Abstract
Bacillus subtilis displays a substrate-inducible decarboxylating activity with the following three phenolic acids: ferulic, p-coumaric, and caffeic acids. Based on DNA sequence homologies between the Bacillus pumilus ferulate decarboxylase gene (fdc) (A. Zago, G. Degrassi, and C. V. Bruschi, Appl. Environ. Microbiol. 61:4484–4486, 1995) and the Lactobacillus plantarum p-coumarate decarboxylase gene (pdc) (J.-F. Cavin, L. Barthelmebs, and C. Diviès, Appl. Environ. Microbiol. 63:1939–1944, 1997), a DNA probe of about 300 nucleotides for the L. plantarum pdc gene was used to screen a B. subtilis genomic library in order to clone the corresponding gene in this bacterium. One clone was detected with this heterologous probe, and this clone exhibited phenolic acid decarboxylase (PAD) activity. The corresponding 5-kb insertion was partially sequenced and was found to contain a 528-bp open reading frame coding for a 161-amino-acid protein exhibiting 71 and 84% identity with the pdc- and fdc-encoded enzymes, respectively. The PAD gene (pad) is transcriptionally regulated by p-coumaric, ferulic, or caffeic acid; these three acids are the three substrates of PAD. The pad gene was overexpressed constitutively in Escherichia coli, and the stable purified enzyme was characterized. The difference in substrate specificity between this PAD and other PADs seems to be related to a few differences in the amino acid sequence. Therefore, this novel enzyme should facilitate identification of regions involved in catalysis and substrate specificity.
Phenolic acids, also called substituted cinnamic acids, are important lignin-related aromatic acids and natural constituents of plant cell walls. These acids (particularly ferulic, p-coumaric, and caffeic acids) bind the complex lignin polymer to the hemicellulose and cellulose in plants (1) or are generally esterified with tartaric acid (for example, in grape must, wine, and cider) and can be released as free acids during wine making by some cinnamoyl esterase activities (9). Most often, free phenolic acids are metabolized by different microorganisms into 4-vinyl derivatives and then are eventually reduced into 4-ethyl derivatives (5, 6). Some of these volatile phenols, particularly vinyl and ethyl guaiacol (generated from ferulic acid), are useful aromatic chemicals (12) or contribute naturally to aroma in wine (10) and other fermented foods and beverages. Other volatile phenols, such as ethyl and vinyl phenols (from p-coumaric acid), are most often considered phenolic off-flavors and are responsible for alterations in organoleptic properties. Previously, only three bacterial phenolic acid decarboxylases (PADs) have been purified and characterized (4, 8, 13). Two of these enzymes have been cloned and sequenced, a ferulate decarboxylase (FDC) from Bacillus pumilus (5) and a p-coumarate decarboxylase (PDC) from Lactobacillus plantarum (17). Although these enzymes exhibit 66% amino acid sequence identity, they differ in structure, biochemical characteristics, and substrate specificity. They are also different from the phenylacrylic decarboxylase cloned from Saccharomyces cerevisiae (7), which exhibited very low activity with ferulic and p-coumaric acids. The substrate specificity of these bacterial decarboxylases (ferulic and p-coumaric acids for FDC and p-coumaric and caffeic acids for PDC) is an obstacle for production of aroma compounds from crude or partially purified substrates, which always contain these two acids. It was our goal to screen new microorganisms in order to isolate decarboxylases with different substrate specificities and to better characterize this enzyme family. A comparison of amino acid sequences should help identify regions that specify substrate specificity and residues essential for catalysis. The results presented here are a first step toward obtaining recombinant enzymes with appropriate substrate specificities for aroma production and toward engineering genetically modified starters for vegetable fermentation and wine making. In the course of our screening, we found that Bacillus subtilis was able to decarboxylate ferulic, p-coumaric, and caffeic acids. We describe the cloning and the results of a transcriptional analysis of a pad gene that encodes a PAD. Purification and characterization of the stable recombinant enzyme overexpressed in Escherichia coli confirmed that B. subtilis PAD can metabolize all three phenolic acids; to our knowledge, this is a novel substrate specificity for an enzyme belonging to the PAD family. The PAD examined, which exhibits extensive similarity to FDC in amino acid sequence and differs from FDC in enzymatic characteristics, should be useful in experiments to determine substrate specificity and in catabolic site characterization studies in which site-directed mutagenesis is used.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
B. subtilis 168 (Institut Pasteur Collection, Paris, France) and E. coli TG1 (11) and I-1111 carrying the pHT315 vector (2) (kindly provided by Didier Lereclus) were kept frozen in 30% (wt/vol) glycerol at −70°C and were grown aerobically in Luria-Bertani (LB) medium (3) or agar medium at 37°C. When appropriate, ampicillin (200 mg/liter) and erythromycin (200 mg/liter) were added to the media used for cloning when plasmids pTZ19R (14) and pHT315 (2), respectively, were used (see Fig. 2). Cells used to determine decarboxylating activity or for DNA extraction were harvested at an A600 of 1 (about 400 mg of dry biomass per liter).
