ABSTRACT
The genus Periweissella was proposed as a novel genus in the Lactobacillaceae in 2022. However, the phylogenetic relationship between Periweissella and other heterofermentative lactobacilli, and the genetic and physiological properties of this genus remain unclear. This study aimed to determine the phylogenetic relationship between Periweissella and the two closest genera, Weissella and Furfurilactobacillus, by the phylogenetic analysis and calculation of (core gene) pairwise average amino acid identity. Targeted genomic analysis showed that fructose bisphosphate aldolase was only present in the genome of Pw. cryptocerci. Mannitol dehydrogenase was found in genomes of Pw. beninensis, Pw. fabaria, and Pw. fabalis. Untargeted genomic analysis identified the presence of flagellar genes in Periweissella but not in other closely related genera. Phenotypes related to carbohydrate fermentation and motility matched the genotypes. Motility genes were organized in a single operon and the proteins shared a high amino acid similarity in the genus Periweissella. The relatively low similarity of motility operons between Periweissella and other motile lactobacilli indicated the acquisition of motility by the ancestral species. Our findings facilitate the phylogenetic, genetic, and phenotypic understanding of the genus Periweissella.
Importance
The genus Periweissella is a heterofermentative genus in the Lactobacillaceae which includes predominantly isolates from cocoa fermentations in tropical climates. Despite the relevance of the genus in food fermentations, genetic and physiological properties of the genus are poorly characterized and genome sequences became available only after 2020. This study characterized strains of the genus by functional genomic analysis, and by determination of metabolic and physiological traits. Phylogenetic analysis revealed that Periweissella is the evolutionary link between rod-shaped heterofermentative lactobacilli and the coccoid Leuconostoc clade with the genera Weissella and Furfurilactobacillus as closest relatives. Periweissella is the only heterofermentative genus in the Lactobacillaceae which comprises predominantly motile strains. The genomic, physiological, and metabolic characterization of Periweissella may facilitate the potential use of strains of the genus as starter culture in traditional or novel food fermentations.
KEYWORDS: Periweissella, Weissella; Furfurilactobacillus; Lactobacillus; heterofermentative lactobacilli; phylogenetic relationship; carbohydrate fermentation; motility
INTRODUCTION
Lactobacillaceae are significant members of human and animal gut microbiota, the plant phyllosphere and are dominant in most food fermentations (1). A taxonomic re-organization of the Lactobacillaceae revealed that the former Leuconostocaceae and Lactobacillaceae belong to the same family (2). However, genome sequence data that were available in August 2019 could not confidently establish the phylogenetic relationship of genera previously classified in the Leuconostocaceae to other lactobacilli (2). The availability of additional whole genome sequences in November 2021 revealed that all heterofermentative lactic acid bacteria form a monophyletic group (3). The inclusion of additional genome sequences also demonstrated that the genus Weissella was no longer monophyletic (3). The genus Periweissella, comprising strains previously classified as Weissella species, was proposed as a novel genus in 2022 (4). Five species, Pw. beninensis, Pw. fabalis, Pw. fabaria, Pw. ghanensis, and Pw. cryptocerci formed a single clade which was closely related to Weissella. Periweissella also shares common morphological characteristics with Weissella, i.e., cells are short rods or cocci. Periweissella spp. were isolated from spontaneously fermented cocoa beans (4), fermented cassava (5), and the gut of insects (6). Weissella species occur in many spontaneous plant fermentations where Periweissella have not been isolated, including fermented kimchi (7), carrot juice (8), and maize (9), spontaneous sourdoughs (10) or daqu (11).
Periweissella is also closely related to the genus Furfurilactobacillus and appeared to form the missing link between the former Leuconostocaceae and other (heterofermentative) Lactobacillaceae, based on a core genome phylogenetic tree of type strains in the family Lactobacillaceae (3). Moreover, one species in the genus Periweissella, Pw. beninensis, was described as motile (4). To date, motility has been characterized biochemically for strains in only seven of the more than 370 species of the Lactobacillaceae (3). Because motility is an exceptional physiological trait in the Lactobacillaceae, it is not routinely assessed when characterizing new strains or taxa, indicating the possibility of additional motile species in genus Periweissella.
The genus Periweissella may form the missing link between the coccoid and rod-shaped lactobacilli, but physiological and genetic traits that differentiate Periweissella from rod-shaped lactobacilli on the one hand and coccoid-shaped on the other are poorly described. It was, therefore, the aim of this study to investigate the genetic and physiological information on Periweissella which can putatively differentiate from closely related Lactobacillaceae. We aimed to determine the phylogenetic position of Periweissella relative to other heterofermentative lactobacilli and Lactiplantibacillus plantarum, the homofermentative type species that is most closely related to heterofermentative lactobacilli. Targeted and untargeted analysis of differentiating genetic traits were performed using genomes of type strains and relevant predicted metabolic or physiological traits were verified by analyses of the carbohydrate metabolism.
MATERIALS AND METHODS
Bacterial strain and growth conditions
Pw. beninensis LMG 25373T (DSM 22752), Pw. fabalis LMG 26217T (DSM 28407), Pw. fabaria LMG 24289T (DSM 21416), Pw. ghanensis LMG 24286T (DSM 19935), Pw. cryptocerci LMG 32586T (KACC 18423), and Liquorilactobacillus vini LMG 23202T were cultured aerobically in de Man-Rogosa-Sharpe (MRS) medium at 30°C overnight. The selection of strains included all strains of Periweissella spp. for which genome sequence data were available in May 2022 when the analyses were completed.
To determine the carbohydrate fermentation profile, strains were cultured at 30°C in the carbohydrate-free medium with supplementation of different carbohydrates (glucose, fructose, arabinose, xylose, glycerol, mannitol, and 1,2-propanediol) at a concentration of 1% (wt/vol). The carbohydrate-free medium contained the following ingredients per liter: peptone, 10 g; Lab-lemco powder, 8 g; yeast extract, 4 g; Tween 80, 1 mL; tri-ammonium citrate, 2 g; sodium acetate, 5 g; MgSO4, 0.2 g; MnSO4, 0.05 g; and K2HPO4, 2 g. The final pH was adjusted to 6.2.
DNA extraction, whole genome sequencing, and assembly
DNA of strains Pw. fabaria LMG 24289T and Pw. ghanensis LMG 24286T was extracted using an automated Maxwell DNA preparation instrument (Promega, Madison, Wisconsin, USA). DNA extracts were treated with RNAse (2 mg/mL, 5 µL/100 µL of extract) and incubated at 37°C for 1 hour. The DNA quality was checked using 1% agarose gel electrophoresis, and DNA quantification was performed using the QuantiFluor ONE dsDNA system and the Quantus fluorometer (Promega, USA). Draft genomes of both strains were sequenced at the MiGS center (Pittsburgh, Philadelphia, USA) using the Illumina NextSeq 2000 (PE150) platform. Quality reports were created with fastp version 0.20.0. Prior to assembly, reads were trimmed (Phred score >Q30) and filtered (length >50 bp) with fastp 0.20.0 (12) with correction option enabled. Assembly was performed with Shovill version 1.1.0, with SPAdes genome assembler 3.14.0 (13) at its core with read error correction disabled and default settings. Contigs shorter than 500 bp were removed from the final assembly. The quality of the final assembly was verified with The Quality Assessment Tool for Genome Analysis [QUAST (14)], which generates summary statistics such as the number of contigs, N50, L50, and the G + C content. Finally, the assemblies were checked for completeness and contamination using CheckM version 1.1.2 (15).
Phylogenetic analysis of type strains genomes
For the phylogenetic analysis of heterofermentative lactobacilli, genome sequences of 67 type strains of the genera Furfurilactobacillus, Periweissella, Weissella, Oenococcus, Convivina, Fructobacillus, and Leuconostoc, and type species of each of the heterofermentative genera in the Lactobacillaceae and the type strain of Lp. plantarum as provided on the List of Prokaryotic names with Standing in Nomenclature (https://lpsn.dsmz.de/) were downloaded from Genbank and re-annotated by Prokka (Table S1) (16). Protein sequences of all genomes were extracted, and an all-against-all blast was performed using BLASTp with the E-value as 10−10. Then single-copy core gene sequences were extracted, trimmed, and concatenated into a new alignment using PGCGAP (17). Phylogenetic trees were computed by IQ-TREE (18) and were visualized by iTOL (19). Bootstrap values were calculated from 1,000 replicates. The final tree was rooted using the clade of the type strain of Lp. plantarum and type species of each of the genera of heterofermentative Lactobacillaceae as outgroup.
