Abstract
In this work we introduce the confluent and various sizes image analysis method (COVASIAM), an automated colony count technique that uses digital imaging technology for detection and separation of confluent microbial colonies and colonies of various sizes growing on petri dishes. The proposed method takes advantage of the optical properties of the surfaces of most microbial colonies. Colonies in the petri dish are epi-illuminated in order to direct the reflection of concentrated light coming from a halogen lamp towards an image-sensing device. In conjunction, a multilevel threshold algorithm is proposed for colony separation and counting. These procedures improved the quantification of colonies showing confluence or differences in size. We tested COVASIAM with a sample set of microorganisms that form colonies with contrasting physical properties: Saccharomyces cerevisiae, Aspergillus nidulans, Escherichia coli, Azotobacter vinelandii, Pseudomonas aeruginosa, and Rhizobium etli. These physical properties range from smooth to hairy, from bright to opaque, and from high to low convexities. COVASIAM estimated an average of 95.47% (ς = 8.55%) of the manually counted colonies, while an automated method based on a single-threshold segmentation procedure estimated an average of 76% (ς = 16.27) of the manually counted colonies. This method can be easily transposed to almost every image-processing analyzer since the procedures to compile it are generically standard.
The growth of microbes in liquid cultures can be monitored by many different methods. To quantify the number of living cells in the culture, both direct or indirect counting methods can be used (5). The plate count or colony-counting method is one of the most popular indirect methods used to measure cell viability because it requires a very small investment in equipment. In the plate count method, a known volume of a liquid culture is spread over the surface of a petri dish, and the number of colonies appearing is counted and expressed as the number of CFU (1, 2). One of the disadvantages of the plate count method is that the growth and distribution of the colonies on the surface of the agar plate are not always homogeneous: colonies may have different diameters, densities, and shapes, and/or they may grow to confluence. These features strongly decrease the accuracy of those methods that sample only small portions of the dish. For a well-trained laboratory technician the above problems are easily solved by manual counting methods. However, in laboratories where the quantitative estimation of viable counts of large numbers of samples is a routine, the use of manual methods becomes tedious and labor-intensive.
Different algorithms and automated systems have been proposed for colony counting (5, 8, 9, 13, 16, 19, 20). However, these methods are not adequate to detect individual colonies growing to confluence, and most of them also assume that the colony size is constant. In this work we introduce the confluent and various sizes image analysis method (COVASIAM), a colony count method designed to detect both isolated and confluent colonies of uniform and nonuniform size. This method takes advantage of the light-reflecting properties of most microbial colonies. We tested COVASIAM with a sample set of microorganisms that form colonies with contrasting physical properties: Saccharomyces cerevisiae, Aspergillus nidulans, Escherichia coli, Azotobacter vinelandii, Pseudomonas aeruginosa, and Rhizobium etli. To evaluate COVASIAM, we compared its performance with manual and automated counting based on a single-threshold segmentation procedure. COVASIAM showed a good performance, and full automation was possible.
MATERIALS AND METHODS
System configuration.
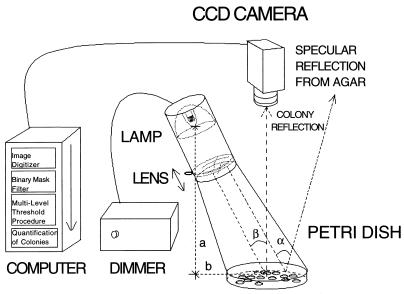
COVASIAM is composed of the elements shown in Fig. 1. The image-sensing device is a monochrome RS-170 charged coupled device TV camera. We used a COHU charged coupled device TV camera, model 6415-2100 with a TV lens (25 mm, 1:1.6, f 25.5) (TAMAON, Japan). Any other equivalent standard TV camera may be useful. Plates were illuminated with a halogen lamp (Decostar 50 W, 12 V, 38°; OSRAM) with an incorporated collecting mirror. The intensity of the light was controlled with a dimmer. To minimize light going to other directions that could produce spurious illumination, the lamp was placed inside a 60-mm-diameter tube. To increase the intensity level of the image and to adjust the size of the reflected spot on the sample, a 95-mm, 1:0.37 biconvex simple positive lens was used. A sliding implement for focusing the lens was used on the tube. For image processing software we used a commercial analyzer (IMAGENIA 2000; Biocom, Les Ulis, France). For object-counting purposes, IMAGENIA 2000 uses an ordinary single-threshold segmentation procedure (6). The macro command files and additional software developed for COVASIAM run straightforwardly in this commercial analyzer (macro files for users of IMAGENIA 2000 are available upon request at the corresponding author’s e-mail address).
