Abstract
Faithful chromosome segregation is fundamentally important for the maintenance of genome integrity and ploidy. By isolating conditional mutants defective in chromosome segregation in the fission yeast Schizosaccharomyces pombe, we identified a role for the essential gene pfs2 in chromosome dynamics. In the absence of functional Pfs2, chromosomal attachment to the mitotic spindle was defective, with consequent chromosome missegregation. Under these circumstances, multiple intracellular foci of spindle checkpoint proteins Bub1 and Mad2 were seen, and deletion of bub1 exacerbated the mitotic defects and the loss of cell viability that resulted from the loss of pfs2 function. Progression from G1 into S phase following release from nitrogen starvation also required pfs2+ function. The product of the orthologous Saccharomyces cerevisiae gene PFS2 is a component of a multiprotein complex required for 3′-end cleavage and polyadenylation of pre-mRNAs and, in keeping with the conservation of this essential function, an S. pombe pfs2 mutant was defective in mRNA 3′-end processing. Mutations in pfs2 were suppressed by overexpression of the putative mRNA 3′-end cleavage factor Cft1. These data suggest unexpected links between mRNA 3′-end processing and chromosome replication and segregation.
Following DNA replication in eukaryotic cells, accurate mitotic chromosome segregation requires the bivalent attachment of the replicated chromosomes to the spindle via sister kinetochores, followed by anaphase movement of the chromosomes to opposite spindle poles. Defects in this process result either in catastrophic failure of mitosis and cell death or in aneuploidy. Spindle checkpoint mechanisms provide protection against these eventualities by imposing a delay over the onset of anaphase until all chromosomes have established symmetrical, bivalent attachments to tense spindle microtubules (14). The molecular basis of this checkpoint is incompletely understood, but many of the proteins involved have been identified and characterized in yeast models and shown to perform analogous checkpoint functions in diverse species, including metazoans and plants. Such proteins include Bub1, Bub3, Mps1, Mad1, Mad2, and Mad3, which are thought to form a heterooligomeric complex at unattached kinetochores (2). During progression through normal, unperturbed mitosis, such complexes would be expected to be short-lived due to the highly dynamic interactions between spindle microtubules and kinetochores. Once all kinetochores have achieved bivalent attachment, the reduction in Mad- and Bub-dependent signaling allows activation of the multisubunit ubiquitin ligase known as the anaphase-promoting complex/cyclosome and subsequent progression into anaphase.
Normal chromosome behavior requires the assembly of specialized protein-DNA complexes at telomeres, centromeres, and origins of replication as well as the establishment and maintenance of cohesion between the sister chromatids following DNA replication. The integrity of these chromosome-associated complexes is potentially threatened by traversal of the chromosomes by DNA replication forks or RNA polymerases. This view first emerged with the identification in the budding yeast Saccharomyces cerevisiae of DNA elements that serve to prevent transcriptional read-through into centromeres or origins of replication (27). Maintenance of heterochromatin at centromeres in the fission yeast Schizosaccharomyces pombe is also required both for the repression of transcription and for accurate chromosome segregation (1). Furthermore, a recent study demonstrated that cohesin, the multiprotein complex which maintains sister chromatid cohesion, accumulates at the 3′ ends of convergent active transcription units (13).
The mutual incompatibility of transcription and centromere function and the capacity of read-through transcription to interfere with the expression of downstream genes emphasize the importance of the appropriate termination of transcription. For RNA polymerase II (Pol II) transcription units, termination is generally coupled to endonucleolytic cleavage of the primary transcript at the site defining the 3′ terminus of the mRNA. Termination typically occurs within a zone that lacks distinctive features at the level of the primary sequence but is situated several hundred bases downstream from the cleavage site. In most cases, cleavage is followed by polyadenylation of the nascent mRNA, with the cleavage and polyadenylation steps being coordinated by multisubunit protein complexes. These complexes are conserved in their overall organization among diverse eukaryotes (23) and comprise a cleavage and polyadenylation specificity factor (CPSF) and a cleavage stimulation factor (CstF) in mammals. In S. cerevisiae, orthologues of many of the subunits of CPSF and CstF have been identified as components of the cleavage and polyadenylation factor (CPF). In the absence of efficient cleavage and polyadenylation, Pol II continues to transcribe beyond the normal termination zone, potentially disrupting the functions of genes and other chromosomal features downstream.
Here we describe the characterization of a fission yeast mutant identified through its inability to execute mitosis faithfully and show that the primary biochemical defect in this mutant is in a component of the cleavage and polyadenylation machinery. These data indicate that the proper regulation of mRNA 3′-end processing is indeed essential for normal chromosome segregation.
MATERIALS AND METHODS
Fission yeast methods.
Conditions for the growth, maintenance, and genetic manipulation of fission yeast were as described previously (15). A complete list of the strains used in this study is given in Table 1. Except where otherwise stated, strains were grown at 30°C in yeast extract (YE) or Edinburgh minimal medium (EMM2) with appropriate supplements. When necessary, gene expression from the nmt1 promoter was repressed by the addition of 5 μM thiamine to the growth medium.
