ABSTRACT
Human norovirus (HuNoV) is the main cause of acute non-bacterial gastroenteritis worldwide. There is no vaccine currently available to prevent HuNoV infection. HuNoV is a highly mutated virus, and its genetic diversity and the lack of cross-protection between different genotypes hinder the broadly protective vaccine development. Among various genotypes, GI.1 is the prototype strain, and GII.4 currently predominates the prevalence of HuNoV. In this work, guided by the structural alignment of GI.1 and GII.4 HuNoV capsid proteins, several chimeric virus-like particles (VLPs) were designed to achieve cross-immunity against these two HuNoV genotypes. The neutralizing epitopes of HuNoV have been identified to be mainly located at the loop regions of VP1 protein exposed on the HuNoV surface. In this study, the exposed loops of GII.4 VP1 protein were grafted into the scaffold of GI.1 genotype to produce the chimeric VLPs. The designed chimeric VLPs were recombinantly expressed by the Hansenula polymorpha expression system developed by our laboratory. Mice were immunized with the chimeric VLPs plus aluminum adjuvant, and then the antibody responses were detected by using the enzyme-linked immunosorbent assay and the histo-blood group antigen-VLP interaction blocking assay. The experimental results show that two of the designed chimeric VLPs induced cross-reactive IgG and cross-blocking antibodies against both the parental GI.1 and GII.4 genotypes of HuNoV. The results also imply that the transplant site design is important to maintain the immunogenicity of foreign epitopes on the scaffold carrier. Our studies may provide a valuable strategy for the development of cross-reactive HuNoV vaccines.
IMPORTANCE
Human norovirus (HuNoV) is highly infectious and can result in severe illnesses in the elderly and children. So far, there is no effective antiviral drug to treat HuNoV infection, and thus, the development of HuNoV vaccines is urgent. However, NoV evolves rapidly, and currently, at least 10 genogroups with numerous genotypes have been found. The genetic diversity of NoV and the lack of cross-protection between different genotypes pose challenges to the development of broadly protective vaccines. In this study, guided by structural alignment between GI.1 and GII.4 HuNoV VP1 proteins, several chimeric-type virus-like particles (VLPs) were designed through surface-exposed loop grafting. Mouse immunization studies show that two of the designed chimeric VLPs induced cross-immunity against both GI.1 and GII.4 HuNoVs. To our knowledge, this is the first designed chimeric VLPs that can induce cross-immune activities across different genogroups of HuNoV, which provides valuable strategies for the development of cross-reactive HuNoV vaccines.
KEYWORDS: human norovirus, chimeric virus-like particle, antigen design, epitope grafting, cross-immune activity
INTRODUCTION
Human norovirus (HuNoV) is the main pathogen causing acute viral gastroenteritis of humans, which is responsible for most of the epidemic outbreaks of acute non-bacterial gastroenteritis worldwide. HuNoV is estimated to result in 19–21 million diarrheal illnesses each year in the United States, in which 1.7–1.9 million require outpatient care, and about 70,000 infected people need hospitalization (1). In China, HuNoV infections continue to increase in recent years, and the outbreak of acute gastroenteritis caused by HuNoV has become a serious public health problem (2, 3). Throughout the world, it is estimated by the World Health Organization that 684 million cases of acute gastroenteritis and 212, 000 deaths were caused by HuNoV per year (4, 5). HuNov can infect both adults and children, in which the elderly (>65 years old) have a higher risk of mortality and the children (<5 years old) with higher morbidity (3, 6). Considering the severe threat of HuNoV to the health of people, it is necessary to develop effective methods to prevent HuNoV infections. Rational design of HuNoV vaccines is one of the important strategies to eliminate the threat of the virus.
According to the phylogenetic analysis, NoV can be classified into ten genogroups including GI, GII, GIII, GIV, GV, GVI, GVII, GVIII, GIX, and GX, of which three genogroups (GI, GII, and GIV) infect humans (7, 8). GI, GII, and GIV HuNoV can be further subdivided into 9, 26, and 2 genotypes, respectively (7, 8). Among these different genotypes of HuNoV, GI.1 is the original Norwalk virus (9), and GII.4 predominated most of the outbreaks of human acute gastroenteritis in the past few decades (7, 10). Many research groups have put efforts into the development of effective vaccines against HuNoV (11). However, HuNoV is a highly variable virus, and its genetic diversity hinders the development of broadly protective vaccines. Many epidemiological investigations have shown that HuNoV evolves rapidly, and every 2–3 years, on average, a new pandemic strain emerges with escape from the host’s immunity elicited by previously circulating HuNoV strains (12, 13). At present, several companies and study groups have developed HuNoV candidate vaccines in the stage of clinical evaluations or pre-clinical trials, in which many vaccines were designed to protect against multiple types of HuNoV with the multivalent combination strategy (11, 14, 15). Our group has also developed a bivalent GI.1/GII.4 recombinant HuNoV virus-like particle (VLP) vaccine that has entered Phase I (NCT04188691) and Phase II (NCT04941261) clinical trials in November 2019 and June 2021, respectively. Besides the multivalent combined vaccines, the construction of chimeric-type VLP is another novel and promising strategy to develop cross-reactive vaccines. However, this strategy is very challenging, and so far, only the chimeric VLPs from different strains within the same genotype or genogroup of HuNoV have been reported to successfully elicit cross-reactive immunity (16, 17). In this study, guided by the tertiary structure analysis, several chimeric VLPs were designed that incorporate the antigenic epitopes from different genogroups, i.e., GI.1 and GII.4, of HuNoV.
The structure of HuNoV capsid is composed of 180 copies of VP1 proteins, which form an icosahedral symmetrical shell. Each capsid protein VP1 can be divided into two domains, namely, S and P domains. The S domain is involved in the assembly of the capsid and constitutes the interior shell. The P domain forms the protrusions on the exterior of the capsid, which is responsible for the virus-host interactions and immune recognitions (18, 19). Many studies have been concerned with the antigenic characteristics of HuNoVs, and multiple blockade epitopes have been identified by experimental and bioinformatic methods (20 – 23). The structural and immunobiological investigations have shown that the most variable regions between the structures of different HuNoV genotypes are the loops on the P domain exposed to the surface of the virion, and most of the identified neutralizing epitopic sites are also located at these loop regions. Similar to HuNoV, the immunogenic epitopes of enterovirus also mainly lie in the loops on the capsid surface (24), and the chimeric VLPs with cross-immune responses have been successfully produced through loop grafting by Zhao et al. (25). In this study, the similar strategy of loop transplanting was used to design the chimeric VLPs for HuNoV.
In this study, based on the structural comparison between GI.1 and GII.4 capsid proteins, eight exposed loop regions on the GII.4 P domain were selected. Aiming at these 8 loops, a total of 15 schemes were designed to graft these exposed loop regions of GII.4 into GI.1 scaffold to construct the recombinant chimeric VLPs. The designed chimeric VLPs were expressed in the Hansenula polymorpha expression system developed by our laboratory and purified by using ion exchange chromatography. Then, BALB/c mice were injected with the chimeric HuNoV VLPs, and the serum from the immunized mice was collected. The enzyme-linked immunosorbent assay (ELISA) and the histo-blood group antigen (HBGA)-VLP interaction blocking assay were performed to evaluate the cross-immunity induced by the chimeric VLPs. The experimental results show that two of the chimeric VLPs can successfully elicit cross-reactive antibodies against both the parental GI.1 and GII.4 HuNoVs. Our studies may provide a valuable strategy for the development of novel vaccines with cross-immune reactions against different genotypes of HuNoV.
