Abstract
The genus Diaporthe (Diaporthaceae, Diaporthales) is a large group of fungi frequently reported as phytopathogens, with ubiquitous distribution across the globe. Diaporthe have traditionally been characterized by the morphology of their ana- and teleomorphic state, revealing a high degree of heterogeneity as soon as DNA sequencing was utilized across the different members of the group. Their relevance for biotechnology and agriculture attracts the attention of taxonomists and natural product chemists alike in context of plant protection and exploitation for their potential to produce bioactive secondary metabolites. While more than 1000 species are described to date, Africa, as a natural habitat, has so far been under-sampled. Several endophytic fungi belonging to Diaporthe were isolated from different plant hosts in Cameroon over the course of this study. Phylogenetic analyses based on DNA sequence data of the internal transcribed spacer region and intervening 5.8S nrRNA gene, and partial fragments of the calmodulin, beta-tubulin, histone and the translation elongation factor 1-α genes, demonstrated that these isolates represent four new species, i.e. D.brideliae, D.cameroonensis, D.pseudoanacardii and D.rauvolfiae. Moreover, the description of D.isoberliniae is here emended, now incorporating the morphology of beta and gamma conidia produced by two of our endophytic isolates, which had never been documented in previous records. Moreover, the paraphyletic nature of the genus is discussed and suggestions are made for future revision of the genus.
Key words: Endophytes, Phomopsis , Sordariomycetes, 4 new taxa
Introduction
The genus Diaporthe (Diaporthaceae, Diaporthales, Sordariomycetes) is a group of fungi attracting considerable interest for its occurrence as plant pathogens, endophytes and saprobes, and its biotechnological potential as producers of secondary metabolites (Udayanga et al. 2011; Gomes et al. 2013; Chepkirui and Stadler 2017; Marin-Felix et al. 2019). Among plant diseases and disease symptoms causally linked to Diaporthe infections are leaf spots, cankers, dieback and fruit rots as well as decays and wilt (Thompson et al. 2011; Udayanga et al. 2011; Guarnaccia and Crous 2017; Guarnaccia et al. 2018). Historically, Diaporthe included teleomorphic species that produced ostiolate ascomata usually immersed in the substrate and often erumpent through a pseudostroma, unitunicate and clavate to cylindrical asci, and hyaline, fusioid, ellipsoid to cylindrical, septate ascospores, sometimes with appendages. On the other hand, the corresponding anamorphs were accommodated within the genus Phomopsis, which was characterized by ostiolate conidiomata and phialidic conidiogenous cells that may produce three types of hyaline conidia, i.e. alpha, beta and gamma (Udayanga et al. 2011). Synonymization following the one-fungus-one name paradigm linked both individual groups together, with the older name Diaporthe recieving priority over Phomopsis (Rossman et al. 2015). Taxonomical classification of Diaporthe relies on its host specificity, disease symptoms and morphological features such as that of ascomata, stroma and spore shapes (Udayanga et al. 2011, Gomes et al. 2013). Nowadays, morphological and ecological traits were shown to exhibit high degrees of homoplasy, as molecular phylogenetic studies over the years demonstrated – a common feature found in rising numbers of fungal groups (Gomes et al. 2013; Jaklitsch et al. 2016). In consequence, recent taxonomical surveys extend to multilocus sequencing, namely ITS, cal, his3, tef1 and tub2, and employ molecular phylogenetic concatenation-based methods for species description and delimitation (Udayanga et al. 2012a). However, most of the over 1000 records are not sequenced (213 species validated by sequence data and typification in Marin-Felix et al. 2019; 293 species and type strains surveyed by Norphanphoun et al. 2022), hence for the future of this genus, it will be critical to recollect and typify old records to bring an expectable high amount of synonyms together. This is the only option to long-term stabilize the taxonomy of Diaporthe (Dissanayake 2017).
Besides rarely occurring infections in immunocompromised human individuals, members of the genus Diaporthe are most well-known as phytopathogens in agriculture (Iriart et al. 2011; Rakita et al. 2017; Marin-Felix et al. 2019). Among the most economically impactful, infections of grapevines, forest trees and plants of ornamental value have to be named, with D.eres and D.ampelina, and more recently D.rudis from apple trees (Martino et al. 2023) being among the most frequently isolated ones in Europe (Pscheidt 1989; Mostert et al. 2001; Guarnaccia and Crous 2017; Guarnaccia et al. 2018; Yang et al. 2018; Manawasinghe et al. 2019). During pathogenesis, secondary metabolites were occasionally described as important virulence factors, ensuring plant infection (Tsurushima et al. 2005). This and their ubiquitous dispersion are conceivably among the main reasons why Diaporthe and the former Phomopsis spp. have been studied extensively for their capability to produce bioactive natural products (Chepkirui and Stadler 2017; Xu et al. 2021). For instance, Goddard et al. 2014 described the isolation of nine Diaporthe strains (described as Phomopsis spp.) from different vine cultivars with and without showing symptoms of esca decline, a plant trunk disease leading to diebacks of vineyards (Mostert et al. 2001). A set of secondary metabolites was subsequently isolated from cultures growing on petri dishes containing potato dextrose agar and tested for their phytotoxicity. Two compounds, namely cytosporone B and phomopsolide B, induced necrosis on leaf discs similar to eutypine, a phytotoxic polyketide from Eutypalata, another noteworthy threat for grape plants (Tey-Rulh et al. 1991). Occurrence in inflorescence and crude sap of infected plants even enabled discerning healthy from infected individuals due to the latter containing eutypine, instrumentalizing the association of fungal secondary metabolites with plant infections for phytopathological surveillance (Mahoney et al. 2005). However, toxin productive capabilities of Diaporthe spp. and esca disease symptoms were shown to not strictly correlate with each other (Goddard et al. 2014). Exploring the ecological impact of the more than 300 to-date described natural products will be an important parameter to study the phytopathogenesis of this group of fungi, as has been highlighted by other authors (Pusztahelyi et al. 2015; Chepkirui and Stadler 2017; Xu et al. 2021).
Further embarking on charting the biodiversity of this genus for biotechnological exploitation, we here aimed to describe species diversity in an almost unstudied habitat, the planta from Cameroon. This paper describes the isolation, morphological and molecular characterization of fungal endophytes that were assigned to the genus Diaporthe.
Material and methods
Taxonomy
Hyphal material (1 mm diam) was scratched from actively growing cultures on YM 6.3 agar (malt extract 10 g/L, yeast extract 4 g/L, D-glucose 4 g/L, agar 20 g/L, pH 6.3 before autoclaving) and transferred onto 9-cm-diam petri dishes containing 2% tap water agar supplemented with sterile pine needles (PNA) (Smith et al. 1996), potato dextrose agar (PDA), oatmeal agar (OA) and malt extract agar (MEA) (Crous et al. 2009). The plates were incubated at 21 °C in darkness. Pigment production and colony characters on PDA, OA and MEA were documented after 15 d. Colony colors were rated with the color chart of The Royal Horticultural Society London (1966). Colony diameters were measured after 5, 10 and 15 d. Cultures were examined periodically for development of ascomata and conidiomata. Morphological characters were examined by mounting fungal structures in clear lactic acid and 30 measurements at x1000 magnification were recorded for each isolate using a Nikon eclipse Ni-U (Nikon Europe BV, Amsterdam, Netherlands) microscope with differential interference contrast. Descriptions, nomenclature and illustrations of taxonomic novelties were deposited in MycoBank (www.MycoBank.org).
DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted using the EZ-10 SPIN column fungal genomic DNA minipreps Kit (Bio Basic Inc. Ontario, Canada) following manufacturer’s instructions. Six different loci were amplified, i.e. the internal transcribed spacer region (ITS), and fragments of the calmodulin (cal), histone 3 (his3), translation elongation factor 1-α (tef1) and beta-tubulin (tub2) genes. The ITS was amplified and sequenced using the primers ITS4 and ITS5 (White et al. 1990), cal with CAL-228 F and CAL-737R (Carbone and Kohn 1999), his3 with CYCH3F and H3-1b (Crous et al. 2004; Glass and Donaldson 1995), tef1 with EF-1-728F and EF-1-986R (Carbone and Kohn 1999) and tub2 with Bt2a and Bt2b (Glass and Donaldson 1995). Amplicons were purified by using an EZ-10 spin column PCR purification Kit (Bio Basic Inc. Ontario, Canada) following the manufacturer’s instructions, and sequenced by employing Sanger sequencing with a commercial provider (Microsynth Seqlab GmbH, Göttingen). Consensus sequences were obtained using Geneious 7.1.9 (http://www.geneious.com, Kearse et al. 2012) and deposited in GenBank (accession numbers in Table 1).
Table 1.
Isolates and reference strains of Diaporthe spp. included in the phylogenetic study. GenBank accession numbers in bold were newly generated in this study. Taxonomic novelties are indicated in bold italic.
1ATCC: American Type Culture Collection, Virginia, USA; BRIP: Queensland Plant Pathology Herbarium, Brisbane, Australia; CAA: Collection of Artur Alves housed at Department of Biology, University of Aveiro, Portugal; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; CECT: Spanish Type Culture Collection at University of Valencia, Valencia, Spain; CFCC: China Forestry Culture Collection Center, Beijing, China; CGMCC: Chinese General Microbiological Culture Collection Center, Beijing, China; CMRP: Taxonline Microbiological Collections of Paraná Network, at the Federal University of Paraná, Brazil; CNUCC: Capital Normal University Culture Collection Center, Beijing, China; COAD: Culture Collection of Octávio de Almeida Drumond. Universidade Federal de Viçosa, Viçosa, Brasil; CPC: Culture collection of Pedro Crous, housed at Westerdijk Fungal Biodiversity Institute; CSUFTCC: Central South University of Forestry and Technology Culture Collection, Hunan, China; DAOM: Plant Research Institute, Department of Agriculture (Mycology), Ottawa, Canada; DAL: strains deposited in fungal collection of the Instituto Agroforestal Mediterráneo–Universitat Politècnica de València, Valencia, Spain; DAOMC: Canadian Collection of Fungal Cultures, Ottawa, Canada; FPH: personal collection of Francesca Peduto Hand, Department of Plant Pathology, The Ohio State University, Columbus; GUCC: Culture Collection at the Department of Plant Pathology, Agriculture College, Guizhou University, China; GZAAS: Herbarium of Guizhou Academy of Agricultural Sciences, Guiyang, China; FAU: Isolates in culture collection of Systematic Mycology and Microbiology Laboratory; FFPRI: the Forestry and Forest Products Research Institute culture collection, Tsukuba, Japan; HKAS: Chinese Academy of Sciences, Kunming, China; HNZZ: Central South University of Forestry and Technology, Changsha, China; ICMP: International Collection of Micro-organisms from Plants, Landcare Research, Private Bag 92170, Auckland, New Zealand; IFRDCC: International Fungal Research and Development Culture Collection, Kunming, China; KUMCC: Kunming Institute of Botany, Kunming, China; JZB: Culture collection of Institute of Plant and Environment Protection, Beijing, China; LC: Working collection of Lei Cai, housed at Institute of Microbiology, Chinese Academy of Sciences, Beijing, China; LGMF, Laboratório de Genética de Microrganismos (LabGeM) culture collection, at the Federal University of Paraná, Brazil; MAFF: Ministry of Agriculture, Forestry and Fisheries, Tokyo, Japan; MFLU: Mae Fah Luang University herbarium, Thailand; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; NCYU: Department of Plant Medicine, National Chiayi University, Chiayi, Taiwan; NTUPPMCC: Department of Plant Pathology and Microbiology, National Taiwan University Culture Collection, PMM: collection of Providence Moyo at the University of Stellenbosch, Stellenbosch, South Africa; PSCG: Personal Culture Collection Y.S. Guo, China; SAUCC: Shandong Agricultural University Culture Collection, Shandong, China; SCHM: Mycological Herbarium of South China Agricultural University, Guangzhou, China; SDBR-CMU: Culture Collection of Sustainable Development of Biological Resources Laboratory at Chiang Mai University, Chiang Mai, Thailand; URM: Culture Collection at the Universidade Federal de Pernambuco, Recife, Brazil; VTCC: Vietnam Type Culture Collection, Center of Biotechnology, Vietnam National University, Hanoi, Vietnam; ZHKUCC: Culture Collection of Zhongkai University of Agriculture and Engineering, Guangzhou, China. T indicates ex-type material. 2ITS: internal transcribed spacers and intervening 5.8S nrDNA; tub2: partial β-tubulin gene; his3: partial histone H3 gene; tef1: partial elongation factor 1-alpha gene; cal: partial calmodulin gene.
Molecular phylogenetic inference
To further put the sampled strains and obtained sequences into their taxonomic context, a molecular phylogenetic inference using the taxon selection and program settings presented by Matio Kemkuignou et al. (2022) was performed. Briefly, five MAFFT alignments (Katoh and Standley 2013) were calculated featuring all surveyed sequences of the respective loci (Table 1) and curated by using Gblocks (Talavera and Castresana 2007; see Suppl. material 1: table S1). Maximum-likelihood (ML) analysis using RAxML (-HPC BlackBox v8.2.12 with default parameters, Stamatakis 2014) as implemented in the CIPRES portal (www.phylo.org) was performed for the combined aligned data, which was obtained concatenating the single locus alignments in SequenceMatrix 1.8 (Vaidya et al. 2011). The phylogenetic tree is shown in Fig. 1. After evaluation of the inferred tree, the alignment was then split into two sections (Fig. 1 shown in reddish-pink and green) and re-aligned using MAFFT. Instead of automatic filtering for conserved positions, alignments were now manually curated, correcting for alignment mistakes and subjected to the earlier described maximum-likelihood molecular phylogenetic inference using IQTree, with the option to approximate Bayesian posterior probability values (-abayes). In addition, single locus trees were calculated and checked visually for congruence with the multi-locus phylogenetic inference among the closest related sequences clustering with the here reported sequences. Support values regarded as significant (bootstrap (bs) >70%; posterior probabilities (pp) >95%) were mapped on the final maximum likelihood tree for each analysis. All alignments are deposited in the supplementary material; all used sequences, as well as the GenBank numbers for the newly generated ones, can be found in Table 1.
Figure 1.
Maximum Likelihood phylogram (lLn = -56509.790498) obtained from the combined ITS, cal, his3, tef1 and tub2 sequences of our strain and reference strains of Diaporthe spp. Diaporthellacorylina CBS 121124 was used as outgroup. Bootstrap support values ≥70 are indicated along branches. Branch lengths are proportional to distance. Figure legend refers to nucleotide substitutions per site.
