Abstract
A characteristic feature of gene expression in eukaryotes is the addition of a 5′-terminal 7-methylguanine cap (m7GpppN) to nascent pre-mRNAs in the nucleus catalyzed by capping enzyme and cap methyltransferase. Small interfering RNA (siRNA) knockdown of cap methyltransferase in HeLa cells resulted in apoptosis as measured by terminal deoxynucleotidyltransferase-mediated dUTP-tetramethylrhodamine nick end labeling assay, demonstrating the importance of mRNA 5′-end methylation for mammalian cell viability. Nuclear localization of cap methyltransferase is mediated by interaction with importin-α, which facilitates its transport and selective binding to transcripts containing 5′-terminal GpppN. The methyltransferase 96-144 region has been shown to be necessary for importin binding, and N-terminal fusion of this sequence to nonnuclear proteins proved sufficient for nuclear localization. The targeting sequence was narrowed to amino acids 120 to 129, including a required 126KRK. Although full-length methyltransferase (positions 1 to 476) contains the predicted nuclear localization signals 57RKRK, 80KKRK, 103KKRKR, and 194KKKR, mutagenesis studies confirmed functional motifs only at positions 80, 103, and the previously unrecognized 126KRK. All three motifs can act as alternative nu clear targeting signals. Expression of N-truncated cap methyltransferase (120 to 476) restored viability of methyltransferase siRNA knocked-down cells. However, an enzymatically active 144-476 truncation mutant missing the three nuclear localization signals was mostly cytoplasmic and ineffective in preventing siRNA-induced loss of viability.
RNA polymerase II (Pol II) gene transcripts are uniquely modified at the 5′ end by addition of the m7GpppN cap structure (3, 22, 25). This modification has been shown to be essential for viability in yeast (14, 24, 28) and in Caenorhabditis elegans (26, 27). It marks transcription start sites and is the earliest mRNA processing event in eukaryotic cells. Although added early in transcription when nascent pre-mRNAs consist of only 20 to 30 nucleotides (nt) (2, 20), caps have multiple effects on later stages of gene expression. These include enhancement of RNA stability, splicing, nucleocytoplasmic transport, and translation initiation facilitated by interaction with distinct nuclear and cytoplasmic cap binding proteins (5, 8, 23).
Capping proceeds in three steps: (i) removal of the 5′-terminal phosphate from nascent Pol II transcripts by RNA triphosphatase (RTP); (ii) addition of GMP from GTP to the resulting diphosphate ends by guanylyltransferase (GT); and (iii) transfer of the methyl group from S-adenosylmethionine to the N7 position of the added guanosine by cap methyltransferase (MT) (4). The first two steps are catalyzed by a bifunctional capping enzyme (CE) in metazoans and by separate polypeptides in yeast (25). CE nuclear localization in mammalian cells is determined by the C-terminal GT domain which binds to the phosphorylated C-terminal domain (P-CTD) of the largest submit of Pol II, probably also accounting for the selective capping of Pol II transcripts (1, 16, 33). Interaction with P-CTD stimulates CE (7, 29), and a similar increase in capping activity resulted from binding to transcription elongation factor SPT5 (29), suggesting a regulatory link between capping and transcription.
Consistent with increasing evidence for regulation of eukaryotic gene expression by an integrated network of protein interactions (13), we also found a functional connection between mRNA cap methylation and intracellular protein transport (30). Human MT was shown to bind selectively to substrate RNA containing 5′-terminal GpppN. This interaction and guanosine N7 methylation were both stimulated by importin-α (Impα), the adapter protein that associates with Impβ to mediate nuclear recruitment of proteins containing a nuclear localization signal (NLS) (6). An N-terminal region in the 476-amino-acid MT was required for Impα binding and nuclear localization, and MT-RNA-Impα complexes formed in vitro were dissociated by Impβ and by the Impα nuclear exporter CAS in the presence of RanGTP (30; also unpublished results), consistent with a classical NLS-based nuclear import pathway of MT (6).
To investigate the role of MT sequences in intracellular targeting and to test if cap methylation in the nucleus is essential in mammalian cells, we have analyzed the localization of MT truncation and fusion proteins. The MT 120-129 sequence, GDGTQNKRKI, including the previously unrecognized 126KRK NLS, proved necessary and sufficient for nuclear recruitment. siRNA knockdown of endogenous MT in HeLa cells resulted in loss of viability, emphasizing the key role of the 7-methylguanosine 5′ cap in gene expression. Consistent with the critical importance of MT function in the nucleus, the N-terminally truncated MT 120-476 mutant localized to the nucleus and prevented the apoptotic effects of siRNA-mediated MT knockdown, while the MT 144-476 truncation mutant did not.
