ABSTRACT
Polycyclic aromatic hydrocarbons (PAHs) are harmful to human health due to their carcinogenic, teratogenic, and mutagenic effects. A thermophilic Hydrogenibacillus sp. strain N12 capable of degrading a variety of PAHs and derivatives was previously isolated. In this study, an aromatic ring-hydroxylating oxygenase, NarA2B2, was identified from strain N12, with substrate specificity including naphthalene, phenanthrene, dibenzothiophene, fluorene, acenaphthene, carbazole, biphenyl, and pyrene. NarA2B2 was proposed to add one or two atoms of molecular oxygen to the substrate and catalyze biphenyl at C-2, 2 or C-3, 4 positions with different characteristics than before. The key catalytic amino acids, H222, H227, and D379, were identified as playing a pivotal role in the formation of the 2-his-1-carboxylate facial triad. Furthermore, we conducted molecular docking and molecular dynamics simulations, notably, D219 enhanced the stability of the iron center by forming two stable hydrogen bonds with H222, while the mutation of F216, T223, and H302 modulated the catalytic activity by altering the pocket’s size and shape. Compared to the wild-type (WT) enzyme, the degradation ratios of acenaphthene by F216A, T223A, and H302A had an improvement of 23.08%, 26.87%, and 29.52%, the degradation ratios of naphthalene by T223A and H302A had an improvement of 51.30% and 65.17%, while the degradation ratio of biphenyl by V236A had an improvement of 77.94%. The purified NarA2B2 was oxygen-sensitive when it was incubated with L-ascorbic acid in an anaerobic environment, and its catalytic activity was restored in vitro. These results contribute to a better understanding of the molecular mechanism responsible for PAHs’ degradation in thermophilic microorganisms.
IMPORTANCE
(i) A novel aromatic ring-hydroxylating oxygenase named NarA2B2, capable of degrading multiple polycyclic aromatic hydrocarbons and derivatives, was identified from the thermophilic microorganism Hydrogenibacillus sp. N12. (ii) The degradation characteristics of NarA2B2 were characterized by adding one or two atoms of molecular oxygen to the substrate. Unlike the previous study, NarA2B2 catalyzed biphenyl at C-2, 2 or C-3, 4 positions. (iii) Catalytic sites of NarA2B2 were conserved, and key amino acids F216, D219, H222, T223, H227, V236, F243, Y300, H302, W316, F369, and D379 played pivotal roles in catalysis, as confirmed by protein structure prediction, molecular docking, molecular dynamics simulations, and point mutation.
KEYWORDS: Hydrogenibacillus, aromatic ring-hydroxylating oxygenase, biodegradation, polycyclic aromatic hydrocarbons
INTRODUCTION
Polycyclic aromatic hydrocarbons (PAHs) are compounds formed by multiple benzene rings. Incomplete combustion of organic molecules at high temperatures (500–800°C) or prolonged exposure to lower temperatures (100–300°C) can produce PAHs, including events such as forest fires and the combustion of fossil fuels (1). Due to their carcinogenic, teratogenic, and mutagenic effects, the US Environmental Protection Agency has designated 16 PAHs as High Priority Pollutants (2).
Over the past few decades, many PAHs-degrading bacteria have been isolated, including Pseudomonas, Sphingomonas, Sphingobium, Mycobacterium, and Rhodococcus (3), with well-studied molecular mechanisms as well as established physiological and biochemical properties. However, the research on PAHs’ degrading bacteria has focused on mesophilic bacteria, and the degradation potential and characteristics of thermophilic microorganisms have rarely been investigated (4). Hydrogenibacillus sp. N12 can grow with naphthalene (NAP) as the only carbon source and energy source and degrade a variety of PAHs and derivatives through co-metabolism, including phenanthrene (PHE), dibenzothiophene (DBT), fluorene (FLN), dibenzofuran, and carbazole (CA) at its optimal growth temperature of 60°C (5). According to previous research, the extraordinary metabolic property of PAHs’ degraders is attributable to multiple degradation enzymes. In particular, the aromatic ring-hydroxylating oxygenase (RHO) may determine the substrate preference of PAHs’ degraders, which catalyzes the first step of microbial degradation of PAHs (6). RHO is a class of multi-component enzyme consisting of a terminal oxygenase with catalytic function as well as an electron transport chain (ETC) with the function of transferring electrons (7). The terminal oxygenase usually occurs as homotrimers (α3) or hexamers (α3β3), with the α-subunit consisting of a Rieske domain responsible for transferring electrons to the active site of the neighboring α-subunit (8). While a significant amount of investigation on the structure and function of RHOs in mesophilic bacteria has been reported, such as NDO-P (the naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4) (9), NDO-R (the naphthalene dioxygenase from Rhodococcus strain NCIMB12038) (10), BPDO-R (the biphenyl dioxygenase from Rhodococcus sp. strain RHA1) (11), NidA3B3 and NidAB (two RHOs of Mycobacterium vanbaalenii PYR-1) (12, 13), PhnI (the RHO from Sphingomonas sp. strain CHY-1) (14, 15), as well as PhnA1B1 and PhnA2B2 (two RHOs from Sphingobium sp. strain SHPJ-2) (16), the degradation mechanisms of RHOs in thermophilic microorganisms have not been reported. Moreover, the degradation pathway and products of PAHs by thermophilic microorganisms may be different from those of moderate-temperature bacteria. This suggests that there may be novel PAHs’ degradation mechanisms found within thermophilic microorganisms. Therefore, identifying and characterizing functional enzymes in thermophilic microorganisms, which degrade PAHs is important in order to clarify bacterial degradation mechanisms.
Here, we identified and characterized a novel RHO NarA2B2 in Hydrogenibacillus sp. N12 (5), which was able to degrade a variety of PAHs and derivatives. The degradation products were detected and identified using gas chromatography-mass spectrometry (GC-MS). Through the prediction of the protein structure and carrying out point mutations to NarA2, we verified some key amino acids involved in the catalysis of NarA2 and explored the mechanism by which NarA2B2 catalyzed PAHs. The in-depth study allows for a better understanding of the PAH degradation mechanism in thermophilic microorganisms, which lays an important foundation for subsequent genetic utilization and bioremediation.
MATERIALS AND METHODS
Chemicals and bacterial strains
PAHs and their derivatives were purchased from J&K Scientific Co., Ltd. and Shanghai Aladdin Biochemical Technology Co., Ltd. N, O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) was purchased from Sigma-Aldrich and used to derivatize metabolites. The purity of all chemicals exceeded 99%. Escherichia coli (E. coli) Top10 was used for plasmid construction, and E. coli BL21(DE3) was used for gene expression. These strains were grown at 37°C on Luria-Bertani (LB) medium supplemented with antibiotics.
Construction of plasmids and purification of proteins
The predicted RHO genes were amplified from the genomic DNA of Hydrogenibacillus sp. strain N12. Chr966 was amplified using primers 966-F1 and 966-R1 (Table 1). Chr967 was amplified using primers 967-F and 967-R (Table 1). Chr966 and Chr967 were cloned into ORF1 and ORF2 of pETDuet, respectively, with a terminator inserted between ORF1 and ORF2, generating the plasmid pETDuet-narA2-Ter-narB2. The ETC components phtAcAd (derived from Mycobacterium vanbaalenii PYR-1) and phdCD (derived from Nocardioides sp. strain KP7) were cloned into pACYCDuet, respectively, and named pACYCDuet-phtAcAd and pACYCDuet-phdCD (12, 17). The phtAcAd was also cloned into the pET28a and named pET28a-phtAcAd for protein expression and purification.
TABLE 1.
