Abstract
Background
Black Americans suffer disparities in risk for cardiometabolic and other chronic diseases. Findings from the Adventist Health Study-2 (AHS-2) cohort have shown associations of plant-based dietary patterns and healthy lifestyle factors with prevention of such diseases. Hence, it is likely that racial differences in metabolic profiles correlating with disparities in chronic diseases are explained largely by diet and lifestyle, besides social determinants of health.
Methods
Untargeted plasma metabolomics screening was performed on plasma samples from 350 participants of the AHS-2, including 171 Black and 179 White participants, using ultrahigh-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) and a global platform of 892 metabolites. Differences in metabolites or biochemical subclasses by race were analyzed using linear regression, considering various models adjusted for known confounders, dietary and/or other lifestyle behaviors, social vulnerability, and psychosocial stress. The Storey permutation approach was used to adjust for false discovery at FDR < 0.05.
Results
Linear regression revealed differential abundance of over 40% of individual metabolites or biochemical subclasses when comparing Black with White participants after adjustment for false discovery (FDR < 0.05), with the vast majority showing lower abundance in Blacks. Associations were not appreciably altered with adjustment for dietary patterns and socioeconomic or psychosocial stress. Metabolite subclasses showing consistently lower abundance in Black participants included various lipids, such as lysophospholipids, phosphatidylethanolamines, monoacylglycerols, diacylglycerols, and long-chain monounsaturated fatty acids, among other subclasses or lipid categories. Among all biochemical subclasses, creatine metabolism exclusively showed higher abundance in Black participants, although among metabolites within this subclass, only creatine showed differential abundance after adjustment for glomerular filtration rate. Notable metabolites in higher abundance in Black participants included methyl and propyl paraben sulfates, piperine metabolites, and a considerable proportion of acetylated amino acids, including many previously found associated with glomerular filtration rate.
Conclusions
Differences in metabolic profiles were evident when comparing Black and White participants of the AHS-2 cohort. These differences are likely attributed in part to dietary behaviors not adequately explained by dietary pattern covariates, besides other environmental or genetic factors. Alterations in these metabolites and associated subclasses may have implications for the prevention of chronic diseases in Black Americans.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12916-023-03101-4.
Keywords: Metabolomics, Cohort, Dietary pattern, Health disparities, Black Americans, Linear regression, Cardiometabolic, Lipids, Creatine
Background
Black or African Americans suffer persistent disparities in cardiovascular and metabolic diseases, particularly higher morbidity and mortality from these diseases, in spite of downward trends in the general US population. For example, while national mortality rates for heart disease and stroke have declined 61% and 70%, respectively, since 1975, rates for African Americans remain higher by roughly 20% and 40%, respectively [1]. Additionally, rates of hypertension [2, 3] and stroke and peripheral arterial disease have been found to be nearly twice as high in Black compared with White Americans, and Black Americans have higher rates of obesity [2]. Furthermore, Blacks have a disproportionate cancer burden, as evidenced by their higher mortality and low survival compared to other racial/ethnic groups, with a higher risk of death for specific cancers including myeloma, stomach cancer, prostate cancer, endometrial cancer, and breast cancer [4]. Moreover, Blacks are 1.5 to 2 times as likely to develop diabetes as Whites [5, 6] and twice as likely to die from diabetes [7].
Reductions in coronary disease progression and death are largely attributed to improvements in risk factors (smoking, hypertension, physical inactivity) [8, 9]. Dietary patterns or habits may play an important role in the prevention and control of chronic diseases. Epidemiologic and experimental studies have linked diets high in plant-based foods to a number of favorable health outcomes, including lower risk of metabolic syndrome, cardiometabolic diseases, and cancer, whereas diets high in non-fish meats and fatty or refined foods are associated with increased risk of these diseases [10–14]. Findings from the Adventist Health Study (AHS)-2 cohort, particularly, have shown strong inverse associations between a vegetarian dietary pattern and diabetes, metabolic syndrome (including lower triglycerides, glucose, blood pressure, waist circumference, and total and low-density lipoprotein (LDL)-cholesterol), and coronary heart disease [15–19]. This reduction in risk of metabolic syndrome and the favorable outcomes for vegetarians are apparent in Black participants as well [15].
Dietary behaviors may have a profound impact on metabolites. We previously reported distinct metabolic signatures for vegans relative to nonvegetarians, with many of the differentially abundant metabolite subclasses implicated in inflammation-related or cardiometabolic conditions [20]. We have also found higher abundance of anti-inflammatory bioactive compounds and more favorable profiles of fatty acids in vegetarians, especially vegans, relative to non-vegetarians [21]. Racial status and culture may strongly influence dietary patterns and food preferences, consequently impacting overall health and disease susceptibility. Besides dietary behaviors, research has shown that social and environmental determinants of health, as opposed to genetic differences, are critical in propagating the burden of chronic disease disparities in African Americans [22–25]. Blacks are over-represented in lower-wage jobs and more likely to live in lower-income neighborhoods disproportionately higher in poverty compared to White neighborhoods [26, 27]. In these neighborhoods, access to health-promoting resources, and quality health care can be problematic. This socioeconomic disadvantage and social vulnerability may promote psychosocial stress, further contributing to disparities in risk of chronic diseases.
Thus, it is therefore necessary to determine if Black relative to White Americans show differences in metabolic profiles, which may be explained by differences in dietary and behavioral factors, or social and environmental influences, besides genetic factors. In the present study, we used an untargeted metabolomics platform to compare plasma metabolic profiles between Black and White participants of the AHS-2 cohort to determine if differences in metabolites and their associated biochemical subclasses, some with implications for disease susceptibility, could be explained by lifestyle, sociodemographic, or other environmental factors.
Methods
Study design/plasma metabolomics profiling
This study included 171 healthy Black (African American, West Indian/Caribbean, African, or other Black) and 179 non-Hispanic White participants from the AHS-2 cohort who previously provided plasma samples in one of three AHS-2 substudies (Calibration, Religion Health Study, Pilot Study) [28–30]. Participants also completed a 204-item food frequency questionnaire (FFQ) at baseline which contained information on dietary habits including consumption of fruits and vegetables, legumes (lentils, soybeans, and other beans), breads and grains, soy foods/drinks/supplements, dairy, eggs, red meats, processed meats, fish, and caffeine/coffee consumption [29]. Participants were subsequently classified by dietary pattern, with vegans never or rarely (< 1/month) consuming animal products including meat, fish, dairy, or eggs; pesco-vegetarians consuming fish at least once per month and other meats < 1/month; and non-vegetarians consuming flesh meats, not only fish ≥ 1/month [29]. The FFQ additionally collected demographic/sociodemographic data including address/zipcode, and other lifestyle behaviors such as exercise, smoking, alcohol drinking, and medication/supplement use.
Plasma samples of 350 study participants were profiled by Metabolon, Inc. (Morrisville, NC) using a global platform (DiscoveryHD4) that provides a comprehensive picture of metabolites within various biochemical classes including amino acid, nucleotide, carbohydrate, lipid, xenobiotic, and microbial classes, further divided into 92 subclasses. Global metabolomics profiling on participant plasma samples was performed in two separate sets (on different days): the first set included samples from 92 participants (pilot), and the second set included a total of 258 samples. Procedures for sample accession and sample preparation including protein precipitation were followed as documented previously [31]. Briefly, samples were prepared using the automated MicroLab STAR® system from Hamilton company, as described previously, and analyzed on four independent ultrahigh-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) platforms. Several quality controls were included — a pooled matrix sample that included a small volume of each experimental sample was used as a technical replicate, water samples were used as process blanks, and a cocktail of quality control internal standards was spiked into every experimental sample to monitor instrument performance. Test samples were randomized across the platform run with QC samples spaced evenly among the injections. Median relative standard deviation (RSD) was calculated for the internal standards added to each sample before injection into the mass spectrometers to determine instrument variability. Process variability was determined by calculating the median RSD for endogenous metabolites present in all of the technical replicates of pooled client samples. The RSD for instrument and process variability was 6% and 8%, respectively. Metabolites were identified by comparison to library entries of purified, authenticated standards or recurrent unknown entities.
A biochemical pathway or “subclass” is annotated for each metabolite within Metabolon’s proprietary reference library of compounds, i.e., lysophospholipids, long-chain monounsaturated fatty acid, long-chain polyunsaturated fatty acid (n3 and n6), xanthine metabolism, creatine metabolism, monoacylglycerol, and diacylglycerol, grouping in accordance with its metabolic function. These subpathways or “subclasses” are grouped into larger/broader pathways or “classes,” i.e., amino acids, lipids, nucleotides, partially characterized molecules (not yet fully characterized), carbohydrates, xenobiotics, drugs, and chemicals.
Statistical analysis
Raw metabolite values representing mass spectrometry peak densities were median scaled (i.e., for each metabolite, individual values were divided by the median for that metabolite) and log-transformed after first imputing undetectable values with the minimum for each metabolite. Metabolites were excluded from analysis if at least 80% were missing, defined as below the limits of detection, yielding a total of 892 metabolites for statistical analysis. Linear regression was performed on individual metabolites or metabolite subclasses using a combined set of n = 350 analytic samples to examine associations of race with log-transformed metabolite abundance, adjusting for relevant covariates or known confounders. Covariates included sex (male vs female), race (Black vs White), age at clinic visit (continuous), batch (continuous), body mass index (BMI, continuous), study (pilot vs non-pilot), substudy (Calibration, Religion Health Study, Pilot), education (high school, some college, college graduate), dietary pattern (vegan, pesco-vegetarian, non-vegetarian), exercise (minutes/week), coffee (ounces/day), use of medications for hypertension or cholesterol or use of aspirin/NSAIDS (yes/no), smoking and alcohol drinking (never, former, current), social vulnerability index (SVI, continuous), psychosocial stress (ordinal), and prevalent chronic diseases including cardiovascular disease, cancer, or diabetes (yes/no).
The Center for Disease Control and Prevention Social Vulnerability Index (SVI) was created by the Agency for Toxic Substances and Disease Registry’s (ATSDR) Geospatial Research, Analysis & Services Program [32], and determined from geocoded addresses for participants in this study. The SVI describes the relative vulnerability of US Census tracts (county subdivisions for which the US Census collects data) based on various social variables, which are grouped into four themes: (1) socioeconomic status, (2) household composition and disability, (3) minority status and language, and (4) housing type and transportation. Each tract received an index or ranking (percentile) for each theme, as well as an overall ranking or combined summary theme, which was used in the current study. Psychosocial stress was defined by two dummy variables — one representing growing up in a single-parent household, defined by divorce of parents or death of a mother or father, “parents divorced” and “mother or father died,” and the other representing personal divorce. Psychosocial stress was a cumulative variable, representing the sum of these two variables.
In the linear regression analysis of differentially abundant metabolites, the adjusted mean with 95% confidence intervals was calculated as the log of the adjusted geometric mean for Black and White participants, and subsequently back-transformed for each metabolite. Fold difference representing the difference in these adjusted means comparing Black with White participants was subsequently calculated.
An adapted Storey et al. permutation approach was used to adjust for false discovery [33], and residualized variables for race were permuted as a means of finding the null distribution of the t scores for metabolite abundance [34], thereby retaining covariances between residualized metabolite abundances. An estimate of the proportion of null metabolites allows for an estimate of the FDR, avoiding the over-conservative Benjamini-Hochberg approach. Consequently, metabolites may be selected with low FDR [33, 35].
For analysis of metabolite subclasses, t-scores were obtained by averaging the respective component metabolite values within a subclass and then dividing by the standard deviation of this average, accounting for the covariances between the metabolites. The metabolite subclass average was regressed on race and other covariates. As a sensitivity analysis, and to identify metabolites and subclasses consistently associated with race, linear regression models examining these associations were generated considering pilot study and non-pilot participants separately. All analyses were conducted in R version 4.0.2.
