ABSTRACT
Developing broad-spectrum vaccines and optimal vaccination strategies is crucial to controlling the COVID-19 pandemic. Here, we generated a chimpanzee adenoviral vector-based COVID-19 vaccine carrying broad-spectrum immunogens, modified full-length spike, and conserved T-cell epitopes of SARS-CoV-2, and assessed its immune response in mice through intramuscular (i.m.), intranasal (i.n.), or combined immunization routes (i.m. + i.n., or i.n. + i.m.). Compared to other vaccination strategies, the two combined regimens elicited higher neutralizing antibody (NAb) responses to all variants. Compared to i.n. + i.m. regimen, the i.m. + i.n. regimen stimulated a stronger secondary GC response, which is more pivotal to high-quality antibody production than the primary GC response. Moreover, the i.m. + i.n. regimen was adept at mediating systemic cellular immunity, while the i.n. + i .m. regimen tended to elicit lung tissue-resident memory T (TRM) cell responses. Overall, the two combined regimens induced comprehensive but distinct immune responses consisting of lgA, lgG, NAbs, GC B cells, long-lived plasma cells, TRM cells, and systemic memory T cells, which conferred complete protection against BA.2 infection in hACE2 transgenic mice, and warranted further investigation as potential universal vaccination strategies.
IMPORTANCE
The development of broad-spectrum SARS-CoV-2 vaccines will reduce the global economic and public health stress from the COVID-19 pandemic. The use of conserved T-cell epitopes in combination with spike antigen that induce humoral and cellular immune responses simultaneously may be a promising strategy to further enhance the broad spectrum of COVID-19 vaccine candidates. Moreover, this research suggests that the combined vaccination strategies have the ability to induce both effective systemic and mucosal immunity, which may represent promising strategies for maximizing the protective efficacy of respiratory virus vaccines.
KEYWORDS: SARS-CoV-2, adenovirus vector, broad-spectrum vaccine, combined immunization routes, systemic immunity, mucosal immunity
INTRODUCTION
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has been a global pandemic since the beginning of 2020 and has severely affected public health. The rapid evolution of SARS-CoV-2 gave rise to numerous variants, and some have been designated as variants of concern/interest (VOC/I) by the World Health Organization. As the dominant strain, the Omicron variant contains over 30 mutations in spike (S) protein that leads to immune evasion and increased transmissibility (1). First-generation COVID-19 vaccines, derived from the spike or RBD of SARS-CoV-2 ancestral strain, have shown waning effectiveness against VOCs, especially Omicron (2).
Both humoral and cellular immune responses are essential in controlling virus infections. Neutralizing antibodies (NAbs) are regarded as the major contributor to host protection against SARS-CoV-2 (3), while SARS-CoV-2-specific T-cell responses play roles in reducing disease acquisition and severity (4). Even when NAbs and memory B cell responses decreased significantly, T-cell responses to diverse variants from Alpha to Omicron were still preserved in most convalescents and vaccines (5). These observations offer a compelling explanation for the effectiveness of first-generation COVID-19 vaccines in protecting against severe disease and death under the condition of impaired or even absent neutralization capacity against Omicron. Nevertheless, ~20% of individuals have a >50% reduction in T-cell reactivity to Omicron spike (6). Given the uncertainty of further evolution, broad-spectrum vaccines, which are capable of providing broad-spectrum humoral and cellular immunity, are urgently required to protect against circulating and emerging variants.
Currently, available vaccines offer limited protection against breakthrough infections, especially for the Omicron sublineage (7). Importantly, SARS-CoV-2 infection primarily targets the respiratory tract, especially the nasal mucosa. The vast majority of first-generation vaccines are administered via the intramuscular route, which is incapable of eliciting IgA secretion at the mucosal surface or tissue-resident memory T (TRM) cells activation in lungs. Both the immune evasive potential of variants and the absence of mucosal immunity are believed to account for their suboptimal effectiveness (8). Hence, the intranasal (i.n.) vaccination, which closely mimics the natural infection route and has the potential to provoke a robust local mucosal immunity may be a promising approach to defending against SARS-CoV-2 infection.
Adenovirus is an attractive vaccine platform owing to its flexibility of administration routes and capacity to induce humoral and cellular immune responses simultaneously (9). First-generation adenoviral vector-based COVID-19 vaccines include ChAdOx1 nCoV-19 (Oxford-AstraZeneca), Ad26.COV2.S (Johnson & Johnson), Sputnik V (Gamaleya Research Institute), and Ad5-nCoV (CanSino Biologics) (10 – 12). Recently, two adenovector-based mucosal vaccines, Salnavak (an intranasal spray version of Sputnik V) and Ad5-nCoV (an inhaled aerosol vaccine developed by CanSino Biologics) have been approved for clinical use (13, 14). Notably, these vaccines were administered through different formulations, including prime-only (i.m. and i.n.) and matched prime-boost (i.m. + i.m. and i.n. + i.n.) regimens, whereas the combined prime-boost vaccination strategies (i.m. + i.n. and i.n. + i.m.) have not been implemented so far. Besides, no head-to-head comparative studies of the aforementioned vaccination strategies are available, which leads to uncertainty surrounding optimal regimens for clinical use.
Herein, we developed an adenoviral vector-based broad-spectrum COVID-19 vaccine carrying modified spike and conserved T-cell epitopes. To grope for optimal regimens, we performed a direct comparison of antibody responses induced by different regimens in mice and found that the two combined regimens were superior in improving antibody responses. Then, we performed in-depth immunophenotype exploration to characterize the mucosal and systemic immune responses induced by them. To our knowledge, this study is the first to comprehensively compare adaptive immune responses of two homogenous combined regimens.
RESULTS
Designing and characterizing the AdC68-vST-vtRBM
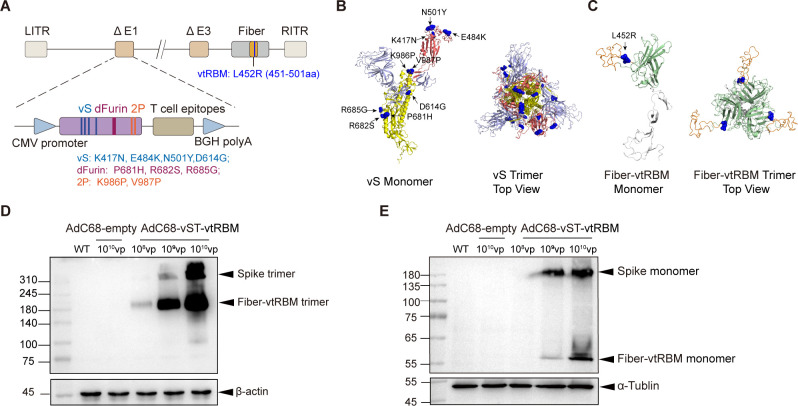
Based on chimpanzee adenovirus serotype-68 (AdC68), we designed a broad-spectrum COVID-19 vaccine AdC68-vST-vtRBM carrying full-length spike with several mutations (vS), numerous T-cell epitopes (TCEs), and truncated receptor-binding motif (RBM) (Fig. 1A). As of February 2021, several VOCs have been described, including Alpha, Beta, and Gamma. These VOCs share N501Y and D614G mutations in spike, which are associated with increased transmissibility (15, 16). The K417N spike mutation in Beta stabilizes a more open spike trimer that achieves greater transmissibility when combined with D614G (17) and also partly resists neutralization by post-vaccination sera (18). The E484K spike mutation shared by Beta and Gamma is a main contributor to neutralization evasion (19, 20). In addition, to stabilize spike in the prefusion conformation, we introduced furin cleavage site mutation (dFurin: 681-PRRAR-685 to 681-HSRAG-685) and two proline substitutions K986P and V987P (2P) (21, 22). Thus, we generated a modified sequence of spike (vS) containing substitutions K417N, E484K, N501Y, D614G, P681H, R682S, R685G, K986P, and V987P. Figure 1B shows its predicted tertiary structure.
Fig 1.
Designing and characterizing the AdC68-vST-vtRBM. (A) Schematic of AdC68-vST-vtRBM construction. TCEs, tandem conserved T-cell epitopes from ORF1, ORF3, and M proteins of ancestral SARS-CoV-2 ; vS, full-length spike with several mutations; dFurin, abolished furin cleavage site; 2P, two proline substitutions; vtRBM, truncated receptor-binding motif containing L452R mutation. (B and C) The sequence of vS (B) or vtRBM-modified fiber (C) was subjected to SWISS-Model server for tertiary structure prediction, and analyzed using PyMol. Ribbon diagrams of monomer and top view trimer were displayed. In (B), S1 subunit is shown in grey with RBD domain in red. S2 subunit is shown in yellow. In (C), knob is shown in green with vtRBM in orange. (D and E) Western blot analysis of vS and vtRBM expression under non-reductive (D) and reductive (E) conditions. HEK293 cells were transduced with 108, 109, and 1010 vp of AdC68-vST-vtRBM. AdC68-empty-transduced (1010 vp) HEK293 cells and untransduced HEK293 cells (WT) were used as controls.
Based on previous studies, the immunogenic and conserved T-cell epitopes from the structural and non-structural proteins of SARS-CoV-2, which have been proven to be involved in T-cell activation to combat COVID-19 (23 – 25), were selected for broadening cellular immunity. Insertion of antigenic epitopes into adenoviral surface protein fiber could induce protective immunity against influenza viruses and Pseudomonas aeruginosa (26, 27). The L452R spike mutation shared by the subsequently emerged variants Kappa and Delta is relevant to immune escape and increased transmissibility (28). To enhance immune responses, the truncated RBM of spike (451–501aa) containing L452R mutation (termed as vtRBM) was incorporated into the HI loop of fiber knob. Both adenovirus fiber and SARS-CoV-2 spike exist as trimers on the surface of native virions. Theoretically, vtRBM-modified fiber would present in a trimeric form on AdC68 virion surface. Figure 1C shows its structural modeling.
Western blot analyses revealed a vaccine dose-dependent expression of vS and vtRBM-modified fiber in vitro (Fig. 1D and E). Due to the lack of furin cleavage site, vS was not cleaved into S1 and S2. Moreover, both vS and vtRBM-modified fiber maintained trimeric structure under non-reducing conditions (Fig. 1D), whereas 2-mercaptoethanol treatment depolymerized them into corresponding monomers (Fig. 1E). These indicated that our vaccine properly expressed vS and vtRBM-modified fiber without blocking their trimerization.
Single-dose intramuscular immunization induced superior systemic T-cell responses over intranasal immunization
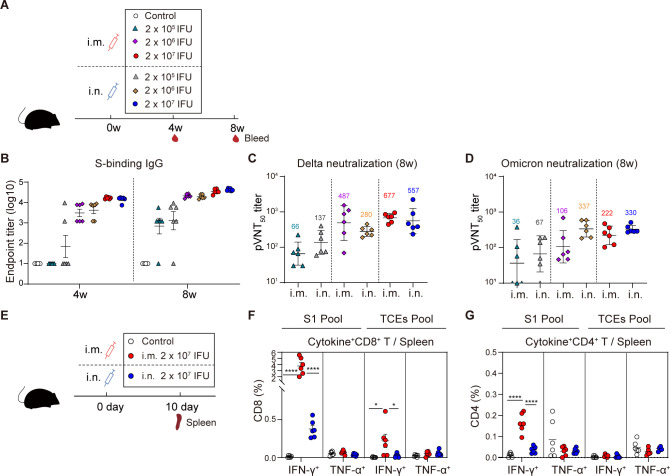
We first assessed the immunogenicity of AdC68-vST-vtRBM as a single-shot intramuscular or intranasal vaccine (Fig. 2A). S-specific IgG was induced in a dose- and time-dependent manner when compared to control mice (Fig. 2B). Consistent with binding antibody (Bab) responses, a dose-dependent neutralizing capacity against Delta and Omicron was observed irrespective of the inoculation route (Fig. 2C and D). AdC68-vST-vtRBM [2 × 107 infectious units (IFU)] induced relatively superior antibody responses and was adopted as the optimal dose in subsequent studies.
Fig 2.
