ABSTRACT
Fecal-oral pathogens encounter constitutively expressed enteric alpha-defensins in the intestine during replication and transmission. Alpha-defensins can be potently antiviral and antibacterial; however, their primary sequences, the number of isoforms, and their activity against specific microorganisms often vary greatly between species, reflecting adaptation to species-specific pathogens. Therefore, alpha-defensins might influence not only microbial evolution and tissue tropism within a host but also species tropism and zoonotic potential. To investigate these concepts, we generated a panel of enteric and myeloid alpha-defensins from humans, rhesus macaques, and mice and tested their activity against group A rotaviruses, an important enteric viral pathogen of humans and animals. Rotaviral adaptation to the rhesus macaque correlated with resistance to rhesus enteric but not myeloid alpha-defensins and sensitivity to human alpha-defensins. Infection by mouse and human rotaviruses was either resistant to or increased by host enteric alpha-defensins, although the effects of cross-species alpha-defensins did not follow an obvious pattern. Because infection by all rotaviruses tested was resistant to or enhanced by enteric alpha-defensins from their hosts, exposure to alpha-defensins may have shaped their evolution. We then used a genetic approach to identify the viral attachment and penetration protein, VP4, as a determinant of alpha-defensin sensitivity. Our results provide a foundation for future studies of the VP4-dependent mechanism of defensin neutralization, highlight the species-specific activities of alpha-defensins, and focus future efforts on a broader range of rotaviruses that differ in VP4 to uncover the potential for enteric alpha-defensins to influence species tropism.
Importance
Rotavirus is a leading cause of severe diarrhea in young children. Like other fecal-oral pathogens, rotaviruses encounter abundant, constitutively expressed defensins in the small intestine. These peptides are a vital part of the vertebrate innate immune system. By investigating the impact that defensins from multiple species have on the infectivity of different strains of rotavirus, we show that some rotaviral infections can be inhibited by defensins. We also found that rotaviruses may have evolved resistance to defensins in the intestine of their host species, and some even appropriate defensins to increase their infectivity. Because rotaviruses infect a broad range of animals and rotaviral infections are highly prevalent in children, identifying immune defenses against infection and how they vary across species and among viral genotypes is important for our understanding of the evolution, transmission, and zoonotic potential of these viruses as well as the improvement of vaccines.
KEYWORDS: defensins, enteric viruses, rotavirus, viral immunity
INTRODUCTION
Mammalian defensins are broadly anti-microbial, cationic peptides that can be subdivided into three families: α, β, and θ (1 – 3). β-Defensins are the most ancestral and are found in many vertebrate species. Many mammals also express α-defensins, while θ-defensins are only expressed in orangutans and some old-world monkeys like baboons and rhesus macaques. The α-defensin subfamily can be further divided into enteric and myeloid, based on expression pattern and gene organization. Enteric α-defensins are constitutively secreted in the small intestine by Paneth cells and are also expressed in the genitourinary tract. Myeloid α-defensins are expressed by neutrophils and are stored in granules which can either be fused with phagolysosomes or secreted during the formation of neutrophil extracellular traps.
Defensins can be both antiviral and antibacterial (4). Although there are numerous examples of bacterial species and enveloped viruses that can be potently inhibited by α-, β-, and θ-defensins, only α-defensins alter non-enveloped viral infections (5 – 7). Thus far, some members of Adenoviridae, Papillomaviridae, Parvoviridae, and Polyomaviridae have been shown to be neutralized by α-defensins. Apart from BK polyomavirus, which is inhibited by viral aggregation prior to entry, α-defensins inhibit non-enveloped viruses by altering uncoating, leading to improper trafficking of the viral genome in the cell.
Although most studies have focused on neutralization of viral infection by α-defensins, some viruses can appropriate α-defensins to enhance their infection. For human adenoviruses (AdVs), serotypes of enteric species are either enhanced or resistant to human enteric α-defensin 5 (HD5), while serotypes of respiratory species are generally neutralized (8 – 11). This paradigm extends to mouse AdVs, where infection by the fecal-orally transmitted mouse AdV serotype 2 is enhanced by mouse enteric α-defensins (12), while the more pantropic mouse AdV serotype 1 is neutralized (13). Because enteric α-defensins are constitutively secreted in the small intestine, fecal-orally transmitted viruses may have evolved resistance to and/or mechanisms to co-opt defensins. The ability of enteric α-defensins to select for the evolution of α-defensin-resistant human AdVs has been verified in vitro (11), demonstrating that this form of viral adaptation is feasible.
Rotavirus is a fecal-oral pathogen and, despite the availability of vaccines, remains one of the leading causes of severe gastroenteritis in young children (14, 15). There are nine species or groups of rotavirus (A–D, F–J) that infect a broad range of animals and pose a concern for both human health and agriculture (16). In humans, group A rotaviruses are, by far, the most epidemiologically relevant and currently are responsible for almost 150,000 deaths annually (15). Two major genotypes, Wa-like and DS-1-like, together account for ~90% of circulating strains (17, 18). Rotavirus has an 11-segment double-stranded RNA genome that is encapsidated in a triple-layered particle (19). VP2 forms the innermost layer and interacts with the RNA-dependent RNA polymerase (VP1) and the capping enzyme (VP3). VP6 constitutes the intermediate layer. The outermost layer consists of the protease-sensitive spike protein (VP4) and calcium ion-stabilized trimers of the glycoprotein (VP7). VP4 matures following trypsin cleavage into VP5* and VP8*. VP5* makes up the foot and the stalk of the spike protein and is vital for membrane penetration. VP8* forms the head of the spike protein and is involved in viral attachment to its receptor, which is often a glycan (20). During entry, VP5*, VP8*, and VP7 disassociate from the viral particle to generate the transcriptionally active double-layered particle. Replication and assembly occur in the cytoplasm within viroplasms, which are formed by NSP2 and NSP5 (21). Because of the segmented nature of the genome, reassortment of gene segments from multiple viruses infecting the same cell can give rise to progeny with mixed genomic compositions, although reassortant formation tends to favor certain gene constellations. This process contributes to rotaviral evolution and provides a powerful approach to investigate rotaviral genetics, particularly prior to the recent development of a tractable reverse genetics system (22).
Rotaviruses are members of the Reoviridae family, and the only study thus far on the effects of defensins on infection by a Reoviridae family member found that reovirus type 3 is resistant to HNP1 (23). Given more recent examples of neutralization of non-enveloped viruses by α-defensins and the potential for enteric α-defensins to influence the evolution of fecal-orally transmitted viruses (5, 6, 11), we examined the effects of human, rhesus macaque, and mouse α-defensins on human (DS-1 and Wa), simian (RRV and SA11), and mouse [epizootic diarrhea of infant mice (EDIM)] group A rotaviruses. We found that α-defensins from different species have distinct antiviral activity, rotaviral adaptation to the rhesus macaque correlates with resistance to rhesus enteric α-defensins, different genogroups of human rotaviruses are differentially affected by human α-defensins, and the sensitivity of rotaviruses to neutralization by α-defensins can be determined by the VP4 capsid protein.
RESULTS
Selection and purification of α-defensins for rotaviral studies
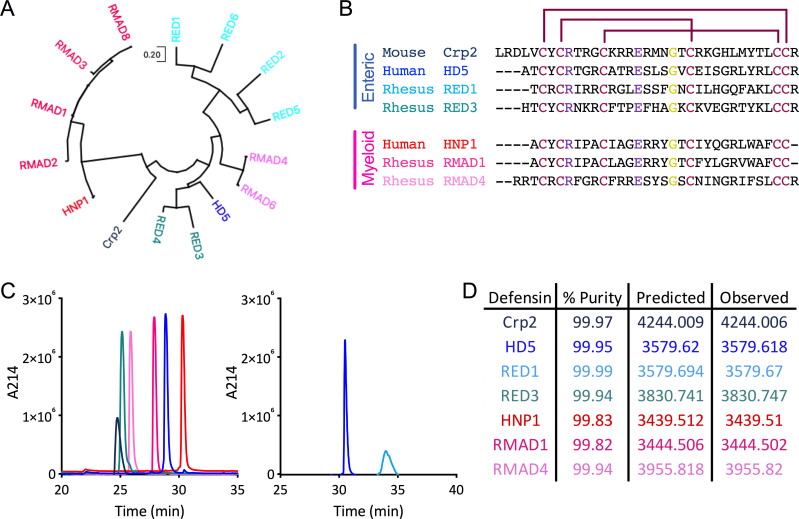
The α-defensin repertoire differs between species at both the genetic and protein levels, and there is considerable variability in the primary sequences of α-defensins despite the conservation of fold, disulfide bonding pattern, a salt bridge, and an invariant glycine (Fig. 1A and B) (1, 2). Humans have three genes that encode myeloid α-defensins (DEFA1, DEFA3, and DEFA4), but four distinct α-defensin proteins have been isolated (HNP1–4). In the human gut and genitourinary tracts, there is a predominant form of the products (HD5 and HD6) of each of the two enteric α-defensin genes (DEFA5 and DEFA6), although additional cleavage products of HD5 have been identified (24). Rhesus macaque myeloid (RMAD) and enteric (RED) α-defensins have not been investigated extensively. However, sequence analysis indicates that the six to nine isoforms that have been described for each subtype of α-defensin can be further subdivided into two groups (Fig. 1A) (25 – 27). The mouse is even more complex, with an absence of intact myeloid α-defensin genes and a large number (>20) of enteric α-defensin genes, which vary by strain, leading to abundant expression of ~6 isoforms (called cryptdins, Crps) in the gut (28).
Fig 1.
