ABSTRACT
Swine acute diarrhea syndrome coronavirus (SADS-CoV) is highly pathogenic to piglets and poses a major threat to the swine industry. SADS-CoV has a wide cell tropism and pathogenic potential in younger animals. Therefore, understanding how SADS-CoV enters cells is essential for curbing its re-emergence and spread. Here, we report that tunicamycin, an N-linked glycoprotein inhibitor, inhibited the attachment of SADS-CoV to host cells, suggesting that the SADS-CoV receptor may be an N-linked glycoprotein but not Neu5Gc or Neu5Ac. Moreover, we found that exogenous trypsin, endogenous serine protease, cathepsin B, cathepsin L, and lysosomal acidification triggered SADS-CoV entry into cells. These findings improve our understanding of the molecular mechanisms underlying SADS-CoV entry and provide insights into the development of potential antiviral targets against SADS-CoV.
IMPORTANCE
Gaining insight into the cell-entry mechanisms of swine acute diarrhea syndrome coronavirus (SADS-CoV) is critical for investigating potential cross-species infections. Here, we demonstrated that pretreatment of host cells with tunicamycin decreased SADS-CoV attachment efficiency, indicating that N-linked glycosylation of host cells was involved in SADS-CoV entry. Common N-linked sugars Neu5Gc and Neu5Ac did not interact with the SADS-CoV S1 protein, suggesting that these molecules were not involved in SADS-CoV entry. Additionally, various host proteases participated in SADS-CoV entry into diverse cells with different efficiencies. Our findings suggested that SADS-CoV may exploit multiple pathways to enter cells, providing insights into intervention strategies targeting the cell entry of this virus.
KEYWORDS: SADS-CoV, entry, N-linked glycoproteins, host proteases
INTRODUCTION
Swine acute diarrhea syndrome coronavirus (SADS-CoV), a novel bat HKU2-related coronavirus belonging to subgenus Rhinacovirus and genus Alphacoronavirus, was first identified in southern China in 2017 (1 – 4). SADS-CoV infection leads to acute diarrhea, vomiting, and high mortality rates among young piglets, particularly those less than 7 days old, but only causes mild or asymptomatic infections in adult swine (2, 4, 5). Recent studies have shown that SADS-CoV has broad cell tropism and high replication efficiency in primary human cells in vitro, suggesting its potential for interspecies transmission (6 – 8). Experimental infection has demonstrated that SADS-CoV can replicate in chicken embryos and young chicks, showing multiple tissue tropisms (9), and induces lethal infection in suckling mice but limited infection and no symptoms in adult mice (10, 11). These results are consistent with the findings in SADS-CoV-infected piglets (12), suggesting that SADS-CoV is highly pathogenic in young animals.
The closely related SADS-CoV (98.48% nucleotide identity at the genome level) was detected in Rhinolophus affinis sampled in 2016, approximately 200 km from a farm experiencing a SADS outbreak (4). In addition, genetically diverse SADS-related CoVs (SADSr-CoVs) have been detected in three other Rhinolophus bats (R. sinicus, R. rex, and R. pusillus) from several provinces in southern China (4, 13, 14). These findings highlight the risk of potential interspecies transmission of SADS-CoV and related CoVs to younger animals. Thus, further characterization of these viruses is urgently required.
Coronavirus receptor binding and membrane fusion are critical first steps in the infection cycle (15 – 17). The coronavirus spike is a member of the class I viral membrane fusion protein family that mediates coronavirus entry into host cells (18, 19). The S1 subunit binds to a receptor on the host cell surface, and the S2 subunit fuses with the viral and host membranes, allowing viral genomes to enter the host cells (20, 21). Coronaviruses exhibit complex receptor-recognition patterns, with some recognizing proteins and others recognizing sugars. For example, severe acute respiratory syndrome virus (SARS)-CoV-1, SARS-CoV-2, and HCoV-NL63 recognize angiotensin-converting enzyme 2; Middle East respiratory syndrome-CoV (MERS-CoV) and HKU4 recognize dipeptidyl peptidase 4; mouse hepatitis virus recognize carcinoembryonic antigen-related cell adhesion molecule 1; HCoV-229E, porcine deltacoronavirus, transmissible gastroenteritis virus, and porcine respiratory coronavirus recognize aminopeptidase N (22 – 28); bovine coronavirus (BCoV) and infectious bronchitis virus recognize N-glycolylneuraminic acid (Neu5Gc); and HCoV-OC43 and HCoV-HKU1 recognize N-acetylneuraminic acid (Neu5Ac) (29 – 31). Moreover, BCoV and HCoV-OC43 may use HLA-1 as the protein receptor (32, 33).
Host proteases that cleave coronavirus spikes also play important roles in viral entry (34, 35) by acting at four stages of the virus infection cycle: (i) proprotein convertases (e.g., furin) during virus packaging; (ii) extracellular proteases (e.g., elastase) and exogenous trypsin after virus release into the extracellular space; (iii) cell surface proteases (e.g., serine protease TMPRSSs) after virus attachment to host cells; and (iv) lysosomal proteases (e.g., cathepsin L and cathepsin B) after virus endocytosis into targeting cells (21, 35). Some coronaviruses, including SARS-CoV-2, infectious bronchitis virus (IBV), and MERS-CoV, possess one or more furin and furin-like protease cleavage sites (35 – 37). Trypsin is a prototype serine endopeptidase that can directly cleave the S proteins of many enteric coronaviruses, and most porcine epidemic diarrhea virus (PEDV) strains are highly dependent on trypsin (38, 39). Cathepsins are typically found in endosomes and lysosomes with both endo- and exopeptidase activities. For example, cathepsin L processes SARS-CoV-1, SARS-CoV-2, MERS-CoV, and HCoV-229E spike proteins, whereas cathepsin B is involved in feline coronavirus and mouse hepatitis virus entry (37, 40 – 42). TMPRSS proteases are type II transmembrane proteins involved in the activation of many coronaviruses, including SARS-CoV-1, SARS-CoV-2, MERS-CoV, HCoV-229E, and PEDV (21, 35, 37, 43).
In vivo and in vitro studies have shown that SADS-CoV has broad cell and tissue tropism in diverse hosts. However, limited information is available on the viral receptors and cell-entry mechanisms of SADS-CoV (44). Accordingly, in this study, we evaluated the effects of tunicamycin on the inhibition of SADS-CoV attachment to cells and the potential for N-linked glycoproteins to act as SADS-CoV receptors. Furthermore, we assessed the roles of trypsin, cathepsin B, cathepsin L, lysosomal acidification, and TMPRSSs in SADS-CoV entry.
RESULTS
Tunicamycin inhibited SADS-CoV infection in cells
SADS-CoV has a wide cell tropism (6 – 8), suggesting that a common molecule is likely to affect viral entry. To assess whether sugars were involved in SADS-CoV infection, we first performed hemagglutination tests. The results showed that SADS-CoV and SADS-CoV S1 proteins could not agglutinate mouse red blood cells (data not shown), implying that O-linked glycosylation did not affect SADS-CoV entry (29).