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| 972 | h− | Laboratory stock |
| 428h | h−ade6-M210 leu1-32 ura4-D18 his7 | Laboratory stock |
| 429h | h+ade6-M216 leu1-32 ura4-D18 his7 | Laboratory stock |
| KZ2 | h+leu1-32 ade6-M210 bub1+-GFP-kanr Ch16 | This study |
| KZ50 | h−ade6-M210 leu1-32 pfs2-3169 bub1+-GFP-kanr | This study |
| KZ52 | h−his2 pfs2-3169 mad2+-GFP-LEU2 bub1+-HA3-kanr | This study |
| KZ67 | h−ade6-M216 leu1-32 ura4-D18 pfs2-3169 bub1::ura4+ | This study |
| pfs2Δ | h+/h− pfs2::ura4+/pfs2+ade6-M210/ade-M216 leu1-32 ura4-D18 | This study |
| pfs2+-GFP | h−pfs2+-GFP::kanrleu1-32 | This study |
| P41-pfs2 | h−pfs2::ura4-nmt41-pfs2 ade6-M210 leu1-32 ura4-D18 his7 | This study |
| P81-pfs2 | h−pfs2::ura4-nmt81-pfs2 leu1-32 ura4-D18 | This study |
| pfs2-11 | h− pfs2-11 | This study |
| bub1Δ | h−bub1::ura4+ade6-M210 leu1-32 ura4-D18 his1 | Jean-Paul Javerzat |
| pfs2-11 bub1Δ | h−pfs2-11 bub1::ura4+ade6-M210 leu1-32 ura4-D18 his1 | This study |
| bub1-GFP | h−bub1+-GFP::kanrleu1-32 ura4-D18 | Laboratory stock |
| pfs2-11 bub1-GFP | h−pfs2-11 bub1+-GFP::kanrleu1-32 ura4-D18 | This study |
| Ch16 | h−ade6-M210 ura4-D18 Ch16 | Laboratory stock |
| pfs2-11 Ch16 | h−pfs2-11 ade6-M210 leu1-32 ura4-D18 Ch16 | This study |
The pfs2-3169 mutant was identified during the course of a large-scale mutagenesis screen to identify essential genes required for accurate chromosome segregation. Parental strain KZ2 (Table 1), which contains minichromosome Ch16 (19) and Bub1-green fluorescent protein (GFP), was mutagenized with nitrosoguanidine as previously described (24), spread on rich yeast extract with five supplements (YE5S) plates, and incubated at 27°C. Colonies were replica plated on YE5S plates at 36°C and YE plates (rich medium lacking exogenously added supplements) at 32°C, on which cells that had lost Ch16 turned red because of the chromosomal ade6-210 mutation. A total of 221 isolates out of 500,000 mutagenized colonies showed both minichromosome loss and temperature-sensitive (ts) phenotypes; furthermore, 39 mutants showed cosegregation of minichromosome loss and ts phenotypes upon backcrossing. Among these isolates, those with mutations in 10 complementation groups (cin1 to cin10) showed hypersensitivity to thiabenzadole compared to a wild-type strain. cin2-3169 is allelic to pfs2. For further analysis of pfs2-3169, strain KZ52, which contains Mad2-GFP and Bub1-hemagglutinin (HA), was used.
Minichromosome loss assays were performed with strains containing the nonessential ade6-M216-marked Ch16 minichromosome derivative of chromosome 3 (19). The ade6-M216 allele complements an unlinked ade6-M210 marker in these strains such that they remain Ade+ as long as the minichromosome is maintained. Chromosome loss was measured in the progeny from a single Ade+ cell after a known number of generations during which selection for adenine prototrophy had been relaxed by growth on YE agar. Rates of chromosome loss per generation were calculated as described elsewhere (16, 28) with the formula 1 − e(1/n)lnRn, where Rn is the proportion of Ade+ cells n generations after the removal of selection. For each strain tested, mean rates were calculated from five independent measurements.
Disruption and modification of pfs2.
One-step gene disruption or modification via homologous recombination was performed following PCR-mediated generation of ura4+ or kanMX selectable cassettes flanked by 80-bp segments from appropriate regions of pfs2+ by using the oligonucleotides described in Table 2. Following transformation of a diploid strain (428h/429h), Ura+ progeny were screened for the desired integration pattern by diagnostic PCRs with primer pairs spanning the presumptive recombination sites (details of the additional primers used for this purpose are available from the authors on request). Meiosis and sporulation were induced by plating on malt extract agar, and tetrad dissection was performed with an MSM micromanipulator (Singer Instruments, Watchet, United Kingdom) as described elsewhere (15). The pfs2+ gene was disrupted by using primers DELA and DELB, construction of the chromosomally GFP-tagged pfs2+ strain (pfs2+-GFP) was accomplished by using primers TAGA and TAGB, and the nmt1 promoter shutoff strains P41-pfs2 and P81-pfs2 were constructed by using primers PSOA and PSOB (Table 2). In order to generate further ts alleles of pfs2, low-fidelity PCRs were performed with primers MUTA and MUTB, which flank the pfs2+ locus, together with genomic S. pombe DNA as a template. The resulting pool of mutagenized pfs2 sequences was used to transform strain P81-pfs2, and transformants were selected by growth at 25°C on YE agar containing thiamine, allowing the growth of cells in which the nmt1 promoter-driven pfs2 allele had been replaced by homologous recombination with the mutagenized PCR product. The resulting colonies were screened for ts growth by replica plating and checked for the loss of the P81-pfs2 allele by PCR, and the sequence change(s) in pfs2 was identified following PCR amplification of the mutant alleles.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence (5′-3′) |
|---|---|
| DELA | TTTAAGTTGGTTTTATTTAAATTAAAGATTGATTTTATTGCTGCTAAATAAGGAATTTCACATCTTTGGGGAATTGGTTACAAAGCGACTATAAGTCAGAAAGTG |
| DELB | GGATGATATTGCATACTCCAAATACCCAAATCATGAGCAAATGGAATTTCAGCAGTAGGGTATAGTAAATAAGAAGACGCAATTCTAAATGCCTTCTGAC |
| TAGA | ACGCTGAGCCGTCAACTCAAAACTCTTTTATCCCAGGTCTCACCTCAAAGTCACAAGATGGGTATCCTCAAAACTATCGACGGATCCCCGGGTTAATTAA |
| TAGB | GCTGCGGTGTGAATTTTTTTTTTTTTAAATCTAATTAATGCCAAGTAAATATGTTTTTCAAATAACAAAGAAATGTATTAGAATTCGAGCTCGTTTAAAC |
| PSOA | CTAGGTGGATTAATTTATTTGGCGTGCCTAATTTGGAACGAAATTAATATCATCTTCAACATCCCACCGATTTACTTATAATACTAATTCTAAATGCCTTCTGAC |
| PSOB | TTTGATAGCCCTGATCCATAGTCCACAGTCCTACGAGTCATAGGCTTTTGAATAATTCTAGCATTTTCAGCTCTTTCCATCAAAGCGACTATAAGTCAGAAAGTG |
| MUTA | ATCAAGCATGGATAGATGATCAAAGATATC |
| MUTB | TCCTCTCGCCGAATCGTAATTTCTTAA |
| URA4F | GTCCCCTGGTATCGGCTTGGATGTTAAAGG |
| URA4R1 | TGCCTTCTGACATAAAACGCCTAG |
| URA4R2 | TGTAGGAGCATGTTTAATAAATTACTATAGCAAATTACTTTTTATT |
| URA4R3 | CATACATCTTTCATTGGCTTTGTAC |
| URA4R4 | GTCTACATGGTATTTTACATTCATC |
| URA4R5 | CCAACACCAATGTTTATAACCAAG |
| URA4R6 | GCTTGTGATATTGACGAAACTTTTTGACATC |
| CDT1F | CTCATCTCGCTCAGATTTTAACCGTATGGC |
| CDT1R | CTCGCGACTTGTTGATTTCTCCAAATTGAG |
| CDC18F | CATATCGTTCAGCAGGAGTCGTGGGTCGTG |
| CDC18R | CGGACTGGGTGAAATGCGACTCTAGTTCAC |
| CDC18F1 | GGTCTTCAACAGAAAGCCATCCTTTGTACG |
| CDC18R1 | GGTACCAATGTCTCCAACAGCTGTAATGAC |
| CDC18R2 | CGGCAAAATTTGAAAACCGTAACGCGTGAA |
| CDC18R3 | CAACCTGAAAAGTTTAAACACCAAGTTGGA |
| CDC18R4 | ACCACTTGAGACTGAACCACGGGTATGCGT |
| CDC18R5 | CCCCAAAATTGAGAAGTGCTTTTGCTACGT |
| CDC18R6 | TTCCGCAGCTTATATGATTACTATGTCGTC |
Microscopy.