Additionally, over-representation analysis was performed for pathway analysis to determine if a pathway was represented more than expected by chance based on the analytical/reference platform of metabolic pathways in KEGG, performing a hypergeometric test using MetaboAnalyst [36]. Importance measures were calculated from the centrality measures (relative betweenness) reflecting the number of shortest paths going through the node) based on pathway topology, with each pathway having a maximum importance of 1.
A random forest classifier was developed to differentiate race according to metabolic profiles. Two testing techniques were implemented to evaluate the robustness of the random forest classifier. One used a 70% training and 30% test set in a cross-validation approach using all samples and all 892 log-transformed metabolites. In this analysis, each of the 50,000 trees learned from a random sample (70%, training set), with the remaining data (30%, test) passed down the tree for class prediction. Mean decreased accuracy was calculated to determine the most influential metabolites, permuting each predictor variable to assess the difference in predictive accuracy. The second approach was a 5-fold cross-validation (also 70% training and 30% test), which was used to evaluate the model. Overall variable importance was calculated based on the goodness of split measures (ascertained by an impurity function) for each variable. Analyses were conducted in R, using randomForest and rpart packages.
Results
Baseline characteristics
The current study included 171 Black and 179 White participants distributed between vegan, pesco-vegetarian, and non-vegetarian diet groups (Table 1). White participants were on average slightly older in years than Blacks (60.4 vs 57.3; p < 0.01), with significantly higher coffee consumption (2.2 oz/day vs 0.3 oz/day; p < 0.001). Blacks had higher SVI ranking (0.65 vs 0.58; p = 0.003), and a greater proportion used blood pressure medication (39% vs 23%; p = 0.004) and had a history of smoking (p = 0.02) and alcohol drinking (p = 0.008). There were additional differences in various dietary components, particularly macronutrients (Additional file 1, Supplementary Table 1). Differences in other characteristics (sex, exercise, diet group, education, disease prevalence) were not statistically significant.
Table 1.
Demographic and lifestyle characteristics of the study populationa,b
| Black | White | P value | |
|---|---|---|---|
| Participants | 171 | 179 | |
| Sex | 0.67 | ||
| Male | 88 (51.5) | 87 (48.6) | |
| Female | 83 (48.5) | 92 (51.4) | |
| Age (years) | 57.3 (11.7) | 60.4 (10.5) | 0.009 |
| BMI (kg/m2) | 27.7 (5.8) | 26.6 (5.5) | 0.07 |
| Exercise (min/week) | 88.8 (99.5) | 92.5 (101.1) | 0.74 |
| Diet group | 0.84 | ||
| Vegan | 63 (36.8) | 62 (34.6) | |
| Pesco-vegetarian | 50 (29.2) | 51 (28.5) | |
| Nonvegetarian | 58 (33.9) | 66 (36.9) | |
| Coffee (oz/day) | 0.3 (1.0) | 2.2 (6.7) | < 0.001 |
| Social vulnerability index (percentile) | 0.65 (0.23) | 0.58 (0.22) | 0.003 |
| Education | 0.18 | ||
| High school | 20 (11.8) | 23 (15.7) | |
| Some college | 76 (45.0) | 63 (37.5) | |
| College graduate | 73 (43.2) | 92 (54.8) | |
| Medication use (yes/no) | |||
| Blood pressure | 55 (39) | 42 (23.3) | 0.004 |
| Cholesterol | 23 (16) | 32 (17.8) | 0.84 |
| Aspirin | 42 (29.8) | 61 (33.9) | 0.51 |
| Smoking | |||
| Ever | 43 (25.6) | 27 (15.2) | |
| Never | 125 (74.4) | 151 (84.8) | 0.02 |
| Alcohol | |||
| Current | 19 (11.4) | 12 (6.7) | 0.008 |
| Former | 62 (37.3) | 46 (25.6) | |
| Never | 85 (51.2) | 121 (67.6) | |
| Prevalent disease | |||
| Cardiovascular | 6 (3.5) | 6 (3.3) | 1 |
| Diabetes | 14 (8.2) | 10 (5.6) | 0.41 |
| Cancer | 13 (7.6) | 14 (7.8) | 1 |
| Psychosocial stress | 0.07 | ||
| No | 145 | 164 | |
| Yes | 26 | 15 | |
aValues presented as n (%) or mean (SD)
bMissing values: smoking, n = 3 (Black), n = 1 (White); alcohol n = 5 (Black), n = 1 (White); education, n = 2 (Black), n = 1 (White)
Linear regression analysis of individual metabolites
In order to obtain a more comprehensive understanding of racial disparities contributing to metabolic differences, we generated a number of statistical models to compare metabolomics profiles between Black and White participants. In the simplest model adjusted for age, sex, race, and study, there were 418 differentially abundant metabolites (Tables 2 and 3, Additional file 1, Supplementary Tables 2 and 3). In the most complete model adjusted additionally for various lifestyle behaviors and environmental stressors (BMI, dietary pattern, exercise, smoking, alcohol drinking, coffee consumption, medication use, diabetes, education, SVI, psychosocial stress, and chronic conditions), there were 404 differentially abundant metabolites, with 56 higher and 348 lower in Blacks. Metabolites most notably higher in Black participants included those of the xenobiotics class, such as the methyl/propyl-parabens, propyl- and methyl-4-hydroxybenzoate sulfate within the benzoate metabolism subclass, and various food component/plant metabolites (Table 4). These metabolites in higher abundance overlapped largely with those in the simpler models (Table 2; Additional file 1, Supplementary Table 2), and the most significant metabolites (< 1.0E−3) were retained. Other compounds with the greatest fold difference increases (above 2-fold) or greatest statistical differences (< 4E−04) in Black relative to White participants included various food component/plant metabolites, particularly the piperine metabolites (derivatives of black pepper), 3-carboxy-4-methyl-5-propyl-2-furanpropanoate (CMPF), some acetylated amino acids (and putative uremic toxins), a tocopherol metabolite, bile acid, chemical/drug, and tobacco metabolite (Tables 2 and 4).
Table 2.
Metabolites (top 40 of 73) present at higher abundance in Black relative to White participants at FDR < 0.05 in simplest modela,b
| Metabolite | Fold difference | Subclass | Major class | FDR (Storey) |
|---|---|---|---|---|
| Propyl 4-hydroxybenzoate sulfate | 7.57 | Benzoate metabolism | Xenobiotics | < 3.6E−04 |
| Methyl-4-hydroxybenzoate sulfate | 4.56 | Benzoate metabolism | Xenobiotics | < 3.6E−04 |
| Piperine | 3.32 | Food component/plant | Xenobiotics | 5.2E−04 |
| 3-Carboxy-4-methyl-5-propyl-2-furanpropanoate (cmpf) | 3.23 | Fatty acid, dicarboxylate | Lipid | 8.8E−04 |
| Umbelliferone sulfate | 2.95 | Food component/plant | Xenobiotics | < 3.6E−04 |
| Sulfate of piperine metabolite c18h21no3 (1) | 2.79 | Food component/plant | Xenobiotics | < 3.6E−04 |
| n-Acetyl-1-methylhistidine | 2.78 | Histidine metabolism | Amino acid | < 3.6E−04 |
| Hydroxy-cmpf | 2.73 | Fatty acid, dicarboxylate | Lipid | 1.9E−03 |
| Thymol sulfate | 2.72 | Food component/plant | Xenobiotics | 9.9E−04 |
| Sulfate of piperine metabolite c16h19no3 (3) | 2.71 | Food component/plant | Xenobiotics | < 3.6E−04 |
| Sulfate of piperine metabolite c16h19no3 (2) | 2.66 | Food component/plant | Xenobiotics | < 3.6E−04 |
| Sulfate of piperine metabolite c18h21no3 (3) | 2.58 | Food component/plant | Xenobiotics | < 3.6E−04 |
| 2-Hydroxyfluorene sulfate | 2.49 | Tobacco metabolite | Xenobiotics | < 3.6E−04 |
| 2-Naphthol sulfate | 2.49 | Chemical | Xenobiotics | < 3.6E−04 |
| n-Acetylalliin | 2.31 | Food component/plant | Xenobiotics | 1.5E−03 |
| Gamma-cehc sulfate | 2.23 | Tocopherol metabolism | Cofactors and vitamins | < 3.6E−04 |
| n-Acetylcitrulline | 2.18 | Urea cycle; arginine and proline metabolism | Amino acid | < 3.6E−04 |
| 2-Piperidinone | 2.04 | Food component/plant | Xenobiotics | < 3.6E−04 |
| Glucuronide of piperine metabolite c17h21no3 (4) | 2.02 | Food component/plant | Xenobiotics | 1.0E−02 |
| Glucuronide of piperine metabolite c17h21no3 (3) | 1.93 | Food component/plant | Xenobiotics | 1.1E−02 |
| Glucuronide of piperine metabolite c17h21no3 (5) | 1.91 | Food component/plant | Xenobiotics | 1.0E−02 |
| Taurodeoxycholic acid 3-sulfate | 1.89 | Secondary bile acid metabolism | Lipid | 5.8E−03 |
| Glutarate (c5-dc) | 1.59 | Fatty acid, dicarboxylate | Lipid | 3.6E−04 |
| Perfluorooctanesulfonate (pfos) | 1.51 | Chemical | Xenobiotics | 1.0E−02 |
| n-Acetylarginine | 1.49 | Urea cycle; arginine and proline metabolism | Amino acid | < 3.6E−04 |
| Serotonin | 1.48 | Tryptophan metabolism | Amino acid | 4.8E−02 |
| n-Acetylglutamine | 1.45 | Glutamate metabolism | Amino acid | < 3.6E−04 |
| Indoleacetylglutamine | 1.44 | Tryptophan metabolism | Amino acid | 3.1E−02 |
| n-Acetylasparagine | 1.42 | Alanine and aspartate metabolism | Amino acid | < 3.6E−04 |
| Hexanoylglutamine | 1.42 | Fatty acid metabolism (acyl glutamine) | Lipid | 1.3E−02 |
| 1-Stearoyl-2-docosahexaenoyl-gpc (18:0/22:6) | 1.41 | Phosphatidylcholine (PC) | Lipid | 3.8E−04 |
| n-Acetyl-2-aminooctanoate | 1.40 | Fatty acid, amino | Lipid | 2.5E−03 |
| 6-Oxopiperidine-2-carboxylate | 1.40 | Lysine metabolism | Amino acid | 9.8E−03 |
| Taurocholenate sulfate | 1.38 | Secondary bile acid metabolism | Lipid | 3.5E−03 |
| Campesterol | 1.37 | Sterol | Lipid | 3.1E−02 |
| Carotene diol (3) | 1.36 | Vitamin A metabolism | Cofactors and vitamins | 5.0E−03 |
| Arginine | 1.35 | Urea cycle; arginine and proline metabolism | Amino acid | < 3.6E−04 |
| Cis-4-decenoylcarnitine (c10:1) | 1.34 | Fatty acid metabolism (acyl carnitine, monounsaturated) | Lipid | 5.0E−04 |
| Picolinate | 1.33 | Tryptophan metabolism | Amino acid | 8.5E−03 |
| 1-(1-Enyl-stearoyl)-2-arachidonoyl-gpe (p-18:0/20:4) | 1.33 | Plasmalogen | Lipid | 2.3E−03 |
aFold difference represents the ratio of geometric means of raw metabolite values of Black relative to White participants
bAdjusted for age, sex, batch, study, and substudy
Table 3.
