Abstract
It is known that metabolic defects in the retinal pigment epithelium (RPE) can cause degeneration of its neighboring photoreceptors in the retina, leading to retinal degenerative diseases such as age-related macular degeneration. However, how RPE metabolism supports the health of the neural retina remains unclear. The retina requires exogenous nitrogen sources for protein synthesis, neurotransmission, and energy metabolism. Using 15N tracing coupled with mass spectrometry, we found human RPE can utilize the nitrogen in proline to produce and export 13 amino acids, including glutamate, aspartate, glutamine, alanine, and serine. Similarly, we found this proline nitrogen utilization in the mouse RPE/choroid but not in the neural retina of explant cultures. Coculture of human RPE with the retina showed that the retina can take up the amino acids, especially glutamate, aspartate, and glutamine, generated from proline nitrogen in the RPE. Intravenous delivery of 15N proline in vivo demonstrated 15N-derived amino acids appear earlier in the RPE before the retina. We also found proline dehydrogenase, the key enzyme in proline catabolism is highly enriched in the RPE but not the retina. The deletion of proline dehydrogenase blocks proline nitrogen utilization in RPE and the import of proline nitrogen–derived amino acids in the retina. Our findings highlight the importance of RPE metabolism in supporting nitrogen sources for the retina, providing insight into understanding the mechanisms of the retinal metabolic ecosystem and RPE-initiated retinal degenerative diseases.
Keywords: retina, retinal pigment epithelium, nitrogen, ammonium, glutamine, aspartate, alanine, leucine, transaminase
The retinal pigment epithelium (RPE) is essential for maintaining metabolic homeostasis of the outer retina. The RPE relies on an active metabolism to transport nutrients from the choroidal blood circulation to the metabolically demanding photoreceptors. It also transports “metabolic waste” from the photoreceptors to the blood, phagocytoses daily-shed photoreceptor tips, and synthesizes ketone bodies and mitochondrial intermediates for photoreceptor utilization. Defects in RPE metabolism could lead to disruption of the metabolic ecosystem in the outer retina, contributing to multiple types of retinal degenerative diseases including age-related macular degeneration (AMD), the leading cause of blindness in the elderly (1, 2, 3).
The RPE is capable of using multiple types of nutrient sources to fuel its metabolism, which spares glucose for the neural retina (4). Our previous research demonstrates that the RPE prefers to use proline, a nonessential amino acid, as a metabolic substrate (5). RNA-sequence data also shows that genes in proline transport and catabolism are highly enriched in human RPE (6). Proline supplementation decreases the effects of oxidative damage in cultured RPE, and dietary proline protects against retinal degeneration induced by oxidative damage of RPE in vivo (7). Aside from being provided by diet, proline can be produced from ornithine, glutamate, or from collagen degradation. Mutations of enzymes in these pathways are known to cause RPE cell death and retinal degeneration (8, 9). For example, mutations of ornithine aminotransferase (OAT) cause gyrate atrophy, an ocular disease characterized by progressive loss of the RPE/choroid (Cho), leading to retinal degeneration. Proline is catabolized through proline dehydrogenase (PRODH) in the mitochondrial matrix into pyrroline-5-carboxylate (P5C), which is further degraded into glutamate, a substrate for the tricarboxylic acid (TCA) cycle. By 13C tracing, we have found the carbon skeleton from proline is catabolized into glutamate and TCA cycle intermediates, which are exported from the retinal side of the RPE to support retinal metabolism (7). However, the fate of the nitrogen groups in proline catabolism remains unclear.
In addition to high metabolic needs for carbon sources such as glucose, the neural retina also requires nitrogen sources to produce ATP, cGMP, glutamate, and other nitrogen-containing metabolites for energy metabolism, phototransduction, and protein synthesis. Our previous studies in both human and mouse explant cultures showed that retinas need substantial amounts of exogenous glutamate and aspartate (6). Glutamate and aspartate are interconvertible through aspartate transaminase. Glutamate is a key neurotransmitter as well as a metabolite substrate for the TCA cycle and many amino acids, such as serine, glycine, glutamine, and γ-aminobutyric acid (GABA). The retinal neurons contain millimolar concentrations of glutamate. However, glutamate and aspartate are the least abundant amino acids in plasma, suggesting that they might be produced locally in the neural retina or the RPE.
In this report, we trace the fate of nitrogen from proline in the RPE and the neural retina by using 15N proline. We found that proline is a nitrogen source for synthesizing 13 amino acids, especially glutamate, aspartate, and glutamine in primary human RPE. However, the neural retina alone cannot utilize the nitrogen directly from proline. Interestingly, neural retina cocultured with RPE imports amino acids that were made by the RPE using nitrogen from proline. In addition, we found the neural retina can utilize nitrogen from proline in vivo to produce glutamate, aspartate, and other amino acids by proline catabolism through PRODH in the RPE. Our results demonstrate that proline catabolism in the RPE is a source of nitrogen for the synthesis of amino acids for the neural retina.
Results
Proline is an important nitrogen source for the synthesis and export of amino acids in human RPE cells
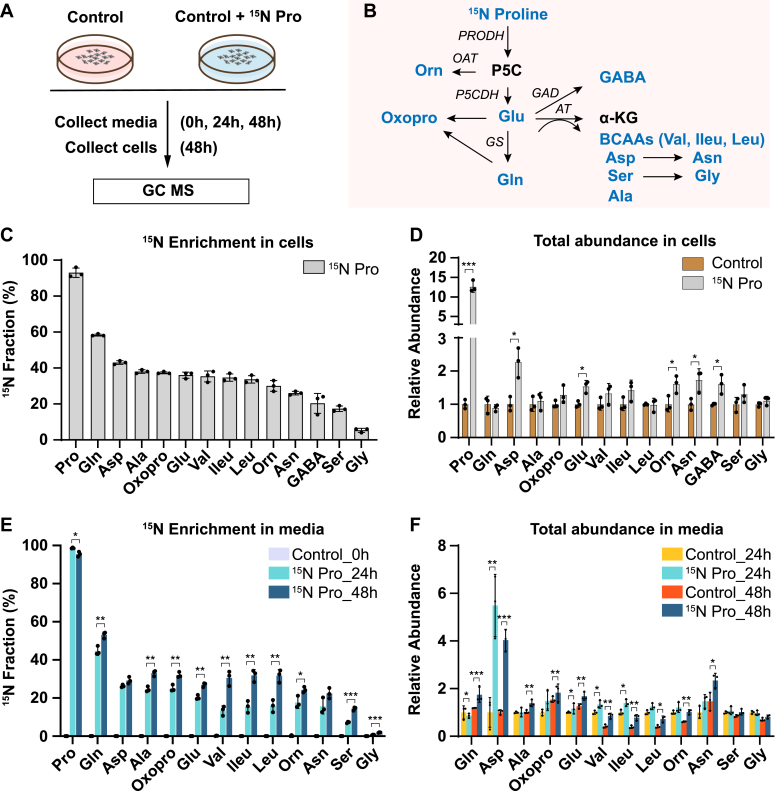
To trace the fate of nitrogen in proline catabolism, we supplemented mature primary human RPE cells in culture with or without 1 mM 15N proline. The physiological concentration of proline in healthy human plasma is 81 to 335 μM (10). We used 1 mM since our previous study showed RPE has a high rate of proline catabolism in regular 2D culture (7). The mature RPE cells formed characteristic cobblestone hexagonal morphology and pigmentation (Fig. S1). The spent media and cells were harvested to analyze 15N-labeled amino acids using GC–MS (Fig. 1A). The nitrogen from proline can be utilized to synthesize 13 types of amino acids (Fig. 1B). Therefore, we targeted these amino acids together with proline in our GC–MS analysis. As expected, human RPE cells actively consumed proline, and 1 mM 15N proline was almost completely used up after incubation for 48 h (Fig. S2). 15N proline was incorporated in all the selected amino acids including glutamine, aspartate, glutamate, alanine, oxoproline, branched-chain amino acids (BCAAs; leucine, valine, and isoleucine), ornithine, asparagine, GABA, serine, and glycine (Fig. 1C). Glutamine and aspartate had the highest enrichment, whereas glycine had the lowest enrichment. Proline also increased the cellular pool sizes of some amino acids, especially aspartate and asparagine (Fig. 1D). The RPE is known to actively transport nutrients to support the neural retina. To investigate whether the RPE can export these proline-derived amino acids out of the cells, we quantified these amino acids in the spent media. Except for GABA, all the other amino acids were labeled by 15N proline in the spend media at 24 h, and the enrichment was further increased at 48 h (Fig. 1E). Similar to the intracellular changes, glutamine, aspartate, alanine, oxoproline, glutamate, and BCAAs were highly enriched by 15N proline in the media. The total amounts of these amino acids were only slightly increased or unchanged except for aspartate, which increased ∼4- to 6-fold by 15N proline (Fig. 1F). These results suggest that proline provides an important nitrogen source for the synthesis of multiple amino acids, which might be exported to support retinal metabolism.
