Abstract
Purpose:
The induction of retinal progenitor cell (RPC) proliferation is a strategy that holds promise for alleviating retinal degeneration. However, the mechanisms that can stimulate RPC proliferation during repair remain unclear. Xenopus tailbud embryos successfully regrow functional eyes within 5 days after ablation, and this process requires increased RPC proliferation. This model facilitates identification of mechanisms that can drive in vivo reparative RPC proliferation. This study assesses the role of the essential H+ pump, V-ATPase, in promoting stem cell proliferation.
Methods:
Pharmacological and molecular loss of function studies were performed to determine the requirement for V-ATPase during embryonic eye regrowth. The resultant eye phenotypes were examined using histology and antibody markers. Misexpression of a yeast H+ pump was used to test whether the requirement for V-ATPase in regrowth is dependent on its H+ pump function.
Results:
V-ATPase inhibition blocked eye regrowth. Regrowth-incompetent eyes resulting from V-ATPase inhibition contained the normal complement of tissues but were much smaller. V-ATPase inhibition caused a significant reduction in reparative RPC proliferation but did not alter differentiation and patterning. Modulation of V-ATPase activity did not affect apoptosis, a process known to be required for eye regrowth. Finally, increasing H+ pump activity was sufficient to induce regrowth.
Conclusions:
V-ATPase is required for eye regrowth. These results reveal a key role for V-ATPase in activating regenerative RPC proliferation and expansion during successful eye regrowth.
Keywords: eye, retina, regeneration, V-ATPase, stem cells, Xenopus
Introduction
Retinal progenitor cells (RPCs) are multipotent stem cells that generate all retinal cell types and the Müller glia of the eye.1 The induction of RPC proliferation is a strategy that holds promise for alleviating retinal degeneration.2,3 However, it has been challenging to identify the molecular mechanisms that are required to stimulate proliferative retinal repair even though vertebrate eye development is well characterized. One reason is that natural stem cell proliferation is typically robust during embryogenesis, whereas studies of retinal regeneration have understandably focused on adult or mature models—environments where RPC proliferation is generally limited. Using a model where robust in vivo RPC proliferation is induced in response to damage can facilitate identification of key mechanisms.
The eye of the clawed frog, Xenopus laevis, is highly homologous to the human eye. Moreover, Xenopus eye development and the corresponding regulatory signaling pathways have been studied comprehensively.4,5 It is also a well-established model for retina, lens, and cornea regeneration.6–8 Xenopus embryos develop externally, and their eye developmental process is similar to humans. These characteristics make the developing Xenopus distinctively suited for eye regeneration studies.
At developmental stage (st.) 27, the Xenopus tailbud embryo eye contains an optic cup and a differentiating lens placode.9 Ablation surgery to remove ∼85% of the embryonic eye tissues (including the lens placode and majority of the optic cup) induced a successful regrowth response that required robust RPC proliferation to rebuild a normal eye within 5 days.10,11 The newly restored eye grown at the injury site is functional and comparable with the control uninjured eye in the same animal.
Using this model, we seek to identify mechanisms that are required for reparative RPC proliferation. Bioelectrical signaling mediated by ion transporters is an understudied yet common early driver of regenerative tissue growth. In particular, the highly conserved multimeric proton (H+) pump, V-ATPase, regulates endogenous membrane voltage and pH in vesicles and plasma membranes of diverse cell types.12 In addition to its essential role in cellular homeostasis, V-ATPase function is required for the maintenance of neural stem cells in mice and Drosophila.13,14 It is also required for appendage regeneration.
In both the Xenopus tadpole tail and adult zebrafish fin, V-ATPase is specifically expressed at the injury site where it acts to initiate regeneration and drives tissue outgrowth events.15,16 Inhibition of V-ATPase function blocked appendage regeneration, whereas ectopic activation of an H+ pump promoted regeneration during nonregenerative states. We asked whether regenerative mechanisms such as V-ATPase can be used reiteratively in diverse tissues.
Here, we show that loss of V-ATPase activity blocked reparative RPC proliferation and resulted in regrowth-incompetent small eyes. Furthermore, misexpression of an H+ pump is sufficient to restore eye regrowth during V-ATPase inhibition. Together, our results show that V-ATPase is required for eye regrowth.
Methods
Embryo culture and surgery
X. laevis were cultured through approved protocols and guidelines (UNLV Institutional Animal Care and Use Committee). Embryos were generated through in vitro fertilization and raised in 0.1 × Marc's Modified Ringer (MMR: 1 mM MgSO4, 2.0 mM KCl, 2 mM CaCl2, 0.1 M NaCl, 5 mM HEPES, pH 7.8) medium.17 The eye ablation surgery and the regrowth assay were performed as described.10,11
At developmental st. 27,9 embryos were anesthetized with tricaine methanesulfonate (Millipore Sigma). Fine surgical forceps (Dumont No. 5) were used to make an incision in the epidermis adjacent to the protruding eyecup and overlying lens placode. The cut was continued around the outline of the eye to enable ablation of the eye tissues. After surgery, embryos were transferred into 0.1 × MMR, washed with medium, allowed to recover, and then cultured at 22°C for up to 5 days.
Assessment of eye regrowth
The regrowth of the operated eyes was compared with either uninjured contralateral eyes or vehicle-treated eyes, and scored using the Regrowth Index (RI) as previously described.10 Four phenotypic categories were used to assess regrowth: full, partial, weak, and none, and a RI score was calculated. Full regrowth indicated that the eye was morphologically the same as a control uninjured eye, whereas no regrowth indicated a missing eye.
The RI ranged from 0 to 300, where 0 indicated no eye regrowth of all embryos under a given condition, and 300 indicated that all embryos in the group showed full regrowth. Raw data from scoring were used to compare the experimental and control conditions. The area of 5-day regrown eyes was calculated by tracing the area of visible eye tissues on a lateral view image of each tadpole. The unoperated eye of the same tadpole was used as control.
Pharmacological exposure and messenger RNA expression
For V-ATPase inhibition, embryos were treated with 20 nM of Concanamycin A (CAS No. 80890-47-7; Cayman Chemical) in 0.1% dimethyl sulfoxide (DMSO) for 24 h after eye surgery. DMSO (0.1%) was used as vehicle control (n > 30). For apoptosis inhibition, 28 μM of M50054 was used (CAS No. 54135-60-3; Millipore; n > 46).