FIG. 2.
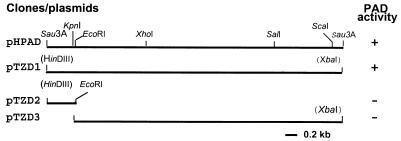
Physical and restriction map of the 5-kb insertion from pHPAD. Fragments that were subcloned in pTZ19R are shown, and the corresponding PAD activities are indicated on the right. Restriction sites in parentheses belong to the multicloning site of the vectors.
DNA manipulation, sequencing, and computer analysis.
Standard molecular procedures described by Sambrook et al. (15) were used for DNA manipulation. Double-stranded DNA from recombinant plasmids was purified with a Qiagen plasmid kit (model Tip 100; Diagen) and was sequenced by the dideoxy chain termination method (16) with a ThermoSequenase cycle sequencing kit (Amersham). Both strands were sequenced with specific synthetic primers (Gibco-BRL). Computer analyses of the sequences were carried out with PC GENE software (Intelligenetics).
Synthesis of a specific probe from the L. plantarum pdc gene by PCR.
PCR were performed with an automated Hybaid DNA thermocycler by using the standard procedure with genomic DNA from L. plantarum as the template and two oligonucleotides, 5′-CACTTGATGACTTTCTCGGCAC-3′ and 5′-CTTCAACCCACTTTGGGAAG-3′, to amplify the first 300 bp of the pdc gene (5). The PCR product was fractionated by agarose gel electrophoresis. The expected band at about 300 bp was recovered by extraction from agarose with a Jet-Sorb kit (Genomed, Bioprobe Systems, Montreuil, France) and was sequenced to check its identity. This PCR product was radiolabeled with [α-32P]dATP (Isotopchim, Ganagobie-Peyruis, France) by random priming (Gibco-BRL kit), and this probe was used to screen the B. subtilis library.
Screening of the B. subtilis genomic library.
A B. subtilis 168 genomic library from the Institut Pasteur (Paris, France), which was constructed in E. coli TG1 by ligation of B. subtilis genomic DNA partially digested with Sau3A and BamHI-digested pHT315, was used. Colony hybridization was carried out at 60°C for 5 h and then at 50°C for 5 h by using standard procedures described previously (5). Clones that hybridized with the pdc probe were detected by exposing membranes to Kodak BIOMAX MS film.
Isolation of total RNA from B. subtilis.
Cells were grown aerobically in 1,000 ml of LB broth to an A600 of 0.7 (280 mg of dry biomass per liter), and the culture was divided into several subcultures that were not induced or were induced with one of the phenolic acids tested at a concentration of 1.2 mM (about 200 mg/liter). These subcultures were incubated for 60 min at 37°C. During this period, 40-ml samples were quickly refrigerated in ice water, and the total RNA was extracted and quantified as described previously (5). The RNA integrity was checked by standard denaturing agarose gel electrophoresis.
Northern blot analysis.
Total RNA from B. subtilis was separated in denaturing formaldehyde agarose gels and transferred to nylon membranes by using the Pharmacia vacuum system. Hybridization was performed at 60°C with a [α-32P]dATP-radiolabeled probe synthesized in a PCR by using plasmid pHPAD carrying the pad gene as a template and primers encompassing the first 300 bp of the pad gene. The sizes of the transcripts were determined by using an RNA ladder (0.24 to 9.5 kb; Gibco-BRL) as the standard.
Primer extension analysis.
