Abstract
The mobility of transposable elements via a cut-and-paste mechanism depends on the elaboration of a nucleoprotein complex known as the synaptic complex. We show here that the Mos1 synaptic complex consists of the two inverted terminal repeats of the element brought together by a transposase tetramer and is designated paired-end complex 2 (PEC2). The assembly of PEC2 requires the formation of a simpler complex, containing one terminal repeat and two transposase molecules and designated single-end complex 2 (SEC2). In light of the formation of SEC2 and PEC2, we demonstrate the presence of two binding sites for the transposase within a single terminal repeat. We have found that the sequence of the Mos1 inverted terminal repeats contains overlapping palindromic and mirror motifs, which could account for the binding of two transposase molecules “side by side” on the same inverted terminal repeat. We provide data indicating that the Mos1 transposase dimer is formed within a single terminal repeat through a cooperative pathway. Finally, the concept of a tetrameric synaptic complex may simply account for the inability of a single mariner transposase molecule to interact at the same time with two kinds of DNA: the inverted repeat and the target DNA.
Mos1 is an autonomous mariner transposable element first isolated from Drosophila mauritiana. It is 1,286 bp long, with 28-bp imperfect inverted terminal repeats (ITR), and contains a single open reading frame encoding a 345-amino-acid transposase (Tnp). On the basis of the sequence of the Tnp and the organization of the element, mariner has been grouped together with the Tc1 and pogo transposable elements to form the Tc1/mariner superfamily (27). One of the main properties of the Tc1/mariner elements is that they do not require host factors for their mobility, because the Tnp is sufficient to promote all the transposition steps. This property accounts for the wide distribution of the Tc1/mariner superfamily among eukaryotic organisms and makes it possible to develop DNA transfer vectors based on Tc1/mariner elements (22).
mariner transposes by a cut-and-paste mechanism, similar to that described for related bacterial insertion sequences (4). Briefly, the two ends of the elements are brought together by transposase oligomerization to form a synaptic complex that triggers cleavages at the transposon ends. This complex is usually designated a paired-end complex (PEC). The Tnp then promotes the integration of the excised transposon at a new target site. Assembling the highly organized synaptic complex is the key of transposition, in which the cleavages progress step by step. The structure of this synaptic complex has been elucidated for several prokaryotic transposons, such as Tn5. In this case, the molecular assembly is dimeric: each double-stranded DNA molecule is bound to both Tnp subunits (8). In other cases, where no crystallographic data are available, the exact stoichiometry of the complex has yet to be defined, as explained for IS911 (21). No complete synaptic complex has so far been crystallized for the eukaryotic transposons, but the structure of Tnp/ITR complexes has been analyzed by a biochemical approach, as in the case of Sleeping Beauty, a Tc1-like element (6, 13). Since mariner elements are related to prokaryotic transposons, their synaptic complexes are thought to contain a dimer of Tnp. This accounts for the currently accepted view of the Mos1 and Himar1 transposition pathways. Two recent reports describe Mos1 Tnp/ITR complexes formed in vitro (1, 9), but it is still difficult to identify the exact organization of the Mos1 synaptic complex. This is the purpose of the work presented here.
We first explored the protein content and intrinsic activities of the Tnp/ITR complexes. Our findings show that the synaptic complex for Mos1 is not as simple as hitherto believed. In fact, this complex consists of paired ITR retaining a tetramer of Tnp's and is thus designated paired-end complex 2 (PEC2). Our results indicate that PEC2 is both the target capture complex and the strand transfer complex. Two non-mutually exclusive pathways are proposed for the assembly of PEC2, both of which would involve the formation of a complex consisting of a single ITR retaining two Tnp molecules and designated single-end complex 2 (SEC2). In light of the formation of SEC2 and PEC2, we demonstrate that there are two binding sites for the Tnp within a single ITR. Indeed, we have found that the sequence of the Mos1 ITR contains overlapping palindromic and mirror motifs that could account for the binding of two Tnp molecules “side by side” on the same ITR. Our data therefore indicate that the Mos1 Tnp binds cooperatively within a single ITR in order to form SEC2. Finally, the occurrence of a tetrameric synaptic complex may account for the inability of the mariner Tnp to interact simultaneously with two kinds of DNA: the ITR and the DNA target. Similarities between Mos1 and two eukaryotic nucleoprotein complexes [CRO and V(D)J] are discussed.
MATERIALS AND METHODS
DNA manipulations.
A double-stranded ITR30 oligonucleotide with BamHI extremities was cloned at the BamHI site of plasmid pBS II SK+ (Stratagene) to produce pBS-3′ITR. This plasmid was subjected to EcoRI/XbaI cleavage, yielding the ITR70 fragment, which was agarose purified. After precipitation, the DNA concentration was estimated on agarose gel. ITR260 was amplified with Taq DNA polymerase from pBS-3′ ITR as recommended by the manufacturer (Promega), by using universal and reverse primers. The PCR product was agarose purified, and the DNA concentration was estimated on agarose gel.
Sequences of ITR30 (corresponding to the right Mos1 ITR) and truncated ITR (Δ-28 or 1-Δ) are shown in Fig. 6. The dinucleotide TT was added at each 5′ extremity so that the double-stranded oligonucleotides could be labeled by the DNA polymerase Klenow fragment after annealing. Briefly, 10 pmol of DNA was filled in by using the DNA polymerase Klenow fragment (Promega), [α-32P]dATP, and unlabeled dCGT. After precipitation with tRNA as a carrier, the probes were resuspended in 50 μl of H2O, with a final concentration of 200 nM.
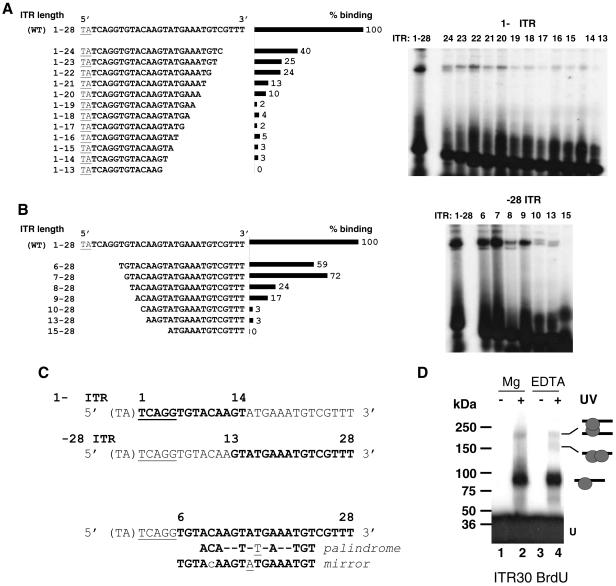
FIG. 6.
Mos1 ITR contains two binding sites for the Tnp. The wild-type ITR (WT) is the full-length sequence (TA + 28 bp). (A) EMSA with ITR truncated at the 3′ end (1-Δ). The sequences of the probes are given on the left. Binding rates obtained with the different 1-Δ ITR, expressed as percentages of that obtained for the WT ITR (taken as 100%), are shown in the center. EMSA data are presented on the right. (B) EMSA with ITR truncated at the 5′ end (Δ-28). The sequences of the probes are given on the left. Binding rates obtained with the different Δ-28 ITR, expressed as percentages of that obtained for the WT ITR (taken as 100%), are shown in the center. EMSA data are presented on the right. (C) Sequence data. The TA dinucleotide flanking the ITR is enclosed in parentheses. The cleavage signal (TCAGG) is underlined. Boldfaced letters represent the shortest sequence retaining a binding signal. The numbers above the top two sequences delineate the minimal truncated ITR that was able to interact with the Tnp. The symmetrical structures (palindrome or mirror) in truncated ITR, which fall between positions 6 and 28, are given at the bottom. The underlined nucleotide represents the center of each symmetrical structure. (D) UV cross-linking assays. SEC2 was assembled by using BrdU ITR30 as a probe with MgCl2 (lanes 1 and 2) or EDTA (lanes 3 and 4). Complexes were exposed to UV light (lanes 2 and 4) or not (lanes 1 and 3) and then loaded onto SDS-PAGE gels with prestained molecular standards. U, unbound DNA. The complexes observed are diagramed on the right.
