Summary
Patient-derived xenografts (PDX) remain valuable models for understanding the biology and for developing novel therapeutics. To expand current PDX models of childhood leukemia, we have developed new PDX models from Hispanic patients, a subgroup with a poorer overall outcome. Of 117 primary leukemia samples obtained, successful engraftment and serial passage in mice were achieved in 82 samples (70%). Hispanic patient samples engrafted at a rate (51/73, 70%) that was similar to non-Hispanic patient samples (31/45, 70%). With a new algorithm to remove mouse contamination in multi-omics datasets including methylation data, we found PDX models faithfully reflected somatic mutations, copy-number alterations, RNA expression, gene fusions, whole-genome methylation patterns, and immunophenotypes found in primary tumor (PT) samples in the first 50 reported here. This cohort of characterized PDX childhood leukemias represents a valuable resource in that germline DNA sequencing has allowed the unambiguous determination of somatic mutations in both PT and PDX.
Subject areas: Cancer, Omics, Transcriptomics
Graphical abstract
Highlights
-
•
Patient-derived xenografts (PDXs) are important preclinical models
-
•
50 pediatric leukemia PDXs are largely derived from Hispanic ethnicity
-
•
Paired patient tumor, PDX, and germline for sequencing analysis
-
•
This cohort uncovers genomic, transcriptomic, and epigenomic features
Cancer; Omics; Transcriptomics
Introduction
Acute leukemia, the most common malignancy in children, accounts for over 25% of all childhood cancers.1 In recent years, significant changes to leukemia management, including improved risk stratification based on molecular and genetic alterations,2,3 new therapeutic approaches,4,5,6,7,8,9 and new comprehensive supportive care measures,10 have improved outcomes for children with lymphoblastic and myeloid leukemias. Cure rates for pediatric leukemias have substantially increased over the past four decades, particularly for those with acute lymphoblastic leukemia (ALL), 90% of whom are predicted to become long-term survivors.11 While acute myeloid leukemia (AML) survival in pediatrics is lower than that of ALL, survival approaching 70% also indicates progress.2
There are still opportunities to improve ALL and AML therapy. A considerable proportion of children with leukemia will not achieve a sustained remission, highlighted by the fact that leukemia remains the second leading cause of childhood cancer deaths.12 It is also recognized that children of Hispanic ethnicity have both a higher incidence and poorer outcome for leukemia when compared to non-Hispanic patients.13,14,15,16,17 Current approaches to therapy have likely reached their maximum benefit. In AML, for instance, the heterogeneity of the leukemia itself has limited biology-driven advances in therapy, and the ability to further intensify cytotoxic chemotherapy has likely been reached.18 Finally, survivors of pediatric leukemia often suffer from long-term chronic health conditions such as osteonecrosis, cardiotoxicity, and peripheral neuropathy due to cytotoxic drug-based therapies. Further advancements to improve outcomes for the pediatric age group are likely to require more biology-based molecular targeted therapy, and this is particularly true for Hispanic patients where few preclinical model systems exist to identify potential therapeutic targets.
Patient-derived xenograft (PDX) models represent a valuable tool to understand tumor evolution and to identify and develop novel therapeutics. PDX models are created by implanting cancer cells or tissues from a patient’s primary tumor into an immunodeficient mouse, simulating human tumor biology in vivo.19,20 Currently, resources in the pediatric PDX community have two major shortcomings. First, most genomic studies in pediatric PDX models do not include germline DNA samples or patient primary tumor samples to delineate somatic from germline genetic alterations.21,22,23,24,25 Second, the majority of published PDX studies have only DNA and RNA sequencing data and fail to explore the epigenomic landscape of either PDX models or matched primary tumors.21,22,23,24,25 Therefore, a comprehensive multi-omics analysis of PDX models including both germline DNA and epigenomics studies are in an urgent need.
An additional prevalent challenge for the PDX field is the lack of proper analysis method to remove mouse contamination from the multi-omics datasets of PDX models, especially DNA methylation data. Resected human PDX samples can exhibit a substantial presence of mouse DNA or RNA, reaching levels as high as 70–80% due to infiltration by murine stromal cells.26 Consequently, contamination from mouse reads could emerge as a critical source of errors during PDX sequencing data analysis. The majority of existing algorithmic frameworks tailored to alleviate mouse contaminant reads from human PDX samples have been designed to accommodate solely DNA sequencing data (e.g., MAPEX algorithm27), or RNA sequencing data (e.g., Xenome algorithm28). Though certain multi-omics solutions, such as XenofilteR,29 have sought to address both DNA and RNA-based contamination, but no published algorithm possess the capacity to effectively remove mouse contaminant from the three major next-generation sequencing (NGS) data modalities: DNA, RNA, and methylation-based sequencing data.
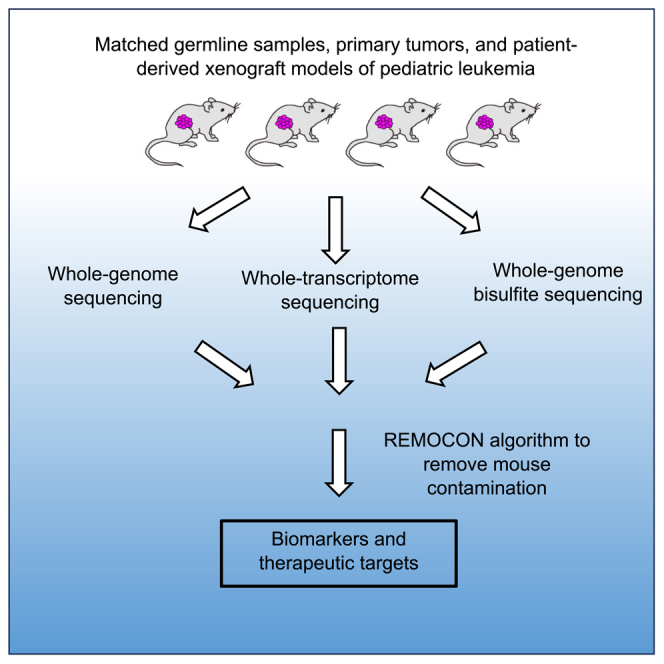
In our cohort described here, we applied whole-exome, whole-transcriptome, and whole-genome bisulfite sequencing (WGBS) technologies to conduct genomic, transcriptomic, and epigenomic analysis on matched germline samples, primary leukemia and PDX models from 50 patients with pediatric leukemia, as well as developed a new algorithm, REMOCON, to remove mouse contaminant from all three major NGS data modalities including genomic, transcriptomic, and epigenomic data. This study represents a new resource of PDX models from pediatric patients with precursor B-cell and T cell ALL, AML, and mixed phenotype ALL samples, including the high fidelity of genomic, transcriptomic and epigenomic landscape of matched leukemia and PDXs from the same patients, as well as genetic features that correlate with PDX growth.
Results
Leukemia patient-derived xenografts were primarily generated from a population enriched for Hispanic ethnicity
Patients with pediatric leukemia with active leukemia were approached for specimen collection at three institutions in Texas. Patients and families were consented and leukemia-containing samples (either bone marrow or peripheral blood) were collected, processed and injected into immunodeficient mice at one institution. Overall, 117 patient samples were injected, and 82 of them successfully engrafted. In this study, the initial set of 50 PDXs with matched germline and primary leukemia samples underwent comprehensive multi-omics sequencing, making them the focus of our analysis. It is important to note that these 50 samples was based solely on their availability and did not involve any other influencing factors. The composition of these 50 PDXs includes 19 cases of standard risk preB ALL, 19 cases of high-risk preB ALL, 6 cases of AML, 5 cases of T cell ALL, and 1 case of mixed phenotype ALL. Whole-exome, whole-transcriptome, and whole-genome bisulfate sequencing were conducted for this cohort. Patient characteristics of these 50 PDXs are highlighted in Table 1, including age at diagnosis, sex, and ethnicity. More detailed demographic and clinical information for each of the pediatric leukemia samples are summarized in Table S1, including initial white blood cell count, end of induction minimal residual disease (MRD) status, clinically relevant cytogenetics, immunophenotype, and relapse status.
Table 1.
Demographic information for the patient leukemia samples utilized for the generation of the patient derived xenografts including age at diagnosis, gender, race, and ethnicity
| Diagnosis | Number of PDXs | Age at diagnosis median in years (range) | Number of female (percentage) | Race (number of white) | Ethnicity | Relapse/refractory |
|---|---|---|---|---|---|---|
| Standard Risk preB ALL | 19 | 4 (1–8) | 10 (52%) | 18 | 15 (79%) | 1 (5%) |
| High Risk preB ALL | 19 | 14 (0–25) | 7 (37%) | 16 | 13 (68%) | 5 (25%) |
| Acute Myeloid Leukemia | 6 | 9 (0.5–10) | 4 (67%) | 6 | 4 (67%) | 4 (67%) |
| T cell Leukemia/Lymphoma | 5 | 10 (4–15) | 1 (20%) | 5 | 4 (80%) | 4 (80%) |
| Mixed Phenotype Acute Leukemia | 1 | 0.67 | 1 (100%) | 1 | 0 | 1 (100%) |
The final column records the percentage of patients who ultimately suffered refractory or relapsed disease.
This PDX cohort is primarily derived from patients with preB cell ALL with an equal distribution between patients designated as standard risk (SR) and high risk (HR) at diagnosis based on the age of the patient and initial total white blood cell count. The percentage of patients with established PDXs who ultimately experienced relapsed/refractory disease was similar to that generally observed in preB ALL (5% for SR-ALL, 25% for HR-ALL), but rates higher than clinically reported30 were noted in patients with AML and T cell ALL (67% and 80% respectively) than (Table 1). Therefore, this collection of PDX models, particularly for AML and T cell ALL, represent a unique resource for relapsed or refractory pediatric leukemia studies.