Primer extension analysis was performed by using two antisense primers, BSD2 (5′-CGTATTCCCATCCGTTTTCATACG-3′) and BSD4 (5′-CGTATAAATCATGTGGCTTCCG-3′), in the 5′ region of the pad gene. Two microliters (10 μg) of RNA was mixed with 10 pmol (0.5 μl) of primer and 10 μl of an extension mixture containing 3 μl of 5× reverse transcriptase buffer, 3 μl of 0.1 M dithiothreitol, 0.3 μl of a solution containing dCTP, dGTP, and dTTP (each at a concentration of 100 mM), 0.3 μl of [α-32P]dATP (Isotopchim), 0.4 μl of RNase inhibitor (40 U/ml; Boehringer Mannheim), and 3.4 μl of distilled water. Denaturation and annealing were performed by incubating the mixture at 90°C for 5 min and at 50°C for 10 min. The mixture was placed at 37°C, and the reverse transcriptase reaction was immediately started by adding 5 U of Moloney murine leukemia virus (M-MuLV) reverse transcriptase (Appligene) and incubating the preparation for 20 min. Then 0.3 μl of dATP (100 mM) was added to the reaction mixture, and the preparation was incubated for an additional 40 min to ensure that complete synthesis of cDNA occurred. Next, 3 μl of loading denaturing buffer (provided in the sequencing kit) was added to 3 μl of the reaction mixture. The mixture was denatured at 80°C for 3 min and loaded onto a sequencing gel, and sequencing reactions were performed with the pad DNA as the template and the same primers.
Preparation of cell extracts and enzyme assays.
Cells of B. subtilis and recombinant E. coli strains grown in LB medium were disrupted with a French press at 1.2 × 108 Pa, and PAD activity was assayed by monitoring the disappearance of absorption peaks of the different substrates and the simultaneous appearance of new peaks corresponding to vinyl derivatives as previously described (4). The total protein concentration was determined with a protein assay kit (Bio-Rad, Richmond, Calif.) by using bovine serum albumin as the standard, and the specific activity was expressed as micromoles of substrate degraded per minute per milligram of protein (units per milligram).
PAGE analysis.
The protein extracts containing PAD activity were resolved by denaturing sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (12% polyacrylamide resolving gel) as previously described (4); molecular mass markers (SDS-PAGE standards, low range; Bio-Rad) were used as standards.
PAD purification from recombinant E. coli.
Crude cell extract from washed and concentrated recombinant E. coli TG1(pHPAD) cells grown in 3 liters of LB medium was obtained with the French press and then fractionated by adding (NH4)2SO4 [30 and 45% (wt/vol) (NH4)2SO4 and saturation at 0°C]. The saturated fraction containing the highest specific activity was dialyzed, applied to a Q-Sepharose ion-exchange chromatography column (16 by 140 mm; Pharmacia LKB, Uppsala, Sweden), and eluted with an NaCl gradient (200 to 500 mM NaCl) in 20 mM Tris buffer (pH 7). Fractions containing PAD activity were pooled, the (NH4)2SO4 concentration was adjusted to 2 M, and the preparation was applied to a hydrophobicity column (16 by 140 mm; Macro-Prep Methyl HIC; Bio-Rad) and eluted with a gradient of (NH4)2SO4 (2 to 0 M) in 20 mM phosphate buffer (pH 7). Then, fractions containing PAD activity were pooled, dialyzed, applied to an anion-exchange chromatography column (10 by 120 mm; DEAE-Sepharose CL-6B; Sigma), and eluted with an NaCl gradient (0 to 400 mM NaCl) in phosphate buffer (pH 7). Finally, the fractions containing PAD activity were pooled, the (NH4)2SO4 concentration was adjusted to 2 M, and the preparation was applied to a smaller hydrophobicity column (16 by 30 mm) and eluted as described above. Fractions containing PAD activity were pooled, dialyzed, and concentrated by spraying flakes of polyethylene glycol 20,000 (Sigma) on the dialysis tube. A Sephacryl HR200 (Pharmacia) size exclusion chromatography column (10 by 100 mm; Bio-Rad) was used to determine the native molecular mass by comparing the elution volume of the enzyme with the elution volumes of three reference proteins (68-kDa bovine serum albumin, 45-kDa egg albumen, and 14.4-kDa lysozyme).
Nucleotide sequence accession number.
The sequence of the 1,200-bp DNA fragment containing the pad gene has been deposited in the GenBank database under accession no. AF017117.
RESULTS AND DISCUSSION
Expression of PAD activity in B. subtilis.
