Abstract
Both the Prp18 protein and the U5 snRNA function in the second step of pre-mRNA splicing. We identified suppressors of mutant prp18 alleles in the gene for the U5 snRNA (SNR7). The suppressors' U5 snRNAs have either a U4-to-A or an A8-to-C mutation in the evolutionarily invariant loop 1 of U5. Suppression is specific for prp18 alleles that encode proteins with mutations in a highly conserved region of Prp18 which forms an unstructured loop in crystals of Prp18. The snr7 suppressors partly restored the pre-mRNA splicing activity that was lost in the prp18 mutants. The close functional relationship of Prp18 and U5 is emphasized by the finding that two snr7 alleles, U5A and U6A, are dominant synthetic lethal with prp18 alleles. Our results support the idea that Prp18 and the U5 snRNA act in concert during the second step of pre-mRNA splicing and suggest a model in which the conserved loop of Prp18 acts to stabilize the interaction of loop 1 of the U5 snRNA with the splicing intermediates.
Pre-mRNA is spliced in two sequential transesterification reactions within the spliceosome (5, 7, 34, 54). The active spliceosome is composed of the U2, U5, and U6 snRNPs together with a dynamic cast of proteins. The U2 and U6 snRNAs appear to form the catalytic core of the spliceosome, while the U5 snRNP is thought to hold the substrate RNA and to align the exons for splicing. We focus here on the second step of splicing, in which the exons are joined to form the product mRNA. The U2, U5, and U6 snRNAs play key roles in the second step, and mutations in each specifically block the second step (17, 35, 43). Six proteins, namely, Prp16, Prp17, Prp18, Prp22, Slu7, and Prp8, function specifically in the second step.
The second step of splicing can be divided into stages based on the different protein and ATP requirements of each stage. After the first transesterification reaction, the DExH-box RNA helicase Prp16 catalyzes an ATP-dependent rearrangement of the spliceosome (49, 50). Prp17 acts at this stage as well, although its function is unknown (33, 47). The Prp16-catalyzed conformational change permits the binding of Slu7, Prp18, and Prp22 to the spliceosome (6, 31); addition of these proteins allows the ATP-independent transesterification reaction to proceed (26, 33, 48). How these three proteins facilitate the second reaction is not known; however, the observations that none of the three is needed for splicing substrates with short branch point-to-3′ splice site distances and that Slu7 affects 3′ splice site choice suggest that the proteins may form a bridge between the branch site and the 3′ splice site (6, 19, 48, 61). Following exon ligation, Prp22, another DExH family member, catalyzes an ATP-dependent conformational change that releases the mRNA (14). The Prp8 protein, which is also required for the first step, appears to have multiple functions during the second step (45; reviewed in references 3 and 13).
The U5 snRNA plays a central role in the second step of splicing, in which it is hypothesized to align the exons for joining (42; reviewed in references 38 and 57). All U5 snRNAs studied have the invariant 9-nucleotide sequence 5′-GCCUUUUAC-3′ within an 11-nucleotide loop, called loop 1 (21). Both genetic and biochemical experiments show that loop 1 interacts with sequences at the ends of the exons, tethering them to the spliceosome. The bases at the 3′ end of exon 1 interact with bases U4 to U6 in loop 1, and the bases at the 5′ end of exon 2 interact with C3 and U4 (37, 39). The bases with which loop 1 interacts in both exons are not conserved, and the basis for this interaction is not well understood. Genetic experiments show that the bases in loop 1 can form base pairs with the pre-mRNA or intermediates and that this pairing can determine splice site selection in some instances (15, 37, 39). Promiscuous base-pairing by uridine residues has been suggested as one mechanism for allowing the interaction of loop 1 bases with the substrate RNAs (39), and proteins are likely to be involved as well. Interaction of exon 1 with loop 1 is established during the first step of splicing and the interaction persists through the second step, while the interaction with exon 2 is not present until the second step (1, 40, 53). In studies using model substrates in vitro, loop 1 was dispensable for the first step (despite the fact that it can alter 5′ splice site choice) and required for the second in yeast extracts (43), but it was dispensable for both steps of splicing in HeLa cell extracts (51).
Proteins are likely to play a role in stabilizing the interaction of loop 1 of U5 with the substrate. The involvement of Prp8 is perhaps the best supported by the evidence. Prp8, a component of the U5 snRNP, cross-links to loop 1 in free U5 snRNP as well as to the ends of both exons during splicing, with kinetics that parallel those of U5 snRNA cross-linking (16, 55; reviewed in references 3 and 57). Other proteins cross-link at or near the 3′ splice site during the second step, but it is not known whether they interact with the exonic sequences or with U5 (36, 58). Genetic studies based on synthetic lethal interactions suggest a network of interactions involving loop 1 of U5 and the second-step proteins Slu7, Prp8, Prp17, and Prp18 (20, 33, 58). Slu7 can affect 3′ splice site choice (19) and may strengthen the binding of exon 1 to the spliceosome (10), but Slu7 has not been shown to interact with U5.
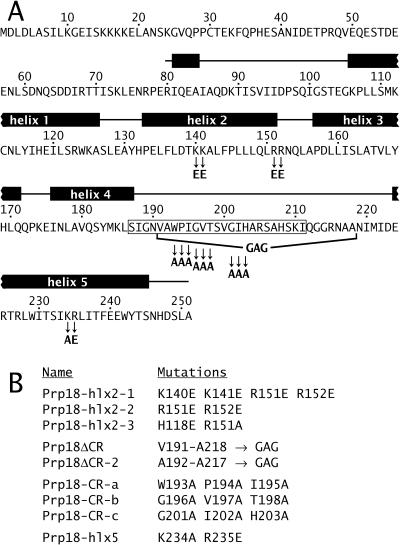
We have focused on understanding the mechanism of action of Prp18. The structure of a fully functional fragment of Saccharomyces cerevisiae Prp18 shows five tightly packed α-helices with an unstructured 36-amino-acid loop between helices 4 and 5 (Fig. 1) (32). This loop is evolutionarily conserved and includes a nearly invariant stretch of 19 amino acids. Only the carboxyl-terminal third of yeast Prp18 is conserved in human Prp18, yet yeast Prp18 can function in human splicing, showing the importance of this region to Prp18 action (28). Mutational analysis based on the structure implies that Prp18 has at least two separable functions. Prp18 interacts with Slu7, and the face of Prp18 opposite the conserved loop binds to Slu7 (2, 61). This interaction is necessary for both proteins to bind stably to the spliceosome (31). Mutant Prp18 proteins lacking their conserved region are partly functional and apparently bind Slu7 and enter the spliceosome normally (2). However, these mutant Prp18 proteins do not support wild-type growth at any temperature. Prp18 is physically associated with the U5 snRNP, although its binding is not tight (22, 27), and we had previously speculated that the conserved loop of Prp18 could interact with U5.
FIG. 1.
Locations of mutations in yeast Prp18. (A) Amino acid sequence of S. cerevisiae Prp18. The positions of α-helices (black boxes labeled 1 through 5) and loops between them (lines connecting the boxes) are shown above the sequence, as determined from the X-ray crystal structure of a large fragment of Prp18 (32). The most conserved region of Prp18 from S187 to I211 is boxed in the sequence. The positions of most of the mutations in Prp18 proteins used in this study are shown. (B) List of Prp18 mutants used; all were described by Bačíková and Horowitz (2).