The pairwise average amino acid identity (AAI) between type strains was calculated by CompareM (https://github.com/dparks1134/CompareM). Sequences of soft-core genes, i.e., genes that are present in more than 90% of the genomes, were also used to determine the average amino acid identity of core proteins (cAAI) between type strains and genera. For determination of the inter-family/inter-genus relatedness, AAI or cAAI values are preferable over ANI values that are most commonly used to determine inter-genus relatedness (2).
In silico genome analysis and analysis of carbohydrate metabolism
The comparative genomic analysis of the genera Weissella, Periweissella, and Furfurilactobacillus was performed with all type strain genomes. Owing to the low number of type strain genomes for the genus Furfurilactobacillus, additional genomes of strains in the genus (Table S2) were included. Annotations from Prokka were used by Roary (20) to produce a gene presence/absence table of Periweissella, Weissella, and Furfurilactobacillus strains. Genome annotation by Prokka is based on curated databases including Swissprot/Uniprot to provide high quality annotations, which facilitates subsequent verification of gene function by wet-lab experimentation (21). The three genera were treated as different phenotype groups and were associated with gene presence/absence patterns using Scoary (Benjamini-Hochberg adjusted P value of < 0.05) (22). Duplicate hits to the same protein, as well as putative and hypothetical proteins were removed from the output results. Only the hits for which four or more Periweissella species were present were chosen. Key enzymes (23) of metabolic pathways for carbohydrate metabolism were downloaded from NCBI (Table S3) and were used as query sequences for BLAST analysis against type strain genomes of Lactobacillaceae (Tables S1 and S4) (3) with an amino acid identity of 40% and a query coverage of more than 70% as cut-offs. Heatmaps depicting the percentage of type species in each genus of the family Lactobacillaceae that harbor genes for metabolic traits were drawn with R software (version 4.1.3, https://www.r-project.org/).
In silico comparison of flagellar operons in Lactobacillaceae
An initial assessment of whether motility operons in Periweissella are functional, the four motility operons in Periweissella were compared to the corresponding operons of seven genome-sequenced species of Lactobacillaceae for which motility was determined by reliable motility assays or by electronmicroscopic observation of flagellar (Table S5). Using the second contig of Pw. ghanensis LMG 24286T as the template, contigs of other draft genomes were re-ordered by Mauve (24) and Benchling Biology Software. A built-in BLASTn with an E-value of 0.01 was performed to determine the % protein identity between homologous proteins in different genomes and multiple alignments of flagellar coding regions were visualized using EasyFig (25).
The amino acid sequences of 45 flagellar-related genes identified from the EasyFig analysis were downloaded from NCBI and were used as query sequences for BLAST analysis following the same procedures as for the analysis of carbohydrate metabolism. A heatmap depicting the presence and absence of genes in each strain was drawn with R software (version 4.1.3, https://www.r-project.org/).
Characterization of motility
Four Periweissella strains, Pw. ghanensis, Pw. fabaria, Pw. fabalis, and Pw. cryptocerci, were inoculated into the center of a tube containing semi-solid MRS medium by stabbing. The tube was incubated at 30°C overnight to observe the spreading growth of strains.
For transmission electron microscopy (TEM), bacterial cells of a one-day-old broth culture were centrifuged at 123 × g for 20min, washed in phosphate buffered saline (PBS) and fixed for 20min in 50 µL of 4% paraformaldehyde at pH 7.3. To prevent clumping of cells, 0.1% Triton-X 100 (Sigma) was added to the fixative. The cells were then adsorbed onto a formvar-coated copper single slot grid for 10 minutes and were rinsed twice in PBS and once in distilled water. Cells were negatively stained with a 2% aqueous solution of uranyl acetate for 10 seconds. The excess fluid was removed with a filter paper and the grid was then air-dried. TEM analysis of Pw. beninensis LMG 25373T, Pw. fabalis LMG 26217T, Pw. fabaria LMG 24289T, Pw. ghanensis LMG 24286T, and Pw. cryptocerci LMG 32586T was performed using a Jeol JEM 1010 transmission electron microscope at 60 kV, equipped with a CCD side-mounted Veleta camera (Emsis, Münster, Germany).
For scanning electron microscopy (SEM) of Pw. beninensis LMG 25373T, 10 µL of a 1-day-old broth culture was spotted on a polycarbonate Whatman Nuclepore Track-Etched Membrane with a pore size of 0.4 µM (Merck, Darmstadt, Germany), dried overnight, and after a short wash in 0.1M sodium cacodylate buffer at pH 7.4 (VWR International, Leuven, Belgium) fixed in freshly prepared fixative (2% paraformaldehyde, VWR), 2.5% glutaraldehyde (VWR) in 0.1 M cacodylate buffer, pH 7.4, at room temperature for 1 hour. The fixative was removed by washing 3 × 5 minutes in 0.1 M cacodylate buffer and samples were then incubated in 2% osmium tetroxide (VWR) in 0.1 M cacodylate buffer for 30 minutes at room temperature. After washing in water for 3 × 5 minutes, the sample was dehydrated using solutions of increasing ethanol concentration (50%, 70%, 85%, 95%, 2 × 100%), for 15 minutes each. Samples were then critical point dried (Leica EM CPD300) and coated with a thin layer of gold or platinum (Quorum Q150T ES) before imaging with a scanning electron microscope (Crossbeam 540, Zeiss, Jena, Germany). Images were taken using a SE2 detector at 1.5 kV.
Carbohydrate fermentation profile
Type strains of Pw. beninensis, Pw. fabalis, Pw. fabaria, and Pw. ghanensis were tested for their ability to metabolise glucose and fructose. Liqorilactobacillus vini LMG 23202T was used as a control strain. Each carbohydrate was added to 10 mL of carbohydrate-free MRS medium at a concentration of 1% (wt/vol) in the case of arabinose, fructose, glucose, and xylose, and of 10 mM in the case of glycerol, mannitol, and 1,2-propanediol. All media were prepared with and without 1% glucose, so that a total of 13 different fermentation conditions were tested. After 48 hours of incubation at 30°C, the cultures were centrifuged at 4,696 × g for 20 minutes and the supernatants were kept at −20°C until further analysis. All incubation tests were performed in triplicate. The concentrations of carbohydrates (arabinose, fructose, glucose, and xylose) and sugar alcohols (arabitol, erythritol, glycerol, mannitol, and sorbitol) were quantified by high-performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD). To this end, 100 µL of cell-free culture supernatant was mixed with 900 µL of deproteinization solution, consisting of 1 L of acetonitrile (Thermo Fisher Scientific, Waltham, Massachusetts, USA) and 0.05 g of fucose as internal standard (IS). Samples were then vortexed for 2 minutes, centrifuged at 19,000 × g for 15 minutes, and filtered using 0.2 µM H-PTFE filters (Millex; Merck). Ethanol, acetic acid, and lactic acid concentrations were quantified using high-performance liquid chromatography with refractive index detection (HPLC-RI) as described previously (26). In this case, the deproteinization step was performed by adding 300 µL of Carrez A solution [36 g/L of K4Fe(CN)6.3H2O] and 300 µL of Carrez B solution (72 g/L of ZnSO4.7H2O) to 600 µL of cell-free culture supernatant. The quantification of 1,2-propanediol was performed by liquid injection gas chromatography with tandem mass spectrometry (LI-GC-MS/MS). In this case, 100 µL of sample were mixed with 900 µL of deproteinization solution, consisting of 1 L of acetone (Merck) with 50 µg of deuterated 3-methyl-1-butanol, 2,3-butanedione, and ethyl decanoate (CDN Isotopes, Pointe-Claire, Quebec, Canada) as IS. The quantification of all compounds mentioned above was performed by external calibration.
RESULTS
Whole genome sequencing
The assembly of the Illumina NextSeq 150 bp paired-end reads resulted in assemblies of 19 (Pw. fabaria LMG 24289T) and 66 (Pw. ghanensis LMG 24286T) contigs with N50 values of 201 Kb and 59 Kb and a total genome size of 1.93 Mb and 1.99 Mb, respectively. Both genomes showed more than 98% completeness and less than 1% contamination as determined using CheckM.
