Abstract
Signaling through the mammalian target of rapamycin (mTOR) controls cell size and growth as well as other functions, and it is a potential therapeutic target for graft rejection, certain cancers, and disorders characterized by inappropriate cell or tissue growth. mTOR signaling is positively regulated by hormones or growth factors and amino acids. mTOR signaling regulates the phosphorylation of several proteins, the best characterized being ones that control mRNA translation. Eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) undergoes phosphorylation at multiple sites. Here we show that amino acids regulate the N-terminal phosphorylation sites in 4E-BP1 through the RAIP motif in a rapamycin-insensitive manner. Several criteria indicate this reflects a rapamycin-insensitive output from mTOR. In contrast, the insulin-stimulated phosphorylation of the C-terminal site Ser64/65 is generally sensitive to rapamycin, as is phosphorylation of another well-characterized target for mTOR signaling, S6K1. Our data imply that it is unlikely that mTOR directly phosphorylates Thr69/70 in 4E-BP1. Although 4E-BP1 and S6K1 bind the mTOR partner, raptor, our data indicate that the outputs from mTOR to 4E-BP1 and S6K1 are distinct. In cells, efficient phosphorylation of 4E-BP1 requires it to be able to bind to eIF4E, whereas phosphorylation of 4E-BP1 by mTOR in vitro shows no such preference. These data have important implications for understanding signaling downstream of mTOR and the development of new strategies to impair mTOR signaling.
There is presently a high level of research interest in signaling through the mammalian target of rapamycin (mTOR). This reflects its key roles in regulating cell and animal growth, the cell cycle, and gene expression (transcription and translation) (16, 28, 60). Recent studies have demonstrated mTOR is essential for cell growth and proliferation (53). Furthermore, rapamycin, a specific inhibitor of mTOR signaling, is in clinical use, or has clinical potential, for graft rejection (45), restenosis after angioplasty (13), certain types of cancer (33), tumor angiogenesis (25, 77), and liver fibrosis (78). Important recent advances have identified new components on this pathway involved in the upstream control of mTOR signaling (e.g., TSC1, TSC2, and the small G-protein Rheb) (20, 40, 47, 50) and in downstream signaling from mTOR, which involves complexes with partner proteins (e.g., raptor [26, 37], GβL [36], and their yeast orthologs [44]). Nonetheless, our overall understanding of the pathway remains far from complete.
The best understood targets of the mTOR pathway are proteins that regulate the translational machinery. One intensively studied target is the translational repressor protein, 4E-BP1 (eukaryotic initiation factor 4E-binding protein 1) (23, 41). Binding of 4E-BP1 to eIF4E prevents the latter protein from engaging with other partners, such as the scaffold eIF4G, and therefore blocks cap-dependent mRNA translation initiation. 4E-BP1 undergoes phosphorylation at multiple sites (Fig. 1A), and phosphorylation at some of them disrupts its ability to bind eIF4E, leading to release of 4E-BP1 and allowing eIF4E to bind eIF4G. Release of 4E-BP1 from eIF4E is generally blocked by rapamycin, indicating an essential role for mTOR signaling.
FIG. 1.
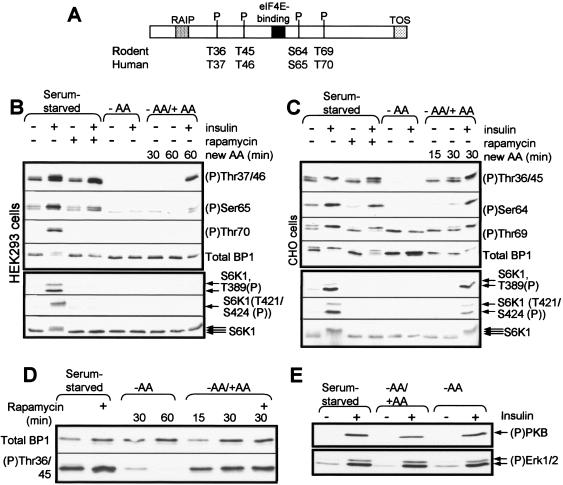
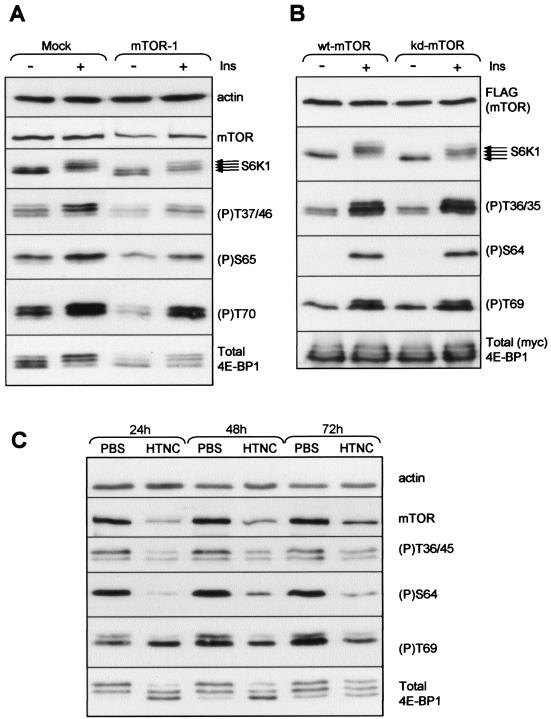
Amino acid withdrawal and rapamycin have distinct effects on 4E-BP1 phosphorylation. (A) The graphic depicts the regulatory RAIP and TOS motifs in 4E-BP1, the eIF4E binding site, and the phosphorylation sites studied here (showing the differences in numbering between human and rodent). (B and C) HEK293 (B) or CHO (C) cells were first starved of serum overnight and then, where indicated, also starved of amino acids (−AA) for 1 h or treated with rapamycin and/or insulin. In some cases (−AA/+AA), amino acids were resupplied to cells that had been deprived of them for 60 min. Times are indicated in minutes. Samples of cell lysate were analyzed by Western blotting with the indicated antisera for 4E-BP1 (total BP1), specific phosphorylation sites in 4E-BP1, or S6K1 (multiple arrows indicate that several differently phosphorylated species are resolved) or an antibody that recognizes S6K1 only when it is phosphorylated at Thr389 or at Thr421 and Ser424 (the two bands correspond to the 70- and 85-kDa species of S6K1). (D) CHO cells were starved of serum overnight and, where indicated, were also starved of amino acids (-AA for times shown) or resupplied with amino acids (−AA/+AA; times shown). Where added, rapamycin was added for 40 min before lysis of the cells. (E) CHO cells, which had been starved of serum and, in some cases, also of amino acids (−AA) or that had been resupplied with amino acids (-AA/+AA), as described above, were in some cases (+) treated with insulin. Samples of cell lysate were analyzed by Western blotting using anti-(P)PKB (Thr308) or anti-(P)Erk, which recognizes the phosphorylated active forms of both Erk1 and Erk2.
Phosphorylation of 4E-BP1 at several sites is stimulated by agents such as insulin, and, in many cell types, this effect requires the presence in the cells' medium of amino acids (60). Phosphorylation of 4E-BP1 is hierarchical, with phosphorylation of Thr36/45 being required for modification of Thr69 (the numbering of residues in human 4E-BP1 is shifted by +1 relative to the rodent orthologs; this site is therefore Thr70 in human 4E-BP1 [PHAS-I]) and Ser64 (21-23, 32, 51, 52). Phosphorylation of other sites, such as Ser101, appears not to be regulated (73).
Which kinase(s) is responsible for the complex phosphorylation of 4E-BP1 in vivo? mTOR can phosphorylate 4E-BP1 in vitro, at least under specific conditions (5, 7, 21, 49, 74). The fact that the C-terminal TOR-signaling (TOS) motif in 4E-BP1 (62) recruits raptor, and thus also mTOR, to 4E-BP1 and plays an important role in the phosphorylation of Ser64/5 and Thr69/70 in vivo appears consistent with the idea that mTOR might indeed directly phosphorylate several sites in 4E-BP1 within cells (3, 9, 55, 63). Rapamycin may disrupt mTOR/raptor complexes (37, 56), which could help explain how rapamycin interferes with the phosphorylation of 4E-BP1. This has given rise to the widely accepted notion that the kinase activity of mTOR directly phosphorylates 4E-BP1 and that inhibition of 4E-BP1 phosphorylation by rapamycin reflects impairment of this activity. However, in vitro, mTOR primarily phosphorylates the N-terminal threonine sites (Thr36 and Thr45 in rodent 4E-BP1 [74]), whose phosphorylation is independent of the TOS motif in vivo (3). In contrast, mTOR only weakly phosphorylates Thr69 and Ser64, the sites which are dependent upon the TOS motif (3, 5, 51). It has also been reported that phosphorylation of some sites in 4E-BP1 is relatively rapamycin insensitive (21). Earlier studies on the mTOR/raptor complex (26) and for the yeast TORC1 complex (which contains the raptor homologue KOG1 [44]) failed to observe an effect of rapamycin on the interactions between these partners.
4E-BP1 also possesses an N-terminal regulatory motif, named the RAIP motif (from the single-letter amino acid code for its sequence), which is required for phosphorylation of many sites in 4E-BP1 in vivo, including the N-terminal threonine residues which are not affected by disruption of the TOS motif (3). In our hands, the RAIP motif does not appear to bind to raptor (3; however, see also reference 9). It also has effects on the phosphorylation of 4E-BP1 in vivo that are different from those of the TOS motif.
Numerous previous studies show that amino acids positively regulate mTOR signaling. Amino acid starvation therefore results in dephosphorylation of proteins such as 4E-BP1 and S6K1 (reviewed in references 1 and 60). However, it is not yet clear how amino acids feed into the regulation of mTOR or whether their withdrawal has effects similar to those of rapamycin.
Here we have studied in detail the site-specific phosphorylation of 4E-BP1 within cells. Our studies reveal new insights into the regulatory events that act downstream of mTOR, which appear to be more complex than previously thought. For example, they indicate that amino acids control a rapamycin-insensitive output from mTOR to 4E-BP1 and that the outputs from mTOR to 4E-BP1 and to S6K1 are distinct.
MATERIALS AND METHODS
Materials.
Chemicals and biochemicals were obtained as described earlier (4). Antisera for specific phosphorylation sites in 4E-BP1 and in S6K1 were from Cell Signaling Technology (Beverly, Mass.). Anti-mTOR was from AbCam (Cambridge, United Kingdom).
Vectors and mutants.