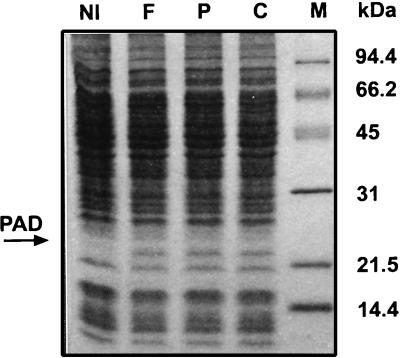
A preliminary experiment revealed that B. subtilis was able to decarboxylate ferulic and p-coumaric acids during growth in LB medium containing these acids (data not shown). To determine whether expression of PAD activity was constitutive or inducible, a 1-liter exponential-phase B. subtilis culture (A600, 0.7) was divided into four subcultures. One subculture was used as a control and was incubated for 1 h at 37°C with no additions, while the other subcultures were supplemented with 1.2 mM ferulic acid, 1.2 mM p-coumaric acid, or 1.2 mM caffeic acid and incubated under the same conditions. Crude cell extracts obtained from these subcultures were tested for PAD activity (see above) with different substrates (Table 1) and were resolved by SDS-PAGE (Fig. 1). No decarboxylase activity was detected in the uninduced cell extract. Each of the three phenolic acids tested was able to induce activity on the three acids. However, cell extracts from caffeic acid-induced cells exhibited lower activity on the three acids, which indicates that caffeic acid could be a less efficient inducer than ferulic acid or p-coumaric acid. The protein electrophoresis patterns revealed that a band at about 22 kDa that was absent in the extract from the uninduced cells was present in the cell extracts from the three induced subcultures. The following other phenolic acids were tested as potential inducers under the same conditions: phenyl acrylic acid (cinnamic acid), 2-hydroxycinnamic acid (o-coumaric acid), 3-hydroxycinnamic acid (m-coumaric acid), 3,4-hydroxy-3-phenylpropionic acid (hydrocaffeic acid), 4-hydroxy-3-phenylpropionic acid (phloretic acid), 2-methoxycinnamic acid, and 3-methoxycinnamic acid. Cell extracts obtained from these cultures were tested for the ability to metabolize these substrates by monitoring the disappearance of absorption peaks for the substrates as previously described (4). None of the compounds tested was metabolized or was able to induce decarboxylase activity. The corresponding SDS-PAGE protein patterns were not different from the uninduced control pattern (data not shown).
TABLE 1.
Expression of PAD activity in B. subtilis
| Inducer | Sp act (μmol min−1 mg of protein−1) in B. subtilis crude cell extracts with the following phenolic acids:
|
||
|---|---|---|---|
| Ferulic acid | p-Coumaric acid | Caffeic acid | |
| None (uninduced control) | <10−4 | <10−4 | <10−4 |
| Ferulic acid | 0.20 | 0.18 | 0.12 |
| p-Coumaric acid | 0.22 | 0.19 | 0.11 |
| Caffeic acid | 0.15 | 0.14 | 0.09 |
FIG. 1.
SDS-PAGE of crude cell extracts from a B. subtilis uninduced culture (lane NI) and from cultures induced with ferulic acid (lane F), p-coumaric acid (lane P), and caffeic acid (lane C). Lane M contained molecular mass standards.
Cloning of the PAD gene from B. subtilis.
Alignment of the PDC gene (pdc) from L. plantarum (5) and the FDC gene (fdc) from B. pumilus (17) revealed the presence of strictly conserved sections that were 12 to 15 bp long (data not shown). On the basis of the hypothesis that the corresponding gene of B. subtilis could be similar, a rapid strategy was used to test whether a DNA probe from one of these genes could be used to screen a B. subtilis genomic library. A preliminary Southern hybridization experiment performed at 50°C showed that a DNA probe encompassing the first 300 bp of the L. plantarum pdc gene hybridized weakly but specifically with one band of B. subtilis total DNA digested with 6-bp restriction site enzymes (data not shown). The probe was synthesized by PCR by using E. coli recombinant plasmid pJPDC1 (5) in order to avoid background noise due to the phylogenetic proximity of L. plantarum and B. subtilis. Then, the same probe synthesized by PCR by using total DNA from L. plantarum as the template was used to screen the B. subtilis genomic library in E. coli. One clone (pHPAD) of the genomic library hybridized clearly with the probe. This clone was found to be able to decarboxylate ferulic, p-coumaric, and caffeic acids at a rate 10-fold higher than the rate observed in the induced B. subtilis strain, without induction by phenolic acid or isopropyl-β-d-thiogalactopyranoside (IPTG). Control strain TG1(pHT315) had no detectable PAD activity. It must be pointed out that the B. subtilis PAD is to our knowledge the first microbial PAD described which is able to metabolize the three phenolic acids. The whole pHPAD insertion and the two subfragments were subcloned in pTZ19R to give plasmids pTZD1, pTZD2, and pTZD3, and the PAD phenotypes of the recombinant E. coli clones were determined (Fig. 2). The results indicate that the PAD-encoding gene is probably in a region that overlaps the pTZD2 and pTZD3 insertions.