The cleavages occurring in the complexes were analyzed by using ITR70 labeled on one strand. For single-strand labeling of the nontransferred strand (yielding ITR70α), the ITR70 fragment was filled in at the EcoRI site by using the DNA polymerase Klenow fragment and [α-32P]dATP without dCGT. For single-strand labeling on the transferred strand (yielding ITR70γ), pBS-3′ITR was digested by XbaI and dephosphorylated by using calf intestine phosphatase (Promega) before EcoRI digestion. After purification, the XbaI/EcoRI fragment was labeled on the dephosphorylated end by using T4 polynucleotide kinase (Promega) and [γ-32P]ATP.
Proteins.
The wild-type Mos1 Tnp protein (amino acids 1 to 345) was amplified by PCR with GoTaq DNA polymerase (Promega) as previously described (1), with minor modifications: 5′-AAGCTTTTATTCAAAGTATTTGCCGTCGC-3′ (Tnp 3′ HindIII) was used as a primer instead of Tnp 3′ BamHI. PCR products were cloned with the pGEM-T Easy system (Promega), sequenced, and subcloned with the pMal-c2 system (New England Biolabs). The Tnp was produced and purified as a fusion protein linked to the maltose-binding protein (MBP), as previously described (1). MBP-Tnp was used instead of Tnp because it has the same specific activity but is much more stable during purification, biochemical assays, and in vitro and in vivo transposition assays (C. Augé-Gouillou et al., unpublished data). The terms “transposase,” “Tnp,” and “Tnp molecule,” used here, refer to a single subunit of Mos1 protein.
Electrophoretic mobility shift assays (EMSA).
Binding reactions were carried out in 50 mM NaCl-0.5 mM dithiothreitol-10 mM Tris (pH 9)-5% glycerol-100 ng of bovine serum albumin (BSA). Each 20-μl reaction mixture contained 0.2 pmol of either 32P-labeled duplex, ITR30 or ITR70, 10 pmol of purified Tnp, and 5 mM EDTA or MgCl2. Mixtures were incubated at either 4 or 30°C for various times. Reaction products were separated by using discontinuous 4 to 6% native polyacrylamide-0.25× TBE gels (30:0.93, acrylamide-bisacrylamide-22 mM Tris-22 mM boric acid-0.6 mM EDTA) containing 5% glycerol. Gels were run at 200 V for 3 h and autoradiographed. Experiments with mixed DNA substrates were performed using ITR30 as a probe, and 0.4 pmol of a 260-bp cold DNA fragment containing the 3′ ITR. Mixtures were incubated for 2 h at 30°C in 5 mM EDTA.
Native polyacrylamide gel electrophoresis (PAGE) was used to determine the molecular sizes of the Tnp/ITR complexes. Samples were prepared by using conditions under which all four complexes were formed (2 h at 30°C; 5 mM EDTA), with ITR30 as a probe, and were run with prestained protein standards (Bio-Rad) on native discontinuous 4 to 7% polyacrylamide-Tris-glycine gels (30:0.8, acrylamide-bisacrylamide). Gels were run at 200 V for 3 h and autoradiographed.
DNA-protein cross-linking. (i) Glutaraldehyde cross-linking.
Complexes were assembled in 15-μl reaction mixtures containing 5 mM EDTA, 0.2 pmol of 32P-labeled ITR30, and 15 pmol of Tnp in 20 mM phosphate buffer (pH 8) instead of Tris buffer, as described above. For the glutaraldehyde binding assays, we took advantage of the presence of methyl groups on the DNA to obtain DNA-protein cross-linking in addition to interprotein cross-linking. By using ITR30 as a probe, complexes were assembled under conditions that allowed a C4 complex (see Fig. 1) to accumulate (overnight at 30°C; 5 mM EDTA). A 1.5-μl volume of 0.5% glutaraldehyde was added for 1 h at 25°C. Reactions were stopped by addition of 3 μl of 6× sodium dodecyl sulfate (SDS)-PAGE loading buffer, and samples were boiled for 10 min. As a control, the same experiment was performed without fixing or boiling the samples.
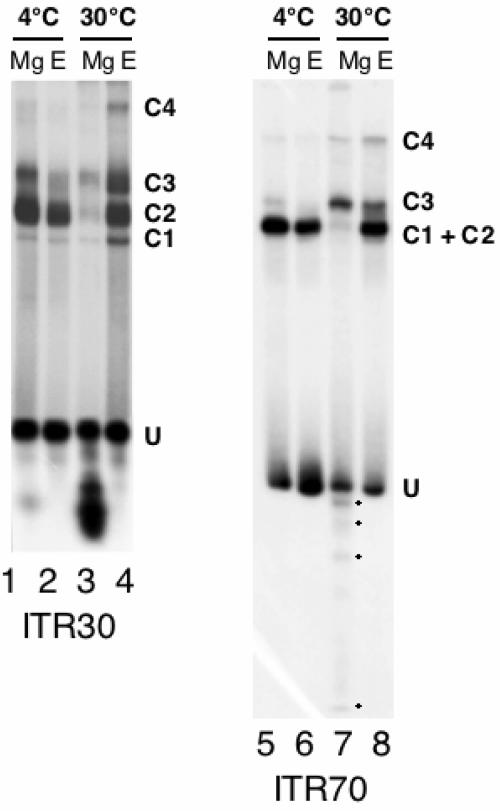
FIG. 1.
Mos1 Tnp forms four complexes with the Mos1 ITR. EMSA were performed using Tnp and ITR30 (lanes 1 to 4) or ITR70 (lanes 5 to 8) as probes. U, unbound DNA; C1, C2, C3, and C4, Tnp/ITR complexes; Mg, incubations in 5 mM MgCl2; E, incubations in 5 mM EDTA. Temperature conditions are indicated above the gels. Putative cleavage products in lane 7 are starred.
(ii) UV cross-linking.
Reactions were performed as described for glutaraldehyde cross-linking, using a modified 32P-labeled ITR30. The upper strand of ITR30 was annealed with the bromodeoxyuridine (BrdU)-containing lower strand, 5′-TTAAACGACAXTXCATACXXGTACACCTGATA-3′ (where X stands for BrdU nucleotides). UV cross-linking assays are currently used to selectively label DNA-binding proteins based on their specific interaction with a DNA recognition site. As a consequence of label transfer, the molecular weight of the DNA-protein complex can be determined rapidly and reliably (5). An important point to consider is that the efficiency of UV cross-linking in usually on the order of 1 to 10%. If a protein binds to its target as a dimer, UV cross-linking will mainly transfer the label to one of the bound molecules. Thus, the observed molecular weight is mainly that of the monomer, since the dimer is only faintly (<1%) linked. For the UV cross-linking assays, mixtures were prepared under conditions that allowed all four complexes to form (2 h at 30°C; 5 mM EDTA) by using either ITR30 or ITR30 BrdU as a probe. The ability of the ITR30-BrdU to engage the four complexes had been confirmed by EMSA previously (data not shown). The mixtures were incubated for 2 h at 30°C, and samples were then exposed to UV irradiation (312 nm) for 30 min at 4°C. Reactions were stopped by addition of 3 μl of 6× SDS-PAGE loading buffer, and samples were boiled for 10 min.
