Abstract
Prior to microtubule capture, sister centromeres resolve from one another, coming to rest on opposite surfaces of the condensing chromosome. Subsequent assembly of sister kinetochores at each sister centromere generates a geometry favorable for equal levels of segregation of chromatids. The holocentric chromosomes of Caenorhabditis elegans are uniquely suited for the study of centromere resolution and subsequent kinetochore assembly. In C. elegans, only two proteins have been identified as being necessary for centromere resolution, the kinase AIR-2 (prophase only) and the centromere protein HCP-4/CENP-C. Here we found that the loss of proteins involved in chromosome cohesion bypassed the requirement for HCP-4/CENP-C but not for AIR-2. Interestingly, the loss of cohesin proteins also restored the localization of HCP-6 to the kinetochore. The loss of the condensin II protein HCP-6 or MIX-1/SMC2 impaired centromere resolution. Furthermore, the loss of HCP-6 or MIX-1/SMC2 resulted in no centromere resolution when either nocodazole or RNA interference (RNAi) of the kinetochore protein KNL-1 perturbed spindle-kinetochore interactions. This result suggests that normal prophase centromere resolution is mediated by condensin II proteins, which are actively recruited to sister centromeres to mediate the process of resolution.
Attachment of a single kinetochore to both mitotic centrosomes (merotelic orientation) is a frequent cause of aneuploidy (8, 46). One safeguard against merotelic orientation is the geometry of sister kinetochores. Sister kinetochores are positioned on opposite surfaces of the mitotic chromosomes so that attachment of one kinetochore to one centrosome results in the reorientation of the chromosome such that the unattached sister kinetochore faces away from the attached centrosome (42). Thus, the chromosome itself prevents attachment of the nonattached sister kinetochore to the same centrosome and promotes attachment of the sister kinetochore to the other centrosome. Sister kinetochores are observed on opposing surfaces of the mitotic chromosomes in early prophase prior to microtubule capture, indicating that this process is intrinsic to the mitotic chromosome (3, 17, 22, 43). Sister kinetochores are assembled at a chromosomal locus, the centromere. Centromeres do not display obvious sequence homologies between species; however, they are functionally similar and contain many conserved proteins (30). One conserved centromere protein, CENP-A, is a histone H3 variant required for recruiting kinetochore proteins (21, 35, 37). During G2/early prophase, centromere-associated proteins, including CENP-A, are observed to resolve into paired sister centromeres, suggesting that centromere resolution is responsible for the positioning of sister kinetochores back-to-back (2, 16, 35).
A process similar to centromere resolution occurs during prophase (52). This process, called sister chromatid resolution, involves functions important for regulating connections between sister chromatids as well as functions that individualize the chromosomes during prophase. Two well-defined multisubunit complexes, the cohesin and the condensin complexes, carry out these respective activities. A complex of four proteins mediates cohesion between sister chromatids, and dissolution of the connections mediated by this complex is necessary for resolution (for a review, see reference 36). This cohesin complex consists of two structural-maintenance-of-chromosome (SMC) proteins, SMC1 and SMC3, with associated non-SMC proteins SCC3 and SCC1/Mcd1/Rad21 (27, 56). Sister chromatid resolution also requires chromosome condensation, which is mediated by the condensin complex (10, 47). The condensin complex is also composed of two SMC proteins, SMC2 and SMC4, along with associated non-SMC proteins, CAP-D2, CAP-G, and CAP-H (18-20, 25, 48). More recently, a second condensin complex, condensin II, which has different non-SMC components and a distinct localization to mitotic chromosomes, was identified (39, 61).
Although centromere resolution and sister chromatid resolution resemble each other, little is known about the mechanisms or proteins involved in centromere resolution. We used the nematode Caenorhabditis elegans to investigate the role of cohesin and condensin proteins in centromere resolution. C. elegans chromosomes are holocentric and differ from monocentric chromosomes in that the kinetochore of holocentric chromosomes is assembled along nearly the entire length of each chromatid (4, 34). Many centromere and kinetochore proteins in C. elegans function similarly to their mammalian counterparts (7, 35, 37), underscoring the conserved natures of the centromere and kinetochore. This makes C. elegans an ideal system for studying aspects of centromere resolution.
Centromere resolution in C. elegans occurs early in the cell cycle when the centromeric histone H3 variant HCP-3 (CENP-A), observed as a single “line” of HCP-3 staining, splits into two lines of staining that are initially close together but that, later in prophase, are further resolved until they are on opposing surfaces of the mitotic chromosome (35). This dynamic rearrangement requires the centromere protein HCP-4 (CENP-C), the C. elegans ortholog of the mammalian centromere protein C (CENP-C). A mitotic cohesin complex composed of homologs of SMC1, SMC3, SCC1, and SCC3 is present in C. elegans and is required for chromatid cohesion in meiosis, but its role in mitosis is less clear (5, 33, 40, 41). Likewise, homologs of the SMC components of both condensin complexes, SMC2 and SMC4, as well as the non-SMC component of condensin II, HCP-6, are required for mitotic chromosome structure in C. elegans (15, 23, 46). Interestingly, these three condensin proteins colocalize with the centromeric histone HCP-3/CENP-A during mitosis, and HCP-6 requires HCP-3/CENP-A for recruitment to the centromere. Here we show that the loss of cohesins via mutation or RNA interference (RNAi) bypasses the requirement for HCP-4/CENP-C in centromere resolution and in the recruitment of the condensin II component HCP-6. We also show the HCP-6 is required for prophase centromere resolution, suggesting that the role of HCP-4/CENP-C in centromere resolution is to dissolve sister centromere cohesion, facilitating the recruitment of HCP-6 during prophase.
MATERIALS AND METHODS
Strains.
The N2 Bristol strain was used as the wild type; other strains used were JM93, which contains an integrated array of the lactose operator sequence LacO; CB879, which contains the him-1(e879) allele; AR1, which contains the hcp-6(mr17) allele; and EU630, which contains the air-2(or207) allele.
RNA interference.
RNAi via feeding was accomplished using strains obtained from MRC Geneservice and performed according to published procedures (13). Double-stranded RNA (dsRNA) for RNAi was produced as described previously by using appropriate oligonucleotides with or without the T7 polymerase promoter sequence 5′-TAATACGACTCACTATAGGG (34). Oligonucleotides used were 5′-GAATTCCATCTCATGGAACTCATGG and 5′-GTCGACCACAGCCATCTTGTCCTGTGC (C. elegans SCC3 [CeSCC3]), 5′-CGCTGCGGTTCATCAGGAGC and 5′-CAATTGCCTTGGCAGCAGTC (CeSMC1), 5′-GAAGCCAGAAGATGCTCCA and CGTCGCCCACTTCTTGCATTCTG (HCP-4/CeCENP-C), 5′-GCTAATGTGAGCCGTCGTG and 5′-CTCTCCAGCGAATCCACTCAGG (CeSCC1), 5′-GTGCTTCTGCCAACAAACGACC and 5′-CGATAGACCAGCTCGTTGTTGGC (CeMCAK), 5′-GAAAGCGTTGTAATCTCGGG and 5′-TTCAACTCTCTTGCTTCGGG (HCP-6), 5′-AAATCAATTCGACAGGGTGC and 5′-GAAAGATGAGCCGCTGAAAG (Mix-1), 5′-GTAGACTCCCACGCACAAG and 5′-TCGTTTCCTAACCGCCACAC (CeSCC2/Y43H11AL.3), and 5′-GTGGCTGAGTTGTTGTCGAA and 5′-TTTCGGAGCGAGAAGACACT (CeTRF4/ZK858.1). RNAi was by soaking L4-stage worms as previously described (35). RNAi with two different dsRNAs were performed with 2.5 mg of each dsRNA per ml for a total dsRNA concentration of 5 mg/ml. Control RNAi experiments showed no phenotypic difference between 2.5 and 5 mg of total dsRNA per ml (data not shown).