Cells fixed with 3.8% formaldehyde were washed with phosphate-buffered saline and stained with 4′,6-diamidino-2-phenylindole (DAPI) before examination by fluorescence microscopy. Images were acquired by using a Zeiss Axioskop microscope equipped with a Planapochromat ×100 objective, an Axiocam cooled charge-coupled device camera, and Axiovision software (Carl Zeiss Ltd., Welwyn Garden City, United Kingdom). Images were assembled by using Adobe Photoshop. In some experiments, living cells growing in EMM2 medium were stained by the addition of 5 μg/ml bis-benzimide (Hoechst 33342; Sigma) before examination by fluorescence microscopy. Visualization of Pfs2-GFP in living cells embedded in 0.6% low-melting-point agarose was performed at room temperature (∼22°C).
Flow cytometry.
Cells fixed with 70% ethanol were rehydrated in 10 mM EDTA (pH 8.0)-0.1 mg of RNase A/ml-1 μM Sytox green and incubated at 37°C for 2 h. Cells were analyzed by using an Epics XL-MCL flow cytometer (Coulter, Fullerton, Calif.).
RT-PCR.
Total RNA was isolated by hot phenol extraction and purified by using an RNeasy kit (Qiagen). A total of 0.5 μg of total S. pombe RNA was reverse transcribed with SuperScript reverse transcriptase (RT) (Invitrogen) and random hexamers according to the manufacturer's instructions. Reverse-transcribed S. pombe cDNA products (2 μl) were PCR amplified as described elsewhere (9) with specific primers (Table 2) and 25 cycles of 95°C for 1 min, 64°C for 1 min, and 72°C for 3 min.
RESULTS
Fission yeast Pfs2 is an essential nuclear protein required for normal chromosome segregation.
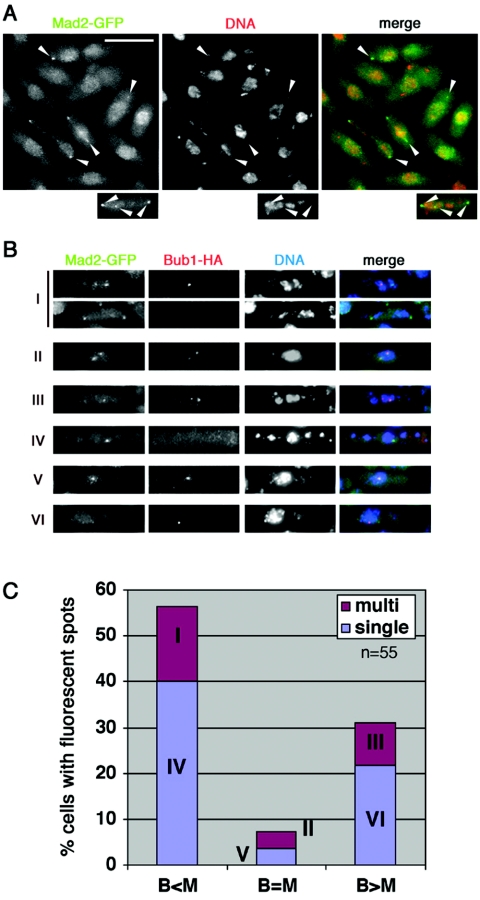
In order to identify novel genes with essential roles in chromosome segregation, we used the fluorescence microscopy-based screen to search a collection of ts S. pombe mutants for strains displaying chromosome missegregation and an unusually high frequency of Mad2 or Bub1 nuclear dots at the restrictive temperature. This screen yielded the cin2-3169 strain, along with a number of other mutant strains (a more complete account of this screen and characterization of other mutants will form the basis of a future report). After asynchronous cin2-3169 cells expressing Mad2-GFP were shifted from 26°C to the restrictive temperature of 36°C, cells containing Mad2-GFP dots accumulated and accounted for 8% of the population by 4 h after the temperature shift (Fig. 1A). The presence of one, two, or occasionally three Mad2-GFP foci in these cells was correlated in most cases with gross defects in chromosome segregation, as judged by DAPI staining (Fig. 1A). Visualization of HA epitope-tagged Bub1 in cin2-3169 cells also expressing Mad2-GFP revealed a variety of combinatorial phenotypes after the shift to 36°C. Most cells with Bub1-HA foci also contained Mad2-GFP spots, although this was not always the case, and in most cases the numbers of Bub1-HA and Mad2-GFP foci within individual cells were not equal (Fig. 1B and C). After incubation for 4 h at 36°C, 85% of the cin2-3169 cells had microtubular arrays typical of interphase, while 10% had postanaphase arrays and 4% contained mitotic spindles. The latter category frequently contained aberrantly segregated chromosomes (data not shown).
FIG. 1.
Chromosome missegregation and accumulation of Mad2 and Bub1 foci in cin2-3169 (pfs2-3169) cells. (A) An asynchronous culture of cin2-3169 mad2+-GFP (KZ52) cells growing at 26°C was shifted to the restrictive temperature of 36°C for 4 h, stained with DAPI to reveal DNA, and examined by fluorescence microscopy. (B) Terminal phenotypes in a cin2-3169 mad2+-GFP bub1+-HA strain (KZ52) after 4 h of incubation at 36°C. Fixed cells were processed for anti-HA immunofluorescence and DAPI staining. Individual cells containing one or more fluorescent spots were assigned to one of six categories: cells containing multiple spots, with a larger number of Mad2-GFP spots than of Bub1-HA spots (I); those with equal numbers of multiple (II) or single (V) spots of each type; those with multiple spots, with a larger number of Bub1-HA spots than of Mad2-GFP spots (III); and those with a single Mad2-GFP spot (IV) or a single Bub1-HA spot (VI). (C) Quantification of the phenotypes indicated in panel B; a total of 55 cells with one or more fluorescent spots were scored.
