Abstract
During spermatogenesis, a large fraction of cellular proteins is degraded as the spermatids evolve to their elongated mature forms. In particular, histones must be degraded in early elongating spermatids to permit chromatin condensation. Our laboratory previously demonstrated the activation of ubiquitin conjugation during spermatogenesis. This activation is dependent on the ubiquitin-conjugating enzyme (E2) UBC4, and a testis-particular isoform, UBC4-testis, is induced when histones are degraded. Therefore, we tested whether there are UBC4-dependent ubiquitin protein ligases (E3s) that can ubiquitinate histones. Indeed, a novel enzyme, E3Histone, which could conjugate ubiquitin to histones H1, H2A, H2B, H3, and H4 in vitro, was found. Only the UBC4/UBC5 family of E2s supported E3Histone-dependent ubiquitination of histone H2A, and of this family, UBC4-1 and UBC4-testis are the preferred E2s. We purified this ligase activity 3,600-fold to near homogeneity. Mass spectrometry of the final material revealed the presence of a 482-kDa HECT domain-containing protein, which was previously named LASU1. Anti-LASU1 antibodies immunodepleted E3Histone activity. Mass spectrometry and size analysis by gel filtration and glycerol gradient centrifugation suggested that E3Histone is a monomer of LASU1. Our assays also show that this enzyme is the major UBC4-1-dependent histone-ubiquitinating E3. E3Histone is therefore a HECT domain E3 that likely plays an important role in the chromatin condensation that occurs during spermatid maturation.
Spermatogenesis is a complex developmental process during which stem cell spermatogonia are transformed into highly differentiated spermatids (12). This transformation can be divided into three phases. The first phase is the proliferative phase, in which spermatogonia undergo successive mitotic divisions. Subsequently, in the meiotic phase, spermatogonia are transformed into spermatocytes in which the genetic material undergoes homologous recombination and two sequential cell divisions, resulting in haploid spermatids. In the final (spermiogenic) phase, the spermatids are transformed into cells that are structurally equipped to reach and fertilize the eggs. During spermiogenesis, each immature spermatid develops an acrosome and a tail, reorganizes its mitochondria, and loses most of its cytoplasm.
Proteolysis plays an important role in these developmental phases. In the first two phases, proteolysis is essential in regulating the cell cycle. Proteolysis also appears to be important for the third (spermiogenic) phase. During this cellular remodeling of haploid spermatids, many proteins are degraded. Histones are among the key proteins that undergo proteolysis (29). Upon degradation in early and late elongated spermatids, histones are replaced by transition proteins, which in turn are replaced by protamines (29). The substitution of histones by protamines is essential to permit the condensation of chromatin into the narrow head of the compact and elongated mature spermatid. The mechanisms underlying the degradation of histones remain unclear. Previous work suggests that many proteins are removed in a cytoplasmic droplet that is phagocytosed by the adjacent Sertoli cells (30, 42). However, the ubiquitin system appears to be involved in this loss of cellular proteins. Indeed, ubiquitinated histones have been detected in the testes of different species (1, 3, 8, 32). During rooster (1) and mouse (3) spermatogenesis, ubiquitinated histone H2As are at the highest levels before the replacement of histones by protamines in late spermatids. During trout germ cell maturation, the levels of monoubiquitinated histone H2B decreased, while those of doubly ubiquitinated histone H2B increased (32). Ubiquitinated H3 was first identified in a study of rat spermatogenesis, with the highest level in elongated spermatids (8). The results from these studies suggest that ubiquitination plays an important role in histone replacement.
The conjugation of ubiquitin to proteins requires the sequential cooperation of three enzymes (reviewed in references 21 and 38). The ubiquitin-activating enzyme (E1) hydrolyzes ATP to convert ubiquitin into an activated form, which is covalently linked at its carboxyl terminus to a cysteine residue of the E1 enzyme via a high-energy thioester linkage. The activated ubiquitin molecule is then transferred to the second enzyme of this pathway, a ubiquitin-conjugating enzyme (E2), and the activated form is maintained through the formation of a thioester linkage with a cysteine residue of the E2 enzyme. The third enzyme in the process, the ubiquitin protein ligase (E3), supports the transfer of ubiquitin to substrates.
E3 is the key to the specificity of ubiquitin conjugation, as it recognizes substrates (reviewed in references 21 and 38). There are two major classes of E3 enzymes. One class is the RING finger-containing E3s. This type of E3 functions as a scaffold protein that binds both the E2 and the substrate, permitting ubiquitin to be transferred from the E2 to the substrate. The RING finger domain chelates two Zn2+ ions and is essential for interaction with the E2. The other class of E3 enzymes is the HECT domain-containing E3s. These E3s support ubiquitination in a process that requires two steps. First, ubiquitin is transferred from E2 to the E3 via a thioester linkage with a cysteine residue in the HECT domain, and then ubiquitin is transferred from the E3 to the substrate. This family of E3s therefore requires a free thiol group for its function.
The E3 covalently links the ubiquitin moiety by its C-terminal residue to the ɛ-amino group of an internal lysine residue of the target protein (21, 38) or in some cases to the α-amino group at the N terminus of the protein (10). Monoubiquitination has important functions as a signal (reviewed in reference 23). Monoubiquitination of membrane proteins signals their internalization to endosomes that traffic to lysosomes for degradation (23, 41). Histone monoubiquitination regulates gene transcription. Monoubiquitination of histone H2B in yeast results in telomeric gene silencing by stimulating the methylation of histone H3 on lysine 4 and lysine 79 (47), while monoubiquitination of histone H3 in yeast results in the activation of transcription (6). For most ubiquitination, though, a polyubiquitin chain is usually formed after the linkage of ubiquitin to the substrate protein (21). When linked via the lysine 48 residue of each ubiquitin, these chains usually target the attached protein to the 26S proteasome for degradation (7). Chains consisting of non-lysine 48 linkages appear to have nonproteolytic functions. For example, lysine 63-linked ubiquitin chains can mediate DNA repair and signaling via the NF-κB pathway (13, 24, 45, 46).
Previous work in our laboratory supports a role for ubiquitination during spermatogenesis. The rate of ubiquitin conjugation increases during the first wave of spermatogenesis that occurs in early postnatal life (39). This increase in conjugation appears to be dependent on the UBC4 family of E2s. UBC4 is induced during spermatogenesis (39). This activation is at least partly due to the induction of UBC4 isoforms. UBC4 is highly expressed in the testis, and it is widely expressed in many different tissues. A particular testis-specific UBC4 isoform, UBC4-testis, is induced in round spermatids and early elongated spermatids (52).
To further explore the role of this UBC4-dependent ubiquitination during male germ cell development, we have been identifying UBC4-dependent E3s that are expressed in the testis. Since histones are known to be ubiquitinated and degraded in early elongated spermatids when UBC4-testis is induced, we hypothesized that histones may be substrates of a UBC4-dependent E3. Therefore, we used 125I-labeled histone H2A as a substrate to biochemically screen for UBC4-dependent E3 activity in testis extracts. Interestingly, a HECT domain E3 was identified to be able to conjugate ubiquitin to different core histones. We now describe the characterization of this new E3.
MATERIALS AND METHODS
Iodination of proteins.
The chloramine-T method was used to label bovine ubiquitin with Na125I to a specific radioactivity of 3,000 cpm/pmol and histones H1, H2A, H3, and H4 (Boehringer Mannheim) to a specific radioactivity of 375,000 cpm/μg. Unincorporated 125I was removed by passing the reaction products over a Sephadex G25 column.
Preparation and quantification of enzymes.
E1 was isolated from rabbit liver by ubiquitin affinity chromatography as previously described (19). Bacterially expressed recombinant UBC4-1, UBC4-testis, E214K, UBC7, UBCH6, UBCH7, and UBCH10 proteins were prepared as described previously (4, 27, 33, 48, 52-54). UBC4-1, UBC4-testis, and E214K enzymatic activities were quantified by measuring the initial release of radioactive pyrophosphate following incubation in the presence of [γ-32P]ATP, ubiquitin, and E1 (20). The remaining E2s were quantified by using a thioester assay (36) (see below). Recombinant glutathione S-transferase (GST) fused to the RING domain of the E3 ARNIP (GST-ARNIP) was expressed in Escherichia coli cells and purified by glutathione-Sepharose beads (5).