Immunofluorescence and immunoblotting.
An EcoRI/SalI DNA fragment was generated via PCR from C. elegans genomic DNA using oligonucleotides 5′-tccgaattcGACATCATTTGTCGGATG-3′ and 5′-tccgtcgacTTCTCCCATTGTCGCCCA (lowercase indicates non-CeSCC3 DNA sequence used for cloning) and cloned into the EcoRI/SalI-digested pET-28a expression vector to generate plasmid pET-28a::Exon3CeSCC3 by standard procedures. Plasmid pET-28a::Exon3CeSCC3 encodes a six-His fusion protein of predicted amino acids 170 to 521 of the open reading frame F18E2.3. The fusion protein was purified by Ni-chelate chromatography (QIAGEN) and used to raise rabbit antisera in New Zealand White rabbits by R&R Research and Development (Stanwood, Wash.). Antibodies were affinity purified (46). The specific reactivity of serum towards CeSCC3 protein was eliminated by preincubation of serum with CeSCC3 protein and was absent in cescc3(RNAi) embryos (see Fig. S2 in the supplemental material).
For immunofluorescence, embryos from Bristol strain N2 or JM93 (14) were prepared, fixed, and stained for immunofluorescence microscopy (34). Primary antibodies were anti-CeSCC3 (this work), anti-HCP-4 (35), anti-HCP-3 (4), anti-LacI (Stratagene), anti-CeMCAK (37), anti-Mix-1 (a gift from R. Chan and B. Meyer), anti-COH-2 (a gift from M. Jantsch and J. Loidl), and anti-HCP-6 (46). Cell cycle stage was determined using the monoclonal antibody (MAb) 414 (12), directed against nuclear pore proteins. For detection of LacO sequences in strain JM93, fixed embryos were incubated for 30 min at room temperature with purified LacI repressor protein at 0.45 μg/ml in blocking solution and washed three times with blocking solution prior to incubation with anti-LacI antibody. No signal was detected in wild-type controls or when LacI protein was omitted, indicating that the observed signals are specific to the integrated LacO array (data not shown). Nocodazole treatment was as previously described (35). Embryos were examined either by a Zeiss Axioscope microscope equipped with a Sensys charge-coupled-device camera (Photometric) or by a Deltavision microscope (Applied Precision). Three-dimensional imaging was accomplished using Volocity (Improvision). Images were first analyzed in Photoshop (Adobe) and then imported into Canvas (Deneba).
For immunoblotting, equal numbers of hermaphrodites were soaked in either RNAi buffer (mock treated) or dsRNA for CeSCC3. After RNAi treatment, worms were washed three times in M9 buffer and the pellet was resuspended EB buffer (50 mM HEPES [7.5], 70 mM potassium acetate, 5 mM magnesium acetate, 0.1% Triton X-100, 20 mM β-glycerol phosphate, 10% glycerol, 0.5 mM dithiothreitol, and complete proteinase inhibitors [Sigma]) and sonicated. Quantitation of total proteins was by DC protein assay (Bio-Rad) and was followed by the addition of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer. Equal amounts of the total protein were used for Western blotting and detected with an ECL colorimetric detection kit (Amersham). For loading controls, proteins bound to the membrane were detected by using Ponceau S (data not shown), and only blots with equal levels of loading were used for Western blotting.
RESULTS
Suppressors of the HCP-4/CENP-C(RNAi) defect in centromere resolution.
Given the importance of cohesin proteins in the cohesion between sister chromatids, we investigated whether the loss of cohesins and proteins likely required for cohesion could suppress the defect in centromere resolution resulting from the loss of HCP-4/CENP-C (35). Centromere resolution was considered to have occurred if any portion of HCP-3/CENP-A staining on the mitotic chromosome was resolved. In wild-type and mock RNAi embryos, the presence of paired lines of HCP-3/CENP-A staining flanking prophase chromosomes indicated that sister centromeres were resolved (Fig. 1A). RNAi of a cohesin, CeSCC3, as expected, did not prevent centromere resolution, as paired sister centromere staining was observed in prophase (Fig. 1B). To test whether the loss of cohesin proteins allowed centromere resolution when HCP-4/CENP-C was absent, we removed both HCP-4/CENP-C and CeSCC3 via RNAi. In cescc3(RNAi) hcp-4/cenp-c(RNAi) embryos, 80.3% (163 of 203) of prophase chromosomes had paired sister centromeres compared to 2.9% (4 of 138) of hcp-4/cenp-c(RNAi) embryos (Fig. 1D and Table 1). Removal of other cohesin proteins, HIM-1/SMC1, or COH-2/SCC1 via RNAi and a genetic mutation of the CeSMC1 gene him-1(e879) (5) also suppressed the need for HCP-4/CENP-C in centromere resolution (Table 1). This suppression appeared specific to the cohesin proteins, as co-RNAi of HCP-4/CENP-C with a kinetochore protein, CeMCAK, or with a condensin II protein, MIX-1/SMC2, did not reinstate centromere resolution (Table 1). We also examined whether the loss of proteins likely to be required for cohesion establishment could also bypass the block in centromere resolution resulting from the loss of HCP-4/CENP-C. Two proteins necessary for cohesion establishment in Saccharomyces cerevisiae are SCC3 and TRF4 (9, 58). We identified C. elegans candidate homologs of these cohesion establishment proteins, CeSCC2 and CeTRF4, based on sequence similarities. The loss of CeSCC2 (Fig. 1E) or CeTRF4 suppressed the failure to resolve sister centromeres in the absence of HCP-4/CENP-C (Table 1). Taken together, these results indicate that the loss of cohesins, and presumably cohesion, suppressed the requirement for HCP-4/CENP-C in centromere resolution.
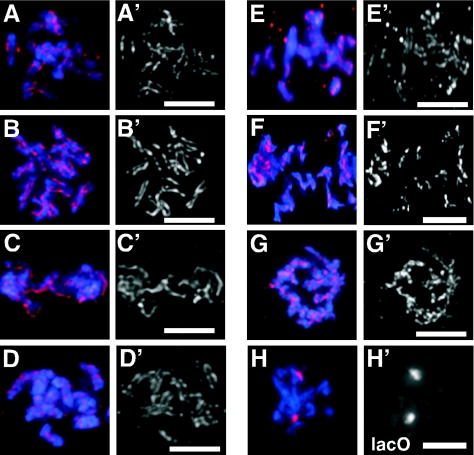
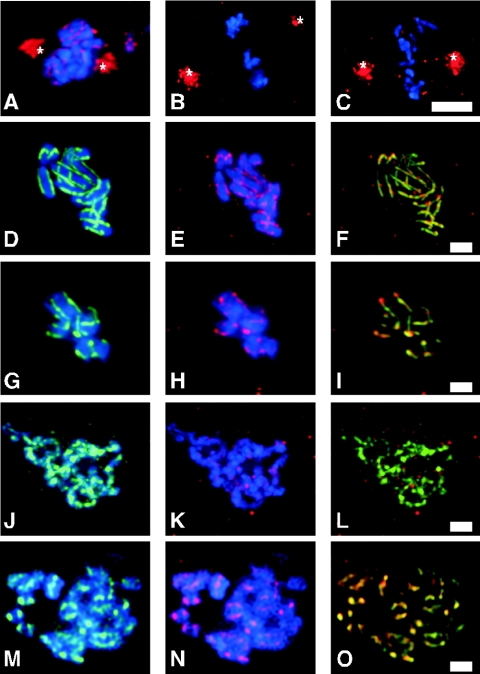
FIG. 1.
