Abstract
Most DNA polymerases incorporate nucleotides opposite template 7,8-dihydro-8-oxoguanine (8-oxoG) lesions with reduced efficiency and accuracy. DNA polymerase (Pol) η, which catalyzes the error-free replication of template thymine-thymine (TT) dimers, has the unique ability to accurately and efficiently incorporate nucleotides opposite 8-oxoG templates. Here we have used pre-steady-state kinetics to examine the mechanisms of correct and incorrect nucleotide incorporation opposite G and 8-oxoG by Saccharomyces cerevisiae Pol η. We found that Pol η binds the incoming correct dCTP opposite both G and 8-oxoG with similar affinities, and it incorporates the correct nucleotide bound opposite both G and 8-oxoG with similar rates. While Pol η incorporates an incorrect A opposite 8-oxoG with lower efficiency than it incorporates a correct C, it does incorporate A more efficiently opposite 8-oxoG than opposite G. This is mainly due to greater binding affinity for the incorrect incoming dATP opposite 8-oxoG. Overall, these results show that Pol η replicates through 8-oxoG without any barriers introduced by the presence of the lesion.
Eukaryotic DNA polymerase (Pol) η is a specialized polymerase that functions in the error-free replication of thymine-thymine (TT) dimers (19, 20), a common DNA lesion induced by UV irradiation. In Saccharomyces cerevisiae, disruption of the RAD30 gene encoding Pol η results in increased sensitivity to UV irradiation and an increase in the number of UV-induced mutations (21, 30, 37). In humans, the lack of Pol η is responsible for the variant form of xeroderma pigmentosum (18, 27), a genetic disorder characterized by increased UV sensitivity and a predisposition for skin cancers.
Both yeast and human Pol η synthesize DNA with low fidelity with error frequencies ranging from 10−2 to 10−3 (19, 28, 44). These enzymes incorporate As opposite the Ts of a template TT dimer—a distorting DNA lesion that blocks classical polymerases—with nearly the same catalytic efficiency (kcat/Km) and accuracy as opposite nondamaged Ts (19, 47). This led to the notion that Pol η has an active site that is unusually tolerant of the distorted geometry of the TT dimer (44). Moreover, the X-ray structure of Pol η has shown that it has an open active site that can readily accommodate this distorted lesion (40).
Pre-steady-state kinetics studies were used to examine the mechanism of nucleotide incorporation opposite a template TT dimer by Pol η (42, 43). With pre-steady-state kinetics analyses, it is possible to directly measure the binding affinity for the DNA and deoxynucleoside triphosphate (dNTP) substrates as well as the first-order rate constants of the individual steps of the nucleotide incorporation reaction (17). Pol η binds dATP with the same affinity opposite the TT dimer as opposite a nondamaged T (43). Similarly, Pol η incorporates the bound nucleotide at the same rate opposite the TT dimer as opposite a nondamaged T (43). Thus, the distorted geometry of the TT dimer presents no barriers to nucleotide incorporation by Pol η.
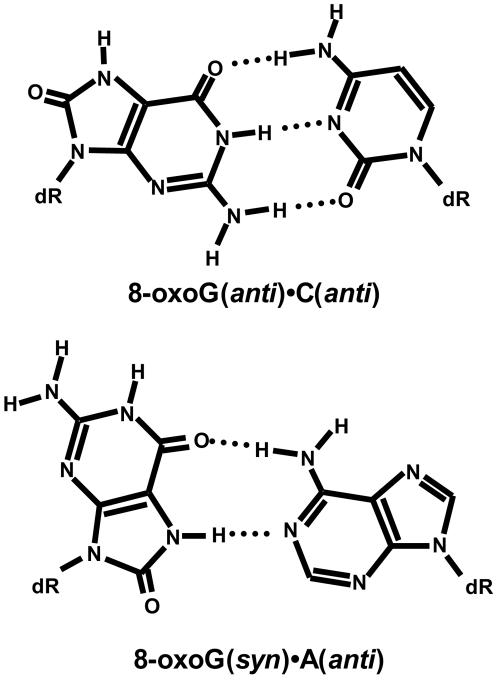
7,8-Dihydro-8-oxoguanine (8-oxoG) is a common form of oxidative damage, with steady-state levels of about 1,000 to 10,000 lesions per mammalian cell (1, 14). This lesion is highly mutagenic giving rise to G · C to T · A transversions (33). This is because 8-oxoG base pairs with both C and A (Fig. 1). In the 8-oxoG · C base pair, the glycosidic bonds of both residues are in the anti configuration, and these bases form normal Watson-Crick hydrogen bonds (25, 34). In the 8-oxoG · A base pair, the glycosidic bond of the 8-oxoG residue is in the syn configuration and it base pairs along its Hoogsteen edge with A (22, 29). With both 8-oxoG · C and 8-oxoG · A base pairs, the DNA adopts the normal B form (22, 25, 29, 34); however, when 8-oxoG is in the anti configuration in the 8-oxoG · C base pair, there are significant structural perturbations of the DNA backbone at the site of the lesion (25).
FIG. 1.
Structures of the 8-oxoG · C and 8-oxoG · A base pairs. The anti and syn designations refer to the relative orientation of the base and sugar around the glycosidic bond.
Classical polymerases typically incorporate nucleotides opposite 8-oxoG with lower catalytic efficiencies than opposite G. For example, Escherichia coli Pol I exo− and Pol II exo− incorporate C opposite 8-oxoG with 30-fold lower efficiencies than opposite G (26). Similar trends have also been observed with bacteriophage T7 DNA polymerase (10), human Pol α (38), and calf thymus Pol δ (6). Moreover, classical polymerases typically incorporate nucleotides opposite 8-oxoG with lower fidelity. E. coli Pol I exo−, human Pol β, and T7 DNA polymerase all incorporate C with slightly higher frequencies (less than fourfold) than A (10, 26, 31), and human Pol α and calf thymus Pol δ incorporate A with slightly higher frequencies than C (6, 38).
Genetic studies with yeast have demonstrated that Pol η functions in the error-free replication of 8-oxoG by preferentially incorporating C opposite G (13). Steady-state kinetics studies have shown that unlike classical polymerases, Pol η replicates through 8-oxoG both efficiently and accurately (13). In fact, Pol η incorporates C opposite 8-oxoG with the same catalytic efficiency as opposite G, and it incorporates C opposite 8-oxoG with a 20-fold higher frequency than A (13).