Conjugation assay.
To measure the conjugation of ubiquitin to histone substrates, the reaction mixture contained the following in a final volume of 20 μl: 10 μl of the fractions from each protein purification step, 50 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol (DTT), 2 mM MgCl2, 2 mM ATP, 0.5 U of pyrophosphatase, 12.5 mM phosphocreatine, 2.5 U of creatine kinase, 50 nM E1, 30 μM 125I-labeled histone H2A, and 250 nM UBC4-1 or UBC4-testis. All ubiquitin conjugation reactions used to screen fractions for E3Histone activity were initiated with 25 μM reductively methylated ubiquitin (RMUb), prepared and quantified as described previously (22), and incubated for 1 h at 37°C. Reaction products were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the results were detected by autoradiography.
For quantification of E3Histone activity, the peak fractions from each purification step were tested in the conjugation assay at 30°C for only 10 min. Under these conditions of shorter time and lower temperature, the assay was linear with respect to the amount of enzyme. E3Histone activity was quantified by adding up the intensity of each band multiplied by the number of ubiquitin moieties linked to histone H2A in that band.
Conjugation assays which tested the ability of different E2s to support E3-mediated ubiquitination of histone H2A contained E2 enzymatic concentrations between 150 and 300 nM. The conjugation assay comparing different E2s or 125I-labeled histone types were carried out under quantification conditions (i.e., 30°C for 10 min).
In the assays of testing the substrate specificity (Fig. 1C) and E2 specificity (Fig. 1D) of E3Histone and testing the different results of E3Histone-mediated ubiquitination of histone H2A with wild-type ubiquitin (Ub) or methylated ubiquitin (MeUb) (Fig. 1B), the E3Histone used was purified with two anion exchange columns, Q-Sepharose and Mono Q columns, and a Superdex 200 gel filtration column.
FIG. 1.
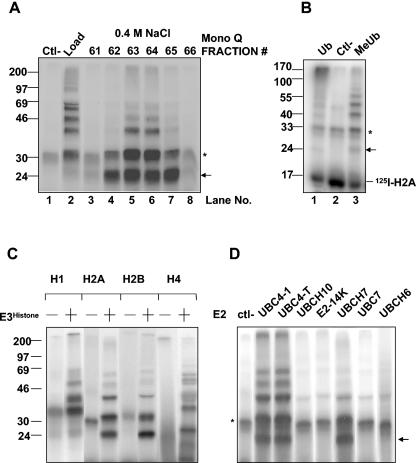
Identification and initial characterization of E3Histone. (A) A soluble testis extract was chromatographed on a Mono Q anion exchange column. The supernatant of the rat testis extract that was centrifuged at 100,000 × g (Load), water as a negative control (Ctl-), and eluted fractions were tested for the ability to support the conjugation of ubiquitin to 125I-labeled histone H2A in the presence of E1 and UBC4. Lanes 3 to 8 are the fractions eluting at approximately 0.4 M NaCl that contained the activity. (B) E3Histone (lanes 1 and 3) or water (lane 2) was used in a ubiquitin conjugation assay of 125I-labeled histone H2A with either wild-type Ub or MeUb. (C) Different 125I-labeled histones were tested in the conjugation assay in the presence (+) or absence (−) of E3Histone. (D) E3Histone-mediated ubiquitination was tested in the presence or absence (ctl-) of the indicated E2s. (A to D) Reaction products were resolved by SDS-PAGE and detected by autoradiography. *, background bands. Arrows show the single monoubiquitinated form of histone H2A. Note that in panels A, C, and D, the free histone substrates were run out of the gels because they created large shadows when the gels were exposed to film with the use of intensifying screens. In panel B, the gel was exposed to a phosphorimager screen, which does not result in the 125I-labeled histone substrate generating a large shadow. (B to D) The E3Histone used was partially purified as described in Materials and Methods.
For the conjugation assay with ARNIP, a RING finger E3, the components and their concentrations were the same as in the assays with E3Histone except that the E3 ARNIP was used at a concentration of 1 μM, 2 μl of 50 μM 125I-labeled ubiquitin was used to start the reaction, and there was no histone H2A. Since there was no exogenous substrate, autoubiquitinating activity was measured. The reaction was incubated at 37°C for 1 h. Reaction products were resolved by SDS-PAGE and detected by autoradiography.
Thioester assay.
The components for a 10-μl reaction were as follows: 1 μl of 10× thioester buffer (0.5 M Tris-HCl [pH 7.5], 0.1 M MgCl2, 20 mM ATP, 5 mM DTT), 1 μl of inorganic pyrophosphatase (0.5 U/μl) (Sigma), and 1 μl of E1 (1 μM). One microliter of 50 μM 125I-labeled ubiquitin was used to start the reaction. The reaction was incubated at 37°C for 1 min. Laemmli sample buffer without 2-mercaptoethanol was used to stop the reaction, and the products were separated by SDS-PAGE at 4°C. The gel was dried, and results were detected by autoradiography.
Preparation of testis extracts.
Testes from Sprague-Dawley rats (200 g; Charles River Laboratories) were sliced and homogenized at 4°C with a Potter-Elvehjem tissue grinder (Fisher) in 5 volumes of ice-cold homogenization buffer (50 mM Tris [pH 7.5], 1 mM DTT). Bovine testes were homogenized in 3 volumes of homogenization buffer with a Waring blender. The homogenate was centrifuged at 10,000 × g for 15 min, and the supernatant was centrifuged at 100,000 × g for 1 h.
Bovine E3Histone purification.
The supernatant that was centrifuged at 100,000 × g was subjected to purification by the following steps. After each step, the fractions were assayed as described above to detect the ability to support UBC4-dependent conjugation of ubiquitin to histone H2A. The peak fractions were pooled for the next step.
The supernatant that was centrifuged at 100,000 × g was loaded onto a 50-mm by 35-cm DEAE-cellulose (Whatman DE-52) column that had been equilibrated with the homogenization buffer. Bound proteins were stepwise eluted with 0.1, 0.2, and 0.3 M NaCl in 50 mM Tris (pH 7.5, 4°C)-1 mM DTT. Most of the E3Histone eluted with buffer containing 0.2 M salt.
The E3Histone activity collected was precipitated by 0 to 50% ammonium sulfate precipitation. The pellet was redissolved in and dialyzed against homogenization buffer.
The dialyzed sample was then loaded onto a 26-mm by 20-cm quaternary amine anion exchange column (Q Sepharose; Pharmacia Amersham Biotech) equilibrated with the homogenization buffer. Bound proteins were eluted with a 0 to 0.5 M NaCl gradient (in 50 mM Tris [pH 7.5]-1 mM DTT).
Peak fractions containing E3Histone activities eluted at ∼0.4 M NaCl. They were pooled and dialyzed against homogenization buffer containing 0.7 M (NH4)2SO4 and loaded onto an 8-mm by 7.5-cm Phenyl 5PW column. Bound proteins were eluted with a 0.7 to 0 M (NH4)2SO4 gradient in 50 mM Tris (pH 7.5)-1 mM DTT.
The peak fractions eluting at ∼0.24 M (NH4)2SO4 were pooled, dialyzed overnight against homogenization buffer, and then loaded onto a 5-mm by 5-cm quaternary amine anion exchange column (Mono Q; Pharmacia Amersham Biotech) equilibrated with the homogenization buffer. Retained proteins were eluted with the same gradient as for the Q Sepharose chromatography noted above.
The peak fractions eluting at ∼0.4 M NaCl were then concentrated by using Centriplus 10 concentrators (Amicon). The concentrated eluate was further purified on a 10-mm by 30-cm gel filtration column (Superdex 200; Pharmacia Amersham Biotech).