Nucleotide and protein sequences.
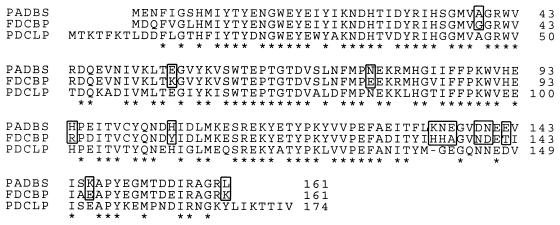
Based on the assumption that the pad gene was located at least partially in the region corresponding to the pTZD2 insertion, sequencing was initiated from the lacZ-proximal end of the pHPAD insertion. An open reading frame (ORF) with a coding capacity of 483 bp was detected (data not shown), and the deduced product of this ORF exhibited extensive similarity to previously described PADs (Fig. 3). This ORF had two 11-nucleotide stretches and one 12-nucleotide stretch with high GC contents, which were identical to the corresponding regions of the L. plantarum pdc gene. These conserved sequences could be responsible for specific cross-hybridization between the DNAs of these two genes. Downstream of the ORF corresponding to the pad gene, a putative ORF transcribed in the opposite direction was partially sequenced and aligned with GenBank sequences. This ORF was found to correspond exactly to the 3′ end of the pnbA gene (accession no. U06089) encoding a p-nitrobenzyl esterase (18). A comparison of the primary structure of the deduced PAD protein sequence (161 amino acids) revealed 84% identity with the FDC sequence of B. pumilus (17) and 71% identity with the PDC sequence of L. plantarum (5) (Fig. 3). The third value obtained in the protein alignment study was less than 28% identity, and no homology was found with other previously described decarboxylases, particularly the phenylacrylic acid decarboxylase (PAD1) of S. cerevisiae (7). A putative Shine-Dalgarno sequence (5′-AAGGAAGA-3′) was observed 12 bp upstream of the ATG initiation codon (position 250). Eight nucleotides beyond the TAA stop codon, a sequence was found that could form a stable stem-loop structure (positions 743 to 783) with an estimated ΔG of −33.8 kcal/mol; this was followed by a stretch of T residues which may function as a rho-independent terminator. A multiple-sequence alignment of B. subtilis PAD with L. plantarum PDC and B. pumilus FDC (Fig. 3) showed that the PAD is similar to the FDC and that the main differences between these two proteins and the PDC are located in the N- and C-terminal parts. If we consider that similar amino acids could not account for the differences in substrate specificity between the B. subtilis PAD and the B. pumilus FDC, the major sequence differences between these two enzymes correspond to only a few amino acids (eight isolated amino acids, one doublet, and one triplet). It is interesting to note that for seven of the amino acid differences, the B. subtilis PAD was identical to the L. plantarum PDC and that these two enzymes are able to metabolize caffeic acid.
FIG. 3.
Comparison of the deduced amino acid sequence encoded by the pad gene of B. subtilis (PADBS) with the sequences of B. pumilus FDC (FDCBP) (accession no. X84815) and L. plantarum PDC (PDCLP) (accession no. U63827). The sequences were aligned by using the Clustal program. Asterisks indicate identical amino acids. The numbers on the right are the amino acid positions in the protein sequences. PAD residues that are neither identical nor similar to FDC residues are enclosed in boxes.
Transcriptional analysis.
