Abstract
We report a newly emerged SARS-CoV-2 Omicron subvariant FY.4 that has mutations Y451H in spike and P42L in open reading frame 3a proteins. FY.4 emergence coincided with increased SARS-CoV-2 cases in coastal Kenya during April–May 2023. Continued SARS-CoV-2 genomic surveillance is needed to identify new lineages to inform COVID-19 outbreak prevention.
Keywords: COVID-19, Omicron, respiratory infections, severe acute respiratory syndrome coronavirus 2, SARS-CoV-2, SARS, coronavirus disease, zoonoses, viruses, coronavirus, FY.4, Kilifi, Kenya
As of August 2023, >340,000 test-confirmed COVID-19 cases and 5,689 COVID-19–related deaths had been reported in Kenya (1). Seropositivity rates in rural and urban populations were high, and vaccine uptake was low (≈28% of the adult population received >1 dose) (2). By August 2022, a total of 69%–81% of rural (Kilifi and Siaya) and 89%–95% of urban (Nairobi and Kisumu) adults in Kenya had IgG against the SARS-CoV-2 spike protein (3).
Genomic surveillance is critical to determine origins of new waves, evolution, and spread patterns of SARS-CoV-2. As of June 2023, a total of 7 distinct waves of SARS-COV-2 infections have been observed in Kenya (1; G. Githinji et al., unpub. data, https://doi.org/10.1101/2022.10.26.22281446). The last 3 waves were dominated by Omicron BA.1-like, BA.5-like, and BQ-like lineages (in that order), associated with increases in SARS-CoV-2 cases because of virus mutations that conferred transmission advantage or escape from preexisting immunity (4).
In coastal Kenya, the Kenya Medical Research Institute Wellcome Trust Research Programme is conducting SARS-CoV-2 genomic surveillance across 5 health facilities within the Kilifi Health and Demographic Surveillance System (KHDSS) (5). Up to 75 respiratory samples are collected weekly from persons of all ages with acute respiratory illness at participating health facilities. SARS-CoV-2 reverse transcription PCR and sequencing are performed on virus-positive samples from KHDSS health facilities and other collaborating health facilities across Kenya.
SARS-CoV-2 positivity rates in KHDSS health facilities increased from 1.2% during the week beginning March 27, 2023, to 42.9% during the week beginning April 24, 2023 (Figure, panel A). After dropping to 23.5% during the first week of May, rates remained at 5.0%–7.7% over the next 3 weeks.
Figure.
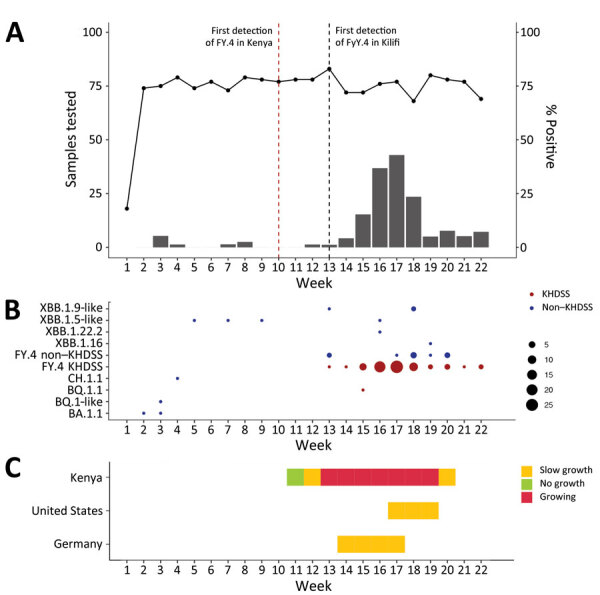
Number of positive samples, distribution, and growth rate for new SARS-CoV-2 Omicron variant with spike protein mutation Y451H, Kilifi, Kenya, March–May 2023. The x axes indicate calendar weeks beginning on January 1, 2023. A) Weekly number of collected samples (black data line) and positive SARS-CoV-2 cases (data bars) in health facilities within the KHDSS during January–May 2023. Vertical dotted lines indicate the weeks when the FY.4 lineage was first detected in Kenya (red) and Kilifi (black). B) Weekly distribution of SARS-CoV-2 lineages observed in samples processed at the Kenya Medical Research Institute Wellcome Trust Research Programme from KHDSS (red dots) and non-KHDSS (blue dots) health facilities during January–May 2023. C) Growth rate estimates for the Omicron FY.4 variant in Kenya relative to those in the United States and Germany. KHDSS, Kilifi Health Demographic Surveillance System.
