Abstract
The Cdc25 family of protein phosphatases positively regulates cell division by activating cyclin-dependent protein kinases (CDKs). In humans and rodents, there are three Cdc25 family members—denoted Cdc25A, Cdc25B, and Cdc25C—that can be distinguished based on their subcellular compartmentalizations, their abundances and/or activities throughout the cell cycle, the CDKs that they target for activation, and whether they are overexpressed in human cancers. In addition, murine forms of Cdc25 exhibit distinct patterns of expression throughout development and in adult tissues. These properties suggest that individual Cdc25 family members contribute distinct biological functions in embryonic and adult cell cycles of mammals. Interestingly, mice with Cdc25C disrupted are healthy, and cells derived from these mice exhibit normal cell cycles and checkpoint responses. Cdc25B−/− mice are also generally normal (although females are sterile), and cells derived from Cdc25B−/− mice have normal cell cycles. Here we report that mice lacking both Cdc25B and Cdc25C are obtained at the expected Mendelian ratios, indicating that Cdc25B and Cdc25C are not required for mouse development or mitotic entry. Furthermore, cell cycles, DNA damage responses, and Cdc25A regulation are normal in cells lacking Cdc25B and Cdc25C. These findings indicate that Cdc25A, or possibly other phosphatases, is able to functionally compensate for the loss of Cdc25B and Cdc25C in mice.
Cell cycle advancement is regulated, in part, by the dephosphorylation and activation of cyclin-dependent protein kinases. Dephosphorylation and activation of cyclin-dependent protein kinases are, in turn, catalyzed by the Cdc25 family of protein phosphatases. Budding and fission yeasts encode a single member of this family, whereas the mammalian genome encodes three family members, designated Cdc25A, Cdc25B, and Cdc25C (19, 43, 45, 51, 52). The unique functional contributions made by individual Cdc25 family members to mammalian cell cycle control and checkpoint control have not yet been defined.
Experiments performed using mammalian tissue culture cells have revealed several distinguishing characteristics of Cdc25 family members. Although cells simultaneously express all three family members, Cdc25A is nuclear, whereas Cdc25B and Cdc25C shuttle in and out of the nucleus throughout interphase, and this activity is dependent on their interactions with 14-3-3 proteins (9-11, 22, 23, 26, 49). Early cell cycle studies indicated that the G1-to-S-phase transition was regulated by Cdc25A (5, 26, 31, 55), whereas the G2-to-M-phase transition was regulated by Cdc25B and Cdc25C. For example, the microinjection of antibodies specific for either Cdc25B or Cdc25C arrests cells in G2, suggesting roles for these proteins at the G2-to-M-phase transition (38, 43). In certain cell types, Cdc25B has been shown to be an unstable protein that accumulates during the S and G2 phases of the cell cycle. In other cell lines, the activity of Cdc25B is regulated such that it is most active during the S and G2 phases of the cell cycle (18, 38, 47). Furthermore, Cdc25B has a Cdk binding site within its C terminus that enables the efficient dephosphorylation and activation of cyclin A/Cdk2 and cyclin B1/Cdk1 complexes in vitro (17, 27, 54). Cdc25B has been proposed to regulate centrosomal microtubule nucleation during mitosis (18). Unlike Cdc25B, the intrinsic phosphatase activity of Cdc25C is low during the S and G2 phases of the cell cycle. Cdc25C is activated in mitosis due to phosphorylation by Cdk1/cyclin B1 and the Polo-like kinase Plk1 (1, 13, 25, 28, 29, 34-36, 38, 50, 60). Cdk1/cyclin B complexes have been proposed to be the primary targets for activation by Cdc25C (14, 20, 25, 39, 58).
Microinjection of antibodies against Cdc25A arrests cells in G1, and the overexpression of Cdc25A accelerates entry of cells into S phase, implicating a role for Cdc25A in regulating the G1-to-S-phase transition (5, 26, 31, 55). In addition, Cdc25A is an E2F target gene, and Cdc25A is required for the efficient induction of S-phase entry by E2F-1 (59). However, recent evidence suggests that Cdc25A also plays a role in regulating the G2-to-M-phase transition (8, 42, 63, 66). Cdc25A is present and active in all stages of the cell cycle, and Cdc25A levels actually rise as cells progress from S phase to mitosis (4, 5, 26, 31, 42, 44, 55). Importantly, Cdc25A has a docking site for cyclin B1/Cdk1 within its C terminus, which is masked by 14-3-3 proteins during interphase but exposed during mitosis (8). Additional evidence includes the observation that Cdc25A overexpression accelerates mitotic entry (44) and the overproduction of phosphatase-dead Cdc25A delays mitotic entry (33).
Cdc25A and Cdc25C are targets of negative regulation by checkpoints that respond to various forms of genotoxic stress. Checkpoint activation maintains Cdc25C in a 14-3-3 protein-bound form, and the overproduction of a mutant of Cdc25C that cannot bind to 14-3-3 proteins causes a partial bypass of both the DNA replication and G2 DNA damage checkpoints (49). Cdc25A stability is regulated as a function of the cell cycle, and Cdc25A is rapidly degraded in a proteasome-dependent manner in cells exposed to UV light or ionizing radiation (IR) (4, 6, 12, 16, 21, 24, 30, 41, 42, 44, 66). Chk1 phosphorylates Cdc25A to target it for proteolysis during an unperturbed cell cycle, and the integrity of the Chk1/Cdc25A pathway is required for cells to delay in the S and G2 phases of the cell cycle following checkpoint activation (57, 63, 66). In addition, the overexpression of Cdc25A bypasses the G1 DNA damage checkpoint and the intra-S-phase checkpoint, resulting in enhanced DNA damage and decreased cell survival (42, 44).
In mice, Cdc25A, Cdc25B, and Cdc25C exhibit overlapping but distinct patterns of expression during development and are expressed in tissue-specific patterns in adult mice (32, 46, 61, 62). This fact suggests that these genes have distinct biological functions in embryonic and adult mice. Mice lacking Cdc25C are viable, develop normally, and do not display any obvious abnormalities (7). Furthermore, the phosphorylation status of Cdk1, the timing of entry into mitosis, and the cellular responses to DNA damage are unperturbed in mouse embryo fibroblasts (MEFs) lacking Cdc25C (7). Mice with Cdc25B disrupted are also viable and healthy, although females are sterile due to a meiotic defect during oogenesis (40). One explanation for the lack of a cell cycle phenotype in mice with either Cdc25B or Cdc25C disrupted is compensation by the other family members and/or other phosphatases. To address this possibility, we generated mice lacking both Cdc25B and Cdc25C. Surprisingly, these mice are obtained at the expected Mendelian ratios and are healthy, demonstrating that Cdc25B and Cdc25C are dispensable for murine embryonic development and for mitotic entry.
MATERIALS AND METHODS
Generation and genotyping of mice with Cdc25B and Cdc25C disrupted.