YCHE78 and PMA1.2 messenger RNA (mRNA) were transcribed in vitro from linearized plasmid constructs using the mMESSAGE Transcription Kit (Thermofisher). Green fluorescent protein (GFP) mRNA was included as a lineage label. mRNA was injected into 1 dorsal blastomere of a 4-cell embryo using a microinjector (Harvard Apparatus) to target 1 side of the embryo. Embryos with GFP fluorescence in the eye region were selected for eye ablation.
The titrated dosages used for injections at the 4-cell stage were as follows: 2.38 ng/embryo for YCHE78 mRNA and 7.74 ng/embryo for PMA1.2 mRNA. These dosages were the highest amount of mRNA that can be injected into tadpoles while allowing for normal embryo development and sensitivity to regeneration events (n > 58).
Embryo sectioning, histology, and immunofluorescence microscopy
MEMFA (100 mM MOPS with a pH 7.4, 2 mM EGTA, 1 mM MgSO4, 3.7% formaldehyde)17 was used to fix samples overnight at 4°C. For histology, paraffin-embedded tissues were sectioned at 10 μm thickness using a Tissue-Tek Accu-Cut Rotary Microtome (n > 6). Alternatively, embryos and tadpoles were embedded in 4%–6% agarose and sectioned into 60 μm slices using a Leica vt1000s vibratome.18
Eye sections were stained with primary antibodies, including Xen1 [pan-neural antibody, clone 3B1, 1:100 dilution, RRID: AB_531871; Developmental Studies Hybridoma Bank (DSHB)], mouse anti-Islet-1 (retinal ganglion cells and the inner nuclear cell layer, clone 40.2D6, 1:200 dilution, RRID: AB_528315; DSHB), mouse anti-Rhodopsin (rod photoreceptor cells, clone 4D2, 1:200 dilution, RRID: AB_10807045; EMD Millipore), rabbit anti-phospho Histone H3 (H3P; mitosis marker, 1:500 dilution, RRID: AB_310177; EMD Millipore), rabbit anti-activated Caspase-3 (C3) antibody (activated C3, 1:300 dilution, RRID: AB_2341188; Cell Signaling), anti-RPE65 antibody (retinal pigment epithelium, 1:500 dilution, RRID: AB_2181003; ThermoFisher Scientific), and anti-PMA1.2 (yeast H+ pump, 1:200 dilution, RRID: AB_1109897; Novus Biologicals). Alexa Fluor conjugated secondary antibodies were used at a 1:1,000 dilution (ThermoFisher). DAPI (4′,6-diamidino-2-phenylindole, dihydrochloride; Sigma-Aldrich) was used to stain DNA (n > 16).
Microscopy/image processing and analysis
Images of whole animals were obtained using a Zeiss Discovery V20 stereomicroscope with an AxioCam MRc camera. Confocal imaging was performed at the UNLV Confocal and Biological Imaging Core using an inverted Nikon A1R-Si confocal laser scanning microscope. ZEN Image Analysis software and/or the open-source FIJI imaging software19 were used to analyze and/or process all acquired images.
Quantification of rod photoreceptor cells and mitotic cells
Quantification of rod photoreceptor cell numbers was performed using agarose sections labeled with an anti-Rhodopsin antibody and DAPI. The number of rod photoreceptor cells was counted per 60 μm section (n > 10 per condition and timepoint). Rod photoreceptor cells expression pattern was measured in pixels along a line drawn across the outer nuclear layer and compared with the overall circumference of the retinal layer from one end of the ciliary marginal zone to the other end of the ciliary marginal zone (n > 5 per timepoint). The ratio of Rhodopsin expression in the retinal layer over the corresponding retinal layer circumference measurement was calculated.
Quantification of mitotic cell numbers was performed using sections labeled with an anti-H3P antibody and the pan-neural Xen1 antibody. H3P-positive (H3P+) cells were counted per 60 μm section (n > 10 tadpoles per condition and timepoint). For each eye, the total area was calculated by measuring the surface area for each eye section and summing the measurements. The total mitotic counts were then normalized for the total area.
Statistical analysis
All statistical analysis was performed using GraphPad Prism (GraphPad Software, San Diego, CA). A comparison of 2 conditions was made using the Mann−Whitney U test for ordinal data with tied ranks, using normal approximation for large sample sizes. Multiple conditions were compared using a Kruskal−Wallis test, with Dunn's Q corrected for tied ranks. All other experiments were analyzed using a Student's t-test.
Results
V-ATPase is required for eye regrowth
To determine the role of V-ATPase in eye regrowth, we first sought to inhibit V-ATPase function. Concanamycin A is a highly specific and established V-ATPase inhibitor (IC50 = 10 nM) that depolarizes tissues,20 and blocks both tadpole tail and zebrafish fin regeneration.15,16 First, we titrated Concanamycin A to identify dosages that allowed normal development and embryo viability. At concentrations of ≤20 nM Concanamycin A, embryos developed normally with no observable abnormalities.
