Abstract
Approximately 10% of cancers overall use alternative lengthening of telomeres (ALT) instead of telomerase to prevent telomere shortening, and ALT is especially common in astrocytomas and various types of sarcomas. The hallmarks of ALT in telomerase-negative cancer cells include a unique pattern of telomere length heterogeneity, rapid changes in individual telomere lengths, and the presence of ALT-associated promyelocytic leukemia bodies (APBs) containing telomeric DNA and proteins involved in telomere binding, DNA replication, and recombination. The ALT mechanism appears to involve recombination-mediated DNA replication, but the molecular details are largely unknown. In telomerase-null Saccharomyces cerevisiae, an analogous survivor mechanism is dependent on the RAD50 gene. We demonstrate here that overexpression of Sp100, a constituent of promyelocytic leukemia nuclear bodies, sequestered the MRE11, RAD50, and NBS1 recombination proteins away from APBs. This resulted in repression of the ALT mechanism, as evidenced by progressive telomere shortening at 121 bp per population doubling, a rate within the range found in telomerase-negative normal cells, suppression of rapid telomere length changes, and suppression of APB formation. Spontaneously generated C-terminally truncated Sp100 that did not sequester the MRE11, RAD50, and NBS1 proteins failed to inhibit ALT. These findings identify for the first time proteins that are required for the ALT mechanism.
The telomeres of human cells contain a linear tandem array of TTAGGG repeats that are bound by telomere-associated proteins and are essential for chromosome stability and genomic integrity (6). The progressive erosion of telomeres that occurs during the proliferation of normal cells leads eventually to the state of replicative senescence, features of which include permanent withdrawal from the cell cycle. Telomere shortening and senescence appear to be a potent tumor suppressor mechanism (14). Most cancer cells escape from the limitation on proliferation that normal telomere shortening imposes by activating a telomere maintenance mechanism, either telomerase (10) or alternative lengthening of telomeres (ALT) (4). Telomerase is active in approximately 85% of cancers (36), and an ALT mechanism is active in many telomerase-negative tumors (3), especially sarcomas and astrocytomas (13, 45). Some tumors utilize more than one telomere maintenance mechanism (3, 13, 45).
Although the detailed molecular mechanisms of ALT are largely unknown, telomerase-null Saccharomyces cerevisiae survivors are dependent on the RAD52 gene (20), which encodes a protein involved in DNA recombination, and there is evidence that ALT in human cells also involves recombination (8, 25). The hallmarks of ALT in all human cell lines examined to date include a unique pattern of telomere length heterogeneity, with telomeres that range from very short to greater than 50 kb long (4). This heterogeneity is generated by a combination of steady telomere attrition at the rate seen in normal telomerase-negative cells and rapid lengthening and shortening events (25, 33).
An additional hallmark of ALT is the presence of promyelocytic leukemia (PML) nuclear bodies containing (TTAGGG)n DNA and telomere-specific binding proteins. PML bodies are found in most cells. They are dynamic structures that respond to cellular stresses, and they have been implicated in a wide variety of cellular functions, including DNA repair, apoptosis, senescence, transcriptional regulation, proteasome degradation, tumor suppression, and response to viruses (reviewed in references 7, 30, and 53). They have variously been proposed to serve as depots for storage of nuclear factors, as sites where proteins are posttranslationally modified, and as macromolecular platforms on which specific nuclear events such as DNA transcription, replication, or repair take place.
PML bodies containing (TTAGGG)n DNA and telomere-specific binding proteins such as TRF1 and TRF2 have only been found in ALT cells, not in mortal cells or telomerase-positive cell lines, so PML bodies with these contents are referred to as ALT-associated PML bodies (APBs) (52). In addition to constitutive components of PML bodies such as PML and Sp100, APBs contain other proteins involved in DNA replication, recombination, and repair including RAD51, RAD52, and RPA (52), RAD51D (41), BLM (38, 51), WRN (15), human RAP1 and BRCA1 (49), MRE11, RAD50, and NBS1 (50, 55), ERCC1 and XPF (54), and human RAD1, RAD9, RAD17, and HUS1 (26). The formation of APBs requires NBS1, which recruits MRE11, RAD50, and BRCA1 into these structures (49).
APBs are found in a minority of cells (approximately 5%) within asynchronously dividing ALT cell populations (52), which has led to the conclusion that they form in a cell cycle-dependent manner (11, 50). The possibility has been considered that APBs act as storage depots of macromolecules involved in the ALT mechanism or that they have a role in processing by-products of this mechanism (52). It has also been suggested that APBs have an integral role in the ALT mechanism (11, 22, 49, 50, 52), and, consistent with this suggestion, inhibition of ALT in some somatic cell hybrids formed by fusion of ALT and telomerase-positive cell lines resulted in a major decrease in APBs (33). Imaging of live cells has revealed a dynamic relationship between telomeres and APBs: an individual telomere may move into and out of contact with an APB (22).
As a step towards understanding the function of APBs, we attempted live cell imaging of PML bodies in ALT cells by transfecting them with an expression plasmid encoding a fusion protein consisting of yellow fluorescent protein (YFP) and Sp100, a core member of PML bodies. Sp100 is a protein of 480 amino acids, a calculated mass of 53 kDa, and an electrophoretic mobility of 100 kDa (with the anomalous mobility being accounted for mainly by its acidic region). It was originally identified as an antigen which reacts with autoantibodies from patients with primary biliary cirrhosis, forming nuclear speckles (40) (hence Sp100, for speckle protein of 100 kDa). Like PML, Sp100 is a dynamic component of PML bodies (48). Sp100 is also found in nuclear foci that do not contain PML; time-lapse fluorescence microscopy showed that these foci are more mobile than PML bodies, which exhibit little movement (48). The region of Sp100 that is responsible for its localization to PML bodies has been mapped to its N terminus (39). Sp100 is thought to have transcription-modulatory activity, primarily through association with factors such as HIPK2, HP1, and ETS-1 (19, 23, 35, 47). Sp100 has also been shown to bind to the DNA recombination and repair protein NBS1 (27).
We found that formation of APBs was suppressed in cells that expressed high levels of Sp100 or the YFP-Sp100 fusion protein. Stable expression of the fusion protein also resulted in progressive telomere shortening for many population doublings, and inhibition of the telomere length fluctuation that is characteristic of ALT cells. Analysis of clones expressing mutant transgenes containing C-terminal truncations of Sp100 demonstrated that suppression of ALT occurred when NBS1, MRE11, and RAD50 were sequestered by the full-length protein. In addition to identifying a protein that can inhibit ALT and demonstrating the close correlation between the presence of APBs and the ALT mechanism, these data identify for the first time proteins that are required for ALT.
MATERIALS AND METHODS
Cells, plasmids, and transfections.
The spontaneously immortalized Li-Fraumeni syndrome fibroblast line IIICF/c (34) and the osteosarcoma cell line Saos-2 (American Type Culture Collection), both of which are ALT+ (3, 4) were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 50 μg of gentamicin per ml in a 5% CO2 humidified atmosphere at 37°C. Cells were growth arrested by withdrawal of methionine for 4 days; methionine-deficient medium was reconstituted from methionine- and cystine-deficient Dulbecco's modified Eagle's medium (Gibco) by adding l-cystine (Gibco).
IIICF/c cells were transfected with the YFP-Sp100 expression plasmid (24) (see Fig. 2C) with Fugene 6 (Roche), 5 μg of DNA per 10-cm dish, and selected with G418 (Roche). From pooled G418-resistant cells that had been cultured for approximately 20 population doublings, individual colonies were isolated by limiting dilution and passaged continuously in medium containing G418 (430 μg/ml). IIICF/c and Saos-2 cells were transfected with plasmid pSG5-Sp100 (39) with Fugene 6 and 0.5 μg of DNA per well in four-well glass slides (Nunc).
