Abstract
Neisseria meningitidis causes invasive meningococcal diseases and has also been identified as a causative agent of sexually transmitted infections, including urethritis. Unencapsulated sequence type 11 meningococci containing the gonococcal aniA-norB locus and belonging to the United States N. meningitidis urethritis clade (US_NmUC) are causative agents of urethral infections in the United States, predominantly among men who have sex with men. We identified 2 subtypes of unencapsulated sequence type 11 meningococci in Japan that were phylogenetically close to US_NmUC, designated as the Japan N. meningitidis urethritis clade (J_NmUC). The subtypes were characterized by PCR, serologic testing, and whole-genome sequencing. Our study suggests that an ancestor of US_NmUC and J_NmUS urethritis-associated meningococci is disseminated worldwide. Global monitoring of urethritis-associated N. meningitidis isolates should be performed to further characterize microbiologic and epidemiologic characteristics of urethritis clade meningococci.
Keywords: Neisseria meningitidis, meningococci, serogroup, invasive meningococcal diseases, urethritis, urethritis clade, whole-genome sequencing, phylogenetic analysis, bacteria, meningitis/encephalitis, sexually transmitted infections, Japan
Neisseria meningitidis causes invasive meningococcal diseases (IMDs), such as meningitis and septicemia. N. meningitidis is classified into 12 defined serogroups; however, most IMDs are associated with the serogroups A, B, C, W, X, and Y (1). Serogrouping is critical for IMD control because meningococcal vaccines have serogroup-specific effects (2). Whole-genome sequencing (WGS)–based typing, such as high-resolution core genome multilocus sequence typing (MLST), is the most powerful method for analyzing meningococcal isolates. However, standard MLST, which identifies sequence types (STs) of isolates according to the unique allelic profiles of 7 housekeeping genes, is still applied to meningococcal epidemiology studies because the most invasive isolates belong to a limited number of clonal complexes (CCs). For example, ST11 and the locus variants comprising CC11 meningococci are well-known hypervirulent N. meningitidis strains that have caused many pandemics (3), including IMD outbreaks that predominately occurred among men who have sex with men (MSM) (4–9).
N. gonorrhoeae is also a human pathogen capable of infecting the urethra, cervix, rectum, and oropharynx. Most gonococcal infections manifest clinically as urethritis in men or cervicitis in women, both of which are sexually transmitted infections (STI). Meningococcus and gonococcus are generally regarded as distinct taxa that cause specific diseases; however, recent findings suggest a greater overlap than was originally reported. N. gonorrhoeae is rarely identified as a causative agent of systemic infection; N. meningitidis has been reported to cause STIs, such as urethritis. An outbreak of meningococcal urethritis predominantly among MSM was reported in multiple cities in the United States (10). Causative agents were identified as CC11 N. meningitidis isolates with several unique features and classified as US N. meningitidis urethritis clade (US_NmUC) (11–14). The capsular polysaccharide (cps) locus in US_NmUC meningococci is disrupted by insertion sequence (IS) 1301 that replaced ccsA, cssB, and cssC genes and part of the csc gene causing loss of encapsulation (12). That genetic mutation also caused the loss of wild-type lipooligosaccharide sialylation, which appeared to increase mucosal surface adherence (15). Moreover, the factor H binding protein (fHbp), which binds to human factor H and inhibits the alternative complement activation pathway in the human immune system (16), was highly expressed in US_NmUC N. meningitidis isolates and might promote evasion from immune responses in the human urogenital tract (12). The most unique feature of US_NmUC meningococci is their acquisition of the N. gonorrhoeae denitrification apparatus that comprises gonococcal alleles encoding nitrate reductase AniA and nitric oxide reductase NorB and the intergenic promoter region, which confers survival in the urogenital tract (12,17).
Most US_NmUC isolates have been recovered from patients with urethritis in the United States. However, 2 US_NmUC meningococci isolates were identified in 2019 in rectal swab samples from MSM in the United Kingdom (18), and 19 US_NmUC meningococci were isolated in Vietnam in 2019 and 2020 (19). US_NmUC meningococci have not yet been reported in other countries. We report the genomic and phenotypic features of 3 unencapsulated ST11 urethritis-associated N. meningitidis strains isolated in Japan that were phylogenetically close to US_NmUC but classified as novel urethritis meningococcus clade subtypes.
