Abstract
The myocyte enhancer factor-2 (MEF2) family of transcription factors plays an important role in regulating cellular programs like muscle differentiation, neuronal survival, and T-cell apoptosis. Multisite phosphorylation is known to control the transcriptional activity of MEF2 proteins, but it is unclear whether other modifications are involved. Here, we report that human MEF2D, as well as MEF2C, is modified by SUMO2 and SUMO3 at a motif highly conserved among MEF2 proteins from diverse organisms. This motif is located within the C-terminal transcriptional activation domain, and its sumoylation inhibits transcription. As a transcriptional corepressor of MEF2, histone deacetylase 4 (HDAC4) potentiates sumoylation. This potentiation is dependent on the N-terminal region but not the C-terminal deacetylase domain of HDAC4 and is inhibited by the sumoylation of HDAC4 itself. Moreover, HDAC5, HDAC7, and an HDAC9 isoform also stimulate sumoylation of MEF2. Opposing the action of class IIa deacetylases, the SUMO protease SENP3 reverses the sumoylation to augment the transcriptional and myogenic activities of MEF2. Similarly, the calcium M kinase and extracellular signal-regulated kinase 5 signaling pathways negatively regulate the sumoylation. These results thus identify sumoylation as a novel regulatory mechanism for MEF2 and suggest that this modification interplays with phosphorylation to promote intramolecular signaling for coordinated regulation in vivo.
How protein function is regulated is a fundamental question relevant to many biological processes. Different mechanisms are involved, and one such mechanism operates through modification at the posttranslational level. Lysine acetylation has recently emerged as an important posttranslational modification and has been shown to regulate functions of histones, about 40 transcription factors, and over 30 other proteins (36, 62, 74). Acetylation of specific lysine residues located at the N-terminal tails of core histones is necessary for controlling chromatin activities in various nuclear processes. Histone deacetylases (HDACs) are the enzymes responsible for reversing the acetylation of histones. Some HDACs also display deacetylase activities towards other proteins. According to sequence similarity and phylogenetic analysis, known HDACs have been grouped into distinct classes (10, 18, 19, 33). Mammalian class II members have been further divided into two subclasses: IIa (HDAC4, -5, -7, and -9) and IIb (HDAC6 and -10) (66, 76).
Class IIa members share a bipartite domain organization and display significant sequence homology in their long N-terminal extensions and C-terminal catalytic domains. A characteristic feature of these deacetylases is dynamic regulation by signal-dependent nucleocytoplasmic trafficking. Ca2+/calmodulin-dependent kinase (CaMK) and protein kinase D phosphorylate-specific serine residues within the N-terminal extensions of class IIa HDACs to promote 14-3-3 association and CRM1-dependent nuclear export (43, 65, 66). Other regulatory mechanisms, such as sumoylation (34, 51, 64), caspase cleavage (40, 50), ubiquitin-dependent proteosomal degradation (26, 38), and mitochondrial targeting (3), have also been reported for some class IIa members. While human HDAC4 is highly sumoylated at Lys559 (34, 64; unpublished observations), substitution of this residue with arginine modestly affects the deacetylase and transcriptional activities of HDAC4, raising the question whether this modification regulates other functions. Related to this, little is known about potential roles of the regions adjacent to the sumoylation site.
HDAC4 and other class IIa members function as signal-responsive transcriptional corepressors for the myocyte enhancer factor-2 (MEF2) family of transcription factors (43). In mammals, there are four MEF2 isoforms: MEF2A, -B, -C, and -D. Originally identified as myocyte enhancer factors, MEF2s have been extensively studied as major transcriptional activators for muscle differentiation (43). Consistent with this, a mutation on the human MEF2A gene plays a potentially causal role in a familial coronary artery disease (71). Recent studies indicate that MEF2s also play important roles in regulating other cellular programs like growth factor responses, neuronal survival, and T-cell apoptosis (8, 23, 42, 77). In addition, human MEF2D is expressed in different tissues (43), and its gene is rearranged in pre-B acute lymphoblastic leukemia patients (78). Class IIa HDACs interact with the DNA-binding domains of MEF2 proteins and convert them from activators to repressors. Upon activation by Ca2+/calmodulin, CaMKs phosphorylate class IIa HDACs and promote nuclear export to relieve transcriptional repression, so CaMKs modify these HDACs to stimulate MEF2-dependent transcription. By contrast, MAP kinases directly modify MEF2s. While p38 phosphorylates MEF2A and MEF2C (9, 21, 22, 80), extracellular signal-regulated kinase 5 (ERK5) phosphorylates MEF2A, -C, and -D (30-32, 73). These phosphorylation events activate transcription, whereas cyclin-dependent kinase 5 (Cdk5)-mediated phosphorylation of MEF2A and -2D inhibits transcription (17). Therefore, MEF2 transcription factors are subject to multisite phosphorylation for the integration of diverse signals in the nucleus.
Different covalent modifications are well known to interplay and regulate functions of histones and transcription factors like the p53 tumor suppressor (1, 13, 48, 79), so we explored whether covalent modifications other than phosphorylation regulate the MEF2 transcriptional activity and, if so, how different modifications may interplay and how class IIa HDACs may be involved. Here, we show that MEF2D, as well as MEF2C, is sumoylated on a single lysine residue located at a consensus sumoylation motif conserved among MEF2 proteins. This modification inhibits transcriptional and myogenic activities. Independent of its deacetylase domain, HDAC4 and other class IIa members potentiate sumoylation, whereas the SUMO protease SENP3 and the ERK5 signal pathway reverse the modification. These results identify sumoylation as a novel regulatory mechanism for MEF2 and suggest the potential interplay of sumoylation with phosphorylation in the control of MEF2-dependent transcription.
MATERIALS AND METHODS
Plasmid constructs.
Expression plasmids for Flag- or hemagglutinin (HA)-tagged human MEF2C, MEF2D, and mutants were generated by PCR with Expand thermostable DNA polymerase (Roche) and subcloned into derivatives of pcDNA3.1 (Invitrogen). DNA sequencing was performed with T7 sequenase 2.0 (Amersham Pharmacia Biotech.). Expression plasmids for HA-tagged SUMO1, SUMO2, and SUMO3 were kindly provided by R. T. Hay (64) and J. Long (41). The Ubc9 expression plasmid was obtained from C. D. Lima (4), and SENP1/-3 expression plasmids were obtained from Leonard I. Zon (5). Green fluorescent protein (GFP) constructs were derived from pEGFP-C2 (BD Biosciences). The luciferase reporter Gal4-tk-luc and expression plasmids for HDAC4 and deletion mutants have been described previously (68-70). Expression plasmids for Myc-tagged HDAC4 and mutant K559R were from R. T. Hay (64), and constructs for MEK5(D) and ERK5 were from J. D. Lee (31).
Cell culture.
HEK293, HeLa, and C3H10T1/2 cells were maintained in Dulbecco's modified eagle medium supplemented with 10% fetal bovine serum (FBS; Sigma), penicillin, and streptomycin. C2C12 cells were cultured in the same medium containing 20% FBS.
In vivo sumoylation assays.
For analyzing the sumoylation of endogenous MEF2D, HEK293 and HeLa cells were washed with phosphate-buffered saline (PBS) and lysed directly in buffer S (15 mM Tris-HCl [pH 6.7], 0.5% sodium dodecyl sulfate, 3% glycerol, 0.8× PBS, 4% NP-40, 0.1% mercaptoethanol, 25 mM N-ethylmaleimide, and protease inhibitors) (12). After a brief sonication (10 s), soluble extracts were used for immunoblotting with anti-MEF2D polyclonal antibody, kindly provided by R. Prywes (23). The extracts were also subject to immunoprecipitation with the anti-MEF2D antibody and immunoblotting with anti-MEF2D, anti-SUMO1, and anti-SUMO2 antibodies, the last two of which were purchased from Zymed Laboratories, Inc., and Chemicon International, respectively. Blots were developed with Supersignal chemiluminescent substrates (Pierce).
For determination of the sumoylation of exogenous proteins in vivo, HEK293 cells were cotransfected with Superfect transfection reagent (QIAGEN). Briefly, expression constructs were transfected into 4 × 105 cells per 10-cm dish at the indicated amounts. For each transfection, the total amount of plasmids was kept constant at 10 μg by supplementation with pKSII(+) (Stratagene), and 20 μl of Superfect was used. At 48 h posttransfection, the cells were washed twice with cold PBS and scrape harvested in 0.5 ml of buffer S. Each cell suspension was sonicated for 10 s, and the soluble extract was subject to affinity purification on M2 agarose beads (Sigma). Beads with bound proteins were washed four times with buffer R (PBS, 5% NP-40, 1% mercaptoethanol, and protease inhibitors), and bound proteins were eluted with Flag peptide (Sigma). Eluted proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane for Western blotting with anti-Flag, anti-HA, or anti-Myc antibody as specified. PBS containing 20% horse serum (Invitrogen) and 0.15% Tween 20 (Sigma) was used for membrane blocking and antibody incubation, and PBS with 0.15% Tween 20 was used for membrane washing. Blots were developed with Supersignal chemiluminescent substrates (Pierce).
Nuclear extract preparation.