Defensin refolding, sequences, and evolutionary relationships. (A) Dendrogram of mature protein sequences of rhesus enteric and myeloid α-defensins, one mouse enteric α-defensin (Crp2), and one each of the human enteric (HD5) and myeloid (HNP1) α-defensins. The evolutionary history was inferred by using the Maximum Likelihood method and JTT matrix-based model (29). The tree with the highest log likelihood is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT model and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGA X (30). (B) Amino acid sequence alignment of the mature α-defensin peptides used in this study. Enteric α-defensin names are written in shades of blue, while myeloid α-defensin names are written in shades of pink. Disulfide bond linkages are indicated by maroon lines. Residues that interact through a conserved salt bridge are colored purple, and the conserved glycine is colored yellow. Dashes represent gaps. For (A and B), accession numbers for defensin protein sequences are NP_066290 (HD5), P59665 (HNP1), AAW51365 (RED1), AAW51366 (RED2), AAW51367 (RED3), AAW51368 (RED4), AAW51369 (RED5), AAW51370 (RED6), AAF06312 (RMAD1), P82317 (RMAD2), AAF06313 (RMAD3), AAF06315 (RMAD4), AAF06316 (RMAD6), AAF06314 (RMAD8), and AAI25549 (Crp2). (C) Peptide samples were analyzed individually by RP-HPLC on a C18 column. Peaks are colored as in (B). Peptides depicted on the same graph were analyzed sequentially on the same day. (D) Percent purity determined by the analyses in (C) as well as predicted and observed molecular masses determined by electrospray ionization are listed for each defensin.
Because an exhaustive analysis of all human, rhesus, and mouse α-defensins was not feasible, we chose a subset for analysis in these studies. HNP1–3 vary by only the first amino acid and are much more abundant in neutrophils than HNP4, so we selected HNP1. HD5 was chosen because it has potent antiviral activity against multiple non-enveloped viruses, unlike HD6. Crp2 was chosen based on our prior analyses with mouse AdVs (12, 13). We chose abundant RMADs that are more (RMAD1) or less (RMAD4) similar in sequence to HNP1. Likewise, we chose REDs that are more (RED3) or less (RED1) similar in sequence to HD5. Due to our focus on an enteric virus, rhesus oral defensins were not included; however, they have comparable sequences to RMAD4 (26).
Two protocols have been described for the oxidative refolding of synthetic human α-defensins, one using guanidine hydrochloride for HNP4, HD5, and HD6 (31) and the other using urea and N,N-dimethylformamide for HNP1-3 (32). We therefore chose a refolding protocol for the REDs, RMADs, and Crp2 based on sequence similarity to the human defensins. Upon purification by semi-preparative reverse-phase high-pressure liquid chromatography (RP-HPLC), analytical RP-HPLC was consistent with high purity and homogeneity (Fig. 1C and D), and the measured masses of the refolded species indicated the formation of the expected three disulfide bonds (Fig. 1D). Although most of the refolded α-defensins had retention times between 25 and 30 min on the analytical C18 RP-HPLC column, we noted an unusually prolonged retention time for RED1, which was consistent with prior studies (27). Thus, we successfully generated purified α-defensins for further study.
Rhesus rotavirus is resistant to rhesus enteric α-defensins but neutralized by rhesus myeloid and human α-defensins
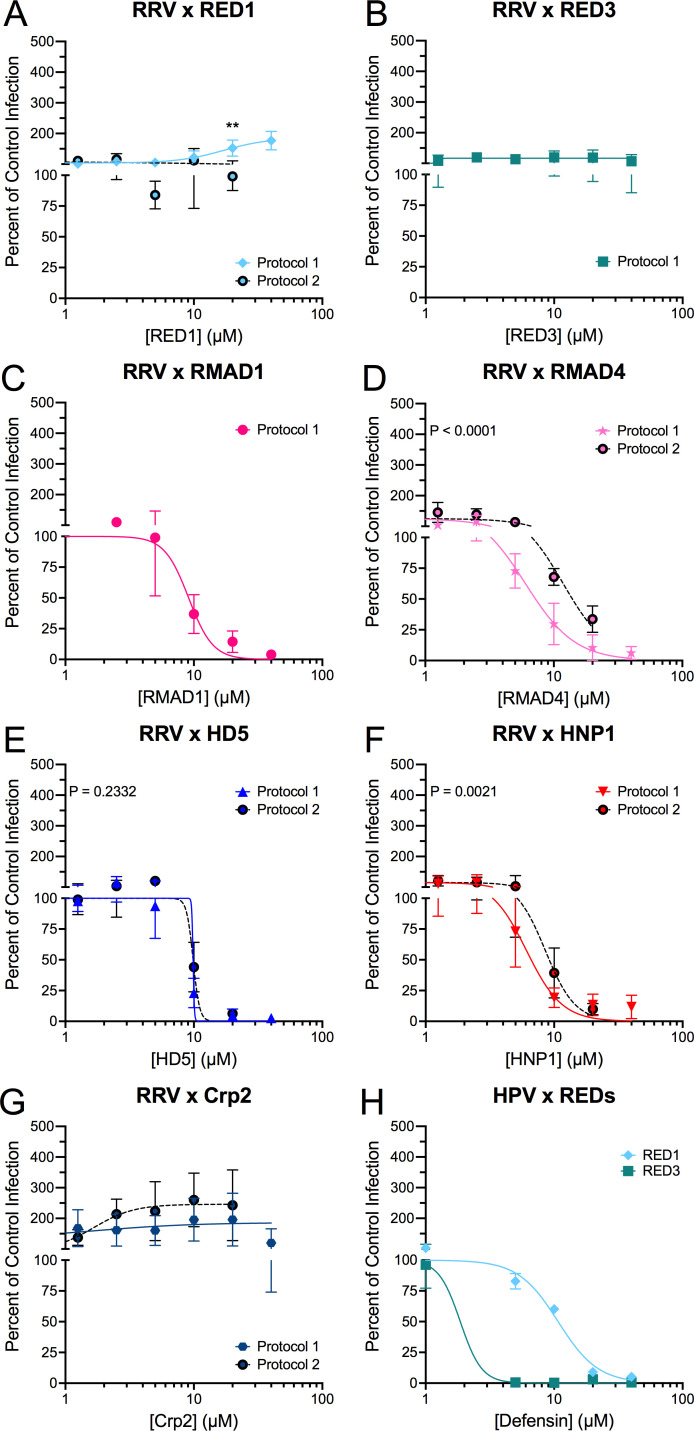
We tested the effects of the panel of purified α-defensins on rhesus rotavirus (RRV) infection starting with the homologous defensins of rhesus macaques (RED1, RED3, RMAD1, and RMAD4). For these experiments, we incubated RRV with each α-defensin on ice before adding the mixture to MA104 cells. Based on our previous studies of α-defensin interactions with mouse and human AdVs (8, 9, 11 – 13, 33 – 35), we defined three phenotypes of defensin effects on infection relative to control infection in the absence of defensin: neutralization (0–49%), resistance (50–199%), and enhancement (≥200%). RRV was potently neutralized by both rhesus myeloid α-defensins, RMAD1 (IC50 = 9.1 µM, 95% CI = 7.4–11.6 µM, Hill slope = −4.6) and RMAD4 (IC50 = 6.0 µM, 95% CI = 5.2–7.0 µM, Hill slope = −2.2), with an almost complete block of infection in the presence of 20 µM defensin (Fig. 2C and D). However, RRV was resistant to both rhesus enteric α-defensins, RED1 and RED3, up to the highest concentration tested (40 µM) (Fig. 2A and B, solid lines).
Fig 2.
RRV is selectively resistant to homologous enteric α-defensins. RRV was incubated with the indicated concentrations of (A) RED1, (B) RED3, (C) RMAD1, (D) RMAD4, (E) HD5, (F) HNP1, or (G) Crp2 before infecting MA104 cells (protocol 1; solid, colored lines). For protocol 2 (dotted, black lines with filled circles), RRV was bound to cells in the cold prior to defensins being added (A and D–G). (H) HPV16 pseudovirus infection was measured in the presence of RED1 and RED3 following protocol 1. Data are normalized to infection in the absence of defensin (control infection) and are the mean ± SD of at least three individual experiments.
To our knowledge, there is only one report describing the bactericidal properties of RED1 and RED3 (27) and none that have investigated their antiviral capabilities. Therefore, to ensure that RED1 and RED3 are functional, we tested their activity against another non-enveloped virus, human papillomavirus 16 (HPV16). Like prior studies with human defensins (33, 36 – 39), HPV16 was potently neutralized by both RED1 (IC50 = 10.7 µM, 95% CI = 9.4–12.1 µM, Hill slope −2.8) and RED3 (IC50 < 5 µM), demonstrating that RED1 and RED3 can be antiviral (Fig. 2H).
Next, we tested the effects of heterologous α-defensins from humans and mice on RRV infection. Although HD5 and RED3 cluster together phylogenetically and are 56% identical (Fig. 1A and B), RRV is completely neutralized by 20 µM HD5 (IC50 = 8.1 µM, lower 95% CI = 7.1 µM, Hill slope = −5.7; Fig. 2E, solid line). Similarly, the heterologous myeloid α-defensin, HNP1, strongly neutralizes RRV (IC50 = 6.1 µM, 95% CI = 5.4–6.9 µM, Hill slope −3.1; Fig. 2F, solid line). Thus, our data are consistent with the hypothesis that selective pressure during fecal-oral transmission among rhesus hosts has pressured RRV to evolve resistance to rhesus enteric α-defensins (RED1 and RED3), but not rhesus myeloid α-defensins or heterologous α-defensins (RMAD1, RMAD4, HD5, and HNP1). The only exception to this trend is the resistance of RRV to mouse Crp2 (up to 40 µM, Fig. 2G).