Phylogenetic analysis of the genera Furfurilactobacillus, Periweissella, Weissella, Oenococcus, Convivina, Fructobacillus, and Leuconostoc
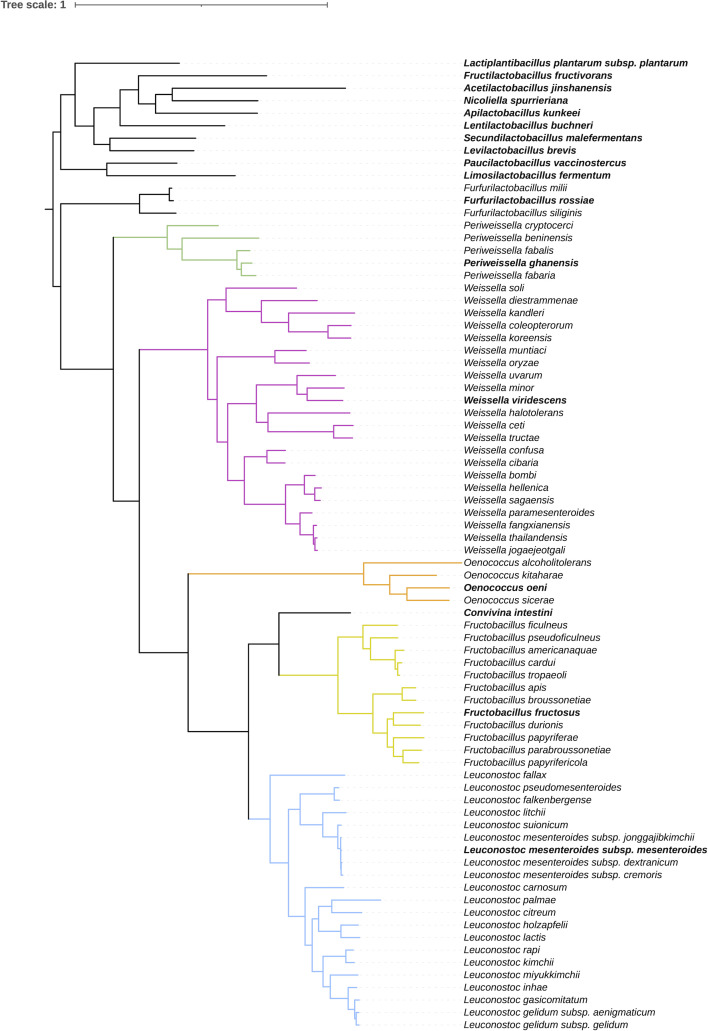
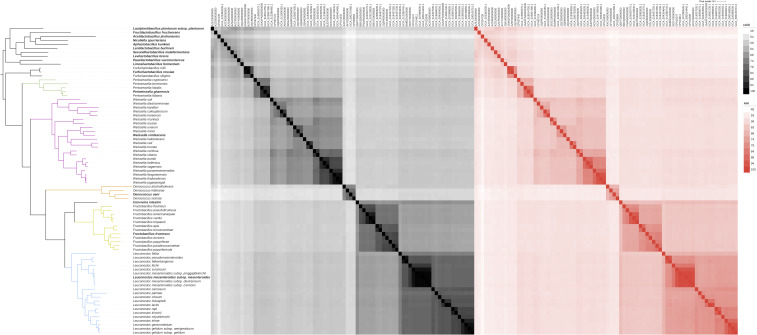
The description of the genus Periweissella was based on phylogenetic comparison with only the genera Weissella, Oenococcus, Convivina, Fructobacillus and Leuconoctoc (4); however, these genera form a monophyletic clade together with other heterofermentative Lactobacillaceae (3). The tree calculated in this study also clearly separated lactobacilli and the former Leuconostocaceae (Fig. 1). The Lactobacillaceae, however, are monophyletic only if the former Leuconostocaceae are included (Fig. 1). We, therefore, extended the phylogenetic analyses of Periweissella to include the genus Furfurilactobacillus and the type species of all 10 other heterofermentative genera using Lactiplantibacillus as outgroup (Fig. 1). Periweissella clustered between Furfurilactobacillus and Weissella and thus provided the phylogenetic link of the coccoid heterofermentative Lactobacillaceae, formerly Leuconostocaceae, to other heterofermentative Lactobacillaceae. The pairwise AAI and cAAI values for the type strains of heterofermentative Lactobacillaceae were also calculated (Fig. 2). The intra-genus AAI and cAAI values of Periweissella were higher than 68% and 74%, respectively (Tables S6 and S7). The genera Periweissella and Weissella were exclusive, i.e., the lowest intra-genus cAAI values were higher than the highest inter-genus cAAI of species in these genera (Fig. S1). The inter-genus AAI values for the genera Periweissella and Furfurilactobacillus were in a range of 57.53%–59.21% (average 58.30%) and the corresponding AAI values of the genera Periweissella and Weissella were in a range of 57.59%–60.00% (average 58.43%), again indicating that the genus Periweissella is about as closely related to Furfurilactobacillus as it is to Weissella.
Fig 1.
Core genome phylogenetic tree of 50 type strains of the genera Furfurilactobacillus, Periweissella, Weissella, Oenococcus, Convivina, Fructobacillus, and Leuconostoc. The type strain of Lactiplantibacillus plantarum and type species of each of the heterofermentative genera in the Lactobacillaceae were used as outgroups. The maximum likelihood tree is based on the concatenated alignment of protein sequences from single-copy core genes and was inferred by IQ-TREE as described in Zheng et al. (2). Species of the same genus are indicated by the same color code for branches and the type species of each genus is printed in bold. The genomes used for the graph are provided in the Table S1.
Fig 2.
Heatmap of the average amino acid identity (AAI) and average amino acid identity of core proteins (cAAI) values between 50 type strains of the genera Furfurilactobacillus, Periweissella, Weissella, Oenococcus, Convivina, Fructobacillus, and Leuconostoc. The type strain of Lactiplantibacillus plantarum and type species of each of the heterofermentative genera in the Lactobacillaceae were used as outgroups. Rows and columns are clustered according to the phylogenetic tree. Values of AAI and cAAI are shown in black and red, respectively. The data used for the graph are provided in Tables S6 and S7.
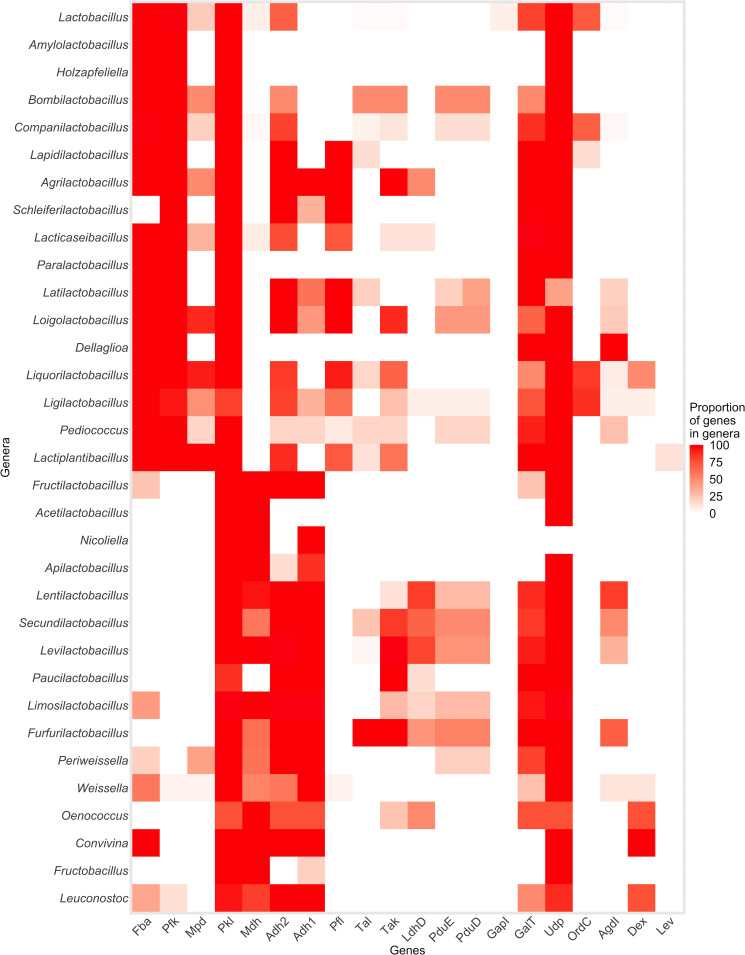
Targeted genomic analysis of the metabolic potential of Periweissella
To investigate the metabolic potential of Periweissella, key enzymes for carbohydrate metabolism (3, 27) in the genus Periweissella as well as in other genera of Lactobacillaceae were identified by protein BLAST (Fig. 3). Fructose bisphosphate aldolase was only present in the genome of Pw. cryptocerci. Periweissella was the only genus in heterofermentative lactobacilli other than Weissella in which mannitol phosphate dehydrogenase was identified (28). The gene coding for mannitol dehydrogenase, which enables the conversion of fructose to mannitol to regenerate reduced co-factors in heterofermentative lactobacilli (27) was found in the genomes of Pw. beninensis, Pw. fabaria, and Pw. fabalis but not in those of Pw. ghanensis and Pw. cryptocerci (Fig. 3). Glycerol dehydratase (28) was present in the genome of Pw. cryptocerci. Pw. cryptocerci does not produce acid from glycerol (29), but its ability to convert glycerol to 1,3 propanediol remains unknown. Glycerol dehydratase (29) was also present in the genomes of several Furfurilactobacillus species but absent in Weissella, Oenococcus, Convivina, Fructobacillus, and Leuconoctoc. Dextransucrases were frequently present in the genomes of Weissella, Oenococcus, Convivina, and Leuconoctoc but absent in Periweissella.