Nuclei were isolated from HEK293 cells according to a previously described procedure (70) and suspended in the hypotonic lysis buffer containing 0.5 M NaCl. After rotation at 4°C for 10 min and high-speed centrifugation, the supernatant was collected as the nuclear extracts, to which 25 mM N-ethylmaleimide was added to minimize desumoylation.
Fluorescence microscopy.
HeLa cells were seeded at 2 × 104 cells per well on glass coverslips in 12-well tissue culture plates. After 16 to 20 h, the cells were transfected with indicated expression constructs with Superfect transfection reagent. About 24 h posttransfection, the cells were rinsed three times with PBS-0.1 mM CaCl2-1 mM MgCl2 and subsequently fixed with PBS-2% paraformaldehyde at room temperature for 15 min. After two rinses with PBS, the coverslips were quenched with PBS-50 mM NH4Cl for 10 min and rinsed twice with PBS. Then, the cells were permeabilized with PBS-0.2% Triton X-100 for 5 min and blocked with PBS-1% bovine serum albumin for 10 min. The coverslips were then incubated with anti-HA antibody for 15 min, rinsed six times with PBS, and incubated with 0.5 μg of Hoechst 53258 (Sigma)/ml and goat anti-mouse immunoglobulin G conjugated with Alexa 488 (Molecular Probes) for 20 min. After six rinses with PBS, the coverslips were then mounted on glass slides with Immu-Mount (Thermo Shandon) and sealed with nail oil. Slides were examined under a Nikon Eclipse TE300 microscope equipped with an epifluorescence unit, a temperature-adjustable platform, and a charge-coupled device camera (Hamamatsu) controlled by a Dell computer running ISee imaging software (Inovision Corp.). Images were recorded and exported for further processing with Adobe Photoshop.
Myogenesis assays.
C3H10T1/2 cells were seeded at 5 × 104 cells per well on polylysine-coated coverslips in 12-well plates and transfected with the indicated plasmids. After 48 h, the cells were washed with PBS and fed with the differentiation medium (Dulbecco's modified eagle medium, 2% horse serum, penicillin, and streptomycin). On day 7, the cells were processed for immunofluorescence microscopy with anti-myosin heavy-chain (anti-MHC) MF-20 antibody (Developmental Studies Hybridoma Bank, Iowa City, Iowa) to detect myotubes. A GFP expression plasmid was cotransfected to normalize the transfection efficiency as previously described (53, 72).
Reporter gene assays.
The assays were performed as previously described (68).
RESULTS
Sumoylation of MEF2C and MEF2D in vivo.
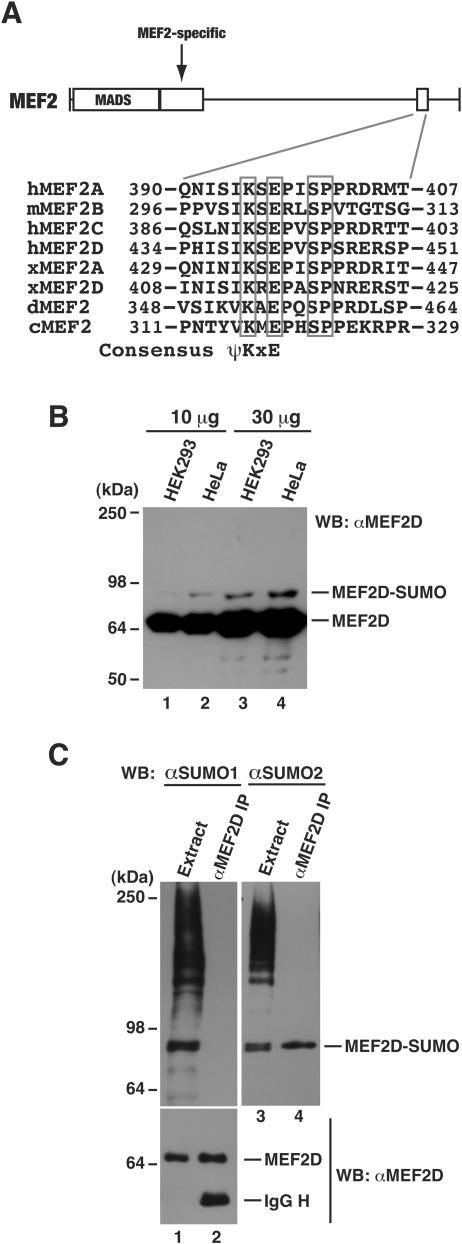
To gain further insight into the function and regulation of MEF2 transcription factors, we inspected their amino acid sequences to identify motifs that may suggest potential covalent modification sites. This inspection revealed a putative sumoylation motif that is conserved among MEF2 proteins from Caenorhabditis elegans, Drosophila melanogaster, Xenopus, and mammals (Fig. 1A). This motif is located at the divergent C-terminal region, away from the highly conserved N-terminal DNA-binding domain. Like C. elegans and Drosophila MEF2 proteins, MEF2B only displays limited sequence similarity to MEF2A, -C, and -D in the C-terminal region, so the conservation of the putative sumoylation motif among MEF2 proteins suggests that it plays an important role. In light of these observations, we investigated whether MEF2 proteins are subject to regulation by this modification. Among mammalian MEF2 isoforms, MEF2D is widely expressed and was thus chosen for most experiments described below. To determine the sumoylation of endogenous proteins, HEK293 and HeLa cell extracts were prepared in buffer S and analyzed by immunoblotting with anti-MEF2D antibody. In addition to regular MEF2 proteins (∼67 kDa), a protein of ∼80 kDa was detected (Fig. 1B). Monosumoylation increases the molecular mass of a protein target by ∼15 kDa, so this 80-kDa species might correspond to sumoylated MEF2D. To confirm this, we performed immunoprecipitation with the anti-MEF2D antibody. The precipitated proteins were analyzed by immunoblotting with anti-SUMO antibodies. Among four mammalian SUMO proteins, SUMO1 is the most divergent and SUMO2 is almost identical to SUMO3 and SUMO4 (7, 20, 46), so we tested anti-SUMO1 and anti-SUMO2 antibodies. As shown in Fig. 1C, unlike anti-SUMO1 antibody, anti-SUMO2 antibody detected an 80-kDa band. Both antibodies could recognize sumoylated HDAC4 (data not shown), which is consistent with previous reports showing that it is modified by both SUMO1 and SUMO2 (34, 64). These results indicate that endogenous MEF2D is modified by SUMO2 addition.
FIG.1.
Sumoylation of endogenous MEF2D. (A) Domain organization of MEF2. The MADS box and MEF2-specific domain (rectangles) are highly conserved, whereas the C-terminal region (solid line) is divergent. The small box in the C-terminal region represents a conserved sequence motif. Shown in detail is the sequence alignment of this motif found on MEF2 proteins from human (h), mouse (m), Xenopus (x), Drosophila (d), and C. elegans (c), with invariant residues boxed. At the bottom of the alignment is the consensus sumoylation motif ψKxE, where ψ is any aliphatic residue and x is any amino acid (55). (B) HEK293 and HeLa extracts were prepared in buffer S and analyzed by Western blotting with anti-MEF2D antibody. (C) HEK293 cells were washed and lysed in buffer S for extract preparation. Extracts were subject to immunoprecipitation with anti-MEF2D antibody and immunoblotting with anti-SUMO and anti-MEF2D antibodies. The band detected by the anti-SUMO2 antibody (lane 4) is specific (see also Fig. 9E).
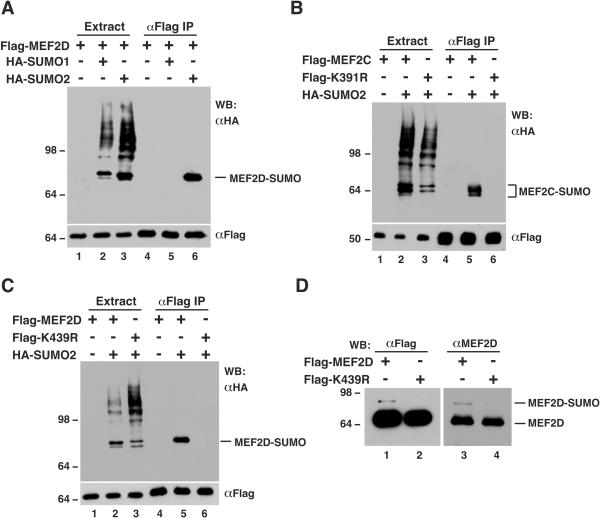
To substantiate this conclusion, in vivo sumoylation assays with HA-tagged SUMO proteins were performed. An expression plasmid for Flag-tagged MEF2D was cotransfected into HEK293 cells along with expression plasmids for HA-tagged SUMO1 and SUMO2. After transfection, cell extracts were subject to affinity purification on anti-Flag M2 agarose, and purified proteins were analyzed by immunoblotting with anti-Flag and anti-HA antibodies. As shown in Fig. 2A, Flag-MEF2D was modified by SUMO2 but not SUMO1. Similar results were obtained with a different cell line, HeLa cells (data not shown). By contrast, HDAC4 was sumoylated by both SUMO isoforms under similar conditions (data not shown). Like MEF2D, MEF2C was sumoylated by SUMO2 (Fig. 2B, lanes 2 and 5). Therefore, both MEF2C and MEF2D are subject to sumoylation in vivo.
FIG. 2.
