulation of cardiac myocyte cell cycle exit.
Acknowledgments
We thank A. Berns for Rblox mice and S. Mao and J. Chen for technical assistance.
This work was supported by gifts from the Laubisch Fund (K.P.R., W.R.M.), as well as grants AHA EIA 0340087N and R01 HL62448 to W.R.M. and grants R01 HL47567 and R01 HL61668 and the M. D. Anderson Foundation professorship to M.D.S.
REFERENCES
- 1.Agah, R., P. A. Frenkel, B. A. French, L. H. Michael, P. A. Overbeek, and M. D. Schneider. 1997. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J. Clin. Investig. 100:169-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agah, R., L. A. Kirshenbaum, M. Abdellatif, L. D. Truong, S. Chakraborty, L. H. Michael, and M. D. Schneider. 1997. Adenoviral delivery of E2F-1 directs cell cycle reentry and p53-independent apoptosis in postmitotic adult myocardium in vivo. J. Clin. Investig. 100:2722-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albert, T., J. Wells, J. O. Funk, A. Pullner, E. E. Raschke, G. Stelzer, M. Meisterernst, P. J. Farnham, and D. Eick. 2001. The chromatin structure of the dual c-myc promoter P1/P2 is regulated by separate elements. J. Biol. Chem. 276:20482-20490. [DOI] [PubMed] [Google Scholar]
- 4.Amati, B., K. Alevizopoulos, and J. Vlach. 1998. Myc and the cell cycle. Front. Biosci. 3:D250-D268. [DOI] [PubMed] [Google Scholar]
- 5.Beltrami, A. P., L. Barlucchi, D. Torella, M. Baker, F. Limana, S. Chimenti, H. Kasahara, M. Rota, E. Musso, K. Urbanek, A. Leri, J. Kajstura, B. Nadal-Ginard, and P. Anversa. 2003. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 114:763-776. [DOI] [PubMed] [Google Scholar]
- 6.Brockes, J. P., and A. Kumar. 2002. Plasticity and reprogramming of differentiated cells in amphibian regeneration. Nat. Rev. Mol. Cell. Biol. 3:566-574. [DOI] [PubMed] [Google Scholar]
- 7.Chen, P. L., D. J. Riley, S. Chen-Kiang, and W. H. Lee. 1996. Retinoblastoma protein directly interacts with and activates the transcription factor NF-IL6. Proc. Natl. Acad. Sci. USA 93:465-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, P. L., D. J. Riley, Y. Chen, and W. H. Lee. 1996. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 10:2794-2804. [DOI] [PubMed] [Google Scholar]
- 9.Clarke, A. R., E. R. Maandag, M. van Roon, N. M. van der Lugt, M. van der Valk, M. L. Hooper, A. Berns, and H. te Riele. 1992. Requirement for a functional Rb-1 gene in murine development. Nature 359:328-330. [DOI] [PubMed] [Google Scholar]
- 10.Classon, M., S. Salama, C. Gorka, R. Mulloy, P. Braun, and E. Harlow. 2000. Combinatorial roles for pRB, p107, and p130 in E2F-mediated cell cycle control. Proc. Natl. Acad. Sci. USA 97:10820-10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cobrinik, D., M. H. Lee, G. Hannon, G. Mulligan, R. T. Bronson, N. Dyson, E. Harlow, D. Beach, R. A. Weinberg, and T. Jacks. 1996. Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev. 10:1633-1644. [DOI] [PubMed] [Google Scholar]
- 12.Field, L. J. 1988. Atrial natriuretic factor-SV40 T antigen transgenes produce tumors and cardiac arrhythmias in mice. Science 239:1029-1033. [DOI] [PubMed] [Google Scholar]
- 13.Flink, I. L., S. Oana, N. Maitra, J. J. Bahl, and E. Morkin. 1998. Changes in E2F complexes containing retinoblastoma protein family members and increased cyclin-dependent kinase inhibitor activities during terminal differentiation of cardiomyocytes. J. Mol. Cell. Cardiol. 30:563-578. [DOI] [PubMed] [Google Scholar]
- 14.Fung, Y. K., A. T'Ang, A. L. Murphree, F. H. Zhang, W. R. Qiu, S. W. Wang, X. H. Shi, L. Lee, B. Driscoll, and K. J. Wu. 1993. The Rb gene suppresses the growth of normal cells. Oncogene 8:2659-2672. [PubMed] [Google Scholar]