RESULTS
Design of chimeric VLPs guided by tertiary structure analysis
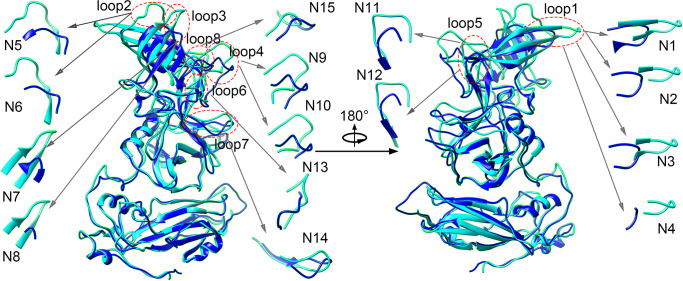
Many studies have shown that the exposed flexible loops on the P domain are the most variable regions between different genotypes of HuNoV, which also serve as the most important neutralizing epitopic sites (18 – 23). In this study, the chimeric VLPs were designed through the grafting of these exposed loops. In order to determine which and how these loop regions are grafted from the GII.4 P domain to replace the corresponding regions in GI.1 HuNoV, the structure alignment was performed between GII.4 and GI.1 VP1 proteins. The three-dimensional structures of GI.1 and GII.4 VP1 proteins with the protein data bank (PDB) accession codes 1ihm and 7k6v (26, 27), respectively, were compared by using UCSF Chimera software (28). The structural alignment is displayed in Fig. 1. It is found that the structure of the S domain is highly conserved, and the major structural divergences between GI.1 and GII.4 are the loops on the P domain. According to the structural comparison, eight loop regions with the most structural variety were chosen for loop grafting to produce the chimeric VLPs, as illustrated in Fig. 1. Many experimental studies have implied that these loops are involved in neutralizing immunogenicity. Taube et al. showed that loop2 and loop3 are the binding sites for neutralizing antibodies for murine NoV that is closely homologous to HuNoV (29). Allen et al. proved by experiments that loop3 and loop4 serve as the epitopic sites involved in antibody binding (30). Lindesmith et al. predicted loop2, loop3, loop4, loop6, and loop8 to be potential epitopic sites by using bioinformatic analysis (20). Cao et al. revealed by X-ray crystallographic experiments that loop1, loop2, and loop5 directly interact with the HBGA receptor (31). Koromyslova et al. and Carmona-Vicente et al. have found that loop5 is involved in the binding of the blockade monoclonal 10E9 and 3C3G3 antibodies (21, 22). In our study, aiming at these 8 loop regions, a total of 15 grafting schemes were designed to produce the chimeric VLPs, in which the loop regions were cut from GII.4 and incorporated into the scaffold GI.1 HuNoV. The designed transplanting schemes are displayed in Table 1. Four grafting schemes were designed for loop1, and two schemes for each of loop2–loop5. As to loop6–loop8, one scheme was designed for each loop.
Fig 1.
Structural alignment between GII.4 (cyan color) and GI.1 (blue color) VP1 proteins by using the UCSF Chimera software (28). Eight exposed loop regions (highlighted by the dotted red ellipses and marked by loop1–loop8 in the figure) with the most structural variety were chosen for loop grafting to produce the chimeric VLPs. Aiming at these 8 loops, a total of 15 chimeric schemes were designed. These 15 transplanting schemes are illustrated by the inserted sub-figures marked with N1–N15, in which the grafting residues of GII.4 are shown in cyan and the substituted residues on GI.1 are shown in blue, respectively. All the figures were drawn with the UCSF Chimera software (28).
TABLE 1.
The designed grafting schemes to construct the GII.4-GI.1 chimeric VLPs
| Scheme no. | Loops | Grafting residues of GII.4 | The residues to be substituted in GI.1 |
|---|---|---|---|
| N1 | loop1 | 336QTTRTDGSTRG346 | 333TQFGHSSQ340 |
| N2 | loop1 | 338TRTDGST344 | 334QFGHSS339 |
| N3 | loop1 | 337TTRTDGST344 | 334QFGHSS339 |
| N4 | loop1 | 338TRTDGS343 | 335FGH337 |
| N5 | loop2 | 367FETDTDRDFEA377 | 361ANGIGS366 |
| N6 | loop2 | 366QFETDTDRDF375 | 361ANGIG365 |
| N7 | loop3 | 290VTHITGSRNYTM301 | 295NGTV298 |
| N8 | loop3 | 291THITGSRNYT300 | 296GT297 |
| N9 | loop4 | 390QDGSTTHRNEP400 | 378PPSHPSGSQV387 |
| N10 | loop4 | 390QDGSTTHRNE399 | 378PPSHPSGSQ386 |
| N11 | loop5 | 440GCSGYPNM447 | 427GPGAY431 |
| N12 | loop5 | 440GCSGYPNMDL449 | 427GPGAYN432 |
| N13 | loop6 | 409SGRNTPNV416 | 396GSSITEAT403 |
| N14 | loop7 | 247EKLFTGPSSAFVVQ260 | 252SMGISPDNVQSV263 |
| N15 | loop8 | 352YTGSADFA359 | 346DTTPDTFV353 |
Expression, purification, and characterization of the designed chimeric VLPs
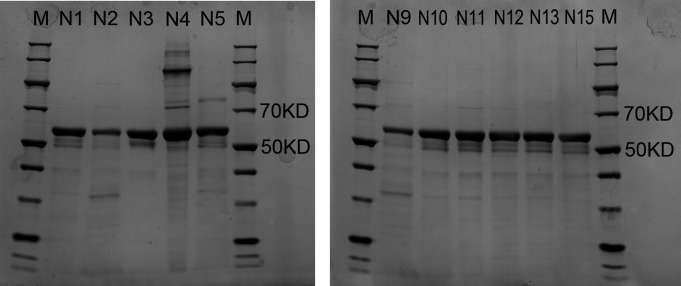
The designed chimeric proteins were expressed by using the Hansenula polymorpha expression platform developed by our laboratory, as described in our previous studies (32). Then, the chimeric VLPs were obtained by purification with chromatography. To validate the expression of the proteins, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out. The SDS-PAGE experimental results show that the specific band with the relative molecular weight (Mr) around 58 kD was observed for 11 protein samples (i.e., the schemes N1, N2, N3, N4, N5, N9, N10, N11, N12, N13, and N15). The experimental Mr value is well consistent with the theoretical value of VP1, which indicates that the chimeric VLPs are successfully expressed and purified for these 11 schemes, as shown in Fig. 2. However, except for these 11 schemes, SDS-PAGE experiments didn’t exhibit the specific band for the rest 4 designed schemes (i.e., N6, N7, N8, and N14). These four failed-expression schemes were removed in the following experiments.
Fig 2.
SDS-PAGE results for the 11 successfully expressed chimeric VLPs, including the N1, N2, N3, N4, N5, N9, N10, N11, N12, N13, and N15 constructs. In the experiment, the same amount of protein samples with 3 µg was loaded per lane. In this figure, lane M represents the molecular mass markers.
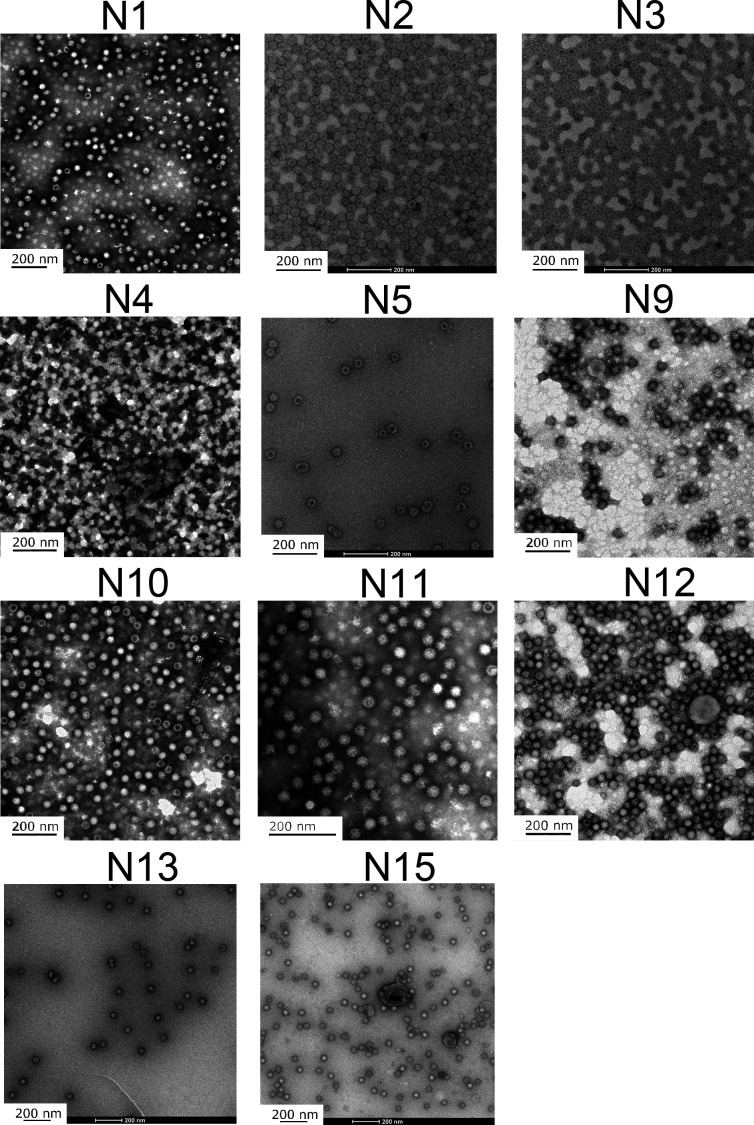
Furthermore, in order to determine whether the designed chimeric proteins successfully self-assemble into VLPs, the morphology of the purified proteins was observed with TEM, as shown in Fig. 3. It is found that all the 11 successfully purified chimeric proteins assemble into particles with uniform sizes and distributions. The diameter and dispersity of the VLPs for the 11 designed proteins were evaluated with the Z-average diameter (ZD) and the polydispersity index (PDI), respectively, by using the dynamic light scattering (DLS) method (33, 34). Three times repeated measurements were performed for each VLP, and the average values of ZD and PDI were calculated, as shown in Table 2. It is found that the diameters of the hydrous particles for the 11 chimeric VLPs are all in the range of 44–60 nm, and the PDI values are less than 0.220, which implies that the particles distribute in a narrow range of sizes. The above experimental results indicate that the 11 designed chimeric proteins can successfully self-assemble into VLPs.