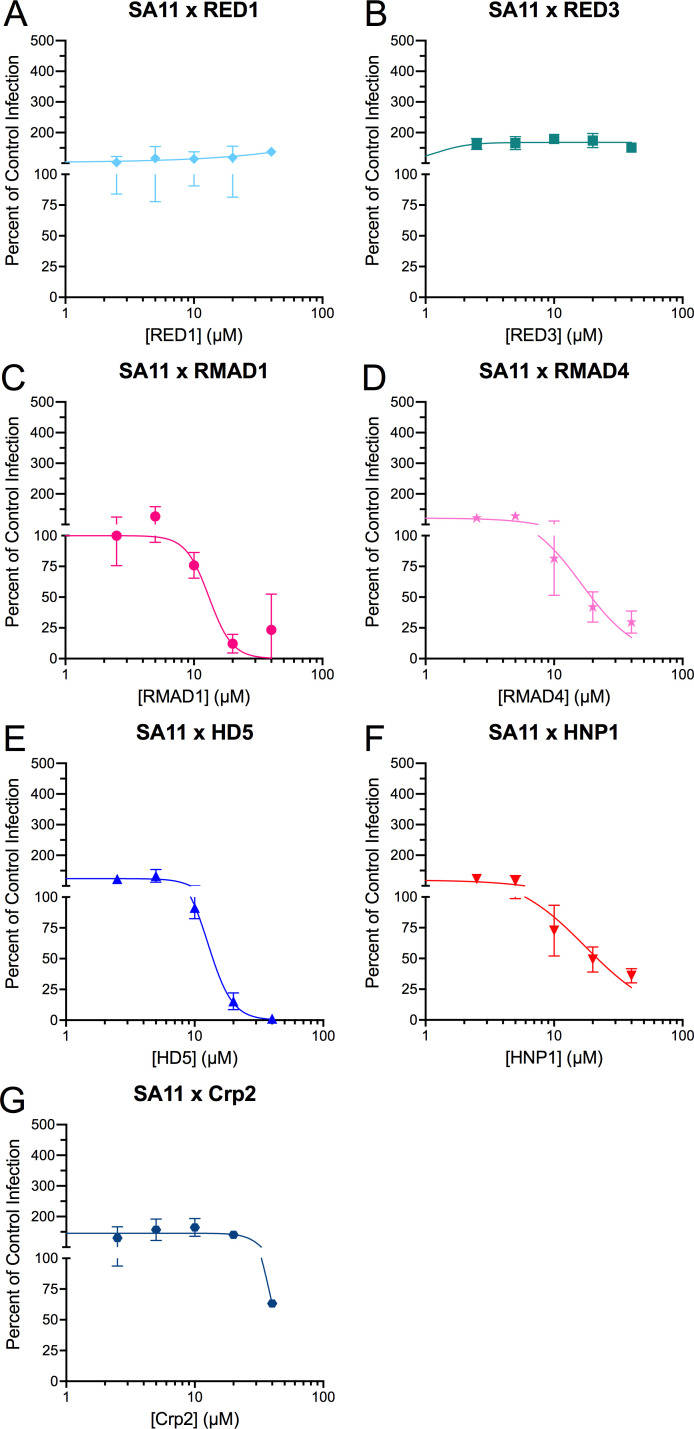
The non-human primate rotavirus SA11 has an infectivity phenotype like that of RRV in the presence of α-defensins
SA11 is a widely studied group A rotavirus strain that was originally isolated from the vervet monkey, Chlorocebus pygerythrus (40). A detailed analysis of the sequences and relative expression of α-defensins of C. pygerythrus, which is estimated to have diverged from macaques 12 million years ago (41), has not been published. Therefore, we did not examine defensin peptides from this species. Nonetheless, we tested the effects of our panel of defensins on SA11 infection. Like RRV, SA11 was resistant to RED1 and RED3 (Fig. 3A and B) and neutralized by RMAD1, RMAD4, HD5, and HNP1 (Fig. 3C through F). However, the IC50 values of RMAD1, RMAD4, HD5, and HNP1 for SA11 infection were 1.3- to 3.0-fold greater than those for RRV. The Hill slopes for RMAD1 and RMAD4 were similar to those for RRV; however, they differed for HD5 (−4.7 for SA11 vs −5.7 for RRV) and HNP1 (−1.5 for SA11 vs −3.1 for RRV). And, unlike the resistance of RRV, Crp2 reduced the infectivity of SA11 at 40 µM, although inhibition was less than 40% (Fig. 3G). Thus, the effects of α-defensins on RRV and SA11 are comparable but not identical, and the high similarity between RRV and SA11 proteins may facilitate future mapping of viral determinants of α-defensin activity.
Fig 3.
SA11 has a similar phenotype to RRV. SA11 was incubated with the indicated concentrations of (A) RED1, (B) RED3, (C) RMAD1, (D) RMAD4, (E) HD5, (F) HNP1, or (G) Crp2 before infecting MA104 cells (protocol 1). Data are normalized to infection in the absence of defensin (control infection) and are the mean ± SD of at least three individual experiments.
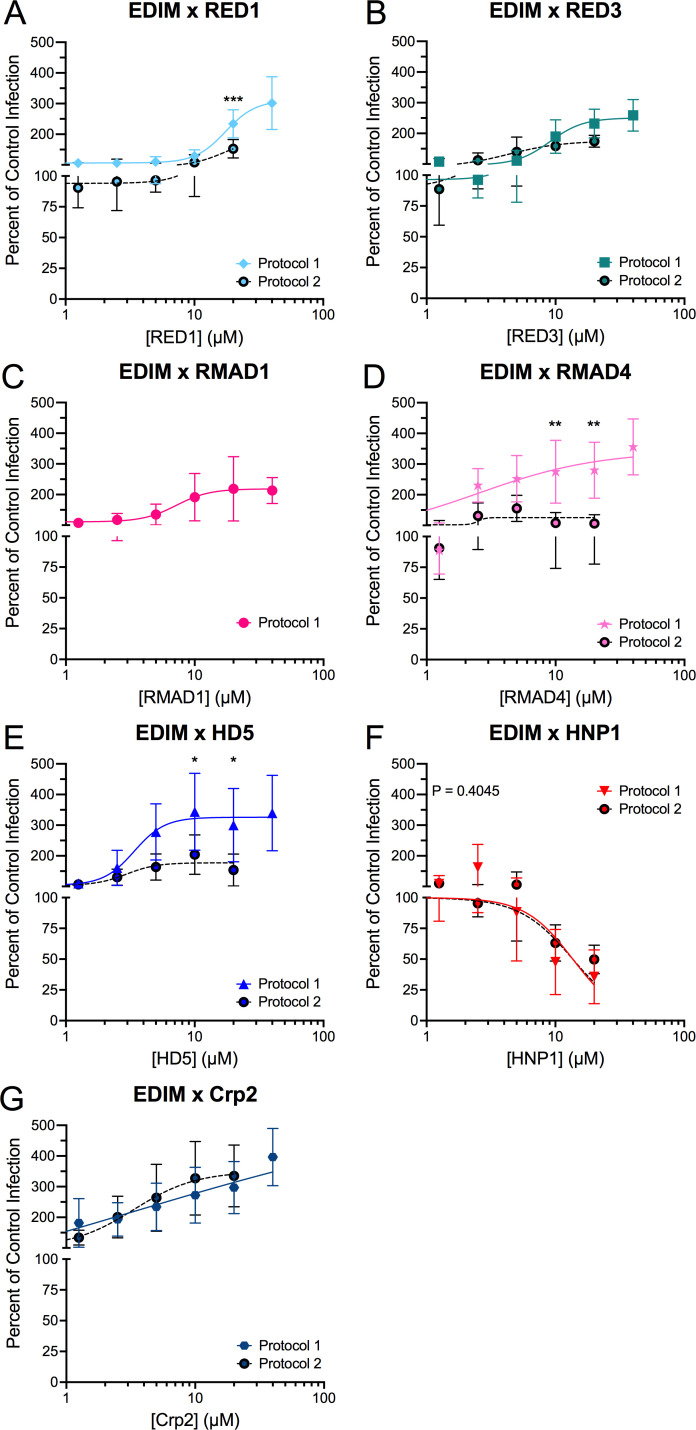
Infectivity of mouse and human rotaviruses is either enhanced by or resistant to homologous enteric alpha-defensins
To investigate the generalizability of our finding that group A rotaviruses are resistant to homologous enteric α-defensins but sensitive to myeloid and heterologous α-defensins, we expanded our analyses to both mouse and human group A rotaviruses. Infection by a mouse rotavirus, EDIM, was enhanced in the presence of the homologous enteric α-defensin, Crp2, with three- to fourfold increased infection in the presence of >20 µM Crp2 (Fig. 4G, solid line) compared to control. This is consistent with the possibility that selective pressure led to EDIM appropriating Crp2 to increase its infectivity. However, among the heterologous α-defensins, EDIM was neutralized by HNP1 (IC50 = 12.4 µM, 95% CI = 7.7–22.8 µM, Hill slope = −2.5) and enhanced two- to fourfold by all other α-defensins (Fig. 4 through F, solid lines).
Fig 4.
EDIM is enhanced by most defensins. EDIM was incubated with the indicated concentrations of (A) RED1, (B) RED3, (C) RMAD1, (D) RMAD4, (E) HD5, (F) HNP1, or (G) Crp2 before infecting MA104 cells (protocol 1; solid, colored lines). For protocol 2 (dotted, black lines with filled circles), EDIM was bound to cells in the cold prior to defensins being added (A, B, and D–G). Data are normalized to infection in the absence of defensin (control infection) and are the mean ± SD of at least three individual experiments.
