Abstract
The Sleeping Beauty (SB) transposon is an emerging tool for transgenesis, gene discovery, and therapeutic gene delivery in mammals. Here we studied 1,336 SB insertions in primary and cultured mammalian cells in order to better understand its target site preferences. We report that, although widely distributed, SB integration recurrently targets certain genomic regions and shows a small but significant bias toward genes and their upstream regulatory sequences. Compared to those of most integrating viruses, however, the regional preferences associated with SB-mediated integration were much less pronounced and were not significantly influenced by transcriptional activity. Insertions were also distinctly nonrandom with respect to intergenic sequences, including a strong bias toward microsatellite repeats, which are predominantly enriched in noncoding DNA. Although we detected a consensus sequence consistent with a twofold dyad symmetry at the target site, the most widely used sites did not match this consensus. In conjunction with an observed SB integration preference for bent DNA, these results suggest that physical properties may be the major determining factor in SB target site selection. These findings provide basic insights into the transposition process and reveal important distinctions between transposon- and virus-based integrating vectors.
Approximately half of the mammalian genome is derived from ancient transposable elements. Although the two general types of transposable elements, (DNA) transposons and retrotransposons, are often regarded as “selfish DNA parasites” or “junk DNA,” their frequent movement in and out of host cell chromosomes has played a significant role in genome diversification and evolution. Members of the Tc1/mariner family of DNA transposons are extremely widespread in nature (44) and can function independently of species-specific host factors (29, 56). Although the vast majority of elements present in vertebrate genomes are nonfunctional (14, 32), an active Tc1-like element called Sleeping Beauty (SB) was recently reconstructed from ancient transposon fossils found within fish genomes (20).
SB elements transpose by a cut-and-paste mechanism that requires the sequence-specific binding of the SB transposase to the transposon ends (25). This transposition process involves the precise excision and reintegration of the transposon from one DNA site to another site, which invariably contains a TA dinucleotide that is duplicated upon insertion. The transfer of DNA strands at the insertion site is mediated by the transposase catalytic core domain, which contains a conserved DDE motif shared by a large group of recombinase proteins, including the V(D)J recombinase and retrovirus integrases (44). SB is capable of efficient transposition in a variety of cell types (24), including human, mouse, and fish cells, and is an emerging tool for genetic research on vertebrates, with potential applications for transgenesis (22), functional genomics (1, 5-9, 11, 12, 16, 18, 19, 21, 33), and human gene therapy (2, 15, 23, 31, 34, 37, 42, 43, 59, 61).
The vast majority of transposable elements do not integrate randomly but display some level of target DNA selectivity. Many factors can substantially influence the spectrum of target sites utilized during genomic integration. In some cases, integration can be heavily skewed toward a particular DNA site, often called a hot spot, because it contains a specific nucleotide sequence that is preferred by integration complexes. In addition, DNA-binding proteins bound to target DNA can also effect target site selection, either negatively, by obstructing the access of integration complexes to target DNA (4, 47), or positively, by promoting structural changes in the target DNA. For instance, DNA bending proteins such as nucleosomes can induce severe target DNA distortions that can bias retroviral integration into these sites (45-47), possibly because these conformational changes enable better recognition by the integration complex. Alternatively, studies of integration targeting by certain yeast transposons (Ty elements in Saccharomyces cerevisiae retrotransposons and Tf elements in Schizosaccharomyces pombe) indicate that target site selection can also be dramatically influenced by physical interactions between locally bound transcription factors and components of the integration complex (49, 63). Finally, although there are some conflicting data (57), most recent reports indicate that that the vast majority of viruses preferentially target integration into active genes (17, 27, 36, 38, 39, 58), suggesting that transcriptional activity can also greatly influence insertion site selection in mammalian cells.
Previous studies of SB transposition have noted a preference for additional nucleotides flanking the TA insertion site (5, 55) and a strong tendency for SB to jump locally into target sites residing on the same chromosome as the donor site (5, 11, 19, 33). Unfortunately, this local transposition phenomenon, which also occurs with the P element in flies (54), introduces strong contextual biases that can significantly complicate studies of SB integration targeting within a host cell genome. To overcome these effects, Vigdal et al. recently studied transposition using a plasmid-based approach and found that SB and other Tc1/mariner transposons share a common preference for insertion sites with a bendable structure (55). However, their study utilized only a limited number of zeocin-selected integration events isolated from a single cell type (HeLa cells) and unfortunately did not rigorously address the mutagenic potential of SB in mammalian cells. Considering the recent adverse events observed with two patients undergoing retrovirus-based gene therapy (13), we believed that it was important to more fully investigate the targeting of SB integration in order to better evaluate the potential for SB-mediated gene transfer in vertebrates.
Here we report a large-scale, genome-wide analysis of SB transposon integration in mammalian cells. We isolated DNA from >1,300 independent SB-mediated integrations in human and mouse cells and then mapped them to their respective genomes. We report that SB insertions are nonuniformly distributed with respect to genes (mostly introns), their upstream regulatory sequences, and numerous repetitive sequence elements. In contrast to most integrating virus-based vectors, however, microarray analyses revealed no correlation between SB integration targeting and transcriptional activity, suggesting that SB might be a safer vector for therapeutic gene delivery. Finally, base composition analysis of SB insertion sites suggests that physical attributes of the target site, such as an inherent distortion of the DNA, rather than sequence-specific preferences, may be the major targeting determinant involved in SB insertion site selection.
MATERIALS AND METHODS
Plasmid construction.
To facilitate the high-throughput analyses described here, we made several modifications to the pT/nori vector described previously (61). First, we removed a unique XbaI site in the plasmid backbone sequence by sequentially treating XbaI-digested pT/nori with T4 DNA polymerase and T4 DNA ligase. We then removed the pUC19 origin of replication from this vector and introduced a unique PmeI restriction enzyme site ∼100 bp outside of the ampicillin gene. This was done by amplifying the ampicillin gene from pUC19 by PCR using primers Amp-1 (5′-GAAAGGGCCTCGTGATACGCCTAT) and Amp-PS (5′-AGTAGCTGGAAGAGCGTTTAAACACTTGGTCTGACAGTTACCAATGC), digesting the PCR product with the AatII and SapI restriction endonucleases, and then ligating it with a 4.1-kb AatII-SapI fragment from pT/nori-ΔXba. Last, we replaced the low-copy-number p15A bacterial origin of replication within the transposon with a 633-bp PCR fragment corresponding to the pUC19 origin of replication by SacII-BstB1 ligation. The resulting vector, called pT/nori-2, contains a unique PmeI site in the plasmid backbone sequence, is resistant to XbaI, NheI, and SpeI restriction enzyme digestion, is amenable to high-copy propagation in bacteria, and supports high-frequency transposition in NIH 3T3 and Huh-7 cells. The pc-SB10 plasmid, used as a source of transposase, has been described previously (60).
Animal studies.
We obtained 6-week-old female C57BL/6 mice from Jackson Laboratory and treated them according to the National Institutes of Health (NIH) Guidelines for Animal Care and the guidelines of Stanford University. We induced transposition in primary mouse hepatocytes by injecting three mice each via the tail vein with 25 μg of the pT/nori-2 vector and 1 μg of the pc-SB10 vector as described previously (61). Mice were euthanized 2 days later under anesthetic, and equal portions of their livers were removed and combined for subsequent isolation of total hepatic DNA.
Cell culture and stable transfections.
We obtained NIH 3T3 mouse fibroblast and Huh-7 human hepatoma cell lines from the American Type Culture Collection. We transfected 5 × 105 cells of each cell line on 6-cm-diameter plates with 1.5 μg of pT/nori-2 and 1.5 μg of pc-SB10 by using Superfect (QIAGEN). For each cell type, we performed multiple transfections to generate independent SB integration libraries. Cells from each transfection were trypsinized 2 days later, diluted onto multiple 10-cm-diameter dishes containing Dulbecco's modified Eagle medium with 600 μg of G418/ml, and selected for growth over a period of 2 weeks. At this time point, the remaining cells were harvested and used to isolate total DNA for recovery of integrated transposon sequences.