Results
The lengths of the fragments of the five loci used in the combined dataset were 458 bp (ITS), 331 bp (cal), 296 bp (his3), 157 bp (tef1) and 510 bp (tub2). The length of the final alignment was 1752 bp. The phylogenetic tree obtained from the RAxML analysis of the combined dataset is shown in Fig. 1. In this tree our endophytic strains isolated from different Cameroonian host plants were located within a clade considered to represent the genus Diaporthe. As the surveyed isolates clustered in two larger clades of Diaporthe, the subsequent molecular phylogenetic inference was split into two to allow for a more accurate and efficient analysis.
The first restricted clade analysis featured 561 bp (ITS), 453 bp (cal), 373 bp (his3), 434 bp (tef1), 820 bp (tub2) for each respective locus, spanning in total 121 taxa and 2641 sites in total (Fig. 2). The second restricted clade analysis, on the other hand, resulted in an alignment featuring 550 bp (ITS), 426 bp (cal), 400 bp (his3), 362 bp (tef1), 719 bp (tub2) for each respective locus, consisting of, in total, 49 taxa and 2457 sites (Fig. 3). The first analysis showed the formation of two large clades in sister position to each other (100 bs / 1 pp), in which the isolates STMA 18289, STMA 18290, CBS 148913 and CBS 148911 clustered within an unsupported smaller one. The former three strains formed a well-supported clade (100 bs /1 pp), while strain CBS 148911 was located in an independent lineage apart from the other Diaporthe spp. The second phylogenetic inference revealed two highly supported clades, in which strains STMA 18291 and STMA 18245, and STMA 18247, STMA 18292 and CBS 148909 nested in, respectively. The first two resolved close to D.isoberliniae (100 bs / 1 pp), while the latter formed a well-supported clade (82 bs / 1 pp) within a cluster formed by D.anacardii, D.macadamiae, D.nebulae and D.velutina. The position of strain CBS 148912 did not receive bootstrap support, but was located in an independent lineage showing a higher nucleotide difference compared to other closely related species.
Figure 2.
Maximum-Likelihood phylogram (lLn = -13511.2844) obtained from the combined ITS, cal, his3, tef1 and tub2 sequences of our strain and related Diaporthe spp. Diaportheamygdali CBS 126679T and D.eres CBS 138594T were used as outgroup. Bootstrap support values ≥ 70/Bayesian posterior probability scores ≥ 0.95 are indicated along branches. Branch lengths are proportional to distance. Novelties are indicated in bold. Type material of the different species is indicated with T. Figure legend refers to nucleotide substitutions per site.
Figure 3.
Maximum Likelihood phylogram obtained (lLn = -10678.2613) from the combined ITS, cal, his3, tef1 and tub2 sequences of our strain and related Diaporthe spp. Diaportheamygdali CBS 126679T and D.eres CBS 138594T were used as outgroup. Bootstrap support values ≥ 70/Bayesian posterior probability scores ≥ 0.95 are indicated along branches. Branch lengths are proportional to distance. Novelties and emended taxa are indicated in bold. Type material of the different species is indicated with T. Figure legend refers to nucleotide substitutions per site.
Taxonomy
. Diaporthe brideliae
L. Schweizer, C. Lamb. & Y. Marín sp. nov.
DBAE24AF-3A20-543B-B97A-0BBE8EAF2E3C
843234
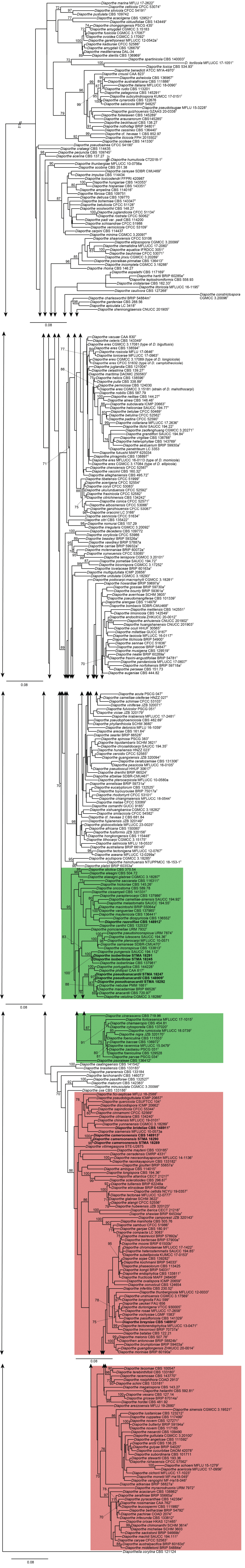
Figure 4.
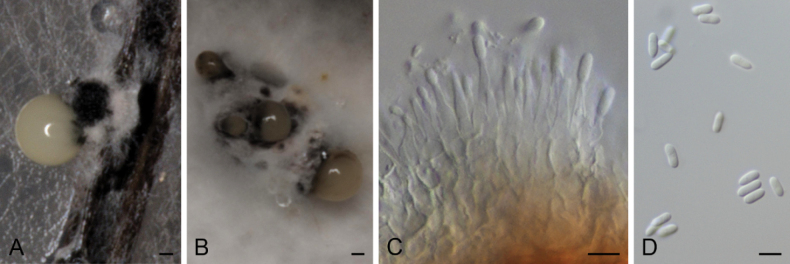
Diaporthebrideliae (ex-type strain CBS 148911) A conidioma in PNAB conidiomata in OAC conidiophores and conidia D alpha and beta conidia. Scale bars: 100 μm (A); 500 μm (B); 5 μm (C, D).
Etymology.
Name refers to the host genus that this fungus was isolated from, Bridelia.
Description.
Conidiomata pycnidial in culture on PNA, globose or irregular, dark brown to black, solitary or in groups, embedded, erumpent, 240–500 μm diam, white to cream or yellow conidial drops exuded from ostioles; conidiomatal wall pale olivaceous green to brown, composed of 1–3 layers, textura angularis. Conidiophores cylindrical to subcylindrical, base pale olivaceous to pale yellow, apex hyaline to subhyaline, straight, densely aggregated, smooth-walled, aseptate or 1(–2) septate, (6–)12–22.5 × 1–3 μm. Conidiogenous cells phialidic, cylindrical, tapering towards the apex, hyaline, mostly terminal, rarely lateral, (7–)8–15.5 × 1–3 μm. Paraphyses not observed. Alpha conidia ovoid to ellipsoidal, hyaline, apex acutely rounded, base acutate, biguttulate, aseptate, (3–)4–6.5 × 1.5–2.5 μm. Beta conidia filiform, curved, tapering towards apex, hyaline, not guttulate, aseptate, 18–32.5 × 1–2 μm. Gamma conidia not observed.
Culture characters.
Colonies on PDA covering the surface of the Petri dish in 2 weeks, grayed white (156B–C) with a grayed orange (174B) ring and grayed orange (163A) margins, velvety to cottony, flat to raised in some zones, margins filamentous to fimbriate; reverse center gray brown (199A) with a yellow orange or grayed orange (167A) zones. Colonies on MEA covering the surface of the Petri dish in 2 weeks, yellow green (153B) with white to grayed yellow (160C) margins, velvety to cottony, flat to raised in some zones, margins filamentous to fimbriate; reverse black (202A) with gray brown (199A) mycelia and yellow green (153B) margins. Colonies on OA covering the surface of the Petri dish in 2 weeks, grayed green (198B) to white mycelium with a yellow green (151B) ring, cottony, flat to raised in some zones, margins filamentous; reverse yellow green (153B) with grayed yellow (161C) margins.
Specimen examined.
Cameroon, Kala Mountain, from Brideliandellensis, 03 Jan. 2019, S.C.N. Wouamba (holotype: CBS H-24921, culture ex-type CBS 148911 = STMA 18286).
Notes.
Diaporthebrideliae is the only report in Bridelia (Phyllanthaceae) from Cameroon. The phylogenetically most related species are D.chinensis, D.siamensis and D.yunnanensis. Diaporthechinensis can be distinguished by the absence of beta conidia, which are produced by the other three species. Diaporthesiamensis is the only species mentioned here that produces gamma conidia (Udayanga et al. 2012b). Diaporthebrideliae can be distinguished from D.yunnanensis by the production of smaller conidiomata (up to 500 μm diam in D.brideliae vs. 880 μm diam in D.yunnanensis).
. Diaporthe cameroonensis
L. Schweizer, C. Lamb. & Y. Marín sp. nov.
7C011D2C-17C4-5063-83A9-9AEB756FC7D2
843235
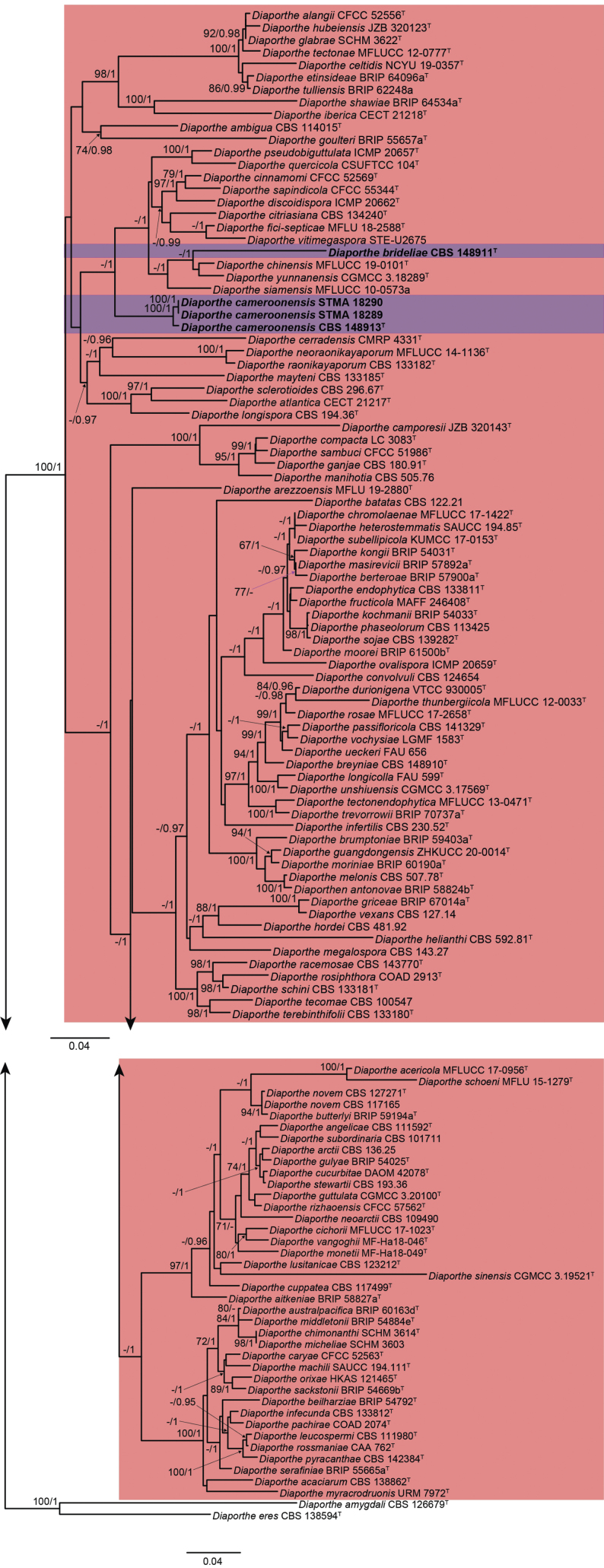
Figure 5.
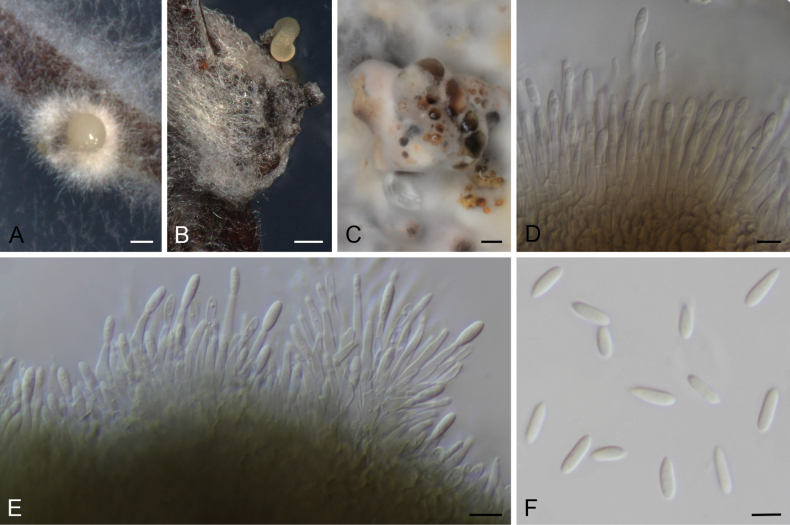
Diaporthecameroonensis (ex-type strain CBS 148913) A conidioma in PNAB conidiomata in OAC conidiophores and conidia D alpha conidia. Scale bars: 100 μm (A); 500 μm (B); 5 μm (C, D).
Etymology.
Named for the country where it was isolated from, Cameroon.
Description.
Conidiomata pycnidial in culture on PNA, globose or irregular, dark brown to black, solitary or in groups, embedded, erumpent, 220–550 μm diam, white to cream conidial drops exuded from ostioles; conidiomatal wall pale olivaceous green to olivaceous brown, composed of 1–3 layers, textura angularis. Conidiophores cylindrical to subcylindrical, tapering towards apex, base subhyaline to pale yellow or pale olivaceous, apex hyaline to subhyaline, straight, densely aggregated, smooth-walled, 1(–3) septate, 12.5–28 × 1–3.5 μm. Conidiogenous cells phialidic, cylindrical to subcylindrical, tapering towards apex, hyaline, terminal, 6–11(–12) × 1.5–3 μm. Paraphyses not observed. Alpha conidia ellipsoidal, hyaline, apex rounded, base rounded to slightly acutate, biguttulate, aseptate, 4.5–6 × (1–)1.5–2.5 μm. Beta and gamma conidia not observed.
Culture characters.