Specific sumoylation of MEF2C and MEF2D. (A to C) In vivo sumoylation assays. Expression plasmids for the indicated proteins were transfected into HEK293 cells. Extracts were prepared in buffer S for immunoprecipitation on M2 agarose, and bound proteins were eluted with Flag peptide and analyzed by Western blotting (WB) with anti-HA (top) or anti-Flag (bottom) antibody. Like HA-SUMO2, HA-SUMO3 was also conjugated to MEF2D (data not shown). (D) Extracts from HEK293 cells expressing Flag-MEF2D or Flag-K439R were used for immunoprecipitation on M2 agarose. Bound proteins were eluted with Flag peptide and subjected to Western blotting analysis with anti-Flag (left) or anti-MEF2D (right) antibody.
Mapping sumoylation sites of MEF2C and MEF2D.
Lys391 of MEF2C is the putative sumoylation site (Fig. 1A), so this residue was replaced by arginine to create the point mutant K391R. This mutant was expressed in HEK293 cells and analyzed for conjugation with HA-SUMO2. As shown in Fig. 2B (lanes 3 and 6), unlike wild-type MEF2C, mutant K391R was not modified by HA-SUMO2, indicating that Lys391 is indeed the sumoylation site of MEF2C. This residue is equivalent to Lys439 of MEF2D. To identify the sumoylation site on MEF2D, Lys439 was changed to arginine to create the point mutant K439R. As shown in Fig. 2C, this substitution abolished the sumoylation, indicating that Lys439 is responsible for the sumoylation of MEF2D.
Mutant K439R was further analyzed to ascertain that the aforementioned 80-kDa species (Fig. 1B) is the sumoylated form of MEF2D. HEK293 cells were transfected with expression plasmids for Flag-tagged MEF2D and K439R. These fusion proteins were affinity purified for subsequent immunoblotting. As shown in Fig. 2D, like the anti-MEF2D antibody, the anti-Flag antibody detected a 80-kDa band in the protein sample affinity purified from cell extracts expressing wild-type MEF2D but not from extracts expressing mutant K439R, further supporting that the endogenous 80-kDa species detected by the anti-MEF2D antibody (Fig. 1B) is indeed the sumoylated form of MEF2D. Therefore, the above results indicate that MEF2D, as well as MEF2C, is sumoylated at a single lysine residue in vivo.
Regulation of the transcriptional and myogenic activities of MEF2D by sumoylation.
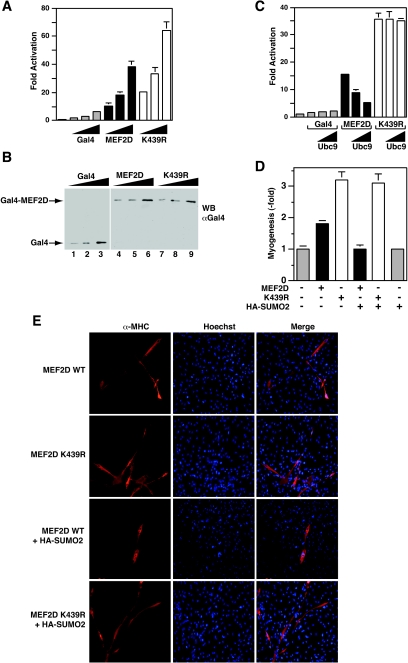
To determine functional consequences of the sumoylation, we compared the subcellular localization, protein stability, and HDAC4-binding ability of wild-type and mutant MEF2D proteins. No significant differences were observed (data not shown). Since sumoylation has been shown to inhibit transcriptional activities of other transcription factors (14, 16, 58, 67) and Lys439 is located within the transcriptional activation domain of MEF2D (Fig. 1A), we tested whether sumoylation affects its transcriptional activity. To avoid potential interference and complication of endogenous MEF2 proteins, MEF2D and mutant K439R were expressed as fusion proteins with the DNA-binding domain of Gal4 for reporter gene assays. As shown in Fig. 3A, the mutant was more active than the wild-type protein. Their expression levels were similar (Fig. 3B), suggesting that sumoylation inhibits the transcriptional activity of MEF2D. To substantiate this, a plasmid expressing the E2 conjugating enzyme Ubc9 was included in the reporter gene assays. In a dose-dependent manner, Ubc9 reduced the transcriptional activity of the wild type but not the mutant MEF2D (Fig. 3C).
FIG. 3.
Effect of sumoylation on MEF2D activities. (A) The luciferase reporter Gal4-tk-Luc (200 ng) was transfected into HEK293 cells along with a β-galactosidase expression plasmid (50 ng) and increasing amounts (25, 50, and 100 ng) of expression plasmids for the Gal4 DNA-binding domain (residues 1 to 147), Gal4-MEF2D, and Gal4-K439R, as indicated. The normalized luciferase activity from the transfection without any effector plasmids was arbitrarily set to 1.0. Average values of at least three independent experiments are shown with standard deviation. (B) Extracts from HEK293 cells transfected as in panel A were prepared for Western blotting with anti-Gal4 antibody (RK5C1; Santa Cruz Biotech). (C) Reporter gene assays were performed as in panel A except that the amounts of expression plasmids for Gal4, Gal4-MEF2D, and Gal4-K439R were kept constant (50 ng) and the amount of the expression plasmid for Ubc9 varied (50 and 200 ng). (D and E) A MyoD expression plasmid (400 ng) was transfected into C3H10T1/2 cells along with the expression plasmid for Flag-MEF2D, Flag-K439R, or HA-SUMO2 (each, 500 ng). Myotubes were detected by indirect immunofluorescence with anti-MHC antibody. Average values of at least three independent experiments are illustrated with standard deviation in panel D, and representative images are shown in panel E.
We then analyzed how sumoylation regulates the myogenic activity of MEF2D. For this, MyoD-dependent myogenic conversion assays were utilized (53). Pluripotent C3H10T1/2 cells were cotransfected with the MyoD expression plasmid, along with expression plasmids for MEF2D, mutant K439R, and SUMO2. As reported, MEF2D increased the myogenic potential of MyoD (Fig. 3D). Compared to wild-type MEF2D, the mutant was more potent in stimulating the myogenic conversion of C3H10T1/2 (Fig. 3D and E). Expression of SUMO2 reduced the ability of wild-type but not mutant MEF2D to potentiate the myogenic conversion (Fig. 3D and E). Without MEF2D, SUMO2 itself had minimal effects (Fig. 3D), suggesting that the effect of SUMO2 is dependent on MEF2D. Collectively, these data indicate that sumoylation negatively regulates the myogenic activity of MEF2D.
Regulation the MEF2 sumoylation by HDAC4.
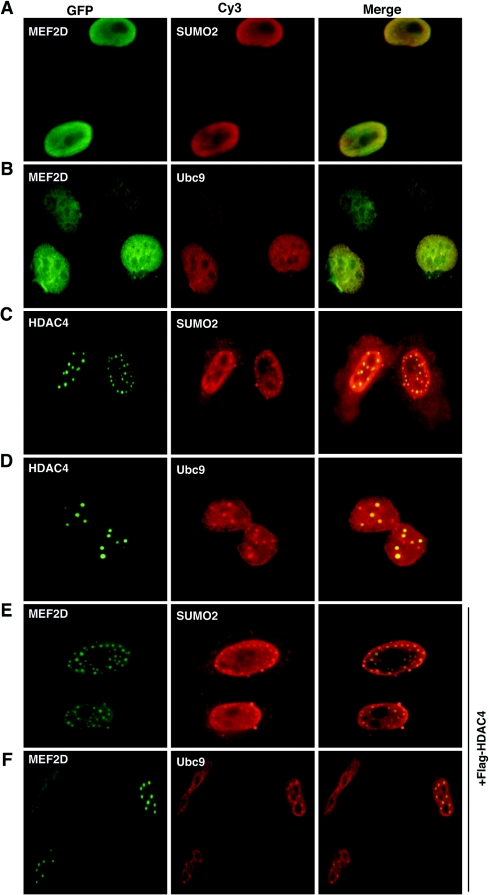
We next examined the subcellular distribution of MEF2D and two components of the sumoylation machinery, SUMO2 and Ubc9. MEF2D colocalized with SUMO2 in nuclear periphery regions (Fig. 4A) and displayed some colocalization with Ubc9 in the nucleus (Fig. 4B). These results are consistent with the conclusion that MEF2D is sumoylated in vivo. Different from MEF2D, HDAC4 colocalized with SUMO2 and Ubc9 in discrete nuclear dots (Fig. 4C and D). MEF2s were reported to associate with HDAC4 in similar nuclear dots (45), so an interesting possibility is that HDAC4 relocates MEF2D, SUMO2 and Ubc9 to nuclear dots. Consistent with this, SUMO1 was reported to be enriched in nuclear dots of HDAC4 (34). To further address this, we analyzed the subcellular distribution of these proteins in the presence of HDAC4 expression. Under this condition, MEF2D colocalized with both SUMO2 and Ubc9 in discrete nuclear dots (Fig. 4E and F), confirming that HDAC4 recruits MEF2D, as well as SUMO2 and Ubc9, to nuclear dots. Nuclear foci of other proteins such as the polycomb protein Pc2 have recently been shown to be sumoylation centers (29, 47, 57). Pc2 and several known SUMO ligases are themselves targets of sumoylation (29, 35, 52, 57). Related to this, HDAC4 is sumoylated by SUMO1 and SUMO2 (34, 64). In addition, this deacetylase interacts with MEF2s and inhibits their transcriptional activity through multiple repression domains (45, 68). These observations led us to ask whether HDAC4 regulates MEF2 sumoylation.