Reduced expression of CeSCC3 suppressed the defect in centromere resolution in hcp-4/cenp-c(RNAi) one-cell embryos. Prophase nuclei from wild-type (A), cescc3(RNAi) (B), hcp-4/cenp-c(RNAi) (C), and hcp-4/cenp-c(RNAi) cescc3(RNAi) (D) embryos are shown. hcp-4/cenp-c(RNAi) cescc2(RNAi) (E), air-2(or207ts) (F), and air-2(or207ts) him-1/smc1(RNAi) (G) hermaphrodites were stained for DNA (blue) and for centromeres (red and apostrophe panels) using an antibody against the centromere-specific histone HCP-3/CENP-A. (H) Embryos from JM93, which contains an integrated array of the lactose operator sequence (LacO), were stained for DNA and the integrated LacO array after CeSCC3 RNAi. Bars, 5 μm.
TABLE 1.
Resolution of sister centromeresa
| Background | % of centromeres resolved (n) |
|---|---|
| Wild-type | 91.4 (210) |
| hcp-4/cenp-c(RNAi) | 2.9 (138) |
| hcp-4/cenp-c(RNAi) Cescc3(RNAi) | 80.3 (203) |
| hcp-4/cenp-c(RNAi) coh-2/scc1(RNAi) | 42.2 (154) |
| hcp-4/cenp-c(RNAi) him-1/smc1(RNAi) | 70.8 (137) |
| hcp-4/cenp-c(RNAi) him-1/smc1(e879) | 63.6 (154) |
| hcp-4/cenp-c(RNAi) Cemcak(RNAi) | 6.4 (202) |
| hcp-4/cenp-c(RNAi) mix-1/smc2(RNAi) | 0.6 (178) |
| hcp-4/cenp-c(RNAi) Cescc2(RNAi) | 75.6 (82) |
| hcp-4/cenp-c(RNAi) CeTRF4(RNAi) | 82.6 (207) |
Embryos of the indicated backgrounds were stained for centromeres using an anti-HCP-3 antibody and for nuclei using MAb 414. Prophase nuclei in early embryos were scored for the presence of resolved sister centromeres. Centromere resolution was considered to have occurred if any portion of HCP-3 staining was resolved. All dsRNAs were at a concentration of 2.5 mg/ml in RNAi experiments. n, total number of chromosomes scored.
Because AIR-2 is necessary for the prophase timing of centromere resolution, we examined whether the loss of cohesins could remove the need for AIR-2 in prophase centromere resolution (23). In air-2(or207ts) embryos, 7.9% (17 of 216) of prophase chromosomes had resolved sister centromeres prior to nuclear envelope breakdown (NEBD) (Fig. 1F). We removed the cohesin Him-1/SMC1 by feeding RNAi in the Air-2(or207) mutant strain. The efficacy of RNAi was monitored at the permissive temperature by determining embryonic lethality (data not shown). Following a shift to the nonpermissive temperature, we examined prophase nuclei in the air-2(or207) him-1/smc1(RNAi) embryos and found that only 2.8% (6 of 216) of prophase chromosomes were resolved (Fig. 1G), indicating that, unlike for HCP-4/CENP-C, the loss of cohesin Him-1/SMC1 did not suppress the effect of AIR-2 loss on centromere resolution. This result indicates that the loss of cohesin suppression is specific to HCP-4/CENP-C involvement in centromere resolution.
Sister chromatids remain linked in cohesin RNAi embryos.
Although the distances separating sister centromeres varied along the chromosome in cohesin RNAi embryos, sister centromeres remained paired and the maximum distance separating sister centromeres was not significantly different from that in the wild type (0.6 ± 0.12 versus 0.58 ± 0.10 μm, respectively). This finding suggested that sister chromatids remained linked. To test whether sister chromatids remained linked in cohesin RNAi embryos, we removed CeSCC3 protein via RNAi and observed lacO arrays in strain JM93, which contains an integrated array of the lactose operator sequence, LacO (14). Using LacI binding and indirect immunofluorescence to detect the arrays, we observed in wild-type nuclei two distinct aggregates corresponding to homologous chromosomes during prophase and four aggregates corresponding to the four spatially separated chromatids at anaphase (see Fig. S1 in the supplemental material). To control for the effectiveness of RNAi and to minimize mis-segregation artifacts, we stained embryos with an antibody specific to CeSCC3 (see below) and scored only one-cell and two-cell embryos that had no detectable CeSCC3 staining. Examination of cescc3(RNAi) embryos showed two aggregates of LacI staining in 51 of 52 prophase nuclei examined (Fig. 1H). These results are consistent with prior observations (5) and indicated that sister chromatids remain linked in cescc3(RNAi) nuclei.
The loss of HCP-4/CENP-C or AIR-2 does not retain cohesins on mitotic chromosomes.
The previous results were confusing in that the loss of cohesins bypassed the need for HCP-4/CENP-C but not AIR-2 in centromere resolution. This result may be explained by a direct involvement of cohesin in inhibiting centromere resolution and may indicate that HCP-4/CENP-C is necessary to remove cohesins from centromeres. To test this idea, we first generated antibodies against CeSCC3 protein that on a Western blot identified a 122-kDa band, close to the predicted size for CeSCC3 protein, and whose intensity was reduced in cescc3(RNAi) extracts (see Fig. S2A in the supplemental material; also data not shown). The CeSCC3 antibody stained nuclei in wild-type embryos, but in cescc3(RNAi) embryos nuclear staining was absent (see Fig. S2B in the supplemental material). This staining pattern is similar to the localization of the cohesin COH-2/SCC1 in C. elegans embryos (33, 41). In vertebrates, cohesin proteins are associated with chromosomes beginning in S phase but are mostly absent by metaphase (28, 50, 57). To determine if C. elegans cohesins are similarly localized, we examined CeSCC3 nuclear staining in individual optical sections by using multiwave capture microscopy at different stages of the cell cycle and observed the overlap between DNA staining and CeSCC3 staining. In interphase and early prophase, nuclear CeSCC3 staining was abundantly detected (Fig. 2A and B). The CeSCC3 staining overlapped with the DNA staining, suggesting that DNA does not exclude the CeSCC3 protein. Following NEBD, CeSCC3 staining was reduced and was not detectably associated with DNA at metaphase (Fig. 2C). Similar results were observed with antibodies against COH-2/SCC1 protein and by different fixation methods (data not shown). These results indicate that cohesins are largely absent from postprophase chromosomes in C. elegans embryos.
