Abstract
The xlnD gene encoding the 85-kDa β-xylosidase was cloned from Aspergillus nidulans. The deduced primary structure of the protein exhibits considerable similarity to the primary structures of the Aspergillus niger and Trichoderma reesei β-xylosidases and some similarity to the primary structures of the class 3 β-glucosidases. xlnD is regulated at the transcriptional level; it is induced by xylan and d-xylose and is repressed by d-glucose. Glucose repression is mediated by the product of the creA gene. Although several binding sites for the pH regulatory protein PacC were found in the upstream regulatory region, it was not clear from a Northern analysis whether PacC is involved in transcriptional regulation of xlnD.
Hydrolysis of xylans is of considerable interest for various biotechnological applications (for reviews see references 7 and 47). Unlike cellulose, xylans are chemically heterogeneous molecules with a characteristic backbone consisting of β-(1,4)-linked d-xylosyl residues replaced with acetyl, l-arabinosyl, and 4-O-methyl-glucuronosyl side chains. Natural xylan degradation by microorganisms occurs through the coordinated action of various enzymes, including the endo-(1,4)-β-xylanases (EC 3.2.1.8), which cleave the β-(1,4) glycosidic bonds between d-xylose residues in the main chain to produce xylooligosaccharides, and β-xylosidase (EC 3.2.1.37), which cleaves xylooligosaccharides to produce xylose.
Filamentous fungi are known to be efficient producers of xylanolytic enzymes, and most commercial xylanolytic preparations are obtained from fermentations of Aspergillus or Trichoderma species. Several genes encoding endo-(1,4)-β-xylanases from these fungal species have been characterized (10, 23, 25, 26, 39, 44), and recently genes encoding β-xylosidases have been cloned from both Aspergillus niger (46) and Trichoderma reesei (32).
Little is currently known about the molecular mechanisms controlling xylanase gene expression in filamentous fungi. The presence of regulatory elements involved in xylan-specific induction in the promoters of the Aspergillus tubingensis and T. reesei xylanase-encoding genes (10, 50) and Cre1-mediated carbon catabolite repression of expression of the T. reesei xln1 gene (31) are the only such data reported so far. The ascomycete Aspergillus nidulans is a model organism for studies of gene regulation due to our extensive knowledge of its genetics and the availability of mutants (1, 9). In recent years the molecular basis of glucose repression by the protein product of the regulatory gene creA has been investigated; it has been found that this protein is a negatively acting transcription factor which binds to a subset of DNA sequence motifs conforming to the consensus sequence 5′-SYGGRG-3′ (8, 24, 28). In addition, studies of mutants (5) disrupted in their responses to external pH (alkaline growth mimic and acidic growth mimic phenotypes) have revealed a regulatory mechanism comprising a signal transduction pathway, encoded by the pal genes, which at alkaline ambient pH results in proteolytic conversion of the PacC transcription factor to its active form. After conversion PacC is able to activate those genes whose expression is appropriate under alkaline conditions and to repress those genes whose expression is suited to acidic ambient pH (3, 12, 33, 34, 43).
When grown in media in which xylan is the only carbon source, A. nidulans produces at least three endo-(1,4)-β-xylanases (17, 36) and one predominantly mycelium-bound β-xylosidase (29). These four enzymes have been purified and kinetically characterized (15–18, 29), and the genes encoding the three endo-β-(1,4)-xylanases (xlnA, xlnB, and xlnC) have been cloned and sequenced (30, 35). In this paper we describe the identification, cloning, and nucleotide sequence of an A. nidulans gene (xlnD) which encodes the previously isolated β-xylosidase (29).
MATERIALS AND METHODS
Strains, plasmids and culture conditions.
Escherichia coli LE392 [e14-(mcrA) hsdR514 supE44 supF58 lacY1 galK2 galT22 metB1 trpR55] and DH5α [endA1 hsdR17 gyrA96 thi-1 relA1 supE44 recA1 ΔlacU169 (φ80 lacZΔM15)] were used as hosts for propagation of bacteriophage λ and plasmids, respectively. A. nidulans biA1 (= CECT2544) was obtained from the Spanish Type Culture Collection and was used as the wild-type strain. A. nidulans G191 (pabaA1 pyrG89 fwA1 uAY9) (4) was used as the host in transformation experiments performed with plasmid pGW635, which contains the A. niger pyrA gene (20) for selection of transformants. A. nidulans creAd30, biA1 was a gift from H. N. Arst, Jr., and strains palA1, biA1, wA3 (a strain which mimics growth at acidic pH), and pacCc14, biA1 (a strain which mimics growth at alkaline pH) were obtained from M. A. Peñalva. A. nidulans mycelia were grown from spores in minimal medium (MM) (37) containing various carbon sources (1%, wt/vol) as indicated below; appropriate supplements were included when necessary. In transfer experiments, MM containing d-fructose (1%, wt/vol) and supplemented with 0.5% (wt/vol) Casamino Acids (Difco Laboratories, Detroit, Mich.) was used to generate mycelial biomass. Buffered media were prepared by adding filter-sterilized sodium phosphate after autoclaving from 1 M stock solutions having pH values of 4.1, 6.0, and 8.0 in order to obtain a final phosphate concentration of 100 mM. In all cases the sodium ion concentration was adjusted to 195 mM by adding 5 M NaCl. Induction media were prepared by replacing d-fructose with d-xylose (1%, wt/vol) from a filter-sterilized stock solution (10%, wt/vol).