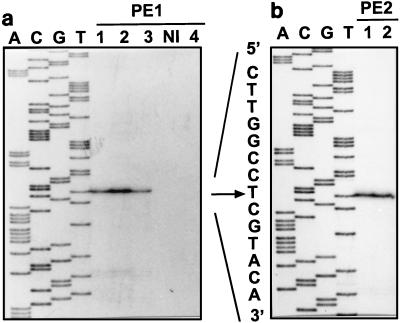
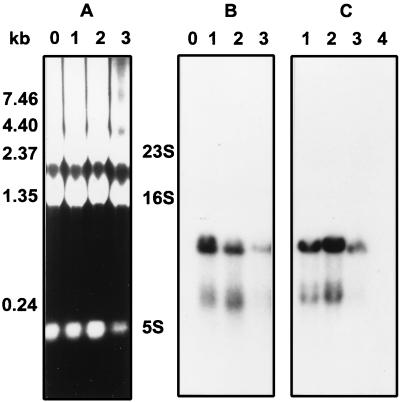
Primer extension experiments were performed with primers BSD2 and BSD4 by using RNA from B. subtilis cultures induced with each of the three phenolic acids 10 min before cells were harvested. Identical results were obtained with both primers, and an A residue (at position 91) located 159 nucleotides upstream from the start codon was identified as the transcription start site (Fig. 4). No primer extension product was detected when RNA from an uninduced culture was used (Fig. 4). Northern blot hybridization with the same templates was performed to determine the sizes and levels of the corresponding mRNA at different sampling times after the inducer was added (Fig. 5A and B). No transcript and no PAD activity (data not shown) were detected in the lane corresponding to the RNA extract and in the protein extract from uninduced cells, respectively. A single transcript of approximately 620 nucleotides (a size corresponding to the size of a DNA fragment from the start site to the 3′ end of the pad gene) was detected in the RNA extract from cells induced by adding ferulic acid, and the level of this transcript was maximal after 10 min of incubation with the inducer. The level of the pad transcript was lower after 30 min of incubation and was very low after 1 h. A smaller band at about 300 bases was detected with the probe, and this band probably corresponded to a degradation product of the pad transcript since it was not detected in total RNA isolated from uninduced cells. Other phenolic acids were tested to determine their inducing abilities under the same conditions (Fig. 5C). The pad transcript was detected only in ferulic acid-, p-coumaric acid-, or caffeic acid-induced samples, which confirmed the results shown in Table 1. Maximal PAD activity was observed after 10 min of induction, after which the substrate was entirely metabolized and the activity started to decrease slowly (data not shown). This decrease in activity could have been due to dilution of the enzyme in dividing cells during the last 50 min. Taken together, these results indicate that the pad gene is transcribed as a monocistronic transcriptional unit and is subjected to transcriptional regulation involving substrate-mediated induction.
FIG. 4.
Mapping of the 5′ end of B. subtilis pad mRNA (arrow) by primer extension analysis. (a) Primer BSD2 (PE1) was used with total RNA from uninduced cells (lane NI) and cells induced by adding 1.2 mM ferulic acid (lane 1), p-coumaric acid (lane 2), caffeic acid (lane 3), and cinnamic acid (lane 4). (b) Primer BSD4 (PE2) was used with RNA from cells induced with ferulic acid (lane 1) and p-coumaric acid (lane 2). The products of the reverse transcriptase reactions were coelectrophoresed with DNA sequencing reaction mixtures (lanes A, C, G, and T) initiated with the same primers on pad template DNA.
FIG. 5.
(A) Denaturing agarose gel electrophoresis of total RNA (10 μg per lane) from B. subtilis uninduced cells (lane 0) and induced cells harvested 10 min (lane 1), 30 min (lane 2), and 60 min (lane 3) after 1.2 mM ferulic acid was added. (B) Corresponding Northern blot analysis. (C) Northern blot analysis of total RNA purified from cells induced for 10 min with ferulic acid (lane 1), p-coumaric acid (lane 2), caffeic acid (lane 3), and cinnamic acid (lane 4). The Northern blot analysis was performed with a [α-32P]dATP-radiolabeled probe that was PCR synthesized by using plasmid pHPAD as a template.
Purification of the recombinant PAD.
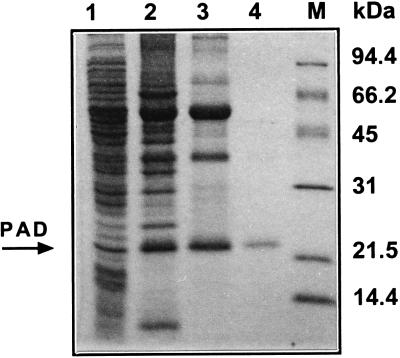
First, we determined that the recombinant PAD expressed in E. coli displayed the same enzymatic characteristics that were observed with the partially purified protein obtained from an induced culture of B. subtilis (data not shown). The two recombinant E. coli TG1 clones (pHPAD and pTZD1) expressed PAD activity at nearly the same level with and without phenolic acid or IPTG inducer in the culture medium (data not shown). A crude extract was obtained from a 3-liter culture of E. coli TG1(pHPAD) as described above, and PAD was purified (Table 2) to apparent SDS-PAGE homogeneity (Fig. 6). About 250 μg of 112-fold-purified PAD with a yield of 8% was obtained and used for further enzymatic characterization.