Colonies on PDA covering the surface of the Petri dish in 2 weeks, grayed yellow (161C–D) with transparent margins and white mycelia, cottony to slightly feathery, flat to raised in some zones, lobate, margins filamentous to fimbriate; reverse grayed yellow (161A–D) with transparent margins. Colonies on MEA covering the surface of the Petri dish in 2 weeks, grayed white (156A–B) with transparent margins and yellow white (158D) mycelia, or grayed-orange (165A) with white mycelia and yellow green (153D) margins, cottony to slightly feathery, flat to raised in some zones, margins filamentous to fimbriate; reverse grayed yellow (161A–D) with transparent margins or grayed orange (165A–B) with yellow green (153D) margins. Colonies on OA covering the surface of the Petri dish in 2 weeks, white with grayed white (156C) patches and grayed green (197D) or gray brown (199D) margins, or yellow green (152B) with brown (200A) patches and yellow-white (158A) mycelia, cottony to slightly feathery, flat to raised in some zones, margins filamentous to fimbriate; reverse grayed green (195A) with yellow green centre (152C) or fully yellow green (152C–D).
Specimens examined.
Cameroon, Kala Mountain, from Atractogynegabonii, 02 Jan. 2019, E. G. M. Anoumedem (holotype CBS H-24922; culture ex-type CBS 148913 = STMA 18288); from Tremaguineensis,11 Apr. 2019, E. G. M. Anoumedem (STMA 18289); from Tremaguineensis, 11 Apr. 2019, E. G. M. Anoumedem (STMA 18290).
Notes.
Different strains belonging to this new species formed a well-supported independent clade (100 bs / 1 pp) apart from all surveyed Diaporthe spp. This species was isolated from Trema (Cannabaceae) and Atractogyne (Rubiaceae). To the best of our knowledge, this is the first Diaporthe species to be isolated from Atractogyne. Diaporthepseudoanacardii, which is introduced further below, has also been isolated from Trema collected in Cameroon. However, both species can easily be distinguished by the length of their conidiogenous cells (12.5–28 μm in D.cameroonensis vs (7.5–)10–45 μm in D.pseudoanacardii) and conidia (4.5–6 μm in D.cameroonensis vs (5–)6–8(–9) μm in D.pseudoanacardii).
. Diaporthe pseudoanacardii
L. Schweizer, C. Lamb. & Y. Marín sp. nov.
F6416B15-FDAE-5E1B-A439-1FD3C73B7427
843236
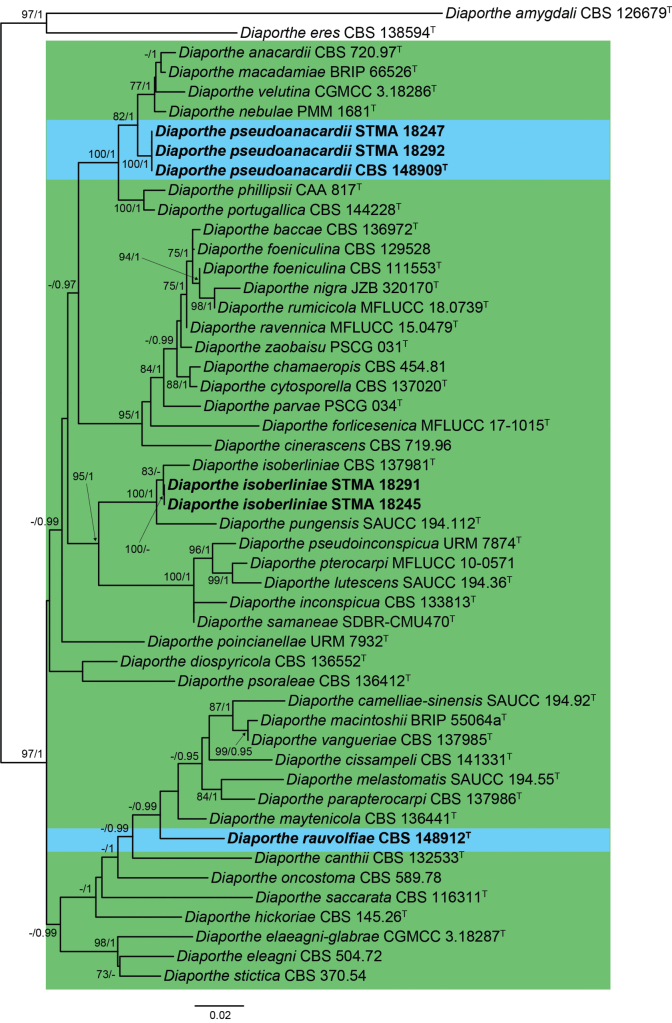
Figure 6.
DiaporthepseudoanacardiiA, B conidioma in PNAC conidiomata in OAD, E conidiophores and conidia F alpha conidia A, C, E, F ex-type strain CBS 148909 B, D STMA 18292. Scale bars: 200 μm (A, B); 1000 μm (C); 5 μm (D–F).
Etymology.
Named after its close phylogenetic relation to Diaportheanacardii.
Description.
Conidiomata pycnidial in culture on PNA, globose or irregular, dark brown to black, solitary or in groups, embedded, erumpent, 190–700(–820) μm diam, white to yellow or cream conidial drops and cirrus exuded from ostioles; conidiomatal wall pale olivaceous to olivaceous brown, composed of 1–2 layers, textura angularis. Conidiophores cylindrical to subcylindrical, base subhyaline to pale yellow or pale olivaceous, apex hyaline to subhyaline, straight, densely aggregated, smooth-walled, 1–2(–3) septate, rarely aseptate, (7.5–)10–45 × 1–3.5(–4) μm. Conidiogenous cells phialidic, cylindrical, tapering towards apex, hyaline to subhyaline, terminal or lateral, 7–28 × 1–3.5(–4) μm. Paraphyses not observed. Alpha conidia ovoid to ellipsoidal, hyaline, apex acutely rounded, base acutate, granular to guttulate, aseptate, (5–)6–8(–9) × 1.5–3 μm. Beta and gamma conidia not observed.
Culture characters.
Colonies on PDA covering the surface of the Petri dish in 2 weeks, white to grayed yellow (162C–D) or grayed white (156A–B), sometimes with transparent margins and white, yellow green (153B–C) and grayed green (195A–B) zones, granulous to cottony or slightly feathery, flat to raised in some zones, margins filamentous to fimbriate; reverse grayed yellow (161C–D or 162D) and brown (200A) or black (202A–B) center, sometimes with transparent margins. Colonies on MEA reaching 59–85 in 2 weeks, white or grayed yellow (161B–C) with normally a white ring, sometimes with grayed green zones (197A–D) and transparent margins, cottony to slightly feathery, lobate, flat to raised in some zones, margins filamentous to fimbriate; reverse grayed green (197A) to brown (200A) with grayed yellow (161B) margins, or grayed green (197A) with grayed yellow (160D) and yellow green (152B) zones and black (202A) margin, or grayed yellow (161 A–B) and transparent margins. Colonies on OA covering the surface of the Petri dish in 2 weeks, grayed green (195A–D) with white margins and yellow (4A–B) or grayed yellow (160D) center, or grayed white (156A–C) with grayed orange (163B–C) center and yellow white (158B–C) margins, cottony to slightly feathery, raised, margins filamentous to fimbriate; reverse yellow green (147B) with gray brown (199B) margins or entire gray brown (199A–B) or grayed green (195A with 198A centre).
Specimens examined.
Cameroon, Kala Mountain, from Tremaguineensis, 11 Apr. 2019, E.G.M. Anoumedem (holotype CBS H-24923; culture ex-type CBS 148909 = STMA 18283); Tonga, West Region, from Pittosporummanii, 19 Jun. 2019, E.G.M. Anoumedem (STMA 18247, STMA 18292).
Notes.
This species resolved in a well-supported clade (82 bs / 1 pp) together with D.anacardii, D.macadamiae, D.nebulae and D.velutina. Diaporthepseudoanacardii can be easily distinguished from all the other species by the absence of beta conidia. All these species are reported from Africa (Gomes et al. 2013; Lesuthu et al. 2019; Wrona et al. 2020), except of D.velutina, which was found in Asia (Gao et al. 2017).
. Diaporthe rauvolfiae
Y. Marín, C. Lamb., Kouam & L. Schweizer sp. nov.
6557A14B-6F0B-5227-8FBD-5F45AECD35CF
843237
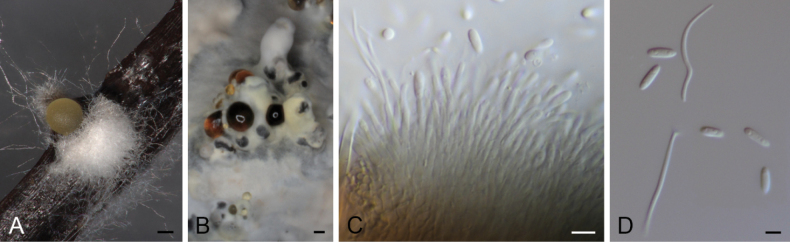
Figure 7.
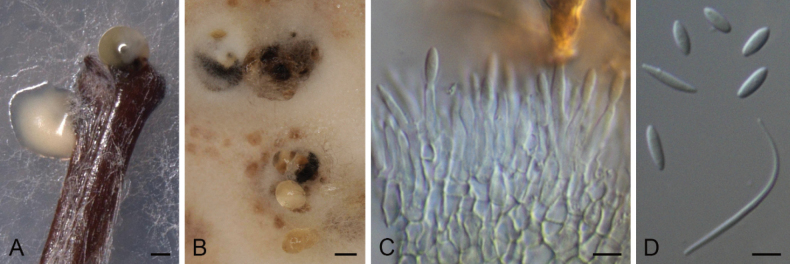
Diaportherauvolfiae (ex-type strain CBS 148912) A conidioma in PNAB conidiomata in OAC conidiophores and conidia D alpha, beta and alpha conidia. Scale bars: 200 μm (A); 500 μm (B); 5 μm (C, D).
Etymology.
Name refers to the host genus that this fungus was isolated from, Rauvolfia.
Description.
Conidiomata pycnidial in culture on PNA, globose or irregular, dark brown to black, solitary or in groups, embedded, erumpent, 210–450(–530) μm diam, white to cream conidial drops exuded from ostioles; conidiomatal wall yellowish brown to olivaceous brown or brown, composed of 1–2 layers, textura angularis. Conidiophores cylindrical to subcylindrical, tapering towards apex, base subhyaline to pale yellow or pale olivaceous, apex hyaline to subhyaline, densely aggregated, smooth-walled, (0–)1–2 septate, 9–19.5 × 1.5–3.5 μm. Conidiogenous cells phialidic, cylindrical to subcylindrical, tapering towards apex, hyaline, mostly terminal, 6.5–13.5 × 1.5–3 μm. Paraphyses not observed. Alpha conidia broadly fusiform to obovoid, hyaline, apex rounded or acute, base acutate, biguttulate to multiguttulate, aseptate, 6.5–9 × 2–3 μm. Beta conidia filiform, curved, tapering towards apex, hyaline, not guttulate, aseptate, 20–36.5 × 1–2 μm. Gamma conidia less frequent, fusiform to obovoid, straight to slightly curved, rarely sinuose, acutate ends or one acutate and other round, hyaline, multiguttulate, aseptate, (8–)9–13 × 1.5–2.5 μm.
Culture characters.
Colonies on PDA reaching 72–76 mm in 2 weeks, grayed yellow (160B–C) with white ring and transparent margins, cottony to slightly feathery, raised, lobate, margins filamentous; reverse grayed yellow (160B–C) with white ring and transparent margins. Colonies on MEA covering the surface of the Petri dish in 2 weeks, white to grayed white (156B), cottony to slightly feathery, raised, margins filamentous; reverse grayed green (197A) to gray brown (199A) with black (202A) center. Colonies on OA covering the surface of the Petri dish in 2 weeks, grayed white (156B–D), cottony to slightly feathery, raised, margins filamentous; reverse gray brown (199A).
Specimen examined.
Cameroon, Tonga, West Region, from Rauvolfiavomitoria, 19 Jun. 2019, E.G.M. Anoumedem (holotype CBS H-24924, culture ex-type CBS 148912 = STMA 18287).
Notes.
Diaportherauvolfiae was located in an independent branch far from other species of Diaporthe (Fig. 3). This species is characterized by the production of alpha, beta and gamma conidia, which were not observed in other species reported from Cameroon except of D.isoberliniae. This latter species differs from D.rauvolfiae in the length of the conidiophores (13–42 μm in D.isoberliniae vs 9–19.5 μm in D.rauvolfiae), beta conidia (11.5–27.5 μm in D.isoberliniae vs 20–36.5 μm in D.rauvolfiae) and gamma conidia (10–18.5(–21) μm in D.isoberliniae vs (8–)9–13 μm in D.rauvolfiae). Both species are not phylogenetically related (Fig. 3).
. Diaporthe isoberliniae
Crous, Persoonia 32: 221. 2014. emend. L. Schweizer, C. Lamb. & Y. Marín
4F731F40-4DE0-527A-841A-A0BD3F50A38A
808909
Figure 8.
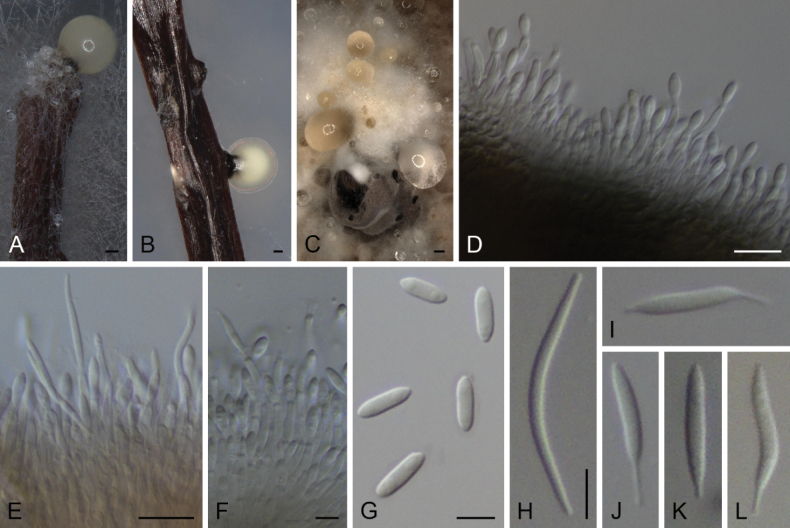
DiaportheisoberliniaeA, B conidioma in PNAC conidiomata in OAD–F conidiophores and conidia G alpha conidia H beta conidia I–L Gamma conidia A, C, D–I STMA 18291 B, J–L STMA 18245. Scale bars: 100 μm (A, B); 500 μm (C), 10 μm (D, E), 5 μm (F–L).
Description.