Methods
N. meningitidis Isolates
Although IMDs are legally notifiable diseases in Japan, STIs caused by N. meningitidis are not. In Japan, meningococcal isolates from patients with STIs are typically collected as part of the countrywide gonococcal surveillance program (headed by M.Y.). Urethral swab samples from male patients suspected of having urethritis and cervical swab samples from female patients suspected of having cervicitis were sent to Sapporo Medical University from ≈100 clinics across Japan. We isolated strains by selective growth on Thayer-Martin medium and analyzed those isolates by using Biotyper matrix-assisted/laser desorption time-of-flight mass spectrometry (Beckman Coulter, https://www.beckmancoulter.com) and commercially available mass spectrometry profiles to identify species. We collected >1,000 gonococcal isolates annually and isolated ≈10 N. meningitidis strains under the gonococcal surveillance program, in which no misidentification of N. meningitidis as N. gonorrhoeae has occurred. We characterized 3 N. meningitidis isolates at the National Institute of Infectious Diseases by using serologic and genetic analyses.
Typing and Antimicrobial Drug Susceptibility Tests
We performed serogrouping by using PCR (20) and slide agglutination tests with meningococcal rabbit antiserum (Remel, http://www.remel.com, or Difco/Becton Dickinson, https://www.bd.com) and a commercial latex agglutination kit (Pastorex Meningitis assay; Bio-Rad Laboratories, https://www.bio-rad.com). We conducted MLST by using the standard method (21). We performed antimicrobial drug susceptibility tests by using E-tests (bioMérieux, https://www.biomerieux.com) and Mueller-Hinton agar with 5% sheep blood (Becton Dickinson), which we interpreted according to the Clinical and Laboratory Standards Institute criteria for agar dilution, as previously described (22).
WGS, Genome Assembly, and Phylogenetic Analysis
We extracted genomic DNA by using the MagMAX DNA Multi-Sample Ultra 2.0 Kit, which we then purified by using the KingFisher Duo Prime Purification System and measured concentrations by using a Qubit dsDNA HS assay kit (all from Thermo Fisher Scientific, https://www.thermofisher.com). We prepared genomic libraries for short read sequencing by using the QIAseq FX DNA Library Kit (QIAGEN, https://www.qiagen.com) and sequenced 300-bp paired-end reads on a MiSeq instrument (Illumina, https://www.illumina.com). For long-read sequencing on a MinION sequencer (Oxford Nanopore Technologies, https://nanoporetech.com), we prepared genomic libraries by using a Rapid Barcoding Kit (Oxford Nanopore Technologies) and sequenced them by using an R9.4.1 flow cell. We basecalled raw data by using Guppy 6.5.7 (23) and removed adaptors before assembly by using Porechop 0.2.3 (https://github.com/rrwick/Porechop). We generated draft genome sequences for both long and short reads by using Unicycler version 0.5.0 in conservative mode (24) and performed annotations of complete genomes and genome assemblies by using the DDBJ Fast Annotation and Submission Tool (https://dfast.ddbj.nig.ac.jp) (25). We used draft genome assemblies for PorA and FetA typing and determining the Meningococcal Deduced Vaccine Antigen Reactivity Index through PubMLST (https://www.pubmlst.org). We performed phylogenetic analyses of N. meningitidis from urethritis patients by using 26 publicly available genomes and constructed core gene alignments by using Roary version 3.12.0 and the -s and -e–mafft options (26), which were subject to SNP-sites version 2.5.1 (27) to extract single-nucleotide variants. We constructed the phylogenetic tree by using IQ-TREE version 2.0.3 (http://www.iqtree.org) with 1,000 ultrafast bootstrap replicates and visualized the tree by using iTOL (28).
Repositories
We deposited the short reads sequence data for NIID835, NIID836, and NIID838 in the DDBJ Sequence Read Archive (https://www.ddbj.nig.ac.jp) under accession nos. DRR494404 (NIID835), DRR494405 (NIID836), and DRR494406 (NIID838) and in the PubMLST database under nos. 135430 (NIID835), 135431 (NIID836), and 135432 (NIID838). The annotated complete genome assemblies of NIID835, NIID836, and NIID838 strains are also available in the GenBank, EMBL (https://www.ebi.ac.uk), and DDBJ databases under accession nos. AP028680 (NIID835), AP028681 and AP028682 (NIID836), and AP028683 (NIID838).
Results
The 3 J_NmUC N. meningitidis strains (NIID835, NIID836, and NIID838) were isolated from 3 men with urethritis that developed 4–5 days after contact with commercial sex workers for oral sexual services (Appendix 1 Table). Although N. meningitidis strains from patients with urethritis in Japan are typically classified as ST11026, which is also isolated from healthy carriers (29), or ST23, which is also isolated from IMD patients and healthy carriers (30), we identified all 3 J_NmUC N. meningitidis strains as ST11 (Appendix 1 Table). To further characterize the 3 J_NmUC N. meningitidis isolates as urethritis clade meningococci, we performed WGS, phylogenetic, and serologic analyses.