MATERIALS AND METHODS
Plasmids.
Chimeras were produced by using the Advantage-HF 2 PCR kit (Clontech, Palo Alto, Calif.) and overlapping primers in a two-step procedure. Chimeric and truncated MT final products were designed to contain a Kozak consensus sequence (12) and BglII restriction site inserted at the N terminus and were cloned into the pcDNA3.1/CT-GFP-TOPO vector (Invitrogen, Carlsbad, Calif.). Full-length MT-green fluorescent protein (GFP) and MT in pET28a (Novagen, Madison, Wis.) have been described (30). For terminal deoxynucleotidyltransferase-mediated dUTP-tetramethylrhodamine nick end labeling (TUNEL) staining, full-length MT and the 120-476 and 144-476 truncation mutants were constructed in the pEGFP-N3 vector (Clontech). Mutagenesis of potential NLSs in all chimeric, truncated, and full-length MT constructs was performed with appropriate primers and the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). All constructs including mutants were confirmed by sequencing. For cross-linking analysis, two copies of simian virus 40 (SV40) NLS were added to pEGFG-N3 at the 3′ end of the GFP coding sequence (pEGFP-N3/2XSV40NLS).
Transfection of mammalian cells.
GFP fusion plasmids were transfected into HeLa S3 cells by using Effectene or PolyFect (QIAGEN, Valencia, Calif.) according to the manufacturer's protocols. Cells were fixed 36 h posttransfection, and nuclei were stained with Hoechst 33258. Samples were also examined by fluorescence microscopy for protein expression.
Preparation and assay of siRNA.
Two 21-nt RNAs containing the nt 317 to 336 and nt 1085 to 1104 coding sequence of MT were transcribed by T7 RNA polymerase, using DNA oligonucleotides as templates (18), to produce short RNAs which were annealed to form MT317 siRNA
5′ GGAACATCCT/GC/GAGTTTCTCTT 3′3′ AACCTTGTAGGA/CG/CTCAAAGGG 5′ and MT1085 siRNA 5′ GGAACATCCACAACACCTTCC 3′3′ GTCCTTGTAGGTGTTGTGGGG 5′ Two-nucleotide mismatch MT317 siRNA was prepared in the same way (mRNA mismatched nucleotides are given in bold after the slash).
HeLa cells, seeded at ∼1 × 105/2-cm2 chamber, were transfected 24 h later with 56 pmol of siRNA, using LipofectAmine 2000 (Invitrogen). Efficacy was tested by Western analysis with rabbit polyclonal anti-human MT and by real-time PCR using an 18S rRNA nt 897 to 1006 fragment as an internal control, LUX fluorogenic primers, Platinum Quantitative PCR Supermix-UDG (Invitrogen), and the Mx4000 Multiplex quantitative PCR system (Stratagene).
TUNEL assay.
Endogenous MT was first reduced in HeLa cells by transfection with MT317 siRNA. After 24 h, pEGFP-N3/MT, pEGFP-N3/MT120-476, or pEGFP-N3/MT144-476 was transfected, and 24 h later, cells were analyzed by TUNEL staining (in situ cell death detection kit, TMR red; Roche Applied Science, Indianapolis, Ind.). The fraction of dead cells was calculated by counting apoptotic (red) cells and dividing by the total number of cells (4′,6′-diamidino-2-phenylindole stained; blue) in a field of ∼1,000 cells.
Protein expression and glutathione S-transferase (GST) pull-down assay.
GST-Impα was expressed and purified as described previously (31). Chimeric, truncated, and full-length MT proteins were synthesized in the TNT Quick Coupled reticulocyte lysate system (Promega, Madison, Wis.). 35S-methionine-labeled, in vitro-translated proteins were incubated with GST-Impα in pull-down assays (29).
Cross-linking and immunoprecipitation.
HeLa cells transfected with pEGFP-N3/2XSV40NLS, pEGFP-N3/MT, pEGFP-N3/MT120-476, or pEGFP-N3/MT120-476(R127I) were collected, washed twice, and incubated with phosphate-buffered saline containing 1 mM dithiobis[succinimidylpropionate] at room temperature for 20 min with gentle agitation. Cells were pelleted, resuspended in 0.1 M Tris (pH 7.5), and lysed in radioimmunoprecipitation buffer by sonication. The clarified lysates were immunoprecipitated with the anti-Pol II antibody 8WG16 (abcam, Cambridge, Mass.), and a GFP monoclonal antibody (Santa Cruz, Santa Cruz, Calif.) was used for Western blot detection.
RESULTS
Human MT sequences target chimeric proteins to the nucleus.