Vectors for rat 4E-BP1 have been described earlier (70). Additional vectors were kindly provided by other investigators as follows: 4E-BP2 (PHAS-II; John Lawrence, University of Virginia, Charlottesville [43]), Rheb (Paul Worley, Johns Hopkins University, Baltimore, Md.), TSC1/TSC2 (Andrew Tee, formerly Harvard University, Boston, Mass., now at the University of Dundee [67]), GST-FKBP12 (Pamela Scott, University of Glasgow, United Kingdom), and PTEN (Peter Downes and Nick Leslie, University of Dundee, United Kingdom [42]). FLAG-mTOR (wild type [WT] or kinase dead) was from Dario Alessi (University of Dundee, United Kingdom), and the vector encoding the myc-Raptor protein was kindly provided by David Sabatini (Boston, Mass.). Point mutations were introduced using the QuikChange (StrataGene) method.
Cell culture, treatment, and lysis.
Chinese hamster ovary (CHO) cells were grown in Ham's F-12 medium supplemented with 10% (vol/vol) fetal calf serum. Human embryonic kidney (HEK293) cells were grown in Dulbecco's modified Eagle medium (DMEM) with 10% (vol/vol) fetal calf serum. Cells were treated as indicated in the figure legends. In all cases, cells were starved of serum overnight before further treatments, such as amino acid deprivation, insulin treatment, etc. For amino acid starvation, cells were transferred for the indicated times to Dulbecco's phosphate-buffered saline containing 10 mM d-glucose. In some experiments, this medium was supplemented with a mixture of amino acids that corresponds to those present in DMEM. Where used, rapamycin was added at 100 nM 40 min before addition of insulin or lysis of the cells. Insulin was used for 30 min at 100 nM. Lysis was performed with our standard lysis buffer (20 mM HEPES [pH 7.5]; 25 mM β-glycerophosphate; 50 mM KCl; 0.2 mM EDTA; 0.5 mM sodium orthovanadate; 0.5% [vol/vol] Triton X-100; 0.1 μM microcystin LR; 15 mM β-mercaptoethanol; 1 μg each of pepstatin, leupeptin, and antipain/ml; 1 mM benzamidine; 1 mM phenylmethylsulfonyl fluoride).
Analytical methods.
Procedures for gel electrophoresis and immunoblotting have been described previously (59, 69), as has the method for the affinity chromatography of eIF4E on m7GTP-Sepharose (73). The ability of mTOR to phosphorylate 4E-BP1 in vitro was assessed as described earlier (70), and recombinant eIF4E and 4E-BP1 were prepared as described previously (64, 70).
In vitro phosphorylation of 4E-BP1 by mTOR.
GST-4E-BP1, FLAG-mTOR, and myc-raptor were expressed in HEK293 cells as described earlier (3). Cells were transfected with 5 μg of pcDNA FLAG-mTOR and a vector encoding the myc-Raptor protein. Cells were grown in medium containing 10% fetal calf serum for 2 days and then were harvested using a detergent-free buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 5 mM EGTA, 20 mM β-glycerophosphate). Cells were lysed by 3 cycles of freeze and thaw.
mTOR/raptor was then immunoprecipitated using protein G coupled to anti-FLAG. The immunoprecipitate was then washed twice with high-salt buffer (50 mM HEPES [pH 7.4], 0.5 M NaCl), twice with low-salt buffer (50 mM HEPES [pH 7.4], 150 mM NaCl), and once in the mTOR kinase buffer (10 mM HEPES, 50 mM β-glycerophosphate, 150 mM NaCl).
Prior to being added to the reaction, equimolar amounts of glutathione S-transferase (GST)-4E-BP1 and eIF4E protein produced in Escherichia coli were incubated together at 30°C for 15 min. Rapamycin (1 μM) and FKBP12 protein (10 μg) were also incubated together at 30°C for 15 min. The reaction was then started by adding 10 mM MnCl2 and 200 μM ATP plus 10 μCi of [γ-32P]ATP at 30°C for 60 min. The reaction was then stopped by adding 4× sodium dodecyl sulfate (SDS) sample buffer, and the products were resolved in an SDS-15% polyacrylamide gel electrophoresis (PAGE) gel. The dried gel was then subjected to autoradiography.
RNA interference.
For siRNA, we exploited HeLa cells from Jacques Pouysségur (Nice, France [2]). Five sets of complementary 21-residue oligoribonucleotides corresponding to different regions of mTOR were synthesized with the Silencer kit from Ambion (Austin, Tex.), using the methods recommended by the manufacturer. The templates were supplied by MWG-Biotech (Ebersberg, Germany). The selected regions were those starting at nucleotides 3572 (mTOR-1, the most effective ones and the only set reported here), 2792, 3073, 3266, and 425, all within the coding region of the mRNA. HeLa cells were grown in DMEM containing 10% (vol/vol) fetal calf serum without antibiotics. When the cells were 30 to 50% confluent, they were transfected using a final concentration of 60 nM each pair of oligoribonucleotides or combinations thereof. After 24 h, cells were retransfected. On the third day, cells were starved of serum for 4 h prior to further treatment or analysis. Cells were scrutinized microscopically. As cells tended to detach from the plate after treatment with the siRNAs directed against mTOR, care was taken to include so-called floating cells in the samples we analyzed.
Targeting of mTOR in embryonic stem (ES) cells.
mTOR was targeted genetically to generate a C-terminally truncated variant exactly as described in reference 53.
RESULTS
Phosphorylation of the N-terminal sites in 4E-BP1 is resistant to rapamycin but decreases upon amino acid withdrawal.
Figure 1A depicts the structures of mammalian 4E-BP1 proteins, showing the phosphorylation sites studied here and other relevant features.
We (8, 72) and others (19, 27, 51) have noted that amino acids modulate the phosphorylation state and the function of 4E-BP1. To study this further, we made use of the phosphospecific antisera that are available for four sites in 4E-BP1. It should be noted that the electrophoretically distinct bands for 4E-BP1 are detected by different sets of phosphospecific antisera. This is because, as documented earlier, phosphorylation of certain sites appears to cause the observed band shifts (21, 22, 51, 52). The most highly phosphorylated topmost band is detected by all phosphospecific antisera used here. Phosphorylation of Thr36/45 (or the corresponding sites in the human protein) in the cells' endogenous 4E-BP1 was readily detectable in lysates from serum-starved HEK293 or CHO cells maintained in medium containing amino acids (Fig. 1B and C). Phosphorylation of these sites was not appreciably increased by insulin, in agreement with our earlier data for both CHO (57) and HEK293 cells (70, 73). Phosphorylation of Thr36/45 (or their equivalents) was not detectably inhibited by rapamycin in CHO or HEK293 cells (Fig. 1B and C); earlier data have also suggested that these sites may be relatively insensitive to rapamycin (22). In contrast, withdrawal of amino acids led to their rapid and complete dephosphorylation in both cell types (Fig. 1B and C). Thus, amino acid withdrawal and rapamycin treatment have strikingly different effects on the phosphorylation of these sites in 4E-BP1. This insensitivity to rapamycin could suggest that Thr36/45 are not phosphorylated by mTOR in vivo. However, we will present further evidence that mTOR is involved in their phosphorylation.
It was also possible that the (putative) mTOR signaling complex that affects Thr36/45 is already formed in amino acid-replete cells, and that rapamycin does not affect this but could block its formation when amino acids are restored. Replenishing amino acids resulted in a rapid and marked increase in the phosphorylation of Thr36/45 in CHO cells (Fig. 1C), but this effect was not blocked by rapamycin (Fig. 1D). This finding argues against the above possibility and reinforces the conclusion that the effects of amino acids and rapamycin are not simply the converse of one another.
In HEK293 cells, re-addition of amino acids after a period of withdrawal did not elicit phosphorylation of 4E-BP1 (Fig. 1B), even though the combination used contains the same mixture and concentrations of amino acids as the medium in which HEK293 cells are grown. The reasons for this are unknown.
In the absence of amino acids, insulin failed to induce phosphorylation of any site in 4E-BP1 in either cell type. Insulin was able to partly restore levels of phosphorylation of these sites in cells to which amino acids had been resupplied (Fig. 1B and C). Because the effects of amino acid deprivation and re-addition are fully reversible in CHO cells, we elected to use them for the remaining experiments on the control of 4E-BP1 phosphorylation by amino acids. Furthermore, we have already extensively characterized the effects of amino acids on mTOR signaling in these cells (8, 57, 72). Our data for HEK293 cells differ in certain respects from those reported earlier (51). A possible reason for this is that while we studied the phosphorylation of the endogenous 4E-BP1, Mothe-Satney et al. (51), for example, only looked at the overexpressed protein. Our earlier data also show that the insulin-induced phosphorylation of the N-terminal threonines in overexpressed rat or human 4E-BP1 is partially blocked by rapamycin (3, 70, 73). The commercially available antibody for (P)Ser65 cross-reacts with an additional site in the human protein (at Ser101 [73]), making it hard to get specific information on the phosphorylation of this site. Given this difficulty, we have generally used rat 4E-BP1 in the transfection experiments described later in this report, because although rat 4E-BP1 contains a serine at the same position, differences in the local sequence mean that it is not recognized by the phosphospecific antibody.
The ability of insulin to elicit phosphorylation of PKB (at Thr308) or Erk (TEY motif) was essentially identical, regardless of whether cells were starved of serum or serum and amino acids or were resupplied with amino acids (Fig. 1E). This confirms that amino acid status does not compromise the ability of CHO cells to respond to insulin.
Regulation of the phosphorylation of Thr69/70 differs between HEK293 and CHO cells.
Thr69/70 can also be phosphorylated, albeit poorly, by mTOR in vitro, but this is reported to be completely eliminated by rapamycin (and also by LY294002 or wortmannin [9, 49]). In vivo, phosphorylation of Thr69/70 is thought to depend on prior phosphorylation of the N-terminal sites and to facilitate phosphorylation of Ser64/65 and, probably, release of eIF4E from 4E-BP1 (22, 53). Phosphorylation of Thr69 in vivo is profoundly decreased by mutation of the TOS motif through which 4E-BP1 binds raptor (3, 9, 55, 63).
In HEK293 cells, the signal for phospho-Thr70 in serum-starved cells is too weak to allow one to examine the effects of amino acid withdrawal or rapamycin (Fig. 1B), but it is strongly increased upon addition of insulin, an effect that is blocked by rapamycin (Fig. 1B). In CHO cells the corresponding signal is much stronger, and amino acid withdrawal has no effect on the level of phosphorylation of this site; neither does insulin (Fig. 1C). The inability of insulin to increase the signal for Thr69 in CHO cells is consistent with our earlier data (57) and similar to the situation reported for 3T3-L1 adipocytes (51). Strikingly, the high level of phosphorylation of Thr69 was not affected at all by rapamycin in these cells (Fig. 1C). This could suggest that, like the N-terminal threonines, Thr69 in CHO cells is regulated by a rapamycin-insensitive output from mTOR. The data also show that, in CHO cells at least, phosphorylation of Thr69 is not tightly coupled to the phosphorylation of the N-terminal threonines.
