Abstract
One of the unique aspects of RNA processing in trypanosomatid protozoa is the presence of a cap 4 structure (m7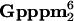 AmpAmpCmpm3Um) at the 5′ end of all mRNAs. The cap 4 becomes part of the mRNA through trans-splicing of a 39-nucleotide-long sequence donated by the spliced leader RNA. Although the cap 4 modifications are required for trans-splicing to occur, the underlying mechanism remains to be determined. We now describe an unconventional nuclear cap binding complex (CBC) in Trypanosoma brucei with an apparent molecular mass of 300 kDa and consisting of five protein components: the known CBC subunits CBP20 and importin-α and three novel proteins that are only present in organisms featuring a cap 4 structure and trans-splicing. Competitive binding studies are consistent with a specific interaction between the CBC and the cap 4 structure. Downregulation of several individual components of the T. brucei CBC by RNA interference demonstrated an essential function at an early step in trans-splicing. Thus, our studies are consistent with the CBC providing a mechanistic link between cap 4 modifications and trans-splicing.
AmpAmpCmpm3Um) at the 5′ end of all mRNAs. The cap 4 becomes part of the mRNA through trans-splicing of a 39-nucleotide-long sequence donated by the spliced leader RNA. Although the cap 4 modifications are required for trans-splicing to occur, the underlying mechanism remains to be determined. We now describe an unconventional nuclear cap binding complex (CBC) in Trypanosoma brucei with an apparent molecular mass of 300 kDa and consisting of five protein components: the known CBC subunits CBP20 and importin-α and three novel proteins that are only present in organisms featuring a cap 4 structure and trans-splicing. Competitive binding studies are consistent with a specific interaction between the CBC and the cap 4 structure. Downregulation of several individual components of the T. brucei CBC by RNA interference demonstrated an essential function at an early step in trans-splicing. Thus, our studies are consistent with the CBC providing a mechanistic link between cap 4 modifications and trans-splicing.
Flagellate protozoa of the family Trypanosomatidae include a large number of diverse organisms, many of which are parasites of great economical and medical importance. The study of the parasitic life style of these organisms has resulted in the original discovery of phenomena of fundamental biological significance, including among others antigenic variation of surface glycoproteins (5), glycosylphosphatidylinositol anchors of membrane proteins (11), and mitochondrial RNA editing (35). The examination of antigenic variation at the gene expression level revealed yet another complexity of eukaryotic biology, namely, polycistronic transcription and trans-splicing of precursor RNAs (20, 30, 43). In particular, transcription of protein coding genes by RNA polymerase II (Pol II) generates polycistronic pre-mRNAs, which are then processed by the coupled action of trans-splicing and polyadenylation to yield monocistronic mature mRNAs (24). trans-splicing transfers the 39-nucleotide (nt)-long capped spliced leader (SL) from the SL RNA to the 5′ end of mRNAs, and this process is mechanistically similar to the removal of intervening sequences (24). At present, there is no evidence for regulation of gene expression at the transcriptional level and, thus, it is very likely that posttranscriptional events play a major role in fine-tuning the output of gene products (7). In this scenario, pre-mRNA and SL RNA cis-acting signals for RNA processing, by virtue of their interaction(s) with components of RNA processing machineries, could be major determinants for regulating gene expression in trypanosomes.
Studies performed mainly in yeast and human cells have shown that the m7GpppN cap structure of nascent Pol II transcripts has key functions in various aspects of RNA processing and that these effects are mediated by two distinct cap binding complexes (CBCs) (17, 22, 39). The predominantly nuclear CBC is comprised of two subunits, termed cap binding proteins 20 (CBP20) (16) and 80 (CBP80) (17, 31). CBP20, in contrast to CBP80, is highly conserved from yeast to humans and contains an RNA binding motif (the RNP domain). Of note is that binding to capped RNA requires an association of the two subunits, with CBP20 directly contacting the cap and CBP80 ensuring high-affinity cap binding (6, 17, 28). Although the CBC subunits are not essential for viability in Saccharomyces cerevisiae (8), they play an active role in both splicing and RNA export (17, 19). CBC augments pre-mRNA processing by increasing the splicing efficiency of cap-proximal 5′ introns. Furthermore, CBC positively affects the efficiency of 3′-end formation (12). In metazoa, where U-snRNP assembly has a cytoplasmic phase, CBC, in cooperation with an adaptor protein named PHAX (for phosphorylated adaptor for RNA export), is required for the nuclear export of U-snRNAs (32). In contrast, export of mRNA does not appear to require CBC, although it accompanies mRNA to the cytoplasm (15, 22, 47), where it is exchanged for eukaryotic initiation factor 4E, the cytoplasmic cap binding protein of eukaryotic translation initiation factor 4F (13).
The mRNA cap in T. brucei has the unusual feature of containing, in addition to 7-methylguanosine, four modified nucleotides making it by definition a cap 4 structure (m7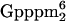 AmpAmpCmpm3Um) (3). Whereas this highly modified cap appears to be conserved throughout the evolution of the family Trypanosomatidae (34), there is at present no evidence that a similar cap structure exists outside this group of organisms. Although permeable cell studies showed that the hypermodified cap is essential for utilization of the SL RNA in trans-splicing (46) and genetic analysis in Leptomonas collosoma established that each of the four nucleotides of the cap 4 is essential for SL RNA function in trans-splicing (27), a specific role for the modified nucleotides has not been elucidated. Nevertheless, these observations reinforce the notion that cap structures have key functions in RNA metabolism. In this paper we have characterized the T. brucei nuclear CBC, which consists of five subunits and is essential for cell viability.
AmpAmpCmpm3Um) (3). Whereas this highly modified cap appears to be conserved throughout the evolution of the family Trypanosomatidae (34), there is at present no evidence that a similar cap structure exists outside this group of organisms. Although permeable cell studies showed that the hypermodified cap is essential for utilization of the SL RNA in trans-splicing (46) and genetic analysis in Leptomonas collosoma established that each of the four nucleotides of the cap 4 is essential for SL RNA function in trans-splicing (27), a specific role for the modified nucleotides has not been elucidated. Nevertheless, these observations reinforce the notion that cap structures have key functions in RNA metabolism. In this paper we have characterized the T. brucei nuclear CBC, which consists of five subunits and is essential for cell viability.
MATERIALS AND METHODS
Cell lines, epitope tagging, and other procedures.
Transfection of procyclic T. brucei cells, epitope tagging, and construction of RNA interference (RNAi) cell lines were done as described previously (2, 9). All experiments described here were carried out with clonal cell lines, and proteins of interests were tagged at the N terminus. Epitope tags used in this study included BB2 (EVHTNQDPLD), hemagglutinin (HA; YPYDVPDYA), FLAG (DYKDDDDK), and the TAPmyc tag, which was modified from the original version (36, 37) and contains a triple myc tag (EQKLISEEDLVEQKL) after the TEV cleavage site to provide a detection method after TEV cleavage. Northern blotting (9) and primer extension (26) analyses were carried out as described elsewhere.