Cross-linked products were separated by using 3 to 8% continuous-gradient NuPage acetate gels (Invitrogen), as recommended by the manufacturer, together with prestained protein standards (Bio-Rad), and gels were autoradiographed.
Analysis of the cleavage products in preformed complexes.
DNA binding reaction mixtures were assembled as described above, using ITR70 (α or γ) as a probe. Samples were scaled up twofold. Incubations were performed in 5 mM MgCl2 for 60 min at 30°C. Complexes were separated as described above, and the wet gel was exposed to film for 1 h at room temperature. Slices containing individual complexes were cut from the gel, immersed in an elution buffer (20 mM Tris [pH 9], 100 mM NaCl, 1 mM dithiothreitol, 5 mM EDTA) and heated to 70°C for 15 min to inactivate the Tnp. Samples were incubated overnight at 37°C, and the DNA eluted was then ethanol precipitated. Products were loaded onto a 9% denaturing Hydrolink (Promega) 0.5× TBE gel containing 7 M urea. Gels were run at 45 W for 2 h and autoradiographed. G+A sequences were prepared from either the ITR70α or the ITR70γ probe by using the standard chemical procedure.
Target capture and strand transfer reactions. (i) Target capture.
The target was a 30-bp duplex oligonucleotide with a single central TA dinucleotide. Complexes were assembled in 20-μl reaction mixtures containing 0.2 pmol of unlabeled ITR30 and 10 pmol of Tnp, without MgCl2 or EDTA, with 100 ng of BSA and 5% glycerol, for 16 h at 30°C. A 0.3-pmol portion of labeled target and 500 ng of sonicated salmon DNA were added, and the samples were incubated at 30°C for a further 2 h. Samples were analyzed by EMSA.
(ii) Strand transfer.
Complexes were assembled in 20-μl reaction mixtures containing 0.2 pmol of labeled duplex ITR30 and 20 pmol of Tnp without MgCl2 or EDTA, with 100 ng of BSA and 5% glycerol, for 16 h at 30°C. A 0.2-pmol portion of a supercoiled plasmid and 5 mM MgCl2 were added, and the mixtures were incubated at 30°C for various times. Reactions were stopped by phenol-chloroform extraction and ethanol precipitation. DNA pellets were resuspended in 15 μl of 1× native loading buffer and were run on a native 9% Hydrolink gel in 1× TBE buffer. The gel was dried and autoradiographed.
RESULTS
Mos1 Tnp forms multiple complexes on 3′ ITR.
As a first step in Mos1 transposition, the Tnp must bind and bridge the two ITR to form the synaptic complex. From what is known about prokaryote-related transposons, two kinds of complexes can be expected. The first type would consist of one Tnp molecule bound to one ITR, yielding a SEC. The second type would be the PEC, which is a dimer of SEC. To check this, we performed a set of comparative analyses, using a 30-bp (ITR30) or a 70-bp (ITR70) probe. Mos1 Tnp/ITR complexes were assembled at either 4 or 30°C for 2 h under various cation conditions (Fig. 1) and were separated on a high-resolution native polyacrylamide gel. Independently of the probe used, up to four complexes were observed when binding assays where performed at 30°C in 5 mM EDTA (Fig. 1, lanes 4 and 8). The lowest complexes (C1 and C2) were always observed at 4°C (lanes 1, 2, 5, and 6) and corresponded to the complexes previously reported (1). The detection of the highest complex (C4) was promoted by a temperature of 30°C (lanes 2 versus 4 and 6 versus 8), and the same temperature was also required to detect the last complex (C3) when ITR70 was used as a probe (compare lanes 5 and 6 to lanes 7 and 8). DNA fragments smaller than the probes were also obtained at 30°C with MgCl2 (Fig. 1, lanes 3 and 7); these may correspond to cleavage products.
Stoichiometry of the Tnp/ITR complexes.
Because we obtained four Tnp/ITR complexes, instead of the two that would be expected if the synaptic complex were dimeric, the protein and DNA contents of these complexes had to be determined. Three different approaches were used to determine the molecular weight and, consequently, the number of Tnps and ITR in each complex: native PAGE (Fig. 2A), UV cross-linking (Fig. 2B), and glutaraldehyde fixation (Fig. 2C) assays. It must be noted that some Tnp-ITR interactions were strong enough to withstand SDS treatment during electrophoresis (Fig. 2C, lane 1). The stability of the Tnp/ITR complexes was confirmed by the observation, in the same lane, of two weaker bands of higher molecular weight. However, the use of SDS-PAGE sometimes led to the detection of a thin band (Fig. 2C, lane 1) that remained inexplicable.
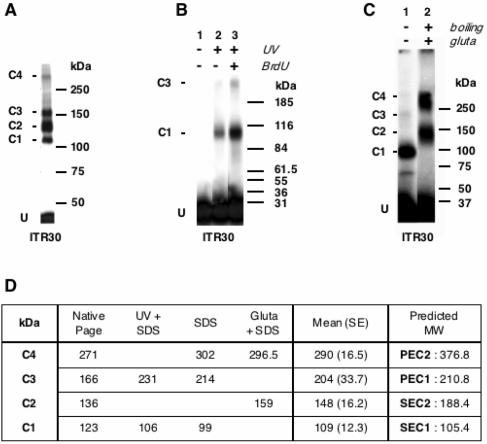
FIG. 2.
Determination of the apparent molecular masses of Tnp/ITR complexes. (A) Native PAGE was performed using ITR30 as a probe and was run with prestained molecular standards. U, unbound DNA; C1, C2, C3, and C4, Tnp/ITR complexes. (B) UV cross-linking assays. PEC2 was assembled using either ITR30 (lanes 1 and 2) or BrdU ITR30 (lane 3) as a probe. Complexes were exposed to UV (lanes 2 and 3) or not (lane 1) and loaded onto SDS-PAGE gels with prestained molecular standards. (C) Glutaraldehyde fixation assays. PEC2s were assembled by using ITR30 as a probe. Complexes were subjected to glutaraldehyde treatment (lane 2) or not (lane 1) and then loaded onto SDS-PAGE gels with prestained molecular standards. (D) The apparent molecular mass of each complex as determined in panels A, B, and C. Values are means of at least three independent assays. The standard error is always less than 15% of the reported mean. The expected molecular mass of each complex is indicated on the right.
The observed molecular masses of the four complexes are reported in Fig. 2D. Molecular masses obtained by using the different methods for each complex were pooled to yield an experimental average. These experimental values were then compared to the predicted values, taking into account MBP-Tnp (83 kDa) and ITR30 (22.4 kDa) counterparts. Thus, a complex consisting of a monomer of Tnp bound to a single ITR would be expected to have a molecular mass of 105.4 kDa, which closely matches the apparent molecular mass of C1 (109 ± 12 kDa). In the same way, a complex consisting of a Tnp dimer bound to two ITR would be expected to have a molecular mass of 210.8 kDa, which closely matches the apparent molecular mass of C3 (204 ± 33.7 kDa). Therefore, C1 and C3 are single-end and paired-end complexes, respectively, as expected from the simple transposition model. They were designated SEC1 and PEC1, respectively.