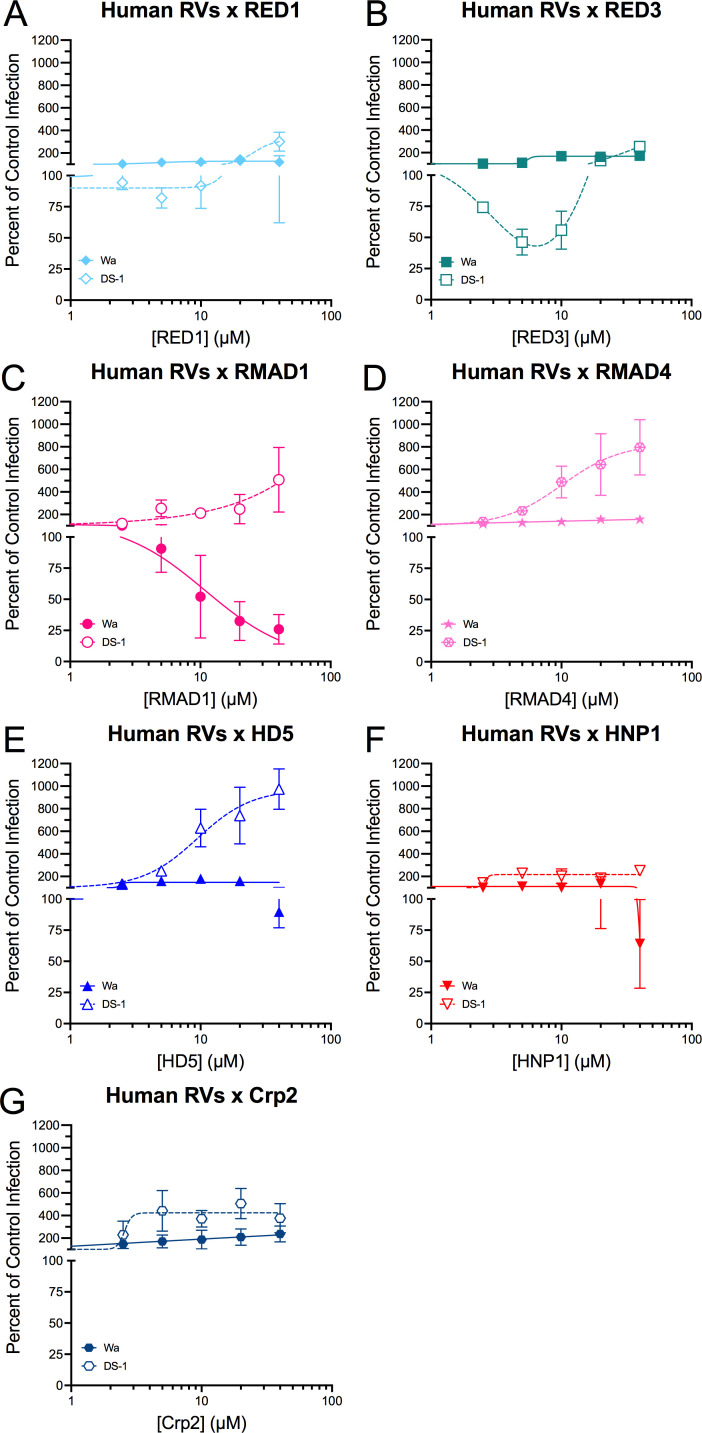
The prototype strains of two of the more prevalent rotavirus A genogroups that infect humans, DS-1 and Wa, had different phenotypes in the presence of α-defensins. DS-1 resembled EDIM in that it was enhanced by all α-defensins to varying degrees (Fig. 5, open symbols with dotted lines). The >9-fold increase in infection in the presence of 40 µM HD5 was particularly striking (Fig. 5), and the bi-phasic effect of RED3 was unusual but reproducible (Fig. 5). In contrast, Wa was resistant to RED1, RED3, RMAD4, HD5, and Crp2 and neutralized by RMAD1 (IC50 = 10.9 µM, Hill slope = −1.3). The infectivity of Wa was reduced by HNP1 at 40 µM, although inhibition was highly variable and less than 50% (Fig. 5, filled symbols and solid lines). Thus, a clear distinction between the activity of homologous and heterologous α-defensins, like we observed for RRV, was not apparent for EDIM, DS-1, or Wa.
Fig 5.
DS-1 and Wa have different sensitivities to defensins. DS-1 (open symbols) and Wa (filled symbols) were incubated with the indicated concentrations of (A) RED1, (B) RED3, (C) RMAD1, (D) RMAD4, (E) HD5, (F) HNP1, or (G) Crp2 before infecting MA104 cells (protocol 1). Data are normalized to infection in the absence of defensin (control infection) and are the mean ± SD of at least three individual experiments.
Order of addition does not substantially alter the effects of α-defensins on rotaviral infection
We previously found that the order of addition of α-defensin and virus to the cell can have a dramatic effect on the outcome of human AdV infection (11). The data in the prior sections were generated by exposing the virus to defensin before the virus was added to cells (protocol 1). To test if the order of addition affects the outcome of the interaction, we re-examined a subset of the α-defensin/rotavirus combinations by binding the virus to MA104 cells before adding α-defensin (protocol 2). Under these conditions, RRV is less sensitive to the myeloid defensins RMAD4 (Fig. 2D, IC50 = 11.8 µM in protocol 2 vs 6.0 µM in protocol 1) and HNP1 (Fig. 2F, IC50 = 8.5 µM in protocol 2 vs 6.1 µM in protocol 1). For the enteric defensins, Crp2 became weakly enhancing (Fig. 2G), while there were only minor differences between protocols 1 and 2 for RRV infection with RED1 and HD5 (Fig. 2A and E).
For human AdV, we found that some enhanced viruses in protocol 1 became neutralized in protocol 2 (11). We therefore compared protocol 1 to protocol 2 for EDIM, which is enhanced by some α-defensins, using most of our α-defensin panel (Fig. 4, black dotted lines). Neutralization of EDIM by HNP1 was protocol-independent (Fig. 4F). In contrast, EDIM was no longer enhanced but became resistant to RED1, RED3, RMAD4, and HD5 under protocol 2 (Fig. 4A, B, D and E). Interestingly, like for RRV, Crp2 enhanced EDIM infection in protocol 2 to an even greater extent than in protocol 1 (Fig. 4G). Therefore, pre-binding the virus to the cell before adding α-defensins only abrogates defensin-dependent enhancement, although Crp2-mediated enhancement was unaffected. We did not observe any protocol-dependent changes from an enhanced to a neutralized phenotype, unlike what was observed for human AdVs.
VP4 is a determinant for α-defensin activity
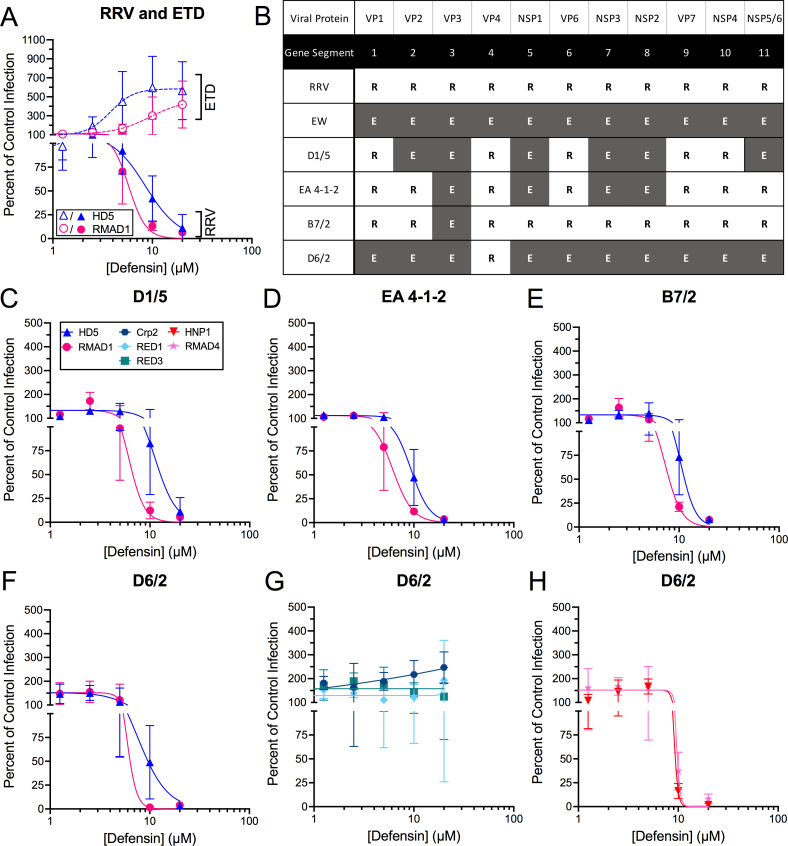
To identify a viral determinant for α-defensin activity, we interrogated a previously characterized panel of reassortant viruses between RRV and EW, a wild-type murine strain that has not been adapted to cell culture (Fig. 6) (42, 43). As a control, we included a cell culture-adapted derivative of EW, ETD. ETD is enhanced by HD5 and RMAD1, while RRV is neutralized by those α-defensins (Fig. 6). The reassortant viruses D1/5, EA 4-1-2, and B7/2 were all neutralized, like RRV, rather than enhanced, like ETD, by both HD5 and RMAD1 (Fig. 6). Because these reassortant viruses have several RRV gene segments in common, we tested an additional virus, D6/2, in which only the VP4 gene segment is derived from RRV. This virus was also neutralized by both HD5 and RMAD1 (Fig. 6); therefore, substituting the RRV VP4 gene segment for that of ETD is sufficient to alter the infection phenotype of the virus in the presence of HD5 and RMAD1. To extend these findings, we tested additional myeloid and enteric α-defensins and found that D6/2 uniformly mimics the RRV phenotype (Fig. 6), although the effect of Crp2 on D6/2 was intermediate between the resistance of RRV and the enhancement of EDIM. In summary, these data strongly support the identification of VP4 as a determinant of neutralization for multiple α-defensins.
