Abstract
The gene coding for Penicillium amagasakiense glucose oxidase (GOX; β-d-glucose; oxygen 1-oxidoreductase [EC 1.1.3.4]) has been cloned by PCR amplification with genomic DNA as template with oligonucleotide probes derived from amino acid sequences of N- and C-terminal peptide fragments of the enzyme. Recombinant Escherichia coli expression plasmids have been constructed from the heat-induced pCYTEXP1 expression vector containing the mature GOX coding sequence. When transformed into E. coli TG2, the plasmid directed the synthesis of 0.25 mg of protein in insoluble inclusion bodies per ml of E. coli culture containing more than 60% inactive GOX. Enzyme activity was reconstituted by treatment with 8 M urea and 30 mM dithiothreitol and subsequent 100-fold dilution to a final protein concentration of 0.05 to 0.1 mg ml−1 in a buffer containing reduced glutathione-oxidized glutathione, flavin adenine dinucleotide, and glycerol. Reactivation followed first-order kinetics and was optimal at 10°C. The reactivated recombinant GOX was purified to homogeneity by mild acidification and anion-exchange chromatography. Up to 12 mg of active GOX could be purified from a 1-liter E. coli culture. Circular dichroism demonstrated similar conformations for recombinant and native P. amagasakiense GOXs. The purified enzyme has a specific activity of 968 U mg−1 and exhibits kinetics of glucose oxidation similar to those of, but lower pH and thermal stabilities than, native GOX from P. amagasakiense. In contrast to the native enzyme, recombinant GOX is nonglycosylated and contains a single isoform of pI 4.5. This is the first reported expression of a fully active, nonglycosylated form of a eukaryotic, glycosylated GOX in E. coli.
Glucose oxidase (GOX; β-d-glucose; oxygen 1-oxidoreductase [EC 1.1.3.4]) is a hydrogen peroxide-generating flavoprotein catalyzing the oxidation of β-d-glucose to d-glucono-1,5-lactone. GOX is used in the food industry for the removal of glucose from powdered eggs, as a source of hydrogen peroxide in food preservation, for gluconic acid production, and in the production of beer and soft drinks, in which its reaction serves an antioxidant function (10, 39, 42). GOX is also used extensively for the quantitative determination of d-glucose in samples such as blood, food, and fermentation products (10, 39, 49). The enzyme has been purified from both Aspergillus niger (45) and Penicillium spp. (33), with A. niger NRRL3 being the most widely used strain for industrial-scale production (11). A problem with utilizing GOX from its native source is the presence of impurities such as catalase, cellulase, and amylase, which may impair some of its applications. To overcome these difficulties and to simplify the stringent purification procedures, which are relatively expensive, A. niger GOX has been cloned and expressed in Saccharomyces cerevisiae as a highly glycosylated form (17).
The most frequent employment of GOX has been in biosensors, in which the biochemical event of glucose oxidation is detected by electrochemical, thermometric, or optical techniques. The most interesting possibilities appear to lie in electron transfer reactions, with artificial electron acceptors or mediators being used to transfer information from the enzyme to the electrode (49). The electrical communication between GOX and the electrode and thereby its biosensor performance are hampered by the protein-bound carbohydrate moiety of the enzyme (1, 15), which most probably impedes electron tunneling through the enzyme (32). Almost complete (24, 27) or partial (15, 32) deglycosylation of GOX is possible, but the procedure is expensive and complicated. A more efficient and effective way of obtaining nonglycosylated GOX would be to express the enzyme in a prokaryotic host. This would also enable the properties and efficiency of GOX to be improved for its use in biosensors by protein engineering techniques (49). As a first step towards this objective, GOX from Penicillium amagasakiense was cloned and expressed in Escherichia coli. GOX from P. amagasakiense was selected since the enzyme has a higher turnover rate and a better affinity for β-d-glucose than its A. niger counterpart (30, 33).
In this study, we describe the cloning and expression of the gene encoding P. amagasakiense GOX and the refolding, purification, and characterization of the nonglycosylated recombinant enzyme. The activity of the recombinant GOX, expressed in the form of insoluble inclusion bodies, was reconstituted, and the active enzyme was shown to possess properties and secondary structure composition similar to those of native P. amagasakiense GOX. This is the first reported expression of a fully active nonglycosylated form of a eukaryotic glycosylated GOX in a prokaryote, which enabled us to demonstrate that in contrast to previous assumptions (4, 9, 47) the protein-bound carbohydrate moiety is not essential for the correct folding of GOX.
MATERIALS AND METHODS
Culture conditions.
P. amagasakiense (ATCC 28686) was grown in potato glucose medium (2) at 25°C and used as a source of genomic DNA. E. coli TG2 (41) was the host strain for the plasmid pCYTEXP1 (5) and the recombinant constructs. Bacterial cells were grown in Luria broth under appropriate selective conditions (41).
PCR.
A 100-μl reaction mixture contained 1 μg of fungal genomic DNA template isolated according to the method of reference 38, 10 μl of 10× polymerase buffer (Perkin-Elmer), 15 μl of 25 mM MgCl2 solution, 2.0 μl of each 10 μM deoxynucleoside triphosphate, 100 pmol each of the N-terminal forward and the C-terminal reverse primers, and 0.5 U of Taq DNA polymerase. A 1.7-kb fragment was obtained following 25 PCR amplification cycles with an annealing temperature of 50°C. The amplified fragment was separated by agarose gel electrophoresis and extracted with the QIAEX kit (Qiagen Inc.). The primers had the following sequences: N-terminal forward primer (N1), 5′ GGCATATGTACTTGCCAGCTCAGCAGATCGACGTTCAGTC 3′; C-terminal reverse primer (C1), 5′ GGGTCGACGAATTCTTAAGCTGACTTAGCATAATCATCCAAGATAGC 3′. The ATG initiation signal, the TAA stop codon, and suitable restriction sites (NdeI to SalI) were introduced for cloning and expression purposes.
Plasmid construction and expression of recombinant GOX.