The two remaining complexes required further analyses. C2 had an apparent molecular mass of 148 ± 16.2 kDa, probably reflecting the presence of one dimer of Tnp bound to a single ITR (expected molecular mass, 188.4 kDa), as previously described (1). Since the experimental molecular mass of C4 (290 ± 16.5 kDa) corresponded exactly to that of a C2 dimer (296 kDa), we concluded that C4 consisted of a Tnp tetramer bound to two ITR. Thus, on the basis of their protein and ITR assignments, C2 and C4 were designated SEC2 and PEC2, respectively. The fact that the predicated molecular masses of SEC2 and PEC2 did not match the observed molecular masses of C2 and C4 might simply be due to a conformation of the complexes that disturbs their electrophoretic mobility.
An alternative procedure to establish the protein content of a complex is the use of tagged protein of different sizes. In contrast to its successful use for the V(D)J synaptic complex (20), this process cannot be applied here. This was mainly due to the fact that we have four complexes (with MBP-Tnp) yielding the detection of at least 12 bands with tagged Tnps of different sizes (2 for SEC1, 3 for SEC2, 3 for PEC1, and 4 for PEC2, plus those due to the isoforms [see Discussion]). Proper analysis of such a complex pattern by EMSA was impossible.
Finally, the data obtained following the use of UV cross-linking assays call for comment. These experiments were done to produce covalent linking of the Tnp molecules to the ITR. From this point of view, SEC1 was to be expected, whereas SEC2 would be detected only if the two Tnp molecules were in close contact with the DNA within this complex. Under the conditions described here (Fig. 2B), this was not the case, but it was possible to detect PEC1. This may indicate that each ITR is bound by both Tnp subunits within this complex, as has previously been shown to occur in the Tn5 synaptic complex (8).
Analysis of complex formation.
To confirm that PEC1 and PEC2 were indeed paired-end complexes, we used experiments involving mixed substrates (Fig. 3A). Incubation of the Tnp with ITR30 alone resulted in the formation of all four complexes (Fig. 3A, lane −). If one of them was a paired-end complex, addition of an unlabeled 260-bp derivative of the 3′ ITR (ITR260) would inevitably lead to the formation of an additional band. As shown in Fig. 3A, lane +, two additional complexes were indeed formed when ITR260 was added. The first complex, which migrated between PEC1 and PEC2, was a PEC1 containing the labeled ITR30 paired to the cold ITR260, as previously shown (9). The second band, which migrated more slowly than PEC2, was a PEC2 containing the labeled ITR30 paired to the cold ITR260. An additional band was detected in both lanes and corresponded to stacking aggregates. These data enabled us to confirm the DNA contents of PEC1 and PEC2.
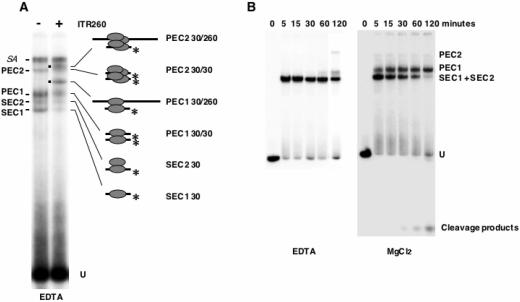
FIG. 3.
Analysis of PEC formation. (A) Demonstration of paired-end complexes. Complexes were assembled with (+) or without (−) a cold 200-bp derivative of the 3′ ITR (ITR260) together with the labeled ITR30 and were separated by EMSA. The molecular composition of each complex is shown on the right. Dots indicate the complexes that contained mixed ITR. An asterisk marks the position of the 32P. SA, stacking aggregates. (B) Time course analysis of complex formation. Complexes were assembled, with ITR70 as a probe, for various lengths of time at 30°C in 5 mM EDTA (left panel) or 5 mM MgCl2 (right panel). The complexes observed (PEC2, PEC1, SEC2, and SEC1) and the unbound DNA (U) are indicated.
The identification of two single-end complexes together with two paired-end complexes raised questions about the relationship between them. In order to find out whether the SECs were precursors of the PECs or not, complexes were assembled over various periods of time at 30°C (Fig. 3B) and were then separated by native gel electrophoresis. In EDTA (Fig. 3B, left panel), SEC2 was formed immediately, whereas PEC1 and PEC2 were detected only after incubation for at least 60 and 120 min, respectively. SEC1 was always poorly represented, regardless of the incubation time used. Furthermore, PEC1 formation was correlated to a decrease in the amount of SEC2. The results found under catalytic conditions (5 mM MgCl2) (Fig. 3B, right panel) were quite different: all four complexes were formed immediately. PEC2 was poorly detected, and its amount did not vary during the time course analyses. The involvement of MgCl2 in complex formation was particularly evident for PEC1, which was detected immediately. These findings are in agreement with those of Dawson and Finnegan (9), who showed that PEC formation was promoted by catalytic conditions. Furthermore, small signals, which may have corresponded to cleavage products, were detected after incubation for at least 30 min, and thereafter increased with time. The increases in cleavage products and amounts of PEC1 were correlated to the decrease in the amount of SEC2. Taken together, these findings strongly suggest that SEC2 is a molecular precursor of PEC1. Because SEC2 was assembled very quickly under our experimental conditions, time course analyses failed to clarify the relationships between SEC1 and SEC2. Furthermore, the data obtained under noncatalytic conditions showed that PEC2 was the last assembly product to be formed.
Analysis of cleavage products in preformed complexes.
The cleavages occurring in each complex were examined to further clarify the sequential steps in the assembly of the Mos1 synaptic complex. As previously pointed out, only low levels of SEC1 were present, even after short incubation times, and this made it impossible to investigate its cleaving properties. We have shown above that SEC2 must be formed quickly from SEC1 solely via the recruitment of a second molecule of Tnp. Since this process took place even under noncatalytic conditions (with EDTA or at 4°C), we consider that the conversion of SEC1 to SEC2 did not involve cleavage.
ITR/Tnp complexes were allowed to assemble under catalytic conditions (5 mM MgCl2), by using either ITR70α (labeled on the nontransferred strand) or ITR70γ (labeled on the transferred strand) as a probe, for 60 min at 30°C. The complexes were then separated by EMSA, and slices containing individual complexes were cut from the gel. The DNA eluted from these slices was analyzed by denaturing gel electrophoresis, together with the G+A sequences of both probes. Representative results for SEC2 and PEC1 are shown in Fig. 4A. At least six cleavages on the nontransferred strand were detected in both complexes. Among these, 15% of the input substrate in SEC2 and 40% of the input substrate in PEC1 were cleaved 2 bases into the ITR (Fig. 4). Similarly, at least three cleavages on the transferred strand were detected in both SEC2 and PEC1. One of these took place just before the TA dinucleotide flanking the ITR (Fig. 4). This cleavage position represented about 5% of the input substrate in SEC2 and 10% of the input substrate in PEC1. Since some of the cleavages detected in SEC2 were increased in PEC1, the hypothesis of a relationship between the two complexes was strengthened.
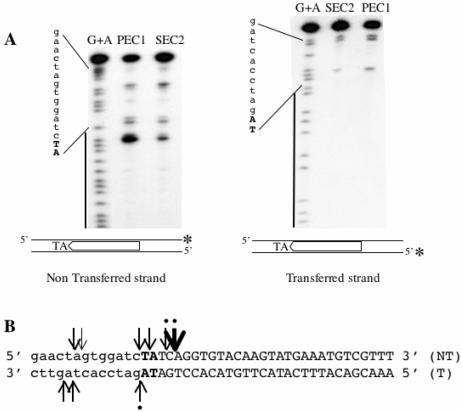
FIG. 4.