Primers used for the construction of plasmids
| Primers | Sequences |
|---|---|
| 966-F1 | 5-CCACAGCCAGGATCCGATGGTCGTTCAAAATGGAATTGTA-3 |
| 966-R1 | 5-CGCCGAGCTCGAATTTTACTCGCTCATCAACTTAGCC-3 |
| 967-F | 5-GAAGGAGATATACATATGGGAGACGCGATCTATTT-3 |
| 967-R | CCAATTGAGATCTGCTTAGAAAAACACCCCGAGATTCT-3 |
| 966-H302A-F | 5-AACGCAGGCACCATCTTTCCCAACATGTC-3 |
| 966-H302A-R | 5-GCCTGCGTTATATGCCAGACGCCG-3 |
| 966-F216A-F | 5-AATGCAGTCGGGGATGCTTATCACACGTT-3 |
| 966-F216A-R | 5-GACTGCATTATCTCCGCCGAGCTT-3 |
| 966-T223A-F | 5-CACGCATTCATGACCCATTATTCTGCTGT-3 |
| 966-T223A-R | 5-GAATGCGTGATAAGCATCCCCGAC-3 |
| 966-D219A-F | 5-GGGGCAGCTTATCACACGTTCATGACCCA-3 |
| 966-D219A-R | 5-AGCTGCCCCGACAAAATTATCTCC-3 |
| 966-H222A-F | 5-TATGCAACGTTCATGACCCATTATTCTGC-3 |
| 966-H222A-R | 5-CGTTGCATAAGCATCCCCGACAAA-3 |
| 966-H227A-F | 5-ACCGCATATTCTGCTGTCCAGTTGAAGAT-3 |
| 966-H227A-R | 5-ATATGCGGTCATGAACGTGTGATA-3 |
| 966-F243A-F | 5-AAGGCAGCGTATTATGGGGTTCACGTATCGTGTGA-3 |
| 966-F243A-R | 5-CGCTGCCTTTGGATCGCCGCTCGG-3 |
| 966-Y300A-F | 5-GCAGCAAACCATGGCACCATCTTTCCCAACATGT-3 |
| 966-Y300A-R | 5-GTTTGCTGCCAGACGCCGGAACGC-3 |
| 966-W316A-F | 5-ACCGCAGCCACGGAAGATGGCAAG-3 |
| 966-W316A-R | 5-GGCTGCGGTGAAGTAACCGAACGACATGTT-3 |
| 966-F369A-F | 5-ACCGCAGGTGCGAGCGGTATGCTG-3 |
| 966-F369A-R | 5-ACCTGCGGTCCGAACAAACGTTTCTCTGGC-3 |
| 966-V236A-F | 5-ATCGCACCGAGCGGCGATCCA-3 |
| 966-V236A-R | 5-CGGTGCGATCTTCAACTGGAC-3 |
| 966-D379A-F | 5-GACGCAGCGGAAATGTGGAGCAACATCAC-3 |
| 966-D379A-R | 5-CGCTGCGTCCTGTTCCAGCATACC-3 |
The plasmids pETDuet-narA2-Ter-narB2 and pET28a-phtAcAd were separately transformed into E. coli BL21(DE3). E. coli BL21(DE3) cells containing recombinant plasmids were cultured in LB medium at 37°C. When the OD600 reached 0.6–0.8, isopropyl-β-D-thiogalactopyranoside was added to the culture at a final concentration of 0.4 mM, and the cells were further incubated at 16°C for 16 h. The cultured cells were harvested (4,800 rpm, 10 min) and resuspended using buffer A (25 mM Tris-HCl, 300 mM NaCl, 20 mM imidazole, and 2 mM 2-mercaptoethanol). The collected cells were broken up using a high-pressure disruptor (800 bar, 50 mL/2 min), and centrifuged at 10,000 rpm for 40 min at 4°C. The recombinant protein was purified with Ni-NTA affinity chromatography and eluted with different concentrations of imidazole buffer. The purified protein was stored in buffer B (15 mM Tris-HCl, 150 mM NaCl, and 2 mM dithiothreitol). A small amount of protein was mixed with an equal volume of 2× SDS loading buffer, identified by SDS-PAGE (12%).
Characterization of the predicted RHO
The predicted RHO was characterized using resting cell biotransformation and enzyme assays.
Resting cell biotransformation
The ETC components pACYCDuet-phtAcAd and pACYCDuet-phdCD were co-transformed into E. coli BL21(DE3) with pETDuet-narA2-Ter-narB2, respectively. The culture and harvest of E. coli BL21(DE3) containing recombinant plasmid were the same as the preparation of purification of protein. The harvested cells were washed three times in PBS buffer. The suspension was starved at 30°C for 3 h without any substrate to prepare resting cells (10 mL, OD600nm = 5). Next, 50 mg/L NAP was added, and the residual NAP concentration was measured after 26 h by highperformance liquid chromatography (HPLC). E. coli BL21(DE3) cells containing pETDuet and pACYCDuet served as a control, with all treatments being the same as the experimental group. All tests above were conducted in triplicate. HPLC was performed using the Agilent 1,200 Infinity with Eclipse XDB-C18 columns, 80% methanol, 20% water (0.1% formic acid), 0.8 mL/min, and 30°C (18).
Enzyme assays
The assay of enzyme activity was performed in the reaction mixtures (1 mL) containing 25 mM Tris-HCl pH 7.5, 10% glycerol, 0.05 mM ferrous ammonium sulfate, 0.2 mM NADH, 0.3 mM L-ascorbic acid, 5.67 µM NarA2B2 complex, and 10 µM PhtAcAd. This mixture was incubated in an anaerobic environment for 20–30 min. Then, PHE was added to a final concentration of 14.24 mg/L to initiate the reaction at 30°C. After 2 h, 20 µL of HCl was added to stop the reaction. The mixture was then extracted with 500 µL of ethyl acetate, and the remaining PHE content was detected using HPLC.
Characterization of the substrate specificity of NarA2B2
Resting cells were supplemented with different substrates, including NAP, PHE, DBT, CA, BP, FLN, and ACE, at a final concentration of 50 mg/L, while PYE was added at a concentration of 10 mg/L. Samples were analyzed after 0, 4, 8, 16, and 24 h. Low molecular weight PAHs were extracted with an equal volume of ethyl acetate for 3 min, and high molecular weight PAHs were extracted with half the volume of ethyl acetate for 6 min. The concentration of residual PAHs was detected by HPLC. The methods for preparing resting cells and detecting by HPLC are the same as described above.
Identification of metabolites by NarA2B2
To detect metabolites, resting cells were re-prepared, and the concentrations of PAHs were the same as described above. A half-volume of ethyl acetate was added and extracted twice at 30°C. The residual water in ethyl acetate was removed, and the extract was condensed 200–300 times. A total of 30 µL of product was then mixed with an equal volume of BSTFA and reacted at 70°C for 30 min before detection by GC-MS (Agilent, GC-7890B; MS-5977B; 30 m × 0.25 mm, 0.25 µm, HP-5MS column). The oven temperature was set at 75°C for 3 min, followed by a ramp-up to 250°C at a rate of 12°C/min, held at 250°C for 1 min, followed by an increase to 300°C at a rate of 10°C/min, and held at 300°C for 10 min (19).
Molecular docking and molecular dynamics simulation
The structure of the NarA2B2 complex was predicted by AlphaFold2 (20). The positions of active site iron and iron-sulfur clusters were determined by SWISS-MODEL (21). The structures of substrates were downloaded from PubChem (22). AutoDock4.2 software was utilized to build the enzyme-substrate complex (23). According to the binding position of active site iron, the cube box of side length 100 Å centered on the iron atom was set and calculated. The Lamarckian genetic algorithm (24) was adopted with search parameters for the stable enzyme-substrate complex with 200 runs. Other parameters were set as default values.
The results of the docking were ranked according to the docking energy, with the structure possessing the lowest energy serving as the initial structure for molecular dynamic (MD) simulations. The substrate parameters were prepared using a two-step restrained electrostatic potential(25) charge-fitting method, facilitated by the Antechamber package. The system was then solvated within an octahedral box of TIP3P water via tleap and subsequently neutralized electrically with either chlorine or sodium ions. Following this, classical MD simulations were executed on the prearranged structures of the eight systems using AMBER20 (26). The energy minimization was conducted in two stages: initially focusing on the water molecules, followed by the remainder of the overall system. Subsequently, the system was gradually heated from 0 to 300 K over a period of 50 ps. Afterward, the system was transitioned to constant pressure and temperature (NPT) and equilibrated for an additional 50 ps to adjust the system to the appropriate density. The equilibrated structure served as the initial structure, and from it, three parallel 50 ns trajectories were collected for further analysis. The root mean square deviation (RMSD) and distances were analyzed using Cpptraj (27). Visualization of the processes was accomplished using PyMOL (28).
Verification of key catalytic sites by point mutation
Mutants were generated using pETDuet-narA2-Ter-narB2 as the template, and primers containing mutation sites with an overlap of 9 bp. A total of 8 µL of PCR product was mixed with 1 µL DpnI and 1 µL CutSmart buffer at 37°C for 1 h. The mix was then directly transformed into E. coli Top10, and the transformed cells were selected for verification. Mutated plasmids were co-transformed into E. coli BL21(DE3) with pACYCDuet-phtAcAd to determine their ability to degrade PAHs (the OD600nm of resting cells was 10). The primers used for the construction of mutants are listed in Table 1.
RESULTS
Identification of predicted aromatic ring-hydroxylating oxygenase NarA2B2
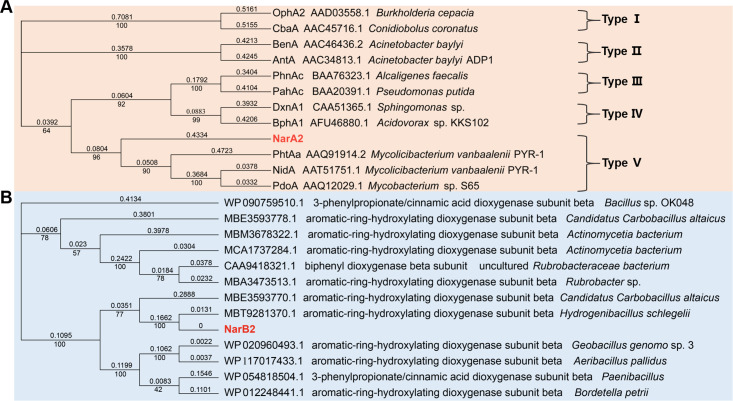
According to previous studies (5), the strain N12 can degrade multiple PAHs and derivatives. Based on genomic annotation and homologous sequence alignment with other reported RHOs, we presumed that Chr966 and Chr967 may be a new RHO in strain N12. Chr966 shares 51.33% amino acid identity with the naphthalene dioxygenase large subunit (NarAa) of Rhodococcus strain NCIMB12038 (10) and contains the conserved sequence of a Rieske-type iron-sulfur cluster, suggesting that Chr966 may be the α-subunit of a novel RHO.