Fig 3.
The particle morphology of the chimeric VLPs observed with TEM. The subfigures display the morphology of the N1, N2, N3, N4, N5, N9, N10, N11, N12, N13 and N15 VLPs, respectively.
TABLE 2.
The values of ZD and PDI for the 11 chimeric VLPs measured with DLS method b
| Scheme no. | Z-Average(d.nm) | PDI | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | Mean ± SD a | 1 | 2 | 3 | Mean ± SD a | |
| N1 | 54.22 | 46.77 | 44.90 | 48.63 ± 4.03 | 0.211 | 0.145 | 0.146 | 0.167 ± 0.031 |
| N2 | 48.30 | 46.63 | 45.31 | 46.75 ± 1.22 | 0.098 | 0.056 | 0.069 | 0.074 ± 0.018 |
| N3 | 42.75 | 42.52 | 43.26 | 42.84 ± 0.31 | 0.019 | 0.062 | 0.020 | 0.034 ± 0.020 |
| N4 | 52.65 | 51.50 | 51.14 | 51.76 ± 0.64 | 0.145 | 0.179 | 0.167 | 0.164 ± 0.014 |
| N5 | 58.36 | 58.32 | 58.43 | 58.37 ± 0.05 | 0.228 | 0.207 | 0.207 | 0.214 ± 0.010 |
| N9 | 45.11 | 45.42 | 44.40 | 44.98 ± 0.43 | 0.136 | 0.126 | 0.129 | 0.130 ± 0.004 |
| N10 | 43.59 | 43.02 | 42.72 | 43.11 ± 0.36 | 0.019 | 0.062 | 0.071 | 0.051 ± 0.023 |
| N11 | 59.13 | 58.85 | 60.98 | 59.65 ± 0.95 | 0.198 | 0.220 | 0.214 | 0.211 ± 0.009 |
| N12 | 58.15 | 58.44 | 57.08 | 57.89 ± 0.58 | 0.197 | 0.200 | 0.212 | 0.203 ± 0.006 |
| N13 | 53.57 | 53.89 | 54.02 | 53.83 ± 0.19 | 0.216 | 0.175 | 0.211 | 0.201 ± 0.018 |
| N15 | 48.34 | 47.24 | 46.96 | 47.51 ± 0.60 | 0.084 | 0.107 | 0.090 | 0.094 ± 0.010 |
SD stands for standard deviation.
Three repeated measurements were performed for each VLP.
Two chimeric VLPs elicit cross-blocking antibody responses against both GI.1 and GII.4 genotypes
For each of the 11 chimeric VLPs, 6 female BALB/c mice (SPF level) were immunized, and another two groups (6 mice per group) were immunized with the parent GI.1 and GII.4 VLPs, respectively, as controls. The parent GI.1 and GII.4 VLPs were produced in our previous studies, and their particle morphology and immunogenicity have been validated (32). The sera were separated from the blood of the immunized mice to evaluate the immune responses induced by these VLPs. Histo-blood group antigens (HBGAs) have been revealed to be important attachment factors for HuNoV entry into the host cells (35 – 41), and therefore, the VLP-HBGA blocking assay that evaluates the ability of the immunized sera to block the binding of GI.1 or GII.4 genotypes to HBGAs has been widely used as a surrogate neutralization detection method (42). In this study, the immune activity of these recombinant VLPs was evaluated by using the HBGA blocking assay.
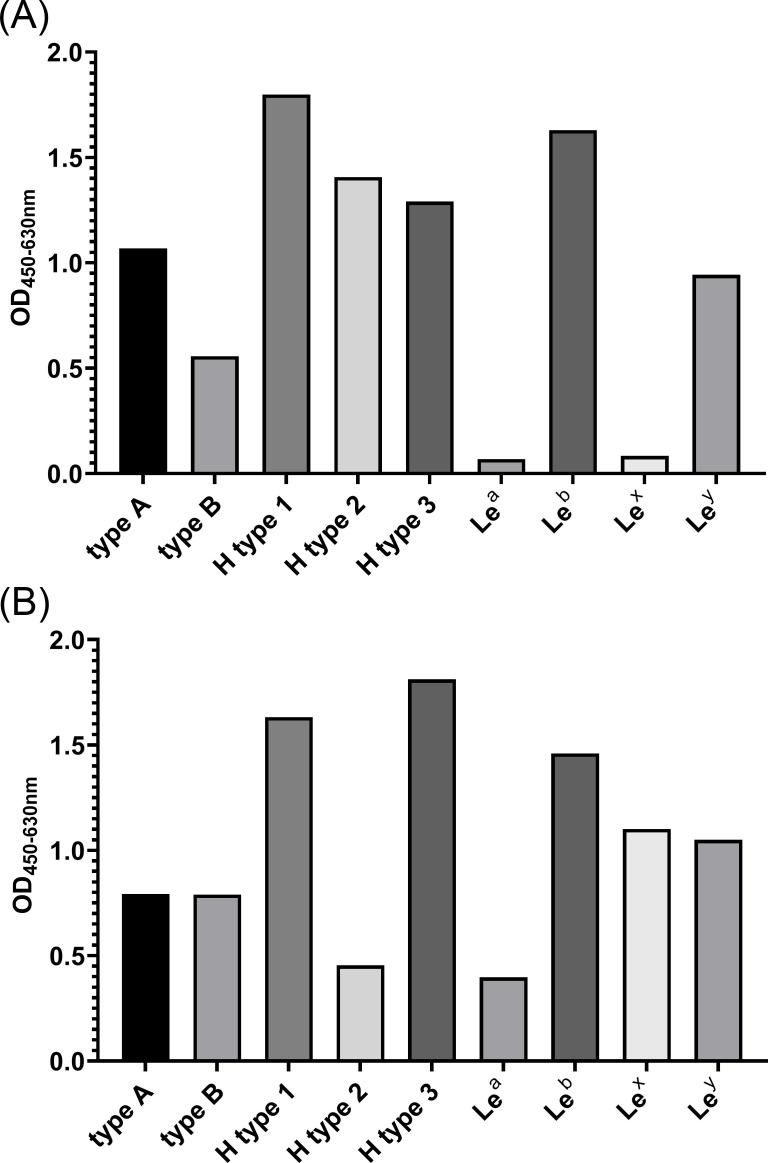
HBGA includes many different types, and several studies have revealed that different HuNoV genotypes prefer to bind with different types of HBGA (38 – 41). Before performing the VLP-HBGA blocking assay, we first detected the binding ability of GI.1 and GII.4 VLPs with different HBGA types, and then, the specific types with strong binding affinity and high detection sensitivity were selected. The results of the VLP-HBGA-binding experiments are shown in Fig. 4. It is found that different types of HGBA have different binding abilities to GI.1 and GII.4 VLPs. H type 1 and Leb type exhibited relatively high binding ability to GI.1 and GII.4 VLPs, and thus, these two types of HBGA were used in the following VLP-HBGA blocking assay for GI.1 and GII.4 VLPs, respectively.
Fig 4.
The binding ability of GI.1 (A) and GII.4 (B) VLPs to nine types of HBGA receptors. The binding ability was evaluated by the value of OD450/630 nm.