Cloning and sequencing procedures.
An A. nidulans genomic library constructed in λ Charon 4A (51) was screened by using hybridization conditions as previously described (35). DNA manipulations were carried out by standard methods as described by Sambrook et al. (40). DNA sequences were determined by the dideoxynucleotide chain termination method (41) with a Sequenase sequencing kit from Amersham-USB used according to the supplier’s instructions; universal, reverse, and gene-specific oligonucleotides were used as primers. The DNA sequences obtained were analyzed with the Genetics Computer Group sequence analysis software package (13).
DNA isolation and RNA isolation.
Fungal high-molecular-weight DNA was isolated as described previously (11). Total RNA was extracted from mycelial tissue by a procedure based on the method of Cathala et al. (6). Mycelia were harvested by filtration and rapidly press dried between sheets of absorbent paper to remove as much liquid as possible. Each mycelial mat was then flash frozen in liquid nitrogen and stored at −70°C. Approximately 100 mg of nitrogen-frozen mycelium was homogenized with a solution containing 600 μl of lysing medium (5 M guanidinium thiocyanate, 10 mM EDTA, 50 mM Tris [pH 7.5]) plus 48 μl of β-mercaptoethanol in a 2-ml screw-cap Eppendorf tube for 45 s at full speed with a Mini-beadbeater (Biospec Products, Bartlesville, Okla.) using five 2-mm-diameter steel balls. The contents reached a temperature of about 50°C. The tube was left at room temperature for 5 min, and the contents were rehomogenized as described above. Mycelial debris was removed by microcentrifugation at 4°C for 15 min. Five hundred microliters of supernatant was removed and added to 1.5 ml of 4 M LiCl, and the preparation was mixed and kept on ice overnight. Precipitated material was collected by microcentrifugation at 4°C for 15 min. The tube was thoroughly drained, and the pelleted material was resolubilized in 500 μl of a solution containing 0.1% (wt/vol) sodium dodecyl sulfate, 1 mM EDTA, and 10 mM Tris (pH 7.5) by homogenization with the Mini-beadbeater for 10 s in the absence of steel balls. The solubilized material was extracted with phenol-chloroform, and the aqueous phase recovered was reextracted with chloroform. Total RNA was precipitated by adding 0.1 volume of 3 M sodium acetate (pH 4.8) and 2.5 volumes of absolute ethanol and then mixing and incubating the preparation at −70°C for 30 to 60 min. RNA was recovered by microcentrifugation at 4°C for 15 min. The tube was drained, and the pellet was dissolved in 100 μl of diethyl pyrocarbonate-treated, MilliQ-filtered water by freezing and thawing.
3-[N-Morpholino]propanesulfonic acid (MOPS) (0.6 M)–formaldehyde gels were used to perform a Northern analysis of total RNA. A 1.9-kb DNA fragment containing the A. niger xlnD gene was generated with oligonucleotides xylos001 and xylos004 and was used as the xlnD-specific probe (46). An 830-bp KpnI-NcoI fragment was used as the actin-specific probe (19). Probes were labelled by the random hexanucleotide primer method (14).
A. nidulans transformations.
Transformation of A. nidulans G191 was carried out as described by Tilburn and coworkers (42) by using plasmids pGW635 (5 μg) and pXDE1 (20 μg); the latter plasmid contained the A. nidulans xlnD gene. Transformants were selected for growth on MM in the absence of uridine and were clonally purified. β-Xylosidase activity was extracted and measured as described previously (29).
Immunoblot analysis.
Mycelia were grown from 2.5 × 108 spores for 14 h at 37°C and 200 rpm in 50 ml of MM containing oat spelt xylan (1%, wt/vol), yeast extract (1%, wt/vol), and Casamino Acids (0.5%, wt/vol) and then extracted with 25 ml of phosphate-buffered saline containing 0.05% Triton X-100 for 24 h at 30°C and 200 rpm. Portions (30 μl) of the mycelial extracts were subjected to sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis and electroblotted onto nitrocellulose membranes. Immunostaining was carried out by the Bio-Rad procedure by using a 1:400 dilution of the A. nidulans β-xylosidase antibody (29), followed by incubation with a 1:3,000 dilution of anti-rabbit immunoglobulin G.
Nucleotide sequence accession number.
The A. nidulans xlnD gene sequence has been deposited in the EMBL nucleotide sequence database under accession no. Y13568.
RESULTS
Cloning of an A. nidulans gene coding for a β-xylosidase.