While steady-state kinetics studies have shown than Pol η efficiently and accurately replicates through 8-oxoG, the mechanism by which Pol η does this remains unclear. This is because in steady-state kinetics, it is not generally possible to directly observe the individual steps of the overall reaction, such as the DNA binding, dNTP binding, and nucleotide incorporation steps. Consequently, in order to examine these individual steps and determine the mechanism of efficient and accurate incorporation by Pol η opposite 8-oxoG, we carried out pre-steady-state kinetics studies of this reaction. We have found that Pol η binds dCTP with the same affinity opposite either 8-oxoG or G, and it incorporates the bound nucleotide at the same rate when bound opposite either template.
MATERIALS AND METHODS
Purification of Pol η.
Yeast Pol η was overexpressed in yeast strain BJ5464 carrying plasmid pR30.175, which encodes Pol η fused in frame with glutathione S-transferase (GST). The GST fusion protein was purified, and the GST portion was removed by treatment with PreScission protease (Amersham BioSciences) as described elsewhere (42). Purified protein was stored at −80°C in 50-μl aliquots. The active concentration of purified protein was determined by active-site titration (see Results).
DNA and nucleotide substrates.
DNA substrates were composed of two synthetic oligonucleotide strands. The same 24-mer primer sequence, 5′-GCC TCG CAG CCG TCC AAC CAA CTC, was used for all incorporation measurements. Two 45-mer templates were used with the sequence 5′-GGA CGG CAT TGG ATC GAC CTG GAG TTG GTT GGA CGG CTG CGA GGC; in one template the underlined G is a nondamaged G, and in the other the underlined base is an 8-oxoG. The oligonucleotide containing the 8-oxoG was synthesized by Midland Certified Reagent Company, Inc. The primer strand was 5′-32P end labeled with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (6,000 Ci/mmol; Amersham BioSciences) at 37°C for 1 h. Labeled primer strands were separated from unreacted [γ-32P]ATP with a Sephadex G-25 spin column (Amersham BioSciences). The labeled primer strands were annealed to template strands in 50 mM Tris Cl and 100 mM NaCl by heating at 90°C for 2 min and slow cooling to room temperature over several hours. Annealed strands were stored at 4°C for up to 2 weeks. Solutions of each dNTP (100 mM, sodium salt) were obtained from New England Biolabs and stored at −80°C.
Pre-steady-state kinetics.
All polymerase reactions were measured in 25 mM TrisCl (pH 7.5), 5 mM MgCl2, 5 mM dithiothreitol, and 10% glycerol at 22°C. The incorporation of A opposite either template G or 8-oxoG was slow enough to be measured by hand. However, the incorporation of C opposite template G or 8-oxoG was much faster and was measured on a rapid chemical quench flow instrument (KinTek Corp.). Preincubated Pol η (33 nM final concentration) and labeled DNA substrate (10 to 100 nM final concentration) in one syringe were mixed rapidly with dCTP (0 to 20 μM final concentration) from the second syringe. Nucleotide incorporation was quenched after various times (0 to 15 s) with 0.2 M EDTA. Extended primer strands (the product) were separated from unextended primer strands (the substrate) on a 15% polyacrylamide sequencing gel containing 8 M urea. The labeled gel bands were quantified with the InstantImager (Packard). Each set of DNA and dNTP concentrations were repeated several times to ensure reproducibility, and the amplitudes and rates obtained from these multiple experiments were in close agreement.
Data analysis.
The amounts of product (P) formed were graphed as a function of time (t), and the data were fit by nonlinear regression (SigmaPlot 8.0) to the burst equation P = A[1 − e−k(obs)t] + vt, where A is the amplitude of the pre-steady-state burst phase, kobs is the first-order rate constant for the burst phase, and v is the velocity of the steady-state phase.
To determine the dissociation constant (Kd) for the nucleotide binding step and the maximum first-order rate constant for nucleotide incorporation (kpol), the observed rate constants for the pre-steady-state phases were graphed as a function of dNTP concentration and the data were fit by nonlinear regression to the hyperbolic equation kobs = (kpol[dNTP])/(Kd + [dNTP]).
To determine the active concentration of Pol η ([Pol η]) and the dissociation constant (Kd) for the DNA binding step, the amplitudes of the pre-steady-state burst phases were graphed as a function of total DNA concentration ([DNA]) and the data were fit to the quadratic equation A = 0.5(Kd + [Pol η] + [DNA])2 − {0.25([Pol η] [DNA])}1/2.
For A incorporation opposite template G and 8-oxoG, the amounts of product (P) formed were graphed as a function of time (t), and the data were fit to a straight line. The observed rate constants (kobs) were obtained from the slopes of the lines divided by the active concentration of Pol η. The kobs values were plotted as a function of [dATP] and fit to the hyperbolic equation to determine the values for Kd for initial dATP binding and the maximum first-order rate constant (kpol) for nucleotide incorporation.
RESULTS
To our knowledge, Pol η is the only polymerase that incorporates nucleotides opposite template 8-oxoG residues with high efficiency and accuracy (13). To understand the mechanistic basis of this efficiency and accuracy, we have carried out a pre-steady-state kinetics analysis of C and A incorporation opposite a nondamaged G template and an 8-oxoG template. We have measured and compared the dissociation constants (Kd) for the nucleotide binding step and the first-order rate constants (kpol) for the nucleotide incorporation step for these four reactions. However, before determining the values of these parameters, we first had to determine whether or not Pol η displayed biphasic kinetics, determine the concentration of active Pol η molecules, and measure the Kd for the DNA binding step.
Biphasic kinetics of C incorporation opposite G and 8-oxoG.
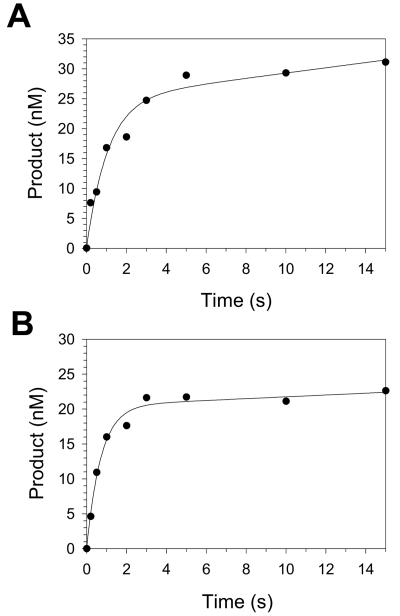
As a necessary first step to examining the mechanisms of nucleotide incorporation opposite G and 8-oxoG, we first examined whether the incorporation of C opposite these templates displayed biphasic kinetics (“burst kinetics”). Using a rapid chemical quench flow instrument, we mixed a solution containing preincubated Pol η (33 nM final concentration) and either template G DNA or template 8-oxoG DNA (100 nM final concentration) with a solution containing dCTP (20 μM final concentration). We rapidly quenched the reactions following time intervals ranging from 0.2 to 15 s. The quenched samples were analyzed by polyacrylamide gel electrophoresis, and the amounts of product formed were graphed as a function of time (Fig. 2). For incorporation opposite both G and 8-oxoG, Pol η clearly displayed two kinetic phases.