An alternative explanation, that mTOR signaling is insensitive to rapamycin in CHO cells, can be ruled out by the data showing that phosphorylation of S6K1 is fully sensitive to rapamycin in CHO cells (Fig. 1C). Possible reasons for this difference between the behavior of Thr69 in CHO cells and the corresponding site in HEK293 cells are explored below.
The phosphorylation of Ser64 is also partially resistant to rapamycin in CHO cells.
The phosphorylation of Ser64 in serum-starved CHO cells is increased by insulin (Fig. 1C). However, both the basal (serum-starved) and insulin-stimulated signals show little, if any, sensitivity to rapamycin, but they are completely eliminated by amino acid withdrawal. The pattern for the phosphorylation of this residue mirrors the appearance and disappearance of the slowest moving γ-species, which is the only one for which a signal for Ser64(P) is seen (Fig. 1C). These data again indicate that the effects of amino acid withdrawal and rapamycin treatment are not identical. Analysis of phosphorylation of Ser65 in human 4E-BP1 in HEK cells is complicated by the cross-reactivity of the commercially available phosphospecific antiserum with Ser101 (73).
Regulation of S6K1 is fully sensitive to rapamycin in both cell types.
The differences in rapamycin sensitivity of specific sites in 4E-BP1 between CHO and HEK293 cells prompted us to test whether phosphorylation of another target for mTOR signaling, S6K1, might also be resistant to rapamycin in CHO cells.
Phosphorylation of Thr389, Thr421, and Ser424 in S6K1 was essentially undetectable in the absence of insulin in both CHO and HEK293 cells but was increased by insulin (Fig. 1B and C, bottom panels). This therefore differs markedly from the situation for the N-terminal sites in 4E-BP1, which are phosphorylated under serum-starved conditions and not stimulated detectably by insulin. Furthermore, while phosphorylation of these sites in 4E-BP1 is not affected by rapamycin, in both types of cells this drug causes a complete block in the phosphorylation of S6K1 that is induced by insulin (Fig. 1B and C, bottom panels) as well as collapsing the ladder of bands seen either in amino acid-replete or insulin-stimulated cells. Rapamycin thus seems to inhibit the phosphorylation of S6K1 at many sites and in both cell types (which is consistent with earlier data [15]).
The rapamycin-insensitive phosphorylation of certain sites in 4E-BP1 is a feature of CHO cells, not of hamster 4E-BP1.
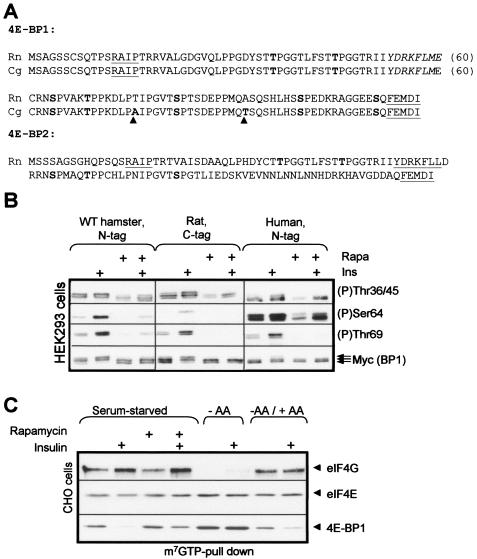
In view of the striking difference in the regulation of specific phosphorylation sites in CHO cells compared to that of the better characterized HEK293 cells, it was possible that hamster 4E-BP1 differed from the human protein in some way that rendered its phosphorylation less sensitive to rapamycin. We therefore cloned cDNAs for hamster 4E-BP1 by reverse transcription-PCR (RT-PCR) using cDNA derived from CHO cells as a template. The sequence of hamster 4E-BP1 is very similar to that of the rat protein, there only being two amino acid differences between them, i.e., Thr76 in rat is replaced by Ala and Ala92 is replaced by Thr (Fig. 2A). All the known phosphorylation sites and both regulatory motifs (RAIP and TOS) are thus fully conserved.
FIG. 2.
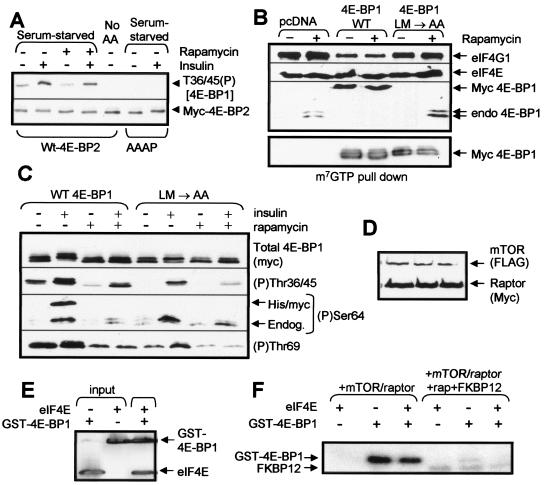
Sequence and regulation of hamster 4E-BP1. (A) Amino acid sequences of rat (Rattus norvegicus [Rn]) and Chinese hamster (Cricetulus griseus [Cg]) 4E-BP1 and of rat 4E-BP2 (C). The RAIP and TOS motifs are underlined, the eIF4E-binding motif is italicized, and phosphorylation sites in 4E-BP1 or the corresponding residues in 4E-BP2 are shown in boldface. Residues that are different in hamster 4E-BP1 compared to those of the rat protein are also in boldface and are highlighted by an arrowhead. (B) HEK293 cells were transfected with vectors for hamster 4E-BP1 or human 4E-BP1, both with N-terminal his/myc tags, or with a vector for rat 4E-BP1 with C-terminal tags. After overnight serum starvation, cells were treated with rapamycin and, where indicated, insulin. Samples of lysate were analyzed by SDS-PAGE and Western blotting using anti-myc (for the total 4E-BP1) or the indicated phosphospecific antisera. (C) CHO cells were treated as indicated (using the notation employed for the other panels), and samples of lysate were subjected to affinity chromatography on m7GTP-Sepharose prior to analysis of the bound material by SDS-PAGE and Western blotting using the indicated antisera. The signal for eIF4E serves as a loading control.
When expressed in HEK293 cells, hamster 4E-BP1 shows a pattern of phosphorylation at Thr69 similar to that of the human protein expressed in HEK293 cells or of the endogenous 4E-BP1. In particular, the low phosphorylation seen in serum-starved cells was increased in response to insulin in a rapamycin-sensitive manner (Fig. 2B; see also Fig. 1B). The phosphorylation of Ser64 in overexpressed 4E-BP1 of rat or hamster origin was blocked by rapamycin (Fig. 2B), consistent with our earlier work on rat 4E-BP1 (3).
These data confirm that the high level of phosphorylation of 4E-BP1 in serum-starved CHO cells is not an intrinsic feature of the hamster protein but a feature of CHO cells. They therefore indicate that the outputs from mTOR to the more C-terminal sites in 4E-BP1 and to S6K1 are distinct, because the former are relatively resistant to rapamycin in CHO cells while phosphorylation of S6K1 is fully sensitive.
Amino acid withdrawal and rapamycin exert different effects on eIF4E-4E-BP1 binding in CHO cells.
The (partial or complete) rapamycin resistance of specific sites in 4E-BP1 in CHO cells prompted us to analyze the effects of different treatments on the association of 4E-BP1 with eIF4E in CHO cells by performing affinity chromatography on m7GTP-Sepharose. In serum-starved CHO cells, as previously noted (72), the level of eIF4G binding to eIF4E is high, even in the absence of insulin (Fig. 2C). Rapamycin failed to block this basal binding of eIF4E to eIF4G. In contrast, amino acid withdrawal led to increased binding of eIF4E to 4E-BP1 and complete loss of its interaction with eIF4G. The changes in the binding of 4E-BP1 to eIF4E correlate closely with the phosphorylation status of Ser64 but not Thr36/45/69, suggesting that Ser64 plays an important role in modulating the eIF4E-4E-BP1 interaction, although there are other published data which argue against this (see, for example, reference 14), and more complicated interpretations are also possible. In contrast, in serum-starved HEK293 cells, binding of eIF4G to eIF4E is almost undetectable, and we have therefore been unable to compare the effects of rapamycin versus those of amino acid withdrawal in them (31, 70).
LY294002 eliminates the phosphorylation of the N-terminal sites in 4E-BP1.
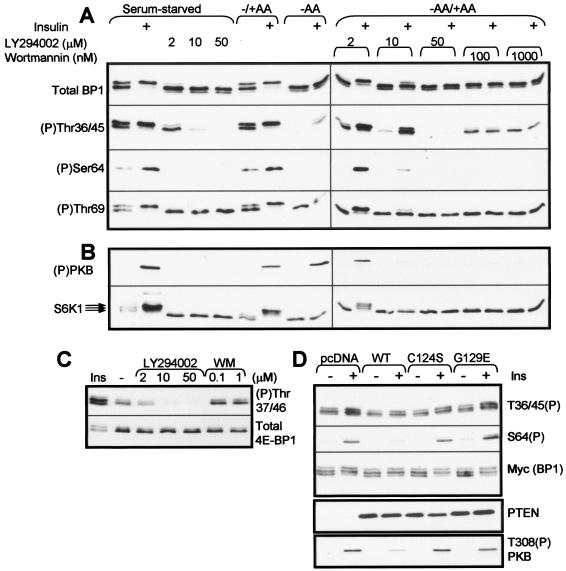
Because phosphorylation of the N-terminal sites in 4E-BP1 is (i) insensitive to rapamycin and (ii) depends on the RAIP motif (which does not appear to bind the mTOR partner raptor [3, 70]), it was important to establish whether their phosphorylation in vivo actually involves mTOR acting in a rapamycin-insensitive manner. To study this, we first made use of the fact that the phosphatidylinositol (PI) 3-kinase inhibitor LY294002 also strongly inhibits the protein kinase activity of mTOR (6, 49). Even at modest concentrations, LY294002 inhibited the phosphorylation of Thr36/45 in CHO cells and caused a shift in the electrophoretic migration of 4E-BP1 to the lower β and α forms (Fig. 3A, top section). When used at 10 μM, LY294002 almost completely eliminated the phosphorylation of Thr36/45 in serum-starved cells or their amino acid-induced rephosphorylation, and it did so completely at the highest concentration tested (50 μM; Fig. 3A). Wortmannin, another PI 3-kinase inhibitor that also inhibits mTOR kinase activity (6, 49), caused a similar shift in the electrophoretic mobility of 4E-BP1, but even at the highest concentration tested it failed to eliminate phosphorylation of Thr36/45 (Fig. 3A). As expected, both inhibitors completely eliminated the insulin-induced phosphorylation of PKB, a downstream effector of PI 3-kinase (Fig. 3B). LY294002 and wortmannin also each decreased the phosphorylation of S6K1 in serum-starved cells (as evidenced by a shift to faster migrating species and by their ability to block completely the insulin-induced phosphorylation of S6K1 in serum-starved or amino acid-replete cells) (Fig. 3B). LY294002 also eliminated the phosphorylation of the N-terminal threonines in 4E-BP1 in HEK293 cells (Fig. 3C). We have previously noted in two other cell types that LY294002 is more effective than wortmannin in blocking 4E-BP1 phosphorylation, while both strongly inhibit S6K1 phosphorylation (66, 71).