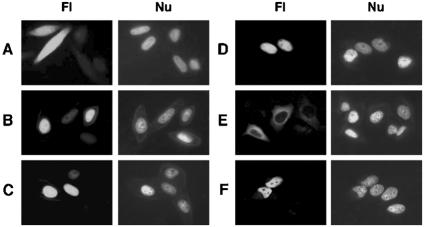
Nuclear localization of mammalian bifunctional CE is specified by the C-terminal GT domain, and the N-terminal RTP expressed as a GFP fusion protein in transfected cells was not compartmentalized (Fig. 1A). MT binds Impα (30) and is thereby recruited to the nucleus (Fig. 1B). Deletion of the first 144 amino acids from MT resulted in loss of both Impα binding and nuclear localization (30). The region required for Impα interaction was narrowed to within positions 96 to 144. This sequence not only is necessary for nuclear localization of MT but also proved sufficient to target RTP to the nuclear compartment (Fig. 1C). The nuclear targeting capacity of the MT 96-144 sequence was confirmed with the 26-kDa GFP, which became nuclearly localized when the MT 96-144 sequence was attached (Fig. 1D). In addition, the ∼98-kDa cytoplasmic RAF1 kinase-GFP fusion protein (Fig. 1E) localized to the nucleus when the MT 96-144 mutant was fused at the N terminus (Fig. 1F). The MT 96-144 nuclear targeting sequence was further delineated, and RTP fused to the MT 120-129 construct was nuclear (Fig. 2A). Nuclear localization was also obtained with RTP fused to the MT 96-129 and 120-144 sequences (data not shown), and in each case, RTP activity was retained in the RTP chimeras as measured with γ-32P-labeled RNA (33) (see Fig. S1 in the supplemental material).
FIG. 1.
Intracellular localization of chimeric GFP-tagged proteins. HeLa cells were transfected with plasmids for expression of RTP-GFP (A), MT-GFP (B), 96-144MT-RTP-GFP (C), 96-144MT-GFP (D), RAF1-GFP (E), or 96-144MT-RAF1-GFP (F) and 36 h later were examined by fluorescence microscopy (Fl) and Hoechst nuclear staining (Nu) as described in Materials and Methods.
FIG. 2.
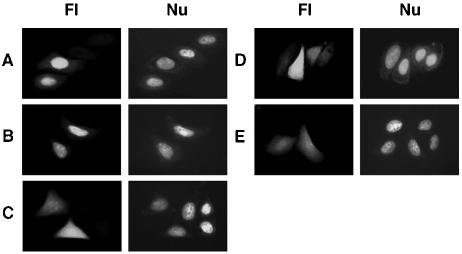
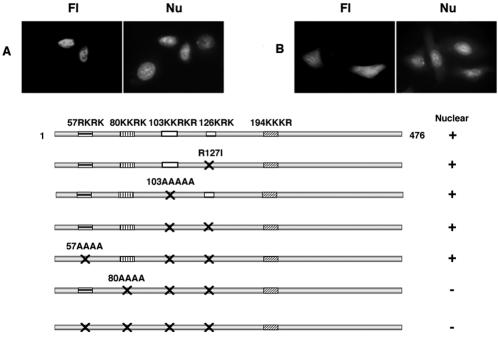
Loss of nuclear targeting by mutation of 126 KRK in MT-RTP-GFP. Nuclear targeting of 120-129MT-RTP-GFP (A) and 96-129MT-RTP-GFP with 103KKRKR mutagenized to all alanines (B). Cytoplasmic localization of 120-129MT-RTP-GFP containing K126A (C), R127I (D), or K128A (E). Fl, fluorescence microscopy; Nu, Hoechst nuclear staining.
MT 126KRK is an NLS in the 120-129 targeting sequence.
The MT 96-129 sequence contains a putative NLS, 103KKRKR, predicted in the PSORTII Prediction Program (http://psort.nibb.ac.jp/form2.html). However, alanine substitution of all five basic residues in the full-length MT (which had no effect on Impα binding [30]) did not alter nuclear localization of the chimeric 96-129MT-RTP (Fig. 2B). Another basic sequence in this region, 126KRK, is not a predicted NLS, but mutation of R127 to isoleucine in the 120-144MT-RTP chimera resulted in loss of nuclear targeting (data not shown). Nuclear localization of RTP fused to the MT 120-129 sequence was similarly lost when amino acid 126, 127, or 128 was mutated (Fig. 2C to E). However, alanine substitutions at other positions (120, 121, 124, and 125) in the MT sequence 120GDGTQNKRKI did not alter nuclear recruitment of RTP chimeras (data not shown), indicating that 126KRK is a previously unrecognized minimum NLS.
Impα binding of MT chimeras.