Fig 3.
Heatmap depicting the percentage of type species in each genus of the family Lactobacillaceae that harbor genes for metabolic traits. Red indicates the gene is present in all type strains of a genus and white indicates the gene is absent in all type strains. Genes used for the graph are provided in Table S3. Genes are indicated as follows: Fba, aldolase; Pfk, phosphofructokinase; Mpd, mannitol-phosphate-dehydrogenase; Pkl, phosphoketolase; Mdh, mannitol dehydrogenase; Adh2, two-domain alcohol dehydrogenase; Adh1, alcohol dehydrogenase; Pfl, pyruvate formate lyase; Tal, transaldolase; Tak, transketolase; LdhD, lactaldehyde dehydrogenase; PduE,; PduD, glycerol dehydratase subunit PduD; GapI, galactose-6-phosphate isomerase; GalT; galactose-1-phosphate-uridyltransferase; Udp, UDP-4-galactose-epimerase; OrdC, ornithine decarboxylase; AgdI, agmatine deiminase; Dex, dextransucrase; Lev, levansucrase.
Untargeted genomic analysis of the metabolic potential of Periweissella
To identify other genes that contribute to the metabolic and ecological traits of Periweissella, Roary and Scoary were used to identify differentially distributed genes in the core genome of Periweissella and the two closest genera, Furfurilactobacillus and Weissella. Around 400 genes were differentially distributed among the three genera (Tables S8 and S9). Of these, 34 genes related to carbohydrate metabolism, drug resistance, and bacterial motility were further selected (Table 1). Motility-related genes were present in four out of five Periweissella strains, but not in Furfurilactobacillus or Weissella. Several ribose metabolism-related genes were present in Periweissella strains but absent from all Weissella strains. Genes coding for multidrug resistance, maltose phosphorylase, and glycerol dehydrogenase were found only in Furfurilactobacillus strains.
TABLE 1.
Gene presence and absence from untargeted genomic analysis of the genera Furfurilactobacillus, Periweissella, and Weissellab
| Annotationa | Gene | Accession no. used for verification | Traits | ||
|---|---|---|---|---|---|
| Furfuri-lactobacillus | Periweissella | Weissella | |||
| Maltose phosphorylase | MalP | QJU50634.1 | + | − | |
| Glycerol dehydrogenase | GldA | ARW49736.1 | + | − | |
| Multidrug efflux system permease | MesP | CAH0417302.1 | + | − | |
| Multidrug resistance protein MdtH | MdtH | GFI63498.1 | + | − | |
| Multidrug resistance protein MdtL | MdtL | QRQ96649.1 | + | − | |
| Multidrug transporter EmrE | EmrE | KRN47543.1 | − | + | − |
| Chemoreceptor glutamine deamidase CheD | CheD | CAH0417903.1 | − | + | − |
| Chemotaxis protein CheA | CheA | GFI63782.1 | − | + | − |
| Chemotaxis protein CheW | CheW | WP_235719183.1 | − | + | − |
| Chemotaxis protein CheY | CheY | KRN88084.1 | − | + | − |
| Chemotaxis protein methyltransferase | CheM | CAH0417905.1 | − | + | − |
| Chemotaxis protein PomA | PomA | CAH0417878.1 | − | + | − |
| CheY P phosphatase CheC | CheC | CAH0417907.1 | − | + | − |
| Flagellar basal body rod protein FlgB | FlgB | CAH0417879.1 | − | + | − |
| Flagellar basal body rod protein FlgC | FlgC | WP_235315572.1 | − | + | − |
| Flagellar basal body rod protein FlgG | FlgG | AUJ33046.1 | − | + | − |
| Flagellar biosynthesis protein FlhA | FlhA | WP_057895838.1 | − | + | − |
| Flagellar biosynthetic protein FlhB | FlhB | BBA81011.1 | − | + | − |
| Flagellar biosynthetic protein FliP | FliP | CAH0417893.1 | − | + | − |
| Flagellar biosynthetic protein FliR | FliR | WP_235315560.1 | − | + | − |
| Flagellar hook basal body complex protein | FliE | WP_235807225.1 | − | + | − |
| Flagellar motor switch protein FliG | FliG | WP_235315569.1 | − | + | − |
| Flagellar motor switch protein FliM | FliM | WP_224288758.1 | − | + | − |
| Flagellar M ring protein | FlmR | CAH0417882.1 | − | + | − |
| Flagellar secretion chaperone FliS | FliS | CAH0417922.1 | − | + | − |
| L asparaginase 1 | Ans | GFI59533.1 | − | + | |
| Ribose import ATP-binding protein RbsA | RbsA | CAH0416239.1 | + | − | |
| Ribose import binding protein RbsB | RbsB | GFI19485.1 | + | − | |
| Ribose import permease protein RbsC | RbsC | CAH0416238.1 | + | − | |
| Multidrug export protein EmrB | EmrB | GFI63251.1 | + | − | |
| Multidrug export protein MepA | MepA | GFI60335.1 | + | + | − |
| L fucose proton symporter | FucP | GFI59483.1 | + | + | − |
Scoary additionally identified ribokinase and ribose-5-phosphate isomerase A as present in Periweissella but absent in Weissella; however, the corresponding genes are present in Weissella but with an amino acid identify of less than 50%.
The presence and absence of genes is indicated by “+” and “−“, respectively.Cells are blank if they are not differentiated between two of the three genera.
Phenotypic carbohydrate fermentation profile of Periweissella
The carbohydrate fermentation profiles of Pw. beninensis, Pw. fabalis, Pw. fabaria, and Pw. ghanensis type strains were tested by detailed quantification of products and metabolites of carbohydrate metabolism. Glucose was metabolized to lactate and acetate or ethanol (Table 2), matching the pattern of metabolites in heterofermentative hexose metabolism via the phosphoketolase pathway (Fig. 3). Xylose and arabinose as the sole carbohydrate sources were not metabolized, in agreement with the absence of the gene for xylose isomerase (XylA) and the gene for L-arabinose isomerase (AraA) in the genomes of all type strains tested. However, these pentoses were metabolized when glucose was also present, especially xylose. None of the strains consumed 1,2-propanediol or glycerol, matching the absence of glycerol dehydratase, glycerol dehydrogenase or the aerobic α-glycerophosphate oxidase in the genomes of the four strains evaluated (Fig. 3). 1,2-Propanediol was not produced by any of the strains, matching the absence of lactaldehyde dehydrogenase. When glucose and fructose were present as substrates, fructose was reduced to mannitol with concomitant formation of acetate instead of ethanol (Table 2). The Pw. fabalis and Pw. fabaria type strains used fructose as carbon source; in these fermentations, fructose was partially reduced to mannitol and partially converted to lactate and acetic acid or ethanol. Despite the presence of mannitol phosphate dehydrogenase in the genomes of the Pw. ghanensis and Pw. fabalis type strains, none was able to use mannitol as carbon source when mannitol was present as the sole carbon source or in combination with glucose.