FIG. 1.
Plate illumination and image acquisition setup.
Illumination of the plates.
For colony detection, this method takes advantage of two important properties. First, most colonies have smooth surfaces that reflect light almost like a mirror. Second, most colonies have convex shapes; according to geometrical optics, a convex spherical mirror produces an image of a far object behind the mirror surface at a distance approximately equal to one-half of the surface radius of curvature; the size of the image is very small (7). Therefore, if we place a light source with a small filament in front of the colonies (epi-illumination), a small bright image of the source will be produced for each colony. The produced image can be considered analogous to a topographic map if the gray levels are regarded as altitudinal levels (Fig. 2b). One advantage of such a method is that, due to the small size of the reflected spot, even colonies that overlap, that grow very close to each other, or that have various sizes appear separated and can be further processed and identified by using the counting algorithm proposed in this work.
FIG. 2.
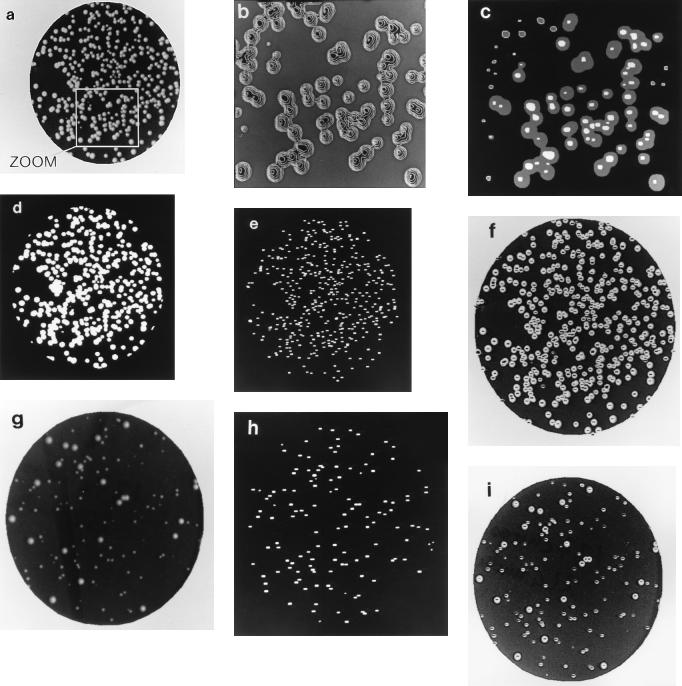
Detection of colony tops of S. cerevisiae by using COVASIAM and a single-threshold segmentation procedure. A plate with highly conglomerated and heterogeneous size colonies is shown. (a) An epi-illuminated plate was digitized and filtered through a binary mask with the proposed method; brilliant white dots are visible in the center of the colonies. Manual count gave 432 CFU. (b) Enhanced image (zoom) of a section of Fig. 2a; the topography of colonies is made clear with the epi-illumination approach. (c) Resulting image of data from Fig. 2b after adding images at T1 and T2 thresholds. (d) Colony segmentation at a single threshold level of data from Fig. 2a; colonies showing confluence are not separated, and small colonies are not detected; only 45% of the colonies in the whole plate were counted. (e) Colony segmentation of data from Fig. 2a by using COVASIAM; colonies showing confluence are well separated. (f) Overlap of Fig. 2a and e for visual inspection of results (detected colonies are seen as black dots). COVASIAM estimated 94.4% of the CFU for the whole plate. (g) Digitized image of S. cerevisiae colonies (strain IBT4) heterogeneous in size. Manual count for the image was 138 CFU. (h) Colony segmentation of data from Fig. 2g by using COVASIAM; small and large colonies are well detected. COVASIAM counted 97.8% of the manual count. (i) Overlap of Fig. 2g and h to aid in visual inspection of results.