Generation of SB integration libraries.
We treated 10 μg of total DNA isolated from mouse liver, NIH 3T3 cells, and Huh-7 cells with PmeI and calf intestinal alkaline phosphatase to minimize recovery of the parental plasmid. We then digested samples with the NheI, SpeI, and XbaI restriction endonucleases, each of which does not cleave within the pT/nori-2 vector but cuts mouse genomic DNA flanking integrated SB transposons. We self-ligated the digested DNA with T4 DNA ligase, transformed 5-μg aliquots of ligated products into ElectroMAX DH10B Escherichia coli (Invitrogen), and selected for kanamycin-resistant (Kanr) growth. Individual Kanr colonies were patched onto Luria-Bertani plates containing either ampicillin (100 μg/ml) or kanamycin (30 μg/ml) and then screened for resistance to ampicillin in order to identify clones representing the parental pT/nori-2 plasmid. In total, we constructed seven sets of NIH 3T3 libraries from seven independent transfections, four sets of Huh-7 libraries from four independent transfections, and a single mouse liver library from three injected mice.
High-throughput analysis of SB integration events.
We isolated plasmid DNA from individual E. coli colonies by using a 96-well format. Each plasmid was digested with HindIII and screened by ethidium gel electrophoresis for bona fide SB transposon plasmids, which had to contain a full-length (3.6-kb) transposon copy to qualify for further analysis. We sequenced plasmids by using primer IR-1 (5′-AGATGTCCTAACTGACTTGCC), which anneals to the 5′ end of the transposon, and obtained ∼1 kb of high-quality DNA sequence in our reads.
Integration site mapping.
We used the BLAT program to map sequences to their relevant genomes (University of California at Santa Cruz [UCSC] Human and Mouse Genome Project working drafts, July and October 2003 freezes, respectively). Insertion sites were considered authentic only if each (i) contained sequence from the nested primer to the end of the 5′ inverted-repeat-direct-repeat (IR/DR) (CTG) sequence, (ii) matched a genomic location starting immediately after the end of the 5′ IR/DR (CTG), (iii) showed ≥95% identity to the genomic sequence over the high-quality sequence region (>100 bp), and (iv) matched no more than one genomic locus with ≥95% identity. We sequenced 892 clones from the mouse liver library, 506 clones from the NIH 3T3 libraries, and 480 clones from the Huh-7 libraries. In total, 1,336 sequences met all the criteria described above and could be mapped to a unique genomic locus. The remaining sequences either were too short to map to any location, were identical to the parental pT/nori-2 plasmid, were consistent with interplasmid transposition events, were duplicate clones recovered from the same integration library, or mapped to multiple locations in the genome. Although interplasmid transposition events were rare, we did recover a total of 41 such events from mouse liver. Recovery of these events was aided in part by the long-term persistence of extrachromosomal target plasmids in the liver following in vivo vector administration and by the inactivation of the plasmid-encoded Ampr gene in these clones, which enabled their recovery in our genetic screen. Inactivation of the Ampr gene appears to have occurred either directly, via transposition into these sequences, or by complete and/or partial loss of the Ampr gene, such as through intraplasmid recombination and/or inadvertent cleavage of the plasmid backbone (i.e., “star activity”) during restriction enzyme treatments. Ultimately, we were able to map 590 integration events from the mouse liver library, 380 integration events from the NIH 3T3 libraries, and 366 integration events from the Huh-7 libraries. Two NIH 3T3 clones (clones 238 and 501) mapped to the identical chromosomal coordinate in the mouse genome but were not considered duplicates because they were recovered from two independent integration libraries. In addition, ∼19% of the total clones analyzed (255 out of 1,336) contained less than 800 bp of total genomic DNA sequence, which enabled analysis of the 3′ end of the element in this subset of clones. These sequences were compared to those contained in the mouse and human genome databases in order to screen for potential chromosomal rearrangements (e.g., translocations, deletions, or insertions) at the insertion site.
Bioinformatics.
We downloaded the coordinates of RefSeq genes, CpG islands, and other annotation tables for the July 2003 human and October 2003 mouse genome freezes from the UCSC genome project website (www.genome.ucsc.edu). We defined an integration as having landed in a gene only if it was between the transcriptional start and transcriptional stop boundaries of one of the 22,753 or 17,958 RefSeq genes mapped to the human or mouse genome, respectively. We determined the AT content for SB integrations in each cell type over a varying window size and analyzed the base composition over a 60-bp region encompassing the target TA dinucleotide. We also analyzed integrations in relation to various genomic repeat elements and in various-size windows around transcriptional start sites, transcriptional stop sites, and CpG islands. In every case, we compared the distribution and content of SB integration sites to those of a set of 10,000 computer-generated random coordinates in order to determine the level of statistical significance.
We assessed the transcriptional activity of each SB-targeted gene by using a publicly available, Web-based microarray gene expression database as previously described (58). We used mouse liver expression databases GSM4659, -4661, and -4669 in the Gene Expression Omnibus (GEO) data repository and GNF Gene Expression Atlas 2 from the Genomics Institute of the Novartis Research Foundation. For the GEO data sets, we filtered all spots by criteria (more than twice the standard deviation of the background, not saturated or irregular) and obtained 9,527 spots, of which 2,272 could be linked to RefSeq genes and used as references for statistical analysis. A total of 23 SB integrations into RefSeq genes were available for the analysis. In the GNF data set, expression data from 169 RefSeq genes that had SB integration within genes and from 56 RefSeq genes that had SB integration within ±5 kb of transcriptional start sites were available.
We analyzed gene expression data for SB-targeted genes in two different ways. First, we compared the expression level of each SB-targeted gene with either the median expression level from all the 61 tissues analyzed (52) or the value from universal control RNA (GEO database). Second, we compared the expression level of each SB-targeted gene with the median expression value from all the genes analyzed in the liver.
Statistical analyses.
We investigated the bias for or against preferred integration into genomic repeats, RefSeq genes, and transcriptional start sites, and in or near CpG islands, by comparing the observed frequency with that from 10,000 random computer-simulated integrations, and we assessed the statistical significance of the bias by using a χ2 test. We determined the probability of any gene of given length being hit at random among all the genes within the human or mouse genome, and we then used this value to calculate P values for each gene of equivalent size being hit n number of times by using a binomial distribution test. For example, the probability of hitting a 28-kb gene such as Ma2a8 at random among all mouse genes (total mRNA size, 7.42 × 108 bases) is equal to 3.77 × 10−5. Using this value in a binomial distribution test, we calculated the probability that Ma2a8 would be not be hit in a total of 380 integration events [P(0) = 0.986], or would be hit once [P(1) = 0.0142] or twice [P(2) = 0.0001], and then subtracted the sum of these values from 1 to obtain the P value for Ma2a8 being hit 3 times out of 380 events in a random-integration model (i.e., P = 0.0003).
To analyze the transcriptional status of SB-targeted genes in the liver, we compared the median expression levels for all SB-targeted genes and those ±5 kb from transcriptional start sites with those of all 13,890 genes on the array by using a nonparametric Mann-Whitney test. To confirm these results, we also analyzed the median expression level of SB-targeted genes by the methods of Schröder et al. as an independent assessment of the data (50). Briefly, we distributed the 13,890 genes on the array into eight equal “bins” by relative expression levels and distributed the 169 genes used as integration targets into these bins based on their expression levels. We then summed each bin and tested for statistical bias using a χ2 test by comparing the observed frequencies to the value that would be expected if one-eighth of all genes analyzed were placed in each bin.