The peak fractions were pooled and centrifuged on a 10 to 40% glycerol gradient in 10 mM Tris-Cl, pH 7.5. The centrifugation was performed at 28,000 rpm in an SW 40Ti rotor (Beckman) for 16 h at 4°C.
Free thiol group requirement test.
To test whether E3Histone might be a HECT domain E3, we assessed whether a free thiol group was required for activity. Following the Superdex 200 chromatography step, some of the partially purified enzyme was treated with 5 mM N-ethylmaleimide (NEM) on ice for 5 min, and then 2.5 mM DTT was added to quench the remaining NEM. The enzyme was then assayed for activity. To confirm the effectiveness of the NEM treatment, 1 pmol of E1 was similarly treated and then subjected to a thioester assay for activity. To confirm that there was sufficient quenching of NEM by DTT, buffer was treated with the same amount of NEM and DTT prior to use in an assay of E1 for thioester activity.
Zn2+ ion requirement test.
To test whether the E3Histone might be a RING domain E3, we assessed whether divalent cations were required for the activity. Glycerol gradient fraction 4 or 20 pmol of recombinant GST-ARNIP was treated with 10 mM EDTA at 4°C overnight. MgCl2 (10 mM) was added to bind the residual EDTA prior to use in a conjugation assay. The treated E3s were then tested in conjugation assays. In an additional test of the requirement of Zn2+, an E3Histone-containing fraction or 20 pmol of recombinant GST-ARNIP was treated with a relatively zinc-specific chelator, N,N,N′,N′-tetrakis-(2-pyridylmethyl)-ethylenediamine (TPEN; 3.2 mM). The assays were then performed as for EDTA-treated enzyme.
Mass spectrometry analysis.
Following glycerol gradient centrifugation, the fractions were analyzed for E3Histone activity and protein composition by both native gel and SDS-PAGE. Prominent bands comigrating with E3Histone activity were excised and analyzed at the Harvard Microchemistry Facility. The proteins in the bands were digested with trypsin, after which the peptides were resolved and analyzed by microcapillary reverse-phase high-pressure liquid chromatography, directly coupled to the nano-electrospray ionization source of an ion trap mass spectrometer (MS). These MS/MS spectra were then correlated with known sequences by using the algorithm Sequest developed at the University of Washington (15) and programs developed at Harvard University (9). MS/MS peptide sequences were then reviewed for consensus with known proteins, and the results were manually confirmed for fidelity.
Antibody design, production, and purification.
A fragment encoding 185 amino acid residues of the N-terminal part of LASU1 was amplified by reverse transcription-PCR using testis mRNA as a template. The amplified nucleotide fragment was cloned into pET15b vector (Novagen) and expressed in E. coli BL21 cells. The expressed protein contained an N-terminal His tag fusion and was purified on a Ni2+ column. Rabbits were injected with the protein in Freund's adjuvant, and after two boosts the serum was collected and the antibody was purified on a column containing GST fused to the protein fragment.
Immunoblotting and immunoprecipitation.
Rabbit sera against LASU1 diluted 1,000 times or purified LASU1 antibody diluted to 2.7 μg/ml was used for immunoblotting on polyvinylidene difluoride membranes (Millipore). Immunoblots were visualized by chemiluminescence using the ECL detection system (Amersham). Antibody against the 20S proteasome (Affiniti) was used to detect the 20S proteasome subunits. To immunoprecipitate LASU1 from E3Histone-containing fractions, an equal volume of LASU1 antiserum or preimmune serum and protein A-Sepharose beads was incubated at 4°C for 2 h as described elsewhere (26). Beads were washed three times in immunoprecipitation buffer (20 mM Tris-HCl [pH 7.5] at 4°C, 1% NP-40, 50 mM NaCl, protease inhibitor cocktail [Roche]) and then incubated with an equal volume of E3Histone-containing fraction at 4°C for 2 h. Beads were washed as before. The pellet and the supernatant were assayed for E3Histone activity as described above.
Nucleotide sequence accession numbers.
The human and mouse LASU1 sequences described here have been deposited in GenBank under accession no. AY929611 and AY929612, respectively.
RESULTS
Identification and characterization of E3Histone.
Since a specific E2, UBC4, is induced in elongating spermatids when histones are removed from spermatids (39), we used 125I-labeled histone H2A as a substrate and UBC4 as the E2 to screen biochemically for a ubiquitin protein ligase that can ubiquitinate histones (see Materials and Methods). When this assay was applied to a rat testis extract (Fig. 1A), multiple high-molecular-weight forms of histone H2A were generated (Fig. 1A, lane 2), compared to the negative control (Fig. 1A, lane 1), which was an assay done in the absence of the testis extract. These results indicated the presence of an E3 in the testis that can ubiquitinate the histone. We carried out the assay with RMUb, as it resulted in easily visible and quantifiable monoubiquitinated forms of H2A. In contrast, the use of wild-type ubiquitin resulted in a high-molecular-weight smear at the top of the gel (Fig. 1B, compare lanes 1 and 3). The presence of multiple bands when RMUb was used must represent monoubiquitination of histone H2A on multiple lysine residues. This result suggests that at least in vitro this ubiquitination is not site specific on histone H2A. Since the substrate of the reaction, 125I-labeled histone H2A, showed a strong signal whose shadow rendered difficult the observation of the 8-kDa-higher monoubiquitinated histone H2A band on film, we typically ran the substrate off the gel. Our more recent assays used a phosphorimager which does not generate a large shadow because of a shorter path (no intensifying screen) to the detection plate (Fig. 1B). To characterize further this E3, the testis extract was fractionated on a Mono Q anion exchange column. The E3 activity eluted at ∼0.4 M salt (Fig. 1A, lanes 5 and 6). This E3 activity was designated E3Histone.
Since all of the different types of histones are degraded during spermatogenesis, we tested whether this E3 can ubiquitinate other histones besides histone H2A (Fig. 1C). Histones H1, H2B, H3, and H4 were radiolabeled and incubated in the presence of E1, UBC4, or RMUb with or without E3Histone. High-molecular-weight forms of the histones were observed for all of the histone types tested, but only in the presence of E3Histone, indicating that E3Histone could ubiquitinate all core histones tested here in vitro.
Since E3s generally interact with specific E2s, we tested the ability of different E2s to support ubiquitination of histone H2A (Fig. 1D). Of the E2s tested, only UBC4-1, UBC4-testis, and UBCH7 supported E3Histone-dependent ubiquitination. These three E2s belong to the UBC4/UBC5 family of E2s.
E3Histone is a HECT domain E3.
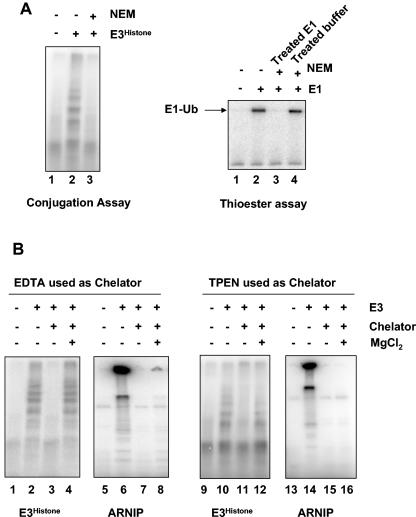
There are two major families of E3s, RING finger E3s and HECT domain E3s. To assess whether E3Histone might be a HECT domain E3, we tested whether E3Histone activity requires the presence of a free thiol group (Fig. 2A, left panel). E3Histone was treated with NEM to inactivate thiol groups. Following addition of DTT to quench the excess NEM, the treated E3Histone was reassayed for E3 activity (Fig. 2A, left panel, lane 3). After the treatment, E3Histone lost its activity, as did E1 when it was similarly treated and tested in a thioester assay as a positive control (Fig. 2A, left and right panels, lane 3). The loss of activity was not due to inactivation of E1 or E2 in the assay from incomplete quenching of NEM by DTT, as identical treatment of a mock sample with NEM and DTT did not affect the activity of subsequently supplemented E1 in a thioester assay (Fig. 2A, right panel, lane 4). Thus, this E3 could require a free thiol group for its function and may therefore be a HECT domain E3. However, at this point we cannot rule out the possibility that under the conditions employed, the NEM may have reacted with other nucleophiles besides thiol groups. Since RING finger motifs chelate Zn2+ ions, we tested E3Histone for the requirement of divalent cations by incubating it with the chelator EDTA or the relatively zinc-specific chelator TPEN (Fig. 2B). Following incubation, the remaining EDTA or TPEN was bound by adding excess Mg2+ to avoid interference by the chelators of the E1-mediated activation of ubiquitin, which requires Mg2+. The treated enzyme was reassayed in the conjugation assay. E3Histone activity was unaffected by EDTA treatment (Fig. 2B, lane 4) and TPEN treatment (Fig. 2B, lane 12). The enzymatic activity of the E3Histone was lower in the TPEN study due to the use of a different fraction. To confirm that this treatment was effective at chelation, we applied the identical protocol to a sample of ARNIP, a known RING finger E3 (5). Indeed, ARNIP autoubiquitination activity was lost upon treatment (Fig. 2B, lanes 8 and 16), indicating that the negative result with E3Histone was valid. These data together suggest that E3Histone is probably a HECT domain E3.