FIG. 2.
Biphasic kinetics of C incorporation opposite G and an 8-oxoG. (A) Preincubated Pol η (33 nM) and the template G DNA substrate (100 nM) were mixed with dCTP (20 μM) by using a rapid chemical quench flow instrument for various reaction times. The amounts of product formed (•) were graphed as a function of time, and the data were fit to the burst equation with an amplitude equal to 25 ± 3 nM and a rate constant for the exponential phase equal to 1.5 ± 0.5 s−1. (B) Preincubated Pol η (33 nM) and the template 8-oxoG DNA substrate (100 nM) were mixed with dCTP (20 μM) for various reaction times. The data were fit to the burst equation with an amplitude equal to 20 ± 1 nM and a rate constant for the exponential phase equal to 1.4 ± 0.2 s−1.
Burst kinetics is observed when the individual step leading to product formation, which for Pol η is the nucleotide incorporation step, occurs earlier along the reaction pathway than the slowest individual step of the overall reaction, which for Pol η is likely the DNA dissociation step. Two kinetic phases are observed because product is formed more rapidly in the first enzyme turnover than it is in subsequent turnovers (the steady-state phase), which are limited by the slow DNA dissociation step. Opposite G, the first phase (the burst phase) had an observed first-order rate constant (kobs) equal to 1.5 s−1, and opposite 8-oxoG the burst phase had a kobs equal to 1.4 s−1.
Active-site titration and Kd for DNA binding.
The presence of a pre-steady-state burst phase allowed us to examine the nucleotide binding step and nucleotide incorporation step in the first enzyme turnover before information regarding these steps was masked by the slow DNA dissociation step that dominates the steady-state phase. Before measuring these steps, however, we next had to carry out an active-site titration to determine the concentration of active Pol η molecules and measure the Kd for the DNA binding step. This was possible because the amplitude of the burst phase reflects the amount of active Pol η-DNA complex formed during the preincubation period (17). Consequently, in order to determine the active Pol η concentration and the Kd for the DNA binding step, we examined the variation of the burst amplitude as a function of total DNA concentration.
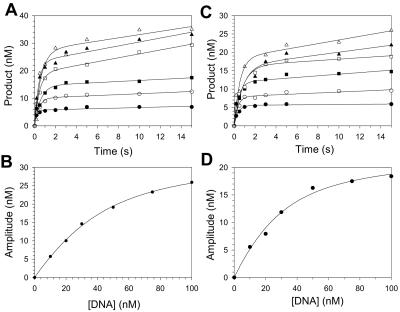
We carried out experiments as described above, except that the DNA final concentration was varied from 10 to 100 nM (Fig. 3). The amplitudes of the burst phases were graphed as a function of total DNA concentration. For incorporation opposite G, the best fit of these data to the quadratic equation gave a concentration of active Pol η equal to 33 nM and a Kd for the DNA binding step equal to 22 nM. For incorporation opposite 8-oxoG, the active Pol η concentration was 23 nM and the Kd was 17 nM. These Pol η concentrations were used for all experiments in the present study, and all subsequent experiments were performed at DNA concentrations about fivefold greater than the Kd to ensure that we were close to saturating conditions. These results also show that the binding affinity for the DNA substrate is approximately the same when G or 8-oxoG is the template base.
FIG. 3.
Active-site titration and Kd for DNA binding. (A) Preincubated Pol η (33 nM) and various concentrations of template G DNA (•, 10 nM; ○, 20 nM; ▪, 30 nM; □, 50 nM; ▴, 75 nM; ▵, 100 nM) were mixed with dCTP (20 μM) for various reaction times. The solid lines represent the best fits to the burst equation. (B) Amplitudes of the exponential phases (•) were graphed as a function of total DNA concentration. The solid line represents the best fit to the quadratic equation, with an active-site concentration equal to 33 ± 1 nM and a Kd for the Pol η-DNA complex equal to 22 ± 2 nM. (C) Preincubated Pol η (33 nM) and various concentrations of template 8-oxoG DNA (•, 10 nM; ○, 20 nM; ▪, 30 nM; □, 50 nM; ▴, 75 nM; ▵, 100 nM) were mixed with dCTP (20 μM) for various reaction times. The solid lines represent the best fits to the burst equation. (D) Amplitudes of the exponential phases (•) were graphed as a function of total DNA concentration. The solid line represents the best fit to the quadratic equation, with an active-site concentration equal to 23 ± 1 nM and a Kd for the Pol η-DNA complex equal to 17 ± 3 nM.
The reproducible one-third reduction in the active Pol η concentration with 8-oxoG relative to G was surprising. However, similar reductions have been seen previously with other polymerases. A 10-fold reduction in the active E. coli Pol II concentration was observed previously with 8-oxoG relative to G (26), a 2-fold reduction in the active human immunodeficiency virus type 1 reverse transcriptase concentration was observed previously with 8-oxoG relative to G (10), and a one-fourth reduction in the active T7 DNA polymerase concentration was observed previously with 8-oxoG relative to G (10). In the case of human immunodeficiency virus type 1 reverse transcriptase, this reduction in active enzyme concentration was attributed to nonproductive substrate binding (11). It is likely that, for reasons unknown, 8-oxoG induces varying degrees of nonproductive substrate binding in many polymerases, including Pol η.
Mechanism of C incorporation opposite G and 8-oxoG.
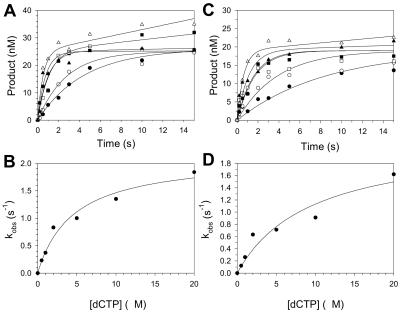
To better understand the mechanistic basis of the efficiency of incorporation opposite 8-oxoG, we determined the mechanism of C incorporation opposite G and 8-oxoG. We examined the variation of the kobs for the burst phase as a function of dCTP concentration. From these experiments it was possible to determine the Kd for the dCTP binding step and the kpol for the nucleotide incorporation step. We carried out experiments as described above with 33 nM Pol η and 100 nM DNA, except that the concentrations of dCTP ranged from 0.5 to 20 μM (Fig. 4). The kobs values for the burst phases were graphed as a function of dCTP concentration, and from the best fit of the data to the hyperbolic equation we obtained a Kd for the dCTP binding step opposite G equal to 4.5 μM and a kpol for the nucleotide incorporation step opposite G equal to 2.1 s−1. We also obtained a Kd for dCTP binding opposite 8-oxoG equal to 9.4 μM and a kpol for incorporation opposite G equal to 2.2 s−1. Thus, dCTP binds 2.1-fold weaker opposite 8-oxoG than G, but nucleotide incorporation occurs at nearly the same rate opposite both templates (Table 1).