FIG. 4.
Colocalization of MEF2D and HDAC4 with SUMO2 and Ubc9. HeLa cells were transfected with expression plasmids for GFP-MEF2D, GFP-HDAC4, HA-SUMO2, and HA-Ubc9 as indicated. Transfected cells were subjected to green fluorescence microscopy to detect GFP. HA-tagged proteins were visualized by immunostaining with anti-HA antibody and Cy3-labeled secondary antibody. In the experiments shown in panels E and F, an expression plasmid for Flag-HDAC4 was cotransfected. Note that expression of SUMO2, Ubc9, or HDAC4 led to uneven distribution of MEF2D, which itself is known to be uniform in the nucleoplasm.
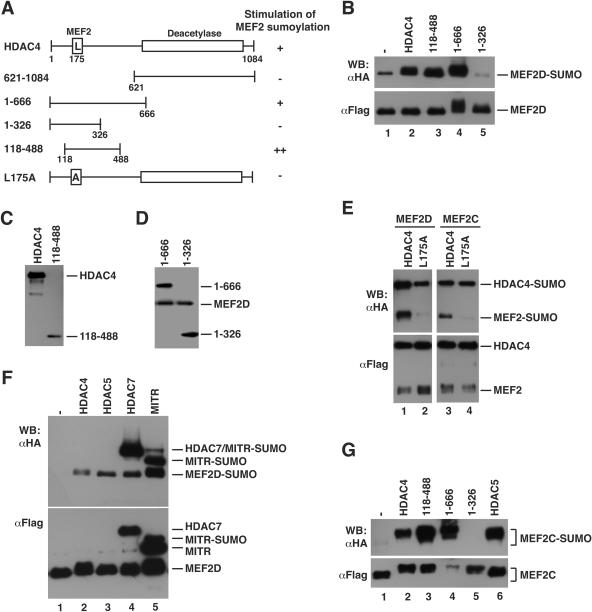
To address this question, sumoylation assays were performed in the presence of exogenous HDAC4. As shown in Fig. 5B (lanes 1 and 2), HDAC4 stimulated the sumoylation of MEF2D. To map which region is responsible, we first tested two deletion mutants, 1-666 and 621-1084 (Fig. 5A). The former, but not the latter, mimicked the effect of full-length HDAC4 (Fig. 5B and data not shown). Consistent with this, trichostatin A treatment did not have any significant effects (data not shown). To further map the region responsible for the stimulation, we tested two smaller deletion mutants, 1-326 and 118-488 (Fig. 5A). Unlike 1-326, mutant 118-488 stimulated the sumoylation of MEF2D (Fig. 5B to D). Compared to full-length HDAC4, this mutant was expressed at a lower level (Fig. 5C), so it may be more potent than wild-type HDAC4. To test whether direct MEF2 binding was required, we tested L175A, an HDAC4 point mutant that is unable to bind MEF2 (70). As shown in Fig. 5E (lanes 1 and 2), this mutant failed to stimulate sumoylation. More interestingly, like mutant 1-326 (Fig. 5B), this point mutant exhibited inhibitory effects on MEF2 sumoylation (Fig. 5E, lanes 1 to 2, and Fig. 5B, lanes 1, 2, and 5). This could be due to dominant negative inhibition. Therefore, through an N-terminal region (residues 118-488) (Fig. 5A and B), HDAC4 stimulates the sumoylation of MEF2D. This region is conserved among HDAC5, -7, and -9, so we analyzed how these HDACs affected MEF2D sumoylation. As shown in Fig. 5F, HDAC5, HDAC7, and the MEF2-interacting transcription repressor MITR exerted effects similar to that of HDAC4, indicating that class IIa members are all able to promote MEF2D sumoylation.
FIG. 5.
Stimulation of MEF2 sumoylation by class IIa HDACs. (A) Schematic representation of HDAC4 and mutants, with the ability to stimulate MEF2 sumoylation shown at right. (B) In vivo sumoylation assays. HEK293 cells were transfected with expression plasmids for HA-SUMO2, Flag-MEF2D, and HDAC4 proteins as indicated. HDAC4 and 118-488 were HA tagged, whereas 1-666 and 1-326 were Flag tagged. Extracts were subjected to immunoprecipitation on M2 agarose, and bound proteins were eluted with Flag peptide for Western blotting analysis with anti-HA (top) or anti-Flag (bottom) antibody. (C) HEK293 cells were transfected with expression plasmids for Flag-MEF2D and HA-tagged HDAC4 and 118-488. Extracts were prepared for Western blotting analysis with anti-HA antibody. (D) As in panel C, except that HDAC4 mutants were Flag tagged and extracts were analyzed by immunoblotting with anti-Flag antibody. (E) As in panel B, except that Flag-tagged HDAC4, mutant L175A, MEF2C, and MEF2D were expressed as indicated. (F) As in panel B, except that different class IIa HDAC members were expressed, with HDAC4 and HDAC5 HA tagged, HDAC7 tagged with both HA and Flag epitopes, and MITR Flag tagged. (G) Same as in panel B, except that Flag-MEF2C was used. Note that the mobility shift of MEF2 proteins (panels B, F, and G) may be due to HDAC-induced phosphorylation.
To examine whether the effect of HDAC4 is specific to MEF2D, we analyzed MEF2C sumoylation. As shown in Fig. 5G (lanes 1 and 2), HDAC4 dramatically stimulated sumoylation of MEF2C. In addition, HDAC4 caused a migration shift of nonsumoylated MEF2C. This shift could be due to phosphorylation, suggesting that HDAC4 also stimulates MEF2 phosphorylation (see Discussion). As shown for MEF2D, mutants 1-666 and 118-488 but not the other deletion mutants or the point mutant L175A promoted the sumoylation of MEF2C (Fig. 5E and G). Therefore, association with class IIa HDACs also potentiates the sumoylation of MEF2C.
Role of HDAC4 sumoylation in regulating MEF2D sumoylation.
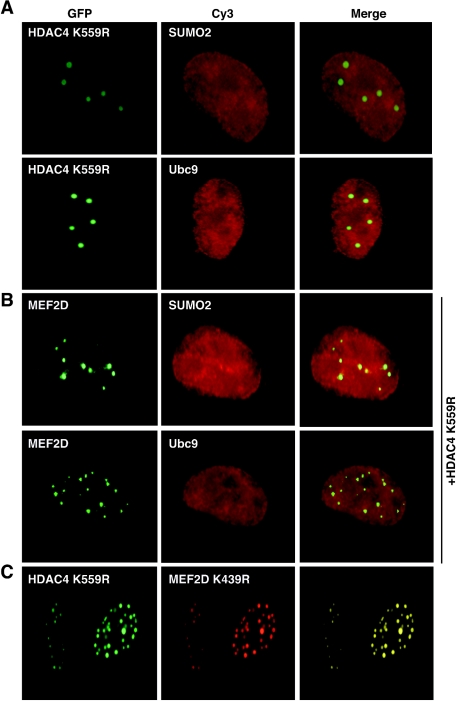
As discussed above, nuclear foci of proteins like Pc2 are sumoylation centers (29, 47, 57), so we wondered whether HDAC4 nuclear dots are novel sumoylation centers. To address this question, we first analyzed the subcellular distribution of K559R, an HDAC4 mutant that is defective in sumoylation (6, 64). As shown in Fig. 6A, this mutant failed to recruit SUMO2 and Ubc9 to nuclear dots. Coexpression of K559R led to the recruitment of MEF2D to nuclear dots, but neither SUMO2 nor Ubc9 was enriched in these dots (compare Fig. 6B with Fig. 4E and F). Moreover, this HDAC4 mutant colocalized with K439R (Fig. 6C), the MEF2D mutant deficient in sumoylation (Fig. 2). Therefore, sumoylation of HDAC4 and MEF2D is dispensable for their colocalization in nuclear dots.
FIG. 6.
Subcellular distribution of MEF2D, HDAC4, and mutants. (A) GFP-K559R was expressed along with HA-SUMO2 or HA-Ubc9 in HeLa cells, followed by fluorescence microscopy to detect GFP. HA-tagged proteins were detected by immunostaining with anti-HA antibody and Cy3-labeled secondary antibody. (B) GFP-MEF2D and Flag-K559R were expressed along with HA-SUMO2 or HA-Ubc9 in HeLa cells. Expressed proteins were detected by immunofluorescence microscopy as in panel A. (C) GFP-K559R and HA-K439R were expressed in HeLa cells and detected as in panel A.
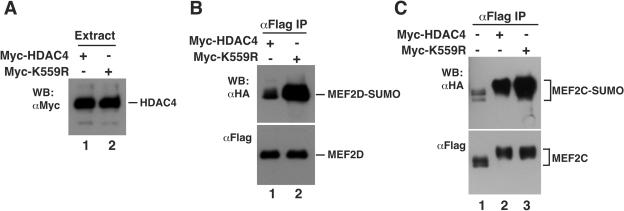
Sumoylation assays were then performed to compare the ability of the wild-type and mutant HDAC4 proteins to stimulate MEF2D sumoylation. As shown in Fig. 7A, both expressed to similar levels. Unexpectedly, mutant K559R was more potent than wild-type HDAC4 in potentiating the sumoylation of MEF2D and MEF2C (Fig. 7B and C), suggesting that sumoylation of HDAC4 exerts negative effects on MEF2 sumoylation. Along with the finding that recruitment of SUMO2 and Ubc9 to HDAC4 nuclear dots was dependent on Lys559 (Fig. 4 and 6), these results indicate that the nuclear dots are not sumoylation centers for MEF2. The biological significance of these dots is presently unclear.