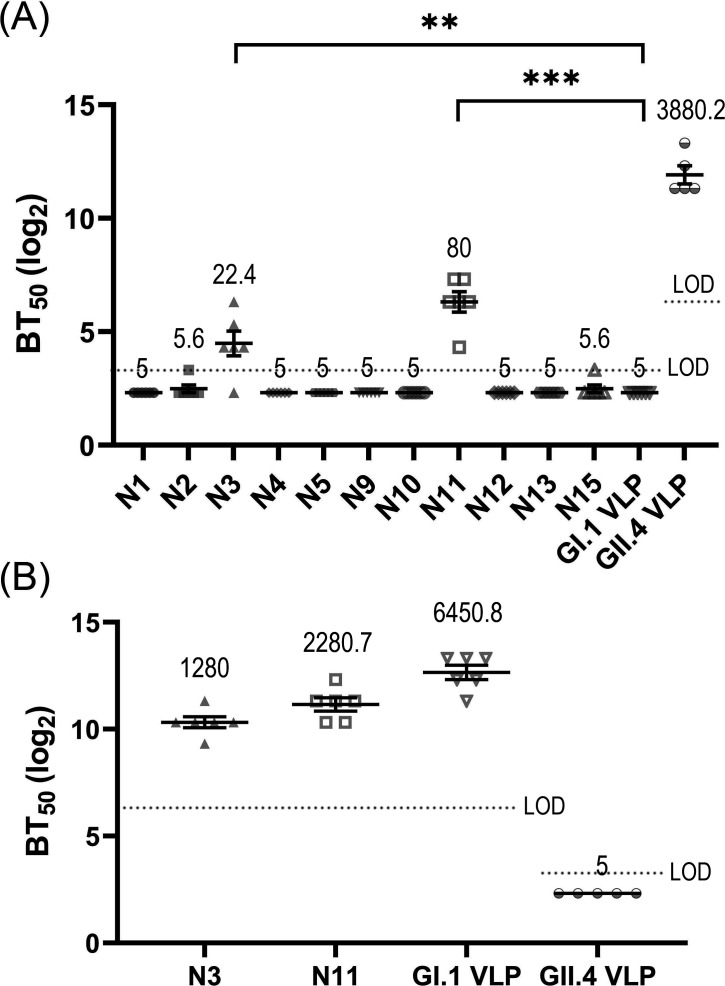
In this study, the exposed loop regions of the GII.4 genotype were grafted into the GI.1 to construct the chimeric VLPs, and we then investigated whether the transplanted sites exhibit HBGA-blockade activities against the GII.4 genotype by using the HBGA-GII.4 blocking assays. The experimental results are displayed in Fig. 5A. Our experiments show that the parent GII.4 VLP can elicit a high level of blocking efficacy against GII.4 genotype, where the geometric mean value of BT50 (GMT) is 3880.2. However, the scaffold GI.1 VLP cannot induce cross-blocking immune responses against the GII.4 genotype with BT50 values less than the detection limit. Many experimental studies have also shown that GI.1 and GII.4 genotypes do not exhibit cross-blocking immune activities (43 – 45), which is consistent with our results. Importantly, among the designed chimeric VLPs, two VLPs, i.e., N3 and N11 schemes, elicit a certain level of blocking ability against the HBGA-GII.4 interactions, in which the GMT values for N3 and N11 VLPs are 22.4 and 80, respectively, as shown in Fig. 5A. Compared with the scaffold GI.1 VLP, the BT50 values of the chimeric N3 and N11 VLPs are statistically significant. These results indicate that unlike the scaffold GI.1 that has no cross-blocking effect, the grafting of GII.4 loop1 in N3 and loop5 in N11 exhibits immunological activities against GII.4 genotype. It should be noted that the schemes N1, N2, and N4 were also designed to graft GII.4 loop1 with various lengths compared to N3, as shown in Table 1. N12 was designed aiming at the same loop (GII.4 loop5) as N11. However, the HBGA-GII.4 blocking experimental results exhibit no HBGA-blockade activities against GII.4 genotype for N1, N2, N4, and N12, as displayed in Fig. 5A. Our experimental results imply that the grafting scheme needs to be well designed to maintain the immunogenicity of the epitopic loops on the scaffold carrier.
Fig 5.
The experimental results of VLP-HBGA blocking assays. (A) The blocking ability against the binding of GII.4 VLP to the receptor Leb-type HBGA for the mice sera immunized with the chimeric VLPs as well as the parent GI.1 and GII.4 VLPs. (B) The blocking ability against GI.1 VLP binding to the receptor H-type1 HBGA for the mice sera immunized with the chimeric N3 and N11 VLPs as well as the parent GI.1 and GII.4 VLPs. The blocking ability was evaluated by the value of BT50, which is calculated as the reciprocal of the highest dilution that blocks 50% VLPs binding to the receptor HBGA. In the VLP-HBGA blocking assays, the detection limit for the serum from the GII.4 VLP immunization group was 10 against GI.1 VLP-HBGA interaction and 80 against GII.4 VLP-HBGA interaction, while the detection limit for the serum from other groups was 80 and 10 against GI.1 and GII.4 VLP-HBGA interactions, respectively. Limit of detection (LOD) was indicated by dashed lines in the figure. Data below the detection limit were set to half of the limit. Data are presented as mean ± SEM. The geometric mean value of BT50 (GMT) is provided in the figure. Abnormal values were obtained for a mouse in GII.4 VLP immunization group, and thus, this mouse was removed from the statistical analyses. Statistically significance of the difference between groups was determined by using one-way ANOVA with Dunnett’s test. **P < 0.01, ***P < 0.001.
For the chimeric N3 and N11, VLPs with positive blocking effects against the GII.4 genotype, we then explored whether the incorporation of the grafted loops has an influence on the immune activities of the scaffold protein. The blocking abilities against the GI.1 genotype for N3 and N11 VLPs were evaluated by using HBGA-GI.1 blocking assay. As a comparison, the blocking activities for the parent GI.1 and GII.4 VLPs were also measured. The experimental results are shown in Fig. 5B. It is found that GI.1 VLP can elicit a high level of blocking activities against GI.1 genotype as expected, whereas the parent GII.4 VLP cannot induce a cross-blocking response against the GI.1 genotype, in which the values of BT50 are less than the detection limit. This is consistent with the experimental results of other research groups (43 – 45). For the chimeric N3 and N11, the blocking abilities against GI.1 genotype were maintained at high levels, in which the GMT values are 1,280 and 2,280.7 for N3 and N11, respectively, as shown in Fig. 5B. Our results imply that the grafted epitopic loops slightly affect the immunobiological properties against GI.1 for N3 and N11. This result is expected because in the chimeric VLPs, only the local region corresponding to the grafted site was replaced in GI.1, and most of the antigenic properties were retained. The above results indicate that the two chimeric schemes, i.e., N3 and N11 VLPs, have cross-immune activity both against GI.1 and GII.4 genotypes. To our knowledge, this is the first designed chimeric VLPs that can induce cross-blocking activity against GI.1 and GII.4 from different genogroups, which provides valuable strategies for the development of cross-reactive vaccines for HuNoV.