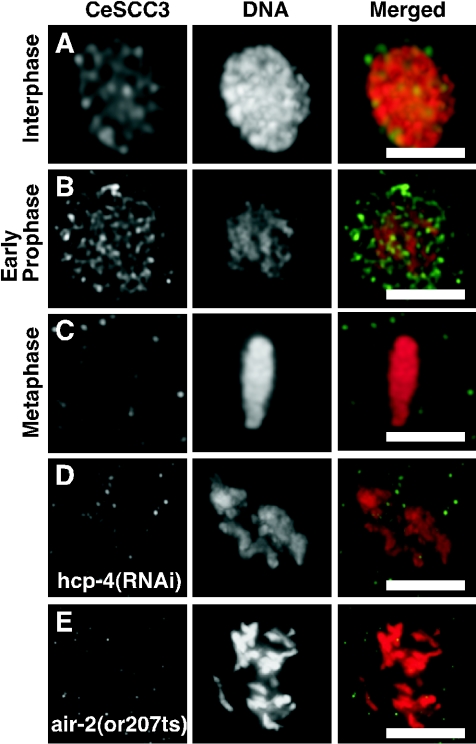
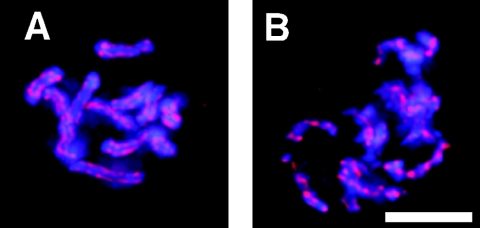
FIG. 2.
CeSCC3 is dynamically excluded from mitotic chromosomes. Images of single optical sections (0.5 μm thick) obtained using multiwave capture from nuclei in wild-type N2 embryos stained for CeSCC3 (green in merged panels) and DNA (red in merged panels) are shown separately and merged. Nuclei were in interphase (A), early prophase (B), or metaphase (C). Post-NEBD nuclei from hcp-4/cenp-c(RNAi) (D) or air-2(or207) (E) embryos were stained as described above. Bars, 5 μm.
Having observed that cohesins are not detectably present on postprophase chromosomes, we tested whether the block in centromere resolution correlated with the retention of cohesins on mitotic chromosomes. We examined mitotic chromosomes from hcp-4/cenp-c(RNAi) embryos stained with CeSCC3 and COH-2/SCC1 antibodies. CeSCC3 and COH-2/SCC1 staining was not observed on postprophase chromosomes in hcp-4/cenp-c(RNAi) embryos (Fig. 2D and data not shown). In metazoans, cohesins are retained on mitotic chromosomes when either Polo kinase or Aurora kinase B is absent (26, 51). In C. elegans, the absence of Air-2 protein did not observably retain cohesins on mitotic chromosomes (Fig. 2E), suggesting that C. elegans chromosomes differ from metazoan chromosomes in cohesin removal during prophase. These results suggest that the blocked or delayed centromere resolution is not caused by the retention of cohesins at the sister centromeres.
Condensin II proteins, HCP-6 and MIX-1/SMC2, are required for centromere resolution.
Because condensin proteins are required for the resolution of sister chromatids, we considered whether condensin proteins were also required for centromere resolution (10, 47). Previously, it was observed that hcp-6(mr17ts) mitotic chromosomes were twisted and contained regions of unresolved sister centromeres (46). One explanation for the poor but observable centromere resolution is that spindle microtubules interacting with the sister kinetochores partially rescue blocked centromere resolution. To test this idea, we inhibited spindle microtubules with nocodazole in wild-type and hcp-6(mr17ts) embryos. Wild-type mitotic chromosomes resolved sister centromeres after treatment with nocodazole, consistent with normal prophase centromere resolution being independent of spindle microtubules (Fig. 3A and reference 35). In hcp-6(mr17ts) embryos, most (83 of 96) mitotic chromosomes had some portion of each sister centromere resolved; however, some regions of the mitotic chromosomes appeared not to have resolved sister centromeres, suggesting that the resolution process was incomplete (Fig. 3B). However, in nocodazole-treated hcp-6(mr17ts) embryos, nearly all (125 of 132) of the mitotic chromosomes had only a single line of centromere staining, characteristic of blocked centromere resolution (Fig. 3C). Similar results were observed when MIX-1/SMC2 was removed or when kinetochore function was compromised via RNAi of the kinetochore protein KNL-1 (Fig. 3D). These results indicated that in the absence of spindle microtubules, centromere resolution required the condensin II proteins HCP-6 and MIX-1/SMC2.
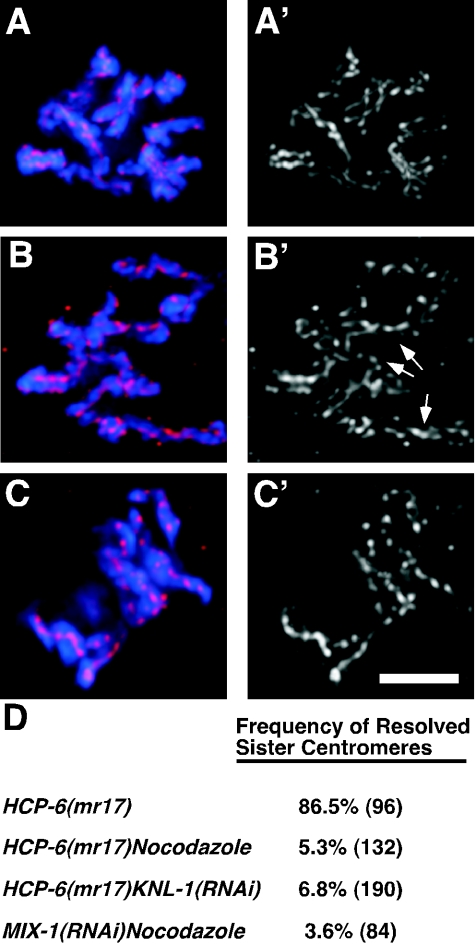
FIG. 3.
HCP-6 and MIX-1/SMC2 are required for centromere resolution. One-cell embryos were stained for DNA (blue) and the centromeric histone HCP-3/CENP-A (red and apostrophe panels). Images are of single post-NEBD nuclei from wild-type embryos treated with nocodazole (A), hcp-6(mr17ts) embryos (B), and hcp-6(mr17ts) embryos treated with nocodazole (C). (D) Quantitative analysis of the incidence of centromere resolution. The numbers of chromosomes examined are in parentheses. Arrows illustrate examples of chromosomes with partial centromere resolution. Bar, 5 μm.
HCP-6 and MIX-1/SMC2 are necessary for cohesin loss to suppress the requirement for HCP-4/CENP-C in centromere resolution.
A requirement for condensin II proteins in prophase centromere resolution may explain the need for AIR-2, which recruits condensin proteins to the centromere (23). To determine whether HCP-4/CENP-C might act upstream of condensins in centromere resolution, we examined whether the loss of cohesins could still bypass the HCP-4/CENP-C requirement in centromere resolution when condensin II proteins are absent. We used RNAi to remove gene function in different genetic backgrounds and quantified the results in Table 2. In a wild-type background, the loss of CeSCC3 bypasses the requirement for HCP-4/CENP-C in centromere resolution in that sister centromeres resolved as in the mock RNAi control (compare Fig. 4A and B). When the same RNAi experiment was performed on hcp-6(mr17ts) embryos, only single linear arrays of centromeric staining were observed on condensed chromosomes (Fig. 4C), indicating a failure to resolve sister centromeres. Furthermore, the HIM-1/SMC1 mutation (e879) bypassed the requirement for HCP-4/CENP-C in centromere resolution, but when HCP-4/CENP-C was removed along with HCP-6 via RNAi in the him-1(e879) mutant, sister centromeres failed to resolve (Fig. 4D and E). These two results indicate the importance of HCP-6 for cohesin loss to suppress the loss of HCP-4/CENP-C in centromere resolution. To test other condensin II proteins, we removed MIX-1/SMC2 along with HCP-4/CENP-C via RNAi, again using the him-1(e879) strain. Similar to the loss of HCP-6, removing HCP-4/CENP-C and MIX-1/SMC2 gene function in the him-1/smc1(e879) strain failed to restore centromere resolution (Fig. 4F). These results indicate that both HCP-6 and MIX-1/SMC2 are required for the loss of cohesin to restore centromere resolution in hcp-4/cenp-c(RNAi) embryos.