We devised a genetic test to look for a functional interaction between Prp18 and loop 1 of the U5 snRNA. Suppressors of prp18 alleles that encode a mutant Prp18 protein lacking its conserved loop were found in the gene for U5 snRNA. The results imply a functional connection between the invariant loop 1 of U5 and the conserved loop of Prp18 and suggest that Prp18 could stabilize the interaction of loop 1 with the splicing intermediates.
MATERIALS AND METHODS
Plasmids.
All PRP18 mutants and plasmids are described in reference 2. The Prp18 mutants used in this study are shown in Fig. 1. A wild-type SNR7 plasmid was made by genomic PCR and cloning of the ClaI-HindIII fragment (44) into pRS316 (52). The snr7 library of U5-loop 1 mutants was obtained from Andy Newman (MRC Laboratory of Molecular Biology, Cambridge, United Kingdom) (37). The six additional point mutations in loop 1 (see Fig. 6, below) were made with the QuikChange mutagenesis kit (Stratagene).
FIG. 6.
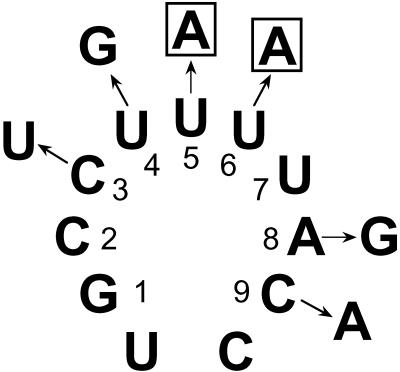
Point mutations in loop 1 of U5 snRNA. The six mutant versions of U5 snRNA shown were expressed in prp18ΔCR yeast. The snr7-U5A and snr7-U6A alleles, whose mutations are boxed in the figure, were synthetic lethal with prp18ΔCR despite the presence of a wild-type SNR7 allele in the yeast.
Strains.
The yeast strains W303-1A (MATa leu2-3,112 his3-11,15 trp1-1 can1-100 ade2-1 ura3-1), W303-1B (MATα), and W303 (diploid) were used (56). The W303-1A prp18::HIS3 strain was made as described previously (27). The W303-1A prp18::KAN strain was made by PCR with pFA6a-kanMX6 (60) replacing the coding sequence for amino acids 117 to 234 in PRP18. These two strains transformed with a pRS314-prp18 plasmid were used as mutant prp18 yeast in all U5 experiments. The W303-1A prp18::URA3 strain described previously (2) was used for making RNA.
SNR7 disruptions were made in diploid W303 by PCR with pFA6a-kanMX6 (41). W303 SNR7/snr7::KAN was transformed with pRS316-SNR7 or pRS316-snr7 and dissected, giving the haploid W303-1B snr7::KAN/pRS316-snr7 (or SNR7) strains. prp18 snr7 strains were made by crossing W303-1A prp18::HIS3 with W303-1B snr7::KAN/pRS316-snr7 and dissecting, yielding the W303-1A prp18::HIS3 snr7::KAN/pRS316-snr7 (or SNR7) strains. These were transformed with pRS314-prp18ΔCR or pRS314-PRP18 for analysis.
Yeast screens and manipulations.
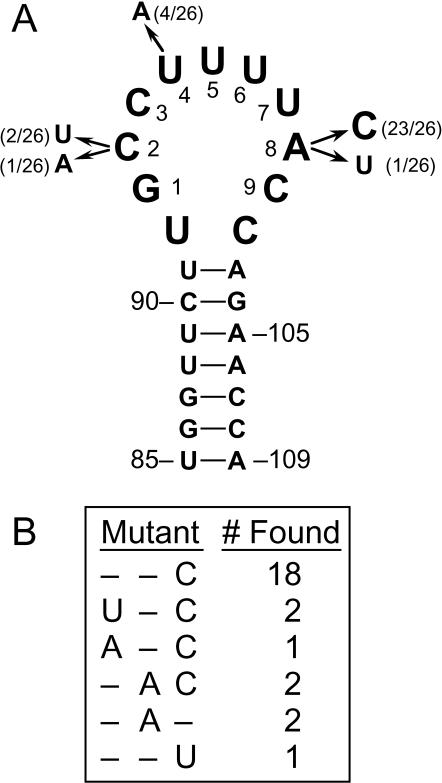
W303-1A prp18::HIS3 yeast were transformed (24) with the pRS314-derived plasmid bearing the prp18-hlx2-1, prp18ΔCR, or prp18-hlx5 alleles. These yeast plus the parent strain were then transformed with the snr7 loop 1 library (in pRS316). Two methods were used to find suppressors. In the first, the transformed yeast were grown at 23°C and replica plated to restrictive temperature. In the second, the transformed yeast were grown for 1 day at 23°C and then shifted to nonpermissive temperature. For the PRP18 knockout strain, no 34°C survivors were found from 35,000 transformants (only replica plating was used). For the prp18-hlx2-1 mutant, 25 of 16,000 (replica plating) and 0 of 9,000 (shifting) colonies grew at 34 or 37°C; all 25 of these grew at 37°C on 5-fluoroorotic acid (5-FOA; which selects for yeast cured of pRS316-snr7 and presumably identifies chromosome-plasmid recombinants that regenerated a wild-type PRP18) and were not considered further. For the prp18-hlx5 mutant, 36 of 40,000 colonies (replica plating) grew at 34°C; all of these grew at 37°C on 5-FOA and were not considered further. For prp18ΔCR yeast, 34 of 6,000 (replica plating) and 20 of 2,000 (shifting) transformants grew at 37°C. Three of these 54 initial candidates grew at 37°C on 5-FOA, and 51 were evaluated further. The U5 library plasmid was isolated from 47 candidates and was retested for suppression of prp18ΔCR. The U5 alleles from the best-scoring 26 candidates were sequenced, and all of these were U5 mutants (Fig. 2).
FIG. 2.
snr7 suppressors of prp18ΔCR. (A) Bases 85 through 109 of S. cerevisiae U5 snRNA are shown in their canonical stem-loop structure (21). The numbering of G93 through C101 as bases 1 to 9, which we use throughout the text, is indicated (37). The positions and changes of the U5 snRNAs transcribed from the suppressors are shown, together with the frequency with which each position was mutated to the nucleotide shown. (B) The sequences of the suppressors are shown, with only positions 2, 4, and 8 displayed, since all other positions are wild type, together with the number of occurrences of each suppressor.
Dominant synthetic lethality of the six mutants shown in Fig. 6 was determined by transformation. W303-1A prp18::KAN yeast bearing either the pRS314-prp18ΔCR or pRS314-PRP18 plasmid were transformed with the pRS316-snr7 plasmids. Several hundred to a couple of thousand transformants were obtained from each plasmid with yeast bearing a wild-type allele for PRP18 and from four of the plasmids (pRS316-snr7-C3U, -U4G, -A8G, and -C9A) in prp18ΔCR yeast. For snr7-U5A and snr7-U6A in prp18ΔCR yeast, no viable colonies were obtained at 26, 30, or 34°C. In some attempts an occasional transformation survivor was seen; the pRS316-snr7 plasmid was recovered from some of these yeast and the snr7 allele was sequenced, but the snr7 allele had been converted to wild type or otherwise mutated in these survivors.
RNA analysis.