Mice with both Cdc25B and Cdc25C disrupted were generated by first mating Cdc25B−/− (40) males with Cdc25C−/− (7) females. F2 double-heterozygous littermates of the strain background C57BL/6;129X1SvJ were then interbred to generate Cdc25B−/− Cdc25C−/− double knockout mice. PCR analysis was carried out on genomic tail DNA of wild-type (WT) mice, Cdc25B−/− knockout mice, Cdc25C−/− knockout mice, and the Cdc25B−/− Cdc25C−/− double knockout mice (BCKO) by using specific primers to confirm the genotypes of the mice. PCR analysis for Cdc25C was achieved by using a three-primer PCR with primer 5′-GGTTCCTTGGATTCATCTGGACC from exon 3, primer 5′-CCTCGTGCTTTACGGTATCGCCG from the neomycin phosphotransferase gene cassette, and primer 5′-CCCTACCATGAGTGCAGGGCACC from the intronic sequence between exons 3 and 4 as described previously (7). Three PCRs were carried out to identify mice lacking Cdc25B. Primers 5′-GCCTGTGAGGCCACCTAC and 5′-CACTGAACACAACAGACTTC amplify a region in the 3′ untranslated region. Primers 5′-TGGAGAGGCTATTCGGCTATGAC and 5′-GGCATCGCCATGGGTCACGACGA amplify a region within the neomycin phosphotransferase gene cassette. Primers 5′-GACTGATCGGAGATTACTCT and 5′-TGGAGAGGCTATTCGGCTATGAC amplify a region between exon 10 and the neomycin phosphotransferase gene cassette.
Histology.
Tissues were fixed in 10% neutral buffered formalin, rinsed in phosphate-buffered saline (PBS), and stored in 70% ethanol. Fixed tissues were embedded in paraffin by using standard procedures. Blocks were sectioned (5 mm) and stained with hematoxylin and eosin.
Animal growth measurements.
Heterozygous crosses were used to generate mice, and postnatal growth curves were obtained by weighing pups at the indicated times by using a Mettler AE 50 scale. Genotyping was performed by PCR analysis of tail cuttings. All recorded weight averages shown in the growth curves were compiled from at least 30 mice per genotype per time point.
Northern blotting.
RNA was isolated from cells by using the QuickPrep total RNA extraction kit (Amersham Pharmacia Biotech) according to the manufacturer's suggestions. RNA was resolved on a 1.2% agarose gel and then transferred to a GeneScreen Plus membrane (NEN). The probes used for screening mouse tissues and cells include a 440-bp HincII/DraIII restriction fragment of the murine Cdc25C (mCdc25C) cDNA, a 427-bp XmnI/PvuI restriction fragment of the mCdc25A cDNA, a 466-bp BglII/SacII restriction fragment of the mCdc25B cDNA, a 600-bp HindIII/EcoRI restriction fragment of GAPDH, and an ∼1.2-kb XbaI/HindIII restriction fragment of the neomycin phosphotransferase gene. Probes were labeled with dCTP (α-32P; NEN) by using the Megaprime DNA labeling system (Amersham). Blots were prehybridized with ExpressHyb solution (Clontech) containing 100 μg of sonicated salmon sperm DNA/ml for 2 to 3 h at 68°C with shaking. Labeled probe was added to 106 cpm/ml, and the blot was hybridized in ExpressHyb solution for 2 h at 68°C. Blots were washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.5% sodium dodecyl sulfate (SDS) at room temperature twice and then washed once with 2× SSC-0.5% SDS for 15 min with shaking at 50°C.
Generation of mouse embryo fibroblasts.
Mouse embryonic fibroblasts were derived from 13.5-day-old embryos. Following the removal of the head and organs, each embryo was rinsed with PBS, minced, and digested with trypsin-EDTA (0.5% trypsin, 0.53 mM EDTA) for 10 min at 37°C, using 1 ml per embryo. Trypsin was inactivated by the addition of Dulbecco's modified Eagle's media (DMEM; Gibco-BRL) containing 10% fetal bovine serum (FBS), 2 mM l-glutamine, a 0.1 mM concentration of nonessential amino acids, 140 mM 2-mercaptoethanol, 100 U of penicillin G/ml, and 100 mg of streptomycin/ml. Cells from single embryos were plated into one 100-mm-diameter tissue culture dish and incubated at 37°C in a 10% CO2-humidified chamber for 3 days and then expanded. Each trypsinization and replating represented one passage, and the split ratio of each passage was 1:3. Early-passage cells (P2 to P4, prior to crisis) were analyzed for cell cycle progression and for cellular responses to DNA damage.
Antibodies.
Fluorescein isothiocyanate (FITC)-conjugated mouse anti-BrdU monoclonal antibody was purchased from BD Biosciences. Mouse monoclonal antibodies specific for Cdc25A were raised against baculovirus-produced glutathione S-transferase (GST)-mCdc25A. Cdk1 was detected with Cdk1-specific rabbit polyclonal antibody (48). β-Catenin mouse monoclonal antibodies were purchased from BD Transduction Laboratories. Phosphohistone H3 antibodies were purchased from Upstate Biotechnology (Lake Placid, N.Y.) and then visualized with FITC-conjugated goat anti-rabbit antibody (Jackson ImmunoResearch, Inc.). Bound primary antibodies were detected with horseradish peroxidase-conjugated rat anti-rabbit antibody (Zymed) or horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) plus IgM antibody (Jackson ImmunoResearch, Inc.), and proteins were visualized by chemiluminescence with the ECL reagent (Amersham Pharmacia Biotech).
S-to-G1-phase progression in absence and presence of IR.
A total of 106 MEFs were seeded onto 100-mm-diameter tissue culture dishes 36 h prior to bromodeoxyuridine (BrdU) labeling. Cells were incubated in 5 ml of culture medium (DMEM, 10% FBS, 2 mM l-glutamine, a 0.1 mM concentration of nonessential amino acids, 140 mM 2-mercaptoethanol, 100 U of penicillin G/ml, and 100 mg of streptomycin/ml) containing 20 mM bromodeoxyuridine (Amersham Pharmacia Biotech) at 37°C for 1 h. Medium was removed and replaced with 10 ml of culture medium, and cells were incubated for the indicated times. Cells were harvested by trypsinization and collected by centrifugation. After the removal of the supernatant by aspiration, cells were washed once in PBS and were then suspended in 0.5 ml of PBS. Cells were fixed by the addition of 5 ml of 70% ethanol at 4°C in the dark. Pelleted cells were resuspended in 1 ml of 0.4 mg of pepsin (Sigma Chemical Co.)/ml in 0.1 N HCl and incubated with rocking for 30 min. Nuclear pellets were suspended in 1 ml of 2 N HCl-0.5% NP-40, and incubated with rocking for 1 h. After neutralization by incubation with 1 ml of 0.1 M sodium borate (pH 8.5) for 5 min, nuclear pellets were suspended in 100 μl of PBS-TB (PBS, 0.5% Tween 20, 1% bovine serum albumin [BSA]) and stained with 5 μl of FITC-conjugated anti-BrdU monoclonal antibody for 1 h in the dark. Nuclei were washed once with 1 ml of PBS-TB and then incubated with 1 ml of PBS-TB containing 30 μg of propidium iodide (PI)/ml and 250 μg of RNase A/ml for 30 min in the dark. Cells were analyzed for DNA content by flow cytometry using a FACSCalibur instrument (Becton Dickinson Instruments). The data were analyzed using CELLQUEST analysis software (Becton Dickinson).
G0-to-S-phase progression.