Figure 1.
Human RPE utilizes nitrogen from proline to generate amino acids and exports them into the media.A, human RPE cells were grown for 20 weeks and switched into DMEM with or without 1 mM 15N proline. The spent media were collected at 0, 24, and 48 h, and cells were collected at 48 h for metabolite analysis with GC–MS. B, a schematic of proline nitrogen metabolism into amino acids (colored in blue) through proline dehydrogenase (PRODH), pyrroline-5-carboxylic acid (P5C) dehydrogenase (P5CDH), ornithine transaminase (OAT), glutamate decarboxylase (GAD), glutamine synthetase (GS), and aminotransferase (AT). These amino acids in proline metabolism were selected for GC–MS analysis. C and D, 15N fraction of metabolites derived from 15N proline in RPE cells and the total abundance of all isotopologs relative to control RPE cells without 15N proline. ∗p < 0.05, ∗∗∗p < 0.001, N = 3. E and F, 15N fraction of metabolites derived from 15N proline in RPE spent media at different time points and the total abundance of all isotopologs relative to control RPE culture media without 15N proline at 24 h or 48 h. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, N = 3. Error bars represent SD. DMEM, Dulbecco's modified Eagle's medium; RPE, retinal pigment epithelium.
RPE, but not the neural retina, is responsible for the amino acid synthesis from proline
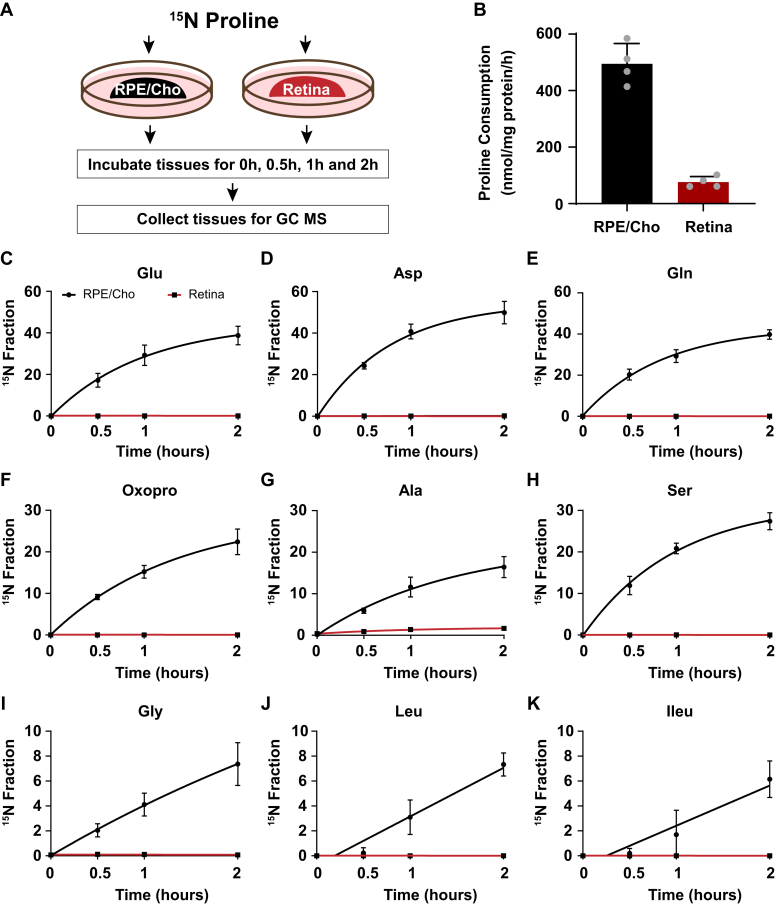
We previously reported that proline is primarily catabolized in the RPE to produce TCA cycle intermediates, but proline catabolism in the neural retina is negligible (7). To investigate the sites of amino acid synthesis from the nitrogen group in proline, we incubated freshly isolated mouse RPE/Cho and the neural retina with 15N proline in the presence of 5 mM glucose in Krebs–Ringer bicarbonate buffer. Then we collected the tissues within 2 h to analyze 15N proline–derived amino acids (Fig. 2A). 15N proline in the media was consumed four to five times faster in the RPE/Cho than the neural retina (Fig. 2B). Consistent with previous findings, 15N from the labeled proline was incorporated rapidly into nine types of amino acids in the RPE/Cho. Similar to human RPE, glutamate, aspartate, glutamine, oxoproline, and alanine were highly enriched with 15N from proline (Fig. 2, C–K). The total abundance of these amino acids remained constant during the 2 h incubation, except for the slight increase of aspartate and decrease of leucine (Fig. S3A). These results further support the previous findings that proline provides important nitrogen sources for amino acid synthesis in the RPE. By contrast, except for a very slight enrichment in alanine, we did not detect enrichment in the other amino acids in the neural retina (Fig. 2, C–K), demonstrating that the neural retina is incapable of utilizing proline as a nitrogen source directly. Proline was not sufficient to maintain the total concentrations of amino acids in the neural retina. In contrast to the RPE/Cho, the total abundance of several amino acids was diminished 20 to 60% after 2 h of incubation (Fig. S3B). These results suggest that while the neural retina requires exogenous amino acids other than proline to support its metabolism, it does not directly utilize proline.
Figure 2.
The synthesis of amino acids from proline occurs mostly in the RPE but not the neural retina ex vivo in mice.A, a schematic of ex vivo incubation of mouse RPE/choroid (RPE/Cho) and retina with 15N proline. Tissues were incubated with 15N proline in Krebs–Ringer bicarbonate buffer (KRB) for 0, 0.5, 1, and 2 h. Tissues and media were collected at these time points for metabolite analysis with GC–MS. B, 15N proline consumption from RPE/Cho and retina. ∗∗∗p < 0.001, N = 4. C–K, 15N fraction of metabolites derived from 15N proline in RPE/Cho and retina at different time points. Error bars represent SD. RPE, retinal pigment epithelium.