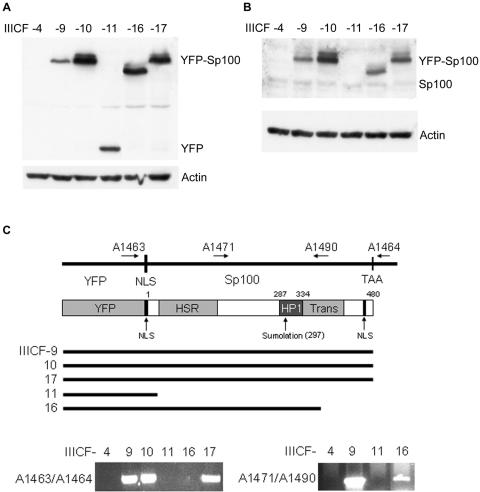
FIG. 2.
Expression levels and mutation profiles of YFP-Sp100 in transfected IIICF/c clones. Clones IIICF-4, -9,-10, -11, -16, and -17 were all analyzed at population doubling 38 to 41. Western blot analyses of YFP-Sp100 (A) and Sp100 (B) with anti-GFP and anti-Sp100 antibodies, respectively, are shown. (C) Schematic representation of YFP-Sp100 and genomic PCR analysis of the integrated YFP-Sp100 plasmid. IIICF-11 and -16 had undergone C-terminal truncations of YFP-Sp100, and no YFP-Sp100 sequence was retained in IIICF-4. NLS, nuclear localization signal; HSR, homogeneous staining region; HP1, heterochromatin protein 1 binding domain; Trans, transactivation domain.
Mutation analysis by PCR.
To determine the integrity of the YFP-Sp100 construct in transfected IIICF/c clones, genomic DNA was subjected to PCR with primers A1463 (5′-TCACATGGTCCTGCTGGAGTTC), and A1464 (5′-AGGTTCAGGGGGAGGTGTGG), which flanked the Sp100 open reading frame (Fig. 2C). Clones IIICF-9, IIICF-10, and IIICF-17 yielded 1.6-kb DNA products that were purified and subjected to automated DNA sequencing. The C terminus of IIICF-16 was determined with primer walking, and a product was ultimately obtained with primers A1471 (5′-GAGGTCTGGCCTCCAACTAAGTC) and A1490 (5′-AATCTGGGGTCGTGAGCAAGTGG). Primer A1490 corresponds to Sp100 amino acid residues 372 to 378 (40).
Antibodies.
The following antibodies were used in this study: mouse anti-NBS1, anti-MRE11, and anti-RAD50 (BD Biosciences); mouse and rabbit anti-green fluorescent protein (GFP) (BD Biosciences); rabbit anti-NBS1 (Ab-1) and anti-MRE11 (Ab-1) (Oncogene Research Products); goat anti-NBS1 (Nibrin, C-19), goat anti-PML (N-19), and mouse anti-PML (Santa Cruz Biotechnology); mouse anti-TRF2 and anti-HP1α (Upstate Biotechnology); rabbit anti-Sp100 (Chemicon); and another rabbit anti-Sp100 antibody (anti-SpGH) described previously (39). Polyclonal anti-TRF1 rabbit serum was raised against a TRF1 peptide, residues 13 to 35.
Immunostaining and fluorescence microscopy.
Cells grown in four-well or two-well chamber slides were fixed for 15 min in 2% paraformaldehyde at room temperature and then permeated with methanol-acetone (1:1) at −20°C for 15 min. In order to clearly visualize NBS1/MRE11/RAD50 foci and other nuclear foci, cells were sometimes treated with a nuclear extraction procedure as described previously (55). Cells were incubated with primary antibodies either for 1 h at room temperature or overnight at 4°C and then incubated with fluorescently conjugated secondary antibodies at room temperature for 30 to 40 min. In some cases, 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma) was included in the secondary incubation to visualize DNA. Finally, the preparations were mounted in antifading medium containing Dabco (Sigma). The secondary antibodies used were as follows: 7-amino-3-methylcoumarin acetic acid (AMCA)-, fluorescein isothiocyanate-, or Texas Red-conjugated goat anti-mouse immunoglobulin, AMCA-, fluorescein isothiocyanate-, or Texas Red-conjugated goat anti-rabbit immunoglobulin, AMCA-conjugated donkey anti-mouse immunoglobulin, fluorescein isothiocyanate-conjugated donkey anti-rabbit immunoglobulin, and Texas Red-conjugated donkey anti-goat immunoglobulin (Jackson ImmunoResearch).
The samples were examined on a Leica DMLB fluorescence microscope. Images were recorded with a Spot cooled charge-coupled device camera (SPOT2; Diagnostic Instruments) and analyzed with Adobe PhotoShop 6.0.
Immunoblotting and coimmunoprecipitation.
For immunoblotting analyses, cell lysates were prepared, electrophoretically separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and electrotransferred to a nylon membrane as described previously (44). Immunoblotting procedures were as recommended by the antibody suppliers. Horseradish peroxidase-conjugated goat anti-mouse or swine anti-rabbit immunoglobulin G (Dako) were used as secondary antibodies.
For coimmunoprecipitation, an equal number of cells for each clone were lysed in Triton X-100 lysis buffer (1% Triton X-100, 25 mM Tris-HCl, [pH 8], 200 mM NaCl, 2 mM EDTA, [pH 8], 1.5 mM MgCl2, 10% glycerol, 0.2 mM dithiothreitol supplemented with Roche complete protease inhibitor and leupeptin). Precleared whole-cell extracts were incubated with the primary antibody-bound protein G-agarose beads (Roche) for 2 h at 4°C. The bound protein complexes were eluted from the beads by boiling with sodium dodecyl sulfate sample buffer and then immunoblotted. For the input control, 10% of whole-cell extracts were used. SYPRO Ruby (Molecular Probes) was used to stain proteins in polyacrylamide gels.
Terminal restriction fragment analysis and telomere repeat amplification protocol analysis.
Telomere length was determined by pulsed-field gel electrophoresis as previously described (4) with 1.5 μg/well of genomic DNA that had been digested with restriction enzymes HinfI and RsaI (Roche) to generate terminal restriction fragments. The gel was exposed to a phosphor screen and scanned with a Storm 860 optical scanner with ImageQuant software (Molecular Dynamics). The molecular weights of telomeric bands were determined by constructing a standard curve from high-molecular-weight DNA markers (Invitrogen) electrophoresed on the same gel. Telomerase activity was assayed with the telomere repeat amplification protocol (16) with the modifications previously described (33).
Telomere fluorescence in situ hybridization analysis and quantitative fluorescence histogram analysis.
Mitotic chromosome preparations were obtained according to standard cytogenetic protocols. Fluorescence in situ hybridization with a Cy3-conjugated telomere-specific peptide nucleic acid probe (PE Biosystems) was performed essentially as described previously (33). Metaphases were evaluated on a Leica DMLB fluorescence microscope with appropriate filter sets, and DAPI and Cy3 images were captured separately as monochromatic 12-bit images with the cooled charge-coupled device camera, merged and pseudo-colored, and further processed for illustrative purposes with Adobe Photoshop 6.0 software.
Fluorescence intensity was measured as described previously (33) with the following modifications. Quantitative histogram analysis was performed with ImagePro Plus 4.0 software (MediaCybernetics) on the unmodified, nonmerged 12-bit monochromatic images captured with exposure times between 0.5 and 2 s with no gamma adjustment. Image bitmap pixel values ranged from 0 (empty scale = black) to 4,095 (full scale = white) following a linear function of a measured intensity with increasing exposure time. Typically, values for the maximum intensities of the short (p) and the long (q) arms of a consistently present and identifiable marker chromosome with an interstitial telomere signal (see Fig. 5A) were recorded for 20 randomly selected metaphase spreads. The maximal values (<4,095) for p- and q-arm telomere fluorescence in situ hybridization intensities of this marker chromosome were corrected for average intensity values of background fluorescence, and the ratios of p-arm to q-arm fluorescence intensities were normalized by the median and plotted in a bar chart (Fig. 5B). The pairwise comparison of statistical significance was made by F test on the variances of normalized ratios.