Phylogenetic analysis has demonstrated that Chr966 is in the same branch as PhtAa (AAQ91914.2) (17), NidA (AAT51751.1) (13), and PdoA (AAQ12029.1) (29), with sequence similarity of 38.9%, 42.92%, and 43.62%, respectively (Fig. 1A). These belong to type V RHOs compatible with a [3Fe-4S]-type ferredoxin and a GR-type reductase (30) (Fig. 1A). The gene with highest sequence similarity to Chr967 is an aromatic-ring-hydroxylating dioxygenase subunit beta of Hydrogenibacillus Schlegelii, followed by an aromatic-ring-hydroxylating dioxygenase subunit beta of Candidatus Carbobacillus altaicus, with similarities of 99.39% and 64.85%, respectively (Fig. 1B).
Fig 1.
Neighbor-joining phylogenetic tree of amino acid sequences of NarA2 (A) and NarB2 (B) in strain N12. The phylogenetic tree of NarAa is based on a novel RHO classification system that classifies RHOs into five types derived from the ETC components (30). The phylogenetic tree of NarB2 is constructed from homologous sequences aligned by the National Center for Biotechnology Information (NCBI).
Functional verification of predicted aromatic ring-hydroxylating oxygenase
As no genes near Chr966 and Chr967 were explicitly annotated as ferredoxin and ferredoxin reductase, several sets of possible ETCs were co-expressed with Chr966 and Chr967, and FLN cannot be degraded (Fig. S1). Exogenous ETCs were introduced to detect the activity of predicted RHOs. According to phylogenetic analysis (Fig. 1A), Chr966 belongs to type V RHO. Therefore, Chr966-967 was co-expressed with phdCD and phtAcAd, respectively, and the activity was detected within resting cell biotransformation. We found that NAP could only be degraded when Chr966-967 was co-expressed with phtAcAd (Fig. S2). Therefore, it was determined that Chr966 and Chr967 are the α- and β-subunits of a novel RHD, named NarA2B2.
Characterization of the substrate specificity of NarA2B2
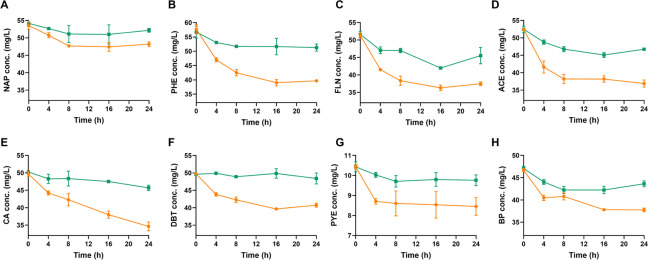
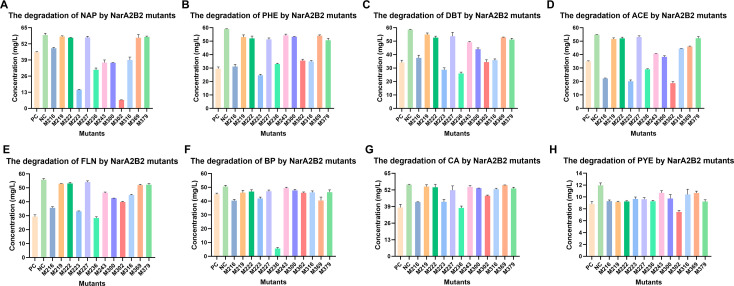
The 14 PAHs were used to identify the substrate specificity of NarA2B2, which refers to the ability of an enzyme to bind and catalyze a specific substrate or group of substrates. As shown, NarA2B2 can degrade NAP (Fig. 2A), PHE (Fig. 2B), FLN (Fig. 2C), ACE (Fig. 2D), CA (Fig. 2E), DBT (Fig. 2F), PYE (Fig. 2G), and BP (Fig. 2H). There is no degradation of the other substrates (Fig. S3). According to the degradation curve, the substrate degraded most rapidly in the first 4 h and outside of CA, the concentration of the substrates remained essentially stable after 16 h. Due to the volatilization, the substrate concentration in the control also decreases slightly over time. However, the degradation of PYE was completed within the first 4 h (Fig. 2). The enzymes may become inactivated more quickly due to the inhibitory effect of PYE. Among all substrates, NarA2B2 has the strongest degradability to CA, and at an initial concentration of 50 mg/L, it can degrade 25.96% of CA. This was followed by DBT, PHE, ACE, and FLN, with degradation ratios of 20.65%, 21.27%, 18.83%, and 20.86%, respectively. The ability to degrade NAP is poor, with degradation ratios of 9.79%. PYE and BP showed intermediate degradation ratios of 12.94% and 12.55%, respectively.
Fig 2.
Detection of substrate specificity of NarA2B2 in E. coli BL21(DE3). (A) NAP (50 mg/L); (B) PHE (50 mg/L); (C) FLN (50 mg/L); (D) ACE (50 mg/L); (E) CA (50 mg/L); (F) DBT (50 mg/L); (G) PYE (10 mg/L); and (H) BP (50 mg/L). The yellow line with circles represents E. coli BL21(DE3) containing pETDuet-narA2-Ter-narB2 and pACYCDuet-phtAcAd, and the green line with squares represents E. coli BL21(DE3) containing pETDuet and pACYCDuet. The concentration in parentheses is the initial concentration. All the tests above were conducted in triplicate. NAP, naphthalene; PHE, phenanthrene; FLN, fluorene; ACE, acenaphthene; CA, carbazole; DBT, dibenzothiophene; PYE, pyrene; and BP, biphenyl.
Intermediates detected with GC-MS
The metabolites degraded by NarA2B2 were detected by GC-MS and structures were determined by comparing them with the National Institute of Standards and Technology (NIST) MS library or MS profiles of the standard compounds. The metabolites are listed in Table 2.
TABLE 2.
GC-MS results of PAHs metabolites produced by NarA2B2a
| Metabolite | Proposed product | Structure | Retention time (min) | Mass spectral characteristics of the product(ion abundances) |
|---|---|---|---|---|
| P1 | 1-naphthol (TMS)b |
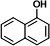
|
11.99 | 201(100), 216(99.73), 185(49.88), 73(36.33), 115(23.8), 217.1(19.57) |
| P2 | 2-naphthol (TMS)b |
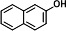
|
12.25 | 201(100), 216(79.72), 145(24.51), 115(21.78), 73(20.59), 127(18.55) |
| P3 | NAP-1,2-dihydrodiol (2TMS)c |
|
13.15 | 191(100), 73(96.08), 147(58.29), 203(29.71), 128(24.85), 192(20.63) |
| P4 | 1,2-dihydroxynaphthalene (2TMS)b,d |
|
14.29 | 73(100), 304(44.75), 216(21.12), 186(12.76), 305(12.25), 74(8.68) |
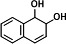
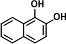
| P5 | PHE-9,10-dihydrodiol (2TMS)c |
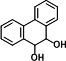
|
16.38 | 147(100), 73(56.52), 266(34.96), 341(29.89), 165(27.07), 356.2(23.99) |
| P6 | Monohydroxy-PHE (TMS)c | __ | 16.75 | 266(100), 235(75.69), 251(61.09), 73(43.33), 267(24.07), 117.7(20.36) |
| P7 | Monohydroxy-PHE (TMS)c | __ | 17.06 | 266(100), 251(62.68), 73(45.43), 235(26.8), 165(24.59), 267(22.84) |
| P8 | PHE-dihydrodiol (2TMS)c | __ | 17.21 | 73(100), 191(81.5), 147(35.9), 165(19.49), 178(19.06), 192(16.28) |
| P9 | Monohydroxy-PHE (TMS)c | __ | 17.29 | 266(100), 251(69.17), 267(24.19), 165(20.45), 73(20.33), 176(18.3) |
| P10 | Monohydroxy-PHE (TMS)c | __ | 17.34 | 266(100), 251(72.61), 235(40.93), 73(30.77), 267(24.15), 165(21.63) |