Proline nitrogen–derived amino acids from the RPE can be utilized by the neural retina in coculture
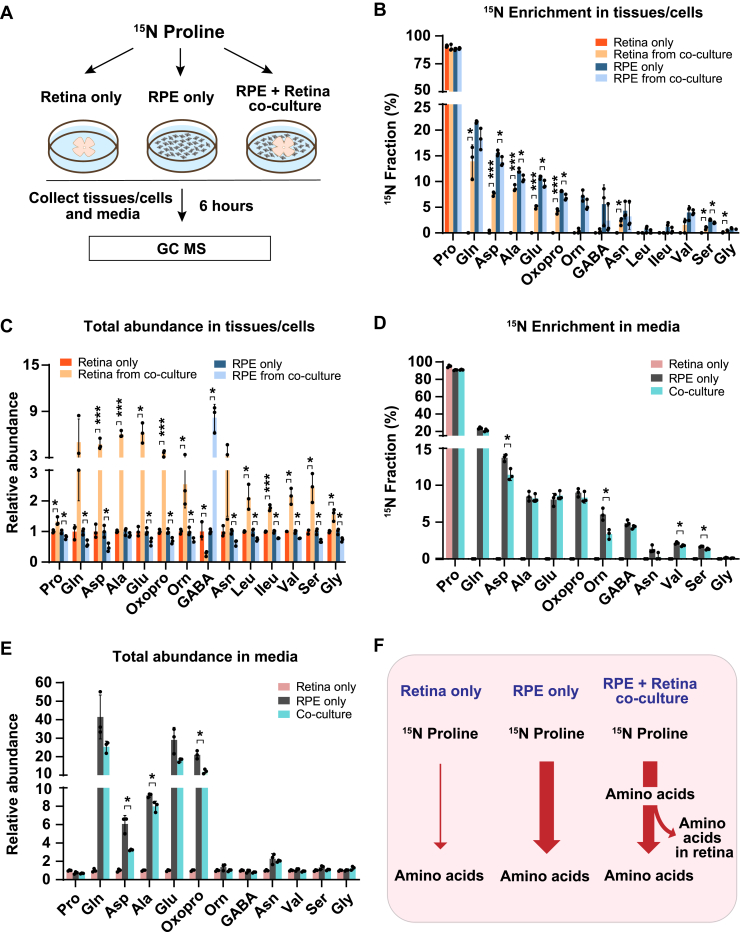
To test whether proline nitrogen–derived amino acids produced from the RPE can be utilized by the neural retina, we cocultured human RPE cells with mouse retina for 6 h in a medium supplemented with 15N proline and cultured human RPE or mouse retina alone under the same conditions as the control groups. The spent medium, RPE cells, and retinas were harvested to analyze proline-derived amino acids by GC–MS (Fig. 3A). As expected, except for 15N proline itself, the neural retina culture alone could not incorporate the 15N from proline into other amino acids. However, cocultured retina had substantial 15N enrichment in multiple amino acids, including glutamine, aspartate, alanine, glutamate, asparagine, valine, serine, and glycine (Fig. 3B). Consistently, compared with RPE culture alone, most of these amino acids showed decreased 15N enrichment in cocultured RPE. Compared with the retina or RPE culture alone, coculturing substantially increased the total abundance (concentrations) of almost all the amino acids except GABA in the retina but conversely, reduced these amino acids in the RPE (Fig. 3C). These results suggest that the retina imports these amino acids produced by the RPE. Similarly, RPE culture alone exported proline-derived amino acids into the media; however, many of these amino acids were reduced in the cocultured media (Fig. 3, D and E). Taken together, we confirm that the neural retina cannot catabolize proline directly, but it can import proline-derived amino acids from the RPE (Fig. 3F).
Figure 3.
The neural retina imports proline-derived amino acids from the RPE.A, human RPE cells and mouse retina were cultured separately or cocultured in DMEM with 1 mM 15N proline for 6 h. Tissues and media were collected for metabolite analysis with GC–MS. B and C, 15N fraction of metabolites derived from 15N proline in RPE cells/retina and the total abundance of all isotopologs in RPE cells/retina from coculture relative to their corresponding control. ∗p < 0.05, ∗∗∗p < 0.001, N = 3. D and E, 15N fraction of metabolites derived from 15N proline in media and the total abundance of all isotopologs in media relative to control media from the retina-only group. ∗p < 0.05, N = 3. F, a schematic of proline nitrogen metabolism in coculture. Retina alone barely utilizes proline for amino acid synthesis, but RPE utilizes proline to produce amino acids to feed the retina. Error bars represent SD. DMEM, Dulbecco's modified Eagle's medium; RPE, retinal pigment epithelium.
RPE exports proline-derived amino acids to the retina in vivo
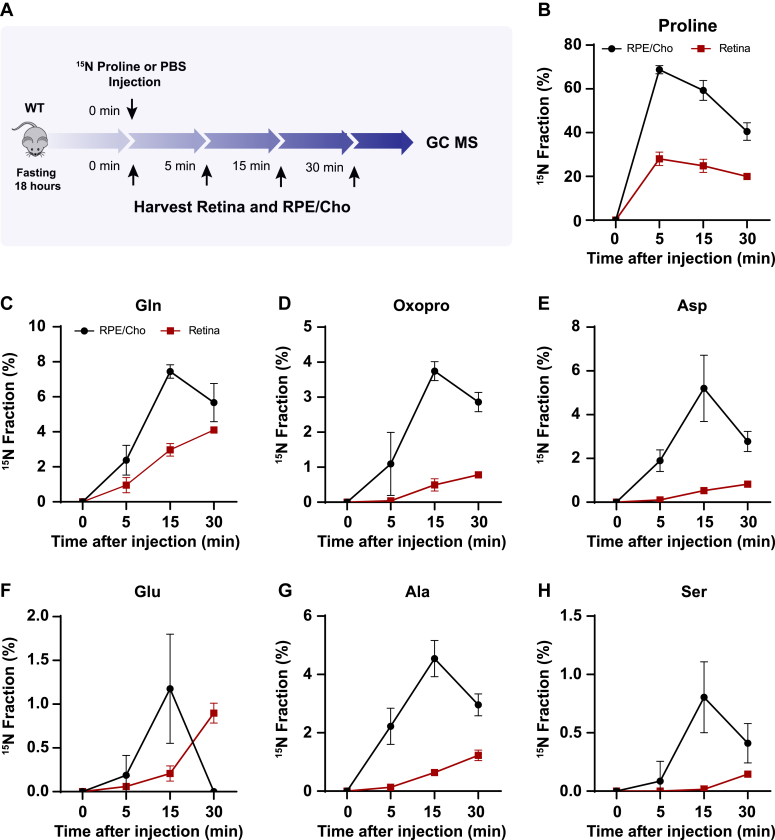
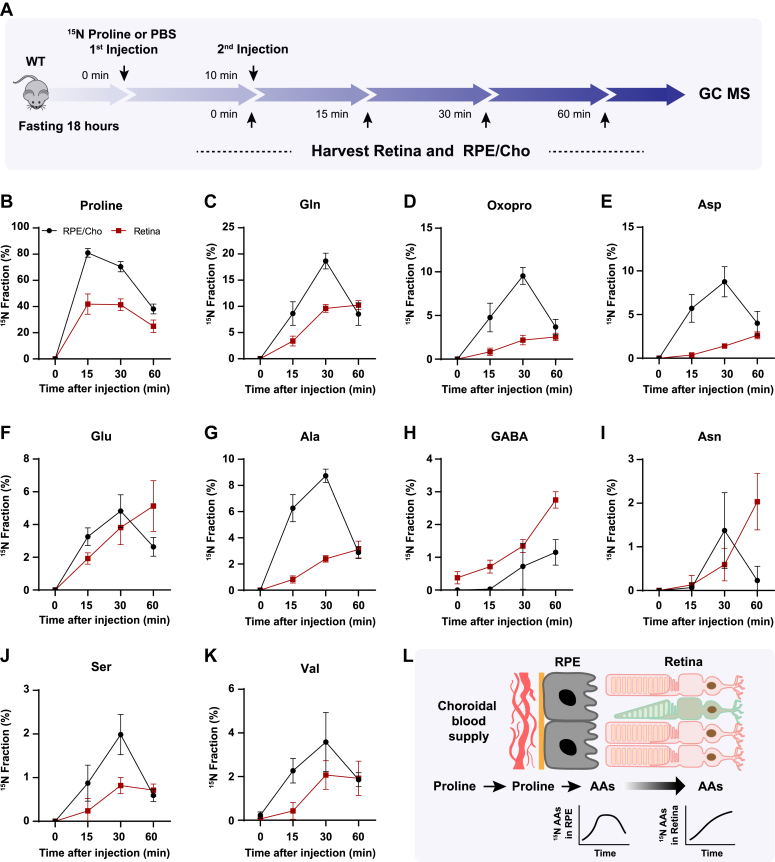
To test whether the retina utilizes proline-derived amino acids from the RPE in vivo, we administered a bolus injection of 15N proline intravenously into WT mice with C57BL/6J background and analyzed 15N-proline-derived amino acids in the retina and RPE/Cho (Fig. 4A). Except for 15N in proline, we detected only six amino acids (glutamine, oxoproline, aspartate, glutamate, alanine, and serine) that were labeled by 15N proline, probably because of the rapid proline catabolism. The enrichment of 15N proline peaked at 5 min after injection in both the retina and RPE/Cho (Fig. 4B). However, the 15N enrichment of other amino acids peaked at 15 min in the RPE/Cho and 30 min in the neural retina, indicating sequential proline catabolism into amino acids from the RPE/Cho to the retina (Fig. 4, C–H). The 15N labeling of all these amino acids in RPE/Cho was higher and earlier than the retina. The RPE/Cho incorporated 15N into the amino acid pool at 5 min, peaked at 15 min, and dropped at 30 min; however, in the retina, there was almost no 15N labeling at 5 min although the labeling appeared at 15 min and was steadily increasing at 30 min. These data strongly suggest the RPE metabolizes proline first before exporting to the retina. Except for transient increases in proline, glutamine, and aspartate, the concentrations of these amino acids were constant in the RPE/Cho and the retina at different time points after injection (Fig. S4, A and B). As the bolus injection had a lower rate of enrichment and labeled fewer amino acids, we administered a second retro-orbital injection of the same dose of 15N proline at 10 min after the first injection and extended the collection time to 60 min (Fig. 5A). Indeed, compared with the bolus injection alone, the double injection increased the number of 15N-labeled amino acids from 6 to 10 and almost doubled the 15N enrichment (Fig. 5, B–K). Similar to the bolus injection, the labeled amino acids appeared earlier with higher enrichment in the RPE/Cho than the retina before 30 min. At 60 min, all the 15N-labeled amino acids, except for GABA, were decreased in the RPE/Cho but continued to rise in the retina (Fig. 5, B–K). These time lags between the RPE and retina further suggest that proline is primarily catabolized in the RPE, and its derived amino acids are utilized by the retina (Fig. 5L). Compared with single injection, double injections have a more transient increase in the concentrations of proline-derived amino acids in the retina (Fig. S5, A and B), suggesting that proline is an important source of synthesis of these amino acids.