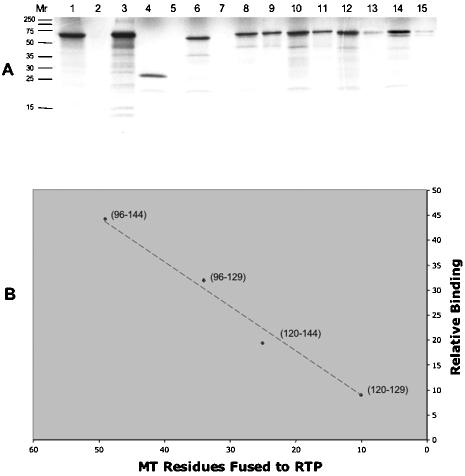
To assess the effect of MT sequences on Impα binding, human MT-RTP-GFP fusion proteins were synthesized in vitro in the presence of [35S]methionine and tested in pull-down assays with GST-Impα. Full-length MT bound to GST-Impα and not to GST (Fig. 3A, lanes 1 to 3). GFP and RTP did not bind to GST-Impα (lanes 4 to 7), but fusion of the MT 96-144 mutant to RTP-GFP resulted in binding that was nearly half the level obtained with full-length MT (compare lanes 8 and 9 with lanes 1 and 3). When the MT sequence was shortened to 34, 24, and 10 amino acids (96 to 129, 120 to 144, and 120 to 129, respectively), binding of the RTP fusion proteins to GST-Impα was retained (Fig. 3A, lanes 10 to 15) but diminished in direct proportion to the number of fused MT residues (Fig. 3B).
FIG. 3.
Binding of MT-RTP-GFP chimeras to GST-Impα. (A) MT input, GST bound, and GST-Impα bound (lanes 1, 2, and 3); input (25% in all cases) and bound are shown in the even and odd lanes, respectively, for GFP (lanes 4 and 5), RTP-GFP (lanes 6 and 7), 96-144MT-RTP-GFP (lanes 8 and 9), 96-129MT-RTP-GFP (lanes 10 and 11), 120-144MT-RTP-GFP (lanes 12 and 13), and 120-129MT-RTP-GFP (lanes 14 and 15). (B) The fraction of each MT-RTP-GFP chimera that bound to GST-Impα was determined by phosphorimager analysis and then normalized relative to the binding of full-length MT-GFP. There was a direct, linear relationship between the relative binding and the number of MT residues fused to RTP-GFP (r2 = 0.9835).
Alternative NLS motifs in MT.
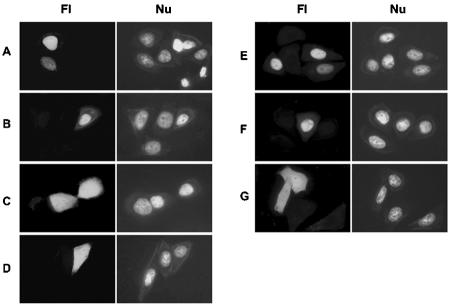
Full-length MT is nuclear (Fig. 1B) and like methyltransferase activity (21, 30), this was little changed by N-terminal truncation to position 96 or 120 (Fig. 4A and B). However, removal of N-terminal residues to position 130 (Fig. 4C) or Ala substitution in the 120-476 construct at position 127 (Fig. 4D) or amino acid 126 or 128 but not 120, 121, or 124 plus 125 (data not shown) resulted in loss of nuclear localization. Nuclear targeting was retained in 96-476MT-GFP with mutations R127I or 103-107 Ala (Fig. 4E and F), but the mutant containing both of these changes lost nuclear localization (Fig. 4G). These results suggest that amino acids 103 to 107 and 126 to 128 can function as alternative NLS motifs.
FIG. 4.
Effects of N-terminal truncation and mutation on MT localization. HeLa cells were examined 36 h after transfection with 96-476MT-GFP (A), 120-476MT-GFP (B), 130-476MT-GFP (C), 120-476(R127A)MT-GFP (D), 96-476(R127I)MT-GFP (E), 96-476(103-107A)MT-GFP (F), or 96-476(103-107A/R127I)MT-GFP (G). Fl, fluorescence microscopy; Nu, Hoechst nuclear staining.