Conidiomata pycnidial in culture on PNA, globose or irregular, dark brown to black, solitary or in groups, embedded, erumpent, 200–460 μm diam, white to cream or yellow conidial drops exuded from ostioles; conidiomatal wall yellowish brown to olivaceous brown or brown, composed of 1–6 layers, textura angularis. Conidiophores cylindrical to subcylindrical, base subhyaline to pale olivaceous, apex hyaline, densely aggregated, smooth-walled, 1–3-septate, 13–42 × 1.5–4 μm. Conidiogenous cells phialidic, cylindrical to subcylindrical, tapering towards apex, hyaline, terminal or lateral, (5.5–)6.5–14 × 1.5–3 μm. Paraphyses not observed. Alpha conidia ellipsoidal to obovoid, or fusoid-ellipsoid, hyaline, apex rounded or subobtuse, base acutate or subtruncate, biguttulate to multiguttulate, aseptate, 5.5–9(–10) × 2–3(–3.5) μm. Beta conidia less frequent, filiform, curved, tapering towards apex, hyaline, not guttulate, aseptate, 11.5–27.5 × 1–2 μm. Gamma conidia less frequent, broadly fusiform, straight to slightly curved, rarely sinuose, apex acutate or filiform, base filiform, hyaline, multiguttulate, aseptate, 10–18.5(–21) × 1.5–2.5 μm.
Culture characters.
Colonies on PDA reaching 63–72 mm or covering the surface of the Petri dish in 2 weeks, white with a grayed yellow (160C) ring and transparent margins, lobate, cottony to slightly feathery, flat to raised in some zones or fully raised, lobate, margins filamentous to fimbriate; reverse grayed yellow (160B–D). Colonies on MEA covering the surface of the Petri dish in 2 weeks, grayed yellow (161A) with a white ring and white to transparent margins, cottony to slightly feathery, flat to raised in some zones or fully raised, margins filamentous to fimbriate; reverse grayed yellow (162A–C) with transparent margins and sometimes with gray brown (199A) center. Colonies on OA covering the surface of the Petri dish in 2 weeks, white to grayed white (156A) with grayed yellow (161A–B) margins or grayed yellow (161C) with brown (200A) dots and white center and margins, cottony to slightly feathery, raised, margins filamentous to fimbriate; reverse grayed green (197B) to/or gray brown (199C–D).
Specimens examined.
Cameroon, Tonga, West Region, from Pittosporummanii, 19 Jun. 2019, E. G. M. Anoumedem (STMA 18245); ibid. STMA 18291.
Notes.
Diaportheisoberliniae was described based on a specimen isolated from Zambia on Isoberliniaangolensis (Fabaceae) (Crous et al. 2014b). To the best of our knowledge, this species had not been recollected since then. We isolated two strains belonging to D.isoberliniae from Cameroon on Pittosporummanii (Pittosporaceae). The description of this species is here emended with beta and gamma conidia, as the shared observations are the first to report on them. The isolate STMA 18245 did not produce beta conidia, but produced gamma conidia, while isolate STMA 18291 produced beta conidia more frequently than gamma conidia. The type strain produced fusoid-ellipsoid alpha conidia of similar sizes, while these are ellipsoid to obovoid in our two Cameroonian strains.
Diaportheisoberliniae is related to D.pungensis. This latter species can be distinguished by the absence of gamma conidia and the production of shorter conidiophores (11–14.5 μm in D.pungensis vs 13–42 μm in D.isoberliniae) (Sun et al. 2021).
Discussion
This study reports on the isolation and assignment of a group of fungi isolated from plant material to the genus Diaporthe. A characterization by sequencing was followed-up with a concatenation-based molecular phylogenetic inference, which afforded heterogenous sequence placements among a phylogenetic dataset featuring DNA sequence data substantially derived from type strains. Taken together with an analysis of the taxon placements in single-locus trees (data not shown), we concluded that the placement pattern among each strain was unique, which combined with the traditional morphological descriptions let us to propose the erection of four new species to accommodate the isolated strains. Secluding species description to either morphology, ecological observations (such as host occurrence or lifestyle) or molecular data alone has been shown to be problematic in Diaporthe, indicating that the commonly observed morphological features that are recorded and observed by taxonomists are not under strict evolutionary selection pressure (Gao et al. 2015, Fan et al. 2018). Additionally, only a limited set of loci are sequenced even for typified Diaporthe spp., which act as a strong limiting factor for comprehensive molecular phylogenetic analysis. That this undersampling may become problematic is best exemplified by highlighting that the ITS region, long treated as unequivocal fungal barcode, is a poor choice for species delimitation and should be utilized with caution. More specifically, multiple copies may occur in fungal genomes potentially showing significant differences, which, if accidentally sequenced independent from each other, may erroneously lead taxonomists to treat specimens, which are in truth only one species, as separate ones. The fact that this scenario is plausible for Diaporthe has recently been reported by Hilário et al. (2021), who found multiple ITS paralogues for a newly sequenced genome of D.novem. A similar finding was already described and discussed for the xylarialean genus Hypoxylon (Stadler et al. 2020). Most interestingly, Hilário et al. (2021) reported indications for a hybrid species, which could, if this was also assumed to be the case for other members of the genus as well, explain the convoluted systematic status of Diaporthe in its current form (Fan et al. 2018). This makes it all the more necessary to treat molecular data cautiously, especially since not all loci commonly used to infer phylogenies are well covered across all described species of Diaporthe. A careful in-depth phylogenetic analysis of species assigned to the D.amygdali “complex”, rigorously applying Genealogical Concordance Phylogenetic Species Recognition (GCPSR) and coalescence based evolutionary principles by Hilário et al. (2021), for example, showed an unexpectedly high genetic heterogeneity. This complex consisted of seven species forming nine statistically supported clades in concatenation-based phylogenies of ITS, cal, his3, tef1 and tub2 – loci also used in this paper – which, however, could not be resolved into reasonably distinct lineages representing individual species in either single-locus trees, or coalescent-based analysis. While this hopefully remains an extreme example inside the systematics of Diaporthe, this finding clearly shows that 1) adding new species based on single-locus sequencing will further destabilize Diaporthe taxonomy and 2) molecular phylogeneticists have to be aware of the possibility that the inferred supermatrix tree may not reflect the evolutionary history of the underlying loci, possibly leading to artificial lineage resolutions. A later study by Norphanphoun et al. (2022) attempted to classify a large collection of Diaporthe species into molecularly distinctly resolving species complexes. They showed that different loci harbored distinct resolution power for members of each complex (Norphanphoun et al. 2022) – a phenomenon that we also observed for our strains (data not shown) – which raises doubt that molecular data alone is enough to justify the classification of species complexes, even if it is just for the mere purpose of easing communication. In our study, we followed a polyphasic strategy combining multi-locus sequencing with morphological characterization, which clearly showed that the collected strains are separable by multiple phenotypic traits – a necessity in this convoluted genus. Given that intraspecific variation is repeatedly shown to be unexpectedly high, this should be complemented by screening for additional well-established discriminative characters, such as secondary metabolite production for chemotaxonomic purposes in the future. Furthermore, aiming for the generation of high-quality genome sequences would enable studies on the genetics governing ecology, lifestyle and speciation. The genomic toolbox to meaningfully embark on this has already been established for other complicated genera such as Penicillium and Aspergillus (Frisvad and Larsen 2015, Kocsubé et al. 2016, Tsang et al. 2018). This would help to find reasons for the frequently reported paraphyly, poor phylogenetic resolution and by consequence, enable the establishment of sound species boundaries for the inevitable revision of the genus (Dissanayake 2017; Gao et al. 2017; Marin-Felix et al. 2019; Hilário et al. 2021). A recent study published by Hongsanan et al. (2023) formalized necessary taxonomic changes with data that is already available today, indicating the huge potential of a more sophisticated follow-up analysis. Lastly, an additional epitypification campaign is imperative to further stabilize the taxonomy of Diaporthe and allies, hopefully enabling species differentiation between saprobes and important phytopathogens for e.g. diagnostic purposes.
Supplementary Material
Acknowledgements
We are grateful to V. Nana (National Herbarium of Cameroon) for the botanical identifications and S.C.N. Wouamba for the isolation of the strain CBS 148911.
Citation
Lambert C, Schweizer L, Matio Kemkuignou B, Anoumedem EGM, Kouam SF, Marin-Felix Y (2023) Four new endophytic species of Diaporthe (Diaporthaceae, Diaporthales) isolated from Cameroon. MycoKeys 99: 319–362. https://doi.org/10.3897/mycokeys.99.110043
Funding Statement
Life-Science Foundation (LSS), Munich
Additional information
Conflict of interest
The authors have declared that no competing interests exist.
Ethical statement
No ethical statement was reported.
Funding
This work was financed by a personal PhD stipend from the German Academic exchange service (DAAD) to B.M.K. (programme ID- 57440921); a stipend granted by the Life-Science Foundation (LSS) located in Munich to C.L.; postdoctoral stipendium from Alexander-von-Humboldt Foundation, Germany, and financial support of the “COPFUN project” (Project-ID 490821847) funded by Deutsche Forschungsgemeinschaft (DFG) to Y.M.F. The World Academy of Sciences (TWAS) (grant 18‐178 RG/CHE/AF/AC_G‐FR 3240303654), and the Alexander von Humboldt Foundation (AvH) through the equipment subsidies (Ref 3.4 - 8151/20 002), the Research Group Linkage (grant IP-CMR-1121341) and the hub project CECANOPROF (3.4-CMR-Hub).
Author contributions
Christopher Lambert: Methodology, Writing – original draft. Lena Schweizer: Methodology. Blondelle Matio Kemkuignou: Methodology. Elodie Gisele M. Anoumedem: Methodology. Simeon F. Kouam: Funding acquisition. Yasmina Marin-Felix: Methodology, Supervision, Writing – original draft and revision.
Author ORCIDs
Christopher Lambert https://orcid.org/0000-0002-1899-8214
Lena Schweizer https://orcid.org/0000-0003-1296-5486
Simeon F. Kouam https://orcid.org/0000-0003-0191-0527
Yasmina Marin-Felix https://orcid.org/0000-0001-8045-4798
Data availability
All of the data that support the findings of this study are available in the main text or Supplementary Information.
Supplementary materials
Phylogenetic study data
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Christopher Lambert, Lena Schweizer, Blondelle Matio Kemkuignou, Elodie Gisèle M. Anoumedem, Simeon F. Kouam, Yasmina Marin-Felix
Data type
docx
References
- Abeywickrama PD, Qian N, Jayawardena RS, Li Y, Zhang W, Guo K, Zhang L, Zhang G, Yan J, Li X, Guo Z, Hyde KD, Peng Y, Zhao W. (2023) Endophytic fungi in green manure crops; friends or foe? Mycosphere: Journal of Fungal Biology 14(1): 1–106. 10.5943/mycosphere/14/1/1 [DOI]
- Allan-Perkins E, Li D-W, Schultes N, Yavuz S, LaMondia J. (2020) The identification of a new species, Diaporthehumulicola, a pathogen causing Diaporthe leaf spot on common hop. Plant Disease 104(9): 2377–2390. 10.1094/PDIS-08-19-1770-RE [DOI] [PubMed] [Google Scholar]
- Ando Y, Masuya H, Aikawa T, Ichihara Y, Tabata M. (2017) Diaporthetoxicodendri sp. nov., a causal fungus of the canker disease on Toxicodendronvernicifluum in Japan. Mycosphere : Journal of Fungal Biology 8(5): 1157–1167. 10.5943/mycosphere/8/5/6 [DOI] [Google Scholar]
- Ariyawansa HA, Tsai I, Wang J-Y, Withee P, Tanjira M, Lin S-R, Suwannarach N, Kumla J, Elgorban AM, Cheewangkoon R. (2021) Molecular phylogenetic diversity and biological characterization of Diaporthe species associated with leaf spots of Camelliasinensis in Taiwan. Plants 10(7): 1434. 10.3390/plants10071434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner K, Fujiyoshi PT, Travadon R, Castlebury LA, Wilcox WF, Rolshausen PE. (2013) Characterization of species of Diaporthe from wood cankers of grape in eastern North American vineyards. Plant Disease 97(7): 912–920. 10.1094/PDIS-04-12-0357-RE [DOI] [PubMed] [Google Scholar]
- Beluzán F, Olmo D, León M, Abad-Campos P, Armengol J. (2021) First report of Diaportheamygdali associated with twig canker and shoot blight of nectarine in Spain. Plant Disease 105(10): 3300. 10.1094/PDIS-10-20-2283-PDN [DOI] [PubMed] [Google Scholar]
- Bhunjun CS, Niskanen T, Suwannarach N, Wannathes N, Chen YJ, McKenzie EHC, Maharachchikumbura SSN, Buyck B, Zhao CL, Fan YG, Zhang JY, Dissanayake AJ, Marasinghe DS, Jayawardena RS, Kumla J, Padamsee M, Chen YY, Liimatainen K, Ammirati JF, Phukhamsakda C, Liu JK, Phonrob W, Randrianjohany É, Hongsanan S, Cheewangkoon R, Bundhun D, Khuna S, Yu WJ, Deng LS, Lu YZ, Hyde KD, Lumyong S. (2022) The numbers of fungi: Are the most speciose genera truly diverse? Fungal Diversity 114(1): 387–462. 10.1007/s13225-022-00501-4 [DOI]
- Cao L, Luo D, Lin W, Yang Q, Deng X. (2022) Four new species of Diaporthe (Diaporthaceae, Diaporthales) from forest plants in China. MycoKeys 91: 25–47. 10.3897/mycokeys.91.84970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone I, Kohn LM. (1999) A method for designing primer sets for the speciation studies in filamentous ascomycetes. Mycologia 91(3): 553–556. 10.1080/00275514.1999.12061051 [DOI] [Google Scholar]
- Chang CQ, Cheng YH, Xiang MM, Jiang ZD. (2005) New species of Phomopsis on woody plants in Fujian Province. Junwu Xuebao 24: 6–11. [Google Scholar]
- Chepkirui C, Stadler M. (2017) The genus Diaporthe: A rich source of diverse and bioactive metabolites. Mycological Progress 16(5): 477–494. 10.1007/s11557-017-1288-y [DOI] [Google Scholar]
- Crous PW, Groenewald JZ, Risede JM, Simoneau P, Hyde KD. (2004) Calonectria species and their Cylindrocladium anamorphs: Species with sphaeropedunculate vesicles. Studies in Mycology 50: 415–430. 10.3114/sim.55.1.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkley GJM, Groenewald JZ, Samson RA. (2009) Fungal Biodiversity. CBS Laboratory Manual Series 1. Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands.