FIG. 2.
E3Histone is probably a HECT domain-containing E3. (A) E3Histone requires a free thiol group for its E3 activity. (Left panel) In the conjugation assay, E3Histone was first treated with NEM or not treated. Excess NEM was then quenched by DTT, and the other components of the conjugation assay were added. Products of the assay were resolved by SDS-PAGE and detected by autoradiography. (Right panel) To confirm that both the NEM treatment and quenching by DTT were adequate, E1 was treated sequentially with NEM and DTT (lane 3) or added subsequently to buffer that had been similarly treated with NEM and DTT (lane 4) and then assayed for the ability to form a thioester bond with 125I-labeled ubiquitin. Reaction products were resolved by SDS-PAGE at 4°C under nonreducing conditions and detected by autoradiography. (B) E3Histone does not require divalent cations for its function. E3Histone was treated with EDTA (lanes 3 and 4) or TPEN (lanes 11 and 12). Remaining chelators were then quenched with excess Mg2+ (lanes 4 and 12) or not quenched (lanes 3 and 11) prior to assays for ubiquitination of histone H2A. To confirm the efficacy of the treatment, the RING finger E3 ARNIP was similarly treated and then assayed for its autoubiquitination ability by incubation with E1, UBC4-1, and 125I-labeled ubiquitin (lanes 5 to 8 and 13 to 16). Products were resolved by SDS-PAGE and detected by autoradiography.
LASU1 is a component of E3Histone.
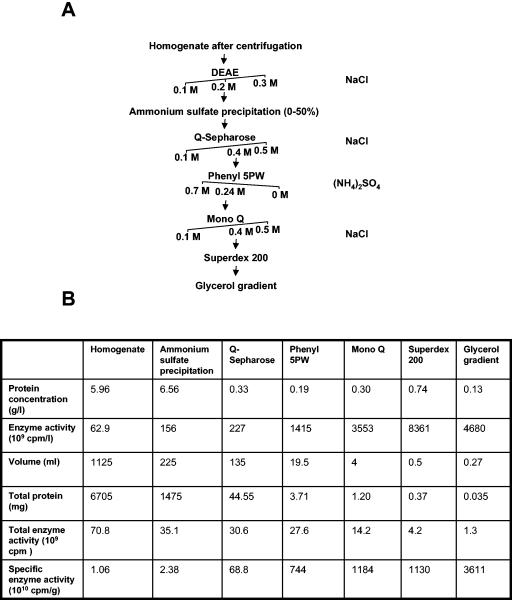
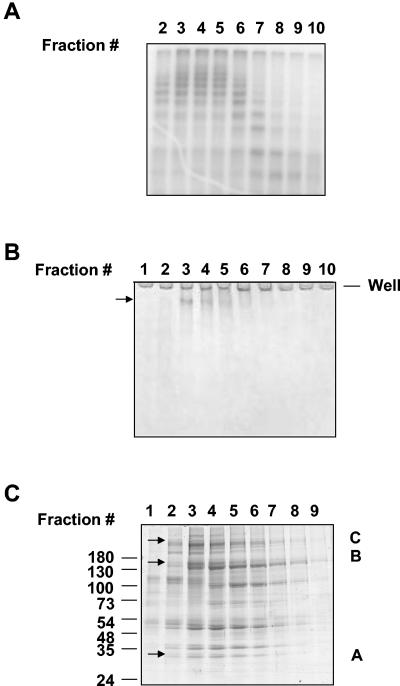
To identify this new E3, we purified it to near homogeneity by the sequential application of different protein purification methods (details are in Materials and Methods). To obtain an adequate amount of purified enzyme, we started with an extract prepared from bovine testes. After the multistep purification (Fig. 3A), the specific enzyme activity was 3,600-fold that of the crude extract (Fig. 3B). The precipitation of the activity by relatively low concentrations of ammonium sulfate suggested that the enzyme was probably large in size. This was indeed confirmed by the gel filtration chromatography step, which suggested a molecular mass of ∼600 kDa for this enzyme (data not shown) (see Fig. 7A). Therefore, we subjected it to a final step of purification by centrifugation on a glycerol gradient. The fractions from this last purification step were assayed for E3Histone activity (Fig. 4A), and the protein components were analyzed by denaturing and native gel electrophoresis (Fig. 4B and C). Native gel electrophoresis revealed a single major band (Fig. 4B). This band was cut and analyzed by tandem MS. SDS-PAGE revealed a number of bands ranging from ∼30 kDa to >180 kDa (Fig. 4C). Several bands on this denaturing gel appeared to comigrate closely with E3Histone activity (Fig. 4A) and so were also cut and analyzed by tandem MS. Interestingly, analysis of the band from native gel electrophoresis (Table 1) identified 68 peptides corresponding to a human 370-kDa HECT domain-containing protein previously named LASU1 (17, 18). This appeared to be the dominant protein in the band and indicated that LASU1 was likely to be E3Histone or a part of E3Histone.
FIG. 3.
Purification of E3Histone from bovine testis extract. (A) E3Histone purification scheme. The salts used to elute proteins in each step are shown on the right of the purification tree. Arrows indicate the salt concentration at which the enzyme eluted. The buffers used are described in the text. (B) Protein purification table. Aliquots of the pooled fractions from each protein purification step were assayed under quantification conditions (see the text) to measure E3Histone activity. The results were detected by phosphorimager and quantified as described in Materials and Methods.
FIG. 7.
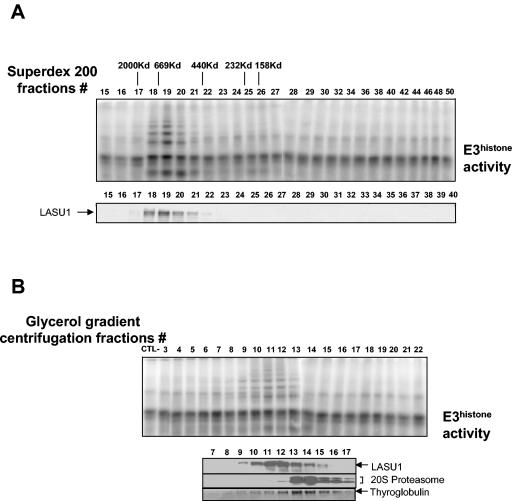
E3Histone appears to be a monomeric protein. (A) Separation of a crude testis extract using a Superdex 200 gel filtration column. (Top panel) Fractions were assayed for E3Histone activity as described in the legend to Fig. 1. Elution positions and molecular weights of the protein standards are shown. (Bottom panel) Western blot of the fractions with the anti-LASU1 antibody. The arrow shows the LASU1 band. (B) Separation of a crude testis extract by glycerol gradient centrifugation. The sample, purified 20S proteasome, or thyroglobulin was applied to a 4 to 40% glycerol gradient and subjected to the centrifugation at 30,000 rpm in an SW 40Ti rotor for 20 h at 4°C. (Top panel) Fractions from 4 to 40% glycerol gradient centrifugation were assayed for E3Histone activity. (Bottom panel) Detection of protein standards and LASU1. 20S proteasome and LASU1 were detected by Western blotting. Thyroglobulin was detected by Coomassie blue staining. (In both panels A and B, the unmodified 125I-labeled E3Histone substrate was run off the bottom of the gels.)