Single-dose intramuscular immunization induced superior systemic T-cell responses over intranasal immunization. (A) Scheme of experiments. Mice (n = 6) were i.m. or i.n. immunized with 2 × 105, 2 × 106, and 2 × 107 IFU of AdC68-vST-vtRBM, respectively. About 100 µL PBS intramuscularly vaccinated mice served as control. Serum was collected at 4 and 8 weeks post-vaccination. (B) Serum anti-spike IgG titers. An ELISA was used to measure anti-spike lgG antibodies at 4 and 8 weeks post-vaccination. (C and D) The neutralizing activity against Delta (C) or Omicron (D) at 8 weeks post-vaccination. (E) Scheme of experiments. Mice (n = 6) were vaccinated with 2 × 107 IFU of AdC68-vST-vtRBM by i.m. or i.n. route, 100 µL PBS intramuscularly vaccinated mice served as control. Mice were euthanized at 10 days post-vaccination. Spleens were harvested for effector T-cell detection. (F and G) Frequencies of splenic CD8+ T cells (F) and CD4+ (G) T cells producing IFN-γ, TNF-α following re-stimulation with peptide pools for S1 and TCEs. Values of geometric mean titer (GMT) were displayed in (C) and (D). Data are represented as GMT ± SD (C and D) or mean ± SEM (B, F and G). One-way ANOVA with Tukey correction was used for multiple comparisons. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
We next carried out intracellular cytokine staining (ICS) assays to evaluate T-cell responses (Fig. 2E). The i.m. vaccination elicited a significant increase of S1-specific IFN-γ-producing CD4+ and CD8+ T cells, along with suboptimal TCEs-specific IFN-γ+ CD8+ T cells. In contrast, i.n. vaccination induced faint responses, with a small quantity of TCEs-specific IFN-γ+ CD8+ T cells activation (Fig. 2F and G). Compared to S1-specific responses, TCEs-specific CD8+ and CD4+ T-cell responses were lower, which may be attributed to the weaker immunogenicity of TCEs. Spike protein contains 1273 amino acids, while TCEs consist of only 208 amino acids. As predicted, CD4+ and CD8+ T cells from phosphate-buffered saline (PBS)-treated mice are not responsive to either peptide pool.
Thus, whether i.m. or i.n. administered, our COVID-19 vaccine showed good immunogenicity in an adjuvant-independent manner. The i.m. vaccination was more effective in eliciting systemic T-cell responses than i.n. vaccination, although both regimens showed equivalent efficiency in inducing systemic antibody responses.
The i.m. + i.n. regimen-induced superior antibody responses
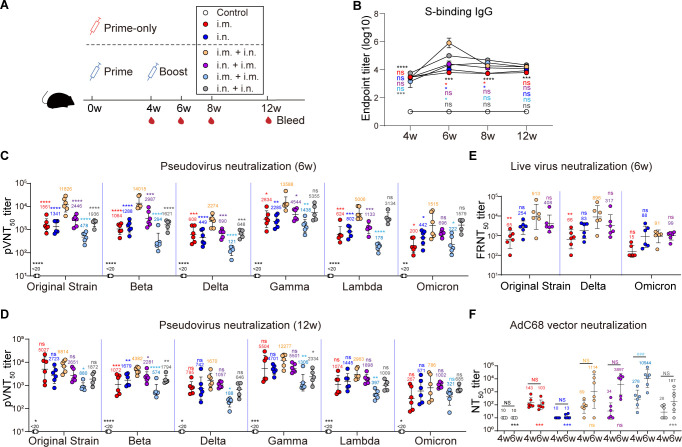
Vaccine effectiveness can be affected by vaccination regimen. To identify the optimal regimen for AdC68-vST-vtRBM, six groups of C57BL/6J mice were immunized using prime-only regimens or homologous prime-boost regimens (Fig. 3A). Compared to the control group, durable BAbs were elicited from 4 weeks post-vaccination (wpv) and maintained at a high level for at least 12 weeks, regardless of the regimen (Fig. 3B). At 6 wpv, i.m. + i.n. regimen provoked a dramatic surge of BAbs that subsequently restored to a comparable level with other regimens.
Fig 3.
The i.m. + i.n. regimen induced superior antibody responses. (A) Scheme of experiments. Mice (n = 6) were immunized in prime-only regimens (i.m., i.n., respectively, at 0 weeks) or prime-boost regimens (i.m. + i.n., i.n. + i.m., i.m. + i.m., and i.n. + i.n., respectively, with an interval of 4 weeks) with 2 × 107 IFU of AdC68-vST-vtRBM. About 100 µL PBS intramuscularly vaccinated mice served as control. Serum samples were collected periodically. (B) Kinetics of serum anti-spike IgG responses. (C and D) Serum pseudovirus neutralizing antibody responses at 6 weeks post-vaccination (C) and 12 weeks post-vaccination (D). (E) Serum live virus neutralizing antibody responses at 6 weeks post-vaccination. (F) Serum neutralizing antibody responses against AdC68 at 4 and 6 weeks post-vaccination. Values of geometric mean titer (GMT) were displayed in (C–F). Data are represented as GMT ± SD (C–F) or mean ± SEM (B). Statistical analysis was conducted using a two-way ANOVA with Sidak’s multiple comparisons test (B and F) or Tukey’s multiple comparisons test or Kruskal-Wallis test with Dunn’s multiple comparisons test according to distribution of data (C–E). Asterisks (*) or “ns” indicate the statistical significance level of each group as compared to i.m. + i.n. group (B–E) or i.m. + i.m. group (F) at the same time point. Pound signs (#) or “NS” indicate the statistical significance level between 4 and 6 wpv in (F). *P ≤ 0.05, **P ≤ 0.01, ***, ### P ≤ 0.001, ****P ≤ 0.0001. NS and ns, not significant.
NAbs induced by all regimens were detectable at 4 wpv and remained stable for at least 12 weeks (Fig. 3C and D; Fig. S1). Our vaccine displayed broad-spectrum neutralizing activity against the original strain, four VOCs [Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (BA.1)] and a VOI [Lambda (C.37)], although NAbs against Omicron were significantly reduced. Consistent with BAbs, the i.m. + i.n. regimen induced the highest NAb titers against all pseudovirus and displayed 6.5-, 12-, 3.5-, 7.0-, 2.7-, and 6.6-fold increase compared to i.m. group at 6 wpv (Fig. 3C). For neutralization activity against the live virus, the i.m. + i.n. regimen also exhibited higher level than the prime-only groups (Fig. 3E).
To understand the impact of vector-specific antibodies, we evaluated the serum neutralizing capacity against AdC68 vector (Fig. 3F). AdC68-NAbs were maintained at baseline levels after priming regardless of vaccination route at 4 wpv (P > 0.05 among all groups). After boosting, especially i.m. boosting (P = 0.0004), an increase in AdC68-NAbs values was observed. The i.m. + i.m. regimen produced the most robust vector-specific antibody responses, perhaps responsible for the impaired neutralizing activity against SARS-CoV-2.
Overall, for adenoviral vector vaccine, the dosage, route, and order of vaccination played important roles in determining the antibody responses.
Stronger secondary GC responses resulted in higher antibody titers
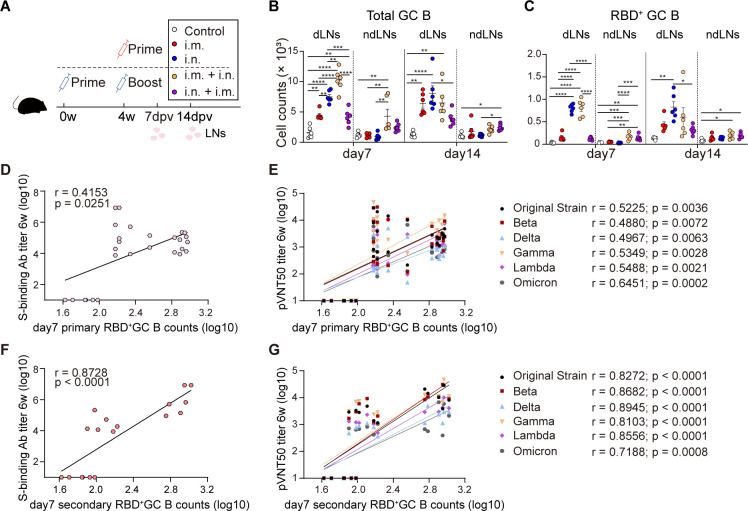
As demonstrated above, both combined prime-boost regimens excelled in antibody induction. In contrast to prime-only or match prime-boost regimens, knowledge about detailed immunological evaluation of two combined regimens was limited. To address the knowledge gap, our next research focused on making comprehensive comparison of adaptive immunity between the two combined regimens. First, we sought to dissect why the i.m. + i.n. regimen was far more potent in inducing SARS-CoV-2-specific antibody responses than the i.n. + i.m. regimen. Germinal centers (GCs) are known to be critical in the development of high-quality antibody responses (29). We aimed to characterize the primary (induced by priming) and secondary (induced by boosting) GC responses induced by two combined regimens in lymph nodes (LNs) and identify their relevance with antibody titers.
Previous studies revealed that GC responses peak between 5 and 14 days post-vaccination (29, 30). We immunized C57BL/6J mice with i.m., i.n., i.m. + i.n., or i.n. + i.m. regimens and harvested LNs at 7 and 14 days post-last vaccination (dpv) (Fig. 4A). The i.n. immunization stimulated robust and comparable total GC B cell response in draining LNs (dLNs) at 7 and 14 dpv (Fig. 4B), as well as RBD+ GC B cell response (Fig. 4C). In contrast with i.n. immunization, the i.m. immunization induced moderate total GC B cell response but minimal RBD+ GC B cell responses, indicating that total GC B cells contained abundant non-RBD+ GC B cells. To determine whether these non-RBD+ GC B cells were vector-specific, we immunized mice with 2 × 107 IFU AdC68-empty via i.m. or i.n. route and collected LNs at seven dpv (Fig. S2A). A significant increase in total GC B cells was observed in dLNs of i.m.- but not i.n.-immunized mice compared to PBS-treated mice (Fig. S2C and D). Thus, for the adenovirus vaccine, vector-specific GC response and antigen-specific GC response seem to be two interacting factors: vector-specific GC response would abate when antigen-specific GC response dominates, while substantial vector-specific GC response impairs antigen-specific GC response to some extent.
Fig 4.
Stronger secondary GC responses resulted in higher antibody titers. (A) Scheme of experiments. Two sets of mouse experiments were performed simultaneously. Mice (n = 6) were immunized in prime-only regimens (i.m., i.n., respectively, at 4 weeks) or prime-boost regimens (i.m. + i.n., i.n. + i.m., respectively, with an interval of 4 weeks) with 2 × 107 IFU of AdC68-vST-vtRBM. About 100 µL PBS intramuscularly vaccinated mice served as control. The i.m. dLNs (pooled popliteal, inguinal, and iliac LNs, the dLNs for i.m. vaccination and also served as the ndLNs for i.n. vaccination) and i.n. dLNs (pooled cervical and mediastinal LNs, the dLNs for i.n. vaccination and also served as the ndLNs for i.m. vaccination) were harvested at 7 and 14 days post-last vaccination. (B and C) Absolute number of total or RBD+ GC B cells. (D) Spearman correlations of S-binding antibody titers and primary RBD+ GC B cell counts. The primary RBD+ GC B counts refer to RBD+ GC B cells induced by i.m. or i.n. priming in dLNs. BAbs here refer to endpoint titers of i.m. + i.m., i.n. + i.n., i.m. + i.n., i.n. + i.m. regimens after boosting. Data of control groups were also contained in all Spearman correlation analysis. (E) Spearman correlations of pVNT50 titers and primary RBD+ GC B cell counts. pVNT50 titers here refer to those of i.m. + i.m., i.n. + i.n., i.m. + i.n., and i.n. + i.m. regimens after boosting. (F) Spearman correlations of S-binding antibody titers and secondary RBD+ GC B cell counts. BAbs here refer to S-binding antibody titers of i.m. + i.n. and i.n. + i.m. regimens after boosting. Secondary RBD+ GC B counts refer to RBD+ GC B cells induced by i.m. + i.n. and i.n. + i.m. regimens in dLNs. (G) Spearman correlations of pVNT50 titers and secondary RBD+ GC B cell counts. pVNT50 titers here refer to those of i.m. + i.n. and i.n. + i.m. regimens after boosting. All data are represented as mean ± SEM and analyzed by one-way ANOVA with Tukey correction.*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001.