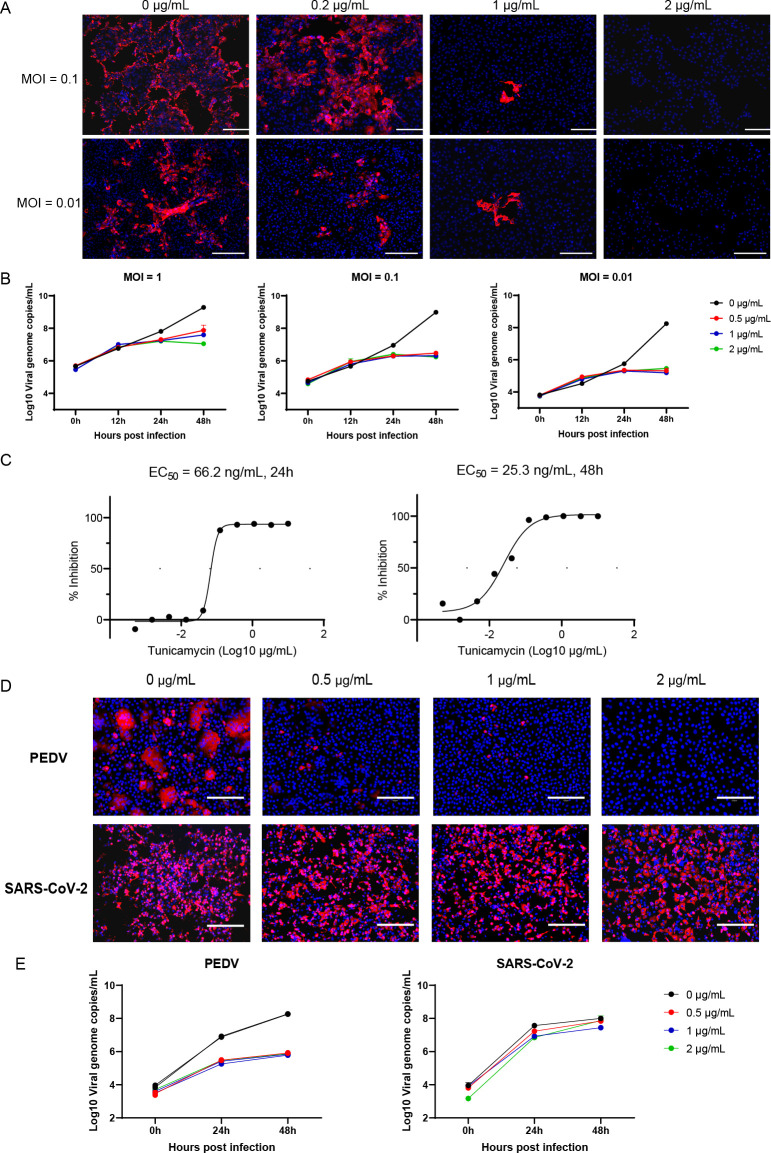
Next, to test whether N-linked glycosylation was involved in SADS-CoV infection, Vero cells were treated with the N-linked glycosylation inhibitor tunicamycin for 24 h, followed by infection with SADS-CoV. We then used quantitative reverse transcription polymerase chain reaction (RT-qPCR) and immunofluorescence assays (IFAs) targeting the nucleocapsid protein (N) to assess SADS-CoV replication. Our results showed that SADS-CoV replication was significantly decreased in Vero cells pretreated with tunicamycin. Indeed, virus replication was barely detected in cells pretreated with 2 µg/mL tunicamycin (Fig. 1A and B). Tunicamycin displayed inhibition against SADS-CoV with half-maximal effective concentration (EC50) values of 66.2 at 24 h post-infection (hpi) and 25.3 ng/mL at 48 hpi, respectively, (Fig. 1C). These results indicated that N-linked sugars were associated with SADS-CoV infection.
Fig 1.
Tunicamycin inhibited SADS-CoV infection. Vero cells were pretreated with tunicamycin for 24 h, washed with phosphate-buffered saline (PBS) three times, and then infected with SADS-CoV (MOI = 0.1). Cells were fixed at 24 hpi and stained with anti-SADS-CoV NP antibodies by IFA (A), and the supernatants were harvested at 0, 12, 24, or 48 hpi for assessment of viral replication dynamics by RT-qPCR (B). (C) Dose-response curves of tunicamycin for SADS-CoV infection, as determined by RT-qPCR. (D) Vero cells were pretreated with tunicamycin for 24 h, washed with PBS three times, and then infected with PEDV or SARS-CoV-2 (MOI = 0.1). Cells were fixed at 24 hpi and stained with anti-PEDV (1:200) or SARS-CoV-2 NP (1:1,000) antibodies by IFA. (E) The supernatants were harvested at 0, 24, or 48 hpi for assessment of viral replication dynamics by qRT-PCR. All data are shown as means ± standard errors of the means (n = 3 biological replicates). Scale bars: 300 µm (A) and 125 µm (D).
To further characterize whether tunicamycin could inhibit other coronavirus infections, Vero cells were pretreated with 1 µg/mL tunicamycin for 24 h to inhibit cell N-linked glycosylation and then infected with PEDV and SARS-CoV-2, and viral replication was assessed using RT-qPCR and IFA (Fig. 1D and E). The results showed that PEDV infection was significantly inhibited by tunicamycin at 24 hpi. By contrast, tunicamycin treatment had no significant effect on SARS-CoV-2 infection. These data implied that tunicamycin was likely an inhibitor of SADS-CoV and PEDV but not SARS-CoV-2.