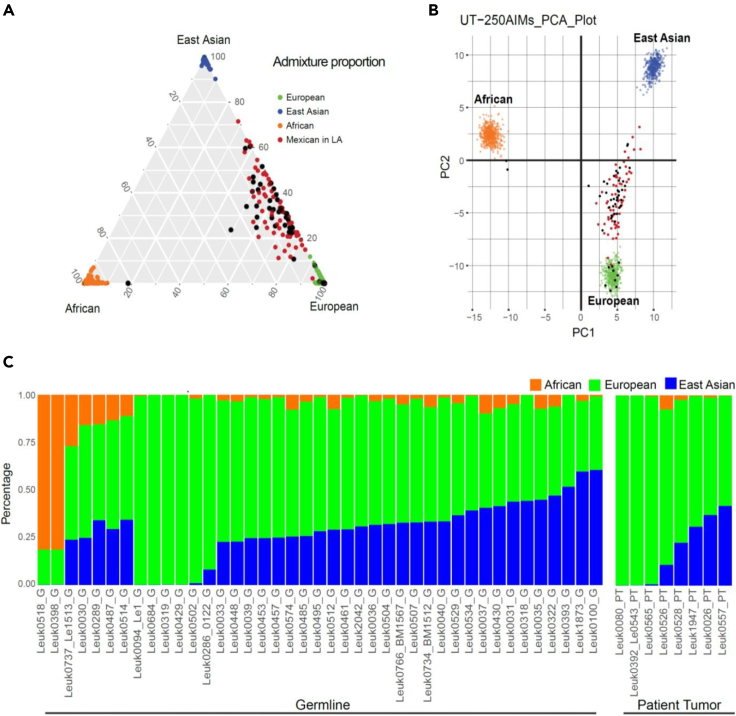
In addition, consistent with the evolving demographics of the population in Texas, the majority of leukemia PDX models in this study were derived from leukemia samples collected from children of Hispanic ethnicity. We previously developed an ancestry informative marker (AIM) panel, called UT-AIM250,31 to infer 3-way genetic admixture from three distinct continental populations (African (AFR), European (EUR), and East Asian (EAS)). In order to validate self-reported ethnicity information in this study, we utilized this marker panel to confirm the Mexican ancestry for the majority of leukemia samples from our cohort (Figure 1).
Figure 1.
Determination of ancestral admixture
(A) Two hundred and fifty (250) ancestral informative markers (UT-AIM250 panels) were extracted from WES to infer 3-way genetic admixtures of 49 samples (41 from germline DNAs, and 8 from tumor DNAs) and plotted as black dots in triangle plots.
(B) PCA plots with 250 genotypes directly. Three continental populations (European, African, and East Asian from the 1000 genome project) are plotted in green, orange, and blue dots, respectively. Samples from Mexicans in LA (also part of the 1000 Genome Project) were plotted in red dots, overlapping with Texas patients in this study.
(C) Bar chart of 49 patients with estimated proportion to each continental population. UT-AIM250 is capable to estimate admixture proportion from 8 tumor samples where germline DNAs were not available.
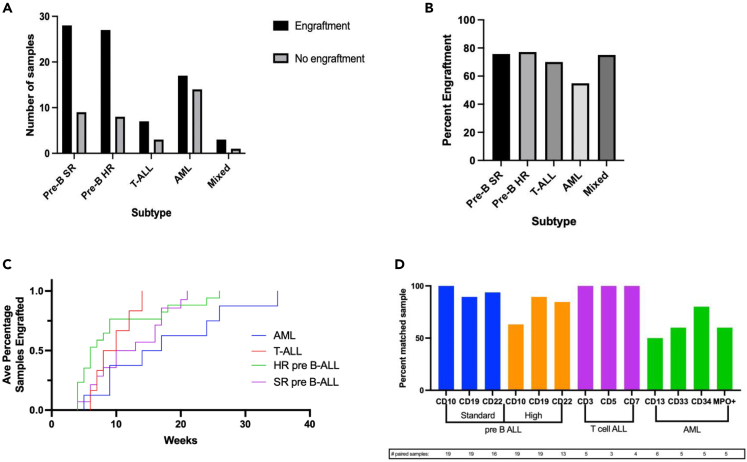
Leukemia patient-derived xenograft models show variability in time to engraftment
Across three different centers, a total of 117 primary leukemia samples were collected for PDX development. The majority of primary leukemia samples collected and injected ultimately engrafted with an overall efficiency of 70%. Figure 2A illustrates the total number of primary leukemias samples of each subtype that were collected and injected and their engraftment status. The percentage of primary leukemia samples that engrafted is presented by subtype in Figure 2B. AML had the poorest overall engraftment efficiency (54.8%) while preB ALL had the highest and showed no statistical difference between SR and HR (75.7% and 77.1% respectively). Because samples were collected at three separate centers in Texas but processed and injected into mice at one site (UT Health San Antonio), we assessed whether time from collection to processing for injection influenced engraftment efficiency. At one distant site (UT Southwestern), samples were collected and shipped with a median time from collection to processing of 2.29 days (range 2–6 days). The vast majority of samples collected at two sites in San Antonio were processed for injection on the day of collection. Delay from collection to injection did not measurably influence engraftment as samples injected in <24 h and those injected at > 24 h engrafted at similar rates [58/79 (73%) versus 24/38 (63%), two-way ANOVA test, p = 0.39]. Delays of >72 h from collection to injection engrafted at similar rates (11/16, 68.8%). Samples from the distant site engrafted at the same overall rate as samples collected at the local site indicating no significant impact of time to injection [40/63 (63%) versus 42/54 (77%), two-way ANOVA test, p = 0.81]. The mean time to engraftment across all 38 preB-cell ALL models was 10.7 weeks (range 4–26), which was similar to 9.5 weeks (range 6–14) for engraftment in T cell ALL models (Figure 2C). AML PDX models displayed an average of 16.5 weeks (range 5–35) to engraft, which was significantly longer than the other the ALL subtypes (Figure 2C). Individual PDX growth and engraftment can be found in Figure S1.
Figure 2.
Leukemia PDX models show variability in time to engraftment and cell surface markers
(A) Number of total leukemia samples collected for PDX collection by subtype detailing engraftment (black) versus no engraftment (gray) after implantation.
(B) Percentage of engraftment samples for each leukemia subtype.
(C) Engraftment rates of leukemia subtypes.
(D) Percentage of maintained cell surface markers on the primary leukemia with its paired patient derived xenograft. Total number of paired samples analyzed for that particular cell surface marker noted later in discussion.
Patient-derived xenograft acute lymphoblastic leukemia models preserved the immunophenotypes in the patient leukemia specimens
Whether PDX models preserve biological features of primary tumor samples is a key issue and sets the foundation for their potential usefulness in preclinical studies.32,33,34 To assess the fidelity between the PDX and the patient’s primary leukemia, we compared the cell surface markers in the engrafted PDX to the paired primary leukemia diagnostic specimen assessed by flow cytometry. Figure 2D shows the percentage of PDX/primary leukemia samples that expressed selected cluster of differentiation (CD) markers. Post-engraftment comparison of cell surface receptors of the PDX to the patient’s primary leukemia demonstrates excellent fidelity in standard risk preB ALL and T cell ALL with a higher proportion of discrepancies in the cell surface markers of AML models. A proportion of high risk preB ALL PDX models lost CD10 and CD19 expression which was present on the patient’s primary diagnostic sample. This contrasts standard risk preB ALL in which CD10 and CD19 expression was preserved. The loss of cell surface marker expression may be notable given the importance of tisagenlecleucel, a CAR-T therapy directed against CD19, for the treatment of refractory and relapsed preB ALL. It should be noted that not all cell surface markers were analyzed in the PDX models, and Figure 2D only represents samples where data was available for both PDX and primary samples. T cell PDX models maintained immunophenotype fidelity across all analyzed cell surface markers, which suggested little evolution between the patient’s primary leukemia and the engrafted PDX. AML models had poor immunophenotypic fidelity when compared to the diagnostic specimens, particularly in CD13 which was maintained in only half of the PDX models. Full immunophenotype information for the PDX models and primary leukemias can be found in Table S1.
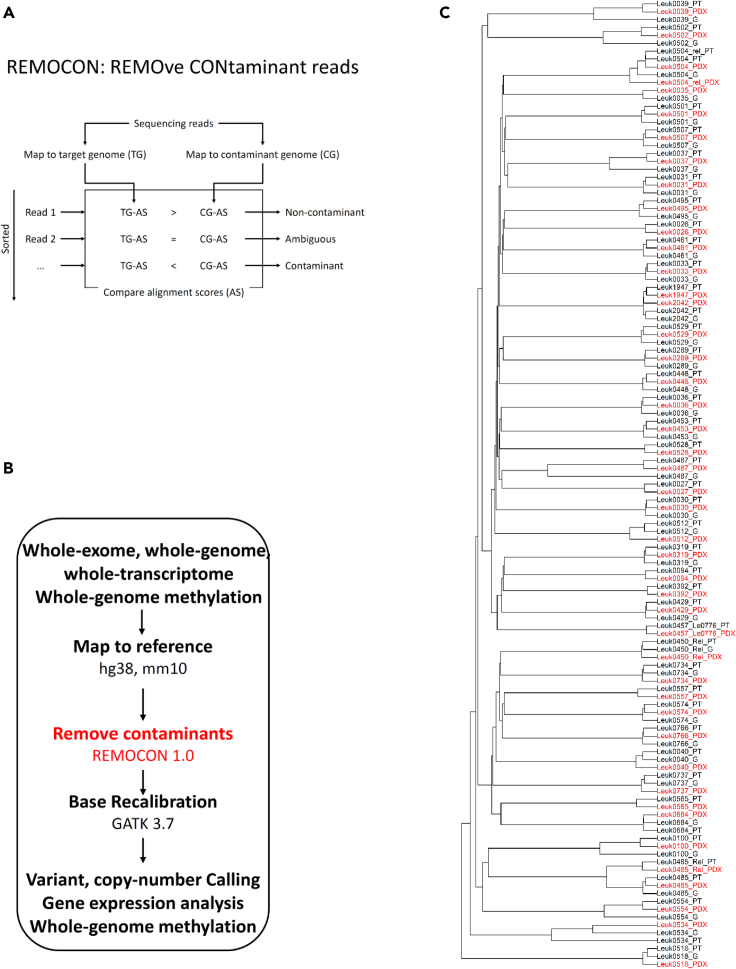
The REMOCON algorithm effectively removes contaminating mouse DNA reads
In addition to evaluating whether cell surface markers were preserved, we examined whether three PDX models maintain genomic, transcriptomic and epigenomic features of the primary leukemia samples. In order to accurately characterize the genomic, transcriptomic, and epigenomic landscape of these PDX models, we developed the REMOCON (REMOve CONtaminant reads) algorithm (Figures 3A and 3B) to remove mouse reads in the next-generation sequencing data.