Complementation of the cin2-3169 mutant with a genomic DNA library (kindly provided by Taro Nakamura and Chikashi Shimoda) yielded 10 transformants that could grow at the restrictive temperature. Of these, five contained a plasmid insert spanning the S. pombe orthologue of the S. cerevisiae PFS2 gene, which we term pfs2+, while three contained an insert spanning cft1+, the S. pombe orthologue of the S. cerevisiae CFT1 gene. Both PFS2 and CFT1 (also known as YHH1) encode components of the S. cerevisiae CPF that have clear counterparts in metazoan CstF and CPSF (Cstf50 and CPSF160, respectively) (23). Backcrossing of the cin2-3169 strain with a strain in which the pfs2+ gene was tagged with GFP (by use of a kanamycin resistance gene as a marker) demonstrated that pfs2 is the gene mutated in cin2-3169 cells, while cft1+ represents an extragenic suppressor. We therefore assigned the designation pfs2-3169 to the pfs2 allele identified in cin2-3169.
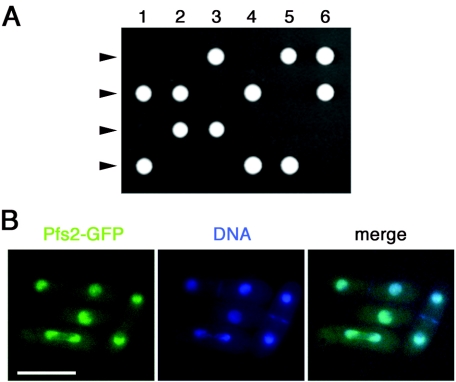
The identification of a ts pfs2 allele suggested that pfs2+ is likely to be essential for cell viability. To address this point, we constructed a ura4-D18/ura4-D18 diploid fission yeast strain in which one of the two pfs2 alleles was disrupted by the ura4+ marker. Following meiosis and tetrad dissection, it was found that only two of each set of four haploid spores were viable, and these were in every case Ura− (Fig. 2A). Thus, pfs2+, like PFS2 in S. cerevisiae, is an essential gene, and pfs2-3169 is a loss-of-function allele.
FIG. 2.
Fission yeast Pfs2 is an essential nuclear protein. (A) Tetrads derived from h+/h− pfs2::ura4+/pfs2+ diploid strains were microdissected onto YE5S agar. Colonies resulting from six such tetrads were photographed after 7 days of growth at 30°C. The genotypes of the segregants were determined by replica plating. (B) Merged images of fluorescence micrographs showing Pfs2-GFP and DNA (Hoechst 33342) localization in living cells. Bar, 10 μm.
To monitor the localization of Pfs2 in living fission yeast, the one-step gene replacement method was used to generate a strain (pfs2+-GFP) with a C-terminally GFP-tagged version of Pfs2. The GFP-tagged protein appeared to be functional, as judged by the full viability of the pfs2+-GFP strain. Examination of living pfs2+-GFP cells by fluorescence microscopy showed that Pfs2-GFP (and, by inference, Pfs2) was localized to the nucleus but was excluded from the nucleolus (Fig. 2B). There were no obvious differences in this pattern among cells at different cell cycle stages.
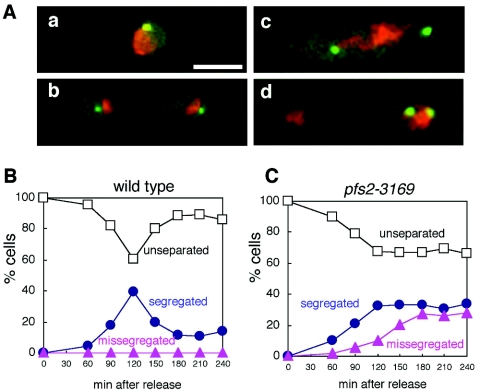
To examine the influence of pfs2 function on progression through mitosis in more detail, pfs2-3169 and control pfs2+ cells expressing GFP localized to the centromere of chromosome 2 (cen2-GFP) were synchronized by arrest in hydroxyurea (HU) at 26°C and then released into HU-free medium at 36°C. Progression through subsequent mitosis then was monitored by fluorescence microscopy to examine cen2-GFP separation and nuclear morphology (Fig. 3). Under these conditions, control pfs2+ cells exhibited uniformly symmetrical segregation of the cen2-GFP signal and daughter nuclei, and a peak of binucleate cells was seen 120 min after release of the HU block; by 180 min, most of these cells had completed mitosis and cytokinesis (Fig. 3A and B). In contrast, approximately 40% of the pfs2-3169 cells initiated cen2-GFP separation, and most of these cells displayed severe chromosome missegregation by 240 min after release of the HU block (Fig. 3A and C). In the remaining 60% of the cells, cen2-GFP remained unseparated. The latter population of cells failed to complete mitosis by 240 min after release of the HU block (Fig. 3C). These data suggested that upon HU block and release, pfs2-3169 cells showed two defects. One was mitotic sister chromatid missegregation, which accompanied the increased frequencies of Mad2 and Bub1 foci, and the other was a block in interphase, resulting in a failure to enter mitosis. Among cells that exhibited cen2-GFP separation, the timing of this event was not significantly altered in the pfs2 mutant, suggesting that it had no gross defect in sister chromatid cohesion.
FIG. 3.
Chromosome segregation defects in pfs2-3169 cells. (A) Segregation patterns of sister centromeres. Wild-type (KZ72) or pfs2-3196 (KZ71) cells carrying cen2-GFP and grown at 26°C were treated with HU for 3 h. After HU was washed away, cultures were shifted to 36°C and incubation was continued. cen2-GFP localization and DAPI staining were visualized by fluorescence microscopy. Representative examples of unseparated (a), equally segregated (b), and missegregated (c and d) centromeres are shown. Bar, 5 μm. (B and C) Quantification of sister centromere segregation patterns following cen2-GFP and DAPI staining of wild-type (B) and pfs2-3169 (C) cells. The percentage of each type of segregation pattern (open squares, unseparated; blue circles, equally segregated; red triangles, missegregated) was plotted at each time point.