Tunicamycin inhibited SADS-CoV attachment to cells
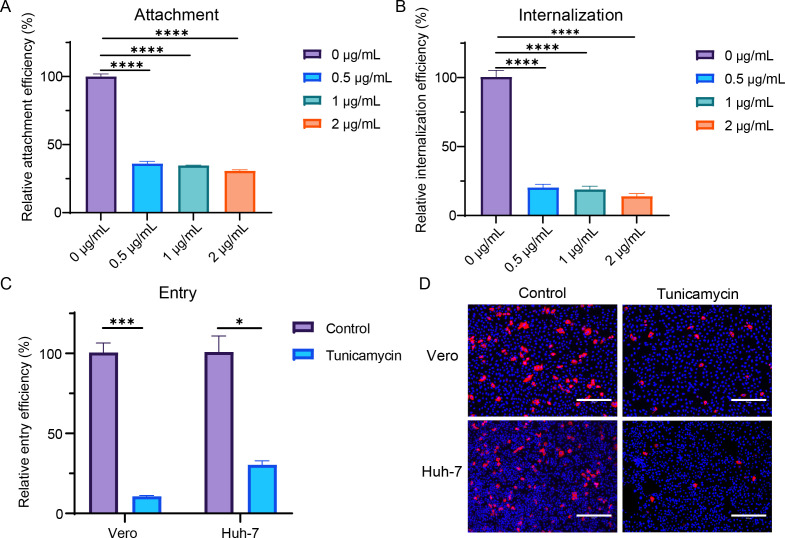
To further determine the effects of tunicamycin on different steps of viral infection, we assessed the attachment, internalization, and entry of SADS-CoV into cells pretreated with different concentrations of tunicamycin (Fig. 2). For viral attachment, SADS-CoV-infected Vero cells [multiplicity of infection (MOI) =10] were incubated on ice for 1 h and then washed with cold PBS three times, and the viral attachment on the cell surface was detected by RT-qPCR. For viral internalization, virus-attached cells were incubated at 37°C for another 1 h, and viral internalization was detected by RT-qPCR. For viral entry, Vero and Huh-7 cells were infected with SADS-CoV (MOI = 1) at 37°C for 6 h. The cells were then harvested, and RNA copies were detected using RT-qPCR. The results showed that viral attachment, internalization, and entry were inhibited in the tunicamycin-treated group, indicating that tunicamycin affected viral attachment and that N-linked glycosylation was likely associated with the SADS-CoV receptor.
Fig 2.
Tunicamycin inhibited viral attachment, internalization, and entry of SADS-CoV. (A) Vero cells were pretreated with tunicamycin for 24 h and then infected with SADS-CoV (MOI = 10) for 1 h on ice. Cells were then washed with cold PBS three times, and viral attachment on the cell surface was detected by RT-qPCR. (B) After viral attachment for 1 h on ice, cells were cultured at 37°C for an additional 1 h, and viral internalization was detected by RT-qPCR. For viral entry, Vero and Huh-7 cells were pretreated with tunicamycin for 24 h and then infected with SADS-CoV (MOI = 1) for 6 h. Cells were lysed, and virus detection was performed using qRT-PCR (C). Additionally, cells were fixed and stained with anti-SADS-CoV NP antibodies for analysis by IFA (D). All data are shown as means ± standard errors of the means (n = 3 biological replicates). Scale bars: 300 µm.
Common N-linked glycans were not involved in SADS-CoV infection
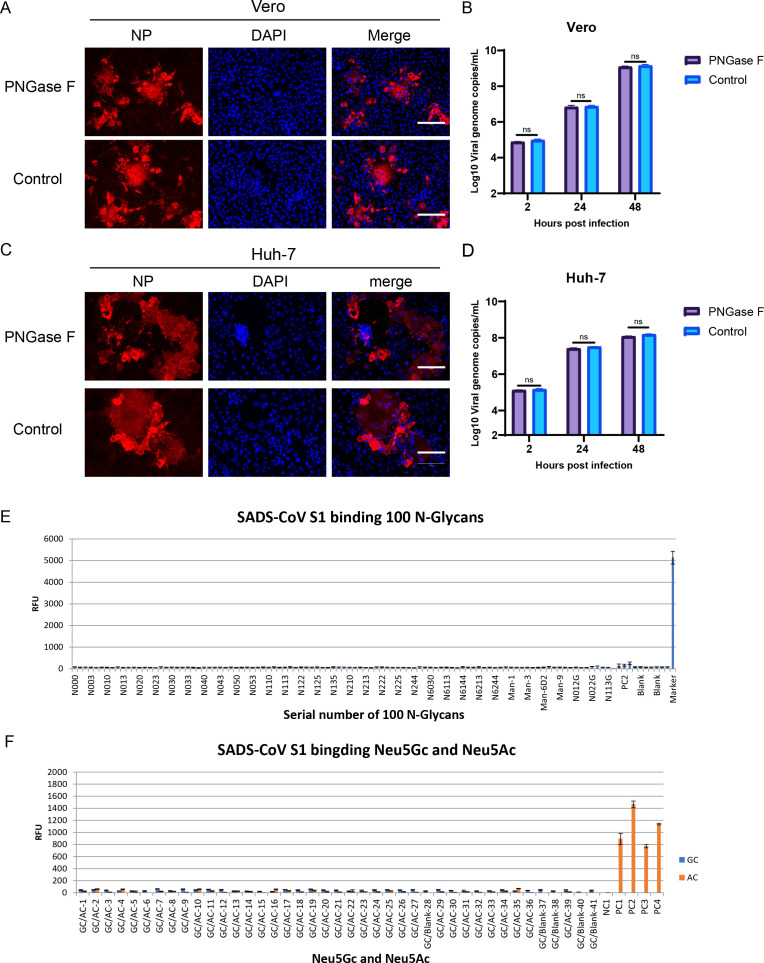
To better elucidate the roles of N-linked glycans in SADS-CoV infection, we first used PNGase F to remove N-linked oligosaccharides from cell surface glycoproteins (Fig. 3A through D). The results showed that viral replication did not significantly change the replication efficiency in the infected Huh-7 or Vero cells between the PNGase F treatment and mock groups. Next, we assessed whether the SADS-CoV S1 protein could bind N-linked glycans by evaluating the binding of the S1 protein with 100 common N-glycans using glycan microarrays. We found no binding between SADS-CoV S1 and the 100 N-glycans (Fig. 3E). These results implied that these N-linked glycans were not involved in SADS-CoV infection. Moreover, we also tested whether the SADS-CoV S1 protein could bind Neu5Gc (an IBV receptor) and Neu5Ac (BCoV and HCoV-OC43 receptor); all tests were negative, indicating that Neu5Gc and Neu5Ac were not associated with SADS-CoV entry (Fig. 3F). Taken together, these results demonstrated that the SADS-CoV receptor was likely an N-linked glycoprotein but not Neu5Gc or Neu5Ac.
Fig 3.
Effects of N-linked glycans on SADS-CoV infection. Vero and Huh-7 cells were pretreated with 2000 U PNGase F for 3 h and then infected with SADS-CoV (MOI = 0.1). Viral replication in Vero cells treated with PNGase F was detected by IFA (A) and qRT-PCR (B). Viral replication in Huh7 cells treated with PNGase F was detected by IFA (C) and qRT-PCR (D). (E) Binding between SADS-CoV S1 and 100 N-glycans was detected using glycan microarrays. (F) Binding of SADS-CoV S1 with Neu5Gc and Neu5Ac was detected using glycan microarrays. All data are shown as means ± standard errors of the means (n = 3 biological replicates). Scale bars: 125 µm.