Metabolites (top 40 of 345) present at lower abundance in Black relative to White participants at FDR < 0.05 in simplest modela,b
| Metabolite | Fold difference | Subclass | Major class | FDR (Storey) |
|---|---|---|---|---|
| Theobromine | 0.26 | Xanthine metabolism | Xenobiotics | 9.2E−04 |
| n-Methylpipecolate | 0.28 | Bacterial/fungal | Xenobiotics | 4.5E−03 |
| Caffeine | 0.29 | Xanthine metabolism | Xenobiotics | 3.6E−03 |
| n6-methyllysine | 0.30 | Lysine metabolism | Amino acid | 2.3E−03 |
| 3-Methylxanthine | 0.32 | Xanthine metabolism | Xenobiotics | 1.2E−04 |
| 5-Acetylamino-6-amino-3-methyluracil | 0.34 | Xanthine metabolism | Xenobiotics | 1.1E−03 |
| Tryptophan betaine | 0.35 | Tryptophan metabolism | Amino acid | 3.8E−04 |
| 7-Methylxanthine | 0.37 | Xanthine metabolism | Xenobiotics | 1.6E−04 |
| 1-Methylxanthine | 0.40 | Xanthine metabolism | Xenobiotics | 1.9E−04 |
| 1,7-Dimethylurate | 0.40 | Xanthine metabolism | Xenobiotics | 1.2E−03 |
| 2-Palmitoleoyl-gpc (16:1) | 0.41 | Lysophospholipid | Lipid | 8.2E−05 |
| Theophylline | 0.44 | Xanthine metabolism | Xenobiotics | 5.3E−03 |
| Galactonate | 0.45 | Fructose, mannose, and galactose metabolism | Carbohydrate | 2.0E−04 |
| 5alpha-androstan-3beta,17alpha-diol disulfate | 0.46 | Androgenic steroids | Lipid | 8.9E−05 |
| 2-Hydroxyacetaminophen sulfate | 0.47 | Drug — analgesics, anesthetics | Xenobiotics | 2.3E−02 |
| Quinate | 0.47 | Food component/plant | Xenobiotics | 5.1E−03 |
| 1,3-Dimethylurate | 0.47 | Xanthine metabolism | Xenobiotics | 7.8E−04 |
| Paraxanthine | 0.48 | Xanthine metabolism | Xenobiotics | 1.4E−02 |
| Palmitoyl-linoleoyl-glycerol (16:0/18:2) [1] | 0.49 | Diacylglycerol | Lipid | 9.8E−05 |
| 3-Aminoisobutyrate | 0.49 | Pyrimidine metabolism, thymine containing | Nucleotide | 5.7E−04 |
| Oleoyl-linoleoyl-glycerol (18:1/18:2) [2] | 0.49 | Diacylglycerol | Lipid | 6.5E−04 |
| Alpha-cehc sulfate | 0.49 | Tocopherol metabolism | Cofactors and vitamins | 6.9E−04 |
| Metabolonic lactone sulfate | 0.50 | Partially characterized molecules | Partially characterized | 2.2E−04 |
| 1-Palmitoleoylglycerol (16:1) | 0.50 | Monoacylglycerol | Lipid | 1.8E−04 |
| 2-Aminophenol sulfate | 0.51 | Food component/plant | Xenobiotics | 2.9E−04 |
| 2-Palmitoleoylglycerol (16:1) | 0.51 | Monoacylglycerol | Lipid | 1.1E−04 |
| 4-Ethylcatechol sulfate | 0.52 | Benzoate metabolism | Xenobiotics | 1.1E−03 |
| 1-Methylurate | 0.52 | Xanthine metabolism | Xenobiotics | 1.8E−04 |
| n6,n6-dimethyllysine | 0.52 | Lysine metabolism | Amino acid | 1.5E−03 |
| Vanillic acid glycine | 0.53 | Food component/plant | Xenobiotics | 1.9E−04 |
| Palmitoloelycholine | 0.53 | Fatty acid metabolism (acyl choline) | Lipid | 4.1E−04 |
| Linoleoyl-linolenoyl-glycerol (18:2/18:3) [2] | 0.54 | Diacylglycerol | Lipid | 9.2E−05 |
| n-Delta-acetylornithine | 0.54 | Urea cycle; arginine and proline metabolism | Amino acid | 2.3E−04 |
| 1,3,7-Trimethylurate | 0.54 | Xanthine metabolism | Xenobiotics | 2.1E−03 |
| Myristoleate (14:1n5) | 0.55 | Long-chain monounsaturated fatty acid | Lipid | 1.3E−02 |
| 3-Methylglutarylcarnitine (2) | 0.55 | Leucine, isoleucine, and valine metabolism | Amino acid | 2.7E−04 |
| Cholate | 0.55 | Primary bile acid metabolism | Lipid | 1.9E−03 |
| Indolebutyrate | 0.55 | Tryptophan metabolism | Amino acid | 2.1E−04 |
| Oleoyl-linoleoyl-glycerol (18:1/18:2) [1] | 0.56 | Diacylglycerol | Lipid | 7.5E−04 |
| Ethylmalonate | 0.57 | Leucine, isoleucine, and valine metabolism | Amino acid | 9.0E−04 |
aFold difference represents the ratio of geometric means of raw metabolite values of Black relative to White participants
bAdjusted for age, sex, batch, study, and substudy
Compound represents structural isomer in the Metabolon spectral library
Table 4.
Metabolites present at higher abundance in Black relative to White participants at FDR < 0.05 in the most completely adjusted modela,b
| Metabolite | Fold difference | Subclass | Major class | FDR (Storey) |
|---|---|---|---|---|
| Propyl 4-hydroxybenzoate sulfate | 7.25 | Benzoate metabolism | Xenobiotics | < 7.6E−05 |
| Methyl-4-hydroxybenzoate sulfate | 4.29 | Benzoate metabolism | Xenobiotics | < 7.6E−05 |
| Piperine | 3.19 | Food component/plant | Xenobiotics | 3.8E−03 |
| n-Acetylalliin | 2.80 | Food component/plant | Xenobiotics | 2.1E−03 |
| Sulfate of piperine metabolite c16h19no3 (3) | 2.67 | Food component/plant | Xenobiotics | 1.1E−03 |
| Sulfate of piperine metabolite c18h21no3 (1) | 2.66 | Food component/plant | Xenobiotics | 1.2E−03 |
| Sulfate of piperine metabolite c18h21no3 (3) | 2.51 | Food component/plant | Xenobiotics | 8.7E−04 |
| Umbelliferone sulfate | 2.49 | Food component/plant | Xenobiotics | 4.8E−03 |
| n-Acetyl-1-methylhistidine | 2.44 | Histidine metabolism | Amino acid | < 7.6E−05 |
| 3-Carboxy-4-methyl-5-propyl-2-furanpropanoate (cmpf) | 2.43 | Fatty acid, dicarboxylate | Lipid | 5.9E−03 |
| Sulfate of piperine metabolite c16h19no3 (2) | 2.41 | Food component/plant | Xenobiotics | 1.0E−03 |
| 2-Hydroxyfluorene sulfate | 2.33 | Tobacco metabolite | Xenobiotics | < 7.6E−05 |
| 2-Naphthol sulfate | 2.32 | Chemical | Xenobiotics | < 7.6E−05 |
| Hydroxy-cmpf | 2.17 | Fatty acid, dicarboxylate | Lipid | 1.0E−02 |
| n-Acetylcitrulline | 2.16 | Urea cycle; arginine and proline metabolism | Amino acid | < 7.6E−05 |
| Thymol sulfate | 2.14 | Food component/plant | Xenobiotics | 5.0E−02 |
| Genistein sulfate | 2.13 | Food component/plant | Xenobiotics | 3.7E−02 |
| Gamma-cehc sulfate | 2.11 | Tocopherol metabolism | Cofactors and vitamins | 1.1E−03 |
| Glycohyocholate | 1.89 | Secondary bile acid metabolism | Lipid | 5.8E−03 |
| 2-Piperidinone | 1.87 | Food component/plant | Xenobiotics | 5.9E−03 |
| Taurodeoxycholic acid 3-sulfate | 1.86 | Secondary bile acid metabolism | Lipid | 1.5E−02 |
| Tauro-beta-muricholate | 1.72 | Primary bile acid metabolism | Lipid | 2.6E−02 |
| Glutarate (c5-dc) | 1.54 | Fatty acid, dicarboxylate | Lipid | 6.8E−03 |
| n-Acetylglutamine | 1.49 | Glutamate metabolism | Amino acid | < 7.6E−05 |
| Taurocholenate sulfate | 1.47 | Secondary bile acid metabolism | Lipid | 6.2E−03 |
| Carotene diol (3) | 1.47 | Vitamin A metabolism | Cofactors and vitamins | 5.0E−03 |
| Spermidine | 1.45 | Polyamine metabolism | Amino acid | 2.0E−02 |
| n-Acetylarginine | 1.45 | Urea cycle; arginine and proline metabolism | Amino acid | 9.6E−04 |
| n-Acetyl-2-aminooctanoate | 1.43 | Fatty acid, amino | Lipid | 1.2E−02 |
| n-Acetylasparagine | 1.38 | Alanine and aspartate metabolism | Amino acid | 2.0E−03 |
| Cis-4-decenoylcarnitine (c10:1) | 1.34 | Fatty acid metabolism (acyl carnitine, monounsaturated) | Lipid | 6.5E−03 |
| Homoarginine | 1.33 | Urea cycle; arginine and proline metabolism | Amino acid | 1.0E−02 |
| 1-(1-Enyl-stearoyl)-2-arachidonoyl-gpe (p-18:0/20:4) | 1.29 | Plasmalogen | Lipid | 1.4E−02 |
| Sphingomyelin (d18:1/20:1, d18:2/20:0) | 1.26 | Sphingomyelins | Lipid | < 7.6E−05 |
| Arginine | 1.26 | Urea cycle; arginine and proline metabolism | Amino acid | 6.8E−03 |
| Creatine | 1.26 | Creatine metabolism | Amino acid | 9.7E−03 |
| Carotene diol (2) | 1.26 | Vitamin A metabolism | Cofactors and vitamins | 3.7E−02 |
| 3-Methoxytyrosine | 1.25 | Tyrosine metabolism | Amino acid | 1.6E−02 |
| Sphingomyelin (d18:2/24:2) | 1.25 | Sphingomyelins | Lipid | 9.6E−04 |
| Guanidinoacetate | 1.25 | Creatine metabolism | Amino acid | 6.9E−03 |
| Succinylcarnitine (c4-dc) | 1.25 | TCA cycle | Energy | 3.5E−02 |
| 2′-o-methyluridine | 1.24 | Pyrimidine metabolism, uracil containing | Nucleotide | 1.5E−02 |
| 2-Hydroxyglutarate | 1.23 | Fatty acid, dicarboxylate | Lipid | 9.3E−03 |
| 1-(1-Enyl-palmitoyl)-2-linoleoyl-gpc (p-16:0/18:2) | 1.22 | Plasmalogen | Lipid | 1.2E−03 |
| 1-(1-Enyl-palmitoyl)-2-arachidonoyl-gpe (p-16:0/20:4) | 1.22 | Plasmalogen | Lipid | 3.5E−02 |
| Sphingomyelin (d18:2/18:1) | 1.21 | Sphingomyelins | Lipid | 2.7E−02 |
| Sphingomyelin (d18:1/20:2, d18:2/20:1, d16:1/22:2) | 1.20 | Sphingomyelins | Lipid | 2.0E−02 |
| 1-(1-Enyl-palmitoyl)-2-arachidonoyl-gpc (p-16:0/20:4) | 1.19 | Plasmalogen | Lipid | 6.2E−03 |
| Sphingomyelin (d18:1/22:2, d18:2/22:1, d16:1/24:2) | 1.18 | Sphingomyelins | Lipid | 6.3E−03 |
| 1-(1-Enyl-palmitoyl)-2-palmitoyl-gpc (p-16:0/16:0) | 1.17 | Plasmalogen | Lipid | 2.0E−03 |
| Gamma-glutamylthreonine | 1.15 | Gamma-glutamyl amino acid | Peptide | 3.7E−02 |
| 5-Methylthioribose | 1.15 | Methionine, cysteine, SAM, and taurine metabolism | Amino acid | 9.4E−03 |
| 1-(1-Enyl-palmitoyl)-2-oleoyl-gpc (p-16:0/18:1) | 1.15 | Plasmalogen | Lipid | 2.1E−02 |
| Sphingomyelin (d18:1/17:0, d17:1/18:0, d19:1/16:0) | 1.13 | Sphingomyelins | Lipid | 2.7E−02 |
| Betaine | 1.12 | Glycine, serine, and threonine metabolism | Amino acid | 3.5E−02 |
| Creatinine | 1.10 | Creatine metabolism | Amino acid | 2.2E−02 |
aFold difference represents the ratio of geometric means of raw metabolite values of Black relative to White participants
bAdjustment for age, sex, batch, substudy, dietary pattern, education, social vulnerability index, exercise, BMI, smoking, alcohol drinking, coffee, medication use, and chronic disease (cardiovascular disease, diabetes, cancer, hypertension)
Among metabolites showing the lowest abundance in Black relative to White participants was the microbial metabolite n-methylpipecolate, along with lysine metabolites (n6-methyllysine, n6,n6-dimethyllysine), various xanthine metabolites, and tryptophan betaine, represented in both the simplest and fullest models (Tables 3 and 5; Additional file 1, Supplementary Table 4). Various types of fatty acids and phospholipids showed the greatest statistical significance among metabolites inversely associated with Black race. Comparing the fully adjusted with simplest model, there was some attenuation of representation of acyl carnitines and secondary bile acids, among other compounds, and greater representation of leucine, isoleucine, and valine metabolites, as well as long-chain polyunsaturated fatty acids, and glycine, serine, threonine metabolites in the fully adjusted model. Overall, results from the fully adjusted model were very similar to other less complex models (Additional file 1, Supplementary Tables 5–8), where across these models at least 58 metabolites were higher, and 345 lower in Black participants, with the most notable difference being attenuation of representation of acyl carnitines in the fullest relative to the simpler models.