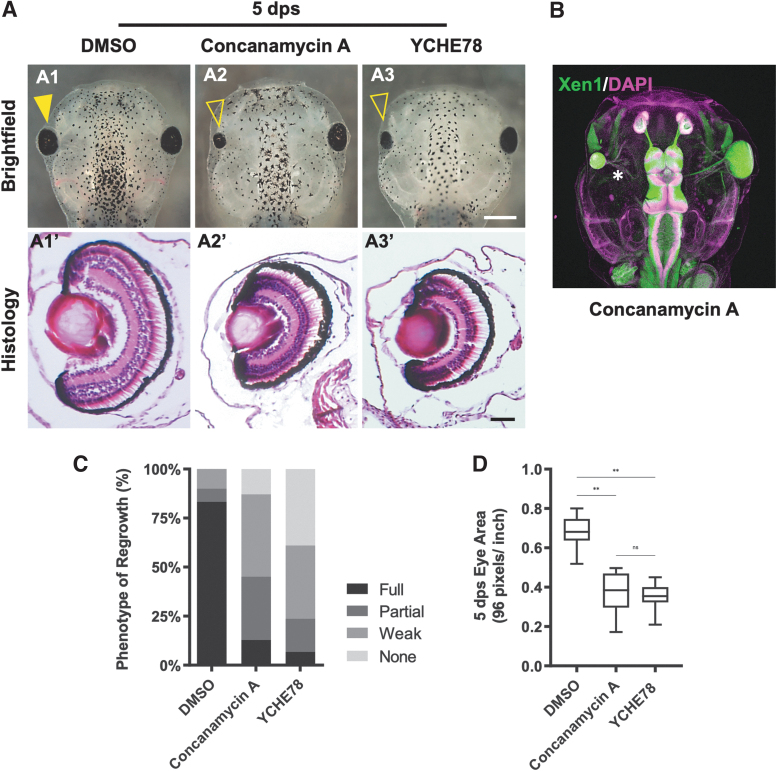
We then performed the eye regrowth assay to assess the effects of pharmacological V-ATPase inhibition. After eye ablation surgery at st. 27, embryos were treated with Concanamycin A for 24 h, and regrowth was examined at 5 days post surgery (dps). Our data showed that Concanamycin treatment for the first 24h gave equivalent inhibition of regrowth as for longer treatment periods. Concanamycin A strongly blocked eye regrowth and resulted in regrowth-incompetent small eyes as compared with vehicle control (Fig. 1A, C, D, P < 0.01). Although V-ATPase inhibition resulted in a small eye, the eye contained an optic nerve that innervated the brain (Fig. 1B, asterisk) similar to the uninjured right eye (Fig. 1B).
FIG. 1.
V-ATPase is required for eye regrowth. (A) (A1–A3) Representative brightfield images of eye regrowth phenotypes for control (DMSO; n = 30), Concanamycin A treatment (n = 31), or YCHE78 expression (n = 59; up = anterior). Closed arrowhead indicates regrowth, whereas open arrowhead indicates blocked regrowth. (A1′–A3′) Show corresponding hematoxylin and eosin-stained eye sections (n = 19, n = 16, and n = 7, respectively; up = dorsal). (B) V-ATPase−inhibited regrowth-incompetent eye at 5 dps is connected by an optic nerve (starred) to the brain. DAPI shows nucleus (magenta) and Xen1 stains neural tissues (green); up = anterior. n = 5. (C) The RI shows the quality of regrowth based on 4 phenotypic categories: full, partial, weak, and no eyes. (D) Area measurements at 5 dps show smaller eyes resulting from Concanamycin A treatment (n = 12) or YCHE78 expression (n = 16) when compared with control DMSO regrown eyes (n = 12). **P < 0.01. Data are means ± SD. Scale bars: (A1–A3) 500 μm, (A1′–A3′) 50 μm, and (B) 200 μm. DAPI, 4′,6-diamidino-2-phenylindole; DMSO, dimethyl sulfoxide; dps, days postsurgery; ns, not significant; RI, Regrowth Index; SD, standard deviation.
To assess the quality of eye regrowth, we calculated the RI (described in the Methods section) for each condition. Control regrown eyes showed an RI of 273 with 83.3% of eyes fully regrown. In contrast, inhibitor-treated eyes showed an RI of 145 with only 12.9% of eyes fully regrown (P < 0.01) (Fig. 1C). Measurements of the eye area at 5 dps indicated that Concanamycin A-treated regrowth-inhibited eyes were on average 55.1% smaller when compared with DMSO-treated regrown eyes (P < 0.01) (Fig. 1D). Together, our data demonstrated that Concanamycin A inhibited eye regrowth.
To confirm the role of V-ATPase in eye regrowth, we sought to molecularly reduce its function by ectopically expressing a well-characterized dominant negative peptide, YCHE78, through mRNA injection targeted to the eye region. The V-ATPase V1 catalytic domain subunit E is required for V-ATPase function. YCHE78 is a truncated subunit E peptide that acts as a dominant negative to inhibit V-ATPase activity when expressed.21 YCHE78 misexpression inhibited tadpole tail regeneration without affecting overall development.15
Consistent with previous studies, we found that injection of 2.4 ng of YCHE78 mRNA into 1 dorsal cell of the 4-cell embryo did not alter normal development. To identify cells expressing YCHE78, YCHE78 mRNA was coinjected with GFP mRNA to facilitate selection of st. 27 embryos that showed GFP expression (and by inference YCHE78) in the eye region. Eye ablation surgeries were performed on the selected embryos, and the embryos were assessed for regrowth at 5 dps.
Consistent with pharmacological V-ATPase inhibition phenotypes, embryos expressing YCHE78 in the eye region showed strong regrowth inhibition with eyes that were on average 52.2% smaller in size as compared with control regrown eyes (RI = 95, P < 0.01; Fig. 1A, A1–A3, C, D). A significant percentage of embryos were missing eyes at the surgery site—likely indicative of a complete absence of regrowth. Histological sections of the regrowth-incompetent small eyes indicated that normal eye morphology was maintained (Fig. 1A, A3′), a similar finding to the Concanamycin A-treated small eyes. Together, the pharmacological and molecular inhibition data revealed an essential role of V-ATPase function in embryonic eye regrowth.
Effects of V-ATPase inhibition on eye regrowth
The regrowth-inhibited small eyes caused by V-ATPase inhibition could be due to defects in eye progenitor cell proliferation and/or defects in cellular differentiation and patterning. We assessed the effects of V-ATPase inhibition by examining the resultant small eyes using histological analyses, cell proliferation assay, and molecular marker characterization of retinal layer patterning.
To characterize the regrowth defects caused by V-ATPase inhibition, we performed histology using hematoxylin and eosin (H&E) staining to examine potential defects in the Concanamycin A-treated or YCHE-expressing regrowth-incompetent eyes at 5 dps. At this timepoint, the tadpole eye is mature and has the structures found in an adult frog eye. Transverse H&E sections through control regrown eyes showed the normal complement of eye tissues, including the lens, cornea, retina, and pigmented epithelium (Fig. 1A, A1′).
Although Concanamycin A-treated regrowth-incompetent eyes or YCHE78 regrowth-inhibited eyes were significantly smaller, both groups of eyes contained similar structures to those found in the control group (Fig. 1A, A2′, A3′). Together, our data indicated that V-ATPase does not act to regulate eye differentiation and patterning during regrowth.