Trypsin was important for SADS-CoV infection in Vero cells but was not essential in Huh-7 cells
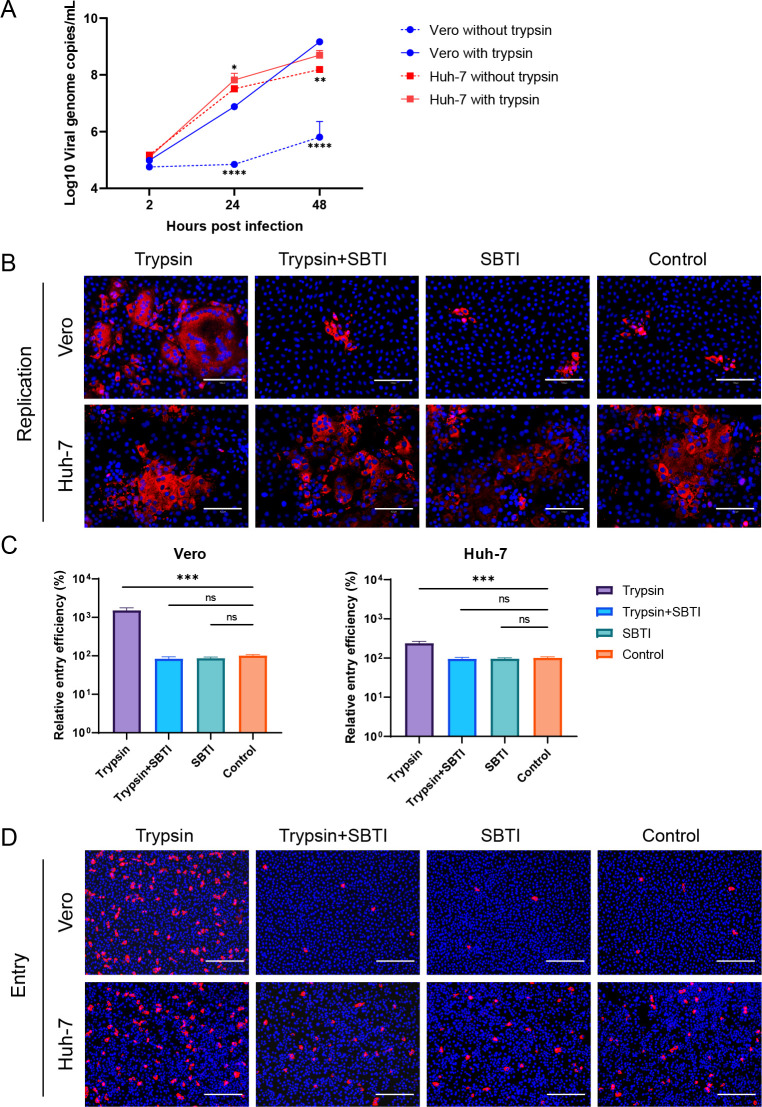
Exogenous trypsin cleavage has been reported to facilitate SADS-CoV infection (6, 8). To assess whether trypsin-mediated viral entry was essential for SADS-CoV infection, we tested viral replication dynamics in cells treated with 4 µg/mL trypsin using an MOI of 0.1 (Fig. 4A). The results showed that trypsin strongly enhanced infectivity in Vero cells, whereas its effect on Huh-7 cells was relatively limited. We then infected Vero and Huh-7 cells with SADS-CoV at an MOI of 0.1, treated the cells with 10 µg/mL of the trypsin inhibitor SBTI and/or 4 µg/mL trypsin, and tested viral replication at 24 hpi using IFA (Fig. 4B). Similar results were observed; that is, viral replication was strongly enhanced in trypsin-treated Vero cells but not in Huh-7 cells. To further explore the effects of trypsin on SADS-CoV entry, Vero and Huh-7 cells were infected with SADS-CoV at an MOI of 1 and then treated with trypsin or SBTI for 6 h. Viral RNA and NP expression in cells were detected (Fig. 4C and D), suggesting that viral entry increased significantly in Vero and Huh-7 cells following trypsin treatment, with stronger entry efficiency observed in Vero cells in the presence of trypsin.
Fig 4.
Effects of trypsin on SADS-CoV infection. (A) For the detection of viral replication dynamics, Vero and Huh-7 cells were infected with SADS-CoV (MOI = 0.1) and treated with or without 4 µg/mL trypsin. The supernatants were harvested at 0, 24, or 48 hpi for the assessment of viral replication dynamics by qRT-PCR. (B) Vero and Huh-7 cells were infected with SADS-CoV (MOI = 0.1) and treated with 4 µg/mL and/or 10 µg/mL TBSI. The cells were fixed at 24 hpi and stained with anti-SADS-CoV NP antibodies by IFA. For viral entry detection, Vero and Huh-7 cells were infected with SADS-CoV (MOI = 1) in the presence of 4 µg/mL trypsin and/or 10 µg/mL TBSI for 6 h. The cells were then lysed, and virus detection was performed using qRT-PCR (C). Alternatively, cells were fixed and stained with anti-SADS-CoV NP antibodies by IFA (D). All data are shown as means ± standard errors of the means (n = 3 biological replicates). Scale bars: 125 µm (B) and 300 µm (D).
Other host protease cleavage enhanced SADS-CoV entry
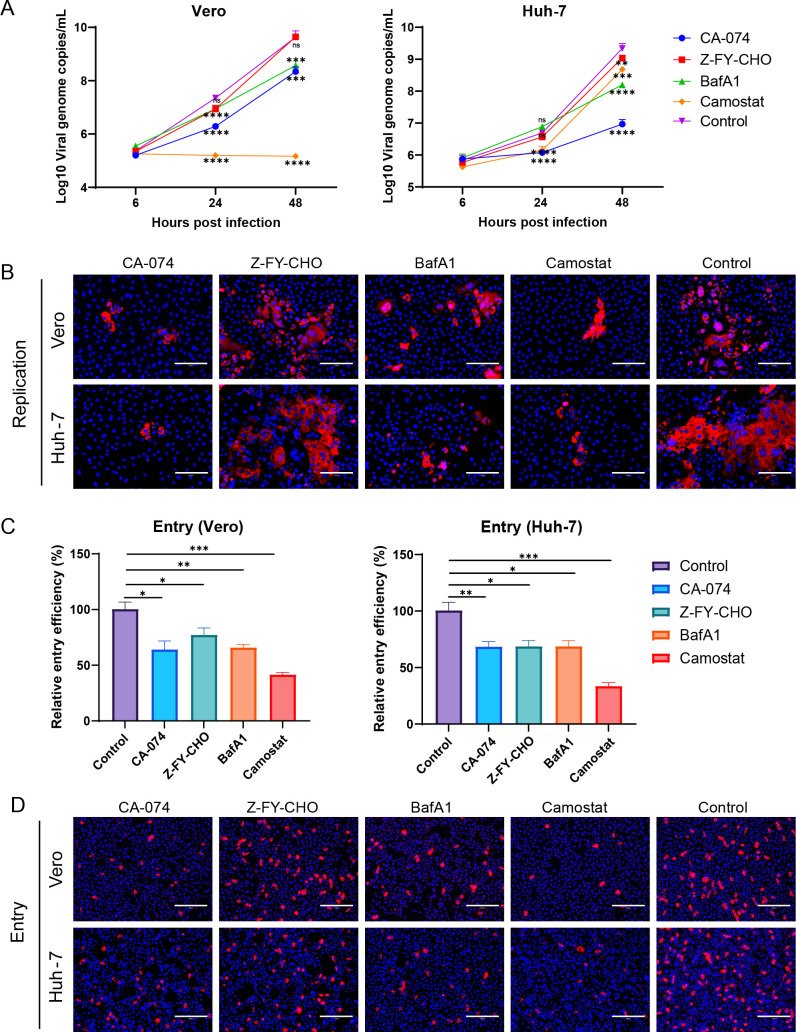
Proteolysis of coronavirus spikes triggers membrane fusion during viral entry. Cleavage of host proteases, including furin, elastase, type II transmembrane serine proteases, and cathepsins, promotes coronavirus entry. To further explore whether another host protease cleavage was involved in SADS-CoV infection, we pretreated Vero and Huh-7 cells with host protease inhibitors (50 µM of the cathepsin B inhibitor CA-074, 50 µM of the cathepsin L inhibitor Z-FY-CHO, 500 nM of the lysosomal acidification inhibitor BafA1, and 50 µM of the serine protease inhibitor camostat) for 3 h and then infected with SADS-CoV (MOI = 0.1). Viral replication was detected at 24 hpi by RT-qPCR and IFA (Fig. 5A and B). Notably, viral replication in Vero cells was significantly inhibited by treatment with Z-FY-CHO, CA-074, and camostat at 24 and 48 hpi, with the most obvious inhibition observed following camostat treatment. These three inhibitors also blocked Huh-7 infection, with CA-074 showing the most significant inhibition.