FIG. 7.
Effect of the HDAC4 mutant K559R on MEF2 sumoylation. (A and B) Expression plasmids for Flag-MEF2D and HA-SUMO2 were transfected into HEK293 cells along with the expression construct for Myc-HDAC4 or Myc-K559R. Extracts were subjected to Western blotting analysis with anti-Myc antibody (A) and immunoprecipitation on M2 agarose (B). For the immunoprecipitation, bound proteins were eluted with Flag peptide and subjected to Western blotting analysis with anti-HA or anti-Flag antibody as indicated. (C) Expression plasmids for Flag-MEF2C and HA-SUMO2 were transfected into HEK293 cells along with the expression construct for Myc-HDAC4 or Myc-K559R. Extracts were prepared for immunoprecipitation and immunoblotting as above.
Regulation of MEF2D sumoylation by SUMO proteases.
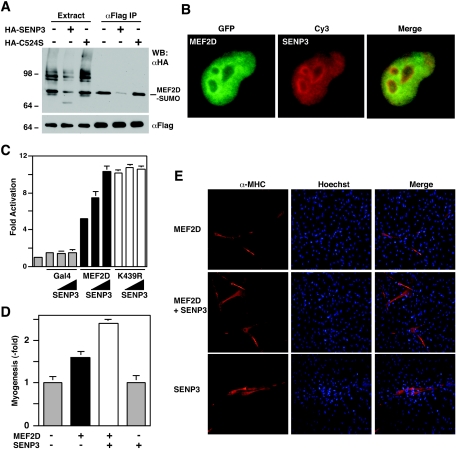
The observation that HDAC4 promoted MEF2 sumoylation (Fig. 5) led us to determine which SUMO protease is involved in desumoylation. A family of SUMO proteases has been identified (44). These proteases display specific subcellular localization and may target different arrays of protein targets. MEF2D was modified by SUMO2 and SUMO3 but not by SUMO1 (Fig. 1C), and SENP3 displays specificity towards SUMO2 and SUMO3 (49, 54), so we tested this SUMO protease. SENP1 was also analyzed for comparison. Both proteases effectively removed SUMO2 and SUMO3 but not SUMO1 from various cellular proteins (data not shown). For desumoylation of MEF2D, SENP3 was more effective than SENP1 (Fig. 8A and data not shown). Moreover, MEF2D and SENP3 displayed partial colocalization when they were coexpressed (Fig. 8B). To test whether the enzymatic activity of SENP3 was directly involved, we replaced Cys524 with serine to create mutant C524S. Cys524 is part of the catalytic center, so this mutation is expected to abrogate the enzymatic activity of SENP3. As shown in Fig. 8A, unlike wild-type SENP3, mutant C524S failed to de-sumoylate MEF2D. These results indicate that SENP3 is able to remove SUMO2 from MEF2D.
FIG. 8.
SENP3 reverses the sumoylation of MEF2D and augments its transcriptional activity. (A) HEK293 cells were transfected with plasmids expressing Flag-MEF2D (2 μg) and HA-SUMO-2 (2 μg), along with HA-SENP3 (6 μg) or its mutant C524S (6 μg). Extracts were used for immunoprecipitation on M2 agarose, and bound proteins were eluted with Flag peptide and subjected to Western blotting analysis with anti-HA (top) or anti-Flag (bottom) antibody. (B) HeLa cells were transfected with expression plasmids for GFP-MEF2D and HA-SENP3, followed by fluorescence microscopy to detect GFP. HA-SENP3 was detected by immunostaining with anti-HA antibody and Cy3-labeled secondary antibody. (C) The luciferase reporter Gal4-tk-Luc (200 ng) and a β-galactosidase expression plasmid (50 ng) were transfected into HEK293 cells along with the expression plasmid for indicated Gal4 proteins (25 ng) and increased amounts of the HA-SENP3 expression plasmid (100 and 200 ng). The normalized luciferase activity from the transfection without any effector plasmids was arbitrarily set to 1.0. Average values of at least three independent experiments are shown with standard deviation. (D and E) The MyoD expression plasmid (400 ng) was transfected into C3H10T1/2 cells along with expression plasmids for Flag-MEF2D (500 ng) and HA-SENP3 (500 ng) as indicated. Myotubes were detected by indirect immunofluorescence with anti-MHC antibody. Average values of at least three independent experiments are illustrated with standard deviation (D) and representative images are shown (E).
Since sumoylation inhibits the transcriptional and myogenic activities of MEF2D (Fig. 3), we determined whether desumoylation reverses the inhibition. In reporter gene assays, SENP3 increased the transcriptional activity of MEF2D in a dose-dependent manner, but minimal effects on mutant K439R were found (Fig. 8C), indicating that SENP3 reverses the inhibitory effect of sumoylation. Similarly, in myogenesis assays, this protease increased the myogenic activity of MEF2D (Fig. 8D and E), further supporting that SENP3 upregulates MEF2-dependent transcription in vivo.
Signal-dependent regulation of MEF2D sumoylation.
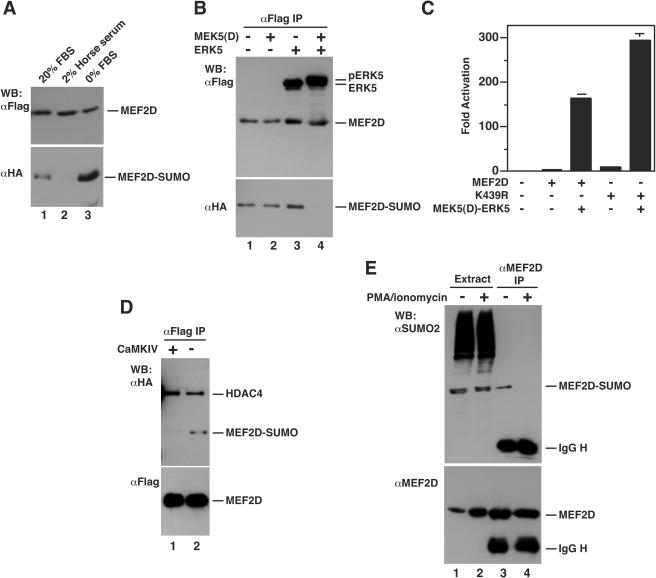
From above, it is clear that the sumoylation of MEF2 is dynamically controlled by HDAC4 and SENP3. In addition, the transcriptional activity of MEF2 is regulated by different signaling pathways (22, 43), so an interesting question is how cell signaling may regulate this modification. To address this, we first tested muscle differentiation conditions and serum starvation. For C2C12 cells, 2% horse serum is known to promote myogenic conversion. As shown in Fig. 9A, this condition inhibited the sumoylation of MEF2D, whereas serum starvation stimulated the modification, suggesting that serum-induced signaling pathways regulate the sumoylation. To identify the pathways involved, we analyzed MEK5 and ERK5, two kinases that are known to act synergistically in stimulating MEF2 transcriptional activity (30-32, 73). As shown in Fig. 9B, activation of this pathway inhibited the sumoylation. Moreover, mutant K439R synergized with activated ERK5 to stimulate the transcriptional activity of MEF2D (Fig. 9C). These results suggest that MEF2 sumoylation is subject to regulation by cell signaling.
FIG. 9.
Signal-dependent sumoylation of MEF2D. (A) Expression plasmids for Flag-MEF2D and HA-SUMO2 were transfected into C2C12 cells. After transfection, cells were washed once with PBS and fed with medium containing 20% FBS (lane 1), 2% horse serum (lane 2), or no serum (lane 3). After 48 h, extracts were prepared in buffer S for immunoprecipitation on M2 agarose. Bound proteins were eluted with Flag peptide and subjected to Western blotting analysis with anti-HA or anti-Flag antibody. (B) Expression plasmids for Flag-MEF2D and HA-SUMO2 were transfected into HEK293 cells along with constructs for MEK5(D) and ERK5 as indicated. Extracts were prepared for immunoprecipitation and immunoblotting as above. Note that phosphorylation by MEK5(D) and ERK5 often retards the migration of MEF2D, but it is not clear on this particular gel. (C) Reporter gene assays were performed as described in the legend to Fig. 3A, except that expression plasmids for Gal4-MEF2D, Gal4-K439R, MEK5(D), and ERK5 (each, 100 ng) were used as indicated. (D) As in panel B, except that an expression plasmid for a constitutively active form of CaMKIV was cotransfected as indicated. Nuclear extracts were prepared for immunoprecipitation and immunoblotting. (E) After treatment with or without PMA (10 ng/ml) and ionomycin (0.5 μM) for 4 h, HEK293 cells were subject to nucleus isolation. Nuclear extracts were then used for immunoprecipitation with anti-MEF2D antibody and immunoblotting with anti-SUMO and anti-MEF2D antibodies.