In keeping with the previous study, we observed that GC formation was limited to dLNs but not non-dLNs (ndLNs) after priming (31). However, at 7 and 14 days post-boosting, modest amounts of RBD+ GC B cells were detected in ndLNs that also served as the dLNs for prime vaccination, suggesting that i.m. or i.n. priming triggered a persistent RBD+ GC response for at least 42 days. After the combined boosting, secondary GC responses were induced in dLNs with a similar kinetic pattern and comparable magnitude to primary GC responses that were elicited via the same immunization route (Fig. 4B and C). In other words, a combined booster did not handicap antigen-specific GC B cell response, and immunophenotype discrepancy between the two combined regimens was mainly attributed to the inherent capability divergence of booster shot in RBD+ GC response induction. Altogether, SARS-CoV-2-specific GC responses to the i.m. + i.n. regimen are characterized by feeble primary GC responses and robust secondary GC responses, which are contrary to that of the i.n. + i.m. regimen. There were weak associations between primary RBD+ GC B cell responses and antibody responses, regardless of NAbs or BAbs (Fig. 4D and E). Intriguingly, the secondary RBD+ GC B cells exhibited robust correlations with the titers of S-specific BAbs and pVNT50 against the original strain and its variants (Fig. 4F and G).
Overall, our data suggested that in terms of homogenous combined regimens, superior secondary GC responses were more important in improving antibody quality than primary GC responses.
The combined boosting broadened primary T-cell responses
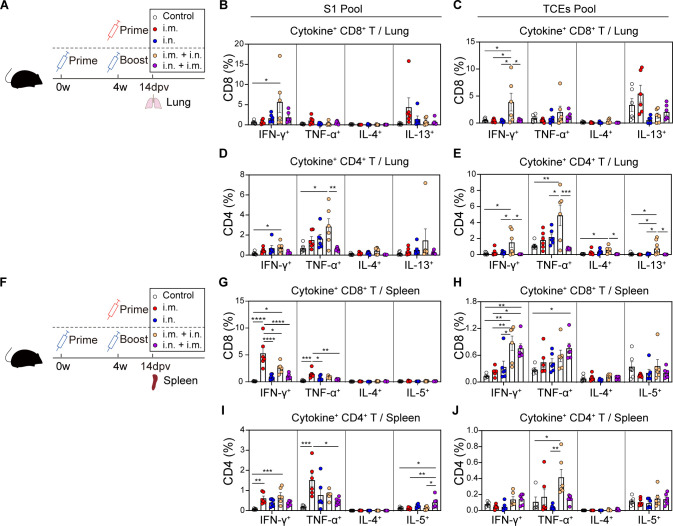
Next, we assessed the phenotype of effector T-cell responses in mice following prime-only or combined prime-boost regimens (Fig. 5A). The effector function of SARS-CoV-2-specific T cells was determined in lungs at 14 dpv by the production of Th1-type cytokines (IFN-γ and TNF-α) or Th2-type cytokines (IL-4 and IL-13) (Fig. 5B through E). We found that single-dose i.m. or i.n. immunization developed poor effector T-cell responses. After combined boosting, the i.m. + i.n., but not i.n. + i.m., regimen induced a prominent increase in both S1- and TCEs-specific T cells, which were characterized by a Th1-biased immune phenotype.
Fig 5.
The combined boosting broadened primary T-cell responses. (A) Scheme of experiments. The immunization schemes were identical to Fig. 4. Lung tissues were harvested at 14 days post-last vaccination. (B–E) Frequencies of pulmonary CD8+ T cells (B and C) and CD4+ T cells (D and E) producing cytokines following re-stimulation with peptide pools for S1 (B and D) and TCEs (C and E). (F) Scheme of experiments. The immunization schemes were identical to Fig. 4. Spleens were harvested at 14 days post-last vaccination. (G–J) Frequencies of splenic CD8+ T cells (G and H) and CD4+ T cells (I and J) producing cytokines following re-stimulation with peptide pools for S1 (G and I) and TCEs (H and J). All data are represented as mean ± SEM and analyzed by one-way ANOVA with Tukey correction. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001.
A parallel experiment was conducted for SARS-CoV-2-specific T-cell immunity evaluation in spleens (Fig. 5F). The i.m.- rather than i.n.-immunized mice developed S1-specific responses with CD8+ T cells dominant over CD4+ T cells, which were not augmented by the combined boosting (Fig. 5G and I). For TCEs-specific responses, the frequency of IFN-γ+ CD8+ T cells was significantly increased in combined prime-boost groups compared with corresponding prime-only groups (Fig. 5H). Consistent with the cellular responses in lungs, S1- and TCEs-specific T cells in spleens exhibited a clear Th1-biased phenotype with prominent IFN-γ and/or TNF-α secretion (Fig. 5I and J).
In summary, the i.m. immunization primed strong effector T-cell responses only in spleens, while SARS-CoV-2-specific CD4+ and CD8+ T cells were activated both in spleens and lungs after i.n. boosting. Meanwhile, the i.n. priming did not elicit appreciable systemic T-cell responses until i.m. boosting. Altogether, the combined boosting could broaden systemic T-cell responses induced by priming.
Two combined regimens triggered both memory mucosal and systemic immune responses
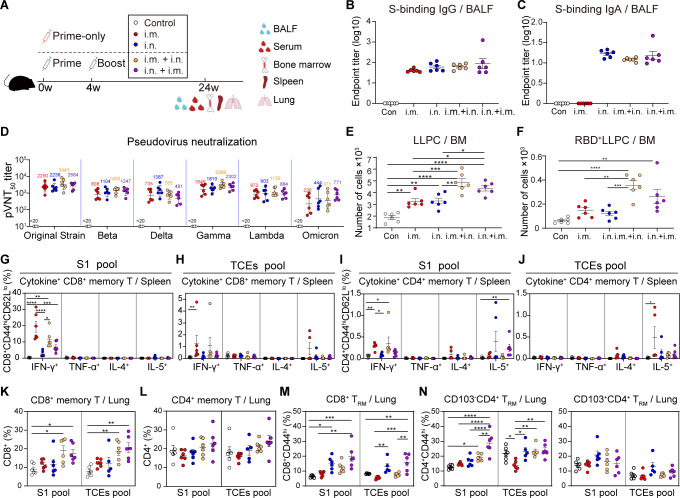
We also investigated whether AdC68-vST-vtRBM induced comprehensive memory immune responses at 24 wpv (Fig. 6A). BAb detection in the bronchoalveolar lavage fluid (BALF) revealed that only the i.m. regimen failed to generate IgA, although the lgG levels were comparable among all groups (Fig. 6B and C). Moreover, serum NAb titer measurements reconfirmed that our vaccine-induced durable and broad-spectrum antibody responses (Fig. 6D).
Fig 6.
Two combined regimens triggered both memory mucosal and systemic immune responses. (A) Scheme of experiments. The immunization scheme was identical to Fig. 3. Serum, spleens, lungs, BALF, and bone marrow were harvested at 24 weeks post-vaccination. All data analyzed at 24 weeks post-vaccination. (B and C) BALF anti-spike IgG (B) and IgA (C) endpoint titers. (D) Serum pseudovirus NAb responses. (E and F) Absolute numbers of total or RBD+ LLPCs population. (G–J) Frequencies of splenic memory CD8+ T cells (G and H) and memory CD4+ T cells (I and J) producing cytokines following re-stimulation with peptide pools for S1 (G and I) and TCEs (H and J). (K and L) Frequencies of pulmonary memory CD8+ T cells (K) and memory CD4+ T cells (L) following re-stimulation with peptide pools for S1 and TCEs. (M and N) Frequencies of lung CD8+ TRM cells (M) and CD4+ TRM cells (N) following re-stimulation with peptide pools for S1 and TCEs. Values of geometric mean titer (GMT) were displayed in (C and D). Data are represented as GMT ± SD (D) or mean ± SEM (B, C, E–N) and analyzed by one-way ANOVA with Tukey correction. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001.
GC B cells eventually polarize into long-lived plasma cells (LLPCs) or memory B cells (MBCs) to provide long-term immunity (32). LLPCs can continuously secrete high-affinity antibodies without pathogen re-stimulation. We found that both i.m. and i.n. immunization induced LLPCs production in bone marrow (BM), which was further enhanced by the combined boosting (Fig. 6E). Accordingly, RBD+ LLPCs were also augmented in combined prime-boost groups compared to prime-only groups (Fig. 6F). Only minimal RBD+ MBCs, IgG1+ RBD+ MBCs, and IgG2a/2b+ RBD+ MBCs were elicited by AdC68-vST-vtRBM vaccination in spleens (Fig. S3C).
Additionally, we found that NAb titers showed positive correlation with total and RBD+ LLPCs (Fig. S3D and E). The amount of total and RBD+ LLPCs was positively associated with total and RBD+ GC B cells, respectively (Fig. S3F and G). The absolute number of RBD+ MBCs was weakly correlated with RBD+ GC B cell counts, and there is no correlation between MBCs and GC B cells (Fig. S3H and I). Altogether, our results indicated that the GC B cells induced by AdC68-vST-vtRBM tend to differentiate into LLPCs eventually, which are responsible for maintenance of long-lasting humoral immunity.
In spleens, the i.m. instead of i.n. vaccination evoked potent S1- and TCEs-specific memory CD8+ T-cell responses that were characterized by IFN-γ production (Fig. 6G and H). SARS-CoV-2-specific memory CD4+ T cells were also triggered by i.m. vaccination, though to a much lesser degree than the CD8+ counterparts (Fig. 6I and J). In keeping with effector T-cell responses, the frequencies of secondary memory T cells remained constant or declined slightly in comparison to corresponding primary counterparts. Distinct from splenic memory T-cell responses, i.m. and i.n. priming had a limited effect on pulmonary T-cell activation, but the combined boosting significantly mobilized them (Fig. 6K and L). Further analysis revealed that the majority of IFN-γ-producing memory T cells were S1-specific but not TCEs-specific (Fig. S3J and K).
Lung TRM cells, which upregulate CD69 expression and reside within specific niches in lungs, are distinguished from splenic and pulmonary memory T cells and are crucial to limiting SARS-CoV-2 transmission (33). The i.n. but not i.m. vaccination induced robust lung CD8+ TRM cell responses to both S1 and TCEs pools, which remained steady after the combined boosting (Fig. 6M). ICS of CD8+ TRM cells showed that all regimens, particularly i.n. + i.m. regimen, induced enrichment of S1-specific IFN-γ+ CD8+ TRM cells (Fig. S3L). Unlike lung CD8+ TRM cells that express typical TRM cell markers (CD69+CD103+), lung CD4+ TRM cells are defined as CD69+ CD103+/− (34). We observed significant increases of SARS-COV-2-specific CD103− CD4+ TRM cells rather than CD103+ CD4+ TRM cells in two combined groups, particularly i.n. + i.m. group (Fig. 6N). Conversely, CD103+ CD4+ TRM cells rather than CD103− CD4+ TRM cells produced substantial IFN-γ upon S1 peptide pool stimulation in all vaccine groups (Fig. S3M).
Collectively, compared with i.n. vaccination, the i.m. vaccination induced comparable systemic humoral responses, along with superior systemic memory cellular responses but faint mucosal immunity. The combined boosting increased the breadth and magnitude of memory humoral responses as well as the diversity of memory T cells, resulting in better well-rounded memory immunity.