Fig 5.
Effects of other protease inhibitors on SADS-CoV infection. For the detection of viral replication dynamics, Vero and Huh-7 cells were pretreated with protease inhibitors (50 µM CA-074, 50 µM Z-FY-CHO, 500 nM BafA1, and 50 µM camostat) for 3 h and then infected with SADS-CoV (MOI = 0.1). The supernatants were harvested at 0, 24, or 48 hpi for the assessment of viral replication dynamics by qRT-PCR (A). Alternatively, cells were fixed at 24 hpi and stained with anti-SADS-CoV NP antibodies for detection by IFA (B). For viral entry detection, Vero and Huh-7 cells were pretreated with protease inhibitors for 3 h and then infected with SADS-CoV (MOI = 0.1) for 6 h. The cells were lysed, and virus detection was performed using qRT-PCR (C). Alternatively, cells were fixed and stained with anti-SADS-CoV NP antibodies for detection by IFA (D). All data are shown as means ± standard errors of the means (n = 3 biological replicates). Scale bars: 125 µm (B) and 300 µm (D).
To further explore the effects of these protease inhibitors on SADS-CoV entry, we pretreated Vero and Huh-7 cells with CA-074, Z-FY-CHO, BafA1, and camostat and then infected the cells with SADS-CoV (MOI = 1) for 6 h. Cells were harvested, and viral RNA and NP expression were detected (Fig. 5C and D). The results showed that viral entry decreased significantly following treatment with CA-074, Z-FY-CHO, BafA1, or camostat in Vero and Huh-7 cells. Furthermore, camostat-treated cells showed the most resistance to SADS-CoV entry.
Taken together, these data suggested that cathepsin B, cathepsin L, lysosomal acidification, and TMPRSSs played important roles in SADS-CoV entry.
DISCUSSION
Here, we evaluated the cell-entry mechanisms of SADS-CoV, a novel bat HKU2-related coronavirus that threatens the swine industry. We found that N-linked glycosylation of host cells played a crucial role in SADS-CoV entry and that Neu5Gc and Neu5Ac, which are receptors of other coronaviruses, were not functional receptors of this virus. Additionally, proteolytic cleavage proteases, including trypsin, TMPRSSs, cathepsin L, and cathepsin B, were found to be involved in viral entry. Our study suggested that the sophisticated cell-entry mechanisms of SADS-CoV may pose major challenges for prophylaxis and therapeutics against emerging diseases caused by these viruses.
Some coronaviruses, including the alphacoronaviruses TGEV and PEDV; the betacoronaviruses MERS-CoV, SARS-CoV-2, BCoV, HCoV-OC43, and HCoV-HKU1; and the gammacoronavirus IBV, recognize sugars as receptors or coreceptors (29, 30, 45 – 52). SADS-CoV has a wide cell tropism and utilizes common molecules, such as sugars, for entry (6 – 8). In this study, we demonstrated for the first time that the N-linked glycosylation inhibitor, tunicamycin, inhibited SADS-CoV infection by blocking viral binding to host cells. However, further PNGase F treatment of SADS-infected cells with PNGase F excluded the involvement of N-linked glycosylation. A glycan microarray assay with 100 common glycans, Neu5Gc and Neu5Ac, further confirmed that these glycans were not functional receptors of SADS-CoV. Thus, the SADS-CoV receptor is likely an N-linked glycoprotein.
Tunicamycin is an antibiotic produced by Streptomyces clavuligerus and Streptomyces lysosuperficus that can interfere with the glycosylation of N-linked glycoproteins and block cell cycle arrest at the G1 phase in human cells (53, 54). Previous studies have suggested that tunicamycin may have applications in the treatment of human colon and prostate cancer by inducing apoptosis in tumor cells (55). In this study, we found that pretreatment with tunicamycin blocked SADS-CoV entry into cells and inhibited PEDV infection, but the infection of SARS-CoV-2 was not affected, which suggested that such inhibition was not due to the cell growth state. These results indicated that tunicamycin could be used as a therapeutic agent in the management of virus-induced acute gastroenteritis in neonatal piglets.
Proteolytic activation of the spike protein by host cell proteases also plays a critical role in coronavirus cell and tissue tropism, host range, and pathogenesis (21, 34, 35). Trypsin is a prototype serine endopeptidase that prefers to cleave arginine (R) and lysine (K) residues (56). Trypsin is primarily a digestive enzyme, with active trypsin found in the small intestine, and has been extensively demonstrated to activate viral glycoprotein cleavage. Most PEDV strains are highly trypsin dependent in Vero cells; trypsin cleavage occurs only after receptor binding and not on free particles (38, 56). In this study, we found that trypsin strongly enhanced infectivity and entry into Vero cells, but its effect on Huh-7 cells was relatively limited, which is similar to the results of other studies (6, 8). Moreover, we characterized how exogenous trypsin facilitates viral entry, even in cells in which trypsin is not essential for replication, such as Huh-7 cells. TMPRSSs are type II transmembrane proteins, and membrane-bound trypsin-like serine proteases are widely expressed in the respiratory tract (21, 35). TMPRSSs are involved in the activation of many coronavirus spikes, including those of MERS-CoV, SARS-CoV-1, SARS-CoV-2, and HCoV-229E (41, 43, 57, 58). We also found that TMPRSSs were involved in SADS-CoV entry into Vero and Huh-7 cells, highlighting the importance of the proteolytic activation of trypsin and TMPRSSs.