TABLE 2.
Purification of PAD from recombinant E. colia
| Purification step | Total protein (mg) | Total activity (U)b | Sp act (U mg−1) | Purifi- cation (fold) | Yield (%) |
|---|---|---|---|---|---|
| Crude extract | 350 | 875 | 2.5 | 1 | 100 |
| (NH4)2SO4 (saturation) | 85 | 637 | 7.5 | 3 | 73 |
| Q-Sepharose (pH 7) | 21 | 504 | 24 | 9.6 | 58 |
| Methyl HIC (pH 7) | 3.3 | 297 | 90 | 36 | 34 |
| DEAE-Sepharose (pH 7) | 0.6 | 123 | 205 | 82 | 14 |
| Methyl HIC (pH 7) | 0.25 | 70 | 280 | 112 | 8 |
For details see the text.
One unit corresponds to 1 μmol of ferulic acid decarboxylated per min.
FIG. 6.
SDS-PAGE of protein extracts obtained during purification of PAD from recombinant E. coli(pHPAD). Lane 1, crude extract; lane 2, (NH4)2PO4-saturated fraction; lane 3, Q-Sepharose fraction; lane 4, second Methyl HIC step fraction (2.5 μg of purified enzyme); lane M, molecular mass standards.
Characterization of the recombinant PAD.
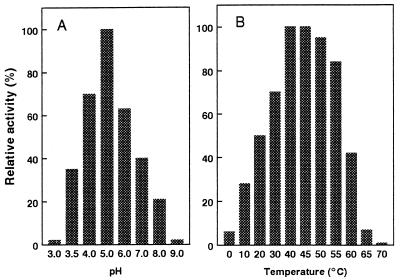
The recombinant PAD had a molecular mass of about 45 kDa, as determined by size exclusion chromatography (data not shown), indicating that it could be a homodimer consisting of two 22-kDa subunits. Maximal enzyme activity was obtained in 25 mM phosphate or Tris buffer (pH 5) without any exogenously added cofactor or metal ion. The enzyme was stable; more that 80% of the activity was conserved in phosphate buffer after 48 h of incubation at 20°C and after repeated freeze-thaw cycles in the same buffer (pH 5 or 6). The activity was completely inhibited by 0.3% (wt/vol) SDS. The purified PAD had Vmax values of 280, 265, and 180 μmol min−1 mg−1 and Km values of 1.1, 1.3, and 2.6 mM for ferulic, p-coumaric, and caffeic acids, respectively. The PAD exhibited relatively high activity within broad pH and temperature ranges around the optimal conditions (pH 5 and 40 to 45°C) (Fig. 7). However, the PAD activity dramatically decreased after 10 min of exposure to pH 3.5 or 65°C (data not shown). This high level of stability of the B. subtilis PAD expressed in E. coli is an advantage for protein engineering since the B. pumilus FDC seemed to be unstable when it was expressed in E. coli (17) and the Pseudomonas fluorescens FDC was active only at restricted pH and temperature ranges (13).
FIG. 7.
Temperature optimum (A) and pH optimum (B) of purified PAD.
The difference in substrate specificity between the PAD of B. subtilis and the FDC of B. pumilus seems to be linked to few differences in the amino acid sequences (Fig. 3). Therefore, site-directed mutagenesis aimed at exchanging these variant residues could allow engineering of a PAD that is not able to metabolize p-coumaric acid, derivatives of which are often considered phenolic off-flavors. Also, this novel PAD sequence should facilitate identification of protein regions and residues involved in catalysis and substrate specificity and, ultimately, development of genetically modified enzymes and microorganisms with desirable phenolic acid specificities. An absence of detectable PAD activity correlated with an absence of detectable corresponding mRNA in B. subtilis uninduced cells was observed previously for the pdc gene in L. plantarum (5). Further studies will be undertaken to characterize this family of phenolic acid-dependent regulatory systems.