FIG. 4.
Analysis of the fractions from the last purification step. (A) Fractions from the glycerol gradient centrifugation were assayed for E3Histone activity. As described in Materials and Methods, MeUb was used to start the reaction. The results were detected by autoradiography. (B) Protein fractions were resolved on a 5% native acrylamide gel and detected by colloidal blue staining. (C) Protein fractions were resolved by 7 to 15% gradient SDS-PAGE and detected by colloidal blue staining. Arrows in panels B and C show the bands cut for tandem mass spectrometric analysis. The presence of proteins in the A-, B-, and C-labeled bands in panel C that were also found in the band from the native gel in panel B is noted in Table 1.
TABLE 1.
MS/MS analysis of the band isolated from the native acrylamide gel in Fig. 4B
| Reference no. | Protein name | Species | No. of peptides | Present in SDS bands from Fig. 4Ca |
|---|---|---|---|---|
| gi 22090626 | HECT domain protein LASU1 | Homo sapiens | 68 | A,B,C |
| gi 26340188 | Unnamed protein product | Mus musculus | 20 | B,C |
| gi 10436857 | Unnamed protein product | Homo sapiens | 3 | A |
| gi 25140230 | Similar to tripeptidyl peptidase II | Homo sapiens | 7 | B |
| gi 1061310 | Valyl-tRNA synthetase | 7 | B | |
| gi 2493459 | Protein kinase C substrate, 60.1-kDa protein, heavy chain (PKCSH) (80K-H protein) | Bostauras | 3 | A |
| gi 28478776 | Similar to elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma) | Mus musculus | 7 | |
| gi 28189194 | Similar to pancreatic tumor-related protein | Bos taurus | 3 | |
| gi 417844 | Valyl-tRNA synthetase (valinetRNA Ligase) (VALRS) | 2 | ||
| gi 27807311 | Vesicle docking protein p115 | Bos taurus | 5 | |
| gi 232037 | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma) | Oryctolagus cuniculus | 3 | |
| gi 12804891 | Similar to tubulin; beta 5 | Homo sapiens | 4 | |
| gi 6015101 | Endoplasmin (94-kDa glucose-regulated protein) (GRP94) | Oryctolagus cuniculus | 5 | |
| gi 25021127 | Similar to transitional endoplasmic reticulum ATPase (EC 3.6.1.) (validated), rat | Mus musculus | 2 | |
| gi 4204880 | Heat shock protein | Homo sapiens | 3 | |
| gi 11265337 | Hypothetical protein DKFZp434K0126.1, human (fragment) | Homo sapiens | 3 | |
| gi 1174593 | Tubulin alpha-2/alpha-4 chain | Patella vulgata | 2 | |
| gi 12847562 | Unnamed protein product | Musmusculus | 2 |
Proteins also detected in the tandem mass spectrometry analysis reports of bands A, B, or C from the denaturing gel (Fig. 4C) are indicated.
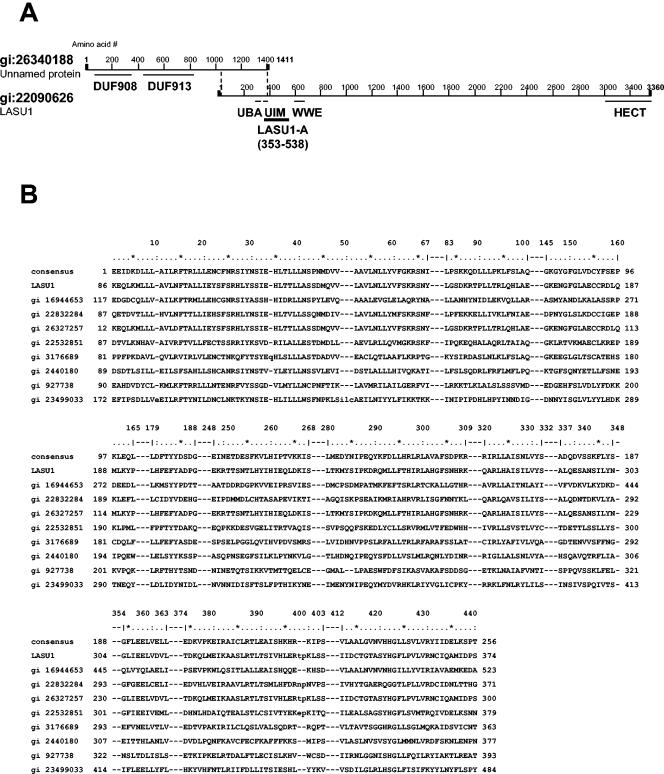
Interestingly, the second-largest number of peptides in the native band corresponded to an unnamed protein (gi 26340188) whose C-terminal 356 amino acids are the same as amino acid residues 41 to 397 in the N-terminal part of LASU1 (Fig. 5A). A search of the expressed sequence tag (EST) database identified other independent sequences of LASU1 in this region and revealed that the first 40 amino acids of LASU1 were miscoded because of a frame shift caused by an extra G at base 266 of the previous DNA sequence of LASU1. Therefore, the C-terminal 396 amino acids of the unnamed protein are exactly the same as the N-terminal 396 amino acids of LASU1. Furthermore, using the LASU1 and unnamed protein sequences to probe the mouse genome database indicated that the sections of DNA encoding these two putative proteins are adjacent to and overlapping each other on the X chromosome. Northern blot analysis of testis mRNA with a probe against the overlapping region detected only a single band that was >10 kb in size (unpublished data). Finally, the cDNA clone 4017134 (Invitrogen) contains the overlapping region of these two proteins and nonoverlapping sequences of the unnamed protein and LASU1 on the N terminus and C terminus, respectively. All of these independent pieces of evidence suggested very strongly that the unnamed protein and LASU1 are parts of the same protein. The complete human LASU1 has 4,374 amino acids, a predicted molecular mass of 481.9 kDa, and a pI of 4.87. Mouse LASU1 has 4,377 amino acids, a predicted molecular mass of 482.7 kDa, and a pI of 4.86.
FIG. 5.
Structure of LASU1. (A) Domain analysis of LASU1 and the unnamed protein. The region (amino acids 353 to 538) used to generate an anti-LASU1 antibody is shown. (B) Alignment of DUF908. (C) Alignment of DUF913. In panels B and C, the conserved residues in the domain are shown. gi 16944653, related to TOM1 protein; gi 22832284, CG8184-PB; gi 26327257, unnamed protein product; gi 22532851, hypothetical protein Y67D8C.5; gi 3176689, contains similarity to ubiquitin carboxyl-terminal hydrolase 14; gi 2440180, SPAC19D5.04; gi 927738, Tom1p; gi 23499033, ubiquitin-protein ligase 1; and gi 8778329, F14J16.10.
Mass spectrometric analysis of the bands from the SDS-PAGE gel (Fig. 4C) revealed that all three bands contained LASU1 and that the two larger ones also contained the unnamed protein. Those bands likely contain degradation fragments of LASU1.
Domain analysis of this protein showed that it has a classical HECT domain at the C-terminal end (Fig. 5A). In addition, a UBA domain and a UIM domain, both known to function in ubiquitin binding, are found in the overlapping region between the unnamed protein and LASU1. A WWE domain was also identified, and this domain has been associated with proteins involved in the regulation of ubiquitin-dependent proteolysis (2). Finally, two domains with unknown function, DUF908 and DUF913, were identified in the N-terminal region located in the unnamed protein (Fig. 5A). Interestingly, most of the other proteins that contain these two domains appear to be related to the 374-kDa Saccharomyces cerevisiae HECT domain ligase TOM1 (14) (Fig. 5B and C). TOM1 and the complete LASU1 share high degrees of similarity in these domains and in the HECT domain but are relatively poorly conserved elsewhere, showing only 17.3% and 34.7% overall amino acid identity and similarity, respectively.