FIG. 4.
Kinetics of dCTP incorporation opposite G and 8-oxoG. (A) Preincubated Pol η (33 nM) and the template G DNA substrate (100 nM) were mixed with various concentrations of dCTP (•, 0.5 μM; ○, 1 μM; ▪, 2 μM; □, 5 μM; ▴, 10 μM; ▴, 20 μM) for various reaction times. The solid lines represent the best fits to the burst equation. (B) Observed rate constants of the exponential phases (•) were graphed as a function of dCTP concentration. The solid line represents the best fit to the hyperbolic equation, with a kpol equal to 2.1 ± 0.2 s−1 and a Kd for the Pol η-DNA-dCTP complex equal to 4.5 ± 1.2 μM. (C) Preincubated Pol η (33 nM) and template 8-oxoG DNA substrate (100 nM) were mixed with various concentrations of dCTP (•, 0.5 μM; ○, 1 μM; ▪, 2 μM; □, 5 μM; ▴, 10 μM; ▵, 20 μM) for various reaction times. The solid lines represent the best fits to the burst equation. (D) Observed rate constants of the burst phases (•) were graphed as a function of dCTP concentration. The solid line represents the best fit to the hyperbolic equation, with a kpol equal to 2.2 ± 0.5 s−1 and a Kd for the Pol η-DNA-dCTP complex equal to 9.4 ± 4.8 μM.
TABLE 1.
Nucleotide incorporation by Pol η on template G and 8-oxoG
| dNTP · template | Kd (μM) | kpol (s−1) | kpol/Kd (μM−1 s−1) | Kd/Kda | kpol/kpolb |
|---|---|---|---|---|---|
| dCTP · G | 4.5 ± 1.2 | 2.1 ± 0.2 | 0.47 | ||
| dCTP · 8-oxoG | 9.4 ± 4.8 | 2.2 ± 0.5 | 0.23 | 2.1 | 0.95 |
| dATP · G | 120 ± 50 | 0.012 ± 0.001 | 0.00010 | 27 | 180 |
| dATP · 8-oxoG | 6.5 ± 1.2 | 0.022 ± 0.001 | 0.0034 | 1.4 | 95 |
Calculated by dividing Kd for dCTP·8-oxoG, dATP·G, or dATP·8-oxoG by Kd for dCTP·G.
Calculated by dividing kpol for dCTP·G by kpol for dCTP·8-oxoG, dATP·G, or dATP·8-oxoG.
Mechanism of A incorporation opposite G and 8-oxoG.
To better understand the mechanistic basis of the accuracy of incorporation opposite 8-oxoG, we determined the mechanism of A incorporation opposite G and 8-oxoG. Unlike C incorporation, A incorporation did not display biphasic kinetics; instead, the amounts of product formed varied linearly with reaction time. The absence of burst kinetics indicates that the slowest step in the overall reaction is the step leading to the formation of product, which in this case is the nucleotide incorporation step. Only one kinetic phase is observed, because the first enzyme turnover and all subsequent enzyme turnovers occur at the same rate.
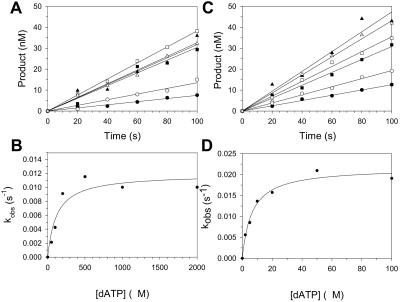
From these experiments, it was possible to determine the Kd for the dATP binding step and the kpol for the incorporation step with G and 8-oxoG by examining the variation of the kobs for the linear phase as a function of dATP concentration. Here, the kobs values were determined by dividing the slopes of the lines by the concentration of active Pol η (33 nM for template G and 23 nM for template 8-oxoG). We carried out experiments as described above, except that the concentrations of dATP ranged from 50 to 2,000 μM for incorporation opposite G and from 2 to 100 μM for incorporation opposite 8-oxoG (Fig. 5). We obtained a Kd for dATP binding opposite G equal to 120 μM and a kpol for incorporation opposite G equal to 0.012 s−1. We also obtained a Kd for dATP binding opposite 8-oxoG equal to 6.5 μM and a kpol for incorporation opposite G equal to 0.022 s−1. Thus, dATP binds significantly weaker (27-fold) opposite G relative to dCTP binding opposite G, but dATP binds with nearly the same affinity opposite 8-oxoG as dCTP binds opposite G (Table 1). In addition, A is incorporated opposite both G and 8-oxoG significantly slower (180- and 95-fold, respectively) than C is incorporated opposite G (Table 1).
FIG. 5.
Kinetics of dATP incorporation opposite G and 8-oxoG. (A) Preincubated Pol η (33 nM) and the template G DNA substrate (100 nM) were mixed with various concentrations of dATP (•, 50 μM; ○, 100 μM; ▪, 200 μM; □, 500 μM; ▴, 1,000 μM; ▵, 2,000 μM) for various reaction times. The solid lines represent the best fits to the linear equation. (B) Observed rate constants (•) were graphed as a function of dATP concentration, and the solid line represents the best fit to the hyperbolic equation, with a kpol of 0.012 ± 0.001 s−1 and a Kd for the Pol η-DNA-dATP complex equal to 120 ± 50 μM. (C) Preincubated Pol η (33 nM) and the template 8-oxoG DNA substrate (100 nM) were mixed with various concentrations of dATP (•, 2 μM; ○, 5 μM; ▪, 10 μM; □, 20 μM; ▴, 50 μM; ▵, 100 μM) for various reaction times. The solid lines represent the best fits to the linear equation. (D) Observed rate constants (•) were graphed as a function of dATP concentration. The solid line represents the best fit to the hyperbolic equation, with a kpol of 0.022 ± 0.001 s−1 and a Kd for the Pol η-DNA-dATP complex equal to 6.5 ± 1.2 μM.