An A. nidulans genomic library (51) was screened by heterologous hybridization by using a DNA probe corresponding to the A. niger xlnD gene, as described above. After two rounds of screening, a positive plaque was detected and purified. The results of a Southern analysis of the phage insert performed with different restriction enzymes correlated well with signals detected previously in Southern blot profiles of restricted A. nidulans wild-type genomic DNA (data not shown). A 4.3-kb EcoRI DNA fragment that hybridized to the A. niger xlnD probe was isolated and subcloned into pUC18, yielding plasmid pXDE1.
Nucleotide sequence of the cloned gene.
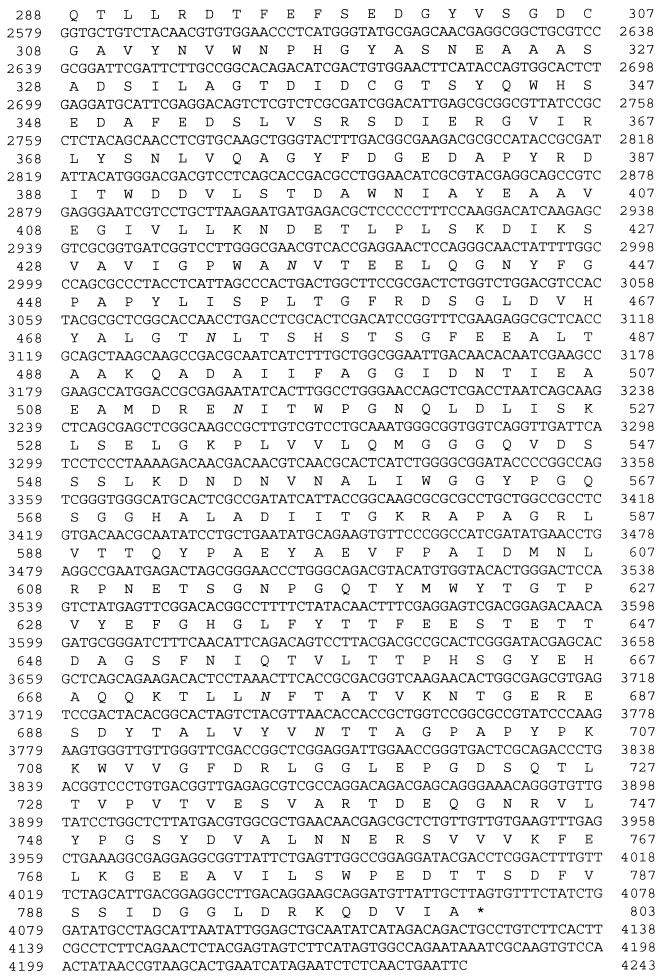
The sequence of the A. nidulans DNA insert (4,243 bp) cloned in plasmid pXDE1 was determined on both strands. A comparison of this sequence with the A. niger xlnD gene sequence revealed a 2,406-bp uninterrupted open reading frame (Fig. 1). The deduced amino acid sequence of the A. nidulans xlnD gene product exhibited high degrees of similarity to the primary structures of the A. niger and T. reesei β-xylosidases (64.3 and 61.9% identity, respectively) (Fig. 2) and also exhibited significant levels of similarity to the primary structures of β-glucosidases belonging to glycosidase family 3 (22). There was a predicted signal peptide cleavage site (49) between amino acid residues 17 and 18. Thus, the mature protein is 785 amino acids long and has a predicted molecular mass of 85,320 Da and an isoelectric point of 4.17.
FIG. 1.
Nucleotide sequence of the A. nidulans xlnD gene and the deduced amino acid sequence of the gene product. Putative CreA and PacC binding sites are underlined and double underlined, respectively. Potential N-glycosylation sites are in italics.
FIG. 2.
Alignment of the β-xylosidase amino acid sequences of A. nidulans, A. niger (46), and T. reesei (34a). Asterisks indicate identical amino acids, and dots indicate conservative changes.
Analysis of the 5′ noncoding sequence of the A. nidulans xlnD gene revealed the presence of sequence elements (Fig. 1) that could be involved in transcriptional initiation (21, 45). One TATA box is present 75 bp upstream of the ATG codon, and two CAAT boxes are present 85 and 100 bp upstream of the ATG codon. The start codon is preceded by the sequence TCACC, which resembles the consensus CCPuCC-ATG sequence found in higher eukaryotes (27). In addition, consensus binding target sequences for the A. nidulans wide domain regulators CreA (8) and PacC (43) are present. An AATAAA polyadenylation motif is present 120 bp downstream of the proposed stop codon (38).
β-Xylosidase overproduction in A. nidulans.