- Crous PW, Summerell BA, Shivas RG, Romberg M, Mel’nik VA, Verkley GJM, Groenewald JZ. (2011a) Fungal Planet description sheets: 92–106. Persoonia 27(1): 130–162. 10.3767/003158511X617561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Shivas RG, Edwards J, Seifert KA, Alfenas AC, Alfenas RF, Burgess TI, Carnegie AJ, Hardy GESJ, Hiscock N, Hüberli D, Jung T, Louis-Seize G, Okada G, Pereira OL, Stukely MJC, Wang W, White GP, Young AJ, McTaggart AR, Pascoe IG, Porter IJ, Quaedvlieg W. (2011b) Fungal Planet description sheets: 69–91. Persoonia 26(1): 108–156. 10.3767/003158511X581723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Swart L, Denman S, Taylor JE, Bezuidenhout CM, Palm ME, Marincowitz S, Groenewald JZ. (2011c) Fungal pathogens of Protaceae. Persoonia 27(1): 20–45. 10.3767/003158511X606239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Shivas RG, Burgess TI, Decock CA, Dreyer LL, Granke LL, Guest DI, Hardy GSJ, Hausbeck MK, Hüberli D, Jung T, Koukol O, Lennox CL, Liew ECY, Lombard L, McTaggart AR, Pryke JS, Roets F, Saude C, Shuttleworth LA, Stukely MJC, Vánky K, Webster BJ, Windstam ST, Groenewald JZ. (2012) Fungal Planet description sheets: 107–127. Persoonia 28(1): 138–182. 10.3767/003158512X652633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Guarro J, Cheewangkoon R, van der Bank M, Swart WJ, Stchigel AM, Cano-Lira JF, Roux J, Madrid H, Damm U, Wood AR, Shuttleworth LA, Hodges CS, Munster M, de Jesús Yáñez-Morales M, Zúñiga-Estrada L, Cruywagen EM, De Hoog GS, Silvera C, Najafzadeh J, Davison EM, Davison PJN, Barrett MD, Barrett RL, Manamgoda DS, Minnis AM, Kleczewski NM, Flory SL, Castlebury LA, Clay K, Hyde KD, Maússe-Sitoe SND, Chen S, Lechat C, Hairaud M, Lesage-Meessen L, Pawłowska J, Wilk M, Śliwińska-Wyrzychowska A, Mętrak M, Wrzosek M, Pavlic-Zupanc D, Maleme HM, Slippers B, Mac Cormack WP, Archuby DI, Grünwald NJ, Tellería MT, Dueñas M, Martín MP, Marincowitz S, de Beer ZW, Perez CA, Gené J, Marin-Felix Y, Groenewald JZ. (2013) Fungal Planet description sheets: 154–213. Persoonia 31(1): 188–296. 10.3767/003158513X675925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Shivas RG, Quaedvlieg W, van der Bank M, Zhang Y, Summerell BA, Guarro J, Wingfield MJ, Wood AR, Alfenas AC, Braun U, Cano-Lira JF, García D, Marin-Felix Y, Alvarado P, Andrade JP, Armengol J, Assefa A, den Breeÿen A, Camele I, Cheewangkoon R, De Souza JT, Duong TA, Esteve-Raventós F, Fournier J, Frisullo S, García-Jiménez J, Gardiennet A, Gené J, Hernández-Restrepo M, Hirooka Y, Hospenthal DR, King A, Lechat C, Lombard L, Mang SM, Marbach PAS, Marincowitz S, Marin-Felix Y, Montaño-Mata NJ, Moreno G, Perez CA, Pérez Sierra AM, Robertson JL, Roux J, Rubio E, Schumacher RK, Stchigel AM, Sutton DA, Tan YP, Thompson EH, Vanderlinde E, Walker AK, Walker DM, Wickes BL, Wong PTW, Groenewald JZ. (2014a) Fungal Planet description sheets: 214–280. Persoonia 32(1): 184–306. 10.3767/003158514X682395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Schumacher RK, Summerell BA, Giraldo A, Gené J, Guarro J, Wanasinghe DN, Hyde KD, Camporesi E, Garethjones EB, Thambugala KM, Malysheva EF, Malysheva VF, Acharya K, Álvarez J, Alvarado P, Assefa A, Barnes CW, Bartlett JS, Blanchette RA, Burgess TI, Carlavilla JR, Coetzee MPA, Damm U, Decock CA, Denbreeÿen A, Devries B, Dutta AK, Holdom DG, Rooney-Latham S, Manjón JL, Marincowitz S, Mirabolfathy M, Moreno G, Nakashima C, Papizadeh M, Shahzadehfazeli SA, Amoozegar MA, Romberg MK, Shivas RG, Stalpers JA, Stielow B, Stukely MJC, Swart WJ, Tan YP, Vanderbank M, Wood AR, Zhang Y, Groenewald JZ. (2014b) Fungal Planet description sheets: 281–319. Persoonia 33(1): 212–289. 10.3767/003158514X685680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Schumacher RK, Wingfield MJ, et al. (2015a) Fungal Systematics and Evolution, FUSE 1. Sydowia 67: 81–118. 10.12905/0380.sydowia67-2015-0081 [DOI] [Google Scholar]
- Crous PW, Wingfield MJ, Le Roux JJ, et al. (2015b) Fungal Planet description sheets: 371–399. Persoonia 35(1): 264–327. 10.3767/003158515X690269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, et al. (2016a) Fungal Planet description sheets: 469–557. Persoonia 37: 218–403. 10.3767/003158516X694499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Richardson DM, Leroux JJ, Strasberg D, Edwards J, Roets F, Hubka V, Taylor PWJ, Heykoop M, Martín MP, Moreno G, Sutton DA, Wiederhold NP, Barnes CW, Carlavilla JR, Gené J, Giraldo A, Guarnaccia V, Guarro J, Hernández-Restrepo M, Kolařík M, Manjón JL, Pascoe IG, Popov ES, Sandoval-Denis M, Woudenberg JHC, Acharya K, Alexandrova AV, Alvarado P, Barbosa RN, Baseia IG, Blanchette RA, Boekhout T, Burgess TI, Cano-Lira JF, Čmoková A, Dimitrov RA, Dyakov MY, Dueñas M, Dutta AK, Esteve-Raventós F, Fedosova AG, Fournier J, Gamboa P, Gouliamova DE, Grebenc T, Groenewald M, Hanse B, Hardy GESTJ, Held BW, Jurjević Ž, Kaewgrajang T, Latha KPD, Lombard L, Luangsa-ard JJ, Lysková P, Mallátová N, Manimohan P, Miller AN, Mirabolfathy M, Morozova OV, Obodai M, Oliveira NT, Ordóñez ME, Otto EC, Paloi S, Peterson SW, Phosri C, Roux J, Salazar WA, Sánchez A, Sarria GA, Shin H-D, Silva BDB, Silva GA, Smith MTH, Souza-Motta CM, Stchigel AM, Stoilova-Disheva MM, Sulzbacher MA, Telleria MT, Toapanta C, Traba JM, Valenzuela-Lopez N, Watling R, Groenewald JZ. (2016b) Fungal Planet description sheets: 400–468. Persoonia 36(1): 316–458. 10.3767/003158516X692185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, Carnegie AJ, Hardy GESJ, Smith D, Summerell BA, Cano-Lira JF, Guarro J, Houbraken J, Lombard L, Martín MP, Sandoval-Denis M, Alexandrova AV, Barnes CW, Baseia IG, Bezerra JDP, Guarnaccia V, May TW, Hernández-Restrepo M, Stchigel AM, Miller AN, Ordoñez ME, Abreu VP, Accioly T, Agnello C, Agustin Colmán A, Albuquerque CC, Alfredo DS, Alvarado P, Araújo-Magalhães GR, Arauzo S, Atkinson T, Barili A, Barreto RW, Bezerra JL, Cabral TS, Camello Rodríguez F, Cruz RHSF, Daniëls PP, da Silva BDB, de Almeida DAC, de Carvalho Júnior AA, Decock CA, Delgat L, Denman S, Dimitrov RA, Edwards J, Fedosova AG, Ferreira RJ, Firmino AL, Flores JA, García D, Gené J, Giraldo A, Góis JS, Gomes AAM, Gonçalves CM, Gouliamova DE, Groenewald M, Guéorguiev BV, Guevara-Suarez M, Gusmão LFP, Hosaka K, Hubka V, Huhndorf SM, Jadan M, Jurjević Ž, Kraak B, Kučera V, Kumar TKA, Kušan I, Lacerda SR, Lamlertthon S, Lisboa WS, Loizides M, Luangsa-ard JJ, Lysková P, Mac Cormack WP, Macedo DM, Machado AR, Malysheva EF, Marinho P, Matočec N, Meijer M, Mešić A, Mongkolsamrit S, Moreira KA, Morozova OV, Nair KU, Nakamura N, Noisripoom W, Olariaga I, Oliveira RJV, Paiva LM, Pawar P, Pereira OL, Peterson SW, Prieto M, Rodríguez-Andrade E, Rojo De Blas C, Roy M, Santos ES, Sharma R, Silva GA, Souza-Motta CM, Takeuchi-Kaneko Y, Tanaka C, Thakur A, Smith MT, Tkalčec Z, Valenzuela-Lopez N, van der Kleij P, Verbeken A, Viana MG, Wang XW, Groenewald JZ. (2017) Fungal Planet description sheets: 625–715. Persoonia 39: 270–467. 10.3767/persoonia.2017.39.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Luangsa-ard JJ, Wingfield MJ, Carnegie AJ, Hernández-Restrepo M, Lombard L, Roux J, Barreto RW, Baseia IG, Cano-Lira JF, Martín MP, Morozova OV, Stchigel AM, Summerell BA, Brandrud TE, Dima B, García D, Giraldo A, Guarro J, Gusmão LFP, Khamsuntorn P, Noordeloos ME, Nuankaew S, Pinruan U, Rodríguez-Andrade E, Souza-Motta CM, Thangavel R, van Iperen AL, Abreu VP, Accioly T, Alves JL, Andrade JP, Bahram M, Baral H-O, Barbier E, Barnes CW, Bendiksen E, Bernard E, Bezerra JDP, Bezerra JL, Bizio E, Blair JE, Bulyonkova TM, Cabral TS, Caiafa MV, Cantillo T, Colmán AA, Conceição LB, Cruz S, Cunha AOB, Darveaux BA, da Silva AL, da Silva GA, da Silva GM, da Silva RMF, de Oliveira RJV, Oliveira RL, De Souza JT, Dueñas M, Evans HC, Epifani F, Felipe MTC, Fernández-López J, Ferreira BW, Figueiredo CN, Filippova NV, Flores JA, Gené J, Ghorbani G, Gibertoni TB, Glushakova AM, Healy R, Huhndorf SM, Iturrieta-González I, Javan-Nikkhah M, Juciano RF, Jurjević Ž, Kachalkin AV, Keochanpheng K, Krisai-Greilhuber I, Li Y-C, Lima AA, Machado AR, Madrid H, Magalhães OMC, Marbach PAS, Melanda GCS, Miller AN, Mongkolsamrit S, Nascimento RP, Oliveira TGL, Ordoñez ME, Orzes R, Palma MA, Pearce CJ, Pereira OL, Perrone G, Peterson SW, Pham THG, Piontelli E, Pordel A, Quijada L, Raja HA, Rosas de Paz E, Ryvarden L, Saitta A, Salcedo SS, Sandoval-Denis M, Santos TAB, Seifert KA, Silva BDB, Smith ME, Soares AM, Sommai S, Sousa JO, Suetrong S, Susca A, Tedersoo L, Telleria MT, Thanakitpipattana D, Valenzuela-Lopez N, Visagie CM, Zapata M, Groenewald JZ. (2018a) Fungal Planet description sheets: 785–867. Persoonia 41(1): 238–417. 10.3767/persoonia.2018.41.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, Hardy GESJ, Gené J, Guarro J, Baseia IG, García D, Gusmão LFP, Souza-Motta CM, Thangavel R, Adamčík S, Barili A, Barnes CW, Bezerra JDP, Bordallo JJ, Cano-Lira JF, de Oliveira RJV, Ercole E, Hubka V, Iturrieta-González I, Kubátová A, Martín MP, Moreau P-A, Morte A, Ordoñez ME, Rodríguez A, Stchigel AM, Vizzini A, Abdollahzadeh J, Abreu VP, Adamčíková K, Albuquerque GMR, Alexandrova AV, Álvarez Duarte E, Armstrong-Cho C, Banniza S, Barbosa RN, Bellanger J-M, Bezerra JL, Cabral TS, Caboň M, Caicedo E, Cantillo T, Carnegie AJ, Carmo LT, Castañeda-Ruiz RF, Clement CR, Čmoková A, Conceição LB, Cruz RHSF, Damm U, da Silva BDB, da Silva GA, da Silva RMF, Santiago ALCMA, de Oliveira LF, de Souza CAF, Déniel F, Dima B, Dong G, Edwards J, Félix CR, Fournier J, Gibertoni TB, Hosaka K, Iturriaga T, Jadan M, Jany J-L, Jurjević Ž, Kolařík M, Kušan I, Landell MF, Leite Cordeiro TR, Lima DX, Loizides M, Luo S, Machado AR, Madrid H, Magalhães OMC, Marinho P, Matočec N, Mešić A, Miller AN, Morozova OV, Neves RP, Nonaka K, Nováková A, Oberlies NH, Oliveira-Filho JRC, Oliveira TGL, Papp V, Pereira OL, Perrone G, Peterson SW, Pham THG, Raja HA. (2018b) Fungal Planet description sheets: 716–784. Persoonia 40(1): 240–393. 10.3767/persoonia.2018.40.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Carnegie AJ, Wingfield MJ, et al. (2019) Fungal Planet description sheets: 868–950. Persoonia 42: 291–473. 10.3767/persoonia.2019.42.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Schumacher RK, Akulov A, Bulgakov TS, Carnegie AJ, Jurjević Ž, Decock C, Denman S, Lombard L, Lawrence DP, Stack AJ, Gordon TR, Bostock RM, Burgess T, Summerell BA, Taylor PWJ, Edwards J, Hou LW, Cai L, Rossman AY, Wöhner T, Allen WC, Castlebury LA, Visagie CM, Groenewald JZ. (2020) New and Interesting Fungi. 3. Fungal Systematics and Evolution 6(1): 157–231. 10.3114/fuse.2020.06.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Hernández-Restrepo M, Schumacher RK, Cowan DA, Maggs-Kölling G, Marais E, Wingfield MJ, Yilmaz N, Adan OCG, Akulov A, Duarte EÁ, Berraf-Tebbal A, Bulgakov TS, Carnegie AJ, de Beer ZW, Decock C, Dijksterhuis J, Duong TA, Eichmeier A, Hien LT, Houbraken JAMP, Khanh TN, Liem NV, Lombard L, Lutzoni FM, Miadlikowska JM, Nel WJ, Pascoe IG, Roets F, Roux J, Samson RA, Shen M, Spetik M, Thangavel R, Thanh HM, Thao LD, van Nieuwenhuijzen EJ, Zhang JQ, Zhang Y, Zhao LL, Groenewald JZ. (2021) New and Interesting Fungi. 4. Fungal Systematics and Evolution 7(1): 255–343. 10.3114/fuse.2021.07.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva RMF, Soares AM, Pádua APSL, Firmino AL, Souza-Motta CM, da Silva GA, Plautz Jr HL, Bezerra JDP, Paiva LM, Ryvarden L, Oliani LC, de Mélo MAC, Magalhães OMC, Pereira OL, Oliveira RJV, Gibertoni TB, Oliveira TGS, Svedese VM, Fan XL. (2019) Mycological Diversity Description II. Acta Botanica Brasílica 33(1): 163–173. 10.1590/0102-33062018abb0411 [DOI] [Google Scholar]