Cleavage positions on preformed SEC2 and PEC1. (A) The DNA contained in SEC2 and PEC1 was eluted from an EMSA gel and analyzed on a sequencing gel. An asterisk marks the position of the 32P. The G+A sequence from each probe was loaded as a marker. A black line on the left indicates the position of the ITR. The flanking TA dinucleotide is shown in bold capital letters. The non-ITR flanking DNA is shown in lowercase letters. (Left panel) Cleavage products obtained by using ITR70α as a probe. (Right panel) Cleavage products obtained by using ITR70γ as a probe. (B) Sequence of the DNA used in cleavage site determination. The cleavage sites are shown with arrows, either on the nontransferred strand (NT) or on the transferred strand (T). Dots indicate cleavages that had been described previously.
The cleavage positions identified in our experiments are summarized in Fig. 4B. The data presented here are in good agreement with what has been reported previously for the cleavage of the nontransferred strand of the Tc1, Himar1, and Mos1 elements (9, 15, 22). However, none of the cleavages of the transferred strand detected here corresponded to the previously reported cleavage that was located right at the end of the ITR, excluding the TA dinucleotide (15, 22). Nonetheless, the main cleavage sites described here do correspond to the additional secondary cleavage sites found by Dawson and Finnegan (9) on the transferred strand of Mos1. Furthermore, we have found that both strands were cleaved both in PEC1 and in SEC2. This is not in accordance with the report of Dawson and Finnegan (9), who detected no cleavages on the transferred strand in SEC2. However, an accurate analysis of their results shows a weak but definite cleavage signal on the transferred strand following SEC formation (Fig. 6, lane 6, in reference 9), which is in agreement with the data presented here. Finally, similar results were reported for the Himar1 transposon (18).
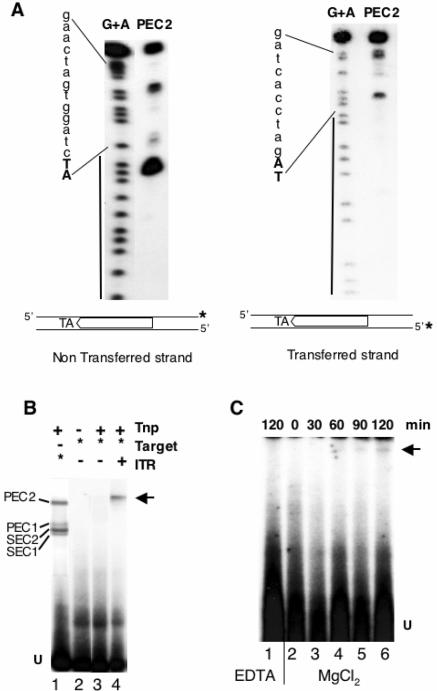
Cleavage sites observed in PEC2 were defined in the same way as for SEC2 and PEC1. A representative result is shown in Fig. 5A. The use of ITR70α as a probe improved the cleavage occurring 2 bases into the ITR; the resulting product was the main product of the reaction. This cleavage accounted for as much as 65% of the initial substrate and accumulated in PEC2 to a greater extent than in PEC1. By use of ITR70γ as a probe, the three cleavage products detected in PEC1 were also detected in PEC2 but were found to be more intense there. However, the proportion of nicked transferred strands in PEC2 accounted for only 15% of the initial substrate. This set of experiments indicated that the cleavages occurred within the synapse and suggested that PEC2 is the final complex, in other words, the target capture complex.
FIG. 5.
PEC2 undergoes target capture. (A) Cleavage positions on preformed PEC2. The DNA contained in PEC2 was eluted from an EMSA gel and analyzed on a sequencing gel. Asterisks mark the position of the 32P. The G+A sequence from each probe was loaded as a marker. Black lines on the left indicate the position of the ITR. The flanking TA dinucleotide is shown in bold capital letters. The non-ITR flanking DNA is shown in lowercase letters. (Left) Cleavage products obtained by using ITR70α as a probe. (Right) Cleavage products obtained by using ITR70γ as a probe. (B) Target capture reactions. EMSA were performed using ITR30 (lane 1) or the target (lanes 2 to 4) as a probe, with or without Tnp. Unlabeled PEC2 was assembled overnight under noncatalytic conditions before addition of the target (lane 4). *, labeled DNA; −, no DNA; +, unlabeled DNA. Arrow indicates the target capture complex. (C) Strand transfer reactions. PEC2 was assembled overnight by using ITR30 as a probe. A supercoiled target was then added after various times, with MgCl2 (lanes 2 to 6) or EDTA (lane 1), and proteins were removed by phenol extraction. Arrow indicates the target labeled by the transferred ITR30.
PEC2 is both the target capture complex and the strand transfer complex.
To ascertain the role of PEC2, we first performed target capture reactions. Unlabeled PEC2 was allowed to assemble under noncatalytic conditions for 16 h at 30°C. Labeled target DNA was then added for another 2 h, and the capture of this piece of DNA was detected by EMSA. The four complexes formed between the labeled ITR30 and the Tnp were used as size indicators (Fig. 5B, lane 1). Tnp alone was unable to directly bind the target DNA (lane 3), in contrast to preformed Tnp/ITR complexes (lane 4). The size of the labeled complex in lane 4 was compatible with target capture by PEC2, yielding a complex with a lower electrophoretic mobility.
We then checked the ability of PEC2 to perform strand transfer reactions. Labeled PEC2 was allowed to assemble under noncatalytic conditions for 16 h at 30°C. A 4-kb supercoiled plasmid was then added for 0 to 120 min, together with MgCl2. If transfer reactions occurred, the target plasmid would have incorporated the labeled ITR and would be detected by autoradiography (Fig. 5C). As expected, at the beginning of the reaction the target was unlabeled (Fig. 5C, lane 2), and the labeling increased with time (lanes 3 to 6). The same reaction, performed in EDTA as a control, showed no target labeling, even after an incubation time of 120 min (lane 1). These findings were in agreement with what is known about the involvement of MgCl2 in strand transfer reactions. Furthermore, they provide direct evidence of the involvement of PEC2 in the Mos1 transposition pathway.
The SEC2 paradox.
The findings discussed in the preceding sections described SEC2 as a single ITR molecule interacting with a dimer of Tnp. From this point of view, two non-mutually exclusive hypotheses can be advanced concerning the organization of the complex. First, the two Tnp molecules could interact directly with the ITR; second, alternatively, just one Tnp molecule could interact with the ITR, the second Tnp being retained within the complex by means of Tnp-Tnp contacts. The first hypothesis would require the presence of two Tnp binding sites within a single ITR, whereas the second would not. The parts of the ITR involved in Tnp binding can be investigated by several techniques, including chemical footprints and binding interference by methylation. None of these tools was able to indicate the number of Tnp molecules retained on the ITR. This was particularly true when the DNA binding domain of the Tnp was bipartite, interacting with two distinct parts of the ITR, as in Tc1, for example (30). Furthermore, footprint analyses have previously shown that the Mos1 ITR (1) is fully protected by the Tnp.