Intranasal immunization alone or in combination with intramuscular immunization provided enhanced protection against BA.2 in vivo
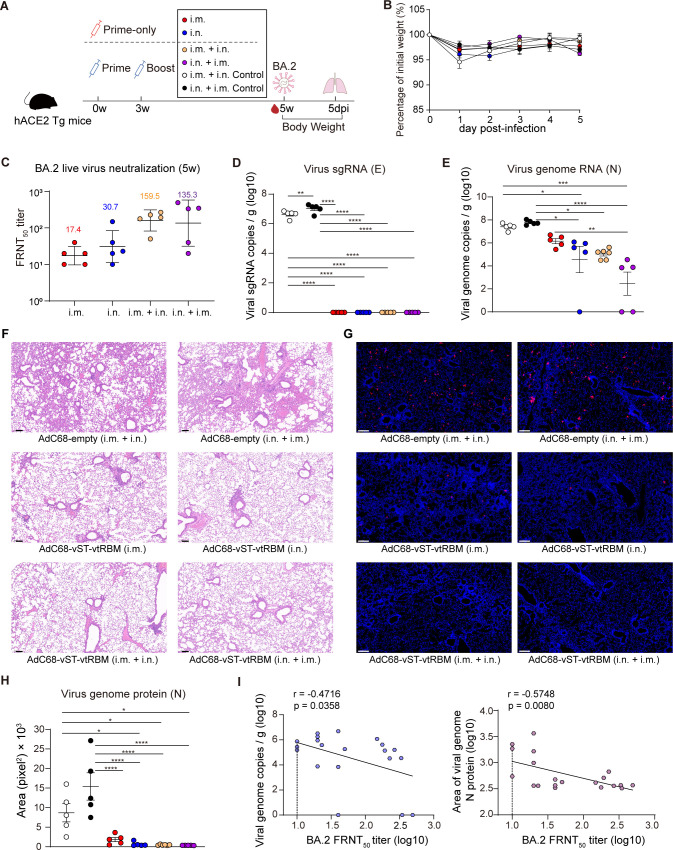
We evaluated the protection potency of AdC68-vST-vtRBM against Omicron BA.2, which can partially evade the neutralization activity of our vaccine and may increase the risk of reinfection. The immunization-challenge schedule is described in Fig. 7A. In agreement with the NAb responses in wild-type C57BL/6J mice, BA.2-NAbs were significantly increased after boosting in hACE2 transgenic mice (derived from the C57BL/6J strain) (Fig. 7C). More specifically, geometric mean FRNT50 titers in the i.m. + i.n. and i.n. + i.m. groups were 8.2- and 3.4-fold higher than corresponding prime-only group.
Fig 7.
Intranasal immunization alone or in combination with intramuscular immunization provided enhanced protection against BA.2 in vivo. (A) Scheme of vaccination and challenge. Human ACE2 transgenic (hACE2 Tg) mice (n = 5–6) were immunized with 2 × 107 IFU of AdC68-vST-vtRBM in prime-only regimens (i.m., i.n., respectively, at 0 weeks) or prime-boost regimens (i.m. + i.n., i.n. + i.m., respectively, with an interval of 3 weeks). Sham control mice were immunized with 2 × 107 IFU of AdC68-empty by i.m. + i.n. and i.n. + i.m. routes, respectively. At 2 weeks post-last vaccination, mice were challenged with 5 × 103 focus-forming units of BA.2 and monitored for clinical signs and weight loss. At 5 days post-infection, mice were euthanized, and lung tissues were collected. (B) Body weight change over 5 days post-infection. (C) Serum BA.2 neutralizing antibody titers at 5 wpv. (D and E) SARS-CoV-2 viral burden in lungs, measured by copies of viral sgRNA (D) and gRNA (E). (F) Hematoxylin and eosin staining for pathological examination, representative magnified images are shown. (G and H) Immunofluorescence staining for viral burden examination. Representative magnified images in (G). Immunofluorescence area of nucleocapsid protein of the complete sections was analyzed using ImageJ software in (H). (I) Spearman correlations of viral gRNA copies (left) or the immunofluorescence area of nucleocapsid protein (right) and BA.2 FRNT50 titers. Values of geometric mean titer (GMT) were displayed in (C). Data are represented as GMT ± SD (C) or mean ± SEM (B, D, E, H and I) and analyzed by one-way ANOVA with Tukey correction. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001. Dotted line represents L.O.D., limit of detection. Scale bars: 100 µm (F) and 200 µm (G).
In keeping with earlier report, there was no significant reduction in body weight over 5 days post-infection (dpi) (35) (Fig. 7B). Subgenomic RNA (SgRNA) loads in lungs ranged from 6.2 to 7.3 log10 copies/g in control groups, but were undetectable in all vaccine groups, indicating that BA.2 virus replication was entirely under control after vaccination (Fig. 7D). Except for the i.m. group, the copy numbers of genomic RNA (gRNA) were significantly reduced in all vaccine groups compared to control groups (Fig. 7E). According to histopathological analysis, negligible lesions were observed in lungs of vaccinated mice, regardless of the regimen (Fig. 7F). The AdC68-empty-immunized mice suffered moderate pulmonary damage at 5 dpi, including congestion, loss of alveolar cavities, and extensive viral antigen expression (Fig. 7G). Consistent with the gRNA quantification, measurement of the fluorescent area of nucleocapsid (N) protein suggested that i.m.-immunized mice underwent more severe viral invasion with 1.8-, 3.0-, and 4.1-fold higher N protein expression than i.n.-, i.m. + i.n.-, and i.n. + i.m.-immunized mice, respectively (Fig. 7H). BA.2-NAb titers showed a weak negative correlation with both gRNA loads and N protein levels in lungs (Fig. 7I).
These data demonstrate that our COVID-19 vaccine can effectively protect mice from BA.2 infection. The intranasal-inclusive regimens were superior to the i.m. regimen in protective efficacy.
DISCUSSION
Here, we developed an adenoviral vector-based broad-spectrum COVID-19 vaccine that can induce NAbs against SARS-CoV-2 and its variants. In our vaccine design, we added several key antigenic mutations targeting multiple variants to the full-length spike gene, which were then linked a cluster of the conserved T-cell epitopes of the SARS-CoV-2 virus. Moreover, the truncated RBM of spike protein was further displayed on adenovirus structural proteins fiber. Such a design has been demonstrated to induce broad-spectrum of cellular and humoral immune responses.
To date, multiple broad-spectrum COVID-19 vaccine candidates based on different platforms including protein and mRNA are in development. Zhao et al. generated a universal Span recombinant protein vaccine by phylogenetically calculating the most frequent mutations in 2675 spike sequences from the NCBI database and found that the Span vaccination of mice elicited broad NAbs against original strain, Beta, Delta, and Omicron (36). Glycosite-deleted spike mRNA or protein vaccine that could expose more conserved epitopes elicited stronger antibody with broader protection against the SARS-CoV-2 variants compared to the unmodified counterparts (37, 38). Wu et al. engineered a chimeric spike protein vaccine termed as STFK1628x, containing NTD from B.1.620 lineage, RBD-S2 from the Gamma variant, and additional RBD mutation patches from the Delta variant, that elicited high titers of broad-spectrum NAbs to protect hamsters from the ancestral SARS-CoV-2, Beta, and BA.1 challenge (39). Most of the broad-spectrum COVID-19 vaccine candidates exclusively target spike or RBD antigen. However, mutations in spike or RBD protein occur most frequently (40), which may enhance viral immune evasion and reduce the efficacy of vaccines. Combining conserved antigens with spike to induce both humoral and cellular immune responses may be a promising strategy that can further improve the broad spectrum of COVID-19 vaccine candidates.
The continued emergence of new variants of SARS-CoV-2 and the waning immunity overtime call for the development of not only broad-spectrum vaccines but also optimal vaccination strategies. Numerous studies have shown that the i.m. or i.m. + i.m. regimen can induce strong systemic humoral and cellular immune responses with the absence of mucosal immunity, whereas the i.n. or i.n. + i.n. regimen fills this gap but falls short of eliciting systemic cellular immunity (41 – 43). Recent studies have increasingly focused on combined regimens, but most of them are limited to heterogenous i.m. + i.n. regimen, such as DNA/vector or mRNA/protein or mRNA/vector regimen (44 – 46). Our findings are consistent with these reports, showing that the i.m. + i.n. regimen can induce both systemic and mucosal immunity. However, the immunokinetic profile of the i.n. + i.m. regimen remains largely unknown. Our study expanded the current understanding of prime-boost regimens, revealing that the i.n. + i.m. regimen also has the potential to induce systemic and mucosal immune responses. Comparing the two combined regimens, we found that the i.n. + i.m. regimen elicited superior TRM cell responses, while the i.m. + i.n. regimen exhibited immunodominance in systemic T-cell responses and antibody responses. Since no parallel comparison between the two combined prime-boost regimens was performed hitherto, the advantage of i.n. + i.m. regimen in triggering mucosal immunity has likely been underestimated.
Shin et al. have defined a “prime and pull” strategy, conventional parenteral vaccination to elicit systemic T-cell responses (prime), followed by intravaginal chemokine treatment (pull) to establish TRM cell barrier at the site of genital herpes simplex virus infection (47). Cuburu et al. ameliorated this pattern and performed an intramuscular-intravaginal regimen of the HPV vaccine. They obtained similar results that the topical boosting could efficiently attract circulating CD8+ T cells into the vaccination site and induced in situ proliferation and differentiation of cognate CD8+ TRM cells (48). Recently, Mao et al. assessed a vaccination strategy that utilizes i.m. priming with mRNA-LNP followed by i.n. boosting with spike proteins, which also elicits robust mucosal humoral and cellular immunity against SARS-CoV-2 infection (45). Theoretically, our homologous i.m. + i.n. regimen is also a prime-pull-amplify approach for the SARS-CoV-2 vaccine and has indeed been proven to evoke mucosal immunity. In terms of the i.n. + i.m. regimen, Khanna et al. showed that heterogenous vector/vector vaccination generated CD4+ T cell-mediated mucosal and systemic immunity, which was related to partial protection against high-dose SIV challenge (49). The mechanisms underlying the differences in immune responses between combined regimens remain to be investigated.
GCs, where antigen-activated B cells undergo somatic hypermutation and positive selection of high-affinity B cell receptor variants, are the foundation for high-quality long-term B cell responses induction (50). Recent studies showed that the mRNA- and protein-based COVID-19 vaccines evoke robust SARS-CoV-2-specific GC responses, which are closely associated with antibody responses (29, 51). For viral vector vaccines, the immunogenicity of the vector per se inevitably has an impact on that of the antigen, which further complicates the adaptive immune responses upon vaccination. Our vaccine was confirmed to promote the generation of SARS-CoV-2-specific GC B cells as well. Further analyses extended this finding by clarifying that secondary GC responses contribute more to antibody response enhancement than primary GC responses, which may be partly explained by the following novel discovery: B cells without primary GC experience overwhelmingly comprise secondary GCs, where redifferentiation of mutant MBCs rarely occurs, meaning that secondary GC responses are characterized by a clonality bottleneck that limits the participation of diverse MBCs induced by priming (52).
In terms of adenovirus vector-based vaccine, pre-existing immunity against adenovirus, including NAbs and specific T cells, dampens the efficacy of adenovirus vectors (53). In our study, we chose the chimpanzee-derived vector AdC68 due to its low seroprevalence in the human population (54, 55). For homologous prime-boost regimens, pre-existing anti-vector immunity provoked by priming may affect the induction of immune responses by boosting. We found that the i.m. + i.m. regimen induced lower NAbs against SARS-CoV-2 but substantially higher vector-NAbs compared with i.m. regimen (Fig. 3C and F), showing the anti-vector pre-existing immunity had inhibited the further enhancement of B-cell responses post-boosting. In addition, the i.m. + i.n. regimen distinctly reduced SARS-CoV-2-specific cytokine production in spleens compared to i.m. regimen (Fig. 5G and 6G through J), indicating that the pre-existing anti-vector immunity also resulted in the suppression of T-cell response elevation. Such inhibition was not observed in lungs, where the cellular response was instead enhanced, which probably due to the absence of vector-specific lung TRM cells (56). Alternating the immunization route could somewhat circumvent pre-existing immunity to adenovirus (57), which may contribute to the stronger immune response induced by the combined prime-boost regimens. In conclusion, taking into account the impact of anti-vector pre-existing immunity will help to further improve viral vector applications.