aniA-norB Locus
We conducted phylogenetic analysis of the 3.5-kb aniA-norB gene sequence (Appendix 2 Figure 1) for 3 J_NmUC N. meningitidis isolates from Japan, 2 N. meningitidis US_NmUC isolates, N. gonorrhoeae FA1090 (GenBank accession no. NC_002946.2), N. meningitidis MC58 (31), and 6 N. meningitidis serogroup C isolates from IMD patients in United States that were genetically very close to US_NmUC (32) (Figure 1). Moreover, we included 3 ST23 N. meningitidis isolates (NM001, NM003, and NIID574) from Japan harboring the gonococcal aniA-norB locus (30), designated as J_NmUC-II (Figure 1). The aniA-norB locus in the 3 ST11 J_NmUC isolates was 100% identical to that in US_NmUC meningococci (12,17), indicating the aniA-norB locus in the 3 J_NmUC strains was of gonococcal origin. In the 3 ST11 J_NmUC and 3 J_NmUC-II isolates, the aniA-norB locus was located between gpxA and NMB1624 genes (Appendix 2 Figure 1), which was identical to that in US_NmUC N. meningitidis strains (12). Collectively, those results indicated that the 3 ST11 J_NmUC isolates acquired the gonococcal aniA-norB locus, similar to US_NmUC meningococci.
Figure 1.
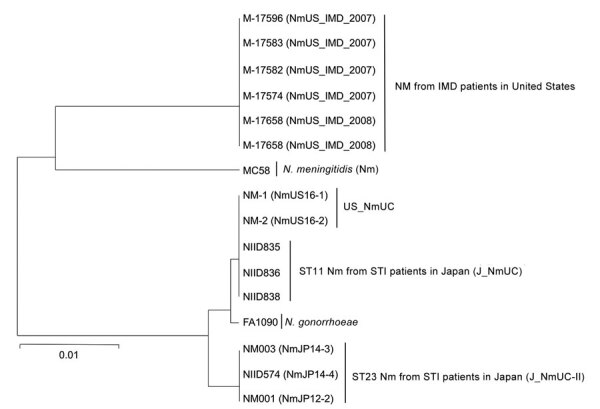
Phylogenetic analysis of the 3.5-kb aniA-norB gene locus of Neisseria spp. isolates in study detecting novel US N. meningitidis urethritis clade subtypes in Japan. Tree was constructed by using the unweighted pair group method with arithmetic mean and 1,000 bootstrap replicates. The gonococcal aniA-norB locus was derived from N. gonorrhoeae FA1090 (GenBank accession no. NC_002946.2); all others are from N. meningitidis isolates. Scale bar indicates nucleotide substitutions per site. IMD, invasive meningococcal disease; Nm, N. meningitidis, ST, sequence type; STI, sexually transmitted infection.
Serogrouping and cps Locus Analysis
Although we initially identified the 3 ST11 J_NmUC N. meningitidis strains as serogroup C meningococci (MenC) by PCR (20), the strains were agglutination negative when we tested with serogroup C–specific antiserum. To clarify this discrepancy, we characterized the cps gene locus (Figure 2). In NIID835 and NIID838 isolates, cssA, cssB, and cssC genes, and part of the csc gene (region A) were deleted and replaced with IS1301, but the ctrABCD gene cluster (region C) was also deleted. In contrast to 2 copies of IS1301 in US_NmUC isolates (12), only 1 copy of IS1301 was found in the cps locus of NIID835 and NIID838 isolates. In the NIID836 J_NmUC isolate, deletions of cssA, cssB, cssC, csc genes were identical to those in NIID835 and NIID838, but the ctrABCD gene cluster remained, containing the pylA, gltS, lipA, and lipB genes, which are typically proximal to the csc and cssE genes. Furthermore, 2 copies of the rfbC, rfbA, and rfbB gene cluster were identified in the NIID836 J_NmUC isolate; only 1 copy was found in NIID835 and NIID838 isolates. Although the cps locus in the 3 J_NmUC meningococcal strains were not identical to that in US_NmUC meningococci, the J_NmUC meningococci were genotypically nongroupable. All of the genetic features within the cps and aniA-norB loci confirmed that the 3 nongroupable ST11 J_NmUC meningococci were classified into the urethritis clade.
Figure 2.