Table 5.
Top 40 (of 348) metabolites present at lower abundance in Black relative to White participants at FDR < 0.05 in the most completely adjusted modela,b
| Metabolite | Fold difference | Subclass | Major class | FDR (Storey) |
|---|---|---|---|---|
| n-Methylpipecolate | 0.27 | Bacterial/fungal | Xenobiotics | 4.8E−03 |
| n6-methyllysine | 0.31 | Lysine metabolism | Amino acid | 2.4E−03 |
| Theobromine | 0.33 | Xanthine metabolism | Xenobiotics | 1.9E−03 |
| Tryptophan betaine | 0.35 | Tryptophan metabolism | Amino acid | 1.8E−04 |
| Myristoleate (14:1n5) | 0.36 | Long-chain monounsaturated fatty acid | Lipid | 1.2E−03 |
| 3-Methylxanthine | 0.37 | Xanthine metabolism | Xenobiotics | 2.5E−04 |
| Palmitoyl-linoleoyl-glycerol (16:0/18:2) [1] | 0.38 | Diacylglycerol | Lipid | 2.3E−04 |
| 5-Acetylamino-6-amino-3-methyluracil | 0.39 | Xanthine metabolism | Xenobiotics | 2.7E−03 |
| 2-Palmitoleoylglycerol (16:1) | 0.40 | Monoacylglycerol | Lipid | 3.0E−04 |
| Metabolonic lactone sulfate | 0.41 | Partially characterized | Partially characterized | 4.8E−04 |
| Alpha-cehc sulfate | 0.41 | Tocopherol metabolism | Cofactors and vitamins | 5.3E−04 |
| Galactonate | 0.42 | Fructose, mannose, and galactose metabolism | Carbohydrate | 4.0E−04 |
| 2-Hydroxyacetaminophen sulfate | 0.42 | Drug — analgesics, anesthetics | Xenobiotics | 2.9E−02 |
| 7-Methylxanthine | 0.43 | Xanthine metabolism | Xenobiotics | 1.5E−03 |
| 1-Palmitoleoylglycerol (16:1) | 0.44 | Monoacylglycerol | Lipid | 2.2E−04 |
| Oleoyl-linoleoyl-glycerol (18:1/18:2) [2] | 0.45 | Diacylglycerol | Lipid | 1.2E−03 |
| Caffeine | 0.45 | Xanthine metabolism | Xenobiotics | 8.2E−03 |
| Linoleoyl-linolenoyl-glycerol (18:2/18:3) [2] | 0.45 | Diacylglycerol | Lipid | 1.6E−04 |
| 1,7-Dimethylurate | 0.46 | Xanthine metabolism | Xenobiotics | 2.6E−03 |
| 1-Methylxanthine | 0.47 | Xanthine metabolism | Xenobiotics | 1.2E−03 |
| Docosatrienoate (22:3n6) | 0.48 | Long-chain polyunsaturated fatty acid (n3 and n6) | Lipid | 2.1E−04 |
| 3-Methylglutarylcarnitine (2) | 0.48 | Leucine, isoleucine, and valine metabolism | Amino acid | 4.0E−04 |
| Palmitoleate (16:1n7) | 0.49 | Long-chain monounsaturated fatty acid | Lipid | 2.0E−04 |
| 2-Palmitoleoyl-gpc (16:1) | 0.49 | Lysophospholipid | Lipid | 4.0E−03 |
| 9-Hydroxystearate | 0.49 | Fatty acid, monohydroxy | Lipid | 8.1E−05 |
| Linoleoyl-linoleoyl-glycerol (18:2/18:2) [2] | 0.50 | Diacylglycerol | Lipid | 8.2E−05 |
| 5alpha-androstan-3beta,17alpha-diol disulfate | 0.50 | Androgenic steroids | Lipid | 1.3E−03 |
| Vanillic acid glycine | 0.51 | Food component/plant | Xenobiotics | 9.6E−04 |
| 3-Aminoisobutyrate | 0.51 | Pyrimidine metabolism, thymine containing | Nucleotide | 4.3E−04 |
| Ceramide (d16:1/24:1, d18:1/22:1) | 0.51 | Ceramides | Lipid | 1.9E−04 |
| Palmitoloelycholine | 0.52 | Fatty acid metabolism (acyl choline) | Lipid | 2.8E−04 |
| 2-Aminophenol sulfate | 0.52 | Food component/plant | Xenobiotics | 2.0E−03 |
| Oleoyl-linoleoyl-glycerol (18:1/18:2) [1] | 0.53 | Diacylglycerol | Lipid | 1.6E−03 |
| 1-Palmitoyl-2-palmitoleoyl-gpc (16:0/16:1) | 0.53 | Phosphatidylcholine (PC) | Lipid | 9.6E−04 |
| Palmitoyl-linoleoyl-glycerol (16:0/18:2) [2] | 0.53 | Diacylglycerol | Lipid | 2.1E−04 |
| n6,n6-dimethyllysine | 0.53 | Lysine metabolism | Amino acid | 8.0E−04 |
| Cystathionine | 0.54 | Methionine, cysteine, SAM, and taurine metabolism | Amino acid | 1.4E−03 |
| 5alpha-androstan-3alpha,17beta-diol monosulfate (1) | 0.54 | Androgenic steroids | Lipid | 4.0E−04 |
aFold difference represents the ratio of geometric means of raw metabolite values of Black relative to White participants
bAdjusted for age, sex, batch, substudy, dietary pattern, education, social vulnerability index, exercise, BMI, coffee, medication use, and chronic disease (cardiovascular disease, diabetes, cancer, hypertension)
Compound represents structural isomer in the Metabolon spectral library
A sensitivity analysis considering only the 258 participants not included in the pilot study [20] showed overall fewer significant metabolites (n = 296), although 95% (281) of metabolites significant in the analysis considering these participants were also significant considering the full dataset (Additional file 1, Supplementary Table 9), including metabolites showing the greatest fold differences or statistical significance. A similar analysis of pilot participants revealed 268 differential metabolites, and again, the majority of these were also differential in the analysis considering the full cohort (75%), and nearly half overlapped with the non-pilot study participants (Additional file 1, Supplementary Tables 10 and 11).
Linear regression analysis of metabolite subclasses
Besides examining associations of race with individual metabolites, we also conducted an analysis of metabolite subclasses by averaging metabolites across their respective biochemical subclasses. Results from the fully adjusted model including adjustment for lifestyle/behavioral variables and environmental stress revealed a total of 38 differentially abundant metabolite subclasses of a total of 92 queried subclasses, comparing the two racial groups (Table 6). Creatine metabolism exclusively showed higher abundance in Blacks (fold change = 1.20). Besides creatine metabolism, subclasses with the strongest significance included long-chain monounsaturated fatty acid, pyrimidine metabolism, ceramides, and several phospho- and glycero-lipid subclasses (notably lysophospholipid, monoacylglycerol, phosphatidylethanolamine, phosphatidylinositol, glycerolipid metabolism, diacylglycerol, monoacylglycerol, phosphatidylethanolamine, glycerolipid metabolism), leucine, isoleucine, and valine metabolism, and lysine metabolism, all showing lower abundance in Blacks. Results from less complex models including only basic covariates (age, sex, substudy, and batch excluding BMI) or basic covariates in addition to dietary pattern, exercise, and/or environmental stress variables showed a similar representation of the most significant subclasses with 44–54 differential metabolic subclasses detected, all represented at lower abundance, with the exception of creatine metabolism (Additional file 1, Supplementary Tables 12–14).
Table 6.