TABLE 2.
Carbohydrate consumption, indicated by (−) and metabolite production, indicated by (+) after 48 hours fermentation in modified MRS mediaa
| Substrates/ metabolites (mM) | Mean consumption/production ± standard deviation (mM) | ||||
|---|---|---|---|---|---|
| Pw. beninensis LMG 25373T | Pw. fabalis LMG 26217T | Pw. fabaria LMG 24289T | Pw. ghanensis LMG 24286T | Lq. vini LMG 23202T | |
| Fructose and glucose | |||||
| Glucose (−) | 69 ± 0 | 70 ± 2 | 57 ± 2 | 69 ± 1 | 21 ± 3 |
| Fructose (−) | 58 ± 0 | 33 ± 0 | 29 ± 7 | 58 ± 0 | 9 ± 4 |
| Mannitol (+) | 39 ± 10 | 39 ± 6 | 9 ± 1 | 46 ± 7 | 0* |
| Lactate (+) | 64 ± 5 | 96 ± 2 | 85 ± 12 | 80 ± 11 | 44 ± 5 |
| Acetate (+) | 23 ± 4 | 34 ± 3 | 21 ± 6 | 38 ± 8 | 0 |
| Ethanol (+) | 11 ± 1 | 58 ± 12 | 38 ± 17 | 13 ± 6 | 0* |
| Fructose | |||||
| Fructose (−) | 23 ± 6 | 53 ± 1 | 48 ± 4 | 19 ± 1 | 9 ± 5 |
| Mannitol (+) | 0 | 22 ± 3 | 16 ± 2 | 0 | 0 |
| Lactate (+) | 20 ± 3 | 37 ± 0 | 43 ± 4 | 22 ± 2 | 35 ± 3 |
| Acetate (+) | 0 | 10 ± 2 | 8 ± 2 | 0 | 0 |
| Ethanol (+) | 7 ± 3 | 8 ± 0 | 10 ± 4 | 9 ± 3 | 0 |
Concentrations are expressed as the difference between the concentrations in the negative control (not inoculated) and in the respective sample ± standard deviation.
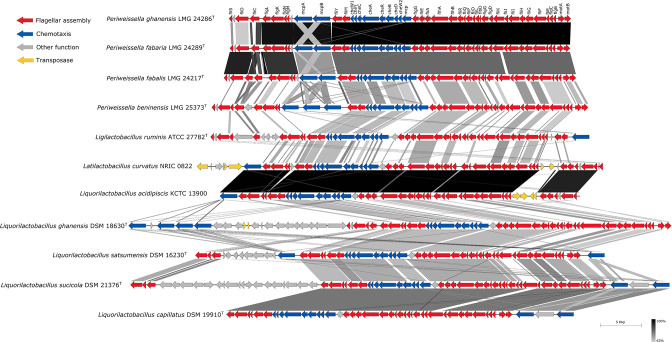
Comparison of flagellar operons of Periweissella as well as all reported motile strains of the family Lactobacillaceae
Prior to the comparison of the motility operons of four Periweissella type strains together with seven other motile lactobacilli (Table S5), the motility genes of the Periweissella species were investigated for their organization into single operons. In Pw. ghanensis LMG 24286T, genes related to motility were located on only one contig, whereas they were distributed on several contigs in all other Periweissella genomes examined. Therefore, the operon of Pw. ghanensis LMG 24286T was used to identify the presence and organization of motility operons in other genomes (Fig. 4). Among the five type strains of Periweissella, Pw. beninensis LMG 25373T was the only strain for which motility was described before (5). The motility operon contained 45 genes involved in flagellar structure, function, and chemotaxis (Fig. 4). Of these 45 genes, 34 genes were also present in Pw. fabalis LMG 26217T and Pw. fabaria LMG 24289T, in a nearly identical organization (Fig. 4). The number and organization of motility genes in Pw. beninensis differs substantially from the other three strains and motility genes were absent in Pw. cryptocerci LMG 32586 (Fig. 4). Since May 2022, the genome of Pw. beninensis 716 became additionally available (Genbank accession number GCA_025211165.1). The genome of Pw. beninensis 716 also encodes for a motility operon that shares 98.96% nucleotide identity with the operon of the type strain and has an identical organization of motility genes (data not shown).
Fig 4.
Comparison of genetic loci coding for flagellar-related proteins in type strains of four Periweissella species as well as seven reported motile lactobacilli strains. Shades of gray of connecting lines represent percent blast identity according to the scale on the right. Periweissella cryptocerci LMG 32586T does not contain any flagellar-related proteins. Pw. beninensis LMG 25373T was re-ordered, and Pw. fabalis LMG 26217T was reversed along with the template of Pw. fabaria LMG 24289T using Mauve (version 2.4.0) (24). Genomes used for the graph are provided in Table S5.
Phenotypic identification of flagellum-mediated motility of Periweissella
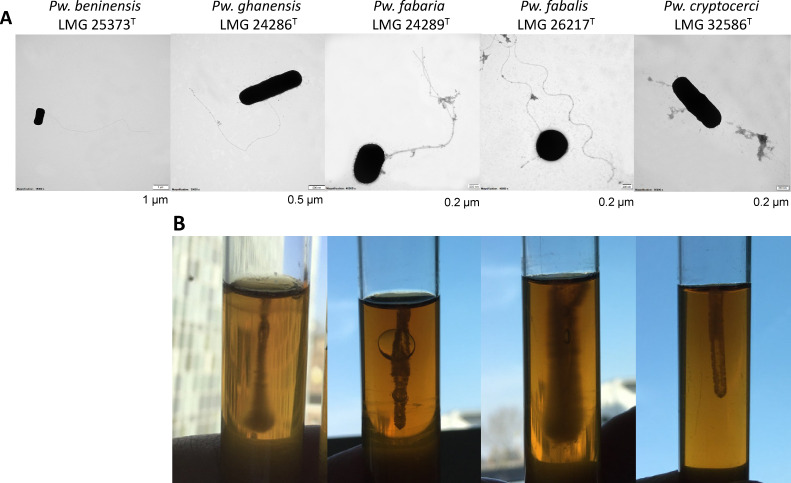
To confirm the motility of the Periweissella type strains, semi-solid agar assays, TEM, and SEM were conducted. The strains Pw. ghanensis LMG 24286T, Pw. fabaria LMG 24289T, and Pw. fabalis LMG 26217T were motile in the semi-solid agar assay and TEM revealed flagellated cells, whereas Pw. cryptocerci LMG 32586T was non-motile and no flagellar structures were detected (Fig. 5). Pw. beninensis LMG 25373T failed to grow under the semi-solid agar conditions, but flagellar filaments of Pw. beninensis LMG 25373T were observed by TEM (Fig. 5) and SEM (data not shown), which was consistent with the presence of motility operons in its genome. TEM analysis showed thin filaments which were positioned along the length of the cells, indicating the swimming motility of these bacteria by the rotation of flagellar filaments (30). Among four motile strains, Pw. fabalis LMG 26217T was considered to have a highly motile ability from its moving at exponential phase in semi-solid MRS medium.
Fig 5.
Flagellar and motility of strains in the genus Periweissella. (A) Transmission electron micrographs of five Periweissella strains. The size of the scale bar at the bottom right is indicated below the images. (B) Semisolid MRS medium assays in tubes for Pw. fabalis LMG 26217T, Pw. fabaria LMG 24289T, Pw. ghanensis LMG 24286T and Pw. cryptocerci LMG 32586T. Pw. beninensis did not grow in the semi-solid MRS medium.
DISCUSSION
This study determined the genetic and physiological properties that may differentiate Periweissella from other heterofermentative Lactobacillaceae by genomic and physiological analyses. The phylogenetic position of Periweissella documents that the genus forms missing link between Leuconostoc, Oenococcus, Convivina, Fructibacillus, and Weissella and other heterofermentative Lactobacillaceae. The genus Periweissella was also found to be only the second predominantly motile genus in addition to Liquorilactobacillus (31–34), thus providing an important physiological trait that distinguishes most Periweissella species from all other heterofermentative lactic acid bacteria (4).