With intranasal-inclusive regimens, our vaccine conferred stronger protection against BA.2 than the i.m. regimen. BA.2-NAb titers showed a weak negative correlation with protective efficacy (Fig. 7I), suggesting that NAb is probably not an exclusive contributor to viral clearance. Compared to the i.m. regimen, intranasal-inclusive regimens induced additional mucosal immunity including lgA and TRM cell responses, which may also be crucial in protective immunity. Their role in combating SARS-CoV-2 has been supported by recent studies (33, 58, 59). It is undeniable that the induction of systemic T-cell responses by the i.m. regimen also contributes to anti-viral protection, but the deficiency of mucosal immunity may lead to limited viral clearance. In addition, there are growing reports supporting the role of Fc effector functions in the efficacy of vaccines and monoclonal antibodies, especially when the neutralizing activity is waning (60, 61). The Fc effector function may also play a role in the protective immunity induced by our vaccines and remains to be examined.
Immunophenotypic identification of the two combined regimens has advanced our understanding of how to effectively orientate the immune response, which can provide a reference for clinical practice. For individuals who have received aerosolized vaccines or have a history of exposure to SARS-CoV-2 (analogous to intranasal inoculation), the i.m. booster may be a promising regimen. Indeed, several reports have demonstrated that COVID-19 convalescents only required a single vaccine dose to achieve more robust humoral immune responses than SARS-CoV-2-naïve individuals who received two doses of i.m. vaccine (62 – 64). As for those who have been vaccinated intramuscularly, the i.n. booster may be a better option.
In summary, our preclinical study lays the groundwork for the further development of broad-spectrum COVID-19 vaccines. The combined vaccination strategies that can induce potent systemic and mucosal immunity simultaneously may represent promising strategies for maximizing the protective efficacy of respiratory virus vaccines.
MATERIALS AND METHODS
Virus and cell lines
HEK-293, 293T, Huh-7 cells, A549, and VeroE6 cells were all cultured in complete Dulbecco’s modified Eagle medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin/streptomycin (NCM Biotech). The cells were maintained at 37°C with 5% CO2 and passaged when confluent. SARS-CoV-2 wild-type SH01 strain (GenBank accession no. MT121215), Delta, and BA.2 were all obtained from the Chinese Center for Disease Control and Prevention following isolation from patients in Shanghai, China. All experiments involving SARS-CoV-2 were performed in the BSL-3 facility of Fudan University according to International Standard Operating Procedures.
Mouse
Age-matched 6- to 8-week-old female C57BL/6J mice and hACE2 transgenic mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China) and Shanghai Model Organisms Center Inc. (Shanghai, China), respectively. C57BL/6J mice studies were conducted under the approval of the Institutional Animal Care and Use Committee of Tianjin Medical University (Tianjin, China). The hACE2 transgenic mice studies were conducted under the approval of the Institutional Animal Care and Use Committee of Fudan University (Shanghai, China). All experiments were performed following the institutional guidelines from the Animal Research and Ethics Board.
Vaccine construction
The full-length spike gene of the Wuhan-Hu-1 strain (YP 009724390.1) was modified by introducing amino acid substitutions (K417N, E484K, N501Y, D614G, P681H, R682S, R685G, K986P, and V987P) to obtain the vS sequence. The vS gene and tandem conserved TCEs from ORF1, ORF3, and M proteins of ancestral SARS-CoV-2 were codon-optimized for human expression and synthesized commercially (Tsingke Biotechnology, Beijing, China). The peptide sequences of TCEs are shown in Table S1. Separated by P2A linker, vS and TCEs gene were incorporated into an open reading frame, which was driven by a cytomegalovirus (CMV) promoter and finally cloned onto E1 region of AdC68-empty using Gibson assembly. Moreover, L452R mutation was introduced into the truncated receptor-binding domain (vtRBM) of spike to produce the vtRBM sequence, which was placed in the HI loop of fiber to produce a fusion sequence.
Western blot
HEK293 cells were transduced with 108, 109, and 1010 vp of AdC68-vST-vtRBM. AdC68-empty-transduced (1010 vp) HEK293 cells and untransduced HEK293 cells were used as controls. Twenty-four hours post-infection, the cells were harvested and split into two fractions. One fraction was boiled at 100°C with 1× reductive loading buffer for 20 min, and the other fraction was ultrasonicated on ice for 1 min and mixed with 1× non-reductive loading buffer. A Western blot was performed against the samples using anti-SARS-CoV-2 RBD antibody (Sino Biological), followed by HRP-conjugated goat anti-rabbit IgG antibody (Abcam). Alpha-tubulin and β-actin were selected as internal control, respectively (Proteintech).
Antibody ELISAs
Spike-specific IgG and IgA antibody levels in mouse serum and BALF were measured by ELISA, as previously described (65). Briefly, ELISA plates were coated overnight at 4°C with ancestral spike protein (Sino Biological). For lgG detection, serum samples were threefold serially diluted with 1:400 starting dilutions, and BALF samples were twofold serially diluted with 1:5 starting dilutions. For IgA detection, BALF samples were twofold serially diluted with an undiluted stock solution as starting dilutions. Following 1-h incubation at 37°C, secondary antibodies were added: HRP-conjugated goat anti-mouse IgG antibody (ab6789, 1:100,000) (Abcam); HRP-conjugated goat anti-mouse IgA (1:5,000) (Southern Biotech). Plates were again incubated for 1 h at 37°C, followed by the addition of TMB substrate (NCM Biotech). Sulfuric acid (2 M H2SO4) solution was used to stop the reaction. Optical density (OD)450–630 was recorded using a microplate reader (Tecan). The binding antibody endpoint titer was determined as the reciprocal of the highest serum dilution that yielded an absorbance greater or equal to 0.1 OD unit above the absorbance of the pre-immune samples.
Production of SARS-CoV-2 pseudoviruses
The backbone plasmid pNL4-3.Luc.R-E and pCAGGS-S-CΔ19 expressing spike of the original strain or its variants were cotransfected into HEK 293T cells by polyethyleneimine (Polysciences). The spike amino acid sequences of original strain and its variants were based on GISAID EPI_ISL_402124 (Wuhan/WIV04/2019), EPI_ISL_712096 (B.1.351variant), EPI_ISL_906075 (P.1variant), EPI_ISL_3023383 (C.37 variant), EPI_ISL_ 2029113 (B.1.617.2 variant), or EPI_ISL_6640916 (BA.1 variant). The supernatant was collected 48 h post-transfection, filtered through 0.45 µm filters, and concentrated overnight with PEG8000 at 4°C.
Pseudovirus neutralization test
To determine the neutralization activity of mouse serum, Huh7 cells were seeded in 96-well plates at a density of 2.5 × 104 cells per well overnight. The heat-inactivated sera were threefold serially diluted with 1:20 starting dilutions and mixed with an equal volume of pseudoviruses. The mixture was incubated at 37°C for 1 h before adding to Huh7 cells. Relative luciferase activity (RLA) was measured using the SteadyGlo Luciferase Assay System (Promega) 48 h later. The 50% pseudovirus neutralization (pVNT50) titer was calculated as the reciprocal of the greatest serum dilution at which RLA was reduced by 50% compared with RLA in virus control wells that were infected with pseudovirus in the absence of mouse serum.
Adenovirus neutralization assay
The adenovirus neutralization assay was performed as previously described (66). An optimized method was performed utilizing AdC68-Luc instead of AdC68-eGFP. Briefly, 5 × 104 cells/well A549 were seeded in 96-well overnight. Sera were threefold serially diluted with 1:20 starting dilutions and mixed with an equal volume of 1 × 107 vp AdC68-Luc. The mixture was incubated at 37°C for 2 h before adding to A549 cells. RLA was measured 48 h later. The 50% adenovirus neutralization (NT50) titer was calculated as the serum dilution at which RLA was reduced by 50% compared with RLA in virus control wells.
Focus reduction neutralization test
Serum samples were threefold serially diluted with an initial dilution of 1:20 and mixed with 0.1 multiplicity of infection (MOI) original strain, 0.002 MOI Delta, or 0.015 MOI BA.2. The mixture was incubated at 37°C for 1 h before transferring to 96-well plates with 4 × 104 VeroE6 cells per well. Plates were incubated at 37°C, 5% CO2 for 48 h, and fixed with 4% paraformaldehyde (PFA) in PBS for 20 min. Plates were washed and incubated with rabbit anti-N antibody (ABclonal), followed by peroxidase-conjugated anti-rabbit IgG (Proteintech) and peroxidase substrate incubation (SeraCare). Virus-infected cell foci were counted on an ImmunoSpot microanalyzer (Cellular Technologies). The 50% focus reduction neutralization (FRNT50) titer was measured as the reciprocal of the greatest serum dilution at which foci were reduced by 50% relative to control wells that were infected with the live virus in the absence of mouse serum.
Immunophenotyping by flow cytometry
All staining steps were performed at 4°C in MACS buffer (DPBS with 0.5% BSA and 50 mM EDTA-2Na). Single-cell suspensions were blocked with anti-CD16/CD32 monoclonal antibody (Thermo Fisher Scientific) and stained with LIVE/DEAD Stain Kit (Invitrogen).
Germinal center B cells
LN cells were stained with eFlour450-IgD, FITC-GL7, PerCP/Cyanine5.5-CD19, PE-Cy7-CD95 (Fas), and Alexa Fluor 647 conjugated RBD for 1 h. The gating strategy is shown in Fig. S2D.
Memory B cells
Splenocytes were stained with BUV395-CD38, eFlour450-IgD, BV605-IgG2a/IgG2b, FITC-GL7, PerCP/Cyanine5.5-CD19, PE-CD138, PE/Dazzle 594-CD45R/B220, PE/Cyanine7- IgG1, APC/Cyanine7-CD3ε, APC/Cyanine7-Ter119, and Alexa Fluor 647 conjugated RBD for 1 h. The gating strategy is shown in Fig. S4A.
Long-lived plasma cells
Bone marrow cells were stained with eFlour450-IgD, PE-CD138, PE/Dazzle 594-CD45R/B220, APC/Cyanine7-CD3ε, and APC/Cyanine7-Ter119 for 1 h. The gating strategy is shown in Fig. S4B.
Excess antibodies were washed away with MACS buffer after staining. All samples were fixed with 2% PFA for 30 min before acquisition, then acquired on a BD LSRFortessa (BD Biosciences) and analyzed using FlowJo v10.
Intracellular cytokine stain assay
Splenocytes or homogenized lung cells were stimulated for 12 h with two pools of S1or TCEs (2 µg/mL) in the presence of GolgiPlug (BD Biosciences). The S1 peptide pool contained 15-mers overlapping by 11 amino acids derived from ancestral strain. The TCEs peptide pool contained 25 epitope peptides in Table S1. After stimulation, the cells were washed with cold MACS buffer and kept at 4°C for the entire staining process. Single-cell suspensions were blocked with anti-CD16/CD32 monoclonal antibody and stained with LIVE/DEAD Stain Kit.
Effector T-cell responses
The cells were stained with PerCP/Cyanine5.5-CD3ε, FITC-CD4, Alexa Fluor 700-CD8a for 30 min and permeabilized with fixation/permeabilization solution (BD Biosciences). Samples were then intracellularly stained for 30 min with cytokine Abs cocktail. For splenocytes, APC-IFN-γ, Brilliant Violet 605-TNF-α, APC-IL-5, and Brilliant Violet 605-IL-4 were used. For lung cells, APC-IFN-γ, Brilliant Violet 605-TNF-α, PE-IL-13, and Brilliant Violet 421-IL-4 were added.
Memory T-cell responses
Splenocytes were stained with PerCP/Cyanine5.5-CD3ε, FITC-CD4, Alexa Fluor 700-CD8a, PE-eflour610-CD44, and eFlour450-CD62L for 1 h. After permeabilizing, the cells were stained with APC-IFN-γ and Brilliant Violet 605-TNF-α cocktail or APC-IL-5 and Brilliant Violet 605-IL-4 cocktail for 30 min. Lung cells were stained with PerCP/Cyanine5.5-CD3ε, APC-Cyanine7-CD4, Alexa Fluor 700-CD8a, FITC-CD44, eFlour450-CD62L, Brilliant Violet 605-CD103, and PE-eFluor 610-CD69 for 1 h, followed by APC-IFN-γ staining for 30 min after permeabilizing. The gating strategy is shown in Fig. S5.