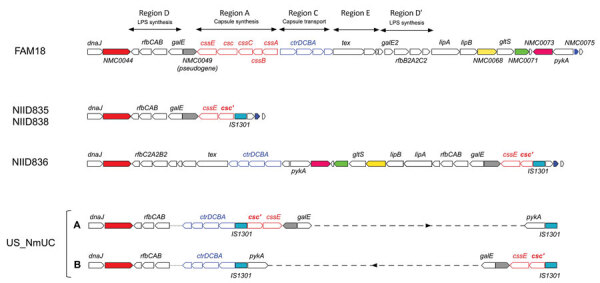
Organization of genes within the cps locus of Neisseria meningitidis isolates in study of detection of novel US N. meningitidis urethritis clade subtypes in Japan. N. meningitidis isolates from Japan (NIID835, NIID836, NIID838) and United States (US_NmUC) were compared with N. meningitidis strain FAM18 (GenBank accession no. AM421808). Open red arrows indicate the cssA, cssB, cssC, csc, and cssE genes in region A responsible for capsule synthesis and open blue arrows the ctrD, ctrC, ctrB, and ctrA genes (in that order) in region C responsible for capsule transport. Insertion sequence IS1301 is indicated. Open reading frames identical to NMC0044 (solid red), NMC0049 (gray), NMC0068 (yellow), NMC0071 (green), NMC0073 (pink), and NMC0075 (blue) in FAM18 are shown for each isolate. Partial deletion is indicated for the csc gene (csc′). The cps locus for US_NmUC had 2 configurations created by a ≈20-kb genome inversion between 2 IS1301 sequences (designated as A and B). Gene alignments in the region between the 2 IS1301 sequences have been omitted and are indicated by the dashed line. Although ctrD, ctrC, ctrB, and ctrA genes were shown to be proximal to dnaJ (12), contigs containing the dnaJ-rfbC, rfbA, and rfbB genes and the ctrD, ctrC, ctrB, and ctrA genes (shown on the left side of A and B), as well as 2 IS1301 and pykA genes (shown on the right side of A and B), were not connected by our analysis because of the absence of US_NmUC long-read sequences. Therefore, unidentified connections of the 2 contigs are indicated by a dotted line.
fHbp Locus
In US_NmUC meningococci, fHbp was speculated to be highly expressed because the fHbp promoter sequence belonged to high fHbp–expressing promoter clade I (33). In the 3 ST11 J_NmUC N. meningitidis isolates, the fHbp promoter sequence, fHbp peptide, and fHbp allele were identical to those in US_NmUC meningococci strains (Appendix 2 Figure 2), suggesting fHbp might also be highly expressed in J_NmUC meningococci (12).
Phylogenetic Analysis by Using WGS
To gain insights into the origin of J_NmUC meningococci, we performed phylogenetic analysis by using WGS to compare 9 ST11 IMD isolates from Japan (29), 1 STI isolate (NmJP12–1) (30), and 7 IMD MenC isolates from the United States that were genetically close to US_NmUC (32) (Figure 3). ST23 J_NmUC-II, ST11 serogroup W meningococci SK001 (NmJP12–1), 8 IMD MenC, and 5 STI MenC (30) isolates were genetically separate from J_NmUC and US_NmUC meningococci; 7 US IMD MenC that were close to US_NmUC (32) were also genetically close to J_NmUC. However, J_NmUC strains were the phylogenetically closest to US_NmUC, eliminating the possibility that J_NmUC was originally derived from MenC strains in Japan.
Figure 3.

Phylogenetic analysis of Neisseria meningitidis from different countries in study of detection of novel US N. meningitidis urethritis clade subtypes in Japan. Strains isolated from patients with IMD (red font) or STI (blue font), serogroup (NG or C), and country of origin are indicated. US_NmUC, J_NmUC, and J_NmUC-II N. meningitidis isolates have detailed profiles (Appendix 1 Table). We included 1 sequence type 11 N. meningitidis strain isolated in Japan from a patient with an STI (SK028) and 4 serogroup C meningococci (MenC) that were phylogenetically close to SK028 (PE5, PE6, PE7, and LNP26948) (29). Moreover, we included 7 MenC phylogenetically close to US_NmUC (IMD strains in the United States) (31), 2 sequence type 11 MenC isolated from IMD patients during 2003–2020 in Japan (NIID647 and NIID716) (28), and 6 MenC phylogenetically close to the 2 MenC from Japan (28). Scale bar indicates nucleotide substitutions per site. C, serogroup C; IMD, invasive meningococcal disease; NG, nongroupable; STI, sexually transmitted infection.
Susceptibility to Antimicrobial Drugs
Antimicrobial resistance in N. meningitidis is considered to be acquired by transmission of genetic material from N. gonorrhoeae, such as the gonococcal aniA-norB locus (14). However, US_NmUC meningococci isolated in the United States were susceptible to the third-generation cephalosporin ceftriaxone, ciprofloxacin, and rifampin, whereas ≈75%–85% of US_NmUC meningococci were nonsusceptible (intermediate susceptibility) to penicillin G (34,35). The 3 J_NmUC meningococci were susceptible to most antimicrobial drugs tested, except the NIID836 strain had intermediate susceptibility to penicillin G, similar to US_NmUC meningococci (34,35). Those results suggest that genetic material related to antimicrobial resistance genes might not be transmitted into J_NmUC N. meningitidis isolates. Of note, the NIID835 strain was susceptible to penicillin G and ceftriaxone despite having the penA327 allele, which typically reduces susceptibility to penicillin G and third-generation cephalosporins (36).