RESULTS
Isolation and mapping of SB integration events.
To better understand transposon target site selection in vertebrate cells, we studied SB transposition in different mammalian cell types using both selective and nonselective conditions. To do this, we transiently transfected female C57BL/6 mouse liver tissue, and stably transfected mouse fibroblast (NIH 3T3) and human hepatoma (Huh-7) cell lines, by using plasmids encoding the SB transposase (pCMV-SB10) and a neomycin-marked SB element (pT/nori-2) that confers G418 drug-resistant growth in mammalian cells and kanamycin resistance in E. coli. For each of the two cell lines, we also performed multiple independent transfections in order to search for potential hot spots for SB insertion in mammalian cells. We then generated transposon integration libraries from each cell type by using a high-throughput plasmid recovery strategy that enriches for integration events via genetic screening in E. coli (Fig. 1). Using this general strategy, we identified a total of 1,877 plasmid clones, each of which corresponds to a potential de novo transposition event. Novel flanking sequences were then identified and mapped within their respective genome by using the BLAT program. Of these 1,877 plasmids, 551 were discarded from consideration because they contained sequences that either were too short to map to any location, were identical to the parental pT/nori-2 plasmid, were duplicate clones recovered from the same integration library, or mapped to multiple locations in the genome. Ultimately, we were able to unambiguously map a total of 1,336 different transposon integrations to unique locations in the mouse and human genomes (Table 1). These integration events were then studied in relation to various host chromosomal features and were compared to 10,000 computer-simulated random integrations used as a control.
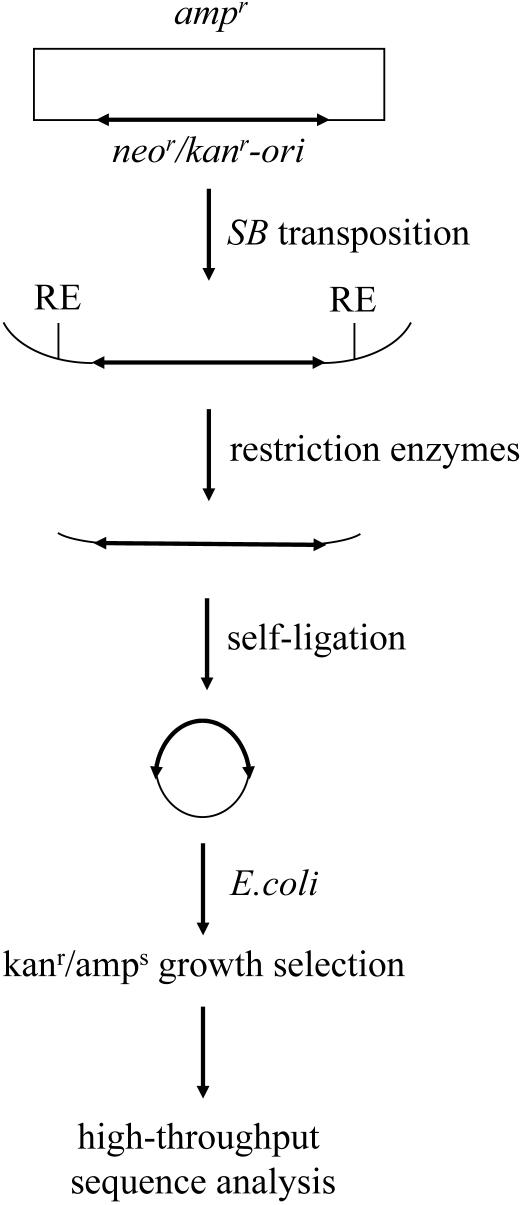
FIG. 1.
Plasmid rescue strategy to isolate transposon insertion sites. SB integration was initiated by cotransfecting different mammalian cells with plasmids encoding the SB transposase and an SB transposon (thick arrow) containing a bacterial origin of replication (ori) and sequences to permit Neor and Kanr growth. In some cases, transfected cells were selected in the antibiotic G418 for stable transposon expression. The genomic sequences flanking integrated elements were recovered by cutting genomic DNA with three compatible restriction enzymes (RE) to minimize the potential for restriction site bias, followed by religation with T4 ligase and transformation into E. coli. Bacteria were selected for Kanr Amps growth and then amplified by using a 96-well format to isolate plasmid DNA. The DNA flanking the recovered transposons was determined by sequence analysis using primers that anneal to the transposon ends and was mapped to its respective genome by using the BLAT program.
TABLE 1.
SB integration libraries
| Source | No. of libraries | Nature of sites | No. of unique sites |
|---|---|---|---|
| Mouse liver | 1 (from 3 mice) | Genomic, unselected | 590 |
| Plasmid, unselected | 21 | ||
| NIH 3T3 cells | 7 | Genomic, selected | 380 |
| Huh-7 cells | 4 | Genomic, selected | 366 |
Chromosomal distribution of transposon insertions.
We investigated the chromosomal distribution pattern for SB integrations by comparing the density of computer-simulated integrations on each mouse chromosome with that of transposon integrations recovered from primary mouse liver tissue, which should contain a normal karyotype. In each of the cell types we studied, there was no Y chromosome present and thus no hits on the Y chromosome were recorded. Although some chromosomes seemed to be somewhat preferred targets for SB integration (e.g., chromosomes 4 and 16), and others appeared to be disfavored (e.g., chromosomes 7, 15, and X), statistical analyses showed that SB insertions were distributed evenly at the chromosomal level (Fig. 2A). Therefore, plasmid-based transposition is significantly more random than chromosomal SB transposition, which has been shown to be heavily biased toward local hopping into closely linked loci (5, 11, 19, 33). In addition, when all SB integrations were mapped in relation to one another, there was no evidence for significant clustering of SB integrations in the mouse genome (Fig. 2B). Indeed, no more than three pairs of transposon integrations mapped within 10 kb of one another in each experimental group. Finally, although there appeared to be potential cold spots for SB integration in the mouse genome, such as the proximal part of chromosome 15 or the central portion of the X chromosome, similar distributional gaps were observed upon three independent control mappings of an equivalent number of random insertions (data not shown). Thus, it is likely that disfavored integration targets cannot be adequately addressed at the chromosomal level, at least at the resolution of 970 events.
FIG. 2.
Genome-wide distribution of SB integrations. (A) Distribution of integration events at the chromosome level. Only insertions isolated from unselected mouse liver tissue (n = 590) were analyzed, because these cells contain a normal karyotype. The distribution of SB integration events was compared to that of 10,000 computer-simulated random integrations to test for statistical significance. (B) SB insertion site mapping in the mouse genome. The relative positions of 970 total independent integration sites for SB (liver plus NIH 3T3 cells) within the mouse genome are shown.
Transposition into extrachromosomal target sequences.
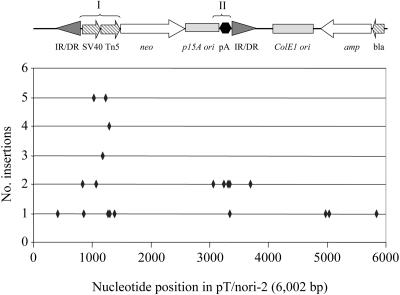
In addition to the 1,336 incidences of genomic integration, we also identified 41 transposition events in our unselected mouse liver library that localized to a new site in the pT/nori-2 target plasmid. These insertion sites were evaluated in greater detail and showed that, although the target plasmid contained a total of 269 potential TA target sites, only 21 were hit by transposons (Fig. 3). In addition, multiple insertions were observed at many of these sites, including two sites, TA1030 and TA1229, each of which was targeted five times. Furthermore, a total of 35 insertion sites (85% of the total) mapped to two small regions of the target plasmid, arbitrarily designated regions I and II, even though these regions together comprised only ∼14% of the plasmid. Region I (nucleotides 834 to 1384) corresponds to the simian virus 40 and Tn5 promoters, whereas region II (nucleotides 3065 to 3346) is located within the polyadenylation signal region.