To obtain this type of illumination a petri dish is positioned over a black mat surface in front of the TV camera at a distance where the whole dish is visualized in the image processing screen. This distance may vary according to which TV camera and lens are used. The halogen lamp is initially placed in vertical position at a horizontal distance (a) of 16 cm from the TV camera-petri dish axis and at a height (b) of 24 cm (the filament of the lamp and the center of the petri dish are taken as a reference) (Fig. 1). Then the lamp is carefully tilted to an angle (β) of 35° to illuminate the petri dish. The best illumination setup for convex microbial species (h/w > 0.15, where h = height of the colony, w = width or diameter of the colony) can be found empirically by changing the β angle to between 30° and 40° from the TV camera axis. In an optimal setup, the light reflected by the colonies is clearly detected without the specular reflection from the agar surface that shades the image of some colonies on the TV camera (α angle). The lamp should need no further reorientation if care is taken to pour the agar media during the preparation of the petri dishes on a horizontal plane-leveled surface.
Under the epi-illumination conditions described above, light reflected by microbial colonies towards the TV camera will be either concentrated or diffuse depending on the optical properties of the colony. Opaque or blurred colonies of S. cerevisiae, A. nidulans, or P. aeruginosa will reflect a larger and more diffuse spot, while species that form more brilliant and transparent colonies like R. etli will reflect a really small and focused spot. However, in other species which also have brilliant surfaces like A. vinelandii and E. coli, a large proportion of the colonies will not reflect the bright spot towards the TV camera at a 35° β angle. We observed that this effect is due to their low degree of convexity (h/w < 0.15). Under the conditions in which they were grown, A. vinelandii and E. coli form colonies whose main body behaves as a plane mirror but whose edges are convex in some parts and flat in other regions. These irregularities in their edges raise the possibility that at β angles near 35°, only a fraction of the colonies reflects the bright spot towards the TV camera (i.e., only when incident light hits the areas with convex edges). This effect can easily be observed if the petri dish is rotated. Upon rotation, the focused and intense spot reflected by some colonies will disappear while it will appear in others. This simple test confirms their morphological irregularities. This problem is easily solved by increasing the β angle up to 60° or more so that the light received by the TV camera is diffuse and not the highly intense light resulting from the reflection of the halogen lamp’s filament. With diffuse reflection, the contrast of the colonies tends to decrease, but the gain in homogeneity of the reflections will compensate for this low contrast effect. We also tested the effect of transillumination instead of epi-illumination. With transillumination, the contrast of A. vinelandii and E. coli colonies decreased even more than if they were epi-illuminated due to their transparency. Therefore, we recommend that for these species and others forming colonies with low convexity and very bright surfaces and with incomplete all-around convex edges, the β angle should be increased up to 60° or more (optimal angle of illumination is found when the reflected bright spot disappears from all colonies).
Image segmentation and counting of colony tops.
With epi-illumination, the semispherical surface of each colony produces a small image of any object placed in front of it. This image is composed of a very bright and small object (the lamp) reflected near the top of the colony and surrounded by less brilliant areas reflected by the adjacent surfaces of the colony. We define the top of the digitized image of a given colony as the most brilliant area of the colony which is aligned with the light source axis (β degrees from the vertical axis). By counting the total number of bright spots over the dark background, the number of colonies is known. An additional problem to solve, however, is that the tops of the colonies might have different gray-level values (or heights) due to a heterogeneous background, differences in illumination, colony size, and/or reflectance. Traditional counters based on image processing that use a single gray-level threshold segmentation procedure (6) detect only the tops with similar heights; thus, undesired manual intervention is needed to select the multiple optimal thresholds.
The multilevel-threshold algorithm that we propose creates first the isodensity contours of the illuminated image (Fig. 2b). The decreasing gray-level range to be scanned goes from the maximum (max_gray) of the image up to the between- class variance (BCV) level obtained with this method (11). Inside this range we can be sure that all the tops will be included in the topographic map eliminating most of the contours corresponding to background. The algorithm makes a polygonal segmentation of the contours in the decreasing range (max_gray, BCV) with a pitch (P) equal to (max_gray − BCV)/N, where N is a constant that defines the height resolution of the topographic map.