Discussion
Meningococcus and gonococcus generally colonize distinct niches in humans causing systemic (meningococcus) and sexually transmitted (gonococcus) disease; few cases exist that identify N. meningitidis as a causative agent for STI (14). Meningococcal urethritis is symptomatically indistinguishable from gonococcal urethritis; one of the main problems in clinical and public health is that meningococcal urethritis cannot be diagnosed by the existing nucleic acid amplification test, a standard method for STI diagnosis (14,34). Urethritis clade meningococci, such as US_NmUC and J_NmUC, have been isolated only from urethritis patients (10), rectal swab samples of asymptomatic MSM (18), and 1 neonatal patient with conjunctivitis (37); virulence was considered equal to gonococci. However, urethritis clade meningococci were also speculated to colonize the upper respiratory tracts of sexual partners of persons who eventually manifested urethritis. No published studies exist regarding carriage of urethritis clade meningococci in the upper respiratory tract; thus, the public health threat of urethritis clade meningococci is unclear, and emergence of this clade should be continuously monitored.
Although deletion of the cps locus or genes within this locus, which results in loss of encapsulation, is a main features of urethritis clade meningococci (12,14), the pattern of deletion within the cps locus was different between J_NmUC and US_NmUC isolates, despite the identical junctions between the csc gene and IS1301 sequences. Because meningococcal loss of encapsulation enhances adherence to human cells (15,38–44), loss of the capsule might promote N. meningitidis–induced urethritis. However, some cases of meningococcal urethritis might be caused by encapsulated N. meningitidis isolates (30). Therefore, the relationship between loss of encapsulation by deletions within the cps locus in N. meningitidis and meningococcal urethritis should be further examined.
Acquisition of the gonococcal aniA-norB locus (12) was another main feature of urethritis clade meningococci (Figure 1). In some N. meningitidis strains, such as M-17541 (Appendix 2 Figure 1), the meningococcal aniA gene was disrupted by an insertion or missense mutation (45,46). Moreover, if the meningococcal aniA gene was intact, expression was lower than that of gonococcal aniA genes (45). However, the gonococcal aniA-norB locus was not detected in some N. meningitidis isolates from patients with meningococcal urethritis (30), suggesting that acquisition of the gonococcal aniA-norB locus was advantageous (12,13,17) but not essential to cause urethritis.
A phylogenetic analysis using WGS data supports the hypothesis that US_NmUC and J_NmUC might be derived from the same ancestor (Figure 3). US_NmUC appears to have originated during 2006–2012 in the United States (32), and the ancestral strain might have been imported into Japan during the same period. However, MenC, serogroup W, and CC11 meningococci have rarely been detected in Japan for >40 years, even in IMD patients (29,47,48). Although CC11 meningococci have never been identified as a causative agent for meningococcal urethritis in Japan (29), J_NmUC meningococci, as well as the ST11 ancestral strain, might be dormant in the urethra or pharynx of persons in Japan. Therefore, further analyses of meningococcal isolates from healthy carriers and patients with urethritis will provide insights into dissemination of the N. meningitidis urethritis clade among the human population in Japan.
In conclusion, few studies have attempted to estimate the prevalence of meningococcal infections, including the urethritis clade. J_NmUC meningococci identified in this study are new subtypes of US_NmUC, and microbiologic characteristics, such as virulence and transmissibility, remain unclear. Continuous monitoring and analyses of J_NmUC meningococci will elucidate more precise features, including transmissibility and pathogenicity. Moreover, detection of J_NmUC in Japan suggests potential dissemination of several types of urethritis clade meningococci (US_NmUC and J_NmUC) worldwide. Global monitoring of urethritis-associated N. meningitidis isolates should be required to reveal further microbiologic and epidemiologic aspects of urethritis clade meningococci and to improve laboratory diagnostic testing for urethritis.
Clinical and genetic isolate profiles for detection of novel US Neisseria meningitidis urethritis clade subtypes in Japan.
Additional information for detection of novel US Neisseria meningitidis urethritis clade subtypes in Japan.
Acknowledgments
This work was supported by a grant from the Japan Agency for Medical Research and Development (grant no. JP21fk0108605) and Ministry of Healthcare and Labour of Japan (grant no. JPMH22HA1007 to H.T.).
This publication made use of the PubMLST website (https://pubmlst.org) developed by Keith Jolley and Martin Maiden (49); the website is located at the University of Oxford. The development of that site has been funded by The Wellcome Trust and European Union.