Here, we performed EMSA using a truncated ITR to delineate the sequence of the ITR involved in binding the Tnp (Fig. 6). Tnp/ITR complexes were assembled for 90 min at 4°C in 5 mM MgCl2. The amount of complex obtained for each truncated ITR was compared to that obtained with full-length ITR30. After removal of bases 15 to 28 (leaving ITR 1-14), a binding signal could still be detected, corresponding to 3% of the signal obtained with ITR30 (Fig. 6A). Similarly, after removal of bases 1 to 12 (leaving ITR 13-28), a binding signal was observed corresponding to 3% of the signal obtained with ITR30 (Fig. 6B). In contrast, after removal of bases 1 to 6 (leaving ITR 7-28), a strong binding signal was observed, corresponding to 70% of the signal obtained with ITR30. The first 6 bases of the ITR are known to contain the cleavage signal, which was, as expected, weakly involved in complex formation. Thus, half of the ITR retained the ability to form complexes with the Tnp, albeit with very low efficiency. This observation is compatible with the first hypothesis, i.e., the presence of two Tnp binding sites within a single ITR. The low binding efficiency of the Tnp within each half-ITR may be due to cooperative binding of the Tnp dimer within the whole ITR (see Discussion).
Because only one ITR binding domain has been attributed to the Mos1 Tnp (1, 31), we speculate that the two binding sites may have related sequences. Excluding the first 6 bases (the cleavage signal), we checked for symmetrical motifs located within the remaining sequence. There are two possible antisymmetrical motifs: a mirror (TGTA.AAGTATGAAATGT), between positions 6 and 23, and a palindrome (ACA..T.T.A..TGT), between positions 9 and 23 (dots stand for irrelevant nucleotides) (Fig. 6C). These motifs are centered on positions A15 and T16 (boldfaced and underlined in sequences), respectively. The data reported here failed to identify the design of the relevant structure but do provide further evidence for the existence of two Tnp binding sites within the ITR. To confirm this, UV cross-linking experiments were performed using ITR30 BrdU as a probe, under conditions that allowed SEC2 to accumulate, despite the fact that small amounts of the other complexes were still formed (30 min at 4°C in either 5 mM MgCl2 or EDTA). Samples were exposed to UV irradiation (312 nm) for 30 min at 4°C before being boiled and loaded onto an SDS-polyacrylamide continuous-gradient gel (3 to 8%) with prestained protein standards (Bio-Rad). In the absence of UV irradiation, we observed only unbound ITR30 (Fig. 6D, lanes 1 and 3). After UV irradiation, two bands were obtained when the incubation was performed in MgCl2 (Fig. 6D, lane 2). Their apparent molecular masses (93 and 216 kDa) indicated that they corresponded to SEC1 and PEC1, respectively. A third band was obtained when the incubation was performed in EDTA (Fig. 6D, lane 4). Its apparent molecular mass (168 kDa) indicated that it corresponded to SEC2. This is strong evidence that two Mos1 Tnp molecules were in direct contact with a single ITR. However, under catalytic conditions, SEC2 was no longer detected following UV cross-linking treatment (Fig. 6D, lane 2), whereas it was still present in the mixture (Fig. 1, lane 1). This may have indicated that only one molecule of Tnp was bound to the ITR, while the second Tnp molecule was retained in the complex only by Tnp-Tnp interactions. Because this kind of conformation frees a DNA binding domain contained in one of the two Tnp molecules, it is tempting to suggest that this particular conformation was more likely to be involved in the formation of PEC1. SEC2, therefore, appeared to be present in two different conformations, depending on whether it was produced under catalytic or noncatalytic conditions (see Discussion). These findings seemed to conflict with those presented in Fig. 2B, where no SEC2 was detected after UV cross-linking performed in EDTA. The main difference between these two experiments was the incubation time: 30 min (Fig. 6D) versus 2 h (Fig. 2B). Time course analyses of complex formation (Fig. 3B, left panel) may account for this apparent paradox. In EDTA, a 30-min incubation time may not have been long enough to allow the conformational change described above to occur. This was supported by the fact that PEC1 was not found, either. In contrast, an incubation time of 120 min did allow the conformational change to occur, and this was reflected by the detection of PEC1. We can therefore conclude that the conformational change in SEC2 appeared to be (i) important for the formation of PEC1 and (ii) promoted by catalytic and/or temperature conditions.
DISCUSSION
The transposition of elements by excision and reinsertion makes it necessary to ensure that only properly coupled events take place. The assembly of the synaptic complex, which prevents or limits nonproductive cleavages, plays this checkpoint role. The formation of these complexes has been extensively studied and elucidated in several prokaryotic transposition systems, such as Mu (19), Tn5 (3), and Tn10 (28), as well as in two eukaryotic systems, Sleeping Beauty (6, 13) and V(D)J (20). Despite the recent description of the cleavages occurring during excision (9), little is known about the formation of synaptic complexes in mariner transposons. Indeed, in a recent publication, Lipkow et al. (18) failed to observe the Himar1 synaptic complex, thus preventing the characterization of this complex, even if they were able to detect its activity. Here, we determined the Tnp content and functional capabilities of Mos1 Tnp/ITR complexes. We found that four distinct complexes can be formed: two are single-end complexes (SEC1 and SEC2), and the others are paired-end complexes (PEC1 and PEC2). SEC2 and PEC1 have the same protein content, corresponding to a Tnp dimer, whereas SEC1 and PEC2 result from monomeric and tetrameric assemblies, respectively. The kinetic relationships between these complexes allow us to propose an alternative model for the mariner transposition pathway (Fig. 7). According to this model, the mariner target capture complex contains a Tnp tetramer.
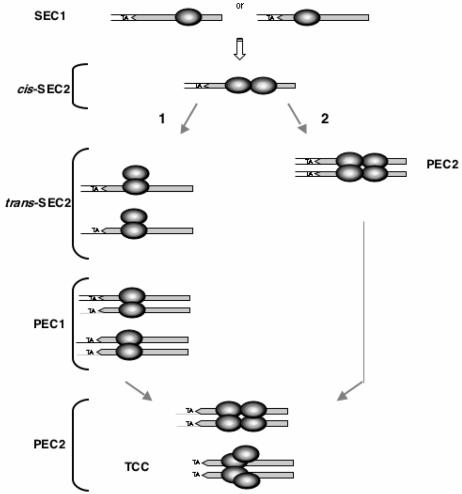
FIG. 7.
Proposed pathway(s) for assembly of the Mos1 synaptic complex. First, a single Tnp molecule binds to the ITR at either half-site (SEC1). A second Tnp is then recruited to form uncleaved cis-SEC2. Two alternative pathways are then proposed for the formation of the target capture complex (TCC). In pathway 1, a conformational change may take place, allowing the formation of trans-SEC2 and the beginning of cleavages. The second ITR is then recruited to form PEC1, and finally two other Tnp molecules are included in the complex to form PEC2. In pathway 2, direct dimerization of cis-SEC2 may allow PEC2 to form and cleavages to take place. In both cases, the TCC contains a Tnp tetramer; two of these Tnp's are bound to the ITR, while the other two are disengaged from the ITR in order to capture the target. Monomers of Tnp are ringed. Double-stranded ITR are shown in gray. The TA dinucleotide flanking the ITR is shown. Flanking DNA is indicated by plain black lines for uncleaved strands and plain gray lines for strands undergoing cleavage.
Assembly of the Mos1 single-end complexes.
Proteins that contain a single HTH motif, like the mariner Tnp (1), have a common mode of DNA binding. Most of them interact as dimers with DNA, as exampled by the bacteriophage lambda Cro repressor (7), which interacts with a 21-bp sequence (the operator) that has an approximate twofold symmetry. Most of the proteins of the Cro family self-associate in the presence of DNA, and the resulting dimers are the binding species (17). Similarly, the POU eukaryotic transcription factors, related to the Cro family, are able to bind to DNA in various dimer configurations (24). Once again, the dimer DNA binding site (designated PORE) presents a twofold symmetry.