Standard methods for DNA manipulation were employed (41). All restriction enzymes and other reagents for recombinant DNA manipulations were obtained from New England Biolabs. The N1C1 PCR fragment of about 1.7 kb was cloned directly into the pCYTEXP1 vector to give the expression plasmid (pPAGOX1). Plasmid pPAGOX1 was used for transformation into the E. coli host strain TG2 by electroporation (18) and selected in the presence of 100 μg of ampicillin μl−1. Cultures were grown at 30°C in Luria broth (41) to an optical density at 600 nm of 0.6, and then expression of recombinant GOX was heat induced at 42°C for 3 h. The cell pellet was resuspended in 10 mM Tris-HCl–50 mM NaCl, pH 8.0, and sonicated on ice for several minutes with pulsed bursts. The soluble supernatant fraction and insoluble pellet were separated by centrifugation at 35,000 × g for 15 min. Both fractions were analyzed by polyacrylamide gel electrophoresis (PAGE), enzyme assay, and Western blotting. The amount of GOX in the inclusion bodies was quantified by densitometric scanning of Coomassie blue-stained sodium dodecyl sulfate (SDS)-polyacrylamide gels.
Solubilization and refolding of insoluble GOX.
The inclusion bodies were washed twice in 20 mM Tris-HCl–100 mM NaCl–1 mM EDTA, pH 8.0, once with and once without Triton X-100. Resuspension of the pellet in 2 M urea led to the solubilization of most of the protein contaminants. GOX was subsequently solubilized in 8 M urea–30 mM dithiothreitol (DTT) and kept on ice for 60 min. The protein concentration of the solubilized sample was determined as described below. The enzyme (5 mg ml−1) was diluted 100-fold in the renaturation buffer (1 mM reduced glutathione [GSH], 1 mM oxidized glutathione [GSSG], 0.05 mM flavin adenine dinucleotide [FAD], 10% [vol/vol] glycerol, 20 mM Tris-HCl, pH 8.0) and left to stand for a week at 10°C.
Purification of reactivated GOX.
The reactivated enzyme was concentrated by diafiltration with a 10-kDa-cutoff membrane (Pall Filtron) and washed with MilliQ water (Millipore). The sample was then mildly acidified by dilution with 20 mM sodium acetate buffer, pH 6.0, and reconcentrated by diafiltration. Aggregated proteins were removed by centrifugation (10,000 × g, 5 min, 4°C) and sterile filtration through a 0.2-μm-pore-size membrane (Sartorius). GOX was then purified by fast protein liquid chromatography on a Q-Sepharose column (2.5 by 8 cm) at 4°C in the same buffer. The proteins were eluted at a flow rate of 1 ml min−1 with a linear gradient of 0 to 1 M NaCl in the same buffer in 40 ml. GOX eluted at 0.7 M NaCl. Fractions containing GOX were pooled, dialyzed against MilliQ water, and analyzed for purity by SDS-PAGE.
Purification and deglycosylation of native P. amagasakiense GOX.
Commercial GOX from P. amagasakiense (Nagase, Osaka, Japan) was purified to electrophoretic homogeneity and deglycosylated by more than 95% with endoglycosidase H as previously described (30). The purified enzyme is referred to as native GOX; the almost completely deglycosylated enzyme is referred to as deglycosylated GOX.
Enzyme assay.
GOX activity was assayed at 420 nm by a standard oxidase assay with 2,2′-azino-di-[3-ethylbenzthiazoline-6-sulfonate] diammonium salt (ABTS) as chromogen (28). Assays were performed under oxygen saturation in 0.1 M sodium acetate buffer, pH 6, at 25°C with 0.1 M glucose as substrate. One unit of GOX is defined as the amount of enzyme that catalyzes the oxidation of 1 μmol of glucose to gluconolactone and H2O2 in 1 min at 25°C.
Protein determination.
Soluble protein concentration was determined by the method of Bradford (7) with the standard assay kit from Bio-Rad (Munich, Germany) with immunoglobulin G as standard. The concentration of insoluble GOX in the inclusion bodies was quantified by densitometric scanning (GT-9000 scanner; Epson) of Coomassie blue-stained SDS-polyacrylamide gels, with the ScanPack program (Epson).
Kinetic parameters.
The Michaelis-Menten kinetic parameters, limiting maximal velocity (Vmax), Km, catalytic constant (kcat), and specificity or apparent second-order constant (kcat/Km), were calculated for glucose and other sugars. The values were calculated for the β-anomers due to their preferential utilization by GOX (13). Activity was determined for each sugar by the standard assay procedure. Lineweaver-Burk plots were used to determine the kinetic parameters on the assumption that simple Michaelis-Menten kinetics were followed. The Kcat values were calculated per mole of native GOX (i.e., per dimer), since only the dimeric form of GOX is active.
Melting temperature.
The temperature at the midpoint of thermal unfolding transition, which is referred to as melting temperature, Tm, was determined from the kinetics of GOX thermal inactivation as described in reference 19.
PAGE.
SDS-PAGE was carried out under reducing conditions by using the buffer of Laemmli (34) and included a 12% polyacrylamide running gel and a 4% polyacrylamide stacking gel. The same buffers without SDS were used for nondenaturing gels, with 7.5 and 3% polyacrylamide running and stacking gels, respectively. The gels were stained with Coomassie brilliant blue R. The isoelectric point of GOX was determined by isoelectric focusing with the Pharmacia Phast System in the pH range 4.0 to 6.5 according to the method described in reference 8.
Carbohydrate analysis.
Protein-associated carbohydrates were detected by transferring the proteins from SDS-polyacrylamide gels onto polyvinylidene difluoride membranes by the Western blot method and staining the proteins for glycoconjugates with the Glycan Detection Kit (Boehringer Mannheim, Mannheim, Germany) according to the manufacturer’s recommendations (6).
Circular dichroism (CD).