Furin and furin-like proteases, which are proprotein convertases, cleave paired basic residues within R/K-(X)0,2,4,6-R/K (X: any amino acid) (59 – 61). Previous studies have reported that furin-induced cleavage of the SADS-CoV S protein is required for cell-cell fusion but not for pseudotyped virus entry (62). Cathepsins, as degradative enzymes, comprise a group of cysteine, serine, and aspartyl proteases and are typically found in endosomes and lysosomes. Cathepsin L is known to process SARS-CoV-1 and SARS-CoV-2 S as well as other coronaviruses, including MERS-CoV, HCoV-229E, and MHV-2 (37, 40, 41, 63). Cathepsin B has also been shown to be involved in the entry of other coronaviruses, such as type II feline coronavirus and MHV-2 (40, 64). In this study, we demonstrated that inhibitors of endogenous serine proteases, cathepsin B, cathepsin L, and lysosomal acidification affected SADS-CoV entry, suggesting that these factors serve as important triggers for SADS-CoV entry into cells. In addition, a recent report showed that SADS-CoV can also enter cells via the clathrin-, caveolae-, and micropinocytosis-mediated endocytosis pathways (65). Similar entry pathways have been reported for PEDV, which can cause diarrhea and death in piglets (66, 67). Taken together, these studies suggest that multiple pathways are likely to participate in viral entry into host cells based on the characteristics of the younger piglet intestinal microenvironment.
In summary, our results demonstrated that N-linked glycoproteins and several cellular proteases were involved in SADS-CoV entry. Importantly, many genetically diverse SADSr-CoVs circulate among Rhinolophus spp. bats in China and elsewhere, highlighting the potential risk of cross-species infections. This study provides evidence for widely employed mechanisms of SADS-CoV entry and illuminates multiple potential prophylactic and therapeutic strategies for these viruses. Our findings suggest that N-linked glycoproteins and host proteases play a role in virus entry, but the mechanism needs further exploration.
MATERIALS AND METHODS
Cells lines and virus
Vero cells (American Type Culture Collection, Manassas, VA, USA; cat. no. CRL-81) and Huh-7 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Life Technologies) and 1% antibiotic-antimycotic (Invitrogen). SADS-CoV was propagated and titrated in Vero cell lines with a maintenance medium containing DMEM, 2% tryptose phosphate broth (Sigma, St. Louis, MO, USA), 5 µg/mL trypsin (Gibco, Carlsbad, CA, USA), and 1% antibiotic-antimycotic (Invitrogen). Viral titration was performed using 10-fold serial dilutions of Vero cells. The 50% tissue culture infective dose was expressed as the reciprocal of the highest dilution showing the cytopathic effect using the Reed-Muench method.
Hemagglutination assays
Each well of a microtiter V plate contained 50 µL PBS; 25 µL of increasing concentrations of SADS-CoV, SADS-CoV S1 protein, PEDV, TGEV, or H1N1; and 25 µL of 4% (vol/vol) mouse erythrocyte suspension. All dilutions were prepared using PBS. Agglutination was allowed to proceed for 60 min at 20°C, and the minimum concentration of these viruses or S1 proteins required for agglutination was determined via serial dilution.
SADS-CoV S1 expression and purification
Codon-optimized SADS-CoV S1 protein was inserted into the pCAGGS vector with an N-terminal S-tag. The constructed plasmids were transiently transfected into HEK293F cells. The supernatant collected for protein purification was purified using an S-tag resin, and the purity and yield were tested using an anti-S-tag monoclonal antibody (generated in-house).
Inhibitors
The N-linked glycosylation inhibitor tunicamycin (T7765-1) was obtained from Sigma-Aldrich (St. Louis, MO, USA). The enzyme for removing N-linked oligosaccharides from glycoproteins, PNGase F, was obtained from New England Biolabs. The trypsin inhibitor SBTI (A51497) was obtained from OKA. The cathepsin B inhibitor CA-074 (HY-103350), cathepsin L inhibitor Z-FY-CHO (HY-128140), serine protease inhibitor camostat mesylate (HY-13512), and lysosomal acidification inhibitor BafA1 (HY-100558) were obtained from MedChem Express.
Virus attachment, internalization, and entry
Vero and Huh-7 cells were seeded in 24-well plates and washed with tunicamycin for 24 h or inhibitors for 3 h. The cells were washed twice with cold PBS and infected with SADS-CoV at an MOI of 10 for 1 h on ice. The supernatants were then removed, and the infected cells were washed three times to eliminate the unbound virus. Cells were lysed with TRIzol reagent (Invitrogen) for viral RNA extraction to determine viral attachment to the cell surface. For virus internalization, cells were cultured with virus (MOI = 10) for 1 h on ice and further cultured at 37°C for another 1 h. The cells were then treated with 1 mg/mL pronase to remove uninternalized virus. After three washes, the cells were harvested for RNA extraction. For viral entry, cells were infected with virus (MOI = 1) for 6 h at 37°C, washed three times with PBS, and lysed with TRIzol reagent for viral RNA extraction or fixed with 4% paraformaldehyde to determine viral entry into cells.
Glycan microarray analysis
In total, 100 N-glycans, sialic acid Neu5Gc- and Neu5Ac-based glycan arrays from Kerixin Biological Technology Co., Ltd. (Nanjing, China), were used to assess the binding of SADS-CoV S1 proteins. The glycan structures in the array are shown in the product manual of Kerixin Biological Technology Co., Ltd. The measurements and data analyses were performed by Kerixin Biological Technology Co., Ltd. Briefly, the SADS-CoV S1 protein was labeled with biotin and diluted to 8 or 1 g/mL with incubation buffer (Kerixin Biological Technology Co., Ltd.). Each labeled protein (200 µL) was added to a subarray and incubated for 1 h in the dark. After removing unbound proteins, the array was washed twice with TBST. Then, 200 µL fluorescein isothiocyanate-conjugated streptavidin was added to a subarray and incubated for 1 h in the dark. After washing and drying, the dried array was scanned using a microarray scanner (LuxScan 10 K/A; CapitalBio Corporation, Beijing, China) and analyzed using LuxScan 3.0.
RNA extraction and RT-qPCR
Viral RNA was extracted from supernatants with a QIAamp 96 Virus QIAcube HT kit (Qiagen, Valencia, CA, USA). RNA was used as a template for RT-qPCR to assess the SADS-CoV-specific genome by targeting the RdRp gene using a HiSxript II One step RT-qPCR SYBR Green Kit (Vazyme), as previously described (8). The average values from duplicates of each gene were used to calculate the viral genomic copies; 10 µL qPCR reaction mixture contained 5 µL 2× One-Step SYBR Green mix, 0.5 µL One-Step SYBR Green Enzyme mix, 0.2 µL (10 µM) of each primer, and 2 µL RNA. Amplification was performed as follows: 50°C for 3 min; 95°C for 30 s; 40 cycles at 95°C for 10 s and 60°C for 30 s; and a default melting curve step using a Bio-Rad Real-Time PCR machine.