ACKNOWLEDGMENTS
We are very grateful to Didier Lereclus and Georges Rapoport (Institut Pasteur, Paris, France) and to the Institut Pasteur for the gift of E. coli I-1111 carrying the pHT315 vector and for the agreement to screen the B. subtilis 168 genomic library. We thank Christophe Mendoza for his help in enzyme purification and Christine Bernard-Rojas for laboratory work.
This study was supported in part by a grant from the Conseil Régional de Bourgogne.
REFERENCES
- 1.Akin D E. Biological structure of lignocellulose and its degradation in the rumen. Anim Feed Sci Technol. 1988;21:215–310. [Google Scholar]
- 2.Arantes O, Lereclus D. Construction of cloning vectors for Bacillus thuringiensis. Gene. 1991;108:115–119. doi: 10.1016/0378-1119(91)90495-w. [DOI] [PubMed] [Google Scholar]
- 3.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;60:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavin J F, Barthelmebs L, Guzzo J, Van Beeumen J, Bart S, Travers J F, Diviès C. Purification and characterization of an inducible p-coumaric acid decarboxylase from Lactobacillus plantarum. FEMS Microbiol Lett. 1997;147:291–295. doi: 10.1128/aem.63.5.1939-1944.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavin J-F, Barthelmebs L, Diviès C. Molecular characterization of an inducible p-coumaric acid decarboxylase from Lactobacillus plantarum: gene cloning, transcriptional analysis, overexpression in Escherichia coli, purification, and characterization. Appl Environ Microbiol. 1997;63:1939–1944. doi: 10.1128/aem.63.5.1939-1944.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatonnet P, Dubourdieu D, Boidron J N, Pons M. The origin of ethylphenols in wines. J Sci Food Agric. 1992;60:165–178. [Google Scholar]
- 7.Clausen M, Lamb C J, Megnet R, Doerner P. PAD1 encodes phenylacrylic acid decarboxylase which confers resistance to cinnamic acid in Saccharomyces cerevisiae. Gene. 1994;142:107–112. doi: 10.1016/0378-1119(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 8.Degrassi G, Polverino De Laureto P, Bruschi C V. Purification and characterization of ferulate and p-coumarate decarboxylase from Bacillus pumilus. Appl Environ Microbiol. 1995;61:326–332. doi: 10.1128/aem.61.1.326-332.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dugelay I, Gunata Z, Sapis J C, Baumes R, Bayonove C. Role of cinnamoyl esterase activities from enzyme preparations on the formation of volatile phenols during winemaking. J Agric Food Chem. 1993;41:2092–2096. [Google Scholar]
- 10.Etiévant P X, Issanchou S, Marie S, Ducruet V, Flanzy C. Sensory impact of volatile phenols on red wine aroma: influence of carbonic maceration and time of storage. Sci Aliment. 1989;9:19–33. [Google Scholar]
- 11.Gibson T J. Studies on the Epstein-Barr virus genome. Ph. D. thesis. Cambridge, United Kingdom: Cambridge University; 1984. [Google Scholar]
- 12.Huang Z, Dostal L, Rosazza J P N. Microbial transformations of ferulic acid by Saccharomyces cerevisiae and Pseudomonas fluorescens. Appl Environ Microbiol. 1993;59:2244–2250. doi: 10.1128/aem.59.7.2244-2250.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Z, Dostal L, Rosazza J P N. Purification and characterization of a ferulic acid decarboxylase from Pseudomonas fluorescens. J Bacteriol. 1994;176:5912–5918. doi: 10.1128/jb.176.19.5912-5918.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rokeach L A, Haselby J A, Hoch S O. Molecular cloning of the cDNA encoding the human Sm-D autoantigen. Proc Natl Acad Sci USA. 1988;85:4832–4836. doi: 10.1073/pnas.85.13.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 16.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zago A, Degrassi G, Bruschi C V. Cloning, sequencing, and expression in Escherichia coli of the Bacillus pumilus gene for ferulic acid decarboxylase. Appl Environ Microbiol. 1995;61:4484–4486. doi: 10.1128/aem.61.12.4484-4486.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zock J, Cantwell C, Swartling J, Hodges R, Pohl T, Sutton K, Rosteck P, Jr, McGilvray D, Queener S. The Bacillus subtilis pnbA gene encoding p-nitrobenzyl esterase: cloning, sequence and high-level expression in Escherichia coli. Gene. 1994;151:37–43. doi: 10.1016/0378-1119(94)90630-0. [DOI] [PubMed] [Google Scholar]