CD spectra were recorded at 20°C on a spectropolarimeter (Jasco J-600). Spectra in the near-UV (300 to 250 nm) and far-UV (250 to 184 nm) ranges were recorded in cells of 0.5- and 1-mm path lengths and at enzyme concentrations of 0.1 and 0.05 mg ml−1, respectively. The secondary structures were predicted by a mathematical calculation of the basis CD spectra with the programs CONTIN (37) and Varselec (25) with a set of CD spectra of proteins with known secondary structures. The fractions of five types of secondary structures (α-helix, antiparallel and parallel β-sheet, β-turn, and “other”) were calculated without the use of constraints for the analysis, i.e., without forcing the sum of the secondary structures to be 1.0. The term “other” refers to random secondary structures which cannot be assigned to the four main conformation classes.
RESULTS
Expression of the GOX PCR fragment in E. coli TG2.
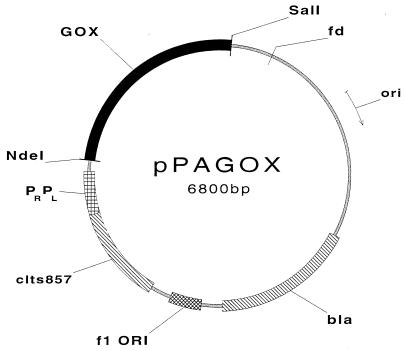
The amino acid sequence deduced from the directly sequenced PCR fragment (50) was completely identical to the amino acid sequence derived for the native P. amagasakiense enzyme by protein sequencing (31). The mature GOX coding region was inserted into the E. coli expression vector pCYTEXP1. When TG2 cells carrying the plasmid pPAGOX1 (Fig. 1) reached mid-logarithmic growth, expression of recombinant GOX was heat induced as described in Materials and Methods. The cells were harvested when the culture reached an optical density at 600 nm of 2.5 to 3.0 and were analyzed by SDS-PAGE.
FIG. 1.
Structure of the plasmid vector for the expression of GOX in E. coli. The plasmid was constructed as described in Materials and Methods. pPAGOX is based on pCYTEXP1 and contains the bacteriophage λ tandem promoters PR and PL preceded by the clts857 repressor gene, the GOX coding region, and the transcription terminator from the bacteriophage fd.
Heat induction resulted in the accumulation of large quantities of a new protein with a molecular mass of 60 kDa which was recognized by monoclonal anti-GOX antibodies. Cell fractionation into soluble and insoluble fractions demonstrated the 60-kDa protein to accumulate in inclusion bodies. Expression experiments with other E. coli strains and under gentler conditions (e.g., lower temperature shift) also led to the expression of GOX in the form of insoluble inclusion bodies, but with much lower quantities (data not shown). Quantification of the expression level of GOX by densitometric scanning of Coomassie blue-stained SDS-polyacrylamide gels demonstrated GOX to represent over 60% of the total insoluble protein fraction. No enzymatic activity was detected in either the soluble or the insoluble fraction.
Refolding and purification of the active recombinant GOX.
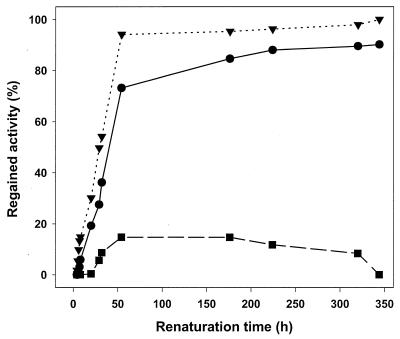
The inclusion bodies obtained from E. coli TG2 were washed with 2 M urea to remove most of the contaminating proteins. GOX was subsequently solubilized in 8 M urea and 30 mM DTT, resulting in a protein concentration of 5 to 10 mg ml−1. Densitometric scanning showed GOX to represent 80% of the solubilized protein. Solubilized GOX was diluted 100-fold to a final protein concentration of 0.05 to 0.1 mg ml−1 in the renaturation buffer (1 mM GSH, 1 mM GSSG, 0.05 mM FAD, 20 mM Tris-HCl [pH 8.0], 10% [vol/vol] glycerol) and left to stand for a week at 10°C. The kinetics of refolding were determined by measuring the increase in enzyme activity after dilution of the protein. The kinetics of GOX reactivation followed an apparent single first-order reaction (Fig. 2). The half-time of GOX reactivation was 19.8 h at 10°C and 30.3 h at 4°C. Maximum reactivation was observed at 10°C. At 20°C, only 14% of the maximal enzyme activity was regained.
FIG. 2.
Reactivation of denatured GOX at various temperatures. The refolding and reactivation conditions were as follows: 0.1 mg of protein ml−1 in 20 mM Tris-HCl–1 mM GSH–1 mM GSSG–0.05 mM FAD–10% glycerol, pH 8.0, incubated at 4°C (•), 10°C (▾), and 20°C (▪). Reactivation was calculated relative to final values, determined after a reactivation time of more than 1,400 h. Reactivation of the completely denatured GOX at 4 and 10°C can be described by a single exponential function yielding half-times of 30.3 and 19.8 h, respectively.
Refolded GOX was concentrated by diafiltration, and excess FAD was removed by washing with MilliQ water. This step resulted in the apparent removal of nearly 85% of the total protein (Table 1). One possible explanation for this large loss of protein through the diafiltration membrane is that most of the protein in the renaturation fraction failed to refold properly and was still in the unfolded state, most probably existing in a nonrandom conformation. Such disordered proteins would be able to pass more readily through the diafiltration membranes than globular proteins of similar molecular mass. With GOX being the main component of the renatured preparation, most of the expressed recombinant GOX appears not to have refolded correctly. However, the possibility that one or more components of the renaturation buffer interfered with the protein assay cannot be excluded.
TABLE 1.