- Dayarathne MC, Jones EBG, Maharachchikumbura SSN, Devadatha B, Sarma VV, Khongphinitbunjong K, Chomnunti P, Hyde KD. (2020) Morpho-molecular characterization of microfungi associated with marine based habitats. Mycosphere: Journal of Fungal Biology 11(1): 1–188. 10.5943/mycosphere/11/1/1 [DOI] [Google Scholar]
- de Silva NI, Maharachchikumbura SSN, Thambugala KM, Bhat DJ, Karunarathna SC, Tennakoon DS, Phookamsak R, Jayawardena RS, Lumyong S, Hyde KD. (2021) Morphomolecular taxonomic studies reveal a high number of endophytic fungi from Magnoliacandolli and M.garrettii in China and Thailand. Mycosphere : Journal of Fungal Biology 11(1): 163–237. 10.5943/mycosphere/12/1/3 [DOI] [Google Scholar]
- de Silva NI, Hyde KD, Lumyong S, Phillips AJL, Bhat DJ, Maharachchikumbura SSN, Thambugala KM, Tennakoon DS, Suwannarach N, Karunarathna SC. (2022) Morphology, phylogeny, host association and geography of fungi associated with plants of Annonaceae, Apocynaceae and Magnoliaceae. Mycosphere: Journal of Fungal Biology 13(1): 955–1076. 10.5943/mycosphere/13/1/12 [DOI] [Google Scholar]
- Dissanayake AJ. (2017) The current status of species in Diaporthe. Mycosphere : Journal of Fungal Biology 8(5): 1106–1156. 10.5943/mycosphere/8/5/5 [DOI] [Google Scholar]
- Dissanayake AJ, Camporesi E, Hyde KD, Zhang W, Yan JY, Li XH. (2017a) Molecular phylogenetic analysis reveals seven new Diaporthe species from Italy. Mycosphere: Journal of Fungal Biology 8(5): 853–877. 10.5943/mycosphere/8/5/4 [DOI] [Google Scholar]
- Dissanayake AJ, Zhang W, Liu M, Hyde KD, Zhao WS, Li XH, Yan JY. (2017b) Diaporthe species associated with peach tree dieback in Hubei, China. Mycosphere: Journal of Fungal Biology 8(5): 533–549. 10.5943/mycosphere/8/5/2 [DOI] [Google Scholar]
- Dissanayake AJ, Chen Y-Y, Liu J-K. (2020) Unravelling Diaporthe species associated with woody hosts from karst formations (Guizhou) in China. Journal of Fungi (Basel, Switzerland) 6(4): 251. 10.3390/jof6040251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doilom M, Dissanayake AJ, Wanasinghe DN, Boonmee S, Liu J-K, Bhat DJ, Taylor JE, Bahkali AH, McKenzie EHC, Hyde KD. (2017) Microfungi on Tectonagrandis (teak) in Northern Thailand. Fungal Diversity 82(1): 107–182. 10.1007/s13225-016-0368-7 [DOI] [Google Scholar]
- Dong Z, Manawasinghe IS, Huang Y, Shu Y, Phillips AJL, Dissanayake AJ, Hyde KD, Xiang M, Luo M. (2021) Endophytic Diaporthe associated with Citrusgrandis cv. Tomentosa in China. Frontiers in Microbiology 11: 3621. 10.3389/fmicb.2020.609387 [DOI] [PMC free article] [PubMed]
- Du Z, Fan XL, Hyde KD, Yang Q, Liang Y-M, Tian C-M. (2016) Phylogeny and morphology reveal two new species of Diaporthe from Betula spp. China. Phytotaxa 269(2): 90–102. 10.11646/phytotaxa.269.2.2 [DOI] [Google Scholar]
- Fan XL, Hyde KD, Udayanga D, Wu X-Y, Tian C-M. (2015) Diaportherostrata, a novel ascomycete from Juglansmandshurica associated with walnut dieback. Mycological Progress 14(10): 82. 10.1007/s11557-015-1104-5 [DOI] [Google Scholar]
- Fan X, Yang Q, Bezerra JDP, Alvarez LV, Tian C-M. (2018) Diaporthe from walnut tree (Juglansregia) in China, with insight of the Diaportheeres complex. Mycological Progress 17(7): 841–853. 10.1007/s11557-018-1395-4 [DOI] [Google Scholar]
- Feng X-X, Chen J-J, Wang G-R, Cao T-T, Zheng Y-L, Zhang C-L. (2019) Diaporthesinensis, a new fungus from Amaranthus sp. in China. Phytotaxa 425(5): 259–268. 10.11646/phytotaxa.425.5.1 [DOI] [Google Scholar]
- Frisvad JC, Larsen TO. (2015) Chemodiversity in the genus Aspergillus. Applied Microbiology and Biotechnology 99(19): 7859–7877. 10.1007/s00253-015-6839-z [DOI] [PubMed] [Google Scholar]
- Gao YH, Sun W, Su YY, Cai Lei. (2014) Three new species of Phomopsis in Gutianshan Nature Reserve in China. Mycological Progress 13: 111–121. 10.1007/s11557-013-0898-2 [DOI] [Google Scholar]
- Gao YH, Su YY, Sun W, Cai L. (2015) Diaporthe species occurring on Lithocarpusglabra in China, with descriptions of five new species. Fungal Biology 119(5): 295–309. 10.1016/j.funbio.2014.06.006 [DOI] [PubMed] [Google Scholar]
- Gao Y, Liu F, Cai L. (2016) Unravelling Diaporthe species associated with Camellia. Systematics and Biodiversity 14(1): 102–117. 10.1080/14772000.2015.1101027 [DOI] [Google Scholar]
- Gao YH, Liu F, Duan W, Crous PW, Cai L. (2017) Diaporthe is paraphyletic. IMA Fungus 8(1): 153–187. 10.5598/imafungus.2017.08.01.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Pan M, Tian C, Fan X. (2021) Cytospora and Diaporthe species associated with hazelnut canker and dieback in Beijing, China. Frontiers in Cellular and Infection Microbiology 11: 664366. 10.3389/fcimb.2021.664366 [DOI] [PMC free article] [PubMed]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61(4): 1323–1330. 10.1128/aem.61.4.1323-1330.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard ML, Mottier N, Jeanneret-Gris J, Christen D, Tabacchi R, Abou-Mansour E. (2014) Differential production of phytotoxins from Phomopsis sp. from grapevine plants showing esca symptoms. Journal of Agricultural and Food Chemistry 62(34): 8602–8607. 10.1021/jf501141g [DOI] [PubMed] [Google Scholar]
- Gomes RR, Glienke C, Videira SIR, Lombard L, Groenewald JZ, Crous PW. (2013) Diaporthe: A genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 31(1): 1–41. 10.3767/003158513X666844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomzhina MM, Gannibal PhB. (2022) Diaporthe species infecting sunflower (Helianthusannuus) in Russia, with the description of two new species. Mycologia 114: 556–574. 10.1080/00275514.2022.2040285 [DOI] [PubMed] [Google Scholar]
- Guarnaccia V, Crous PW. (2017) Emerging citrus diseases in Europe caused by Diaporthe spp. IMA Fungus 8(2): 317–334. 10.5598/imafungus.2017.08.02.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V, Crous PW. (2018) Species of Diaporthe on Camellia and Citrus in the Azores Islands. Phytopathologia Mediterranea 57: 307–319. 10.14601/Phytopathol_Mediterr-23254 [DOI] [Google Scholar]
- Guarnaccia V, Groenewald JZ, Woodhall J, Armengol J, Cinelli T, Eichmeier A, Ezra D, Fontaine F, Gramaje D, Gutierrez-Aguirregabiria A, Kaliterna J, Kiss L, Larignon P, Luque J, Mugnai L, Naor V, Raposo R, Sándor E, Váczy KZ, Crous PW. (2018) Diaporthe diversity and pathogenicity revealed from a broad survey of grapevine diseases in Europe. Persoonia 40(1): 135–153. 10.3767/persoonia.2018.40.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YS, Crous PW, Bai Q, Fu M, Yang MM, Wang XH, Du YM, Hong N, Xu WX, Wang GP. (2020) High diversity of Diaporthe species associated with pear shoot canker in China. Persoonia 45(1): 132–162. 10.3767/persoonia.2020.45.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilário S, Amaral IA, Gonçalves MFM, Lopes A, Santos L, Alves A. (2020) Diaporthe species associated with twig blight and dieback of Vacciniumcorymbosum in Portugal, with description of four new species. Mycologia 112(2): 293–308. 10.1080/00275514.2019.1698926 [DOI] [PubMed] [Google Scholar]
- Hilário S, Santos L, Alves A. (2021) Diaportheamygdali, a species complex or a complex species? Fungal Biology 125(7): 505–518. 10.1016/j.funbio.2021.01.006 [DOI] [PubMed]
- Hongsanan S, Norphanphoun C, Senanayake IC, Jayawardena RS, Manawasinghe IS, Abeywickrama PD, Khuna S, Suwannarach N, Senwanna C, Monkai J, Hyde KD, Gentekaki E, Bhunjun CS. (2023) Annotated notes on Diaporthe species. Mycosphere : Journal of Fungal Biology 14(1): 918–1189. [Google Scholar]
- Hu DM, Cai L, Hyde KD. (2012) Three new ascomycetes from freshwater in China. Mycologia 104(6): 1478–1489. 10.3852/11-430 [DOI] [PubMed] [Google Scholar]
- Huang F, Hou X, Dewdney MM, Fu Y, Chen G, Hyde KD, Li H. (2013) Diaporthe species occurring on citrus in China. Fungal Diversity 61(1): 237–250. 10.1007/s13225-013-0245-6 [DOI] [Google Scholar]
- Huang F, Udayanga D, Wang X, Hou X, Mei X, Fu Y, Hyde KD, Li H. (2015) Endophytic Diaporthe associated with Citrus, a phylogenetic reassessment with seven new species from China. Fungal Biology 119(5): 331–347. 10.1016/j.funbio.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Huang S, Xia J, Zhang X, Sun W. (2021) Morphological and phylogenetic analyses reveal three new species of Diaporthe from Yunnan, China. MycoKeys 78: 49–77. 10.3897/mycokeys.78.60878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde KD, Hongsanan S, Jeewon R, et al. (2016) Fungal diversity notes 367–492, taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 80: 1–270. 10.1007/s13225-016-0373-x [DOI] [Google Scholar]
- Hyde KD, Chaiwan N, Norphanphoun C, et al. (2018) Mycosphere notes 169–224. Mycosphere: Journal of Fungal Biology 9(2): 271–430. 10.5943/mycosphere/9/2/8 [DOI] [Google Scholar]
- Hyde KD, Tennakoon DS, Jeewon R, Bhat DJ, Maharachchikumbura SSN, Rossi W, Leonardi M, Lee HB, Mun HY, Houbraken J, Nguyen TTT, Jeon SJ, Frisvad JC, Wanasinghe DN, Lücking R, Aptroot A, Cáceres MES, Karunarathna SC, Hongsanan S, Phookamsak R, de Silva NI, Thambugala KM, Jayawardena RS, Senanayake IC, Boonmee S, Chen J, Luo Z-L, Phukhamsakda C, Pereira OL, Abreu VP, Rosado AWC, Bart B, Randrianjohany E, Hofstetter V, Gibertoni TB, Soares AMS, Plautz Jr HL, Sotão HMP, Xavier WKS, Bezerra JDP, de Oliveira TGL, de Souza-Motta CM, Magalhães OMC, Bundhun D, Harishchandra D, Manawasinghe IS, Dong W, Zhang S-N, Bao D-F, Samarakoon MC, Pem D, Karunarathna A, Lin C-G, Yang J, Perera RH, Kumar V, Huang S-K, Dayarathne MC, Ekanayaka AH, Jayasiri SC, Xiao Y, Konta S, Niskanen T, Liimatainen K, Dai Y-C, Ji X-H, Tian X-M, Mešić A, Singh SK, Phutthacharoen K, Cai L, Sorvongxay T, Thiyagaraja V, Norphanphoun C, Chaiwan N, Lu Y-Z, Jiang H-B, Zhang J-F, Abeywickrama PD, Aluthmuhandiram JVS, Brahmanage RS, Zeng M, Chethana T, Wei D, Réblová M, Fournier J, Nekvindová J, do Nascimento Barbosa R, dos Santos JEF, de Oliveira NT, Li G-J, Ertz D, Shang Q-J, Phillips AJL, Kuo C-H, Camporesi E, Bulgakov TS, Lumyong S, Jones EBG, Chomnunti P, Gentekaki E, Bungartz F, Zeng X-Y, Fryar S, Tkalčec Z, Liang J, Li G, Wen T-C, Singh PN, Gafforov Y, Promputtha I, Yasanthika E, Goonasekara ID, Zhao R-L, Zhao Q, Kirk PM, Liu J-K, Yan JY, Mortimer PE, Xu J, Doilom M. (2019) Fungal diversity notes 1036–1150: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 96(1): 1–242. 10.1007/s13225-019-00429-2 [DOI] [Google Scholar]
- Hyde KD, Dong Y, Phookamsak R, Jeewon R, Bhat DJ, Jones EBG, Liu N-G, Abeywickrama PD, Mapook A, Wei D, Perera RH, Manawasinghe IS, Pem D, Bundhun D, Karunarathna A, Ekanayaka AH, Bao D-F, Li J, Samarakoon MC, Chaiwan N, Lin C-G, Phutthacharoen K, Zhang S-N, Senanayake IC, Goonasekara ID, Thambugala KM, Phukhamsakda C, Tennakoon DS, Jiang H-B, Yang J, Zeng M, Huanraluek N, Liu J-KJ, Wijesinghe SN, Tian Q, Tibpromma S, Brahmanage RS, Boonmee S, Huang S-K, Thiyagaraja V, Lu Y-Z, Jayawardena RS, Dong W, Yang E-F, Singh SK, Singh SM, Rana S, Lad SS, Anand G, Devadatha B, Niranjan M, Sarma VV, Liimatainen K, Aguirre-Hudson B, Niskanen T, Overall A, Alvarenga RLM, Gibertoni TB, Pfliegler WP, Horváth E, Imre A, Alves AL, da Silva Santos AC, Tiago PV, Bulgakov TS, Wanasinghe DN, Bahkali AH, Doilom M, Elgorban AM, Maharachchikumbura SSN, Rajeshkumar KC, Haelewaters D, Mortimer PE, Zhao Q, Lumyong S, Xu J, Sheng J. (2020) Fungal diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100(1): 5–277. 10.1007/s13225-020-00439-5 [DOI] [Google Scholar]