A total of 106 MEFs were seeded onto 100-mm-diameter tissue culture dishes. The following day, cells were incubated in 10 ml of medium (DMEM, 2 mM l-glutamine, a 0.1 mM concentration of nonessential amino acids, 100 U of penicillin G/ml, and 100 μg of streptomycin/ml) containing 0.1% FBS for 96 h. Cells were reseeded at 106 in medium containing 10% FBS and pulsed with 20 μM BrdU for 1 h prior to harvest. Harvesting, fixing, staining, and analysis of cells were carried out as described above.
IR-induced G1-phase checkpoint in MEFs.
Cells were plated and serum starved as for G1 analysis with the following changes. After serum starvation, 2 × 106 cells were seeded onto 100-mm-diameter tissue culture dishes and either mock irradiated or exposed to 20 Gy of IR from a 60Co source. Medium containing 10% FBS and 20 μM BrdU was added, and cells were harvested 24 h later. Cells were fixed, stained, and analyzed by flow cytometry as described above.
IR-induced G1-phase checkpoint in thymocytes.
Eight- to 10-week-old mice were irradiated with 10 Gy of IR, and injected intraperitoneally with 1 ml of 10 mM BrdU-1 mM fluorodeoxyuridine (Amersham Pharmacia Biotech) at 2 h postirradiation (3). Mice were sacrificed 1 h after the injection, and thymocytes were isolated by crushing thymi between glass slides. The prepared thymocytes were fixed, stained, and analyzed by flow cytometry as outlined above.
IR-induced S-phase checkpoint.
MEFs were labeled with 10 nCi of 14C-thymidine (NEN Life Science Products)/ml for 24 h to control for total DNA content between samples. Cells were then washed and incubated in culture medium for 24 h. Cells were either mock irradiated or exposed to 5, 10, or 20 Gy of IR, incubated for the indicated times, and then pulse labeled with 2.5 μCi of [3H]thymidine (NEN Life Science Products)/ml for 15 min. Cells were harvested, washed twice in PBS, and then incubated in 100% methanol for 5 min, followed by incubation in 10% trichloroacetic acid for 5 min and then 0.3 N NaOH. After neutralization with HCl, radioactivity was determined by liquid scintillation counting. The resulting ratios of 3H counts per minute to 14C counts per minute, corrected for counts per minute that resulted from channel crossover, were measures of DNA synthesis.
IR-induced G2-phase checkpoint.
MEFs were either mock irradiated or exposed to 6 Gy of IR. After 40 min of incubation, 100 ng of nocodazole/ml was added to trap cells in mitosis, and samples were harvested by trypsinization 1 h 20 min later. A total of 5 × 105 cells were fixed by the addition of 5 ml of 70% ethanol at 4°C for as long as 24 h. After fixation, cells were washed twice with PBS, suspended in 1 ml of 0.25% Triton X-100 in PBS, and incubated at 4°C with rocking for 15 min. After centrifugation, cell pellets were suspended in 100 μl of PBS containing 1% BSA and 0.75 μg of anti-histone H3 antibody and incubated for 1.5 h at room temperature. Cells were rinsed with PBS containing 1% BSA and incubated with FITC-conjugated goat anti-rabbit IgG antibody diluted at a ratio of 1:1,200 in PBS containing 1% BSA. After a 30-min incubation at room temperature in the dark, cells were washed again and incubated with 500 μl of PBS with 1% BSA containing 30 μg of PI/ml and 250 μg of RNase A/ml for 30 min, also in the dark. Cells were analyzed by flow cytometry as described above.
Synchronization of MEFs and analysis of Cdk1 phosphorylation.
A total of 5 × 105 MEFs were seeded onto 100-mm-diameter tissue culture dishes. The following day, cells were incubated in 10 ml of culture medium containing 0.1% FBS for 48 h. Cells were then incubated in culture medium containing 15% FBS and 1 μg of aphidicolin/ml (Calbiochem) for 20 h. Cells were released from the aphidicolin block by being washed twice with 10 ml of PBS and then being incubated in culture medium containing 15% FBS. Cells were harvested prior to release (time zero) or at 3, 6, 9, or 12 h after release by trypsinization and collected by centrifugation. After the removal of the supernatant by aspiration, cells were washed once in PBS and then were suspended in 0.5 ml of PBS. Cells were fixed by the addition of 5 ml of 70% ethanol at 4°C. Cells were washed once with 1 ml of PBS-1% FBS and then incubated with 1 ml of PBS-1% FBS containing 30 μg of PI/ml and 250 μg of RNase A/ml for 30 min. Cells were analyzed for DNA content by flow cytometry using a FACSCalibur instrument (Becton Dickinson). The data were analyzed by using CELLQUEST analysis software (Becton Dickinson). For monitoring Cdk1 phosphorylation, cells were lysed in MCLB {50 mM Tris-HCl, pH 8.0, 2 mM dithiothreitol, 5 mM EDTA, 0.5% Nonidet P-40, 100 mM NaCl, 1 μM sodium orthovanadate, phosphatase inhibitor cocktail [containing 25 μM (−)-p-bromotetramisole oxalate, 5 mM cantharidin, and 5 mM microcystin LR; Calbiochem] and protease inhibitor cocktail [containing 1.04 mM 4-(2-aminoethyl) benzenesulfonyl fluoride (AEBSF), 15 μM pepstatin A, 14 μM E-64, 40 μM bestatin, 20 μM leupeptin, and 800 nM aprotinin; Sigma] } for 15 min. at 4°C. Cell lysates containing 2 mg of total cellular protein were incubated with 50 μl of packed p13suc1 agarose (Upstate Biotechnology). After incubation at 4°C for 3 h, precipitates were washed four times with 1 ml of MCLB. Proteins were resolved by SDS-polyacrylamide gel electrophoresis on a 12% SDS gel. Cdk1 phosphorylation status was monitored by immunoblotting.
Monitoring Cdc25A levels.
Asynchronously growing populations of MEFs were lysed in MCLB. Lysates were resolved by SDS-polyacrylamide gel electrophoresis on an 8% SDS gel, transferred to nitrocellulose, and incubated with mouse ascites specific for mCdc25A. For synchronization studies, 6 × 105 MEFs were seeded onto 100-mm-diameter tissue culture dishes and serum starved in the same manner as for the Cdk1 studies. Cells were harvested prior to release (time zero) or at 3, 6, 9, or 12 h after release, analyzed for DNA content by flow cytometry, and processed to monitor Cdc25A levels by Western blotting.
RESULTS AND DISCUSSION
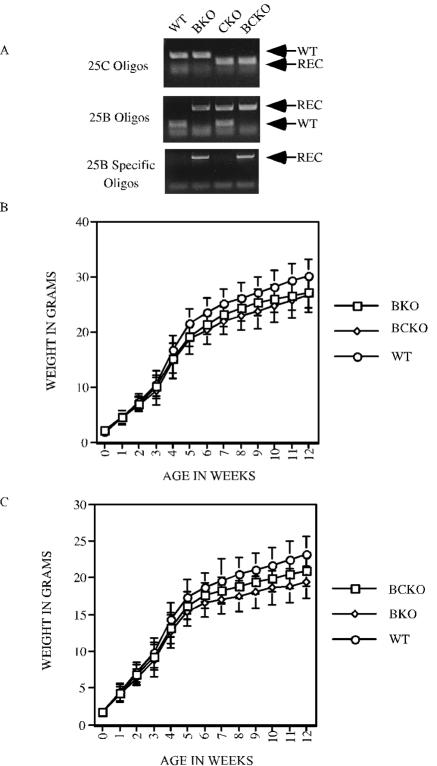
Mice heterozygous for both Cdc25B (40) and Cdc25C (7) were bred to generate mice lacking both phosphatases. PCR analysis was carried out on genomic tail DNA by using specific primers to confirm the genotypes of all mice (Fig. 1A). The cumulative genotyping of heterozygous crosses resulted in the expected ratio of each genotype (Table 1). Given that mice lacking both Cdc25B and Cdc25C were obtained at the expected Mendelian ratios, this outcome indicates that Cdc25B and Cdc25C are not required for mouse development or mitotic entry. All major organs from BCKO mice were examined histologically after hematoxylin-eosin staining and appeared normal (data not shown). Fluorescence-activated cell-sorting (FACS) analysis using the T-cell markers CD4 and CD8 revealed the expected percentages of single-positive (CD4+CD8− and CD4−CD8+) and double-negative (CD4−CD8−) T cells in the peripheral blood of BCKO animals (data not shown).