Figure 3.
Development and application of REMOCON algorithm to analyze whole-exome, whole-transcriptome, and whole-genome methylation datasets from germline, leukemia, and PDX samples from the same patients with leukemia
(A) A schematic strategy for the REMOCON algorithm to remove contaminant reads, in which reads are mapped to the human genome (target genome; TG) and mouse genome (contaminant genome; CG). The read which alignment score (AS) to the mouse genome is greater than that to the human genome is defined as contaminant read.
(B) A Pipeline to analyze whole-exome, whole-genome, whole-transcriptome, and whole-genome methylation datasets in this cohort by removing contaminant reads.
(C) Applying whole-exome data to build phylogenetic relations among germline (G for short), primary leukemia (PT for short) and PDX samples from the same patients after REMOCON analysis. To facilitate visualization, all PDX samples are labeled red.
We directly compared whole-exome sequencing (WES) data before and after applying REMOCON. We focused on the unsupervised clustering based on WES data of paired primary leukemia and PDX samples. Before applying REMOCON (Figure S2), only 25 of 50 paired primary leukemia and PDX samples clustered together using an unsupervised approach. In contrast, after applying REMOCON, all 50 paired primary leukemia and PDX samples from the same patient did cluster together (Figure 3C) (Fisher exact test, p < 0.001), indicating that REMOCON effectively removes mouse contaminant reads. This step would be critical to reveal the biological patterns possibly masked by contaminating mouse nucleic acid sequencing reads.
In addition, we further conducted analyses to compare experimentally measured and REMOCON-estimated mouse contamination in the same PDX models. Firstly, we utilized lactate dehydrogenase (LDH) isoenzyme assays to experimentally estimate the level of mouse contamination in each of 50 PDX samples (see STAR Methods). Secondly, we employed REMOCON to independently calculate the level of mouse contamination in the same set of 50 PDX models. Because all 50 PDXs have WES data but other omics data are missing in some of 50 PDXs, we utilized WES data for estimating mouse contamination to maximize the number of samples available for REMOCON analysis. As shown in Figure S3A, we compared the REMOCON-estimated mouse contamination (y axis) with the experimentally measured mouse contamination (x axis). We observed a significantly positive correlation between these two measures (Pearson correlation coefficient r = 0.53, p-value = 0.0457). This correlation provides strong evidence supporting the reliability of REMOCON as a tool for detecting mouse contamination in PDX models. Furthermore, we also applied another published method for removing mouse contamination reads to conduct the same analysis (Figure S3B) and observed the same trend but less significant correlation (Pearson correlation coefficient r = 0.47, p-value = 0.1294).
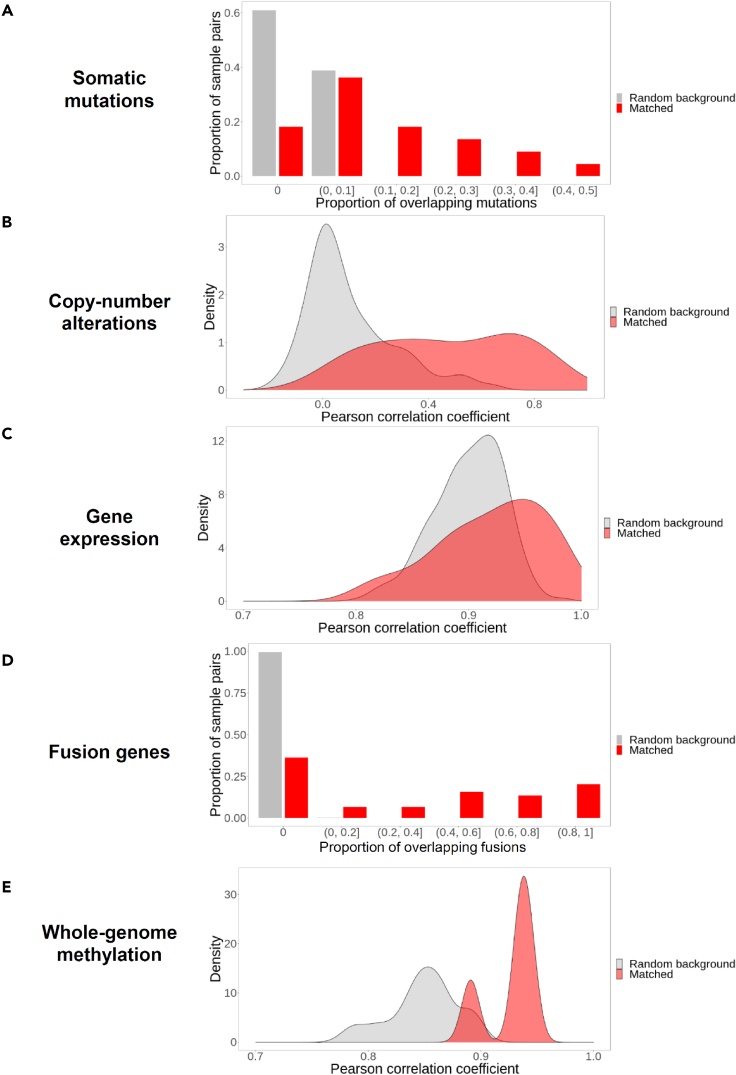
Gene alterations and the transcriptome are conserved in patient-derived xenografts models of leukemia
To continue exploring the conservation between matched pediatric leukemia samples and PDXs in our cohort, we compared somatic mutations (Figure 4A), copy-number variations (Figure 4B), gene expression (Figure 4C), presence of fusion genes (Figure 4D), and whole-genome methylation (Figure 4E) in matched leukemia-PDX pairs. In our analyses, we compared the matched leukemia-PDX pairs from the same patients (termed as “matched pairs” for short, red color in Figure 4) to the randomly chosen leukemia-PDX pairs taken from the different patients (termed as “random background” for short, gray color in Figure 4). To avoid trivial bias, we did not allow random matching between ALL and AML specimens.
Figure 4.
Conservation between matched pediatric leukemia samples and PDXs (red color) on genomic, transcriptomic, and epigenomic landscape in comparison with randomly chosen pairs of leukemia samples and PDXs (gray color)
This analysis contains mutation (A), copy-number alterations (B), gene expression (C), fusion genes (D), and genome-wide methylation states (E).
First of all, as shown in Figure 4A, for random background leukemia-PDX pairs defined above (gray color), as shown in the first gray bar from the left in Figure 4A, 61% of them (from y axis) do not share any somatic mutations (from x axis), and the rest 39% (from y axis) only have 0 to 10% (from x axis) somatic mutations shared between them (the second gray bar from the left in Figure 4A). None of the random background leukemia-PDX pairs have more than 10% somatic mutations shared between them, which is consistent with the lack of conservation between random chosen leukemia-PDX pairs from different patients. However, for all the matched leukemia-PDX pairs from the same patient, only 18% of them do not share somatic mutations (the first red bar from the left in Figure 4A), and 82% of them have shared somatic mutation(s) (the rest of red bars, Figure 4A). In summary, this analyses showed that 82% of matched leukemia-PDX pairs from the same patient shared somatic mutations as compared to only 39% of randomly chosen leukemia-PDX pairs (Wilcoxon rank-sum test, p < 1 X 10–15, Figure 4A), which supports a statistically significant conservation of somatic mutation landscape in matched leukemia-PDX pairs in comparison to random expectation.
Secondly, we used the same rationale to analyze the conservation of copy-number alterations between random background and matched pairs. We calculated the Pearson correlation coefficients of the gene copy-number alterations between leukemia samples and PDX samples, as shown in x axis in Figure 4B. 0 represents no correlation and 1 represents the highest correlation between leukemia samples and PDX samples. For random background leukemia-PDX pairs, the majority of leukemia-PDX pairs has no correlation (Pearson correlation coefficients equal to 0) on their gene copy-number alterations (gray color, Figure 4B). However, in the matched leukemia-PDX pairs, Pearson correlation coefficients are not centered on 0 (red color, Figure 4B) and is significantly higher than those in the random pairs (red vs. gray, Wilcoxon rank-sum test, p = 2.3 X 10-8) (Figure 4B), which supports a notable degree of similarity in the copy-number alterations among matched leukemia-PDX pairs in comparison to random expectation.
Thirdly, in order to explore the conservation of gene expression pattern, we calculated the Pearson correlation coefficients of the gene expression level between leukemia samples and PDX samples in random and matched pairs. We found that Pearson correlation coefficients of the gene expression is also higher in matched leukemia-PDX pairs than in the randomly chosen pairs (Wilcoxon rank-sum test, p = 0.0066) (Figure 4C), indicating conserved gene expression pattern in matched leukemia-PDX pairs from the same patients.
Fourthly, in order to explore the conservation of gene fusion pattern, we calculated the proportion of overlapping fusion genes between leukemia samples and PDX samples in random and matched pairs. We found that two-thirds of matched leukemia-PDX pairs showed expression of the same fusion genes, a finding does not present in any randomly chosen pairs (Wilcoxon rank-sum test, p < 1 X 10–15, Figure 4D). Lastly, we observed that the methylation level in all protein-coding genes’ promoter and coding regions was more similar in the tumor and PDX samples pairs (Wilcoxon rank-sum test, p = 0.0044) (Figure 4E).
To further display detailed transcriptome pattern of matched leukemia-PDX pairs, we focused on 715 known cancer genes reported in the COSMIC database35 to remove non-cancer transcription noise signals and then performed clustering analysis (Figure S4A). We observed that all the samples are divided into three major clusters (from left to right) that are enriched in AML, T cell leukemia, and preB ALL subtypes, respectively. We repeated the clustering analysis using 24 known pediatric leukemia genes (Figure S4B). We found that these 24 pediatric leukemia genes are clearly separated into two clusters (from top to bottom): the top-panel cluster (Figure S4B) represents a group of genes highly expressed in most high-risk preB ALL samples and a small portion of standard risk preB ALL samples, while the bottom-panel cluster (Figure S4B) represents genes highly expressed in most of standard risk preB ALL and AML samples. These genes suggest that these known cancer genes, especially known pediatric leukemia genes, have the potential as biomarkers to separate pediatric leukemia subtypes.