Protein purifications.
Whole-cell and S100 extracts were prepared as described previously (10). For gel filtration chromatography, 500 μl of an S100 extract (the equivalent of about 500 ml of cells at a density of 6 × 106 cells/ml) was fractionated on a Superdex-200 column (Amersham Biosciences) as described elsewhere (10).
Tandem affinity purification (TAP) was carried out according to the methods described in reference 36. Purified protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for silver staining with the SilverQuest kit (Invitrogen). Individual protein bands (with apparent molecular masses of 30, 55, 75, and 110 kDa) were cut out and digested with trypsin, and the resulting peptides were then subjected to nano-liquid chromatography and tandem mass spectrometry (MS) analysis at the Protein Chemistry Core Facility at Columbia University. The output file from this procedure was then used to search a T. brucei database. The following matches were predicted: TbCBP30 with the 30-kDa protein MS data with an expect value of 3.1 × 10−9; T. brucei importin-α with the 55-kDa protein MS data with an expect value of 0.10; TbCBP66 with the 75-kDa protein MS data with an expect value of 0.43; TbCBP110 with the 110-kDa protein MS data with an expect value of 8.1 × 10−5.
Immunoprecipitations and affinity selection.
The following antibodies were used: mouse monoclonal anti-BB2 antibodies were raised against 10 amino acids from the immunologically well-characterized major structural protein of the S. cerevisiae Ty1 virus-like particle in Keith Gull's laboratory (4); horseradish peroxidase-conjugated rat anti-HA antibodies (Roche); mouse monoclonal anti-FLAG antibodies (Sigma); mouse monoclonal anti-myc antibodies (a generous gift from Susan Baserga); and rabbit polyclonal anti-protein A antibodies (Sigma). Immunoprecipitations were performed as described elsewhere (41).
For m7GTP-Sepharose pull-down experiments, 15 μl of m7GTP-Sepharose beads (Amersham Biosciences) was washed three times in NET-2 buffer (150 mM NaCl, 50 mM Tris-HCl [pH 7.5], 0.05% NP-40) and then mixed with 150 μl of S100 extract, 150 μl of NET-2 buffer, and 33 μl of yeast tRNA (10 mg/ml). The mixture was rotated at 4°C for 30 min, then the beads were spun down and washed five times with NET-2 buffer, and the affinity-selected material was subjected to Western blot analysis.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were carried out essentially as described previously (18). The binding reaction mixture contained 1.6 μg of yeast tRNA/μl, 1 U of RNasin (Roche)/μl, and 103 cpm of labeled SL RNA probe/μl (5.1 fmol/μl) in BB buffer (10 mM potassium phosphate [pH 8.0], 0.25 M KCl, 2 mM EDTA, and 5% glycerol). The total volume of the samples was 10 μl. Reaction mixtures were incubated at 28°C for 30 min and then on ice for 10 min. Next, each sample was mixed with 3 μl of sample buffer (5% Ficoll, 20% glycerol, and 0.05% bromophenol blue in BB buffer) and loaded onto a 5% polyacrylamide gel (40:1 acrylamide-bis-acrylamide). Gels were run at room temperature at 4 V/cm in 0.5× Tris-borate-EDTA for 6 h. Following drying, the gels were exposed to a phosphorimager screen, and data were acquired on a Cyclone PhosphorImager (Packard). Quantitation of bands was carried out using the Optiquant software (Packard).
TbCBC used in band shift assays (except for the results shown in Fig. 4, below) was purified from an S100 extract of cells expressing TAPmyc-CBP20 up to the TEV cleavage step. The concentration of TbCBC was determined by Western blot analysis, using as a standard a myc tag-containing protein (Positope; Invitrogen). The recombinant human CBC sample was a generous gift from Kristin Wilson and Richard Cerione, and its concentration was determined by SDS-PAGE followed by Coomassie blue staining, using bovine serum albumin as a standard.
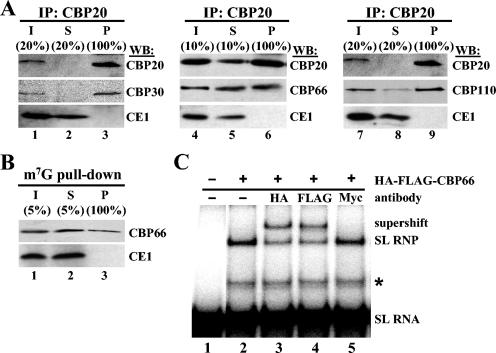
FIG. 4.
(A) S100 extracts from cells expressing BB2-tagged CBP20 and HA-tagged CBP30 (lanes 1 to 3), BB2-tagged CBP20 and HA-tagged CBP66 (lanes 4 to 6), or BB2-tagged CBP20 and HA-tagged CBP110 (lanes 7 to 9) were immunoprecipitated (IP) with anti-BB2 antibodies. The input (I; lanes 1, 4, and 7), supernatant (S; lanes 2, 5, and 8), and immunoprecipitate (P; lanes 3, 6, and 9) were then subjected to Western blot analysis (WB) with anti-BB2 antibodies to detect CBP20, anti-HA antibodies to detect CBP30, CBP66, or CBP110, and anti-capping enzyme 1 (CE1) (42) antibodies to control for specificity. (B) An S100 extract from cells expressing HA-FLAG-tagged CBP66 (CBP66) was affinity selected with m7GTP-Sepharose beads, and the input (lane 1), supernatant (lane 2), and affinity-selected material (lane 3) were subjected to Western blot analysis with anti-HA antibodies (CBP66) and anti-CE1 antibodies to control for specificity. (C) Supershift analysis of CBP66 binding to the SL RNA. 32P-labeled SL RNA (lane 1) was incubated with HA-FLAG-tagged CBP66, partially purified by fractionation on a Superdex-200 column, in the absence of antibodies (lane 2) or in the presence of anti-HA (lane 3), anti-FLAG (lane 4), and anti-myc (lane 5) antibodies. The supershift is marked. A second band shift, described in the legend of Fig. 2B, is indicated with an asterisk.
32P-labeled SL RNA was prepared in vivo in a T. brucei permeable cell system supplemented with [α-32P]UTP (45). Labeled total RNA was separated on a 6% acrylamide-8 M urea gel, and the SL RNA was cut out from the gel and recovered by soaking the gel piece in water (25). Similarly, unlabeled competitor SL RNA was prepared using the permeable cell system without the addition of [α-32P]UTP. The concentration of the unlabeled SL RNA was determined by Northern blotting using in vitro-transcribed SL RNA as a standard.
For in vitro RNA synthesis, the T7 Ampliscribe kit (Epicentre) was used. To prepare unlabeled, uncapped RNA, XhoI-digested pBluescript II KS was used as a template to yield a 96-nt-long RNA. Unlabeled m7G-capped RNA was synthesized in the presence of 6 mM m7GpppG and 1.5 mM GTP.