Fig 6.
VP4 is a determinant of defensin neutralization. (A) ETD, a culture-adapted strain of murine rotavirus EW, (open symbols) and RRV (closed symbols) infection in the presence of HD5 or RMAD1. Note that the RRV data are independent of the data in Fig. 2. (B) Genetic composition of reassortant viruses between RRV (R, white boxes) and the murine rotavirus, EW (E, gray boxes). (C–F) Infection of the indicated reassortant viruses in the presence of HD5 or RMAD1. (G) D6/2 infection in the presence of enteric α-defensins, Crp2, RED1, or RED3. (H) D6/2 infection in the presence of myeloid α-defensins, HNP1, or RMAD4. All viruses were incubated with defensins before infecting MA104 cells (protocol 1). Defensins in panels (C–H) are denoted by the key in panel (C). Data are normalized to infection in the absence of defensin (control infection) and are the mean ± SD of at least three individual experiments.
DISCUSSION
Enteric α-defensins are important for host immunity against enteric pathogens and influence the composition of the intestinal microbiome (44). Comparative genetic studies suggest that differences in the primary sequences and number of isoforms of defensins between species reflect adaptation to pathogens during evolution (2); however, studies that directly compare the effects of heterologous and homologous α-defensins on viral infection are limited. Our approach to investigate a panel of α-defensins from multiple host species led to several important insights. We found that RED1, RED3, RMAD1, and RMAD4 are all capable of inhibiting the infection of at least one type of non-enveloped virus. Prior studies of the antiviral capabilities of rhesus α-defensins identified RMAD4 as having anti-HIV properties while RMAD3 did not (45); however, both RMAD1, which differs from RMAD3 by only two residues (L22R and G23R), and RMAD4 were largely equivalent in our assays, with the exception of Wa being neutralized by RMAD1 and resistant to RMAD4. They were also both comparable to HNP1 for RRV and SA11; however, both RMADs enhanced EDIM/ETD to varying degrees, while HNP1 was neutralizing, and both RMADs were much more enhancing than HNP1 for DS-1. The similar activity of these myeloid α-defensins is remarkable given that RMAD4 differs substantially in linear sequence from RMAD1 and HNP1 (36.7% identical to each), which are themselves quite closely related (86.7% identical). Among the enteric α-defensins, HD5, and RED3 (56% identical), have the same effect on Wa but different effects on RRV, SA11, and DS-1. Conversely, both REDs were largely equivalent for all viruses despite being only 48.4% identical, and HD5 and Crp2 are 46.9% identical but similarly enhance EDIM infection. Thus, α-defensin antiviral activity cannot be easily predicted by sequence nor the degree of positive charge; however, the activities identified here inform the design of α-defensin mutants to dissect the features of α-defensins that dictate their effects on rotaviral infection.
A key finding of our study is that VP4, the viral attachment, and membrane penetrating protein, can dictate the neutralizing activity of α-defensins on rotaviral infection. Although we have not formally excluded a cellular target for α-defensin activity that determines the outcome of infection, the importance of VP4 strongly suggests that defensin binding to the virus and not the cell is critical. Such a mechanism would also be consistent with defensin-dependent mechanisms that we and others have determined for a wide range of non-enveloped viruses (5 – 7). Moreover, like for other non-enveloped viruses, our data show that α-defensins impact rotaviral entry upstream of VP6 translation, which we used to quantify infection. Mechanistically, α-defensins could directly compete with VP4 for receptor binding, although receptor competition has not been documented for other non-enveloped viruses. However, our use of rotaviruses with differing receptor specificities (20), which is mediated by VP8*, argues against receptor competition as a mechanism. Alternatively, defensin-mediated aggregation, although not inherently neutralizing, could have the effect of reducing the total number of cells that are infected; however, this is an unlikely mechanism for neutralization of rotavirus, since binding RRV to cells prior to the addition of RMAD4, HD5, or HNP1 (protocol 2) did not circumvent aggregation and rescue infection. α-Defensins could also modulate membrane penetration mediated by VP5*, either directly or indirectly, thereby blocking infection. Because VP4 determines the cellular entry pathway and subcellular compartment from which rotavirus escapes into the cytosol (46), defensin-bound VP4 may direct the virus into a non-permissive cellular pathway that does not trigger VP5*-mediated membrane penetration. Alternatively, defensin interactions with the host membrane in a rotavirus-containing compartment could disrupt cellular factors needed to trigger uncoating, such as calcium levels (46, 47). This would effectively convert a permissive pathway into a non-permissive pathway. More directly, α-defensins could perturb the conformational changes in VP5* or block access of VP5* loops to the membrane that are required for membrane penetration (19).
Although our use of viral strains is somewhat broad, one caveat of our studies is that they are limited to MA104 cells. Rotaviral entry remains incompletely understood, cell- and strain-specific entry mechanisms have been reported, and the nature of the membranous compartment that is penetrated by VP5* has not been definitively identified (19, 46). Thus, confirming these findings in other cell types, particularly more relevant intestinal cells, is needed. At the same time, the study of α-defensins as novel inhibitors may provide valuable tools to dissect entry mechanisms. Prior studies of α-defensin interactions with non-enveloped viruses have focused on DNA viruses, where we and others have observed alterations in intracellular trafficking in the presence of defensins that preclude the genomes of AdVs (11, 34, 35, 48), polyomaviruses (49), and papillomaviruses (36, 38, 39) from reaching the nucleus. Rotavirus is the first non-enveloped virus that replicates in the cytoplasm to be shown to be neutralized by α-defensins. The intracellular routes of RNA viral genomes to reach their replicative niche are different than those of DNA viruses, and additional studies of rotavirus may reveal novel defensin-dependent mechanisms.
Although our data strongly point to VP4 as a critical determinant for neutralization, we considered other possible mechanisms. The steepness of the Hill slopes of the inhibition curves suggests cooperative binding, which differs between defensin/virus combinations and may reflect the involvement of additional binding partners on the viral capsid. VP7 makes up the facets of the outer layer of the capsid, which become destabilized during internalization in part due to the loss of Ca2+ (50). Thus, α-defensins could block infection by crosslinking or stabilizing VP7 trimers, as has been observed for neutralizing antibodies (50), or by bridging a VP4-VP7 interaction, thereby constraining VP5 and preventing uncoating and membrane penetration. Because there is some evidence that α-defensins function as lectins (1, 6) and because VP7 is a glycoprotein, a mechanism that involves both a protein-protein interaction with VP4 and a lectin-glycan interaction with VP7 is plausible. EDIM and RRV VP7 differ not only in sequence but also in N-linked glycosylation. However, the one N-linked glycosylation site on RRV VP7 (N69) is conserved between them and is not surface accessible in intact virus. Thus, the involvement of VP7 in the VP4-dependent neutralization mechanism through strain-specific glycans is still formally possible but unlikely.
A role for VP4 as the determinant for enhancement is less clear. A likely mechanism for enhanced infection is that the defensin/virus interaction leads to enhanced cell binding. This would in turn lead to increased uptake and infection if the virus enters a cellular pathway conducive to uncoating and membrane penetration. We have previously reported enhanced cell binding of both mouse and human AdVs, even for defensin/virus combinations that result in neutralized infection (8, 11, 34). Increased cell binding mediated by α-defensins is consistent with the marked loss of enhancement when EDIM is bound to cells prior to the addition of RMAD4 and HD5 (protocol 2) compared to when defensin/virus complexes are added to cells (protocol 1). However, we note that Crp2 still potently enhances EDIM under protocol 2, perhaps due to Crp2 reducing the off-rate of the virus-receptor interaction. Increased association of the virus and cell in the presence of defensin need not be mediated by VP4-receptor interactions but could be a result of defensins bridging the virus and cell through binding to cellular proteins, glycans, or the lipid bilayer. For example, DS-1 has a surface-exposed glycosylation site on VP7 (N146), located in the middle of the VP7 trimer, that is not present in the other viruses studied in this paper. A defensin dimer could bind atop the VP7 trimer and bridge defensin-specific receptor interactions. Alternatively, the α-defensin could augment VP5*-dependent membrane penetration, leading to more efficient escape into the cytosol. The nature of α-defensin interactions with cells that enhance viral and bacterial infection remains elusive, and further studies of enhanced rotaviral infection may provide critical insight. Moreover, the mechanisms for enhancement and neutralization are not mutually exclusive. Thus, resistant viruses may contain VP4 proteins to which specific α-defensins cannot bind or could reflect a balance between these putative enhancement mechanisms and one or more of the neutralization mechanisms described above.
Based on our prior studies of mouse and human AdVs and consistent with early studies of α-defensin interactions with non-enveloped viruses, we hypothesized that the resistance of fecal-orally transmitted viruses reflects evolution driven by defensins. By this process, the virus evolves to be resistant to or enhanced by the constitutively expressed enteric α-defensins of its host while remaining susceptible to neutralization by myeloid α-defensins (11, 12). Our analysis of RRV, and to some extent EDIM, DS-1, and Wa, supports this hypothesis. Furthermore, the differential effects of HD5 on Wa (G1P[8]), DS-1 (G2P[4]), and animal rotaviruses in combination with variation in HD5 expression could potentially contribute to the epidemiology of human rotaviruses. While human Paneth cells develop during gestation (51), the global variance in the level of HD5 expression is not known, and differences in HD5 mRNA expression in specific populations because of nutritional state, body mass index, and enteropathy have been described (52 – 54). Because the relative prevalence of Wa-like versus DS-1-like genotypes varies over time and location in the human population, it is difficult to determine if the resistance of P[8]-containing viruses to HD5 versus enhanced infection of P[4]-containing viruses by HD5 is consequential. In addition, circulating P[8] (and presumably P[4]) strains have evolved compared to the original Wa P[8] that we studied and may no longer be just resistant HD5 (55). Furthermore, the variation in HD5 expression combined with the differential effects of HD5 on animal strains could modulate susceptibility to zoonotic rotaviral infection. Finally, HD5 may impact vaccine efficacy. The Rotarix vaccine and one component of the RotaTeq vaccine both contain G1P[8]. The effect of HD5 on the P[5] VP4 of the remaining four components of the RotaTeq vaccine remains to be determined. Therefore, a more expansive analysis of strains differing by VP4, including clinical isolates, is warranted.