In Xenopus, retinogenesis is completed over 2 days (st. 24–40).22 It was previously shown that chemically blocking cell cycle progression in RPCs resulted in small eyes with mostly normal retinal cell layers,23 indicating that retinal differentiation and proliferation can be separated, and that retinogenesis can be initiated independent of eye field size. Our previous work demonstrated that eye regrowth required an increase in cell proliferation coupled with a concurrent postponement in retinal differentiation as compared with normal eye development.24
During regrowth, retinogenesis is delayed by 1 day to allow for RPC proliferation, and yet was still completed within a 2-day period once it was initiated. Our histological data indicated that overall retinal differentiation appeared to be unaffected by V-ATPase inhibition (Fig. 1A, A1′, A3′). We asked whether V-ATPase inhibition of regrowth could alter the timing of retinal differentiation after eye ablation.
We examined this process at 3 timepoints (1, 2, and 3 dps) using antibody markers.24 First, we compared eye development in control (vehicle-treated) and V-ATPase−inhibited (Concanamycin A) embryos without performing eye ablations, and observed the same spatiotemporal patterns (data not shown). This result confirmed that the reduction of V-ATPase activity due to 20 nM Concanamycin A exposure allowed for normal overall development.
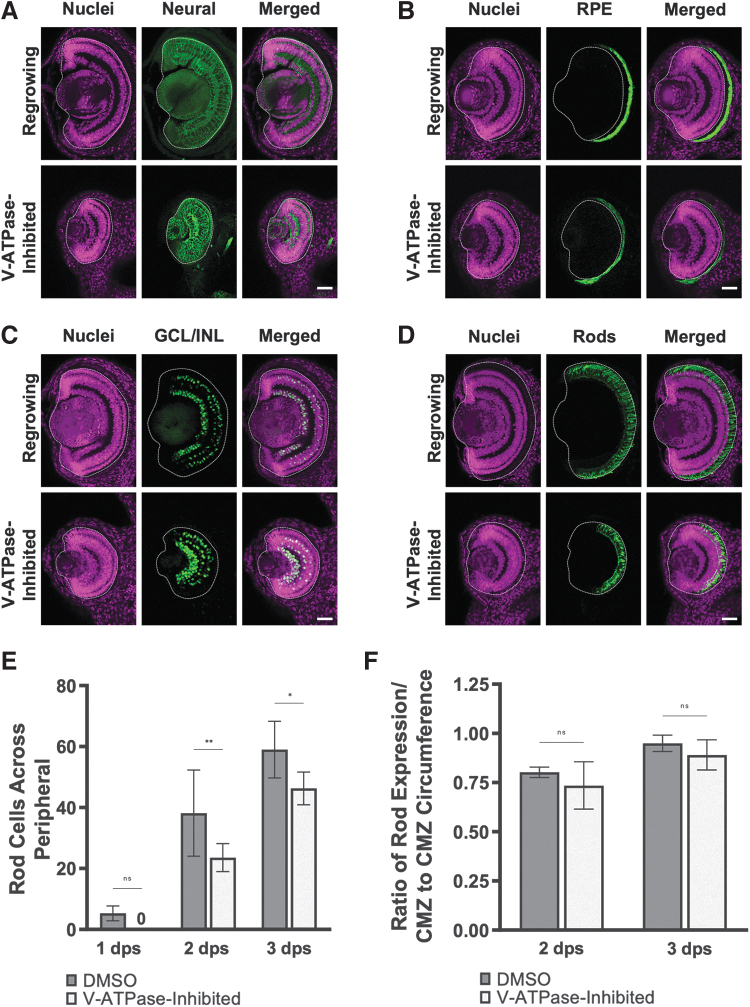
The Xen1 antibody recognizes neural tissues and enables the assessment of retinal layer formation.25 In the control (DMSO-treated) regrowing eye, the initial retinal layering was visible by 1 dps and completed by 3 dps. The V-ATPase−inhibited nonregrowing eye, although smaller in size, showed a similar pattern (Fig. 2A). Using established markers for retinal pigmented epithelium (Fig. 2B), retinal ganglion cells (Fig. 2C), and rod photoreceptors (Fig. 2D), we observed that the temporal differentiation patterns in the V-ATPase−inhibited regrowth-incompetent eyes were indistinguishable from control regrowing eyes.
FIG. 2.
V-ATPase−inhibited nonregrowing eyes showed normal structure and patterning. Transverse eye sections at 3 dps. (A) Pan-neural marker Xen1 (green) outlines neural patterning. (B) Anti-RPE65 antibody (green) shows the pigmented epithelium. (C) Anti-Islet-1 antibody (green) shows retinal ganglion cells and subsets of amacrine, bipolar, and horizontal cells. (D) Anti-Rhodopsin antibody (green) shows rod photoreceptors. (A–D) White dashed lines outline each eye. Cell nuclei (DAPI) are shown in magenta. n > 10 per timepoint and condition. Up = dorsal. Scale bars = 50 μm. (E) Quantification of rod photoreceptor cells in control or V-ATPase−inhibited eyes. The number of rod photoreceptor cells was counted across the retinal periphery. *P < 0.05, **P < 0.01 (n > 10 per timepoint and condition). (F) The ratio of Rhodopsin expression over the retinal layer circumference measurements is shown (n > 5 per timepoint and condition). Data are means ± SD. CMZ, ciliary marginal zone.
We further examined the rod photoreceptor differentiation process. Consistent with the small eyes that resulted from V-ATPase inhibition of regrowth, the number of rod photoreceptors was less than their control counterparts (Fig. 2E). However, when the photoreceptor counts were normalized to the corresponding periphery length for each timepoint, the ratio was similar between the V-ATPase−inhibited eye and the control regrowing eye (Fig. 2F). Together, our data indicated that retinal formation was unaffected during regrowth inhibition, and that V-ATPase likely does not play a main role in regulating differentiation of Xenopus eye tissues.
If V-ATPase acts to regulate cell proliferation during regrowth, then this function would be consistent with our observations that differentiation remains unaffected when V-ATPase activity is blocked. We hypothesized that V-ATPase regulates stem cell proliferation during eye regrowth. To test this hypothesis, we assayed cell cycle progression in RPCs during the first 24 h of regrowth using an established mitotic marker, the anti-H3P antibody.10,26 We counted the number of H3P+ cells in the eye region and normalized the counts for area.