FIG. 3.
Effects of PI 3-kinase inhibitors or PTEN on the phosphorylation of specific sites in 4E-BP1. (A and B) CHO cells were starved of serum and then, in some cases (−AA), were also starved of amino acids (60 min). Amino acids were then added back to some plates for 30 min (−AA/+AA). Where indicated, cells were treated with LY294002 or wortmannin (WM) (at the indicated concentrations, for 40 min) and then, where indicated, with insulin (Ins). Samples of lysates were analyzed by Western blotting using the indicated antisera for 4E-BP1 or specific phosphorylation sites in 4E-BP1 (A) or for (P)PKB (Thr308) or S6K1 (B), as indicated. (C) Serum-starved HEK293 cells were treated with insulin or with PI 3-kinase inhibitors as described for panels A and B for CHO cells. (D) HEK293 cells were transfected with a vector encoding rat 4E-BP1, plus vectors for wild-type PTEN, the indicated PTEN point mutants, or the empty vector (pcDNA). Samples of lysate (20 μg of protein) were analyzed by SDS-PAGE and Western blotting using antisera for 4E-BP1, specific sites in 4E-BP1, PTEN, or phosphorylated PKB, as indicated.
In contrast, neither LY294002 nor wortmannin affected the phosphorylation state of Thr69 in CHO cells, even when used at relatively high concentrations. These data strongly suggest that the phosphorylation of Thr69 in vivo is not directly catalyzed by mTOR. The data also therefore cast doubt on the notion that the TOS motif acts to recruit mTOR to phosphorylate 4E-BP1 directly at this site.
In principle, the effects of LY294002 or wortmannin on the phosphorylation of the N-terminal sites in 4E-BP1 could be due to their ability to block PI 3-kinase rather than an effect upon mTOR itself. This seemed unlikely, given that there is no detectable phosphorylation of PKB, a well-studied effector of PI 3-kinase, in unstimulated cells (Fig. 1E); insulin induces its phosphorylation in HEK293 cells (Fig. 3D, bottom part). To study this issue further, we made use of the lipid phosphatase, PTEN, which dephosphorylates lipids at the 3 position and thus reverses the action of PI 3-kinase. As expected, expression of WT PTEN decreased the insulin-induced phosphorylation of PKB (Fig. 3D). As negative controls, we used mutants of PTEN that are catalytically inactive (C124S) or lack lipid phosphatase activity but still possess protein phosphatase activity (G129E) (42, 54). As expected, neither affected the ability of insulin to increase the phosphorylation of PKB.
Because expression of active PTEN did not affect phosphorylation of the N-terminal sites in 4E-BP1, it seems that the effects of LY294002 on the phosphorylation of Thr37/46 are not due to its effect on PI 3-kinase activity and are consistent with the idea that they reflect the ability of LY294002 to inhibit mTOR kinase activity. For example, these sites may indeed be phosphorylated by mTOR but in a rapamycin-insensitive manner. The expression of WT PTEN, but not the mutants, caused the almost complete inhibition of the insulin-induced increase in S65 phosphorylation (Fig. 3A), indicating that insulin-induced phosphorylation of Ser65 requires PI 3-kinase signaling.
TSC1/2 and Rheb affect the phosphorylation of several sites in 4E-BP1.
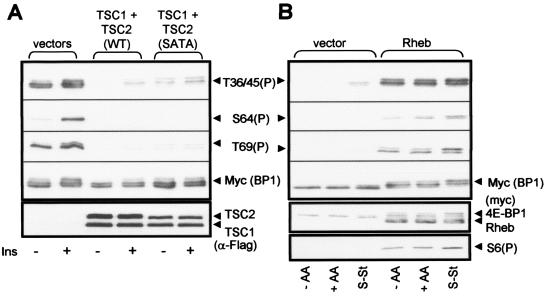
The above data are consistent with the idea that the N-terminal sites in 4E-BP1 are targets for a rapamycin-insensitive output from mTOR. We wished to test this idea further. Recent studies have identified the TSC1/TSC2 complex (TSC1/2) as an important negative regulator of mTOR signaling, with TSC2 acting as a GTPase-activator protein for the small G-protein Rheb, a positive regulator of mTOR signaling (for reviews see references 47 and 50). Phosphorylation of TSC2 by PKB alleviates the inhibition of mTOR signaling. We therefore expressed wild-type TSC1 and TSC2, or TSC1 and a nonphosphorylatable mutant of TSC2 (SATA) (48), as FLAG-tagged proteins in HEK293 cells, and we studied the effect on the phosphorylation of 4E-BP1. Expression levels of TSC1/2 were determined by Western analysis using anti-FLAG. Expression of TSC1/2 greatly decreased the phosphorylation of Thr36/45 in 4E-BP1 (Fig. 4A). The greater effect of WT TSC2 compared to that of the SATA mutant likely reflects the higher expression levels of the former. TSC1/2 also prevented the phosphorylation of Ser64 and Thr69 (Fig. 4A).
FIG. 4.
Effects of proteins that regulate mTOR signaling on the phosphorylation of 4E-BP1. (A) HEK293 cells were transfected with vectors for rat 4E-BP1 and TSC1 and TSC2, TSC1 and the SATA mutant of TSC2, or the corresponding empty vectors. Cells were starved of serum overnight and then treated with insulin (Ins). Samples of cell lysate were analyzed by SDS-PAGE and Western blotting with the indicated antisera (anti-FLAG was used to detect overexpressed TSC1 and TSC2). (B) HEK293 cells were transfected with vectors for rat 4E-BP1 and either Rheb or the corresponding empty vector. Cells were starved of serum overnight (S-St), deprived of amino acids (−AA), or transferred to the medium used for amino acid starvation but containing amino acids (+AA). Samples of cell lysate were analyzed with the indicated antisera (anti-myc was used to detect both 4E-BP1 and Rheb). S6(P) indicates an antibody that detects rpS6 when it is phosphorylated at Ser235.
Other data have shown that the increased expression of Rheb enhances the phosphorylation of 4E-BP1 (20, 34, 47, 68), and the data in Fig. 4B demonstrate that Rheb clearly promotes the phosphorylation of the N-terminal threonine residues in 4E-BP1. This was true even when the cells had previously been starved of amino acids. Expression of Rheb also promoted the phosphorylation of the other sites in 4E-BP1 and of S6, confirming its ability to activate mTOR signaling (Fig. 4B).
Taken together, the data for the effects of the PI 3-kinase inhibitors, and of the expression of PTEN, TSC1/2, or Rheb, support the conclusion that phosphorylation of the N-terminal threonines in 4E-BP1 does involve signaling via mTOR but reflects an output from mTOR which is insensitive to rapamycin.
Evaluation of additional approaches to studying the role of mTOR in the phosphorylation of 4E-BP1.
The above data suggest that the phosphorylation of the N-terminal sites in 4E-BP1 reflects a rapamycin-insensitive output from mTOR. We sought to employ additional approaches to study this. The first was to use siRNA to try to decrease cellular levels of mTOR by RNA interference (RNAi). We performed extensive experiments using five different siRNAs to try to decrease cellular levels of mTOR and to examine the effect of this on 4E-BP1 phosphorylation. The five siRNAs were used singly, pairwise, or in triple combinations in at least three experiments in every case. Some degree of reduction in mTOR levels was seen by 48 h, especially using siRNA 1 (Fig. 5A). Although, in some cases, longer-term exposure to certain siRNAs (or combinations) did seem to result in a further reduction in mTOR levels, the cells rounded up and tended to become detached from the dishes, showed signs of cell death, and responded poorly to insulin. We did not, therefore, have confidence in data obtained using such cells. For example, we have previously shown that mTOR signaling is inactivated well before cells enter an apoptotic program (69). At the 48-h time point, the greatest reduction in mTOR levels we saw was about 50 to 60%. However, this had no detectable effect on the phosphorylation of S6K1 (another mTOR target), as judged by the ladder of bands that corresponds to differentially phosphorylated forms of the protein. It is also apparent that the overall level of S6K1 is decreased (compared to that of the actin control; Fig. 5A). In the case of 4E-BP1, overall levels were also decreased, and, taking this into account, there was no clear reduction in the real level of phosphorylation of Thr37/46, Ser65, or Thr70 (Fig. 5A). This indicates that this siRNA approach does not decrease mTOR levels to a point where downstream signaling is impaired detectably. These data are consistent with those of Murakami et al. (53). That recent study, which appeared while this paper was under revision, used cells in which mTOR was genetically knocked out or modified. Their data showed that even an 80 to 90% decrease in mTOR levels did not interfere with downstream signaling to 4E-BP1. It is therefore not surprising that the 50 to 60% decrease we achieved also did not suffice to do so.
FIG. 5.
Approaches to impair mTOR signaling. (A) HeLa cells were transfected with 60 nM siRNA directed against mTOR (mTOR-1) or were mock transfected. Forty-eight hours after the first transfection, cells were starved of serum for 4 h and, in some cases, were then treated with insulin (Ins) (100 nM, 30 min). Lysates were then analyzed by SDS-PAGE and Western blotting for actin (normalization control), mTOR, S6K1, total 4E-BP1, or the indicated phosphorylation sites in 4E-BP1. (B) HEK293 cells were transfected with vectors encoding wild-type or kinase-dead FLAG-mTOR along with rat 4E-BP1. Twenty-four hours later, cells were starved of serum overnight and, in some cases, were then treated with insulin (100 nM, 30 min). Lysates were analyzed as described for panel A (but not here for actin levels). (C) Targeted ES cells (53) were treated with HTNC or (as negative control) phosphate-buffered saline (PBS). After the times indicated, lysates were prepared and analyzed by SDS-PAGE and Western blotting using the indicated antisera.
The second approach was to use a kinase-dead form of mTOR which might in principle act as a dominant negative. Although kinase-dead mTOR has previously been shown to be functionally inactive (75), it has not, to our knowledge, been reported to act in a dominant-negative fashion. Nonetheless, we expressed either wild-type or kinase-dead mTOR in HEK 293 cells together in each with rat 4E-BP1 (because interpretations of the data for this species are easier due to lack of cross-reactivity between the anti-Ser64/65 phosphoantibody and other sites, which complicates analysis of the human protein, as explained elsewhere in this paper). Equal expression was confirmed using the FLAG tag. However, expression of kinase-dead mTOR caused no change in the phosphorylation of S6K1 or any of the sites in 4E-BP1 tested (Fig. 5B). This indicates that kinase-dead mTOR does not act in a dominant-negative (interfering) manner. Therefore, unfortunately, neither of the above approaches provides further insight into the role of mTOR in regulating phosphorylation of the N-terminal sites in 4E-BP1.