Refolding and purification of recombinant GOX derived from a 1-liter E. coli culture
| Purification step | Total protein (mg) | Total activity (U) | Sp act (U mg−1) | Yield (%) |
|---|---|---|---|---|
| Solubilizationa | 248 | 0 | 0 | 0 |
| Renaturationb | 203 | 14,680 | 73 | 100 |
| Concnc | 32 | 12,950 | 407 | 88 |
| Acidificationd | 14 | 12,400 | 898 | 84 |
| Q-Sepharose | 12 | 11,860 | 968 | 81 |
Solubilization of the apoenzyme in 8 M urea and 30 mM DTT. The total protein concentration is the amount of protein determined after solubilization of the inclusion bodies.
Renaturation in refolding medium. The maximum activity reconstituted after renaturation was used to represent the initial GOX activity and corresponded to 100% yield.
Concentration of the refolding medium by diafiltration.
Acidification of the medium to pH 6.0.
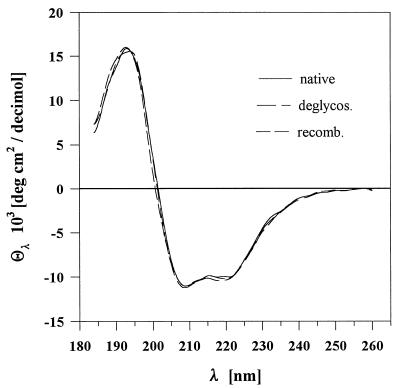
Mild acidification of the sample by dilution with 20 mM acetate buffer, pH 6.0, caused aggregation of most of the remaining contaminants, which were removed by centrifugation and sterile filtration. The enzyme was ultimately purified to electrophoretic homogeneity by anion-exchange chromatography on a Q-Sepharose column. The purification steps and yields of GOX are summarized in Table 1. Inclusion bodies (200 to 250 mg) from a 1-liter E. coli culture yielded 10 to 12 mg of GOX. The CD spectrum of the purified recombinant enzyme was identical to those of native and deglycosylated P. amagasakiense GOXs (Fig. 3), demonstrating similar configurations of the three enzymes and implying correct folding of the active recombinant GOX (12). The fraction of residues in a given conformation class was very similar for native, deglycosylated, and recombinant P. amagasakiense GOXs (Table 2). The secondary structure composition estimated from the CD spectra of P. amagasakiense GOX was in good agreement with the values obtained from X-ray data for the α-helices (0.28) and β-sheets (0.18) of A. niger GOX (22).
FIG. 3.
CD spectra of GOXs. Spectra of recombinant (recomb.), deglycosylated (deglycos.), and native GOXs were recorded at 20°C on a Jasco J-600 spectropolarimeter in 0.5- and 1-mm cuvettes at enzyme concentrations of 0.1 and 0.05 mg ml−1, respectively.
TABLE 2.
Comparison of secondary structure compositions of recombinant, native, and deglycosylated P. amagasakiense GOXs estimated from CD spectraa
| Secondary structure | Recombinant | Native | Deglycosylated |
|---|---|---|---|
| α-Helix | 0.31 | 0.32 | 0.35 |
| Antiparallel β-sheet | 0.14 | 0.16 | 0.16 |
| Parallel β-sheet | 0.03 | 0.04 | 0.03 |
| β-Turn | 0.14 | 0.14 | 0.16 |
| Otherb | 0.21 | 0.24 | 0.24 |
The values are presented as the fractions of residues in a given conformation class without using constraints for the total.
The term “other” refers to random secondary structure elements, such as long, mostly unordered loops, that cannot be unequivocally assigned to the four conformation classes.
Characterization of recombinant GOX.
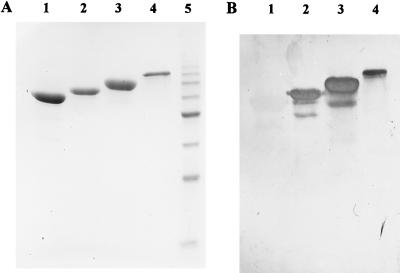
SDS-PAGE demonstrated the recombinant enzyme to have a lower molecular mass than both native and deglycosylated GOXs from P. amagasakiense (Fig. 4A). Recombinant GOX migrated as a single protein band with a molecular mass of 60 kDa (SDS-PAGE) and 120 kDa (native PAGE). These values are in good agreement with the molecular mass of 64 kDa estimated from the amino acid sequence analysis of the native enzyme (31) and deduced from the DNA sequence of the GOX gene (50). The higher molecular masses of the native (30% higher) and deglycosylated (10% higher) P. amagasakiense GOXs may be attributed to the protein-bound carbohydrates (30). The N-terminal sequences and the digestion patterns of the recombinant and native GOXs were identical (data not shown). However, as expected for E. coli-produced proteins and in contrast to native and deglycosylated P. amagasakiense GOXs, the recombinant enzyme contained no detectable protein-bound carbohydrates (Fig. 4B). Isoelectric focusing under native conditions revealed four bands of pH 4.37, 4.42, 4.46, and 4.51 for native and deglycosylated GOXs (30). In contrast, recombinant GOX has a single protein band of pI 4.5, which is identical to the pI of native P. amagasakiense GOX which has been purified to isoelectric homogeneity (26, 28) and then deglycosylated and crystallized (24).
FIG. 4.
Analysis of protein-bound carbohydrates by SDS-PAGE (A) and Western blotting (B). Purified native, deglycosylated, and recombinant GOXs were separated by SDS-PAGE and transferred by electroblotting onto a polyvinylidene difluoride membrane. Proteins were visualized in SDS gels by staining with Coomassie blue. Protein-bound carbohydrates were detected on the membrane with the Glycan Detection Kit (Boehringer Mannheim). Lanes 1, recombinant GOX; lanes 2, deglycosylated GOX; lanes 3, native GOX; lanes 4, transferrin (control); lane 5, molecular mass markers (10-kDa protein ladder from Gibco BRL).