- 15.Furukawa, Y., S. Iwase, J. Kikuchi, M. Nakamura, H. Yamada, and M. Matsuda. 1999. Transcriptional repression of the E2F-1 gene by interferon-alpha is mediated through induction of E2F-4/pRB and E2F-4/p130 complexes. Oncogene 18:2003-2014. [DOI] [PubMed] [Google Scholar]
- 16.Gaussin, V., T. Van de Putte, Y. Mishina, M. C. Hanks, A. Zwijsen, D. Huylebroeck, R. R. Behringer, and M. D. Schneider. 2002. Endocardial cushion and myocardial defects after cardiac myocyte-specific conditional deletion of the bone morphogenetic protein receptor ALK3. Proc. Natl. Acad. Sci. USA 99:2878-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu, W., J. W. Schneider, G. Conderstil, S. Kaushai, V. Mahdavi, and B. Nadal-Ginard. 1993. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell 72:309-324. [DOI] [PubMed] [Google Scholar]
- 18.Jacks, T., A. Fazeli, E. M. Schmitt, R. T. Bronson, M. A. Goodell, and R. A. Weinberg. 1992. Effects of an Rb mutation in the mouse. Nature 359:295-300. [DOI] [PubMed] [Google Scholar]
- 19.Jahn, L., J. Sadoshima, A. Greene, C. Parker, K. G. Morgan, and S. Izumo. 1996. Conditional differentiation of heart- and smooth muscle-derived cells transformed by a temperature-sensitive mutant of SV40 T antigen. J. Cell Sci. 109:397-407. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, Z., E. Zacksenhaus, B. L. Gallie, and R. A. Phillips. 1997. The retinoblastoma gene family is differentially expressed during embryogenesis. Oncogene 14:1789-1797. [DOI] [PubMed] [Google Scholar]
- 21.Katz, E. B., M. E. Steinhelper, J. B. Delcarpio, A. I. Daud, W. C. Claycomb, and L. J. Field. 1992. Cardiomyocyte proliferation in mice expressing alpha-cardiac myosin heavy chain-SV40 T-antigen transgenes. Am. J. Physiol. 262:H1867-H1876. [DOI] [PubMed] [Google Scholar]
- 22.Kirshenbaum, L. A., and M. D. Schneider. 1995. Adenovirus E1A represses cardiac gene transcription and reactivates DNA synthesis in ventricular myocytes, via alternative pocket protein- and p300-binding domains. J. Biol. Chem. 270:7791-7794. [DOI] [PubMed] [Google Scholar]
- 23.Kusek, J. C., R. M. Greene, and M. M. Pisano. 2001. Expression of the E2F and retinoblastoma families of proteins during neural differentiation. Brain Res. Bull. 54:187-198. [DOI] [PubMed] [Google Scholar]
- 24.LeCouter, J. E., B. Kablar, P. F. Whyte, C. Ying, and M. A. Rudnicki. 1998. Strain-dependent embryonic lethality in mice lacking the retinoblastoma-related p130 gene. Development 125:4669-4679. [DOI] [PubMed] [Google Scholar]
- 25.Lee, E. Y. H. P., C.-Y. Chang, N. Hu, Y.-C. J. Wang, C.-C. Lai, K. Herrup, W.-H. Lee, and A. Bradley. 1992. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 359:288-294. [DOI] [PubMed] [Google Scholar]
- 26.Lee, M. H., B. O. Williams, G. Mulligan, S. Mukai, R. T. Bronson, N. Dyson, E. Harlow, and T. Jacks. 1996. Targeted disruption of p107: functional overlap between p107 and Rb. Genes Dev. 10:1621-1632. [DOI] [PubMed] [Google Scholar]
- 27.Leone, G., R. Sears, E. Huang, R. Rempel, F. Nuckolls, C. H. Park, P. Giangrande, L. Wu, H. I. Saavedra, S. J. Field, M. A. Thompson, H. Yang, Y. Fujiwara, M. E. Greenberg, S. Orkin, C. Smith, and J. R. Nevins. 2001. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol. Cell 8:105-113. [DOI] [PubMed] [Google Scholar]
- 28.Liao, H. S., P. M. Kang, H. Nagashima, N. Yamasaki, A. Usheva, B. Ding, B. H. Lorell, and S. Izumo. 2001. Cardiac-specific overexpression of cyclin-dependent kinase 2 increases smaller mononuclear cardiomyocytes. Circ. Res. 88:443-450. [DOI] [PubMed] [Google Scholar]
- 29.Lipinski, M. M., and T. Jacks. 1999. The retinoblastoma gene family in differentiation and development. Oncogene 18:7873-7882. [DOI] [PubMed] [Google Scholar]
- 30.MacLellan, W. R., and M. D. Schneider, unpublished results.