TABLE 2.
Condensin II epistasis
| Background | % of centromeres resolved (no. of chromosomes scored):
|
|
|---|---|---|
| In prophase | Post-NEBD | |
| Cescc3(RNAi) hcp-4/cenp-c(RNAi) | 62.3 (138) | 82 (144) |
| hcp-6(mr17) Cescc3(RNAi) hcp-4/cenp-c(RNAi) | 2.7 (270) | 10.5 (84) |
| hcp-6(RNAi) hcp-4/cenp-c(RNAi) | 2.7 (258) | 11.5 (96) |
| him-1/smc1(e879) hcp-6(RNAi) hcp-4/cenp-c(RNAi) | 5.0 (240) | 8.3 (132) |
| mix-1/smc2(RNAi) hcp-4/cenp-c(RNAi) | 2.1 (192) | 5 (120) |
| him-1/smc1(e879) mix-1/smc2(RNAi) hcp-4/cenp-c(RNAi) | 1.7 (180) | 6.7 (120) |
Embryos were fixed and stained with anti-HCP-3/CENP-A antibody and MAb 414 as described in Materials and Methods. Staining embryos with antibodies against HCP-6, Mix-1/SMC2, HCP-4/CENP-C, and CeSCC3 in parallel monitored RNAi efficiency. Nuclei from one-cell embryos with visibly condensed chromosomes were scored for resolved sister centromeres. Centromere resolution occurred if any portion of HCP-3/CENP-A staining was resolved.
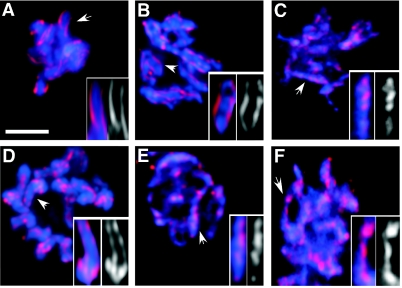
FIG. 4.
Condensin II components required for centromere resolution. Mitotic nuclei from mock RNAi embryos (A), hcp-4/cenp-c(RNAi) cescc3(RNAi) in wild-type embryos (B), and hcp-4/cenp-c(RNAi) cescc3(RNAi) in hcp-6(mr17ts) embryos (C) after the adult hermaphrodites had been shifted to the nonpermissive temperature for 2 h were stained for centromeric histone, HCP-3/CENP-A (red), MAb 414 (not shown), and DNA (blue). HIM-1/SMC1(e879) post-NEBD nuclei from hcp-4/cenp-c(RNAi) (D), hcp4/cenp-c(RNAi) hcp-6(RNAi) (E), and hcp-4/cenp-c(RNAi) mix-1/smc2(RNAi) (F) embryos was stained as described above. Insets show a closer view of marked (arrow) chromosomes along with centromere staining. Bar, 5 μm.
The loss of cohesins restores HCP-6 localization.
The previous result supports a role for HCP-4/CENP-C acting upstream of condensins to promote centromere resolution. In addition to its role in centromere resolution, HCP-4/CENP-C is also necessary for the recruitment of different proteins to the centromere or kinetochore (35, 37). To determine if the recruitment of these proteins was also suppressed by the loss of cohesins, we stained cescc3(RNAi), hcp-4/cenp-c(RNAi), and cescc3(RNAi) hcp-4/cenp-c(RNAi) embryos with antibodies against known kinetochore components, CeMCAK, HCP-1/CENP-F, and CeBUB1. Localization of CeMCAK to centrosomes and kinetochores was not affected by the loss of CeSCC3 (Fig. 5A), but CeMCAK kinetochore localization was affected by the loss of HCP-4/CENP-C (Fig. 5B) as previously observed (37). When we removed both HCP-4/CENP-C and CeSCC3 via RNAi, CeMCAK kinetochore localization was not restored, although its centrosome localization was unaffected (Fig. 5C). Likewise the kinetochore localization of HCP-1/CENP-F or CeBUB1 was not restored (data not shown). These results indicate that centromere resolution is not sufficient to enable the assembly of all kinetochore components.
FIG. 5.
HCP-6 localization to the centromere is restored along with centromere resolution. Nuclei from cescc3(RNAi) (A) or hcp-4/cenp-c(RNAi) (B) and cescc3(RNAi) hcp-4/cenp-c(RNAi) (C) embryos were stained for DNA (blue) and CeMCAK (red). (A) Four-cell embryo; (B and C) two-cell embryos. Centrosomes are indicated by asterisks. The intensity of CeMCAK centrosome staining was used for normalization. The second centrosome in panel B was not completely imaged, so only the fully imaged centrosome was used for normalization, Bar, 5 μm. Nuclei from the wild-type (D to F), cescc3(RNAi) (G to I), hcp-4/cenp-c(RNAi) (J to L), or hcp-4/cenp-c(RNAi) cescc3(RNAi) (M to O) embryos were stained for DNA (blue), centromeric histone, HCP-3/CENP-A (green), and HCP-6 (red). (F, I, L, O) Merged HCP-3/CENP-A and HCP-6 staining in yellow. Bar, 2 μm.
Another protein that requires HCP-4/CENP-C for its recruitment to the centromere or kinetochore is HCP-6 (46). In wild-type embryos, HCP-6 was faintly present at the centromere or kinetochore prior to centromere resolution but became abundantly present after sister centromeres were resolved (Fig. 5D to F). This localization of HCP-6 was not dependent on cohesin since RNAi of CeSCC3 or CeSMC1 still showed colocalization between HCP-6 and the centromeric histone HCP-3/CENP-A in 42 of 45 embryos examined (Fig. 5G to I and data not shown). HCP-6 centromere localization, however, was dependent on HCP-4/CENP-C (Fig. 5J to L) (46). In contrast to the kinetochore protein results, we found that in CeSCC3 HCP-4/CENP-C RNAi embryos, HCP-6 colocalization with HCP-3/CENP-A was restored along with centromere resolution in 80% (24 of 30) of the embryos (Fig. 5M to O). The restored localization of HCP-6 was also observed when CeSMC1 was removed via RNAi or by using the genetic him-1(e879) mutant (data not shown), indicating that the loss of cohesins suppresses the requirement for HCP-4/CENP-C in both HCP-6 localization and centromere resolution.
HCP-4/CENP-C is not required for MIX-1/SMC2 recruitment.
The requirement for MIX-1/SMC1 in centromere resolution raised the question of whether HCP-4/CENP-C was responsible for MIX-1/SMC2 recruitment. MIX-1/SMC2 is centromere localized on mitotic chromosomes independently of HCP-3/CENP-A (15). Consistent with centromere localization, we observed MIX-1/SMC2 localization on mitotic chromosomes (Fig. 6A). In hcp-4/cenp-c(RNAi) embryos, MIX-1/SMC2 localized to mitotic chromosomes as a single linear array that resembled the single linear aggregate observed with HCP-3/CENP-A staining (Fig. 6B). This result indicates that unlike HCP-6, MIX-1/SMC2 recruitment to mitotic chromosomes does not depend on HCP-4/CENP-C.
FIG. 6.
MIX-1 recruitment is independent of HCP-4/CENP-C. Mitotic nuclei from mock RNAi (A) or hcp-4/cenp-c(RNAi) (B) embryos stained for MIX-1/SMC2 (red) and DNA (blue) are shown.