Since HDAC4 association is required for its ability to stimulate MEF2 sumoylation (Fig. 5), cell signaling may act through class IIa HDACs and regulate MEF2 sumoylation. CaMKs are known to upregulate the transcriptional activity of MEF2 by phosphorylating class IIa members and stimulating their nuclear export (43), so we examined whether CaMK pathways regulate the sumoylation. Considering that nuclear and cytoplasmic proteins become mixed during cell lysis, we performed sumoylation assays with extracts from isolated nuclei (70). As shown in Fig. 9D, expression of a constitutively active form of CaMKIV (61, 72) inhibited the sumoylation of MEF2D. We also asked how nuclear export of class IIa HDACs may affect the sumoylation of endogenous MEF2D. For this, HEK293 cells were treated with phorbol myristate acetate (PMA) and ionomycin. This treatment is known to promote the nuclear export of class IIa HDACs (11, 65). Nuclear extracts were prepared for immunoprecipitation and immunoblotting to detect MEF2D and its sumoylated form. As shown in Fig. 9E, the treatment inhibited the sumoylation, further supporting that nuclear localization of class IIa HDACs is required for stimulating the sumoylation of MEF2D.
DISCUSSION
Sumoylation inhibits the transcriptional activity of MEF2.
Herein, we have demonstrated that MEF2D, as well as MEF2C, is sumoylated on a single lysine residue located within the transcriptional activation domain (Fig. 1 and 2). This lysine residue is conserved among MEF2 proteins (Fig. 1A), suggesting that other MEF2 proteins are similarly modified. Among four human SUMO isoforms, SUMO2, -3, and -4 are almost identical, whereas SUMO1 is the most divergent (7, 20, 46). Numerous proteins have been reported to be modified by SUMO1, but fewer targets have been found for the other three isoforms. SUMO2 and SUMO3 could be conjugated to MEF2D (Fig. 1 and 2 and data not shown), whereas SUMO1 addition was undetectable under the experimental conditions that we tested (Fig. 1 and 2). By contrast, these three SUMO proteins are all effective in modifying HDAC4 under similar conditions (34, 51, 64; unpublished observations). Addition of SUMO2 and SUMO3 has been shown to exert distinct effects from SUMO1 (2, 25, 39). It is presently unclear whether modification of MEF2 by specific SUMO isoforms is to impose unique functional effects.
Sumoylation has been shown to regulate protein function in different manners (14, 16, 58, 67). First, sumoylation and ubiquitination may create a gridlock at the same lysine residue. This mode of action has been reported for IκB, whose sumoylation blocks poly ubiquitination and subsequent ubiquitin-dependent proteasomal degradation (12). In addition, the same lysine residue can be subject to monoubiquitination, which may stimulate transcription (14). Mutation of Lys439 did not inhibit the ubiquitination of MEF2D or affect its stability (data not shown), suggesting that this residue is not ubiquitinated. Second, sumoylation may regulate protein-protein interactions. Sumoylation of MEF2D exhibited minimal effects on HDAC4 binding (Fig. 6C and data not shown). It is unclear whether the modification affects the association of HDAC4 with other proteins. Third, sumoylation has been shown to affect subcellular distribution of several proteins, including promyelocytic leukemia protein (PML), MEK1, and Sp3 (28, 47, 56, 60). Mutant K439R displayed localization patterns similar to that of wild-type MEF2D (data not shown), so sumoylation might not affect the subcellular localization of MEF2D. Fourth, the sumoylation site within MEF2 proteins is similar to the synergy control motif found in other transcription factors (27, 63). Emerging evidence indicates that sumoylation negatively regulates the transcriptional activity of various transcription factors (14, 16, 58, 67). Consistent with this theme, sumoylation of MEF2D inhibited its transcriptional and myogenic activities (Fig. 3).
How does sumoylation exert its inhibitory effect? MEF2 proteins synergize with transcriptional activators, such as MyoD, NF-AT, GATA, thyroid hormone receptor, and SMAD proteins (43). Transcriptional coactivators such as p300 also associate with MEF2 to activate transcription, so it is possible that sumoylation of MEF2 regulates the interaction with transcriptional activators and coactivators. Alternatively, sumoylation may create a docking site for transcriptional corepressors. Related to this, SUMO1 modification of p300 and Elk-1 creates docking sites for HDAC6 and HDAC2, respectively (15, 74). Further investigation is needed to elucidate how modification by SUMO2, SUMO3, and perhaps SUMO4 inhibits the transcriptional activity of MEF2.
HDAC4 upregulates the sumoylation of MEF2.
PIAS (protein inhibitor of activated STAT) proteins, RanBP2, and Pc2 have recently been found to possess SUMO ligase activity (29, 35, 52, 57). These proteins act as nonenzymatic adaptors to stimulate sumoylation of specific targets. We tested RanBP2, Pc2, and a PIAS protein and found that none of them were able to stimulate the sumoylation of MEF2D (data not shown). Instead, class IIa HDACs stimulated the sumoylation of MEF2C and MEF2D (Fig. 5). This stimulation was dependent on residues 118 to 488 but not the C-terminal catalytic domain of HDAC4 (Fig. 5B and G), so the deacetylase activity is not required. Consistent with this conclusion, trichostatin A treatment did not appear to affect MEF2 sumoylation (data not shown). Moreover, mutant L175A failed to potentiate the sumoylation of MEF2 (Fig. 5E), indicating that the physical association is critical. Like this mutant, the HDAC4 mutant 1-326 exhibited dominant-negative effects (Fig. 5). Nuclear export of class IIa HDACs by activated CaMKIV, or by treatment with PMA and ionomycin, abolished the sumoylation of exogenous and endogenous MEF2D proteins (Fig. 9D and E). HDAC4 had no obvious stimulatory effects on the sumoylation of HDAC1 and Pc2 (data not shown). Thus, these results clearly establish that class IIa deacetylases are able to promote MEF2 sumoylation (Fig. 5 and 10). Related to this unexpected and intriguing finding, the class IIb member HDAC6 binds to ubiquitin and may possess ubiquitin ligase activity (26, 37, 59).
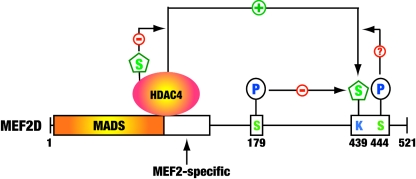
FIG. 10.
Cartoon showing how sumoylation of MEF2 is regulated. Class IIa deacetylases, such as HDAC4, associate with MEF2 to potentiate the sumoylation, whereas the phosphorylation-dependent nuclear export of these HDACs precludes them from upregulating MEF2 sumoylation. This modification is also regulated by phosphorylation at distant and adjacent sites. For example, sumoylation of human MEF2D at Lys439 is inhibited by ERK5-mediated phosphorylation of Ser179. The underlying mechanisms remain elusive. Whether phosphorylation at Ser444 by Cdk5 positively regulates the sumoylation of MEF2 is an interesting question awaiting further exploration. P, phosphorylation; S, sumoylation.
Pc2 recruits the E1A C-terminal-binding protein CtBP and the E2 conjugating enzyme Ubc9 to polycomb group bodies (29), and PIASy sequesters LEF1, SUMO, and Ubc9 to nuclear bodies (35, 57), so it has been proposed that that polycomb protein Pc2 and PIASy bodies function as sumoylation centers to promote the association of substrates with the SUMO machinery. Since HDAC4 forms nuclear dots in vivo, we asked if these dots are sumoylation centers. The HDAC4 mutant K559R could not recruit SUMO2 and Ubc9 to the nuclear dots (Fig. 6) but was more potent than the wild type in potentiating the sumoylation of MEF2 (Fig. 7), indicating that the nuclear dots are not sumoylation centers. This raises the interesting question of how class IIa HDAC4 promotes MEF2 sumoylation. One possibility is that HDAC4 possesses E3-like SUMO ligase activity. Alternatively, it could act through other mechanisms. Related to this, we considered whether HDAC4 reverses the potential acetylation at the sumoylation site. This is not so likely, since the deacetylase domain is dispensable (Fig. 5A). Intriguingly, HDAC4 and other class IIa members caused a mysterious mobility shift of MEF2C and MEF2D (Fig. 5B, F, and G and 7C). This shift could be due to phosphorylation, raising the interesting possibility that these HDACs promote phosphorylation of MEF2 to upregulate the sumoylation (see below).
Interplay between the sumoylation and phosphorylation of MEF2.
The SUMO protease SENP3 reversed the sumoylation of MEF2D (Fig. 8), confirming that this modification is dynamic in vivo, so an interesting question is how cell signaling regulates this modification. Upon activation by Ca2+/calmodulin, CaMKs phosphorylate class IIa HDACs and promote their nuclear export (43). Since physical association with MEF2 was important for HDAC4 to potentiate sumoylation (Fig. 5E), cytoplasmic retention was expected to downregulate MEF2 sumoylation. Indeed, CaMKIV inhibited this modification (Fig. 9D). In addition, MEF2D sumoylation decreased when a muscle differentiation medium was used (Fig. 9A). ERK5, which is known to phosphorylate Ser179 of MEF2D in vivo (30-32, 73), inhibited sumoylation (Fig. 9B), suggesting that phosphorylation of Ser179 promotes long-range interplay with the sumoylation (Fig. 10). Adjacent to the sumoylation site of MEF2D is Ser444 (Fig. 1A), a residue that is known to be phosphorylated by Cdk5 (17). This serine residue is highly conserved among MEF2 proteins and adjacent to the sumoylation site (Fig. 1A). Related to this, sumoylation of HSF1 is stimulated by phosphorylation of a serine residue at the equivalent position (24), so an interesting possibility is that phosphorylation of MEF2D at Ser444 affects the sumoylation. Therefore, sumoylation at the conserved motif of MEF2 proteins (Fig. 1A) may be regulated by phosphorylation events occurring at neighboring and distant sites (Fig. 10) (75). In light of these observations, we propose that the sumoylation motif and adjacent phosphorylation site (Fig. 1A) constitute a novel “modification cassette” for coordinated regulation in vivo (Fig. 10) (13). Of relevance, this cassette is part of a small transferable repression domain that was recently identified (81).