In summary, we examined the effects of α-defensins—homologous and heterologous, enteric and myeloid—on rotaviral infections. All of the group A rotaviruses that we tested are resistant to or enhanced by enteric α-defensins from their host species. Although the exact mechanisms of enhancement and neutralization are yet to be uncovered, identification of VP4 as a determinant for defensin-mediated neutralization and elucidation of the differential activities of α-defensins will focus future efforts to understand the molecular properties of both partners that dictate the outcome of the interaction.
MATERIALS AND METHODS
Cells and viruses
Monkey kidney epithelial MA104 cells were a kind gift from Monica McNeal of Cincinnati Children’s Hospital Medical Center (Cincinnati, OH, USA) and HeLa cells were purchased from the American Type Culture Collection. Cells were propagated in DMEM supplemented with 0.1 mM non-essential amino acids, 100 units/mL penicillin, 100 µg/mL streptomycin, 4 mM L-glutamine, and 10% fetal bovine serum (complete DMEM). HPV16 pseudovirus (PsV) encapsidating an eGFP reporter plasmid was produced via transfection and maturation as previously described (38). RRV, simian rotavirus (SA11), and EDIM were kind gifts from Monica McNeal. Human rotavirus isolate Wa was obtained from ATCC, while DS-1 was generously gifted by Mary Estes of Baylor College of Medicine (Houston, TX, USA). ETD and reassortant viruses D1/5, EA 4-1-2, B7/2, and D6/2 were previously described (42, 43).
To create stocks of rotaviruses, the inoculum was trypsin-activated by incubation for 45–60 min at 37°C with 10 µg/mL type IX-S EDTA-free porcine trypsin (Sigma-Aldrich) and then added to a monolayer of MA104 cells. After incubation for 1 h at 37°C, the inoculum was removed, and the media was replaced with serum-free DMEM containing 0.5 µg/mL type IX-S EDTA-free porcine trypsin. The culture was harvested after incubation for 3–7 days at 37°C, upon the appearance of complete cytopathic effect. Cells were lysed by three freeze-thaw cycles and centrifuged to remove cell debris. Viral lysate was stored in aliquots at −80°C. The identity of each virus stock was verified by isolating viral genomic RNA (Thermo Scientific GeneJet Viral DNA and RNA Purification Kit), amplifying gene segment 9 (VP7) for all viruses as well as gene segment 4 (VP4) for the reassortant viruses and DS-1 by RT-PCR (Promega GoScript Reverse Transcriptase) using sequence-specific primers followed by end point PCR, and sequencing. Primers used for amplification and sequencing are listed in Table 1. DS-1, Wa, and SA11 were also verified by whole-genome sequencing.
TABLE 1.
Primers used to verify the identity of rotaviruses
| Viruses | Segment | RT and PCR primers | Sequencing primers |
|---|---|---|---|
| B7/2, D1/5, D6/2, EA 4-1-2 | 4 | GGCTATAAAATGGCTTCGCTCATTTATAG, GGTCACATCCTCTAGAAATTGCTTACAG | GCTAATGCTTCCCAAACACAATGG |
| DS-1 | 4 | TATGCTCCAGTTAACTGGGGAC, GGCTGATAATGACCTAACATACACC | TATGCTCCAGTTAACTGGGGAC |
| EDIM | 4 | GGCTATAAAATGGCTTCACTCATTTATAGAC, GGTCACATCCTCTAGACACTGC | CGTACCTACACATTGTTCGG, CAGAGATCTCTTTCAAACCAGCG, GTGCCGTCCAACGATAACTATC, AGCGTCAGAGAAGTTCATTCCG, GAAGAGTAGTATGCCTGACCTGC |
| ETD | 4 | GGCTATAAAATGGCTTCACTCATTTATAGAC, GGTCACATCCTCTAGACACTGC | CGTACCTACACATTGTTCGG |
| RRV | 4 | GGCTATAAAATGGCTTCGCTCATTTATAG, GGTCACATCCTCTAGAAATTGCTTACAG | GCTAATGCTTCCCAAACACAATGG, CGTATACACGAGATGGAGAGGAGG, AACGACAGTTGGGTGAACTTAGAG, GTCACTGAAGCTTCAGAGAAGTTC, GTAGTACTTAGTGGTCACATTCGG |
| B7/2, D1/5, EA 4-1-2, RRV | 9 | GGCTTTAAAAGAGAGAATTTC, GGTCACATCATACAATTCTAA | TTCCGTCTGGCTAGCGGTT |
| D6/2, EDIM, ETD | 9 | GGCTTTAAAAGAGAGAATTTC, GGTCACATCATACAGCTGTAA | TTCCGTTTGGCTAGCGGTT |
| DS-1 | 9 | GGCTTTAAAAGAGAGAATTTC, GGTCACATCATACAAATCTGA | TTCCGTCTGGCTAGCGGTT |
| SA11 | 9 | GGCTTTAAAAGAGAGAATTTC, GGTCACATCATACAATTCTAA | TTCCGTTTGGCTAGCGGTT |
| Wa | 9 | GGCTTTAAAAGAGAGAATTTC, GGTCACATCATACAATTCTAA | TTCCGTCTGGCTAACGGTT |
α-Defensin peptides
Peptides (see Fig. 1B for sequences) were synthesized by CPC Scientific (Sunnyvale, CA, USA). Most peptides (HD5, Crp2, RMAD4, RED1, and RED3) were initially dissolved at 2 mg/mL in 8 M GuHCl, 12 mM reduced glutathione, and 1.2 mM oxidized glutathione in water and were then diluted with 0.25 M NaHCO3 in water to a final concentration of 0.5 mg/mL peptide, 2 M guanidine hydrochloride, 3 mM reduced glutathione, 0.3 mM oxidized glutathione, and 0.19 M NaHCO3, pH 8.3. However, HNP1 and RMAD1 were initially dissolved at 0.5 mg/mL in 50% N,N-dimethylformamide (DMF), 4 M urea, 6 mM reduced glutathione, and 0.6 mM oxidized glutathione in water and were then diluted with 0.2 M NaHCO3 in water to a final concentration of 0.25 mg/mL in 25% N,N-dimethylformamide (DMF), 2 M urea, 3 mM reduced glutathione, 0.3 mM oxidized glutathione, and 0.1 M NaHCO3, pH 8.1. Note that the ratio of reduced to oxidized glutathione was inadvertently adjusted to 5:1 for one preparation each of HNP1 and RMAD1, although this did not affect the final product. After overnight incubation at room temperature (RT), peptides were purified by RP-HPLC over an acetonitrile gradient using a Hichrom Prevail C18 column (5 µm, 22 × 250 mm) or a Waters Delta-Pak C18 column (5 µm, 19 × 300 mm, 300 Å). Fractions containing the correctly folded species were lyophilized, resuspended in water, quantified by UV absorbance at 280 nm using calculated molar extinction coefficients (56), adjusted to approximately 1 mM with water, and stored in aliquots at −80°C. For all peptides except RED1, the correctly folded species was found in a distinct peak with a shorter retention time than unfolded or partially folded intermediates. However, the RED1 chromatogram was more complex, and the correctly folded species was identified in a peak with an intermediate retention time. Purity was assessed by analytical RP-HPLC performed with an Agilent Zorbax 300SB-C18 column (5 µm, 2.1 × 250 mm) or a Thermo Scientific Acclaim 120 C18 column (5 µm, 4.6 × 100 mm, 120 Å). Formation of three disulfide bonds was verified by electrospray ionization mass spectrometry in high-resolution mode on a Thermo Scientific LTQ Orbitrap or Orbitrap Elite mass spectrometer. Average yield of purified defensins from 20 to 59 mg of partially purified (81.0–86.3%), synthesized peptide was ~15%. Defensin purification and analysis were performed by the Proteomics & Metabolomics Core Shared Resource of the Fred Hutch/University of Washington Cancer Consortium. Additional mass spectrometry analysis was performed by the Mass Spectrometry Center of the Department of Medicinal Chemistry of the University of Washington School of Pharmacy.