- 31.MacLellan, W. R., and M. D. Schneider. 2000. Genetic dissection of cardiac growth control pathways. Annu. Rev. Physiol. 62:289-319. [DOI] [PubMed] [Google Scholar]
- 32.MacLellan, W. R., G. Xiao, M. Abdellatif, and M. D. Schneider. 2000. A novel Rb- and p300-binding protein inhibits transactivation by MyoD. Mol. Cell. Biol. 20:8903-8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macleod, K. F., Y. Hu, and T. Jacks. 1996. Loss of Rb activates both p53-dependent and independent cell death pathways in the developing mouse nervous system. EMBO J. 15:6178-6188. [PMC free article] [PubMed] [Google Scholar]
- 34.MacPherson, D., J. Sage, D. Crowley, A. Trumpp, R. T. Bronson, and T. Jacks. 2003. Conditional mutation of Rb causes cell cycle defects without apoptosis in the central nervous system. Mol. Cell. Biol. 23:1044-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marino, S., M. Vooijs, G. H. van Der, J. Jonkers, and A. Berns. 2000. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 14:994-1004. [PMC free article] [PubMed] [Google Scholar]
- 36.Minamino, T., V. Gaussin, F. J. DeMayo, and M. D. Schneider. 2001. Inducible gene targeting in postnatal myocardium by cardiac-specific expression of a hormone-activated Cre fusion protein. Circ. Res. 88:587-592. [DOI] [PubMed] [Google Scholar]
- 37.Moberg, K. H., T. J. Logan, W. A. Tyndall, and D. J. Hall. 1992. Three distinct elements within the murine c-myc promoter are required for transcription. Oncogene 7:411-421. [PubMed] [Google Scholar]
- 38.Novitch, B. G., G. J. Mulligan, T. Jacks, and A. B. Lassar. 1996. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J. Cell Biol. 135:441-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novitch, B. G., D. B. Spicer, P. S. Kim, W. L. Cheung, and A. B. Lassar. 1999. pRb is required for MEF2-dependent gene expression as well as cell-cycle arrest during skeletal muscle differentiation. Curr. Biol. 9:449-459. [DOI] [PubMed] [Google Scholar]
- 40.Oh, H., S. B. Bradfute, T. D. Gallardo, T. Nakamura, V. Gaussin, Y. Mishina, J. Pocius, L. H. Michael, R. R. Behringer, D. J. Garry, M. L. Entman, and M. D. Schneider. 2003. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc. Natl. Acad. Sci. USA 100:12313-12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poss, K. D., L. G. Wilson, and M. T. Keating. 2002. Heart regeneration in zebrafish. Science 298:2188-2190. [DOI] [PubMed] [Google Scholar]
- 42.Robanus-Maandag, E., M. Dekker, M. Van de Valk, M. L. Carrozza, J. C. Jeanny, J. H. Dannenberg, A. Berns, and H. te Riele. 1998. p107 is a suppressor of retinoblastoma development in pRb-deficient mice. Genes Dev. 12:1599-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossant, J., and A. McMahon. 1999. “Cre”-ating mouse mutants—a meeting review on conditional mouse genetics. Genes Dev. 13:142-145. [DOI] [PubMed] [Google Scholar]
- 44.Russo, G. L., and M. Russo. 2003. Ins and outs of apoptosis in cardiovascular diseases. Nutr. Metab. Cardiovasc. Dis. 13:291-300. [DOI] [PubMed] [Google Scholar]
- 45.Sage, J., A. L. Miller, P. A. Perez-Mancera, J. M. Wysocki, and T. Jacks. 2003. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature 424:223-228. [DOI] [PubMed] [Google Scholar]