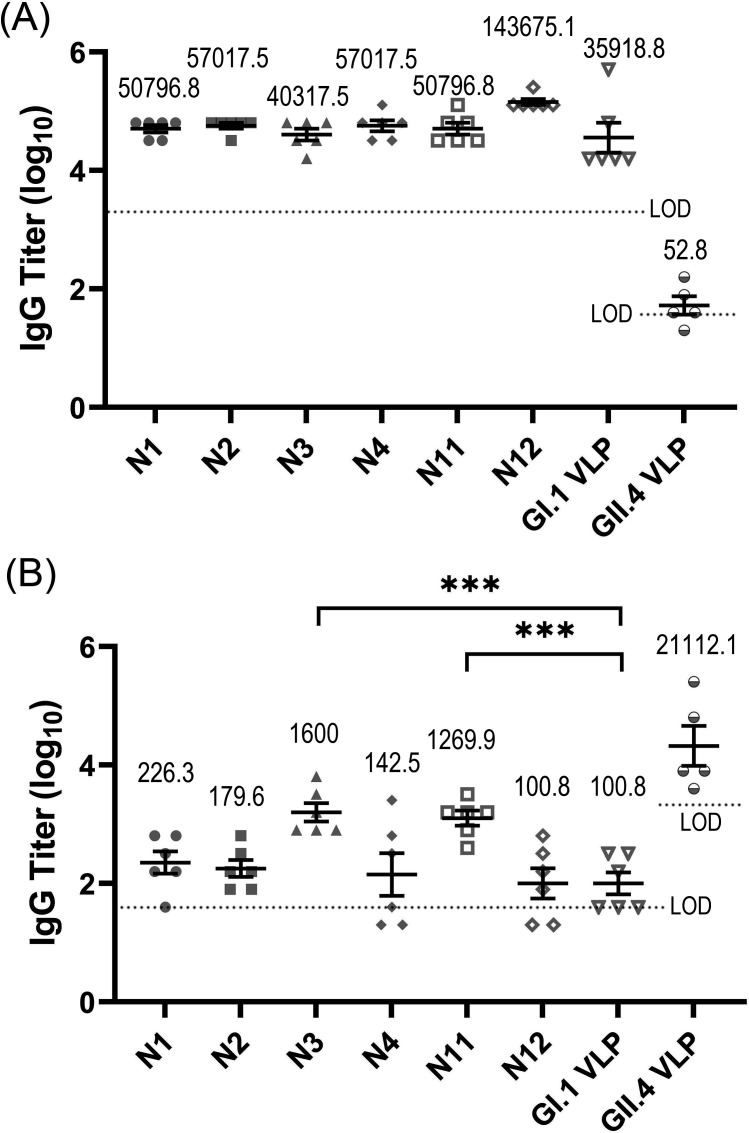
The titers of specific IgG antibodies against GI.1 and GII.4 genotypes elicited by N3 and N11 chimeric VLPs
The VLP-HBGA blocking assays have shown that the chimeric N3 and N11 VLPs can elicit cross-blocking activities against both GI.1 and GII.4 genotypes. We then evaluated the IgG antibody levels elicited by N3 and N11 VLPs by using the ELISA. As a comparison, the levels of IgG antibodies induced by the parent GI.1 and GII.4 VLPs, as well as the chimeric schemes N1, N2, N4 (the chimeric schemes aiming at the grafting of the same loop as N3) and N12 (the chimeric scheme aiming at the grafting of the same loop as N11), with negative cross-blocking capability were also evaluated by ELISA. The titers of the specific IgG antibodies against GI.1 and GII.4 genotypes in the immunized sera for these VLPs were measured, and the results are displayed in Fig. 6. The experimental results show that GI.1 VLP induced high titers of IgG antibodies specific to GI.1 genotype but induced very low titers specific to GII.4 genotype, as shown in Fig. 6. Similarly, the IgG antibodies induced by GII.4 only specifically interacts with GII.4 genotype, but has negligible interactions with GI.1 genotype, as shown in Fig. 6. These results indicate that individual GI.1 and GII.4 VLPs can elicit antibodies only specifically interacted with the corresponding immunized antigens, but the elicited antibodies have very weak cross-interactions with the other type VLPs. This agrees with the results obtained by the VLP-HBGA blocking experiments, where the individual GI.1 and GII.4 do not exhibit cross-blocking activities.
Fig 6.
The titers of IgG antibodies specific to GI.1 (A) and GII.4 (B) genotypes in the mice immunized with the chimeric N1, N2, N3, N4, N11, and N12 VLPs, as well as the parent GI.1 and GII.4 VLPs. In these assays, the detection limit of GI.1-specific antibodies was 40 for the serum samples immunized with GII.4 VLP and 2,000 for the serum samples from the other groups, respectively. For the GII.4-specific antibodies, the detection limit was 2,000 for the sera from the GII.4 VLP immunization group and 40 for the sera from the other groups, respectively. Limit of detection (LOD) was indicated by dashed lines in the figure. Data less than the detection limit were set to half of the limit. Data are presented as mean ± SEM. The geometric mean value of BT50 (GMT) is provided in the figure. Abnormal values were obtained for a mouse in GII.4 VLP immunization group, and thus, this mouse was removed from the statistical analyses. Statistical significance of the difference between groups was determined by using one-way ANOVA with Dunnett’s test. ***P < 0.001.
For the chimeric N3 and N11 VLPs, high levels of anti-GI.1 antibodies were elicited, where the titers of the GI.1-specific IgG antibodies induced by these two chimeric VLPs are comparable to that elicited by the GI.1 VLP, as shown in Fig. 6A. As discussed above, this result is not surprising because GI.1 is the scaffold of the designed chimeric VLPs, and most of the epitopic sites are retained. More importantly, N3 and N11 chimeric VLPs also elicit an obvious level of antibodies against GII.4 genotype, as shown in Fig. 6B. The geometric mean titers (GMTs) of the GII.4-specific IgG antibodies elicited by these two chimeric VLPs are 1,600 and 1,269.9, respectively, which are obviously higher than that of the control group (i.e., the GI.1VLP). This result implies that the grafting of GII.4 loop1 in N3 and loop5 in N11 exhibits immunological activity in the chimeric VLPs. The above results indicate that these two chimeric schemes, i.e., N3 and N11, induced cross-immune activities against both GI.1 and GII.4 HuNoVs, which is consistent with the results obtained by VLP-HBGA blocking assays discussed in the previous section. However, for the chimeric N1, N2, N4, and N12 VLPs with negative cross-blocking capability, high levels of antibodies specific to the GI.1 genotype were induced but very low levels of antibodies specific to the GII.4 were elicited, as displayed in Fig. 6. This result implies that the transplanted loops lost their immunogenicity in these chimeric VLPs. Therefore, the construction of chimeric VLP is a challenging task, where the grafting schemes need to be well designed.
DISCUSSION
HuNoV is a highly infectious pathogen, which is the leading cause for most of the outbreaks of acute gastroenteritis worldwide. The spread of HuNoV is a serious threat to human health, especially for the elderly and children, which also results in a heavy financial burden. It is urgent to develop effective vaccines to prevent the spread of HuNoV. However, NoV is a virus with a high rate of mutations. At present, 10 genogroups with numerous genotypes of NoV have been found, among which GI.1 is the prototype strain and GII.4 is the most prevalent genotype in human. Each HuNoV genotype also includes many strains, and on average, a new strain emerges per 2–3 years. The genetic diversity and high variability of HuNoV require the development of effective vaccines with cross protections. Constructing chimeric VLP that incorporates the epitopes from different virus strains is a promising but challenging strategy to develop cross-protective vaccines. At present, for HuNoV only the chimeric VLPs for different strains within the same genotype or genogroup have been successfully designed. To our knowledge, it has not been reported to develop the chimeric HuNoV VLPs across different genogroups.
In this study, guided by the structural analysis, several chimeric VLPs across GI and GII genogroups were designed. In our strategy, the surface-exposed loops of GII.4 HuNoV, where the neutralizing epitopes are mainly located, were grafted onto the GI.1 scaffold. The designed chimeric VLPs were expressed with the Hansenula polymorpha expression platform constructed by our laboratory. The SDS-PAGE experiments confirmed the expressions of 11 out of 15 designed proteins. The observations with TEM and the measurements of DLS showed that these 11 designed proteins are successfully assembled into VLPs with uniform sizes. ELISA assays exhibited that two of the 11 chimeric VLPs, i.e., N3 and N11, elicit high levels of IgG antibodies specific to both GI.1 and GII.4 HuNoVs. Upon loop transplantation, the GII.4-specific IgG antibody GMTs reached 1,600 and 1,269.9 for N3 and N11, respectively, which were significantly higher than the value of 100.8 for the parent GI.1VLP. HGBA-VLP blocking assays displayed that the two positive chimeric VLPs elicited cross-blocking response against both GI.1 and GII.4 HuNoVs. After loop grafting, the HBGA-blocking antibody GMT against the GII.4 genotype was improved from less than the detection limit to 22.4 and 80 for N3 and N11, respectively. The results also imply that the transplant site design is important to maintain the immunogenicity of foreign epitopes on the scaffold carrier.
It should be mentioned that GII.4 HuNoV is currently more prevalent than the GI.1 genotype (7, 10), and thus, it may be better to use the GII.4 VP1 protein as the scaffold to incorporate the epitopes from GI.1. In fact, we have also designed several schemes that graft the GI.1 epitopes onto the GII.4 backbone, but preliminary results show that no VLP or no cross-immunological activity was obtained for the designed schemes. These negative schemes using GII.4 as the scaffold are also provided in Table S1 in the supplemental material for reference. Further studies are ongoing in our laboratory.
The attachment of HuNoV to HBGAs is an essential process in the infection of host cells, and the blocking antibodies that inhibit this critical step may serve as neutralizing antibodies. Estes group has proved that the HBGA-blocking antibody titer is highly correlated with the neutralization level in adults receiving a candidate bivalent HuNoV vaccine (46). Experimental human challenge studies showed that HBGA-blocking antibody titers correlated with protection against HuNoV infection and associated illness (47, 48). The cross-blocking ability may indicate potential cross-neutralizing activity of our designed chimeric VLPs against both GI.1 and GII.4 HuNoVs. Our studies may provide a valuable strategy for the possible development of cross-reactive HuNoV vaccines. Certainly, blocking the attachment of HuNoV to HBGA is not the only way to neutralize virus infection. One of the limitations of our study is that the cross-neutralizing reactivity of the chimeric VLP constructs needs to be verified by using live virus neutralization assay. In recent years, Estes group has developed human intestinal stem cell-derived enteroids (HIEs) for the in vitro cultivation of HuNoVs (49, 50), which provides the possibility to test the neutralization against live virus. We will evaluate the neutralizing activity of the constructed chimeric VLPs in the future studies.