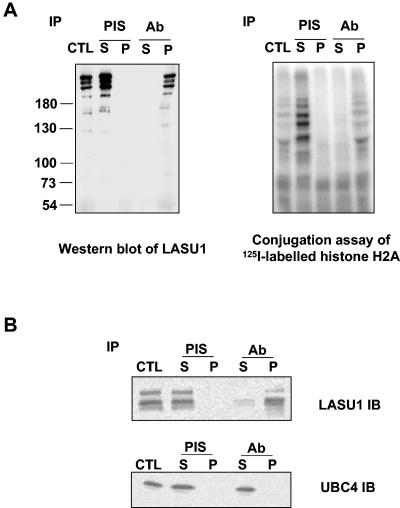
To confirm that E3Histone indeed contains LASU1, an antibody was raised against a His-tagged fragment of LASU1 containing residues 353 to 538 of the LASU1 sequence (residues 1367 to 1552 of the complete protein). The antibody was affinity purified on a column bearing the fragment fused to GST. This antibody was tested for its ability to immunoprecipitate E3Histone (Fig. 6A). The antibody, but not the preimmune serum, bound E3Histone activity, confirming that LASU1 is a part of E3Histone. Since UBC4 appears to be the cognate E2 for E3Histone, we tested for the presence of UBC4 in the immunoprecipitated LASU1 (Fig. 6B). UBC4 was not detectable in the pellet, suggesting that its interaction with E3Histone is transient, which is consistent with UBC4-Ub thioester acting as a substrate in the reaction.
FIG. 6.
E3Histone contains HECT domain protein LASU1. (A) E3Histone was immunoprecipitated (IP) with preimmune serum (PIS) or anti-LASU1 antiserum (Ab). (Left panel) Western blot of the indicated sample with anti-LASU1 antibody. (Right panel) Samples were assayed for E3Histone activity. (B) UBC4 was not coimmunoprecipitated with E3Histone. Crude testis extract was immunoprecipitated with the LASU1 antibody, and then the supernatants and pellets were subjected to immunoblotting with anti-LASU1 antibody or anti-UBC4 antibody. Multiple bands in the LASU1 blot likely represent degradation products of LASU1 during the procedure. CTL, the same amount of sample before IP; S, supernatant; P, pellet.
E3Histone appears to be a monomeric protein.
To determine whether E3Histone is a monomeric protein or a multisubunit complex, we estimated the molecular mass of this E3. Since purifying the protein resulted in significant degradation, we chromatographed a crude testis extract on a Superdex 200 gel filtration column (Fig. 7A). The peak of E3Histone eluted at fraction 19. When compared to the elution positions of protein standards, this corresponded to a molecular mass of ∼600 kDa. Immunoblotting revealed that LASU1 protein comigrated as expected with E3Histone activity.
Since gel filtration chromatography is not a precise method of sizing such large proteins, density gradient centrifugation was also used to estimate the molecular mass of E3Histone. Glycerol gradient centrifugation was performed on E3Histone and also on thyroglobulin (667 kDa) and the 20S proteasome (700 kDa), which were used as standards (Fig. 7B). The peak of thyroglobulin was in fraction 13 and the peak of the 20S proteasome was in fractions 13 and 14, while that of E3Histone was detected in fraction 11. Thus, E3Histone is smaller than the 20S proteasome (700 kDa) and thyroglobulin (667 kDa) but larger than the ferritin standard used on gel filtration, suggesting a size between 440 and 667 kDa.
To determine whether E3Histone contains other subunits besides LASU1, the anti-LASU1 antibody was covalently linked to protein A beads by DMP, and the covalently linked antibody was used to immunoprecipitate E3Histone from a crude rat testis extract. The proteins that were immunoprecipitated by the antibody were resolved by SDS-PAGE. Several protein bands that were in the pellet that was immunoprecipitated by the antibody but not by the rabbit immunoglobulin G control were cut and analyzed by MS (data not shown). There was only one protein, heat shock protein 70 (HSP70), which was present in the proteins identified in the E3Histone band on native gel electrophoresis (Fig. 4B). However, in both cases, the numbers of peptides ascribed to HSP70 were small, and to date we have not been able to confirm this finding by Western blot analysis using anti-HSP70 antibody of the E3Histone that was immunoprecipitated by use of anti-LASU1 antibody (data not shown). On the other hand, all of LASU1 immunoreactivity comigrated with E3Histone activity (Fig. 7A and B). Taken together, these data suggest that E3Histone probably consists simply of the 482-kDa protein LASU1.
E3Histone is the major UBC4-1-dependent E3 that ubiquitinates histone H2A in the testis.
So far, two different RING domain E3s, Np95 (11) and BRCA1/BARD1 (28), have been found to ubiquitinate histone H2A in vitro by cooperation with UBCH5B, the human homolog of UBC4-1. Therefore, we tested whether other UBC4-dependent E3s besides E3Histone are present in the testis. We fractionated a testis extract on the Superdex 200 gel filtration column (Fig. 7A) and glycerol gradient (Fig. 7B) and screened fractions for their ability to ubiquitinate histone H2A in the presence of UBC4-1. In both cases, we did not find any significant activity in fractions other than the ones containing E3Histone. Therefore, E3Histone appears to be the major UBC4-1-dependent E3 in the testis that ubiquitinates histone H2A.
DISCUSSION
During spermatid maturation, several chromatin rearrangements occur. Initially, the histones are replaced by transition proteins which in turn are finally replaced by protamines (34). Histones are still degraded when genes encoding transition proteins are inactivated in the mouse, indicating that the loss of histones is not simply due to displacement by the transition proteins (56). Ubiquitination of several histones is increased just prior to their degradation (29), suggesting that ubiquitin-dependent proteolysis plays an important role in histone replacement. In this paper, we identify and characterize a new testis ubiquitin protein ligase, E3Histone, which is an excellent candidate ligase for mediating the ubiquitination of histones. It ubiquitinates all core histones in vitro (Fig. 1A and C). With wild-type ubiquitin, E3Histone produced much-higher-molecular-weight forms of histone than when MeUb was used (Fig. 1B), indicating that E3Histone can form polyubiquitin chains on histones, thus conferring the potential for recognition and degradation by the proteasome. Its preferred interaction with UBC4 would also be consistent with such a role, as UBC4 isoforms are induced in early elongating spermatids (39) when histones begin to be replaced. Indeed, the induction of UBC4 may be a mechanism by which histone ubiquitination and degradation are initiated and regulated.
Mass spectrometric analysis identified E3Histone as a HECT domain-containing ligase, previously named LASU1. This was consistent with our observations that E3Histone activity was abolished by the thiol reactive compound N-ethylmaleimide but not by the divalent cation chelators EDTA or TPEN (Fig. 2). In addition, our EST sequence analysis corrected a frameshift-inducing sequencing error present in the previous sequence of LASU1, thus resulting in a much longer N terminus that includes what was hitherto a protein of unknown function. Remarkably, mass spectrometric coupled to bioinformatics analyses of this bovine enzyme was able to identify 88 peptides matching the human LASU1 or mouse unnamed protein sequence. This result suggests a significant degree of conservation of sequence among these species, which was confirmed by the observation of 97.7% identity and 98.5% similarity between mouse and human sequences present in the EST databases. All other proteins identified in the analyses had fewer than seven matching peptides, suggesting that LASU1 was the dominant protein in this enzyme and that these other proteins were likely contaminants. Indeed, they were not detected in MS analyses of protein bands in immunoprecipitates of E3Histone. Also, none of the other identified proteins appeared to be a potential ligase. The predicted molecular mass of 482 kDa was reasonably close to the estimated molecular mass of E3Histone of ∼600 kDa, considering the limited precision of mass estimation by chromatography for such a large protein and the assumption of globular enzymes in these estimations. The enzyme bound to anion exchange columns, as would be expected for the predicted pI of 4.87. All of these observations together argue strongly that LASU1 is E3Histone. The latter findings also suggest that E3Histone functions as a monomeric protein, but the mass determinations are not precise enough to rule out the presence of an additional small subunit(s), particularly nonstoichiometrically in subsets of the enzyme molecules.