In summary, we observed that matched leukemia-PDX pairs from the same patients (red color, Figure 4) have a significantly higher correlation than random expectation (gray color, Figure 4), suggesting that in our cohort the PDX samples preserve most of the genomic, transcriptomic, and epigenomic features from matched pediatric leukemia samples and therefore can faithfully model the disease.
Landscape of somatic mutations, homozygous copy-number deletions, and loss of heterogeneity in matched pediatric leukemia samples and patient-derived xenografts
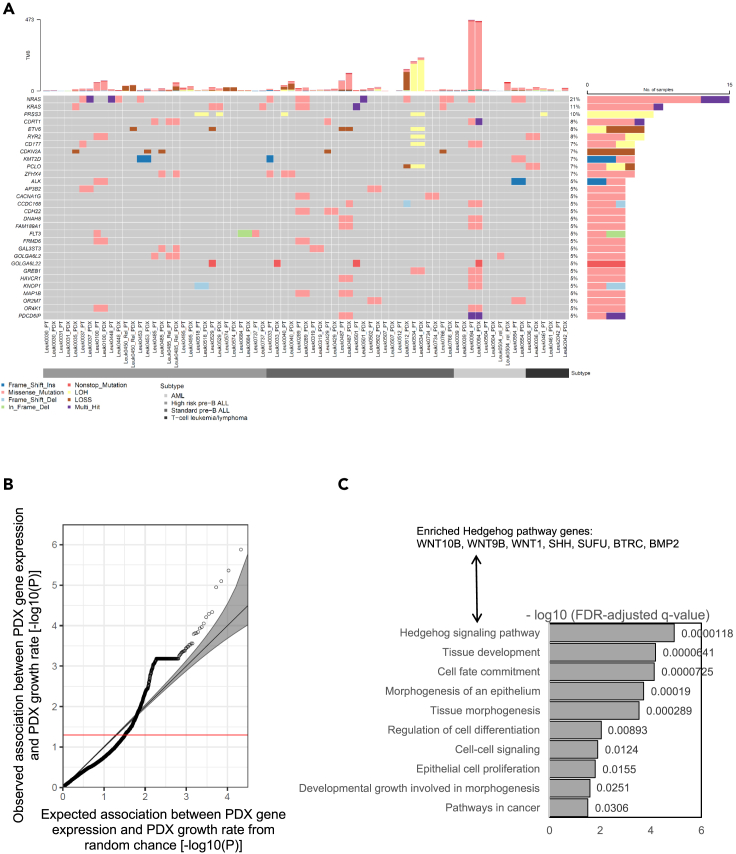
After confirming the general concordance between patient and PDX leukemia specimens, we next defined the somatic mutations, homozygous gene copy-number deletions, and loss of heterogeneity (LOH) from whole-exome sequencing data. Figure 5A describes the top 30 genes that showed alterations in at least 5% of samples in our collection. We also clustered samples based on subtypes highlighted in Table 1. In summary, NRAS harbored the most frequently recurrent alterations (21% of samples) followed by KRAS (11% of samples), both of which are known oncogenes in childhood AML36 and ALL.37,38 Besides these two established oncogenes, our analysis identified multiple known pediatric leukemia driver events, including deletion and LOH of ETV639 (8% of samples), deletion of CDKN2A40 (7% of samples), as well as frameshift mutations and missense mutations in KMT2D41 (6% of samples). We also manually collected a list of previously reported fusion genes in pediatric leukemia and compared them with the fusion genes detected in our cohort. We identified two previously reported pediatric leukemia fusion genes in our cohort: ETV6-RUNX1 fusion42 in one PDX model (UHS0487 in Table S1), and TCF3-PBX1 fusion43,44 in two PDX models (UHS0518 and UHS0528 in Table S1).
Figure 5.
PDX mutation landscape and phenotype-transcriptome association
(A) Mutation, homozygous copy-number deletion (LOSS), and loss of heterogeneity (LOH) landscape of the top 30 mutated genes in matched pediatric leukemia samples and PDXs from the same patients. TMB, total mutational burden, which is defined as the total number of somatic mutations per coding area of a genome.
(B) The quantile-quantile (Q-Q) plot of the association test between PDX gene expression and PDX growth rate. Each circle in the plot represents one gene. The expected p value in a Q-Q plot is calculated as the following way: If no gene is associated with the trait (growth rate here), the p values we get from tests should follow a uniform (0,1) distribution. Suppose there are n genes in the plot, and the observed p values are sorted from smallest to largest. The expected p value for the i th gene is i/n.
(C) Pathway enrichment analysis for PDX genes whose expression significantly associated with PDX growth rate in (A) (FDR-adjusted p < 0.05).
In order to utilize this new PDX resource (Figure 1) to study Hispanic-specific genetic alterations in pediatric leukemia, we re-analyzed the above mutations and copy-number pattern in Hispanic and non-Hispanic groups independently (Figure S5). Among the 30 most frequent mutations and copy-number alterations in our cohort, 12 found only in PDXs from the Hispanic population included deletion and LOH of ETV6 and mutations in KMT2D (Figure S5). However, due to a small sample size the enrichment of these mutations in leukemias from the Hispanic population was not statistically significant (Fisher exact test, p > 0.05). A re-examination on Hispanic-specific mutations and copy-number alterations in pediatric leukemia is warranted in our future study with a larger cohort.
Association between patient-derived xenograft gene expression and patient-derived xenograft growth rate
In order to elucidate factors that might influence PDX growth, we sought genes in which a positive correlation between PDX gene expression and PDX growth rate (i.e., elevated PDX gene expression correlated with increased PDX growth, or decreased PDX gene expression correlated with decreased PDX growth). In studying all 50 PDX models, we identified 201 genes (FDR-adjusted p < 0.05) whose expression positively correlated with PDX growth (Figure 5B). The ALL- or AML-specific subsets lacked statistical power to detect associated genes because of the small sample size. Furthering this analysis, we identified that multiple GO or KEGG pathways significantly correlated with PDX growth rate (Figure 5C). The hedgehog pathway, known for regulating cell proliferation and tumor growth,45,46 had the most significant enrichment (FDR-adjusted q-value = 1.18 X 10−5); seven genes overlapping with the KEGG hedgehog pathway term were identified to significantly associate with PDX growth rate (Figure 5C). The contributions of the Hedgehog signaling pathway, and other identified pathways such as cell-cell signaling and cell fate commitment (Figure 5C), to leukemia PDX engraftment and growth will need further functional evaluation in these models.
Discussion
Novel pediatric PDX models are a critical research resource to evaluate the efficacy of new therapies. Pediatric PDX models which mimic the disease heterogeneity and patient ethnic diversity seen clinically have to date been limited. We report in this study on a comprehensive resource of whole-exome, whole-transcriptome, and whole-genome methylation sequencing datasets characterizing 50 new pediatric leukemia PDX models (Table S1). All PDX models and related next-generation sequencing (NGS) datasets in our study are freely available for the cancer research community, as described in the Data Availability section. This large-scale PDX resource provides further tools for the pediatric leukemia research community. By establishing a cohort of pediatric leukemia PDXs covering multiple subtypes, these PDX models can complement other pediatric leukemia PDXs20,47 and more accurately recapitulate the underlying etiology of pediatric leukemia and assist in efficient drug development.
It is established that the risk of leukemia in Hispanic children is higher than in the overall population, and these patients have poorer outcomes.16,17 The majority of the leukemia samples collected in this cohort were from Hispanic patients and PDX development was equivalent in samples from Hispanic children compared to non-Hispanic patients. In the cohort of 50 analyzed here, the majority of PDX were from patients of Hispanic ethnicity. For standard risk 15 of 19 PDXs established (79%) and for high-risk preB ALL 13 of 19 (68%) were derived from Hispanic patients. Similarly, 4 of 6 AML and 4 of 5 T cell ALL were samples from Hispanic patients, although the sample number is low (n = 6 and 5, respectively). This cohort represents a novel, Hispanic-predominant set.
Our work also includes the development of a competitive bioinformatics algorithm, REMOCON, to remove mouse contaminant reads from DNA-based, RNA-based, and methylation-based sequencing data, which is also freely available. Given PDX samples can harbor up to 70–80% mouse DNA or RNA due to the infiltration of murine stromal cells,26 mouse read contamination is an important source of errors in PDX sequencing data analysis and needs to be addressed prior to downstream analyses. Existing algorithm packages to remove mouse contaminant reads from human PDX samples have focused on either DNA sequencing data (e.g., MAPEX algorithm27) or RNA sequencing data (e.g., Xenome algorithm28), or both (e.g., XenofilteR29). However, these previously published algorithms cannot process mouse contaminant reads from all three major NGS data types, including DNA-based, RNA-based, or methylation-based sequencing data. We demonstrated that REMOCON recovers the correct biological patterns of leukemia PDX models masked by mouse contaminant reads. In addition, we demonstrated a faithful recapitulation of pediatric leukemia disease in these PDX models through analysis of somatic mutations, copy number alterations, RNA expression, gene fusions, and whole-genome methylation patterns.
Recently Ben-David et al.48 reported that PDX copy number patterns display significant divergence from the primary tumors that these PDXs originate from, and therefore questioned whether genetic evolution in PDXs can reflect the genomic conservation of primary tumors of the same origin, or as a consequence of mouse-specific selective pressures. However, other studies have not confirmed this report.49,50,51,52 This is an important debate because the conclusion could impact the capacity of PDXs to faithfully model patient treatment response. In our PDX samples, we demonstrate significant fidelity in somatic mutations, copy number alterations, RNA expression, gene fusions, and whole-genome methylation patterns from matched leukemia samples, suggesting the high fidelity of PDX samples in our cohort.