Figure 4.
Proline-derived amino acids appear later in the retina than the RPE/Cho.A, a schematic of in vivo tracing of 15N proline. Mice were fasted for 18 h and received a single injection of 15N proline (150 mg/kg) or PBS. Retina and RPE/Cho were harvested at 0, 5, 15, and 30 min after injection for metabolite analysis with GC–MS. B–H, 15N fraction of proline and metabolites derived from 15N proline in RPE/Cho and retina at different time points. N = 4. Error bars represent SD. Cho, choroid; RPE, retinal pigment epithelium.
Figure 5.
There is a delayed synthesis of amino acids from15N proline in the retina compared with the RPE/Cho.A, a schematic of in vivo tracing of 15N proline with double injection. Mice were fasted for 18 h and received two injections of 15N proline (150 mg/kg) or PBS at 0 min and 10 min, respectively. Retina and RPE/Cho were harvested at 0, 5, 15, and 30 min after the second injection for metabolite analysis with GC–MS. B–K, 15N fraction of proline and metabolites derived from 15N proline in RPE/Cho and retina at different time points. N = 8. L, a schematic of proline catabolism into amino acids in RPE to be exported into the neural retina. Error bars represent SD. Cho, choroid; RPE, retinal pigment epithelium.
Loss of PRODH blocks the utilization of proline-derived amino acids in the retina from the RPE/Cho
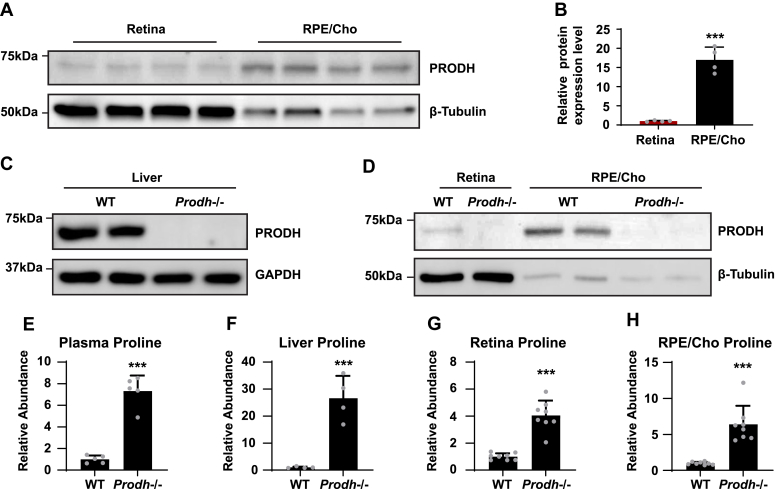
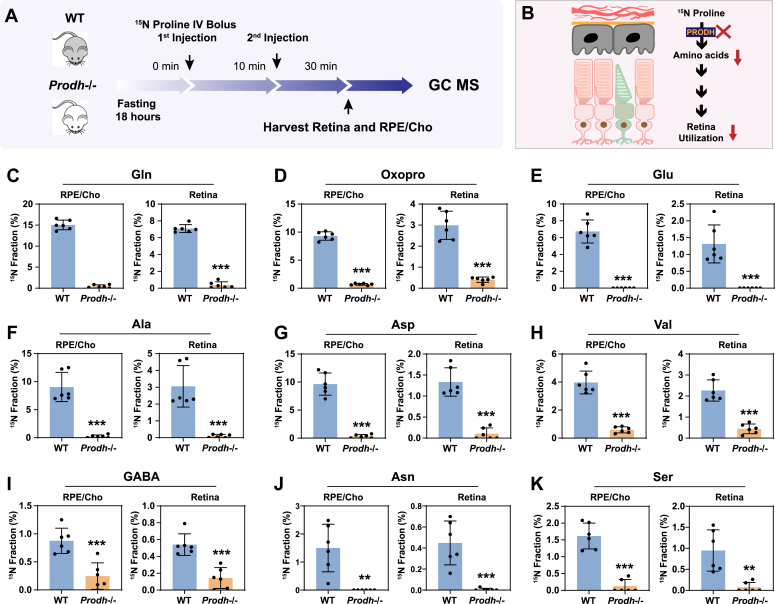
To investigate whether proline catabolism in the RPE is critical for the amino acid supply in the retina, we tested the protein expression of PRODH, the key enzyme in proline catabolism. Immunoblotting showed that PRODH protein expression in RPE/Cho is 15- to 20-fold higher than the neural retina (Fig. 6, A and B, Fig. S6), which is consistent with our previous results that 15N proline is primarily catabolized in the RPE/Cho but not the retina. The inbred mouse line PRO/ReJ (Prodh−/−) carries a G -> T substitution 135 bp upstream of the native terminal codon in the PRODH gene, which leads to premature translational termination (11). Because of the lack of PRODH activity, this mouse line has hyperprolinemia (12). We validated that the PRODH protein expression was not detectable in the liver, retina, and RPE/Cho of Prodh−/− mice (Fig. 6, C and D, Fig. S6). Consistently, loss of PRODH caused substantial accumulation of proline in the liver, plasma, retina, and RPE/Cho (Fig. 6, E–H). To study whether deletion of PRODH can block the synthesis and export of proline-derived amino acids from the RPE to the retina, we administered a double injection of 15N proline as mentioned before and collected the retina and RPE/Cho 30 min after the first injection (Fig. 7, A and B). Similarly, we detected nine 15N-labeled amino acids in both the retina and RPE/Cho in the WT mice with 15N enrichment ranging from 1 to 15%. RPE/Cho had higher enrichment of all the amino acids when compared with the retina. However, in the Prodh−/− mice, the 15N enrichment of these amino acids was significantly diminished to less than 1% (Fig. 7, C–K), demonstrating that proline catabolism in the RPE is required for the utilization of proline-derived amino acids in the retina. Compared with the WT mice, the Prodh−/− mice had lower concentrations of glutamate, alanine, and aspartate in the RPE/Cho (Fig. S7A), suggesting that proline is important to maintain the availability of these amino acids. Alanine also was lower in the retina of the Prodh−/− mice. However, several amino acids including glutamine, oxoproline, and asparagine were increased in Prodh−/− retina (Fig. S7B), indicating there might be compensatory pathways for the synthesis of these amino acids.