Metabolite subclasses associated with race (Black vs White) at FDR < 0.05 in the most completely adjusted modela,b
| Subclass | Fold difference (95% CI) | n total metabolites | n significant metabolites | n > 1 | n < 1 | FDR |
|---|---|---|---|---|---|---|
| Creatine metabolism | 1.20 (1.11, 1.29) | 3 | 3 | 3 | 0 | < 9.0E−05 |
| Long-chain monounsaturated fatty acid | 0.61 (0.49, 0.75) | 7 | 7 | 0 | 7 | 9.5E−05 |
| Pyrimidine metabolism, thymine containing | 0.73 (0.63, 0.83) | 2 | 1 | 0 | 1 | 1.2E−04 |
| Ceramides | 0.74 (0.65, 0.85) | 9 | 8 | 0 | 8 | 1.4E−04 |
| Lysophospholipid | 0.75 (0.67, 0.84) | 29 | 24 | 0 | 24 | 1.6E−04 |
| Monoacylglycerol | 0.65 (0.54, 0.79) | 16 | 13 | 0 | 13 | 1.6E−04 |
| Phosphatidylethanolamine (PE) | 0.70 (0.60, 0.83) | 12 | 10 | 0 | 10 | 1.8E−04 |
| Phosphatidylinositol (PI) | 0.75 (0.67, 0.83) | 6 | 6 | 0 | 6 | 2.4E−04 |
| Glycerolipid metabolism | 0.77 (0.68, 0.87) | 2 | 2 | 0 | 2 | 2.6E−04 |
| Tryptophan metabolism | 0.84 (0.77, 0.92) | 20 | 11 | 0 | 11 | 3.3E−04 |
| Diacylglycerol | 0.57 (0.47, 0.68) | 11 | 10 | 0 | 10 | 4.7E−04 |
| Pyrimidine metabolism, orotate containing | 0.79 (0.70, 0.90) | 3 | 2 | 0 | 2 | 7.1E−04 |
| Long-chain polyunsaturated fatty acid (n3 and n6) | 0.72 (0.61, 0.86) | 17 | 14 | 0 | 14 | 7.7E−04 |
| Leucine, isoleucine, and valine metabolism | 0.87 (0.80, 0.94) | 28 | 15 | 0 | 15 | 7.8E−04 |
| Lysine metabolism | 0.83 (0.75, 0.92) | 16 | 6 | 0 | 6 | 9.5E−04 |
| Fatty acid metabolism (acyl choline) | 0.72 (0.60, 0.87) | 9 | 8 | 0 | 8 | 9.8E−04 |
| Xanthine metabolism | 0.49 (0.32, 0.74) | 13 | 12 | 0 | 12 | 1.5E−03 |
| Ascorbate and aldarate metabolism | 0.84 (0.75, 0.93) | 6 | 5 | 0 | 5 | 2.2E−03 |
| Phosphatidylcholine (PC) | 0.85 (0.77, 0.94) | 19 | 10 | 0 | 10 | 2.4E−03 |
| Fatty acid, dihydroxy | 0.85 (0.76, 0.94) | 5 | 3 | 0 | 3 | 2.9E−03 |
| Fructose, mannose, and galactose metabolism | 0.79 (0.68, 0.92) | 4 | 1 | 0 | 1 | 3.3E−03 |
| Guanidino and acetamido metabolism | 0.73 (0.60, 0.89) | 2 | 1 | 0 | 1 | 3.4E−03 |
| Pentose metabolism | 0.85 (0.76, 0.95) | 6 | 4 | 0 | 4 | 4.7E−03 |
| Long-chain saturated fatty acid | 0.82 (0.72, 0.95) | 7 | 5 | 0 | 5 | 7.0E−03 |
| Dihydroceramides | 0.79 (0.66, 0.94) | 2 | 2 | 0 | 2 | 7.4E−03 |
| Pyrimidine metabolism, cytidine containing | 0.84 (0.74, 0.96) | 5 | 2 | 0 | 2 | 1.3E−02 |
| Eicosanoid | 0.75 (0.59, 0.95) | 3 | 2 | 0 | 2 | 1.7E−02 |
| Chemical | 0.87 (0.78, 0.98) | 20 | 9 | 1 | 8 | 1.9E−02 |
| Fatty Acid, monohydroxy | 0.90 (0.82, 0.98) | 19 | 5 | 0 | 5 | 2.0E−02 |
| Androgenic steroids | 0.80 (0.66, 0.97) | 21 | 8 | 0 | 8 | 2.1E−02 |
| Pantothenate and CoA metabolism | 0.78 (0.63, 0.98) | 2 | 1 | 0 | 1 | 3.0E−02 |
| Gamma-glutamyl amino acid | 0.93 (0.86, 1.00) | 15 | 9 | 1 | 8 | 3.4E−02 |
| Nicotinate and nicotinamide metabolism | 0.86 (0.75, 0.99) | 5 | 2 | 0 | 2 | 3.6E−02 |
| Sphingosines | 0.88 (0.78, 1.00) | 2 | 1 | 0 | 1 | 3.6E−02 |
| Purine metabolism, adenine containing | 0.94 (0.88, 1.00) | 5 | 2 | 0 | 2 | 4.0E−02 |
| Fatty acid metabolism (acyl carnitine, dicarboxylate) | 0.86 (0.73, 1.01) | 4 | 2 | 0 | 2 | 4.6E−02 |
| Sphingolipid synthesis | 0.89 (0.79, 1.00) | 3 | 2 | 0 | 2 | 4.6E−02 |
| Phospholipid metabolism | 0.92 (0.84, 1.00) | 6 | 2 | 0 | 2 | 4.7E−02 |
aLinear regression analysis based on composite t-statistics derived by dividing by the standard deviation of the averaged, log-transformed metabolites
bAdjustment for age, sex, batch, substudy, dietary pattern, education, social vulnerability index, exercise, BMI, smoking, alcohol drinking, coffee, medication use, and chronic conditions (CVD, diabetes, hypertension, cancer)
For many of the differentially represented subclasses, the vast majority of component metabolites were significantly differential (names of component metabolites in Additional file 1, Supplementary Table 15), and showed lower abundance in Blacks, as determined from linear regression analysis of individual metabolites. Examples are the long-chain monounsaturated fatty acid, monoacylglycerol, phosphatidylethanolamine, ceramides, lysophospholipid, diacylglycerol, long-chain polyunsaturated fatty acid, and xanthine metabolism subclasses (Table 6). Several of the top subclasses were replicated, both in analyses excluding or including only pilot participants, with many in common between both sub-cohorts. (Additional file 1, Supplementary Table 16). Pathway analysis of metabolic pathways in KEGG revealed an over-representation of biosynthesis of unsaturated fatty acids pathway (FDR = 0.02), consistent with results from subclass analyses (Additional file 1, Supplementary Table 17).
In light of the finding of several differentially abundant subclasses of lipids, we performed additional analyses to adjust for dietary fat, including mono-, poly-, and saturated fat consumption. Results from the linear regression analysis of individual metabolites were overall similar to those from other models (Additional file 1, Supplementary Table 18). However, far fewer lipid/fatty acid and other subclasses were statistically differential after adjustment for dietary fat, although several were retained, including long-chain monounsaturated fatty acids among others (Additional file 1, Supplementary Table 19). Additionally, given the relevance of several differentially abundant metabolites or biochemical subclasses related to kidney function, namely, creatine metabolism and acetylated amino acids, we also compared metabolic profiles among a subset of individuals with plasma creatinine measurements (mg/dL) adjusting for estimated glomerular filtration rate (eGFR). Adjustment for eGFR (mL/min/m2) resulted in far fewer metabolites showing higher abundance in Blacks, notably many acetylated amino acids, which were no longer significant, although creatine was still present in higher abundance. Interestingly, the inclusion of eGFR in analyses of differential abundance of biochemical subclasses yielded a higher number of differential subclasses, with some showing stronger statistical significance, most notably, lysine metabolism (Additional file 1, Supplementary Tables 20–23). Creatine metabolism, however, was no longer significant after adjustment for eGFR.
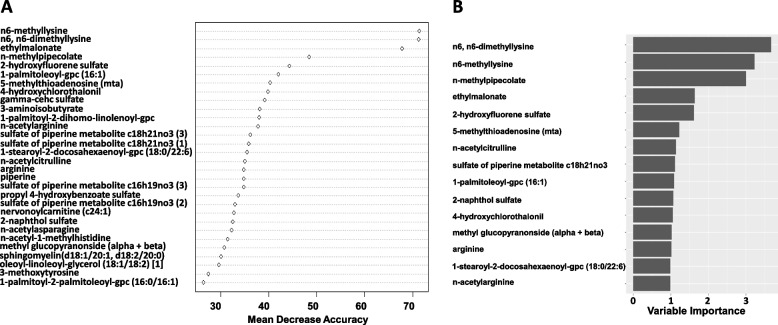
Random forest
Random forest analysis was performed to identify metabolic profiles differentiating race, reasoning that metabolites showing the greatest importance in distinguishing race might be more reflective of biological differences. Random forest classification showed a predictive accuracy in the test set of 81.3% and revealed the greatest discriminating potential for n6,n6-dimethyllysine, n6-methyllysine, n-methylpipecolate, ethylmalonate, and 2-hydroxyfluorene sulfate (Fig. 1A). These metabolites were among those found to be most differential in the linear regression analysis. Random forest was also performed using 5-fold cross-validation, with a predictive accuracy of 87.9%, and overlap of the top 5 metabolites showing the greatest variable importance (Fig. 1B), thereby providing evidence of the robustness of the random forest classifier and validating the discriminating ability of the most influential metabolites.
Fig. 1.
Random forest analysis showing A mean decrease accuracy representing an average decrease in accuracy after permutation of each respective metabolite and B importance plot using 5-fold cross-validation, classifying Black and White participants
Discussion
Differences in metabolite abundance may account for racial disparities in susceptibility to chronic diseases [37–39]. Using an untargeted metabolomics platform, we investigated differences in plasma metabolites with roles in various biological pathways with relevance to chronic diseases. We noted differences in a number of xenobiotic and lipid metabolite subclasses — particularly lysophospholipids, mono- and diacylglycerols, ceramides, and related lipids.
We hypothesized that inter-related lifestyle and socioeconomic factors largely contribute to racial differences in metabolic profiles. However, expanded models adjusting for these variables did not appreciably alter results, neither did adjustment for lifestyle (dietary pattern, exercise, etc.) and social stress variables, as the number of differential metabolites was not diminished, although differences in some xenobiotic metabolites are likely explained by differences in lower caffeine consumption (as many xanthine metabolites are caffeine derivatives), and lower medication use (i.e., drugs/analgesics) by Black participants. Additionally, the differences we noted in consumption of select foods or nutrients may partially explain differences in lipids and other metabolites not evident with adjustment for dietary pattern alone. For example, two of the most differentially abundant or discriminating metabolites from linear regression or random forest analyses, n-methylpipecolate and tryptophan betaine (lower in Blacks) have been associated with consumption of certain types of legumes [40–42], and legumes were among foods consumed at higher amounts among Black participants.
Dietary habits and food preferences could have a major impact on the types/distribution on lipid metabolites found to be differential between Black and White participants, for example, phospholipids or diacylglycerols, besides triglycerides in plasma (again lower in Blacks). Synthesis of diacylglycerols and phospholipids is heavily influenced by diet. Indeed, we noted differences in dietary intakes of saturated and unsaturated fatty acids, which helped explain differences in biochemical subclasses or pathways relevant to lipid biosynthesis or metabolism. Fatty acids from the diet diffuse in micelles from the gut lumen into enterocytes and are converted to phosphatidic acid, and subsequently diacylglycerols or phospholipids, and later secreted as vesicles into plasma in chylomicrons or very-low-density lipoprotein, and transported to other tissues [43]. Lysophoshospholipids are intermediates of other phospholipids, generated by enzymatic activity resulting in the removal of an acyl group. Variations in the diet (consumption of oleic or palmitic acids, and related oils) could mean variations in phospholipid profiles, as well as the fatty acids incorporated into the C1 and C2 positions of diacylglycerols, which are continually in flux [44]. Hence differences in some of these lipid types reflect differences in food preferences, which are culturally influenced. Accordingly, Black Americans within AHS-2 were shown to have higher consumption of certain types of carbohydrates and “soul” or Caribbean foods such as macaroni and cheese, red/pinto beans, rice and beans, black-eyed peas, okra, and other foods [45], which could contribute to differences in lipid metabolism. This is consistent with the higher consumption of carbohydrates in Black participants in the current study.
A high carbohydrate diet might increase de novo lipogenesis and consequently phospholipids or triglycerides, as glucose impacts this process by providing acetyl-coA, and can induce expression or production of lipogenic genes or enzymes [46]. Induction of lipogenic genes is also evident in the context of obesity. Lysophospholipid metabolism is also altered with obesity, and in turn impacts obesity-related diseases [47]. Besides differences in diet or obesity, the other possible explanation for differences in phospholipids and other fatty acids relates to ethnic differences in triglycerides. Black Americans have been found to have lower triglycerides in spite of higher rates of several cardiometabolic diseases, termed “the triglyceride paradox” [48], although the reason is not clear. It is not clear from the current study how levels of plasma triglycerides differed between the two racial groups. But there were lower levels of long-chain fatty acids and monoacylglycerols, which are products of triglyceride digestion by pancreatic lipase. Thus, it is possible that lower levels of triglycerides correlate with lower products of triglyceride digestion (i.e., lower monoacylglycerols and monounsaturated/long-chain fatty acids) in Blacks. As glycerol and glycerol-3-phosphate (comprising the glycerophospholipids subclass) are precursors for phospholipids and di- and triacylglycerols, it might be expected that these would be significantly lower in Blacks.
No lipid metabolite subclasses showed higher abundance in Black participants. There was, however, a proportionately greater representation of sphingomyelins and plasmalogens in Black participants when considering individual metabolites. Both plasmalogens and sphingomyelins have important roles in cell structure and signaling. While their pathophysiological roles are not completely clear, dysregulation may be associated with neurodegenerative disorders, cancer, cardiovascular disease, diabetes, and other metabolic diseases [49, 50] which could explain higher rates of these diseases in Blacks.
Lysine and leucine metabolism showed significant and consistent differences comparing Black and White participants. Products of leucine breakdown such as 3-methylglutaconate and ethylmalonate, among others, have been previously associated with single nucleotide polymorphisms (SNPs) [51], which could partly explain the differences. Ethylmalonate is a branched fatty acid that has been associated with a protein-altering variant in acyl-CoA dehydrogenase (ACAD), which may affect fatty acid oxidation [51, 52]. This is consistent with the differences observed in fatty acid and lipid metabolism in the current study, as leucine and lysine catabolism both yield acetyl CoA to regulate fatty acid synthesis. It was interesting that methylated lysines, particularly, showed the strongest ability to differentiate Black and White participants. These metabolites are especially interesting because of their potential involvement in regulation of gene expression. For instance, histone lysine methylation may be associated with activation or silencing of genes depending on position [53, 54], and these metabolites could have been present in plasma as a result of histone degradation. It is also possible that these lysine residues are nonhistone derived. Nonhistone lysine methylation has implications for oncogenesis and cancer progression [55]. As methylation of lysine is regulated by lysine methyltransferase, it would be interesting to determine in future studies if such enzymes show differential activity in Black and White Americans in the context of both health and disease.