FIG. 1.
Genotyping and growth rates of mice with Cdc25B and Cdc25C disrupted. (A) PCR analysis of mouse tail DNA from WT, Cdc25B−/− (BKO), Cdc25C−/− (CKO), and Cdc25B−/−C−/− (BCKO) mice, using the indicated oligonucleotides. REC, recombinant. (B and C) WT, Cdc25B−/− (BKO), and Cdc25B−/−C−/− (BCKO) male (B) and female (C) mice were weighed beginning at birth and at weekly intervals up to 12 weeks. Weights (in grams) represent averages from at least 30 mice per genotype per time point. Standard deviations are shown as error bars along the y axis.
TABLE 1.
Actual and expected genotypes of double-heterozygous mating
| Parameter (n) | Result (%) for indicated genotypea
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| W/W | W/H | W/K | H/W | H/H | H/K | K/W | K/H | K/K | |
| Expected ratio | 1 (6.25) | 2 (13) | 1 (6.25) | 2 (13) | 4 (26) | 2 (13) | 1 (6.25) | 2 (13) | 1 (6.25) |
| No. of mice | |||||||||
| Males (398) | 32 (8.04) | 47 (11.81) | 29 (7.29) | 46 (11.56) | 93 (23.37) | 48 (12.06) | 25 (6.28) | 53 (13.32) | 25 (6.28) |
| Females (412) | 25 (6.07) | 68 (16.50) | 33 (8.01) | 51 (12.38) | 95 (23.06) | 44 (10.68) | 25 (6.07) | 40 (9.71) | 31 (7.52) |
| No. of MEFs (236) | 19 (8.05) | 31 (13.14) | 14 (5.93) | 24 (10.17) | 80 (33.89) | 22 (9.32) | 7 (2.97) | 24 (10.17) | 15 (6.36) |
| Total no. of mice and MEFs (1,046) | 76 (7.26) | 146 (13.96) | 76 (7.27) | 121 (11.57) | 268 (25.62) | 114 (10.89) | 57 (5.45) | 117 (11.19) | 71 (6.79) |
The first genotype represents Cdc25B, and the second genotype represents CDc25C. W, wild type; H, heterozygous; K, knockout.
The weights of individual mice from birth to 3 months of age were also determined (Fig. 1B and C). As previously reported, weights of WT and Cdc25C−/− mice were indistinguishable (data not shown) (7). In contrast, Cdc25B−/− and BCKO mice were 8 to 14% and 6 to 10% smaller than their WT littermates, respectively, beginning at 4 weeks of age. The basis for the differences in weights is unknown at this time.
Cell cycle progression is normal in cells lacking Cdc25B and Cdc25C.
Fibroblasts derived from wild-type and knockout mouse embryos were analyzed for their ability to traverse from G0 into S phase. MEFs were synchronized by serum starvation, and at various times after the addition of complete media, cells were incubated with BrdU for 1 h to specifically label cells that were undergoing DNA replication. Cells were stained for DNA content with PI and for replicative DNA synthesis with anti-BrdU antibody and analyzed by flow cytometry. By gating on BrdU-positive cells, the timing of S-phase entry from G0 could be monitored. As seen in Fig. 2A, similar percentages of WT and BCKO MEFs were undergoing DNA replication at each time point tested, indicating that the progression from G0 into S phase is not perturbed in the absence of Cdc25B and Cdc25C.
FIG. 2.
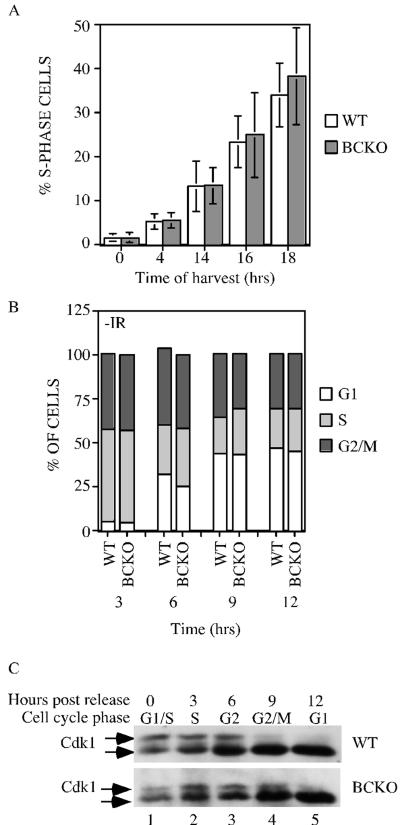
Cell cycle analysis of cells lacking Cdc25B and Cdc25C. (A) WT and BCKO MEFs (passages 4 to 6) were serum starved for 96 h to synchronize cells in G0. After the addition of serum, cells were incubated with BrdU for 1 h prior to harvest. Cells were stained with PI and for BrdU and were analyzed by flow cytometry. Each experiment was performed six times in duplicate with independent MEF strains, and standard deviations are shown as error bars along the y axis. Differences between WT and BCKO MEFs were not statistically different at any of the time points (0 h [P = 0.936], 4 h [P = 0.7374], 14 h [P = 0.9441], 16 h [P = 0.6278], and 18 h [P = 0.2667]). (B) MEFs at passages 4 to 6 were pulse labeled with BrdU for 1 h. Cells were harvested at the indicated times (hours) and stained with PI and for BrdU. The cellular DNA content of BrdU-positive cells was analyzed by flow cytometry. Graphs show the averages from six independent experiments. (C) Early-passage MEFs prepared from wild-type and BCKO mice were synchronized in early S phase. Cells were harvested prior to release (time zero) or at various times (hours) after release. Cdk1 precipitates were resolved on a 12% SDS gel, and Cdk1 was detected by Western blotting. The arrows indicate two electrophoretic forms of Cdk1.
Wild-type and BCKO MEFs were also analyzed for their ability to traverse from S phase through G2 and mitosis and into G1. Cells were pulse labeled with BrdU to mark S-phase cells. Cells were harvested at various times after the incubation and processed for analysis by flow cytometry. By gating on BrdU-positive cells, cell cycle progression from S phase through G2/M phase and into the G1 phase of the cell cycle could be monitored. At the time of the pulse, 17% of WT cells and 19% of BCKO cells incorporated BrdU. As seen in Fig. 2B, the percentages of BrdU-positive wild-type and BCKO cells in each phase of the cell cycle were similar. By 3 h, 42.3 and 42.9% of BrdU-positive wild-type and BCKO cells, respectively, had progressed to the G2/M phase of the cell cycle, and by 6 h, 31.9 and 25.2% of BrdU-positive wild-type and BCKO cells, respectively, had progressed to the G1 phase of the cell cycle. These findings indicate that the time required for cells to progress from S to G1 phases of the cell cycle is unaltered in murine cells lacking Cdc25B and Cdc25C.