As part of an initially independent investigation of pfs2+ function, we used targeted homologous recombination to replace the endogenous pfs2+ promoter with either of two attenuated versions of the thiamine-repressible nmt1 promoter, P41 and P81 (3, 4). The resulting strains, P41-pfs2 and P81-pfs2, grew healthily in the absence of thiamine but were unable to grow when the attenuated nmt1 promoters were repressed by the addition of thiamine to the medium (data not shown). The availability of these promoter shutoff strains made possible the generation of further ts alleles of pfs2, following targeted recombination of randomly mutated chromosomal pfs2 sequences and simultaneous restoration of the pfs2+ promoter (see Materials and Methods). This approach gave rise to a second tightly ts allele, pfs2-11. These cells ceased to divide within 1 h after being shifted to the restrictive temperature of 36°C and, like pfs2-3169 cells, displayed chromosome segregation defects at later time points (Fig. 4 and data not shown). These data indicate that the phenotypes associated with both alleles resulted from similar losses of pfs2 function. The connection between pre-mRNA 3′-end processing and chromosome segregation suggested by our data for pfs2-3169 cells is therefore not based on unrepresentative, allele-specific effects.
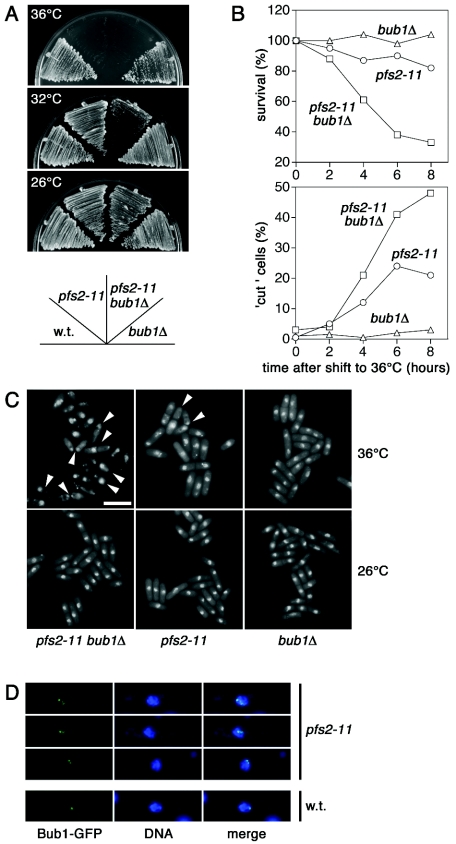
FIG. 4.
The spindle checkpoint is required for cell survival after the inactivation of pfs2. (A) Wild-type strain 972 and the other strains indicated were streaked in parallel on YE5S agar plates and photographed after incubation for 3 to 5 days at the indicated temperatures. (B) At 2-h intervals, samples from cultures of pfs2-11, bub1Δ, and pfs2-11 bub1Δ cells incubated at the restrictive temperature of 36°C were taken to score the percentage of aberrant mitosis in each culture. One thousand cells at the indicated times were plated on YES agar plates to assess cell survival. Colonies were counted after 5 days of growth at 26°C. (C) Fluorescence micrographs of DAPI-stained pfs2-11, bub1Δ, and pfs2-11 bub1Δ cells grown at the permissive temperature of 26°C or after a shift to 36°C for 6 h. Bar, 10 μm. (D) Merged images of fluorescence micrographs showing wild-type or pfs2-11 cells carrying Bub1-GFP. Cells were grown at 36°C for 2 h, fixed with methanol, and stained with DAPI.
The spindle checkpoint is required for cell survival after inactivation of pfs2.
Despite the chromosome segregation defects described above, pfs2 mutants retained viability during protracted incubation at the restrictive temperature (Fig. 4B). The increased frequencies of Mad2 and Bub1 foci in these cells suggested an involvement of at least some aspects of the spindle checkpoint machinery in the events following the loss of pfs2 function. To investigate this notion further, we tested spindle checkpoint-defective strains for genetic interactions with pfs2-11 cells. Strains with a deletion of bub1 (bub1Δ) were not inherently ts, but the combination of bub1Δ and pfs2-11 resulted in a reduction in the restrictive temperature relative to that seen with pfs2-11 alone (Fig. 4A). This genetic interaction was reflected in a more rapid loss of viability of the double mutant compared with that of each of the single mutants after a shift of the cells to 36°C (Fig. 4B). Loss of viability was correlated with the appearance of “cut” cells, in which septation had occurred in the absence of nuclear division (Fig. 4B and C). Consistent with the results obtained with pfs2-3169 cells (Fig. 1), cells with multiple Bub1-GFP foci were observed by 2 h after pfs2-11 cells were shifted to 36°C (Fig. 4D).
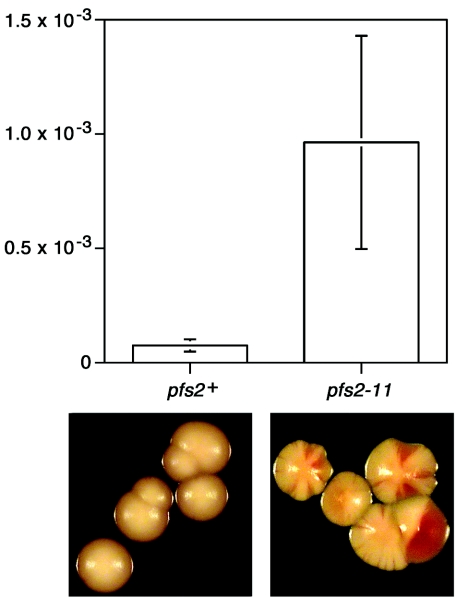
The failure of chromosome segregation seen after pfs2 mutants were shifted to the restrictive temperature suggested that even a partial loss of pfs2 function might severely compromise the fidelity of chromosome transmission. pfs2+ (control) and pfs2-11 cells containing a nonessential minichromosome (Ch16) carrying an adenine biosynthetic marker were therefore used to measure rates of chromosome loss during growth at the semipermissive temperature of 32°C (Fig. 5). This assay showed that the spontaneous loss of the minichromosome was 13-fold more frequent in pfs2-11 cells than in control pfs2+ cells. We conclude that the proper functioning of pfs2+ makes an important contribution to the maintenance of genome stability in S. pombe.
FIG. 5.
Increased chromosome segregation failure rate in pfs2-11 cells. (Upper panel) Rates of minichromosome loss were calculated for pfs2+ and pfs2-11 cells grown at the semipermissive temperature of 32°C. Mean loss rates were 9.6 × 10−4 and 7.5 × 10−5 per generation for the pfs2-11 cells and the control pfs2+ cells, respectively. (Lower panels) Examples of pfs2+ colonies (left) and Ade− pfs2-11 colonies indicative of chromosome loss (right).