DISCUSSION
Steady-state kinetics studies have been carried out with a wide range of DNA polymerases to quantify the efficiency and accuracy of nucleotide incorporation opposite 8-oxoG templates. Most DNA polymerases, including E. coli Pol I and Pol II (26), Bacillus stearothermophilus Pol I (15), bacteriophage T7 DNA polymerase (10), human Pol α (38), and fetal calf thymus Pol δ (6), incorporate nucleotides opposite 8-oxoG with both low efficiency (i.e., the efficiency of C incorporation opposite 8-oxoG is more than fivefold lower than opposite G) and low accuracy (i.e., the efficiency of C incorporation is less than fivefold greater than A incorporation). By contrast, human Pol β incorporates nucleotides opposite 8-oxoG with high efficiency (C incorporation opposite 8-oxoG is only threefold lower than opposite G) but low accuracy (C incorporates only twofold better than A) (31). In addition, bacteriophage Rb69 DNA polymerase (9) and human Pol ι (41) incorporate nucleotides opposite 8-oxoG with low efficiency but high accuracy (C incorporation is preferred over A by 20- and 30-fold, respectively). To our knowledge, only Pol η incorporates nucleotides opposite 8-oxoG with both high efficiency (the efficiency of C incorporation opposite G and 8-oxoG is the same) and high accuracy (C incorporation is preferred over A by 20-fold) (13).
Such steady-state kinetics studies are useful for measuring the efficiency and accuracy of nucleotide incorporation by DNA polymerases (4). However, in steady-state kinetics, one only observes the slowest step in the overall reaction, which for DNA polymerases is usually a step following the nucleotide incorporation step along the reaction pathway (3, 6, 8, 12, 24, 35, 36, 43, 45, 46, 48). For this reason, steady-state kinetics does not generally provide information regarding the mechanism of nucleotide incorporation by polymerases. This requires pre-steady-state kinetics, in which it is possible in principle to observe all the individual steps of the nucleotide incorporation reaction (16, 17). For DNA polymerases, these steps include at least the following: (i) DNA binding to the enzyme (E) to form the E-DNA binary complex, (ii) dNTP binding to the E-DNA binary complex to form the E-DNA-dNTP ternary complex, (iii) the incorporation of the bound nucleotide onto the 3′ end of the primer strand, (iv) release of the PPi product, and (v) dissociation of the DNA to regenerate the free enzyme. While it is likely that some of these steps can be divided into further steps, including conformational changes in the binary and ternary complexes (5, 7, 32, 42, 49), this simple, minimal mechanism is sufficient for our purpose here.
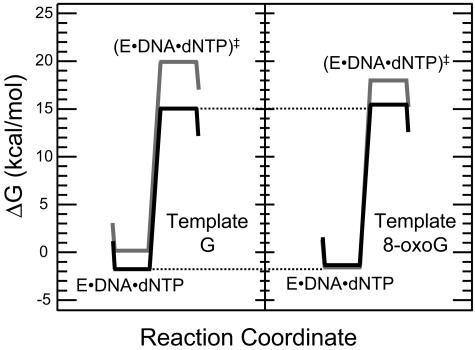
To understand the mechanistic basis of the efficiency of nucleotide incorporation opposite 8-oxoG by Pol η, we have carried out a pre-steady-state kinetics study. For the incorporation of C opposite G and 8-oxoG, we have measured the Kd values for dNTP binding (step 2) and the kpol values for nucleotide incorporation (step 3). From these values, we generated free energy diagrams for portions of these two nucleotide incorporation reactions (Fig. 6). The left panel shows the incorporation of C opposite G, and the right panel shows the incorporation of C opposite 8-oxoG. Here we have found that at the nucleotide binding step (step 2), Pol η binds dCTP 2.1-fold tighter when the template residue is a G than when it is an 8-oxoG. This difference is very small and corresponds to a ΔΔG equal to 0.43 kcal/mol. We have also found that at the nucleotide incorporation step (step 3), the bound nucleotide is incorporated by Pol η at nearly the same rate when the template residue is a G or an 8-oxoG, and the ΔΔG‡ corresponds to only −0.03 kcal/mol. These results show that the 8-oxoG lesion presents no barrier to Pol η at either the nucleotide binding step or nucleotide incorporation step.
FIG. 6.
Free energy diagrams of a portion of the nucleotide incorporation reaction. The left panel represents the incorporation of C (black) and A (gray) opposite template G, and the right panel represents the incorporation of C and A opposite template 8-oxoG. The ground state of the Pol η-DNA-dNTP complex is labeled E-DNA-dNTP, and the transition state of the nucleotide incorporation step is labeled (E-DNA-dNTP)‡. To calculate these values, we have assumed arbitrary concentrations of DNA equal to 100 nM and dNTP equal to 100 μM.
Pre-steady-state kinetics studies have also been carried out with several DNA polymerases that incorporate nucleotides opposite 8-oxoG with low efficiencies. These include E. coli Pol I and Pol II (26) and the bacteriophage T7 DNA polymerase (10). For both E. coli Pol I and Pol II, the efficiencies of C incorporation opposite 8-oxoG are about 30-fold lower than those opposite G. Interestingly, this similar decrease in efficiency results from distinct mechanisms. In the case of Pol I, the 8-oxoG impacts both the nucleotide binding step and the nucleotide incorporation step. The dCTP binds sixfold weaker with a template 8-oxoG, and the nucleotide is incorporated sixfold slower with a template 8-oxoG (26). In the case of Pol II, the 8-oxoG impacts primarily the nucleotide binding step. The dCTP binds 11-fold weaker with a template 8-oxoG, but the nucleotide is incorporated only 2-fold slower with a template 8-oxoG (26). For the bacteriophage T7 DNA polymerase, the incorporation of C opposite 8-oxoG is about 140-fold less efficient than opposite G. In this case, the 8-oxoG impacts primarily the nucleotide incorporation step. The dCTP binds 4-fold weaker with a template 8-oxoG, but the nucleotide is incorporated 50-fold slower with a template 8-oxoG (10). Thus, for other polymerases, the 8-oxoG lesion is able to create barriers at the nucleotide binding step, the nucleotide incorporation step, or both steps.