DISCUSSION
HCP-4/CENP-C removes cohesion from sister centromeres.
One of the aims of this study was to investigate whether cohesins and condensins, proteins involved in sister chromatid resolution, affected centromere resolution. It was previously shown that sister centromeres are resolved from one another to opposite surfaces of the chromosome prior to microtubule capture and that RNAi of the centromere protein HCP-4/CENP-C results in a failure of sister centromeres to resolve (35). We used this observation to look for genetic interactions between HCP-4/CENP-C and cohesin proteins. RNAi of cohesins COH-2/SCC1, CeSCC3, and HIM-1/SMC1 or a viable loss-of-function mutation, him-1(e879), enabled sister centromeres to resolve in the absence of HCP-4/CENP-C. The presence of two resolved sister centromeres in these suppression experiments supports the previous conclusion that two fully duplicated but unresolved sister centromeres are present on mitotic chromosomes in hcp-4/cenp-c(RNAi) embryos.
The suppression of the requirement for HCP-4/CENP-C in centromere resolution by the loss of cohesins or candidate cohesion establishment genes suggests that the role of HCP-4/CENP-C is to remove cohesion. There are two possible locations for this cohesion: one is the cohesion between sister chromatids, and the other is the cohesion between sister centromeres. We observed that when cohesins were reduced via RNAi or mutation, sister chromatids remain close, suggesting that sister chromatid cohesion was maintained. However, centromere resolution was restored by the loss of cohesins when HCP-4/CENP-C was also absent, further suggesting that it is cohesion between sister centromeres that is affected by HCP-4/CENP-C. This suggestion is consistent with HCP-4/CENP-C localization to sister centromeres. Although the suppression results support a role for cohesins in inhibiting centromere resolution, we did not observe cohesins on hcp-4/cenp-c(RNAi) mitotic chromosomes. This failure to observe the retention of cohesins at the centromere in hcp-4/cenp-c(RNAi) embryos may result from only very low levels of cohesin being necessary for cohesion. In Schizosaccharomyces pombe, low levels of the cohesin protein SCC1/RAD21 are viable and therefore sufficient to maintain cohesion until anaphase (55). However, our experiments showing that the condensin II proteins HCP-6 and MIX-1/SMC2 are necessary for cohesin loss to bypass the requirement for HCP-4/CENP-C suggests that it is not the simple retention of cohesins at sister centromeres that holds sister centromeres together. Given these results, we favor the idea that the function of HCP-4/CENP-C in centromere resolution is to remove residual cohesion present at sister centromeres (Fig. 7).
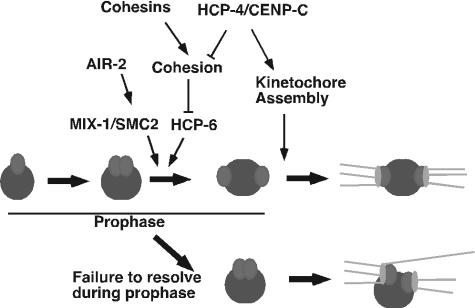
FIG. 7.
Diagram of centromere resolution and kinetochore assembly pathways. See the text for details. The resolution of the centromeres of a mitotic chromosome is depicted. Initially sister centromeres are close. Following the recruitment of condensin II via both the AIR-2 and HCP-4/CENP-C pathways, sister centromeres resolve from one another and assemble sister kinetochores on opposing surfaces of the mitotic chromosome. In the absence of condensin II proteins, such as HCP-6, centromere resolution fails, but kinetochore assembly allows functional kinetochores to assemble. However, inappropriate merotelic attachments (46) result in a “twisting” of the mitotic chromosomes and chromosome mis-segregation.
Cohesin dynamics of holocentric chromosomes.
The observation that cohesins are not retained on mitotic chromosomes suggests that holocentric chromosomes differ from monocentric chromosomes in their cohesin dynamics. We observed that cohesin staining overlapped with DNA staining, suggesting that cohesins are associated with DNA in interphase and early prophase. Furthermore, the loss of cohesins affects meiotic chromosome cohesion and mitotic chromosome segregation, suggesting that cohesins function in C. elegans similarly to the way they function in other organisms (40, 41). However, cohesin proteins are largely absent from mitotic holocentric chromosomes. This observation argues that for holocentric chromosomes, the accumulation of cohesins near centromeres is not necessary to establish or maintain sister kinetochore bi-orientation. A possible reason for the difference in cohesin retention on mitotic chromosomes is the difference in spindle microtubule dynamics. Poleward flux of spindle microtubules is thought to generate tension at sister kinetochores (29, 32, 60). Centromeric heterochromatin and even the areas surrounding yeast centromeres are enriched in cohesin, suggesting that cohesin may be required to oppose tension generated at sister kinetochores (1, 31, 53-55, 59). In C. elegans embryos, spindle microtubules do not undergo significant poleward flux, and consequently little tension is present between sister kinetochores (24). With little tension generated by sister kinetochores, there would be less of a need for cohesin to be retained on C. elegans chromosomes. Despite this reduced need for cohesin, some small amount of cohesin may still be required for mitotic chromosome segregation, as the C. elegans separase gene (Sep-1) is still necessary for the separation of sister chromatids during mitosis (44).
Chan et al. (5) observed that sister chromatids in cohesin RNAi embryos remained closely juxtaposed even after the more stringent denaturation steps necessary for in situ hybridization. We also found that the loss of cohesin proteins in C. elegans did not result in a precocious dissociation of sister chromatids during mitosis. In vertebrates, sister chromatids often remain in close proximity in the absence of cohesins, suggesting that a second form of cohesion may also be present (45). Topological links between sister DNA strands may provide another form of cohesion, and the idea that the dissolution of these DNA-mediated linkages is one requirement for sister chromatid resolution has been proposed (26). The persistence of such linkages may explain our and other's findings that the loss of cohesins does not cause precocious dissociation of sister chromatids during mitosis in the C. elegans embryo.
Condensin II is required for centromere resolution.
We showed that in the absence of spindle-kinetochore forces, the loss of HCP-6 or MIX-1/SMC2 led to a failure to resolve sister centromeres, suggesting that it is the condensin II complex that is required for centromere resolution. Previous results suggested that the loss of HCP-6 did not prevent centromere resolution; instead, mitotic chromosomes were twisted (46). We extended these results by observing that often only portions of mitotic chromosomes actually resolved sister centromeres; furthermore, the presence of twisting may result from unresolved topological linkages between sister centromeres. This possibility is further supported by the observation that spindle-kinetochore forces, which favor the removal of topological linkages between DNA strands, are sufficient to partially drive the resolution of some portions of sister centromeres. In yeast, condensin is thought to resolve cohesin-independent linkages either directly or by recruiting topoisomerase II (11, 49). Furthermore, condensin II helps to resolve cohesin-independent linkages during meiosis in C. elegans (6). These results suggest that the condensin II complex drives prophase centromere resolution by the resolution of DNA linkages.
Our work on holocentric chromosomes in C. elegans suggests that the condensin II complex plays a unique role in organizing the centromere. Recently, it was shown that condensin II colocalized with the inner kinetochore plate and that the deletion of condensins affected the back-to-back geometry of sister kinetochores in vertebrate chromosomes (38). The similar localizations and the requirement for condensin II in monocentric and holocentric chromosomes' kinetochore geometric organization further indicates that the centromere structure is highly conserved across phylogenies. This conservation of centromere biology supports the idea that the main difference between holocentric and monocentric chromosomes is the percentage of the mitotic chromosomes encompassed by the centromere.