Plasmid pXDE1, which contains the A. nidulans xlnD gene plus 1,660 bp of upstream sequence, was introduced into A. nidulans G191 by cotransformation with plasmid pGW635. Uridine prototrophs were selected and analyzed by Southern hybridization. Five cotransformants were tested for β-xylosidase overexpression by direct growth in MM containing oat spelt xylan as the carbon source. Samples (5 ml) were collected in duplicate after 14, 24, and 36 h of incubation, and the β-xylosidase activities in extracted mycelia were measured (Table 1). In all of the strains analyzed, β-xylosidase activity was lower in the 36-h samples than in the 14-h samples due to the presence of protease activity (data not shown). All of the cotransformants exhibited higher levels of β-xylosidase activity than the nontransformed strain. β-Xylosidase overexpression was greatest in cotransformants TXD1.4 and TXD1.10. These two cotransformants and A. nidulans G191 were also grown for 17 h in MM containing d-fructose and Casamino Acids as carbon and nitrogen sources, respectively, and mycelia were then transferred to d-xylose-containing induction medium. Samples (5 ml) were collected in duplicate after 2, 4, and 8 h of posttransfer incubation, and the β-xylosidase activities in extracted mycelia were measured (Table 2). Activity in the nontransformed strain was detected 2 h after transfer and then decreased, and the minimum activity was reached after 8 h. The cotransformants had similar activity profiles but higher absolute levels at all time points after transfer.
TABLE 1.
Time course of β-xylosidase production during growth of A. nidulans G191 and five xlnD cotransformants on MM containing 1% oat spelt xylan as the sole carbon source
| Strain | β-Xylosidase activity (U/g of mycelial dry wt) after growth for:
|
||
|---|---|---|---|
| 14 h | 24 h | 36 h | |
| G191 | 6.28 | 2.13 | 0.46 |
| TXD1.1 | 158.57 | 5.42 | 2.90 |
| TXD1.3 | 131.42 | 7.85 | 23.33 |
| TXD1.4 | 617.14 | 62.85 | 20.33 |
| TXD1.6 | 70.00 | 13.57 | 2.90 |
| TXD1.10 | 442.85 | 100.71 | 28.33 |
TABLE 2.
Time course of β-xylosidase production by A. nidulans G191 and two xlnD cotransformants during incubation of washed, d-fructose-grown mycelia in d-xylose-containing induction media
| Strain | β-Xylosidase activity (U/g of mycelial dry wt) after growth for:
|
|||
|---|---|---|---|---|
| 0 h | 2 h | 4 h | 8 h | |
| G191 | 1.60 | 27.71 | 12.33 | 4.91 |
| TXD1.4 | 3.25 | 83.14 | 80.11 | 63.75 |
| TXD1.10 | 3.60 | 183.57 | 156.22 | 141.08 |
Western blot analysis of the overproduced β-xylosidase.
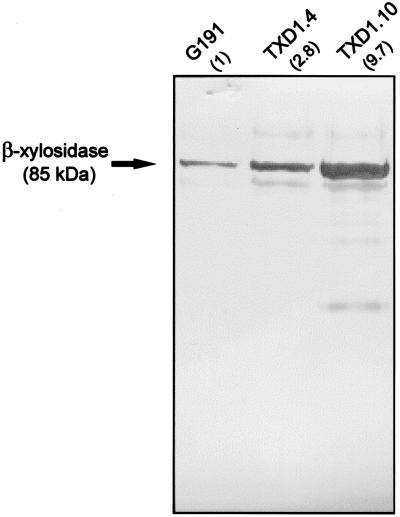
In previous work in our laboratory Kumar and Ramón isolated and characterized an 85-kDa β-xylosidase from A. nidulans (29). Antibodies raised against this enzyme (29) were used to probe a Western blot of mycelial extracts prepared from G191 (the untransformed strain) and the overexpressing transformants TXD1.4 and TXD1.10. Figure 3 shows that in the extracts from the overproducing transformants there were increased levels of a specific band detected by the A. nidulans β-xylosidase antibody whose mobility was identical to the mobility of a band found in the untransformed strain extract. The intensities of staining of the bands corresponded to the levels of β-xylosidase activity measured in the mycelial extracts.
FIG. 3.
Western blot. Mycelial extracts of G191 (an untransformed strain) and transformed β-xylosidase overproducers TXD1.4 and TXD1.10 were probed with an antibody raised against the A. nidulans 85-kDa β-xylosidase (29). The numbers in parentheses are the relative (compared to nontransformed extract) β-xylosidase activities in the mycelial extracts.
Transcriptional regulation of xlnD.
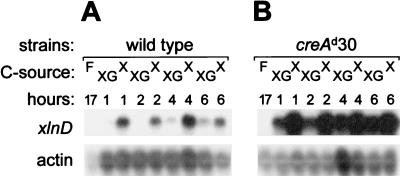
The levels of expression of xlnD were investigated by performing a Northern blot analysis of transfer cultures and in all cases were compared to the levels of actin transcripts as an internal control. Mycelial biomass was grown from spores for 17 h in d-fructose-containing MM supplemented with Casamino Acids and then transferred to d-xylose-containing induction medium. Mycelial samples taken 1, 2, 4, and 6 h after the transfer were used to prepare total RNA. No xlnD expression was detected after growth of the A. nidulans wild-type strain in d-fructose-containing medium. However, within 1 h of transfer to inducing conditions a strong xlnD transcript signal was detected, the level of which remained high throughout the time course analyzed (Fig. 4A, lanes X).