Thirdly, we exploited the recently available ability to decrease cellular mTOR activity by using the genetic approach described by Murakami et al. (53). This involved using a targeting vector designed to result in deletion of the extreme C terminus (the last six amino acids) of mTOR, which is essential for kinase activity (58, 65), in targeted ES cells. To achieve the necessary recombination event, ES cells were treated with a fusion protein comprising a hexahistidine tag, the cell-permeant Tat peptide, the simian virus 40 nuclear localization signal, and the Cre recombinase (referred to as HTNC [53]). This also results in decreased levels of mTOR expression (53). It was shown previously that targeting mTOR in this way decreases the level of phosphorylation of 4E-BP1, as judged by its mobility on SDS-PAGE (53). However, the state of phosphorylation of specific sites in 4E-BP1 was not examined in that study. In particular, it was important to know whether decreasing mTOR levels affected Thr36/45, the sites we find to be insensitive to rapamycin. As shown in Fig. 5C (upper section), treatment of the ES cells with HTNC substantially reduces mTOR expression (as reported previously [53]). It also caused a marked loss of phosphorylation of the N-terminal sites that are insensitive to rapamycin without altering overall levels of 4E-BP1 (Fig. 5C, bottom section). The observation that their phosphorylation was not completely eliminated likely reflects the fact that these cells do contain some residual mTOR protein. HTNC treatment also greatly reduced the level of phosphorylation of Ser64, in line with the sensitivity of this site to rapamycin. In contrast, HTNC treatment had almost no effect on phosphorylation of Thr69. As already noted, this site is insensitive to rapamycin in CHO cells.
The RAIP motif mediates amino acid signaling to 4E-BP2.
The above data show that 4E-BP1 is basally phosphorylated at the N-terminal threonine residues in HEK293 or CHO cells, and this requires the presence of amino acids in the medium. Our earlier work showed that the N-terminal RAIP motif plays a key role in maintaining phosphorylation of these sites in serum-starved cells, while mutating the TOS motif had almost no effect on this (3). This strongly suggests that the RAIP motif may mediate the amino acid input into their phosphorylation. To test this, we exploited the facts that (i) an identical motif is present in 4E-BP2 (Fig. 2A) and (ii) that the sequences around the N-terminal sites are similar in the two proteins, making it possible that the phosphospecific antisera might cross-react. The anti-4E-BP1 (P)Thr36/45 antibody also detects 4E-BP2 when it is overexpressed in HEK293 cells (Fig. 6A). The signal was lost when cells were starved of amino acids, but it was not affected by rapamycin. This behavior is identical to that of 4E-BP1 (Fig. 1B and C). To test the role of the RAIP motif in 4E-BP2, we mutated two residues within the motif to alanine, thus creating 4E-BP2(AAAP). Similar mutations in 4E-BP1 eliminate phosphorylation of the N-terminal threonines (3, 70). Strikingly, the 4E-BP2(AAAP) mutant does not show any detectable phosphorylation at the N-terminal threonine sites, even in the presence of amino acids (Fig. 6A), consistent with the idea that the RAIP motif shared by 4E-BP1 and 4E-BP2 serves to mediate an amino acid-dependent input that is required for the phosphorylation of these residues.
FIG. 6.
Features of 4E-BP2 and 4E-BP1 required for efficient phosphorylation in vivo. (A) HEK293 cells were transfected with vectors for wild-type 4E-BP2 or a mutant in which the RAIP motif was altered to AAAP. Cells were starved of serum overnight and, in some cases, also of amino acids (60 min). Where indicated, cells were treated with rapamycin or insulin. Samples were analyzed by SDS-PAGE and Western blotting using anti-myc (to detect 4E-BP2) or anti-4E-BP1 (Thr36/45 phosphospecific antibody). (B and C) HEK293 cells were transfected with vectors for WT 4E-BP1 or the LM/AA mutant, each as N-terminally his/myc-tagged proteins. After serum starvation overnight, cells were treated with rapamycin or insulin as indicated. (B) Samples of lysate were subjected to affinity chromatography on m7GTP-Sepharose prior to analysis of the bound material by SDS-PAGE and Western blotting using the indicated antisera. (C) Samples of lysate were analyzed by SDS-PAGE and Western blotting using anti-myc and the indicated phosphospecific antisera for 4E-BP1. For (P)Ser64, both the endogenous (endog.; serves as a positive control) and recombinant (his/myc) 4E-BP1 proteins can be seen. (D to F) HEK293 cells were transfected with vectors encoding FLAG-tagged mTOR and myc-raptor. (D) Western blot demonstrating that mTOR and myc-raptor are efficiently expressed. (E) GST-4E-BP1 and eIF4E were mixed and pulled down on glutathione-Sepharose (pull down), and the bound material was analyzed by SDS-PAGE and Western blotting using antisera for eIF4E and 4E-BP1. Purified eIF4E and GST-4E-BP1 (input) were run as controls. (F) Recombinant 4E-BP1 was incubated with immunoprecipitated mTOR/raptor in the presence of [γ-32P]ATP, MnCl2, and, where indicated, recombinant eIF4E and/or GST-FKBP12 plus rapamycin.
Interaction with eIF4E is required for regulated phosphorylation of several sites in 4E-BP1.
The above data raise important questions about the inputs that lead to phosphorylation of the different sites in 4E-BP1. It has recently been suggested that the hierarchical nature of the phosphorylation of 4E-BP1 arises from a gradual loosening of its complex with eIF4E, which allows increased access to the relevant kinases (24). If this were so, one would anticipate that a form of 4E-BP1 that could not be sequestered by eIF4E would be phosphorylated more efficiently in vivo than one that could not bind eIF4E. Free 4E-BP1 is thought to lack any folded structure in solution (18) and can be phosphorylated by several kinases (e.g., Erk) in vitro (29), although they do not appear to play a direct role in its phosphorylation in vivo. In fact, binding of 4E-BP1 to eIF4E blocks its ability to be phosphorylated by Erk in vitro (11). An important issue is whether its regulated hierarchical phosphorylation is an intrinsic feature of 4E-BP1 itself or whether it is related to the structure that is apparently induced upon binding to eIF4E. To test this, we generated a mutant of rat 4E-BP1 in which two residues (LM) within the eIF4E-binding site (Fig. 2A) were converted to alanines (LM/AA mutant). As expected, this variant cannot bind to eIF4E (Fig. 6B). When expressed in HEK293 cells under serum-starved conditions, the level of phosphorylation of this mutant at both the N-terminal sites and at Thr69 (Fig. 6C) is markedly reduced relative to that of the wild-type protein, and the phosphorylation of Ser64 is eliminated. This suggests that binding to eIF4E is required for the efficient phosphorylation of 4E-BP1 in vivo, although we cannot formally exclude the possibility that these mutations affect its phosphorylation via other mechanisms.
If mTOR is the kinase responsible for phosphorylating 4E-BP1 in vivo, one would thus expect it to phosphorylate 4E-BP1 complexed with eIF4E better than the free protein. We therefore studied the ability of mTOR complex to phosphorylate purified recombinant 4E-BP1 in the absence or presence of recombinant eIF4E. As raptor is thought to recruit mTOR to 4E-BP1 (3, 9, 55, 63), we coexpressed FLAG-tagged mTOR with myc-raptor (Fig. 6D). Care was taken to add similar molar quantities of GST-4E-BP1 and, where present, eIF4E. The binding of the recombinant proteins to one another was confirmed in a GST pull-down assay (Fig. 6E). As shown in Fig. 6F, mTOR phosphorylated 4E-BP1 to similar extents, whether or not eIF4E was present. If anything, there was a slight but consistent inhibition. These data also show that the phosphorylation of 4E-BP1 by mTOR is blocked by rapamycin/FKBP12. This experiment does not, therefore, match the simple explanation that association of 4E-BP1 with eIF4E enhances its efficacy as a substrate for direct phosphorylation by mTOR. The kinases that phosphorylate 4E-BP1 in vivo presumably preferentially phosphorylate it when it is complexed with eIF4E.
DISCUSSION
Implications of the present data for understanding mTOR signaling.
A generally accepted view of mTOR signaling is that the kinase domain of mTOR directly phosphorylates targets such as 4E-BP1 and S6K1, that its ability to do so is stimulated by amino acids or hormones in a similar manner through alleviation of the inhibitory effects of TSC1/2, and that these outputs are susceptible to inhibition by rapamycin, although control of protein phosphatases may also play a role here (1). The data we present here question several of these ideas and indicate that a reassessment of this simple model is required. In particular, our data show that (i) the effects of rapamycin and amino acid withdrawal are not equivalent, e.g., in the case of the phosphorylation of the N-terminal threonines in 4E-BP1; (ii) the regulation of 4E-BP1 and S6K1 involves distinct inputs, e.g., while the phosphorylation of S6K1 is completely blocked by rapamycin in CHO or HEK293 cells, phosphorylation of the N-terminal sites in 4E-BP1 is not; (iii) amino acids and insulin appear to stimulate different outputs from mTOR to 4E-BP1; and (iv) the insensitivity in CHO cells of the phosphorylation of Thr69 to LY294002, which inhibits mTOR kinase activity, indicates that mTOR does not directly phosphorylate this site in vivo. Our data also imply that, within cells, 4E-BP1 is phosphorylated as a complex with eIF4E, which differs from the characteristics of the phosphorylation of 4E-BP1 by mTOR in vitro.
The N-terminal sites are regulated by amino acids but not by rapamycin.
The insensitivity of the phosphorylation of the N-terminal sites in 4E-BP1 to rapamycin could suggest that the sites are not actually regulated by mTOR signaling in vivo. However, several pieces of our data argue in favor of a role for mTOR signaling in their phosphorylation: (i) phosphorylation of Thr36/45 is blocked by LY294002, which inhibits mTOR kinase activity, independently of its effect on PI 3-kinase; (ii) expression of TSC1 and TSC2 (negative regulators of mTOR signaling) suppresses their phosphorylation; (iii) overexpression of Rheb, a positive regulator of mTOR, increases their phosphorylation; (iv) starvation of cells for amino acids, which is well known to impair mTOR signaling, decreases their phosphorylation; and (v) their phosphorylation is also decreased by deletion of the final (essential) six amino acids from mTOR. Taken together, our data suggest that phosphorylation of these N-terminal threonyl residues in 4E-BP1 represents a rapamycin-insensitive output from mTOR. However, we cannot formally exclude, e.g., the theoretical possibility that Rheb and TSC1/2 also stimulate other relevant signaling events, although there is little evidence that they do.
A potentially important point here is that certain processes that are sensitive to PI 3-kinase inhibitors but not to rapamycin may actually involve signaling through mTOR rather than PI 3-kinase. Caution must therefore be exercised in interpreting such data.
The role of the RAIP motif.