- Iantas J, Savi DC, Schibelbein Rd S, Noriler SA, Assad BM, Dilarri G, Ferreira H, Rohr J, Thorson JS, Shaaban KA, Glienke C. (2021) Endophytes of brazilian medicinal plants with activity against phytopathogens. Frontiers in Microbiology 12: 714750. 10.3389/fmicb.2021.714750 [DOI] [PMC free article] [PubMed]
- Iriart X, Binois R, Fior A, Blanchet D, Berry A, Cassaing S, Amazan E, Papot E, Carme B, Aznar C, Couppié P. (2011) Eumycetoma caused by Diaporthephaseolorum (Phomopsisphaseoli): A case report and a mini-review of Diaporthe/Phomopsis spp invasive infections in humans. Clinical Microbiology and Infection 17(10): 1492–1494. 10.1111/j.1469-0691.2011.03568.x [DOI] [PubMed] [Google Scholar]
- Jaklitsch WM, Gardiennet A, Voglmayr H. (2016) Resolution of morphology-based taxonomic delusions. Acrocordiella, Basiseptospora, Blogiascospora, Clypeosphaeria, Hymenopleella, Lepteutypa, Pseudapiospora, Requienella, Seiridium and Strickeria. Persoonia 37(1): 82–105. 10.3767/003158516X690475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Voglmayr H, Piao C-G, Li Y. (2021) Two new species of Diaporthe (Diaporthaceae, Diaporthales) associated with tree cankers in the Netherlands. MycoKeys 85: 31–56. 10.3897/mycokeys.85.73107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software v. 7: Improvements in performance and usability. Molecular Biology and Evolution 30(4): 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Mentjies P, Drummond A. (2012) Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics (Oxford, England) 28(12): 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocsubé S, Perrone G, Magistà D, Houbraken J, Varga J, Szigeti G, Hubka V, Hong SB, Frisvad JC, Samson RA. (2016) Aspergillus is monophyletic: Evidence from multiple gene phylogenies and extrolites profiles. Studies in Mycology 85(1): 199–213. 10.1016/j.simyco.2016.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence DP, Travadon R, Baumgartner K. (2015) Diversity of Diaporthe species associated with wood cankers of fruit and nut crops in northern California. Mycologia 107(5): 926–940. 10.3852/14-353 [DOI] [PubMed] [Google Scholar]
- Lesuthu P, Mostert L, Spies CFJ, Moyo P, Regnier T, Halleen F. (2019) Diaporthenebulae sp. nov. and first report of D.cynaroidis, D.novem, and D.serafiniae on grapevines in South Africa. Plant Disease 103(5): 808–817. 10.1094/PDIS-03-18-0433-RE [DOI] [PubMed] [Google Scholar]
- Li WJ, McKenzie EHC, Liu JK, Bhat DJ, Dai D-Q, Camporesi E, Tian Q, Maharachchikumbura SSN, Luo Z-L, Shang Q-J, Zhang J-F, Tangthirasunun N, Karunarathna SC, Xu J-C, Hyde KD. (2020) Taxonomy and phylogeny of hyaline-spored coelomycetes. Fungal Diversity 100(1): 279–801. 10.1007/s13225-020-00440-y [DOI] [Google Scholar]
- Lin S, Taylor NJ, Peduto Hand F. (2018) Identification and characterization of fungal pathogens causing fruit rot of deciduous holly. Plant Disease 102(12): 2430–2445. 10.1094/PDIS-02-18-0372-RE [DOI] [PubMed] [Google Scholar]
- Liu JK, Hyde KD, Jones EBG, Ariyawansa HA, Bhat DJ, Boonmee S, Maharachchikumbura SSN, McKenzie EHC, Phookamsak R, Phukhamsakda C, Shenoy BD, Abdel-Wahab MA, Buyck B, Chen J, Chethana KWT, Singtripop C, Dai DQ, Dai YC, Daranagama DA, Dissanayake AJ, Doilom M, D’souza MJ, Fan XL, Goonasekara ID, Hirayama K, Hongsanan S, Jayasiri SC, Jayawardena RS, Karunarathna SC, Li WJ, Mapook A, Norphanphoun C, Pang KL, Perera RH, Peršoh D, Pinruan U, Senanayake IC, Somrithipol S, Suetrong S, Tanaka K, Thambugala KM, Tian Q, Tibpromma S, Udayanga D, Wijayawardene NN, Wanasinghe D, Wisitrassameewong K, Zeng XY, Abdel-Aziz FA, Adamčík S, Bahkali AH, Boonyuen N, Bulgakov T, Callac P, Chomnunti P, Greiner K, Hashimoto A, Hofstetter V, Kang JC, Lewis D, Li XH, Liu XZ, Liu ZY, Matsumura M, Mortimer PE, Rambold G, Randrianjohany E, Sato G, Sri-Indrasutdhi V, Tian CM, Verbeken A, von Brackel W, Wang Y, Wen TC, Xu JC, Yan JY, Zhao RL, Camporesi E. (2015) Fungal diversity notes 1–110: Taxonomic and phylogenetic contributions to fungal species. Fungal Diversity 72(1): 1–197. 10.1007/s13225-015-0324-y [DOI] [Google Scholar]
- Lombard L, van Leeuwen GCM, Guarnaccia V, Polizzi G, Rijswick PCJ, Rosendahl KCHM, Gabler J, Crous PW. (2014) Diaporthe species associated with Vaccinium, with specific reference to Europe. Phytopathologia Mediterranea 53(2): 287–299. 10.14601/PHYTOPATHOL_MEDITERR-14034 [DOI] [Google Scholar]
- Long H, Zhang Q, Hao Y-Y, Shao X-Q, Wei X-X, Hyde KD, Wang Y, Zhao D-G. (2019) Diaporthe species in south-western China. MycoKeys 57: 113–127. 10.3897/mycokeys.57.35448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QT, Zhang JY, Sun YR, Tang X, Lu YZ, Zhang Z. (2022) Diaportheorixae sp. nov., an endophytic species isolated from Orixajaponica in southern China. Phytotaxa 544(1): 37–51. 10.11646/phytotaxa.544.1.3 [DOI] [Google Scholar]
- Machingambi NM, Dreyer LL, Oberlander KC, Roux J, Roets F. (2015) Death of endemic Virgiliaoroboides trees in South Africa caused by Diaporthevirgiliae sp. nov. Plant Pathology 64(5): 1149–1156. 10.1111/ppa.12341 [DOI] [Google Scholar]
- Mahoney N, Molyneux RJ, Smith LR, Schoch TK, Rolshausen PE, Gubler WD. (2005) Dying-arm disease in grapevines: Diagnosis of infection with Eutypalata by metabolite analysis. Journal of Agricultural and Food Chemistry 53(21): 8148–8155. 10.1021/jf0510236 [DOI] [PubMed] [Google Scholar]
- Manawasinghe IS, Dissanayake AJ, Li X, Liu M, Wanasinghe DN, Xu J, Zhao W, Zhang W, Zhou Y, Hyde KD, Brooks S, Yan J. (2019) High genetic diversity and species complexity of Diaporthe associated with grapevine dieback in China. Frontiers in Microbiology 10: 1936. 10.3389/fmicb.2019.01936 [DOI] [PMC free article] [PubMed]
- Manawasinghe IS, Jayawardena RS, Li HL, Zhou YY, Zhang W, Phillips AJL, Wanasinghe DN, Dissanayake AJ, Li XH, Li YH, Hyde KD, Yan JY. (2021) Microfungi associated with Camelliasinensis: A case study of leaf and shoot necrosis on Tea in Fujian, China. Mycosphere: Journal of Fungal Biology 12(1): 430–518. 10.5943/mycosphere/12/1/6 [DOI] [Google Scholar]
- Mapook A, Hyde KD, McKenzie EHC, Jones EBG, Bhat DJ, Jeewon R, Stadler M, Samarakoon MC, Malaithong M, Tanunchai B, Buscot F, Wubet T, Purahong W. (2020) Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaenaodorata (Siam weed). Fungal Diversity 101(1): 1–175. 10.1007/s13225-020-00444-8 [DOI] [Google Scholar]
- Marin-Felix Y, Hernández-Restrepo M, Wingfield MJ, Akulov A, Carnegie AJ, Cheewangkoon R, Gramaje D, Groenewald JZ, Guarnaccia V, Halleen F, Lombard L, Luangsa-ard J, Marincowitz S, Moslemi A, Mostert L, Quaedvlieg W, Schumacher RK, Spies CFJ, Thangavel R, Taylor PWJ, Wilson AM, Wingfield BD, Wood AR, Crous PW. (2019) Genera of phytopathogenic fungi: GOPHY 2. Studies in Mycology 92(1): 47–133. 10.1016/j.simyco.2018.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino I, Agustí-Brisach C, Nari L, Gullino ML, Guarnaccia V. (2023) Characterization and pathogenicity of fungal species associated with dieback of apple trees in Northern Italy. Plant Disease PDIS-04-23-0645-RE. 10.1094/PDIS-04-23-0645-RE [DOI] [PubMed]
- Matio Kemkuignou B, Schweizer L, Lambert C, Anoumedem EGM, Kouam SF, Stadler M, Marin-Felix Y. (2022) New polyketides from the liquid culture of Diaporthebreyniae sp. nov. MycoKeys 90: 85–118. 10.3897/mycokeys.90.82871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matio Kemkuignou B, Lambert C, Stadler M, Kouam SF, Marin-Felix Y. (2023) Unprecedented antimicrobial and cytotoxic polyketides from cultures of Diaportheafricana sp. nov. Journal of Fungi (Basel, Switzerland) 9(7): 781. 10.3390/jof9070781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milagres CA, Belisário R, Silva MA, Lisboa DO, Pinho DB, Furtado GQ. (2018) A novel species of Diaporthe causing leaf spot in Pachiraglabra. Tropical Plant Pathology 43(5): 460–467. 10.1007/s40858-018-0242-0 [DOI] [Google Scholar]
- Monkai J, Hongsanan S, Bhat DJ, Dawoud TM, Lumyong S. (2023) Integrative taxonomy of novel Diaporthe species associated with medicinal plants in Thailand. Journal of Fungi (Basel, Switzerland) 9(6): 603. 10.3390/jof9060603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostert L, Crous PW, Kang J-C, Phillips AJL. (2001) Species of Phomopsis and a Libertella sp. occurring on grapevines with specific reference to South Africa: Morphological, cultural, molecular and pathological characterization. Mycologia 93(1): 146–167. 10.1080/00275514.2001.12061286 [DOI] [Google Scholar]
- Noriler SA, Savi DC, Ponomareva L, Rodrigues R, Rohr J, Thorson JS, Glienke C, Shaaban KA. (2019) Vochysiamides A and B: Two new bioactive carboxamides produced by the new species Diaporthevochysiae. Fitoterapia 138: 104–273. 10.1016/j.fitote.2019.104273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norphanphoun C, Gentekaki E, Hongsanan S, Jayawardena R, Senanayake IC, Manawasinghe IS, Abeywickrama PD, Bhunjun CS, Hyde KD. (2022) Diaporthe: Formalizing the species-group concept. Mycosphere: Journal of Fungal Biology 13(1): 752–819. 10.5943/mycosphere/13/1/9 [DOI] [Google Scholar]
- Ozawa K, Mochizuki K, Takagi D, Ishida K, Sunada A, Ohkusu K, Kamei K, Hashimoto A, Tanaka K. (2019) Identification and antifungal sensitivity of two new species of Diaporthe isolated. Journal of Infection and Chemotherapy 25(2): 96–103. 10.1016/j.jiac.2018.10.008 [DOI] [PubMed] [Google Scholar]
- Pereira C, Ferreira B, Aucique-Perez C, Barreto R. (2021) Diaportherosiphthora sp. nov.: Yet another rose dieback fungus. Crop Protection 139: 105365. 10.1016/j.cropro.2020.105365 [DOI]
- Perera RH, Hyde KD, Dissanayake AJ, Jones EB, Liu JK, Wei D, Liu ZY. (2018) Diaporthecollariana sp. nov., with prominent collarettes associated with Magnoliachampaca fruits in Thailand. Studies in Fungi 3(1): 141–151. 10.5943/sif/3/1/16 [DOI] [Google Scholar]
- Perera RH, Hyde KD, Maharachchikumbura SSN, Jones EBG, McKenzie EHC, Stadler M, Lee HB, Samarakoon MC, Ekanayaka AH, Camporesi E, Liu JK, Liu ZY. (2020) Fungi on wild seeds and fruits. Mycosphere: Journal of Fungal Biology 11(1): 2108–2480. 10.5943/mycosphere/11/1/14 [DOI] [Google Scholar]
- Petrović K, Riccioni L, Dordević V, Tubic SB, Miladinović J, Ceran M, Rajković D. (2018) Diaporthepseudolongicolla - the new pathogen on soybean seed in Serbia. Ratarstvo i Povrtarstvo 55(2): 103–109. 10.5937/ratpov55-18582 [DOI] [Google Scholar]
- Phukhamsakda C, McKenzie EHC, Phillips AJL, Jones EBG, Bhat DJ, Stadler M, Bhunjun CS, Wanasinghe DN, Thongbai B, Camporesi E, Ertz D, Jayawardena RS, Perera RH, Ekanayake AH, Tibpromma S, Doilom M, Xu J, Hyde KD. (2020) Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Diversity 102(1): 1–203. 10.1007/s13225-020-00448-4 [DOI] [Google Scholar]
- Pscheidt J. (1989) Time of infection and control of Phomopsis fruit rot of grape. Plant Disease 73(10): 829–833. 10.1094/PD-73-0829 [DOI] [Google Scholar]
- Pusztahelyi T, Holb IJ, Pócsi I. (2015) Secondary metabolites in fungus-plant interactions. Frontiers in Plant Science 6: 1–23. 10.3389/fpls.2015.00573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakita RM, O’Brien KD, Bourassa L. (2017) Diaporthe soft tissue infection in a heart transplant patient. Transplant Infectious Disease 19(3): 1–3. 10.1111/tid.12680 [DOI] [PubMed] [Google Scholar]
- Rossman AY, Adams GC, Cannon PF, Castlebury LA, Crous PW, Gryzenhout M, Jaklitsch WM, Mejia LC, Stoykov D, Udayanga D, Voglmayr H, Walker DM. (2015) Recommendations of generic names in Diaporthales competing for protection or use. IMA Fungus 6(1): 145–154. 10.5598/imafungus.2015.06.01.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JM, Correia VG, Phillips AJL. (2010) Primers for mating-type diagnosis in Diaporthe and Phomopsis, their use in teleomorph induction in vitro and biological species definition. Fungal Biology 114(2–3): 255–270. 10.1016/j.funbio.2010.01.007 [DOI] [PubMed] [Google Scholar]