Next, the phosphorylation status of Cdk1 was monitored in early passage fibroblasts derived from wild-type and BCKO mouse embryos. Cdk1 is proposed to be a key target of Cdc25B and Cdc25C in vivo. The phosphorylation of Cdk1 varies as a function of the cell cycle, and changes in the electrophoretic mobility of Cdk1 can be used as a specific indicator of Cdk1 phosphorylation status and cell cycle position (2, 56). Wild-type and BCKO MEFs were arrested in early S phase, by first being cultured in medium containing low serum and then being released into complete medium containing aphidicolin. Cells released from G1/S arrest were then analyzed for DNA content by flow cytometry and were processed to monitor Cdk1 phosphorylation status (Fig. 2C). Wild-type and BCKO cells were in S phase by 3 h after release, in G2 phase by 6 h after release, and in the M and G1 phases of the cell cycle by 9 and 12 h after release, respectively. As seen in Fig. 2C, two electrophoretic forms of Cdk1 were present in cells arrested at the G1/S border (lane 1), as well as cells in the S (lane 2) and G2 (lane 3) phases of the cell cycle. As expected, a loss of the slower electrophoretic form of Cdk1 with a concomitant increase in the fastest electrophoretic form of Cdk1 was observed as cells entered mitosis (lane 4) and moved into G1 (lane 5). The slight delay in the dephosphorylation of Cdk1 seen in BCKO cells at the 9-h time point is a consistent finding. However, the cell cycle analysis did not reveal significant differences between wild-type and BCKO cells in their abilities to traverse through the cell cycle after release from the G1/S-phase arrest. These findings demonstrate that sufficient quantities of active Cdk1 were generated in BCKO cells to drive cells into mitosis.
Cells lacking Cdc25B and Cdc25C respond normally to ionizing radiation.
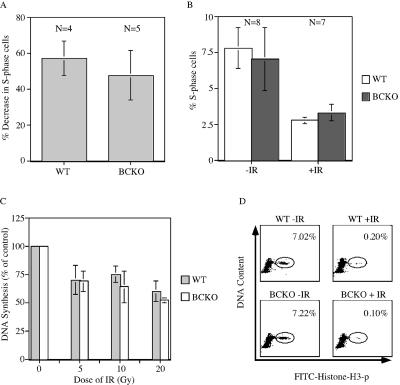
Ionizing radiation activates the IR-induced DNA damage checkpoint to induce cell cycle delays. This checkpoint monitors the integrity of the genome and arrests cells in G1 before DNA replication (termed the IR-induced G1-phase checkpoint), in S phase (the IR-induced S-phase checkpoint), or in G2 before mitosis (the IR-induced G2-phase checkpoint). Checkpoints target the Cdc25A and Cdc25C regulatory pathways to elicit cell cycle delays following ionizing radiation exposure (15, 37, 41, 42, 44, 49, 66). To monitor the IR-induced G1-phase checkpoint in wild-type and BCKO cells, early passage MEFs were synchronized by serum starvation, released into complete medium containing BrdU, and immediately mock irradiated or gamma irradiated. Cells harvested 24 h after release were stained for DNA content with PI and for replicative DNA synthesis with anti-BrdU antibody. As seen in Fig. 3A, wild-type and BCKO cells showed 57 and 48% reductions, respectively, in the numbers of S-phase cells relative to the numbers of mock-irradiated control cells at the 24 h time point. The difference was not statistically significant (P = 0.2909), indicating that the ability of cells to delay entry into S phase is intact in cells lacking Cdc25B and Cdc25C.
FIG. 3.
The IR-induced DNA damage checkpoint is intact in BCKO cells. (A) Early-passage MEFs were serum starved for 96 h. Arrested cells were mock irradiated or gamma irradiated and then incubated in BrdU-containing complete medium. Cells were stained with PI and for BrdU and analyzed by flow cytometry 24 h later. The percent decreases in S-phase cells following irradiation are indicated. Differences between WT and BCKO cells were not significant (P = 0.2909) (B) WT and BCKO mice were mock irradiated (−IR) or gamma irradiated (+IR) with 10 Gy of IR and injected intraperitoneally with BrdU 2 h later. One hour after the BrdU injection, thymocytes were prepared and analyzed by flow cytometry. The percentages of S-phase cells are indicated. Standard deviations are shown as error bars along the y axis. (C) Radioresistant DNA synthesis was assessed 1 h after the exposure of early-passage MEFs to various doses of IR. Experiments were repeated four times. Differences between WT and BCKO MEFs were not statistically different at any dose of IR (5 Gy [P = 0.9035], 10 Gy [P = 0.2095], and 20 Gy [P = 0.1333]). Standard deviations are shown as error bars along the y axis. (D) Early-passage MEFs were mock irradiated or gamma irradiated and incubated in nocodazole 40 min later. Cells were costained for DNA content and histone H3 phosphorylation (p) and were analyzed by flow cytometry 2 h after IR. The percentages of mitotic cells are indicated. The panels are representative results obtained using WT (n = 2) and BCKO (n = 3) MEFs.
The IR-induced G1-phase checkpoint in mice was also monitored (3). Animals were mock irradiated or irradiated with 10 Gy of IR and injected with BrdU 2 h postirradiation. At 1 h postinjection, mice were sacrificed and their thymi were dissected. Thymocytes were analyzed by flow cytometry. As seen in Fig. 3B, irradiated wild-type and BCKO mice exhibited 48 and 51% reductions, respectively, in the numbers of thymocytes undergoing DNA replication compared with the numbers of their mock-irradiated counterparts. The difference was not statistically significant (P = 0.1638), indicating that the ability of thymocytes to delay entry into S phase is intact in animals lacking Cdc25B and Cdc25C.
To determine whether the absence of Cdc25B and Cdc25C perturbed the IR-induced S-phase checkpoint, [3H]thymidine incorporation into DNA was monitored 1 h after exposure of cells to various doses of IR (Fig. 3C). Both wild-type and BCKO cells had an intact S-phase checkpoint, as indicated by the dose-dependent decrease in [3H]thymidine incorporation into newly synthesized DNA. Finally, the ability of G2 cells in cultures of wild-type and BCKO MEFs to delay in G2 within 2 h post-IR was monitored (Fig. 3D). Cells were costained with PI to assess DNA content and with anti-phosphohistone H3 antibody to identify mitotic cells (64, 65). This experimental paradigm determines the percentages of G2 cells that move into mitosis in the absence and presence of double-strand DNA breaks (64, 65). There was a 96% decrease in the number of wild-type MEFs that moved from G2 into mitosis following exposure to IR. A similar decrease in the percentage of mitotic cells was measured for cells lacking both Cdc25B and Cdc25C, which is indicative of an intact IR-induced G2-phase checkpoint.
Analysis of Cdc25A in MEFs lacking Cdc25B and Cdc25C.