Recombinant GOX was optimally active at pH 5.2 to 6.2 and exhibited more than 80% of the maximum activity between pH 4.5 to 5.2 and 6.2 to 6.5. Outside this range, the enzyme activity decreased rapidly. Two pKa values (pK1 = 3.95 ± 0.15; pK2 = 7.03 ± 0.11) were determined upon calculation of the data with the GraFit program (35). The temperature dependence profile of the recombinant enzyme was virtually identical to those of native and deglycosylated GOXs from P. amagasakiense (20, 36), with a broad temperature optimum at 28 to 40°C (about 1,200 U mg−1). Activity increased twofold with an increase in temperature from 15 to 28°C and decreased rapidly above 40°C to less than 200 U mg−1 at 67°C. In comparison, A. niger GOX exhibited optimum activity at 55°C (29).
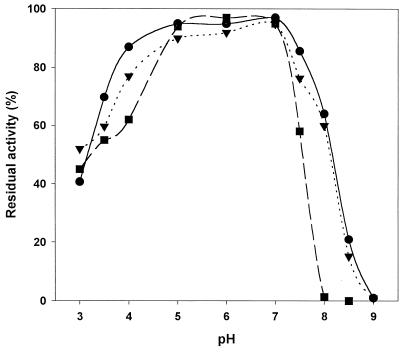
Optimal stability was observed at pH 5 to 7, with more than 90% of the residual activity being retained after 72 h of incubation at room temperature (Fig. 5). However, recombinant GOX was slightly less stable above pH 7.0 than native and deglycosylated GOXs (Fig. 5). The recombinant enzyme was stable up to 40°C. Above 40°C, its stability decreased rapidly with increases in temperature (Table 3). Thermal inactivation followed first-order kinetics. The thermal stability was affected by the medium pH, with the enzyme being more stable at pH 5 than pH 6. Recombinant GOX demonstrated considerably lower stability than native and deglycosylated P. amagasakiense GOXs (Table 3), the latter being only slightly less thermostable than the native enzyme. A melting temperature, Tm, of 58°C at pH 5 and 54°C at pH 6 was estimated for the recombinant enzyme, in comparison with a Tm of 61°C (pH 5) and 58°C (pH 6) for the native GOX from P. amagasakiense. The activation energy for the destruction reaction, Ed, which was only marginally affected by the medium pH, was lower for the recombinant enzyme (92.3 kcal/mol at pH 5 and 93.8 kcal/mol at pH 6) than for native GOX (110.3 and 109.4 kcal/mol at pH 5 and 6, respectively). In contrast to native and deglycosylated GOXs, the thermal stability of recombinant GOX was dependent on the protein concentration, with the enzyme being more stable at lower protein concentrations. Consequently, at 40°C the half-life decreased from 105 h for a diluted sample (0.74 μg ml−1) to 12 h for a 100-fold-more concentrated solution (74 μg ml−1).
FIG. 5.
Effect of pH on GOX stability. The residual activities of recombinant (▪), native (•), and deglycosylated (▾) GOXs were measured in 0.1 M sodium acetate buffer, pH 6.0, after 72 h of storage at room temperature in sodium citrate (pH 3.0 to 4.0), sodium acetate (pH 5.0 to 6.0), or Tris-HCl (pH 7.0 to 9.0) (all 50 mM).
TABLE 3.
Effect of temperature and pH on the stability of GOX
| Incubation temp (°C) |
t1/2a (min) at pH:
|
|||||
|---|---|---|---|---|---|---|
| 5.0
|
6.0
|
|||||
| Recombb | Deglycosc | Native | Recombb | Deglycosc | Native | |
| 50 | 781 | 3,219 | 3,984 | 96 | 918 | 1,110 |
| 55 | 78 | 228 | 582 | 14 | 80 | 109 |
| 60 | 12 | 29 | 40 | 2 | 8 | 9 |
| 65 | 2.6 | 3.2 | 3.3 | 1.3 | 1.5 | 1.7 |
t1/2, half-life of GOX (0.74 μg ml−1) in 50 mM sodium acetate, pH 5.0 or 6.0. Data are the means of three independent measurements.
Recomb, recombinant GOX.
Deglycos, deglycosylated GOX.
The substrate specificity of recombinant GOX was similar to that of native GOX, with β-d-glucose being the preferred substrate, showing at least a 5-fold-higher turnover rate and a 30-fold-higher specificity constant than those for other sugars. The kinetic parameters of monosaccharide oxidation by the recombinant enzyme are summarized in Table 4. The kinetic parameters of glucose oxidation by recombinant GOX were very similar to those of native and deglycosylated P. amagasakiense GOXs and considerably better than those of A. niger GOX (Table 5).
TABLE 4.
Kinetic parameters of recombinant GOX for different sugarsa
| Substrate | Vmax (U mg−1) | Km (mM) | kcatb (s−1) | kcat/Km (s−1 mM−1) |
|---|---|---|---|---|
| d-Glucose | 926 ± 9 | 6.2 ± 0.5 | 2,003 | 323.0 |
| 2-Deoxy-d-glucose | 38 ± 1 | 8.3 ± 0.7 | 83 | 10.0 |
| d-Mannose | 186 ± 6 | 106 ± 8 | 401 | 3.8 |
| d-Galactose | 73 ± 5 | 952 ± 9 | 159 | 0.2 |
| d-Xylose | 33 ± 1 | 384 ± 27 | 72 | 0.2 |
Initial reaction velocities of GOX were determined for each sugar at 25°C under oxygen saturation in 0.1 M sodium acetate, pH 6.0. Data are the means (± standard deviations for Vmax and Km) of at least three independent measurements.
The kcat values were calculated per mole of native GOX, since only the dimeric form of the enzyme exhibits activity.
TABLE 5.
Kinetic parameters for β-d-glucoses of recombinant, native, and deglycosylated P. amagasakiense GOXs and native A. niger GOX
| GOX | Kinetic parametera
|
|||
|---|---|---|---|---|
| Vmax (U mg−1) | Km (mM) | kcatc (s−1) | kcat/Km (s−1 mM−1) | |
| P. amagasakiense | ||||
| Recombinant | 926 ± 9 | 6.2 ± 0.5 | 2,003 | 323 |
| Native | 925 ± 8 | 5.7 ± 0.6 | 2,001 | 351 |
| Deglycosylated | 901 ± 12 | 6.3 ± 0.5 | 1,949 | 309 |
| A. nigerb | 458 | 30 | 920 | 31 |
Initial reaction velocities were determined at 25°C under oxygen saturation in 0.1 M sodium acetate, pH 6.0. Data are the means (± standard deviations, where shown) of at least three independent measurements.