Figure 6.
PRODH is more abundant in the RPE/Cho, and deletion of Prodh causes proline accumulation.A, RPE/Cho has higher PRODH protein expression than RPE/Cho. β-tubulin is used as loading control. B, relative protein expression level of PRODH in retina and RPE/Cho. The relative protein level is the protein expression relative to the retina after normalized to the expression of tubulin. ∗∗∗p < 0.001, N = 4 (replicate lanes from four independent samples). C and D, Prodh mutation causes the depletion of PRODH proteins in liver, retina, and RPE/Cho in Prodh−/− mice. E–H, total abundance of proline in liver, plasma, RPE/Cho, and retina from Prodh−/− mice relative to those tissues from WT mice. ∗∗∗p < 0.001, N ≥ 4. Error bars represent SD. Cho, choroid; PRODH, proline dehydrogenase; RPE, retinal pigment epithelium.
Figure 7.
Depletion of PRODH blocks the synthesis of amino acids from proline in the RPE and its export into the retina.A, a schematic of in vivo tracing of 15N proline. Mice were fasted for 18 h and received two injections of 15N proline (150 mg/kg) at 0 min and 10 min, respectively. Retina and RPE/Cho were harvested at 30 min after the second injection for metabolite analysis with GC–MS. B, Prodh−/− blocks the synthesis of amino acids from 15N proline in the RPE and its export into the retina. C–K, 15N fraction of metabolites derived from 15N proline in RPE/Cho and retina. N = 6. Error bars represent SD. Cho, choroid; PRODH, proline dehydrogenase; RPE, proline dehydrogenase.
Discussion
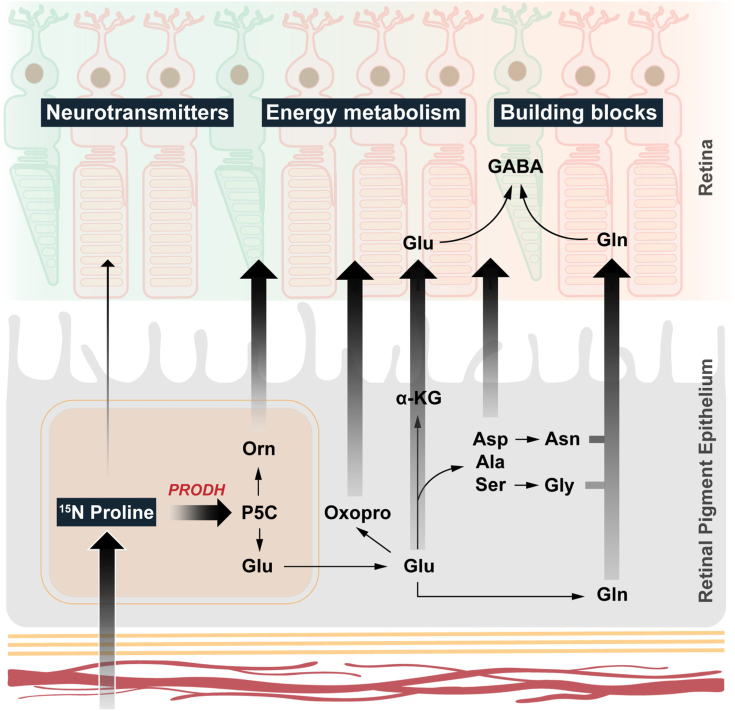
In this study, we found that proline is a nitrogen source for synthesizing 13 types of amino acids in human RPE in vitro. More importantly, these proline-derived amino acids can be exported from the RPE to be used by the neural retina. The retina itself is unable to directly catabolize proline (Fig. 8). Our findings demonstrate that in addition to its classical function as a nutrient transporter between the Cho and the outer retina, the RPE serves as a nutrient-manufacturing center that supports retinal metabolism.
Figure 8.
RPE synthesizes and export amino acids to the retina using nitrogen from proline. RPE uptakes and catabolizes proline through PRODH to produce P5C, which can be converted into ornithine, glutamate, and multiple amino acids. All these amino acids can be exported to be utilized by the neural retina. P5C, pyrroline-5-carboxylate; PRODH, proline dehydrogenase; RPE, retinal pigment epithelium.
Photoreceptors in the outer retina are among the most energy-demanding cells in the human body, and their metabolic needs depend on their neighboring RPE. Because of the lack of a direct blood supply, photoreceptors must take up nutrients including glucose, amino acids, fatty acids, vitamins, and minerals from the choriocapillaris through the RPE. Deletion of the glucose transporter in the RPE is sufficient to cause photoreceptor degeneration. Besides directly transporting nutrients to the outer retina, the RPE also serves as a nutrient reservoir and manufacturer to support retinal metabolism. Transcriptomics data show that genes involved in glucose storage, proline metabolism, and serine metabolism are highly upregulated in human RPE (6). Cultured RPE can store glucose as glycogen and oxidize glucose as mitochondrial intermediates to export them to the retinal side (5, 13). RPE can recycle photoreceptor-derived fatty acids and succinate into ketone bodies and malate to support the metabolic needs of the retina. Similar to the catabolism of its carbon skeleton into mitochondrial intermediates (5), proline feeds its nitrogen into multiple amino acids in the RPE to export them for use by the retina. A common theme of these findings is the idea that active RPE metabolism is essential for producing nutrients necessary to support the metabolically demanding neural retina.
Most of the enzymes that contribute to proline nitrogen metabolism are located in the mitochondria including PRODH, P5C dehydrogenase, OAT, and mitochondrial isoforms of transaminases. Inhibition of mitochondrial respiration can block proline catabolism and the function of nutrient release in RPE (14), suggesting that the supply of amino acids from proline catabolism to the retina requires healthy RPE mitochondrial metabolism. Mitochondrial metabolic defects in the RPE are known to cause RPE dysfunction including dedifferentiation and lipid deposit formation, resulting in subsequent retinal degeneration and AMD in animal models and human donors (1, 15, 16). However, the mechanisms by which RPE mitochondrial dysfunction leads to photoreceptor degeneration remain unclear. Interestingly, dedifferentiated RPE loses the ability to utilize proline (7), and the expression of transcripts that encode proteins involved in proline transport and catabolism is significantly diminished in dedifferentiated RPE (17). We speculate that a consequence of mitochondrial defects in the RPE may diminish nutrient production including proline catabolism. This could result in decreased export of amino acids and other nutrients to the neural retina, causing metabolic stress in photoreceptors and eventual degeneration.
Photoreceptors have a high metabolic need for amino acids. Every day, ∼10% of the photoreceptor outer segment material is shed and resynthesized. Rhodopsin makes up more than 90% of outer segment proteins (18), and human rhodopsin protein consists of 9.2% alanine, 23.8% BCAAs, and 11.2% serine and glycine (19). Photoreceptors have extremely high energy expenditure with each photoreceptor consuming 1 to 1.5 × 108 ATP/s (20). To achieve phototransduction, photoreceptors require the availability of a large amount of cGMP from GTP. The synthesis of ATP, GTP, and cGMP requires nitrogen from aspartate, glutamine, and glycine. Retinal cells including photoreceptors and glial cells need to maintain high concentrations of glutamate, aspartate, GABA, glutamine, and glutathione for neurotransmission, the transfer of reducing equivalents and for activities that counter the effects of oxidative stress (21, 22, 23). By unbiasedly analyzing the nutrient consumption of retinas from media that are nutrient rich, we reported that glutamate and aspartate are specifically consumed by human retinas, mouse retinas, and mature human retinal organoid but not by immature organoids (6). Those observations are consistent with our finding in this study that proline is an important source of nitrogen for synthesis in the RPE of multiple amino acids that are used by the neural retina.