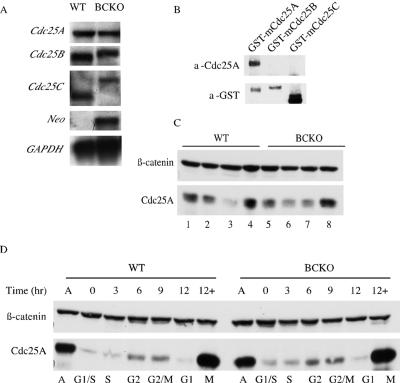
One potential explanation for the lack of an apparent phenotype in mice and cells with Cdc25B and Cdc25C disrupted is compensation by the remaining Cdc25 family member, Cdc25A. The levels of Cdc25A mRNA in wild-type and BCKO MEFs were monitored by Northern blotting, and the levels of Cdc25A protein were monitored by Western blotting (Fig. 4). As seen in Fig. 4A, the levels of Cdc25A mRNA in BCKO cells were not detectably altered. The larger RNA species detected in Northern blots probed for Cdc25B- and Cdc25C-specific mRNAs are due to the insertion of the neomycin phosphotransferase gene after homologous recombination. Importantly, Cdc25B and Cdc25C proteins are not produced in BCKO cells (7, 40). A mouse monoclonal antibody raised against recombinant GST-mCdc25A was used to examine the levels of Cdc25A protein in various primary MEF strains. To test the specificity of the monoclonal antibody, GST-tagged Cdc25A, Cdc25B, and Cdc25C were blotted with the monoclonal antibody and an antibody against GST (Fig. 4B). Only GST-mCdc25A was recognized by the monoclonal antibody, indicating that it does not cross-react with mCdc25B or mCdc25C. Differences in total cellular levels of Cdc25A proteins of various MEF strains were observed, but a consistent pattern of elevated levels of Cdc25A in BCKO-derived MEFs was not seen (Fig. 4C). Next, Cdc25A regulation as a function of the cell cycle was monitored because the abundance of Cdc25A is regulated in a cell cycle-specific manner by ubiquitin-mediated proteolysis (4, 6, 12, 26, 30, 41, 42, 44). As seen in Fig. 4D, Cdc25A levels rose as wild-type and BCKO MEFs progressed from S phase through G2 and into mitosis and then fell as cells exited mitosis and entered G1. A substantial increase in Cdc25A was observed in cells arrested with nocodazole in mitosis. Thus, the regulation of Cdc25A turnover during the cell cycle was normal in cells lacking Cdc25B and Cdc25C.
FIG. 4.
Regulation of Cdc25A is normal in cells lacking Cdc25B and Cdc25C. (A) RNA isolated from WT and BCKO MEFs was processed for Northern blot analysis using probes specific for mCdc25A, mCdc25B, mCdc25C, neomycin phosphotransferase (Neo), and GAPDH. (B) GST-tagged mouse Cdc25 proteins purified from overproducing insect cells (7) were analyzed for reactivity with the mouse Cdc25A (a-Cdc25A) monoclonal antibody and GST (a-GST) antibody. (C) Lysates derived from early-passage WT and BCKO MEFs were analyzed for Cdc25A and β-catenin levels by Western blotting. (D) Asynchronous early-passage MEFs (A) were synchronized at the G1/S border. G1/S-arrested cells (0) were released from the block and processed for Western blotting and flow cytometry at the indicated time points. +, samples that were incubated with nocodazole at the time of release.
Concluding remarks.
Mice lacking two members of the Cdc25 family of protein phosphatases, Cdc25B and Cdc25C, are viable, develop normally, and do not display any obvious abnormalities with the exception that females lacking both phosphatases are sterile, as are females lacking Cdc25B alone. Importantly, these findings challenge the paradigm that entry into mitosis requires Cdc25B and Cdc25C. It is unknown at this time whether Cdc25A, the remaining member of the Cdc25 family, functionally compensates for the lack of Cdc25B and Cdc25C or whether other compensatory pathways are involved. Importantly, Cdc25A has a docking site for Cdk1/cyclin B1 within its C terminus, indicating that Cdc25A is fully capable of activating Cdk1 to regulate mitotic entry (8). Mice that can have Cdc25A, Cdc25B, and Cdc25C conditionally deleted are currently being generated. In this way, Cdc25 phosphatases can be disrupted individually and in combination to determine if “adaptation” during development accounts for the lack of observed phenotype in the BCKO animals, as has been seen in other germ line knockout models (53).
Acknowledgments
A.M.F. was supported by an award from the American Heart Association. This work was supported by a grant from the National Institutes of Health.
H.P.-W. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Abrieu, A., T. Brassac, S. Galas, D. Fisher, J. C. Labbe, and M. Doree. 1998. The Polo-like kinase Plx1 is a component of the MPF amplification loop at the G2/M-phase transition of the cell cycle in Xenopus eggs. J. Cell Sci. 111:1751-1757. [DOI] [PubMed] [Google Scholar]
- 2.Atherton-Fessler, S., F. Liu, B. Gabrielli, M. S. Lee, C.-Y. Peng, and H. Piwnica-Worms. 1994. Cell cycle regulation of the p34cdc2 inhibitory kinases. Mol. Biol. Cell 5:989-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barlow, C., K. D. Brown, C. X. Deng, D. A. Tagle, and A. Wynshaw-Boris. 1997. Atm selectively regulates distinct p53-dependent cell-cycle checkpoint and apoptotic pathways. Nat. Genet. 17:453-456. [DOI] [PubMed] [Google Scholar]
- 4.Bernardi, R., D. A. Lieberman, and B. Hoffman. 2000. Cdc25A stability is controlled by the ubiquitin-proteasome pathway during cell cycle progression and terminal differentiation. Oncogene 19:2447-2454. [DOI] [PubMed] [Google Scholar]
- 5.Blomberg, I., and I. Hoffman. 1999. Ectopic expression of Cdc25A accelerates the G1/S transition and leads to premature activation of cyclin E- and cyclin A-dependent kinases. Mol. Cell. Biol. 19:6183-6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busino, L., M. Donzelli, M. Chiesa, D. Guardavaccaro, D. Ganoth, N. V. Dorrello, A. Hershko, M. Pagano, and G. F. Draetta. 2003. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature 426:87-91. [DOI] [PubMed] [Google Scholar]
- 7.Chen, M.-S., J. Hurov, L. S. White, T. Woodford-Thomas, and H. Piwnica-Worms. 2001. Absence of apparent phenotype in mice lacking Cdc25C protein phosphatase. Mol. Cell. Biol. 21:3853-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, M.-S., C. E. Ryan, and H. Piwnica-Worms. 2003. Chk1 kinase negatively regulates mitotic function of Cdc25A phosphatase through 14-3-3 binding. Mol. Cell. Biol. 23:7488-7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conklin, D. S., K. Galaktionov, and D. Beach. 1995. 14-3-3 proteins associate with cdc25 phosphatases. Proc. Natl. Acad. Sci. USA 92:7892-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalal, S. N., C. M. Schweitzer, J. Gan, and J. Decaprio. 1999. Cytoplasmic localization of human Cdc25C during interphase requires an intact 14-3-3 binding site. Mol. Cell. Biol. 19:4465-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davezac, N., V. Baldin, B. Gabrielli, A. Forrest, N. Theis-Febvre, M. Yashida, and B. Ducommun. 2000. Regulation of CDC25B phosphatases' subcellular localization. Oncogene 19:2179-2185. [DOI] [PubMed] [Google Scholar]