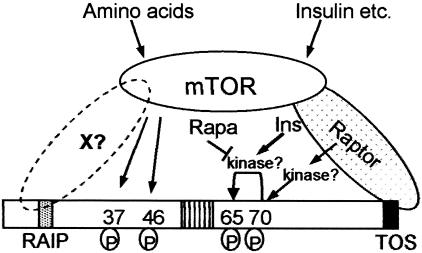
The phosphorylation of the two N-terminal threonine residues in 4E-BP1 (70) or 4E-BP2 (this study) is dependent upon the N-terminal RAIP motif. Because the present data show that their phosphorylation also requires amino acids to be present, it is logical to conclude that the amino acid input to 4E-BP1 phosphorylation operates via the RAIP motif. Although this motif has been reported to be required for interaction of 4E-BP1 with raptor (9), we were unable to demonstrate such an interaction (3), consistent with the fact that the two motifs play different roles in regulating 4E-BP1. Thus, it is tempting to suggest that the RAIP motif interacts with a different partner, which may be part of a rapamycin-insensitive, amino acid-regulated mTOR complex (Fig. 7; also see below). Much of our data is consistent with the notion that mTOR may directly phosphorylate these N-terminal threonine residues, although it remains puzzling that this phosphorylation is so slow in vitro, especially in the absence of high concentrations of Mn2+ ions. Perhaps the efficient phosphorylation of the N-terminal sites by mTOR requires additional components that have yet to be identified and that interact with the RAIP motif (Fig. 7). The finding that, in vitro, mTOR fails to phosphorylate the 4E-BP1/eIF4E complex better than free 4E-BP1 (in contrast to the behavior of 4E-BP1 phosphorylation in vivo) also suggests that the events that couple phosphorylation of the N-terminal sites to mTOR are rather complex. The reduced phosphorylation of the nonbinding mutant may perhaps arise because binding of the normally unstructured (18) 4E-BP1 to eIF4E induces a structure that is recognized by the relevant kinase(s) or because the kinases preferentially recognize the eIF4E/4E-BP1 complex. This clearly contrasts with the phosphorylation of 4E-BP1 by mTOR in vitro, where binding to eIF4E does not enhance phosphorylation. This could be interpreted as suggesting that the kinase that is responsible for phosphorylation of 4E-BP1 in vivo is not mTOR itself but an enzyme whose activity or activation requires mTOR kinase activity.
FIG. 7.
Model for regulation of the phosphorylation of 4E-BP1, integrating the findings of the present report. Our data are consistent with mTOR directly phosphorylating Thr37/46. This requires the RAIP motif in 4E-BP1, which may interact with a (putative) partner protein, X, that could also recruit mTOR to 4E-BP1. The kinase acting at Thr70 is unknown but appears unlikely to be mTOR itself, based on insensitivity to LY294002 and the lack of effect of deletion of the most C-terminal residues of mTOR. The identity of the kinase acting at Ser65 is also unknown, but this phosphorylation event is stimulated by insulin (Ins) and blocked by rapamycin (rapa). The kinase might be mTOR itself. In cells where the phosphorylation of Thr70 is low under serum-starved conditions and stimulated by insulin, this effect too is blocked by rapamycin. This suggests rapamycin may interfere with signaling from mTOR to 4E-BP1 rather than with their direct phosphorylation by mTOR. Amino acids primarily influence the phosphorylation of Thr37/46. The requirement for binding of 4E-BP1 to eIF4E for its efficient phosphorylation in vivo is not shown, for reasons of clarity.
Insulin-induced phosphorylation of Ser64/65 requires PI 3-kinase signaling.
Our data for the phosphorylation of Ser65 in HEK293 cells seem straightforward. Its phosphorylation is stimulated by insulin, and this effect is mediated via PI 3-kinase signaling: it is blocked by moderate concentrations of PI 3-kinase inhibitors or by PTEN. This is consistent with the present model for the control of mTOR signaling that involves TSC1/2 and Rheb (Fig. 7). In cells, phosphorylation of this site also requires amino acids. Given the hierarchical nature of the phosphorylation of 4E-BP1 and the fact that Ser65 is late in this hierarchy, it is likely that this requirement is a secondary effect of the positive influence of amino acids on the phosphorylation of, e.g., Thr37/46/70.
Phosphorylation of Ser65 is highly dependent upon the TOS motif and, thus, presumably on raptor binding. Taken together with the present data, this could suggest that mTOR acts directly to phosphorylate Ser65. However, mTOR actually only phosphorylates this site feebly, if at all, in vitro (this seems to depend upon the antibody used to immunoprecipitate mTOR) (see, e.g., reference 74). However, phosphorylation here is clearly dependent upon intact mTOR function, as shown by the drastic reduction in the phosphorylation of this site in cells in which the last six residues of mTOR were deleted. In CHO cells, neither the basal nor the insulin-stimulated phosphorylation of Ser64 is eliminated by rapamycin, although rapamycin does potently inhibit the direct phosphorylation of this site by mTOR in vitro (49).
The insulin-induced phosphorylation of this site does not seem to be an automatic consequence of phosphorylation of sites that are considered to be earlier elements in the hierarchical phosphorylation of 4E-BP1, i.e., Thr36/45/69. This is indicated both by the fact that Ser64 is not constitutively phosphorylated in CHO cells (where the other sites are fully phosphorylated in the absence of insulin) and by the behavior of the 4E-BP1 mutant that cannot bind to eIF4E. Although the other sites are phosphorylated in this mutant, Ser64 is not. Thus, phosphorylation of Ser64 requires an additional, insulin-stimulated input and probably the binding of 4E-BP1 to eIF4E (Fig. 7). Also, phosphorylation of this site therefore does not, as argued previously (24), merely reflect a successive loosening of the 4E-BP1-eIF4E interaction, allowing increased access by kinases to the phosphorylation sites. Thus, the identity of the kinase that acts here remains unclear. It is likely to be stimulated by insulin in a rapamycin-sensitive manner.
What role does mTOR play in the regulation of Thr69/70?
Thr69 is highly phosphorylated in CHO cells. Previous work has shown (i) that phosphorylation of Thr69/70 is dependent upon the TOS motif (3, 9) and (ii) that immunoprecipitated mTOR can directly phosphorylate this site in a rapamycin-sensitive manner (see, e.g., reference 49). However, our data for the endogenous 4E-BP1 in CHO cells show that phosphorylation here is not inhibited by LY294002 or rapamycin, even though, as noted above, both agents do completely block the phosphorylation of S6K1. Furthermore, deletion of the C terminus of mTOR has very little effect on phosphorylation of this site. These data suggest (i) that Thr69 is not a direct target for mTOR and, thus, (ii) that the function of the TOS motif is not simply to recruit mTOR to 4E-BP1 to allow direct phosphorylation by mTOR of Thr69. The TOS motif may serve to allow phosphorylation of Thr69 by a further kinase (which could be associated with raptor/mTOR [Fig. 7]).
Models for signaling downstream of mTOR.
The observation that TSC1/2 and Rheb influence the phosphorylation of all the sites in 4E-BP1 we have tested indicates that they regulate outputs from mTOR that (i) can be either sensitive or insensitive to rapamycin and (ii) involve both the RAIP and TOS motifs in 4E-BP1. Thus, although all the effectors of mTOR that we have studied are affected by TSC1/2 and Rheb, the control of the S6 kinases and of the different sites in 4E-BP1 actually reflect different outputs from the mTOR complex. The phosphorylation and activation of S6K1 (8) require inputs from amino acids and insulin and are completely eliminated by rapamycin. In contrast, the situation for 4E-BP1 is more complicated than has previously been thought. The N-terminal sites are regulated in response to amino acids in a rapamycin-insensitive manner, and this output is not appreciably activated by insulin in serum-starved cells. Our earlier work showed that loss of the TOS motif has much less effect on the phosphorylation of Thr36/45 than on Thr69 (3). This conflicts with the idea (9) that the TOS motif may recruit mTOR to directly phosphorylate 4E-BP1 at Thr36/45. In fact, as discussed above, it seems likely that the amino acid input, which maintains the phosphorylation of Thr36/45 (and the corresponding residues in 4E-BP2) in serum-starved cells, is mediated via the RAIP motif.
Because S6K1 remains fully sensitive to rapamycin (and LY294002) in CHO cells while phosphorylation of, e.g., Thr69 in 4E-BP1 is not, the outputs from mTOR to 4E-BP1 and to S6K1 must be distinct. In serum-starved HEK293 cells, where phosphorylation of Thr70 is low and increases in response to insulin, rapamycin does block its insulin-induced phosphorylation. It seems possible that rapamycin may block the activation of signaling downstream of mTOR rather than the function of mTOR itself (Fig. 7). After all, its complex with FKBP12 does not bind directly to the kinase domain of mTOR. Consistent with this, other data indicate that rapamycin may disrupt the interaction between raptor and mTOR (37, 56). However, the data of Jacinto et al. (35) argue against this and suggest rapamycin may directly inhibit mTOR kinase activity. The reasons why phosphorylation of this site in CHO cells is insensitive to rapamycin are unclear. Perhaps, in CHO cells, assembly of the mTOR complex involved in its phosphorylation is constitutive, whereas the complexes involved in this in HEK293 cells are only assembled in response to insulin and in a rapamycin-sensitive manner. Other interpretations are also possible.
Taken together, our data point to the operation of multiple links between mTOR and its downstream targets, 4E-BP1 and S6K1. They provide evidence that multiple inputs link mTOR to the control of this single target protein. The possible existence of such outputs has been hinted at previously. For example, it has been known for some time that TOR2, one of two yeast mTOR homologs, performs a rapamycin-insensitive function (30, 39, 76). While this report was under review, a similar complex was identified in mammalian cells (35, 61). Fingar et al. (17) have recently concluded, on the basis of quite different experiments, that mTOR may also possess rapamycin-insensitive functions.
Other recent data have pointed to a role for a rapamycin-insensitive input from mTOR to 4E-BP1 in cells infected with cytomegalovirus (38) and in the trafficking of the amino transporter component 4F2hc (12). The present data show that rapamycin-sensitive and -insensitive inputs can converge on a single target protein, 4E-BP1. Thus, rapamycin cannot be regarded as reliably reporting either the functions of mTOR or the consequences of amino acid deprivation for mTOR signaling. Interestingly, the phosphorylation of S6K1 remained rapamycin sensitive in cytomegalovirus-infected cells (38).