We have also used pre-steady-state kinetics studies to examine the mechanistic basis of the accuracy of nucleotide incorporation opposite 8-oxoG by Pol η. The free energy diagrams for the incorporation of A opposite G (left panel) and 8-oxoG (right panel) are shown in Fig. 6. We found that the mechanistic basis for the discrimination against A incorporation was quite different opposite G and 8-oxoG. For template G, Pol η binds dCTP 27-fold tighter than dATP, corresponding to a ΔΔG equal to 1.93 kcal/mol, and Pol η incorporates the bound C 180-fold faster than A, corresponding to a ΔΔG‡ equal to 3.02 kcal/mol. For 8-oxoG, Pol η binds dCTP with approximately the same affinity as dATP, corresponding to a ΔΔG equal to 0.21 kcal/mol. However, Pol η incorporates the bound C 100-fold faster than A, corresponding to a ΔΔG‡ equal to 2.70 kcal/mol. We conclude from this that the accuracy of incorporation opposite 8-oxoG by Pol η comes from a barrier to incorporation of A at mainly the nucleotide incorporation step.
It is curious and interesting that, in the case of Pol η, the binding affinity of dCTP and dATP opposite 8-oxoG is approximately the same. This is different from what has been observed with other polymerases. For example, in the cases of E. coli Pol I (26) and T7 DNA polymerase (10), dATP binding opposite 8-oxoG is 10- and 14-fold weaker, respectively, than dCTP binding. This difference is likely because the active site of Pol η is larger than the active sites of other DNA polymerases, allowing the 8-oxoG to easily interconvert between the syn and anti configurations in the Pol η-DNA binary complex. A rapid equilibrium between the syn and anti states would allow dCTP and dATP to bind with nearly the same affinity. However, despite this, the strong kinetic barrier for incorporating the A bound opposite 8-oxoG still allows Pol η to replicate through this lesion with greater accuracy than these other polymerases.
High-resolution structures have been determined recently for four DNA polymerases bound to DNA substrates containing 8-oxoG lesions (2, 9, 15, 23), and these structures reveal how these polymerases might avoid a potential steric clash between the O8 atom on the 8-oxoG and the sugar moiety when the 8-oxoG is in the anti configuration. Several high-resolution structures of B. stearothermophilus Pol I (which incorporates opposite 8-oxoG with low efficiency and low accuracy) in complex with DNA containing 8-oxoG provide a series of snapshots of the incorporation of C and A opposite 8-oxoG (15). However, structures of ternary complexes with incoming dCTP or dATP opposite 8-oxoG—complexes that are most critical to understanding the structural basis of the efficiency and accuracy of nucleotide incorporation opposite this lesion—were not among these. Structures were determined for the binary complexes following the incorporation of C or A opposite 8-oxoG. When C pairs with 8-oxoG in the “postinsertion site” (i.e., the primer-terminal base pair), the 8-oxoG is in the anti configuration. In this configuration, a potential steric clash between the O8 of the 8-oxoG and the O4′ of the sugar is avoided by a change in relative orientation of the sugar and base, leading to a distortion of the template strand in the vicinity of the lesion. This distortion resembles what has been observed previously with incorrect base pairs (15), and this may be responsible for the substantially decreased ability of Pol I to incorporate C opposite 8-oxoG.
For human Pol β (which incorporates opposite 8-oxoG with high efficiency but low accuracy), a structure of a ternary complex with incoming dCTP opposite 8-oxoG was determined (23). A structure with A opposite 8-oxoG was also determined, but the incoming dATP was hydrolyzed to dAMP, the 8-oxoG was in the anti rather than syn configuration, and the polymerase was in the inactive, open conformation (23); thus, it is unclear how to interpret this particular structure. However, the structure of the ternary complex with incoming dCTP opposite 8-oxoG provided excellent insight into how this lesion is accommodated in the anti configuration. To accommodate the O8 of the 8-oxoG, the single-stranded region of the template strand on the 5′ side of the 8-oxoG is flipped in the active site of the polymerase relative to the manner in which Pol β binds nondamaged DNA. This flip results from a 180° rotation of the α torsional angle along the covalent bond connecting the O5′ and the 5′ P atoms (23). This dramatic change in the conformation of the DNA substrate is apparently well tolerated by this enzyme, as demonstrated by its high efficiency of incorporation of nucleotides opposite 8-oxoG.
For bacteriophage T7 DNA polymerase (which incorporates opposite 8-oxoG with low efficiency and low accuracy), several high-resolution structures were determined with DNA containing 8-oxoG, including structures of the ternary complex with incoming dCTP bound opposite 8-oxoG as well as structures following the incorporation step in which either C or A opposite 8-oxoG is in the postinsertion site (2). In addition, for Rb69 DNA polymerase (which incorporates opposite 8-oxoG with low efficiency but high accuracy), a structure was determined for the ternary complex with dCTP bound opposite 8-oxoG (2, 9). Interestingly, in both of these polymerases, the large bend in the primer strand between the template residue (the 8-oxoG) and the adjacent residue on its 5′ side avoids any potential steric clash with the O8 on the 8-oxoG in the anti configuration. In fact, no structural differences were observed between the conformations of the bound DNA containing 8-oxoG and bound nondamaged DNA (2, 9). It therefore remains unclear why these polymerases do not incorporate nucleotides opposite this lesion more efficiently.
Based upon these structural studies, there appear to be three modes by which the 8-oxoG lesion is accommodated by DNA polymerases to avoid steric problems: (i) the relative positions of the sugar and base change to create a distortion in the geometry of the template strand, (ii) the single-stranded region of the template strand rotates by 180° between the template residue and the adjacent 5′ residue, and (iii) the DNA is bound with a very large bend between the template residue and the adjacent 5′ residue. Although it is possible that Pol η utilizes any one of these modes to accommodate the 8-oxoG lesion, we believe that the first one is the most likely. The second and third modes, for example, would require that the structural basis for the efficient and accurate replication through 8-oxoG lesions and TT dimers be very different. Consider the case when the first T of the TT dimer is the template residue. Because the two bases of the TT dimer are covalently cross-linked to one another, it is not possible to introduce either a large bend or a 180° rotation between the template residue and the next residue. Instead, based upon the high-resolution structure of Pol η (40), pre-steady-state kinetics studies of nucleotide incorporation opposite the TT dimer (43), and nucleotide incorporation studies opposite modified N3-methyl-TT dimers (39), a compelling case has been made that both bases of the TT dimer are bound within the active site of Pol η without substantial bends or rotations. Because of the unusual tolerance of Pol η for distorted template geometries, Pol η directly incorporates As opposite the two Ts of the TT dimer without any barriers introduced by the distorted geometry of this template lesion (43).