The proposal of the genus Periweissella was based on the phylogenetic analysis of coccoid lactobacilli (4). By including other heterofermentative lactobacilli in the analysis, the phylogenetic tree as well as cAAI and AAI values of the present study showed that the phylogenetic relationship of the new genus Periweissella to Furfurilactobacillus species is as close as the relationship to Weissella species. Addition of the genomes of Periweissella species to the core genome phylogenetic tree of the Lactobacillaceae also demonstrated that all heterofermentative lactic acid bacteria form a monophyletic clade among the Lactobacillaceae (3), indicating that the transition from homofermentation to heterofermentation was a unique and irreversible event in the evolution of lactic acid bacteria.
Periweissella have the shape of short, coccoid rods or of cocci. In the Lactobacillaceae, the coccoid or coccus-shaped genera Leuconostoc, Oenococcus, Weissella, and Periweissella each form a monophyletic clade. The genus Pediococcus and the species Lapidilactobacillus dextrinicus, Loigolactobacillus coryniformis, and Limosilactobacillus equigenerosi are also monophyletic and have been described as cocci or as short coccoid rods (2). Analyses of the orders Bacilli and Erysipelotrichia indicated that the transition of rod to coccus is essentially irreversible (35). This assumption is only partially congruent with cell morphology in the Lactobacillaceae. Most clades with coccoid morphology have a rod-shaped ancestor; however, the rod-shaped genera Fructobacillus and Convivina appear to have coccus-shaped ancestors (Fig. 1) (3), thus indicating the possibility of a coccus-to-rod transition.
The phylogenetic position of a genus in the Lactobacillaceae family often relates to its physiological and metabolic potential as well as its ecology (23, 36), but only limited information is available on differentiating ecological and metabolic properties of the genus Periweissella (4). This study analyzed the metabolic potential with a genome data set that was rarefied to include only type strains and additional genome sequences of furfurilactobacilli. With very few exceptions, genome sequences are available for the over 340 type strains in the Lactobacillaceae, but for most species, the type strain is the only strain with genome sequence data. Less than 30 species, e.g., Lactiplantibacillus plantarum, Levilactobacillus brevis, W. cibaria, and W. confusa, account for more than half of the 5,000 genomes of strains in the Lactobacillaceae that are available on NCBI. Rarefaction of the genome data set is necessary to have equal representation of each species (23). Rarefaction by type strains, however, also prevents elucidation of the full species-level metabolic diversity. Periweissella species are heterofermentative and follow the common genus-level trait proposed by Zheng et al. (23). The phosphoketolase gene is present in all Periweissella species, indicating carbohydrate metabolism via the phosphoketolase pathway (27), which matches the phenotype that all tested Periweissella strains can grow with glucose as the sole carbon source and produce lactate and ethanol.
Most heterofermentative Lactobacillaceae express mannitol dehydrogenase to use fructose as electron acceptor if other hexoses or disaccharides are present (3, 23). In contrast, mannitol phosphate dehydrogenase, which supports the use of mannitol as carbon source, was detected exclusively in homofermentative Lactobacillaceae (3, 23, 37). The presence of mannitol phosphate dehydrogenase in Periweissella is thus a feature that distinguishes Periweissella from all other heterofermentative Lactobacillaceae, including the genera Weissella and Furfurilactobacillus. Acid production from mannitol by resting cells has been described for Pw. beninensis and Pw. fabaria; however, genes coding for mannitol-phophate-dehydrogenase are present in Pw. ghanensis and Pw. fabalis (5, 38, 39). Our study demonstrates, however, that mannitol does not support growth of these two species when offered as sole carbon source (Fig. 3; Table 2). The analysis of carbohydrate fermentation by lactobacilli can provide different results when conversion by resting cells or growth is compared (40). Periweissella and the closely related genera Furfurilactobacillus and Weissella are among the five exceptional genera of heterofermentative Lactobacillaceae that include type strains that lack mannitol dehydrogenase (3, 23) (Fig. 3). Accordingly, in the presence of fructose as the sole carbon source, only the type strains of Pw. fabalis and Pw. fabaria converted fructose into lactate, acetate, and mannitol via the phosphoketolase pathway and mannitol dehydrogenase (Table 2). The strain Pw. beninensis LMG 25373T did not produce mannitol when fructose was present as the sole carbon source; however, mannitol was produced when both glucose and fructose were available as substrates. Remarkably, Pw. ghanensis LMG 24286T also produced mannitol when glucose as well as fructose were present as substrate, although mannitol dehydrogenase was not identified in the genome of Pw. ghanensis LMG 24286T (Fig. 3; Table 2) when using the biochemically characterized proteins from Limosilactobacillus reuteri (41) or Fructilactobacillus sanfranciscensis (42) as BLAST query sequences. Pw. ghanensis LMG 24286T encodes for mannitol-phosphate dehydrogenase, which converts fructose-6-phosphate into mannitol 1-phosphate (Fig. 3). Mannitol 1-phosphate is converted into mannitol by a phosphatase without recovery of the chemical energy of the phosphate bond, which is energetically unfavorable (28, 43). Although the phenotype for carbohydrate fermentation in strains of Periweissella largely matched the genotype, the fate of fructose in those Periweissella strains that encode both mannitol dehydrogenase and mannitol 1-phosphate dehydrogenase (Pw. fabalis LMG 26217T) or only mannitol 1-phosphate dehydrogenase (Pw. ghanensis LMG 24286T) remains subject of future studies.
Pangenomes of Periweissella and the two closest genera, Weissella and Furfurilactobacillus, were compared with the aim to elucidate the genes that are differentially distributed between these genera. Flagellar-related genes are exclusive to Periweissella; the L-asparaginase gene is exclusive to Weissella. Genes coding for the multidrug export protein MepA and the L-fucose proton symporter are present in both Furfurilactobacillus and Periweissella but are absent in Weissella. Considering the mismatching of motility genes and the motile phenotype in Ligilactobacillus ruminis (44) and the disruption of transposase in the motility operon in Lactobacillus hordei (45), the motility of four strains of Periweissella were further investigated by the semi-soft agar and TEM analyses. Four Periweissella strains were shown to be motile and / or to express flagella, which is consistent with the presence of flagellar operons.
The high similarity of motility operons in Latilactobacillus curvatus and Ligilactobacillus acidipiscis as well as the presence of mobile genetic elements flanking the Lt. curvatus NRIC 0822 motility operon suggested recent acquisition of motility in these two species (45). In contrast, our analysis provided no indication for the presence of mobile genetic elements in motility operons in the genera Periweissella and Liquorilactobacillus, or differences between the G + C content of the motility operon and the entire genome. Bioinformatic analyses indicated that the loss of motility is much more likely than acquisition of motility (35). Taken together, motility was likely acquired independently by the ancestral species of the genera Periweissella to Liquorilactobacillus but lost again in some of the progeny.
The comparison of free-living and host-associated bacteria has demonstrated that loss of motility is associated with the transition from a free-living to a host-associated lifestyle (35). In addition, the expression of motility genes by intestinal microorganisms was reduced by mucosal anti-flagellin antibodies (46). To date, several species in four genera of the Lactobacillaceae have been shown to be motile. Motile strains are frequent in the genera Periweissella and Liquorilactobacillus, much less frequent in the genus Ligilactobacillus and only one strain of Lt. curvatus has been described as motile (5, 31–34, 38, 45, 47–52). Several of these references, however, relate to species new descriptions without providing detail on methods and results. This study in conjunction with past studies clearly establishes that motility of lactobacilli is a physiological trait that relates to phylogeny and ecology of the organisms (5, 31–34, 38, 45, 47–52). The vertebrate gut adapted genera Lactobacillus and Limosilactobacillus are not known to include motile species or strains. Of the four motile genera, only Ligilactobacillus consists predominantly but not exclusively of vertebrate host-adapted species; Latilactobacillus and those species of Liquorilactobacillus for which sufficient information is available were designated as free-living organisms (36). The lifestyle of species in the genus Periweissella is unknown and most isolates were obtained from fermented plant foods, which suggests a free-living lifestyle in association with plants (5, 32, 38, 48, 53). The source of isolation of motile Periweissella species partially overlaps with the source of isolation of Liquorilactobacillus (31, 32, 34). Conversely, Periweissella does not share the source of isolation with closely related heterofermentative lactobacilli including Leuconostoc, Weissella, Furfurilactobacillus, or Levilactobacillus. The similarity of sources for motile Periweissella species and Liquorilactobacillus species is indicative of the importance of flagellum-mediated motility and the advantages that likely accompany this trait, such as niche colonization or biofilm formation (44).