- 46.Santoni-Rugiu, E., J. Falck, N. Mailand, J. Bartek, and J. Lukas. 2000. Involvement of myc activity in a G1/S-promoting mechanism parallel to the pRb/E2F pathway. Mol. Cell. Biol. 20:3497-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider, J. W., W. Gu, L. Zhu, V. Mahdavi, and B. Nadal-Ginard. 1994. Reversal of terminal differentiation mediated by p107 in Rb−/− muscle cells. Science 264:1467-1471. [DOI] [PubMed] [Google Scholar]
- 48.Schneider, M. D., P. A. Payne, H. Ueno, M. B. Perryman, and R. Roberts. 1986. Dissociated expression of c-myc and a fos-related competence gene during cardiac myogenesis. Mol. Cell. Biol. 6:4140-4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soonpaa, M. H., and L. J. Field. 1994. Assessment of cardiomyocyte DNA synthesis during hypertrophy in adult mice. Am. J. Physiol. 266:H1439-H1445. [DOI] [PubMed] [Google Scholar]
- 50.Soonpaa, M. H., and L. J. Field. 1997. Assessment of cardiomyocyte DNA synthesis in normal and injured adult mouse hearts. Am. J. Physiol. 272:H220-H226. [DOI] [PubMed] [Google Scholar]
- 51.Soonpaa, M. H., K. K. Kim, L. Pajak, M. Franklin, and L. J. Field. 1996. Cardiomyocyte DNA synthesis and binucleation during murine development. Am. J. Physiol. 271:H2183-H2189. [DOI] [PubMed] [Google Scholar]
- 52.Soonpaa, M. H., G. Y. Koh, L. Pajak, S. Jing, H. Wang, M. T. Franklin, K. K. Kim, and L. J. Field. 1997. Cyclin D1 overexpression promotes cardiomyocyte DNA synthesis and multinucleation in transgenic mice. J. Clin. Investig. 99:2644-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tam, S. K. W. Gu, V. Mahdavi, and B. Nadal-Ginard. 1995. Cardiac myocyte terminal differentiation. Potential for cardiac regeneration. Ann. N. Y. Acad. Sci. 752:72-79. [DOI] [PubMed] [Google Scholar]
- 54.Thalmeier, K., H. Synovzik, R. Mertz, E. L. Winnacker, and M. Lipp. 1989. Nuclear factor E2F mediates basic transcription and trans-activation by E1a of the human MYC promoter. Genes Dev. 3:527-536. [DOI] [PubMed] [Google Scholar]
- 55.Ueno, H., M. B. Perryman, R. Roberts, and M. D. Schneider. 1988. Differentiation of cardiac myocytes after mitogen withdrawal exhibits three sequential states of the ventricular growth response. J. Cell Biol. 107:1911-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vooijs, M., M. Van de Valk, H. te Riele and A. Berns. 1998. Flp-mediated tissue-specific inactivation of the retinoblastoma tumor suppressor gene in the mouse. Oncogene 17:1-12. [DOI] [PubMed] [Google Scholar]
- 57.Weinmann, A. S., S. M. Bartley, T. Zhang, M. Q. Zhang, and P. J. Farnham. 2001. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 21:6820-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao, G., S. Mao, G. Baumgarten, J. Serrano, M. C. Jordan, K. P. Roos, M. C. Fishbein, and W. R. MacLellan. 2001. Inducible activation of c-Myc in adult myocardium in vivo provokes cardiac myocyte hypertrophy and reactivation of DNA synthesis. Circ. Res. 89:1122-1129. [DOI] [PubMed] [Google Scholar]
- 59.Young, A. P., and G. D. Longmore. 2004. Differences in stability of repressor complexes at promoters underlie distinct roles for Rb family members. Oncogene 23:814-823. [DOI] [PubMed] [Google Scholar]
- 60.Yu, B. D., M. Becker-Hapak, E. L. Snyder, M. Vooijs, C. Denicourt, and S. F. Dowdy. 2003. Distinct and nonoverlapping roles for pRB and cyclin D:cyclin-dependent kinases 4/6 activity in melanocyte survival. Proc. Natl. Acad. Sci. USA 100:14881-14886. [DOI] [PMC free article] [PubMed] [Google Scholar]