To calculate the apparent disassociation constant (Kd), 5.1 fmol of labeled SL RNA/μl was used, and increasing amounts of CBCs were used in EMSAs. With Kaleidagraph software, the Kd was calculated by fitting the collected data as a function of protein concentration and using the equation θ = [protein]/([protein] + Kd), where θ is the fraction of bound RNA.
For competition assays, we used a concentration of CBCs that resulted in 10 to 15% of the labeled SL RNA forming an RNP. A total of 5.1 fmol of probe/μl was mixed with various amounts of cold competitor, and then 5.0 fmol of TbCBC/μl or 5.2 fmol of HsCBC/μl was added to assemble the EMSA reaction mixture. The mixtures were processed as described above. Using Kaleidagraph software, Kd1/2 was calculated by fitting the data to the equation Y = bottom + (top - bottom)/(1 + 10 exp(log [competitor] - log [Kd1/2]), where top and bottom are the Y values at the top and bottom plateau of the curve (http://www.curvefit.com).
For supershift analysis, 100 ng of antibody was added to the EMSA reaction mixture after the 10-min ice incubation step, and then the samples were kept on ice for another 30 min, which was followed by gel electrophoresis and data collection as described for the EMSA.
RESULTS
Identification of a T. brucei CBP20 homolog.
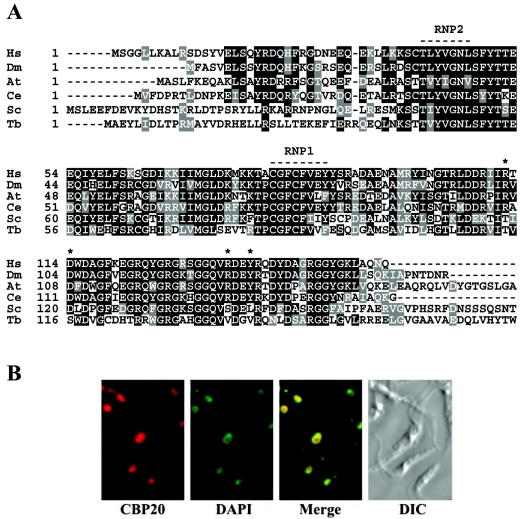
A search of the T. brucei genome database revealed a putative homolog of the 20-kDa subunit of the nuclear CBC, which we named TbCBP20 (Sanger accession no. Tb06.28P18.1290) (Fig. 1A). However, we were not able to identify the partner of CBP20 in the nuclear CBC, namely CBP80, even when using very relaxed search parameters. TbCBP20 is a single-copy gene and encodes a protein with a predicted molecular mass of 21 kDa which is predominantly localized in the nucleus (Fig. 1B). Similar to other CBP20s, the T. brucei protein contains an RNA binding domain (RNP) of about 100 amino acids, including the highly conserved RNP2 and RNP1 sequences (Fig. 1A). Overall, TbCBP20 exhibits 47% identity and 70% similarity with its human counterpart and 42% identity and 68% similarity with yeast CBP20. This homology is most evident within the RNP domain, which is the cap binding region of CBP20.
FIG. 1.
(A) ClustalW alignment of CBP20 homologs. Hs, Homo sapiens (NP_031388); Dm, Drosophila melanogaster (NP_524396); At, Arabidopsis thaliana (AAD29697); Ce, Caenorhabditis elegans (NP_492130); Sc, S. cerevisiae (AAF21454); Tb, T. brucei (TP06.28P18.1190). Two RNA binding domains (RNP) are indicated, and the asterisks highlight four nonconserved residues in the T. brucei sequence that have been shown to directly contact the m7G cap. (B) Immunofluorescence analysis of T. brucei CBP20. TAPmyc-CBP20 was immunostained with anti-protein A antibody (CBP20), and DNA was stained with 4′,6′-diamidino-2-phenylindole (DAPI). The merged picture and the differential interference contrast (DIC) image of the field are included.
To gather experimental evidence that TbCBP20 is a component of the nuclear CBC, we first generated different epitope-tagged versions of the protein in vivo by homologous recombination with PCR-generated cassettes (40). For this, we knocked out one allele of TbCBP20 by inserting the Blasticidin (BSR) resistance gene and then tagged the second allele at the N terminus either with a BB2 epitope tag (BB2-CBP20) or a modified TAP tag containing a myc epitope (TAP-CBP20). The expression of both tagged proteins was verified by Western blot analysis with anti-BB2 or anti-myc monoclonal antibodies (data not shown). Next, we incubated S100 extracts from cells expressing BB2-CBP20 with an affinity matrix containing m7GTP coupled to Sepharose. To control for specificity, we also prepared S100 extracts from two cell lines expressing BB2-tagged proteins not expected to bind to m7GTP-Sepharose, namely, an RNA methyltransferase (MT40 [reference 1]) and a cleavage and polyadenylation specificity factor involved in pre-mRNA 3′-end formation (CPSF73). Equal amounts of the three S100 extracts were mixed and, following affinity selection, pulled-down proteins were analyzed by Western blotting with anti-BB2 antibodies (Fig. 2A). This showed that TbCBP20 was specifically selected with m7G-Sepharose beads, while the two control proteins were not pulled down at a detectable level.
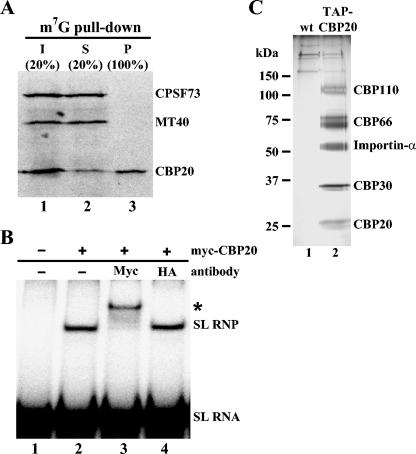
FIG. 2.
(A) m7GTP-Sepharose pull-down experiment. S100 extracts made from cells expressing either BB2-tagged CBP20 (CBP20), BB2-tagged CPSF73 (CPSF73), or BB2-tagged MT40 (MT40) were mixed together and subjected to affinity selection with m7GTP-Sepharose beads. The input (lane 1), supernatant (lane 2), and affinity-selected material (lane 3) were subjected to Western blot analysis with anti-BB2 antibodies. (B) Supershift analysis of CBP20 binding to the SL RNA. 32P-labeled SL RNA (lane 1) was incubated with TAPmyc-tagged CBP20, partially purified by the TAP method, in the absence of antibodies (lane 2) or in the presence of anti-myc (lane 3) and anti-HA (lane 4) antibodies. The supershift is indicated by an asterisk. (C) Affinity purification of the T. brucei CBC. S100 extracts from cells expressing untagged (wild type [wt], lane 1) or TAPmyc-tagged CBP20 (TAP-CBP20, lane 2) were subjected to the TAP procedure, and eluates were separated by SDS-PAGE. Gel slices containing the indicated proteins were excised, digested with trypsin, and analyzed by MS sequencing.