The creatine metabolism subclass was exclusively higher in Black participants, with all three component metabolites statistically higher in the fully adjusted model. Higher creatine was the top metabolite associated positively with estimated glomerular filtration rate (eGFR) in African American participants of the Atherosclerosis Risk in Communities (ARIC) study without chronic kidney disease at baseline [56]. In the current study, race was associated with creatine metabolism after adjustment for social, demographic, and environmental factors, and creatine (though not creatine metabolism) showed differential abundance even after adjustment for eGFR in a sub-analysis. Our finding of higher abundance of creatine or the creatine metabolism subclass in Blacks may highlight clinically relevant racial distinctions, as creatinine is the principal determinant of eGFR [57]. Because even healthy Black or African Americans have shown higher levels of creatinine, various equations have been generated to calculate eGFR with adjustment for race. But the use of such equations is controversial as they often yield higher estimates of GFR in Blacks, leading to a greater number of undiagnosed cases. Recently, a new race-free calculation of eGFR was developed with either creatinine or another filtration marker called cystatin-C [58]. Given the apparently complex role of race in creatine metabolism, metabolomics analysis will likely provide increased discriminatory power to GFR estimation in the future.
The higher abundance of creatine-related metabolites was paralleled by higher abundance of arginine, a precursor for creatine that has been found to be higher in other individuals of African descent [59]. Even more interestingly, though not directly related to creatine metabolism, a number of n-acetylated metabolites, which have relevance to kidney function, showed significant differences in the current study, though in both directions. A number of these metabolites have been found inversely associated with eGFR in African Americans [56, 60], many also associated with SNPs more common in African Americans [51, 60], and particularly mapping to loci related to acetylated amino acids, consequently impacting enzyme-metabolite interactions. Ten of 12 recently identified metabolites with novel validated inverse associations with creatinine glomerular filtration rate in novel subpathways (as discovered among participants of the ARIC and Bogalusa Heart Study (BHS) cohorts), were significantly lower in Black participants in the current study, including some metabolites showing greatest fold changes or discriminating ability, such as n-methylpipecolate, and 3-amino-isobutyrate, among others. Consequently, the observed racial differences in abundance of acetylated amino acids which are putative uremic solutes, and particularly those in higher abundance in Black participants, may have important implications for kidney health disparities as Blacks present with significantly higher kidney function diseases.
Metabolites of methyl- and propyl- parabens, common preservatives in foods, cosmetics (hair, skin, etc.), and medications, were markedly higher (~ 6-fold) in Blacks. African or Black Americans have shown higher levels of parabens in urine in other studies [61], likely attributable to hair products [62]. Particularly hair products have been found to contain endocrine-disrupting chemicals and have been linked with breast cancer (due to parabens or other chemicals) [13, 62–67], and are used more commonly by Black women [67, 62, 68]. Though parabens are generally considered safe, there are some questions about toxicity given some reports of oxidative DNA damage upon light irradiation in dermal tissues [69]. Our findings warrant further investigation of these metabolites in studies with Black participants and biospecimens.
The minimal impact of other lifestyle factors, and socioeconomic or psychosocial stressors (which were not notably different between Black and White participants in this study) on metabolic profiles might be explained by the overall healthier lifestyles among Black AHS-2 participants when compared to the general population. AHS-2 participants have healthier lifestyles, diminishing some of the social disparities that exist between Black and White Americans, and translating into better health outcomes for Black participants. The AHS-2 cohort consists of a large proportion of individuals following vegetarian or plant-based diets, with very little tobacco smoking or alcohol drinking, consistent with the religious doctrine. Thus these participants are more alike in such areas that would otherwise contribute to confounding, yet with variety in dietary habits/patterns and other lifestyle factors. While these are unique features or strengths of this cohort, there are inevitable limitations in comparability with other populations. An external cohort of non-Adventists will have notably different lifestyles and behaviors which impact health outcomes and biological pathways impacting disease. AHS-2 participants, including Blacks, have lower overall cancer incidence and lower all-cause and/or cancer mortality compared to the National Longitudinal Mortality Study (NLMS) population and its Surveillance, Epidemiology, and End Results (SEER) substudy, representing US census populations [70]. The religious engagement and church activity also may favorably impact health and mortality. Such findings highlight the relevance of healthy lifestyles, behaviors, and social experiences (healthy diets, absence of smoking, alcohol drinking, religion, management of stress) in controlling racial disparities. Consequently, there are limitations in terms of external validity.
It is not clear how the differential metabolic profiles might relate to susceptibility to metabolic diseases, and it is challenging to disentangle the biological and social effects in explaining racial differences, which may be inherently intertwined. For example, social stress may interact with biological factors and particularly genetic or epigenetic factors that influence metabolite abundance. The metabolic profile is attributable to various biological and environmental factors, i.e., diet, genetic polymorphisms, the gut microbial community, physical activity, and stress, among others. The statistical models generated were not comprehensive with regard to covariates, and there is a likelihood of residual confounding, particularly in light of the strong relationship between diet and metabolites. However, we have attempted to examine the contributions of lifestyle and social factors by generating and comparing models of varying complexity. Hence, the observed racial differences in metabolic profiles should be interpreted with caution. Nonetheless, many of the differentially abundant lipid subclasses have important physiological roles, and consequently implications for health and disease. Lysophospholipids and other signaling lipids have roles in cardiometabolic and neurological health, and inflammatory responses [71, 72], besides obesity [47]. Besides dietary components/nutrients, it is possible that the lower abundance of certain metabolites reflects differences in body composition that were unaddressed with adjustment for BMI. The potential contribution of genetic polymorphisms also should not be ignored, which might impact kidney health or function and disease risk. Interestingly, metabolomic lactone sulfate which was markedly lower in Black participants, has been associated with a polymorphism in a CYP3A gene and poor cardiometabolic health. An association of such biomarkers with a lower risk of cardiometabolic and other diseases in Black Americans may highlight, all the more, racial disparities in the progression of chronic diseases, and the critical contributions of social determinants of health.
This study has notable strengths, including the approximately equal numbers of Black and White participants with many lifestyle similarities, and comprehensive data collected on study participants, allowing for considerable model complexity (inclusion of data on social vulnerability, dietary patterns, and other lifestyle behaviors and environmental stress). One issue, however, is that the Black participants in this cohort may not reflect the general population as far as socioeconomic or -cultural characteristics, which could be seen as a strength or a limitation. The Black AHS-2 participants overall have a higher level of education and experience less socioeconomic disadvantage. Another limitation is the somewhat small sample size for the current study precluding more comprehensive comparisons considering aspects of diet or stratified analyses, although there was more than enough power to detect statistical differences in plasma metabolites by race, and we were able to repeat analyses comparing two sub-samples.
Conclusions
Black and White participants show distinct metabolic profiles, most notably differences in phospholipid metabolism and related lipid subclasses, besides creatine and lysine metabolism, and metabolites with relevance to kidney function. Differences in some of these metabolites and subclasses are likely a reflection of unique dietary behaviors, not simply dietary patterns, while other differences may be attributed to genetic influences. Many differences in xenobiotic metabolites probably relate to lifestyle choices, such as the use of medications and caffeinated beverages. Whether attributed to dietary, genetic, or other factors, these differential metabolic profiles may have implications for cardiometabolic and renal health in Blacks, driving various health disparities outcomes.
Supplementary Information
Additional file 1: Supplementary Table 1. Dietary intakes of study participants. Supplementary Table 2. All metabolites present at higher abundance in Black relative to White participants at FDR < 0.05 in simplest model. Supplementary Table 3. All metabolites present at lower abundance in Black relative to White participants at FDR < 0.05 in simplest model. Supplementary Table 4. Metabolites (n = 348) present at lower abundance in Black relative to White participants at FDR < 0.05. Supplementary Table 5. Metabolites present at higher abundance in Black relative to White participants at FDR < 0.05 (with additional adjustment for BMI and dietary pattern). Supplementary Table 6. Metabolites present at lower abundance in Black relative to White participants at FDR < 0.05 (with additional adjustment for BMI and dietary pattern). Supplementary Table 7. Metabolites present at higher abundance in Black relative to White participants at FDR < 0.05 (with additional adjustment for BMI, dietary pattern, exercise, social vulnerability index, and psychosocial stress). Supplementary Table 8. Metabolites present at lower abundance in Black relative to White participants at FDR < 0.05 (with additional adjustment for BMI, dietary pattern, exercise, social vulnerability index, and psychosocial stress). Supplementary Table 9. Metabolites present at lower abundance in Black relative to White participants at FDR < 0.05 excluding pilot study participants. Supplementary Table 10. Metabolites present at lower abundance in Black relative to White participants at FDR < 0.05 in pilot study participants. Supplementary Table 11. Differentially abundant metabolites in common between analyses with pilot (n = 93) and non-pilot (n = 258) study participants at FDR<0.05. Supplementary Table 12. Metabolite subclasses associated with race (Black vs White) at FDR in simplest model < 0.05. Supplementary Table 13. Metabolite subclasses associated with race (Black vs White) at FDR < 0.05 (adjusted additionally for BMI and dietary pattern). Supplementary Table 14. Metabolite subclasses associated with race (Black vs White) at FDR < 0.05 (adjusted additionally for BMI, dietary pattern, social vulnerability index, and psychosocial stress). Supplementary Table 15. Names of statistically significant component metabolites (based on linear regression analysis) contained within each subclass differentially abundant at FDR < 0.05. Supplementary Table 16. Differentially abundant subclasses in common between analyses with pilot (n = 93) and non-pilot (n = 258) study participants at FDR<0.05. Supplementary Table 17. Results from pathway analysis of metabolic pathways over-represented comparing Black and White participants (at P ≤ 0.5). Supplementary Table 18. Metabolites differentially abundant in Black relative to White participants at FDR < 0.05 (adjusted additionally for dietary pattern, dietary fat (polyunsaturated, monounsaturated, and saturated), education, social vulnerability index, exercise, BMI, coffee, medication use, chronic disease cardiovascular disease, diabetes, hypertension). Supplementary Table 19. Metabolite subclasses associated with race (Black vs White) at FDR < 0.05 (adjusted additionally for BMI, dietary pattern, social vulnerability index, psychosocial stress, and dietary fat). Supplementary Table 20. Metabolites present at higher abundance in Black relative to White participants at FDR < 0.05 in n = 140 study participants with plasma creatinine measurements (and adjustment for eGFR). Supplementary Table 21. Metabolites present at higher abundance in Black relative to White participants at FDR < 0.05 in n = 140 study participants with plasma creatinine measurements. Supplementary Table 22. Metabolite subclasses associated with race (Black vs White) in subset of study participants with plasma creatinine (and adjustment for eGFR) at FDR < 0.05. Supplementary Table 23. Metabolite subclasses associated with race (Black vs White) in subset of study participants with plasma creatinine at FDR < 0.05.
Acknowledgements
The authors would like to acknowledge Dr. Penelope Duerksen-Hughes for her contributions in the design and implementation of the Metabolomics Pilot project, which preceded the current study.