| P11 | Monohydroxy-PHE (TMS)c | __ | 17.58 | 266(100), 251(89.01), 73(29.91), 267(23.94), 165(21.67), 252(20.12) |
| P12 | PHE-dihydrodiol (2TMS)c | __ | 17.79 | 191(100), 73(73.74), 147(31.89), 192(22.9), 253(21.13), 356(7.43) |
| P13 | Dihydroxyl-PHE (2TMS)c | __ | 19.36 | 73(100), 354(49.75), 266(17.45), 355(16.22), 236(14.77), 45(10.76) |
| P14 | Monohydroxy-DBT (TMS)c | __ | 16.86 | 272(100), 241(66.43), 257(53.33), 73(52.39), 273(22.04), 57(20.73) |
| P15 | Dibenzothiophene-S-oxideb |
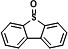
|
17.29 | 171(100), 200(91.11), 172(60.73), 184(51.88), 139(28.82), 168(15.58) |
| P16 | 4-phenylphenol (TMS)b |
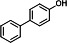
|
12.53 | 211(100), 227(79.29), 242(49.77), 57.1(31.55), 71(22.1), 152(21.47) |
| P17 | 3-hydroxybiphenyl (TMS)b |
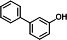
|
13.84 | 227(100), 242(77.9), 211(56.14), 152(27.26), 228(20.65), 113.5(19.97) |
| P18 | 2,2-biphenyldiol (2TMS)b |
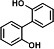
|
14.77 | 73(100), 330(17.15), 74(8.88), 331(5.22), 75(4.92), 212(4.7) |
| P19 | Monodroxy-CA (TMS)c | __ | 16.78 | 255(100), 239(61.79), 224(57.64), 240(56.11), 209(29.74), 256(25.31) |
| P20 | PYE-4,5-dihydrodiol (2TMS)c |
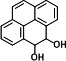
|
18.87 | 147(100), 73(70.43), 290(49.38), 202(28.76), 291(28.73), 380(18.64) |
| P21 | Monodroxy-PYE (TMS)c | __ | 20.21 | 290(100), 73(36.07), 291(25.45), 275(22.52), 189(15.62), 259(12.71) |
| P22 | 1‐acenaphthenoneb,d |
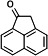
|
13.32 | 140(100), 168(87.06), 139(85.76), 70(14.82), 141(12.03), 169(11.98) |
| P23 | 1-acenaphthenolc |
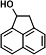
|
14.216 | 242(100), 152(83.56), 75(80.37), 241(59.27), 153(52.77), 73(29.59) |
| P24 | 1,2-dihydroxyacenaphthene (2TMS) (cis or trans)c,d |
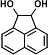
|
15.764 | 147(100), 73(74.44), 168(27.02), 330(23.48), 152(19.87), 241(18.43) |
| P25 | 1,2-dihydroxyacenaphthene (2TMS) (cis or trans)c,d |
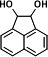
|
15.868 | 73(100), 147(97.35), 168(30.19), 330(23.41), 241(19.82), 152(18.18) |
| P26 | 1,2-dihydroxyacenaphthylene (2TMS)b,d |
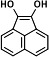
|
16.324 | 73(100), 328(65.69), 75(26.66), 240(25.1), 329(19.68), 117(16.84) |
| P27 | 9-fluorenoneb |
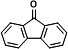
|
14.107 | 180(100), 152(60.24), 151(33.55), 150(21.24), 181(19.22), 76(14.73) |
| P28 | 9-fluorenolc |
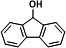
|
14.534 | 165(100), 254(63.01), 253(31.67), 163(20.98), 166(20.48), 164(19.71) |
| P29 | Monodroxy-FLN (TMS)c | __ | 15.493 | 254(100), 223(96.55), 239(45.48), 73(39.27), 255(22.74), 165(22.54) |
| P30 | Monodroxy-FLN (TMS)c | __ | 15.597 | 165(100), 254(95.73), 239(48.42), 73(32.33), 255(21.27), 152(18.28) |
| P31 | Monodroxy-FLN (TMS)c | __ | 15.822 | 254(100), 165(70.34), 239(70), 73(34.01), 255(21.86), 152(16.88) |
| P32 | 2‐fluorenol (TMS)c |
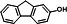
|
16.001 | 254(100), 239(74.86), 165(31.14), 255(23.04), 73(19.45), 240(16.29) |
| P33 | Dihydroxy-FLN (2TMS)c | __ | 17.087 | 342(100), 253(77.63), 73(74.73), 341(35.39), 343(32.47), 254(21.55) |
| P34 | 2-hydroxy-9-fluorenone (TMS)b |
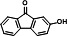
|
17.134 | 253(100), 268(71.41), 73(25.15), 254(22.35), 269(16.59), 139(14.21) |
| P35 | Dihydroxy-FLN (2TMS)c | __ | 17.670 | 73(100), 342(64.24), 343(20.16), 254(19.36), 253(17.96), 74(8.63) |
TMS: trimethylsilyl derivatized compound; 2TMS: ditrimethylsilyl derivatized compound. The metabolites whose structure was not determined were indicated by "__."
The structure of the metabolites was determined by comparing them with the NIST MS library.
The structure of the metabolites was determined by comparing them with the published literatures.
The metabolites were only found in the samples for 24 h.
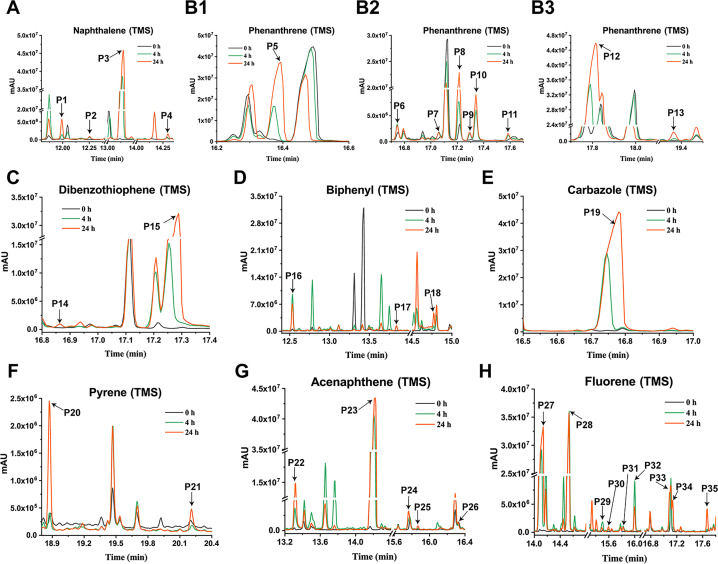
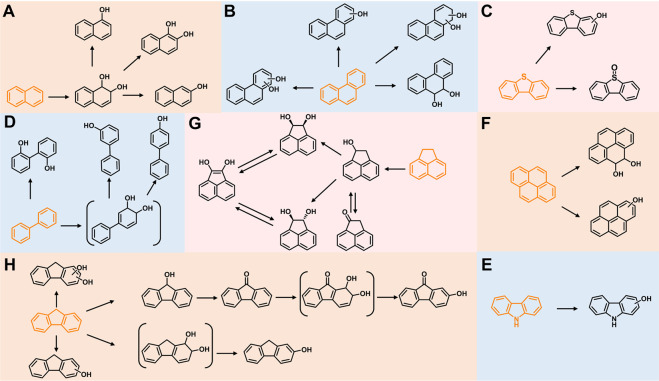
Four metabolites were detected in the 24-h degradation sample of NAP, namely 1-naphthol (TMS), 2-naphthol (TMS), NAP-1,2-dihydrodiol (2TMS), and 1,2-dihydroxynaphthalene (2TMS), with retention times of 11.99, 12.25, 13.15, and 14.29 min, respectively. However, 1,2-dihydroxynaphthalene was not detected in the 4-h degradation sample. 1-Naphthol and 2-naphthol may be products of unstable spontaneous dehydration of NAP-1,2-dihydrodiol. Subsequently, the metabolic pathway of NAP branches from 1,2-dihydroxynaphthalene into the meta-cleavage pathway and the ortho-cleavage pathway (31) (Fig. 3A and 4A).
Fig 3.
GC-MS profile of degradation samples of PAHs and their derivatives by E. coli BL21(DE3) with heterologous expression of NarA2B2 and PhtAcAd. The sample at 0 h was used as the control, and the samples at 4 and 24 h were the experimental group. (A) Naphthalene (P1: 1-naphthol, TMS; P2: 2-naphthol, TMS; P3: NAP-1,2-dihydrodiol, 2TMS; P4: 1,2-dihydroxynaphthalene, 2TMS). (B1–B3) Phenanthrene (P6, P7, P9, P10, P11: monohydroxy-PHE, TMS; P5: PHE-9,10-dihydrodiol, 2TMS; P8, P12: PHE-dihydrodiol, 2TMS; P13: dihydroxyl-PHE, 2TMS). (C) Dibenzothiophene (P14: monohydroxy-DBT, TMS; P15: dibenzothiophene-S-oxide). (D) Biphenyl (P16: 4-phenylphenol, TMS; P17: 3-hydroxybiphenyl, TMS; P18: 2,2-biphenyldiol, 2TMS). (E) Carbazole (P19: monodroxy-CA, TMS). (F) Pyrene (P20: PYE-4,5-dihydrodiol, 2TMS; P21: monodroxy-PYE, TMS). (G) Acenaphthene (P22: 1-acenaphthenone; P23: 1-acenaphthenol, TMS; P24–25: 1,2-dihydroxyacenaphthene, 2TMS; P26: 1,2-dihydroxyacenaphthylene, 2TMS). (H) Fluorene (P27: 9-fluorenone; P28: 9-fluorenol, TMS; P29-31: monodroxy-FLN, TMS; P32: 2-fluorenol, TMS; P33, P35: dihydroxy-FLN, 2TMS; P34: 2-hydroxy-9-fluorenone, TMS).