Pfs2 is required to prevent transcriptional read-through.
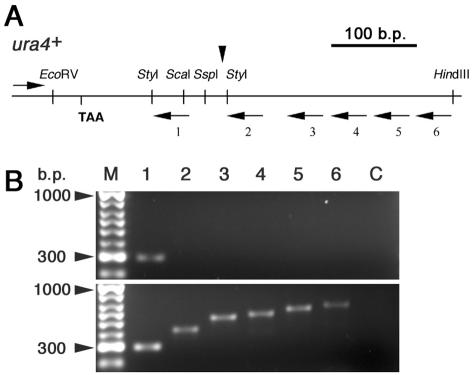
Since Pfs2 was previously implicated in pre-mRNA cleavage and polyadenylation in S. cerevisiae, it seemed likely that the essential function of pfs2+ in S. pombe might also involve pre-mRNA processing. To investigate this possibility, steady-state RNA from the well-characterized downstream region of the ura4+ gene was examined by RT-PCR (Fig. 6). In keeping with the results of an earlier study of cleavage and polyadenylation in this gene (9), transcripts extending beyond the 3′ end of the mature ura4+ mRNA were undetectable in control pfs2+ cells grown for 3 h at 36°C (Fig. 6B, upper panel). In contrast, steady-state ura4+ transcripts extended at least 300 bases beyond the normal 3′ end in pfs2-11 cells grown under the same conditions (Fig. 6B, lower panel). These data suggested that in S. pombe, as in S. cerevisiae, pfs2+ function is required for the normal 3′-end processing of pre-mRNA.
FIG. 6.
Pfs2 is required for transcriptional termination. (A) Schematic representation of the 547-bp region encompassed by the EcoRV-HindIII fragment downstream from the ura4+ gene. The positions of the translation stop codon (TAA) and the major site of pre-mRNA 3′-end cleavage and polyadenylation (vertical arrowhead) are indicated, along with the oligonucleotides used for RT-PCR analysis (horizontal arrows). (B) Electrophoretic separation of RT-PCR products with total RNA isolated from cultures of wild-type (upper panel) and pfs2-11 (lower panel) cells grown at the restrictive temperature of 36°C for 3 h. RT-PCR products were separated on a 1.5% agarose gel. Lane numbers correspond to the 3′ oligonucleotides (URA4R1 to URA4R6) used for RT and PCR amplification; all reactions included a common 5′ primer (URA4F), as indicated in panel A. In the negative control reactions (lane C), no reverse transcriptase was added to the RT-PCR. Lane M, size markers.
Pfs2 is required for entry into S phase.
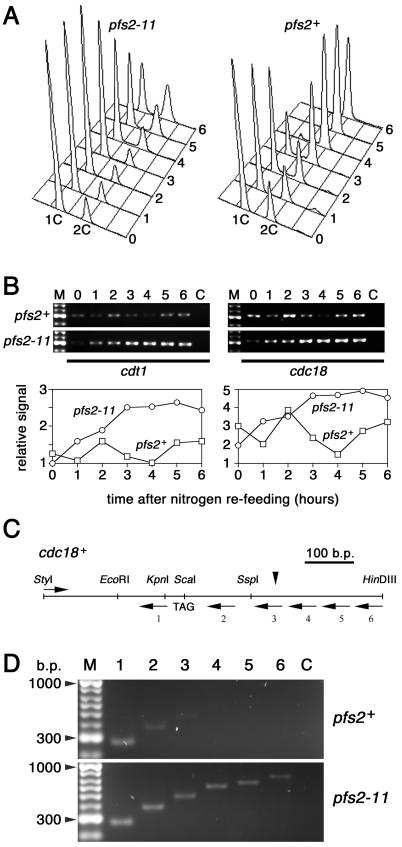
Although defective chromosome segregation was the most striking aspect of the pfs2 mutant phenotype at the cellular level, 85% of the cells were arrested in interphase when pfs2 was inactivated in asynchronous populations (data not shown). Similarly, 60% of pfs2-3169 cells released from an HU (early-S-phase) block at 36°C were unable to progress into mitosis (Fig. 3). This interphase function of pfs2+ was investigated by using nitrogen starvation to arrest pfs2-11 and pfs2+ cells in G1 at 26°C and then releasing the starved cells by nitrogen refeeding at 36°C (Fig. 7). Flow cytometry indicated that the pfs2+ population had entered S phase within 2 h of nitrogen refeeding and had completed DNA replication by 4 h (Fig. 7A). In contrast, very few of the pfs2-11 cells had left G1 by the 4-h time point, and substantial numbers of G1-arrested cells were still present after 6 h. Thus, pfs2+ function is required for transit at the normal rate from G1 phase into S phase. The likely requirement of Pfs2 for general pre-mRNA processing might suggest that nitrogen-starved cells lacking pfs2+ function simply fail to achieve the level of macromolecular biosynthesis needed for commitment to the mitotic cell cycle at the start. This interpretation was, however, not supported by measurements of RNA transcripts from cdt1+ and cdc18+ (Fig. 7B), two genes that are transcribed in a start-dependent manner (8). RT-PCR analysis indicated that the levels of transcripts from each of these genes peaked at 2 h after nitrogen refeeding in control pfs2+ cells; in pfs2-11 cells, the transcript levels increased with similar kinetics up to 2 h but continued to rise, and transcripts were maintained at high levels until at least 6 h after release. These data suggested that, under these conditions, pfs2-11 cells were substantially delayed in cell cycle progression at a point beyond the start and close to the initiation of S phase. Although start-dependent transcription was activated in these cells, our findings with the ura4+ gene (Fig. 6) suggested that the 3′-end processing of these transcripts might be defective. We were able to address this notion for transcripts from cdc18+, since the 3′ end of a cDNA has been mapped for this gene (11). RT-PCR indicated that steady-state cdc18+ transcripts extended at least 200 bases beyond the normal mRNA 3′ end in pfs2-11 cells released from nitrogen starvation for 2 h at 36°C (Fig. 7C and D).
FIG. 7.