The finding that MT 96-476 mutated at both sites lost nuclear localization (Fig. 4G) while mutation of 103KKRKR to all alanines or 126KRK to KIK, or both, did not alter nuclear localization of the full-length MT (data not shown) suggested that other sequences in the MT 1-96 region operate as alternative NLS(s). MT sequences 57RKRK, 80KKRK, 103KKRKR, and 194KKKR are predicted to be NLSs by the PSORTII Prediction Program. Full-length MT containing mutated 103 and 126 motifs and also 57RKR converted to all alanines retained nuclear targeting (Fig. 5A). However, when the additional change was conversion of 80KKRK to four alanines, nuclear localization was lost (Fig. 5B). Thus, the 57RKRK sequence is apparently not a functional NLS, while the 80KKRK motif targets MT-GFP to the nucleus when the 103-107 and 126-128 NLS sites are inactivated, as summarized in Fig. 5C. Our findings indicate that MT contains motifs that function as alternative NLSs at positions 80 and 103, as predicted by PSORTII, and at position 126, which is not listed in either PSORTII or the Predict NLS server. The predicted NLS motifs at positions 57 and 194 do not act as NLSs in full-length MT (Fig. 5B) and MT144-476 (30), respectively.
FIG. 5.
Nuclear targeting of MT NLS mutants. MT triple mutants MT(57-60A, 103-107A, R127I)-GFP (A) and MT(80-83A, 103-107A, R127I)-GFP (B) were expressed in transfected HeLa cells. (C) Summary showing that MT sequence 126KRK is an NLS in addition to two other putative NLS motifs annotated by PSORTII Prediction, 80KKRK and 103KKRKR, while the predicted 57RKRK and 194KKKR motifs are not NLSs. Fl, fluorescence microscopy; Nu, Hoechst nuclear staining.
Induction of apoptosis by MT siRNA and complementation by nuclearly localized MT truncations.
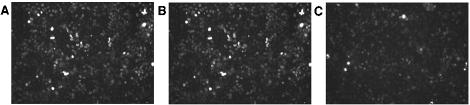
Twenty potential siRNAs targeting different regions of the MT mRNA coding sequence were synthesized and screened for efficacy in decreasing the expression of both endogenous MT and exogenously expressed MT-GFP fusion proteins. Two siRNAs, MT317 and MT1085 (corresponding to nt 317 to 336 and 1085 to 1104 in the coding region), showed high levels of inhibitory activity, while the others had little or no siRNA activity. For example, as shown in Fig. 6, expression of MT-GFP (A) was little affected by transfected MT317 mismatch RNA (B) but markedly decreased by MT317 siRNA (C). The siRNA effect was confirmed in 293T cells by real-time PCR; at 48 h, MT317 and MT317 mismatch siRNA decreased MT mRNA by 84% ± 15% and 17% ± 9%, respectively. By Western assays, MT decreases at 48 h were 74 and 17% for MT317 and MT317 mismatch siRNA, respectively.
FIG. 6.
MT siRNA effect on MT-GFP expression. HeLa cells were transfected with MT-GFP alone (A) or also with MT317 mismatch siRNA (B) and MT317 siRNA (C) and examined by fluorescence microscopy.
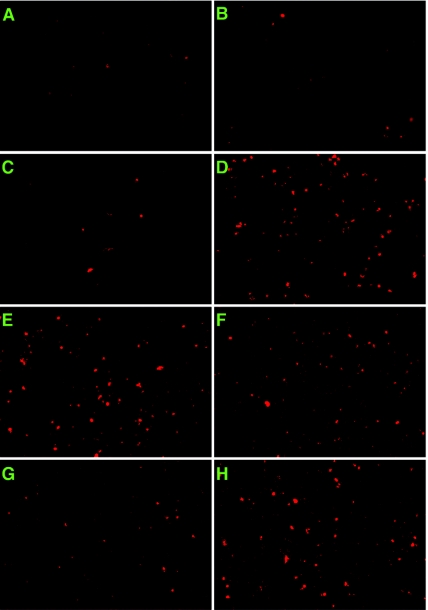
Knockdown of MT in HeLa cells induced apoptosis as measured by TUNEL staining (Fig. 7). Untreated, mock-transfected, and mismatch siRNA-treated cultures contained low levels of TUNEL positive cells (Fig. 7A to C), which increased to 25 to 27% of the culture after transfection with MT1085 siRNA (Fig. 7D). Consistent with TUNEL staining, caspase 3 was activated by MT317 and MT1085 siRNA transfection, similar to the case with staurosporin-treated cells, as measured by Western blotting with cleaved caspase 3 antibody (Cell Signaling Technology, Beverly, Mass.) (data not shown). Apoptosis was similarly induced by MT317 and 1085 siRNAs in the p53-defective non-small-cell lung cancer line H1299 (ATCC CRL-5803), suggesting p53 independence (data not shown).
FIG. 7.
Prevention of apoptosis by nuclearly localized MT-GFP constructs. TUNEL staining of untreated HeLa cells (A), mock-transfected cells (B), or cells transfected with MT317 mismatch (C), MT1085 (D), or MT317 (E), followed by transfection of full-length MT (F), the nuclear-targeted MT 120-476 truncation mutant (G), or the cytoplasmically localized MT 144-476 mutant (H).