Quantification of viral infection
Rotavirus was activated with 10 µg/mL type IX-S EDTA-free porcine trypsin for 1 h at 37°C. Rotavirus or HPV PsV was then diluted to a concentration that was predetermined to yield 50–80% of maximal signal for inhibition studies and 10–25% of maximal infection for enhancement studies in the absence of defensin. For protocol 1, rotavirus or HPV PsV was incubated with defensin on ice for 45 min in serum-free DMEM (SFM). The mixture (35 or 50 µL) was then added to a well containing a confluent monolayer of MA104 (for rotavirus) or HeLa (for HPV PsV) cells in clear bottom, black wall 96-well plates (PerkinElmer) that had been washed twice with SFM. For protocol 2, 35 or 50 µL of RV was added to a well of MA104 cells and incubated while rocking for 1 h at 4°C. The cells were then washed two times with cold SFM, 35 or 50 µL defensin in SFM was added, and samples were incubated while rocking for 45 min at 4°C. For both protocols, samples were then incubated while rocking for 2 h at 37°C, washed with SFM, and incubated for an additional 20–24 h with 100 µL/well complete DMEM. For HPV PsV samples, the total monolayer fluorescence of each well was quantified with a Typhoon 9400 (GE Healthcare) variable mode imager. Rotavirus samples were fixed with 2% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min at RT, washed two times with PBS, and then permeabilized and quenched with 20 mM glycine, 0.5% Triton X-100 in PBS for 20 min at RT. Cells were stained with a 1,000-fold dilution of a primary anti-rotavirus antibody in 0.05% Tween-80/1% bovine serum albumin (BSA) in PBS for 1 h at RT. This antibody was produced in rabbits using an inoculum of mostly double-layered Wa, SA11, and ST3 particles and was a kind gift from Monica McNeal (57). Cells were washed three times with 0.05% Tween-80 in PBS and stained with a 1,000-fold dilution of Alexa Fluor 488-conjugated goat anti-rabbit antibody (Fisher Scientific) in 0.05% Tween-80/1% BSA in PBS for 1 h at RT. Cells were washed once with 0.05% Tween-80 in PBS and then two times with PBS. Total monolayer fluorescence of each well was quantified with either a Typhoon 9400 (GE Healthcare) or Sapphire (Azure) variable mode image. For all samples, Fiji (version 2.1.0/1.53c) was used to quantify background-subtracted total monolayer fluorescence (58). Data are shown as a percent of control infection in the absence of defensin.
Statistical analysis
Statistical analysis and non-linear regression of log-transformed data were performed using GraphPad Prism 9.2.0. Hill slopes are best fit values. For enhanced or resistant infection data, results of ordinary two-way analysis of variance with Sídák’s multiple comparisons test, with individual variances computed for each concentration, comparing protocol 1 and protocol 2 from 1.25 to 20 µM defensin are indicated by asterisks: *, P = 0.01–0.05; **, P = 0.001– 0.01; ***, P = 0.0001–0.001; ****, P < 0.0001. Unmarked comparisons within the specified analysis range are not significant (P > 0.05). For neutralized infection data, log IC50 values between protocol 1 and protocol 2 were compared using the extra sum-of-squares F test, and the P value of the comparison is given.
ACKNOWLEDGMENTS
The authors thank Youngmee Sul and Beth Bromme for technical assistance with HPV experiments. The authors are extremely grateful to Daniel Pfalmer and Alexander L. Greninger, and the NGS core lab at the University of Washington’s Clinical Virology Lab for whole-genome sequencing of Wa, DS-1, and SA11.
This work was supported by R01 AI104920 (to J.G.S.), F30 AI140620 (to K.D.), and R01 AI125249 (to H.B.G.) from the National Institute of Allergy and Infectious Diseases (www.niaid.nih.gov) and by the Office of the Director, National Institutes of Health (www.nih.gov/institutes-nih/nih-office-director) under Award Number S10 OD026741 (to J.G.S.). C.T.H. and K.D. were also supported by Public Health Service, National Research Service Award T32 AI083203 from the National Institute for Allergy and Infectious Diseases. H.B.G. was also supported by Merit Grant GRH0022 from the U.S. Department of Veterans Affairs (www.va.gov). Additional support to J.G.S. in the form of pilot awards and subsidized core service was provided by NIH grants P51 OD010425 from the Office of Research Infrastructure Programs (orip.nih.gov), UL1 TR000423 from the National Center for Advancing Translational Sciences (ncats.nih.gov), and P30 CA015704 from the National Cancer Institute (www.cancer.gov). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Jason G. Smith, Email: jgsmith2@uw.edu.
Stacey Schultz-Cherry, St. Jude Children's Research Hospital, Memphis, Tennessee, USA .
REFERENCES
- 1. Lehrer RI, Lu W. 2012. alpha-defensins in human innate immunity. Immunol Rev 245:84–112. doi: 10.1111/j.1600-065X.2011.01082.x [DOI] [PubMed] [Google Scholar]
- 2. Das S, Nikolaidis N, Goto H, McCallister C, Li J, Hirano M, Cooper MD. 2010. Comparative genomics and evolution of the alpha-defensin multigene family in primates. Mol Biol Evol 27:2333–2343. doi: 10.1093/molbev/msq118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Selsted ME, Ouellette AJ. 2005. Mammalian defensins in the antimicrobial immune response. Nat Immunol 6:551–557. doi: 10.1038/ni1206 [DOI] [PubMed] [Google Scholar]
- 4. Xu D, Lu W. 2020. Defensins: a double-edged sword in host immunity. Front Immunol 11:764. doi: 10.3389/fimmu.2020.00764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Holly MK, Diaz K, Smith JG. 2017. Defensins in viral infection and pathogenesis. Annu Rev Virol 4:369–391. doi: 10.1146/annurev-virology-101416-041734 [DOI] [PubMed] [Google Scholar]
- 6. Wilson SS, Wiens ME, Smith JG. 2013. Antiviral mechanisms of human defensins. J Mol Biol 425:4965–4980. doi: 10.1016/j.jmb.2013.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brice DC, Diamond G. 2020. Antiviral activities of human host defense peptides. Curr Med Chem 27:1420–1443. doi: 10.2174/0929867326666190805151654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smith JG, Silvestry M, Lindert S, Lu W, Nemerow GR, Stewart PL, Sherry B. 2010. Insight into the mechanisms of adenovirus capsid disassembly from studies of defensin neutralization. PLoS Pathog 6:e1000959. doi: 10.1371/journal.ppat.1000959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Holly MK, Smith JG. 2018. Adenovirus infection of human enteroids reveals interferon sensitivity and preferential infection of goblet cells. J Virol 92:e00250-18. doi: 10.1128/JVI.00250-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tartaglia LJ, Badamchi-Zadeh A, Abbink P, Blass E, Aid M, Gebre MS, Li Z, Pastores KC, Trott S, Gupte S, Larocca RA, Barouch DH. 2019. Alpha-defensin 5 differentially modulates adenovirus vaccine vectors from different serotypes in vivo. PLoS Pathog 15:e1008180. doi: 10.1371/journal.ppat.1008180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diaz K, Hu CT, Sul Y, Bromme BA, Myers ND, Skorohodova KV, Gounder AP, Smith JG. 2020. Defensin-driven viral evolution. PLoS Pathog 16:e1009018. doi: 10.1371/journal.ppat.1009018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wilson SS, Bromme BA, Holly MK, Wiens ME, Gounder AP, Sul Y, Smith JG. 2017. Alpha-defensin-dependent enhancement of enteric viral infection. PLoS Pathog 13:e1006446. doi: 10.1371/journal.ppat.1006446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gounder AP, Myers ND, Treuting PM, Bromme BA, Wilson SS, Wiens ME, Lu W, Ouellette AJ, Spindler KR, Parks WC, Smith JG. 2016. Defensins potentiate a neutralizing antibody response to enteric viral infection. PLoS Pathog 12:e1005474. doi: 10.1371/journal.ppat.1005474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Crawford SE, Ramani S, Tate JE, Parashar UD, Svensson L, Hagbom M, Franco MA, Greenberg HB, O’Ryan M, Kang G, Desselberger U, Estes MK. 2017. Rotavirus infection. Nat Rev Dis Primers 3:17083. doi: 10.1038/nrdp.2017.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Troeger C, Khalil IA, Rao PC, Cao S, Blacker BF, Ahmed T, Armah G, Bines JE, Brewer TG, Colombara DV, Kang G, Kirkpatrick BD, Kirkwood CD, Mwenda JM, Parashar UD, Petri WA, Riddle MS, Steele AD, Thompson RL, Walson JL, Sanders JW, Mokdad AH, Murray CJL, Hay SI, Reiner RC. 2018. Rotavirus vaccination and the global burden of rotavirus diarrhea among children younger than 5 years. JAMA Pediatr 172:958–965. doi: 10.1001/jamapediatrics.2018.1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Martella V, Bányai K, Matthijnssens J, Buonavoglia C, Ciarlet M. 2010. Zoonotic aspects of rotaviruses. Vet Microbiol 140:246–255. doi: 10.1016/j.vetmic.2009.08.028 [DOI] [PubMed] [Google Scholar]
- 17. Hungerford D, Vivancos R, EuroRotaNet network members, Read JM, Pitzer VE, Cunliffe N, French N, Iturriza-Gómara M. 2016. In-season and out-of-season variation of rotavirus genotype distribution and age of infection across 12 European countries before the introduction of routine vaccination, 2007/08 to 2012/13. Euro Surveill 21. doi: 10.2807/1560-7917.ES.2016.21.2.30106 [DOI] [PubMed] [Google Scholar]
- 18. Leshem E, Lopman B, Glass R, Gentsch J, Bányai K, Parashar U, Patel M. 2014. Distribution of rotavirus strains and strain-specific effectiveness of the rotavirus vaccine after its introduction: a systematic review and meta-analysis. Lancet Infect Dis 14:847–856. doi: 10.1016/S1473-3099(14)70832-1 [DOI] [PubMed] [Google Scholar]