If we define the colony tops as the contours having an Euler’s number equal to 1 (one connected contour with no more holes inside), the detection of tops is achieved by eliminating in each polygonal segmentation those contours that are external and conserving the holes (the more inward ones). This is done as follows: take as an example those polygonal contours generated at the sequential thresholds T1 and T2, where T1 is max_gray − P and T2 is T1 − P. If we fill the polygonal contours generated at T1 with a constant gray-level value of G and the contours at T2 with the same level G but in a separate image, we can make the arithmetic addition of both obtained images I(T1) + I(T2), where the resulting image will contain the tops that will have a gray-level value of 2G (Fig. 2c). The tops will be defined as the contours with a gray-level standard deviation equal to zero. Those contours not having this null standard deviation will be eliminated (external contours having holes), while contours corresponding to single holes will be saved in a temporary image. In this way, the tops recursively detected in each segmentation procedure will be updated in the temporary image. The final binarized image will contain every separated colony, and counting will be possible.
In addition to this procedure, the digitized image of the petri dish should be filtered with a binary mask in order to eliminate its periphery since the plastic borders of the petri dish may reflect parasite illuminations. The mask is composed of a black image with a white circle encompassing 87% of the area of the petri dish (i.e., for a petri dish with an internal diameter of 8.8 cm, the digital mask should have a diameter of 8.2 cm). To process the whole image with the mask, the “and” operator should be used. When a digital mask is not used, a considerable amount of artifacts arise affecting the accuracy of the system. The introduced error may vary up to 100% for petri dishes with 30 colonies and 10% for dishes with 300 colonies. Since the digital mask encompasses 87% of the total agar surface, to obtain the total number of CFU in the plate COVASIAM’s count should be divided by a factor of 0.87.
Microorganisms and growth media.
S. cerevisiae strain W303a-LEU+ (15) and strain IBT4 (4) were grown at 25°C in YPD medium (1% Bacto yeast extract, 2% Bacto Peptone, 1% dextrose) supplemented with 40 mg of adenine sulfate per ml. A. nidulans RMS011 (17) was grown at 37°C in YPD medium. E. coli JM109 (21) was grown at 37°C in Luria-Bertani (LB) medium (1% Bacto tryptone, 0.5% Bacto yeast extract, 1% NaCl, adjusted to pH 7.0). A. vinelandii ATCC 9046 was grown at 30°C in modified BS media (3). R. etli CE3 (10) was grown at 30°C in PY media (2% Bacto Peptone, 1% Bacto yeast extract, 0.4 mg of CaCl2 per ml). P. aeruginosa IGB83 (12) was grown at 30°C in LB medium. In all cases, solid media were prepared in 2% Bacto agar.
RESULTS
We first implemented COVASIAM in the counting of colonies of the yeast S. cerevisiae. These colonies are highly convex with smooth and opaque surfaces. Figure 2a shows a digitized image of a plate containing colonies of S. cerevisiae (strain W303a) obtained after epi-illumination and binary-mask filtering. In this plate colonies have moderate differences in size and a high level of conglomeration. The manual count for this image was 432 CFU. Figure 2d shows the segmentation of Figure 2a using an automated single-threshold segmentation procedure (6), routinely used in most commercial image processing analyzers (see Materials and Methods). Colonies showing confluence were not separated, and this method estimated 195 CFU, representing only 45% of the CFU obtained by manual counting. Figure 2e shows the segmentation result of Fig. 2a by using COVASIAM. White dots correspond to the detected tops. COVASIAM gave an estimate of 408 CFU for this image, this number representing 94.4% of the manual count. Figure 2f displays the superimposed images of Fig. 2a and e to aid in the visual evaluation of the results.
To evaluate COVASIAM’s performance in populations containing large differences in colony size, we used cultures of S. cerevisiae (strain IBT4) with a small-colony phenotype. The small-colony phenotype of IBT4 is very unstable: at a very large frequency some cells give rise to colonies of a wild-type size (4). Figure 2g shows the digitized image of strain IBT4 colonies after epi-illumination and filtering. The manual count for this image was 138 CFU. Figure 2h shows the segmentation result of Fig. 2g, and Fig. 2i shows the superimposed images of Fig. 2g and h. COVASIAM gave 135 CFU, representing 97.8% of the manual counts. Clearly, COVASIAM detects each colony, isolated or confluent, large or small.