There are multiple lines of evidence that proline metabolism is uniquely important in retinal health and disease. First, proline is the only amino acid whose consumption is linked to RPE differentiation (7, 9). Proline stimulates RPE mitochondrial function. Proline, along with its derivatives such as hydroxyproline and glycine, constitutes the primary components of the extracellular matrix. Both mitochondrial function and extracellular matrix turnover play crucial roles in RPE function and the pathogenesis of AMD (9, 24). Second, mutations of multiple genes involved in proline metabolism, including OAT, P5C dehydrogenase, and P5C reductase, cause inherited retinal degenerations and RPE atrophy in patients and animal models (9, 25, 26, 27). Third, the proline transporter, SLC6A20, is a signature RPE gene that is highly enriched in both human and mouse RPE (28, 29). Its expression correlates with proline consumption during RPE differentiation (7). Mutations in SLC6A20 are associated with AMD and macular telangiectasia (30, 31). Finally, the level of proline in aqueous humor and plasma is perturbed in patients with AMD, diabetic retinopathy, and glaucoma in multiple metabolomics studies (9).
Healthy RPE can flexibly utilize amino acids including proline, glutamine, glutamate, aspartate, BCAAs, and alanine when they are given as the sole nitrogen source. However, under regular enriched RPE culture media containing all the amino acids, human RPE utilizes proline at a much faster rate than other amino acids. In addition, RPE produces more glutamate and alanine than it consumes. The catabolism of glutamine can generate free ammonia that can be toxic to retinal neurons (32). Compared with aspartate and BCAAs, proline has high solubility, and its catabolism can transfer electrons directly to FAD to form FADH2 (9). The RPE is known to have abundant FAD and its precursor riboflavin for FAD-dependent metabolism, including succinate and fatty acid oxidation (33, 34, 35). Riboflavin deficiency can deplete FAD, resulting in RPE dysfunction and retinal degeneration (36). Compared with other amino acids, proline catabolism is more energy efficient as it generates both FADH2 and NADH2 without producing free ammonia. Dietary proline supplementation in young pigs increases protein synthesis in a dose-dependent manner while decreasing concentrations of urea in plasma (37).
Different from RPE, the neural retina resides in a low-oxygen environment so ubiquinol (QH2) produced by mitochondria complex I accumulates in the retina and transfers its electrons to produce succinate (34, 38, 39). The PRODH reaction also generates QH2. We hypothesize that the production of QH2 from complex I and the very low amounts of PRODH enzyme in the retina severely limit the oxidation of proline in the retina. Our results confirm that the retina does not catabolize proline. Furthermore, gene expression data show significantly lower expression levels of proline catabolism genes in the human retina compared with RPE (6). We found proline can be a nitrogen source for 13 types of amino acids, covering 50% of proteogenic amino acids. The amounts of glutamate, aspartate, and alanine are diminished in Prodh−/− mice, supporting the idea that proline can be an important nitrogen donor in retinal protein synthesis and metabolism.
Surprisingly, the amounts of glutamine, oxoproline, and asparagine are increased in Prodh−/− retinas, suggesting compensatory utilization of other alternative nitrogen donors. Glutamine and asparagine are sensitive to the changes of ammonia (40), and oxoproline can be spontaneously formed by glutamine. Blocking nitrogen utilization from proline may activate ammonia production from the deamination of amino acids and nucleosides, leading to an increase in the donation of glutamine and asparagine to other amino acids (41). Further studies are needed to identify these compensatory mechanisms in diseased RPE and retina.
Among proline nitrogen–derived amino acids, GABA is the only amino acid with a decreased concentration in the cocultured retinas but increased in the cocultured RPE (Fig. 3C). These results suggest a GABA efflux from the retina to the RPE. Consistently, in the RPE from 15N proline–injected mice, the percentage of 15N labeling drops at 60 min for all the amino acids except for GABA (Fig. 5). However, the concentration of retinal GABA remains constant or slightly increased after injection. One possibility is that there is a rapid rate of GABA synthesis and recycling between the RPE and retina in vivo. GABA can also be produced through its transaminase from succinic semialdehyde, which is not in the culture media. GABA is located mostly in ganglion cells, amacrine cells, horizontal cells, and Müller glial cells (42, 43). Ganglion cells and other inner neurons are easily stressed and damaged after transection of the optic nerve in the retinal explant culture. Another possibility for the decrease of GABA content in vitro is that it may be caused by the stimulation and damage of these inner retinal neurons after isolation.
In conclusion, we demonstrate that the neural retina needs nitrogen to maintain its amino acid pools and that proline is an important source of nitrogen. However, the retina by itself is nearly unable to catabolize proline directly. Instead, it relies on the RPE to extract the nitrogen from proline. The RPE actively catabolizes proline to synthesize multiple amino acids that are then exported to support retinal metabolism. These findings highlight the importance of RPE metabolism in supporting nitrogen utilization in the neural retina, providing insight on a potential mechanism of RPE-initiated retinal degenerative diseases.
Experimental procedures
Reagents
All the reagents and resources are detailed in the key resources table (Table S1).
Animals
Both genders of ∼2-month-old WT mice (C57 B6/J background, stock no.: 000664) and Prodh−/− mice (C57BL/6J and 129P1/ReJ background, stock no.: 001137) were purchased from the Jackson laboratory. Mouse experiments were performed in accordance with the National Institutes of Health guidelines, and the protocol was approved by the Institutional Animal Care and Use Committee of West Virginia University.
For the tracer study in vivo, mice were deprived of food for 18 h and received bolus or double intravenous injections of 15N proline retro-orbitally at 150 mg/kg under anesthesia with isoflurane. Mouse retina and RPE/Cho were quickly harvested and snap-frozen in liquid nitrogen and stored at −80 °C before use.
Human RPE cell culture
Human RPE cells were isolated from specimens of fetal (78–98 days) human globes, obtained from the Birth Defects Research Laboratory at the University of Washington with ethics board approval. This study was performed in accordance with ethical and legal guidelines of the University of Washington institutional review board, and experimental protocols follow ethical guidelines from the Declaration of Helsinki.
Human RPE cells were cultured as previously described (5, 7). Briefly, human fetal RPE cells were passaged using 0.25% trypsin and plated in 5% fetal bovine serum (FBS; v/v) in minimum essential medium-alpha supplemented with N1, nonessential amino acid, 1% pencillin/streptomycin (v/v), and a mix of taurine, hydrocortisone, and triiodothyronine with rock inhibitor for the first week. The media are then changed to the same recipe with reduced 1% (v/v) FBS in minimum essential medium-alpha media for maintenance. Human fetal RPE was grown in 12-well plates on Matrigel for 20 weeks for experiment. Upon experiment, media were changed to clear Dulbecco's modified Eagle's medium (no glucose, sodium pyruvate, glutamine, or phenol red) and then supplemented with 5.5 mM glucose and 1 mM unlabeled or 1 mM 15N-proline with 1% FBS and penicillin/streptomycin (v/v). Media were collected at 0, 24, and 48 h of culturing for metabolomic analysis. Human RPE cells were washed with 154 mM NaCl solution and collected on dry ice after 48 h of culturing for the same purpose.
Retina and RPE/Cho explant culture
Mouse retina and RPE/Cho were quickly isolated in 200 μl of cold Hank's balanced salt solution as previously described (7, 40). The retina and RPE/Cho were transferred into 200 μl of preincubated Krebs–Ringer bicarbonate buffer (7) with 15N proline in the presence of 5 mM glucose, followed by a 2 h incubation at 37 °C in a CO2 incubator. The tissues were collected and snap-frozen in liquid nitrogen and stored at −80 °C before use.
RPE and retina coculture
Human RPE cells were grown for 12 to 16 weeks in 12-well plates coated with Matrigel before being used for coculture experiments. RPE was equilibrated in unlabeled proline media for approximately 1 h before starting the coculture. Murine retinas were harvested from 6- to 8-week-old C57Bl/6J mice and allowed to equilibrate in unlabeled proline media for about 1 h. After equilibration, the cells alone or cocultured with the retina were switched to 15N proline media and incubated for 6 h. After 6 h, media were harvested to a separate tube from the tissue. The retinas were quickly removed from hfRPE and placed into a cold 154 mM NaCl solution, spun down, liquid removed, and snap frozen. RPE cells were washed with the same saline solution and then harvested on ice with cold 80% methanol and stored frozen at −80 °C until processing.