All samples were fixed with 2% PFA for 30 min before the acquisition, then acquired on a BD LSRFortessa (BD Biosciences) and analyzed using FlowJo v10.
SARS-CoV-2 viral burden determination
Half a fraction of mouse lung tissues were weighed and homogenized. Virus RNA was isolated and Reverse transcription PCR (RT-PCR) assays were performed using HiScript II One Step qRT-PCR Probe Kit (Vazyme). Two sets of primers were used to detect the N gene of the viral genome and the E gene of sgRNA utilizing real-time PCR.
N qPCR Forward: 5′-GACCCCAAAATCAGCGAAAT-3′;
N qPCR Reverse: 5′-CTGGTTACTGCCAGTTGAATCTG-3′;
N-probe: FAM-ACCCCGCATTACGTTTGGTGGACC-TAMRA;
E qPCR Forward: 5′-CGATCTCTTGTAGATCTGTTCTC-3′;
E qPCR Reverse: 5′-ATATTGCAGCAGTACGCACACA-3′;
E-probe: FAM-ACACTAGCCATCCTTACTGCGCTTCG-TAMRA.
Viral loads were expressed on a log10 scale as viral copies per gram after being calculated with a standard curve.
Hematoxylin-eosin/immunofluorescence stain
Hematoxylin-eosin (HE)/immunofluorescence staining was performed for another half fraction of lung tissues. Tissues were fixed in 4% paraformaldehyde, dehydrated, embedded in paraffin, and sectioned. Sections were deparaffinized in xylene and stained with HE for pathological examination. For immunofluorescence staining, after deparaffinization, peroxidase blocking, and antigen retrieval, sections were covered with rabbit anti-SARS-CoV-2 nucleocapsid antibody (1:1,000) (Sino Biological) overnight at 4°C, followed by Cy3-conjugated goat anti-rabbit IgG antibody (1:300) (Servicebio) incubation at room temperature for 50 min in the dark. DAPI (Servicebio) was used for nuclear counterstaining at room temperature for 10 min. Finally, sections were mounted using an anti-fade mounting medium (Servicebio) and scanned using Pannoramic MIDI (3DHISTECH). Images were processed by CaseViewer software (3DHISTECH). To quantify the abundance of Cy3 labeled nucleocapsid protein, the immunofluorescence area of the complete sections was analyzed using the ImageJ software.
Statistical analysis
All data are presented as means ± SEM unless otherwise stated, asterisks in the figures indicate the level of statistical significance (*P < 0.05, **P < 0.01, ***, ### P < 0.001, and ****P < 0.0001). Groups were compared via one-way ANOVA with Tukey’s multiple comparisons test or Kruskal-Wallis test with Dunn’s multiple comparisons test according to distribution of data. Analysis of binding IgG and adenoviral vector neutralization over time were performed using two-way ANOVA with Sidak’s post-test for multiple comparisons. Correlations were determined using Spearman’s rank coefficient with a 95% CI. All tests were performed using GraphPad Prism 8 software.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (32070926) and the Key Project of the Natural Science Foundation of Tianjin (20JCZDJC00090) to D.Z. and supported by the National Key Research and Development Program (2021YFA1300803) and Shanghai Municipal Science and Technology Major Project (ZD2021CY001) to Q.C.
D.Z., Q.C., J.X., and M.X. conceived the project and designed the experiments. M.X., Y.W., X.W., J.L., G.H., F.H., W.D., Q.Z., Y.L., L.S., Y.W., S.D., C.P., and Z.H. performed the experiments. D.Z., Q.C., J.X., M.X., and X.Z. analyzed the data. M.X. and Y.W. wrote the manuscript. D.Z. revised the manuscript.
The authors declare no competing interests.
Contributor Information
Jianqing Xu, Email: xujianqing@fudan.edu.cn.
Qiliang Cai, Email: qiliang@fudan.edu.cn.
Dongming Zhou, Email: zhoudongming@tmu.edu.cn.
Tom Gallagher, Loyola University Chicago, Maywood, Illinois, USA .
DATA AVAILABILITY
All data from this study are included within this manuscript/Supplementary Material and are available from the Lead Contact (Dongming Zhou, zhoudongming@tmu.edu.cn) upon request.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.00724-23.
Supplemental Information includes five figures and one table.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Jian F, Yu Y, Song W, Yisimayi A, Yu L, Gao Y, Zhang N, Wang Y, Shao F, Hao X, Xu Y, Jin R, Wang Y, Xie XS, Cao Y. 2022. Further humoral immunity evasion of emerging SARS-CoV-2 BA.4 and BA.5 subvariants. Lancet Infect Dis 22:1535–1537. doi: 10.1016/S1473-3099(22)00642-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nasreen S, Chung H, He S, Brown KA, Gubbay JB, Buchan SA, Fell DB, Austin PC, Schwartz KL, Sundaram ME, Calzavara A, Chen B, Tadrous M, Wilson K, Wilson SE, Kwong JC, Canadian Immunization Research Network (CIRN) Provincial Collaborative Network (PCN) Investigators . 2022. Effectiveness of COVID-19 vaccines against symptomatic SARS-CoV-2 infection and severe outcomes with variants of concern in Ontario. Nat Microbiol 7:379–385. doi: 10.1038/s41564-021-01053-0 [DOI] [PubMed] [Google Scholar]
- 3. Khoury DS, Cromer D, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, Subbarao K, Kent SJ, Triccas JA, Davenport MP. 2021. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med 27:1205–1211. doi: 10.1038/s41591-021-01377-8 [DOI] [PubMed] [Google Scholar]
- 4. Tan AT, Linster M, Tan CW, Le Bert N, Chia WN, Kunasegaran K, Zhuang Y, Tham CYL, Chia A, Smith GJD, Young B, Kalimuddin S, Low JGH, Lye D, Wang L-F, Bertoletti A. 2021. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep 34:108728. doi: 10.1016/j.celrep.2021.108728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tarke A, Coelho CH, Zhang Z, Dan JM, Yu ED, Methot N, Bloom NI, Goodwin B, Phillips E, Mallal S, Sidney J, Filaci G, Weiskopf D, da Silva Antunes R, Crotty S, Grifoni A, Sette A. 2022. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from alpha to Omicron. Cell 185:847–859. doi: 10.1016/j.cell.2022.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Naranbhai V, Nathan A, Kaseke C, Berrios C, Khatri A, Choi S, Getz MA, Tano-Menka R, Ofoman O, Gayton A, Senjobe F, Zhao Z, St Denis KJ, Lam EC, Carrington M, Garcia-Beltran WF, Balazs AB, Walker BD, Iafrate AJ, Gaiha GD. 2022. T cell reactivity to the SARS-CoV-2 Omicron variant is preserved in most but not all individuals. Cell 185:1259. doi: 10.1016/j.cell.2022.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuhlmann C, Mayer CK, Claassen M, Maponga T, Burgers WA, Keeton R, Riou C, Sutherland AD, Suliman T, Shaw ML, Preiser W. 2022. Breakthrough infections with SARS-CoV-2 Omicron despite mRNA vaccine booster dose. Lancet 399:625–626. doi: 10.1016/S0140-6736(22)00090-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Azzi L, Dalla Gasperina D, Veronesi G, Shallak M, Ietto G, Iovino D, Baj A, Gianfagna F, Maurino V, Focosi D, Maggi F, Ferrario MM, Dentali F, Carcano G, Tagliabue A, Maffioli LS, Accolla RS, Forlani G. 2022. Mucosal immune response in BNT162b2 COVID-19 vaccine recipients. EBioMedicine 75:103788. doi: 10.1016/j.ebiom.2021.103788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tatsis N, Ertl HCJ. 2004. Adenoviruses as vaccine vectors. Mol Ther 10:616–629. doi: 10.1016/j.ymthe.2004.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Falsey AR, Sobieszczyk ME, Hirsch I, Sproule S, Robb ML, Corey L, Neuzil KM, Hahn W, Hunt J, Mulligan MJ, McEvoy C, DeJesus E, Hassman M, Little SJ, Pahud BA, Durbin A, Pickrell P, Daar ES, Bush L, Solis J, Carr QO, Oyedele T, Buchbinder S, Cowden J, Vargas SL, Guerreros Benavides A, Call R, Keefer MC, Kirkpatrick BD, Pullman J, Tong T, Brewinski Isaacs M, Benkeser D, Janes HE, Nason MC, Green JA, Kelly EJ, Maaske J, Mueller N, Shoemaker K, Takas T, Marshall RP, Pangalos MN, Villafana T, Gonzalez-Lopez A. 2021. Phase 3 safety and efficacy of AZD1222 (ChAdOx1 nCoV-19) COVID-19 vaccine. N Engl J Med 385:2348–2360. doi: 10.1056/NEJMoa2105290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Halperin SA, Ye L, MacKinnon-Cameron D, Smith B, Cahn PE, Ruiz-Palacios GM, Ikram A, Lanas F, Lourdes Guerrero M, Muñoz Navarro SR, Sued O, Lioznov DA, Dzutseva V, Parveen G, Zhu F, Leppan L, Langley JM, Barreto L, Gou J, Zhu T, CanSino COVID-19 Global Efficacy Study Group . 2022. Final efficacy analysis, interim safety analysis, and immunogenicity of a single dose of recombinant novel coronavirus vaccine (adenovirus type 5 vector) in adults 18 years and older: an international, multicentre, randomised, double-blinded, placebo-controlled phase 3 trial. Lancet 399:237–248. doi: 10.1016/S0140-6736(21)02753-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Logunov DY, Dolzhikova IV, Shcheblyakov DV. 2021. Data discrepancies and substandard reporting of interim data of Sputnik V phase 3 trial REPLY. Lancet 397:1883–1884. doi: 10.1016/S0140-6736(21)00894-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tukhvatulin AI, Gordeychuk IV, Dolzhikova IV, Dzharullaeva AS, Krasina ME, Bayurova EO, Grousova DM, Kovyrshina AV, Kondrashova AS, Avdoshina DV, Gulyaev SA, Gulyaeva TV, Moroz AV, Illarionova VV, Zorkov ID, Iliukhina AA, Shelkov AY, Botikov AG, Erokhova AS, Shcheblyakov DV, Esmagambetov IB, Zubkova OV, Tokarskaya EA, Savina DM, Vereveyko YR, Ungur AS, Naroditsky BS, Ishmukhametov AA, Logunov DY, Gintsburg AL. 2022. Immunogenicity and protectivity of intranasally delivered vector-based heterologous prime-boost COVID-19 vaccine Sputnik V in mice and non-human primates. Emerg Microbes Infect 11:2229–2247. doi: 10.1080/22221751.2022.2119169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li JX, Wu SP, Guo XL, Tang R, Huang BY, Chen XQ, Chen Y, Hou LH, Liu JX, Zhong J, Pan HX, Shi FJ, Xu XY, Li ZP, Zhang XY, Cui LB, Tan WJ, Chen W, Zhu FC, Grp C-S. 2022. Safety and immunogenicity of heterologous boost immunisation with an orally administered aerosolised Ad5-nCoV after two-dose priming with an Inactivated SARS-CoV-2 vaccine in Chinese adults: a randomised, open-label, single-centre trial. Lancet Respir Med 10:739–748. doi: 10.1016/S2213-2600(22)00087-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou B, Thao TTN, Hoffmann D, Taddeo A, Ebert N, Labroussaa F, Pohlmann A, King J, Steiner S, Kelly JN, Portmann J, Halwe NJ, Ulrich L, Trüeb BS, Fan X, Hoffmann B, Wang L, Thomann L, Lin X, Stalder H, Pozzi B, de Brot S, Jiang N, Cui D, Hossain J, Wilson MM, Keller MW, Stark TJ, Barnes JR, Dijkman R, Jores J, Benarafa C, Wentworth DE, Thiel V, Beer M. 2021. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 592:122–127. doi: 10.1038/s41586-021-03361-1 [DOI] [PubMed] [Google Scholar]