The pre-steady-state kinetics analyses with 8-oxoG presented in the present study provide strong evidence that Pol η is utilizing the exact same mechanism for replicating through 8-oxoG lesions as for replicating through TT dimers. If this is correct, then the DNA containing the 8-oxoG lesion is bound by Pol η in the anti configuration when base pairing with an incoming dCTP without significant bends or rotations between the template 8-oxoG and the adjacent residue in the template strand. The steric problems between the 8-oxoG in this configuration and the sugar moiety are resolved by altering the relative positions of the sugar and base. This creates a distortion in the template strand on both sides of the lesion, probably of a magnitude similar to or less than the distortion caused by the TT dimer. However, because the active site of Pol η can hold two template residues and is large enough to accommodate such an altered DNA backbone conformation, Pol η directly incorporates C opposite the 8-oxoG without any barriers introduced by the distorted template geometry.
Acknowledgments
We thank Christine M. Kondratick and Craig A. Howell for discussions and for reading a draft of the manuscript. We also thank Louise Prakash and Satya Prakash for providing the plasmid for overexpressing yeast Pol η.
REFERENCES
- 1.Beckman, K. B., and B. N. Ames. 1997. Oxidative decay of DNA. J. Biol. Chem. 272:19633-19636. [DOI] [PubMed] [Google Scholar]
- 2.Brieba, L. G., B. F. Eichman, R. J. Kokoska, S. Doublie, T. A. Kunkel, and T. Ellenberger. 2004. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 23:3452-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capson, T. L., J. A. Peliska, B. F. Kaboord, M. W. Frey, C. Lively, M. Dahlberg, and S. J. Benkovic. 1992. Kinetic characterization of the polymerase and exonuclease activities of the gene 43 protein of bacteriophage T4. Biochemistry 31:10984-10994. [DOI] [PubMed] [Google Scholar]
- 4.Creighton, S., L. B. Bloom, and M. F. Goodman. 1995. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 262:232-256. [DOI] [PubMed] [Google Scholar]
- 5.Dahlberg, M. E., and S. J. Benkovic. 1991. Kinetic mechanism of DNA polymerase I (Klenow fragment): identification of a second conformational change and evaluation of the internal equilibrium constant. Biochemistry 30:4835-4843. [DOI] [PubMed] [Google Scholar]
- 6.Einolf, H. J., and F. P. Guengerich. 2001. Fidelity of nucleotide insertion at 8-oxo-7,8-dihydroguanine by mammalian DNA polymerase δ. Steady-state and pre-steady state kinetic analysis. J. Biol. Chem. 276:3764-3771. [DOI] [PubMed] [Google Scholar]
- 7.Fiala, K. A., and Z. Suo. 2004. Mechanism of DNA polymerization catalyzed by Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry 43:2116-2125. [DOI] [PubMed] [Google Scholar]
- 8.Fiala, K. A., and Z. Suo. 2004. Pre-steady state kinetic studies of the fidelity of Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry 43:2106-2115. [DOI] [PubMed] [Google Scholar]
- 9.Freisinger, E., A. P. Grollman, H. Miller, and C. Kisker. 2004. Lesion (in)tolerance reveals insights into DNA replication fidelity. EMBO J. 23:1494-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furge, L. L., and F. P. Guengerich. 1997. Analysis of nucleotide insertion and extension at 8-oxo-7,8-dihydroguanine by replicative T7 polymerase exo− and human immunodeficiency virus-1 reverse transcriptase using steady state and pre-steady state kinetics. Biochemistry 36:6475-6487. [DOI] [PubMed] [Google Scholar]
- 11.Furge, L. L., and F. P. Guengerich. 1999. Explanation of pre-steady state kinetics and decreased burst amplitude of HIV-1 reverse transcriptase at sites of modified DNA bases with an additional, nonproductive enzyme-DNA-nucleotide complex. Biochemistry 38:4818-4825. [DOI] [PubMed] [Google Scholar]
- 12.Graves, S. W., A. A. Johnson, and K. A. Johnson. 1998. Expression, purification, and initial kinetic characterization of the large subunit of the human mitochondrial DNA polymerase. Biochemistry 37:6050-6058. [DOI] [PubMed] [Google Scholar]
- 13.Haracska, L., S. L. Yu, R. E. Johnson, L. Prakash, and S. Prakash. 2000. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nat. Genet. 25:458-461. [DOI] [PubMed] [Google Scholar]
- 14.Helbock, H. J., K. B. Beckman, M. K. Shigenaga, P. B. Walter, A. A. Woodall, H. C. Yeo, and B. N. Ames. 1998. DNA oxidation matters: the HPLC-electrochemical detection assay of 8-oxodeoxyguanosine and 8-oxoguanine. Proc. Natl. Acad. Sci. USA 95:288-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu, G. W., M. Ober, T. Carell, and L. S. Beese. 2004. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature 431:217-221. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, K. A. 1992. Transient-state kinetic analysis of enzyme reaction pathway. Enzymes XX:1-61. [Google Scholar]
- 17.Johnson, K. A. 1995. Rapid quench kinetic analysis of polymerases, adenosine triphosphatases, and enzyme intermediates. Methods Enzymol. 249:38-61. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, R. E., C. M. Kondratick, S. Prakash, and L. Prakash. 1999. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285:263-265. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, R. E., M. T. Washington, S. Prakash, and L. Prakash. 2000. Fidelity of human DNA polymerase η. J. Biol. Chem. 275:7447-7450. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, R. E., S. Prakash, and L. Prakash. 1999. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη. Science 283:1001-1004. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, R. E., S. Prakash, and L. Prakash. 1999. Requirement of DNA polymerase activity of yeast Rad30 protein for its biological function. J. Biol. Chem. 274:15975-15977. [DOI] [PubMed] [Google Scholar]
- 22.Kouchakdjian, M., B. Verraiah, S. Shibutani, M. Eisenberg, F. Johnson, A. P. Grollman, and D. J. Patel. 1991. NMR structural studies of the ionizing radiation adduct 7-hydroxy-8-oxodeoxyguanosine (8-oxo-tH-dG) opposite deoxyadenosine in a DNA duplex. 8-oxo-7H-dG(syn) · dA(anti) alignment at lesion site. Biochemistry 30:1403-1412. [DOI] [PubMed] [Google Scholar]