The Mos1 Tnp, which interacts as a dimer with a 22-bp sequence having an approximate twofold symmetry shares this mode of DNA binding. Indeed, the mariner ITR contains a palindrome and/or an overlapping mirror (Fig. 6C) that may be organized as two half-ITR, each of which contains a binding site for the Tnp. This configuration would allow the Tnp bound to the ITR to be arranged side by side, but facing in opposite directions, on the same side of the DNA double helix. This proposal is sustained by the recent work of Lipkow et al. (18). They have described two patches of the Himar1 ITR protected by the Tnp and centered on positions 10 and 20. As remarked by the authors, these patches are in phase with the helical repeat of DNA.
In light of what is known about the DNA binding of proteins that contain a single HTH motif, the following schema can be proposed (Fig. 7). The Mos1 Tnp interacts with the ITR, within one of the two binding sites, yielding SEC1. SEC1 is rapidly converted into SEC2, solely as a result of the recruitment of a second molecule of Tnp. This hypothesis is reinforced by the fact that SEC2 is formed independently of the cations present and of the temperature conditions (Fig. 1). Assays using the deletion mutant ITR (Fig. 6) showed that separated half-ITR are only poor binding sites. This observation may account for the cooperative homodimerization of the Tnp on the ITR, as proven for the neural POU proteins (26). However, the direct dimerization of the Mos1 Tnp (in solution) cannot be ruled out, even if the detection of SEC1 in EMSA seems to argue against this proposal.
Finally, our UV cross-linking data (Fig. 6D) indicate that Tnp molecules in SEC2 exist in two conformations: in the “cis” position (cis-SEC2), where the two Tnp molecules are bound side by side to the ITR, or in the “trans” position (trans-SEC2), where only one Tnp molecule is bound to the ITR and the other is retained in the complex by means of protein-protein interactions (Fig. 7). The occurrence of trans-SEC2 in Himar1 Tnp/ITR complexes has been suggested recently (18).
Mos1 paired-end complexes.
As the time course analyses suggest, SEC2 may be the molecular precursor of PEC1 (Fig. 3). In light of the fact that MgCl2 speeded up the formation of PEC1, and in light of the discussion in the preceding section, we therefore propose that PEC1 is simply produced from trans-SEC2 by the addition of a second ITR. Indeed, this is in agreement with the fact that SEC2 and PEC1 have the same Tnp content (Fig. 2). As far as we are aware, a synaptic complex assembly pathway of this type has never been reported for prokaryotic transposons, since the PEC is the molecular species mainly and quickly detected by EMSA, using the full-length Tn5 or Tn10 Tnp (12, 25). However, the assembly of the eukaryotic PEC formed in V(D)J recombination has recently been elucidated. In this case, the PEC was simply obtained by adding a second complementary signal (RSS) to SEC2, the RAG1/RAG2 content of SEC2 being the same as that of the PEC (20). If a transposon like Mos1 was the progenitor of the V(D)J recombination, as suggested recently by Dawson and Finnegan (9), it would not be surprising to observe similarities in the pathways by which these two synaptic complexes are assembled. The ultimate step in the assembly of the Mos1 synaptic complex is the conversion of PEC1 into PEC2 by the recruitment of two molecules of Tnp, although the formation of PEC2 as a result of SEC2 dimerization cannot be completely ruled out. Finally, the mariner synaptic complex contains a tetramer of catalytic units. This is also true of the V(D)J synaptic complex, which contains a RAG1 tetramer.
Cleavages and synapsis.
Time course analyses (Fig. 4) show that cleavages start in SEC2, mainly on the nontransferred strand, and continue in PEC1 and PEC2. The same observations have been carried out for V(D)J recombination, where SEC1 and SEC2 undergo single-strand nicking in MgCl2 (20). The ability of SEC2 to undergo cleavages (9, 18; this study) is unexpected, because it provides an opportunity for deleterious breakage to occur as a result of protein assembly on a single ITR. In many transposition pathways, this is prevented by the fact that both DNA ends must be present before the proteins required for catalysis can be assembled. This is due to cleavage in trans, where a Tnp subunit carries out cleavage and strand transfer, not on the DNA molecule to which it is bound, but rather in the ITR opposite. In the present study, we describe two possible conformations for SEC2: cis and trans. In trans-SEC2, the second Tnp molecule is released from the ITR and takes up a position that could account for the catalytic activity detected in SEC2. The same concept can be applied to the V(D)J recombination pathway, because SEC1 and SEC2 contain a dimer and a tetramer of RAG1 (the equivalent of Tnp), respectively. Although this has not been established, RAG1 may occupy a trans position relative to the RSS in both SECs.
Our findings also indicate that there is no obvious qualitative difference between the catalytic activities of PEC1 and PEC2. However, the fact that there are two paired-end complexes raises the question of their respective roles. The fact that neither of the transferred strands is totally cleaved in either PEC1 or PEC2 suggests that PEC1 may be simply a step in the assembly of the target capture complex. However, this step can be bypassed by the direct dimerization of SEC2, since uncleaved PEC2 can undergo cleavages, target capture, and strand transfer (Fig. 5B). We therefore propose that PEC2 is the target capture complex. These findings lead us to consider a model for the assembly of the Mos1 synaptic complexes (Fig. 7) but provide no information about how the target enters the synaptic complexes. We do not know whether the target is captured after the complete excision of the element. Further investigation will be required to clarify this point.
The final comment about our data is that, as has already been shown, cleavages in the mariner family and the Tc1-related transposons are not at all precisely targeted and therefore give rise to a collection of excision products containing variable ends. On the one hand, imprecise cleavages may account for the lesions that are sometimes generated by the excision of Tc1/mariner elements and that have recently been reported for both mariner (2) and Sleeping Beauty (10, 14). On the other hand, some of the mariner excision products described to date could be abortive, as has been suggested for the transposons Ac/Ds (11), Tc1 (23), and Sleeping Beauty (14).
Biochemical relevance of the tetramer.
Some of the transposons studied so far involve a tetramer in the assembly of their synaptic complexes. This is true of Sleeping Beauty, a member of the eukaryotic Tc1/mariner superfamily (6, 13). Nevertheless, Sleeping Beauty has long terminal repeats (about 230 bp), in which the Tnp binding sites are repeated twice in a direct orientation. Other members of the Tc1/mariner family do not share this feature; they generally have short terminal repeats of about 30 bp. Recently, Lipkow et al. (18) have described a tetrameric structure containing the Himar1 Tnp. Since they were unable to detect paired-end complexes, they concluded that the Tnp tetramer mediated overproduction inhibition. These data have to be interpreted in light of what is known about the in vitro activities of the Himar1 and Mos1 Tnps. The Himar1 Tnp undergoes aggregation for concentrations higher than 10 nM, yielding overproduction inhibition (16), whereas the Mos1 Tnp does not (29). This difference may account for the apparent divergence between the data of Lipkow et al. (18) and our present data. As a consequence, the PECs involving Mos1 Tnp were detectable by EMSA, whereas Tnp aggregates were the main species obtained with the Himar1 Tnp. However, the data presented here have clearly established that a Tnp tetramer is involved in mariner transposition. This raises the question of the role of the two “supernumerary” Tnps. As currently accepted, transposition calls for three pieces of DNA: the two ITR and the target site, containing a TA dinucleotide. We propose that in the target capture complex, two molecules of Tnp remain bound to the ITR, whereas the other two are involved in target binding and cleavage. The requirement for four Tnp molecules probably reflects the inability of a single Mos1 Tnp to interact at the same time with two different DNA sequences, probably because the Tnp contains only one DNA binding motif. Although this is an intellectually satisfying model, the target capture pathway in mariner transposition remains to be further elucidated.