FIG. 4.
Northern blots of total RNAs extracted from wild-type mycelia (A) and the creAd30 mutant (B) after growth in the presence of d-fructose (lanes F) for 17 h and transfer to inducing conditions (1% d-xylose) (lanes X) and inducing-repressing conditions (1% d-xylose plus 1% d-glucose) (lanes XG).
The carbon catabolite repressibility of xlnD expression was investigated with the severe carbon catabolite repression mutant creAd30 (2). Mycelial biomasses from the wild type and a creAd30 mutant were each divided into two halves and transferred to induction medium and induction medium supplemented with d-glucose. In the wild-type mycelial samples, xlnD transcript levels were considerably reduced at the early time points in the presence of d-glucose (Fig. 4A, lanes XG). In the case of creAd30, elevated levels of the xlnD transcript were observed in the presence of glucose at all of the time points examined, although the transcript levels never attained the levels seen in the samples that were induced and derepressed (samples containing d-xylose but not d-glucose) (Fig. 4B).
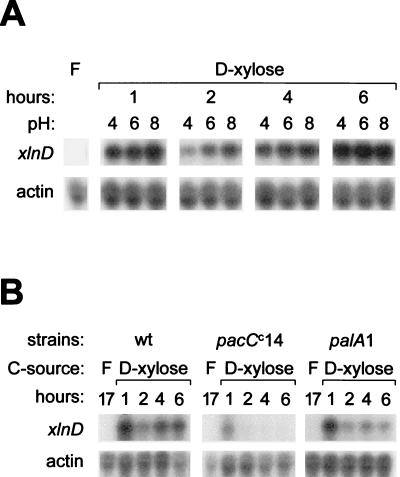
The influence of pH on xlnD expression was investigated by the following two techniques: (i) by examining the consequences of transferring wild-type mycelial biomass to acidic (pH 4), neutral (pH 6), and alkaline (pH 8) buffered induction media, and (ii) by analyzing the expression of xlnD in A. nidulans pH regulatory mutants. Transfer of d-fructose-grown wild-type mycelium to sodium phosphate-buffered induction media revealed no apparent influence of pH at any of the time points analyzed (Fig. 5A). The pH of each medium was measured at the time of harvest to ensure that the buffering capacity was adequate throughout the experiment, and no significant pH shifts were detected (data not shown). xlnD expression in both the acid mimic mutant palA1 and the alkaline mimic mutant pacCc14 exhibited induction patterns similar to the pattern observed for the wild-type strain after mycelial biomass was transferred to nonbuffered induction media, although induction in the pacCc14 mutant seemed to be partially reduced (Fig. 5B). The transcript levels appeared to decrease at later time points in the mutants, whereas the transcript level remained essentially the same in the wild type.
FIG. 5.
(A) Northern blots of total RNA extracted from wild-type mycelia after growth in the presence of d-fructose (lane F) for 17 h and transfer to inducing conditions in media buffered with sodium phosphate to pH 4, 6, and 8. (B) Similar experiment performed with mycelia from the wild type (wt) and the pacCc14 and palA1 mutants after fructose-grown biomass was transferred to nonbuffered inducing conditions (1% d-xylose) for 1, 2, 4, and 6 h.
DISCUSSION
An A. nidulans 4.3-kb EcoRI genomic DNA fragment subcloned in pXDE1 harbors a functional β-xylosidase gene (designated xlnD) since A. nidulans multicopy transformants exhibit significant β-xylosidase overexpression (10- to 100-fold greater expression) compared to the wild type. Western blot analysis performed with a polyclonal antibody raised against the A. nidulans 85-kDa β-xylosidase revealed elevated levels of production of this enzyme in overexpressing transformants. Taken together, these data show that the cloned sequences present in pXDE1 encode the previously characterized (29) 85-kDa β-xylosidase.
The nucleotide sequence of xlnD has been determined. The coding region of the gene consists of an uninterrupted 2,406-bp open reading frame which encodes an 802-amino-acid protein. The deduced molecular mass of the mature protein (85.3 kDa) corresponds closely to the molecular mass previously determined for the purified A. nidulans β-xylosidase (29). This implies that although a number of potential N-glycosylation sites occur within the primary structure, the protein is either not glycosylated to a high degree or not glycosylated at all. The vast majority of enzymic activity (>90%) appears to be cell wall associated (data not shown). The basis of this association might be a consequence of the enzyme’s molecular size since the purified activity has been found to be a dimer (29) and this might result in its capture within the cell wall, a situation analogous to the situation which has been described for the A. niger glucose oxidase (48). The deduced primary structure of the cloned β-xylosidase exhibits a high degree of similarity to the primary structures of other previously characterized fungal β-xylosidases (Fig. 2), confirming that the cloned gene encodes the A. nidulans 85-kDa β-xylosidase.