Limitations of the study
First of all, the majority of our PDX models are derived from patients with B cell ALL with fewer T cell and AML models generated, limiting the power of our genomic analyses in AML and T cell ALL more globally. We are continuing to expand this valuable leukemia PDX resource to include more T cell and AML models. The ability to generate patient derived xenografts from multiple centers spread across a significant geographic distance is critical in rare diseases such as pediatric cancer. The breakdown of our sample collection is reflective of the incidence of pediatric leukemia but is unique in the focus on patients of Hispanic ethnicity. Second, the role of genomic DNA methylation in the development of pediatric cancer is increasingly notable.53,54,55 However, it is cost prohibitive in large scale studies at this time. Our future endeavors include covering more samples in our cohort with multi-omics analysis including genomic DNA methylation sequencing. Third, we have observed the loss of certain surface markers (e.g., CD13) in the PDX models. However, the timeline of this shift in immunophenotype remains obscure. Further investigation on the time course of immunophenotype drift could provide valuable information to help understanding the related limitations and guide how to use these PDX models accordingly. Our forthcoming research endeavors are poised to delve comprehensively into this facet, thereby advancing our understanding of this immunophenotype drift process.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-mouse CD45-FITC antibody | BD Biosciences, San Jose, CA, USA | Cat. No. 103108 |
| anti-human CD45-APC antibody | BD Biosciences, San Jose, CA, USA | Cat. No. 20-0459 |
| Biological samples | ||
| Patient-derived xenografts (PDX) | University of Texas Southwestern Medical Center/Children’s Health (Dallas, TX), the Greehey Children’s Cancer Research Institute UT Health San Antonio (San Antonio, TX), or Methodist Children’s Hospital (San Antonio, TX) | https://datacommons.swmed.edu/cce/ppdxe/ |
| Critical commercial assays | ||
| lactate dehydrogenase (LDH) isoenzyme assay | Helena Laboratories | Cat. No. 3538T |
| KAPA HyperPrep kit | Roche | Cat#5190-6210 |
| TruSeq mRNA Stranded Library Prep Kit | Illumina | Cat#20020595 |
| Deposited data | ||
| Raw WES and RNAseq data | This paper | EGAS00001006710 |
| PDX information | This paper | https://datacommons.swmed.edu/cce/ppdxe/ |
| Software and algorithms | ||
| REMOCON algorithm | This paper | https://github.com/jiwoongbio/REMOCON |
| Burrows-Wheeler Aligner (BWA, v0.7.17) | Li et al.56 | https://github.com/lh3/bwa |
| Picard (2.21.3) | NA | https://broadinstitute.github.io/picard |
| Genome Analysis Toolkit (GATK, 4.1.4.0) | McKenna et al.57 | https://gatk.broadinstitute.org/ |
| TopHat package | Kim et al.58 | http://ccb.jhu.edu/software/tophat/index.shtml |
| DESeq2 R Bioconductor package | Gentleman et al.59 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| CN-fusion | NA | https://github.com/jiwoongbio/CN-fusion |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contacts Peter J. Houghton (HoughtonP@uthscsa.edu).
Materials availability
This study did not generate unique reagents and is not part of a clinical trial.
Experimental model and study participant details
Newly diagnosed or relapsed patient with leukemia younger than 26 years at the time of presentation were eligible and approached for enrollment on IRB approved institutional biobanking protocols. Informed consent was obtained prior to any study procedures including specimen collection. Patient specimens and demographic information were collected for consented patients at either the University of Texas Southwestern Medical Center/Children’s Health (Dallas, TX), the Greehey Children’s Cancer Research Institute UT Health San Antonio (San Antonio, TX), or Methodist Children’s Hospital (San Antonio, TX). Risk stratification and diagnosis were based on National Cancer Institute (NCI) criteria60 and World Health Organization (WHO) classification.61 Peripheral blood and bone marrow specimens of patients with active disease were collected in antibiotic containing RPMI 1640 Medium and stored at 4°C until implantation. Samples collected at UTSW/Children’s Health were shipped overnight to UTHSCSA using cold shipping containers to preserve specimens. DNA and RNA were extracted from Ficoll processed whole blood and bone marrow samples using Qiagen DNeasy Blood and Tissue Kit (Cat #69504) or Qiagen AllPrep DNA/RNA/protein kits (Cat#80004) respectively. Saliva was processed for germline DNA using prepIT.L2P (DNA Genotek). Patient germline sample from either whole blood or saliva was collected once marrow remission was clinically confirmed by flow cytometry. Ethnicity information was obtained by self-report from patient family. In our geographic area those reporting ethnicity are primarily of Mexican ancestry.
Method details
Establishment of leukemia PDX models
Patient-derived xenografts (PDX) were generated as described62 with slight modifications. NOD.Cg-Prkdc Il2rg/SzJ (NSG) mice (Jackson Laboratory, Bar Harbor, ME, USA) were used to establish patient-derived leukemia xenografts. Bone marrow and peripheral blood samples were collected from patients who provided written informed consent. This study was approved by the Institutional Review Board and the Institutional Animal Care and Use Committees of UTSW and UTHSA.
Peripheral blood mononuclear cells were obtained through density gradient centrifugation (Histopaque®-1077 solution, Sigma-Aldrich, United Kingdom). Mononuclear cells (1 × 106 to 5 × 106) isolated from fresh BM or PB samples of primary leukemia patients were intravenously injected in tail of NSG mice. Assessment of engraftment of human leukemic cells in mice peripheral blood started from 3-4 weeks of implantation and analyzed over a period of 3-6 months. Time of being in the experiment depended on 1) the number and quality of viable cells, and 2) the type of leukemia injected in NSG mice. About 3-4 weeks after injection, leukemia progression was monitored in the peripheral blood of mice every 2-3 weeks by flow cytometry analyses using BD LSRFortessa X-20 Cell Analyzer (BD Biosciences, San Jose, CA, USA) with anti-mouse CD45-FITC and anti-human CD45-APC antibodies (BD Biosciences, San Jose, CA, USA). As shown in the Figure S1, we reported the percentage of human CD45+ cells from a flow cytometry analysis of a gated plot (hCD45+ vs. mCD45+). In addition, (hCD45+ vs. mCD45+) plot was determined from the live cells in the plot (viability vs. SSC), which preceded by doublets exclusion plots (FSC-A vs. FSC-H) and (FCS vs. SSC) respectively. We collected about 50 uL of blood (3 drops) from retro-orbital sinus with a sterile hematocrit capillary tube and under isoflurane anesthesia according Institutional IACUC protocol.
We sacrificed the mice with the carbon dioxide asphyxiation or by cervical dislocation at the first indication of morbidity (≥20% weight loss, lethargy, riffled fur) according the Institutional IACUC protocol and/or when the proportion of human CD45+ cells in the peripheral blood exceeded 50%. To make sure that the maximum of xenograft mononuclear cells would be collected, we harvested and purified them from bone marrow of femurs/tibias and spleen cells by density gradient centrifugation using Histopaque 1077.
Purified tumor cells had been collected and cryopreserved in freeze-down sterile solution of 90% FBS and 10% DMSO for later use. Cryovials were kept at -80°C freezer for 24 h in Nalgene Cryo 1°C Freezing Containers and then transferred to liquid nitrogen for long storage. Aliquots a small number of cells kept frozen and used for downstream analysis. Human cells enrichment in the xenograft samples were evaluated by flow cytometry using the same antibody panel as for patient samples. PDX samples were analyzed by flow cytometry post-engraftment for expression of cell surface markers and compared to the patient primary leukemia sample flow cytometry completed at diagnosis. Bone marrow and spleen from PDX samples were collected at the stage of mortality or the highest level of engraftment. Xenograft identity was verified by DNA fingerprinting by STR analysis performed at the McDermott Center Sequencing Core, UT Southwestern Medical Center, Dallas, TX.
Human/mouse LDH isozyme assay for testing of leukemia samples
With lactate dehydrogenase (LDH) isoenzyme assays (Helena Laboratories, Cat. No. 3538T), we quantitatively measured the levels of human/mouse lactate dehydrogenase (LDH) isoenzymes in the xenograft leukemia samples using agarose gel electrophoresis on the QuickGel Chamber from Helena Laboratories, Cat. No. 3538T. Lactate dehydrogenase (LDH) is a tetrameric enzyme that in vertebrates exists in five electrophoretically distinguishable forms known as isoenzymes (LD1-LD5). Each isoenzyme is designated by a number which is related to its electrophoretic mobility. LDH distribution patterns of mice and human are different. The isoenzymes of LDH have been determined by various methods. Electrophoresis provides far more information than the other methods because it allows complete separation of all five isoenzymes according to their electrophoretic mobility on agarose and visualization of the differences of patterns of distribution. After separation, each isoenzyme was detected colorimetrically: a tetrazolium salt is reduced with the formation of a colored formazan dye. A high quality scanning densitometer the QuickScan 2000 (Cat. No. 1660) was used to scan the gels for quantitative results with further analysis by ImageJ. A human control sample (HeLa, ATCC® CCL-2 cells) and mouse control sample (skeletal muscle tissue of normal mice) were included in each agarose gel. Fresh and frozen leukemia cells (1-5 X106) shows the same efficiency in this assay.
Flow cytometry analysis
To characterize human leukemia cells from mouse cells, multicolor panels were designed for flow cytometry analysis, and each fluorochrome-conjugated antibody was titrated for optimal staining.
Briefly, 50 μl of total peripheral blood or purified mononuclear cells (<106) were incubated 30 min at 4°C in the dark with fluorochrome labeled antibodies. The following monoclonal antibodies were used: FITC anti-human CD34, PE anti-human CD13, PE/Cy5 anti-human CD33, APC/Cyanine7 anti-human HLA-DR, PerCP anti-human CD61, PE Mouse Anti-Human Myeloperoxidase (MPO), PE/Cy5 anti-human CD3, PE anti-human CD5, PE/Cy7 anti-human CD7, PE/Cy7 anti-human CD10, APC/Cyanine7 anti-mouse CD19, PE anti-human CD22, Alexa Fluor® 700 anti-human CD34, Alexa Fluor® 700 anti-human CD38, Brilliant Violet 711™ anti-human CD117 (c-kit), FITC Myeloperoxidase Monoclonal Antibody (8E6), Alexa Fluor® 594 anti-human CD79a, PE/Cy5 anti-human CD11b. Purified Rat Anti-Mouse CD16/CD32 (Mouse BD Fc Block™) Clone 2.4G2 (BioLegend, San Diego, CA, USA). After incubation, cells were washed with 5 ml of PBS and centrifuged at 380 × g (4°C) for 5 min. Red blood cells were removed by incubation with 1 ml of RBC lysis buffer (BioLegend, San Diego, CA, USA). Finally, cells were washed in PBS, resuspended in flow cytometry staining buffer (FCSB) (eBioscience, San Diego, CA, USA), and immediately analyzed by flow cytometry analysis.