Interestingly, LASU1 was found to be expressed in many different tissues at both mRNA and protein levels and more highly expressed in the testis, brain, lung, and kidney (unpublished data). E3Histone could therefore have other important roles in different tissues besides its function on chromatin condensation in elongated spermatids. It is possible that it may play a role in the ubiquitination of histones in other cells. However, in most cells, ubiquitination of histones appears to modify function rather than target for degradation. In eukaryotic cells, 10 to 15% of total histone H2A is ubiquitinated (51), while 10% of histone H2B is ubiquitinated in yeast (40) and 1.5% of cellular histone H2B is monoubiquitinated in higher eukaryotes (51). The function of this ubiquitination remains largely unknown except for a few examples. Monoubiquitination of histone H2B in S. cerevisiae leads to methylation of histone H3 on its lysine 4 and lysine 79 residues, which in turn causes telomeric gene silencing (6, 47). This ubiquitination is mediated by the RING domain-containing E3, Bre1, which relies on Rad6p as its cognate E2 (25, 40, 55). Inactivation of a mouse homolog of Rad6p leads to defective spermatogenesis but does not appear to prevent histone degradation. Furthermore, monoubiquitination of histone H1 in drosophila by TAFII250, another RING finger-containing protein, results in transcriptional activation (37). These results taken together support a link between histone ubiquitination and gene transcription. In addition, two other ubiquitin protein ligases belonging to the RING domain E3 family, BRCA1/BARD1 (28) and Np95 (11), have been recently shown to promote ubiquitination of histone in vitro.
In this paper, we identify for the first time a HECT domain-containing E3 that ubiquitinates histones. Besides a role in mediating histone degradation, this ubiquitination could also be involved in transcriptional regulation. Indeed, previous studies with the rat homolog of LASU1 suggested that it may function as a DNA-binding transcriptional regulator (17, 18). The most similar other known E3s are S. cerevisiae Tom1 and E3s closely related to Tom1. Tom1 mutants are pleiotropic and include defects in transcription that appear to be mediated through ADA coactivator proteins (43) as well as defects in the export of mRNA (14, 44, 49). Suppressors of tom1 mutants include STO1/G4p2/STM1, a basic protein partially homologous to histone H1 that can bind to G4 nucleic acids (16, 31, 50). Thus, Tom1 appears to interact with basic nucleic acid binding proteins and in that way is similar to the demonstrated ability of LASU1 to interact with and ubiquitinate histones.
LASU1 is here demonstrated to be one of the largest E3s to be characterized to date. Like other large E3s, such as the 374-kDa Tom1 (50) and 300-kDa EDD (35), the large size likely permits it to interact with a variety of substrates and to exert a large number of functions which remain to be defined.
Acknowledgments
This work was supported by Canadian Institutes of Health Research grant MT12121 to S.S.W. Z.L. is the recipient of a McGill Major Fellowship. R.O. is the recipient of a McGill Major Fellowship and a Molson Foundation Fellowship. S.S.W. is the recipient of a Chercheur National salary award from the Fonds de la Recherche en Santé du Québec.
REFERENCES
- 1.Agell, N., M. Chiva, and C. Mezquita. 1983. Changes in nuclear content of protein conjugate histone H2A-ubiquitin during rooster spermatogenesis. FEBS Lett. 155:209-212. [DOI] [PubMed] [Google Scholar]
- 2.Aravind, L. 2001. The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem. Sci. 26:273-275. [DOI] [PubMed] [Google Scholar]
- 3.Baarends, W. M., J. W. Hoogerbrugge, H. P. Roest, M. Ooms, J. Vreeburg, J. H. Hoeijmakers, and J. A. Grootegoed. 1999. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev. Biol. 207:322-333. [DOI] [PubMed] [Google Scholar]
- 4.Bastians, H., L. M. Topper, G. L. Gorbsky, and J. V. Ruderman. 1999. Cell cycle-regulated proteolysis of mitotic target proteins. Mol. Biol. Cell 10:3927-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beitel, L. K., Y. A. Elhaji, R. Lumbroso, S. S. Wing, V. Panet-Raymond, B. Gottlieb, L. Pinsky, and M. A. Trifiro. 2002. Cloning and characterization of an androgen receptor N-terminal-interacting protein with ubiquitin-protein ligase activity. J. Mol. Endocrinol. 29:41-60. [DOI] [PubMed] [Google Scholar]
- 6.Briggs, S. D., T. Xiao, Z. W. Sun, J. A. Caldwell, J. Shabanowitz, D. F. Hunt, C. D. Allis, and B. D. Strahl. 2002. Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418:498. [DOI] [PubMed] [Google Scholar]
- 7.Chau, V., J. W. Tobias, A. Bachmair, D. Marriott, D. J. Ecker, D. K. Gonda, and A. Varshavsky. 1989. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243:1576-1583. [DOI] [PubMed] [Google Scholar]
- 8.Chen, H. Y., J. M. Sun, Y. Zhang, J. R. Davie, and M. L. Meistrich. 1998. Ubiquitination of histone H3 in elongating spermatids of rat testes. J. Biol. Chem. 273:13165-13169. [DOI] [PubMed] [Google Scholar]
- 9.Chittum, H. S., W. S. Lane, B. A. Carlson, P. P. Roller, F. D. Lung, B. J. Lee, and D. L. Hatfield. 1998. Rabbit beta-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry 37:10866-10870. [DOI] [PubMed] [Google Scholar]
- 10.Ciechanover, A., and R. Ben-Saadon. 2004. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 14:103-106. [DOI] [PubMed] [Google Scholar]
- 11.Citterio, E., R. Papait, F. Nicassio, M. Vecchi, P. Gomiero, R. Mantovani, P. P. Di Fiore, and I. M. Bonapace. 2004. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol. Cell. Biol. 24:2526-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clermont, Y., R. Oko, and L. Hermo. 1993. Cell biology of mammalian spermatogenesis. Oxford University Press, New York, N.Y.
- 13.Deng, L., C. Wang, E. Spencer, L. Yang, A. Braun, J. You, C. Slaughter, C. Pickart, and Z. J. Chen. 2000. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103:351-361. [DOI] [PubMed] [Google Scholar]
- 14.Duncan, K., J. G. Umen, and C. Guthrie. 2000. A putative ubiquitin ligase required for efficient mRNA export differentially affects hnRNP transport. Curr. Biol. 10:687-696. [DOI] [PubMed] [Google Scholar]
- 15.Eng, J., A. L. McCormick, and J. R. Yates, Jr. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 16.Frantz, J. D., and W. Gilbert. 1995. A yeast gene product, G4p2, with a specific affinity for quadruplex nucleic acids. J. Biol. Chem. 270:9413-9419. [DOI] [PubMed] [Google Scholar]
- 17.Gu, J., R. Dubner, A. J. Fornace, Jr., and M. J. Iadarola. 1995. UREB1, a tyrosine phosphorylated nuclear protein, inhibits p53 transactivation. Oncogene 11:2175-2178. [PubMed] [Google Scholar]
- 18.Gu, J., S. G. Irving, and M. J. Iadarola. 1997. URE, an initiator (Inr)-like site, suppresses the promoter of the rat dynorphin gene. Biochem. Biophys. Res. Commun. 231:172-177. [DOI] [PubMed] [Google Scholar]
- 19.Haas, A. L., and P. M. Bright. 1988. The resolution and characterization of putative ubiquitin carrier protein isozymes from rabbit reticulocytes. J. Biol. Chem. 263:13258-13267. [PubMed] [Google Scholar]
- 20.Haas, A. L., and I. A. Rose. 1982. The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. J. Biol. Chem. 257:10329-10337. [PubMed] [Google Scholar]
- 21.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 22.Hershko, A., and H. Heller. 1985. Occurrence of a polyubiquitin structure in ubiquitin-protein conjugates. Biochem. Biophys. Res. Commun. 128:1079-1086. [DOI] [PubMed] [Google Scholar]
- 23.Hicke, L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann, R. M., and C. M. Pickart. 1999. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96:645-653. [DOI] [PubMed] [Google Scholar]
- 25.Hwang, W. W., S. Venkatasubrahmanyam, A. G. Ianculescu, A. Tong, C. Boone, and H. D. Madhani. 2003. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 11:261-266. [DOI] [PubMed] [Google Scholar]
- 26.King, R. W., J. M. Peters, S. Tugendreich, M. Rolfe, P. Hieter, and M. W. Kirschner. 1995. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell 81:279-288. [DOI] [PubMed] [Google Scholar]
- 27.Lin, H., and S. S. Wing. 1999. Identification of rabbit reticulocyte E217K as a UBC7 homologue and functional characterization of its core domain loop. J. Biol. Chem. 274:14685-14691. [DOI] [PubMed] [Google Scholar]
- 28.Mallery, D. L., C. J. Vandenberg, and K. Hiom. 2002. Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J. 21:6755-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meistrich, M. L. 1989. Histones and other basic nuclear proteins. CRC Press, Boca Raton, Fla.