Fig 4.
Proposed metabolic characteristics of PAHs and derivatives catalyzed by E. coli BL21(DE3) with heterologous expression of NarA2B2 and PhtAcAd. (A) NAP; (B) PHE; (C) DBT; (D) BP; (E) CA; (F) PYE; (G) ACE; and (H) FLN. The metabolites in parentheses were inferred from the reported pathway and were not detected in this experiment.
In the dagradation sample of PHE, nine metabolites were detected. Compared to published studies, they were determined to be five isomers of monohydroxy-PHE (TMS), three kinds of PHE-dihydrodiol (2TMS), and a dihydroxy-PHE (2TMS). Of the three types of PHE-dihydrodiol, one was identified as PHE-9,10-dihydrodiol (16). We speculate that there are at least two pathways for NarA2B2 to degrade PHE, one of which begins at the C-9,10 positions. Due to the lack of available information, the structure of other metabolites was not identified (Fig. 3B and 4B).
In the case of DBT, a kind of monohydroxy-DBT (TMS) and dibenzothiophene-S-oxide were detected (Fig. 3C and 4C). Among them, NarA2B2 degraded DBT more easily to produce the latter. Monohydroxy-DBT (TMS) was determined by comparison with previous studies (32). Based upon the DBT metabolic pathway in N12 resting cells (5), monohydroxy-DBT may be 1-hydroxydibenzothiophene or 2-hydroxydibenzothiophene, formed by dehydration of DBT-1,2-dihydrodiol.
In the degradation sample of BP, three metabolites, 3-hydroxybiphenyl (TMS), 4-phenylphenol (TMS), and 2,2-dihydroxybiphenyl (2TMS), were detected. In general, the terminal oxygenase introduced activated molecular oxygen to the biphenyl at the C-2,3 positions (33–36). However, the catalytic positions of NarA2B2 for BP are C-2,2 positions and C-3,4 positions (Fig. 3D and 4D). NarA2B2 can also catalyze CA, and monohydroxy-CA (TMS) has been detected (Fig. 3E and 4E). In the degradation of PYE, two products were detected, monohydroxy-PYE and PYE-4,5-dihydrodiol (Fig. 3F and 4F).
Five intermediates were detected in the degraded samples of ACE for 24 h, namely 1-acenaphthenone, 1-acenaphthenol (TMS), 1,2-dihydroxyacenaphthylene (2TMS), and two kinds of 1,2-dihydroxyacenaphthene (2TMS). Of these, only 1-acenaphthenol was detected at 4 h. NarA2B2 can continuously catalyze ACE. ACE is transformed to 1-acenaphthenol via a one-step monooxygenation, then converted to 1-acenaphthenone via an endogenous dehydrogenase. Simultaneously, 1-acenaphthenol is altered to 1,2-dihydroxyacenaphthene and 1,2-dihydroxyacenaphthylene by the catalysis of NarA2B2 (Fig. 3G and 4G) (31).
Nine metabolites were detected from the FLN sample, including 9-fluorenone, 2-hydroxy-9-fluorenone (TMS), 9-fluorenol (TMS), 2-fluorenol (TMS), three kinds of monohydroxy-FLN (TMS), and two kinds of dihydroxy-FLN (2TMS). Akin to ACE, we hypothesize that NarA2B2 can perform continuous catalysis of FLN. First, NarA2B2 adds one oxygen atom at the C-9 position to form 9-fluorenol as fluorene monooxygenase, and 9-fluorenol is then converted to 9-fluorenone under the catalysis of an endogenous dehydrogenase. Subsequently, NarA2B2 catalyzes 9-fluorenone to produce 9-fluorenone-1,2-dihydrodiol, which spontaneously dehydrates to form 2-hydroxy-9-fluorenone (Fig. 3H and 4H) (31). At the same time, NarA2B2 catalyzes FLN to FLN-dihydrodiol (31), and dihydroxy-FLN was formed under the action of endogenous dehydrogenase.
The predicted structure of NarA2B2
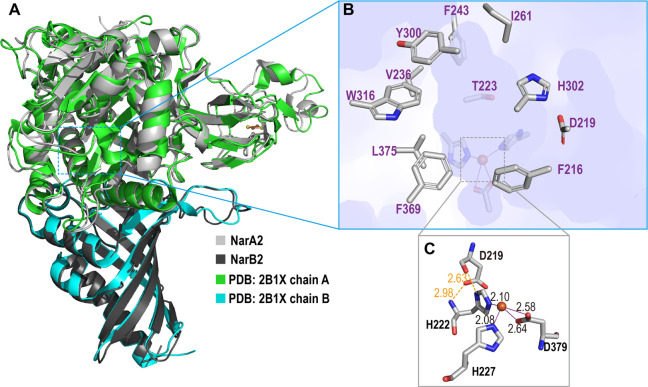
According to the complex structure of NarA2B2 predicted by AlphaFold2, NarA2 and NarB2 are well-combined as shown in Fig. 5A. Compared to the most similar crystal structure, a naphthalene 1,2-dioxygenase from Rhodococcus sp. (PDB: 2B1X, sequence similarity 45%) (10), NarA2B2 has a high structure similarity with an RMSD of 0.531 Å. The high conservation of both sequence and structure indicates that the predicted structure of NarA2B2 is reliable and comparable. Both active site iron and iron-sulfur clusters are bound to the α-subunit. The active site iron coordinating with H222, H227, and D379 has the characteristics of the classic 2-His-1-carboxylate facial triad (Fig. 5C) (37). It is notable that D219 forms two stable hydrogen bonds with H222 (H222N---D219OD1: 2.98 Å, H222Nε---D219O: 2.63 Å), essentially strengthening the iron center. The substrate pocket is assumed to be located around the vacant site of the iron octahedral. As shown in Fig. 5B, key residues surrounding the pocket (F216, D219, T223, V236, F243, I261, Y300, H302, W316, F369, L375) are selected within 4 Å as a docked substrate, indicating π-π potential interactions for PAHs are provided by aromatic rings.
Fig 5.
Overall structure of NarA2B2. (A) NarA2B2 predicted by AlphaFold2 with aligned naphthalene 1,2-dioxygenase from Rhodococcus sp. (PDB: 2B1X); the α-subunit NarA2 is colored in light grey (and green in 2B1X) and the β-subunit NarB2 is dark gray (and cyan in 2B1X). (B) The substrate pocket of NarA2B2, with the surface of NarA2B2 colored in light purple, and the side chain of residues around the active site are shown in sticks. (C) The active site iron center of NarA2B2, distances are given in Å.
According to the results of protein structure prediction, we speculate that F216, D219, H222, T223, H227, V236, F243, Y300, H302, W316, F369, and D379 have involvement in the catalysis of NarA2B2. Therefore, we performed point mutations to detect the degradation of PAHs. Mutants D219A, H222A, H227A, F369A, or D379A were found to have lost the ability to degrade NAP, PHE, DBT, FLN, and CA. F243A and Y300A have a greater influence on the degradation of the substrates with three-fused rings. F243A basically loses its ability to degrade PHE, DBT, BP, and CA. Y300A cannot degrade PHE, BP, and CA; and W316A has little effect on catalytic capacity and mainly inhibits the degradation of ACE, FLN, CA, and PYE. While T223A and H302A enhanced the degradation ability of ACE and NAP, and F216A increased the degradation ratio of ACE. Compared to the wild-type enzyme, the degradation ratios of ACE by F216A, T223A, and H302A had an improvement of 23.08%, 26.87%, and 29.52%, respectively; the degradation ratios of NAP by T223A and H302A had an improvement of 51.30% and 65.17%. The ability of V236A to degrade BP was significantly enhanced with an improvement of 77.94% (Fig. 6).
Fig 6.
The degradation of PAHs and derivatives by NarA2B2 mutants in E. coli BL21(DE3) at 24 h. (A) NAP; (B) PHE; (C) DBT; (D) ACE; (E) FLN; (E) BP; (G) CA; and (H) PYE. PC: BL21(DE3) containing pETDuet-narA2-Ter-narB2 and pACYCDuet-phtAcAd without mutations; NC: BL21(DE3) containing pETDuet and pACYCDuet; M216: F216A; M219: D219A; M222: H222A; M223: T223A; M227: H227A; M236: V236A; M243: F243A; M300: Y300A; M302: H302A; M316: W316A; M369: F369A; and M379: D379A.