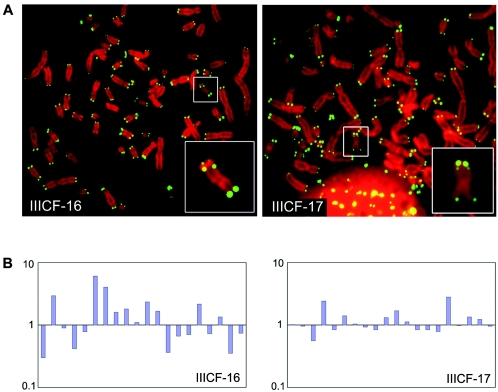
FIG. 5.
Inhibition of telomere length fluctuation in IIICF/c cells overexpressing full-length YFP-Sp100. (A) Telomere fluorescence in situ hybridization analysis, with a peptide nucleic acid telomere probe, on individual metaphase nuclei from clones IIICF-16 and -17 at population doubling 27. The insets show the marker chromosome (with an interstitial telomere signal) that was used for the quantitative analysis. (B) Ratios of p-arm to q-arm telomere fluorescence in situ hybridization signals on the marker chromosome from 20 metaphase spreads for each clone were measured as described in the text. Each bar represents the ratio for an individual chromosome, normalized by the median. The standard deviation of the ratios for IIICF-17 (0.55) was significantly lower than that for IIICF-16 (1.45; F test, P = 0.0001).
RNA interference.
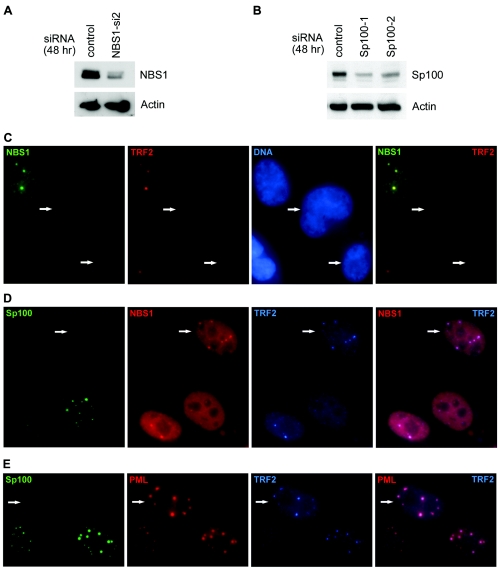
Small interfering RNAs (siRNAs) were designed and synthesized for the target sequence of NBS1 cDNA: 5′-AAGAAGCAGCCTCCACAAATT-3′ (NBS1-si2), and target sequences of Sp100 cDNA: 5′-CAGGAAATTATGATAAACTCA-3′ (Sp100-1) and 5′-AACCATGGAATCCAAATTAAT-3′ (Sp100-2; Qiagen). The nonsilencing control siRNA was purchased from Qiagen. IIICF/c and Saos-2 cells were transfected with siRNA with RNAiFect (Qiagen). For Western analysis, 400 pmol of siRNA was used for each well of a six-well plate; 48 h after transfection, cells were harvested for protein isolation. For immunofluorescence studies, 80 or 160 pmol of siRNA was used for each well of a four- or two-well chamber slide, respectively. Cells were growth arrested 48 h after transfection and fixed and immunostained 4 days later.
RESULTS
APBs are suppressed by overexpression of Sp100 in ALT cells.
As is characteristic of human cell lines that maintain their telomeres with the ALT mechanism, the IIICF/c spontaneously immortalized Li-Fraumeni syndrome fibroblast line has telomeres of very heterogeneous lengths (34) and forms APBs (Fig. 1A). In this study, APBs were mostly detected by visualizing TRF1 or TRF2 within a PML body, i.e., by double immunostaining to detect colocalization of PML protein with one or the other of these telomere binding proteins in a bright nuclear focus of characteristic morphology. Immunostaining for TRF1 or TRF2 to detect APBs produced equivalent results (TRF1 foci completely colocalized with TRF2 foci in >99% of cells under the conditions used in these experiments; data not shown), so antibodies against these two proteins were functionally interchangeable, with the choice being determined by the species of origin of the other antibodies used in colocalization analyses. For example, rabbit anti-TRF1 antibody was used in combination with mouse anti-PML, and mouse anti-TRF2 was used together with rabbit anti-Sp100 (Fig. 1).
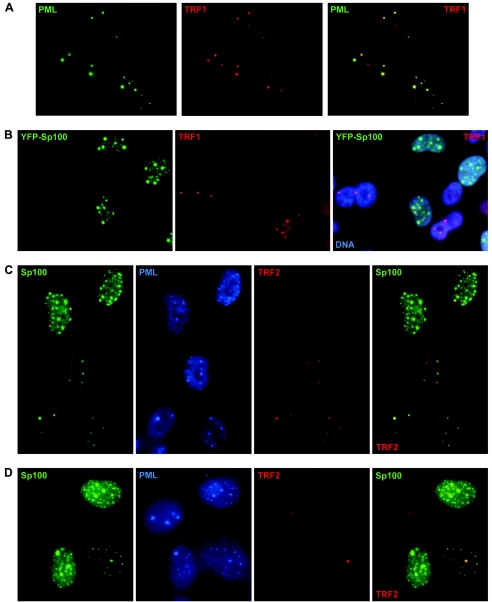
FIG. 1.
Suppression of ALT-associated PML bodies (APBs) by overexpression of Sp100. APBs were visualized in growth-arrested cells with antibodies against PML and either TRF1 or TRF2. Sp100 was detected by fluorescence (YFP-Sp100) or immunostaining (Sp100). (A) Most untransfected control IIICF/c cells exhibited APBs, visualized here as prominent TRF1 foci within PML bodies. (B) APB formation was inhibited in transfected IIICF/c cells overexpressing YFP-Sp100 but not in cells with undetectable levels of transgene expression. Similarly, APB formation was suppressed by overexpression of Sp100 in both IIICF/c (C) and Saos-2 cells (D).
APBs are typically somewhat larger than the PML bodies seen in APB-negative cells, and the quantity of telomere binding proteins (and of telomeric DNA) that they contain is often greater than the amount present at a telomere, so with appropriate microscope settings, APBs may often be seen as large, bright TRF1 or TRF2 foci when individual telomeres are not bright enough to be visualized. To facilitate observation of APBs, the cells were subjected to growth arrest by withdrawal of methionine from the medium, which increased the proportion of cells containing APBs to 50 to 60%, in contrast to ≈5% in exponentially dividing populations.
IIICF/c cells were transfected transiently with expression plasmids encoding either YFP-Sp100 fusion protein or Sp100, and it was observed that there was an inverse correlation between the presence of APBs and expression of these proteins. PML bodies in general were unaffected, but APBs were essentially absent from IIICF/c cells expressing high levels of YFP-Sp100 and present in cells expressing either low levels of or no YFP-Sp100 (Fig. 1B). Similarly, when IIICF/c cells were transfected with a plasmid encoding full-length Sp100 without the YFP tag, APBs were not detected in cells expressing high levels of Sp100 (Fig. 1C), showing that Sp100 has the same effect as YFP-Sp100 and therefore that the fusion protein does not suppress APBs in a dominant negative manner. Similar results were obtained with a second ALT cell line, Saos-2, indicating that the inverse correlation between APB formation and overexpression of Sp100 is not unique to IIICF/c cells (Fig. 1D).