FIG. 3.
Distribution of SB transposon insertions in the pT/nori-2 target plasmid. The number of insertions per TA dinucleotide is shown relative to the nucleotide map of the target plasmid. Locations of the functional domains of the plasmid, as well as the two preferred regions (I and II), are shown. Arrows indicate promoters and genes, shaded triangles represent transposon inverted repeats, and the polyadenylation signal region is shown in black.
Transposon insertion into intragenic regions.
We studied transposon insertions in relation to transcriptional units to determine the mutagenic potential associated with SB integration under various experimental conditions. Of the 366 transposon integrations isolated from Huh-7 cells, 143 (39%) mapped within at least 1 of the 21,804 human RefSeq genes, a frequency significantly higher than that observed for 10,000 computer-simulated random integrations (33%) (P = 0.02 by the χ2 test) (Table 2). This SB-RefSeq integration value was virtually identical to the 38% gene-targeting frequency recently reported for the avian sarcoma-leukosis virus (ASLV) (36) but significantly lower than those previously reported for integration by the avian sarcoma virus (ASV), murine leukemia virus (MLV), and human immunodeficiency virus type 1 (HIV-1) (41, 50, 58). Moreover, 184 of the 590 integrations in mouse liver (31%) and 125 of the 380 insertions in NIH 3T3 cells (33%) mapped within at least 1 of the 18,090 mouse RefSeq genes. When compared to the expected frequency of 26% for random integration, these data reveal an even higher statistical preference for SB insertion into mouse genes (P = 8 × 10−5 for combined data sets by the χ2 test) (Table 3), suggesting that SB target site selection may differ somewhat between species. Even so, however, the frequency at which mouse RefSeq genes were targeted during SB transposition was much lower than that recently reported for adeno-associated virus (AAV) (39), suggesting that SB integration targets mammalian genes much less frequently than most commonly used integrating viral vectors.
TABLE 2.
Frequencies of SB integration events within intragenic regions of human cellsa
| Genomic location | % of integrationsb
|
|||||
|---|---|---|---|---|---|---|
| Random | SBc | ASVd | MLVde | HIV-1df | ASLVg | |
| In RefSeq genes | 33.2 | 39.1h | 53.0h | 50.7h | 83.4h | 38.2h |
| 5 kb upstream of genes | 2.9 | 3.8 | 5.1h | 13.0h | 3.4 | ND |
| ±5 kb from transcription start site | 5.4 | 8.5h | 8.9h | 21.4h | 11.4h | ND |
| ±5 kb from CpG islands | 8.3 | 11.2h | ND | ND | ND | ND |
| ±1 kb from CpG islands | 1.9 | 2.5 | 2.8 | 15.2h | 1.9 | 3.2 |
Compared with integration frequencies of ASV, MLV, HIV-1, and ASLV. Results from the present study are boldfaced.
Values shown for ASV, MLV, HIV-1, and ASLV represent the expected integration frequencies for each vector after the values originally reported were normalized according to the random integration frequencies shown. ND, not determined.
SB integrations were compared to 10,000 computer-simulated random integrations and analyzed by using a χ2 test for statistical significance.
Adjusted values from the work of Narezkina et al. (41).
Adjusted values from the work of Wu et al. (58).
Adjusted values from the work of Schröder et al. (50).
Adjusted values from the work of Mitchell et al. (36).
Values are distinguishable from those for random integration (P ≤ 0.03).
TABLE 3.
Frequencies of SB integration events within intragenic regions of mouse cellsa
| Genomic location | % of integrationsb
|
||||
|---|---|---|---|---|---|
| Random |
SB
|
AAVf | |||
| In mouse liverc | In NIH 3T3 cellsd | Alle | |||
| In RefSeq genes | 26.0 | 31.2 | 32.9 | 31.9 | 53.1 |
| 5 kb upstream of genes | 3.2 | 5.1 | 6.1 | 5.5 | 25.8 |
| ±5 kb from transcription start site | 6.4 | 10.5 | 9.2 | 10.0 | 43.9 |
| ±5 kb from CpG islands | 6.4 | 9.5 | 8.7 | 9.2 | 49.3 |
Compared with frequencies of AAV integrations in a recent study.
Integrations were compared to 10,000 computer-simulated random integrations and were analyzed by using a χ2 test for statistical significance.
All values in this group are distinguishable from random integration (P ≤ 0.01).
All values in this group (except for ±5 kb from CpG islands [P = 0.07]) are distinguishable from random integration (P ≤ 0.03).
The mouse liver and NIH 3T3 data sets were combined. All values in this group are distinguishable from random integration (P < 0.001).
From the work of Nakai et al. (39). All values in this group are distinguishable from random integration (P < 0.001).
Among the 452 total integrations that occurred in genes, which were distributed evenly along the transcript, 435 (96%) mapped within intron sequences. Since an identical frequency of hitting introns was observed in the random control group, the bias toward introns is probably due to the fact that they are much larger than most exons, thereby presenting a much larger target into which a transposon can integrate. Among the 17 remaining transposon insertions that mapped within genes, 9 were found in exons and 8 mapped to the 3′ untranslated region (3′ UTR) (data not shown). Surprisingly, all seven of the 3′ UTR insertions found in mouse liver were in the same orientation as the target gene. This was vastly different from the completely random orientation of inserts observed for SB in introns and exons, and differed from the single 3′ UTR insertion isolated from NIH 3T3 cells, which was in the opposite orientation from the gene. Further studies will be needed to determine whether the orientational bias observed in mouse liver genes is significant or not.
Based on the regional preferences noted in our interplasmid group (Fig. 3), we also investigated whether SB integration showed any bias toward important regulatory sequences, as recently reported for recombinant MLV- and AAV-based vectors (36, 38, 39, 58). Compared to the frequency predicted from random integrations, SB showed an average 1.8-fold-higher tendency to insert into a 5-kb region upstream of mouse RefSeq genes. In addition, integrations were also biased toward the 10-kb region encompassing known CpG islands and the transcriptional start sites of mouse RefSeq genes (Table 3). In general, these regional preferences were much less pronounced in human cells (Table 2), thus further suggesting that SB target site selection may differ for different cell types. Furthermore, while there were slight variations in the actual degree of integration bias observed among selected and unselected populations of mouse cells, the overall preference observed in each case was essentially the same. Therefore, the use of experimental selective-enrichment protocols does not appear to greatly skew SB insertion site selection in mammalian cells.
Although there was no obvious bias toward integration into a particular functional gene category, we found eight genes (Dmd, Gfra1, 3632451006Rik, Stk10, DLEC1, C14orf127, C20orf44, and Ma2a8), ranging from 28 to 431 kb, that were recurrently targeted by SB (the average sizes of 18,090 mouse and 21,804 human RefSeq genes are 41 and 58 kb, respectively). The Ma2a8 gene was hit a total of three times in NIH 3T3 cells, whereas each of the other seven genes was targeted twice in one of the cell types examined. We calculated the probability of hitting a similarly sized gene among all the RefSeq genes in the human and mouse genomes and then determined the statistical significance of each targeted gene being hit n number of times by using a binomial distribution. Results showed a highly significant bias toward each of the eight genes (0.0003 < P < 0.04), with the smallest of these genes, Ma2a8 (28 kb), showing the most integrations (n = 3) in NIH 3T3 cells. Remarkably, two of the three Ma2a8 insertions, represented by clones 5-238 and 7-501, mapped to the same target TA dinucleotide. Importantly, these two events were isolated from two separate SB integration libraries and thus must represent independent events. These data suggest that SB integration may exhibit cell type-specific biases toward a subset of genes and/or genomic regions.