This model postulates the existence of novel, i.e., so far unidentified, components (Fig. 7). For example, what accounts for the role of the RAIP motif (a binding partner, by analogy with the TOS motif and raptor)? Which kinases phosphorylate Thr69 and Ser64 in 4E-BP1? Rictor (also called mAvo3, on account of its similarity to the yeast TOR partner, Avo3p) binds to mTOR and mediates a rapamycin-insensitive output to the actin cytoskeleton, analogous to the role of the rapamycin-insensitive TORC2 complex in yeast (35, 44, 61). It could, in principle, also mediate the rapamycin-insensitive regulation of 4E-BP1 at the N-terminal sites. However, (i) siRNA against Rictor (mAvo3) does not affect these sites (E. Jacinto, personal communication), and (ii) the mTOR/mAvo3 complex does not phosphorylate these sites in 4E-BP1 in vitro (35). It is therefore probable that their phosphorylation is mediated by mTOR independently of Rictor (mAvo3) and raptor. Importantly, these findings point to the possible existence of a third type of mTOR complex (Fig. 7).
A range of studies indicate a key role for eIF4E, and the regulation of its availability, in cell transformation and the dysregulation of cell proliferation (10, 46). Targeting the signaling events that control the phosphorylation on 4E-BP1 therefore seems a promising avenue for developing new anti-cancer therapies. It is therefore important to achieve a full understanding of the upstream control of 4E-BP1, e.g., of the signaling molecules (scaffolds or kinases) that link mTOR to the control of the phosphorylation of 4E-BP1. It may then be possible to design agents that inhibit one arm of the pathway (e.g., phosphorylation of the N-terminal sites in 4E-BP1) without affecting others (phosphorylation of S6K1 and other sites in 4E-BP1). Such agents may be valuable in cancer therapy.
Acknowledgments
This work was supported by funding from the Wellcome Trust and AstraZeneca.
We are grateful to a number of collaborators for providing reagents, as detailed in the text, and to Josep Parra-Palau and Richard Wilson (both from Dundee) for help in creating the vectors for mutants of 4E-BP2.
REFERENCES
- 1.Avruch, J., C. Belham, Q. Weng, K. Hara, and K. Yonezawa. 2001. The p70 S6 kinase integrates nutrient and growth signals to control translational capacity. Prog. Mol. Subcell. Biol. 26:115-154. [DOI] [PubMed] [Google Scholar]
- 2.Berra, E., E. Benizri, A. Ginouves, V. Volmat, D. Roux, and J. Pouyssegur. 2003. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 22:4082-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beugnet, A., X. Wang, and C. G. Proud. 2003. The TOR-signaling and RAIP motifs play distinct roles in the mTOR-dependent phosphorylation of initiation factor 4E-binding protein 1 in vivo. J. Biol. Chem. 278:40722. [DOI] [PubMed] [Google Scholar]
- 4.Browne, G. J., and C. G. Proud. 2004. A novel mTOR-regulated phosphorylation site in elongation factor 2 kinase modulates the activity of the kinase and its binding to calmodulin. Mol. Cell. Biol. 24:2986-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunn, G. J., C. C. Hudson, A. Sekulic, J. M. Williams, H. Hosoi, P. J. Houghton, J. C. Lawrence, and R. T. Abraham. 1997. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 277:99-101. [DOI] [PubMed] [Google Scholar]
- 6.Brunn, G. J., J. Williams, C. Sabers, G. Weiderrecht, J. C. Lawrence, and R. T. Abraham. 1996. Direct inhibition of the signalling functions of the mammalian target of rapamycin by the phosphoinositide 3-kinase inhibitors, wortmannin and LY294002. EMBO J. 15:5256-5267. [PMC free article] [PubMed] [Google Scholar]
- 7.Burnett, P. E., R. K. Barrow, N. A. Cohen, S. H. Snyder, and D. M. Sabatini. 1998. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. USA 95:1432-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, L. E., X. Wang, and C. G. Proud. 1999. Nutrients differentially modulate multiple translation factors and their control by insulin. Biochem. J. 344:433-441. [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, K.-M., L. P. McMahon, and J. C. Lawrence. 2003. Two motifs in the translational repressor PHAS-I required for efficient phosphorylation by mammalian target of rapamycin and for recognition by raptor. J. Biol. Chem. 278:19667-19673. [DOI] [PubMed] [Google Scholar]
- 10.De Benedetti, A., and J. R. Graff. 2004. eIF-4E expression and its role in malignancies and metastases. Oncogene 23:3189-3199. [DOI] [PubMed] [Google Scholar]
- 11.Diggle, T. A., S. K. Moule, M. B. Avison, A. Flynn, E. J. Foulstone, C. G. Proud, and R. M. Denton. 1996. Both rapamycin-sensitive and -insensitive pathways are involved in the phosphorylation of the initiation factor 4E binding protein (4E-BP1) in response to insulin in rat epididymal fat cells. Biochem. J. 316:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edinger, A. L., C. M. Linardic, G. G. Chiang, C. B. Thompson, and R. T. Abraham. 2003. Differential effects of rapamycin on mammalian target of rapamycin signaling functions in mammalian cells. Cancer Res. 63:8451-8460. [PubMed] [Google Scholar]
- 13.Fattori, R., and T. Piva. 2003. Drug-eluting stents in vascular intervention. Lancet 361:247-249. [DOI] [PubMed] [Google Scholar]
- 14.Ferguson, G., I. Mothe-Satney, and J. C. Lawrence, Jr. 2003. Ser-64 and Ser-111 in PHAS-I are dispensable for insulin-stimulated dissociation from eIF4E. J. Biol. Chem. 278:47459-47465. [DOI] [PubMed] [Google Scholar]
- 15.Ferrari, S., R. B. Pearson, M. Siegmann, S. C. Kozma, and G. Thomas. 1993. The immunosuppressant rapamycin induces inactivation of p70s6k through dephosphorylation of a novel set of sites. J. Biol. Chem. 268:16091-16094. [PubMed] [Google Scholar]
- 16.Fingar, D. C., and J. Blenis. 2004. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 23:3151-3171. [DOI] [PubMed] [Google Scholar]
- 17.Fingar, D. C., C. J. Richardson, A. R. Tee, L. Cheatham, C. Tsou, and J. Blenis. 2004. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 24:200-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fletcher, C. M., A. M. McGuire, A.-C. Gingras, H. Li, H. Matsuo, N. Sonenberg, and G. Wagner. 1998. 4E binding proteins inhibit the translation factor eIF4E without folded structure. Biochemistry 37:9-15. [DOI] [PubMed] [Google Scholar]
- 19.Fox, H. L., P. T. Pham, S. R. Kimball, L. S. Jefferson, and C. J. Lynch. 1998. Amino acid effects on translational repressor 4E-BP1 are mediated primarily by L-leucine in isolated adipocytes. Am. J. Physiol. 44:C1232—C1238. [DOI] [PubMed] [Google Scholar]
- 20.Garami, A., F. J. Zwartkruis, T. Nobukuni, M. Joaquin, M. Roccio, H. Stocker, S. C. Kozma, E. Hafen, J. L. Bos, and G. Thomas. 2003. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell 11:1457-1466. [DOI] [PubMed] [Google Scholar]
- 21.Gingras, A.-C., S. P. Gygi, B. Raught, R. D. Polakiewicz, R. T. Abraham, M. F. Hoekstra, R. Aebersold, and N. Sonenberg. 1999. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13:1422-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gingras, A.-C., B. Raught, S. P. Gygi, A. Niedzwieka, M. Miron, S. K. Burley, R. D. Polakiewicz, A. Wyslouch-Cieczyska, R. Aebersold, and N. Sonenberg. 2001. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 15:2852-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gingras, A.-C., B. Raught, and N. Sonenberg. 2001. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 15:807-826. [DOI] [PubMed] [Google Scholar]
- 24.Gross, J. D., N. J. Moerke, H. T. von der, A. A. Lugovskoy, A. B. Sachs, J. E. McCarthy, and G. Wagner. 2003. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115:739-750. [DOI] [PubMed] [Google Scholar]
- 25.Guba, M., P. von Breitenbuch, M. Steinbauer, G. Koehl, S. Flegel, M. Hornung, C. J. Bruns, C. Zuelke, S. Farkas, M. Anthuber, K. W. Jauch, and E. K. Geissler. 2002. Rapamycin inhibits primary and metastatic tumor growth by antiangiogenesis: involvement of vascular endothelial growth factor. Nat. Med. 8:128-135. [DOI] [PubMed] [Google Scholar]
- 26.Hara, K., Y. Maruki, X. Long, K. Yoshino, N. Oshiro, S. Hidayat, C. Tokunaga, J. Avruch, and K. Yonezawa. 2002. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110:177-189. [DOI] [PubMed] [Google Scholar]
- 27.Hara, K., K. Yonezawa, Q.-P. Weng, M. T. Kozlowski, C. Belham, and J. Avruch. 1998. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF4E BP1 through a common effector mechanism. J. Biol. Chem. 273:14484-14494. [DOI] [PubMed] [Google Scholar]
- 28.Harris, T. E., and J. C. Lawrence. 2003. TOR signaling. Science STKE 2003:15. [DOI] [PubMed] [Google Scholar]
- 29.Haystead, T. A. J., C. M. M. Haystead, C. Hu, T. A. Lin, and J. C. Lawrence. 1994. Phosphorylation of PHAS-I by MAP kinase. Identification of a site phosphorylated by MAP kinase in vitro and in response to insulin in rat adipocytes. J. Biol. Chem. 269:23185-23191. [PubMed] [Google Scholar]
- 30.Helliwell, S. B., P. Wagner, J. Kunz, M. Deuter-Reinhard, R. Henriquez, and M. N. Hall. 1994. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol. Biol. Cell 5:105-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herbert, T. P., G. R. Kilhams, I. H. Batty, and C. G. Proud. 2000. Distinct signalling pathways mediate insulin and phorbol ester-stimulated eIF4F assembly and protein synthesis in HEK 293 cells. J. Biol. Chem. 275:11249-11256. [DOI] [PubMed] [Google Scholar]
- 32.Herbert, T. P., A. R. Tee, and C. G. Proud. 2002. The extracellular signal-regulated kinase pathway regulates the phosphorylation of 4E-BP1 at multiple sites. J. Biol. Chem. 277:11591-11596. [DOI] [PubMed] [Google Scholar]
- 33.Houghton, P. J., and S. Huang. 2004. mTOR as a target for cancer therapy. Curr. Topics Microbiol. Immunol. 279:339-359. [DOI] [PubMed] [Google Scholar]
- 34.Inoki, K., Y. Li, T. Xu, and K. L. Guan. 2003. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17:1829-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacinto, E., R. Loewith, A. Schmidt, S. Lin, M. A. Ruegg, A. Hall, and M. N. Hall. 2004. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6:1122-1128. [DOI] [PubMed] [Google Scholar]