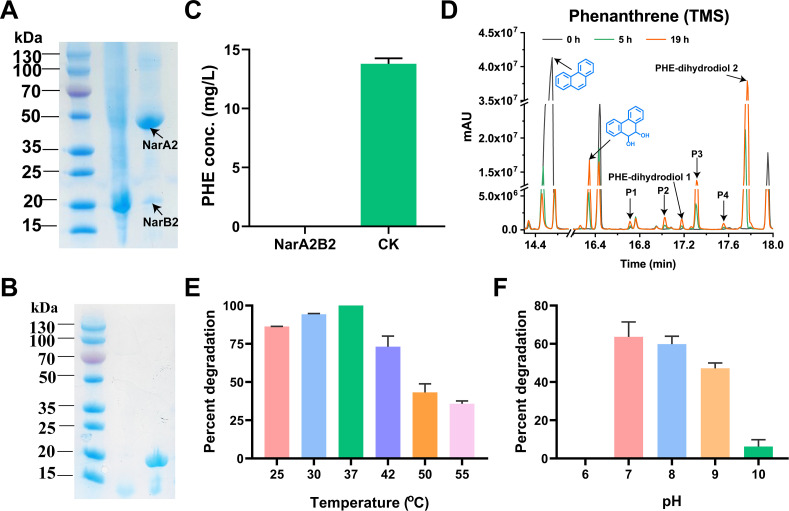
Purification and properties of purified NarA2B2
As NarA2B2 showed strong degradation ability towards PHE during the identification of the substrate specificity of NarA2B2 by resting cell biotransformation, PHE was chosen as the substrate to test the activity of purified NarA2B2 in vitro. We purified NarA2B2 and PhtAcAd by Ni-NTA affinity chromatography. The elution solutions of both recombinant proteins appeared dark brown in color. According to the SDS-PAGE results, we found that the purity of recombinant proteins exceeded 90% (Fig. 7A and B). NarA2B2 contains two subunits with molecular weights close to 50 and 20 kDa (Fig. 7A), consistent with the molecular masses of the polypeptides deduced from Chr966 and Chr967. Within 2 h, 5.67 × 10−3 µmol NarA2B2 could completely convert 14.24 µg of PHE (Fig. 7C), indicating that NarA2B2 has the catalytic ability in vitro. Furthermore, we further detected the catalytic products of purified NarA2B2 on PHE by GC-MS, which were basically consistent with those of resting cells. Also, 9,10-PHE-dihydrodiol, two unknown structures of PHE-dihydrodiol, and four monohydroxy-PHE (P1-P4) were detected (Fig. 7D). Meanwhile, we tested the optimal catalytic temperature and pH value of NarA2B2 and found that under the premise of using PhtAcAd as the ETC, the optimal catalytic temperature of NarA2B2 is 37°C, and NarA2B2 also exhibits catalytic activity at 55°C (Fig. 7E). The optimal catalytic pH value of NarA2B2 is 7, and it has no catalytic activity in acidic environments. The catalytic activity of NarA2B2 is stronger in weak alkaline environments. (Fig. 7F).
Fig 7.
The purification and characterization of NarA2B2. (A) The purification of NarA2B2. (B) The purification of PhtAcAd. (C) The degradation of PHE by purified NarA2B2. NarA2B2: the reaction mixture contained purified NarA2B2 and PhtAcAd; CK: the reaction mixture did not contain purified NarA2B2 and PhtAcAd. (D) GC-MS profile of PHE degraded by purified NarA2B2. The sample at 0 h was used as the control, and the samples at 5 and 19 h were the experimental group (P1–P4: monohydroxy-PHE, TMS); (E) The catalytic temperature range suitable for NarA2B2 when PHE was used as the substrate; (F) The catalytic pH range suitable for NarA2B2 when PHE was used as the substrate.
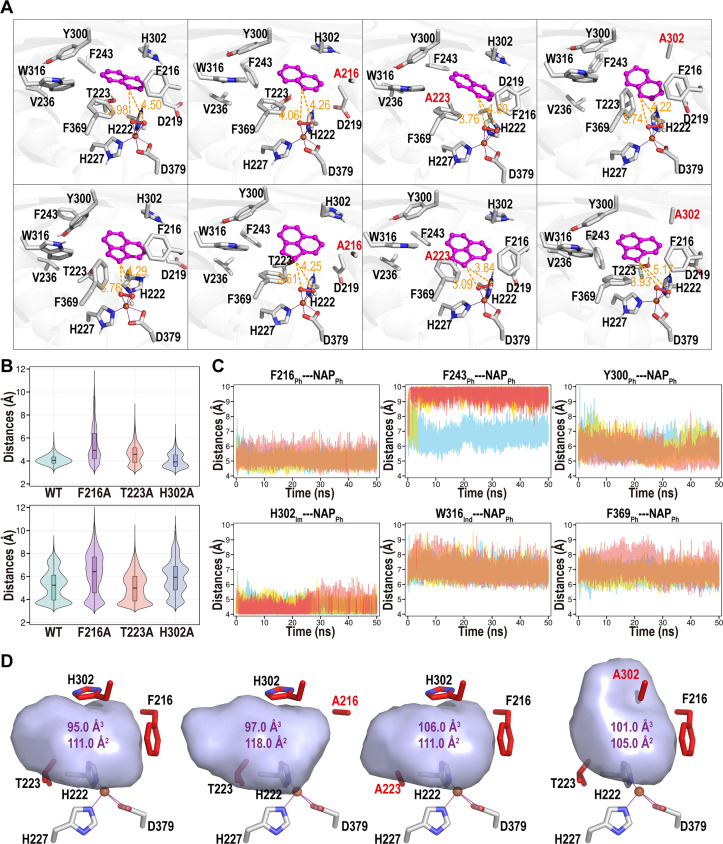
Interaction analysis of structure binding domain
Molecular docking and MD simulations were further developed to examine the structure of NarA2B2, as shown in Fig. 8 (Fig. S4 and S5). Two representative substrates, NAP and ACE (selected due to their notable alterations between the wild type and mutants), were subjected to simulations with NarA2B2. The clustering structure and attack distances, analyzed in Fig. 8A and B, clearly indicate that the NarA2B2 wild type and all three mutants (F216A, T223A, and H302A) can configure a near-attack conformation. The clustering structures in this scenario reflect a sequence of structures rather than a single static structure. The attack distance of clustering structures varies between 3.09 and 4.06 Å (Fig. 8A), but that of all the structures exhibit significant fluctuations (Fig. S5). The attack distances have a minimum of 2.92 Å (WT-NAP), 2.84 Å (F216A-NAP), 2.81 Å (T223A-NAP), 2.76 Å (H302A-NAP), 2.91 Å (WT-ACE), 2.78 Å (F216A-ACE), 2.90 Å (T223A-ACE), 2.97 Å (H302A-ACE), and a concentration below 4 Å (Fig. 8B). Hence, both the WT and mutants can interact with NAP and ACE.
Fig 8.
Structural analysis of NarA2B2 complex with representative substrate. (A) Clustering structures of NarA2B2 and its mutations with NAP (top row) and ACE (bottom row). Active site residues are displayed in stick format, along with the substrate and OOH moiety, shown in stick and ball formats, respectively. Substrates (NAP and ACE) are highlighted in magenta. Attacking distances between the substrate and the OOH moiety are indicated in orange, with distances in Å. (B) Violin density plot illustrating the attacking distances during MD simulations between NarA2B2 and its mutations with either NAP (top) or ACE (bottom). The attacking distances are determined by the minimum distance between the carbon atom of the substrate and the oxygen atom bound to iron. (C) Potential π interaction distances between residues and NAP within the WT-NAP system complex. Distances are measured from the benzene ring’s center of mass within residues and NAP. The terms “Ph,” “Im,” and “Ind” denote phenyl, imidazole, and Indole rings, respectively. Data includes three parallel MD simulations, represented by red, yellow, and blue lines. (D) Active site pocket structures of NarA2B2 and its mutations, with calculated volumes (in Å3) and areas (in Å2).
Given the lack of a hydrogen bond donor or acceptor between the substrate and NarA2B2 (as PAHs typically lack oxygen or nitrogen atoms), it is challenging for NarA2B2 to stabilize a substrate via a hydrogen bond network. The differential performance of the mutants on various substrates is primarily due to the substrate pocket and π interactions (encompassing sandwich, T-shaped, and parallel-displaced π interactions) within the aromatic or imidazole ring of NarA2B2. Figure 8C shows that NAP can form stable π interactions with F216, Y300, and H302 (with center-of-mass distances between two rings of 5.09 ± 0.34 Å, 5.81 ± 0.53 Å, and 4.51 ± 0.36 Å). It also exhibits weak interactions with W316 and F369 and a random interaction with F243. H302A’s lack of π interactions boosts ACE degradation by increasing substrate mobility, thereby improving the likelihood of oxygen contact.
Furthermore, the mutants F216A and H302A expand the substrate pocket as shown in Fig. 8D. The removal of the hydroxyl group in F216A and T223A mutants enhances the hydrophobic environment, which is advantageous for PAHs. To summarize, while the loss of a phenyl ring decreases π-π interactions, it provides additional space for high-molecular-weight PAHs. Simultaneously, the absence of a hydroxyl group augments the hydrophobic environment. Variations in the pocket’s volume and modifications in interactions can significantly alter the catalytic efficiency. The divergent performance of mutants with various substrates can be attributed to a combination of these factors.