As in A. niger, expression of xlnD is specifically induced by oat spelt xylan, as well as by d-xylose (46). Transcription of xlnD does not appear to be influenced by the external pH. No significant differences were found in xlnD transcript levels after d-xylose induction at pH 4, 6, and 8. This finding is consistent with the expression data obtained with pH regulatory mutants, although the level of transcription in the pacCc14 alkaline mimic mutant seems to be somewhat lower than the level of transcription in the wild type. In contrast, xlnD expression is subject to carbon catabolite repression by d-glucose, indicating that the glucose repression of β-xylosidase activity observed previously (29) occurs at the level of mRNA transcription; a transcript is observed upon induction by d-xylose but not in the presence of d-glucose. xlnD is, however, transcribed in the presence of d-glucose in the creAd30 mutant, from which it can be concluded that carbon catabolite repression of the gene is, at least in part, controlled by CreA. Carbon catabolite repression mediated by CreA has also been observed in other fungal genes encoding xylanolytic enzymes (10, 31, 36). Nine putative CreA binding sites are located in the xlnD upstream sequences. Deletion analysis of these sites is in progress in order to determine their in vivo function.
ACKNOWLEDGMENTS
We thank H. N. Arst, Jr., and M. A. Peñalva for kindly providing the A. nidulans mutant strains used in this work and M. Penttilä for providing the revised sequence of the T. reesei β-xylosidase.
This work was supported by grant BIOTECH BIO2-CT93-0174 from D.G. XII of the European Commission and by grant CICYT ALI93-0809 from the Spanish Government. A.P.M. was the recipient of EC Biotechnology Programme fellowship BIO2-CT94-8136.
REFERENCES
- 1.Arst H N, Jr, Scazzocchio C. Formal genetics and molecular biology of the control of gene expression in Aspergillus nidulans. In: Bennet J W, Lasure L L, editors. Gene manipulations in fungi. New York, N.Y: Academic Press; 1985. pp. 309–343. [Google Scholar]
- 2.Arst H N, Jr, Tollervey D, Dowzer C E A, Kelly J M. An inversion truncating the creA gene of Aspergillus nidulans results in carbon catabolite derepression. Mol Microbiol. 1990;4:851–854. doi: 10.1111/j.1365-2958.1990.tb00656.x. [DOI] [PubMed] [Google Scholar]
- 3.Arst H N, Jr, Bignell E, Tilburn J. Two new genes involved in signalling ambient pH in Aspergillus nidulans. Mol Gen Genet. 1994;245:787–790. doi: 10.1007/BF00297286. [DOI] [PubMed] [Google Scholar]
- 4.Ballance D J, Turner G. Development of a high frequency transforming vector for Aspergillus nidulans. Gene. 1985;36:321–331. doi: 10.1016/0378-1119(85)90187-8. [DOI] [PubMed] [Google Scholar]
- 5.Caddick M X, Brownlee A G, Arst H N., Jr Regulation of gene expression by pH of the growth medium in Aspergillus nidulans. Mol Gen Genet. 1986;203:346–353. doi: 10.1007/BF00333978. [DOI] [PubMed] [Google Scholar]
- 6.Cathala G, Savouret J F, Mendez B, West B L, Karin M, Martial J A, Baxter J D. A method for isolation of intact, translationally active ribonucleic acid. DNA. 1983;2:329–335. doi: 10.1089/dna.1983.2.329. [DOI] [PubMed] [Google Scholar]
- 7.Coughlan M P, Hazlewood G P. β-1,4-d-Xylan-degrading enzyme systems: biochemistry, molecular biology and applications. Biotechnol Appl Biochem. 1993;17:259–289. [PubMed] [Google Scholar]
- 8.Cubero B, Scazzocchio C. Two different, adjacent and divergent zinc finger binding sites are necessary for CREA-mediated carbon catabolite repression in the proline gene cluster of Aspergillus nidulans. EMBO J. 1994;13:407–415. doi: 10.1002/j.1460-2075.1994.tb06275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis M A, Hynes M J. Regulatory circuits in Aspergillus nidulans. In: Bennet J W, Lasure L L, editors. More gene manipulations in fungi. New York, N.Y: Academic Press; 1991. pp. 151–189. [Google Scholar]
- 10.de Graaff L H, van den Broeck H, van Ooijen A J J, Visser J. Regulation of the xylanase-encoding xlnA gene of Aspergillus tubingensis. Mol Microbiol. 1994;12:479–490. doi: 10.1111/j.1365-2958.1994.tb01036.x. [DOI] [PubMed] [Google Scholar]
- 11.de Graaff L H, van den Broeck H, Visser J. Isolation and transformation of the pyruvate kinase gene of Aspergillus nidulans. Curr Genet. 1988;13:315–321. doi: 10.1007/BF00424425. [DOI] [PubMed] [Google Scholar]
- 12.Denison S H, Orejas M, Arst H N., Jr Signalling of ambient pH in Aspergillus involves a cysteine protease. J Biol Chem. 1995;270:28519–28522. doi: 10.1074/jbc.270.48.28519. [DOI] [PubMed] [Google Scholar]