Condensin II recruitment pathways.
An interesting result of this study was the observation that the localization of HCP-6, a condensin II component, was restored along with centromere resolution. Stear and Roth (46) showed that HCP-6 required HCP-3/CENP-A and HCP-4/CENP-C for localization. The restoration of HCP-6 localization when HCP-4/CENP-C was absent via cohesin RNAi suggests that HCP-6 recruitment is inhibited by cohesin and cohesion. We were unable to detect any interactions between HCP-4/CENP-C and cohesins, suggesting that the requirement for HCP-4/CENP-C in HCP-6 recruitment is to remove cohesion (Fig. 7). HCP-4/CENP-C is required for both centromere resolution and kinetochore assembly (35, 37). Yet only centromere resolution is restored in the absence of HCP-4/CENP-C by the loss of cohesion, suggesting that the loss of cohesion is not a prerequisite for recruiting kinetochore proteins.
Condensin II components MIX-1/SMC2 and HCP-6 differ in their levels of dependence on HCP-4/CENP-C for localization to mitotic chromosomes. Recently, Chan et al. (6) showed that HCP-6 and MIX-1/SMC2 were not interdependent for their recruitment during mitosis but were codependent during meiosis, suggesting that the holocomplex is not the target for recruitment during mitosis. Because both HCP-6 and MIX-1/SMC2 are required for centromere resolution, it is likely that an active holocomplex is sequentially assembled at the centromere. One pathway depends on HCP-4/CENP-C to remove cohesion and enable the recruitment of HCP-6. It will be interesting to determine if other non-SMC components of condensin II are likewise dependent on HCP-4/CENP-C for recruitment. A second pathway may require the AIR-2 kinase, which is required for the localization of condensin II proteins to the centromere in C. elegans and vertebrates (23, 38). AIR-2 and condensins act separately during prophase with respect to chromosome condensation (23). However, AIR-2 is required for centromere resolution during prophase, as is condensin MIX-1/SMC2, supporting the idea that AIR-2 involvement in centromere resolution is through the recruitment of MIX-1/SMC2 and CeSMC4 to the centromere. Neither HCP-4/CENP-C nor AIR-2 affects the others' localization, supporting the idea that there are at least two distinct pathways for recruiting condensin proteins to the centromere (37). Future work is needed on the mechanism of condensin II protein recruitment to centromeres to better understand the significance of this novel process of condensin complex assembly.
Supplementary Material
Acknowledgments
We thank T. Record and M. Capp for providing LacI repressor protein, J. McGhee for providing tagged nematode strain JM93, and anonymous reviewers for their comments during the preparation of the manuscript. Other strains used here were provided by the Caenorhabditis Genetics Center.
This work was supported by National Institutes of Health grant GM48435 to M.B.R and ACS grant IRG-72-001-30-IRG to L.L.M.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Blat, Y., and N. Kleckner. 1999. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98:249-259. [DOI] [PubMed] [Google Scholar]
- 2.Brenner, S., D. Pepper, M. W. Berns, E. Tan, and B. R. Brinkley. 1981. Kinetochore structure, duplication, and distribution in mammalian cells: analysis by human autoantibodies from scleroderma patients. J. Cell Biol. 91:95-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinkley, B. R., and E. Stubblefield. 1970. Ultrastructure and interaction of the kinetochore and centriole in mitosis and meiosis. Adv. Cell Biol. 1:119-185. [Google Scholar]
- 4.Buchwitz, B. J., K. Ahmad, L. L. Moore, M. B. Roth, and S. Henikoff. 1999. A histone-H3-like protein in C. elegans. Nature 401:547-548. [DOI] [PubMed] [Google Scholar]
- 5.Chan, R. C., A. Chan, M. Jeon, T. F. Wu, D. Pasqualone, A. E. Rougvie, and B. J. Meyer. 2003. Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature 423:1002-1009. [DOI] [PubMed] [Google Scholar]
- 6.Chan, R. C., A. F. Severson, and B. J. Meyer. 2004. Condensin restructures chromosomes in preparation for meiotic divisions. J. Cell Biol. 167:613-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheeseman, I. M., S. Niessen, S. Anderson, F. Hyndman, J. R. Yates III, K. Oegema, and A. Desai. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18:2255-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cimini, D., B. Howell, P. Maddox, A. Khodjakov, F. Degrassi, and E. D. Salmon. 2001. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J. Cell Biol. 153:517-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciosk, R., M. Shirayama, A. Shevchenko, T. Tanaka, A. Toth, and K. Nasmyth. 2000. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 5:243-254. [DOI] [PubMed] [Google Scholar]
- 10.Coelho, P. A., J. Queiroz-Machado, and C. E. Sunkel. 2003. Condensin-dependent localisation of topoisomerase II to an axial chromosomal structure is required for sister chromatid resolution during mitosis. J. Cell Sci. 116:4763-4776. [DOI] [PubMed] [Google Scholar]
- 11.D'Amours, D., F. Stegmeier, and A. Amon. 2004. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell 117:455-469. [DOI] [PubMed] [Google Scholar]
- 12.Davis, L. I., and G. Blobel. 1986. Identification and characterization of a nuclear pore complex protein. Cell 45:699-709. [DOI] [PubMed] [Google Scholar]
- 13.Fraser, A. G., R. S. Kamath, P. Zipperlen, M. Martinez-Campos, M. Sohrmann, and J. Ahringer. 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408:325-330. [DOI] [PubMed] [Google Scholar]
- 14.Fukushige, T., M. J. Hendzel, D. P. Bazett-Jones, and J. D. McGhee. 1999. Direct visualization of the elt-2 gut-specific GATA factor binding to a target promoter inside the living Caenorhabditis elegans embryo. Proc. Natl. Acad. Sci. USA 96:11883-11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagstrom, K. A., V. F. Holmes, N. R. Cozzarelli, and B. J. Meyer. 2002. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 16:729-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He, D., and B. R. Brinkley. 1996. Structure and dynamic organization of centromeres/prekinetochores in the nucleus of mammalian cells. J. Cell Sci. 109:2693-2704. [DOI] [PubMed] [Google Scholar]
- 17.Heneen, W. K. 1975. Ultrastructure of the prophase kinetochore in cultured cells of rat-kangaroo (Potorous tridactylis). Hereditas 79:209-220. [PubMed] [Google Scholar]
- 18.Hirano, T. 2000. Chromosome cohesion, condensation, and separation. Annu. Rev. Biochem. 69:115-144. [DOI] [PubMed] [Google Scholar]
- 19.Hirano, T., R. Kobayashi, and M. Hirano. 1997. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 89:511-521. [DOI] [PubMed] [Google Scholar]
- 20.Hirano, T., and T. J. Mitchison. 1994. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell 79:449-458. [DOI] [PubMed] [Google Scholar]
- 21.Howman, E. V., K. J. Fowler, A. J. Newson, S. Redward, A. C. MacDonald, P. Kalitsis, and K. H. Choo. 2000. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA 97:1148-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jokelainen, P. T. 1967. The ultrastructure and spatial organization of the metaphase kinetochore in mitotic rat cells. J. Ultrastruct. Res. 19:19-44. [DOI] [PubMed] [Google Scholar]
- 23.Kaitna, S., P. Pasierbek, M. Jantsch, J. Loidl, and M. Glotzer. 2002. The Aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous chromosomes during meiosis. Curr. Biol. 12:798-812. [DOI] [PubMed] [Google Scholar]