W303-1A prp18::URA3 yeast transformed with pRS314, pRS314-PRP18, or one of the pRS314-prp18 plasmids were grown in SD-Trp at 26°C to an A600 of 0.5 for harvesting or at 26°C to an A600 of 0.25 and then shifted to 37°C for 2 h before harvesting. Cell pellets were frozen at −70°C. For the U5 suppressors, overnight cultures of W303-1A prp18::KAN yeast bearing either pRS314-PRP18 or pRS314-prp18ΔCR and pRS316-SNR7 or one of the pRS316-snr7 plasmids were grown in SD-Trp-Ura and were used to inoculate cultures in yeast extract-peptone-dextrose which were grown at 30 or 34°C to an A600 of 0.5 for harvesting or at 30°C to an A600 of 0.25 and then for 30 min or 2 h at 37°C before harvesting (11). Alternatively, W303-1A prp18::HIS3 yeast were used as above, except that SD-Trp-Ura was used throughout.
RNA was prepared by hot phenol extraction, essentially according to the method described in reference 12. Six micrograms of total RNA per lane was run in agarose-formaldehyde gels (46), and blots were probed according to the method of Cheng and Abelson (9). DNA for making probes was obtained by cloning appropriate PCR products of yeast genomic DNA into Bluescript KS(−). Probes for mRNAs were made by random priming of gel-purified restriction fragments (18), and the oligonucleotide probe for SCR1 RNA was obtained from Tharun Sundaresan (Uniformed Services University of the Health Sciences, Bethesda, Md.). Probes specific for introns were made by 35 cycles of reactions of 2 ng of intronic DNA fragment, 2 pmol of antisense primer, 50 μCi of [α-32P]dCTP, and Taq DNA polymerase in 20 μl to generate single-stranded, full-length probe. Blots were quantitated with a Molecular Dynamics PhosphorImager.
RESULTS
Experimental rationale and design.
We surmised that Prp18 played a role in stabilizing the interaction of loop 1 of U5 with the splicing intermediates or products. This conjecture was based on the association of Prp18 with the U5 snRNP (27), the specific requirement of loop 1 of U5 for the second step of splicing (43), and the synthetic lethal interaction of the prp18-1 allele with mutations in the part of the U5 snRNA gene (SNR7) that corresponds to loop 1 (20). Cross-linking results suggest that Prp8 is involved in this stabilization (16, 55), and we have used a complementary genetic approach to look for evidence of a functional interaction between Prp18 and loop 1 of U5. We reasoned that splicing of some pre-mRNAs in prp18 yeast could be enhanced by mutations in loop 1 of U5 that strengthened its base-pairing with these substrate RNAs. Thus, we sought suppressors of four prp18 alleles in yeast bearing a wild-type copy of the U5 snRNA gene (SNR7) together with a copy in which the bases corresponding to loop 1 had been randomly mutated (37).
We used three functionally distinct mutants of Prp18 plus a PRP18 knockout strain for our suppressor search (2). Two of these are multiple point mutants (shown in Fig. 1): the Prp18-hlx2-1 protein has four mutations in helix 2 that disrupt its interaction with Slu7, and the Prp18-hlx5 protein has two mutations in helix 5 that may interfere with the interaction of Prp18 with another, unidentified splicing factor. In the third mutant protein, Prp18ΔCR, 28 of the 36 amino acids that comprise the conserved loop between helices 4 and 5 have been deleted. Previous work provided strong evidence that the three mutant proteins fold properly (2). In particular, prp18ΔCR is dominant negative at all temperatures when highly expressed; the Prp18ΔCR protein apparently enters the spliceosome but is not fully functional.
Isolation of suppressors.
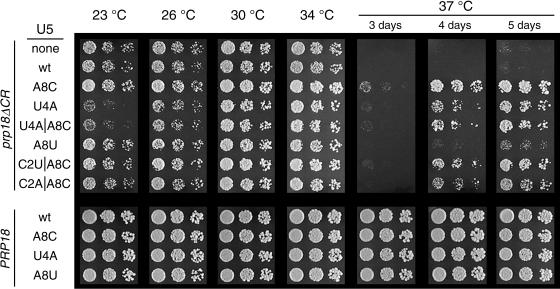
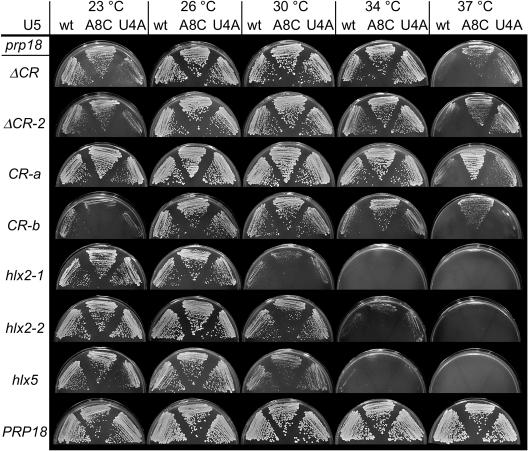
Suppressors of the three prp18 alleles described above and of a prp18 knockout allele were sought in the snr7 library. Yeast with a disrupted PRP18 gene were transformed with a prp18-bearing plasmid followed by a plasmid with an snr7 allele from the library of loop 1 mutants (37). A total of 8,000 to 40,000 candidates for each prp18 allele (depending on the transformation efficiency) from the randomly mutagenized library of U5 mutants were screened by replica plating or by temperature shifting of the plates to the lowest reliably nonpermissive temperature for each prp18 allele. For three of the prp18 alleles (prp18-hlx2-1, prp18-hlx5, and a prp18 knockout), no suppressors were found. For prp18ΔCR, 54 high-temperature (37°C) suppressors were found, of which the best-scoring 26 were analyzed further. Figure 2 shows the mutations in loop 1 of the U5 snRNA from the snr7 suppressors projected on a secondary structure diagram (panel A) and a tabulation of the sequences (panel B). Figure 3 shows the growth of prp18ΔCR and wild-type yeast with the isolated suppressors over a range of temperatures. The suppressors, which were isolated in yeast with both a wild-type and a mutant allele of SNR7, appear to be dominant.
FIG. 3.
Growth of prp18ΔCR and wild-type yeast bearing U5-loop 1 suppressors. Yeast with a HIS3-disrupted PRP18 were transformed with a plasmid bearing prp18ΔCR or PRP18 (indicated at the left) plus a plasmid with a mutant of SNR7 (U5 snRNA) (shown under the U5 heading immediately left of the photographs). Yeast were spotted in fivefold dilutions and grown for 5 days at 23°C, 4 days at 26°C, 3 days at 30 or 34°C, or 3 to 5 days at 37°C, as indicated at the top. The yeast shown here had a wild-type, chromosomal copy of SNR7.
The suppressors fall into two classes. We named the snr7 suppressors by appending the loop 1 mutation(s) to snr7: hence, for example, snr7-A8C. The major class has a mutation at position A8 (24 of 26 suppressors). snr7-A8C itself accounts for 18 of 26 suppressors and appears to be the strongest of the suppressors. Three suppressors have mutations at C2 in addition to A8C, but these were not found without A8C and did not improve suppression. snr7-A8U was also found, but it is the weakest of the six suppressors (Fig. 3). The second class of suppressor has the mutation U4A, which was found by itself and with A8C. The suppressors all have one or two mutations in loop 1, in contrast to the multiply mutant sequences found by Newman and Norman (37), who had selected for splicing of a specific mutant message in a wild-type strain, using the same library.