Pfs2 is required for entry into S phase. (A) pfs2+ (strain 972) and pfs2-11 cells were arrested in G1 by nitrogen starvation and released into nitrogen-rich medium at the nonpermissive temperature of 36°C. Cells harvested at hourly intervals were processed for DNA staining and flow cytometry. (B) (Upper panels) Total RNA samples from the cultures shown in panel A were isolated at the indicated times. RT-PCR analysis was performed with primers specific for coding sequences of cdt1+ (CDT1F and CDT1R; left panels) and cdc18+ (CDC18F and CDC18R; right panels), and products were separated on a 1.5% agarose gel. (Lower panels) Normalized levels of RT-PCR products. (C and D) Transcriptional read-through of cdc18+ in pfs2-11 cells. (C) Schematic representation of the 743-bp region containing the StyI-HinDIII fragment encompassing the 3′ end of the cdc18+ gene. The positions of the translation stop codon (TAG) and the site of pre-mRNA 3′-end cleavage and polyadenylation (vertical arrowhead) (11) are indicated, along with the oligonucleotides used for RT-PCR analysis (horizontal arrows). (D) RT-PCR products obtained with total RNA isolated from cultures of wild-type (upper panel) and pfs2-11 (lower panel) cells harvested 2 h after nitrogen refeeding as in panel A.Lane numbers correspond to the 3′ oligonucleotides (CDC18R1 to CDC18R6) used for RT and PCR amplification; all reactions included a common 5′ primer (CDC18F1), as indicated in panel C. Negative control reactions (lane C) in panels B and D contained no reverse transcriptase.
DISCUSSION
The S. pombe pfs2+ gene described here encodes a 509-amino-acid protein containing seven WD (β-transducin) repeats. Previous studies identified CstF50, the human counterpart of Pfs2, through its involvement in pre-mRNA 3′-end processing and S. cerevisiae Pfs2p as a component of the CPF complex (22, 29). Mutation of pfs2 in S. pombe caused the accumulation of transcripts extending beyond the 3′ ends of mature mRNAs of the ura4+ and cdc18+ genes (Fig. 6 and 7). While it is formally possible that this finding represents the stabilization of read-through RNAs that are normally present only at low levels, a more straightforward explanation would be that pfs2 function is required for pre-mRNA cleavage and polyadenylation. The identification of cft1+ as a multicopy suppressor adds further weight to this idea.
This study has revealed unexpected connections between pfs2 and cell cycle progression. The first point in the cycle at which pfs2 function appears to be critical is before the onset of S phase but after the activation of start-dependent transcription (Fig. 7). The start-dependent transcripts that accumulated in pfs2-11 cells released from nitrogen starvation at the restrictive temperature did not appear to be appropriately processed (Fig. 7C and D). The impaired function of these transcripts may well have contributed to the observed cell cycle arrest, since both Cdt1 and Cdc18 are required for the assembly of functional prereplicative complexes at chromosomal origins of replication (10, 11, 18). The observation that cells released from G1 arrest by nitrogen refeeding were able to progress beyond the start (as judged by the activation of cdt1+/cdc18+ transcription) in the absence of pfs2 function is surprising, since efficient pre-mRNA 3′-end processing would be expected to be a prerequisite for the gene expression needed for cell cycle commitment. It is possible that nitrogen-starved cells retain a stable stockpile of mature mRNAs that facilitate G1 progression when the nutritional status improves. Alternatively, a loss of pfs2 function may affect the processing of different transcripts to different degrees, and the efficient expression of genes required for G1 transit may be comparatively independent of pfs2. Further experiments will be required to distinguish among these possibilities. Defects in the cell cycle machinery, as opposed to defects in cell growth, generally lead to cell elongation in S. pombe. Cells lacking pfs2 function (either as a result of the shift of a ts strain to its restrictive temperature or after promoter shutoff) did not become appreciably elongated, however. This observation suggested that general macromolecular synthesis was at least partially compromised under these circumstances, arguing against a highly specific involvement of pfs2 in the regulated expression of cell cycle-related genes.
Chromosome segregation was a second cell cycle stage during which the pfs2 mutant phenotype was manifested. Two general explanations may account for the defective mitosis seen after the loss of pfs2 function. One realistic possibility is that a loss of transcriptional termination has a direct effect on some aspect of chromosome structure or kinetochore function. The recent observation that cohesin clusters at sites of convergent transcription (13) suggests an intimate relationship between sister chromatid cohesion and the distribution of active Pol II complexes. In addition, centromere function in S. pombe may well be perturbed by deregulated transcriptional read-through, as it is in S. cerevisiae (27). Alternatively, the impact of a loss of pfs2 function on mitosis may be indirect, through impaired expression of one or more genes required for faithful chromosome segregation. This situation would be analogous to a phenotype previously described in S. cerevisiae, where mutation of pre-mRNA splicing factors was seen to cause spindle checkpoint activation as a secondary consequence of defective tubulin gene expression (5). These two general interpretations are not mutually exclusive.
The chromosome segregation defects in pfs2 mutant cells resulted in the accumulation of multiple foci of Mad2 and Bub1, presumably marking unattached kinetochores (Fig. 1 and 4). Many of the cells with multiple Mad2 or Bub1 foci had proceeded into an anaphase-like state, suggesting that the spindle checkpoint was not properly enforced after pfs2 inactivation. Nonetheless, the severity of the mitotic abnormalities in and the loss of viability of pfs2 mutant cells were both accentuated by the deletion of bub1 (Fig. 4). These observations suggested that the spindle checkpoint was only partially activated in pfs2 mutant cells with mitotic abnormalities, a conclusion supported by the observation that Mad2 and Bub1 were not always colocalized to presumptive unattached kinetochores (Fig. 1).
Additional connections between RNA processing and chromosome segregation were suggested by the characterization of S. pombe mutants defective in the 5′-3′ exoribonuclease encoded by dhp1 (26). These mutants accumulated polyadenylated RNA in their nuclei and exhibited chromosome segregation defects similar to those reported here for pfs2 mutants. A further link specifically between pre-mRNA cleavage and polyadenylation and chromosome segregation was recently established by affinity purification of cleavage and polyadenylation complexes from S. cerevisiae and S. pombe (6, 7, 17, 25). In both yeasts, the serine-threonine protein phosphatase Glc7/Dis2 was identified as a component of these complexes after purification with a variety of tagged subunits, including Cft1 and Pfs2. In S. pombe, the dis2+ gene was first identified through a screen for mutants defective in mitotic chromosome separation (21). The same screen identified dis3+, which encodes a protein homologous to the S. cerevisiae protein Dis3p/Rrp44p, a component of the 3′-5′ exoribonuclease termed the exosome (12, 20, 21). It will be interesting to determine whether the chromosome segregation defects of dis2 and dis3 mutants, like those of pfs2 mutants, are associated with aberrant RNA processing.
A partial loss of pfs2 function in fission yeast gave rise to a dramatic elevation in the rate of chromosome loss (Fig. 5). It will be interesting to determine whether this scenario also occurs in mammals; in that case, defects in the cleavage and polyadenylation machinery could contribute to the chromosomal instability that characterizes most cancer cells.