The kinetic data for native A. niger GOX were taken from reference 29.
The kcat values were calculated per mole of native GOX since only the dimeric form of the enzyme is active.
DISCUSSION
Cloning of the P. amagasakiense GOX gene by PCR amplification with genomic DNA as template has enabled us to express the enzyme in E. coli. Although A. niger GOX has previously been cloned and expressed in yeast (17) and Aspergillus spp. (48), this is the first reported expression of a fully active, nonglycosylated form of the eukaryotic, glycosylated GOX in a prokaryote. Expression of the recombinant GOX in E. coli led to the formation of insoluble inclusion bodies. Accumulation of GOX as the major protein component in the inclusion bodies was advantageous for the purification of the enzyme, since inclusion bodies could be rapidly removed from the total cell lysate by centrifugation and the inactive enzyme could be reactivated by a relatively simple renaturation step. Consequently, 10% of the total aggregated GOX was retrieved in an active form, resulting in the final recovery of 12 mg of pure P. amagasakiense GOX from a 1-liter E. coli culture.
Although numerous procedures are known for the folding and refolding of proteins (3, 14, 19), many tests had to be performed before the presently acceptable refolding conditions were found for GOX. Renaturation of GOX was difficult since the enzyme is an FAD-dependent dimer which contains intramolecular disulfide bridges (22, 31). Optimal reactivation of GOX was observed at pH 8 in a renaturation medium containing not only the cofactor FAD and excess DTT but also the redox system GSH-GSSG. These results imply the presence of at least one disulfide bridge in the native enzyme. This is not surprising, since the highly homologous A. niger GOX contains one disulfide bridge per subunit (22). Protein concentration was also critical, with the best renaturation results being obtained at low protein concentrations (0.05 to 0.1 mg ml−1), in accordance with previous observations (3, 14, 19). A lower limit of protein concentration in the refolding experiments was also found, which is a strong indication for dimerization. In vitro refolding of proteins lacking disulfide bridges normally takes only milliseconds and with disulfide bridges hours or perhaps days (21, 43). In the case of GOX, the refolding process required a week. An alternative procedure, using compounds such as DsbA (16) to mediate disulfide bond formation, could accelerate the rate of refolding and lower the cost of this system.
The slow renaturation process is most probably due to two interdependent processes: FAD incorporation and dimer formation. Dimerization is possible only after FAD incorporation and correct folding of GOX (44, 46), due to the proximity of the FAD-binding domain, which is located at the dimer interface and is covered by the FAD covering lid (22, 31). The FAD covering lid, which is a short contiguous region formed by 24 amino acids at the dimer interface, couples FAD binding with dimer formation (31). This lid is in an open configuration in the monomeric form of GOX (22). Thus, a precise sequence of events is essential for the correct refolding of GOX. The cofactor must first bind to the FAD-binding domain to enable the lid to close. Closure of the lid is accompanied by the formation of the dimer interface and the enclosure of the FAD moiety (22).
The carbohydrate moiety of various glycoproteins is believed to play an important role in the folding of proteins into a specific configuration (4, 9, 47). Although recombinant GOX contains no protein-bound carbohydrates, CD analysis of its structural properties showed the enzyme to have a secondary structure virtually identical to that of native GOX, implying correct folding of the nonglycosylated enzyme. Hence, the protein-bound carbohydrate moiety does not appear to be essential for the correct folding of GOX. The carbohydrate moiety also does not affect the kinetic properties of GOX, with the nonglycosylated enzyme exhibiting kinetics of sugar oxidation similar to those of native and deglycosylated GOXs (29, 30). The protein-bound carbohydrate does, however, appear to contribute to the high thermostability of GOX, with the recombinant enzyme being less thermostable than native GOX. This is not surprising since the carbohydrate chain at Asn-89 in A. niger (22) and Asn-93 in P. amagasakiense (31) GOX is situated near the dimer interface and links the FAD-binding lid of one subunit with the second subunit of the dimer (22), thereby providing extra stability to GOX. The integrity of this carbohydrate chain is maintained in the enzymatically deglycosylated GOX (22, 31), which exhibits thermostability similar to that of native GOX (29, 30), presumably due to the inaccessibility of this glycosylation site to the glycohydrolases (22).
Reactivated recombinant GOX exhibited not only biochemical properties very similar to but also electrophoretic behavior comparable to and CD spectra identical to those of the native enzyme. Since the apo- and holoforms of GOX show differences in their CD spectra (12), the identical CD spectra of recombinant and native P. amagasakiense GOXs indicate that the recombinant enzyme has been correctly refolded. However, confirmation of these results is possible only by a comparison of the tertiary structures of the native and recombinant enzymes. The crystal structure of native P. amagasakiense GOX is currently being solved; preliminary crystallization experiments with the recombinant GOX have been initiated, and minicrystals have been obtained. Crystals of GOX suitable for X-ray analysis can be grown only with isoelectrically homogeneous preparations of deglycosylated GOX (24, 27). Thus, the expression of a single isoform of a nonglycosylated GOX eliminates the expensive and elaborate steps which are essential for the crystallization of native P. amagasakiense GOX (24, 27, 28).
The renaturation and purification method described here for recombinant GOX may be a useful tool for improving enzyme-electrode contacts. A major limitation of GOX, and of most redox enzymes used in the biosensor technology, is its inability to transfer electrons directly from its embedded redox site to an electrode (23, 49). Several methods have been developed to improve electrical communication between redox proteins and electrode surfaces, including the site-specific positioning of electron-mediating units in GOX (40). This procedure, which involves modification of the FAD removed from native GOX followed by reconstitution of the apoenzyme with the modified cofactor, could be significantly simplified and improved by refolding the recombinant enzyme in the presence of the modified FAD. In addition, partially or almost completely deglycosylated GOX has been observed to exhibit improved electron transfer properties (1, 15), probably due to increased hydrogen tunneling (32). Thus, the availability of nonglycosylated recombinant GOX should make it possible to improve further the biosensor performance of GOX (15) and may even circumvent the need for artificial electron acceptors or mediators by enabling direct electron transfer from the active site of GOX to the enzyme electrode (1, 49).