The U5-loop 1 suppressors have a clear salutary effect on growth of prp18ΔCR yeast, but they do not fully restore wild-type growth (Fig. 3). The suppressors are graded in their effect, with snr7-A8C being the strongest and snr7-A8U the weakest. Yeast bearing the prp18ΔCR allele grow more slowly than wild-type yeast at 23 to 34°C (Fig. 3 and reference 2), and snr7-A8C suppressed the prp18ΔCR phenotype at all temperatures. In contrast, both suppressor alleles with a U4A mutation exacerbated the slow-growth phenotype of prp18ΔCR yeast at 23 and 26°C. None of the suppressors had a discernible effect when expressed in wild-type cells (Fig. 3).
Genetic characterization of the suppressors.
The U5-loop 1 suppressors are specific for prp18 alleles that encode Prp18 proteins with mutations in their conserved regions. We tested two representative alleles, snr7-A8C and snr7-U4A, for suppression of seven different prp18 mutant alleles plus the original conserved-region deletion allele (Fig. 4). A second conserved-region deletion mutant, prp18ΔCR-2, which is very similar to prp18ΔCR, was suppressed by both snr7-A8C and snr7-U4A. Three point-mutant alleles that encode Prp18 proteins in which three consecutive invariant amino acids in the conserved region have been replaced by alanines (Fig. 1) were suppressed well by snr7-A8C, but only one of these alleles, prp18-CR-a, was clearly suppressed by snr7-U4A (prp18-CR-a and prp18-CR-b are shown in Fig. 4; prp18-CR-c is not). Three other prp18 alleles, including the prp18-hlx2-1 and prp18-hlx5 alleles that we had used in the initial suppressor screen plus the allele prp18-hlx2-2 (Fig. 1) that is less temperature sensitive than prp18-hlx2-1, were not detectably suppressed by either of the snr7 alleles. We conclude that suppression is specific for mutations within the conserved loop of Prp18 and that snr7-A8C is a more general suppressor than snr7-U4A.
FIG. 4.
Allele specificity of U5-loop 1 suppressors. Assays of seven prp18 alleles with two representative suppressors are shown. Yeast with a HIS3-disrupted PRP18 were transformed with a plasmid bearing the prp18 mutant (or wild-type) allele, indicated at the left under the prp18 heading and with a second plasmid bearing the SNR7 (U5 snRNA) allele indicated at the top of the photographs. The prp18 alleles used are shown in Fig. 1. Yeast were streaked and grown at the temperature indicated at the top for 4 days at 37°C and as stated in the legend to Fig. 3 at other temperatures.
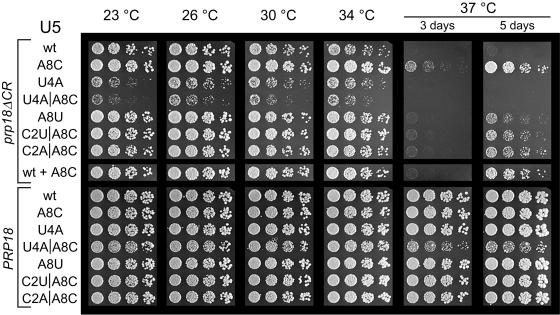
Only the suppressors that have the A8C mutation are effective when there is no wild-type copy of the U5 snRNA gene. We tested the U5-loop 1 suppressors, which had been isolated in strains with a chromosomal, wild-type SNR7 gene, in yeast that had only a mutant copy of SNR7 (Fig. 5). The suppression of the prp18ΔCR growth phenotype by snr7-A8C was independent of the presence of SNR7 (Fig. 5, compare A8C with wild type plus A8C). Likewise, suppression by the snr7 alleles with A8C combined with C2U and C2A or by A8U did not depend on the wild-type SNR7. In contrast, the snr7-U4A allele and, more dramatically, the snr7-U4A A8C allele, slowed the growth of prp18ΔCR strains at 23 to 34°C (below the already slow growth rate of the prp18ΔCR strain) and did not suppress the temperature sensitivity conferred by prp18ΔCR in the absence of a wild-type SNR7. This finding implies that in a prp18ΔCR strain some pre-mRNAs are spliced more efficiently with the U4A U5 snRNAs, while the rest are more efficiently spliced with wild-type U5 snRNA. In an snr7 knockout strain that was otherwise wild type, five of the six snr7 suppressor alleles had no apparent effect on growth, whereas the snr7-U4A A8C double mutant conferred a slow-growth phenotype at high temperature (Fig. 5).
FIG. 5.
Growth of yeast with a single mutant allele of SNR7 (U5 snRNA gene) with a prp18ΔCR or wild-type PRP18 allele. Haploid snr7::KAN prp18::HIS3 yeast carrying a mutant or wild-type SNR7 plasmid were transformed with a plasmid bearing prp18ΔCR or PRP18 as indicated at the left. The yeast were spotted in fivefold dilutions and grown as described in the legend to Fig. 3. Yeast whose U5 is indicated as wt + A8C had both a chromosomal wild-type and plasmid-borne snr7-A8C for comparison with Fig. 3.
To determine whether other SNR7 point mutations that we did not find in our screen would also suppress prp18ΔCR, we made six additional single mutations in SNR7 in bases corresponding to loop 1 of U5 and assayed them for suppression of the temperature sensitivity of a prp18ΔCR SNR7 strain. Three of the snr7 alleles, A8G, C9A, and U4G (Fig. 6), had no effect; snr7-C3U was a weak suppressor at 37°C (slightly weaker than snr7-A8U). Unexpectedly, both the snr7-U5A and snr7-U6A mutations were dominant synthetic lethal at all temperatures in a prp18ΔCR SNR7 strain (that is, they killed the yeast despite the presence of a wild-type gene for U5) (data not shown). None of the six snr7 alleles had a discernible effect on the growth of wild-type yeast.
Effect of mutations in PRP18 on splicing in vivo.
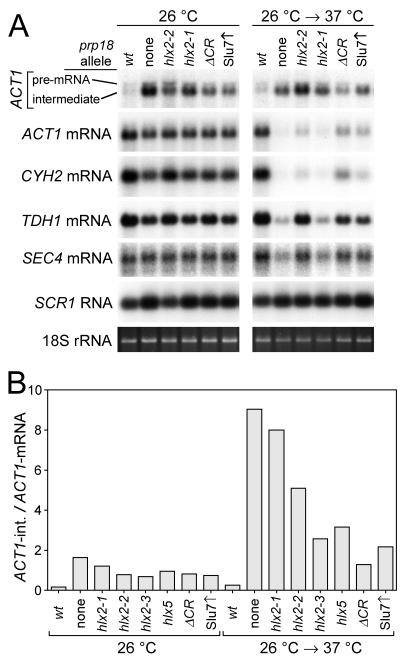
As a prerequisite to understanding the effect of the snr7 suppressors on splicing, we evaluated the splicing defects of representative prp18 yeast. Four spliced mRNAs, ACT1, CYH2, POP8, and RP51a, were assayed using Northern blotting, but splicing intermediates could be reliably quantitated only for ACT1 (Fig. 7A). Three intronless RNAs, the TDH1 and SEC4 mRNAs and the SCR1 RNA, were also assayed by blotting. Samples were loaded by comparison of A260 values, which reliably agreed with rRNA levels measured by staining (Fig. 7A).
FIG. 7.