Transcriptional status of targeted genes.
We tested whether SB's preference for intragenic regions could be explained in part by transcriptional activation or repression of these genomic loci. To do this, we analyzed the transcriptional status of SB-targeted genes within the mouse liver data set by using multiple sources from publicly available microarray databases. Comparisons of the median expression signals of the 169 SB-targeted genes and the 56 genes targeted within ±5 kb of their transcriptional start sites showed no significant difference from those of the 13,890 RefSeq genes available in the GNF Gene Expression Atlas 2 database (average difference values of 177 and 142 versus 184 for all genes; P > 0.62 by the Mann-Whitney test). In addition, analysis of three independent sources present in the GEO data repository revealed that the transcriptional activity of SB-targeted genes was not significantly different from that of the entire population (0.06 < P < 0.44 by the Mann-Whitney test). Taken together, these data suggest that, in sharp contrast to the vast majority of integrating viral vectors (17, 27, 36, 38, 58), SB integration does not preferentially target actively transcribed genes.
Integration into endogenous repeat elements.
We also analyzed the distribution of SB insertions with respect to various genomic repeat elements and detected additional unforeseen integration biases (Table 4). Most obviously, in all three cell types, SB integration targeted short (2- to 6-bp) TA-containing microsatellite repeats with a 10-fold-higher frequency than random integration (P < 0.0001 by the χ2 test). It is not clear at present whether these biases are caused solely by the presence of multiple TA target dinucleotides in these arrays, or whether additional factors inherent in these sequences also contribute to their frequent usage. In addition to these preferences, our results indicate that SB integration avoids certain types of long terminal repeat (LTR) elements and shows particular disfavor for mouse LINE-L1 repeats, even though the latter are the most abundant repeat element in the mouse genome. A similar trend against both classes of repeat elements was also observed in human Huh-7 cells, but the overall bias was generally weaker than in primary and cultured mouse cells. Such observed differences could theoretically be due in part to large differences in the copy number and/or global distribution of each class of repetitive element either in different species or in different cell types. In addition, SB integration in human cells showed a marked preference for MIR-type SINE elements but was also 10-fold less likely than random integration to target human Alu repeats (P < 10−8). The latter tendency contrasts sharply with the profound insertional bias HIV-1 shows for Alu repeats in human SupT1 cells (50) and could indicate that the preferred targeting sequence for SB is particularly rare in these elements.
TABLE 4.
SB and random integration frequencies into genomic repeat elements
| Targeted regionb | % of integrations (P values)a
|
||||
|---|---|---|---|---|---|
| Mouse
|
Human
|
||||
| Random | Liver | NIH 3T3 cells | Random | Huh-7 cells | |
| All genomic repeats | 43.1 | 37.3 (0.009) | 36.8 (0.02) | 48.7 | 41.0 (0.005) |
| DNA element | 0.8 | 1.2 (0.35) | 1.8 (0.04) | 3.4 | 3.6 (0.91) |
| LINE (L1) | 20.1 | 11.2 (<0.0001) | 7.6 (<0.0001) | 16.7 | 13.1 (0.08) |
| LTRc | 8.7 | 5.3 (0.005) | 3.4 (0.0004) | 3.7 | 1.6 (0.04) |
| SINE | |||||
| Alu | 2.1 | 2.0 (0.95) | 2.1 (0.96) | 11.1 | 1.6 (<0.0001) |
| MIR | 2.3 | 1.0 (0.26) | 4.7 (0.66) | 2.6 | 6.3 (0.0001) |
| Microsatellited | 0.5 | 5.4 (<0.0001) | 7.1 (<0.0001) | 0.2 | 2.5 (<0.0001) |
SB integrations were compared to 10,000 computer-simulated random integrations and were analyzed by using a χ2 test. A P value of <0.05 indicates a significant difference between SB integration into the indicated cell type and random integration.
LINE, long interspersed nuclear element; SINE, short interspersed nuclear element; MIR, mammalian interspersed repeat.
Endogenous retrovirus-K and mammalian LTR retrotransposon types only (others showed no deviation from random integration).
Only microsatellites containing at least one TA dinucleotide were considered.
Insertion site sequence preferences.
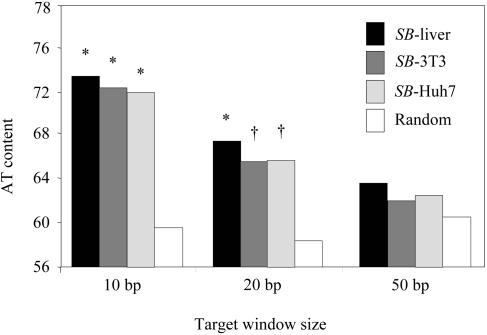
We analyzed the overall AT content of transposon insertion sites and compared it to that of the control group. For each of the three cell types we studied, SB integration showed a strong preference for localized regions of higher AT content, especially over a 10-bp window (Fig. 4). Based on these results, we aligned all the junction sequences and compared them to that of the control group to determine if these local preferences could be attributed to a consensus targeting sequence for the transposase. Although no nucleotides other than the target TA dinucleotide were strictly required, the nucleotides at ±3 and ±1 bp relative to the target TA were approximately 70 and 48% conserved, respectively. In addition, we detected a 14-bp consensus containing a symmetric palindromic core sequence [5′-RCAYA(TA)TRTGY-3′] centered at the insertion site (Table 5), which is consistent with previously reported base pair preferences for both SB (5, 55) and Tc1 (28). Nevertheless, when we examined the most frequently used sites from the plasmid and NIH 3T3 groups, we found that most of the preferred sites showed only limited homology with the consensus sequence (Table 6). For instance, one of the most frequently used sites identified in our interplasmid transposition group, TA1229, had the flanking sequence TGGGC(TA)TCTGG, which had only 3 nucleotides out of 10 in common with the consensus sequence. These results suggest that the primary sequence, although important, may not be the most significant determinant in SB target site selection.
FIG. 4.
Genomic AT contents of SB integrations in three different cell types. The total AT contents of SB insertion sites were analyzed by using variable window sizes and were compared to those of 10,000 random integrations. Significant differences were determined by a χ2 test. *, P < 0.0001; †, P < 0.02.
TABLE 5.
Target site analysis for 1,309 unique genomic SB insertionsa
| Nucleotide | No. of insertion sites with the indicated nucleotide at position:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −5 | −4 | −3 | −2 | −1 | T | A | +1 | +2 | +3 | +4 | +5 | +6 | +15 | |
| A | 461 | 284 | 904 | 214 | 615 | 0 | 1,309 | 194 | 553 | 228 | 426 | 288 | 463 | 379 |
| C | 230 | 350 | 68 | 327 | 226 | 0 | 0 | 257 | 230 | 111 | 260 | 344 | 223 | 240 |
| G | 337 | 233 | 120 | 227 | 247 | 0 | 0 | 230 | 325 | 51 | 341 | 214 | 262 | 225 |
| T | 282 | 442 | 217 | 541 | 221 | 1,309 | 0 | 628 | 201 | 919 | 282 | 463 | 361 | 465 |
| Consensusb | R | C | A | Y | A | T | A | T | R | T | G | Y | A | T |
The base composition of the 60-bp region encompassing the TA target dinucleotide (underlined) was determined and compared to that of 10,000 randomly selected genomic coordinates. The random nucleotide frequencies at each position were identical to the genome average: 30% for A, 20% for C, 20% for G, and 30% for T. Only positions at which significant deviation between the two groups was observed (P < 0.01 by the χ2 test) are shown.
R stands for A or G; Y stands for C or T.