Acknowledgments
We are grateful to Nick Proudfoot and members of his group for valuable discussions, to Paul Russell for bringing our attention to the reannotation of the pfs2 and cid14 genes, to Stephen Kearsey for comments on the manuscript, to Jean-Paul Javerzat for providing the bub1 deletion strain, and to Taro Nakamura and Chikashi Shimoda for providing the genomic DNA library.
This work was supported by Cancer Research UK, the Human Frontiers Science Program (research grant to T.T.), the Association for International Cancer Research, and the Wellcome Trust (research career development award to S.-W.W.).
REFERENCES
- 1.Allshire, R. C., E. R. Nimmo, K. Ekwall, J. P. Javerzat, and G. Cranston. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9:218-233. [DOI] [PubMed] [Google Scholar]
- 2.Amon, A. 1999. The spindle checkpoint. Curr. Opin. Genet. Dev. 9:69-75. [DOI] [PubMed] [Google Scholar]
- 3.Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 4.Basi, G., E. Schmid, and K. Maundrell. 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123:131-136. [DOI] [PubMed] [Google Scholar]
- 5.Dahan, O., and M. Kupiec. 2002. Mutations in genes of Saccharomyces cerevisiae encoding pre-mRNA splicing factors cause cell cycle arrest through activation of the spindle checkpoint. Nucleic Acids Res. 30:4361-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dichtl, B., D. Blank, M. Sadowski, W. Hubner, S. Weiser, and W. Keller. 2002. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J. 21:4125-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 8.Hofmann, J. F., and D. Beach. 1994. cdt1 is an essential target of the Cdc10/Sct1 transcription factor: requirement for DNA replication and inhibition of mitosis. EMBO J. 13:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humphrey, T., C. E. Birse, and N. J. Proudfoot. 1994. RNA 3′ end signals of the S. pombe ura4 gene comprise a site determining and efficiency element. EMBO J. 13:2441-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kearsey, S. E., S. Montgomery, K. Labib, and K. Lindner. 2000. Chromatin binding of the fission yeast replication factor mcm4 occurs during anaphase and requires ORC and cdc18. EMBO J. 19:1681-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly, T. J., G. S. Martin, S. L. Forsburg, R. J. Stephen, A. Russo, and P. Nurse. 1993. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell 74:371-382. [DOI] [PubMed] [Google Scholar]
- 12.Kinoshita, N., M. Goebl, and M. Yanagida. 1991. The fission yeast dis3+ gene encodes a 110-kDa essential protein implicated in mitotic control. Mol. Cell. Biol. 11:5839-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lengronne, A., Y. Katou, S. Mori, S. Yokobayashi, G. P. Kelly, T. Itoh, Y. Watanabe, K. Shirahige, and F. Uhlmann. 2004. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430:573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Millband, D. N., L. Campbell, and K. G. Hardwick. 2002. The awesome power of multiple model systems: interpreting the complex nature of spindle checkpoint signaling. Trends Cell Biol. 12:205-209. [DOI] [PubMed] [Google Scholar]
- 15.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 16.Murakami, S., M. Yanagida, and O. Niwa. 1995. A large circular minichromosome of Schizosaccharomyces pombe requires a high dose of type II DNA topoisomerase for its stabilization. Mol. Gen. Genet. 246:671-679. [DOI] [PubMed] [Google Scholar]
- 17.Nedea, E., X. He, M. Kim, J. Pootoolal, G. Zhong, V. Canadien, T. Hughes, S. Buratowski, C. L. Moore, and J. Greenblatt. 2003. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′ ends. J. Biol. Chem. 278:33000-33010. [DOI] [PubMed] [Google Scholar]
- 18.Nishitani, H., Z. Lygerou, T. Nishimoto, and P. Nurse. 2000. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature 404:625-628. [DOI] [PubMed] [Google Scholar]
- 19.Niwa, O., T. Matsumoto, and M. Yanagida. 1986. Construction of a minichromosome by deletion and its mitotic and meiotic behaviour in fission yeast. Mol. Gen. Genet. 203:397-405. [Google Scholar]
- 20.Noguchi, E., N. Hayashi, Y. Azuma, T. Seki, M. Nakamura, N. Nakashima, M. Yanagida, X. He, U. Mueller, S. Sazer, and T. Nishimoto. 1996. Dis3, implicated in mitotic control, binds directly to Ran and enhances the GEF activity of RCC1. EMBO J. 15:5595-5605. [PMC free article] [PubMed] [Google Scholar]
- 21.Ohkura, H., Y. Adachi, N. Kinoshita, O. Niwa, T. Toda, and M. Yanagida. 1988. Cold-sensitive and caffeine-supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. EMBO J. 7:1465-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohnacker, M., S. M. Barabino, P. J. Preker, and W. Keller. 2000. The WD-repeat protein Pfs2p bridges two essential factors within the yeast pre-mRNA 3′-end-processing complex. EMBO J. 19:37-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Proudfoot, N., and J. O'Sullivan. 2002. Polyadenylation: a tail of two complexes. Curr. Biol. 12:R855-R857. [DOI] [PubMed] [Google Scholar]
- 24.Radcliffe, P., D. Hirata, D. Childs, L. Vardy, and T. Toda. 1998. Identification of novel temperature-sensitive lethal alleles in essential beta-tubulin and nonessential alpha 2-tubulin genes as fission yeast polarity mutants. Mol. Biol. Cell 9:1757-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roguev, A., A. Shevchenko, D. Schaft, H. Thomas, and A. F. Stewart. 2004. A comparative analysis of an orthologous proteomic environment in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Mol. Cell Proteomics 3:125-132. [DOI] [PubMed] [Google Scholar]
- 26.Shobuike, T., K. Tatebayashi, T. Tani, S. Sugano, and H. Ikeda. 2001. The dhp1+ gene, encoding a putative nuclear 5′-3′ exoribonuclease, is required for proper chromosome segregation in fission yeast. Nucleic Acids Res. 29:1326-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snyder, M., R. J. Sapolsky, and R. W. Davis. 1988. Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 8:2184-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart, E., C. R. Chapman, F. Al-Khodairy, A. M. Carr, and T. Enoch. 1997. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 16:2682-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takagaki, Y., and J. L. Manley. 1992. A human polyadenylation factor is a G protein beta-subunit homologue. J. Biol. Chem. 267:23471-23474. [PubMed] [Google Scholar]