- 19. Herrmann T, Torres R, Salgado EN, Berciu C, Stoddard D, Nicastro D, Jenni S, Harrison SC. 2021. Functional refolding of the penetration protein on a non-enveloped virus. Nature 590:666–670. doi: 10.1038/s41586-020-03124-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ramani S, Hu L, Venkataram Prasad BV, Estes MK. 2016. Diversity in rotavirus-host glycan interactions: a "Sweet" spectrum. Cell Mol Gastroenterol Hepatol 2:263–273. doi: 10.1016/j.jcmgh.2016.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Borodavka A, Desselberger U, Patton JT. 2018. Genome packaging in multi-segmented dsRNA viruses: distinct mechanisms with similar outcomes. Curr Opin Virol 33:106–112. doi: 10.1016/j.coviro.2018.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kanai Y, Komoto S, Kawagishi T, Nouda R, Nagasawa N, Onishi M, Matsuura Y, Taniguchi K, Kobayashi T. 2017. Entirely plasmid-based reverse genetics system for rotaviruses. Proc Natl Acad Sci U S A 114:2349–2354. doi: 10.1073/pnas.1618424114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Daher KA, Selsted ME, Lehrer RI. 1986. Direct inactivation of viruses by human granulocyte defensins. J Virol 60:1068–1074. doi: 10.1128/JVI.60.3.1068-1074.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ghosh D, Porter E, Shen B, Lee SK, Wilk D, Drazba J, Yadav SP, Crabb JW, Ganz T, Bevins CL. 2002. Paneth cell trypsin is the processing enzyme for human defensin-5. Nat Immunol 3:583–590. doi: 10.1038/ni797 [DOI] [PubMed] [Google Scholar]
- 25. Tang YQ, Yuan J, Miller CJ, Selsted ME. 1999. Isolation, characterization, cDNA cloning, and antimicrobial properties of two distinct subfamilies of alpha-defensins from rhesus macaque leukocytes. Infect Immun 67:6139–6144. doi: 10.1128/IAI.67.11.6139-6144.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vasudevan S, Yuan J, Osapay G, Tran P, Tai K, Liang W, Kumar V, Selsted ME, Cocco MJ. 2008. Synthesis, structure, and activities of an oral mucosal alpha-defensin from rhesus macaque. J Biol Chem 283:35869–35877. doi: 10.1074/jbc.M806915200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tanabe H, Yuan J, Zaragoza MM, Dandekar S, Henschen-Edman A, Selsted ME, Ouellette AJ. 2004. Paneth cell alpha-defensins from rhesus macaque small intestine. Infect Immun 72:1470–1478. doi: 10.1128/IAI.72.3.1470-1478.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shanahan MT, Tanabe H, Ouellette AJ. 2011. Strain-specific polymorphisms in Paneth cell alpha-defensins of C57BL/6 mice and evidence of vestigial myeloid alpha-defensin pseudogenes. Infect Immun 79:459–473. doi: 10.1128/IAI.00996-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. doi: 10.1093/bioinformatics/8.3.275 [DOI] [PubMed] [Google Scholar]
- 30. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu Z, Ericksen B, Tucker K, Lubkowski J, Lu W. 2004. Synthesis and characterization of human alpha-defensins 4-6. J Pept Res 64:118–125. doi: 10.1111/j.1399-3011.2004.00179.x [DOI] [PubMed] [Google Scholar]
- 32. Wu Z, Powell R, Lu W. 2003. Productive folding of human neutrophil alpha-defensins in vitro without the pro-peptide. J Am Chem Soc 125:2402–2403. doi: 10.1021/ja0294257 [DOI] [PubMed] [Google Scholar]
- 33. Tenge VR, Gounder AP, Wiens ME, Lu W, Smith JG. 2014. Delineation of interfaces on human alpha-defensins critical for human adenovirus and human papillomavirus inhibition. PLoS Pathog 10:e1004360. doi: 10.1371/journal.ppat.1004360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gounder AP, Wiens ME, Wilson SS, Lu W, Smith JG. 2012. Critical determinants of human alpha-defensin 5 activity against non-enveloped viruses. J Biol Chem 287:24554–24562. doi: 10.1074/jbc.M112.354068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith JG, Nemerow GR. 2008. Mechanism of adenovirus neutralization by human alpha-defensins. Cell Host Microbe 3:11–19. doi: 10.1016/j.chom.2007.12.001 [DOI] [PubMed] [Google Scholar]
- 36. Buck CB, Day PM, Thompson CD, Lubkowski J, Lu W, Lowy DR, Schiller JT. 2006. Human alpha-defensins block papillomavirus infection. Proc Natl Acad Sci U S A 103:1516–1521. doi: 10.1073/pnas.0508033103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gulati NM, Miyagi M, Wiens ME, Smith JG, Stewart PL. 2019. Alpha-defensin HD5 stabilizes human papillomavirus 16 capsid/core interactions. Pathog Immun 4:196–234. doi: 10.20411/pai.v4i2.314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wiens ME, Smith JG. 2017. Α-defensin HD5 inhibits human papillomavirus 16 infection via capsid stabilization and redirection to the lysosome. mBio 8:e02304-16. doi: 10.1128/mBio.02304-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wiens ME, Smith JG. 2015. Alpha-defensin HD5 inhibits furin cleavage of human papillomavirus 16 L2 to block infection. J Virol 89:2866–2874. doi: 10.1128/JVI.02901-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Malherbe H, Harwin R. 1963. The cytopathic effects of vervet monkey viruses. S Afr Med J 37:407–411. [PubMed] [Google Scholar]
- 41. Kumar S, Stecher G, Suleski M, Hedges SB. 2017. Timetree: a resource for timelines timetrees, and divergence times. Mol Biol Evol 34:1812–1819. doi: 10.1093/molbev/msx116 [DOI] [PubMed] [Google Scholar]
- 42. Broome RL, Vo PT, Ward RL, Clark HF, Greenberg HB. 1993. Murine rotavirus genes encoding outer capsid proteins VP4 and VP7 are not major determinants of host range restriction and virulence. J Virol 67:2448–2455. doi: 10.1128/JVI.67.5.2448-2455.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Feng N, Yasukawa LL, Sen A, Greenberg HB. 2013. Permissive replication of homologous murine rotavirus in the mouse intestine is primarily regulated by VP4 and NSP1. J Virol 87:8307–8316. doi: 10.1128/JVI.00619-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Salzman NH, Bevins CL. 2013. Dysbiosis--a consequence of Paneth cell dysfunction. Semin Immunol 25:334–341. doi: 10.1016/j.smim.2013.09.006 [DOI] [PubMed] [Google Scholar]
- 45. Tanabe H, Ouellette AJ, Cocco MJ, Robinson WE. 2004. Differential effects on human immunodeficiency virus type 1 replication by alpha-defensins with comparable bactericidal activities. J Virol 78:11622–11631. doi: 10.1128/JVI.78.21.11622-11631.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Arias CF, López S. 2021. Rotavirus cell entry: not so simple after all. Curr Opin Virol 48:42–48. doi: 10.1016/j.coviro.2021.03.011 [DOI] [PubMed] [Google Scholar]
- 47. Salgado EN, Garcia Rodriguez B, Narayanaswamy N, Krishnan Y, Harrison SC. 2018. Visualization of calcium ion loss from rotavirus during cell entry. J Virol 92:e01327-18. doi: 10.1128/JVI.01327-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nguyen EK, Nemerow GR, Smith JG. 2010. Direct evidence from single-cell analysis that human {alpha}-defensins block adenovirus uncoating to neutralize infection. J Virol 84:4041–4049. doi: 10.1128/JVI.02471-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zins SR, Nelson CDS, Maginnis MS, Banerjee R, O’Hara BA, Atwood WJ. 2014. he human alpha defensin HD5 neutralizes JC polyomavirus infection by reducing endoplasmic reticulum traffic and stabilizing the viral capsid. J Virol 88:948–960. doi: 10.1128/JVI.02766-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Aoki ST, Settembre EC, Trask SD, Greenberg HB, Harrison SC, Dormitzer PR. 2009. Structure of rotavirus outer-layer protein VP7 bound with a neutralizing Fab. Science 324:1444–1447. doi: 10.1126/science.1170481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Heida FH, Beyduz G, Bulthuis MLC, Kooi EMW, Bos AF, Timmer A, Hulscher JBF. 2016. Paneth cells in the developing gut: when do they arise and when are they immune competent? Pediatr Res 80:306–310. doi: 10.1038/pr.2016.67 [DOI] [PubMed] [Google Scholar]
- 52. Dhaliwal W, Shawa T, Khanam M, Jagatiya P, Simuyandi M, Ndulo N, Bevins CL, Sanderson IR, Kelly P. 2010. Intestinal antimicrobial gene expression: impact of micronutrients in malnourished adults during a randomized trial. J Infect Dis 202:971–978. doi: 10.1086/655903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dhaliwal W, Bajaj-Elliott M, Kelly P. 2003. Intestinal defensin gene expression in human populations. Mol Immunol 40:469–475. doi: 10.1016/s0161-5890(03)00156-1 [DOI] [PubMed] [Google Scholar]
- 54. Wehkamp J, Salzman NH, Porter E, Nuding S, Weichenthal M, Petras RE, Shen B, Schaeffeler E, Schwab M, Linzmeier R, Feathers RW, Chu H, Lima H, Fellermann K, Ganz T, Stange EF, Bevins CL. 2005. Reduced Paneth cell alpha-defensins in ileal Crohn's disease. Proc Natl Acad Sci U S A 102:18129–18134. doi: 10.1073/pnas.0505256102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Velasquez DE, Jiang B. 2019. Evolution of P[8], P[4], and P[6] VP8* genes of human rotaviruses globally reported during 1974 and 2017: possible implications for rotavirus vaccines in development. Hum Vaccin Immunother 15:3003–3008. doi: 10.1080/21645515.2019.1619400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci 4:2411–2423. doi: 10.1002/pro.5560041120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McNeal MM, Broome RL, Ward RL. 1994. Active immunity against rotavirus infection in mice is correlated with viral replication and titers of serum rotavirus IgA following vaccination. Virology 204:642–650. doi: 10.1006/viro.1994.1579 [DOI] [PubMed] [Google Scholar]
- 58. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]