RNA levels in prp18 mutants. (A) RNAs were detected by Northern blotting of denaturing agarose gels of total RNA extracted from a wild-type strain, four prp18 mutant strains, and a prp18 knockout strain in which Slu7 was overexpressed, as indicated at the top of blots. The prp18 mutants are shown in Fig. 1. Yeast were grown either at 26°C, a permissive temperature for all the strains, or at 26°C with a 2-h shift to 37°C, a restrictive temperature, as indicated at the top of the panel. For ACT1, the pre-mRNA (1,750 bases) and the lariat-exon 2 intermediate (1,600 bases) were detected with an intron-specific probe, and the mRNA (1,450 bases) was detected with a full-length probe (59). The faint band for the 309-base released intron is not shown. CYH2, TDH1, and SEC4 mRNAs were detected with full-length probes, and SCR1 RNA was detected with an oligonucleotide probe. 18S rRNA was detected by staining with ethidium bromide. (B) The ratio of ACT1 intermediate to mRNA was determined from quantitation of Northern blots of RNA extracted from the indicated prp18 mutant strains grown at 26°C or shifted to 37°C. The prp18 mutants are shown in Fig. 1. The units on the y axis are arbitrary and do not represent the actual i/m ratio. The ACT1 lariat-intermediate runs just above the mRNA, and the intermediate would be visible in the section of the blot shown in panel A if there were enough to see.
We examined the amounts of splicing intermediates and products in prp18 mutant yeast (Fig. 7A). At 26°C, a permissive temperature for all the mutants, ACT1 splicing intermediates accumulated in amounts well-correlated with the severity of the prp18 mutation. After a 2-h shift to 37°C, the amounts of accumulated intermediates could not be as readily interpreted: some amounts declined (perhaps from a decline in transcription), and the highest levels were found in yeast with the weakest ts alleles (e.g., prp18-hlx2-2). The amounts of the spliced ACT1 and CYH2 mRNAs were reduced less than 2-fold by prp18 mutations at 26°C and were sharply reduced (up to 15-fold) following a 2-h shift to 37°C. The behavior of the RP51A and POP8 mRNAs was similar to that of the ACT1 and CYH2 mRNAs (data not shown). The amount of accumulated intermediate was only a small fraction of the amount of mRNA; presumably, the intermediates that are not spliced rapidly are degraded (4, 25).
The amounts of the intronless mRNAs for TDH1 and SEC4 varied in the prp18 strains (Fig. 7A). At 26°C, the level of SEC4 mRNA is relatively constant (11), but the level of TDH1 mRNA changed in parallel with the levels of ACT1 and CYH2. At 37°C, the level of SEC4 mRNA varied up to 2.5-fold and that of TDH1 mRNA varied up to 4-fold. The SCR1 RNA, a polymerase III transcript that has been used as a standard (8), varied about 1.6-fold and was higher in prp18 yeast than in wild-type yeast. These variations in the levels of intronless mRNAs, even at permissive temperature, indicate that transcription and/or RNA stability is affected in the prp18 mutants and they complicate the interpretation of the levels of the spliced mRNAs. The four spliced mRNAs we examined were affected substantially more than the intronless mRNAs at 37°C, but not at 26°C.
The ratio of splicing intermediates to mRNA (termed the i/m ratio) may provide the best relative measure of severity of Prp18 mutation (Fig. 7B). If the yeast are at steady state, then the i/m ratio is directly proportional to the rate constant for the second step (using the kinetic formulation of Frank and Guthrie [19]). Comparison of different prp18 strains using the i/m ratio is independent of transcription rates and of the decay rates of intermediates, but it is sensitive to mRNA decay rates. Using the i/m ratio for comparison has the practical advantage that normalized comparisons of absolute amounts of RNA are avoided. The prp18 knockout strain had the highest i/m ratio at both assay temperatures (Fig. 7B); the sixfold increase in the ratio between 26 and 37°C shows that the second step is slowing at high temperature, as expected from in vitro results (27). In the three prp18-hlx2 strains that are temperature sensitive to different extents, the i/m ratio tracked the severity of the alleles at both 26 and 37°C (Fig. 7B). The prp18-hlx5 mutant gave an i/m ratio at 26°C that was apparently in good accord with its severity (between that of prp18-hlx2-1 and prp18-hlx2-2), but the ratio increased only half as much at 37°C as those of the prp18-hlx2 mutants.
The prp18ΔCR allele behaved differently from the other prp18 alleles. At 26°C its i/m ratio was comparable to those of the other strains; however, on shifting to 37°C the ratio increased only 1.6-fold, apparently as a result of a decrease in mRNA, not an increase in intermediates (based on the A260 normalization). In addition, at 37°C the level of the intronless TDH1 mRNA declined almost as much as that of ACT1 mRNA. The chemistry of the Prp18ΔCR protein may explain the differences between its behavior and that of the other Prp18 mutants. The Prp18ΔCR protein binds to the spliceosome at all temperatures but does not function correctly, and its functional defects may be exacerbated at high temperature (2). The mutations in the other Prp18 proteins interfere with interactions of Prp18 with the spliceosome, perhaps blocking or inhibiting its entry into the spliceosome at nonpermissive temperature, leading to a more pronounced effect on splicing upon temperature shift. Overexpression of Slu7 partially compensates for loss of Prp18 (33, 61), and we suggested that the splicing phenotype of prp18 knockout strains in which Slu7 was overexpressed would be similar to that of prp18ΔCR yeast (2), and the data generally supported this notion (Fig. 7).
The results support the idea that the i/m ratio is a reliable method for comparing splicing defects. Its utility appears to extend to yeast that have been shifted to nonpermissive temperature, although these cannot be strictly at steady state. We were only able to measure the ratio for ACT1 RNAs, and we cannot tell whether splicing of ACT1 pre-mRNA is directly affected or is reporting the status of splicing in general, as might be the case if splicing factors are sequestered in inactive complexes.
Effect of suppressors on splicing in prp18ΔCR mutants.
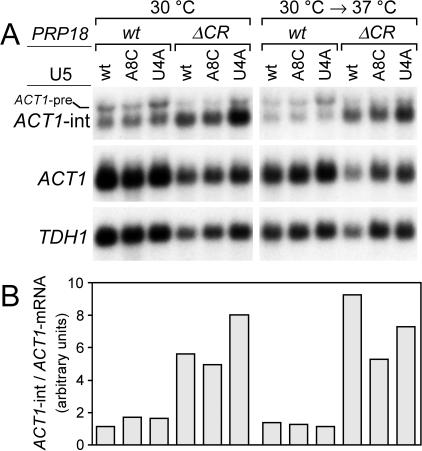
Assays of splicing in a prp18ΔCR strains showed that the snr7 suppressor alleles do suppress the prp18ΔCR splicing phenotype, although the suppression effects were relatively modest. As described above, the ratio of intermediates to products is a kinetically interpretable measurement of splicing efficiency and is well-suited to measuring small differences because it does not depend on absolute comparisons between samples. Levels of ACT1 splicing intermediates and mRNA were measured at 30 to 37°C in prp18ΔCR SNR7 strains bearing a plasmid with either the SNR7, snr7-A8C, or snr7-U4A allele.