The results described in the above section indicated that TbCBP20 or its associated protein(s) had cap binding activity. To extend this observation, an S100 extract, prepared from cells expressing BB2-tagged CBP20, was fractionated on a Superdex-200 gel filtration column and individual fractions were assayed by Western blotting with anti-BB2 antibodies. This revealed that TbCBP20 fractionated with an apparent molecular mass of approximately 300 kDa (Fig. 3A, CBP20). Next, a selected number of fractions were tested for cap binding activity in EMSAs. For this analysis, 32P-labeled SL RNA, containing the cap 4 structure, was incubated with aliquots from the Superdex-200 fractions, and the resulting complexes were separated by nondenaturing PAGE (Fig. 3B). Two major retarded protein-RNA complexes were detected in this assay: one of them cofractionated with TbCBP20, whereas a second one did not coincide with fractions containing TbCBP20 and was not pursued further. To corroborate the binding of CBP20 to the SL RNA, we performed supershift assays (Fig. 2B). Addition of antibodies recognizing the CBP20 epitope tag resulted in a slower-migrating, antibody-protein-RNA supershift complex (lane 3) which was not seen with control antibodies (lane 4), indicating that the formation of the shifted complex was CBP20 specific. Taken together, our results so far are consistent with TbCBP20 being a component of an approximately 300-kDa complex with cap binding activity.
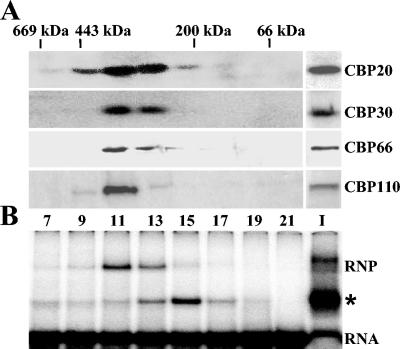
FIG. 3.
(A) Gel filtration chromatography. S100 extracts from cells expressing either BB2-CBP20 (CBP20), HA-FLAG-CBP30 (CBP30), HA-FLAG-CBP66 (CBP66), or TAPmyc-CBP110 (CBP110) were fractionated on a Superdex-200 gel filtration column, and the indicated fractions were subjected to Western blotting with the respective antibodies. The elution positions of bovine serum albumin (66 kDa), β-amylase (200 kDa), apoferritin (443 kDa), and thyroglobulin (669 kDa) are indicated. (B) EMSAs were performed with 32P-labeled SL RNA and aliquots from the BB2-CBP20 fractionation (CBP20 in panel A). A second band shift, indicated by an asterisk, did not cofractionate with the proteins shown in panel A and was not pursued further.
Purification of the TbCBC revealed a known CBC-associated protein, importin-α, and three novel proteins.
To get a handle on the protein component(s) associated with TbCBP20, we used the TAP method (see Materials and Methods). A modified TAP tag consisting of two protein A binding peptides, a TEV protease cleavage site, a triple myc tag, and a calmodulin binding peptide was inserted at the N terminus of TbCBP20 in a strain where the second allele was replaced by the BSR resistance marker. Since TbCBP20 is encoded by an essential gene (see below), this strategy ensured that the TAP-tagged protein was functional. We monitored TAP-CBP20 during the purification by Western blotting with anti-myc antibodies and verified complex integrity and its associated cap binding activity by gel shift assays (data not shown). In parallel, an equivalent amount of S100 extract from wild-type cells was processed to control for specificity. Purified samples were separated alongside on an SDS-PAGE gel and silver stained (Fig. 2C). Protein bands that were present in both the control and sample lanes were considered contaminants and were not pursued further. In addition to the band corresponding to CBP20, four proteins with approximate molecular masses of 35, 55, 75, and 110 kDa were specifically and consistently present. The four protein bands were excised from the gel and subjected to MS analysis, which led to the identification of the corresponding genes. The 55-kDa protein was tentatively identified as importin-α, or karyopherin-α, a bona fide component of the nuclear CBC (15), and was not pursued further in this study.
The most interesting result from the MS data was the identification of three novel proteins associated with the TbCBC, namely, TbCBP30, TbCBP66, and TbCBP110 (see the supplemental material for further information). We named the proteins migrating with approximate molecular masses of 35 and 75 kDa in SDS-PAGE gels (Fig. 2C) TbCBP30 and TbCBP66, respectively, to reflect their predicted molecular masses. Most intriguingly, BLAST searches of all three proteins revealed that these proteins appeared to be restricted to members of the Trypanosomatidae family, i.e., homologs were only found in Leishmania major, Trypanosoma cruzi, Trypanosoma congolense, and Trypanosoma vivax, identifying proteins with unknown functions. Additional searches of domain and motif databases were surprisingly negative for TbCBP30 and TbCBP110 and thus failed to provide us with any clues to their function. In contrast, TbCBP66 was found to contain an unusual zinc finger motif (CCCH) present in a diverse range of RNA binding proteins and an RNA recognition motif (RRM) close to the N terminus (see the supplemental material for further information).
TbCBP30, TbCBP66, and TbCBP110 are components of the TbCBC.
To verify that TbCBP30, TbCBP66, and TbCBP110 were indeed subunits of the 300-kDa CBC, we generated a series of cell lines expressing epitope-tagged versions of the three proteins and used a combination of m7G pull-down experiments, gel filtration chromatography, coimmunoprecipitations, EMSAs, and supershifts (see Materials and Methods). In a first set of experiments, S100 extracts from cells expressing either epitope-tagged CBP30, CBP66, or CBP110 were fractionated on a Superdex-200 gel filtration column in parallel with an S100 extract from cells expressing BB2-tagged CBP20 (Fig. 3A). Western blotting with the appropriate antibodies showed a similar fractionation profile for all four proteins, underscoring that TbCBP20, TbCBP30, TbCBP66, and TbCBP110 are part of an approximately 300-kDa complex. We further showed that fractions containing the epitope-tagged protein bound the SL RNA by using an EMSA (Fig. 3B). This result was corroborated by affinity selections with m7G-Sepharose, which revealed that CBP30, CBP66, and CBP110 were pulled down by the affinity matrix (Fig. 4B and data not shown). Next, we examined whether TbCBP30, TbCBP66, and TbCBP110 could be coimmunoprecipitated with TbCBP20. For this, we inserted an epitope tag at the N terminus of either CBP30, CBP66, or CBP110 in the background of cells expressing BB2-tagged CBP20. Whole-cell extracts of each cell line were then immunoprecipitated with anti-BB2 antibodies (CBP20) and subjected to Western blot assays (Fig. 4A). This showed that CBP30, CBP66, and CBP110 were coimmunoprecipitated with CBP20, whereas a control protein (CE1 [reference 42]) remained in the supernatant, indicating that each protein was in a complex with CBP20.
Since the MS data for CBP66 were not as convincing as those for CBP30 and CBP110 (see Materials and Methods), we felt it critical to add one additional experiment to confirm that CBP66 was indeed part of the complex binding the SL RNA. Thus, we performed supershift assays (Fig. 4C), which revealed a supershift with antibodies against epitopes present on CBP66 (lanes 3 and 4) but not with control antibodies (lane 5). Taken together with the TAP-tag purification results, our data provide convincing evidence that TbCBP30, TbCBP66, and TbCBP110 are components of the T. brucei nuclear CBC.