MATERIALS AND METHODS
Design of chimeric VLPs guided by structural analysis
The tertiary structures of GI.1 and GII.4 VP1s were aligned with UCSF Chimera software (28). Guided by the structural alignments, chimeric VLPs were designed through exposed loop grafting, in which the surface-exposed loops of GII.4 were transplanted into the scaffold GI.1 VLP. Experimental studies have indicated that these loops serve as the most important neutralizing epitopic sites. According to the structural alignments, 8 loops were selected and a total of 15 schemes were designed to graft these exposed loop regions of GII.4 into GI.1 scaffold to construct the chimeric VLPs. The location of these loops and the designed chimeric schemes are displayed in Fig. 1; Table 1.
The expression, purification, and identification of the recombinant VP1 of the designed VLPs
The designed chimeric VLPs, were expressed with the Hansenula polymorpha yeast expression platform constructed by our laboratory and purified with the ion exchange chromatography method. The expression of the VP1 proteins was confirmed by using the SDS-PAGE experiments. In the SDS-PAGE experiments, 12% SDS-PAGE gel was used, and the specific bands corresponding to the expressed proteins were observed. According to the location of the appeared bands, the relative molecular weight of the proteins was evaluated and compared with the theoretical values.
Morphological detection of the designed chimeric VLPs by using TEM and DLS
In order to determine whether the designed proteins successfully assemble into VLPs, the morphology of the designed VLPs was observed with a transmission electron microscope (TEM), and the size distributions were tested with the dynamic light scattering (DLS) method. In TEM experiments, the VLP solutions were dropped on the copper carrier covered with carbon. After 5 minutes absorption and then negative staining with phosphotungstic acid, the morphology of the VLPs was observed with TEM. In DLS experiments, the VLP solutions were pre-filtrated with a microfilter of 0.22 µm, and then, the particle diameters and the polydispersity index (PDI), which reflects the size distribution of the particles, were measured. For each protein sample, the measurement was repeated three times, and then, the mean value was calculated for the particle diameter and PDI.
Mice immunization
The SPF level of BALB/c female mice, aged 6–8 weeks and weighted 18–22 g, were used in the immune experiments. For each chimeric VLP, six mice were immunized by intraperitoneal injections. A total of three doses (25 µg antigen mixed with 250 µg aluminum adjuvant for each dose) were immunized on week 0, week 2, and week 4, respectively. Another two groups of mice (six mice per group) were immunized with three doses of the parent GI.1 and GII.4 VLPs plus aluminum adjuvant, respectively, as controls. On week 6, blood samples were harvested from the orbital veniplex of the immunized mice, and sera were extracted from the blood samples and used to evaluate the immune effects. All the mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. All the animal operating procedures were approved by the Institutional Animal Care and Use Committee of the National Vaccine and Serum Institute (NVSI).
VLP-HBGA binding and blocking assays
HBGAs have been revealed to be important attachment factors for HuNoV entry into the host cells (35 – 41). In this study, the immune activity of the recombinant VLPs was evaluated by using VLP-HBGA interaction blocking assay, which measured the antibody titers in the sera of the immunized mice to block the binding of GI.1 or GII.4 VLPs to HBGAs. HBGA includes many different types (35 – 41), and different HuNoV genotypes prefer to bind with different types of HBGA (38 – 41). We first determined the specific type of HBGA with strong binding affinity and high detection sensitivity for GI.1 and GII.4 HuNoV VLPs, respectively, by using VLP-HBGA binding assay. Nine kinds of biotinylated HBGA were used in our experiments including H type 1-PAA-biotin, H type 2-PAA-biotin, H type 3-PAA-biotin, Lea-PAA-biotin, Leb-PAA-biotin, Lex-PAA-biotin, Ley-PAA-biotin, type A-PAA-biotin, and type B-PAA-biotin (purchased from Glycotech). In the binding assay, the biotinylated HBGAs were diluted to 2.5 µg/mL and coated onto the well walls of the 96-well plate with 100 µL per well, after which the plate was incubated at 25℃ for 1 hour and washed four times with 100 mM phosphate buffer (PB) at pH6.4. GI.1 and GII.4 VLP samples were diluted to 1.6 µg/mL and 0.8 µg/mL, respectively. The diluted VLP sample were added to the well with 100 µL per well and incubated at 4℃ for 2 hours, followed by washing four times with 100 mM PB at pH6.4. The GI.1- and GII.4-specific IgG labeled by HRP (purchased from Beijing Qinbang Biotechnology Co., Ltd) were diluted to working concentrations (1:8,000 for GI.1 and 1:16,000 for GII.4, respectively), which were added into the well of the plate with 100 µL per well and incubated at 4℃ for 1 hour. After washing the plate four times, the color was developed for 5 minutes by adding peroxidase liquid substrate. Then, the color reaction was stopped with sulfuric acid, and the value of the optical density at 450/630 nm (OD450/630 nm) was obtained with the ELISA microplate reader. Based on the experimental results of the VLP-HBGA binding assays, the H type 1 and Leb type of HBGA were chosen to be used in the following GI.1 and GII.4 VLP-HBGA blocking assays, respectively.
In the VLP-HBGA blocking assay, the biotinylated HBGAs were diluted and coated onto the well of the plate as described in the above VLP-HBGA binding assay. Serum of the mice from the GII.4 VLP immunization group was first diluted to 1:5 for the GI.1 VLP-HBGA blocking assay and 1:40 for the GII.4 VLP-HBGA blocking assay. While serum from the other groups was first diluted to 1:40 for the GI.1 VLP-HBGA blocking assay and 1:5 for the GII.4 VLP-HBGA blocking assay. Subsequently, twofold serial dilutions of the serum sample were prepared. Then, the VLP sample was diluted to working concentrations, i.e., 3.2 µg/mL for GI.1 and 1.6 µg/mL for GII.4 VLPs, respectively. The diluted VLP sample was mixed with the diluted serum in equal volume and incubated at 37°C for 1 hour. The VLP-serum mixture was added into the well of the plate with 100 µL per well and incubated at 4°C for 2 hours. Subsequently, the plate was washed with 100 mM PB at PH6.4 for four times. The GI.1- and GII.4-specific IgG antibodies marked by HRP were diluted to working concentrations and added into the well as described in the above VLP-HBGA binding assay. After performing the color reaction, the value of OD450/630 nm was obtained. The titer of the blocking antibodies in the serum was evaluated by the value of BT50, which is defined as the highest dilution that blocks 50% of VLP binding with HBGA. In the VLP-HBGA blocking assays, the detection limit for the serum from the GII.4 VLP immunization group was 10 against GI.1 VLP-HBGA interaction and 80 against GII.4 VLP-HBGA interaction, while the detection limit for the serum from other groups was 80 and 10 against GI.1 and GII.4 VLP-HBGA interactions, respectively. The value of BT50 below the detection limit was set to half of the limit.
Enzyme-linked immunosorbent assay
The tilter of GI.1- and GII.4-specific IgG antibodies in the sera from the mice immunized with the chimeric VLPs were evaluated by ELISA. In the experiment, the VLP solution was diluted to 3 µg/mL, and each well of the 96-well plate was coated with 100 µL of the sample at 4°C for 8 hours, followed by washing three times with phosphate-buffered saline with Tween 20 (PBST). Then, the plate was blocked by phosphate-buffered saline (PBS) solution with 1% bovine serum albumin (BSA) and incubated at 37°C for 2 hours. The serum samples from the mice immunized with the GII.4 VLP were first diluted 40 and 2,000 times for the GI.1- and GII.4-specific antibody detections, respectively. While the sera from the other groups were first diluted 2,000 and 40 times for the detection of IgG antibodies specific to GI.1 and GII.4 genotypes, respectively. After that, twofold serial dilutions were carried out. The diluted serum sample was coated onto the well of the 96-well plate with 100 µL per well at 37°C for 1 hour. Then, the goat anti-mice IgG labeled by HRP (purchased from Beijing Zhongshan Jinqiao Biotechnology Co., Ltd) at 1:40,000 was added into the well with 100 µL per well followed by incubation at 37°C for 1 hour. After washing the plate three times with PBST, the color was then developed for 5 minutes by adding peroxidase liquid substrate and stopped by adding sulfuric acid. The value of OD450/630 nm was read by the ELISA microplate reader. If OD450/630 nm is larger than a cutoff value, the reaction is considered to be positive. Otherwise, the reaction is negative. The cutoff value for ELISA was usually set as two or three times the mean absorbance value of the negative controls (51, 52). Here, the cutoff value was set empirically as 2.1 times the OD450/630 nm mean value of the adjuvant control group. The titer for the specific IgG antibodies was determined as the highest dilution that yields a positive reaction. The detection limit of GI.1-specific antibodies was 40 for the serum samples immunized with GII.4 VLP and 2,000 for the serum samples from the other groups, respectively. For the GII.4-specific antibodies, the detection limit was 2,000 for the sera from the GII.4 VLP immunization group and 40 for the sera from the other groups, respectively. If the IgG antibody was below the detection limit, the titer was set to half of the limit.