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique, the Ministère de l'Education, de la Recherche et de la Technologie, the Groupement de Recherche CNRS 2157, the Institut Fédératif de Recherche 136, the Ligue Nationale contre le Cancer, and the Association Contre le Cancer (grant 7684). B. Brillet is funded by a Ph.D. grant from the Région Centre.
M. Ghosh revised the English version of this paper.
REFERENCES
- 1.Augé-Gouillou, C., M. H. Hamelin, M. V. Demattei, G. Periquet, and Y. Bigot. 2001. The ITR binding domain of the Mariner Mos1 transposase. Mol. Genet. Genomics 265:58-65. [DOI] [PubMed] [Google Scholar]
- 2.Bessereau, J. L., A. Wright, D. C. Williams, K. Schuske, M. W. Davis, and E. M. Jorgensen. 2001. Mobilization of a Drosophila transposon in the Caenorhabditis elegans germ line. Nature 413:70-74. [DOI] [PubMed] [Google Scholar]
- 3.Bhasin, A., I. Y. Goryshin, M. Steiniger-White, D. York, and W. S. Reznikoff. 2000. Characterization of a Tn5 pre-cleavage synaptic complex. J. Mol. Biol. 302:49-63. [DOI] [PubMed] [Google Scholar]
- 4.Chandler, M., and J. Mahillon. 2002. Insertion sequences revisited, p. 305-366. In N. L. Craig et al. (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 5.Chodosh, L. A. 2000. UV cross-linking of proteins to nucleic acids, p. 12.5.1-12.5.6. In F. M. Ausubel et al. (ed.), Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 6.Cui, Z., A. M. Geurts, G. Liu, C. D. Kaufman, and P. B. Hackett. 2002. Structure-function analysis of the inverted terminal repeats of the Sleeping Beauty transposon. J. Mol. Biol. 318:1221-1235. [DOI] [PubMed] [Google Scholar]
- 7.Darling, P. J., J. M. Holt, and G. K. Ackers. 2000. Coupled energetics of λ cro repressor self-assembly and site-specific DNA operator binding. II. Cooperative interactions of cro dimers. J. Mol. Biol. 302:625-638. [DOI] [PubMed] [Google Scholar]
- 8.Davies, D. R., I. Y. Goryshin, W. S. Reznikoff, and I. Rayment. 2000. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289:77-85. [DOI] [PubMed] [Google Scholar]
- 9.Dawson, A., and D. J. Finnegan. 2003. Excision of the Drosophila mariner transposon Mos1: comparison with bacterial transposition and V(D)J recombination. Mol. Cell 11:225-235. [DOI] [PubMed] [Google Scholar]
- 10.Fischer, S. E. J., E. Wienholds, and R. H. A. Plasterk. 2001. Regulated transposition of a fish transposon in the mouse germ line. Proc. Natl. Acad. Sci. USA 98:6759-6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorbunova, V., and A. A. Levy. 1997. Circularized Ac/Ds transposons: formation, structure and fate. Genetics 145:1161-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haniford, D. B. 2002. Transposon Tn10, p. 457-483. In N. L. Craig et al. (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 13.Izsvak, Z., D. Khare, J. Behlke, U. Heinemann, R. H. Plasterk, and Z. Ivics. 2002. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J. Biol. Chem. 277:34581-34588. [DOI] [PubMed] [Google Scholar]
- 14.Izsvak, Z., E. E. Stüwe, D. Fiedler, A. Katzer, P. A. Jeggo, and Z. Ivics. 2004. Healing the wounds inflicted by Sleeping Beauty transposition by double-strand break repair in mammalian somatic cells. Mol. Cell 13:279-290. [DOI] [PubMed] [Google Scholar]
- 15.Lampe, D. J., M. E. A. Churchill, and H. M. Robertson. 1996. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 15:5470-5479. [PMC free article] [PubMed] [Google Scholar]
- 16.Lampe, D. J., T. E. Grant, and H. M. Robertson. 1998. Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics 149:179-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lefevre, K. R., and M. H. Cordes. 2003. Retroevolution of lambda Cro toward a stable monomer. Proc. Natl. Acad. Sci. USA 100:2345-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipkow, K., N. Buisine, D. Lampe, and R. Chalmers. 2004. Early intermediates of mariner transposition: catalysis without synapsis of the transposon ends suggests a novel architecture of the synaptic complex. Mol. Cell. Biol. 24:8301-8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizuuchi, M., T. A. Baker, and K. Mizuuchi. 1992. Assembly of the active form of the transposase-Mu DNA complex: a critical control point in Mu transposition. Cell 70:303-311. [DOI] [PubMed] [Google Scholar]
- 20.Mundy, C. L., N. Patenge, A. G. W. Matthews, and M. A. Oettinger. 2002. Assembly of the RAG1/RAG2 synaptic complex. Mol. Cell. Biol. 22:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Normand, C., G. Duval-Valentin, L. Haren, and M. Chandler. 2001. The terminal inverted repeats of IS911: requirements for synaptic complex assembly and activity. J. Mol. Biol. 308:853-871. [DOI] [PubMed] [Google Scholar]
- 22.Plasterk, R. H. A., and H. G. A. M. Van Luenen. 2002. The Tc1/mariner family of transposable elements, p. 519-532. In N. L. Craig et al. (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 23.Radice, A. D., and S. W. Emmons. 1993. Extra chromosomal circular copies of the transposon Tc1. Nucleic Acids Res. 21:2663-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reményi, A., A. Tomilin, E. Pohl, K. Lins, A. Philippsen, R. Reinhold, H. R. Schöler, and M. Wilmanns. 2001. Differential dimer activities of the transcription factor Oct-1 by DNA induced interface swapping. Mol. Cell 8:569-580. [DOI] [PubMed] [Google Scholar]
- 25.Reznikoff, W. S. 2002. Tn5 transposition, p. 403-422. In N. L. Craig et al. (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 26.Rhee, J. M., C. A. Gruber, M. Trieu, and E. E. Turner. 1998. Highly cooperative homodimerization is a conserved property of neural POU proteins. J. Biol. Chem. 273:34169-34205. [DOI] [PubMed] [Google Scholar]
- 27.Robertson, H. M. 1995. The Tc1-mariner superfamily of transposons in animals. J. Insect Physiol. 41:99-105. [Google Scholar]
- 28.Sakai, J., R. M. Chalmers, and N. Kleckner. 1995. Identification and characterization of a pre-cleavage synaptic complex that is an early intermediate in Tn10 transposition. EMBO J. 14:4374-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tosi, L. R. O., and S. M. Beverley. 2000. cis and trans factors affecting Mos1 mariner evolution and transposition in vitro, and its potential for functional genomics. Nucleic Acids Res. 28:784-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vos, J. C., and R. H. Plasterk. 1994. Tc1 transposase of Caenorhabditis elegans is an endonuclease with a bipartite DNA binding domain. EMBO J. 13:6125-6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, L., A. Dawson, and D. J. Finnegan. 2001. DNA binding activity and subunit interactions of the mariner transposase. Nucleic Acids Res. 29:3566-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]