Binding specificity of the T. brucei CBC.
The presence of novel proteins in the T. brucei CBC might be a reflection of a critical interaction with the hypermodified cap 4 structure. To begin to address this issue, we used binding titration and competition experiments coupled with an EMSA. We examined two parameters of the SL RNA complex formation, namely, binding affinity and specificity, and compared them with those of the human CBC under identical assay conditions. The rationale for using the human CBC was that it will bind the cap 4 through recognition of the 5′-terminal m7G, but that the binding specificity will not be affected by the additional modifications. In contrast, we speculated that in the case of the T. brucei CBC the interaction with the SL RNA will not only be through the m7G, but also that the cap 4 structure will play a role.
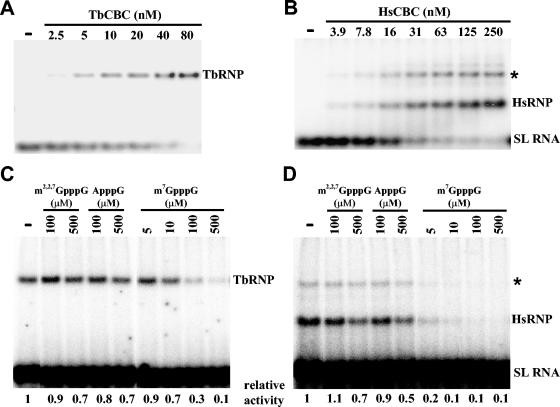
First, equilibrium binding titration assays were performed to determine the apparent disassociation constants (Kd). For this, increasing amounts of the T. brucei or human CBC were added to a constant amount of 32P-labeled SL RNA, which carries the cap 4 structure at the 5′ end (Fig. 5A and B). Whereas only one RNP complex was formed for the T. brucei CBC, two distinct complexes were detected for the human CBC, indicating that complexes of different oligomerization states were formed. Indeed, due to the presence of a glutathione S-transferase tag at the N terminus of CBP20, the human CBC has a tendency to dimerize (Kristin Wilson, personal communication). For the results reported here, both shifted bands were quantitated with a phosphorimager. In the experiment shown, the trypanosome CBC bound the SL RNA substrate with a Kd of 26 ± 5 nM (mean ± error), while the human CBC bound the SL RNA substrate with a Kd of 31 ± 4 nM.
FIG. 5.
Binding of T. brucei and human CBCs to RNA. (A and B) 32P-labeled cap 4-containing SL RNA was incubated without protein (-), with partially purified T. brucei CBC (A), or with recombinant human CBC sample (B) and analyzed by EMSA. The SL RNA concentration in panels A and B was 5.1 fmol/μl, and the protein concentrations were as indicated. (C and D) Inhibition of SL RNA binding by different analogs. Aliquots of 5.1 fmol of 32P-labeled cap 4 SL RNA/μl were mixed with various amounts of cold competitor as indicated, and then 5.0 fmol of TbCBC/μl (C) or 5.2 fmol HsCBC/μl (D) was added and processed for EMSA. The asterisk denotes a dimer that forms with the human CBC. Complex formation is expressed as relative activity with respect to the no-competitor control (-).
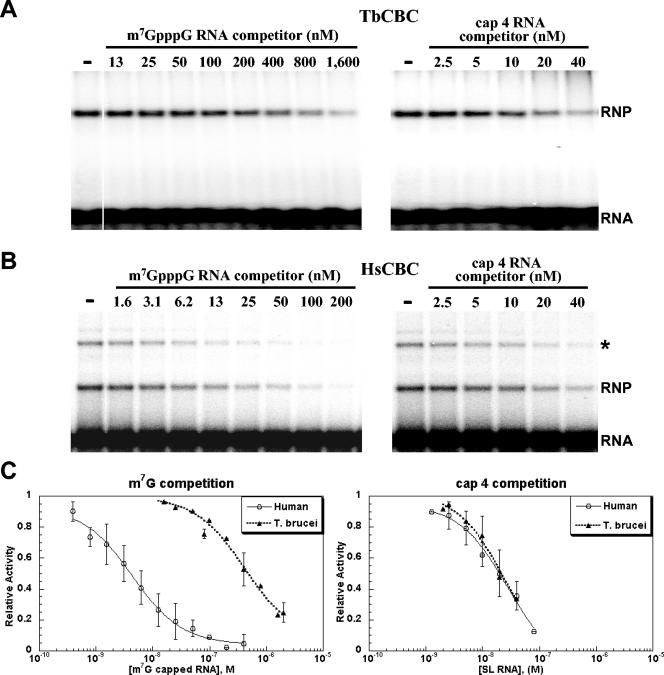
To assess the binding specificity of the RNP complexes shown in Fig. 5A and B, we carried out competition experiments. 32P-labeled cap 4 RNA substrate was mixed with increasing amounts of unlabeled competitor and then incubated with either T. brucei or human CBC at nonsaturating binding concentrations. As expected, two cap analogs, m2,2,7GpppG and ApppG, did not compete efficiently for binding of either the T. brucei or human CBC with the SL RNA (Fig. 5C and D). In contrast, m7GpppG was an efficient competitor for both CBCs. Interestingly, binding of the human complex was reduced about fivefold at the lowest concentration tested (5 μM), whereas the T. brucei complex was not affected at this concentration, and 20 times more m7GpppG was needed to obtain a competition similar to the human one.
To extend the above observation, we next performed competition experiments to examine the T. brucei and human CBCs for their ability to discriminate between m7G-capped and cap 4-containing RNA (Fig. 6). In particular, we determined the amount of competitor needed to dissociate 50% of the RNP complex (Kd1/2). In the case of human CBC, the Kd1/2 values were 4.4 and 29 nM for m7G and cap 4 RNA, respectively. On the other hand, for the trypanosome CBC, the Kd1/2 values were 410 and 27 nM for m7G and cap 4 RNA, respectively. Thus, for the trypanosome CBC, significantly more m7G competitor was needed than cap 4 competitor, while for the human CBC more cap 4 competitor was needed than m7G competitor. Finally, we calculated the relative binding affinities, using the equation Krel = [Kd1/2(m7G)]/[Kd1/2(cap 4)]. For the human CBC the ratio was 0.15, while the value for the T. brucei CBC was 15, which translates into an approximately 100-fold increase in specificity. Thus, it appeared from our experiments that the trypanosome CBC can distinguish between a cap 4 and m7G structure and that it has a much higher preference for a cap 4 substrate.
FIG. 6.
(A) TbCBC (5.0 fmol/μl) was incubated with 5.1 fmol of cap 4 SL RNA/μl in the presence of increasing amounts of m7GpppG-capped RNA (left panel) or cap 4 SL RNA (right panel). (B) HsCBC (5.2 fmol/μl) was incubated with 5.1 fmol of cap 4 SL RNA/μl in the presence of increasing amounts of m7GpppG-capped RNA (left panel) or cap 4 SL RNA (right panel). (C) Representative analysis of the competition titrations for T. brucei and human CBCs with the indicated RNA competitors. Data were plotted as the fraction of relative RNP formation with respect to the level with no competitor. Each set of data was a collection of at least two independent experiments.