IFA
The infected cells were fixed with 4% paraformaldehyde for 24 h, and an indirect IFA was performed to detect the SADS-CoV antigen, as previously described (8). The cells were permeabilized with PBS/0.01% Triton X-100 for 10 min and then blocked with 5% bovine serum albumin at room temperature for 1 h, followed by overnight incubation at 4°C with rabbit anti-SADS-CoV N protein polyclonal antibody (1:500, made in-house), rabbit anti-PEDV N protein polyclonal antibody (1:200, made in-house), or rabbit anti-SARS-CoV-2 N protein polyclonal antibody (1:1,000, made in-house). The slides were then incubated with Cy3-conjugated goat anti-rabbit IgG (1:200; Abcam, Cambridge, MA, USA; cat. no. ab6939). After washing with PBS, slides were stained with DAPI (1:100; Beyotime). Images were captured using a fluorescence microscope.
Statistical analysis
All experiments were independently repeated at least three times. Statistical analysis was performed using PRISM 8.0.2 for Windows (GraphPad). Two-tailed Student’s t-tests were performed to determine significant differences between the two experimental groups.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31830096 to Z.L.S.) and National Key R&D Program of China (2021YFC2300901 to P.Z.). We thank Juan Min and Ding Gao from Center for Instrumental Analysis and Metrology of the Wuhan Institute of Virology for their technical support.
Conceptualization, Z.L.S.; Methodology, Y.C., J.N.Z., P.Z., and R.J.T.; Investigation, Y.C., X.L., L.J.Y., Y.L., Y.L.Y., M.Q.L., H.F.L., and Y.T.H.; Resources, Y.C., X.L., and B.H.; Writing-Original Draft, Y.C.; Writing-Review & Editing, Z.L.S.; Funding Acquisition, Z.L.S. and P.Z.
The authors declare no conflict of interest.
Contributor Information
Zheng-Li Shi, Email: zlshi@wh.iov.cn.
Tom Gallagher, Loyola University Chicago, Maywood, Illinois, USA .
REFERENCES
- 1. Gong L, Li J, Zhou Q, Xu Z, Chen L, Zhang Y, Xue C, Wen Z, Cao Y. 2017. A new bat-HKU2-like coronavirus in swine. Emerg Infect Dis 23:1607–1609. doi: 10.3201/eid2309.170915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pan Y, Tian X, Qin P, Wang B, Zhao P, Yang YL, Wang L, Wang D, Song Y, Zhang X, Huang YW. 2017. Discovery of a novel swine enteric alphacoronavirus (SeAcoV) in Southern China. Vet Microbiol 211:15–21. doi: 10.1016/j.vetmic.2017.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fu X, Fang B, Liu Y, Cai M, Jun J, Ma J, Bu D, Wang L, Zhou P, Wang H, Zhang G. 2018. Newly emerged porcine enteric alphacoronavirus in Southern China: identification, origin and evolutionary history analysis. Infect Genet Evol 62:179–187. doi: 10.1016/j.meegid.2018.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhou P, Fan H, Lan T, Yang X-L, Shi W-F, Zhang W, Zhu Y, Zhang Y-W, Xie Q-M, Mani S, Zheng X-S, Li B, Li J-M, Guo H, Pei G-Q, An X-P, Chen J-W, Zhou L, Mai K-J, Wu Z-X, Li D, Anderson DE, Zhang L-B, Li S-Y, Mi Z-Q, He T-T, Cong F, Guo P-J, Huang R, Luo Y, Liu X-L, Chen J, Huang Y, Sun Q, Zhang X-L-L, Wang Y-Y, Xing S-Z, Chen Y-S, Sun Y, Li J, Daszak P, Wang L-F, Shi Z-L, Tong Y-G, Ma J-Y. 2018. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556:255–258. doi: 10.1038/s41586-018-0010-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sun Y, Cheng J, Luo Y, Yan XL, Wu ZX, He LL, Tan YR, Zhou ZH, Li QN, Zhou L, Wu RT, Lan T, Ma JY. 2019. Attenuation of a virulent swine acute diarrhea syndrome coronavirus strain via cell culture passage. Virology 538:61–70. doi: 10.1016/j.virol.2019.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang YL, Qin P, Wang B, Liu Y, Xu GH, Peng L, Zhou J, Zhu SJ, Huang YW. 2019. Broad cross-species infection of cultured cells by bat HKU2-related swine acute diarrhea syndrome coronavirus and identification of its replication in murine dendritic cells in vivo highlight its potential for diverse interspecies transmission. J Virol 93:e01448-19. doi: 10.1128/JVI.01448-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edwards CE, Yount BL, Graham RL, Leist SR, Hou YJ, Dinnon KH III, Sims AC, Swanstrom J, Gully K, Scobey TD, Cooley MR, Currie CG, Randell SH, Baric RS. 2020. Swine acute diarrhea syndrome coronavirus replication in primary human cells reveals potential susceptibility to infection. Proc Natl Acad Sci U S A 117:26915–26925. doi: 10.1073/pnas.2001046117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Luo Y, Chen Y, Geng R, Li B, Chen J, Zhao K, Zheng X-S, Zhang W, Zhou P, Yang X-L, Shi Z-L. 2021. Broad cell tropism of SADS-CoV in vitro implies its potential cross-species infection risk. Virol Sin 36:559–563. doi: 10.1007/s12250-020-00321-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mei XQ, Qin P, Yang YL, Liao M, Liang QZ, Zhao Z, Shi FS, Wang B, Huang YW. 2022. First evidence that an emerging mammalian alphacoronavirus is able to infect an avian species. Transbounding Emerging Dis 69. doi: 10.1111/tbed.14535 [DOI] [PubMed] [Google Scholar]
- 10. Chen Y, Jiang R-D, Wang Q, Luo Y, Liu M-Q, Zhu Y, Liu X, He Y-T, Zhou P, Yang X-L, Shi Z-L, Subbarao K. 2022. Lethal swine acute diarrhea syndrome coronavirus infection in suckling mice. J Virol 96. doi: 10.1128/jvi.00065-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Duan Y, Yuan C, Suo X, Li Y, Shi L, Cao L, Kong X, Zhang Y, Zheng H, Wang Q. 2023. Bat-origin swine acute diarrhea syndrome coronavirus is lethal to neonatal mice. J Virol 97:e0019023. doi: 10.1128/jvi.00190-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Y, Jiang R-D, Wang Q, Luo Y, Liu M-Q, Zhu Y, Liu X, He Y-T, Zhou P, Yang X-L, Shi Z-L, Subbarao K. 2022. Lethal swine acute diarrhea syndrome coronavirus infection in suckling mice. J Virol 96. doi: 10.1128/jvi.00065-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fan Y, Zhao K, Shi ZL, Zhou P. 2019. Bat coronaviruses in China. Viruses 11:210. doi: 10.3390/v11030210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lau SKP, Woo PCY, Li KSM, Huang Y, Wang M, Lam CSF, Xu H, Guo R, Chan K-H, Zheng B-J, Yuen K-Y. 2007. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology 367:428–439. doi: 10.1016/j.virol.2007.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Siddell S, Wege H, Ter Meulen V. 1983. The biology of coronaviruses. J Gen Virol 64 (Pt 4):761–776. doi: 10.1099/0022-1317-64-4-761 [DOI] [PubMed] [Google Scholar]