Spontaneous mutations of the YFP-Sp100 transgene.
To determine whether suppression of APB formation correlated with inhibition of ALT-mediated telomere length maintenance, stable clones of YFP-Sp100-transfected cells were generated by limiting dilution of the pooled G418-selected culture. Although the cells were maintained continuously on G418 selection, the yield of YFP-positive clones was low (<10% of total clones). This suggested that overexpression of the YFP-Sp100 transgene was disadvantageous for the proliferation or survival of these cells. Of a total of 13 YFP-positive clones, only two (designated IIICF-10 and IIICF-17) exhibited repression of APB formation.
A panel of six clones (IIICF-4, -9, -10, -11, -16, and -17) were chosen for more detailed analysis. All clones were negative for telomerase activity, as assessed by the telomere repeat amplification protocol assay (data not shown). Western blotting with anti-GFP antibodies (which detect YFP also) showed that only two of the clones, IIICF-10 and -17, expressed high levels of full-length YFP-Sp100 (Fig. 2A and B). IIICF-9 expressed lower levels of full-length YFP-Sp100, while no YFP-Sp100 was detected in IIICF-4. IIICF-11 expressed a band of ≈35 kDa that was detected by the anti-GFP but not anti-Sp100 antibodies, whereas IIICF-16 expressed a fusion protein of reduced size (Fig. 2A and B). Two other clones that were not included in the panel expressed fusion proteins that were very similar in size to the fusion protein in IIICF-16 (data not shown).
Analysis of the integrated transgene by genomic PCR (Fig. 2C) showed that the YFP-Sp100 coding sequence was intact in each of the clones (IIICF-9, -10, and -17) which expressed the full-length fusion protein. Also consistent with the Western data, IIICF-16 had undergone a deletion of the C terminus of the Sp100 sequence. No point mutations were detected by sequencing the transgenes of IIICF-9, -10, and -17 (data not shown). Fluorescence microscopy (Fig. 3) showed that IIICF-4 had no detectable expression of YFP-Sp100 (consistent with the absence of a detectable protein by Western blotting). IIICF-11 had diffuse low-level nuclear fluorescence; taken together with the Western and PCR analyses, this indicates that the transgenic fusion protein in this clone consisted mostly of YFP together with a nuclear localization signal and a small portion of the Sp100 N terminus. The remaining four clones expressed abundant nuclear foci of YFP-Sp100.
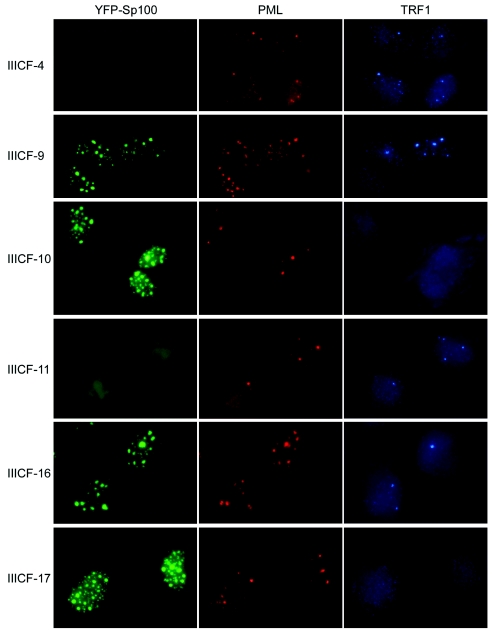
FIG. 3.
Suppression of APB formation in IIICF/c clones overexpressing full-length YFP-Sp100. YFP fluorescence and double immunostaining of growth-arrested IIICF-4, -9, -10, -11, -16, and -17 cells (population doubling 30 to 33) with anti-TRF1 and anti-PML antibodies are shown. APB formation was suppressed in IIICF-10 and -17, which express high levels of YFP-Sp100, whereas they formed normally in cells that had no residual expression of the transgene (IIICF-4), expressed a C-terminally truncated transgene (IIICF-11 and -16), or expressed full-length YFP-Sp100 at a lower level (IIICF-9).
Correlation between status of the YFP-Sp100 transgene and repression of APB formation.
Immunostaining for PML showed that essentially all of the cells in each of the IIICF/c clones contained PML bodies and that most of the YFP-Sp100 in IIICF-9 was contained within PML bodies. In the clones that expressed high levels of YFP-Sp100, however, some of the YFP-Sp100 foci did not contain detectable amounts of PML protein (Fig. 3), in agreement with a previous report indicating that overexpression of Sp100 results in nuclear aggregates that are separate from PML bodies (29).
Double immunostaining for PML and TRF1 showed that APBs formed normally in clones IIICF-4, -9, -11, and -16, whereas there was almost complete suppression of APBs in IIICF-10 and -17, which expressed high levels of full-length YFP-Sp100 (Fig. 3). Under the conditions used here, approximately 50% of the cells in the clones that expressed either no fusion protein or a truncated protein contained APBs, whereas only 2 to 3% of the cells in clones IIICF-10 and -17 formed APBs (Table 1). The effect of YFP-Sp100 on APB formation was not due to a generalized disruption of TRF1 or TRF2 localization or stability; Western blotting showed that the total levels of TRF1 and TRF2 were similar in the six clones, and immunostaining of cytospun metaphase chromosomes and of interphase nuclei for these proteins showed no difference between IIICF-16 (APB+) and IIICF-17 (APB−) apart from lack of APBs in IIICF-17 (data not shown), indicating that association of these proteins with the telomere was not disrupted. The results for the panel of clones are therefore consistent with the results of the transient transfection analyses and indicate that overexpression of full-length YFP-Sp100 is associated with suppression of APB formation.
TABLE 1.
Proportion of APB-positive cells in YFP-Sp100-transfected IIICF/c cell lines
| Cell line | YFP-Sp100 statusa | No. of APB+ cells/no. examined (% positive)b | Telomere shorteningc |
|---|---|---|---|
| IIICF-4 | Loss | 62/133 (46.6) | No |
| IIICF-9 | Full length (low level) | 53/137 (38.7) | No |
| IIICF-10 | Full length (high level) | 4/134 (3.0) | Yes |
| IIICF-11 | YFP retained with small Sp100 N-terminus | 74/143 (51.7) | No |
| IIICF-16 | C-terminal truncation | 68/140 (48.6) | No |
| IIICF-17 | Full length (high level) | 3/152 (2.0) | Yes |
As assessed by Western blotting and analysis of the integrated YFP-Sp100 transgene (Fig. 2).
APBs were detected by immunostaining of cells (all at doubling 30 to 33) for TRF1 and PML after 4 days of growth arrest. Examples are shown in Fig. 3.
As assessed by Southern analysis of Terminal Restriction Fragments (Fig. 4).
Inactivation of the ALT mechanism in cells with suppressed formation of APBs.