- 16. Zhao S, Lou J, Cao L, Zheng H, Chong MKC, Chen Z, Chan RWY, Zee BCY, Chan PKS, Wang MH. 2021. Quantifying the transmission advantage associated with N501Y substitution of SARS-CoV-2 in the UK: an early data-driven analysis. J Travel Med 28:taab011. doi: 10.1093/jtm/taab011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wrobel AG, Benton DJ, Roustan C, Borg A, Hussain S, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ. 2022. Evolution of the SARS-CoV-2 spike protein in the human host. Nat Commun 13:1178. doi: 10.1038/s41467-022-28768-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kuzmina A, Khalaila Y, Voloshin O, Keren-Naus A, Boehm-Cohen L, Raviv Y, Shemer-Avni Y, Rosenberg E, Taube R. 2021. SARS-CoV-2 spike variants exhibit differential infectivity and neutralization resistance to convalescent or post-vaccination sera. Cell Host Microbe 29:522–528. doi: 10.1016/j.chom.2021.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Widera M, Wilhelm A, Hoehl S, Pallas C, Kohmer N, Wolf T, Rabenau HF, Corman VM, Drosten C, Vehreschild M, Goetsch U, Gottschalk R, Ciesek S. 2021. Limited neutralization of authentic severe acute respiratory syndrome coronavirus 2 variants carrying E484K in vitro. J Infect Dis 224:1109–1114. doi: 10.1093/infdis/jiab355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Noh JY, Jeong HW, Shin E-C. 2021. SARS-CoV-2 mutations, vaccines, and immunity: implication of variants of concern. Signal Transduct Target Ther 6:203. doi: 10.1038/s41392-021-00623-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, Graham BS, McLellan JS. 2020. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367:1260–1263. doi: 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bos R, Rutten L, van der Lubbe JEM, Bakkers MJG, Hardenberg G, Wegmann F, Zuijdgeest D, de Wilde AH, Koornneef A, Verwilligen A, van Manen D, Kwaks T, Vogels R, Dalebout TJ, Myeni SK, Kikkert M, Snijder EJ, Li Z, Barouch DH, Vellinga J, Langedijk JPM, Zahn RC, Custers J, Schuitemaker H. 2020. Ad26 vector-based COVID-19 vaccine encoding a prefusion-stabilized SARS-CoV-2 spike immunogen induces potent humoral and cellular immune responses. NPJ Vaccines 5:91. doi: 10.1038/s41541-020-00243-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saini SK, Hersby DS, Tamhane T, Povlsen HR, Amaya Hernandez SP, Nielsen M, Gang AO, Hadrup SR. 2021. SARS-CoV-2 genome-wide T cell epitope mapping reveals immunodominance and substantial CD8+ T cell activation in COVID-19 patients. Sci Immunol 6:eabf7550. doi: 10.1126/sciimmunol.abf7550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nathan A, Rossin EJ, Kaseke C, Park RJ, Khatri A, Koundakjian D, Urbach JM, Singh NK, Bashirova A, Tano-Menka R, Senjobe F, Waring MT, Piechocka-Trocha A, Garcia-Beltran WF, Iafrate AJ, Naranbhai V, Carrington M, Walker BD, Gaiha GD. 2021. Structure-guided T cell vaccine design for SARS-CoV-2 variants and sarbecoviruses. Cell 184:4401–4413. doi: 10.1016/j.cell.2021.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nelde A, Bilich T, Heitmann JS, Maringer Y, Salih HR, Roerden M, Lübke M, Bauer J, Rieth J, Wacker M, Peter A, Hörber S, Traenkle B, Kaiser PD, Rothbauer U, Becker M, Junker D, Krause G, Strengert M, Schneiderhan-Marra N, Templin MF, Joos TO, Kowalewski DJ, Stos-Zweifel V, Fehr M, Rabsteyn A, Mirakaj V, Karbach J, Jäger E, Graf M, Gruber L-C, Rachfalski D, Preuß B, Hagelstein I, Märklin M, Bakchoul T, Gouttefangeas C, Kohlbacher O, Klein R, Stevanović S, Rammensee H-G, Walz JS. 2021. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat Immunol 22:74–85. doi: 10.1038/s41590-020-00808-x [DOI] [PubMed] [Google Scholar]
- 26. Tang X, Yang Y, Xia X, Zhang C, Yang X, Song Y, Dai X, Wang M, Zhou D. 2017. Recombinant adenoviruses displaying matrix 2 ectodomain epitopes on their fiber proteins as universal influenza vaccines. J Virol 91:e02462-16. doi: 10.1128/JVI.02462-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sharma A, Krause A, Xu Y, Sung B, Wu W, Worgall S, Wu M. 2013. Adenovirus-based vaccine with epitopes incorporated in novel fiber sites to induce protective immunity against Pseudomonas aeruginosa. PLoS One 8:e56996. doi: 10.1371/journal.pone.0056996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tchesnokova V, Kulasekara H, Larson L, Bowers V, Rechkina E, Kisiela D, Sledneva Y, Choudhury D, Maslova I, Deng K, Kutumbaka K, Geng H, Fowler C, Greene D, Ralston J, Samadpour M, Sokurenko E. 2021. Acquisition of the L452R mutation in the ACE2-binding interface of spike protein triggers recent massive expansion of SARS-CoV-2 variants. J Clin Microbiol 59:e0092121. doi: 10.1128/JCM.00921-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lederer K, Castaño D, Gómez Atria D, Oguin TH, Wang S, Manzoni TB, Muramatsu H, Hogan MJ, Amanat F, Cherubin P, Lundgreen KA, Tam YK, Fan SHY, Eisenlohr LC, Maillard I, Weissman D, Bates P, Krammer F, Sempowski GD, Pardi N, Locci M. 2020. SARS-CoV-2 mRNA vaccines foster potent antigen-specific germinal center responses associated with neutralizing antibody generation. Immunity 53:1281–1295. doi: 10.1016/j.immuni.2020.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang C, Hart M, Chui C, Ajuogu A, Brian IJ, de Cassan SC, Borrow P, Draper SJ, Douglas AD. 2016. Germinal center B cell and T follicular helper cell responses to viral vector and protein-in-adjuvant vaccines. The Journal of Immunology 197:1242–1251. doi: 10.4049/jimmunol.1502472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kumamoto Y, Hirai T, Wong PW, Kaplan DH, Iwasaki A. 2016. CD301B+ dendritic cells suppress T follicular helper cells and antibody responses to protein antigens. Elife 5:e17979. doi: 10.7554/eLife.17979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Quast I, Tarlinton D. 2021. B cell memory: understanding COVID-19. Immunity 54:205–210. doi: 10.1016/j.immuni.2021.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grau-Expósito J, Sánchez-Gaona N, Massana N, Suppi M, Astorga-Gamaza A, Perea D, Rosado J, Falcó A, Kirkegaard C, Torrella A, Planas B, Navarro J, Suanzes P, Álvarez-Sierra D, Ayora A, Sansano I, Esperalba J, Andrés C, Antón A, Ramón Y Cajal S, Almirante B, Pujol-Borrell R, Falcó V, Burgos J, Buzón MJ, Genescà M. 2021. Peripheral and lung resident memory T cell responses against SARS-CoV-2. Nat Commun 12:3010. doi: 10.1038/s41467-021-23333-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mueller SN, Mackay LK. 2016. Tissue-resident memory T cells: local specialists in immune defence. Nat Rev Immunol 16:79–89. doi: 10.1038/nri.2015.3 [DOI] [PubMed] [Google Scholar]
- 35. Halfmann PJ, Iida S, Iwatsuki-Horimoto K, Maemura T, Kiso M, Scheaffer SM, Darling TL, Joshi A, Loeber S, Singh G, Foster SL, Ying B, Case JB, Chong Z, Whitener B, Moliva J, Floyd K, Ujie M, Nakajima N, Ito M, Wright R, Uraki R, Warang P, Gagne M, Li R, Sakai-Tagawa Y, Liu Y, Larson D, Osorio JE, Hernandez-Ortiz JP, Henry AR, Ciuoderis K, Florek KR, Patel M, Odle A, Wong L-Y, Bateman AC, Wang Z, Edara V-V, Chong Z, Franks J, Jeevan T, Fabrizio T, DeBeauchamp J, Kercher L, Seiler P, Gonzalez-Reiche AS, Sordillo EM, Chang LA, van Bakel H, Simon V, Consortium Mount Sinai Pathogen Surveillance (PSP) study group, Douek DC, Sullivan NJ, Thackray LB, Ueki H, Yamayoshi S, Imai M, Perlman S, Webby RJ, Seder RA, Suthar MS, García-Sastre A, Schotsaert M, Suzuki T, Boon ACM, Diamond MS, Kawaoka Y. 2022. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature 603:687–692. doi: 10.1038/s41586-022-04441-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao Y, Ni W, Liang S, Dong L, Xiang M, Cai Z, Niu D, Zhang Q, Wang D, Zheng Y, Zhang Z, Zhou D, Guo W, Pan Y, Wu X, Yang Y, Jing Z, Jiang Y, Chen Y, Yan H, Zhou Y, Xu K, Lan K. 2023. Vaccination with Span, an antigen guided by SARS-CoV-2 S protein evolution, protects against challenge with viral variants in mice. Sci Transl Med 15:eabo3332. doi: 10.1126/scitranslmed.abo3332 [DOI] [PubMed] [Google Scholar]
- 37. Huang H-Y, Liao H-Y, Chen X, Wang S-W, Cheng C-W, Shahed-Al-Mahmud M, Liu Y-M, Mohapatra A, Chen T-H, Lo JM, Wu Y-M, Ma H-H, Chang Y-H, Tsai H-Y, Chou Y-C, Hsueh Y-P, Tsai C-Y, Huang P-Y, Chang S-Y, Chao T-L, Kao H-C, Tsai Y-M, Chen Y-H, Wu C-Y, Jan J-T, Cheng T-J, Lin K-I, Ma C, Wong C-H. 2022. Vaccination with SARS-CoV-2 spike protein lacking glycan shields elicits enhanced protective responses in animal models. Sci Transl Med 14:eabm0899. doi: 10.1126/scitranslmed.abm0899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu C-Y, Cheng C-W, Kung C-C, Liao K-S, Jan J-T, Ma C, Wong C-H. 2022. Glycosite-deleted mRNA of SARS-CoV-2 spike protein as a broad-spectrum vaccine. Proc Natl Acad Sci U S A 119:e2119995119. doi: 10.1073/pnas.2119995119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu Y, Wang S, Zhang Y, Yuan L, Zheng Q, Wei M, Shi Y, Wang Z, Ma J, Wang K, Nie M, Xiao J, Huang Z, Chen P, Guo H, Lan M, Xu J, Hou W, Hong Y, Chen D, Sun H, Xiong H, Zhou M, Liu C, Guo W, Guo H, Gao J, Gan C, Li Z, Zhang H, Wang X, Li S, Cheng T, Zhao Q, Chen Y, Wu T, Zhang T, Zhang J, Cao H, Zhu H, Yuan Q, Guan Y, Xia N. 2022. Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine. Cell Host Microbe 30:1732–1744. doi: 10.1016/j.chom.2022.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van Dorp L, Acman M, Richard D, Shaw LP, Ford CE, Ormond L, Owen CJ, Pang J, Tan CCS, Boshier FAT, Ortiz AT, Balloux F. 2020. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect Genet Evol 83:104351. doi: 10.1016/j.meegid.2020.104351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Afkhami S, D’Agostino MR, Zhang A, Stacey HD, Marzok A, Kang A, Singh R, Bavananthasivam J, Ye G, Luo X, Wang F, Ang JC, Zganiacz A, Sankar U, Kazhdan N, Koenig JFE, Phelps A, Gameiro SF, Tang S, Jordana M, Wan Y, Mossman KL, Jeyanathan M, Gillgrass A, Medina MFC, Smaill F, Lichty BD, Miller MS, Xing Z. 2022. Respiratory mucosal delivery of next-generation COVID-19 vaccine provides robust protection against both ancestral and variant strains of SARS-CoV-2. Cell 185:896–915. doi: 10.1016/j.cell.2022.