Statistical analysis of the experimental results
The difference between groups was analyzed by using one-way ANOVA with Dunnett’s test, and the difference with P < 0.05 is considered to be statistically significant.
ACKNOWLEDGMENTS
We thank Xiangfeng Cong for his help in statistical analysis.
This work was supported in part by the Beijing Municipal Science and Technology Project (Z221100007922048).
We declare no competing interests.
Contributor Information
Jing Zhang, Email: zhangj791@hotmail.com.
Qi Ming Li, Email: liqiming189@163.com.
Rebecca Ellis Dutch, University of Kentucky College of Medicine, Lexington, Kentucky, USA .
DATA AVAILABILITY
The data that support the findings of this study have been provided in the main text and the supplemental material. The sequences of the designed HuNoV chimeric VP1 proteins reported in this study are available from GenBank under accession numbers OR412756-OR412770 for the N1-N15 constructs, respectively.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.00938-23.
GII.4 VP1 scaffold-based schemes and experimental results.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Grant LR, O’Brien KL, Weatherholtz RC, Reid R, Goklish N, Santosham M, Parashar U, Vinjé J. 2017. Norovirus and sapovirus epidemiology and strain characteristeics among navajo and apache infants. PLoS ONE 12:e0169491. doi: 10.1371/journal.pone.0169491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xue Y, Pan H, Hu J, Wu H, Li J, Xiao W, Zhang X, Yuan Z, Wu F. 2015. Epidemiology of norovirus infections among diarrhea outpatients in a diarrhea surveillance system in Shanghai, China: a cross-sectional study. BMC Infect Dis 15:183. doi: 10.1186/s12879-015-0922-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou HL, Zhen SS, Wang JX, Zhang CJ, Qiu C, Wang SM, Jiang X, Wang XY. 2017. Burden of acute gastroenteritis caused by norovirus in China: a systematic review. J Infect 75:216–224. doi: 10.1016/j.jinf.2017.06.004 [DOI] [PubMed] [Google Scholar]
- 4. Shioda K, Barclay L, Becker-Dreps S, Bucardo-Rivera F, Cooper PJ, Payne DC, Vinjé J, Lopman BA. 2017. Can use of viral load improve Norovirus clinical diagnosis and disease attribution? Open Forum Infect Dis 4:fx131. doi: 10.1093/ofid/ofx131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pires SM, Fischer-Walker CL, Lanata CF, Devleesschauwer B, Hall AJ, Kirk MD, Duarte ASR, Black RE, Angulo FJ. 2015. Aetiology-specific estimates of the global and regional incidence and mortality of diarrhoeal diseases commonly transmitted through food. PLoS ONE 10:e0142927. doi: 10.1371/journal.pone.0142927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hall AJ, Lopman BA, Payne DC, Patel MM, Gastañaduy PA, Vinjé J, Parashar UD. 2013. Norovirus disease in the United States. Emerg Infect Dis 19:1198–1205. doi: 10.3201/eid1908.130465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cannon JL, Bonifacio J, Bucardo F, Buesa J, Bruggink L, Chan MCW, Fumian TM, Giri S, Gonzalez MD, Hewitt J, Lin JH, Mans J, Muñoz C, Pan CY, Pang XL, Pietsch C, Rahman M, Sakon N, Selvarangan R, Browne H, Barclay L, Vinjé J. 2021. Global trends in norovirus genotype distribution among children with acute gastroenteritis. Emerg Infect Dis 27:1438–1445. doi: 10.3201/eid2705.204756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chhabra P, de Graaf M, Parra GI, Chan MC-W, Green K, Martella V, Wang Q, White PA, Katayama K, Vennema H, Koopmans MPG, Vinjé J. 2019. Updated classification of norovirus genogroups and genotypes. J Gen Virol 100:1393–1406. doi: 10.1099/jgv.0.001318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Glass RI, Parashar UD, Estes MK. 2009. Norovirus gastroenteritis. N Engl J Med 361:1776–1785. doi: 10.1056/NEJMra0804575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Winder N, Gohar S, Muthana M. 2022. Norovirus: an overview of virology and preventative measures. Viruses 14:2811. doi: 10.3390/v14122811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cortes-Penfield NW, Ramani S, Estes MK, Atmar RL. 2017. Prospects and challenges in the development of a norovirus vaccine. Clin Ther 39:1537–1549. doi: 10.1016/j.clinthera.2017.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ahmed SM, Lopman BA, Levy K, Vespignani A. 2013. A systematic review and meta-analysis of the global seasonality of norovirus. PLoS ONE 8:e75922. doi: 10.1371/journal.pone.0075922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cannon JL, Barclay L, Collins NR, Wikswo ME, Castro CJ, Magaña LC, Gregoricus N, Marine RL, Chhabra P, Vinjé J. 2017. Genetic and epidemiologic trends of norovirus outbreaks in the United States from 2013 to 2016 demonstrated emergence of novel GII.4 recombinant viruses. J Clin Microbiol 55:2208–2221. doi: 10.1128/JCM.00455-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lucero Y, Vidal R, O’Ryan GM. 2018. Norovirus vaccines under development. Vaccine 36:5435–5441. doi: 10.1016/j.vaccine.2017.06.043 [DOI] [PubMed] [Google Scholar]
- 15. Mattison CP, Cardemil CV, Hall AJ. 2018. Progress on norovirus vaccine research: public health considerations and future directions. Expert Rev Vaccines 17:773–784. doi: 10.1080/14760584.2018.1510327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Debbink K, Lindesmith LC, Donaldson EF, Swanstrom J, Baric RS. 2014. Chimeric GII.4 norovirus virus-like-particle-based vaccines induce broadly blocking immune responses. J Virol 88:7256–7266. doi: 10.1128/JVI.00785-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huo Y, Ma J, Zheng L, Liu J, Yang Z, Wang C, Zhao Q. 2022. Expression of chimeric proteins based on a backbone of the GII.4 norovirus VP1 and their application in the study of a GII.6 norovirus-specific blockade epitope. Arch Virol 167:819–827. doi: 10.1007/s00705-022-05362-5 [DOI] [PubMed] [Google Scholar]
- 18. Prasad BVV, Hardy ME, Dokland T, Bella J, Rossmann MG, Estes MK. 1999. X-ray crystallographic structure of the norwalk virus capsid. Nature 286:287–290. doi: 10.1126/science.286.5438.287 [DOI] [PubMed] [Google Scholar]
- 19. Hardy ME. 2005. Norovirus protein structure and function. FEMS Microbiol Lett 253:1–8. doi: 10.1016/j.femsle.2005.08.031 [DOI] [PubMed] [Google Scholar]
- 20. Lindesmith LC, Beltramello M, Donaldson EF, Corti D, Swanstrom J, Debbink K, Lanzavecchia A, Baric RS. 2012. Immunogenetic mechanisms driving norovirus GII.4 antigenic variation. PLoS Pathog 8:e1002705. doi: 10.1371/journal.ppat.1002705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koromyslova AD, Morozov VA, Hefele L, Hansman GS. 2019. Human norovirus neutralized by a monoclonal antivody targeting the histo-blood group antigen packet. J Virol 93:e02174-18. doi: 10.1128/JVI.02174-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carmona-Vicente N, Vila-Vicent S, Allen D, Gozalbo-Rovira R, Iturriza-Gómara M, Buesa J, Rodríguez-Díaz J. 2016. Characterization of a novel conformational GII.4 norovirus epitope: implications for norovirus-host interactions. J Virol 90:7703–7714. doi: 10.1128/JVI.01023-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lindesmith LC, Brewer-Jensen PD, Mallory ML, Debbink K, Swann EW, Vinjé J, Baric RS. 2018. Antigenic characterization of a novel recombinant GII.P16-GII.4 Sydney norovirus strain with minor sequence variation leading to antibody escape. J Infect Dis 217:1145–1152. doi: 10.1093/infdis/jix651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rossmann MG, He Y, Kuhn RJ. 2002. Picornavirus-receptor interactions. Trends Microbiol 10:324–331. doi: 10.1016/s0966-842x(02)02383-1 [DOI] [PubMed] [Google Scholar]