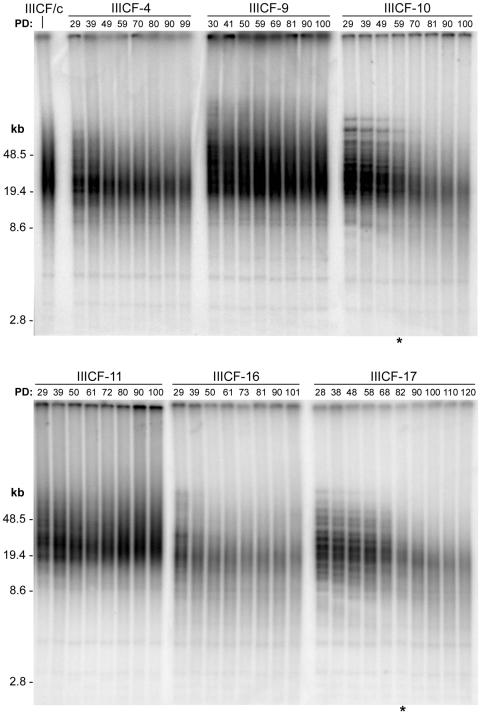
To determine whether YFP-Sp100-induced suppression of APB formation was accompanied by inactivation of the ALT mechanism, we analyzed terminal restriction fragment lengths in these six clones (Fig. 4) over a period of at least 70 to 90 population doublings from the earliest time points possible (population doubling 28 to 29, where population doubling 0 was defined as the point at which the pooled selected population was subcloned by limiting dilution). Densitometric analysis of the invariant bands, which correspond to interstitial (TTAGGG)n DNA sequences, confirmed equal sample loading (variation among lanes was < 30%). In each of the APB-positive clones (IIICF-4, -9, -11, and -16), faint higher-molecular-weight telomeric bands were visible at early population doublings, which reduced in size and then disappeared into the terminal restriction fragment smear pattern (Fig. 4). Analyses of G418-selected clones (i.e., expressing no transgene other than that for neomycin resistance) revealed the same pattern of terminal restriction fragment dynamics (data not shown). This effect in clonally derived populations of IIICF/c cells presumably results from the brief persistence of terminal restriction fragment bands corresponding to individual telomeres, which undergo steady attrition as in other telomerase-negative cells as well as stochastic lengthening and shortening events in cells within the population (25), resulting in relatively rapid dispersal of the bands.
FIG. 4.
Progressive telomere shortening in clones overexpressing YFP-Sp100. Terminal restriction fragment length was analyzed by Southern blotting of genomic DNA from parental IIICF/c cells and IIICF-4, -9,-10, -11, -16, and -17 cells at the population doubling (PD) indicated. The asterisk indicates the population doubling levels at which small patches of revertant cells appeared within the cultures.
In contrast, clones IIICF-10 and -17, in which APBs were suppressed, had much more distinct higher-molecular -weight bands at early population doublings that underwent progressive shortening with further cellular proliferation and remained visible until population doublings 60 and 80. The rate of shortening of individual bands was calculated for IIICF-17 by reference to the molecular weight markers and was found to be very similar for each of the bands (see Fig. S1 in the supplemental material). The mean rate of shortening was 121 nucleotides per population doubling, which is within the range reported for normal telomerase-negative human cells (37) and for somatic cell hybrids in which ALT has been repressed (33). The persistence for so many population doublings of distinct, albeit shortening, terminal restriction fragment bands corresponding to individual telomeres in the clonally derived IIICF-10 and -17 cultures indicates that the stochastic telomere lengthening and shortening events characteristic of ALT were substantially reduced or abolished in these cells.
When the terminal restriction fragment bands became blurred (approximately population doubling a 60 and 80 in IIICF-10 and -17, respectively; Fig. 4), small patches of revertant cells containing APBs started to appear in the cultures. From the reversion points onwards, the total YFP-Sp100 protein levels decreased gradually (see Fig. S2A in the supplemental material), accompanied by the gradual reappearance of APBs (Fig. S2B in the supplemental material). There was thus a close correlation between telomere shortening and repression of APB formation.
To confirm that the ALT mechanism had been repressed, we performed quantitative telomere fluorescence in situ hybridization analysis to determine whether the fluctuations in telomere length characteristic of ALT cells (33) had been inhibited. We measured the ratio of the telomere signals on the p and q arms of a marker chromosome that is present in both IIICF-16 and -17 cells. The marker chromosome was unambiguously identifiable by its size and characteristic DAPI staining pattern and also by the presence of an interstitial telomere signal (Fig. 5A). We note here that IIICF-17 cells, like IIICF-16 and all other ALT cell lines examined to date, have some chromosome ends with no detectable telomeric signal (Fig. 5A). It is thus very unlikely that this clone was selected by the experimental conditions for its ability to survive prolonged repression of ALT due to a preexisting lack of short telomeres. The marker chromosome p-arm:q-arm telomere fluorescence in situ hybridization intensity ratios showed relatively low variation from metaphase to metaphase in the IIICF-17 cell population, whereas they showed a high level of fluctuation in IIICF-16 cells; this difference was highly significant (F test, P = 0.0001 [Fig. 5B]). These results indicate that ALT activity was suppressed in IIICF-17 cells expressing full-length YFP-Sp100 but not in IIICF-16 cells harboring mutant YFP-Sp100, which is consistent with the results of the terminal restriction fragment length and APB analyses.
Sequestration of NBS1 together with MRE11 and RAD50 by YFP-Sp100.
Although the heterochromatin protein family, HP 1α, -β, and -γ, are Sp100-binding proteins, it seemed unlikely that their binding to overexpressed Sp100 or YFP-Sp100 accounted for inhibition of ALT, because the C-terminally truncated YFP-Sp100 expressed by IIICF-16 retained the whole HP1 binding domain (Fig. 2C), and ALT remained fully active in these cells. In confirmation of the mutational analysis, immunostaining showed that HP1α colocalized with both full-length YFP-Sp100 in IIICF-17 and truncated YFP-Sp100 in IIICF-16 cells (see Fig. S3 in the supplemental material).
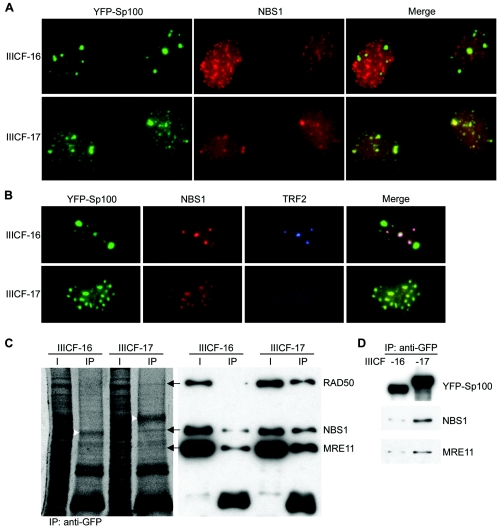
It has been reported that NBS1 physically interacts with Sp100 (27). Immunostaining of IIICF-16 and -17 cells revealed a differential pattern of NBS1 distribution. NBS1 colocalized with the aggregates of full-length YFP-Sp100 in IIICF-17 cells and colocalized to a much lesser extent with the aggregates of truncated YFP-Sp100 in IIICF-16 cells (Fig. 6A). Visualization of YFP-Sp100, NBS1, and TRF2 within the same cells showed that NBS1 colocalized with TRF2 in APBs but was largely excluded from the aggregates of truncated YFP-Sp100 in the IIICF-16 cells. In contrast, NBS1 colocalized with the aggregates of full-length YFP-Sp100 in IIICF-17 cells, in which APBs were repressed (Fig. 6B).
FIG. 6.
Sequestration of NBS1 by YFP-Sp100 in cells with suppression of ALT activity. (A) NBS1 (detected by immunostaining) colocalized with YFP-Sp100 (detected by YFP fluorescence) in IIICF-17 cells but to a much lesser extent with the truncated YFP-Sp100 in IIICF-16 cells. (B) NBS1 colocalized much more extensively with APBs (visualized as foci of TRF2 immunostaining) than with the truncated YFP-Sp100 aggregates in IIICF-16 cells (population doubling 40). In contrast, in IIICF-17 cells (population doubling 39), NBS1 was sequestered in aggregates of full-length YFP-Sp100 and APBs were suppressed. (C) Coimmunoprecipitation analyses showed decreased association of the MRE11/RAD50/NBS1 complex with C-terminally truncated YFP-Sp100. Anti-GFP antibody was used to immunoprecipitate (IP) proteins from the total protein extracts of growth-arrested IIICF-16 and -17 cells (population doubling 33). Proteins separated by gel electrophoresis were either stained with SYPRO Ruby (left panel) or probed with the indicated antibodies (right panel). The arrows (left panel) indicate the bands corresponding to RAD50, NBS1, and MRE11 (detected by immunoblotting in the right panel). The arrowheads indicate the YFP-Sp100 bands. (D) Coimmunoprecipitation of NBS1 and MRE11 was performed as in panel C except that the protein was extracted from cells that were growing asynchronously (population doubling 39). For panels C and D, equal numbers of cells were used from each clone, and 10% of the lysate was used for the input lanes (I). For panels A and B, cells were subjected to nuclear extraction before fixation. Fluorescent images of each channel were acquired and processed in an identical manner for pairwise comparisons in panels A and B.