To test if the MT 126KRK NLS sequence is required for MT to function in vivo, HeLa cells were transfected first with MT317 to knock down endogenous MT and then with full-length MT-GFP, 120-476MT-GFP, or 144-476MT-GFP constructs. 120-476MT-GFP and 144-476MT-GFP do not contain the sequences complementary to siRNA MT317, and consequently their expression was not decreased, unlike endogenous MT and full-length MT-GFP (data not shown). In comparison to the low number of TUNEL-positive cells observed in untreated (1 to 3%), mock-transfected (1 to 3%), and mismatch MT317-transfected cells (6%), MT317 siRNA treatment significantly increased the percentage (Fig. 7E) (18 to 19%). MT-GFP contains the MT317 complementary sequences and, as expected, did not reverse the siRNA-induced apoptosis (Fig. 7F) (17%). By contrast, 120-476MT-GFP was very effective in restoring MT activity and maintaining viability (Fig. 7G) (7%). 144-476MT-GFP, which does not contain any NLS and was mostly cytoplasmic, also did not prevent apoptosis induced by MT317 (Fig. 7H) (28%).
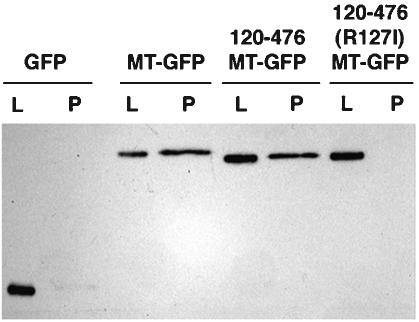
Consistent with restoration of cell viability and the functional activity of the 120-476MT-GFP protein in the nucleus, immunoprecipitation of cross-linked cell lysates with Pol II antibody yielded MT-containing complexes, as with the full-length MT-GFP (Fig. 8). By contrast, 120-476MT-GFP containing the R127I mutation, which was mostly cytoplasmic, did not cross-link to Pol II, nor did the control, nuclearly localized GFP (Fig. 8).
FIG. 8.
Association of nuclearly localized MT with Pol II. HeLa cells were transfected with the indicated constructs, cross-linked with dithiobis[succinimidylpropionate], and immunoprecipitated with anti-Pol II. Cell lysates (L) and precipitates (P) were analyzed by Western blotting with anti-GFP.
Previously, MT 144-476 was shown to retain methyltransferase activity (30). However, to assure that the inability of 144-476MT-GFP to prevent apoptosis was a consequence of cytoplasmic localization rather than loss of enzymatic activity, GFP fusions of full-length MT, MT 120-144, and MT 144-476 were affinity purified from transfected HeLa cells by binding to beads containing GFP monoclonal antibody. The proteins were assayed with radiolabeled GpppG-ended RNA as described previously (19). Conversion to m7GpppG termini was almost complete in each case (data not shown). The results indicate that RNA cap methylation is essential for HeLa cell viability and that the 126KRK NLS sequence has a critical role in localizing MT to the nucleus.
DISCUSSION
The diversity of nuclear import signals and the existence of multiple protein import pathways make it difficult to identify NLS motifs in a primary sequence. In addition, not all NLSs that contain a stretch of basic amino acids are utilized in the classical import pathway (10, 15, 17). We tested directly the putative NLSs in MT at positions 57, 80, 103, and 194 and a previously unrecognized NLS at position 126 for nuclear targeting of full-length MT. Only the sequences at positions 80, 103, and 126 functioned as NLSs. Although 57RKRK and 194KKKR are predicted NLSs and have one more basic amino acid than 126KRK, these motifs failed to nuclearly target MT-GFP when other NLS sites were removed or mutagenized (Fig. 5C). The motifs at positions 57 and 126 both contain KRK, but only the latter was active as NLS, showing that the context of the basic residues is important for function. This is also illustrated by the prototype NLS in SV40 large T antigen, where PKKKRKV is functional while PKTKRKV is not (11). In addition to sequence, the relative positioning of NLS motifs in three-dimensional structures likely influences function by modulating availability for interactions with nuclear adapter/importer proteins, e.g., as described for the bipartite NLS sequences in Smad proteins (32).
Our studies showed that the 96-144 region of MT, the Impα binding region, fused N-terminally to nonnuclear proteins, including GFP and RTP-GFP as well as RAF1 kinase-GFP, can target them to the nucleus (Fig. 1). The same results were obtained with several chimeric proteins in transfected mouse and human cells, including the 3T3, 293, 293T, and 293H lines (data not shown). Thus, the MT 96-144 region can facilitate nuclear localization of proteins unrelated to the mRNA capping machinery and may contain a universal nuclear targeting sequence.