Previously we showed that proliferation was highest during the first day of regrowth.11 Thus 4 early timepoints were examined: 6, 12, 18, and 24 h postsurgery (hps) (Fig. 3). At 6 hps, H3P+ cells were found in control (DMSO-treated) regrowing eyes (5.3 mitoses per 10−4 μm2 area) but reduced in V-ATPase−inhibited eyes (2.7 mitoses per 10−4 μm2 area, P < 0.01). At 12 hps, mitotic activity in control regrowing eyes was the highest of the timepoints assayed (8.5 mitoses per 10−4 μm2 area).
FIG. 3.
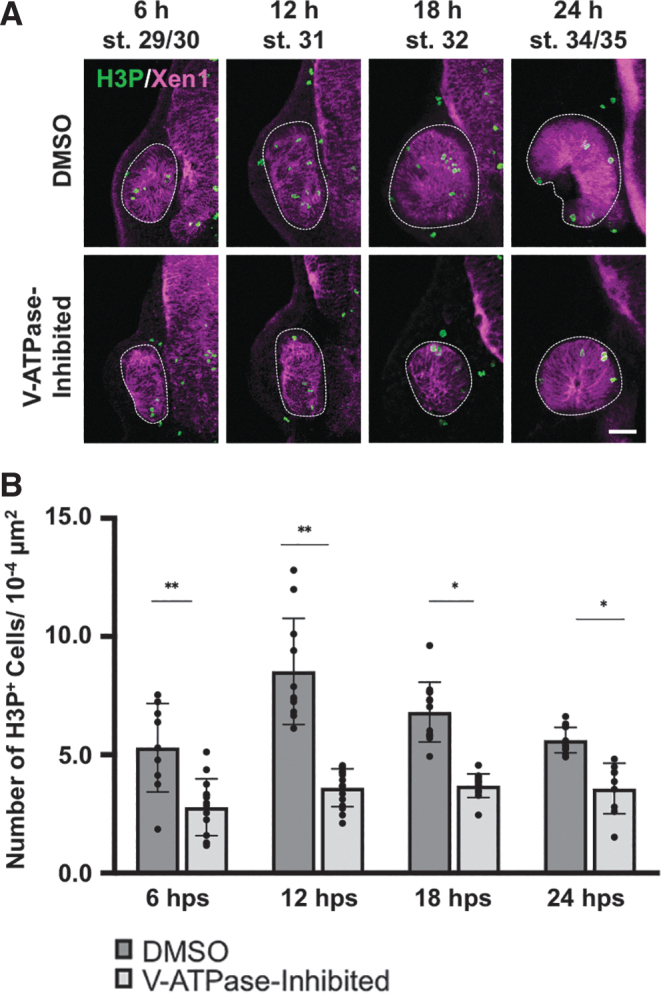
V-ATPase regulates cell proliferation in regrowing eyes. (A) Representative fluorescence images of eye sections stained with anti-H3P antibody to identify mitotic cells (green). Magenta color indicates neural tissues (Xen1). White dashed lines outline each eye. Sample size ranges from n = 9 to 12 per condition and timepoint. Up = dorsal. Scale bar = 50 μm. (B) Quantification of mitoses in control DMSO regrowing and V-ATPase−inhibited eyes. Graph shows the number of H3P-positive cells. *P < 0.05, **P < 0.01 (n = ≥9 per timepoint and condition). Data are means ± SD.
In contrast, V-ATPase−inhibited eyes showed reduced numbers of H3P+ cells (3.6 mitoses per 10−4 μm2 area, P < 0.01). At 18 hps, control regrowing eyes also contained more H3P+ cells (6.8 mitoses per 10−4 μm2 area) than V-ATPase−inhibited eyes (3.7 mitoses per 10−4 μm2 area, P < 0.05). At 24 hps, V-ATPase−inhibited eyes showed a 63% reduction in the number of mitoses (3.5 mitoses per 10−4 μm2 area) as compared with control regrowing eyes (5.6 mitoses per 10−4 μm2 area, P < 0.05). At each timepoint, the number of mitotic cells in V-ATPase−inhibited eyes was significantly and consistently lower than time-matched control eyes. Together, our data demonstrated that inhibition of V-ATPase acts to significantly reduce the levels of RPC proliferation during eye regrowth.
Increased H+ pump activity is insufficient to rescue regrowth failure due to apoptotic inhibition
Our previous studies showed that apoptosis is required for eye regrowth, and that apoptosis is a regrowth-specific mechanism.11,24 We asked whether V-ATPase and apoptosis can interact to regulate eye regrowth. To test this hypothesis, we first examined apoptotic activity during V-ATPase inhibition using Concanamycin A treatment. During the first 24 h of regrowth, apoptosis is upregulated in the regrowing eye24 (Fig. 4A).
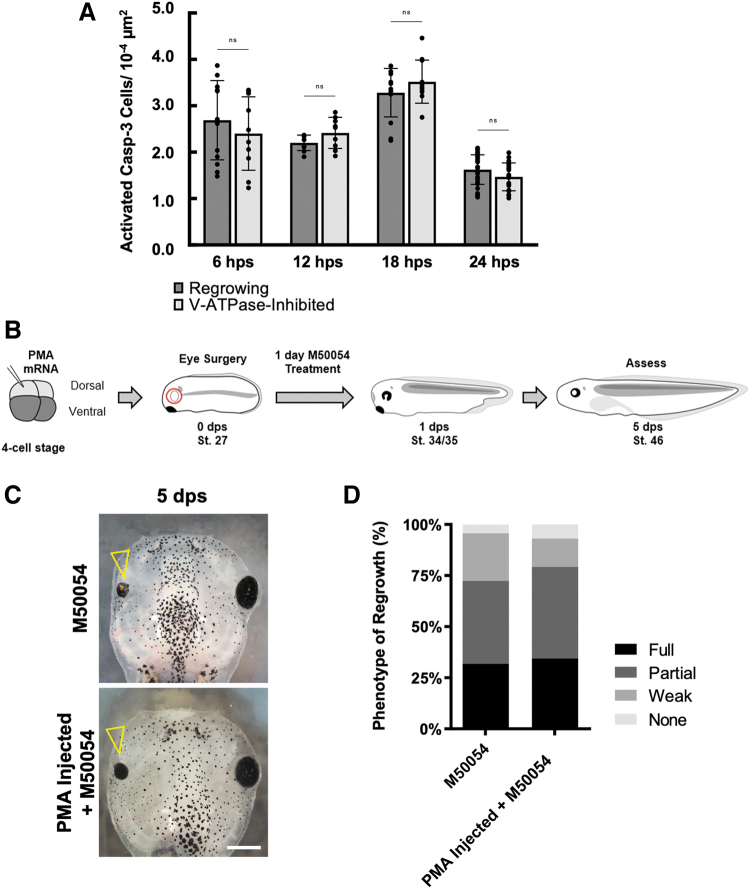
FIG. 4.