The effects of the two snr7 alleles tested were not the same. The snr7-A8C allele reduced the i/m ratio by 1.6-fold ± 0.2-fold at 37°C and had a smaller effect at 34 and 30°C (Fig. 8). The change in the ratio appeared to result primarily from an increase in the amount of ACT1 mRNA, not from a decrease in splicing intermediates, based on the A260 normalization. The level of the intronless TDH1 mRNA also increased in the suppressed strains, so that there was no relative change in the ratio of ACT1 to TDH1 mRNAs at any temperature. The snr7-U4A allele caused a small decrease, 1.3-fold ± 0.2-fold, in the intermediates to products ratio at 37°C but caused a 1.4-fold ± 0.1-fold increase in the ratio at 30°C, consistent with the deleterious effect of snr7-U4A on growth of prp18ΔCR yeast at 30°C (Fig. 5). In snr7-U4A strains, the amounts of intermediates and both ACT1 and TDH1 mRNAs were apparently larger than in unsuppressed strains at all temperatures (although this conclusion rests on the accuracy of our normalization scheme). The suppression effects of both snr7 alleles as quantified by their effects on the intermediates to products ratio were small—only 1.6-fold at their largest. The magnitudes of these effects are similar to the change seen on shifting prp18ΔCR yeast from permissive to restrictive temperature. The observed suppression effect is consistent with the idea that the snr7 suppressors reverse the splicing phenotype of the prp18ΔCR strain, although the size of the effects does not allow us to draw a definitive conclusion. Models in which other steps of splicing are affected to suppress indirectly are disfavored by our results.
FIG. 8.
Splicing in prp18ΔCR yeast with snr7 suppressors. (A) ACT1 lariat intermediate (ACT1-int), ACT1 mRNA, and TDH1 mRNA were assayed by Northern blotting of total RNA isolated from PRP18 (wild-type) or prp18ΔCR yeast with a wild-type SNR7 or suppressor snr7-A8C or snr7-U4A allele on a plasmid together with a chromosomal wild-type SNR7 gene. The PRP18 allele, the plasmid-carried form of U5, and the temperature of growth are indicated at the top. Yeast were shifted to 37°C for 2 h. The three RNA species, plus the ACT1 pre-mRNA (ACT1-pre), are indicated in the three panels, which show sequential probings of one blot with ACT1 intron, ACT1 full-length, and TDH1 full-length probes. (B) The ratios of ACT1 lariat intermediate to ACT1 mRNA, determined from quantitation of blots in panel A, are shown. The histogram is aligned with panel A and shares the key at the top of the figure. The units on the y axis are arbitrary and do not represent the actual i/m ratio. Representative data from one experiment are shown here; in the text averages from at least three measurements are used.
DISCUSSION
We report the isolation and characterization of suppressors of prp18 alleles in the SNR7 (U5 snRNA) gene. Based on previous genetic and biochemical evidence that connected Prp18 with the U5 snRNP, we specifically sought suppressors of mutant PRP18 alleles in a library of snr7 alleles with mutations in bases corresponding to loop 1 of the U5 snRNA, as explicated at the beginning of Results. Two U5 suppressors of the prp18ΔCR allele, snr7-U4A and snr7-A8C, were identified. The snr7-A8C allele is the stronger of the two suppressors and is dominant, whereas snr7-U4A only suppresses in the presence of a wild-type SNR7 allele. Suppression is specific to prp18 alleles that encode Prp18 proteins with mutations in their conserved regions, suggesting that the evolutionarily invariant loop 1 of U5 and conserved loop of Prp18 function together during the second step of splicing. The interdependence of the functions of Prp18 and U5 is emphasized by the finding of a dominant synthetic lethal interaction between two snr7 loop 1 mutations and the prp18ΔCR allele. Analysis of mRNAs from prp18ΔCR strains showed that the splicing defect of prp18ΔCR strains was partly compensated by both of the snr7 suppressor alleles, consistent with the notion that the suppressors restore the lost function(s) to the spliceosome. Our results show that the conserved loop of Prp18 interacts genetically with loop 1 of U5 and suggest a direct functional interaction between them. We suggest that Prp18 acts to stabilize the interactions between loop 1 and the splicing intermediates during the second step of splicing. Previous structural and mutational studies of Prp18 showed that the face of Prp18 that is opposite the conserved loop interacts with the Slu7 protein, and the results here suggest that Prp18 forms a bridge between U5 and Slu7.
The suppression results provide important information about the roles of Prp18 and the U5 snRNA in splicing. The suppressors display some allele specificity in that they only suppress mutations within the conserved loop of Prp18, but because they suppress a deletion of the conserved region they do not imply a direct physical interaction in the way that a true allele-specific suppressor would (23). The suppressors' specificity instead appears to be for one function of Prp18, and the suppressors must then replace that function, essentially acting as bypass suppressors. The measurement of the effect of the suppressors on splicing, which is described in more detail in Results, supports the idea that the defect of Prp18ΔCR in splicing has been overcome by the suppressors, but the specific mechanism cannot be inferred. We envision two general types of mechanistic models for the suppression. In the first type, the suppressors restore the Prp18ΔCR-affected process, implying a close functional connection between the conserved loop of Prp18 and loop 1 of U5 snRNA. In the second type, the suppressors bypass the need for the process affected by Prp18ΔCR, perhaps by interfering with a checkpoint or proofreading step that would slow or halt splicing in prp18ΔCR strains; no proofreading steps are known at or after the second step. On balance, we think that the evidence favors a mechanism in which the Prp18ΔCR-affected process is restored by the snr7 suppressors.
Our conclusions considerably extend earlier results concerning the interaction of Prp18 and U5. Two previous studies addressed the connection of Prp18 and U5. Frank et al. (20) found a synthetic lethal interaction between prp18-1 and two snr7 alleles with mutations in the loop 1 region. Horowitz and Abelson (27) found that Prp18 is associated with the U5 snRNP by coimmunoprecipitation. Our study provides new information in two ways. First, our results imply that the interaction of Prp18 with U5 specifically involves the conserved loop of Prp18 and, second, our suppression results suggest a specific functional relationship between Prp18 and loop 1 of U5. Suppressors provide much stronger evidence of a close functional connection than synthetic lethals (20, 23, 30).
The roles and interactions of the bases in loop 1 of U5 have been investigated, and the current model suggests how our suppressor U5 snRNAs could function. The snr7-U4A allele, our weaker suppressor, is easier to interpret within the framework of known U5 actions. Base U4 of loop 1 interacts with the 3′-terminal base in exon 1 as well as the 5′-terminal base in exon 2; these interactions can occur by base-pairing, although strict base complementarity cannot be required (39). The 3′-terminal base of exon 1 interacts with U4 during both steps of splicing while interaction with exon 2 occurs after the first step of splicing, but it is not known whether U4 could interact with both exons simultaneously (40, 53). We imagine that mutating U4 to A could strengthen the interaction(s), perhaps through base-pairing, with some transcripts, thereby facilitating their splicing. Obviously, interactions with other transcripts would be weakened, inhibiting their splicing. This view is sustained by the observation that in a prp18ΔCR strain the snr7-U4A allele only works well in the presence of a wild-type SNR7 allele. That is, in the SNR7/snr7-U4A prp18ΔCR strains, there is a mandatory division of labor, with each U5 snRNA splicing a subset of the pre-mRNAs optimally. In a PRP18 wild-type strain, snr7-U4A works fine (i.e., an snr7-U4A strain grows normally), consistent with the idea that Prp18 acts to stabilize interactions of loop 1 with the substrate RNA. Our results on the effect of the snr7-U4A allele on ACT1 splicing do not lead to an unequivocal conclusion, with splicing inhibited at 30°C and improved at 37°C. A simple base-pairing model would not explain improvement of splicing of ACT1 pre-mRNA (exon 1 ends with TCTG-3′ and exon 2 begins with 5′-AGG), suggesting either that there is a different type of interaction or that ACT1 is reporting the status of splicing in general, as described in Results.