TbCBP20, TbCBP30, and TbCBP110 are essential genes.
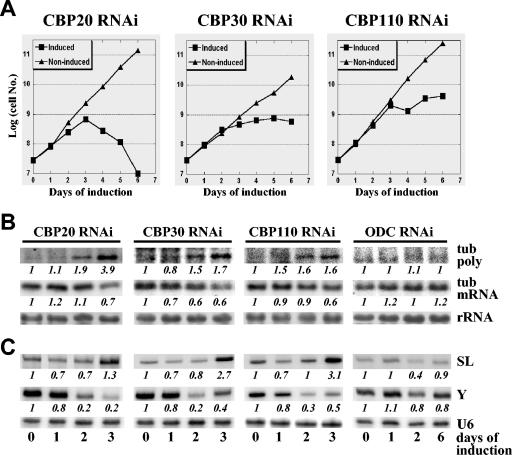
To begin to understand the function of the CBC in T. brucei, TbCBP20, TbCBP30, and TbCBP110 were downregulated one at a time by RNAi. To do this, appropriate constructs expressing hairpin-style double-stranded RNAs under the control of a tetracycline-inducible promoter were stably integrated in the nontranscribed rRNA spacer region, and clonal cell lines were established. Upon RNAi induction with tetracycline, production of double-stranded RNA and degradation of the target mRNA were verified by Northern blotting and/or reverse transcription-PCR (data not shown). Monitoring cell growth during RNAi induction revealed that all three cell lines stopped dividing after 3 days and eventually died (Fig. 7A), demonstrating that TbCBP20, TbCBP30, and TbCBP110 are essential genes.
FIG. 7.
RNAi silencing of TbCBC components. (A) Growth curves of CBP20, CBP30, and CBP110 RNAi cell lines in the absence (noninduced) or presence (induced) of tetracycline. (B) The cell lines were induced for the indicated number of days, and total RNA was subjected to Northern blotting to detect tubulin polycistronic RNA (tub poly) and tubulin mRNA (tub mRNA). RNAi of ODC, an essential gene in a posttranslational modification pathway, was used as a control for unspecific RNAi effects, and rRNA staining with methane blue served as a loading control (rRNA). The levels of tubulin polycistronic RNA and tubulin mRNA are presented as the factor increase and decrease, respectively, with respect to the amount present at day zero and were normalized to the level of rRNA (italic numbers). (C) RNA samples as described for panel B were subjected to primer extension analysis to monitor the SL RNA (SL) and Y-structure intermediate (Y). Primer extension analysis of the U6 snRNA was used to control for RNA input levels (U6). The levels of the SL RNA and Y-structure intermediate are presented as the factor increase and decrease, respectively, with respect to the amount present at day zero and were normalized to the level of U6 snRNA (italic numbers).
RNAi silencing of TbCBC subunits inhibits trans-splicing.
To examine whether the T. brucei CBC had a role in pre-mRNA processing like its yeast and mammalian counterparts (21, 22), TbCBP20, TbCBP30, or TbCBP110 was downregulated by RNAi and total RNA was prepared 1, 2, and 3 days after induction. A combination of Northern blotting and primer extension analysis was then applied to monitor the processing of tubulin RNAs. In trypanosomatids, pre-mRNAs are polycistronic and processed by trans-splicing the 39-nt-long cap 4-containing SL sequence from the SL RNA to the 5′ end of the mature mRNA (7). Like cis-splicing, trans-splicing can be described as a two-step reaction. However, since this is an intermolecular reaction, the branched intermediate is in a Y configuration and is analogous to the lariat structure of cis-splicing. We chose to monitor the maturation of tubulin mRNAs, which are encoded by a cluster of about 20 alternating units of α- and β-tubulin genes and have been the subject of previous studies (44). As shown in Fig. 7B, downregulation of TbCBP20, TbCBP30, or TbCBP110 over a period of 3 days revealed an increase of polycistronic RNA spanning two tubulin gene units. Concomitantly, the levels of mature α-tubulin mRNA decreased in CBP20 and CBP30 RNAi cell lines, but this was less obvious in CBP110 RNAi cells. No change of these RNA levels was seen in control cells, where an essential gene not involved in RNA metabolism (ornithine decarboxylate cyclase [ODC] [48]) was downregulated, and RNA samples were subjected to the same analysis.
To corroborate the involvement of the T. brucei CBC in pre-mRNA processing, the SL RNA and the Y-structure intermediate were assayed by primer extension analysis (Fig. 7C). Targeting either TbCBP20, TbCBP30, or TbCBP110 with RNAi increased the amount of the SL RNA over the 3-day induction period, with a concomitant decrease of the Y-structure intermediate. This effect was specific for the CBC-silenced cells, since the levels of both RNAs did not significantly change in ODC-silenced cells. Of note is that downregulation of CBC components did not affect trimethylguanosine-capped RNAs, like the U1 and U2 snRNAs (data not shown). Taken together, our data showed that downregulation of TbCBC components results in an accumulation of trans-splicing substrates (tubulin polycistronic RNA and the SL RNA) and in a decrease of a trans-splicing intermediate (Y-structure intermediate), as well as of the final product (tubulin mRNA), which is consistent with the TbCBC being required before the first step of the trans-splicing reaction.
DISCUSSION
In eukaryotes, the m7G cap structure on Pol II transcripts serves important functions in mRNA biogenesis pathways (22). In the nucleus, the cap is bound by a heterodimeric CBC whose subunits are not essential for viability in S. cerevisiae (8). We have shown here that T. brucei, an early divergent eukaryote, has a rather complex CBC consisting of five subunits. Silencing individual subunits by RNAi revealed that they are essential for cell viability. A striking novel finding is the existence of three subunits, namely, TbCBP30, TbCBP66, and TbCBP110, that appear to be unique to members of the Trypanosomatidae family and are not found in any other eukaryotic organism. Until now, studies in a variety of organisms have documented the nuclear CBC as consisting of a small (CBP20) and a large (CBP80) subunit (17, 18, 31). One surprising finding from high-resolution structure studies was that only the CBP20 subunit has a direct role in binding m7G-capped RNA and that the CBP80 subunit does not directly bind to or cover the cap binding site, despite the well-documented requirement of this subunit for ensuring high-affinity cap binding (6, 28, 29). The structural data correlate with the high sequence conservation of CBP20, which is reflected in the T. brucei protein: it exhibits 42% sequence identity with its human counterpart, and most of the 14 residues directly contacting the m7G cap are conserved (Fig. 1A). There are nevertheless four variable positions, namely, R112, D114, R135, and Y138, in the human protein. Except D114, the other three variations have been documented in either S. cerevisiae, Schizosaccharomyces pombe, or Encephalitozoon cuniculi (29). Although D114 interacts with the N-1 position of the guanosine base and partially ensures guanosine specificity, a D114A mutation only slightly reduced RNA binding and retained m7G base specificity (28). Thus, the T. brucei CBP20 appears to maintain the highly conserved mode of m7G binding noted throughout eukaryotic evolution.