V-ATPase modulation does not alter apoptotic regulation of regrowth. (A) Quantification of apoptotic cell numbers during regrowth using the activated Caspase-3 antibody. P > 0.05 (n = ≥9 per timepoint). Data are means ± SEM. (B) Experimental strategy for rescue of the apoptotic inhibition phenotype by expression of the yeast H+ pump, PMA1.2. (C) Representative brightfield images of the outcomes from (B). Open arrowheads indicate regrowth-incompetent eyes. Scale bar = 500 μm. (D) Eyes were scored for the RI and quantified for regrowth quality (n = 47 M50054, n = 58 PMA injection+M50054). SEM, standard error of the mean.
We assessed apoptosis using an antibody that specifically detected activated C3, an effector caspase that induces apoptosis,11,27,28 and examined activated C3 levels at 4 different timepoints: 6, 12, 18, and 24 hps. For each timepoint, the number of activated C3-positive cells was similar between control regrowing eyes and V-ATPase−inhibited regrowth-incompetent eyes (P > 0.8 for each timepoint) (Fig. 4A), suggesting that V-ATPase does not act upstream of apoptosis.
As V-ATPase is a multimeric complex, increasing its activity by simultaneous overexpression of multiple subunits is challenging. We tested if increasing H+ pump activity through misexpression of a single peptide non-V-ATPase type H+ pump to mimic V-ATPase activity can restore eye regrowth during apoptotic inhibition (Fig. 4B). PMA1.2 is a single peptide yeast H+ pump that, when overexpressed, is sufficient to substitute for the function of V-ATPase in driving tadpole tail regeneration.15,29 M50054 is a well-characterized inhibitor of C3 activity and has been shown to block tissue regeneration.11,30
Consistent with previous results, M50054 treatment after eye ablation greatly reduced regrowth11 (RI = 198) (Fig. 4C, D). Overexpression of PMA1.2 in the eye region failed to restore regrowth in embryos treated with M50054 after eye ablation (RI = 207 P > 0.05) (Fig. 4C, D). This result showed that an increase in H+ pump activity is insufficient to rescue eye regrowth when apoptosis is blocked. Together, our data indicate that V-ATPase and apoptosis likely act in separate pathways to regulate eye regrowth.
The H+ pump function of V-ATPase is sufficient to induce regrowth
Based on previous studies, the requirement of V-ATPase in eye regrowth is likely due to its H+ pump function. However, another possibility is that the accessory subunits of the multimeric V-ATPase complex may interact with other proteins and signaling pathways. To test if the H+ pump function is sufficient to induce regrowth, we asked if overexpression of a non-V-ATPase H+ pump could restore eye growth in the presence of V-ATPase inhibition. As a single peptide yeast H+ pump, PMA1.2 does not interact with the V-ATPase inhibitor Concanamycin A. Control embryos treated with Concanamycin A after eye ablation surgery showed poor regrowth and small eye sizes (RI = 134) (Fig. 5A, B).
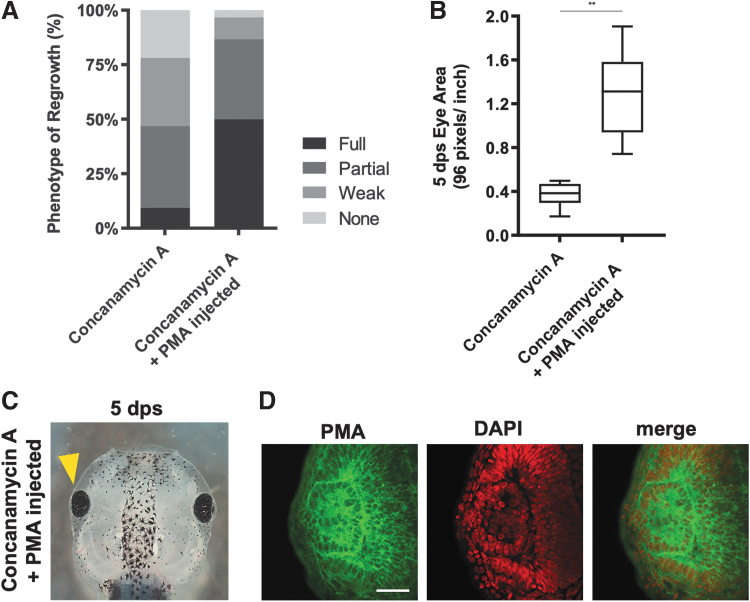
FIG. 5.
Increased H+ pump activity restored eye regrowth. (A) Eyes were scored for the RI and quantified for regrowth quality. The data show that PMA1.2 expression is sufficient to rescue eye regrowth during V-ATPase inhibition. (B) Measurements of 5 dps eye size show PMA1.2 expression restored the V-ATPase−inhibited small eye phenotype (n = 22) to normal as compared with regrowth-incompetent eyes treated with Concanamycin A only (n = 12). **P < 0.01. Data are means ± SD. (C) Restoration of eye size by PMA1.2 expression (yellow arrowhead; n = 30). (D) An eye section showing membrane expression of PMA1.2 (green). Nuclei are shown by DAPI (red). n = 5. Scale bar = 50 μm.
In contrast, overexpression of PMA1.2 was sufficient to induce eye regrowth in embryos treated with Concanamycin A after surgery (RI = 233, P < 0.01) (Fig. 5C). Measurements of regrown eyes expressing PMA1.2 showed that the average size of this group was >3 times the size of Concanamycin A-treated regrowth-incompetent eyes (P < 0.01) (Fig. 5B). There was a significant increase in the full eye phenotype in PMA1.2-expressing Concanamycin A-treated tadpoles (50%) as compared with control Concanamycin A-treated tadpoles (9.3%) (Fig. 5A, C).