During January–May, a total of 120 (7.4%) of 1,612 samples collected from KHDSS health facilities were positive for SARS-CoV-2. We sequenced 96 (80%) of 120 samples that had PCR cycle threshold values <35 by using GridION (Oxford Nanopore Technologies, https://www.nanoporetech.com) (n = 35) or MiSeq (Illumina, https://www.illumina.com) (n = 61) instruments; we recovered 76 (79%) genomes with >70% coverage. We also received 39 SARS-CoV-2–positive samples from health facilities outside the KHDSS and sequenced 32 (82%) of them, yielding 25 (78%) genomes (Appendix Table).
We assigned the 76 genomes from KHDSS health facilities to 2 lineages: BQ.1.1 (n = 1) and FY.4 (n = 75). The SARS-CoV-2 infection rate increase observed in late March coincided with detection of the new Omicron FY.4 lineage (Figure, panel B). In Kenya, FY.4 was first observed in Lamu County (n = 6) on March 10, 2023 (Figure, panel A); in KHDSS, FY.4 was first identified from samples collected on March 27 and, in April and May, it became the dominant lineage, representing 98% of all SARS-CoV-2 cases. By May 31, according to the GISAID database (https://www.gisaid.org), FY.4 had been detected in 4 other counties: Mombasa (n = 2), Narok (n = 2), Nairobi (n = 7), and Kiambu (n = 31). The FY.4 subvariant has since been detected in Austria, Belgium, Germany, Italy, India, Sweden, Canada, France, China, Australia, Spain, the United Kingdom, and the United States (6).
In KHDSS health facilities, patients infected with FY.4 had cough (98%), fever (78%), and nasal discharge (74%); 7% had difficulty breathing (Table). Only 13 (16%) participants reported receiving >1 dose of a COVID-19 vaccine. A serosurveillance study (February–June 2022) found that 67% of unvaccinated KHDSS health facility residents had SARS-CoV-2 IgG, indicating a high percentage of this population might have been naturally infected (3).
Table. Clinical signs and symptoms for patients infected with SARS-CoV-2 Omicron FY.4 variant with spike protein mutation Y451H, Kilifi Health Demographic Surveillance System, Kilifi, Kenya, January–May 2023*.
| Signs/symptoms | No. (%) patients |
|---|---|
| Fever | |
| Yes | 57 (78.1) |
| No |
16 (21.9) |
| Cough | |
| Yes | 72 (98.6) |
| No |
1 (1.4) |
| Nasal discharge | |
| Yes | 54 (74.0) |
| No |
19 (26.0) |
| Difficulty breathing | |
| Yes | 5 (6.8) |
| No |
68 (93.2) |
| Sore throat | |
| Yes | 28 (38.4) |
| No |
45 (61.6) |
| Body malaise | |
| Yes | 25 (34.2) |
| No |
48 (65.8) |
| Conscious level | |
| Alert |
73 (100.0) |
| COVID-19 vaccination status | |
| Yes | 12 (16.4) |
| No | 60 (82.2) |
| No data |
1 (1.4) |
| COVID19 vaccine doses | |
| 1 | 3 (4.1) |
| 2 | 7 (9.6) |
| No data | 63 (86.3) |
*Total number of patients was 73.
Compared with other Omicron lineages, FY.4 had 2 amino acid mutations, Y451H in the spike protein and P42L in the open reading frame (ORF) 3a protein. The effect of the Y451H change is unknown; however, an L452R mutation in the receptor-binding domain of the spike protein (near the Y451H site) increased virus infectivity and fusogenicity by enhancing spike stability and cleavage (7). Changes within ORF3a epitopes can cause complete loss of CD8+ T-cell recognition of ancestral SARS-CoV-2 lineages and Alpha variant (8).
We applied a Bayesian hierarchical model (9) to estimate the growth rate of FY.4-like lineages in Kenya. Growth estimates serve as an epidemic warning system for lineages that have consistent increases in frequency for >2 consecutive weeks. We compared growth rate estimates from Kenya with those from Germany and the United States, the only other countries reporting FY.4 cases for >2 consecutive weeks during the last weeks of May 2023 (Figure, panel C). The model warned of a high epidemic potential in Kenya from March 26 through May, suggesting continued increases in cases attributed to the FY.4 lineage.
In conclusion, we detected emergence of a new Omicron lineage with unique spike and ORF3a gene mutations in coastal Kenya by using SARS-CoV-2 genomic surveillance. FY.4 subvariant detection coincided with an increase in SARS-CoV-2 cases in Kilifi. FY.4 was also detected in other parts of Kenya, and growth estimates suggest potential for continued spread of the FY.4 subvariant. Continued SARS-CoV-2 genomic surveillance is critical for identifying new lineages to inform COVID-19 prevention measures.
Appendix. Additional information for new SARS-CoV-2 Omicron variant with spike protein mutation Y451H, Kilifi, Kenya, March–May 2023.