This finding was confirmed by coimmunoprecipitation analyses of protein extracts from growth-arrested cells with anti-GFP antibodies. There were clear differences between IIICF-16 and -17 in the intensities of the three bands corresponding to NBS1, MRE11, and RAD50 (Fig. 6C). Analysis by densitometry showed that NBS1 and MRE11 binding in IIICF-16 cells was reduced by 4.4- and 3.5-fold, respectively, compared to IIICF-17; the decrease in RAD50 binding was greater and not accurately quantifiable. These proteins were coimmunoprecipitated more strongly by full-length YFP-Sp100 than by the C-terminally truncated version. Similar results were obtained with exponentially growing cell cultures (Fig. 6D). Immunostaining of cytospun metaphase chromosomes revealed no difference between the localization of NBS1 to the telomeres of IIICF-16 and -17 cells (data not shown). The results indicate that inactivation of NBS1 (rather than its removal from telomeres) by binding to YFP-Sp100 is associated with inhibition of the ALT mechanism.
Depletion of NBS1 (but not Sp100) by small interfering RNA prevents the formation of APBs.
To determine the effect of disrupting NBS1 or Sp100 function on the formation of APBs, we used 21-nucleotide siRNAs for silencing NBS1 (NBS1-si2 siRNA) and Sp100 (Sp100-1 or Sp100-2 siRNA). Forty-eight hours after transfection with siRNAs, the level of NBS1 or Sp100 in IIICF/c cells was dramatically reduced compared to the nonsilencing control (Fig. 7A and B). Double immunostaining of the NBS1-si2-transfected IIICF/c cells for NBS1 and TRF2 showed that APB formation was reduced by more than 90% in cells where NBS1 expression was undetectable (Fig. 7C; Table 2), indicating that NBS1 is required for the the formation of APBs. Similar results were obtained by immunostaining NBS1-depeleted cells for TRF1 or detecting telomeric DNA with a fluorescently labeled peptide nucleic acid probe (Table 2). However, APB formation was not inhibited by depletion of Sp100 by either of the Sp100 siRNAs (Fig. 7D and E). TRF2 foci colocalized with PML bodies as well as NBS1 foci in Sp100-depleted cells, suggesting that Sp100 is dispensable for the formation of APBs. Similar results were obtained with another ALT cell line, Saos-2 (data not shown).
FIG. 7.
APB formation requires NBS1 but not Sp100. (A) The NBS1 protein level was downregulated in IIICF/c cells 48 h after NBS1 siRNA (NBS1-si2) transfection. NBS1 and β-actin were detected by immunoblotting. (B) Sp100 siRNAs (Sp100-1 and Sp100-2) downregulated Sp100 expression in IIICF/c cells 48 h after transfection. The blot was probed with the indicated antibodies. (C) APB formation was suppressed in cells that lack NBS1, whereas cells lacking Sp100 formed APBs normally, as demonstrated by colocalization of TRF2 with NBS1 (D) and PML (E). The arrows indicate cells lacking NBS1 in panel C or Sp100 in panels D and E. For panels C to E, IIICF/c cells were growth arrested 48 h after siRNA transfection. The Sp100-1 and Sp100-2 siRNAs were used in panels D and E, respectively.
TABLE 2.
Proportion of APB-positive IIICF/c cells after siRNA treatment
| siRNA treatmenta | Total no. of cells examinedb | No. of cells APB+ (%) | APB detection methodc |
|---|---|---|---|
| NBS1-si2 | 113 | 4 (3.5) | PNA, PML |
| NBS1-si2 | 110 | 3 (2.7) | TRF1, PML |
| NBS1-si2 | 121 | 5 (4.1) | TRF1, TRF2 |
| Sp100-1 | 127 | 75 (59.1) | TRF2, PML |
| No siRNA | 144 | 78 (54.2) | TRF2, PML |
Cells were treated with siRNA (sequence specified in Materials and Methods) for 2 days and then subjected to growth arrest for 4 days.
Only cells that were negative by immunostaining for the knocked-down protein (NBS1 or Sp100) were examined for APBs.
APBs were detected by colocalization of the specified proteins as assessed by dual immunostaining or colocalization of PML protein with (TTAGGG)n DNA visualized by hybridization with a labeled peptide nucleic acid (PNA) probe.
DISCUSSION
We have shown that increased expression of either Sp100 or YFP-Sp100 is able to inhibit the formation of APBs. The YFP-tagged protein therefore has wild-type function in this regard and does not exhibit dominant negative activity. Consistent with this conclusion, knockdown of Sp100 expression by siRNA did not prevent APB formation. Suppression of APBs was accompanied by other evidence that ALT was inhibited, including progressive telomere shortening and significant dampening of the telomere length fluctuation that is characteristic of ALT cells. This is the first time any protein or gene that consistently produces a sustained inhibition of ALT has been identified.
Inhibition of ALT has been obtained previously by somatic cell hybridization approaches only. Fusion of ALT cells with normal cells or with single chromosomes from normal cells resulted in repression of ALT, indicating that ALT is activated by loss of one or more repressor genes (17, 28, 32). When ALT cells were fused with telomerase-positive cells, in some cases ALT was repressed, and this was manifested by a rapid reduction in mean telomere length over the first 20 population doublings, followed by a prolonged period in which the telomeres shortened at a rate similar to that in telomerase-negative normal cells before the telomere length stabilized due to telomerase activity (33). Inhibition of ALT in these hybrid cells was presumably due to the same unidentified repressor(s) present in telomerase-negative normal cells and was not the result of telomerase activity because, except in rare cases, ALT and telomerase activity can coexist within the same cells (5, 9, 12, 33).
YFP-Sp100 was able to cause a sustained inhibition of ALT for up to 80 population doublings, during which time the cells proliferated at about 80% of the rate of untransfected cells (data not shown) and underwent progressive telomere shortening at the rate of 121 bp/population doubling. This is remarkable, given that ALT cells characteristically contain some chromosomes that already have ends with so little telomeric sequence that it is undetectable by fluorescence in situ hybridization. IIICF-10 and -17 are no exception in this regard (see Fig. 5), which indicates that their prolonged survival did not result from selection of clones that happened to have sufficient telomeric sequence on all chromosome ends. It has been shown that recombination-mediated telomere maintenance acts preferentially on the shortest telomeres of the yeasts Saccharomyces cerevisiae (42) and Kluyveromyces lactis (21). Interestingly, although the proportion of APB-containing cells was reduced by more than 90%, the suppression of APBs was not complete. It seems possible, therefore, that ALT activity was not completely repressed in the cells expressing YFP-Sp100 and that this residual activity was sufficient to maintain the shortest telomeres.
There are several lines of evidence indicating that YFP-Sp100 inhibits ALT by sequestering NBS1. The full-length protein that caused inhibition of ALT-mediated telomere maintenance and inhibited APB formation was found to sequester NBS1 into YFP-Sp100 aggregates, whereas the truncation mutant that did not inhibit ALT was ineffective in this regard. We showed by knockdown of NBS1 expression that NBS1 is essential for formation of APBs, in agreement with previous results (49). Furthermore, the three bands that were differentially coimmunoprecipitated with the full-length and truncated YFP-Sp100 proteins were identified by Western blotting as NBS1 and the other two members of the MRN complex, MRE11 and RAD50.