Metabolite analysis with GC–MS
Metabolites from tissue, cells, and media were extracted in 80% cold methanol as described previously (5). Metabolite extracts were transferred into glass inserts containing internal standard (myristic acid d27 in 1 mg/ml) and dried in a speedvac. Dried samples were derivatized by methoxyamine and N-tertbutyldimethylsilyl-N-methyl trifluoroacetamide and analyzed by the Agilent 7890B/5977B GC–MS system with DB-5MS column (30 m × 0.25 mm × 0.25 μm film) as we described in detail before (5, 40). Mass spectra were collected from 80 to 600 m/z under SIM mode. Table S2 lists the detailed parameters including monitored ions for the measured metabolites. The data were analyzed by Agilent MassHunter Quantitative Analysis Software by extracting ion abundance for each monitored ion in Table S2. Raw total ion abundance for isotopologs, the purity of the tracers, and derivatization reagents were imported in ISOCOR software (https://isocor.readthedocs.io/en/latest/) to calculate the percentage of labeling by correcting natural abundance and tracer purity (40). The percentage of labeled ions or enrichment was presented as 15N fraction, which is the ratio of 15N-labeled ion abundance with the total abundance. The total abundance was the total ion abundance from all isotopologs. The protein concentrations in the pellet after metabolite extraction were determined by bicinchoninic acid assay to normalize the metabolite data as reported (PMID: 26358904).
Western blot
Western blot was performed as previously described (44). Briefly, proteins were extracted from tissues using radioimmunoprecipitation assay buffer and determined for protein concentration by the bicinchoninic acid protein assay kit. Protein samples (20 μg) were boiled and loaded onto SDS-PAGE gels. The gels were transferred to nitrocellulose membranes and blocked with 5% nonfat milk in 1× Tris-buffered saline containing 0.1% Tween-20. The membranes were incubated with primary antibodies (see details in Table S1) at 4 °C overnight. After washing with 1× PBS containing 0.1% Tween-20, the membranes were incubated with a rabbit or a mouse secondary antibody conjugated with horseradish peroxidase (1:2000 dilution) for 1 h, followed by three washes with 1× PBS containing 0.1% Tween-20. A chemiluminescence reagent kit was used to visualize protein bands with horseradish peroxidase secondary antibodies. The original immunoblot images were converted to 8 bit format and underwent minor adjustments in brightness and contrast using ImageJ (NIH, https://imagej.nih.gov/ij/download.html), ensuring that the overall visualization of the bands remained unchanged.
Statistics
The significance of differences between means was determined by unpaired two-tailed t tests or one-way ANOVA with Bonferroni post hoc test using GraphPad Prism 9.0 (GraphPad Software, Inc). Only p < 0.05 was considered to be significant. All data are presented as the mean ± SD.
Ethical approval
The mouse experiments were performed in accordance with the National Institutes of Health guidelines and the protocol approved by the Institutional Animal Care and Use Committee of West Virginia University.
The use of human RPE cells were reviewed by the University of Washington Institutional Review Board, and experimental protocols follow ethical guidelines from the Declaration of Helsinki.
Data availability
GC–MS data generated in this study are provided in the supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This work was supported by the National Institute of General Medical Sciences P20GM144230 (Visual Sciences Center of Biomedical Research Excellence), National Institute of Child Health and Human Development grant R24HD000836 (The Birth Defects Research Laboratory from the University of Washington), Research to Prevent Blindness unrestricted grant (University of Washington), and funds for Core facilities P20 GM103434 (to W. V. INBRE grant).
Author contributions
J. R. C. and J. D. conceptualization; S. Z., R. X., A. L. E., Y. W., R. M., J. B. H., J. R. C., and J. D. investigation; S. Z., R. X., A. L. E., J. B. H., J. R. C., and J. D. writing–original draft; J. R. C. and J. D. supervision; J. R. C. and J. D. funding acquisition.
Funding and additional information
This work was supported by the National Eye Institute grant EY034364 (to J. R. C. and J. D.), EY031324 (to J. D.), EY032462 (to J. D.), Bright Focus Foundation M2020141 (to J. D.), MD2020217 (to J. R. C.), the Retina Research Foundation (to J. D.).
Reviewed by members of the JBC Editorial Board. Edited by Kirill Martemyanov
Contributor Information
Jennifer R. Chao, Email: jrchao@uw.edu.
Jianhai Du, Email: jianhai.du@hsc.wvu.edu.
Supporting information
References
- 1.Ferrington D.A., Fisher C.R., Kowluru R.A. Mitochondrial defects drive degenerative retinal diseases. Trends Mol. Med. 2020;26:105–118. doi: 10.1016/j.molmed.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin H., Xu H., Liang F.Q., Liang H., Gupta P., Havey A.N., et al. Mitochondrial DNA damage and repair in RPE associated with aging and age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2011;52:3521–3529. doi: 10.1167/iovs.10-6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hurley J.B. Retina metabolism and metabolism in the pigmented epithelium: a busy intersection. Annu. Rev. Vis. Sci. 2021;7:665–692. doi: 10.1146/annurev-vision-100419-115156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanow M.A., Giarmarco M.M., Jankowski C.S., Tsantilas K., Engel A.L., Du J., et al. Biochemical adaptations of the retina and retinal pigment epithelium support a metabolic ecosystem in the vertebrate eye. Elife. 2017;6 doi: 10.7554/eLife.28899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao J.R., Knight K., Engel A.L., Jankowski C., Wang Y., Manson M.A., et al. Human retinal pigment epithelial cells prefer proline as a nutrient and transport metabolic intermediates to the retinal side. J. Biol. Chem. 2017;292:12895–12905. doi: 10.1074/jbc.M117.788422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li B., Zhang T., Liu W., Wang Y., Xu R., Zeng S., et al. Metabolic features of mouse and human retinas: rods versus cones, macula versus periphery, retina versus RPE. iScience. 2020;23 doi: 10.1016/j.isci.2020.101672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yam M., Engel A.L., Wang Y., Zhu S., Hauer A., Zhang R., et al. Proline mediates metabolic communication between retinal pigment epithelial cells and the retina. J. Biol. Chem. 2019;294:10278–10289. doi: 10.1074/jbc.RA119.007983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kennaway N.G., Stankova L., Wirtz M.K., Weleber R.G. Gyrate atrophy of the choroid and retina: characterization of mutant ornithine aminotransferase and mechanism of response to vitamin B6. Am. J. Hum. Genet. 1989;44:344–352. [PMC free article] [PubMed] [Google Scholar]
- 9.Du J., Zhu S., Lim R.R., Chao J.R. Proline Metabolism and Transport in Retinal Health and Disease. Amino acids. 2021;53:1789–1806. doi: 10.1007/s00726-021-02981-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toyoshima K., Nakamura M., Adachi Y., Imaizumi A., Hakamada T., Abe Y., et al. Increased plasma proline concentrations are associated with sarcopenia in the elderly. PLoS One. 2017;12 doi: 10.1371/journal.pone.0185206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gogos J.A., Santha M., Takacs Z., Beck K.D., Luine V., Lucas L.R., et al. The gene encoding proline dehydrogenase modulates sensorimotor gating in mice. Nat. Genet. 1999;21:434–439. doi: 10.1038/7777. [DOI] [PubMed] [Google Scholar]
- 12.Blake R.L., Russell E.S. Hyperprolinemia and prolinuria in a new inbred strain of mice, PRO-Re. Science. 1972;176:809–811. doi: 10.1126/science.176.4036.809. [DOI] [PubMed] [Google Scholar]
- 13.Senanayake P., Calabro A., Hu J.G., Bonilha V.L., Darr A., Bok D., et al. Glucose utilization by the retinal pigment epithelium: evidence for rapid uptake and storage in glycogen, followed by glycogen utilization. Exp. Eye Res. 2006;83:235–246. doi: 10.1016/j.exer.2005.10.034. [DOI] [PubMed] [Google Scholar]