Biography
Dr. Takahashi is the chief of the urogenital infection laboratory, Department of Bacteriology I, National Institute of Infectious Diseases, Japan. His research interests focus on meningococcal epidemiology and molecular pathogenesis as well as the gonococcal equivalents.
Footnotes
Suggested citation for this article: Takahashi H, Morita M, Yasuda M, Ohama Y, Kobori Y, Kojima M, et al. Detection of novel US Neisseria meningitidis urethritis clade subtypes in Japan. Emerg Infect Dis. 2023 Nov [date cited]. https://doi.org/10.3201/eid2911.231082
References
- 1.Acevedo R, Bai X, Borrow R, Caugant DA, Carlos J, Ceyhan M, et al. The Global Meningococcal Initiative meeting on prevention of meningococcal disease worldwide: Epidemiology, surveillance, hypervirulent strains, antibiotic resistance and high-risk populations. Expert Rev Vaccines. 2019;18:15–30. 10.1080/14760584.2019.1557520 [DOI] [PubMed] [Google Scholar]
- 2.Taha MK, Martinon-Torres F, Köllges R, Bonanni P, Safadi MAP, Booy R, et al. Equity in vaccination policies to overcome social deprivation as a risk factor for invasive meningococcal disease. Expert Rev Vaccines. 2022;21:659–74. 10.1080/14760584.2022.2052048 [DOI] [PubMed] [Google Scholar]
- 3.Mustapha MM, Marsh JW, Harrison LH. Global epidemiology of capsular group W meningococcal disease (1970-2015): Multifocal emergence and persistence of hypervirulent sequence type (ST)-11 clonal complex. Vaccine. 2016;34:1515–23. 10.1016/j.vaccine.2016.02.014 [DOI] [PubMed] [Google Scholar]
- 4.Schmink S, Watson JT, Coulson GB, Jones RC, Diaz PS, Mayer LW, et al. Molecular epidemiology of Neisseria meningitidis isolates from an outbreak of meningococcal disease among men who have sex with men, Chicago, Illinois, 2003. J Clin Microbiol. 2007;45:3768–70. 10.1128/JCM.01190-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marcus U, Vogel U, Schubert A, Claus H, Baetzing-Feigenbaum J, Hellenbrand W, et al. A cluster of invasive meningococcal disease in young men who have sex with men in Berlin, October 2012 to May 2013. Euro Surveill. 2013;18:20523. 10.2807/1560-7917.ES2013.18.28.20523 [DOI] [PubMed] [Google Scholar]
- 6.Kratz MM, Weiss D, Ridpath A, Zucker JR, Geevarughese A, Rakeman J, et al. Community-based outbreak of Neisseria meningitidis serogroup C infection in men who have sex with men, New York City, New York, USA, 2010–2013. Emerg Infect Dis. 2015;21:1379–86. 10.3201/eid2108.141837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taha MK, Claus H, Lappann M, Veyrier FJ, Otto A, Becher D, et al. Evolutionary events associated with an outbreak of meningococcal disease in men who have sex with men. PLoS One. 2016;11:e0154047. 10.1371/journal.pone.0154047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nanduri S, Foo C, Ngo V, Jarashow C, Civen R, Schwartz B, et al. Outbreak of serogroup C meningococcal disease primarily affecting men who have sex with men—Southern California, 2016. MMWR Morb Mortal Wkly Rep. 2016;65:939–40. 10.15585/mmwr.mm6535e1 [DOI] [PubMed] [Google Scholar]
- 9.Folaranmi TA, Kretz CB, Kamiya H, MacNeil JR, Whaley MJ, Blain A, et al. Increased risk for meningococcal disease among men who have sex with men in the United States, 2012–2015. Clin Infect Dis. 2017;65:756–63. 10.1093/cid/cix438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bazan JA, Peterson AS, Kirkcaldy RD, Briere EC, Maierhofer C, Turner AN, et al. Notes from the field: increase in Neisseria meningitidis–associated urethritis among men at two sentinel clinics—Columbus, Ohio, and Oakland County, Michigan, 2015. MMWR Morb Mortal Wkly Rep. 2016;65:550–2. 10.15585/mmwr.mm6521a5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toh E, Gangaiah D, Batteiger BE, Williams JA, Arno JN, Tai A, et al. Neisseria meningitidis ST11 complex isolates associated with nongonococcal urethritis, Indiana, USA, 2015–2016. Emerg Infect Dis. 2017;23:336–9. 10.3201/eid2302.161434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tzeng YL, Bazan JA, Turner AN, Wang X, Retchless AC, Read TD, et al. Emergence of a new Neisseria meningitidis clonal complex 11 lineage 11.2 clade as an effective urogenital pathogen. Proc Natl Acad Sci U S A. 2017;114:4237–42. 10.1073/pnas.1620971114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bazan JA, Stephens DS, Turner AN. Emergence of a novel urogenital-tropic Neisseria meningitidis. Curr Opin Infect Dis. 2021;34:34–9. 