- 23.Krahn, J. M., W. A. Beard, H. Miller, A. P. Grollman, and S. H. Wilson. 2003. Structure of DNA polymerase β with the mutagenic DNA lesion 8-oxodeoxyguanine reveals structural insights into its coding potential. Structure 11:121-127. [DOI] [PubMed] [Google Scholar]
- 24.Kuchta, R. D., V. Mizrahi, P. A. Benkovic, K. A. Johnson, and S. J. Benkovic. 1987. Kinetic mechanism of DNA polymerase I (Klenow). Biochemistry 26:8410-8417. [DOI] [PubMed] [Google Scholar]
- 25.Lipscomb, L. A., M. E. Peek, M. L. Morningstar, S. M. Verghis, E. M. Miller, A. Rich, J. M. Essigmann, and L. D. Williams. 1995. X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine. Proc. Natl. Acad. Sci. USA 92:719-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowe, L. G., and F. P. Guengerich. 1996. Steady state and pre-steady state kinetic analysis of dNTP insertion opposite 8-oxo-7,8-dihydroguanine by Escherichia coli polymerases I exo− and II exo−. Biochemistry 35:9840-9849. [DOI] [PubMed] [Google Scholar]
- 27.Masutani, C., R. Kusumoto, A. Yamada, N. Dohmae, M. Yokoi, M. Yuasa, M. Araki, S. Iwai, K. Takio, and F. Hanaoka. 1999. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature 399:700-704. [DOI] [PubMed] [Google Scholar]
- 28.Matsuda, T., K. Bebenek, C. Matsutani, F. Hanaoka, and T. A. Kunkel. 2000. Low fidelity of DNA synthesis by human DNA polymerase η. Nature 404:1011-1013. [DOI] [PubMed] [Google Scholar]
- 29.McAuley-Hecht, K. E., G. A. Leonard, N. J. Gibson, J. B. Thomson, W. P. Watson, W. N. Hunter, and T. Brown. 1994. Crystal structure of a DNA duplex containing 8-hydrodeoxyguanine-adenine base pairs. Biochemistry 33:10266-10270. [DOI] [PubMed] [Google Scholar]
- 30.McDonald, J. P., A. S. Levine, and R. Woodgate. 1997. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics 147:1557-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, H., R. Prasad, S. H. Wilson, F. Johnson, and A. P. Grollman. 2000. 8-Oxo-dGTP incorporation by DNA polymerase β is modified by active-site residue Asn279. Biochemistry 39:1029-1033. [DOI] [PubMed] [Google Scholar]
- 32.Mizrahi, V., R. N. Henrie, J. F. Marlier, K. A. Johnson, and S. J. Benkovic. 1985. Rate-limiting steps in the DNA polymerase I reaction pathway. Biochemistry 24:4010-4018. [DOI] [PubMed] [Google Scholar]
- 33.Moriya, M. 1993. Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-oxoguanine in DNA induced targeted G · C→T · A transversions in simian kidney cells. Proc. Natl. Acad. Sci. USA 90:1122-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oda, Y., S. Uesugi, M. Ikehara, S. Nishimura, Y. Kawase, H. Ishikawa, H. Inoue, and E. Ohtsuda. 1991. NMR studies of a DNA containing 8-hydroxydeoxyguanosine. Nucleic Acid Res. 19:1407-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel, S. S., I. Wong, and K. A. Johnson. 1991. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry 30:511-525. [DOI] [PubMed] [Google Scholar]
- 36.Roettger, M. P., K. A. Fiala, S. Sompalli, Y. Dong, and Z. Suo. 2004. Pre-steady state kinetic studies of the fidelity of human DNA polymerase μ. Biochemistry 43:13827-13838. [DOI] [PubMed] [Google Scholar]
- 37.Roush, A. A., M. Suarez, E. C. Friedberg, M. Radman, and W. Siede. 1998. Deletion of the Saccharomyces cerevisiae gene RAD30 encoding an Escherichia coli DinB homolog confers UV radiation sensitivity and altered mutability. Mol. Gen. Genet. 257:686-692. [DOI] [PubMed] [Google Scholar]
- 38.Shibutani, S., M. Takeshita, and A. P. Grollman. 1991. Insertions of specific bases during DNA synthesis past the oxidation-damaged base 8-oxo-dG. Nature 349:431-434. [DOI] [PubMed] [Google Scholar]
- 39.Sun, L., K. Zhou, P. Hohler, E. T. Kool, F. Yuan, Z. Wang, and J. S. Taylor. 2003. Yeast Pol η holds a cis-syn thymine dimer loosely in the active site during elongation opposite the 3′-T of the dimer, but tightly opposite the 5′-T. Biochemistry 42:9431-9437. [DOI] [PubMed] [Google Scholar]
- 40.Trincao, J., R. E. Johnson, C. R. Escalante, S. Prakash, L. Prakash, and A. K. Aggarwal. 2001. Structure of the catalytic core of S. cerevisiae DNA polymerase η: implications for translesion DNA synthesis. Mol. Cell 8:417-426. [DOI] [PubMed] [Google Scholar]
- 41.Vaisman, A., and R. Woodgate. 2001. Unique misinsertion specificity of polι may decrease the mutagenic potential of deaminated cytosines. EMBO J. 20:6520-6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Washington, M. T., L. Prakash, and S. Prakash. 2001. Yeast DNA polymerase η utilizes an induced-fit mechanism of nucleotide incorporation. Cell 107:917-927. [DOI] [PubMed] [Google Scholar]
- 43.Washington, M. T., L. Prakash, and S. Prakash. 2003. Mechanism of nucleotide incorporation opposite a thymine-thymine dimer by yeast DNA polymerase η. Proc. Natl. Acad. Sci. USA 100:12093-12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Washington, M. T., R. E. Johnson, S. Prakash, and L. Prakash. 1999. Fidelity and processivity of Saccharomyces cerevisiae DNA polymerase η. J. Biol. Chem. 274:36835-36838. [DOI] [PubMed] [Google Scholar]
- 45.Washington, M. T., R. E. Johnson, L. Prakash, and S. Prakash. 2003. The mechanism of nucleotide incorporation by human DNA polymerase η differs from that of the yeast enzyme. Mol. Cell. Biol. 23:8316-8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Washington, M. T., R. E. Johnson, L. Prakash, and S. Prakash. 2004. Human DNA polymerase ι utilizes different nucleotide incorporation mechanisms dependent upon the template base. Mol. Cell. Biol. 24:936-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Washington, M. T., R. E. Johnson, S. Prakash, and L. Prakash. 2000. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc. Natl. Acad. Sci. USA 97:3094-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Werneburg, B. G., J. Ahn, X. Zhong, R. J. Hondal, V. S. Kraynov, and M. D. Tsai. 1996. DNA polymerase β: pre-steady-state kinetic analysis and roles of arginine-283 in catalysis and fidelity. Biochemistry 35:7041-7050. [DOI] [PubMed] [Google Scholar]
- 49.Zhong, X., S. S. Patel, B. G. Werneburg, and M. D. Tsai. 1997. DNA polymerase β: multiple conformational changes in the mechanism of catalysis. Biochemistry 36:11891-11900. [DOI] [PubMed] [Google Scholar]