- 13.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feinberg B P, Vogelstein B. A technique for radiolabelling DNA restriction fragments to high specific activity. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 15.Fernández-Espinar M T, Piñaga F, de Graaff L H, Visser J, Ramón D, Vallés S. Purification, characterization and regulation of the synthesis of an Aspergillus nidulans acidic xylanase. Appl Microbiol Biotechnol. 1994;42:555–562. [Google Scholar]
- 16.Fernández-Espinar M T, Piñaga F, Sanz P, Ramón D, Vallés S. Purification and characterization of a neutral endoxylanase from Aspergillus nidulans. FEMS Microbiol Lett. 1993;113:223–228. [Google Scholar]
- 17.Fernández-Espinar M T, Ramón D, Piñaga F, Vallés S. Xylanase production by Aspergillus nidulans. FEMS Microbiol Lett. 1992;91:91–96. doi: 10.1111/j.1574-6968.1994.tb06622.x. [DOI] [PubMed] [Google Scholar]
- 18.Fernández-Espinar M T, Vallés S, Piñaga F, Pérez-González J A, Ramón D. Construction of an Aspergillus nidulans multicopy transformant for the xlnB gene and its use to purify the minor X24 xylanase. Appl Microbiol Biotechnol. 1996;45:338–341. [Google Scholar]
- 19.Fidel S, Doonan J H, Morris N R. Aspergillus nidulans contains a single actin gene which has unique intron locations and encodes γ-actin. Gene. 1988;70:283–293. doi: 10.1016/0378-1119(88)90200-4. [DOI] [PubMed] [Google Scholar]
- 20.Goosen T, Bloemheuvel G, Gysler C, de Bie D A, van den Broek H W J, Swart K. Transformation of Aspergillus niger using the homologous orotidine-5-phosphate-decarboxylase gene. Curr Genet. 1987;13:499–503. doi: 10.1007/BF00384612. [DOI] [PubMed] [Google Scholar]
- 21.Gurr S J, Unkles S E, Kinghorn J R. The structure and organization of nuclear genes of filamentous fungi. In: Kinghorn J R, editor. Gene structure in eukaryotic microbes. Oxford, United Kingdom: IRL Press; 1987. pp. 93–139. [Google Scholar]
- 22.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hessing J G M, van Rotterdam C, Verbakel J M A, Roza M, Maat J, van Gorcom R F M, van den Hondel C A M J J. Isolation and characterization of a 1,4-β-endoxylanase gene of A. awamori. Curr Genet. 1994;26:228–232. doi: 10.1007/BF00309552. [DOI] [PubMed] [Google Scholar]
- 24.Hintz W E, Lagosky P A. A glucose-derepressed promoter for expression of heterologous products in the filamentous fungus Aspergillus nidulans. Bio/Technology. 1993;11:815–818. doi: 10.1038/nbt0793-815. [DOI] [PubMed] [Google Scholar]
- 25.Ito K, Ikesamu T, Ishikawa T. Cloning and sequencing of the xynA gene encoding xylanase A of Aspergillus kawachii. Biosci Biotechnol Biochem. 1992;56:906–912. doi: 10.1271/bbb.56.906. [DOI] [PubMed] [Google Scholar]
- 26.Ito K, Iwashita K, Iwano K. Cloning and sequencing of the xynC gene encoding acid xylanase of Aspergillus kawachii. Biosci Biotechnol Biochem. 1992;56:1338–1340. doi: 10.1271/bbb.56.1338. [DOI] [PubMed] [Google Scholar]
- 27.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 28.Kulmburg P, Mathieu M, Dowzer C, Kelly J, Felenbok B. Specific binding sites in the alcR and alcA promoters of the ethanol regulon for the CREA suppressor mediating carbon catabolite repression in Aspergillus nidulans. Mol Microbiol. 1993;7:847–857. doi: 10.1111/j.1365-2958.1993.tb01175.x. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S, Ramón D. Purification and regulation of the synthesis of a β-xylosidase from Aspergillus nidulans. FEMS Microbiol Lett. 1996;135:287–293. [Google Scholar]
- 30.MacCabe A P, Fernández-Espinar M T, de Graaff L H, Visser J, Ramón D. Identification, isolation and sequence of the Aspergillus nidulans xlnC gene encoding the 34-kDa xylanase. Gene. 1996;175:29–33. doi: 10.1016/0378-1119(96)00116-3. [DOI] [PubMed] [Google Scholar]
- 31.Mach R L, Strauss J, Zeilinger S, Schindler M, Kubicek C P. Carbon catabolite repression of xylanase I (xyn1) gene expression in Trichoderma reesei. Mol Microbiol. 1996;21:1273–1281. doi: 10.1046/j.1365-2958.1996.00094.x. [DOI] [PubMed] [Google Scholar]
- 32.Margolles-Clark E, Tenkanen M, Nakari-Setälä T, Penttilä M. Cloning of genes encoding α-l-arabinofuranosidase and β-xylosidase from Trichoderma reesei by expression in Saccharomyces cerevisiae. Appl Environ Microbiol. 1996;62:3840–3846. doi: 10.1128/aem.62.10.3840-3846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Negrete-Urtasun S, Denison S, Arst H N., Jr Characterization of the pH signal transduction pathway gene palA of Aspergillus nidulans and identification of possible homologs. J Bacteriol. 1997;179:1832–1835. doi: 10.1128/jb.179.5.1832-1835.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orejas M, Espeso E A, Tilburn J, Sarkar S, Arst H N, Jr, Peñalva M A. Activation of the Aspergillus PacC transcription factor in response to alkaline ambient pH requires proteolysis of the carboxy-terminal moiety. Genes Dev. 1995;9:1622–1632. doi: 10.1101/gad.9.13.1622. [DOI] [PubMed] [Google Scholar]
- 34a.Penttilä, M. Personal communication.