In summary, we have demonstrated that MEF2 is sumoylated on a single lysine residue located at a conserved sumoylation motif. This modification negatively modulates the transcriptional and myogenic activities of MEF2. While class IIa HDACs are able to upregulate the sumoylation, SENP3 acts as the SUMO protease to reverse the modification. By controlling the phosphorylation of class IIa HDACs and MEF2, different signaling pathways dynamically regulate the sumoylation of MEF2. Therefore, this study identifies sumoylation as a novel regulatory mechanism for MEF2 proteins and suggests that this modification interplays with phosphorylation and acetylation to effectively control the biological activity of these transcription factors in vivo.
Acknowledgments
We thank R. T. Ray, R. Prywes, J. D. Lee, L. I. Zon, R. St.-Arnaud, J. Long and Z. Wu for their generosity in providing antibodies, plasmids, and cell lines.
This work was supported by funds from the Canadian Cancer Society through the National Cancer Institute of Canada and from the Canada Foundation for Innovation (to X.-J.Y.).
REFERENCES
- 1.Appella, E., and C. W. Anderson. 2001. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268:2764-2772. [DOI] [PubMed] [Google Scholar]
- 2.Ayaydin, F., and M. Dasso. 2004. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol. Biol. Cell 15:5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakin, R. E., and M. O. Jung. 2004. Cytoplasmic sequestration of HDAC7 from mitochondrial and nuclear compartments upon initiation of apoptosis. J. Biol. Chem. 279:51218-51225. [DOI] [PubMed] [Google Scholar]
- 4.Bernier-Villamor, V., D. A. Sampson, M. J. Matunis, and C. D. Lima. 2002. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108:345-356. [DOI] [PubMed] [Google Scholar]
- 5.Best, J. L., S. Ganiatsas, S. Agarwal, A. Changou, P. Salomoni, O. Shirihai, P. B. Meluh, P. P. Pandolfi, and L. I. Zon. 2002. SUMO-1 protease-1 regulates gene transcription through PML. Mol. Cell 10:843-855. [DOI] [PubMed] [Google Scholar]
- 6.Bischof, O., O. Kirsh, M. Pearson, K. Itahana, P. G. Pelicci, and A. Dejean. 2002. Deconstructing PML-induced premature senescence. EMBO J. 21:3358-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bohren, K. M., V. Nadkarni, J. H. Song, K. H. Gabbay, and D. Owerbach. 2004. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J. Biol. Chem. 279:27233-27238. [DOI] [PubMed] [Google Scholar]
- 8.Clarke, N., N. Arenzana, T. Hai, A. Minden, and R. Prywes. 1998. Epidermal growth factor induction of the c-jun promoter by a Rac pathway. Mol. Cell. Biol. 18:1065-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox, D. M., M. Du, M. Marback, E. C. Yang, J. Chan, K. W. Siu, and J. C. McDermott. 2003. Phosphorylation motifs regulating the stability and function of myocyte enhancer factor 2A. J. Biol. Chem. 278:15297-15303. [DOI] [PubMed] [Google Scholar]
- 10.Cress, W. D., and E. Seto. 2000. Histone deacetylases, transcriptional control, and cancer. J. Cell. Physiol. 184:1-16. [DOI] [PubMed] [Google Scholar]
- 11.Dequiedt, F., H. Kasler, W. Fischle, V. Kiermer, M. Weinstein, B. G. Herndier, and E. Verdin. 2003. HDAC7, a thymus-specific class II histone deacetylase, regulates Nur77 transcription and TCR-mediated apoptosis. Immunity 18:687-698. [DOI] [PubMed] [Google Scholar]
- 12.Desterro, J. M., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell 2:233-239. [DOI] [PubMed] [Google Scholar]
- 13.Fischle, W., Y. Wang, and C. D. Allis. 2003. Binary switches and modification cassettes in histone biology and beyond. Nature 425:475-479. [DOI] [PubMed] [Google Scholar]
- 14.Gill, G. 2004. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 18:2046-2059. [DOI] [PubMed] [Google Scholar]
- 15.Girdwood, D., D. Bumpass, O. A. Vaughan, A. Thain, L. A. Anderson, A. W. Snowden, E. Garcia-Wilson, N. D. Perkins, and R. T. Hay. 2003. p300 transcriptional repression is mediated by SUMO modification. Mol. Cell 11:1043-1054. [DOI] [PubMed] [Google Scholar]
- 16.Girdwood, D. W., M. H. Tatham, and R. T. Hay. 2004. SUMO and transcriptional regulation. Semin. Cell Dev. Biol. 15:201-210. [DOI] [PubMed] [Google Scholar]
- 17.Gong, X., X. Tang, M. Wiedmann, X. Wang, J. Peng, D. Zheng, L. A. Blair, J. Marshall, and Z. Mao. 2003. Cdk5-mediated inhibition of the protective effects of transcription factor MEF2 in neurotoxicity-induced apoptosis. Neuron 38:33-46. [DOI] [PubMed] [Google Scholar]
- 18.Gregoretti, I. V., Y. M. Lee, and H. V. Goodson. 2004. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 338:17-31. [DOI] [PubMed] [Google Scholar]
- 19.Grozinger, C. M., and S. L. Schreiber. 2002. Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem. Biol. 9:3-16. [DOI] [PubMed] [Google Scholar]
- 20.Guo, D., M. Li, Y. Zhang, P. Yang, S. Eckenrode, D. Hopkins, W. Zheng, S. Purohit, R. H. Podolsky, A. Muir, et al. 2004. A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nat. Genet. 36:837-841. [DOI] [PubMed] [Google Scholar]
- 21.Han, J., Y. Jiang, Z. Li, V. V. Kravchenko, and R. J. Ulevitch. 1997. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature 386:296-299. [DOI] [PubMed] [Google Scholar]
- 22.Han, J., and J. D. Molkentin. 2000. Regulation of MEF2 by p38 MAPK and its implication in cardiomyocyte biology. Trends Cardiovasc. Med. 10:19-22. [DOI] [PubMed] [Google Scholar]
- 23.Han, T.-H., and R. Prywes. 1995. Regulatory role of MEF2D in serum induction of the c-jun promoter. Mol. Cell. Biol. 15:2907-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hietakangas, V., J. K. Ahlskog, A. M. Jakobsson, M. Hellesuo, N. M. Sahlberg, C. I. Holmberg, A. Mikhailov, J. J. Palvimo, L. Pirkkala, and L. Sistonen. 2003. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol. Cell. Biol. 23:2953-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmstrom, S., M. E. Van Antwerp, and J. A. Iniguez-Lluhi. 2003. Direct and distinguishable inhibitory roles for SUMO isoforms in the control of transcriptional synergy. Proc. Natl. Acad. Sci. USA 100:15758-15763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hook, S. S., A. Orian, S. M. Cowley, and R. N. Eisenman. 2002. Histone deacetylase 6 binds polyubiquitin through its zinc finger (PAZ domain) and copurifies with deubiquitinating enzymes. Proc. Natl. Acad. Sci. USA 99:13425-13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iniguez-Lluhi, J. A., and D. Pearce. 2000. A common motif within the negative regulatory regions of multiple factors inhibits their transcriptional synergy. Mol. Cell. Biol. 20:6040-6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss III, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kagey, M. H., T. A. Melhuish, and D. Wotton. 2003. The polycomb protein Pc2 is a SUMO E3. Cell 113:127-137. [DOI] [PubMed] [Google Scholar]
- 30.Kasler, H. G., J. Victoria, O. Duramad, and A. Winoto. 2000. ERK5 is a novel type of mitogen-activated protein kinase containing a transcriptional activation domain. Mol. Cell. Biol. 20:8382-8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato, Y., V. V. Kravchenko, R. I. Tapping, J. Han, R. J. Ulevitch, and J.-D. Lee. 1997. BMK1/ERK5 regulate serum-induced early gene expression through transcription factor MEF2C. EMBO J. 16:7054-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato, Y., M. Zhao, A. Morikawa, T. Sugiyama, D. Chakravortty, N. Koide, T. Yoshida, R. I. Tapping, Y. Yang, T. Yokochi, et al. 2000. Big mitogen-activated kinase regulates multiple members of the MEF2 protein family. J. Biol. Chem. 275:18534-18540. [DOI] [PubMed] [Google Scholar]
- 33.Khochbin, S., A. Verdel, C. Lemercier, and D. Seigneurin-Berny. 2001. Functional significance of histone deacetylase diversity. Curr. Opin. Genet. Dev. 11:162-166. [DOI] [PubMed] [Google Scholar]
- 34.Kirsh, O., J. S. Seeler, A. Pichler, A. Gast, S. Muller, E. Miska, M. Mathieu, A. Harel-Bellan, T. Kouzarides, F. Melchior, et al. 2002. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 21:2682-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kotaja, N., U. Karvonen, O. A. Janne, and J. J. Palvimo. 2002. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 22:5222-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kouzarides, T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kovacs, J. J., C. Hubbert, and T. P. Yao. 2004. The HDAC complex and cytoskeleton. Novartis Found. Symp. 259:170-177. [PubMed] [Google Scholar]