DISCUSSION
In this study, we identified a novel RHO, NarA2B2 in Hydrogenibacillus sp. N12, which could degrade multiple PAHs and heterocyclic derivatives, including NAP, PHE, FLN, ACE, CA, BP, DBT, and PYE. The metabolites of PAHs and derivatives catalyzed by NarA2B2 were identified through GC-MS, and the metabolic characteristics of NarA2B2 were analyzed. NarA2B2 could add one or two atoms of molecular oxygen to substrates. In particular, it was found that the degradation of BP differed from previous reports through the absence of the addition of two oxygen atoms from the C2 and C-3 positions. The catalytic position of BP by NarA2B2 is instead at two C-2 positions or C-3, C-4 positions. Through protein structure prediction and point mutations, we identified key functional sites of the iron coordinating site and active pocket involved in dioxygen activation as well as substrate binding. Considering the detected intermediate metabolites involve one or two oxygen atoms, NarA2 may act as both monooxygenase and dioxygenase. According to the catalytic cycle of Rieske non-heme iron enzymes reported in previous studies (38–40), the mechanism of NarA2 begins with the dioxygen activation intermediate, a ferric (hydro) peroxo species, with hydroxylated product formation with the mononuclear ferric reduction by an external electron.
As NarA2B2 is derived from the thermophilic microorganism Hydrogenibacillus sp. N12, we speculate that the enzyme may have unique physiological and biochemical properties compared to mesophilic bacteria. Currently, few RHOs are purified and characterized, such as phthalate dioxygenase, naphthalene dioxygenase, and biphenyl dioxygenase (41). We purified the oxygenase and ETC components through Ni-NTA affinity chromatography. The function of ETC components was detected by potassium ferricyanide; when PhtAcAd was mixed with potassium ferricyanide and NADH, the buffer changed from brownish yellow to colorless in a short time (data not presented), it indicates that ETC has the function of transferring electrons. We used the reported method to detect the enzyme assay of NarA2B2 (41), but we did not obtain the catalytic activity of purified NarA2B2 in vitro. We also optimized the catalytic conditions, such as different catalytic temperatures, addition of glycerol to the reaction mixtures, using NADH or NADPH as electron donors, addition of L-ascorbic acid or L-ascorbic acid sodium to reduce Fe3+, and even used thrombin to cleave the 6× His-Tag from the recombinant protein, but none of these optimizations enabled us to obtain the catalytic activity of NarA2B2 in vitro (Fig. S6 to S9). We also found that E. coli (DE3) expressing NarA2B2 and PhtAcAd and strain N12 did not exhibit the ability to degrade PAHs after cell lysis (Fig. S10 and S11.). We speculate that NarA2B2 may lose catalytic function due to its inability to maintain a three-dimensional structure outside the cell, or the catalysis requires more specific conditions. We find that the naphthalene dioxygenase PhnI and ETC components of Sphingomonas CHY-1 were purified under an environment with O2 < 2 ppm (41). This inspired us that NarA2B2 may also need to be purified under anaerobic conditions to maintain the catalytic center Fe2+ from oxidation. Therefore, we attempted to incubate NarA2B2 with L-ascorbic acid under anaerobic conditions to restore its activity. Through the optimization of experimental detail, we successfully obtained the catalytic activity of NarA2B2 in vitro. Under the premise of using PhtAcAd as the ETC, the optimal catalytic temperature of NarA2B2 is 37°C, and the optimal pH is 7. The optimal catalytic temperature and pH value of NarA2B2 may vary when it is adapted to different ETCs. When NarA2B2 was compatible with the ETC components of strain N12, its optimal catalytic temperature may be higher than 37°C, and it may also exhibit stronger catalytic activity. Moreover, since strain N12 lacks genetic manipulation tools, we are unable to knock out the gene narA2B2, which makes it difficult for us to determine the interaction of NarA2B2 and NarAaAb in strain N12 (5).
Conclusions
A novel RHO NarA2B2 from Hydrogenibacillus sp. N12 was identified and characterized, which is capable of degrading a variety of PAHs and their derivatives. Depending on the degradation characteristics, NarA2B2 adds one or two atoms of molecular oxygen to the aromatic ring. Through protein structure prediction, molecular simulation docking, and point mutation, we identified several key amino acids involved in catalysis and explored the catalytic mechanism of NarA2B2. We successfully obtained the in vitro catalytic activity of NarA2B2 by incubating it with L-ascorbate under anaerobic conditions.
ACKNOWLEDGMENTS
This work was supported by grants from the National Key R&D Program of China (2021YFA0909500), the National Natural Science Foundation of China (32030004), and the Shanghai Excellent Academic Leaders Program (20XD1421900).
L.G. and H.T. conceived and designed the experiments. L.G., X.O., and X.Q. performed the experiments. W.W. and H.T. received projects and contributed reagents and materials. L.G. and H.T. wrote the paper. L.G., X.O., Y.H., X.Q., W.W., H.T., and P.X. discussed and revised the manuscript. All authors commented on the manuscript before submission. All authors read and approved the final manuscript.
The authors declare no competing financial or non-financial interest.
Contributor Information
Weiwei Wang, Email: oudigouzai@sjtu.edu.cn.
Hongzhi Tang, Email: tanghongzhi@sjtu.edu.cn.
Haruyuki Atomi, Kyoto University, Kyoto, Japan.
DATA AVAILABILITY
The whole-genome sequence of Hydrogenibacillus sp. strain N12 was deposited into the NCBI database under the accession number CP080953. The protein IDs of PhtAc and PhtAd were AAQ91918.1 and AAQ91919.1, respectively.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.00865-23.
Supplemental figures.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Haritash AK, Kaushik CP. 2011. Seasonal and spatial occurrence and distribution of respirable particulate-bound atmospheric polycyclic aromatic hydrocarbons in Hisar city (India) and their potential health-risks. Asian J Water Environ Pollut 8:73–80. [Google Scholar]
- 2. Menzie CA, Potocki BB, Santodonato J. 1992. Exposure to carcinogenic PAHs in the environment. Environ Sci Technol 26:1278–1284. doi: 10.1021/es00031a002 [DOI] [Google Scholar]
- 3. Gupta S, Pathak B, Fulekar MH. 2015. Molecular approaches for biodegradation of polycyclic aromatic hydrocarbon compounds: a review. Rev Environ Sci Biotechnol 14:241–269. doi: 10.1007/s11157-014-9353-3 [DOI] [Google Scholar]
- 4. Müller R, Antranikian G, Maloney S, Sharp R. 1998. Thermophilic degradation of environmental pollutants, p 155–169. In Advances in biochemical engineering/biotechnology. doi: 10.1007/BFb0102286 [DOI] [Google Scholar]
- 5. Qiu X, Wang W, Zhang L, Guo L, Xu P, Tang H. 2022. A thermophile Hydrogenibacillus sp. strain efficiently degrades environmental pollutants polycyclic aromatic hydrocarbons. Environ Microbiol 24:436–450. doi: 10.1111/1462-2920.15869 [DOI] [PubMed] [Google Scholar]
- 6. Gibson DT, Parales RE. 2000. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr Opin Biotechnol 11:236–243. doi: 10.1016/s0958-1669(00)00090-2 [DOI] [PubMed] [Google Scholar]
- 7. Singleton DR, Hu J, Aitken MD. 2012. Heterologous expression of polycyclic aromatic hydrocarbon ring-hydroxylating dioxygenase genes from a novel pyrene-degrading betaproteobacterium. Appl Environ Microbiol 78:3552–3559. doi: 10.1128/AEM.00173-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Larkin MJ, Allen CC, Kulakov LA, Lipscomb DA. 1999. Purification and characterization of a novel naphthalene dioxygenase from Rhodococcus sp. J Bacteriol 181:6200–6204. doi: 10.1128/JB.181.19.6200-6204.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kauppi B, Lee K, Carredano E, Parales RE, Gibson DT, Eklund H, Ramaswamy S. 1998. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 6:571–586. doi: 10.1016/s0969-2126(98)00059-8 [DOI] [PubMed] [Google Scholar]
- 10. Gakhar L, Malik ZA, Allen CCR, Lipscomb DA, Larkin MJ, Ramaswamy S. 2005. Structure and increased thermostability of Rhodococcus sp. naphthalene 1,2-dioxygenase. J Bacteriol 187:7222–7231. doi: 10.1128/JB.187.21.7222-7231.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Furusawa Y, Nagarajan V, Tanokura M, Masai E, Fukuda M, Senda T. 2004. Crystal structure of the terminal oxygenase component of biphenyl dioxygenase derived from Rhodococcus sp. J Mol Biol 342:1041–1052. doi: 10.1016/j.jmb.2004.07.062 [DOI] [PubMed] [Google Scholar]