In the intracytoplasmic cell markers staining assay, Fixation/Permeabilization Working Solution (eBioscience, San Diego, CA, USA) was used. For flow cytometry acquisition (BD LSRFortessa X-20 Cell Analyzer (BD Biosciences, San Jose, CA, USA)), cells were resuspended in a final volume of 500 μl of FCSB. Data analysis was performed using FlowJo version 10.0.7 (Tree Star, CA). To define the best gating strategy to be applied, compensation was done with unstained, and “fluorescence minus one” (FMO) samples.
Patient leukemia cells underwent diagnostic flow cytometry evaluation as part of routine, standard of care. Flow cytometry results were collected and cell surface markers which were identified as >dim + were noted. Patient derived xenograft leukemia cells were isolated and underwent flow cytometry as above. Paired samples were assessed for clusters of differentiation (CD) cell surface markers which were in common. Not all paired samples underwent the same, full analysis. CD markers assessed in both patient derived and PDX samples were identified as positive (>dim or >10%) or negative (no expression or dim) and the percentage in common calculated.
REMOCON algorithm
In order to remove mouse contaminant reads, we developed an algorithm called REMOCON (short for REMOve CONtaminant reads) which is publicly accessible at https://github.com/jiwoongbio/REMOCON. REMOCON is a series of PERL Script to be able to implement existing alignment tools, including BWA (v0.7.17)56 and SAMtools (http://www.htslib.org). Detailed installation and implementation protocols are provided at https://github.com/jiwoongbio/REMOCON. REMOCON utilizes the differential mappability of mouse reads onto the human and mouse reference genomes. REMOCON removes reads that are mapped only to the mouse reference genome or mapped with higher confidence to the mouse than the human reference genome. Figure 3B displays our integrated analysis pipeline by leveraging REMOCON to remove mouse contaminant reads from DNA-based, RNA-based, and methylation-based sequencing data, and then performs downstream analyses, including but not limited to variant calling, copy-number calling, gene expression analysis, and whole-genome methylation measurements.
Whole exome sequencing and data analysis
DNA libraries for whole-exome sequencing was constructed using KAPA HyperPrep kit. Approximately 250-500ng genomic DNA were sheared with Covaris S220 Ultra Sonicator to an average of 200-400bp fragments for DNA-seq library preparation. Then DNA-seq libraries were quantified and pooled together to go through two rounds of hybridization to enrich the DNA fragments of exome regions by using IDT xGen Exome Research Panel (V1 and V2). The final library was amplified, quantified, and loaded for 100bp paired end sequencing with Genome Sequencing Facility at UTHSA. An average of 60M reads were generated per sample. Trim Galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) was used for quality and adapter trimming. The reference genome sequences of human (hg38) and mouse (mm10) were downloaded from Illumina iGenomes (https://support.illumina.com/sequencing/sequencing_software/igenome.html). The sequencing reads were aligned to human and mouse genome sequences using Burrows-Wheeler Aligner (BWA, v0.7.17),56 and contamination reads from mouse DNA were removed using REMOCON. Picard (2.21.3) (https://broadinstitute.github.io/picard) was used to remove PCR duplicates and Genome Analysis Toolkit (GATK, 4.1.4.0)57 was used to recalibrate base qualities. Calling variants and genotyping were performed using GATK HaplotypeCaller and low-quality variant calls were excluded by the following filtering thresholds: QD (Variant Confidence/Quality by Depth) < 2, FS (Phred-scaled p-value using Fisher's exact test to detect strand bias) > 60, MQ (RMS Mapping Quality) < 40, DP (Approximate read depth) < 3, GQ (Genotype Quality) < 7. A custom Perl script (https://github.com/jiwoongbio/Annomen) was used to annotate variants with RefSeq63 human transcripts and proteins, dbSNP (build 151),64 Genome Aggregation Database (gnomAD, r3.0),65 Catalogue Of Somatic Mutations In Cancer (COSMIC, v90)35 (cancer.sanger.ac.uk). Jaccard distances between samples were calculated by point mutations (i.e., single nucleotide variation) from WES data, and then used to generate hierarchical clustering of samples. Somatic mutations were identified by GATK Mutect2 and somatic copy number alterations were identified by DEFOR.66 Oncoplot was generated using maftools67 with somatic mutations selected by the following criteria: variant-supporting reads in tumor sample ≥3, variant allele frequency (VAF) in tumor sample ≥0.1, variant-supporting reads in normal sample <3, VAF in normal sample <0.1, and also (dbSNP allele frequency ≤0.01, or gnomAD allele frequency ≤0.01, or COSMIC occurrence ≥3). Loss-of-heterozygosity (LOH) variants selected by the following criteria: VAF in tumor sample ≥0.7 and gnomAD homozygous individuals ≤3. The clustering analysis was based on Jaccard distances among mutations identified from WES data.
RNA sequencing and data analysis
The quality of Total RNA was checked by Agilent Fragment Analyzer (Agilent Technologies, Santa Clara, CA), and only RNAs with RQN >7 were used for subsequent mRNA-seq library preparation and sequencing. Approximately 500ng Total RNA was used for RNA-seq library preparation by following the Illumina TruSeq stranded mRNA sample preparation guide. After RNA-seq libraries were subjected to quantification process, pooled for cBot amplification and subsequent 100bp paired read sequencing run with Illumina HiSeq 3000 platform. An average of 80M reads were obtained per sample. The reads were aligned to human and mouse transcript sequences using Bowtie (v2.3.4.3)68 within the TopHat package,58 and mouse contamination and ambiguous reads were removed using REMOCON. The quality of sample libraries and strand-specificity were estimated based on the alignments. SAMtools (v1.9) was employed to sort the alignments. HTSeq Python package69 was employed to count reads per gene and DESeq R Bioconductor package59 was used to normalize read. SpliceFisher70 (https://github.com/jiwoongbio/SpliceFisher) was used to identify differential alternative splicing events and calculate PSI (percent spliced-in) values. Fusion genes were identified by a custom Perl script (https://github.com/jiwoongbio/CN-fusion).
Whole genome bisulfite sequencing and data analysis
Whole genome bisulfite sequencing (WGBS) was done with Zymo Methylation-Gold Kit for DNA bisulfite conversion and SwiftBiosciences Accel-NGS® Methyl-Seq kit for library preparation, using approximately 100ng gDNA as starting material. After WGBS libraries were subjected to quantification process, pooled for cBot amplification and subsequent 100bp paired read sequencing run with Illumina HiSeq 3000 platform. An average of 250-300M reads were obtained per sample. The reads were aligned to human and mouse genome sequences using Bismark (v0.22.3),71 and REMOCON was used to calculate alignment scores and remove contamination reads. The alignments were deduplicated and the methylation sites were called using Bismark pipeline.
Quantification and statistical analysis
Copy number alterations and somatic mutations are discontinuous variables and therefore their associations with other variables are conducted by Fisher’s exact test. Gene expression values are continuous variables and therefore their associations with other features are conducted by Wilcoxon rank sum test. For each sample in Figure 5B, the growth curve data was fitted using linear regression model to estimate the growth rate. Correlation analysis was performed as Spearman rank-order correlation with a two-tailed P value, and Spearman Rho was calculated. In all statistical tests, nominal p-values were corrected for multiple testing using the Benjamini–Hochberg method. A Benjamini-Hochberg adjusted P value of <0.05 was considered as statistically significant.
PDX model availability
All PDX models are available after completing the request form on the PDX Explorer website under an MTA from UTHSA.
Acknowledgments
This work was supported by the following funding: the Cancer Prevention and Research Institute of Texas (CPRIT) (RP180319 and RP160732), UTSW Harold C. Simmons Comprehensive Cancer Center from the National Cancer Institute (CA142543) (to S.X.S); the Rally Foundation (USA), Children’s Cancer Fund (Dallas, USA), the CPRIT (RP180319, RP200103, RP180805, RP160732, and RP220599), NIH-NIDDK R01 DK127037, NIH-NCI R01CA263079, NIH-NCI R21CA259771, and NIH-NHGRI UM1-011996 (to L.X.); Mays Cancer Center Support Grant P30 CA054174, CPRIT RP160732 and RP220599 (to P.J.H., Y.C., G.E.T.). Z.L. is supported by NIH NCI R50CA265339. The Sequencing data used in the study were generated at The Greehey Children’s Cancer Research Institute (GCCRI) Genome Sequencing Facility (GSF). GSF is a Mays Cancer Center Next Generation Sequencing Shared Resource (NGSSR) and is supported by NIH-NCI P30 CA054174, NIH Shared Instrument grants S10OD021805 and S10OD030311 (Z.L.), and CPRIT Core Facility Awards RP160732, RP220599, and RP220662 (Y.C.). The resources of the high-performance computing environment from Quantitative Biomedical Research Center (QBRC) and BioHPC at UT Southwestern Medical Center, as well as the Texas Advanced Computing Center (TACC) at The University of Texas at Austin, are gratefully acknowledged.
Author contributions
A.R., L.J.K., E.B., Y.C., Y.X., G.E.T., R.T.K., S.X.S., P.J.H., and L.X. conceived of and designed the study. S.X.S., P.J.H., and L.X. supervised the investigation. Y.Z., Z.L., S.X.S., P.J.H., and L.X. acquired the data. A.R., L.J.K., E.B., J.K., H.Z., X.X., L.G., Q.Z., Y.C., and L.X. performed the analysis. A.R., L.J.K., E.B., S.X.S., P.J.H., and L.X. wrote the article, which was reviewed and edited by all other co-authors.
Declaration of interests
The authors declare no conflicts of interest.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: October 10, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108171.
Contributor Information
Stephen X. Skapek, Email: Stephen.Skapek@UTSouthwestern.edu.
Peter J. Houghton, Email: HoughtonP@uthscsa.edu.
Lin Xu, Email: Lin.Xu@UTSouthwestern.edu.
Supplemental information
Data and code availability
-
•
Raw sequencing data are available at European Genome-Phenome Archive under the accession number EGAS00001006710.