We found that formation of APBs was not impeded by siRNA-mediated knockdown of Sp100 expression. In other cells, it has previously been observed that Sp100 is required for recruiting NBS1 into PML bodies (27); this result was obtained in NT2 cells (non-ALT) that do not usually express detectable levels of either PML or Sp100, which possibly accounts for the different findings.
While there is an excellent correlation between the presence of APBs and ALT, in the absence of data regarding function, it has previously been difficult to determine whether APBs are necessary for ALT. The ALT mechanism appears to involve recombination-mediated DNA replication (8). APBs are a hallmark of ALT cells, contain telomeric DNA and proteins involved in recombination and DNA repair (15, 26, 38, 41, 49-52, 54, 55), and are sites of DNA synthesis (26, 49, 50). Imaging of live cells has revealed that individual telomeres are dynamically associated with APBs (22). Our demonstration here that YFP-Sp100 represses both APBs and ALT-mediated telomere maintenance supports the notion that there is a functional link between APBs and ALT.
In addition to being present in APBs, the MRE11/RAD50/NBS1 complex has been shown to be associated with telomeres (55). Although we could find no effect of YFP-Sp100 overexpression on localization of NBS1 to the telomere, we cannot definitively exclude the possibility that YFP-Sp100 overexpression may inhibit ALT by interfering with the function of these proteins at the telomere. RAD50 and MRE11 are present in TRF2 immunocomplexes and located at telomeres throughout interphase, and they are joined by NBS1 in S phase (55). NBS1 has also been reported to bind to TRF1 (50).
The MRE11/RAD50/NBS1 complex may have nonhomologous end-joining activity and is thought to have a major role in homologous recombination in mammalian cells. It has been suggested that MRE11 and RAD50 may be required to stabilize telomere loop formation (which is essentially a recombinational event) throughout most of the cell cycle and that the transient recruitment of NBS1 in S phase may be required for access of the DNA replication machinery (55). The analogous complex in S. cerevisiae (MRE11/RAD50/XRS2) has a key role in telomere maintenance and recombinational DNA repair (1, 2, 31, 46). There is genetic evidence that RAD50 is part of a pathway that is involved in survival of telomerase-null S. cerevisiae (18). Moreover, of the two types of telomerase-null yeast survivor mechanisms, type II most closely resembles human ALT and is dependent on the RAD50 gene (42, 43).
Identification of the essential role of the MRE11/RAD50/NBS1 proteins in the ALT mechanism suggests possible targets for the development of ALT inhibitors, although our finding that cells in which ALT is repressed can continue to proliferate for up to 80 population doublings indicates that ALT inhibitors may be ineffective as anticancer agents unless combined with other therapies.
Supplementary Material
Acknowledgments
We thank David Spector for the YFP-Sp100 plasmid and Hans Will for the pSG5-Sp100 plasmid and anti-Sp100 antibody.
This work was supported by project grants from the Cancer Council NSW and the National Health and Medical Research Council of Australia (NHMRC), an NHMRC postgraduate scholarship, and an Australian Postgraduate Award.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Boulton, S. J., and S. P. Jackson. 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17:1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bressan, D. A., B. K. Baxter, and J. H. J. Petrini. 1999. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7681-7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryan, T. M., A. Englezou, L. Dalla-Pozza, M. A. Dunham, and R. R. Reddel. 1997. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 3:1271-1274. [DOI] [PubMed] [Google Scholar]
- 4.Bryan, T. M., A. Englezou, J. Gupta, S. Bacchetti, and R. R. Reddel. 1995. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 14:4240-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerone, M. A., J. A. Londono-Vallejo, and S. Bacchetti. 2001. Telomere maintenance by telomerase and by recombination can coexist in human cells. Hum. Mol. Genet. 10:1945-1952. [DOI] [PubMed] [Google Scholar]
- 6.de Lange, T. 2002. Protection of mammalian telomeres. Oncogene 21:532-540. [DOI] [PubMed] [Google Scholar]
- 7.Dellaire, G., and D. P. Bazett-Jones. 2004. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. BioEssays 26:963-977. [DOI] [PubMed] [Google Scholar]
- 8.Dunham, M. A., A. A. Neumann, C. L. Fasching, and R. R. Reddel. 2000. Telomere maintenance by recombination in human cells. Nat. Genet. 26:447-450. [DOI] [PubMed] [Google Scholar]
- 9.Ford, L. P., Y. Zou, K. Pongracz, S. M. Gryaznov, J. W. Shay, and W. E. Wright. 2001. Telomerase can inhibit the recombination-based pathway of telomere maintenance in human cells. J. Biol. Chem. 276:32198-32203. [DOI] [PubMed] [Google Scholar]
- 10.Greider, C. W., and E. H. Blackburn. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405-413. [DOI] [PubMed] [Google Scholar]
- 11.Grobelny, J. V., A. K. Godwin, and D. Broccoli. 2000. ALT-associated PML bodies are present in viable cells and are enriched in cells in the G2/M phase of the cell cycle. J. Cell Sci. 113:4577-4585. [DOI] [PubMed] [Google Scholar]
- 12.Grobelny, J. V., M. Kulp-McEliece, and D. Broccoli. 2001. Effects of reconstitution of telomerase activity on telomere maintenance by the alternative lengthening of telomeres (ALT) pathway. Hum. Mol. Genet. 10:1953-1961. [DOI] [PubMed] [Google Scholar]
- 13.Hakin-Smith, V., D. A. Jellinek, D. Levy, T. Carroll, M. Teo, W. R. Timperley, M. J. McKay, R. R. Reddel, and J. A. Royds. 2003. Alternative lengthening of telomeres and survival in patients with glioblastoma multiforme. Lancet 361:836-838. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100:57-70. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, F. B., R. A. Marciniak, M. McVey, S. A. Stewart, W. C. Hahn, and L. Guarente. 2001. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 20:905-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, N. W., M. A. Piatyszek, K. R. Prowse, C. B. Harley, M. D. West, P. L. C. Ho, G. M. Coviello, W. E. Wright, S. L. Weinrich, and J. W. Shay. 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 266:2011-2015. [DOI] [PubMed] [Google Scholar]
- 17.Kumata, M., M. Shimizu, M. Oshimura, M. Uchida, and T. Tsutsui. 2002. Induction of cellular senescence in a telomerase negative human immortal fibroblast cell line, LCS-AF. 1-3, by human chromosome 6. Int. J. Oncol. 21:851-856. [DOI] [PubMed] [Google Scholar]
- 18.Le, S., J. K. Moore, J. E. Haber, and C. W. Greider. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehming, N., A. Le Saux, J. Schuller, and M. Ptashne. 1998. Chromatin components as part of a putative transcriptional repressing complex. Proc. Natl. Acad. Sci. USA 95:7322-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73:347-360. [DOI] [PubMed] [Google Scholar]
- 21.McEachern, M. J., and S. Iyer. 2001. Short telomeres in yeast are highly recombinogenic. Mol. Cell 7:695-704. [DOI] [PubMed] [Google Scholar]