- 12. Kim S-J, Kweon O, Freeman JP, Jones RC, Adjei MD, Jhoo J-W, Edmondson RD, Cerniglia CE. 2006. Molecular cloning and expression of genes encoding a novel dioxygenase involved in low- and high-molecular-weight polycyclic aromatic hydrocarbon degradation in Mycobacterium vanbaalenii PYR-1. Appl Environ Microbiol 72:1045–1054. doi: 10.1128/AEM.72.2.1045-1054.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kweon O, Kim S-J, Freeman JP, Song J, Baek S, Cerniglia CE. 2010. Substrate specificity and structural characteristics of the novel rieske nonheme iron aromatic ring-hydroxylating oxygenases NidAB and NidA3B3 from Mycobacterium vanbaalenii PYR-1. mBio 1:e00135–10. doi: 10.1128/mBio.00135-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Demanèche S, Meyer C, Micoud J, Louwagie M, Willison JC, Jouanneau Y. 2004. Identification and functional analysis of two aromatic-ring-hydroxylating dioxygenases from a Sphingomonas strain that degrades various polycyclic aromatic hydrocarbons. Appl Environ Microbiol 70:6714–6725. doi: 10.1128/AEM.70.11.6714-6725.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jakoncic J, Jouanneau Y, Meyer C, Stojanoff V. 2007. The catalytic pocket of the ring-hydroxylating dioxygenase from Sphingomonas CHY-1. Biochem Biophys Res Commun 352:861–866. doi: 10.1016/j.bbrc.2006.11.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang L, Liu H, Dai J, Xu P, Tang H. 2022. Unveiling degradation mechanism of PAHs by a Sphingobium strain from a microbial consortium. mLife 1:287–302. doi: 10.1002/mlf2.12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stingley RL, Khan AA, Cerniglia CE. 2004. Molecular characterization of a phenanthrene degradation pathway in Mycobacterium vanbaalenii PYR-1. Biochem Biophys Res Commun 322:133–146. doi: 10.1016/j.bbrc.2004.07.089 [DOI] [PubMed] [Google Scholar]
- 18. Zhang L, Qiu X, Huang L, Xu J, Wang W, Li Z, Xu P, Tang H. 2021. Microbial degradation of multiple PAHs by a microbial consortium and its application on contaminated wastewater. J Hazard Mater 419:126524. doi: 10.1016/j.jhazmat.2021.126524 [DOI] [PubMed] [Google Scholar]
- 19. Krivobok S, Kuony S, Meyer C, Louwagie M, Willison JC, Jouanneau Y. 2003. Identification of pyrene-induced proteins in Mycobacterium sp. strain 6PY1: evidence for two ring-hydroxylating dioxygenases. J Bacteriol 185:3828–3841. doi: 10.1128/JB.185.13.3828-3841.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. doi: 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schwede T, Kopp J, Guex N, Peitsch MC. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31:3381–3385. doi: 10.1093/nar/gkg520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, Zaslavsky L, Zhang J, Bolton EE. 2021. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res 49:D1388–D1395. doi: 10.1093/nar/gkaa971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goodsell DS, Morris GM, Olson AJ. 1996. Automated docking of flexible ligands: applications of autodock. J Mol Recognit 9:1–5. doi: [DOI] [PubMed] [Google Scholar]
- 24. Fuhrmann J, Rurainski A, Lenhof H-P, Neumann D. 2010. A new Lamarckian genetic algorithm for flexible ligand‐receptor docking. J Comput Chem 31:1911–1918. doi: 10.1002/jcc.21478 [DOI] [PubMed] [Google Scholar]
- 25. Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc 117:5179–5197. doi: 10.1021/ja00124a002 [DOI] [Google Scholar]
- 26. Case DA, Belfon K, Ben-Shalom IY, Brozell SR, Cerutti DS, Cheatham TE, Cruzeiro VWD, Darden TA, Duke RE, Giambasu G, Gilson MK, Gohlke H, Goetz AW, Harris R, Izadi S, Izmailov SA, Jin C, Kasavajhala K, Kaymak MC, King E, Kovalenko A, Kurtzman T, Lee TS, LeGrand S, Li P, Lin C, Liu J, Luchko T, Luo R, Machado M, Man V, Manathunga M, Merz KM, Miao Y, Mikhailovskii O, Monard G, Nguyen H, O’Hearn KA, Onufriev A, Pan F, Pantano S, Qi R, Rahnamoun A, Roe DR, Roitberg A, Sagui C, Schott-Verdugo S, Shen J, Simmerling CL, Skrynnikov NR, Smith J, Swails J, Walker RC, Wang J, Wei H, Wolf RM, Wu X, Xue Y, York DM, Zhao S, Kollman PA. 2020. Amber. University of California, San Francisco. [Google Scholar]
- 27. Roe DR, Cheatham TE. 2013. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput 9:3084–3095. doi: 10.1021/ct400341p [DOI] [PubMed] [Google Scholar]
- 28.PyMOL. The PyMOL molecular graphics system, version 2.0. Schrödinger, LLC. https://pymol.org/ [Google Scholar]
- 29. Sho M, Hamel C, Greer CW. 2004. Two distinct gene clusters encode pyrene degradation in Mycobacterium sp. strain S65. FEMS Microbiol Ecol 48:209–220. doi: 10.1016/j.femsec.2004.01.011 [DOI] [PubMed] [Google Scholar]
- 30. Kweon O, Kim S-J, Baek S, Chae J-C, Adjei MD, Baek D-H, Kim Y-C, Cerniglia CE. 2008. A new classification system for bacterial Rieske non-heme iron aromatic ring-hydroxylating oxygenases. BMC Biochem 9:11. doi: 10.1186/1471-2091-9-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mallick S, Chakraborty J, Dutta TK. 2011. Role of oxygenases in guiding diverse metabolic pathways in the bacterial degradation of low-molecular-weight polycyclic aromatic hydrocarbons: a review. Crit Rev Microbiol 37:64–90. doi: 10.3109/1040841X.2010.512268 [DOI] [PubMed] [Google Scholar]
- 32. Gai Z, Yu B, Li L, Wang Y, Ma C, Feng J, Deng Z, Xu P. 2007. Cometabolic degradation of dibenzofuran and dibenzothiophene by a newly isolated carbazole-degrading Sphingomonas sp. strain. Appl Environ Microbiol 73:2832–2838. doi: 10.1128/AEM.02704-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Furukawa K, Suenaga H, Goto M. 2004. Biphenyl dioxygenases: functional versatilities and directed evolution. J Bacteriol 186:5189–5196. doi: 10.1128/JB.186.16.5189-5196.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pieper DH, Seeger M. 2008. Bacterial metabolism of polychlorinated biphenyls. J Mol Microbiol Biotechnol 15:121–138. doi: 10.1159/000121325 [DOI] [PubMed] [Google Scholar]
- 35. Haddock JD, Horton JR, Gibson DT. 1995. Dihydroxylation and dechlorination of chlorinated biphenyls by purified biphenyl 2, 3-dioxygenase from Pseudomonas sp strain LB400. J Bacteriol 177:20–26. doi: 10.1128/jb.177.1.20-26.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seeger M, Cámara B, Hofer B. 2001. Dehalogenation, denitration, dehydroxylation, and angular attack on substituted biphenyls and related compounds by a biphenyl dioxygenase. J Bacteriol 183:3548–3555. doi: 10.1128/JB.183.12.3548-3555.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bruijnincx PCA, van Koten G, Klein Gebbink RJM. 2008. Mononuclear non-heme iron enzymes with the 2-His-1-carboxylate facial triad: recent developments in enzymology and modeling studies. Chem Soc Rev 37:2716–2744. doi: 10.1039/b707179p [DOI] [PubMed] [Google Scholar]
- 38. Zhu L, Zhou J, Zhang R, Tang X, Wang J, Li Y, Zhang Q, Wang W. 2020. Degradation mechanism of biphenyl and 4-4'-dichlorobiphenyl cis-dihydroxylation by non-heme 2,3 dioxygenases BphA: a QM/MM approach. Chemosphere 247:125844. doi: 10.1016/j.chemosphere.2020.125844 [DOI] [PubMed] [Google Scholar]
- 39. Szaleniec M, Wojtkiewicz AM, Bernhardt R, Borowski T, Donova M. 2018. Bacterial steroid hydroxylases: enzyme classes, their functions and comparison of their catalytic mechanisms. Appl Microbiol Biotechnol 102:8153–8171. doi: 10.1007/s00253-018-9320-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang Z, Chi X, Funabashi M, Baba S, Nonaka K, Pahari P, Unrine J, Jacobsen JM, Elliott GI, Rohr J, Van Lanen SG. 2011. Characterization of LipL as a non-heme, Fe (II)-dependent α-ketoglutarate: UMP dioxygenase that generates uridine-5′-aldehyde during A-90289 biosynthesis. J Biol Chem 286:7885–7892. doi: 10.1074/jbc.M110.203562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jouanneau Y, Meyer C, Jakoncic J, Stojanoff V, Gaillard J. 2006. Characterization of a naphthalene dioxygenase endowed with an exceptionally broad substrate specificity toward polycyclic aromatic hydrocarbons. Biochemistry 45:12380–12391. doi: 10.1021/bi0611311 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figures.
Data Availability Statement
The whole-genome sequence of Hydrogenibacillus sp. strain N12 was deposited into the NCBI database under the accession number CP080953. The protein IDs of PhtAc and PhtAd were AAQ91918.1 and AAQ91919.1, respectively.