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Feng L, Wang Q, Shan C, Yang C, Feng Y, Wu J, Liu X, Zhou Y, Jiang R, Hu P, Liu X, Zhang F, Li P, Niu X, Liu Y, Zheng X, Luo J, Sun J, Gu Y, Liu B, Xu Y, Li C, Pan W, Zhao J, Ke C, Chen X, Xu T, Zhong N, Guan S, Yuan Z, Chen L. 2020. An adenovirus-vectored COVID-19 vaccine confers protection from SARS-CoV-2 challenge in rhesus macaques. Nat Commun 11:4207. doi: 10.1038/s41467-020-18077-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tang J, Zeng C, Cox TM, Li C, Son YM, Cheon IS, Wu Y, Behl S, Taylor JJ, Chakaraborty R, Johnson AJ, Shiavo DN, Utz JP, Reisenauer JS, Midthun DE, Mullon JJ, Edell ES, Alameh MG, Borish L, Teague WG, Kaplan MH, Weissman D, Kern R, Hu H, Vassallo R, Liu S-L, Sun J. 2022. Respiratory mucosal immunity against SARS-CoV-2 following mRNA vaccination. Sci Immunol 7:eadd4853. doi: 10.1126/sciimmunol.add4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lapuente D, Fuchs J, Willar J, Vieira Antão A, Eberlein V, Uhlig N, Issmail L, Schmidt A, Oltmanns F, Peter AS, Mueller-Schmucker S, Irrgang P, Fraedrich K, Cara A, Hoffmann M, Pöhlmann S, Ensser A, Pertl C, Willert T, Thirion C, Grunwald T, Überla K, Tenbusch M. 2021. Protective mucosal immunity against SARS-CoV-2 after heterologous systemic prime-mucosal boost immunization. Nat Commun 12:6871. doi: 10.1038/s41467-021-27063-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mao T, Israelow B, Peña-Hernández MA, Suberi A, Zhou L, Luyten S, Reschke M, Dong H, Homer RJ, Saltzman WM, Iwasaki A. 2022. Unadjuvanted intranasal spike vaccine elicits protective mucosal immunity against sarbecoviruses. Science 378:eabo2523. doi: 10.1126/science.abo2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vesin B, Lopez J, Noirat A, Authié P, Fert I, Le Chevalier F, Moncoq F, Nemirov K, Blanc C, Planchais C, Mouquet H, Guinet F, Hardy D, Vives FL, Gerke C, Anna F, Bourgine M, Majlessi L, Charneau P. 2022. An intranasal lentiviral booster reinforces the waning mRNA vaccine-induced SARS-CoV-2 immunity that it targets to lung mucosa. Mol Ther 30:2984–2997. doi: 10.1016/j.ymthe.2022.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shin H, Iwasaki A. 2012. A vaccine strategy that protects against genital herpes by establishing local memory T cells. Nature 491:463–467. doi: 10.1038/nature11522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Çuburu N, Kim R, Guittard GC, Thompson CD, Day PM, Hamm DE, Pang Y-Y, Graham BS, Lowy DR, Schiller JT. 2019. A prime-pull-amplify vaccination strategy to maximize induction of circulating and genital-resident intraepithelial CD8+ memory T cells. J Immunol 202:1250–1264. doi: 10.4049/jimmunol.1800219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Khanna M, Jackson RJ, Alcantara S, Amarasena TH, Li Z, Kelleher AD, Kent SJ, Ranasinghe C. 2019. Mucosal and systemic SIV-specific cytotoxic CD4+ T cell hierarchy in protection following intranasal/Intramuscular recombinant pox-viral vaccination of pigtail macaques. Sci Rep 9:5661. doi: 10.1038/s41598-019-41506-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. De Silva NS, Klein U. 2015. Dynamics of B cells in germinal centres. Nat Rev Immunol 15:137–148. doi: 10.1038/nri3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alameh M-G, Tombácz I, Bettini E, Lederer K, Sittplangkoon C, Wilmore JR, Gaudette BT, Soliman OY, Pine M, Hicks P, Manzoni TB, Knox JJ, Johnson JL, Laczkó D, Muramatsu H, Davis B, Meng W, Rosenfeld AM, Strohmeier S, Lin PJC, Mui BL, Tam YK, Karikó K, Jacquet A, Krammer F, Bates P, Cancro MP, Weissman D, Luning Prak ET, Allman D, Locci M, Pardi N. 2021. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity 54:2877–2892. doi: 10.1016/j.immuni.2021.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mesin L, Schiepers A, Ersching J, Barbulescu A, Cavazzoni CB, Angelini A, Okada T, Kurosaki T, Victora GD. 2020. Restricted clonality and limited germinal center reentry characterize memory B cell reactivation by boosting. Cell 180:92–106. doi: 10.1016/j.cell.2019.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sumida SM, Truitt DM, Kishko MG, Arthur JC, Jackson SS, Gorgone DA, Lifton MA, Koudstaal W, Pau MG, Kostense S, Havenga MJE, Goudsmit J, Letvin NL, Barouch DH. 2004. Neutralizing antibodies and CD8+ T lymphocytes both contribute to immunity to adenovirus serotype 5 vaccine vectors. J Virol 78:2666–2673. doi: 10.1128/jvi.78.6.2666-2673.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ersching J, Hernandez MIM, Cezarotto FS, Ferreira JDS, Martins AB, Switzer WM, Xiang Z, Ertl HCJ, Zanetti CR, Pinto AR. 2010. Neutralizing antibodies to human and simian adenoviruses in humans and New-World monkeys. Virology 407:1–6. doi: 10.1016/j.virol.2010.07.043 [DOI] [PubMed] [Google Scholar]
- 55. Zhang S, Huang W, Zhou X, Zhao Q, Wang Q, Jia B. 2013. Seroprevalence of neutralizing antibodies to human adenoviruses type-5 and type-26 and chimpanzee adenovirus type-68 in healthy Chinese adults. J Med Virol 85:1077–1084. doi: 10.1002/jmv.23546 [DOI] [PubMed] [Google Scholar]
- 56. Fausther-Bovendo H, Kobinger GP. 2014. Pre-existing immunity against ad vectors: humoral, cellular, and innate response, what’s important? Hum Vaccin Immunother 10:2875–2884. doi: 10.4161/hv.29594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Croyle MA, Patel A, Tran KN, Gray M, Zhang Y, Strong JE, Feldmann H, Kobinger GP. 2008. Nasal delivery of an adenovirus-based vaccine bypasses pre-existing immunity to the vaccine carrier and improves the immune response in mice. PLoS One 3:e3548. doi: 10.1371/journal.pone.0003548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Havervall S, Marking U, Svensson J, Greilert-Norin N, Bacchus P, Nilsson P, Hober S, Gordon M, Blom K, Klingström J, Åberg M, Smed-Sörensen A, Thålin C. 2022. Anti-spike mucosal IgA protection against SARS-CoV-2 Omicron infection. N Engl J Med 387:1333–1336. doi: 10.1056/NEJMc2209651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gilbert PB, Montefiori DC, McDermott AB, Fong Y, Benkeser D, Deng W, Zhou H, Houchens CR, Martins K, Jayashankar L, Castellino F, Flach B, Lin BC, O’Connell S, McDanal C, Eaton A, Sarzotti-Kelsoe M, Lu Y, Yu C, Borate B, van der Laan LWP, Hejazi NS, Huynh C, Miller J, El Sahly HM, Baden LR, Baron M, De La Cruz L, Gay C, Kalams S, Kelley CF, Andrasik MP, Kublin JG, Corey L, Neuzil KM, Carpp LN, Pajon R, Follmann D, Donis RO, Koup RA, Immune Assays Team§, Moderna, Inc. Team§, Coronavirus Vaccine Prevention Network (CoVPN)/Coronavirus Efficacy (COVE) Team§, United States Government (USG)/CoVPN Biostatistics Team§ . 2022. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science 375:43–50. doi: 10.1126/science.abm3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tauzin A, Gong SY, Beaudoin-Bussières G, Vézina D, Gasser R, Nault L, Marchitto L, Benlarbi M, Chatterjee D, Nayrac M, Laumaea A, Prévost J, Boutin M, Sannier G, Nicolas A, Bourassa C, Gendron-Lepage G, Medjahed H, Goyette G, Bo Y, Perreault J, Gokool L, Morrisseau C, Arlotto P, Bazin R, Dubé M, De Serres G, Brousseau N, Richard J, Rovito R, Côté M, Tremblay C, Marchetti GC, Duerr R, Martel-Laferrière V, Kaufmann DE, Finzi A. 2022. Strong humoral immune responses against SARS-CoV-2 spike after BNT162b2 mRNA vaccination with a 16-week interval between doses. Cell Host Microbe 30:97–109. doi: 10.1016/j.chom.2021.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ullah I, Prévost J, Ladinsky MS, Stone H, Lu M, Anand SP, Beaudoin-Bussières G, Symmes K, Benlarbi M, Ding S, Gasser R, Fink C, Chen Y, Tauzin A, Goyette G, Bourassa C, Medjahed H, Mack M, Chung K, Wilen CB, Dekaban GA, Dikeakos JD, Bruce EA, Kaufmann DE, Stamatatos L, McGuire AT, Richard J, Pazgier M, Bjorkman PJ, Mothes W, Finzi A, Kumar P, Uchil PD. 2021. Live imaging of SARS-CoV-2 infection in mice reveals that neutralizing antibodies require FC function for optimal efficacy. Immunity 54:2143–2158. doi: 10.1016/j.immuni.2021.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mazzoni A, Di Lauria N, Maggi L, Salvati L, Vanni A, Capone M, Lamacchia G, Mantengoli E, Spinicci M, Zammarchi L, Kiros ST, Rocca A, Lagi F, Colao MG, Parronchi P, Scaletti C, Turco L, Liotta F, Rossolini GM, Cosmi L, Bartoloni A, Annunziato F, COVID-19 Research Group . 2021. First-dose mRNA vaccination is sufficient to reactivate immunological memory to SARS-CoV-2 in subjects who have recovered from COVID-19. J Clin Invest 131:e149150. doi: 10.1172/JCI149150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Goel RR, Apostolidis SA, Painter MM, Mathew D, Pattekar A, Kuthuru O, Gouma S, Hicks P, Meng W, Rosenfeld AM, Dysinger S, Lundgreen KA, Kuri-Cervantes L, Adamski S, Hicks A, Korte S, Oldridge DA, Baxter AE, Giles JR, Weirick ME, McAllister CM, Dougherty J, Long S, D’Andrea K, Hamilton JT, Betts MR, Luning Prak ET, Bates P, Hensley SE, Greenplate AR, Wherry EJ. 2021. Distinct antibody and memory B cell responses in SARS-CoV-2 naive and recovered individuals following mRNA vaccination. Sci Immunol 6:eabi6950. doi: 10.1126/sciimmunol.abi6950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tejedor Vaquero S, de Campos-Mata L, Ramada JM, Díaz P, Navarro-Barriuso J, Ribas-Llaurado C, Rodrigo Melero N, Carolis C, Cerutti A, Gimeno R, Magri G. 2021. The mRNA-1273 vaccine induces cross-variant antibody responses to SARS-CoV-2 with distinct profiles in individuals with or without pre-existing immunity. Front Immunol 12:737083. doi: 10.3389/fimmu.2021.737083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu J, Xu K, Xing M, Zhuo Y, Guo J, Du M, Wang Q, An Y, Li J, Gao P, Wang Y, He F, Guo Y, Li M, Zhang Y, Zhang L, Gao GF, Dai L, Zhou D. 2021. Heterologous prime-boost immunizations with chimpanzee adenoviral vectors elicit potent and protective immunity against SARS-CoV-2 infection. Cell Discov 7:123. doi: 10.1038/s41421-021-00360-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang X, Xing M, Zhang C, Yang Y, Chi Y, Tang X, Zhang H, Xiong S, Yu L, Zhou D. 2014. Neutralizing antibody responses to enterovirus and adenovirus in healthy adults in China. Emerg Microbes Infect 3:e30. doi: 10.1038/emi.2014.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information includes five figures and one table.
Data Availability Statement
All data from this study are included within this manuscript/Supplementary Material and are available from the Lead Contact (Dongming Zhou, zhoudongming@tmu.edu.cn) upon request.