- 12.Donzelli, M., M. Squatrito, D. Ganoth, A. Hershko, M. Pagano, and G. F. Draetta. 2002. Dual mode of degradation of Cdc25 A phosphatase. EMBO J. 21:4875-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ducommun, B., G. Draetta, P. Young, and D. Beach. 1990. Fission yeast cdc25 is a cell cycle regulated protein. Biochem. Biophys. Res. Commun. 167:301-309. [DOI] [PubMed] [Google Scholar]
- 14.Dunphy, W. G., and A. Kumagai. 1991. The cdc25 protein contains an intrinsic phosphatase activity. Cell 67:189-196. [DOI] [PubMed] [Google Scholar]
- 15.Falck, J., N. Mailand, R. G. Syljuasen, J. Bartek, and J. Lukas. 2001. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410:842-847. [DOI] [PubMed] [Google Scholar]
- 16.Falck, J., J. H. Petrini, B. R. Williams, J. Lukas, and J. Bartek. 2002. The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nat. Genet. 30:290-294. [DOI] [PubMed] [Google Scholar]
- 17.Gabrielli, B. G., J. M. Clark, A. K. McCormack, and K. A. Ellem. 1997. Hyperphosphorylation of the N-terminal domain of Cdc25 regulates activity toward cyclin B1/Cdc2 but not cyclin A/Cdk2. J. Biol. Chem. 272:28607-28614. [DOI] [PubMed] [Google Scholar]
- 18.Gabrielli, B. G., C. P. C. De Souza, I. D. Tonks, J. M. Clark, and N. K. Hayward. 1996. Cytoplasmic accumulation of cdc25B phosphatase in mitosis triggers centrosomal microtubule nucleation in HeLa cells. J. Cell Sci. 109:1081-1093. [DOI] [PubMed] [Google Scholar]
- 19.Galaktionov, K., and D. Beach. 1991. Specific activation of cdc25 tyrosine phosphatases by B-type cyclins: evidence for multiple roles of mitotic cyclins. Cell 67:1181-1194. [DOI] [PubMed] [Google Scholar]
- 20.Gautier, J., M. J. Solomon, R. N. Booher, J. F. Bazan, and M. W. Kirschner. 1991. cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2. Cell 67:197-211. [DOI] [PubMed] [Google Scholar]
- 21.Goloudina, A., H. Yamaguchi, D. B. Chervyakova, E. Appella, A. J. Fornace, and D. V. Bulavin. 2003. Regulation of human Cdc25A stability by serine 75 phosphorylation is not sufficient to activate a S-phase checkpoint. Cell Cycle 2:473-478. [PubMed] [Google Scholar]
- 22.Graves, P. R., C. M. Lovly, G. L. Uy, and H. Piwnica-Worms. 2001. Localization of human Cdc25C is regulated both by nuclear export and 14-3-3 binding. Oncogene 20:1839-1851. [DOI] [PubMed] [Google Scholar]
- 23.Graves, P. R., L. Yu, J. K. Schwarz, J. Gales, E. A. Sausville, P. M. O'Connor, and H. Piwnica-Worms. 2000. The Chk1 protein kinase and the Cdc25C regulatory pathway are targets of the anticancer agent UCN-01. J. Biol. Chem. 275:5600-5605. [DOI] [PubMed] [Google Scholar]
- 24.Hassepass, I., R. Voit, and I. Hoffmann. 2003. Phosphorylation at serine-75 is required for UV-mediated degradation of human Cdc25A phosphatase at the S-phase checkpoint. J. Biol. Chem. 278:29824-29829. [DOI] [PubMed] [Google Scholar]
- 25.Hoffmann, I., P. R. Clarke, M. J. Marcote, E. Karsenti, and G. Draetta. 1993. Phosphorylation and activation of human cdc25-C by cdc2-cyclin B and its involvement in the self-amplification of MPF at mitosis. EMBO J. 12:53-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann, I., G. Draetta, and E. Karsenti. 1994. Activation of the phosphatase activity of human cdc25A by a cdk2-cyclin E dependent phosphorylation at the G1/S transition. EMBO J. 13:4302-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honda, R., Y. Ohba, A. Nagata, H. Okayama, and H. Yasuda. 1993. Dephosphorylation of human p34cdc2 kinase on both Thr-14 and Tyr-15 by human cdc25B phosphatase. FEBS Lett. 318:331-334. [DOI] [PubMed] [Google Scholar]
- 28.Izumi, T., and J. L. Maller. 1993. Elimination of cdc2 phosphorylation sites in the cdc25 phosphatase blocks initiation of M-phase. Mol. Biol. Cell 4:1337-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izumi, T., D. H. Walker, and J. M. Maller. 1992. Periodic changes in phosphorylation of the Xenopus cdc25 phosphatase regulate its activity. Mol. Biol. Cell 3:927-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin, J., T. Shirogane, L. Xu, G. Nalepa, J. Qin, S. J. Elledge, and J. W. Harper. 2003. SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 17:3062-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jinno, S., K. Suto, A. Nagata, M. Igarashi, Y. Kanaoka, H. Nojima, and H. Okayama. 1994. Cdc25A is a novel phosphatase functioning early in the cell cycle. EMBO J. 13:1549-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kakizuka, A., B. Sebastian, U. Borgmeyer, I. Hermans-Borgmeyer, J. Bolado, T. Hunter, M. F. Hoekstra, and R. M. Evans. 1992. A mouse cdc25 homolog is differentially and developmentally expressed. Genes Dev. 6:578-590. [DOI] [PubMed] [Google Scholar]
- 33.Kim, S. H., L. Chuan, and J. L. Maller. 1999. A maternal form of the phosphatase Cdc25A regulates early embryonic cell cycles in Xenopus laevis. Dev. Biol. 212:381-391. [DOI] [PubMed] [Google Scholar]
- 34.Kuang, J., C. L. Ashorn, M. Gonzalez-Kuyvenhoven, and J. E. Penkala. 1994. cdc25 is one of the MPM-2 antigens involved in the activation of maturation-promoting factor. Mol. Biol. Cell 5:135-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumagai, A., and W. G. Dunphy. 1996. Purification and molecular cloning of Plx1, a cdc25-regulatory kinase from Xenopus egg extracts. Science 273:1377-1380. [DOI] [PubMed] [Google Scholar]
- 36.Kumagai, A., and W. G. Dunphy. 1992. Regulation of the cdc25 protein during the cell cycle in Xenopus extracts. Cell 70:139-151. [DOI] [PubMed] [Google Scholar]
- 37.Kumagai, A., Z. Guo, K. Emami, S. X. Wang, and W. G. Dunphy. 1998. The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. J. Cell Biol. 142:1559-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lammer, C., S. Wagerer, R. Saffrich, D. Mertens, W. Ansorge, and I. Hoffmann. 1998. The cdc25B phosphatase is essential for the G2/M phase transition in human cells. J. Cell Sci. 111:2445-2453. [DOI] [PubMed] [Google Scholar]