COVASIAM is largely based on the optical properties of convex colonies. In order to test if some other inherent physical features, such as degree of convexity, brightness, and texture, may influence colony detection, we selected other species in which these parameters contrast to those of S. cerevisiae: A. nidulans, E. coli, A. vinelandii, P. aeruginosa, and R. etli. Colonies of the filamentous fungi A. nidulans are of medium convexity with hairy (mycelial) and opaque surfaces. E. coli colonies have a low convexity with smooth and bright surfaces. A. vinelandii colonies have low to medium convexity with smooth and bright surfaces. R. etli colonies have high convexity with smooth and very bright surfaces. P. aeruginosa colonies have a very low convexity on their external part with a more convex central body, with smooth and a somewhat opaque surface. The counts obtained for all these species were as accurate as with S. cerevisiae (Fig. 3), providing that proper adjustments are made for the illumination as described in Materials and Methods.
FIG. 3.
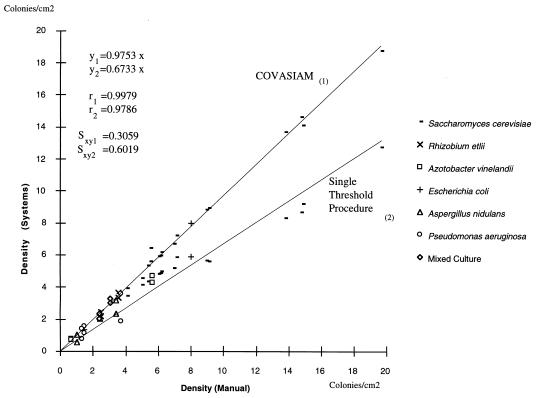
Correlation between the colony density estimates obtained by manual counting with the colony densities estimates obtained by using COVASIAM and a single-threshold segmentation procedure. A total of 31 plates of different species with different colony types, densities, sizes, and contrasted optical properties were used. Mixed culture is a combination of S. cerevisiae, A. nidulans, and E. coli colonies.
Figure 3 shows the relationship of the colony density estimates obtained with manual counting to the colony densities estimates obtained by using COVASIAM and an automated single-threshold segmentation procedure. Plated cultures of S. cerevisiae, A. nidulans, E. coli, A. vinelandii, R. etli, P. aeruginosa and a mixture of S. cerevisiae, A. nidulans, and E. coli were used. Regression analysis revealed a high correlation between COVASIAM and manual counting (regression equation 1 is y1 = 0.9753x, Pearson coefficient 1 [r1] is 0.9979, and standard error 1 [syx1] is 0.3059). Moreover, COVASIAM estimated an average of 95.47% (ς = 8.55%) of the manually counted colonies, showing that it provides a good estimate of the number of colonies. The automated commercial analyzer estimated an average of 76% (ς = 16.27) of the manually counted colonies (regression equation 2 is y2 = 0.6733x, Pearson coefficient 2 [r2] is 0.9786, standard error 2 [syx2] is 0.6019).
It should be emphasized that in this work we evaluated plates beyond the recommended maximum colony density of 300 CFU/plate, for plates 9 cm in external diameter (1, 2). This was done only to prove the method’s performance in the most extreme conditions (i.e., to obtain a higher proportion of confluent colonies and to decrease the colony size).
DISCUSSION
Figures 2f and i demonstrate that most of the colonies were separated and counted by COVASIAM. Only colonies reflecting the incident light towards the TV camera with gray levels within the counting range established are detected (BCV gray-level threshold; see Materials and Methods). Low contrasted colonies are easily misdetected since their gray-level value can be very close to the mean background’s gray-level value and therefore out of the counting range. Moreover, colonies that do not direct the bright incident light towards the TV camera are not detected. For a few confluent colonies this was the case because the particular region on their surfaces that could reflect the incident light towards the TV camera was shielded by a neighbor. In other cases, we observed that the epi-illuminated side of most undetected colonies was flatter than that of their detected neighbors. To prove this, the lamp can be rotated around the camera keeping a constant β angle, and most undetected colonies will be detectable to the detriment of some initially detected colonies.