Analyses of binding of MT chimeras to GST-Impα provided some insights into their interactions with the adapter protein. RTP-GFP fused to MT 96-144 bound GST-Impα, and a linear relationship was found between the fraction of input bound and the number of fused residues, consistent with Impα binding-dependent nuclear transport of MT chimeras in vivo (Fig. 3B). Binding was diminished by N-terminal truncation of MT as well as mutation of NLS motifs in MT and MT-RTP-GFP (data not shown), but in vitro binding to transporter did not directly correlate with NLS function. For example, constructs120-129(R127I)MT-RTP-GFP, 120-476(R127A)MT-GFP,and 96-476(103-7A/R127I)MT-GFP all bound GST-Impα, although less than the nonmutated counterparts (data not shown), but were not nuclearly localized (Fig. 2D and 4D and G). This is not entirely surprising, since nuclear transport is a regulated shuttling mechanism involving a threshold level and a balance of multiple proteins including Impα, Impβ, RanGDP, RanGTP, and CAS nuclear export factor.
Studies of the MT mutant constructs showed that one of the motifs at positions 80, 103, and 126 can act as an alternative NLS when the other two sites become unavailable (Fig. 5C), assuring the targeting of MT to the nucleus, where its function is essential for viability. GST-Impα binding assays indicated that when the NLSs at 103KKRKR and 126KRK are available, as in 96-144MT-RTP-GFP and 96-129MT-RTP-GFP, binding was ∼49 and ∼30% of that obtained with the full-length MT, respectively (Fig. 3). When only one NLS (126KRK) was available, as in 120-144MT-RTP-GFP and 120-129MT-RTP-GFP, binding was significantly reduced (Fig. 3). The presence of a second NLS increased Impα interaction in vitro and apparently also enhanced nuclear localization. In nuclearly targeted fusion proteins, increased cytoplasmic fluorescence was consistently observed with chimeras containing a single NLS, notably 120-129MT-RTP-GFP, 120-476MT-GFP, and 96-476(103-7A)MT-GFP (Fig. 2A and 4B and F), compared to those with two or more NLSs. Although one NLS suffices, the presence of two apparently results in more cargo binding to the nuclear transporter complex. This increased binding likely differs from bipartite NLS binding because the NLSs within MT are probably too far apart to constitute a bipartite motif consisting of two basic stretches separated by a 10-residue spacer (9). There could also be preferential binding of Impα to one NLS over another, as well as NLSs that differ in import mechanism, as in the case of the glucocorticoid receptor NLSs (10).
mRNA capping is functionally conserved from yeast to humans, and our data indicate that mRNA capping, as in yeast and C. elegans, is also required for the viability of mammalian cells. This requirement depends on nuclear localization of MT. Its presence at nuclear functional sites is apparently assured by alternative NLSs and interaction with NLS receptor Impα (30), which networks with adaptor molecules, including carrier protein Impβ and RanGTP. Enzymatically active 144-476MT-GFP, expressed in an endogenous MT knocked-down background, was not nuclearly localized and failed to prevent apoptosis. By contrast, 120-476MT-GFP was nuclear and was effective in reversing the loss of viability induced by MT siRNA. The results clearly demonstrate the importance of the 126KRK NLS in translocating MT into the nuclear compartment to access and methylate the 5′ ends of capped nascent Pol II transcripts.