The snr7-A8C suppressor is difficult to interpret mechanistically because of the paucity of data about the function of A8. Newman and Norman (39) found that an A8C mutation within a multiply mutant loop 1 had complex effects on splicing of a model pre-mRNA with a disabling G-to-A mutation at the first base of the intron, but A8C did not have a determinative role in splice site choice. snr7-A8C is a stronger suppressor of prp18ΔCR than snr7-U4A. snr7-A8C is dominant, and it suppresses the splicing phenotype of prp18ΔCR more persuasively than does the snr7-U4A allele. snr7-A8U is a weak suppressor of prp18ΔCR, but snr7-A8G is not. We suggest three possible mechanisms for snr7-A8C suppression. First, A8 could interact with some pre-mRNAs, and A8C could restore splicing by base-pairing interaction. No evidence suggests that A8 interacts with substrate RNA in the pre-mRNAs that have been studied, although base-pairing has been suggested for the adjacent base U7 (39), and there may be some flexibility in the way that loop 1 interacts with pre-mRNAs (15, 42). Second, A8C could change the conformation of loop 1, indirectly affecting or enhancing the ability of loop 1 to interact with the substrate RNA. We view both of these mechanisms as restoration-of-function suppressors. Third, A8C could bypass the role of the conserved loop of Prp18, perhaps by disabling a checkpoint, as described above. From on our results we do not favor any one of these models over another.
We found that the snr7-U5A and snr7-U6A alleles were both dominant synthetic lethal with prp18ΔCR, killing the yeast despite the presence of a wild-type allele of SNR7. Both the U5 and U6 bases pair with the 3′ end of exon 1, and mutation at either position can activate cryptic 5′ splice sites. Yeast with snr7-U6A as their only version of SNR7 are temperature sensitive at 37°C, and snr7-U6A is synthetic lethal with the prp18-1 mutant (20, 41). The dominant synthetic lethality that we observe can be explained either as a general inhibition of splicing or as a specific effect on a small number of transcripts. In the SNR7/snr7 prp18ΔCR strains, half the spliceosomes (those with the mutant U5 snRNA) could be inactive. The remaining splicing activity could be insufficient, or those spliceosomes could sequester splicing factors, ultimately blocking splicing more completely. However, spliceosomes blocked at the second step are rapidly degraded (4, 25); in addition, yeast tolerate equal amounts of wild-type and ATPase-defective versions of the RNA helicase Prp16 (29), suggesting that blocking half the spliceosomes is not lethal. Alternatively, the U5A or U6A versions of the U5 snRNA could be preferentially recruited to some transcripts by base pairing, inhibiting or affecting the splicing of selected pre-mRNAs. Wild-type yeast tolerate considerable variation in the sequence of loop 1 (41). Our finding that prp18ΔCR yeast cannot cope with some loop 1 sequences even in the presence of a wild-type U5 underscores the close functional relationship of the conserved loop of Prp18 and loop 1 of U5.
We analyzed pre-mRNA splicing in six prp18 strains that represented the three classes of Prp18 mutant that we had identified previously (2). All the prp18 strains showed defects in the second step of splicing; our best quantitative assessment of the splicing defects, using the ratio of splicing intermediates to mRNA, showed a good correlation between the severities of the temperature sensitivities and of the second-step splicing defects. While this result does not rule out other functions for Prp18, it is consistent with the idea that the only function of Prp18 is in splicing. Using microarrays, Clark et al. (11) found that the levels of the vast majority of spliced mRNAs are not significantly changed compared to intronless mRNAs in a prp18 knockout strain at 26°C (the levels of less than 10% of the spliced mRNAs changed by more than 50% compared to reference intronless mRNAs). We observed parallel declines in spliced and intronless mRNAs, in substantial agreement with the microarray results.
The studies we report here imply a close functional relationship between the conserved loop of Prp18 and loop 1 of the U5 snRNA, and we suggest that Prp18 may act to stabilize the complex interaction of loop 1 with the splicing intermediates. Previous work has suggested a role for Prp8 in this stabilization (16, 55), and the two proteins could act together in this function. Combining earlier structural and mutational analysis of Prp18 with the work presented here yields a picture of Prp18 in which Prp18 is bound to the spliceosome by interaction of helices 1 and 2 with Slu7, and perhaps by interaction of helix 5 with another component of the spliceosome, positioning the conserved loop to interact with loop 1 of U5. Understanding the precise mechanism by which Prp18 acts will require biochemical analysis of these processes.
Acknowledgments
We thank Tharun Sundaresan for providing the SCR1 RNA probe, Ken Gable for assistance with data analysis, and Carl Mann and Javier Cáceres for comments on the manuscript.
This work was supported by National Institutes of Health grant GM57267 to D.S.H.
REFERENCES
- 1.Alvi, R. K., M. Lund, and R. T. O'Keefe. 2001. ATP-dependent interaction of yeast U5 snRNA loop 1 with the 5′ splice site. RNA 7:1013-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bačíková, D., and D. S. Horowitz. 2002. Mutational analysis identifies two separable roles of the Saccharomyces cerevisiae splicing factor Prp18. RNA 8:1280-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beggs, J. D., S. Teigelkamp, and A. J. Newman. 1995. The role of PRP8 protein in nuclear pre-mRNA splicing in yeast. J. Cell Sci. 19(Suppl.):101-105. [DOI] [PubMed] [Google Scholar]
- 4.Bousquet-Antonelli, C., C. Presutti, and D. Tollervey. 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102:765-775. [DOI] [PubMed] [Google Scholar]
- 5.Brow, D. A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36:333-360. [DOI] [PubMed] [Google Scholar]
- 6.Brys, A., and B. Schwer. 1996. Requirement for SLU7 in yeast pre-mRNA splicing is dictated by the distance between the branchpoint and the 3′ splice site. RNA 2:707-717. [PMC free article] [PubMed] [Google Scholar]
- 7.Burge, C. B., T. H. Tuschl, and P. A. Sharp. 1999. Splicing of precursors to mRNAs by the spliceosomes, p. 525-560. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), RNA World II. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 8.Caponigro, G., D. Muhlrad, and R. Parker. 1993. A small segment of the MATα1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell. Biol. 13:5141-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, S.-C., and J. Abelson. 1987. Spliceosome assembly in yeast. Genes Dev. 1:1014-1027. [DOI] [PubMed] [Google Scholar]
- 10.Chua, K., and R. Reed. 1999. The RNA splicing factor hSlu7 is required for correct 3′ splice-site choice. Nature 402:207-210. [DOI] [PubMed] [Google Scholar]
- 11.Clark, T. A., C. W. Sugnet, and M. Ares, Jr. 2002. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science 296:907-910. [DOI] [PubMed] [Google Scholar]
- 12.Collart, M. A., and S. Oliviero. 2000. Preparation of yeast RNA by extraction with hot acidic phenol, p. 13.12.1-13.12.2. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 13.Collins, C. A., and C. Guthrie. 2000. The question remains: is the spliceosome a ribozyme? Nat. Struct. Biol. 7:850-854. [DOI] [PubMed] [Google Scholar]
- 14.Company, M., J. Arenas, and J. Abelson. 1991. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature 349:487-493. [DOI] [PubMed] [Google Scholar]