TABLE 6.
Alignment of core consensus with preferred target sites
| Integration site(s) | Proportion of total integrations | Sequencea at:
|
||
|---|---|---|---|---|
| Left end | TA | Right end | ||
| pTA1030 | 5/41 | ttATt | TA | TGcag |
| pTA1229 | 5/41 | tgggc | TA | TcTGg |
| pTA1286 | 4/41 | GggCt | TA | cATGg |
| pTA1173 | 3/41 | ACACg | TA | gAaag |
| NIH 3T3 clones 238 and 501 | 2/380 | GttTA | TA | TATcC |
| Consensus | RCAYA | TA | TRTGY | |
Capitalized nucleotides match the consensus.
DISCUSSION
In this work, we studied transposon target site selection in mammalian cells by characterizing more than 1,300 de novo SB transposon insertions. This is the largest collection of somatic transposition events ever described for a Tc1/mariner transposon member in mammals. In the process, we have found that although widely distributed, SB integration is inherently nonrandom, showing clear target site preferences both for and against many different chromosomal features.
Most significantly, SB integration showed a statistical bias toward transcriptional units and their upstream regulatory sequences irrespective of whether insertions were selected for or not. Transposon insertion also occurred recurrently in a subset of genes in both selected and unselected mammalian cells, a finding that further suggests some level of selectivity in SB integration. Even so, the overall tendency for SB to target these intragenic regions in human cells is still relatively weak compared to that of other integrating vector systems (Table 2) (17, 27, 36, 38, 41, 58), which could explain why similar preferences were not noted previously in two much smaller studies of SB target site selection (5, 55). Interestingly, the P element has also been shown to favor the 5′ ends of transcriptional units in flies, although its integration is biased predominantly toward the 5′ UTR (51), which was not the case for SB. In addition, although recombinant MLV-based vectors also favor the transcriptional start sites of genes, their regional bias is much stronger than that seen with SB, appearing to be restricted to a much narrower, 1-kb window around the start site (58). Therefore, despite a common integration bias near the 5′ ends of genes, each of these integrating elements appears to target distinct chromosomal regions.
Our results further demonstrate that SB elements frequently target microsatellite DNA during genomic integration. These highly abundant simple sequence repeats reside predominantly in noncoding DNA (10), and their expansion during replication slippage can cause repeat instability, increased recombination rates, and numerous human diseases (10). Recent work with worms indicates that Tc1 transposons also accumulate preferentially in regions with high recombination rates (48), suggesting that these transposable elements may utilize the recombination machinery during genomic integration. Furthermore, the finding that Micropan-4 transposons are enriched within (AT)n microsatellites in rice (53) suggests that these and other transposable elements may have evolved integration mechanisms to target these chromosomal regions. These preferences may have been selected for in nature, not only because these repeats are both nonessential and abundant but also because their disruption would likely enhance the overall stability of these large arrays, thus providing some potential long-term benefits to the host. Alternatively, however, it may be that the introduction of transposon IR sequences by themselves causes enhanced recombination rates at the target site. If this is true, then the disruption and stabilization of these large microsatellite arrays could potentially serve as a means to maintain a genetic status quo by counterbalancing the negative effects of transposon insertion. Future studies should therefore make use of various sensitive reporter-based systems to better clarify what effect(s) transposon excision and/or integration has on microsatellite instability in mammalian cells.
We also found that SB integration is significantly biased toward AT-rich DNA. In agreement with these preferences, we have defined a consensus target site sequence for SB that is AT rich and palindromic. Notably, this consensus sequence is virtually identical to that previously defined for the Tc1 element in worms (28), which has been experimentally shown to adopt a bendable DNA structure (55). Therefore, our data indicate that SB and Tc1 probably employ similar or related pathways for selecting a target site, one that appears to include a twofold dyad symmetry at the insertion site. Consequently, we propose a model for the integration of Tc1-like transposable elements in which each externally bound transposase subunit interacts with sequences immediately flanking the essential target TA dinucleotide. These interactions likely help ensure the proper positioning of the transposon termini prior to strand transfer into a target DNA molecule, but they also introduce a higher-order selectivity, such as sequence accessibility or a bias toward a defined DNA structure.
In support of the theory that DNA structure plays a role in SB target site selection, we have shown that SB strongly favors integration into AT-rich palindromes, which are particularly susceptible to local melting and have been experimentally shown to adopt a bendable DNA structure (55). Moreover, a recent report by Vigdal et al. demonstrating that SB and other Tc1/mariner transposon insertion sites possess common physical properties, including a tendency to be highly bendable (55), is also consistent with these notions. In theory, these structures might be preferred targets for integration because they favor the formation and/or stability of the integration complex or, alternatively, because they promote SB-mediated cleavage, such as through distortion or twisting of the target DNA. Indeed, the nonsymmetric preferences for A at position +6 and T at position +15 downstream from the canonical TA target site could reflect a tendency for SB to target sites capable of DNA distortion at one end. Finally, recent three-dimensional modeling of SB target sites also indicates that there may be a preference for sites containing an inherent geometric deformation (30).
One additional possibility is that SB integration might be influenced to some degree by regionally bound host cell factors. Such a targeting mechanism has been shown to be utilized by a variety of integrating yeast retrotransposons (49, 63) and is generally suspected to play a role in the nonuniform integration pattern exhibited by certain retroviruses (3). In agreement with this notion, SB shows a weak bias toward promoter regions that cannot otherwise be explained by localized remodeling of these regions during transcriptional activation. Nevertheless, we have been unable to find any transcription factor binding sites common to the chromosomal regions preferentially targeted by SB (data not shown). Even so, we note that at least two ubiquitous DNA-binding proteins, HMG-B1 and Ku, are capable of physically interacting with SB in mammalian cells (26, 62) and that neither protein binds DNA with any obvious sequence specificity. This suggests that SB may bind these and/or other factors, including nucleosomes, and thus carry out integration locally. Of course, none of these proposed models are mutually exclusive, and it seems likely that more than one mechanism contributes to SB target site selection.
In addition to providing basic insights into SB target site selection, our study also indicates that SB-mediated integration might be safer for therapeutic in vivo gene delivery than most viruses currently used in the clinic. For instance, although the integration of recombinant AAV (rAAV) vectors can be a rare event in transduced tissues (40), rAAV integration has been shown to be frequently associated with various chromosomal abnormalities, including chromosomal translocations and the deletion of ≥1 Mb of host DNA at insertion sites (35, 39). In marked contrast, no alteration of target chromosomes other than the 2-bp TA duplication was observed during SB integration, suggesting an important qualitative advantage over rAAV-based vectors. In addition, although SB integration appears to be somewhat biased toward genes, its intragenic targeting frequency is significantly lower than that of every integrating vector system described to date (17, 27, 36, 38, 39, 41, 58), with the notable exception of those based on ASLV (36) (Tables 2 and 3). Of course, as is true for virtually any vector that frequently targets intronic regions of the genome, it will still be necessary to carefully design and test any clinical-grade transposon vectors in future work so as to avoid any unwanted side effects on RNA processing (i.e., alternative splicing) at the target site. Furthermore, although the chemistry of SB integration shares many similarities with that of retroviral integration, our observation that SB integrates independently of transcriptional activity suggests that the molecular mechanism(s) used to select a target site is probably distinct among these integrating elements. This property of SB integration may prove important for certain ex vivo and in vivo gene therapy applications, especially in instances where transgene expression in corrected cells provides a strong growth advantage over the general population (13, 37).
Acknowledgments
We thank J. Park for critical reading of the manuscript.
This work was supported by NIH grant AR44012 to M.A.K. and by a Walter V. Berry Fellowship to S.R.Y.