- 30.Morales, C., and L. Hermo. 1985. Nature and function of endocytosis in Sertoli cells of the rat. Am. J. Anat. 173:203-217. [DOI] [PubMed] [Google Scholar]
- 31.Nelson, L. D., M. Musso, and M. W. Van Dyke. 2000. The yeast STM1 gene encodes a purine motif triple helical DNA-binding protein. J. Biol. Chem. 275:5573-5581. [DOI] [PubMed] [Google Scholar]
- 32.Nickel, B. E., S. Y. Roth, R. G. Cook, C. D. Allis, and J. R. Davie. 1987. Changes in the histone H2A variant H2A.Z and polyubiquitinated histone species in developing trout testis. Biochemistry 26:4417-4421. [DOI] [PubMed] [Google Scholar]
- 33.Nuber, U., S. Schwarz, P. Kaiser, R. Schneider, and M. Scheffner. 1996. Cloning of human ubiquitin-conjugating enzymes UbcH6 and UbcH7 (E2-F1) and characterization of their interaction with E6-AP and RSP5. J. Biol. Chem. 271:2795-2800. [DOI] [PubMed] [Google Scholar]
- 34.Oliva, R., and G. H. Dixon. 1991. Vertebrate protamine genes and the histone-to-protamine replacement reaction. Prog. Nucleic Acid Res. Mol. Biol. 40:25-94. [DOI] [PubMed] [Google Scholar]
- 35.Oughtred, R., N. Bedard, O. A. Adegoke, C. R. Morales, J. Trasler, V. Rajapurohitam, and S. S. Wing. 2002. Characterization of rat100, a 300-kilodalton ubiquitin-protein ligase induced in germ cells of the rat testis and similar to the Drosophila hyperplastic discs gene. Endocrinology 143:3740-3747. [DOI] [PubMed] [Google Scholar]
- 36.Oughtred, R., N. Bedard, A. Vrielink, and S. S. Wing. 1998. Identification of amino acid residues in a class I ubiquitin-conjugating enzyme involved in determining specificity of conjugation of ubiquitin to proteins. J. Biol. Chem. 273:18435-18442. [DOI] [PubMed] [Google Scholar]
- 37.Pham, A. D., and F. Sauer. 2000. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science 289:2357-2360. [DOI] [PubMed] [Google Scholar]
- 38.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 39.Rajapurohitam, V., C. R. Morales, M. El-Alfy, S. Lefrancois, N. Bedard, and S. S. Wing. 1999. Activation of a UBC4-dependent pathway of ubiquitin conjugation during postnatal development of the rat testis. Dev. Biol. 212:217-228. [DOI] [PubMed] [Google Scholar]
- 40.Robzyk, K., J. Recht, and M. A. Osley. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287:501-504. [DOI] [PubMed] [Google Scholar]
- 41.Rotin, D., O. Staub, and R. Haguenauer-Tsapis. 2000. Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J. Membr. Biol. 176:1-17. [DOI] [PubMed] [Google Scholar]
- 42.Russell, L., and Y. Clermont. 1976. Anchoring device between Sertoli cells and late spermatids in rat seminiferous tubules. Anat. Rec. 185:259-278. [DOI] [PubMed] [Google Scholar]
- 43.Saleh, A., M. Collart, J. A. Martens, J. Genereaux, S. Allard, J. Cote, and C. J. Brandl. 1998. TOM1p, a yeast hect-domain protein which mediates transcriptional regulation through the ADA/SAGA coactivator complexes. J. Mol. Biol. 282:933-946. [DOI] [PubMed] [Google Scholar]
- 44.Sasaki, T., A. Toh-e, and Y. Kikuchi. 2000. Extragenic suppressors that rescue defects in the heat stress response of the budding yeast mutant tom1. Mol. Gen. Genet. 262:940-948. [DOI] [PubMed] [Google Scholar]
- 45.Spence, J., R. R. Gali, G. Dittmar, F. Sherman, M. Karin, and D. Finley. 2000. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell 102:67-76. [DOI] [PubMed] [Google Scholar]
- 46.Spence, J., S. Sadis, A. L. Haas, and D. Finley. 1995. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell. Biol. 15:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun, Z. W., and C. D. Allis. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104-108. [DOI] [PubMed] [Google Scholar]
- 48.Townsley, F. M., A. Aristarkhov, S. Beck, A. Hershko, and J. V. Ruderman. 1997. Dominant-negative cyclin-selective ubiquitin carrier protein E2-C/UbcH10 blocks cells in metaphase. Proc. Natl. Acad. Sci. USA 94:2362-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Utsugi, T., A. Hirata, Y. Sekiguchi, T. Sasaki, A. Toh-e, and Y. Kikuchi. 1999. Yeast tom1 mutant exhibits pleiotropic defects in nuclear division, maintenance of nuclear structure and nucleocytoplasmic transport at high temperatures. Gene 234:285-295. [DOI] [PubMed] [Google Scholar]
- 50.Utsugi, T., A. Toh-e, and Y. Kikuchi. 1995. A high dose of the STM1 gene suppresses the temperature sensitivity of the tom1 and htr1 mutants in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1263:285-288. [DOI] [PubMed] [Google Scholar]
- 51.West, M. H., and W. M. Bonner. 1980. Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res. 8:4671-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wing, S. S., N. Bedard, C. Morales, P. Hingamp, and J. Trasler. 1996. A novel rat homolog of the Saccharomyces cerevisiae ubiquitin-conjugating enzymes UBC4 and UBC5 with distinct biochemical features is induced during spermatogenesis. Mol. Cell. Biol. 16:4064-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wing, S. S., F. Dumas, and D. Banville. 1992. A rabbit reticulocyte ubiquitin carrier protein that supports ubiquitin-dependent proteolysis (E214k) is homologous to the yeast DNA repair gene RAD6. J. Biol. Chem. 267:6495-6501. [PubMed] [Google Scholar]
- 54.Wing, S. S., and P. Jain. 1995. Molecular cloning, expression and characterization of a ubiquitin conjugation enzyme (E2(17)kB) highly expressed in rat testis. Biochem. J. 305:125-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wood, A., N. J. Krogan, J. Dover, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, A. Golshani, Y. Zhang, J. F. Greenblatt, M. Johnston, and A. Shilatifard. 2003. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11:267-274. [DOI] [PubMed] [Google Scholar]
- 56.Yu, Y. E., Y. Zhang, E. Unni, C. R. Shirley, J. M. Deng, L. D. Russell, M. M. Weil, R. R. Behringer, and M. L. Meistrich. 2000. Abnormal spermatogenesis and reduced fertility in transition nuclear protein 1-deficient mice. Proc. Natl. Acad. Sci. USA 97:4683-4688. [DOI] [PMC free article] [PubMed] [Google Scholar]