- 14.Zhang R., Engel A.L., Wang Y., Li B., Shen W., Gillies M.C., et al. Inhibition of mitochondrial respiration impairs nutrient consumption and metabolite transport in human retinal pigment epithelium. J. Proteome Res. 2021;20:909–922. doi: 10.1021/acs.jproteome.0c00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao C., Yasumura D., Li X., Matthes M., Lloyd M., Nielsen G., et al. mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J. Clin. Invest. 2011;121:369–383. doi: 10.1172/JCI44303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felszeghy S., Viiri J., Paterno J.J., Hyttinen J.M.T., Koskela A., Chen M., et al. Loss of NRF-2 and PGC-1alpha genes leads to retinal pigment epithelium damage resembling dry age-related macular degeneration. Redox Biol. 2019;20:1–12. doi: 10.1016/j.redox.2018.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boles N.C., Fernandes M., Swigut T., Srinivasan R., Schiff L., Rada-Iglesias A., et al. Epigenomic and transcriptomic changes during human RPE EMT in a stem cell model of epiretinal membrane pathogenesis and prevention by nicotinamide. Stem Cell Reports. 2020;14:631–647. doi: 10.1016/j.stemcr.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filipek S., Stenkamp R.E., Teller D.C., Palczewski K. G protein-coupled receptor rhodopsin: a prospectus. Annu. Rev. Physiol. 2003;65:851–879. doi: 10.1146/annurev.physiol.65.092101.142611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nathans J., Hogness D.S. Isolation and nucleotide sequence of the gene encoding human rhodopsin. Proc. Natl. Acad. Sci. U. S. A. 1984;81:4851–4855. doi: 10.1073/pnas.81.15.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingram N.T., Fain G.L., Sampath A.P. Elevated energy requirement of cone photoreceptors. Proc. Natl. Acad. Sci. U. S. A. 2020;117:19599–19603. doi: 10.1073/pnas.2001776117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winkler B.S., Giblin F.J. Glutathione oxidation in retina: effects on biochemical and electrical activities. Exp. Eye Res. 1983;36:287–297. doi: 10.1016/0014-4835(83)90013-1. [DOI] [PubMed] [Google Scholar]
- 22.Kalloniatis M., Marc R.E., Murry R.F. Amino acid signatures in the primate retina. J. Neurosci. 1996;16:6807–6829. doi: 10.1523/JNEUROSCI.16-21-06807.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinamaki A.A., Muhonen A.S., Piha R.S. Taurine and other free amino acids in the retina, vitreous, lens, iris-ciliary body, and cornea of the rat eye. Neurochem. Res. 1986;11:535–542. doi: 10.1007/BF00965323. [DOI] [PubMed] [Google Scholar]
- 24.Phang J.M., Liu W., Zabirnyk O. Proline metabolism and microenvironmental stress. Annu. Rev. Nutr. 2010;30:441–463. doi: 10.1146/annurev.nutr.012809.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolthuis D.F., van Asbeck E., Mohamed M., Gardeitchik T., Lim-Melia E.R., Wevers R.A., et al. Cutis laxa, fat pads and retinopathy due to ALDH18A1 mutation and review of the literature. Eur. J. Paediatr. Neurol. 2014;18:511–515. doi: 10.1016/j.ejpn.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Liang S.T., Audira G., Juniardi S., Chen J.R., Lai Y.H., Du Z.C., et al. Zebrafish carrying pycr1 gene deficiency display aging and multiple behavioral abnormalities. Cells. 2019;8:453. doi: 10.3390/cells8050453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elnahry A.G., Elnahry G.A. Gyrate atrophy of the choroid and retina: a review. Eur. J. Ophthalmol. 2022;32:1314–1323. doi: 10.1177/11206721211067333. [DOI] [PubMed] [Google Scholar]
- 28.Liao J.L., Yu J., Huang K., Hu J., Diemer T., Ma Z., et al. Molecular signature of primary retinal pigment epithelium and stem-cell-derived RPE cells. Hum. Mol. Genet. 2010;19:4229–4238. doi: 10.1093/hmg/ddq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strunnikova N.V., Maminishkis A., Barb J.J., Wang F., Zhi C., Sergeev Y., et al. Transcriptome analysis and molecular signature of human retinal pigment epithelium. Hum. Mol. Genet. 2010;19:2468–2486. doi: 10.1093/hmg/ddq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao X.R., Huang H., Kim H. Genome-wide association analyses identify 139 loci associated with macular thickness in the UK Biobank cohort. Hum. Mol. Genet. 2019;28:1162–1172. doi: 10.1093/hmg/ddy422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonelli R., Jackson V.E., Prasad A., Munro J.E., Farashi S., Heeren T.F.C., et al. Identification of genetic factors influencing metabolic dysregulation and retinal support for MacTel, a retinal disorder. Commun. Biol. 2021;4:274. doi: 10.1038/s42003-021-01788-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose C., Michalak A., Rao K.V., Quack G., Kircheis G., Butterworth R.F. L-ornithine-L-aspartate lowers plasma and cerebrospinal fluid ammonia and prevents brain edema in rats with acute liver failure. Hepatology. 1999;30:636–640. doi: 10.1002/hep.510300311. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y., Grenell A., Zhong F., Yam M., Hauer A., Gregor E., et al. Metabolic signature of the aging eye in mice. Neurobiol. Aging. 2018;71:223–233. doi: 10.1016/j.neurobiolaging.2018.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hass D.T., Bisbach C.M., Robbings B.M., Sadilek M., Sweet I.R., Hurley J.B. Succinate metabolism in the retinal pigment epithelium uncouples respiration from ATP synthesis. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.110917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reyes-Reveles J., Dhingra A., Alexander D., Bragin A., Philp N.J., Boesze-Battaglia K. Phagocytosis-dependent ketogenesis in retinal pigment epithelium. J. Biol. Chem. 2017;292:8038–8047. doi: 10.1074/jbc.M116.770784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinha T., Ikelle L., Makia M.S., Crane R., Zhao X., Kakakhel M., et al. Riboflavin deficiency leads to irreversible cellular changes in the RPE and disrupts retinal function through alterations in cellular metabolic homeostasis. Redox Biol. 2022;54 doi: 10.1016/j.redox.2022.102375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu G., Bazer F.W., Burghardt R.C., Johnson G.A., Kim S.W., Knabe D.A., et al. Proline and hydroxyproline metabolism: implications for animal and human nutrition. Amino acids. 2011;40:1053–1063. doi: 10.1007/s00726-010-0715-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spinelli J.B., Rosen P.C., Sprenger H.G., Puszynska A.M., Mann J.L., Roessler J.M., et al. Fumarate is a terminal electron acceptor in the mammalian electron transport chain. Science. 2021;374:1227–1237. doi: 10.1126/science.abi7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bisbach C.M., Hass D.T., Robbings B.M., Rountree A.M., Sadilek M., Sweet I.R., et al. Succinate can Shuttle reducing Power from the Hypoxic retina to the O(2)-rich pigment epithelium. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu R., Ritz B.K., Wang Y., Huang J., Zhao C., Gong K., et al. The retina and retinal pigment epithelium differ in nitrogen metabolism and are metabolically connected. J. Biol. Chem. 2020;295:2324–2335. doi: 10.1074/jbc.RA119.011727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang T., Steel G., Milam A.H., Valle D. Correction of ornithine accumulation prevents retinal degeneration in a mouse model of gyrate atrophy of the choroid and retina. Proc. Natl. Acad. Sci. U. S. A. 2000;97:1224–1229. doi: 10.1073/pnas.97.3.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lam D.M. The biosynthesis and content of gamma-aminobutyric acid in the goldifsh retina. J. Cell Biol. 1972;54:225–231. doi: 10.1083/jcb.54.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolz J., Frumkes T., Voigt T., Wassle H. Action and localization of gamma-aminobutyric acid in the cat retina. J. Physiol. 1985;362:369–393. doi: 10.1113/jphysiol.1985.sp015684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu S., Huang J., Xu R., Wang Y., Wan Y., McNeel R., et al. Isocitrate dehydrogenase 3b is required for spermiogenesis but dispensable for retinal viability. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2022.102387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GC–MS data generated in this study are provided in the supporting information.