In conclusion, this study provides a more comprehensive insight into the phylogenetic and physiological properties of Periweissella and builds a link to connect Periweissella and other heterofermentative lactobacilli. Periweissella is proposed as the second predominantly motile genus among lactobacilli. These findings will promote the understanding and industrial application of strains of this new genus.
ACKNOWLEDGMENTS
L.D.V. and C.M. acknowledge their financial support from the Research Council of the Vrije Universiteit Brussel (SRP7 and IOF342 projects). P.V., M.C., and E.D. acknowledge the TEM facility of the Nematology Research Unit, member of the UGent TEM-Expertise center (life sciences). M.G. acknowledges financial support from the Natural Sciences and Engineering Research Council of Canada and the Canada Research Chairs Program. N.Q. acknowledges scholarship support from the China Scholarship Council.
Contributor Information
Michael G. Gänzle, Email: mgaenzle@ualberta.ca.
Danilo Ercolini, Universita degli Studi di Napoli Federico II, Naples, Italy.
DATA AVAILABILITY
The raw reads and annotated assemblies of Pw. fabaria LMG 24289T and Pw. ghanensis LMG 24286T have been publicly deposited under the BioProject accession number PRJEB48651 and the GenBank accession numbers CAKKNS000000000 and CAKKNT000000000, respectively.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.01034-23.
Figures S1 and S2 and supplemental table legends
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Table S7
Table S8
Table S9
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Marco ML, Sanders ME, Gänzle M, Arrieta MC, Cotter PD, De Vuyst L, Hill C, Holzapfel W, Lebeer S, Merenstein D, Reid G, Wolfe BE, Hutkins R. 2021. The International scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on fermented foods. Nat Rev Gastroenterol Hepatol 18:196–208. doi: 10.1038/s41575-020-00390-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zheng J, Wittouck S, Salvetti E, Franz C, Harris HMB, Mattarelli P, O’Toole PW, Pot B, Vandamme P, Walter J, Watanabe K, Wuyts S, Felis GE, Gänzle MG, Lebeer S. 2020. A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int J Syst Evol Microbiol 70:2782–2858. doi: 10.1099/ijsem.0.004107 [DOI] [PubMed] [Google Scholar]
- 3. Qiao N, Wittouck S, Mattarelli P, Zheng J, Lebeer S, Felis GE, Gänzle MG. 2022. After the storm - Perspectives on the taxonomy of Lactobacillaceae. JDS Commun 3:222–227. doi: 10.3168/jdsc.2021-0183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bello S, Rudra B, Gupta RS. 2022. Phylogenomic and comparative genomic analyses of Leuconostocaceae species: identification of molecular signatures specific for the genera Leuconostoc, Fructobacillus and Oenococcus and proposal for a novel genus Periweissella gen. nov. Int J Syst Evol Microbiol 72:05284. doi: 10.1099/ijsem.0.005284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Padonou SW, Schillinger U, Nielsen DS, Franz C, Hansen M, Hounhouigan JD, Nago MC, Jakobsen M. 2010. Weissella beninensis sp. nov., a motile lactic acid bacterium from submerged cassava fermentations, and emended description of the genus Weissella. Int J Syst Evol Microbiol 60:2193–2198. doi: 10.1099/ijs.0.014332-0 [DOI] [PubMed] [Google Scholar]
- 6. Heo J, Hamada M, Cho H, Weon H-Y, Kim J-S, Hong S-B, Kim S-J, Kwon S-W. 2019. Weissella cryptocerci sp. nov., isolated from gut of the insect Cryptocercus kyebangensis. Int J Syst Evol Microbiol 69:2801–2806. doi: 10.1099/ijsem.0.003564 [DOI] [PubMed] [Google Scholar]
- 7. Jung JY, Lee SH, Lee HJ, Seo HY, Park WS, Jeon CO. 2012. Effects of Leuconostoc mesenteroides starter cultures on microbial communities and metabolites during kimchi fermentation. Int J Food Microbiol 153:378–387. doi: 10.1016/j.ijfoodmicro.2011.11.030 [DOI] [PubMed] [Google Scholar]
- 8. Wuyts S, Van Beeck W, Oerlemans EFM, Wittouck S, Claes IJJ, De Boeck I, Weckx S, Lievens B, De Vuyst L, Lebeer S. 2018. Carrot juice fermentations as man-made microbial ecosystems dominated by lactic acid bacteria. Appl Environ Microbiol 84:e00134-18. doi: 10.1128/AEM.00134-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pswarayi F, Gänzle MG. 2019. Composition and origin of the fermentation microbiota of mahewu, a Zimbabwean fermented cereal beverage. Appl Environ Microbiol 85:e03130-18. doi: 10.1128/AEM.03130-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gobbetti M, Minervini F, Pontonio E, Di Cagno R, De Angelis M. 2016. Drivers for the establishment and composition of the sourdough lactic acid bacteria biota. Int J Food Microbiol 239:3–18. doi: 10.1016/j.ijfoodmicro.2016.05.022 [DOI] [PubMed] [Google Scholar]
- 11. Wang X, Du H, Zhang Y, Xu Y, Björkroth J. 2018. Environmental microbiota drives microbial succession and metabolic profiles during Chinese liquor fermentation. Appl Environ Microbiol 84:e02369-17. doi: 10.1128/AEM.02369-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen S, Zhou Y, Chen Y, Gu J. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. doi: 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 17. Liu H, Xin B, Zheng J, Zhong H, Yu Y, Peng D, Sun M. 2020. Build a bioinformatics analysis platform and apply it to routine analysis of microbial genomics and comparative genomics. Protoc Exch. [Google Scholar]
- 18. Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:2461. doi: 10.1093/molbev/msaa131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Letunic I, Bork P. 2021. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. doi: 10.1093/nar/gkab301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ruiz-Perez CA, Conrad RE, Konstantinidis KT. 2021. MicrobeAnnotator: a user-friendly, comprehensive functional annotation pipeline for microbial genomes. BMC Bioinformatics 22:11. doi: 10.1186/s12859-020-03940-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. 2016. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol 17:262. doi: 10.1186/s13059-016-1132-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zheng J, Ruan L, Sun M, Gänzle M. 2015. A genomic view of lactobacilli and pediococci demonstrates that phylogeny matches ecology and physiology. Appl Environ Microbiol 81:7233–7243. doi: 10.1128/AEM.02116-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. De Roos J, De Vuyst L. 2018. Acetic acid bacteria in fermented foods and beverages. Curr Opin Biotechnol 49:115–119. doi: 10.1016/j.copbio.2017.08.007 [DOI] [PubMed] [Google Scholar]
- 27. Gänzle MG. 2015. Lactic metabolism revisited: metabolism of lactic acid bacteria in food fermentations and food spoilage. Curr Opin in Food Sci 2:106–117. doi: 10.1016/j.cofs.2015.03.001 [DOI] [Google Scholar]
- 28. Nguyen T, Kim T, Ta HM, Yeo WS, Choi J, Mizar P, Lee SS, Bae T, Chaurasia AK, Kim KK. 2019. Targeting mannitol metabolism as an alternative antimicrobial strategy based on the structure-function study of mannitol-1-phosphate dehydrogenase in Staphylococcus aureus. mBio 10:e02660-18. doi: 10.1128/mBio.02660-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Doi Y. 2019. Glycerol metabolism and its regulation in lactic acid bacteria. Appl Microbiol Biotechnol 103:5459. doi: 10.1007/s00253-019-09930-9 [DOI] [PubMed] [Google Scholar]