REFERENCES
- 1.Balciunas, D., A. E. Davidson, S. Sivasubbu, S. B. Hermanson, Z. Welle, and S. C. Ekker. 2004. Enhancer trapping in zebrafish using the Sleeping Beauty transposon. BMC Genomics 5:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belur, L. R., J. L. Frandsen, A. J. Dupuy, D. H. Ingbar, D. A. Largaespada, P. B. Hackett, and R. Scott McIvor. 2003. Gene insertion and long-term expression in lung mediated by the Sleeping Beauty transposon system. Mol. Ther. 8:501-507. [DOI] [PubMed] [Google Scholar]
- 3.Bushman, F. D. 2003. Targeting survival: integration site selection by retroviruses and LTR-retrotransposons. Cell 115:135-138. [DOI] [PubMed] [Google Scholar]
- 4.Bushman, F. D. 1994. Tethering human immunodeficiency virus 1 integrase to a DNA site directs integration to nearby sequences. Proc. Natl. Acad. Sci. USA 91:9233-9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson, C. M., A. J. Dupuy, S. Fritz, K. J. Roberg-Perez, C. F. Fletcher, and D. A. Largaespada. 2003. Transposon mutagenesis of the mouse germline. Genetics 165:243-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark, K. J., A. M. Geurts, J. B. Bell, and P. B. Hackett. 2004. Transposon vectors for gene-trap insertional mutagenesis in vertebrates. Genesis 39:225-233. [DOI] [PubMed] [Google Scholar]
- 7.Davidson, A. E., D. Balciunas, D. Mohn, J. Shaffer, S. Hermanson, S. Sivasubbu, M. P. Cliff, P. B. Hackett, and S. C. Ekker. 2003. Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon. Dev. Biol. 263:191-202. [DOI] [PubMed] [Google Scholar]
- 8.Dupuy, A. J., K. Clark, C. M. Carlson, S. Fritz, A. E. Davidson, K. M. Markley, K. Finley, C. F. Fletcher, S. C. Ekker, P. B. Hackett, S. Horn, and D. A. Largaespada. 2002. Mammalian germ-line transgenesis by transposition. Proc. Natl. Acad. Sci. USA 99:4495-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupuy, A. J., S. Fritz, and D. A. Largaespada. 2001. Transposition and gene disruption in the male germline of the mouse. Genesis 30:82-88. [DOI] [PubMed] [Google Scholar]
- 10.Ellegren, H. 2004. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 5:435-445. [DOI] [PubMed] [Google Scholar]
- 11.Fischer, S. E., E. Wienholds, and R. H. Plasterk. 2001. Regulated transposition of a fish transposon in the mouse germ line. Proc. Natl. Acad. Sci. USA 98:6759-6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grabher, C., T. Henrich, T. Sasado, A. Arenz, J. Wittbrodt, and M. Furutani-Seiki. 2003. Transposon-mediated enhancer trapping in medaka. Gene 322:57-66. [DOI] [PubMed] [Google Scholar]
- 13.Hacein-Bey-Abina, S., C. Von Kalle, M. Schmidt, M. P. McCormack, N. Wulffraat, P. Leboulch, A. Lim, C. S. Osborne, R. Pawliuk, E. Morillon, R. Sorensen, A. Forster, P. Fraser, J. I. Cohen, G. de Saint Basile, I. Alexander, U. Wintergerst, T. Frebourg, A. Aurias, D. Stoppa-Lyonnet, S. Romana, I. Radford-Weiss, F. Gross, F. Valensi, E. Delabesse, E. Macintyre, F. Sigaux, J. Soulier, L. E. Leiva, M. Wissler, C. Prinz, T. H. Rabbitts, F. Le Deist, A. Fischer, and M. Cavazzana-Calvo. 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302:415-419. [DOI] [PubMed] [Google Scholar]
- 14.Hartl, D. L., A. R. Lohe, and E. R. Lozovskaya. 1997. Modern thoughts on an ancyent marinere: function, evolution, regulation. Annu. Rev. Genet. 31:337-358. [DOI] [PubMed] [Google Scholar]
- 15.He, C. X., D. Shi, W. J. Wu, Y. F. Ding, D. M. Feng, B. Lu, H. M. Chen, J. H. Yao, Q. Shen, D. R. Lu, and J. L. Xue. 2004. Insulin expression in livers of diabetic mice mediated by hydrodynamics-based administration. World J. Gastroenterol. 10:567-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heggestad, A. D., L. Notterpek, and B. S. Fletcher. 2004. Transposon-based RNAi delivery system for generating knockdown cell lines. Biochem. Biophys. Res. Commun. 316:643-650. [DOI] [PubMed] [Google Scholar]
- 17.Hong, B. K., B. Calmels, H. Hanawa, J. Gray, R. E. Donahue, D. A. Persons, A. W. Nienhuis, P. Hematti, and C. E. Dunbar. 2004. Retroviral insertion site analysis in rhesus macaques transplanted with CD34+ hematopoietic stem cells transduced with a simian immunodeficiency virus-based lentiviral vector. Mol. Ther. 9:S269. [Google Scholar]
- 18.Horie, K., A. Kuroiwa, M. Ikawa, M. Okabe, G. Kondoh, Y. Matsuda, and J. Takeda. 2001. Efficient chromosomal transposition of a Tc1/mariner-like transposon Sleeping Beauty in mice. Proc. Natl. Acad. Sci. USA 98:9191-9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horie, K., K. Yusa, K. Yae, J. Odajima, S. E. Fischer, V. W. Keng, T. Hayakawa, S. Mizuno, G. Kondoh, T. Ijiri, Y. Matsuda, R. H. Plasterk, and J. Takeda. 2003. Characterization of Sleeping Beauty transposition and its application to genetic screening in mice. Mol. Cell. Biol. 23:9189-9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivics, Z., P. B. Hackett, R. H. Plasterk, and Z. Izsvak. 1997. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91:501-510. [DOI] [PubMed] [Google Scholar]
- 21.Ivics, Z., and Z. Izsvak. 2004. Transposable elements for transgenesis and insertional mutagenesis in vertebrates: a contemporary review of experimental strategies. Methods Mol. Biol. 260:255-276. [DOI] [PubMed] [Google Scholar]
- 22.Ivics, Z., C. D. Kaufman, H. Zayed, C. Miskey, O. Walisko, and Z. Izsvak. 2004. The Sleeping Beauty transposable element: evolution, regulation and genetic applications. Curr. Issues Mol. Biol. 6:43-55. [PubMed] [Google Scholar]
- 23.Izsvak, Z., and Z. Ivics. 2004. Sleeping Beauty transposition: biology and applications for molecular therapy. Mol. Ther. 9:147-156. [DOI] [PubMed] [Google Scholar]
- 24.Izsvak, Z., Z. Ivics, and R. H. Plasterk. 2000. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J. Mol. Biol. 302:93-102. [DOI] [PubMed] [Google Scholar]
- 25.Izsvak, Z., D. Khare, J. Behlke, U. Heinemann, R. H. Plasterk, and Z. Ivics. 2002. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J. Biol. Chem. 277:34581-34588. [DOI] [PubMed] [Google Scholar]
- 26.Izsvak, Z., E. E. Stuwe, D. Fiedler, A. Katzer, P. A. Jeggo, and Z. Ivics. 2004. Healing the wounds inflicted by Sleeping Beauty transposition by double-strand break repair in mammalian somatic cells. Mol. Cell 13:279-290. [DOI] [PubMed] [Google Scholar]
- 27.Kang, Y., T. E. Scheetz, C. J. Moressi, D. T. Tran, L. Xia, B. L. Davidson, T. L. Casavant, and P. B. McCray. 2004. In vitro and in vivo analysis of feline immunodeficiency virus-based lentiviral vector integration. Mol. Ther. 9:S2. [Google Scholar]
- 28.Korswagen, H. C., R. M. Durbin, M. T. Smits, and R. H. Plasterk. 1996. Transposon Tc1-derived, sequence-tagged sites in Caenorhabditis elegans as markers for gene mapping. Proc. Natl. Acad. Sci. USA 93:14680-14685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lampe, D. J., M. E. Churchill, and H. M. Robertson. 1996. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 15:5470-5479. [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, G., A. M. Geurts, K. Yae, A. R. Srinivasan, S. C. Fahrenkrug, D. A. Largaespada, J. Takeda, K. Horie, W. K. Olson, and P. B. Hackett. 2005. Target-site preferences of Sleeping Beauty transposons. J. Mol. Biol. 346:161-173. [DOI] [PubMed] [Google Scholar]