Abbreviations
- ACAD
Acyl-CoA dehydrogenase
- AHS-2
Adventist Health Study-2
- ATSDR
Agency for Toxic Substances and Disease Registry
- BMI
Body mass index
- NLMS
National Longitudinal Mortality Study population
- SEER
Surveillance, Epidemiology, and End Results
- SNP
Single nucleotide polymorphism
- UPLC-MS/MS
Ultrahigh-performance liquid chromatography-tandem mass spectroscopy
Authors’ contributions
FMB, GEF, ROM, CAC, SM, and JWL wrote or edited the manuscript, with FMB taking the lead role. FMB and GEF designed and conceptualized the study. GEF and FMB conducted statistical analyses, with GEF taking the lead role. JU and SAW performed data curating, processing, and management. All authors contributed to the interpretation of results and read and approved the final manuscript.
Funding
This research was funded by the NIH National Institute on Minority Health and Health Disparities K01MD015194. Additionally, the research was supported by Basic Science and Translational Research Pilot Funds, and the Ardmore Institute of Health (“The Adventist Health Study: The next generation in transformational health knowledge”). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Availability of data and materials
The datasets used and/or analyzed during the current study are not publicly available due to data use restrictions but will be available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Participants in this study from whom data was collected provided informed consent. This study was conducted following the rules of the Declaration of Helsinki of 1975, revised in 2013. The project was approved by the Loma Linda University institutional review board, protocol #5190502.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bonow RO, Grant AO, Jacobs AK. The cardiovascular state of the union: confronting healthcare disparities. Circulation. 2005;111(10):1205–1207. doi: 10.1161/01.CIR.0000160705.97642.92. [DOI] [PubMed] [Google Scholar]
- 2.Carnethon MR, Pu J, Howard G, Albert MA, Anderson CAM, Bertoni AG, et al. Cardiovascular health in African Americans: a scientific statement from the American Heart Association. Circulation. 2017;136(21):e393–e423. doi: 10.1161/CIR.0000000000000534. [DOI] [PubMed] [Google Scholar]
- 3.Thomas SJ, Booth JN, 3rd, Dai C, Li X, Allen N, Calhoun D, et al. Cumulative incidence of hypertension by 55 years of age in Blacks and Whites: the CARDIA study. J Am Heart Assoc. 2018;7(14):e007988. doi: 10.1161/JAHA.117.007988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giaquinto AN, Miller KD, Tossas KY, Winn RA, Jemal A, Siegel RL. Cancer statistics for African American/Black people 2022. CA Cancer J Clin. 2022;72(3):202–229. doi: 10.3322/caac.21718. [DOI] [PubMed] [Google Scholar]
- 5.Howard G, Safford MM, Moy CS, Howard VJ, Kleindorfer DO, Unverzagt FW, et al. Racial differences in the incidence of cardiovascular risk factors in older Black and White adults. J Am Geriatr Soc. 2017;65(1):83–90. doi: 10.1111/jgs.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menke A, Casagrande S, Geiss L, Cowie CC. Prevalence of and trends in diabetes among adults in the United States, 1988–2012. JAMA. 2015;314(10):1021–1029. doi: 10.1001/jama.2015.10029. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham TJ. Vital signs: racial disparities in age-specific mortality among Blacks or African Americans - United States, 1999–2015. MMWR Morb Mortal Wkly Rep. 2017;66(18):490. doi: 10.15585/mmwr.mm6618a11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease C, Prevention Decline in deaths from heart disease and stroke–United States, 1900–1999. MMWR Morb Mortal Wkly Rep. 1999;48(30):649–56. [PubMed] [Google Scholar]
- 9.Ford ES, Ajani UA, Croft JB, Critchley JA, Labarthe DR, Kottke TE, et al. Explaining the decrease in US deaths from coronary disease, 1980–2000. New Engl J Med. 2007;356(23):2388–2398. doi: 10.1056/NEJMsa053935. [DOI] [PubMed] [Google Scholar]
- 10.Bouvard V, Loomis D, Guyton KZ, Grosse Y, Ghissassi FE, Benbrahim-Tallaa L, et al. Carcinogenicity of consumption of red and processed meat. Lancet Oncol. 2015;16(16):1599–1600. doi: 10.1016/S1470-2045(15)00444-1. [DOI] [PubMed] [Google Scholar]
- 11.McMacken M, Shah S. A plant-based diet for the prevention and treatment of type 2 diabetes. J Geriatr Cardiol. 2017;14(5):342–354. doi: 10.11909/j.issn.1671-5411.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olfert MD, Wattick RA. Vegetarian diets and the risk of diabetes. Curr Diab Rep. 2018;18(11):101. doi: 10.1007/s11892-018-1070-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan A, Sun Q, Bernstein AM, Schulze MB, Manson JE, Willett WC, et al. Red meat consumption and risk of type 2 diabetes: 3 cohorts of US adults and an updated meta-analysis. Am J Clin Nutr. 2011;94(4):1088–1096. doi: 10.3945/ajcn.111.018978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romagnolo DF, Selmin OI. Mediterranean diet and prevention of chronic diseases. Nutr Today. 2017;52(5):208–222. doi: 10.1097/NT.0000000000000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser G, Katuli S, Anousheh R, Knutsen S, Herring P, Fan J. Vegetarian diets and cardiovascular risk factors in Black members of the Adventist Health Study-2. Public Health Nutr. 2015;18(3):537–545. doi: 10.1017/S1368980014000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rizzo NS, Sabate J, Jaceldo-Siegl K, Fraser GE. Vegetarian dietary patterns are associated with a lower risk of metabolic syndrome: the adventist health study 2. Diabetes Care. 2011;34(5):1225–1227. doi: 10.2337/dc10-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tonstad S, Stewart K, Oda K, Batech M, Herring RP, Fraser GE. Vegetarian diets and incidence of diabetes in the Adventist Health Study-2. Nutr Metab Cardiovasc Dis. 2013;23(4):292–299. doi: 10.1016/j.numecd.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tharrey M, Mariotti F, Mashchak A, Barbillon P, Delattre M, Fraser GE. Patterns of plant and animal protein intake are strongly associated with cardiovascular mortality: the Adventist Health Study-2 cohort. Int J Epidemiol. 2018;47(5):1603–1612. doi: 10.1093/ije/dyy030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alshahrani SM, Fraser GE, Sabate J, Knutsen R, Shavlik D, Mashchak A, et al. Red and processed meat and mortality in a low meat intake population. Nutrients. 2019;11(3):622. doi: 10.3390/nu11030622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miles FL, Orlich MJ, Mashchak A, Chandler PD, Lampe JW, Duerksen-Hughes P, et al. The biology of veganism: plasma metabolomics analysis reveals distinct profiles of vegans and non-vegetarians in the Adventist Health Study-2 Cohort. Nutrients. 2022;14(3):709. doi: 10.3390/nu14030709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miles FL, Lloren JIC, Haddad E, Jaceldo-Siegl K, Knutsen S, Sabate J, et al. Plasma, urine, and adipose tissue biomarkers of dietary intake differ between vegetarian and non-vegetarian diet groups in the Adventist Health Study-2. J Nutr. 2019;149(4):667–675. doi: 10.1093/jn/nxy292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill-Briggs F, Ephraim PL, Vrany EA, Davidson KW, Pekmezaris R, Salas-Lopez D, et al. Social determinants of health, race, and diabetes population health improvement: Black/African Americans as a population exemplar. Curr Diab Rep. 2022;22(3):117–128. doi: 10.1007/s11892-022-01454-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mensah GA, Brown AGM, Pratt CA. Nutrition disparities and cardiovascular health. Curr Atheroscler Rep. 2020;22(4):15. doi: 10.1007/s11883-020-0833-3. [DOI] [PubMed] [Google Scholar]
- 24.Williams DR, Mohammed SA, Shields AE. Understanding and effectively addressing breast cancer in African American women: unpacking the social context. Cancer. 2016;122(14):2138–2149. doi: 10.1002/cncr.29935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yelton B, Friedman DB, Noblet S, Lohman MC, Arent MA, Macauda MM, et al. Social determinants of health and depression among African American adults: a scoping review of current research. Int J Environ Res Public Health. 2022;19(3):1498. doi: 10.3390/ijerph19031498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mensah GA. Cardiovascular diseases in African Americans: fostering community partnersnips to stem the tide. Am J Kidney Dis. 2018;72(5):S37–S42. doi: 10.1053/j.ajkd.2018.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reardon SF, Fox L, Townsend J. Neighborhood income composition by household race and income, 1990–2009. Ann Am Acad Polit Ss. 2015;660(1):78–97. doi: 10.1177/0002716215576104. [DOI] [Google Scholar]
- 28.Chan J, Knutsen SF, Sabate J, Haddad E, Yan R, Fraser GE. Feasibility of running clinics to collect biological specimens in a nationwide cohort study–Adventist Health Study-2. Ann Epidemiol. 2007;17(6):454–457. doi: 10.1016/j.annepidem.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 29.Jaceldo-Siegl K, Knutsen SF, Sabate J, Beeson WL, Chan J, Herring RP, et al. Validation of nutrient intake using an FFQ and repeated 24 h recalls in Black and White subjects of the Adventist Health Study-2 (AHS-2) Public Health Nutr. 2010;13(6):812–819. doi: 10.1017/S1368980009992072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JW, Morton KR, Walters J, Bellinger DL, Butler TL, Wilson C, et al. Cohort profile: the biopsychosocial religion and health study (BRHS) Int J Epidemiol. 2009;38(6):1470–1478. doi: 10.1093/ije/dyn244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mesnage R, Teixeira M, Mandrioli D, Falcioni L, Ducarmon QR, Zwittink RD, et al. Use of shotgun metagenomics and metabolomics to evaluate the impact of glyphosate or Roundup MON 52276 on the gut microbiota and serum metabolome of Sprague-Dawley rats. Environ Health Perspect. 2021;129(1):17005. doi: 10.1289/EHP6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flanagan BE, Gregory EW, Hallisey EJ, Heitgerd JL, Lewis B. A social vulnerability index for disaster management. J Homel Secur Emerg Manag. 2011;8(1):0000102202154773551792. [Google Scholar]
- 33.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freedman D, Lane D. A nonstochastic interpretation of reported significance levels. J Bus Econ Stat. 1983;1(4):292–298. [Google Scholar]
- 35.Miles F, Fraser, GF, Mashchak A. Differences in DNA methylation patterns between vegans and non-vegetarians in the AHS-2 cohort. Poster presented at: American Society for Nutrition, 2019. Poster theater flash session: nutrigenomics in health and disease. Curr Dev Nutr. 2019;1(4):292–98.
- 36.Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49(W1):W388–W96. doi: 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corwin E, Dunlop AL, Fernandes J, Li S, Pearce B, Jones DP. Metabolites and metabolic pathways associated with glucocorticoid resistance in pregnant African-American women. Compr Psychoneuroendocrinol. 2020;1–2:100001. doi: 10.1016/j.cpnec.2020.100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duran AM, Beeson WL, Firek A, Cordero-MacIntyre Z, De Leon M. Dietary omega-3 polyunsaturated fatty-acid supplementation upregulates protective cellular pathways in patients with type 2 diabetes exhibiting improvement in painful diabetic neuropathy. Nutrients. 2022;14(4):761. doi: 10.3390/nu14040761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vantaku V, Donepudi SR, Piyarathna DWB, Amara CS, Ambati CR, Tang W, et al. Large-scale profiling of serum metabolites in African American and European American patients with bladder cancer reveals metabolic pathways associated with patient survival. Cancer. 2019;125(6):921–932. doi: 10.1002/cncr.31890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baxter BA, Oppel RC, Ryan EP. Navy beans impact the stool metabolome and metabolic pathways for colon health in cancer survivors. Nutrients. 2018;11(1):28. doi: 10.3390/nu11010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Souza RJ, Shanmuganathan M, Lamri A, Atkinson SA, Becker A, Desai D, et al. Maternal diet and the serum metabolome in pregnancy: robust dietary biomarkers generalizable to a multiethnic birth cohort. Curr Dev Nutr. 2020;4(10):144. doi: 10.1093/cdn/nzaa144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia-Aloy M, Ulaszewska M, Franceschi P, Estruel-Amades S, Weinert CH, Tor-Roca A, et al. Discovery of intake biomarkers of lentils, chickpeas, and white beans by untargeted LC-MS metabolomics in serum and urine. Mol Nutr Food Res. 2020;64(13):e1901137. doi: 10.1002/mnfr.201901137. [DOI] [PubMed] [Google Scholar]
- 43.Hodson L, Fielding BA. Trafficking and partitioning of fatty acids: the transition from fasted to fed state. Clin Lipidol. 2010;5(1):131–144. doi: 10.2217/clp.09.72. [DOI] [Google Scholar]
- 44.Meisenberg GaS, William. The metabolism of fatty acids and triglycerides. 2012.