-
•
The open-source REMOCON PERL package is available at GitHub: https://github.com/jiwoongbio/REMOCON.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Ward E., DeSantis C., Robbins A., Kohler B., Jemal A. Childhood and adolescent cancer statistics, 2014. CA. Cancer J. Clin. 2014;64:83–103. doi: 10.3322/caac.21219. [DOI] [PubMed] [Google Scholar]
- 2.Elgarten C.W., Aplenc R. Pediatric acute myeloid leukemia: updates on biology, risk stratification, and therapy. Curr. Opin. Pediatr. 2020;32:57–66. doi: 10.1097/MOP.0000000000000855. [DOI] [PubMed] [Google Scholar]
- 3.Conneely S.E., Stevens A.M. Acute Myeloid Leukemia in Children: Emerging Paradigms in Genetics and New Approaches to Therapy. Curr. Oncol. Rep. 2021;23:16. doi: 10.1007/s11912-020-01009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schultz K.R., Bowman W.P., Aledo A., Slayton W.B., Sather H., Devidas M., Wang C., Davies S.M., Gaynon P.S., Trigg M., et al. Improved early event-free survival with imatinib in Philadelphia chromosome-positive acute lymphoblastic leukemia: a children's oncology group study. J. Clin. Oncol. 2009;27:5175–5181. doi: 10.1200/jco.2008.21.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathew N.R., Baumgartner F., Braun L., O'Sullivan D., Thomas S., Waterhouse M., Müller T.A., Hanke K., Taromi S., Apostolova P., et al. Sorafenib promotes graft-versus-leukemia activity in mice and humans through IL-15 production in FLT3-ITD-mutant leukemia cells. Nat. Med. 2018;24:282–291. doi: 10.1038/nm.4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maude S.L., Laetsch T.W., Buechner J., Rives S., Boyer M., Bittencourt H., Bader P., Verneris M.R., Stefanski H.E., Myers G.D., et al. Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. N. Engl. J. Med. 2018;378:439–448. doi: 10.1056/NEJMoa1709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollard J.A., Guest E., Alonzo T.A., Gerbing R.B., Loken M.R., Brodersen L.E., Kolb E.A., Aplenc R., Meshinchi S., Raimondi S.C., et al. Gemtuzumab Ozogamicin Improves Event-Free Survival and Reduces Relapse in Pediatric KMT2A-Rearranged AML: Results From the Phase III Children's Oncology Group Trial AAML0531. J. Clin. Oncol. 2021;39:3149–3160. doi: 10.1200/jco.20.03048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunsmore K.P., Winter S.S., Devidas M., Wood B.L., Esiashvili N., Chen Z., Eisenberg N., Briegel N., Hayashi R.J., Gastier-Foster J.M., et al. Children's Oncology Group AALL0434: A Phase III Randomized Clinical Trial Testing Nelarabine in Newly Diagnosed T-Cell Acute Lymphoblastic Leukemia. J. Clin. Oncol. 2020;38:3282–3293. doi: 10.1200/jco.20.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown P.A., Ji L., Xu X., Devidas M., Hogan L.E., Borowitz M.J., Raetz E.A., Zugmaier G., Sharon E., Bernhardt M.B., et al. Effect of Postreinduction Therapy Consolidation With Blinatumomab vs Chemotherapy on Disease-Free Survival in Children, Adolescents, and Young Adults With First Relapse of B-Cell Acute Lymphoblastic Leukemia: A Randomized Clinical Trial. JAMA. 2021;325:833–842. doi: 10.1001/jama.2021.0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown P., Inaba H., Annesley C., Beck J., Colace S., Dallas M., DeSantes K., Kelly K., Kitko C., Lacayo N., et al. Pediatric Acute Lymphoblastic Leukemia, Version 2.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2020;18:81–112. doi: 10.6004/jnccn.2020.0001. [DOI] [PubMed] [Google Scholar]
- 11.Hunger S.P., Lu X., Devidas M., Camitta B.M., Gaynon P.S., Winick N.J., Reaman G.H., Carroll W.L. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: a report from the children's oncology group. J. Clin. Oncol. 2012;30:1663–1669. doi: 10.1200/JCO.2011.37.8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horn S.R., Stoltzfus K.C., Mackley H.B., Lehrer E.J., Zhou S., Dandekar S.C., Fox E.J., Rizk E.B., Trifiletti D.M., Rao P.M., Zaorsky N.G. Long-term causes of death among pediatric patients with cancer. Cancer. 2020;126:3102–3113. doi: 10.1002/cncr.32885. [DOI] [PubMed] [Google Scholar]
- 13.Conneely S.E., McAtee C.L., Gupta R., Lubega J., Scheurer M.E., Rau R.E. Association of race and ethnicity with clinical phenotype, genetics, and survival in pediatric acute myeloid leukemia. Blood Adv. 2021;5:4992–5001. doi: 10.1182/bloodadvances.2021004735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shoag J.M., Barredo J.C., Lossos I.S., Pinheiro P.S. Acute lymphoblastic leukemia mortality in Hispanic Americans. Leuk. Lymphoma. 2020;61:2674–2681. doi: 10.1080/10428194.2020.1779260. [DOI] [PubMed] [Google Scholar]
- 15.Barrington-Trimis J.L., Cockburn M., Metayer C., Gauderman W.J., Wiemels J., McKean-Cowdin R. Rising rates of acute lymphoblastic leukemia in Hispanic children: trends in incidence from 1992 to 2011. Blood. 2015;125:3033–3034. doi: 10.1182/blood-2015-03-634006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcotte E.L., Domingues A.M., Sample J.M., Richardson M.R., Spector L.G. Racial and ethnic disparities in pediatric cancer incidence among children and young adults in the United States by single year of age. Cancer. 2021;127:3651–3663. doi: 10.1002/cncr.33678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadan-Lottick N.S., Ness K.K., Bhatia S., Gurney J.G. Survival variability by race and ethnicity in childhood acute lymphoblastic leukemia. JAMA. 2003;290:2008–2014. doi: 10.1001/jama.290.15.2008. [DOI] [PubMed] [Google Scholar]
- 18.Mendez L.M., Posey R.R., Pandolfi P.P. The Interplay Between the Genetic and Immune Landscapes of AML: Mechanisms and Implications for Risk Stratification and Therapy. Front. Oncol. 2019;9:1162. doi: 10.3389/fonc.2019.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houghton J.A., Houghton P.J., Webber B.L. Growth and characterization of childhood rhabdomyosarcomas as xenografts. J. Natl. Cancer Inst. 1982;68:437–443. [PubMed] [Google Scholar]
- 20.Lee E.M., Bachmann P.S., Lock R.B. Xenograft models for the preclinical evaluation of new therapies in acute leukemia. Leuk. Lymphoma. 2007;48:659–668. doi: 10.1080/10428190601113584. [DOI] [PubMed] [Google Scholar]
- 21.Bissig-Choisat B., Kettlun-Leyton C., Legras X.D., Zorman B., Barzi M., Chen L.L., Amin M.D., Huang Y.H., Pautler R.G., Hampton O.A., et al. Novel patient-derived xenograft and cell line models for therapeutic testing of pediatric liver cancer. J. Hepatol. 2016;65:325–333. doi: 10.1016/j.jhep.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy A.J., Chen X., Pinto E.M., Williams J.S., Clay M.R., Pounds S.B., Cao X., Shi L., Lin T., Neale G., et al. Forty-five patient-derived xenografts capture the clinical and biological heterogeneity of Wilms tumor. Nat. Commun. 2019;10:5806. doi: 10.1038/s41467-019-13646-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nanni P., Landuzzi L., Manara M.C., Righi A., Nicoletti G., Cristalli C., Pasello M., Parra A., Carrabotta M., Ferracin M., et al. Bone sarcoma patient-derived xenografts are faithful and stable preclinical models for molecular and therapeutic investigations. Sci. Rep. 2019;9:12174. doi: 10.1038/s41598-019-48634-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rokita J.L., Rathi K.S., Cardenas M.F., Upton K.A., Jayaseelan J., Cross K.L., Pfeil J., Egolf L.E., Way G.P., Farrel A., et al. Genomic Profiling of Childhood Tumor Patient-Derived Xenograft Models to Enable Rational Clinical Trial Design. Cell Rep. 2019;29:1675–1689.e9. doi: 10.1016/j.celrep.2019.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J., Li Q., Noureen N., Fang Y., Kurmasheva R., Houghton P.J., Wang X., Zheng S. PCAT: an integrated portal for genomic and preclinical testing data of pediatric cancer patient-derived xenograft models. Nucleic Acids Res. 2021;49:D1321–D1327. doi: 10.1093/nar/gkaa698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneeberger V.E., Allaj V., Gardner E.E., Poirier J.T., Rudin C.M. Quantitation of Murine Stroma and Selective Purification of the Human Tumor Component of Patient-Derived Xenografts for Genomic Analysis. PLoS One. 2016;11:e0160587. doi: 10.1371/journal.pone.0160587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mannakee B.K., Balaji U., Witkiewicz A.K., Gutenkunst R.N., Knudsen E.S. Sensitive and specific post-call filtering of genetic variants in xenograft and primary tumors. Bioinformatics. 2018;34:1713–1718. doi: 10.1093/bioinformatics/bty010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conway T., Wazny J., Bromage A., Tymms M., Sooraj D., Williams E.D., Beresford-Smith B. Xenome--a tool for classifying reads from xenograft samples. Bioinformatics. 2012;28:i172–i178. doi: 10.1093/bioinformatics/bts236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kluin R.J.C., Kemper K., Kuilman T., de Ruiter J.R., Iyer V., Forment J.V., Cornelissen-Steijger P., de Rink I., Ter Brugge P., Song J.Y., et al. XenofilteR: computational deconvolution of mouse and human reads in tumor xenograft sequence data. BMC Bioinf. 2018;19:366. doi: 10.1186/s12859-018-2353-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dores G.M., Devesa S.S., Curtis R.E., Linet M.S., Morton L.M. Acute leukemia incidence and patient survival among children and adults in the United States, 2001-2007. Blood. 2012;119:34–43. doi: 10.1182/blood-2011-04-347872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L.J., Zhang C.W., Su S.C., Chen H.I.H., Chiu Y.C., Lai Z., Bouamar H., Ramirez A.G., Cigarroa F.G., Sun L.Z., Chen Y. An ancestry informative marker panel design for individual ancestry estimation of Hispanic population using whole exome sequencing data. BMC Genom. 2019;20:1007. doi: 10.1186/s12864-019-6333-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woiterski J., Ebinger M., Witte K.E., Goecke B., Heininger V., Philippek M., Bonin M., Schrauder A., Röttgers S., Herr W., et al. Engraftment of low numbers of pediatric acute lymphoid and myeloid leukemias into NOD/SCID/IL2Rcgammanull mice reflects individual leukemogenecity and highly correlates with clinical outcome. Int. J. Cancer. 2013;133:1547–1556. doi: 10.1002/ijc.28170. [DOI] [PubMed] [Google Scholar]