- 22.Molenaar, C., K. Wiesmeijer, N. P. Verwoerd, S. Khazen, R. Eils, H. J. Tanke, and R. W. Dirks. 2003. Visualizing telomere dynamics in living mammalian cells using PNA probes. EMBO J. 22:6631-6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moller, A., H. Sirma, T. G. Hofmann, H. Staege, E. Gresko, K. S. Ludi, E. Klimczak, W. Droge, H. Will, and M. L. Schmitz. 2003. Sp100 is important for the stimulatory effect of homeodomain-interacting protein kinase-2 on p53-dependent gene expression. Oncogene 22:8731-8737. [DOI] [PubMed] [Google Scholar]
- 24.Muratani, M., D. Gerlich, S. M. Janicki, M. Gebhard, R. Eils, and D. L. Spector. 2001. Metabolic-energy-dependent movement of PML bodies within the mammalian cell nucleus. Nat. Cell Biol. 4:106-110. [DOI] [PubMed] [Google Scholar]
- 25.Murnane, J. P., L. Sabatier, B. A. Marder, and W. F. Morgan. 1994. Telomere dynamics in an immortal human cell line. EMBO J. 13:4953-4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nabetani, A., O. Yokoyama, and F. Ishikawa. 2004. Localization of hRad9, hHus1, hRad1 and hRad17, and caffeine-sensitive DNA replication at ALT (alternative lengthening of telomeres)-associated promyelocytic leukemia body. J. Biol. Chem. 279:25849-25857. [DOI] [PubMed] [Google Scholar]
- 27.Naka, K., K. Ikeda, and N. Motoyama. 2002. Recruitment of NBS1 into PML oncogenic domains via interaction with SP100 protein. Biochem. Biophys. Res. Commun. 299:863-871. [DOI] [PubMed] [Google Scholar]
- 28.Nakabayashi, K., T. Ogata, M. Fujii, H. Tahara, T. Ide, R. Wadhwa, S. C. Kaul, Y. Mitsui, and D. Ayusawa. 1997. Decrease in amplified telomeric sequences and induction of senescence markers by introduction of human chromosome 7 or its segments in SUSM-1. Exp. Cell Res. 235:345-353. [DOI] [PubMed] [Google Scholar]
- 29.Negorev, D., A. M. Ishov, and G. G. Maul. 2001. Evidence for separate ND10-binding and homo-oligomerization domains of Sp100. J. Cell Sci. 114:59-68. [DOI] [PubMed] [Google Scholar]
- 30.Negorev, D., and G. G. Maul. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234-7242. [DOI] [PubMed] [Google Scholar]
- 31.Nugent, C. I., G. Bosco, L. O. Ross, S. K. Evans, A. P. Salinger, J. K. Moore, J. E. Haber, and V. Lundblad. 1998. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8:657-660. [DOI] [PubMed] [Google Scholar]
- 32.Perrem, K., T. M. Bryan, A. Englezou, T. Hackl, E. L. Moy, and R. R. Reddel. 1999. Repression of an alternative mechanism for lengthening of telomeres in somatic cell hybrids. Oncogene 18:3383-3390. [DOI] [PubMed] [Google Scholar]
- 33.Perrem, K., L. M. Colgin, A. A. Neumann, T. R. Yeager, and R. R. Reddel. 2001. Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells. Mol. Cell. Biol. 21:3862-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogan, E. M., T. M. Bryan, B. Hukku, K. Maclean, A. C. M. Chang, E. L. Moy, A. Englezou, S. G. Warneford, L. Dalla-Pozza, and R. R. Reddel. 1995. Alterations in p53 and p16INK4 expression and telomere length during spontaneous immortalization of Li-Fraumeni syndrome fibroblasts. Mol. Cell. Biol. 15:4745-4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seeler, J.-S., A. Marchio, D. Sitterlin, C. Transy, and A. Dejean. 1998. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc. Natl. Acad. Sci. USA 95:7316-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shay, J. W., and S. Bacchetti. 1997. A survey of telomerase activity in human cancer. Eur. J. Cancer. 33:787-791. [DOI] [PubMed] [Google Scholar]
- 37.Smogorzewska, A., and T. de Lange. 2004. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73:177-208. [DOI] [PubMed] [Google Scholar]
- 38.Stavropoulos, D. J., P. S. Bradshaw, X. Li, I. Pasic, K. Truong, M. Ikura, M. Ungrin, and M. S. Meyn. 2002. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum. Mol. Genet. 11:3135-3144. [DOI] [PubMed] [Google Scholar]
- 39.Sternsdorf, T., K. Jensen, B. Reich, and H. Will. 1999. The nuclear dot protein Sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J. Biol. Chem. 274:12555-12566. [DOI] [PubMed] [Google Scholar]
- 40.Szostecki, C., H. H. Guldner, H. J. Netter, and H. Will. 1990. Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J. Immunol. 145:4338-4347. [PubMed] [Google Scholar]
- 41.Tarsounas, M., P. Munoz, A. Claas, P. G. Smiraldo, D. L. Pittman, M. A. Blasco, and S. C. West. 2004. Telomere maintenance requires the RAD51D recombination/repair protein. Cell 117:337-347. [DOI] [PubMed] [Google Scholar]
- 42.Teng, S.-C., J. Chang, B. McCowan, and V. A. Zakian. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell 6:947-952. [DOI] [PubMed] [Google Scholar]
- 43.Teng, S.-C., and V. A. Zakian. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toouli, C. D., L. I. Huschtscha, A. A. Neumann, J. R. Noble, L. M. Colgin, B. Hukku, and R. R. Reddel. 2002. Comparison of human mammary epithelial cells immortalized by simian virus 40 T-antigen or by the telomerase catalytic subunit. Oncogene 21:128-139. [DOI] [PubMed] [Google Scholar]
- 45.Ulaner, G. A., H. Y. Huang, J. Otero, Z. Zhao, L. Ben-Porat, J. M. Satagopan, R. Gorlick, P. Meyers, J. H. Healey, A. G. Huvos, A. R. Hoffman, and M. Ladanyi. 2003. Absence of a telomere maintenance mechanism as a favorable prognostic factor in patients with osteosarcoma. Cancer Res. 63:1759-1763. [PubMed] [Google Scholar]
- 46.Usui, T., T. Ohta, H. Oshiumi, J. Tomizawa, H. Ogawa, and T. Ogawa. 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95:705-716. [DOI] [PubMed] [Google Scholar]
- 47.Wasylyk, C., S. E. Schlumberger, P. Criqui-Filipe, and B. Wasylyk. 2002. Sp100 interacts with ETS-1 and stimulates its transcriptional activity. Mol. Cell. Biol. 22:2687-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiesmeijer, K., C. Molenaar, I. M. Bekeer, H. J. Tanke, and R. W. Dirks. 2002. Mobile foci of Sp100 do not contain PML: PML bodies are immobile but PML and Sp100 proteins are not. J. Struct. Biol. 140:180-188. [DOI] [PubMed] [Google Scholar]
- 49.Wu, G., X. Jiang, W. H. Lee, and P. L. Chen. 2003. Assembly of functional ALT-associated promyelocytic leukemia bodies requires Nijmegen breakage syndrome 1. Cancer Res. 63:2589-2595. [PubMed] [Google Scholar]
- 50.Wu, G., W.-H. Lee, and P. L. Chen. 2000. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies bodies during late S/G2 phases in immortalized telomerase-negative cells: Implication of NBS1 in alternative lengthening of telomeres. J. Biol. Chem. 275:30618-30622. [DOI] [PubMed] [Google Scholar]
- 51.Yankiwski, V., R. A. Marciniak, L. Guarente, and N. F. Neff. 2000. Nuclear structure in normal and Bloom syndrome cells. Proc. Natl. Acad. Sci. USA 97:5214-5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yeager, T. R., A. A. Neumann, A. Englezou, L. I. Huschtscha, J. R. Noble, and R. R. Reddel. 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 59:4175-4179. [PubMed] [Google Scholar]
- 53.Zhong, S., P. Salomoni, and P. P. Pandolfi. 2000. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2:E85-E90. [DOI] [PubMed] [Google Scholar]
- 54.Zhu, X. D., L. Niedernhofer, B. Kuster, M. Mann, J. H. Hoeijmakers, and T. de Lange. 2003. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell 12:1489-1498. [DOI] [PubMed] [Google Scholar]
- 55.Zhu, X.-D., B. Kuster, M. Mann, J. H. J. Petrini, and T. de Lange. 2000. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 25:347-352. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.