- 45.Akbar JA, Jaceldo-Siegl K, Fraser G, Herring RP, Yancey A. The contribution of soul and Caribbean foods to nutrient intake in a sample of Blacks of US and Caribbean descent in the Adventist Health Study-2: a pilot study. Ethn Dis. 2007;17(2):244–249. [PubMed] [Google Scholar]
- 46.Ameer F, Scandiuzzi L, Hasnain S, Kalbacher H, Zaidi N. De novo lipogenesis in health and disease. Metabolism. 2014;63(7):895–902. doi: 10.1016/j.metabol.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 47.Del Bas JM, Caimari A, Rodriguez-Naranjo MI, Childs CE, Paras Chavez C, West AL, et al. Impairment of lysophospholipid metabolism in obesity: altered plasma profile and desensitization to the modulatory properties of n-3 polyunsaturated fatty acids in a randomized controlled trial. Am J Clin Nutr. 2016;104(2):266–279. doi: 10.3945/ajcn.116.130872. [DOI] [PubMed] [Google Scholar]
- 48.Yu SS, Castillo DC, Courville AB, Sumner AE. The triglyceride paradox in people of African descent. Metab Syndr Relat Disord. 2012;10(2):77–82. doi: 10.1089/met.2011.0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bozelli JC, Jr, Azher S, Epand RM. Plasmalogens and chronic inflammatory diseases. Front Physiol. 2021;12:730829. doi: 10.3389/fphys.2021.730829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kikas P, Chalikias G, Tziakas D. Cardiovascular implications of sphingomyelin presence in biological membranes. Eur Cardiol. 2018;13(1):42–45. doi: 10.15420/ecr.2017:20:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Long T, Hicks M, Yu HC, Biggs WH, Kirkness EF, Menni C, et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat Genet. 2017;49(4):568–578. doi: 10.1038/ng.3809. [DOI] [PubMed] [Google Scholar]
- 52.Yousri NA, Fakhro KA, Robay A, Rodriguez-Flores JL, Mohney RP, Zeriri H, et al. Whole-exome sequencing identifies common and rare variant metabolic QTLs in a Middle Eastern population. Nat Commun. 2018;9(1):333. doi: 10.1038/s41467-017-01972-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thienpont B, Aronsen JM, Robinson EL, Okkenhaug H, Loche E, Ferrini A, et al. The H3K9 dimethyltransferases EHMT1/2 protect against pathological cardiac hypertrophy. J Clin Invest. 2017;127(1):335–348. doi: 10.1172/JCI88353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang QJ, Chen HZ, Wang L, Liu DP, Hill JA, Liu ZP. The histone trimethyllysine demethylase JMJD2A promotes cardiac hypertrophy in response to hypertrophic stimuli in mice. J Clin Invest. 2011;121(6):2447–2456. doi: 10.1172/JCI46277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carlson SM, Gozani O. Nonhistone lysine methylation in the regulation of cancer pathways. Cold Spring Harb Perspect Med. 2016;6(11):a026435. doi: 10.1101/cshperspect.a026435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu B, Zheng Y, Nettleton JA, Alexander D, Coresh J, Boerwinkle E. Serum metabolomic profiling and incident CKD among African Americans. Clin J Am Soc Nephrol. 2014;9(8):1410–1417. doi: 10.2215/CJN.11971113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delanaye P, Cavalier E, Cristol JP, Delanghe JR. Calibration and precision of serum creatinine and plasma cystatin C measurement: impact on the estimation of glomerular filtration rate. J Nephrol. 2014;27(5):467–475. doi: 10.1007/s40620-014-0087-7. [DOI] [PubMed] [Google Scholar]
- 58.Inker LA, Eneanya ND, Coresh J, Tighiouart H, Wang D, Sang Y, et al. New creatinine- and cystatin C-based equations to estimate GFR without race. N Engl J Med. 2021;385(19):1737–1749. doi: 10.1056/NEJMoa2102953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Valkengoed IGM, Argmann C, Ghauharali-van der Vlugt K, Aerts J, Brewster LM, Peters RJG, et al. Ethnic differences in metabolite signatures and type 2 diabetes: a nested case-control analysis among people of South Asian, African and European origin. Nutr Diabetes. 2017;7(12):300. doi: 10.1038/s41387-017-0003-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo SY, Feofanova EV, Tin A, Tung S, Rhee EP, Coresh J, et al. Genome-wide association study of serum metabolites in the African American Study of Kidney Disease and Hypertension. Kidney Int. 2021;100(2):430–439. doi: 10.1016/j.kint.2021.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Calafat AM, Ye X, Wong LY, Bishop AM, Needham LL. Urinary concentrations of four parabens in the U.S. population: NHANES 2005–2006. Environ Health Perspect. 2010;118(5):679–85. doi: 10.1289/ehp.0901560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stiel L, Adkins-Jackson PB, Clark P, Mitchell E, Montgomery S. A review of hair product use on breast cancer risk in African American women. Cancer Med-US. 2016;5(3):597–604. doi: 10.1002/cam4.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Darbre PD, Harvey PW. Parabens can enable hallmarks and characteristics of cancer in human breast epithelial cells: a review of the literature with reference to new exposure data and regulatory status. J Appl Toxicol. 2014;34(9):925–938. doi: 10.1002/jat.3027. [DOI] [PubMed] [Google Scholar]
- 64.Eberle CE, Sandler DP, Taylor KW, White AJ. Hair dye and chemical straightener use and breast cancer risk in a large US population of Black and White women. Int J Cancer. 2020;147(2):383–391. doi: 10.1002/ijc.32738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Helm JS, Nishioka M, Brody JG, Rudel RA, Dodson RE. Measurement of endocrine disrupting and asthma-associated chemicals in hair products used by Black women. Environ Res. 2018;165:448–458. doi: 10.1016/j.envres.2018.03.030. [DOI] [PubMed] [Google Scholar]
- 66.Llanos AAM, Rabkin A, Bandera EV, Zirpoli G, Gonzalez BD, Xing CY, et al. Hair product use and breast cancer risk among African American and White women. Carcinogenesis. 2017;38(9):883–892. doi: 10.1093/carcin/bgx060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rao R, McDonald JA, Barrett ES, Greenberg P, Teteh DK, Montgomery SB, et al. Associations of hair dye and relaxer use with breast tumor clinicopathologic features: findings from the Women’s circle of Health Study. Environ Res. 2022;203:111863. doi: 10.1016/j.envres.2021.111863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Teteh D, Ericson M, Monice S, Dawkins-Moultin L, Bahadorani N, Clark P, et al. The Black identity, hair product use, and breast cancer scale. PLoS One. 2019;14(12):e0225305. doi: 10.1371/journal.pone.0225305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Okamoto Y, Hayashi T, Matsunami S, Ueda K, Kojima N. Combined activation of methyl paraben by light irradiation and esterase metabolism toward oxidative DNA damage. Chem Res Toxicol. 2008;21(8):1594–1599. doi: 10.1021/tx800066u. [DOI] [PubMed] [Google Scholar]
- 70.Fraser GE, Cosgrove CM, Mashchak AD, Orlich MJ, Altekruse SF. Lower rates of cancer and all-cause mortality in an Adventist cohort compared with a US Census population. Cancer. 2020;126(5):1102–1111. doi: 10.1002/cncr.32571. [DOI] [PubMed] [Google Scholar]
- 71.Hao Y, Guo M, Feng Y, Dong Q, Cui M. Lysophospholipids and their G-coupled protein signaling in Alzheimer’s disease: from physiological performance to pathological impairment. Front Mol Neurosci. 2020;13:58. doi: 10.3389/fnmol.2020.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tan ST, Ramesh T, Toh XR, Nguyen LN. Emerging roles of lysophospholipids in health and disease. Prog Lipid Res. 2020;80:101068. doi: 10.1016/j.plipres.2020.101068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Table 1. Dietary intakes of study participants. Supplementary Table 2. All metabolites present at higher abundance in Black relative to White participants at FDR < 0.05 in simplest model. Supplementary Table 3. All metabolites present at lower abundance in Black relative to White participants at FDR < 0.05 in simplest model. Supplementary Table 4. Metabolites (n = 348) present at lower abundance in Black relative to White participants at FDR < 0.05. Supplementary Table 5. Metabolites present at higher abundance in Black relative to White participants at FDR < 0.05 (with additional adjustment for BMI and dietary pattern). Supplementary Table 6. Metabolites present at lower abundance in Black relative to White participants at FDR < 0.05 (with additional adjustment for BMI and dietary pattern). Supplementary Table 7. Metabolites present at higher abundance in Black relative to White participants at FDR < 0.05 (with additional adjustment for BMI, dietary pattern, exercise, social vulnerability index, and psychosocial stress). Supplementary Table 8. Metabolites present at lower abundance in Black relative to White participants at FDR < 0.05 (with additional adjustment for BMI, dietary pattern, exercise, social vulnerability index, and psychosocial stress). Supplementary Table 9. Metabolites present at lower abundance in Black relative to White participants at FDR < 0.05 excluding pilot study participants. Supplementary Table 10. Metabolites present at lower abundance in Black relative to White participants at FDR < 0.05 in pilot study participants. Supplementary Table 11. Differentially abundant metabolites in common between analyses with pilot (n = 93) and non-pilot (n = 258) study participants at FDR<0.05. Supplementary Table 12. Metabolite subclasses associated with race (Black vs White) at FDR in simplest model < 0.05. Supplementary Table 13. Metabolite subclasses associated with race (Black vs White) at FDR < 0.05 (adjusted additionally for BMI and dietary pattern). Supplementary Table 14. Metabolite subclasses associated with race (Black vs White) at FDR < 0.05 (adjusted additionally for BMI, dietary pattern, social vulnerability index, and psychosocial stress). Supplementary Table 15. Names of statistically significant component metabolites (based on linear regression analysis) contained within each subclass differentially abundant at FDR < 0.05. Supplementary Table 16. Differentially abundant subclasses in common between analyses with pilot (n = 93) and non-pilot (n = 258) study participants at FDR<0.05. Supplementary Table 17. Results from pathway analysis of metabolic pathways over-represented comparing Black and White participants (at P ≤ 0.5). Supplementary Table 18. Metabolites differentially abundant in Black relative to White participants at FDR < 0.05 (adjusted additionally for dietary pattern, dietary fat (polyunsaturated, monounsaturated, and saturated), education, social vulnerability index, exercise, BMI, coffee, medication use, chronic disease cardiovascular disease, diabetes, hypertension). Supplementary Table 19. Metabolite subclasses associated with race (Black vs White) at FDR < 0.05 (adjusted additionally for BMI, dietary pattern, social vulnerability index, psychosocial stress, and dietary fat). Supplementary Table 20. Metabolites present at higher abundance in Black relative to White participants at FDR < 0.05 in n = 140 study participants with plasma creatinine measurements (and adjustment for eGFR). Supplementary Table 21. Metabolites present at higher abundance in Black relative to White participants at FDR < 0.05 in n = 140 study participants with plasma creatinine measurements. Supplementary Table 22. Metabolite subclasses associated with race (Black vs White) in subset of study participants with plasma creatinine (and adjustment for eGFR) at FDR < 0.05. Supplementary Table 23. Metabolite subclasses associated with race (Black vs White) in subset of study participants with plasma creatinine at FDR < 0.05.
Data Availability Statement
The datasets used and/or analyzed during the current study are not publicly available due to data use restrictions but will be available from the corresponding author on reasonable request.