- 15.Cortes, J. J., E. J. Sontheimer, S. D. Seiwert, and J. A. Steitz. 1993. Mutations in the conserved loop of human U5 snRNA generate use of novel cryptic 5′ splice sites in vivo. EMBO J. 12:5181-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dix, I., C. S. Russell, R. T. O'Keefe, A. J. Newman, and J. D. Beggs. 1998. Protein-RNA interactions in the U5 snRNP of Saccharomyces cerevisiae. RNA 4:1675-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabrizio, P., and J. Abelson. 1990. Two domains of yeast U6 small nuclear RNA required for both steps of nuclear precursor messenger RNA splicing. Science 250:404-409. [DOI] [PubMed] [Google Scholar]
- 18.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabelling DNA restriction fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 19.Frank, D., and C. Guthrie. 1992. An essential splicing factor, SLU7, mediates 3′ splice site choice in yeast. Genes Dev. 6:2112-2124. [DOI] [PubMed] [Google Scholar]
- 20.Frank, D., B. Patterson, and C. Guthrie. 1992. Synthetic lethal mutations identify interactions between U5 snRNA and four proteins required for the second step of splicing. Mol. Cell. Biol. 12:5197-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frank, D. N., H. Roiha, and C. Guthrie. 1994. Architecture of the U5 small nuclear RNA. Mol. Cell. Biol. 14:2180-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottschalk, A., G. Neubauer, J. Banroques, M. Mann, R. Lührmann, and P. Fabrizio. 1999. Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6 52 U5] tri-snRNP. EMBO J. 18:4535-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guarente, L. 1993. Synthetic enhancement in gene interaction: a genetic tool come of age. Trends Genet. 9:362-366. [DOI] [PubMed] [Google Scholar]
- 24.Hill, J., K. A. Ian, G. Donald, and D. E. Griffiths. 1991. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 19:5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hilleren, P. J., and R. Parker. 2003. Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol. Cell. 12:1453-1465. [DOI] [PubMed] [Google Scholar]
- 26.Horowitz, D. S., and J. Abelson. 1993. Stages in the second reaction of pre-mRNA splicing: the final step is ATP independent. Genes Dev. 7:320-329. [DOI] [PubMed] [Google Scholar]
- 27.Horowitz, D. S., and J. Abelson. 1993. A U5 small nuclear ribonucleoprotein particle protein involved only in the second step of splicing in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:2959-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horowitz, D. S., and A. R. Krainer. 1997. A human protein required for the second step of pre-mRNA splicing is functionally related to a yeast splicing factor. Genes Dev. 11:139-151. [DOI] [PubMed] [Google Scholar]
- 29.Hotz, H. R., and B. Schwer. 1998. Mutational analysis of the yeast DEAH-Box splicing factor prp16. Genetics 149:807-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huffaker, T. C., M. A. Hoyt, and D. Botstein. 1987. Genetic analysis of the yeast cytoskeleton. Annu. Rev. Genet. 21:259-284. [DOI] [PubMed] [Google Scholar]
- 31.James, S. A., W. Turner, and B. Schwer. 2002. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA 8:1068-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang, J., D. S. Horowitz, and R.-M. Xu. 2000. Crystal structure of the functional domain of the splicing factor Prp18. Proc. Natl. Acad. Sci. USA 97:3022-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones, M. H., D. N. Frank, and C. Guthrie. 1995. Characterization and functional ordering of Slu7p and Prp17p during the second step of pre-mRNA splicing in yeast. Proc. Natl. Acad. Sci. USA 92:9687-9691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12:5-14. [DOI] [PubMed] [Google Scholar]
- 35.McPheeters, D. S., and J. Abelson. 1992. Mutational analysis of the yeast U2 snRNA suggests a structural similarity to the catalytic core of group I introns. Cell 71:819-831. [DOI] [PubMed] [Google Scholar]
- 36.McPheeters, D. S., and P. Muhlenkamp. 2003. Spatial organization of protein-RNA interactions in the branch site-3′ splice site region during pre-mRNA splicing in yeast. Mol. Cell. Biol. 23:4174-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newman, A., and C. Norman. 1991. Mutations in yeast U5 snRNA alter the specificity of 5′ splice-site cleavage. Cell 65:115-123. [DOI] [PubMed] [Google Scholar]
- 38.Newman, A. J. 1997. The role of U5 snRNP in pre-mRNA splicing. EMBO J. 16:5797-5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newman, A. J., and C. Norman. 1992. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell 68:743-754. [DOI] [PubMed] [Google Scholar]
- 40.Newman, A. J., S. Teigelkamp, and J. D. Beggs. 1995. snRNA interactions at 5′ and 3′ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA 1:968-980. [PMC free article] [PubMed] [Google Scholar]
- 41.O'Keefe, R. T. 2002. Mutations in U5 snRNA loop I influence the splicing of different genes in vivo. Nucleic Acids Res. 30:5476-5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Keefe, R. T., and A. J. Newman. 1998. Functional analysis of the U5 snRNA loop 1 in the second catalytic step of yeast pre-mRNA splicing. EMBO J. 17:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Keefe, R. T., C. Norman, and A. J. Newman. 1996. The invariant U5 snRNA Loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell 86:679-689. [DOI] [PubMed] [Google Scholar]
- 44.Patterson, B., and C. Guthrie. 1987. An essential yeast snRNA with a U5-like domain is required for splicing in vivo. Cell 49:613-624. [DOI] [PubMed] [Google Scholar]
- 45.Query, C. C., and M. M. Konarska. 2004. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol. Cell 14:343-354. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Sapra, A. K., Y. Arava, P. Khandelia, and U. Vijayraghavan. 2004. Genome-wide analysis of pre-mRNA splicing: intron features govern the requirement for the second-step factor Prp17 in Saccharomyces cerevisiae and Schizosaccharomyces pombe. J. Biol. Chem. 279:52437-52446. [DOI] [PubMed] [Google Scholar]
- 48.Schwer, B., and C. H. Gross. 1998. Prp22, a DExH RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 17:2086-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwer, B., and C. Guthrie. 1992. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 11:5033-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwer, B., and C. Guthrie. 1991. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature 349:494-499. [DOI] [PubMed] [Google Scholar]
- 51.Ségault, V., C. L. Will, M. Polycarpou-Schwarz, I. W. Mattaj, C. Branlant, and R. Lührmann. 1999. Conserved loop I of U5 small nuclear RNA is dispensable for both catalytic steps of pre-mRNA splicing in HeLa nuclear extracts. Mol. Cell. Biol. 19:2782-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sontheimer, E. J., and J. A. Steitz. 1993. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science 262:1989-1996. [DOI] [PubMed] [Google Scholar]
- 54.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92:315-326. [DOI] [PubMed] [Google Scholar]
- 55.Teigelkamp, S., A. J. Newman, and J. D. Beggs. 1995. Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J. 14:2602-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 57.Turner, I. A., C. M. Norman, M. J. Churcher, and A. J. Newman. 2004. Roles of the U5 snRNP in spliceosome dynamics and catalysis. Biochem. Soc. Trans. 32:928-931. [DOI] [PubMed] [Google Scholar]
- 58.Umen, J. G., and C. Guthrie. 1995. Prp16p, Slu7p, and Prp8p interact with the 3′ splice site in two distinct stages during the second catalytic step of pre-mRNA splicing. RNA 1:584-597. [PMC free article] [PubMed] [Google Scholar]
- 59.Vijayraghavan, U., M. Company, and J. Abelson. 1989. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev. 3:1206-1216. [DOI] [PubMed] [Google Scholar]
- 60.Wach, A. 1996. PCR synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, X., and B. Schwer. 1997. Functional and physical interaction between the yeast splicing factors Slu7 and Prp18. Nucleic Acids Res. 25:2146-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]