- 25. Zhao H, Li H-Y, Han J-F, Deng Y-Q, Zhu S-Y, Li X-F, Yang H-Q, Li Y-X, Zhang Y, Qin E-D, Chen R, Qin C-F. 2015. Novel recombinant chimeric virus-like particle is immunogenic and protective against both enterovirus 71 and coxsackievirus A16 in mice. Sci Rep 5:7878. doi: 10.1038/srep07878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Prasad BVV, Hardy ME, Dokland T, Bella J, Rossmann MG, Estes MK. 1999. X-ray crystallographic structure of the norwalk virus capsid. Science 286:287–290. doi: 10.1126/science.286.5438.287 [DOI] [PubMed] [Google Scholar]
- 27. Hu L, Salmen W, Chen R, Zhou Y, Neill F, Crowe JE, Atmar RL, Estes MK, Prasad BVV. 2022. Atomic structure of the predominant GII.4 human norovirus capsid reveals novel stability and plasticity. Nat Commun 13:1241. doi: 10.1038/s41467-022-28757-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 29. Taube S, Rubin JR, Katpally U, Smith TJ, Kendall A, Stuckey JA, Wobus CE. 2010. High-resolution X-ray structure and functional analysis of the murine norovirus 1 capsid protein protruding domain. J Virol 84:5695–5705. doi: 10.1128/JVI.00316-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Allen DJ, Noad R, Samuel D, Gray JJ, Roy P, Iturriza-Gómara M. 2009. Characterisation of a GII-4 norovirus variant-specific surface-exposed site involved in antibody binding. Virol J 6:150. doi: 10.1186/1743-422X-6-150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cao S, Lou Z, Tan M, Chen Y, Liu Y, Zhang Z, Zhang XC, Jiang X, Li X, Rao Z. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J Virol 81:5949–5957. doi: 10.1128/JVI.00219-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ma Z, Tang F, Zhang X, Zhang J, Hou J, Chen S, Li Q. 2016. Evaluation of immune effects of recombinant Norovirus GI.1 and GII.4 virus-like particles. Chin J Microbiol Immunol 36:930–934. [Google Scholar]
- 33. Murphy RM. 1997. Static and dynamic light scattering of biological macromolecules: what can we learn? Curr Opin Biotechnol 8:25–30. doi: 10.1016/s0958-1669(97)80153-x [DOI] [PubMed] [Google Scholar]
- 34. Lorber B, Fischer F, Bailly M, Roy H, Kern D. 2012. Protein analysis by dynamic light scattering: methods and techniques for students. Biochem Mol Biol Educ 40:372–382. doi: 10.1002/bmb.20644 [DOI] [PubMed] [Google Scholar]
- 35. Harrington PR, Lindesmith L, Yount B, Moe CL, Baric RS. 2002. Binding of norwalk virus-like particles to ABH histo-blood group antigens is blocked by antisera from infected human volunteers or experimentally vaccinated mice. J Virol 76:12335–12343. doi: 10.1128/jvi.76.23.12335-12343.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Harrington PR, Vinjé J, Moe CL, Baric RS. 2004. Norovirus capture with histo-blood group antigens reveals novel virus-ligand interactions. J Virol 78:3035–3045. doi: 10.1128/jvi.78.6.3035-3045.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marionneau S, Ruvoën N, Le Moullac-Vaidye B, Clement M, Cailleau-Thomas A, Ruiz-Palacois G, Huang P, Jiang X, Le Pendu J. 2002. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 122:1967–1977. doi: 10.1053/gast.2002.33661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Donaldson EF, Lindesmith LC, Lobue AD, Baric RS. 2010. Viral shape-shifting: norovirus evasion of the human immune system. Nat Rev Microbiol 8:231–241. doi: 10.1038/nrmicro2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. de Rougemont A, Ruvoen-Clouet N, Simon B, Estienney M, Elie-Caille C, Aho S, Pothier P, Le Pendu J, Boireau W, Belliot G. 2011. Qualitative and quantitative analysis of the binding of GII.4 norovirus variants onto human blood group antigens. J Virol 85:4057–4070. doi: 10.1128/JVI.02077-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shirato-Horikoshi H, Ogawa S, Wakita T, Takeda N, Hansman GS. 2007. Binding activity of norovirus and sapovirus to histo-blood group antigens. Arch Virol 152:457–461. doi: 10.1007/s00705-006-0883-z [DOI] [PubMed] [Google Scholar]
- 41. Singh BK, Leuthold MM, Hansman GS. 2015. Human noroviruses’ fondness for histo-blood group antigens. J Virol 89:2024–2040. doi: 10.1128/JVI.02968-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van Loben Sels JM, Meredith LW, Sosnovtsev SV, de Graaf M, Koopmans MPG, Lindesmith LC, Baric RS, Green KY, Goodfellow IG. 2021. A luciferase-based approach for measuring HBGA blockade antibody titers against human norovirus. J Virol Methods 297:114196. doi: 10.1016/j.jviromet.2021.114196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Debbink K, Lindesmith LC, Donaldson EF, Baric RS. 2012. Norovirus immunity and the great escape. PLoS Pathog 8:e1002921. doi: 10.1371/journal.ppat.1002921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Malm M, Tamminen K, Vesikari T, Blazevic V. 2016. Type-specific and cross-reactive antibodies and T cell responses in Norovirus VLP immunized mice are targeted both to conserved and variable domains of capsid VP1 protein. Mol Immun 78:27–37. doi: 10.1016/j.molimm.2016.08.009 [DOI] [PubMed] [Google Scholar]
- 45. Lindesmith LC, Donaldson E, Leon J, Moe CL, Frelinger JA, Johnston RE, Weber DJ, Baric RS. 2010. Heterotypic humoral and cellular immune responses following norwalk virus infection. J Virol 84:1800–1815. doi: 10.1128/JVI.02179-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Atmar RL, Ettayebi K, Ayyar BV, Neill FH, Braun RP, Ramani S, Estes MK. 2020. Comparison of microneutralization and histo-blood group antigen-blocking assays for functional norovirus antibody detection. J Infect Dis 221:739–743. doi: 10.1093/infdis/jiz526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reeck A, Kavanagh O, Estes MK, Opekun AR, Gilger MA, Graham DY, Atmar RL. 2010. Serological correlate of protection against norovirus-induced gastroenteritis. J Infect Dis 202:1212–1218. doi: 10.1086/656364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Atmar RL, Bernstein DI, Lyon GM, Treanor JJ, Al-Ibrahim MS, Graham DY, Vinjé J, Jiang X, Gregoricus N, Frenck RW, Moe CL, Chen WH, Ferreira J, Barrett J, Opekun AR, Estes MK, Borkowski A, Baehner F, Goodwin R, Edmonds A, Mendelman PM. 2015. Serological correlates of protection against a GII.4 norovirus. Clin Vaccine Immunol 22:923–929. doi: 10.1128/CVI.00196-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ettayebi K., Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng X-L, Qu L, Kou B, Opekun AR, Burrin D, Graham DY, Ramani S, Atmar RL, Estes MK. 2016. Replication of human noroviruses in stem cell-derived human enteroids. Science 353:1387–1393. doi: 10.1126/science.aaf5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ettayebi K, Tenge VR, Cortes-Penfield NW, Crawford SE, Neill FH, Zeng X-L, Yu X, Ayyar BV, Burrin D, Ramani S, Atmar RL, Estes MK, Esstman SMM. 2021. New insights and enhanced human norovirus cultivation in human intestinal enteroids. mSphere 6:e01136-20. doi: 10.1128/mSphere.01136-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lardeux F, Torrico G, Aliaga C. 2016. Calculation of the ELISA’s cut-off based on the change-point analysis method for detection of trypanosoma cruzi infection in bolivian dogs in the absence of controls. Mem Inst Oswaldo Cruz 111:501–504. doi: 10.1590/0074-02760160119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Frey A, Di Canzio J, Zurakowski D. 1998. A statistically defined endpoint titer determination method for immunoassays. J Immunol Methods 221:35–41. doi: 10.1016/s0022-1759(98)00170-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GII.4 VP1 scaffold-based schemes and experimental results.
Data Availability Statement
The data that support the findings of this study have been provided in the main text and the supplemental material. The sequences of the designed HuNoV chimeric VP1 proteins reported in this study are available from GenBank under accession numbers OR412756-OR412770 for the N1-N15 constructs, respectively.