10.1097/QCO.0000000000000697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burns BL, Rhoads DD. Meningococcal urethritis: old and new. J Clin Microbiol. 2022;60:e0057522. 10.1128/jcm.00575-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartley SN, Tzeng YL, Heel K, Lee CW, Mowlaboccus S, Seemann T, et al. Attachment and invasion of Neisseria meningitidis to host cells is related to surface hydrophobicity, bacterial cell size and capsule. PLoS One. 2013;8:e55798. 10.1371/journal.pone.0055798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yee WX, Barnes G, Lavender H, Tang CM. Meningococcal factor H-binding protein: implications for disease susceptibility, virulence, and vaccines. Trends Microbiol. 2023;31:805–15. 10.1016/j.tim.2023.02.011 [DOI] [PubMed] [Google Scholar]
- 17.Tzeng YL, Sannigrahi S, Berman Z, Bourne E, Edwards JL, Bazan JA, et al. Acquisition of gonococcal AniA-NorB pathway by the Neisseria meningitidis urethritis clade confers denitrifying and microaerobic respiration advantages for urogenital adaptation. Infect Immun. 2023;91:e0007923. 10.1128/iai.00079-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brooks A, Lucidarme J, Campbell H, Campbell L, Fifer H, Gray S, et al. Detection of the United States Neisseria meningitidis urethritis clade in the United Kingdom, August and December 2019 - emergence of multiple antibiotic resistance calls for vigilance. Euro Surveill. 2020;25:2000375. 10.2807/1560-7917.ES.2020.25.15.2000375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen HT, Phan TV, Tran HP, Vu TTP, Pham NTU, Nguyen TTT, et al. Outbreak of sexually transmitted nongroupable Neisseria meningitidis–associated urethritis, Vietnam. Emerg Infect Dis. 2023;29:2130–4. 10.3201/eid2910.221596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taha MK. Simultaneous approach for nonculture PCR-based identification and serogroup prediction of Neisseria meningitidis. J Clin Microbiol. 2000;38:855–7. 10.1128/JCM.38.2.855-857.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95:3140–5. 10.1073/pnas.95.6.3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito R, Nakajima J, Prah I, Morita M, Mahazu S, Ota Y, et al. Penicillin- and ciprofloxacin-resistant invasive Neisseria meningitidis isolates from Japan. Microbiol Spectr. 2022;10:e0062722. 10.1128/spectrum.00627-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wick RR, Judd LM, Holt KE. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 2019;20:129. 10.1186/s13059-019-1727-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 2017;13:e1005595. 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanizawa Y, Fujisawa T, Nakamura Y. DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics. 2018;34:1037–9. 10.1093/bioinformatics/btx713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3. 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, Keane JA, et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genom. 2016;2:e000056. 10.1099/mgen.0.000056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–6. 10.1093/nar/gkab301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi H, Morita M, Kamiya H, Fukusumi M, Sunagawa M, Nakamura-Miwa H, et al. Genomic characterization of Japanese meningococcal strains isolated over a 17-year period between 2003 and 2020 in Japan. Vaccine. 2023;41:416–26. 10.1016/j.vaccine.2022.10.083 [DOI] [PubMed] [Google Scholar]
- 30.Ma KC, Unemo M, Jeverica S, Kirkcaldy RD, Takahashi H, Ohnishi M, et al. Genomic characterization of urethritis-associated Neisseria meningitidis shows that a wide range of N. meningitidis strains can cause urethritis. J Clin Microbiol. 2017;55:3374–83. 10.1128/JCM.01018-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tettelin H, Saunders NJ, Heidelberg J, Jeffries AC, Nelson KE, Eisen JA, et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–15. 10.1126/science.287.5459.1809 [DOI] [PubMed] [Google Scholar]