Supplementary Material
Acknowledgments
We thank A. Rabson and Y. Wen for plasmids and D. Perez and P. Srinivasan for advice and assistance.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Cho, E. J., T. Takagi, C. R. Moore, and S. Buratowski. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 11:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coppola, J. A., A. S. Field, and D. A. Luse. 1983. Promoter-proximal pausing by RNA polymerase II in vitro: transcripts shorter than 20 nucleotides are not capped. Proc. Natl. Acad. Sci. USA 80:1251-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furuichi, Y., and A. J. Shatkin. 2000. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 55:135-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furuichi, Y., S. Muthukrishnan, J. Tomasz, and A. J. Shatkin. 1976. Mechanism of formation of reovirus mRNA 5′-terminal blocked and methylated sequence, m7GpppGmpC. J. Biol. Chem. 251:5043-5053. [PubMed] [Google Scholar]
- 5.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 6.Görlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 7.Ho, C. K., and S. Shuman. 1999. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell 3:405-411. [DOI] [PubMed] [Google Scholar]
- 8.Izaurralde, E., J. Lewis, C. McGuigan, M. Jankowska, E. Darzynkiewicz, and I. W. Mattaj. 1994. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78:657-668. [DOI] [PubMed] [Google Scholar]
- 9.Jans, D. A., and S. Hubner. 1996. Regulation of protein transport to the nucleus: central role of phosphorylation. Physiol. Rev. 76:651-685. [DOI] [PubMed] [Google Scholar]
- 10.Kaffman, A., and E. K. O'Shea. 1999. Regulation of nuclear localization: a key to a door. Annu. Rev. Cell Dev. Biol. 15:291-339. [DOI] [PubMed] [Google Scholar]
- 11.Kalderon, D., B. Roberts, W. D. Richardson, and A. E. Smith. 1984. A short amino acid sequence able to specify nuclear location. Cell 39:499-509. [DOI] [PubMed] [Google Scholar]
- 12.Kozak, M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15:8125-8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 14.Mao, X., B. Schwer, and S. Shuman. 1995. Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol. Cell. Biol. 15:4167-4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265-303. [DOI] [PubMed] [Google Scholar]
- 16.McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, S. Shuman, and D. L. Bentley. 1997. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:3306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677-690. [DOI] [PubMed] [Google Scholar]
- 18.Paddison, P. J., A. A. Caudy, and G. J. Hannon. 2002. Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl. Acad. Sci. USA 99:1443-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pillutla, R. C., Z. Yue, E. Maldonado, and A. J. Shatkin. 1998. Recombinant human mRNA cap methyltransferase binds capping enzyme/RNA polymerase IIo complexes. J. Biol. Chem. 273:21443-21446. [DOI] [PubMed] [Google Scholar]
- 20.Rasmussen, E. B., and J. T. Lis. 1993. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc. Natl. Acad. Sci. USA 90:7923-7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saha, N., B. Schwer, and S. Shuman. 1999. Characterization of human, Schizosaccharomyces pombe, and Candida albicans mRNA cap methyltransferases and complete replacement of the yeast capping apparatus by mammalian enzymes. J. Biol. Chem. 274:16553-16562. [DOI] [PubMed] [Google Scholar]
- 22.Shatkin, A. J. 1976. Capping of eukaryotic mRNAs. Cell 9:645-653. [DOI] [PubMed] [Google Scholar]
- 23.Shatkin, A. J., and J. L. Manley. 2000. The ends of the affair: capping and polyadenylation. Nat. Struct. Biol. 7:838-842. [DOI] [PubMed] [Google Scholar]
- 24.Shibagaki, Y., N. Itoh, H. Yamada, S. Nagata, and K. Mizumoto. 1992. mRNA capping enzyme. Isolation and characterization of the gene encoding mRNA guanylytransferase subunit from Saccharomyces cerevisiae. J. Biol. Chem. 267:9521-9528. [PubMed] [Google Scholar]
- 25.Shuman, S. 2001. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 66:1-40. [DOI] [PubMed] [Google Scholar]
- 26.Srinivasan, P., F. Piano, and A. J. Shatkin. 2003. mRNA capping enzyme requirement for Caenorhabditis elegans viability. J. Biol. Chem. 278:14168-14173. [DOI] [PubMed] [Google Scholar]
- 27.Takagi, T., A. K. Walker, C. Sawa, F. Diehn, Y. Takase, T. K. Blackwell, and S. Buratowski. 2003. The Caenorhabditis elegans mRNA 5′-capping enzyme. In vitro and in vivo characterization. J. Biol. Chem. 278:14174-14184. [DOI] [PubMed] [Google Scholar]
- 28.Tsukamoto, T., Y. Shibagaki, S. Imajoh-Ohmi, T. Murakoshi, M. Suzuki, A. Nakamura, H. Gotoh, and K. Mizumoto. 1997. Isolation and characterization of the yeast mRNA capping enzyme beta subunit gene encoding RNA 5′-triphosphatase, which is essential for cell viability. Biochem. Biophys. Res. Commun. 239:116-122. [DOI] [PubMed] [Google Scholar]
- 29.Wen, Y., and A. J. Shatkin. 1999. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 13:1774-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen, Y., and A. J. Shatkin. 2000. Cap methyltransferase selective binding and methylation of GpppG-RNA are stimulated by importin-α. Genes Dev. 14:2944-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wen, Y., Z. Yue, and A. J. Shatkin. 1998. Mammalian capping enzyme binds RNA and uses protein tyrosine phosphatase mechanism. Proc. Natl. Acad. Sci. USA 95:12226-12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao, Z., R. Latek, and H. F. Lodish. 2003. An extended bipartite nuclear localization signal in Smad4 is required for its nuclear import and transcriptional activity. Oncogene 22:1057-1069. [DOI] [PubMed] [Google Scholar]
- 33.Yue, Z., E. Maldonado, R. Pillutla, H. Cho, D. Reinberg, and A. J. Shatkin. 1997. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc. Natl. Acad. Sci. USA 94:12898-12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.