Transverse sections through PMA1.2-expressing V-ATPase−inhibited regrown eyes showed that overall eye structure is normal. To confirm that PMA1.2 is expressed in the eye region, we used a PMA-specific antibody and detected its expression in the plasma membrane of retinal cells (Fig. 5D). Together, our data showed that expression of an H+ pump rescued the Concanamycin A phenotype and significantly improved eye regrowth. Thus, the H+ pump function of V-ATPase is needed to induce eye regrowth.
Discussion
In this study, we showed that V-ATPase induces reparative RPC proliferation during eye regrowth while maintaining proper patterning and differentiation. During regrowth, V-ATPase acts to extend the endogenous developmental RPC multipotency period to generate the needed cell numbers for the full restoration of eye. This finding is consistent with previous studies showing that V-ATPase can promote neural stem cell proliferation.13,14,31
The required role of V-ATPase in Xenopus tadpole tail regeneration is well characterized.15 Similarly, our findings demonstrate that V-ATPase is required in eye regrowth, and that inhibition of V-ATPase greatly reduced regenerative proliferation. Thus V-ATPase appears to be a conserved mechanism for tissue repair as it is required for both appendage regeneration and eye regrowth. Further studies are needed to identify the similarities and differences of V-ATPase in the 2 tissues.
V-ATPase is widely expressed on cellular membranes as it is an essential ion pump with homeostatic functions.32 The pharmacological and molecular reagents that were used to inhibit V-ATPase in this study are well established and confirmed in their high specificity.15,16,20
Moreover, the inhibitor effect is dosage dependent. During appendage regeneration, V-ATPase is highly upregulated.15,16 To further investigate the role of V-ATPase, it would be informative to perform transcriptomics profiling of eye regrowth to characterize regrowth-dependent gene expression changes and identify potential increased expression of V-ATPase subunits.
The function of V-ATPase in the eye is beginning to be explored.33 How V-ATPase regulates RPC proliferation is an important area of investigation. One hypothesis is that loss of V-ATPase restricts the number of RPCs. Alternatively, V-ATPase may act to regulate RPC cell cycle entry and/or progression. A recent study reported a role for a V-ATPase subunit in zebrafish eye development.34 Disruption of V-ATPase is also implicated in eye diseases such as age-related macular degeneration.33,34 In addition, ocular channelopathies can lead to vision loss.35 Thus, understanding the pathways regulated by V-ATPase in the eye is important.
Our work here showed that V-ATPase is unlikely to interact with apoptosis—a process that is separately required for eye regrowth. Notably, V-ATPase has been shown to regulate the activities of key signaling transduction pathways including Notch and Wnt.36–39 For example, a reduction in V-ATPase activity decreased Notch signaling.38 As Notch and Wnt pathways are involved in regulating regeneration of several tissue types, it would be informative to examine whether a similar mechanism is utilized for eye regrowth.
In this work, we have largely focused on retinal regrowth. It should be noted that the lens, retinal pigmented epithelium, and other eye structures are also restored after embryonic eye ablation. Due to its ease of accessibility and well-understood developmental processes, the embryonic regrowth model holds considerable promise for systematically and efficiently identifying mechanisms that successfully stimulate multitissue repair in the eye. Moreover, the potential of ocular repair through pharmacological modulation of ion fluxes represents a worthwhile direction for future investigation.
Acknowledgments
We thank members of Tseng lab and Dr. Hong Sun for their helpful discussions. Confocal imaging was performed at the UNLV Confocal and Biological Imaging Core, with assistance of Sophie Choe. Several antibodies used in this study were obtained from the DSHB, a resource created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology (Iowa City, IA).
Authors' Contributions
C.X.K. and I.N. performed investigation, analysis, visualization, and writing. K.A-S.T. contributed to conceptualization, supervision, and writing.
Author Disclosure Statement
No conflicts of interest.
Funding Information
This study was supported by grants from the National Science Foundation (1726925), National Institutes of Health (R16GM146672), University of Nevada, Las Vegas (Doctoral Graduate Research Assistantship, K.A-S.T.; and The President's UNLV Foundation Graduate Research Fellowship, C.X.K.), and research fellowships from the Nevada NASA Space Grant Consortium (80NSSC20M0043, C.X.K. and I.N.) and NSF (IIA-1301726, I.N.).
References
- 1. Wetts R, Fraser SE. Multipotent precursors can give rise to all major cell types of the frog retina. Science 1988;239:1142–1145. [DOI] [PubMed] [Google Scholar]
- 2. Oswald J, Baranov P. Regenerative medicine in the retina: From stem cells to cell replacement therapy. Ther Adv Ophthalmol 2018;10:2515841418774433; doi: 10.1177/2515841418774433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang Y, Tang Z, Gu P.. Stem/progenitor cell-based transplantation for retinal degeneration: A review of clinical trials. Cell Death Dis 2020;11:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zuber ME. Eye field specification in Xenopus laevis. Curr Top Dev Biol 2010;93:29–60. [DOI] [PubMed] [Google Scholar]
- 5. Chow RL, Lang RA. Early eye development in vertebrates. 2001;17:255–296; doi: 10.1146/annurev.cellbio.17.1.255 [DOI] [PubMed] [Google Scholar]
- 6. Vergara MN, Del Rio-Tsonis K.. Retinal regeneration in the Xenopus laevis tadpole: A new model system. Mol Vis 2009;15:1000–1013. [PMC free article] [PubMed] [Google Scholar]
- 7. Viczian AS. Advances in retinal stem cell biology. J Ophthalmic Vis Res 2013;8:147–159. [PMC free article] [PubMed] [Google Scholar]
- 8. Beck CW. Studying Regeneration in Xenopus. In: Retinal Development. (Gautier J-C. ed.) Humana Press: Totowa, NJ, 2012; pp. 525–539. [DOI] [PubMed] [Google Scholar]