- 35.Pérez-González J A, de Graaff L H, Visser J, Ramón D. Molecular cloning and expression in Saccharomyces cerevisiae of two Aspergillus nidulans xylanase genes. Appl Environ Microbiol. 1996;62:2179–2182. doi: 10.1128/aem.62.6.2179-2182.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piñaga F, Fernández-Espinar M T, Vallés S, Ramón D. Xylanase production in Aspergillus nidulans: induction and carbon catabolite repression. FEMS Microbiol Lett. 1994;115:319–324. doi: 10.1111/j.1574-6968.1994.tb06622.x. [DOI] [PubMed] [Google Scholar]
- 37.Pontecorvo G, Roper J A, Hemmons L J, MacDonald K D, Bufton A W J. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 38.Proudfoot N, Brownlee G G. 3′ non-coding region sequences in eukaryotic mRNA. Nature. 1976;263:211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- 39.Saarelainen R, Paloheimo M, Fagerström R, Suominen P L, Nevalainen K M H. Cloning, sequencing, and enhanced expression of the Trichoderma reesei endoxylanase II (pI 9) gene xln2. Mol Gen Genet. 1993;241:497–503. doi: 10.1007/BF00279891. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tilburn J, Scazzocchio C, Taylor G G, Zabicky-Zissman J H, Lockington R A, Davies R W. Transformation by integration in Aspergillus nidulans. Gene. 1983;26:205–221. doi: 10.1016/0378-1119(83)90191-9. [DOI] [PubMed] [Google Scholar]
- 43.Tilburn J, Sarkar S, Widdick D A, Espeso E A, Orejas M, Mungroo J, Peñalva M A, Arst H N., Jr The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 1995;14:779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Törrönen A, Mach R L, Messner R, González R, Kalkkinen N, Harkki A, Kubicek C P. The two major xylanases from Trichoderma reesei: characterization of both enzymes and genes. Bio/Technology. 1992;10:1461–1465. doi: 10.1038/nbt1192-1461. [DOI] [PubMed] [Google Scholar]
- 45.Unkles S E. Gene organization in industrial filamentous fungi. In: Kinghorn J R, editor. Applied molecular genetics of filamentous fungi. London, United Kingdom: Chapman and Hall; 1992. pp. 28–53. [Google Scholar]
- 46.van Peij N N, Brinkmann J, Vrsanská M, Visser J, de Graaff L H. β-Xylosidase activity, encoded by xlnD, is essential for complete hydrolysis of xylan by Aspergillus niger but not for induction of the xylanolytic enzyme spectrum. Eur J Biochem. 1997;245:164–173. doi: 10.1111/j.1432-1033.1997.00164.x. [DOI] [PubMed] [Google Scholar]
- 47.Visser J, Beldman G, Kusters-van Someren M A, Voragen A G J. Xylans and xylanases. Amsterdam, The Netherlands: Elsevier; 1992. [Google Scholar]
- 48.Visser J, Bussink H J, Witteveen C. Gene expression in filamentous fungi: expression of pectinases and glucose oxidase in A. niger. In: Smith A, editor. Gene expression in recombinant microorganisms. New York, N.Y: Marcel Dekker Inc.; 1994. pp. 241–308. [PubMed] [Google Scholar]
- 49.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeillinger S, Mach R L, Schindler M, Herzog P, Kubicek C P. Different inducibility of expression on the two xylanase genes xyn1 and xyn2 in Trichoderma reesei. J Biol Chem. 1996;271:25624–25629. doi: 10.1074/jbc.271.41.25624. [DOI] [PubMed] [Google Scholar]
- 51.Zimmermann C R, Orr W C, Leclerc R F, Barnard E C, Timberlake W E. Molecular cloning and selection of genes regulated in Aspergillus development. Cell. 1980;21:709–715. doi: 10.1016/0092-8674(80)90434-1. [DOI] [PubMed] [Google Scholar]