In addition to the errors introduced by the misdetection of some colonies due to the illumination procedures and/or the performance of the proposed multilevel segmentation algorithm, another source of error comes from the fact that COVASIAM is a sampling method. The impending use of a digital mask avoids the analysis of 13% of the plate border area. As with all sampling methods, the uneven distribution of the colonies in the plate will influence the accuracy of the method. Since COVASIAM samples 87% of the whole plate, the impact of the sampling error introduced in the final count is highly diminished. Altogether, COVASIAM estimated an average of 95.47% (ς = 8.55%) of colonies scored by manual counting, proving to be very effective in detecting and counting different kinds of colonies regardless of their size and degree of confluence (Fig. 2).
The single-threshold segmentation procedure estimated only an average of 76% (ς = 16.27%) of colonies scored manually, showing this procedure to be less reliable than COVASIAM. The estimation error might be due to two different causes: the confluent colonies are considered as a single object or not counted if a size-form criterion is applied to eliminate artifacts (this error becomes larger as colony density increases) and/or small colonies are not counted since they are not included under the single threshold. COVASIAM avoids both kinds of errors.
Other methods for the separation of confluent colonies are based on the finding of the colony watersheds (14, 18). At low resolution, watershed methods do not resolve confluent colonies (i.e., 10 by 10 pixels or fewer per colony, with a 512 by 480 image resolution and petri dishes 9 cm in diameter). Low resolution of colonies is the consequence of counting the whole petri dish with standard video equipment. An alternative and simple method for colony counting is to measure the total area occupied by the colonies and divide it by the average colony area (8, 20). However, in those cases where colonies of different size are mixed in the plate, this approach cannot offer accurate results. Moreover, the alternative procedure of eroding the image repeatedly until reduction to one single point per colony faces another problem (18). In this case, the low resolution of confluent colonies provokes the conglomerate to be reduced to one single point. Therefore, the iterative erosion procedure does not separate confluent colonies under these particular conditions.
COVASIAM proved to be effective for the counting of colonies with different degrees of convexity and brilliance from species like S. cerevisiae, A. nidulans, E. coli, A. vinelandii, P. aeruginosa, and R. etli (Fig. 3). The proposed method could be effective in the counting of mixed colony types providing that each colony type can be counted effectively in pure cultures under the same illumination conditions (i.e., by using the same β angle). We evaluated COVASIAM in a mixture of A. nidulans, E. coli, and S. cerevisiae. The accuracy of the counting was comparable to the ones obtained with pure cultures of these organisms (Fig. 3).
As in most automated methods, spreaders are not treated differently than nonspreaders, and therefore each colony formed by a spreader has the potential to be counted as an individual. Only spreaders that arise on the periphery of the plate are avoided by COVASIAM, because they are not present in the 87% of the sampling area.
Epi-illumination as described in this work has proved to be very suitable for microbial colony counting, and more specifically for confluent colonies and colonies of various sizes. Transillumination can also be compatible with COVASIAM, providing that the input image gray-level scale is inverted (seen as the negative). In plates with a very small number of confluent colonies or none, transillumination works as well as epi-illumination with the exception of colonies that show a very poor contrast with the background (i.e., colonies with a high degree of transparency as A. vinelandii and E. coli growing on BS or LB media, respectively). In plates with a large number of confluent colonies having a very poor translucency (i.e., A. nidulans) the accuracy of COVASIAM using transillumination decreases.
Most of the image processing functions required for the implementation of the proposed algorithm in COVASIAM are generically standard and are available with their respective specific syntax in most commercial image analyzers. Therefore, COVASIAM can be readily transposed to practically every image processing analyzer (macro files for users of IMAGENIA 2000 are available upon request at corresponding author’s e-mail address).
ACKNOWLEDGMENTS
This work was supported partially by grants from Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica-UNAM (IN202795), and CONACYT-México (3461-N9310) to J.N.-S.
We thank J. Sergio Casas, Leticia Vega Alvarado, and Mario González Cardel for their technical assistance. We also thank Guadalupe Espín, Gloria Soberón, Olivia Sánchez, and Carmen Quinto for kindly providing cultures of A. vinelandii, P. aeruginosa, A. nidulans, and R. etli, respectively.