- 9. Nieuwkoop PD, Faber J. Normal Table of. Xenopus laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilized Egg Till the End of Metamorphosis. 2nd ed. Garland Publishing: New York; 1994. [Google Scholar]
- 10. Kha CX, Guerin DJ, Tseng KA-S. Studying In Vivo Retinal Progenitor Cell Proliferation in Xenopus laevis. In: Retinal Development: Methods in Molecular Biology. Methods in Molecular Biology. (Mao C-A. ed.) Humana: New York, NY, 2020; pp. 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kha CX, Son PH, Lauper J, et al. A model for investigating developmental eye repair in Xenopus laevis. Exp Eye Res 2018;169:38–47. [DOI] [PubMed] [Google Scholar]
- 12. Gluck S. V-ATPases of the plasma membrane. J Exp Biol 1992;172:29–37. [DOI] [PubMed] [Google Scholar]
- 13. Wissel S, Harzer H, Bonnay F, et al. Time-resolved transcriptomics in neural stem cells identifies a v-ATPase/Notch regulatory loop. J Cell Biol 2018;217:3285–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lange C, Prenninger S, Knuckles P, et al. The H(+) vacuolar ATPase maintains neural stem cells in the developing mouse cortex. Stem Cells Dev 2011;20:843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Adams DS, Masi A, Levin M.. H+ pump-dependent changes in membrane voltage are an early mechanism necessary and sufficient to induce Xenopus tail regeneration. Development 2007;134:1323–1335. [DOI] [PubMed] [Google Scholar]
- 16. Monteiro J, Aires R, Becker JD, et al. V-ATPase proton pumping activity is required for adult zebrafish appendage regeneration. PLoS One 2014;9:e92594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sive HL, Grainger RM, Harland RM. Early Development of Xenopus laevis: A Laboratory Manual. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY; 2010. [Google Scholar]
- 18. Blackiston D, Vandenberg LN, Levin M.. High-throughput Xenopus laevis immunohistochemistry using agarose sections. Cold Spring Harb Protoc 2010;2010:pdb.prot5532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: An open-source platform for biological-image analysis. Nature Methods 2012;9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huss M, Ingenhorst G, König S, et al. Concanamycin A, the specific inhibitor of V-ATPases, binds to the V(o) subunit c. J Biol Chem 2002;277:40544–40548. [DOI] [PubMed] [Google Scholar]
- 21. Lu M, Vergara S, Zhang L, et al. The amino-terminal domain of the E subunit of vacuolar H(+)-ATPase (V-ATPase) interacts with the H subunit and is required for V-ATPase function. J Biol Chem 2002;277:38409–38415. [DOI] [PubMed] [Google Scholar]
- 22. Wong LL, Rapaport DH. Defining retinal progenitor cell competence in Xenopus laevis by clonal analysis. Development 2009;136:1707–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harris WA, Hartenstein V.. Neuronal determination without cell division in Xenopus embryos. Neuron 1991;6:499–515. [DOI] [PubMed] [Google Scholar]
- 24. Kha CX, Guerin DJ, Tseng KA-S. Using the Xenopus developmental eye regrowth system to distinguish the role of developmental versus regenerative mechanisms. Front Physiol 2019;10:502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ruiz i Altaba A. Planar and vertical signals in the induction and patterning of the Xenopus nervous-system. Development 1992;116:67–80. [DOI] [PubMed] [Google Scholar]
- 26. Smith JC, Saka Y.. Spatial and temporal patterns of cell division during early Xenopus embryogenesis. Dev Biol 2001;229:307–318. [DOI] [PubMed] [Google Scholar]
- 27. Tseng A-S, Levin M.. Tail regeneration in Xenopus laevis as a model for understanding tissue repair. J Dent Res 2008;87:806–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sîrbulescu RF, Zupanc GKH. Dynamics of caspase-3-mediated apoptosis during spinal cord regeneration in the teleost fish, Apteronotus leptorhynchus. Brain Res 2009;1304:14–25. [DOI] [PubMed] [Google Scholar]
- 29. Masuda CA, Montero-Lomelí M.. An NH2-terminal deleted plasma membrane H+-ATPase is a dominant negative mutant and is sequestered in endoplasmic reticulum derived structures. Biochem Cell Biol 2000;78:51–58. [PubMed] [Google Scholar]
- 30. Tseng A-S, Adams DS, Qiu D, et al. Apoptosis is required during early stages of tail regeneration in Xenopus laevis. Dev Biol 2007;301:62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li L, Yang S, Zhang Y, et al. ATP6V1H regulates the growth and differentiation of bone marrow stromal cells. Biochem Biophys Res Commun 2018;502:84–90. [DOI] [PubMed] [Google Scholar]
- 32. Pamarthy S, Kulshrestha A, Katara GK, et al. The curious case of vacuolar ATPase: Regulation of signaling pathways. Mol Cancer 2018;17:41; doi: 10.1186/s12943-018-0811-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shine L, Kilty C, Gross J, et al. Vacuolar ATPases and Their Role in Vision. In: Retinal Degenerative Diseases. Advances in Experimental Medicine and Biology, Vol. 801 (Ash J, Grimm C, Hollyfield J, et al. eds.). Springer: New York, NY; 2014; doi: 10.1007/978-1-4614-3209-8_13 [DOI] [PubMed] [Google Scholar]
- 34. Nuckels RJ, Ng A, Darland T, et al. The vacuolar-ATPase complex regulates retinoblast proliferation and survival, photoreceptor morphogenesis, and pigmentation in the zebrafish eye. Invest Ophthalmol Vis Sci 2009;50:893–905. [DOI] [PubMed] [Google Scholar]
- 35. Kabra M, Pattnaik BR. Sensing through non-sensing ocular ion channels. Int J Mol Sci 2020;21:6925; doi: 10.3390/ijms21186925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yan Y, Denef N, Schüpbach T.. The vacuolar proton pump, V-ATPase, is required for notch signaling and endosomal trafficking in Drosophila. Dev Cell 2009;17:387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vaccari T, Duchi S, Cortese K, et al. The vacuolar ATPase is required for physiological as well as pathological activation of the Notch receptor. Development 2010;137:1825–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kobia F, Duchi S, Deflorian G, et al. Pharmacologic inhibition of vacuolar H+ ATPase reduces physiologic and oncogenic Notch signaling. Mol Oncol 2014;8:207–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cruciat C-M, Ohkawara B, Acebron SP, et al. Requirement of prorenin receptor and vacuolar H+-ATPase-mediated acidification for Wnt signaling. Science 2010;327:459–463. [DOI] [PubMed] [Google Scholar]