- 30. Wadhwa N, Berg HC. 2022. Bacterial motility: machinery and mechanisms. Nat Rev Microbiol 20:161–173. doi: 10.1038/s41579-021-00626-4 [DOI] [PubMed] [Google Scholar]
- 31. Chao S-H, Tomii Y, Sasamoto M, Fujimoto J, Tsai Y-C, Watanabe K. 2008. Lactobacillus capillatus sp. nov., a motile bacterium isolated from stinky tofu brine. Int J Syst Evol Microbiol 58:2555–2559. doi: 10.1099/ijs.0.65834-0 [DOI] [PubMed] [Google Scholar]
- 32. Nielsen DS, Schillinger U, Franz C, Bresciani J, Amoa-Awua W, Holzapfel WH, Jakobsen M. 2007. Lactobacillus ghanensis sp. nov., a motile lactic acid bacterium isolated from Ghanaian cocoa fermentations. Int J Syst Evol Microbiol 57:1468–1472. doi: 10.1099/ijs.0.64811-0 [DOI] [PubMed] [Google Scholar]
- 33. Endo A, Okada S. 2005. Lactobacillus satsumensis sp. nov., isolated from mashes of shochu, a traditional Japanese distilled spirit made from fermented rice and other starchy materials. Int J Syst Evol Microbiol 55:83–85. doi: 10.1099/ijs.0.63248-0 [DOI] [PubMed] [Google Scholar]
- 34. Irisawa T, Okada S. 2009. Lactobacillus sucicola sp. nov., a motile lactic acid bacterium isolated from oak tree (Quercus sp.) sap. Int J Syst Evol Microbiol 59:2662–2665. doi: 10.1099/ijs.0.006478-0 [DOI] [PubMed] [Google Scholar]
- 35. El Baidouri F, Venditti C, Humphries S. 2016. Independent evolution of shape and motility allows evolutionary flexibility in Firmicutes bacteria. Nat Ecol Evol 1:9. doi: 10.1038/s41559-016-0009 [DOI] [PubMed] [Google Scholar]
- 36. Duar RM, Lin XB, Zheng J, Martino ME, Grenier T, Pérez-Muñoz ME, Leulier F, Gänzle M, Walter J. 2017. Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol Rev 41:S27–S48. doi: 10.1093/femsre/fux030 [DOI] [PubMed] [Google Scholar]
- 37. Wisselink HW, Weusthuis RA, Eggink G, Hugenholtz J, Grobben GJ. 2002. Mannitol production by lactic acid bacteria: a review. Int Dairy J 12:151–161. doi: 10.1016/S0958-6946(01)00153-4 [DOI] [Google Scholar]
- 38. De Bruyne K, Camu N, De Vuyst L, Vandamme P. 2010. Weissella fabaria sp. nov., from a Ghanaian cocoa fermentation. Int J Syst Evol Microbiol 60:1999–2005. doi: 10.1099/ijs.0.019323-0 [DOI] [PubMed] [Google Scholar]
- 39. Fanelli F, Montemurro M, Chieffi D, Cho G-S, Franz C, Dell’Aquila A, Rizzello CG, Fusco V. 2022. Novel insights into the phylogeny and biotechnological potential of Weissella species. Front Microbiol 13:914036. doi: 10.3389/fmicb.2022.914036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao X, Gänzle MG. 2018. Genetic and phenotypic analysis of carbohydrate metabolism and transport in Lactobacillus reuteri. Int J Food Microbiol 272:12–21. doi: 10.1016/j.ijfoodmicro.2018.02.021 [DOI] [PubMed] [Google Scholar]
- 41. Sasaki Y, Laivenieks M, Zeikus JG. 2005. Lactobacillus reuteri ATCC 53608 MDH gene cloning and recombinant mannitol dehydrogenase characterization. Appl Microbiol Biotechnol 68:36–41. doi: 10.1007/s00253-004-1841-x [DOI] [PubMed] [Google Scholar]
- 42. Korakli M, Vogel RF. 2003. Purification and characterisation of mannitol dehydrogenase from Lactobacillus sanfranciscensis. FEMS Microbiol Lett 220:281–286. doi: 10.1016/S0378-1097(03)00129-0 [DOI] [PubMed] [Google Scholar]
- 43. Calmes B, Guillemette T, Teyssier L, Siegler B, Pigné S, Landreau A, Iacomi B, Lemoine R, Richomme P, Simoneau P. 2013. Role of mannitol metabolism in the pathogenicity of the necrotrophic fungus Alternaria brassicicola. Front Plant Sci 4:131. doi: 10.3389/fpls.2013.00131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Neville BA, Forde BM, Claesson MJ, Darby T, Coghlan A, Nally K, Ross RP, O’Toole PW. 2012. Characterization of pro-inflammatory flagellin proteins produced by Lactobacillus ruminis and related motile lactobacilli. PLoS One 7:e40592. doi: 10.1371/journal.pone.0040592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cousin FJ, Lynch SM, Harris HMB, McCann A, Lynch DB, Neville BA, Irisawa T, Okada S, Endo A, O’Toole PW. 2015. Detection and genomic characterization of motility in Lactobacillus curvatus: confirmation of motility in a species outside the Lactobacillus salivarius clade. Appl Environ Microbiol 81:1297–1308. doi: 10.1128/AEM.03594-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cullender TC, Chassaing B, Janzon A, Kumar K, Muller CE, Werner JJ, Angenent LT, Bell ME, Hay AG, Peterson DA, Walter J, Vijay-Kumar M, Gewirtz AT, Ley RE. 2013. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe 14:571–581. doi: 10.1016/j.chom.2013.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sharpe ME, Latham MJ, Garvie EI, Zirngibl J, Kandler O. 1973. Two new species of Lactobacillus isolated from the bovine rumen, Lactobacillus ruminis sp.nov. and Lactobacillus vitulinus sp.nov. J Gen Microbiol 77:37–49. doi: 10.1099/00221287-77-1-37 [DOI] [PubMed] [Google Scholar]
- 48. Snauwaert I, Papalexandratou Z, De Vuyst L, Vandamme P. 2013. Characterization of strains of Weissella fabalis sp. nov. and Fructobacillus tropaeoli from spontaneous cocoa bean fermentations. Int J Syst Evol Microbiol 63:1709–1716. doi: 10.1099/ijs.0.040311-0 [DOI] [PubMed] [Google Scholar]
- 49. Puertas AI, Arahal DR, Ibarburu I, Elizaquível P, Aznar R, Dueñas MT. 2014. Lactobacillus sicerae sp. nov., a lactic acid bacterium isolated from Spanish natural cider. Int J Syst Evol Microbiol 64:2949–2955. doi: 10.1099/ijs.0.059980-0 [DOI] [PubMed] [Google Scholar]
- 50. Mañes-Lázaro R, Song J, Pardo I, Cho J-C, Ferrer S. 2009. Lactobacillus aquaticus sp. nov., isolated from a Korean freshwater pond. Int J Syst Evol Microbiol 59:2215–2218. doi: 10.1099/ijs.0.008276-0 [DOI] [PubMed] [Google Scholar]
- 51. Kaneuchi C, Seki M, Komagata K. 1988. Taxonomic study of Lactobacillus mali Carr and Davis 1970 and related strains: validation of Lactobacillus mali Carr and Davis 1970 over Lactobacillus yamanashiensis Nonomura 1983. Int J Syst Bacteriol 38:269–272. doi: 10.1099/00207713-38-3-269 [DOI] [Google Scholar]
- 52. Kajikawa A, Midorikawa E, Masuda K, Kondo K, Irisawa T, Igimi S, Okada S. 2016. Characterization of flagellins isolated from a highly motile strain of Lactobacillus agilis. BMC Microbiol 16:49. doi: 10.1186/s12866-016-0667-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. De Bruyne K, Camu N, Lefebvre K, De Vuyst L, Vandamme P. 2008. Weissella ghanensis sp. nov., isolated from a Ghanaian cocoa fermentation. Int J Syst Evol Microbiol 58:2721–2725. doi: 10.1099/ijs.0.65853-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 and S2 and supplemental table legends
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Table S7
Table S8
Table S9
Data Availability Statement
The raw reads and annotated assemblies of Pw. fabaria LMG 24289T and Pw. ghanensis LMG 24286T have been publicly deposited under the BioProject accession number PRJEB48651 and the GenBank accession numbers CAKKNS000000000 and CAKKNT000000000, respectively.