- Santos L, Phillips AJL, Crous PW. (2017) Diaporthe species on Rosaceae with descriptions of D.pyracanthae sp. nov. and D.malorum sp. nov. Mycosphere: Journal of Fungal Biology 8(5): 485–511. 10.5943/mycosphere/8/5/1 [DOI] [Google Scholar]
- Senanayake IC, Crous PW, Groenewald JZ, Maharachchikumbura SSN, Jeewon R, Philips AJL, Bhat JD, Perera RH, Li QR, Ji WJ, Tangthirasunun N, Noprhanphoun C, Karunarathna SC, Camporesi E, Manawasighe IS, Al-Sadi AM, Hade KD. (2017) Families of Diaporthales based on morphological and phylogenetic evidence. Studies in Mycology 86: 217–296. 10.1016/j.simyco.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si YZ, Li DW, Zhong J, Huang L, Zhu LH. (2022) Diaporthesapindicola sp. nov. causes leaf spots of Sapindusmukorossi in China. Plant Disease 106: 1105–1113. 10.1094/PDIS-04-21-0777-RE [DOI] [PubMed] [Google Scholar]
- Smith H, Wingfield MJ, Crous PW, Coutinho TA. (1996) Sphaeropsissapinea and Botryosphaeriadothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. South African Journal of Botany 62(2): 86–88. 10.1016/S0254-6299(15)30596-2 [DOI] [Google Scholar]
- Stadler M, Lambert C, Wibberg D, Kalinowski J, Cox RJ, Kolařík M, Kuhnert E. (2020) Intragenomic polymorphisms in the ITS region of high-quality genomes of the Hypoxylaceae (Xylariales, Ascomycota). Mycological Progress 19(3): 35–245. 10.1007/s11557-019-01552-9 [DOI] [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England) 30(9): 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Huang S, Xia J, Zhang X, Li Z. (2021) Morphological and molecular identification of Diaporthe species in south-western China, with description of eight new species. MycoKeys 77: 65–95. 10.3897/mycokeys.77.59852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology 56(4): 564–577. 10.1080/10635150701472164 [DOI] [PubMed] [Google Scholar]
- Tan YP, Shivas RG. (2022) Nomenclatural novelties. Index of Australian Fungi 2: 1–12. [Google Scholar]
- Tan YP, Shivas RG. (2023) Nomenclatural novelties. Index of Australian Fungi 6: 1–17. 10.5281/zenodo.7894166 [DOI] [Google Scholar]
- Tan YP, Edwards J, Grice KRE, Shivas RG. (2013) Molecular phylogenetic analysis reveals six new Diaporthe species from Australia. Fungal Diversity 61(1): 251–260. 10.1007/s13225-013-0242-9 [DOI] [Google Scholar]
- Tanney JB, McMullin DR, Green BD, Miller JD, Seifert KA. (2016) Production of antifungal and antiinsectan metabolites by the Picea endophyte Diaporthemaritima sp. nov. Fungal Biology 120(11): 1448–1457. 10.1016/j.funbio.2016.05.007 [DOI] [PubMed] [Google Scholar]
- Tennakoon DS, Kuo CH, Maharachchikumbura SSN, Thambugala KM, Gentekaki E, Phillips AJL, Bhat DJ, Wanasinghe DN, de Silva NI, Promputtha I, Hyde KD. (2021) Taxonomic and phylogenetic contributions to Celtisformosana, Ficusampelas, F.septica, Macarangatanarius and Morusaustralis leaf litter inhabiting microfungi. Fungal Diversity 108(1): 1–215. 10.1007/s13225-021-00474-w [DOI] [Google Scholar]
- Tey-Rulh P, Philippe I, Renaud JM, Tsoupras G, de Angelis P, Fallot J, Tabacchi R. (1991) Eutypine, a phytotoxin produced by Eutypalata the causal agent of dying-arm disease of grapevine. Phytochemistry 30(2): 471–473. 10.1016/0031-9422(91)83707-R [DOI] [Google Scholar]
- The Royal Horticultural Society London (1966) R.H.S. Colour Chart. London, UK.
- Thompson SM, Tan YP, Young AJ, Neate SM, Aitken EAB, Shivas RG. (2011) Stem cankers on sunflower (Helianthusannuus) in Australia reveal a complex of pathogenic Diaporthe (Phomopsis) species. Persoonia 27(1): 80–89. 10.3767/003158511X617110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SM, Tan YP, Shivas RG, Neate SM, Morin L, Bissett A, Aitken EAB. (2015) Green and brown bridges between weeds and crops reveal novel Diaporthe species in Australia. Persoonia 35(1): 39–49. 10.3767/003158515X687506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SM, Tan YP, Shivas RG. (2023) Nomenclatural novelties. Index of Australian Fungi 7: 1–7. 10.5281/zenodo.7972551 [DOI] [Google Scholar]
- Tibpromma S, Hyde KD, Bhat JD, Mortimer PE, Xu JC, Promputtha I, Doilom M, Yang JB, Tang AMC, Karunarathna SC. (2018) Identification of endophytic fungi from leaves of Pandanaceae based on their morphotypes and DNA sequence data from southern Thailand. MycoKeys 33: 25–67. 10.3897/mycokeys.33.23670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toghueo RMK, Vázquez de Aldana BR, Zabalgogeazcoa I. (2023) Diaporthe species associated with the maritime grass Festucarubrasubsp.pruinosa. Frontiers in Microbiology 14: 1105299. 10.3389/fmicb.2023.1105299 [DOI] [PMC free article] [PubMed]
- Tsang CC, Tang JYM, Lau SKP, Woo PCY. (2018) Taxonomy and evolution of Apsergillus, Penicillium and Talaromyces in the omics era – Past, present and future. Computational and Structural Biotechnology Journal 16: 197–210. 10.1016/j.csbj.2018.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurushima T, Don LD, Kawashima K, Murakami J, Nakayashiki H, Tosa Y, Mayama S. (2005) Pyrichalasin H production and pathogenicity of Digitaria-specific isolates of Pyriculariagrisea. Molecular Plant Pathology 6(6): 605–613. 10.1111/j.1364-3703.2005.00309.x [DOI] [PubMed] [Google Scholar]
- Udayanga D, Liu X, McKenzie EHC, Chukeatirote E, Bahkali AHA, Hyde KD. (2011) The genus Phomopsis: Biology, applications, species concepts and names of common phytopathogens. Fungal Diversity 50(1): 189–225. 10.1007/s13225-011-0126-9 [DOI] [Google Scholar]
- Udayanga D, Liu X, Crous PW, McKenzie EHC, Chukeatirote E, Hyde KD. (2012a) A multi-locus phylogenetic evaluation of Diaporthe (Phomopsis). Fungal Diversity 56(1): 157–171. 10.1007/s13225-012-0190-9 [DOI] [Google Scholar]
- Udayanga D, Liu XZ, Mckenzie EHC, Chukeatirote E, Hyde KD. (2012b) Multi-locus phylogeny reveals three new species of Diaporthe from Thailand. Cryptogamie. Mycologie 33(3): 295–309. 10.7872/crym.v33.iss3.2012.295 [DOI] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Chukeatirote E, Hyde KD. (2014a) Insights into the genus Diaporthe: Phylogenetic species delimitation in the D.eres species complex. Fungal Diversity 67(1): 203–229. 10.1007/s13225-014-0297-2 [DOI] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Hyde KD. (2014b) Species limits in Diaporthe: Molecular re-assessment of D.citri, D.cytosporella, D.foeniculina and D.rudis. Persoonia 32(1): 83–101. 10.3767/003158514X679984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Chukeatirote E, Hyde KD. (2015) The Diaporthesojae species complex, phylogenetic re-assessment of pathogens associated with soybean, cucurbits and other field crops. Fungal Biology 119(5): 383–407. 10.1016/j.funbio.2014.10.009 [DOI] [PubMed] [Google Scholar]
- Vaidya G, Lohmann DJ, Meier R. (2011) SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27(2): 171–180. 10.1111/j.1096-0031.2010.00329.x [DOI] [PubMed] [Google Scholar]
- Van Rensburg JCJ, Lamprecht SC, Groenewald JZ, Castlebury LA, Crous PW. (2006) Characterization of Phomopsis spp. associated with die-back of rooibos (Aspalathuslinearis) in South Africa. Studies in Mycology 55: 65–74. 10.3114/sim.55.1.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanasinghe DN, Phukhamsakda C, Hyde KD, Jeewon R, Lee HB, Gareth Jones EB, Tibpromma S, Tennakoon DS, Dissanayake AJ, Jayasiri SC, Gafforov Y, Camporesi E, Bulgakov TS, Ekanayake AH, Perera RH, Samarakoon MC, Goonasekara ID, Mapook A, Li W-J, Senanayake IC, Li J, Norphanphoun C, Doilom M, Bahkali AH, Xu J, Mortimer PE, Tibell L, Tibell S, Karunarathna SC. (2018) Fungal diversity notes 709–839: Taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Diversity 89(1): 1–236. 10.1007/s13225-018-0395-7 [DOI] [Google Scholar]
- Wang X, Guo Y, Du Y, Yang Z, Huang X, Hong N, Xu W, Wang G. (2021) Characterization of Diaporthe species associated with peach constriction canker, with two novel species from China. MycoKeys 80: 77–90. 10.3897/mycokeys.80.63816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal genes for phylogenetics. In: Gelfand M, Sninsky JI, White TJ. (Eds) PCR protocols: a guide to methods and applications.Academic, New York, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Wrona CJ, Mohankumar V, Schoeman MH, Tan YP, Shivas RG, Jeff-Ego OS, Akinsanmi A. (2020) Phomopsis husk rot of macadamia in Australia and South Africa caused by novel Diaporthe species. Plant Pathology 69(5): 911–921. 10.1111/ppa.13170 [DOI] [Google Scholar]
- Xu TC, Lu YH, Wang JF, Song Z-Q, Hou T-G, Liu S-S, Liu C-S, Wu S-H. (2021) Bioactive secondary metabolites of the genus Diaporthe and anamorph Phomopsis from terrestrial and marine habitats and endophytes: 2010–2019. Microorganisms 9(2): 1–49. 10.3390/microorganisms9020217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Fan XL, Du Z, Tian CM. (2017a) Diaporthejuglandicola sp. nov. (Diaporthales, Ascomycetes) evidenced by morphological characters and phylogenetic analysis. Mycosphere: Journal of Fungal Biology 8(5): 817–826. 10.5943/mycosphere/8/5/3 [DOI] [Google Scholar]
- Yang Q, Fan XL, Du Z, Liang Y-M, Tian C-M. (2017b) Diaporthecamptothecicola sp. nov. on Camptothecaacuminata in China. Mycotaxon 132(3): 591–601. 10.5248/132.591 [DOI] [Google Scholar]
- Yang Q, Fan XL, Du Z, Tian C-M. (2017c) Diaporthe species occurring on Sennabicapsularis in southern China, with descriptions of two new species. Phytotaxa 302(2): 145–155. 10.11646/phytotaxa.302.2.4 [DOI] [Google Scholar]
- Yang Q, Fan X-L, Guarnaccia V, Tian C-M. (2018) High diversity of Diaporthe species associated with dieback diseases in China, with twelve new species described. MycoKeys 39: 97–149. 10.3897/mycokeys.39.26914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Jiang N, Tian C-M. (2020) Three new Diaporthe species from Shaanxi Province, China. MycoKeys 67: 1–18. 10.3897/mycokeys.67.49483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Jiang N, Tian C-M. (2021a) New species and records of Diaporthe from Jiangxi Province, China. MycoKeys 77: 41–64. 10.3897/mycokeys.77.59999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Tang J, Zhou GY. (2021b) Characterization of Diaporthe species on Camelliaoleifera in Hunan Province, with descriptions of two new species. MycoKeys 84: 15–33. 10.3897/mycokeys.84.71701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata M, Palma MA, Aninat MJ, Piontelli E. (2020) Polyphasic studies of new species of Diaporthe from native forest in Chile, with descriptions of Diaporthearaucanorum sp. nov., Diaporthefoikelawen sp. nov. and Diaporthepatagonica sp. nov. International Journal of Systematic and Evolutionary Microbiology 70(5): 3379–3390. 10.1099/ijsem.0.004183 [DOI] [PubMed] [Google Scholar]
- Zhou H, Hou C-L. (2019) Three new species of Diaporthe from China based on morphological characters and DNA sequence data analyses. Phytotaxa 422(2): 23. 10.11646/phytotaxa.422.2.3 [DOI] [Google Scholar]
- Zhu Y-Q, Ma C-Y, Xue H, Piao C-G, Li Y, Jiang N. (2023) Two new species of Diaporthe (Diaporthaceae, Diaporthales) in China. MycoKeys 95: 209–228. 10.3897/mycokeys.95.98969 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic study data
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Christopher Lambert, Lena Schweizer, Blondelle Matio Kemkuignou, Elodie Gisèle M. Anoumedem, Simeon F. Kouam, Yasmina Marin-Felix
Data type
docx
Data Availability Statement
All of the data that support the findings of this study are available in the main text or Supplementary Information.