- 39.Lee, M. S., S. Ogg, M. Xu, L. L. Parker, D. J. Donoghue, J. L. Maller, and H. Piwnica-Worms. 1992. cdc25+ encodes a protein phosphatase that dephosphorylates p34cdc2. Mol. Biol. Cell 3:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lincoln, A. J., D. Wickramasinghe, P. Stein, R. M. Schultz, M. E. Palko, A. P. De Miguel, L. Tessarollo, and P. J. Donovan. 2002. Cdc25b phosphatase is required for resumption of meiosis during oocyte maturation. Nat. Genet. 30:446-449. [DOI] [PubMed] [Google Scholar]
- 41.Mailand, N., J. Falck, C. Lukas, R. G. Syljuasen, M. Welcker, J. Bartek, and J. Lukas. 2000. Rapid destruction of human Cdc25A in response to DNA damage. Science 288:1425-1429. [DOI] [PubMed] [Google Scholar]
- 42.Mailand, N., A. V. Podtelejnikov, A. Groth, M. Mann, J. Bartek, and J. Lukas. 2002. Regulation of G2/M events by Cdc25A through phosphorylation-dependent modulation of its stability. EMBO J. 21:5911-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Millar, J. B. A., J. Blevitt, L. Gerace, K. Sadhu, C. Featherstone, and P. Russell. 1991. p55cdc25 is a nuclear protein required for the initiation of mitosis in human cells. Proc. Natl. Acad. Sci. USA 88:10500-10504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molinari, M., C. Mercurio, J. Dominguez, F. Goubin, and G. F. Draetta. 2000. Human Cdc25A inactivation in response to S phase inhibition and its role in preventing premature mitosis. EMBO Rep. 1:71-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagata, A., M. Igarashi, S. Jinno, K. Suto, and H. Okayama. 1991. An additional homolog of the fission yeast cdc25+ gene occurs in humans and is highly expressed in some cancer cells. New Biol. 3:959-968. [PubMed] [Google Scholar]
- 46.Nargi, J. L., and T. A. Woodford-Thomas. 1994. Cloning and characterization of a cdc25 phosphatase from mouse lymphocytes. Immunogenetics 39:99-108. [DOI] [PubMed] [Google Scholar]
- 47.Nishijima, H., H. Nishitani, T. Seki, and T. Nishimoto. 1997. A dual-specificity phosphatase Cdc25B is an unstable protein and triggers p34(cdc2)/cyclin B activation in hamster BHK21 cells arrested with hydroxyurea. J. Cell Biol. 138:1105-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parker, L. L., S. Atherton-Fessler, M. S. Lee, S. Ogg, F. L. Falk, K. I. Swenson, and H. Piwnica-Worms. 1991. Cyclin promotes the tyrosine phosphorylation of p34cdc2 in a wee1+ dependent manner. EMBO J. 10:1255-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng, C.-Y., P. R. Graves, R. S. Thoma, Z. Wu, A. Shaw, and H. Piwnica-Worms. 1997. Mitotic- and G2-checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine 216. Science 277:1501-1505. [DOI] [PubMed] [Google Scholar]
- 50.Qian, Y. W., E. Erikson, F. E. Taieb, and J. L. Maller. 2001. The polo-like kinase Plx1 is required for activation of the phosphatase Cdc25C and cyclin B-Cdc2 in Xenopus oocytes. Mol. Biol. Cell 12:1791-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Russell, P., and P. Nurse. 1986. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell 45:145-153. [DOI] [PubMed] [Google Scholar]
- 52.Sadhu, K., S. I. Reed, H. Richardson, and P. Russell. 1990. Human homolog of fission yeast cdc25 mitotic inducer is predominantly expressed in G2. Proc. Natl. Acad. Sci. USA 87:5139-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sage, J., A. L. Miller, P. A. Perez-Mancera, J. M. Wysocki, and T. Jacks. 2003. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature 424:223-228. [DOI] [PubMed] [Google Scholar]
- 54.Sebastian, B., A. Kakizuka, and T. Hunter. 1993. Cdc25M2 activation of cyclin-dependent kinases by dephosphorylation of threonine-14 and tyrosine-15. Proc. Natl. Acad. Sci. USA 90:3521-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sexl, V., J. Diehl, C. Sherr, R. Ashmun, D. Beach, and M. F. Roussel. 1999. A rate limiting function of Cdc25A for S phase entry inversely correlates with tyrosine dephosphorylation of Cdk2. Oncogene 18:573-582. [DOI] [PubMed] [Google Scholar]
- 56.Solomon, M. J., T. Lee, and M. W. Kirschner. 1992. Role of phosphorylation in p34cdc2 activation: identification of an activating kinase. Mol. Biol. Cell 3:13-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sorensen, C. S., R. G. Syluasen, J. Falck, T. Schroeder, L. Ronnstrand, K. K. Khanna, B.-B. Zhou, J. Bartek, and J. Lukas. 2003. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell 3:247-258. [DOI] [PubMed] [Google Scholar]
- 58.Strausfeld, U., J. C. Labbe, D. Fesquet, J. C. Cavadore, A. Picard, K. Sadhu, P. Russell, and M. Doree. 1991. Dephosphorylation and activation of a p34cdc2/cyclin B complex in vitro by human cdc25 protein. Nature 351:242-245. [DOI] [PubMed] [Google Scholar]
- 59.Vigo, E., H. Muller, E. Properini, G. Hatevoer, P. Cartwright, M. C. Moroni, and K. Helin. 1999. CDC25A phosphatase is a target of E2F and is required for efficient E2F-induced S phase. Mol. Cell. Biol. 19:6379-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Villa-Moruzzi, E. 1993. Activation of the cdc25C phosphatase in mitotic HeLa cells. Biochem. Biophys. Res. Commun. 196:1248-1254. [DOI] [PubMed] [Google Scholar]
- 61.Wickramasinghe, D., S. Becker, M. K. Ernst, J. L. Resnick, J. M. Centanni, L. Tessarollo, L. B. Grabel, and P. J. Donovan. 1995. Two CDC25 homologues are differentially expressed during mouse development. Development 121:2047-2056. [DOI] [PubMed] [Google Scholar]
- 62.Wu, S., and D. J. Wolgemuth. 1995. The distinct and developmentally regulated patterns of expression of members of the mouse Cdc25 gene family suggest differential functions during gametogenesis. Dev. Biol. 170:195-206. [DOI] [PubMed] [Google Scholar]
- 63.Xiao, Z., Z. Chen, A. H. Gunasekera, T. J. Sowin, S. H. Rosenberg, S. Fesik, and H. Zhang. 2003. Chk1 mediates S and G2 arrests through Cdc25A degradation in response to DNA-damaging agents. J. Biol. Chem. 278:21767-21773. [DOI] [PubMed] [Google Scholar]
- 64.Xu, B., S.-T. Kim, and M. B. Kastan. 2001. Involvement of Brcai in S-phase and G2-phase checkpoints after ionizing radiation. Mol. Cell. Biol. 21:3445-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu, B., S.-T. Kim, D.-S. Lim, and M. B. Kastan. 2002. Two molecularly distinct G2/M checkpoints are induced by ionizing radiation. Mol. Cell. Biol. 22:1049-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao, H., J. L. Watkins, and H. Piwnica-Worms. 2002. Disruption of the checkpoint kinase 1/cell division cycle 25A pathway abrogates ionizing radiation-induced S and G2 checkpoints. Proc. Natl. Acad. Sci. USA 99:14795-14800. [DOI] [PMC free article] [PubMed] [Google Scholar]