- 32.Retchless AC, Kretz CB, Chang HY, Bazan JA, Abrams AJ, Norris Turner A, et al. Expansion of a urethritis-associated Neisseria meningitidis clade in the United States with concurrent acquisition of N. gonorrhoeae alleles. BMC Genomics. 2018;19:176. 10.1186/s12864-018-4560-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biagini M, Spinsanti M, De Angelis G, Tomei S, Ferlenghi I, Scarselli M, et al. Expression of factor H binding protein in meningococcal strains can vary at least 15-fold and is genetically determined. Proc Natl Acad Sci U S A. 2016;113:2714–9. 10.1073/pnas.1521142113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sukhum KV, Jean S, Wallace M, Anderson N, Burnham CA, Dantas G. Genomic characterization of emerging bacterial uropathogen Neisseria meningitidis, which was misidentified as Neisseria gonorrhoeae by nucleic acid amplification testing. J Clin Microbiol. 2021;59:e01699–20. 10.1128/JCM.01699-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bazan JA, Tzeng YL, Bischof KM, Satola SW, Stephens DS, Edwards JL, et al. Antibiotic susceptibility profile for the US Neisseria meningitidis urethritis clade. Open Forum Infect Dis. 2023;10:ofac661. [DOI] [PMC free article] [PubMed]
- 36.Willerton L, Lucidarme J, Walker A, Lekshmi A, Clark SA, Walsh L, et al. Antibiotic resistance among invasive Neisseria meningitidis isolates in England, Wales and Northern Ireland (2010/11 to 2018/19). PLoS One. 2021;16:e0260677. 10.1371/journal.pone.0260677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kretz CB, Bergeron G, Aldrich M, Bloch D, Del Rosso PE, Halse TA, et al. Neonatal conjunctivitis caused by Neisseria meningitidis US urethritis clade, New York, USA, August 2017. Emerg Infect Dis. 2019;25:972–5. 10.3201/eid2505.181631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Virji M, Makepeace K, Ferguson DJ, Achtman M, Sarkari J, Moxon ER. Expression of the Opc protein correlates with invasion of epithelial and endothelial cells by Neisseria meningitidis. Mol Microbiol. 1992;6:2785–95. 10.1111/j.1365-2958.1992.tb01458.x [DOI] [PubMed] [Google Scholar]
- 39.Stephens DS, Spellman PA, Swartley JS. Effect of the (alpha 2—>8)-linked polysialic acid capsule on adherence of Neisseria meningitidis to human mucosal cells. J Infect Dis. 1993;167:475–9. 10.1093/infdis/167.2.475 [DOI] [PubMed] [Google Scholar]
- 40.McNeil G, Virji M, Moxon ER. Interactions of Neisseria meningitidis with human monocytes. Microb Pathog. 1994;16:153–63. 10.1006/mpat.1994.1016 [DOI] [PubMed] [Google Scholar]
- 41.Kolb-Mäurer A, Unkmeir A, Kämmerer U, Hübner C, Leimbach T, Stade A, et al. Interaction of Neisseria meningitidis with human dendritic cells. Infect Immun. 2001;69:6912–22. 10.1128/IAI.69.11.6912-6922.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hill DJ, Griffiths NJ, Borodina E, Virji M. Cellular and molecular biology of Neisseria meningitidis colonization and invasive disease. Clin Sci (Lond). 2010;118:547–64. 10.1042/CS20090513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sutherland TC, Quattroni P, Exley RM, Tang CM. Transcellular passage of Neisseria meningitidis across a polarized respiratory epithelium. Infect Immun. 2010;78:3832–47. 10.1128/IAI.01377-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi H, Kim KS, Watanabe H. Differential in vitro infectious abilities of two common Japan-specific sequence-type (ST) clones of disease-associated ST-2032 and carrier-associated ST-2046 Neisseria meningitidis strains in human endothelial and epithelial cell lines. FEMS Immunol Med Microbiol. 2008;52:36–46. 10.1111/j.1574-695X.2007.00342.x [DOI] [PubMed] [Google Scholar]
- 45.Stefanelli P, Colotti G, Neri A, Salucci ML, Miccoli R, Di Leandro L, et al. Molecular characterization of nitrite reductase gene (aniA) and gene product in Neisseria meningitidis isolates: is aniA essential for meningococcal survival? IUBMB Life. 2008;60:629–36. 10.1002/iub.95 [DOI] [PubMed] [Google Scholar]
- 46.Barth KR, Isabella VM, Clark VL. Biochemical and genomic analysis of the denitrification pathway within the genus Neisseria. Microbiology (Reading). 2009;155:4093–103. 10.1099/mic.0.032961-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi H, Kuroki T, Watanabe Y, Tanaka H, Inouye H, Yamai S, et al. Characterization of Neisseria meningitidis isolates collected from 1974 to 2003 in Japan by multilocus sequence typing. J Med Microbiol. 2004;53:657–62. 10.1099/jmm.0.45541-0 [DOI] [PubMed] [Google Scholar]
- 48.Fukusumi M, Kamiya H, Takahashi H, Kanai M, Hachisu Y, Saitoh T, et al. National surveillance for meningococcal disease in Japan, 1999-2014. Vaccine. 2016;34:4068–71. 10.1016/j.vaccine.2016.06.018 [DOI] [PubMed] [Google Scholar]
- 49.Jolley KA, Maiden MCJ. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical and genetic isolate profiles for detection of novel US Neisseria meningitidis urethritis clade subtypes in Japan.
Additional information for detection of novel US Neisseria meningitidis urethritis clade subtypes in Japan.