- 38.Li, X., S. Song, Y. Liu, S. H. Ko, and H. Y. Kao. 2004. Phosphorylation of the histone deacetylase 7 modulates its stability and association with 14-3-3 proteins. J. Biol. Chem. 279:34201-34208. [DOI] [PubMed] [Google Scholar]
- 39.Li, Y., H. Wang, S. Wang, D. Quon, Y. W. Liu, and B. Cordell. 2003. Positive and negative regulation of APP amyloidogenesis by sumoylation. Proc. Natl. Acad. Sci. USA 100:259-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, F., M. Dowling, X.-J. Yang, and G. D. Kao. 2004. Caspase-mediated specific cleavage of human histone deacetylase 4. J. Biol. Chem. 279:34537-34546. [DOI] [PubMed] [Google Scholar]
- 41.Long, J., G. Wang, D. He, and F. Liu. 2004. Repression of Smad4 transcriptional activity by SUMO modification. Biochem. J. 379:23-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao, Z., A. Bonni, F. Xia, M. Nadal-Vicens, and M. E. Greenberg. 1999. Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science 286:785-790. [DOI] [PubMed] [Google Scholar]
- 43.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2002. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 27:40-47. [DOI] [PubMed] [Google Scholar]
- 44.Melchior, F., M. Schergaut, and A. Pichler. 2003. SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem. Sci. 28:612-618. [DOI] [PubMed] [Google Scholar]
- 45.Miska, E. A., C. Karlsson, E. Langley, S. J. Nielsen, J. Pines, and T. Kouzarides. 1999. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 18:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muller, S., C. Hoege, G. Pyrowolakis, and S. Jentsch. 2001. SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell Biol. 2:202-210. [DOI] [PubMed] [Google Scholar]
- 47.Muller, S., M. J. Matunis, and A. Dejean. 1998. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nathan, D., D. E. Sterner, and S. L. Berger. 2003. Histone modifications: now summoning sumoylation. Proc. Natl. Acad. Sci. USA 100:13118-13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishida, T., H. Tanaka, and H. Yasuda. 2000. A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur. J. Biochem. 267:6423-6427. [DOI] [PubMed] [Google Scholar]
- 50.Paroni, G., M. Mizzau, C. Henderson, G. Del Sal, C. Schneider, and C. Brancolini. 2004. Caspase-dependent regulation of histone deacetylase 4 nuclear-cytoplasmic shuttling promotes apoptosis. Mol. Biol. Cell 15:2804-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petrie, K., F. Guidez, L. Howell, L. Healy, S. Waxman, M. Greaves, and A. Zelent. 2003. The histone deacetylase 9 gene encodes multiple protein isoforms. J. Biol. Chem. 278:16059-16072. [DOI] [PubMed] [Google Scholar]
- 52.Pichler, A., A. Gast, J. S. Seeler, A. Dejean, and F. Melchior. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108:109-120. [DOI] [PubMed] [Google Scholar]
- 53.Puri, P. L., V. Sartorelli, X.-J. Yang, Y. Hamamori, L. Kedes, A. Graessmann, Y. Nakatani, and M. Levrero. 1997. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell 1:35-45. [DOI] [PubMed] [Google Scholar]
- 54.Reverter, D., and C. D. Lima. 2004. A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2-SUMO complex. Structure (Cambridge) 12:1519-1531. [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez, M. S., C. Dargemont, and R. T. Hay. 2001. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276:12654-12659. [DOI] [PubMed] [Google Scholar]
- 56.Ross, S., J. L. Best, L. I. Zon, and G. Gill. 2002. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol. Cell 10:831-842. [DOI] [PubMed] [Google Scholar]
- 57.Sachdev, S., L. Bruhn, H. Sieber, A. Pichler, F. Melchior, and R. Grosschedl. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15:3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seeler, J. S., and A. Dejean. 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 4:690-699. [DOI] [PubMed] [Google Scholar]
- 59.Seigneurin-Berny, D., A. Verdel, S. Curtet, C. Lemercier, J. Garin, S. Rousseaux, and S. Khochbin. 2001. Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Mol. Cell. Biol. 21:8035-8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sobko, A., H. Ma, and R. A. Firtel. 2002. Regulated SUMOylation and ubiquitination of DdMEK1 is required for proper chemotaxis. Dev. Cell 2:745-756. [DOI] [PubMed] [Google Scholar]
- 61.Soderling, T. R. 2000. CaM-kinases: modulators of synaptic plasticity. Curr. Opin. Neurobiol. 10:375-380. [DOI] [PubMed] [Google Scholar]
- 62.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanian, L., M. D. Benson, and J. A. Iniguez-Lluhi. 2003. A synergy control motif within the attenuator domain of CCAAT/enhancer-binding protein alpha inhibits transcriptional synergy through its PIASy-enhanced modification by SUMO-1 or SUMO-3. J. Biol. Chem. 278:9134-9141. [DOI] [PubMed] [Google Scholar]
- 64.Tatham, M. H., E. Jaffray, O. A. Vaughan, J. M. Desterro, C. H. Botting, J. H. Naismith, and R. T. Hay. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276:35368-35674. [DOI] [PubMed] [Google Scholar]
- 65.Vega, R. B., B. C. Harrison, E. Meadows, C. R. Roberts, P. J. Papst, E. N. Olson, and T. A. McKinsey. 2004. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol. Cell. Biol. 24:8374-8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Verdin, E., F. Dequiedt, and H. G. Kasler. 2003. Class II histone deacetylases: versatile regulators. Trends Genet. 19:286-293. [DOI] [PubMed] [Google Scholar]
- 67.Verger, A., J. Perdomo, and M. Crossley. 2003. Modification with SUMO: a role in transcriptional regulation. EMBO Rep. 4:137-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang, A. H., N. R. Bertos, M. Vezmar, N. Pelletier, M. Crosato, H. H. Heng, J. Th'ng, J. Han, and X.-J. Yang. 1999. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol. Cell. Biol. 19:7816-7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, A. H., M. J. Kruhlak, J. Wu, N. R. Bertos, M. Vezmar, B. I. Posner, D. P. Bazett-Jones, and X.-J. Yang. 2000. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol. Cell. Biol. 20:6904-6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, A. H., and X.-J. Yang. 2001. Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol. Cell. Biol. 21:5992-6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang, L., C. Fan, S. E. Topol, E. J. Topol, and Q. Wang. 2003. Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Science 302:1578-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu, Q., L. Yu, L. Liu, C. F. Cheung, X. Li, S. K. Yee, X.-J. Yang, and Z. Wu. 2002. p38 mitogen-activated protein kinase-, calcium-calmodulin-dependent protein kinase-, and calcineurin-mediated signaling pathways transcriptionally regulate myogenin expression. Mol. Biol. Cell 13:1940-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang, C. C., O. I. Ornatsky, J. C. McDermott, T. F. Cruz, and C. A. Prody. 1998. Interaction of myocyte enhancer factor 2 (MEF2) with a mitogen-activated protein kinase, ERK5/BMK1. Nucleic Acids Res. 26:4771-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang, X. J. 2004. Lysine acetylation and the bromodomain: a new partnership for signaling. BioEssays 26:1076-1087. [DOI] [PubMed] [Google Scholar]
- 75.Yang, X.-J. 2005. Multisite protein modification and intramolecular signaling. Oncogene 10:1653-1662. [DOI] [PubMed]
- 76.Yang, X.-J., and E. Seto. 2003. Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr. Opin. Genet. Dev. 13:143-153. [DOI] [PubMed] [Google Scholar]
- 77.Youn, H. D., L. Sun, R. Prywes, and J. O. Liu. 1999. Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2. Science 286:790-793. [DOI] [PubMed] [Google Scholar]
- 78.Yuki, Y., I. Imoto, M. Imaizumi, S. Hibi, Y. Kaneko, T. Amagasa, and J. Inazawa. 2004. Identification of a novel fusion gene in a pre-B acute lymphoblastic leukemia with t(1;19)(q23;p13). Cancer Sci. 95:503-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang, Y. 2003. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 17:2733-2740. [DOI] [PubMed] [Google Scholar]
- 80.Zhao, M., L. New, V. V. Kravchenko, Y. Kato, H. Gram, F. D. Padova, E. N. Olson, R. J. Ulevitch, and J. Han. 1998. Regulation of the MEF2 family of transcription factors by p38. Mol. Cell. Biol. 19:21-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu, B., and T. Gulick. 2004. Phosphorylation and alternative pre-mRNAsplicing converge to regulate myocyte enhancer factor 2C activity. Mol. Cell. Biol. 24:8264-8275. [DOI] [PMC free article] [PubMed] [Google Scholar]