- 31.Liu, L., S. Sanz, A. D. Heggestad, V. Antharam, L. Notterpek, and B. S. Fletcher. 2004. Endothelial targeting of the Sleeping Beauty transposon within lung. Mol. Ther. 10:97-105. [DOI] [PubMed] [Google Scholar]
- 32.Lohe, A. R., E. N. Moriyama, D. A. Lidholm, and D. L. Hartl. 1995. Horizontal transmission, vertical inactivation, and stochastic loss of mariner-like transposable elements. Mol. Biol. Evol. 12:62-72. [DOI] [PubMed] [Google Scholar]
- 33.Luo, G., Z. Ivics, Z. Izsvak, and A. Bradley. 1998. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 95:10769-10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mikkelsen, J. G., S. R. Yant, L. Meuse, Z. Huang, H. Xu, and M. A. Kay. 2003. Helper-independent Sleeping Beauty transposon-transposase vectors for efficient nonviral gene delivery and persistent gene expression in vivo. Mol. Ther. 8:654-665. [DOI] [PubMed] [Google Scholar]
- 35.Miller, D. G., E. A. Rutledge, and D. W. Russell. 2002. Chromosomal effects of adeno-associated virus vector integration. Nat. Genet. 30:147-148. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell, R. S., B. F. Beitzel, A. R. Schroder, P. Shinn, H. Chen, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2:E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montini, E., P. K. Held, M. Noll, N. Morcinek, M. Al-Dhalimy, M. Finegold, S. R. Yant, M. A. Kay, and M. Grompe. 2002. In vivo correction of murine tyrosinemia type I by DNA-mediated transposition. Mol. Ther. 6:759-769. [DOI] [PubMed] [Google Scholar]
- 38.Nakai, H., E. Montini, S. Fuess, T. A. Storm, M. Grompe, and M. A. Kay. 2003. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat. Genet. 34:297-302. [DOI] [PubMed] [Google Scholar]
- 39.Nakai, H., X. Wu, S. Fuess, T. A. Storm, D. Munroe, E. Montini, S. Burgess, M. Grompe, and M. A. Kay. Large-scale molecular characterization of adeno-associated virus vector integration in mouse liver. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 40.Nakai, H., S. R. Yant, T. A. Storm, S. Fuess, L. Meuse, and M. A. Kay. 2001. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J. Virol. 75:6969-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narezkina, A., K. D. Taganov, S. Litwin, R. Stoyanova, J. Hayashi, C. Seeger, A. M. Skalka, and R. A. Katz. 2004. Genome-wide analyses of avian sarcoma virus integration sites. J. Virol. 78:11656-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohlfest, J. R., P. D. Lobitz, S. G. Perkinson, and D. A. Largaespada. 2004. Integration and long-term expression in xenografted human glioblastoma cells using a plasmid-based transposon system. Mol. Ther. 10:260-268. [DOI] [PubMed] [Google Scholar]
- 43.Ortiz-Urda, S., Q. Lin, S. R. Yant, D. Keene, M. A. Kay, and P. A. Khavari. 2003. Sustainable correction of junctional epidermolysis bullosa via transposon-mediated nonviral gene transfer. Gene Ther. 10:1099-1104. [DOI] [PubMed] [Google Scholar]
- 44.Plasterk, R. H., Z. Izsvak, and Z. Ivics. 1999. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 15:326-332. [DOI] [PubMed] [Google Scholar]
- 45.Pruss, D., F. D. Bushman, and A. P. Wolffe. 1994. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc. Natl. Acad. Sci. USA 91:5913-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pruss, D., R. Reeves, F. D. Bushman, and A. P. Wolffe. 1994. The influence of DNA and nucleosome structure on integration events directed by HIV integrase. J. Biol. Chem. 269:25031-25041. [PubMed] [Google Scholar]
- 47.Pryciak, P. M., and H. E. Varmus. 1992. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell 69:769-780. [DOI] [PubMed] [Google Scholar]
- 48.Rizzon, C., E. Martin, G. Marais, L. Duret, L. Segalat, and C. Biemont. 2003. Patterns of selection against transposons inferred from the distribution of Tc1, Tc3 and Tc5 insertions in the mut-7 line of the nematode Caenorhabditis elegans. Genetics 165:1127-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandmeyer, S. 2003. Integration by design. Proc. Natl. Acad. Sci. USA 100:5586-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schröder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 51.Spradling, A. C., D. M. Stern, I. Kiss, J. Roote, T. Laverty, and G. M. Rubin. 1995. Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc. Natl. Acad. Sci. USA 92:10824-10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su, A. I., T. Wiltshire, S. Batalov, H. Lapp, K. A. Ching, D. Block, J. Zhang, R. Soden, M. Hayakawa, G. Kreiman, M. P. Cooke, J. R. Walker, and J. B. Hogenesch. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 101:6062-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Temnykh, S., G. DeClerck, A. Lukashova, L. Lipovich, S. Cartinhour, and S. McCouch. 2001. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 11:1441-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tower, J., G. H. Karpen, N. Craig, and A. C. Spradling. 1993. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics 133:347-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vigdal, T. J., C. D. Kaufman, Z. Izsvak, D. F. Voytas, and Z. Ivics. 2002. Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J. Mol. Biol. 323:441-452. [DOI] [PubMed] [Google Scholar]
- 56.Vos, J. C., I. De Baere, and R. H. Plasterk. 1996. Transposase is the only nematode protein required for in vitro transposition of Tc1. Genes Dev. 10:755-761. [DOI] [PubMed] [Google Scholar]
- 57.Weidhaas, J. B., E. L. Angelichio, S. Fenner, and J. M. Coffin. 2000. Relationship between retroviral DNA integration and gene expression. J. Virol. 74:8382-8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749-1751. [DOI] [PubMed] [Google Scholar]
- 59.Yant, S. R., A. Ehrhardt, J. G. Mikkelsen, L. Meuse, T. Pham, and M. A. Kay. 2002. Transposition from a gutless adeno-transposon vector stabilizes transgene expression in vivo. Nat. Biotechnol. 20:999-1005. [DOI] [PubMed] [Google Scholar]
- 60.Yant, S. R., and M. A. Kay. 2003. Nonhomologous-end-joining factors regulate DNA repair fidelity during Sleeping Beauty element transposition in mammalian cells. Mol. Cell. Biol. 23:8505-8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yant, S. R., L. Meuse, W. Chiu, Z. Ivics, Z. Izsvak, and M. A. Kay. 2000. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat. Genet. 25:35-41. [DOI] [PubMed] [Google Scholar]
- 62.Zayed, H., Z. Izsvak, D. Khare, U. Heinemann, and Z. Ivics. 2003. The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping Beauty transposition. Nucleic Acids Res. 31:2313-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu, Y., J. Dai, P. G. Fuerst, and D. F. Voytas. 2003. Controlling integration specificity of a yeast retrotransposon. Proc. Natl. Acad. Sci. USA 100:5891-5895. [DOI] [PMC free article] [PubMed] [Google Scholar]