REFERENCES
- 1.Alvarez-Icaza M, Kalisz H M, Hecht H J, Aumann K D, Schomburg D, Schmid R D. The design of enzyme sensors based on the enzyme structure. Biosens Bioelectron. 1995;10:735–742. doi: 10.1016/0956-5663(95)96964-z. [DOI] [PubMed] [Google Scholar]
- 2.American Type Culture Collection. Catalogue of fungi/yeasts. 17th ed. Rockville, Md: American Type Culture Collection; 1987. p. 415. [Google Scholar]
- 3.Anfinsen C B. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 4.Barbarić S, Mrša V, Ries B, Mildner P. Role of carbohydrate part of yeast acidic phosphatase. Arch Biochem Biophys. 1984;234:567–575. doi: 10.1016/0003-9861(84)90305-9. [DOI] [PubMed] [Google Scholar]
- 5.Belev T N, Singh M, McCarthy J E G. A fully modular vector system for the optimization of gene expression in Escherichia coli. Plasmid. 1991;26:147–150. doi: 10.1016/0147-619x(91)90056-3. [DOI] [PubMed] [Google Scholar]
- 6.Boehringer Mannheim GmbH. DIG Glycan Detection Kit instructions. Mannheim, Germany: Boehringer Mannheim GmbH; 1997. [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Butcher L A, Tomkins J K. A comparison of silver staining methods for detecting proteins in ultrathin polyacrylamide gels on support film after isoelectric focussing. Anal Biochem. 1985;148:384–388. doi: 10.1016/0003-2697(85)90243-x. [DOI] [PubMed] [Google Scholar]
- 9.Chu F K, Trimble R B, Maley F. The effect of carbohydrate depletion on the properties of yeast external invertase. J Biol Chem. 1978;253:8691–8693. [PubMed] [Google Scholar]
- 10.Crueger A, Crueger W. Carbohydrates. In: Rehm H-J, Reed G, editors. Biotechnology. 6a. Weinheim, Germany: Verlag Chemie; 1984. pp. 421–457. [Google Scholar]
- 11.Crueger A, Crueger W. Glucose transforming enzymes. In: Fogarty W M, Kelly C T, editors. Microbial enzymes and biotechnology. 2nd ed. Weinheim, Germany: Verlag Chemie; 1990. pp. 177–226. [Google Scholar]
- 12.D’Anna J A, Jr, Tollin G. Studies of flavin-protein interaction in flavoproteins using protein fluorescence and circular dichroism. Biochemistry. 1972;11:1073–1080. doi: 10.1021/bi00756a020. [DOI] [PubMed] [Google Scholar]
- 13.Feather M S. A nuclear magnetic resonance study of the glucose oxidase reaction. Biochim Biophys Acta. 1970;220:127–128. doi: 10.1016/0005-2744(70)90237-8. [DOI] [PubMed] [Google Scholar]
- 14.Fischer B, Sumner I, Goodenough P. Isolation, renaturation and formation of disulfide bonds of eukaryotic proteins expressed in E. coli as inclusion bodies. Biotechnol Bioeng. 1993;41:3–13. doi: 10.1002/bit.260410103. [DOI] [PubMed] [Google Scholar]
- 15.Fraser D M, Zakeeruddin S M, Grätzel M. Mediation of glycosylated and partially-deglycosylated glucose oxidase of Aspergillus niger by a ferrocene-derivatised detergent. Biochim Biophys Acta. 1992;1099:91–101. [PubMed] [Google Scholar]
- 16.Frech C, Schmid F X. DsbA mediated disulfide bond formation and catalyzed prolyl isomerization in oxidative protein folding. J Biol Chem. 1995;270:5367–5374. doi: 10.1074/jbc.270.10.5367. [DOI] [PubMed] [Google Scholar]
- 17.Frederick K R, Tung J, Emerick R S, Masiarz F R, Chamberlain S H, Vasavada A, Rosenberg S, Chakraborty S, Schopfer L M, Massey V. Glucose oxidase from Aspergillus niger. J Biol Chem. 1990;265:3793–3802. [PubMed] [Google Scholar]
- 18.Fromm M, Taylor L P, Walbot V. Expression of genes transferred into monocot and dicot plant cells by electroporation. Proc Natl Acad Sci USA. 1985;82:5824–5828. doi: 10.1073/pnas.82.17.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghélis C, Yon J. Protein folding. New York, N.Y: Academic Press, Inc.; 1982. [Google Scholar]
- 20.Gibson Q H, Swoboda B E P, Massey V. Kinetics and mechanism of action of glucose oxidase. J Biol Chem. 1964;239:3927–3934. [PubMed] [Google Scholar]
- 21.Gilbert H F. The formation of native disulfide bonds. In: Pain R H, editor. Mechanisms of protein folding. Oxford, United Kingdom: Oxford University Press; 1994. pp. 104–136. [Google Scholar]
- 22.Hecht H-J, Kalisz H M, Hendle J, Schmid R D, Schomburg D. Crystal structure of glucose oxidase from Aspergillus niger refined at 2.3Å resolution. J Mol Biol. 1993;229:153–172. doi: 10.1006/jmbi.1993.1015. [DOI] [PubMed] [Google Scholar]
- 23.Heller A. Electrical connection of enzyme redox contacts to electrodes. J Phys Chem. 1992;96:3579–3587. [Google Scholar]
- 24.Hendle J, Hecht H-J, Kalisz H M, Schmid R D, Schomburg D. Crystallization and preliminary X-ray diffraction studies of a deglycosylated glucose oxidase from Penicillium amagasakiense. J Mol Biol. 1992;223:1167–1169. doi: 10.1016/0022-2836(92)90267-n. [DOI] [PubMed] [Google Scholar]