REFERENCES
- 1.Brock T D, Madigan M T. Biology of microorganisms. 5th ed. Englewood Cliffs, N.J: Prentice-Hall; 1988. Growth and its control; p. 318. [Google Scholar]
- 2.Busta F F, Peterson E H, Adams D M, Johnson M G. Colony count methods. In: Speck M L, editor. Compendium of methods for the microbiological examination of foods. Washington, D.C: American Public Health Association; 1984. pp. 62–83. [Google Scholar]
- 3.Campos M E, Martínez-Salazar J, Lloret L, Moreno S, Núñez C, Espín G, Soberón-Chavez G. Characterization of the gene coding for GDP-mannose dehydrogenase (algD) from Azotobacter vinelandii. J Bacteriol. 1996;178:1793–1799. doi: 10.1128/jb.178.7.1793-1799.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Folch-Mallol, J. L., L. M. Martínez-Mejía, and J. Nieto-Sotelo. Unpublished data.
- 5.García-Armesto M R, Prieto M, García-Lopez M L, Otero A, Moreno B. Modern microbiological methods for foods: colony count and direct count methods. A review. Microbiología. 1993;9(1):1–13. [PubMed] [Google Scholar]
- 6.Haralick R M, Shapiro L G. Computer and robot vision. I. New York, N.Y: Addison-Wesley Publishing Company, Inc.; 1992. Binary machine vision (thresholding and segmentation) pp. 13–23. [Google Scholar]
- 7.Hecht E. Optics. 2nd ed. New York, N.Y: Addison-Wesley; 1989. Spherical mirrors; pp. 158–163. [Google Scholar]
- 8.Mukherjee D P, Pal A, Sarma S E, Majumder D D. Bacterial colony counting using distance transform. Int J Biomed Comput. 1995;38(2):131–140. doi: 10.1016/0020-7101(94)01043-z. [DOI] [PubMed] [Google Scholar]
- 9.Nishijima S, Namura S, Mitsuya K, Asada Y. An evaluation, using computerized image analysis, of antimicrobial efficacy of an automatic hand washing machine with ultrasonic wave spraying. J Dermatol. 1983;20(10):654–656. doi: 10.1111/j.1346-8138.1993.tb01357.x. [DOI] [PubMed] [Google Scholar]
- 10.Noel K D, Sánchez A, Fernández L, Leemans S, Ceballos M A. Rhizobium phaseoli symbiotic mutants with transposon Tn5. J Bacteriol. 1984;158:148–155. doi: 10.1128/jb.158.1.148-155.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern SMC- 1979;9:62–66. [Google Scholar]
- 12.Palmeros B, Güereca L, Alagón A, Soberón-Chávez G. Biochemical characterization of the lipolytic activity of Pseudomonas aeruginosa IGB 83. Process Biochem. 1994;29:207–212. [Google Scholar]
- 13.Pickett D A, Welch D F. Evaluation of the automated Bact-Alert system for pediatric blood culturing. Am J Clin Pathol. 1995;103(3):320–323. doi: 10.1093/ajcp/103.3.320. [DOI] [PubMed] [Google Scholar]
- 14.Russ J C. Computer assisted microscopy: the measurement and analysis of images. New York, N.Y: Plenum Press; 1990. Binary image editing; pp. 153–160. [Google Scholar]
- 15.Sánchez Y, Lindquist S L. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 16.Spadinger I, Palcic B. Cell survival measurements at low doses using an automated image cytometry device. Int J Radiat Biol. 1993;63(2):183–189. doi: 10.1080/09553009314550241. [DOI] [PubMed] [Google Scholar]
- 17.Stringer M A, Dean R A, Sewall T C, Timberlake W E. Rodletless, a new Aspergillus developmental mutant induced by directed gene inactivation. Genes Dev. 1991;5:1161–1171. doi: 10.1101/gad.5.7.1161. [DOI] [PubMed] [Google Scholar]
- 18.Vincent L, Soille P. Watersheds in digital spaces: an efficient algorithm based on immersion simulations. IEEE Trans Pattern Machine Intelligence. 1991;13(6):583–598. [Google Scholar]
- 19.Walsh L R. Established antimicrobial susceptibility testing methods with a new twist—points to consider and a glimpse of the future. Adv Exp Med Biol. 1994;349:97–105. doi: 10.1007/978-1-4757-9206-5_9. [DOI] [PubMed] [Google Scholar]
- 20.Wilson I G. Use of the IUL Countermat automatic colony counter for spiral plated total viable counts. Appl Environ Microbiol. 1995;61:3158–3160. doi: 10.1128/aem.61.8.3158-3160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]