- 36.Kim, D. H., D. Sarbassov, S. M. Ali, R. R. Latek, K. V. Guntur, H. Erdjument-Bromage, P. Tempst, and D. Sabatini. 2002. GβL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 11:895-904. [DOI] [PubMed] [Google Scholar]
- 37.Kim, D. H., D. Sarbassov, S. M. Ali, J. E. King, R. R. Latek, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163-175. [DOI] [PubMed] [Google Scholar]
- 38.Kudchodkar, S. B., Y. Yu, T. G. Maguire, and J. C. Alwine. 2004. Human cytomegalovirus infection induces rapamycin-insensitive phosphorylation of downstream effectors of mTOR kinase. J. Virol. 78:11030-11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kunz, J., R. Henriquez, U. Schneider, M. Deuter-Reinhard, N. R. Movva, and M. N. Hall. 1993. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73:585-596. [DOI] [PubMed] [Google Scholar]
- 40.Kwiatkowski, D. J. 2003. Tuberous sclerosis: from tubers to mTOR. Ann. Hum. Genet. 67:87-96. [DOI] [PubMed] [Google Scholar]
- 41.Lawrence, J. C., and R. T. Abraham. 1997. PHAS/4E-BPs as regulators of mRNA translation and cell proliferation. Trends Biochem. Sci. 22:345-349. [DOI] [PubMed] [Google Scholar]
- 42.Leslie, N. R., A. Gray, I. Pass, E. A. Orchiston, and C. P. Downes. 2000. Analysis of the cellular functions of PTEN using catalytic domain and C-terminal mutations: differential effects of C-terminal deletion on signalling pathways downstream of phosphoinositide 3-kinase. Biochem. J. 346(Pt. 3):827-833. [PMC free article] [PubMed] [Google Scholar]
- 43.Lin, T. A., and J. C. Lawrence. 1996. Control of the translational regulators PHAS-I and PHAS-II by insulin and cAMP in 3T3-L1 adipocytes. J. Biol. Chem. 271:30199-30204. [DOI] [PubMed] [Google Scholar]
- 44.Loewith, R., E. Jacinto, S. Wullschleger, A. Lorberg, J. L. Crespo, D. Bonenfant, W. Oppliger, P. Jenoe, and M. N. Hall. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10:457-468. [DOI] [PubMed] [Google Scholar]
- 45.MacDonald, A. S. 2003. Rapamycin in combination with cyclosporine or tacrolimus in liver, pancreas, and kidney transplantation. Transplant. Proc. 35:201S—208S. [DOI] [PubMed] [Google Scholar]
- 46.Mamane, Y., E. Petroulakis, L. Rong, K. Yoshida, L. W. Ler, and N. Sonenberg. 2004. eIF4E-from translation to transformation. Oncogene 23:3172-3179. [DOI] [PubMed] [Google Scholar]
- 47.Manning, B. D., and L. C. Cantley. 2003. Rheb fills a GAP between TSC and TOR. Trends Biochem. Sci. 28:573-576. [DOI] [PubMed] [Google Scholar]
- 48.Manning, B. D., A. R. Tee, M. N. Logsdon, J. Blenis, and L. C. Cantley. 2002. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol. Cell 10:151-162. [DOI] [PubMed] [Google Scholar]
- 49.McMahon, L. P., K. M. Choi, T. A. Lin, R. T. Abraham, and J. C. Lawrence, Jr. 2002. The rapamycin-binding domain governs substrate selectivity by the mammalian target of rapamycin. Mol. Cell. Biol. 22:7428-7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McManus, E. J., and D. R. Alessi. 2002. TSC1-TSC2: a complex tale of PKB-mediated S6K regulation. Nat. Cell Biol. 4:E214—E216. [DOI] [PubMed] [Google Scholar]
- 51.Mothe-Satney, I., G. J. Brunn, L. P. McMahon, C. T. Capaldo, R. T. Abraham, and J. C. Lawrence. 2000. Mammalian target of rapamycin-dependent phosphorylation of PHAS-1 in four (S/T)P sites detected by phospho-specific antibodies. J. Biol. Chem. 275:33836-33843. [DOI] [PubMed] [Google Scholar]
- 52.Mothe-Satney, I., D. Yang, P. T. Fadden, A. J. Haystead, and J. C. Lawrence. 2000. Multiple mechanisms control phosphorylation of PHAS-I in five (S/T)P sites that govern translational repression. Mol. Cell. Biol. 20:3558-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murakami, M., T. Ichisaka, M. Maeda, N. Oshiro, K. Hara, F. Edenhofer, H. Kiyama, K. Yonezawa, and S. Yamanaka. 2004. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol. Cell. Biol. 24:6710-6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Myers, M. P., I. Pass, I. H. Batty, K. J. Van der, J. P. Stolarov, B. A. Hemmings, M. H. Wigler, C. P. Downes, and N. K. Tonks. 1998. The lipid phosphatase activity of PTEN is critical for its tumor suppressor function. Proc. Natl. Acad. Sci. USA 95:13513-13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nojima, H., C. Tokunaga, S. Eguchi, N. Oshiro, S. Hidayat, K. Yoshino, K. Hara, J. Tanaka, J. Avruch, and K. Yonezawa. 2003. The mTOR partner, raptor, binds the mTOR substrates, p70 S6 kinase and 4E-BP1, through their TOS (TOR signaling) motifs. J. Biol. Chem. 278:15461-15464. [DOI] [PubMed] [Google Scholar]
- 56.Oshiro, N., K. Yoshino, S. Hidayat, C. Tokunaga, K. Hara, S. Eguchi, J. Avruch, and K. Yonezawa. 2004. Dissociation of raptor from mTOR is a mechanism of rapamycin-induced inhibition of mTOR function. Genes Cells 9:359-366. [DOI] [PubMed] [Google Scholar]
- 57.Patel, J., X. Wang, and C. G. Proud. 2001. Glucose exerts a permissive effect on the regulation of the initiation factor 4E binding protein 4E-BP1. Biochem. J. 358:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peterson, R. T., P. A. Beal, M. J. Comb, and S. L. Schreiber. 2000. FKBP12-rapamycin-associated protein (FRAP) autophosphorylates at serine 2481 under translationally repressive conditions. J. Biol. Chem. 275:7416-7423. [DOI] [PubMed] [Google Scholar]
- 59.Price, N. T., S. F. Nakielny, S. J. Clark, and C. G. Proud. 1989. The two forms of the beta-subunit of initiation factor-2 from reticulocyte lysates arise from proteolytic degradation. Biochim. Biophys. Acta 1008:177-182. [DOI] [PubMed] [Google Scholar]
- 60.Proud, C. G. 2002. Regulation of mammalian translation factors by nutrients. Eur. J. Biochem. 269:5338-5349. [DOI] [PubMed] [Google Scholar]
- 61.Sarbassov, D., S. M. Ali, D. H. Kim, D. A. Guertin, R. R. Latek, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2004. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14:1296-1302. [DOI] [PubMed] [Google Scholar]
- 62.Schalm, S. S., and J. Blenis. 2002. Identification of a conserved motif required for mTOR signaling. Curr. Biol. 12:632-639. [DOI] [PubMed] [Google Scholar]
- 63.Schalm, S. S., D. C. Fingar, D. M. Sabatini, and J. Blenis. 2003. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr. Biol. 13:797-806. [DOI] [PubMed] [Google Scholar]
- 64.Stern, B. D., M. Wilson, and R. Jagus. 1993. Use of nonreducing SDS-PAGE for monitoring renaturation of recombinant protein synthesis initiation factor, eIF-4 alpha. Protein Expr. Purif. 4:320-327. [DOI] [PubMed] [Google Scholar]
- 65.Takahashi, T., K. Hara, H. Inoue, Y. Kawa, C. Tokunaga, S. Hidayat, K. Yoshino, Y. Kuroda, and K. Yonezawa. 2000. Carboxyl-terminal region conserved among phosphoinositide-kinase-related kinases is indispensable for mTOR function in vivo and in vitro. Genes Cells 5:765-775. [DOI] [PubMed] [Google Scholar]
- 66.Tang, X., L. Wang, C. G. Proud, and C. P. Downes. 2003. Muscarinic receptor-mediated activation of p70 S6 kinase 1 (S6K1) in 1321N1 astrocytoma cells: permissive role of phosphoinositide 3-kinase. Biochem. J. 374:137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tee, A. R., D. C. Fingar, B. D. Manning, D. J. Kwiatkowski, L. C. Cantley, and J. Blenis. 2002. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc. Natl. Acad. Sci. USA 99:13571-13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tee, A. R., B. D. Manning, P. P. Roux, L. C. Cantley, and J. Blenis. 2003. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 13:1259-1268. [DOI] [PubMed] [Google Scholar]
- 69.Tee, A. R., and C. G. Proud. 2000. DNA damage causes inactivation of translational regulators linked to mTOR signalling. Oncogene 19:3021-3031. [DOI] [PubMed] [Google Scholar]
- 70.Tee, A. R., and C. G. Proud. 2002. Caspase cleavage of initiation factor 4E-binding protein 1 yields a dominant inhibitor of cap-dependent translation and reveals a novel regulatory motif. Mol. Cell. Biol. 22:1674-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang, L., and C. G. Proud. 2002. Ras/Erk signalling is required for regulation of translation factors linked to mTOR and protein synthesis by hypertrophic agents in cardiomyocytes. Circ. Res. 91:821-829. [DOI] [PubMed] [Google Scholar]
- 72.Wang, X., L. E. Campbell, C. M. Miller, and C. G. Proud. 1998. Amino acid availability regulates p70 S6 kinase and multiple translation factors. Biochem. J. 334:261-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang, X., W. Li, J.-L. Parra, A. Beugnet, and C. G. Proud. 2003. The C-terminus of initiation factor 4E-binding protein 1 contains multiple regulatory features that influence its function and phosphorylation. Mol. Cell. Biol. 23:1546-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang, D., G. J. Brunn, and J. C. Lawrence. 1999. Mutational analysis of sites in the translational regulator, PHAS-I, that are selectively phosphorylated by mTOR. FEBS Lett. 453:387-390. [DOI] [PubMed] [Google Scholar]
- 75.Yonezawa, K., K. I. Yoshino, C. Tokunaga, and K. Hara. 2004. Kinase activities associated with mTOR. Curr. Top. Microbiol. Immunol. 279:271-282. [DOI] [PubMed] [Google Scholar]
- 76.Zheng, X. F., D. Florentino, J. Chen, G. R. Crabtree, and S. L. Schreiber. 1995. TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell 82:121-130. [DOI] [PubMed] [Google Scholar]
- 77.Zhong, H., K. Chiles, D. Feldser, E. Laughner, C. Hanrahan, M. M. Georgescu, J. W. Simons, and G. L. Semenza. 2000. Modulation of hypoxia-inducible factor 1α expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer Res. 60:1541-1545. [PubMed] [Google Scholar]
- 78.Zhu, J., J. Wu, E. Frizell, S. L. Liu, R. Bashey, R. Rubin, P. Norton, and M. A. Zern. 1999. Rapamycin inhibits hepatic stellate cell proliferation in vitro and limits fibrogenesis in an in vivo model of liver fibrosis. Gastroenterology 117:1198-1204. [DOI] [PubMed] [Google Scholar]