Acknowledgments
We thank the laboratories, hospitals, and organizations that shared specimens for sequencing at the Kenya Medical Research Institute Wellcome Trust Research Programme and submitting laboratories that generated and shared genetic sequence data via the GISAID Initiative (https://www.gisaid.org), on which this research is based. Submitting and originating laboratories that generated the GISAID data used in this study are listed at https://doi.org/10.55876/gis8.230627zw. Genome sequences generated in this study are available in the GISAID’s EpiCoV database. The dataset and analysis scripts used are available in the Harvard Dataverse repository (https://doi.org/10.7910/DVN/ZMGR5P). The whole-genome sequencing study protocol was reviewed and approved by the Scientific and Ethics Review Committee at the Kenya Medical Research Institute headquarters in Nairobi, Kenya (SERU no. 4035).
This work was supported by the New Variant Assessment Programme, a UK Health Security Agency program funded by the UK Department of Health and Social Care as a global initiative to strengthen genomic surveillance for pandemic preparedness and response to emerging and priority infectious diseases; the UK National Institute for Health and Care Research (project references 17/63/82 and 16/136/33) that uses aid from the UK government to support global health research; UK Foreign, Commonwealth and Development Office, and The Wellcome Trust, UK (grant nos. 220985/Z/20/Z and 226002/A/22/Z).
The views expressed in this publication are those of the author(s) and not necessarily those of National Institute for Health and Care Research, Department of Health and Social Care, or the Foreign Commonwealth and Development Office, Africa Centers for Disease Control, World Health Organization Africa, or African Society for Laboratory Medicine.
Biography
Mr. Mwanga is a research assistant at Kenya Medical Research Institute Wellcome Trust Research Programme. His research focuses on leveraging epidemiologic and genomic sequence data to investigate virus evolution, transmission, and spread in human populations to inform effective control and intervention strategies.
Footnotes
Suggested citation for this article: Mwanga MJ, Lambisia AW, Morobe JM, Murunga N, Moraa E, Ndwiga L, et al. New SARS-CoV-2 Omicron variant with spike protein mutation Y451H, Kilifi, Kenya, March–May 2023. Emerg Infect Dis. 2023 Nov [date cited]. https://doi.org/10.3201/eid2911.230894
These authors contributed equally to this article.
References
- 1.Our World in Data. Coronavirus pandemic (COVID-19) [cited 2023 Aug 24]. https://ourworldindata.org/coronavirus
- 2.World Health Organization. WHO coronavirus (COVID-19) vaccination dashboard. 2023. [cited 2023 Aug 24]. https://www.afro.who.int/health-topics/coronavirus-covid-19/vaccines
- 3.Kagucia EW, Ziraba AK, Nyagwange J, Kutima B, Kimani M, Akech D, et al. SARS-CoV-2 seroprevalence and implications for population immunity: Evidence from two Health and Demographic Surveillance System sites in Kenya, February-December 2022. Influenza Other Respir Viruses. 2023;17:e13173. 10.1111/irv.13173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He Q, Wu L, Xu Z, Wang X, Xie Y, Chai Y, et al. An updated atlas of antibody evasion by SARS-CoV-2 Omicron sub-variants including BQ.1.1 and XBB. Cell Rep Med. 2023;4:100991. 10.1016/j.xcrm.2023.100991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott JAG, Bauni E, Moisi JC, Ojal J, Gatakaa H, Nyundo C, et al. Profile: The Kilifi Health and Demographic Surveillance System (KHDSS). Int J Epidemiol. 2012;41:650–7. 10.1093/ije/dys062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22:30494. 10.2807/1560-7917.ES.2017.22.13.30494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Motozono C, Toyoda M, Zahradnik J, Saito A, Nasser H, Tan TS, et al. ; Genotype to Phenotype Japan (G2P-Japan) Consortium. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. 2021;29:1124–1136.e11. 10.1016/j.chom.2021.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Silva TI, Liu G, Lindsey BB, Dong D, Moore SC, Hsu NS, et al. ; COVID-19 Genomics UK (COG-UK) Consortium; ISARIC4C Investigators. The impact of viral mutations on recognition by SARS-CoV-2 specific T cells. iScience. 2021;24:103353. 10.1016/j.isci.2021.103353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guzmán-Rincón LM, Hill EM, Dyson L, Tildesley MJ, Keeling MJ. Bayesian estimation of real-time epidemic growth rates using Gaussian processes: local dynamics of SARS-CoV-2 in England. J R Stat Soc Ser C Appl Stat. 2023;qlad056. 10.1093/jrsssc/qlad056 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix. Additional information for new SARS-CoV-2 Omicron variant with spike protein mutation Y451H, Kilifi, Kenya, March–May 2023.