- 25.Hennessey J P, Jr, Johnson W C., Jr Information content in the circular dichroism of proteins. Biochemistry. 1981;20:1085–1094. doi: 10.1021/bi00508a007. [DOI] [PubMed] [Google Scholar]
- 26.Kalisz H M, Schmid R D. Separation of glucose oxidase isozymes from Penicillium amagasakiense by ion-exchange chromatography. ACS Symp Ser. 1993;529:102–111. [Google Scholar]
- 27.Kalisz H M, Hecht H-J, Schomburg D, Schmid R D. Crystallization and preliminary x-ray diffraction studies of a deglycosylated glucose oxidase from Aspergillus niger. J Mol Biol. 1990;213:207–209. doi: 10.1016/S0022-2836(05)80179-2. [DOI] [PubMed] [Google Scholar]
- 28.Kalisz H M, Hendle J, Schmid R D. Purification of the glycoprotein glucose oxidase from Penicillium amagasakiense. J Chromatogr. 1990;521:245–250. doi: 10.1016/0021-9673(90)85049-2. [DOI] [PubMed] [Google Scholar]
- 29.Kalisz H M, Hecht H-J, Schomburg D, Schmid R D. Effects of carbohydrate depletion on the structure, stability and activity of glucose oxidase from Aspergillus niger. Biochim Biophys Acta. 1991;1080:138–142. doi: 10.1016/0167-4838(91)90140-u. [DOI] [PubMed] [Google Scholar]
- 30.Kalisz H M, Hendle J, Schmid R D. Structural and biochemical properties of glycosylated and deglycosylated glucose oxidase from Penicillium amagasakiense. Appl Microbiol Biotechnol. 1997;47:502–507. doi: 10.1007/s002530050963. [DOI] [PubMed] [Google Scholar]
- 31.Kieß M, Hecht H-J, Kalisz H M. Glucose oxidase from Penicillium amagasakiense: primary structure and comparison with other glucose-methanol-choline (GMC) oxidoreductases. Eur J Biochem. 1998;252:90–99. doi: 10.1046/j.1432-1327.1998.2520090.x. [DOI] [PubMed] [Google Scholar]
- 32.Kohen A, Jonsson T, Klinman J P. Effects of protein glycosylation on catalysis: changes in hydrogen tunneling and enthalpy of activation in the glucose oxidase reaction. Biochemistry. 1997;36:2603–2611. doi: 10.1021/bi962492r. [DOI] [PubMed] [Google Scholar]
- 33.Kusai K, Sekuzu I, Hagihara B, Okunuki K, Yamauchi S, Nakai M. Crystallization of glucose oxidase from Penicillium amagasakiense. Biochim Biophys Acta. 1960;40:555–557. doi: 10.1016/0006-3002(60)91406-2. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Leatherbarrow R J. GraFit Version 3.0. Staines, United Kingdom: Erithacus Software; 1992. [Google Scholar]
- 36.Nakamura S, Ogura Y. Action mechanism of glucose oxidase of Aspergillus niger. J Biochem. 1968;63:308–316. [PubMed] [Google Scholar]
- 37.Provencher S W, Glöckner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 38.Raeder U, Broda P. Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol. 1985;1:17–20. [Google Scholar]
- 39.Richter G. Glucose oxidase. In: Godfrey T, Reichelt J, editors. Industrial enzymology. New York, N.Y: Macmillan; 1983. pp. 428–436. [Google Scholar]
- 40.Riklin A, Katz E, Willner I, Stocker A, Bückmann A F. Improving enzyme-electrode contacts by redox modification of cofactors. Nature. 1995;376:672–675. doi: 10.1038/376672a0. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 42.Scott D. Applications of glucose oxidase. In: Reed G, editor. Enzymes in food processing. 2nd ed. New York, N.Y: Academic Press, Inc.; 1975. pp. 519–547. [Google Scholar]
- 43.Sosnick T R, Mayne L, Hiller R, Englander S W. The barriers in protein folding. Nature Struct Biol. 1994;1:149–156. doi: 10.1038/nsb0394-149. [DOI] [PubMed] [Google Scholar]
- 44.Swoboda B E P. The relationship between molecular conformation and the binding of flavin-adenine dinucleotide in glucose oxidase. Biochim Biophys Acta. 1969;175:365–379. doi: 10.1016/0005-2795(69)90014-2. [DOI] [PubMed] [Google Scholar]
- 45.Swoboda B E P, Massey V. Purification and properties of the glucose oxidase from Aspergillus niger. J Biol Chem. 1965;240:2209–2215. [PubMed] [Google Scholar]
- 46.Tsuge H, Mitsuda H. Reconstitution of flavin-adenine dinucleotide in the apoenzyme of glucose oxidase. J Vitaminol. 1971;17:24–31. doi: 10.5925/jnsv1954.17.24. [DOI] [PubMed] [Google Scholar]
- 47.Wang F F C, Hirs C H W. Influences of the heterosaccharides in porcine pancreatic ribonuclease on the conformation and stability of the protein. J Biol Chem. 1977;252:8358–8364. [PubMed] [Google Scholar]
- 48.Whittington H, Kerry-Williams S, Bidgood K, Dodsworth N, Peberdy J, Dobson M, Hinchliffe E, Ballance D J. Expression of the Aspergillus niger glucose oxidase gene in A. niger, A. nidulans and Saccharomyces cerevisiae. Curr Genet. 1990;18:531–536. doi: 10.1007/BF00327024. [DOI] [PubMed] [Google Scholar]
- 49.Wilson R, Turner A P F. Glucose oxidase: an ideal enzyme. Biosens Bioelectron. 1992;7:165–185. [Google Scholar]
- 50.Witt S, Kalisz H M, Singh M. DNA sequence of the glucose oxidase gene from Penicillium amagasakiense. EMBL accession no. AF012277. 1997. [Google Scholar]