The conservation of the amino acid sequence of CBP20 proteins contrasts with a relatively low overall conservation of CBP80s. Indeed, it has been argued that CBP80 has diverged through coevolution with its other protein binding partners and that one of its main functions is to provide a platform for binding to proteins participating in different steps of the biogenesis and transport of capped RNAs (14, 28, 29). Our efforts to identify subunits of the T. brucei nuclear CBC, either biochemically or through database mining, have so far not revealed a credible CBP80 homolog. In addition to the well-characterized CBP20 and CBP80, there are reports of several other proteins specifically associated with capped RNA. Using different cross-linking techniques, proteins of 89, 115, and 120 kDa have been identified (33, 38), but so far no function has been ascribed to these proteins and they remain poorly characterized. Furthermore, a fraction of importin-α was found in a nuclear complex with S. cerevisiae or Xenopus laevis CBC (15). Thus, we were not too surprised to recover importin-α as a subunit of the T. brucei CBC. One corollary of our finding is that the T. brucei complex most likely shuttles between the nucleus and the cytoplasm and is imported into the nucleus through importin-α, similar to what has been described for the yeast and human CBCs (15). However, at present we do not have evidence to support this view.
In addition to CBP20 and importin-α, the T. brucei CBC has subunits of 30, 66, and 110 kDa. As mentioned above, none of these proteins appears to be a homolog of CBP80, but further studies on the architecture and subunit interactions of TbCBC will be required to clarify this point. Nevertheless, our studies exposed a striking difference in the composition of the T. brucei CBC compared to other CBCs: three subunits are only found in trypanosomatid protozoa that feature cap 4 structures and trans-splicing. Our experiments further demonstrated that TbCBP30, TbCBP66, and TbCBP110 (Fig. 7A and data not shown), as well as TbCBP20, are essential genes and that RNAi-induced silencing results in trans-splicing inhibition. The fact that trans-splicing substrates accumulated and that at least one intermediate decreased suggested to us that these subunits, and most likely TbCBC, are essential at an early step of trans-splicing. Previous studies have shown that yeast and human CBCs are not absolutely essential for cis-splicing but that they enhance the assembly of an early splicing complex, the commitment complex (21, 23). Although the precise role of the T. brucei CBC in trans-splicing needs to be elucidated, it can nevertheless be speculated that TbCBC plays an essential role in assembling an early trans-spliceosome. One possibility could be that TbCBC is bringing the SL RNA to the spliceosome. In this scenario, TbCBP30, TbCBP66, and/or TbCBP110 could provide a platform to interact with components of the trans-spliceosome, or they could even be bona fide trans-splicing factors. Regrettably, the rarity of recognizable domains in their amino acid sequences makes it at present impossible to shed light on these possibilities.
One goal of the studies presented here was to extend our previous investigations on the requirement of the cap 4 structure in trans-splicing. We now have characterized a novel T. brucei CBC that binds with a 15-fold-higher affinity to cap 4-containing RNA than to m7G-capped RNA. We would predict that initial cap 4 binding involves recognition of the 5′-terminal m7G residue by CBP20 (see above). The increased specificity is then brought about by an interaction of one or more of the additional subunits with the hypermodified cap structure. Taken together with a requirement for TbCBC in trans-splicing, this complex offers an attractive link between the cap 4 structure and trans-splicing. Our results provide a starting point for functional and structural studies aimed at understanding how TbCBC interacts with the cap 4 structure, as well as with the trans-splicing machinery.
Supplementary Material
Acknowledgments
This work was supported by Public Health Service grant AI43594 from the National Institute of Allergy and Infectious Diseases to C.T.
We are grateful to Kristin Wilson and Richard Cerione for providing us with recombinant human CBC. We thank Susan Baserga, Keith Gull, and Paul Englund for their generous supply of reagents and Susan Baserga, Tony Koleske, Elisabetta Ullu, Kristin Wilson, and Sandy Wolin for critical comments on the manuscript.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Arhin, G. K., S. Shen, H. Irmer, E. Ullu, and C. Tschudi. 2004. Role of a 300-kilodalton nuclear complex in the maturation of Trypanosoma brucei initiator methionyl-tRNA. Eukaryot. Cell 3:893-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arhin, G. K., S. Shen, E. Ullu, and C. Tschudi. 2004. A PCR-based method for gene deletion and protein tagging in Trypanosoma brucei. Methods Mol. Biol. 270:277-286. [DOI] [PubMed] [Google Scholar]
- 3.Bangs, J. D., P. F. Crain, T. Hashizume, J. A. McCloskey, and J. C. Boothroyd. 1992. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 267:9805-9815. [PubMed] [Google Scholar]
- 4.Bastin, P., Z. Bagherzadeh, K. R. Matthews, and K. Gull. 1996. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 77:235-239. [DOI] [PubMed] [Google Scholar]
- 5.Borst, P., and S. Ulbert. 2001. Control of VSG gene expression sites. Mol. Biochem. Parasitol. 114:17-27. [DOI] [PubMed] [Google Scholar]
- 6.Calero, G., K. F. Wilson, T. Ly, J. L. Rios-Steiner, J. C. Clardy, and R. A. Cerione. 2002. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat. Struct. Biol. 9:912-917. [DOI] [PubMed] [Google Scholar]
- 7.Clayton, C. E. 2002. Life without transcriptional control? From fly to man and back again. EMBO J. 21:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colot, H. V., F. Stutz, and M. Rosbash. 1996. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev. 10:1699-1708. [DOI] [PubMed] [Google Scholar]
- 9.Djikeng, A., S. Shen, C. Tschudi, and E. Ullu. 2004. Analysis of gene function in Trypanosoma brucei using RNA interference. Methods Mol. Biol. 265:73-83. [DOI] [PubMed] [Google Scholar]
- 10.Djikeng, A., H. Shi, C. Tschudi, S. Shen, and E. Ullu. 2003. An siRNA ribonucleoprotein is found associated with polyribosomes in Trypanosoma brucei. RNA 9:802-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferguson, M. A., S. W. Homans, R. A. Dwek, and T. W. Rademacher. 1988. Glycosyl-phosphatidylinositol moiety that anchors Trypanosoma brucei variant surface glycoprotein to the membrane. Science 239:753-759. [DOI] [PubMed] [Google Scholar]
- 12.Flaherty, S. M., P. Fortes, E. Izaurralde, I. W. Mattaj, and G. M. Gilmartin. 1997. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc. Natl. Acad. Sci. USA 94:11893-11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fortes, P., T. Inada, T. Preiss, M. W. Hentze, I. W. Mattaj, and A. B. Sachs. 2000. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol. Cell 6:191-196. [PubMed] [Google Scholar]