- 33.Wang K., Sanchez-Martin M., Wang X., Knapp K.M., Koche R., Vu L., Nahas M.K., He J., Hadler M., Stein E.M., et al. Patient-derived xenotransplants can recapitulate the genetic driver landscape of acute leukemias. Leukemia. 2017;31:151–158. doi: 10.1038/leu.2016.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richter-Pechańska P., Kunz J.B., Bornhauser B., von Knebel Doeberitz C., Rausch T., Erarslan-Uysal B., Assenov Y., Frismantas V., Marovca B., Waszak S.M., et al. PDX models recapitulate the genetic and epigenetic landscape of pediatric T-cell leukemia. EMBO Mol. Med. 2018;10:e9443. doi: 10.15252/emmm.201809443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E., et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berman J.N., Gerbing R.B., Alonzo T.A., Ho P.A., Miller K., Hurwitz C., Heerema N.A., Hirsch B., Raimondi S.C., Lange B., et al. Prevalence and clinical implications of NRAS mutations in childhood AML: a report from the Children's Oncology Group. Leukemia. 2011;25:1039–1042. doi: 10.1038/leu.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang D.C., Chen S.H., Liu H.C., Yang C.P., Yeh T.C., Jaing T.H., Hung I.J., Hou J.Y., Lin T.H., Lin C.H., Shih L.Y. Mutational status of NRAS, KRAS, and PTPN11 genes is associated with genetic/cytogenetic features in children with B-precursor acute lymphoblastic leukemia. Pediatr. Blood Cancer. 2018;65 doi: 10.1002/pbc.26786. [DOI] [PubMed] [Google Scholar]
- 38.Jerchel I.S., Hoogkamer A.Q., Ariës I.M., Steeghs E.M.P., Boer J.M., Besselink N.J.M., Boeree A., van de Ven C., de Groot-Kruseman H.A., de Haas V., et al. RAS pathway mutations as a predictive biomarker for treatment adaptation in pediatric B-cell precursor acute lymphoblastic leukemia. Leukemia. 2018;32:931–940. doi: 10.1038/leu.2017.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cavé H., Cacheux V., Raynaud S., Brunie G., Bakkus M., Cochaux P., Preudhomme C., Laï J.L., Vilmer E., Grandchamp B. ETV6 is the target of chromosome 12p deletions in t(12;21) childhood acute lymphocytic leukemia. Leukemia. 1997;11:1459–1464. doi: 10.1038/sj.leu.2400798. [DOI] [PubMed] [Google Scholar]
- 40.Carrasco Salas P., Fernández L., Vela M., Bueno D., González B., Valentín J., Lapunzina P., Pérez-Martínez A. The role of CDKN2A/B deletions in pediatric acute lymphoblastic leukemia. Pediatr. Hematol. Oncol. 2016;33:415–422. doi: 10.1080/08880018.2016.1251518. [DOI] [PubMed] [Google Scholar]
- 41.Zhang H., Wang H., Qian X., Gao S., Xia J., Liu J., Cheng Y., Man J., Zhai X. Genetic mutational analysis of pediatric acute lymphoblastic leukemia from a single center in China using exon sequencing. BMC Cancer. 2020;20:211. doi: 10.1186/s12885-020-6709-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun C., Chang L., Zhu X. Pathogenesis of ETV6/RUNX1-positive childhood acute lymphoblastic leukemia and mechanisms underlying its relapse. Oncotarget. 2017;8:35445–35459. doi: 10.18632/oncotarget.16367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamps M.P. E2A-Pbx1 induces growth, blocks differentiation, and interacts with other homeodomain proteins regulating normal differentiation. Curr. Top. Microbiol. Immunol. 1997;220:25–43. doi: 10.1007/978-3-642-60479-9_2. [DOI] [PubMed] [Google Scholar]
- 44.Jia M., Hu B.F., Xu X.J., Zhang J.Y., Li S.S., Tang Y.M. Clinical features and prognostic impact of TCF3-PBX1 in childhood acute lymphoblastic leukemia: A single-center retrospective study of 837 patients from China. Curr. Probl. Cancer. 2021;45:100758. doi: 10.1016/j.currproblcancer.2021.100758. [DOI] [PubMed] [Google Scholar]
- 45.Duman-Scheel M., Weng L., Xin S., Du W. Hedgehog regulates cell growth and proliferation by inducing Cyclin D and Cyclin E. Nature. 2002;417:299–304. doi: 10.1038/417299a. [DOI] [PubMed] [Google Scholar]
- 46.Agathocleous M., Locker M., Harris W.A., Perron M. A general role of hedgehog in the regulation of proliferation. Cell Cycle. 2007;6:156–159. doi: 10.4161/cc.6.2.3745. [DOI] [PubMed] [Google Scholar]
- 47.Gopalakrishnapillai A., Kolb E.A., Dhanan P., Bojja A.S., Mason R.W., Corao D., Barwe S.P. Generation of Pediatric Leukemia Xenograft Models in NSG-B2m Mice: Comparison with NOD/SCID Mice. Front. Oncol. 2016;6:162. doi: 10.3389/fonc.2016.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ben-David U., Ha G., Tseng Y.Y., Greenwald N.F., Oh C., Shih J., McFarland J.M., Wong B., Boehm J.S., Beroukhim R., Golub T.R. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat. Genet. 2017;49:1567–1575. doi: 10.1038/ng.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bruna A., Rueda O.M., Greenwood W., Batra A.S., Callari M., Batra R.N., Pogrebniak K., Sandoval J., Cassidy J.W., Tufegdzic-Vidakovic A., et al. A Biobank of Breast Cancer Explants with Preserved Intra-tumor Heterogeneity to Screen Anticancer Compounds. Cell. 2016;167:260–274.e22. doi: 10.1016/j.cell.2016.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeRose Y.S., Wang G., Lin Y.C., Bernard P.S., Buys S.S., Ebbert M.T.W., Factor R., Matsen C., Milash B.A., Nelson E., et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat. Med. 2011;17:1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li S., Shen D., Shao J., Crowder R., Liu W., Prat A., He X., Liu S., Hoog J., Lu C., et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013;4:1116–1130. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woo X.Y., Giordano J., Srivastava A., Zhao Z.M., Lloyd M.W., de Bruijn R., Suh Y.S., Patidar R., Chen L., Scherer S., et al. Conservation of copy number profiles during engraftment and passaging of patient-derived cancer xenografts. Nat. Genet. 2021;53:86–99. doi: 10.1038/s41588-020-00750-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nordlund J., Bäcklin C.L., Wahlberg P., Busche S., Berglund E.C., Eloranta M.L., Flaegstad T., Forestier E., Frost B.M., Harila-Saari A., et al. Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol. 2013;14:r105. doi: 10.1186/gb-2013-14-9-r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peneder P., Stütz A.M., Surdez D., Krumbholz M., Semper S., Chicard M., Sheffield N.C., Pierron G., Lapouble E., Tötzl M., et al. Multimodal analysis of cell-free DNA whole-genome sequencing for pediatric cancers with low mutational burden. Nat. Commun. 2021;12:3230. doi: 10.1038/s41467-021-23445-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lietz C.E., Newman E.T., Kelly A.D., Xiang D.H., Zhang Z., Luscko C.A., Lozano-Calderon S.A., Ebb D.H., Raskin K.A., Cote G.M., et al. Genome-wide DNA methylation patterns reveal clinically relevant predictive and prognostic subtypes in human osteosarcoma. Commun. Biol. 2022;5:213. doi: 10.1038/s42003-022-03117-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gaynon P.S., Angiolillo A.L., Carroll W.L., Nachman J.B., Trigg M.E., Sather H.N., Hunger S.P., Devidas M., Children's Oncology Group Long-term results of the children's cancer group studies for childhood acute lymphoblastic leukemia 1983-2002: a Children's Oncology Group Report. Leukemia. 2010;24:285–297. doi: 10.1038/leu.2009.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sabattini E., Bacci F., Sagramoso C., Pileri S.A. WHO classification of tumours of haematopoietic and lymphoid tissues in 2008: an overview. Pathologica. 2010;102:83–87. [PubMed] [Google Scholar]
- 62.Morton C.L., Papa R.A., Lock R.B., Houghton P.J. Preclinical chemotherapeutic tumor models of common childhood cancers: solid tumors, acute lymphoblastic leukemia, and disseminated neuroblastoma. Curr. Protoc. Pharmacol. 2007;Chapter 14:Unit14.8. doi: 10.1002/0471141755.ph1408s39. [DOI] [PubMed] [Google Scholar]
- 63.O'Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sherry S.T., Ward M., Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. [PubMed] [Google Scholar]
- 65.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang H., Zhan X., Brugarolas J., Xie Y. DEFOR: depth- and frequency-based somatic copy number alteration detector. Bioinformatics. 2019;35:3824–3825. doi: 10.1093/bioinformatics/btz170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mayakonda A., Lin D.C., Assenov Y., Plass C., Koeffler H.P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Han T., Goralski M., Gaskill N., Capota E., Kim J., Ting T.C., Xie Y., Williams N.S., Nijhawan D. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science. 2017;356:eaal3755. doi: 10.1126/science.aal3755. [DOI] [PubMed] [Google Scholar]
- 71.Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw sequencing data are available at European Genome-Phenome Archive under the accession number EGAS00001006710.
-
•
The open-source REMOCON PERL package is available at GitHub: https://github.com/jiwoongbio/REMOCON.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.