- 24.Labbe, J. C., E. K. McCarthy, and B. Goldstein. 2004. The forces that position a mitotic spindle asymmetrically are tethered until after the time of spindle assembly. J. Cell Biol. 167:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lavoie, B. D., K. M. Tuffo, S. Oh, D. Koshland, and C. Holm. 2000. Mitotic chromosome condensation requires Brn1p, the yeast homologue of Barren. Mol. Biol. Cell 11:1293-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Losada, A., M. Hirano, and T. Hirano. 2002. Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes Dev. 16:3004-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Losada, A., M. Hirano, and T. Hirano. 1998. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 12:1986-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Losada, A., T. Yokochi, R. Kobayashi, and T. Hirano. 2000. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J. Cell Biol. 150:405-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maddox, P., A. Straight, P. Coughlin, T. J. Mitchison, and E. D. Salmon. 2003. Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J. Cell Biol. 162:377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maney, T., L. M. Ginkel, A. W. Hunter, and L. Wordeman. 2000. The kinetochore of higher eucaryotes: a molecular view. Int. Rev. Cytol. 194:67-131. [DOI] [PubMed] [Google Scholar]
- 31.Megee, P. C., C. Mistrot, V. Guacci, and D. Koshland. 1999. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol. Cell 4:445-450. [DOI] [PubMed] [Google Scholar]
- 32.Mitchison, T. J., and E. D. Salmon. 1992. Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J. Cell Biol. 119:569-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mito, Y., A. Sugimoto, and M. Yamamoto. 2003. Distinct developmental function of two Caenorhabditis elegans homologs of the cohesin subunit Scc1/Rad21. Mol. Biol. Cell 14:2399-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore, L. L., M. Morrison, and M. B. Roth. 1999. HCP-1, a protein involved in chromosome segregation, is localized to the centromere of mitotic chromosomes in Caenorhabditis elegans. J. Cell Biol. 147:471-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore, L. L., and M. B. Roth. 2001. HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J. Cell Biol. 153:1199-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nasmyth, K. 2001. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35:673-745. [DOI] [PubMed] [Google Scholar]
- 37.Oegema, K., A. Desai, S. Rybina, M. Kirkham, and A. A. Hyman. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ono, T., Y. Fang, D. L. Spector, and T. Hirano. 2004. Spatial and temporal regulation of condensins I and II in mitotic chromosome assembly in human cells. Mol. Biol. Cell 15:3296-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ono, T., A. Losada, M. Hirano, M. P. Myers, A. F. Neuwald, and T. Hirano. 2003. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 115:109-121. [DOI] [PubMed] [Google Scholar]
- 40.Pasierbek, P., M. Fodermayr, V. Jantsch, M. Jantsch, D. Schweizer, and J. Loidl. 2003. The Caenorhabditis elegans SCC-3 homologue is required for meiotic synapsis and for proper chromosome disjunction in mitosis and meiosis. Exp. Cell Res. 289:245-255. [DOI] [PubMed] [Google Scholar]
- 41.Pasierbek, P., M. Jantsch, M. Melcher, A. Schleiffer, D. Schweizer, and J. Loidl. 2001. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15:1349-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rieder, C. L., and E. D. Salmon. 1998. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 8:310-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roos, U. P. 1973. Light and electron microscopy of rat kangaroo cells in mitosis. II. Kinetochore structure and function. Chromosoma 41:195-220. [DOI] [PubMed] [Google Scholar]
- 44.Siomos, M. F., A. Badrinath, P. Pasierbek, D. Livingstone, J. White, M. Glotzer, and K. Nasmyth. 2001. Separase is required for chromosome segregation during meiosis I in Caenorhabditis elegans. Curr. Biol. 11:1825-1835. [DOI] [PubMed] [Google Scholar]
- 45.Sonoda, E., T. Matsusaka, C. Morrison, P. Vagnarelli, O. Hoshi, T. Ushiki, K. Nojima, T. Fukagawa, I. C. Waizenegger, J. M. Peters, W. C. Earnshaw, and S. Takeda. 2001. Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev. Cell 1:759-770. [DOI] [PubMed] [Google Scholar]
- 46.Stear, J. H., and M. B. Roth. 2002. Characterization of HCP-6, a C. elegans protein required to prevent chromosome twisting and merotelic attachment. Genes Dev. 16:1498-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steffensen, S., P. A. Coelho, N. Cobbe, S. Vass, M. Costa, B. Hassan, S. N. Prokopenko, H. Bellen, M. M. Heck, and C. E. Sunkel. 2001. A role for Drosophila SMC4 in the resolution of sister chromatids in mitosis. Curr. Biol. 11:295-307. [DOI] [PubMed] [Google Scholar]
- 48.Strunnikov, A. V., E. Hogan, and D. Koshland. 1995. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev. 9:587-599. [DOI] [PubMed] [Google Scholar]
- 49.Sullivan, M., T. Higuchi, V. L. Katis, and F. Uhlmann. 2004. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell 117:471-482. [DOI] [PubMed] [Google Scholar]
- 50.Sumara, I., E. Vorlaufer, C. Gieffers, B. H. Peters, and J. M. Peters. 2000. Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell Biol. 151:749-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sumara, I., E. Vorlaufer, P. T. Stukenberg, O. Kelm, N. Redemann, E. A. Nigg, and J. M. Peters. 2002. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol. Cell 9:515-525. [DOI] [PubMed] [Google Scholar]
- 52.Sumner, A. T. 1991. Scanning electron microscopy of mammalian chromosomes from prophase to telophase. Chromosoma 100:410-418. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka, T., M. P. Cosma, K. Wirth, and K. Nasmyth. 1999. Identification of cohesin association sites at centromeres and along chromosome arms. Cell 98:847-858. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka, T., J. Fuchs, J. Loidl, and K. Nasmyth. 2000. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2:492-499. [DOI] [PubMed] [Google Scholar]
- 55.Tomonaga, T., K. Nagao, Y. Kawasaki, K. Furuya, A. Murakami, J. Morishita, T. Yuasa, T. Sutani, S. E. Kearsey, F. Uhlmann, K. Nasmyth, and M. Yanagida. 2000. Characterization of fission yeast cohesin: essential anaphase proteolysis of rad21 phosphorylated in the S phase. Genes Dev. 14:2757-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toth, A., R. Ciosk, F. Uhlmann, M. Galova, A. Schleiffer, and K. Nasmyth. 1999. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13:320-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waizenegger, I. C., S. Hauf, A. Meinke, and J. M. Peters. 2000. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 103:399-410. [DOI] [PubMed] [Google Scholar]
- 58.Wang, Z., I. B. Castano, A. De Las Penas, C. Adams, and M. F. Christman. 2000. Pol kappa: a DNA polymerase required for sister chromatid cohesion. Science 289:774-779. [DOI] [PubMed] [Google Scholar]
- 59.Watanabe, Y., and P. Nurse. 1999. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 400:461-464. [DOI] [PubMed] [Google Scholar]
- 60.Waters, J. C., T. J. Mitchison, C. L. Rieder, and E. D. Salmon. 1996. The kinetochore microtubule minus-end disassembly associated with poleward flux produces a force that can do work. Mol. Biol. Cell 7:1547-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yeong, F. M., H. Hombauer, K. S. Wendt, T. Hirota, I. Mudrak, K. Mechtler, T. Loregger, A. Marchler-Bauer, K. Tanaka, J. M. Peters, and E. Ogris. 2003. Identification of a subunit of a novel Kleisin-beta/SMC complex as a potential substrate of protein phosphatase 2A. Curr. Biol. 13:2058-2064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.