- 14.Gibson, W., G. Kanmogne, and M. Bailey. 1995. A successful backcross in Trypanosoma brucei. Mol. Biochem. Parasitol. 69:101-110. [DOI] [PubMed] [Google Scholar]
- 15.Gorlich, D., R. Kraft, S. Kostka, F. Vogel, E. Hartmann, R. A. Laskey, I. W. Mattaj, and E. Izaurralde. 1996. Importin provides a link between nuclear protein import and U snRNA export. Cell 87:21-32. [DOI] [PubMed] [Google Scholar]
- 16.Izaurralde, E., J. Lewis, C. Gamberi, A. Jarmolowski, C. McGuigan, and I. W. Mattaj. 1995. A cap-binding protein complex mediating U snRNA export. Nature 376:709-712. [DOI] [PubMed] [Google Scholar]
- 17.Izaurralde, E., J. Lewis, C. McGuigan, M. Jankowska, E. Darzynkiewicz, and I. W. Mattaj. 1994. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78:657-668. [DOI] [PubMed] [Google Scholar]
- 18.Izaurralde, E., J. Stepinski, E. Darzynkiewicz, and I. W. Mattaj. 1992. A cap binding protein that may mediate nuclear export of RNA polymerase II-transcribed RNAs. J. Cell Biol. 118:1287-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarmolowski, A., W. C. Boelens, E. Izaurralde, and I. W. Mattaj. 1994. Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol. 124:627-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laird, P. W., J. C. Zomerdijk, D. de Korte, and P. Borst. 1987. In vivo labelling of intermediates in the discontinuous synthesis of mRNAs in Trypanosoma brucei. EMBO J. 6:1055-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis, J. D., D. Gorlich, and I. W. Mattaj. 1996. A yeast cap binding protein complex (yCBC) acts at an early step in pre-mRNA splicing. Nucleic Acids Res. 24:3332-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis, J. D., and E. Izaurralde. 1997. The role of the cap structure in RNA processing and nuclear export. Eur. J. Biochem. 247:461-469. [DOI] [PubMed] [Google Scholar]
- 23.Lewis, J. D., E. Izaurralde, A. Jarmolowski, C. McGuigan, and I. W. Mattaj. 1996. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 10:1683-1698. [DOI] [PubMed] [Google Scholar]
- 24.Liang, X. H., A. Haritan, S. Uliel, and S. Michaeli. 2003. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell 2:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mair, G., E. Ullu, and C. Tschudi. 2000. Cotranscriptional cap 4 formation on the Trypanosoma brucei spliced leader RNA. J. Biol. Chem. 275:28994-28999. [DOI] [PubMed] [Google Scholar]
- 26.Mandelboim, M., S. Barth, M. Biton, X. H. Liang, and S. Michaeli. 2003. Silencing of Sm proteins in Trypanosoma brucei by RNA interference captured a novel cytoplasmic intermediate in spliced leader RNA biogenesis. J. Biol. Chem. 278:51469-51478. [DOI] [PubMed] [Google Scholar]
- 27.Mandelboim, M., C. L. Estrano, C. Tschudi, E. Ullu, and S. Michaeli. 2002. On the role of exon and intron sequences in trans-splicing utilization and cap 4 modification of the trypanosomatid Leptomonas collosoma SL RNA. J. Biol. Chem. 277:35210-35218. [DOI] [PubMed] [Google Scholar]
- 28.Mazza, C., M. Ohno, A. Segref, I. W. Mattaj, and S. Cusack. 2001. Crystal structure of the human nuclear cap binding complex. Mol. Cell 8:383-396. [DOI] [PubMed] [Google Scholar]
- 29.Mazza, C., A. Segref, I. W. Mattaj, and S. Cusack. 2002. Large-scale induced fit recognition of an m7GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 21:5548-5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy, W. J., K. P. Watkins, and N. Agabian. 1986. Identification of a novel Y branch structure as an intermediate in trypanosome mRNA processing: evidence for trans splicing. Cell 47:517-525. [DOI] [PubMed] [Google Scholar]
- 31.Ohno, M., N. Kataoka, and Y. Shimura. 1990. A nuclear cap binding protein from HeLa cells. Nucleic Acids Res. 18:6989-6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohno, M., A. Segref, A. Bachi, M. Wilm, and I. W. Mattaj. 2000. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101:187-198. [DOI] [PubMed] [Google Scholar]
- 33.Patzelt, E., D. Blaas, and E. Kuechler. 1983. CAP binding proteins associated with the nucleus. Nucleic Acids Res. 11:5821-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perry, K. L., K. P. Watkins, and N. Agabian. 1987. Trypanosome mRNAs have unusual “cap 4” structures acquired by addition of a spliced leader. Proc. Natl. Acad. Sci. USA 84:8190-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pollard, V. W., S. P. Rohrer, E. F. Michelotti, K. Hancock, and S. L. Hajduk. 1990. Organization of minicircle genes for guide RNAs in Trypanosoma brucei. Cell 63:783-790. [DOI] [PubMed] [Google Scholar]
- 36.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 37.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 38.Rozen, F., and N. Sonenberg. 1987. Identification of nuclear cap specific proteins in HeLa cells. Nucleic Acids Res. 15:6489-6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shatkin, A. J. 1985. mRNA cap binding proteins: essential factors for initiating translation. Cell 40:223-224. [DOI] [PubMed] [Google Scholar]
- 40.Shen, S., G. K. Arhin, E. Ullu, and C. Tschudi. 2001. In vivo epitope tagging of Trypanosoma brucei genes using a one step PCR-based strategy. Mol. Biochem. Parasitol. 113:171-173. [DOI] [PubMed] [Google Scholar]
- 41.Shi, H., A. Djikeng, C. Tschudi, and E. Ullu. 2004. Argonaute protein in the early divergent eukaryote Trypanosoma brucei: control of small interfering RNA accumulation and retroposon transcript abundance. Mol. Cell. Biol. 24:420-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva, E., E. Ullu, R. Kobayashi, and C. Tschudi. 1998. Trypanosome capping enzymes display a novel two-domain structure. Mol. Cell. Biol. 18:4612-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sutton, R. E., and J. C. Boothroyd. 1986. Evidence for trans splicing in trypanosomes. Cell 47:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ullu, E., K. R. Matthews, and C. Tschudi. 1993. Temporal order of RNA-processing reactions in trypanosomes: rapid trans splicing precedes polyadenylation of newly synthesized tubulin transcripts. Mol. Cell. Biol. 13:720-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ullu, E., and C. Tschudi. 1990. Permeable trypanosome cells as a model system for transcription and trans-splicing. Nucleic Acids Res. 18:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ullu, E., and C. Tschudi. 1991. trans splicing in trypanosomes requires methylation of the 5′ end of the spliced leader RNA. Proc. Natl. Acad. Sci. USA 88:10074-10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Visa, N., E. Izaurralde, J. Ferreira, B. Daneholt, and I. W. Mattaj. 1996. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J. Cell Biol. 133:5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, Z., J. C. Morris, M. E. Drew, and P. T. Englund. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275:40174-40179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.