- 16. Cui J, Li F, Shi ZL. 2019. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17:181–192. doi: 10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tyrrell DAJ, Myint SH. 1996. Coronaviruses. In Baron S (ed), Medical microbiology. The University of Texas medical branch at Galveston, Galveston (TX). [PubMed] [Google Scholar]
- 18. Li F. 2012. Evidence for a common evolutionary origin of coronavirus spike protein receptor-binding subunits. J Virol 86:2856–2858. doi: 10.1128/JVI.06882-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harrison SC. 2008. Viral membrane fusion. Nat Struct Mol Biol 15:690–698. doi: 10.1038/nsmb.1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kirchdoerfer RN, Cottrell CA, Wang N, Pallesen J, Yassine HM, Turner HL, Corbett KS, Graham BS, McLellan JS, Ward AB. 2016. Pre-fusion structure of a human coronavirus spike protein. Nature 531:118–121. doi: 10.1038/nature17200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li F. 2016. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol 3:237–261. doi: 10.1146/annurev-virology-110615-042301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li WH, Moore MJ, Vasilieva N, Sui JH, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454. doi: 10.1038/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen L, Lin YL, Peng GQ, Li F. 2012. Structural basis for multifunctional roles of mammalian aminopeptidase N. Proc Natl Acad Sci U S A 109:17966–17971. doi: 10.1073/pnas.1210123109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raj VS, Mou H, Smits SL, Dekkers DHW, Müller MA, Dijkman R, Muth D, Demmers JAA, Zaki A, Fouchier RAM, Thiel V, Drosten C, Rottier PJM, Osterhaus ADME, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254. doi: 10.1038/nature12005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tan K, Zelus BD, Meijers R, Liu J, Bergelson JM, Duke N, Zhang R, Joachimiak A, Holmes KV, Wang J. 2002. Crystal structure of murine sCEACAM1a 1,4: a coronavirus receptor in the CEA family. EMBO J 21:2076–2086. doi: 10.1093/emboj/21.9.2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 588:270–273. doi: 10.1038/s41586-020-2951-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu K, Li W, Peng G, Li F. 2009. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc Natl Acad Sci U S A 106:19970–19974. doi: 10.1073/pnas.0908837106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li W, Hulswit RJG, Kenney SP, Widjaja I, Jung K, Alhamo MA, van Dieren B, van Kuppeveld FJM, Saif LJ, Bosch B-J. 2018. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc Natl Acad Sci U S A 115:E5135–E5143. doi: 10.1073/pnas.1802879115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang X, Dong W, Milewska A, Golda A, Qi Y, Zhu QK, Marasco WA, Baric RS, Sims AC, Pyrc K, Li W, Sui J. 2015. Human coronavirus HKU1 spike protein uses O-acetylated Sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J Virol 89:7202–7213. doi: 10.1128/JVI.00854-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hulswit RJG, Lang Y, Bakkers MJG, Li W, Li Z, Schouten A, Ophorst B, van Kuppeveld FJM, Boons G-J, Bosch B-J, Huizinga EG, de Groot RJ. 2019. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc Natl Acad Sci U S A 116:2681–2690. doi: 10.1073/pnas.1809667116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Winter C, Schwegmann-Weßels C, Cavanagh D, Neumann U, Herrler G. 2006. Sialic acid is a receptor determinant for infection of cells by avian infectious bronchitis virus. J Gen Virol 87:1209–1216. doi: 10.1099/vir.0.81651-0 [DOI] [PubMed] [Google Scholar]
- 32. Collins AR. 1993. HLA class I antigen serves as a receptor for human coronavirus OC43. Immunol Invest 22:95–103. doi: 10.3109/08820139309063393 [DOI] [PubMed] [Google Scholar]
- 33. Szczepanski A, Owczarek K, Bzowska M, Gula K, Drebot I, Ochman M, Maksym B, Rajfur Z, Mitchell JA, Pyrc K. 2019. Canine respiratory coronavirus, bovine coronavirus, and human coronavirus OC43: receptors and attachment factors. Viruses 11:328. doi: 10.3390/v11040328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Klenk HD, Garten W. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol 2:39–43. doi: 10.1016/0966-842x(94)90123-6 [DOI] [PubMed] [Google Scholar]
- 35. Millet JK, Whittaker GR. 2015. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res 202:120–134. doi: 10.1016/j.virusres.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peng G, Sun D, Rajashankar KR, Qian Z, Holmes KV, Li F. 2011. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc Natl Acad Sci U S A 108:10696–10701. doi: 10.1073/pnas.1104306108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu C, Ma Y, Yang Y, Zheng Y, Shang J, Zhou Y, Jiang S, Du L, Li J, Li F. 2016. Cell entry of porcine epidemic diarrhea coronavirus is activated by lysosomal proteases. J Biol Chem 291:24779–24786. doi: 10.1074/jbc.M116.740746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Park J-E, Cruz DJM, Shin H-J. 2011. Receptor-bound porcine epidemic diarrhea virus spike protein cleaved by trypsin induces membrane fusion. Arch Virol 156:1749–1756. doi: 10.1007/s00705-011-1044-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang YL, Meng F, Qin P, Herrler G, Huang YW, Tang YD. 2020. Trypsin promotes porcine deltacoronavirus mediating cell-to-cell fusion in a cell type-dependent manner. Emerg Microbes Infect 9:457–468. doi: 10.1080/22221751.2020.1730245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qiu Z, Hingley ST, Simmons G, Yu C, Das Sarma J, Bates P, Weiss SR. 2006. Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry. J Virol 80:5768–5776. doi: 10.1128/JVI.00442-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kawase M, Shirato K, Matsuyama S, Taguchi F. 2009. Protease-mediated entry via the endosome of human coronavirus 229E. J Virol 83:712–721. doi: 10.1128/JVI.01933-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. 2020. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A 117:11727–11734. doi: 10.1073/pnas.2003138117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280. doi: 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Q, Luo Y, Shang W, Shi Z, Xiao G, Zhang L. 2021. Comprehensive interactome analysis of the spike protein of swine acute diarrhea syndrome coronavirus. Biosaf Health 3:156–163. doi: 10.1016/j.bsheal.2021.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schultze B, Cavanagh D, Herrler G. 1992. Neuraminidase treatment of avian infectious bronchitis coronavirus reveals a hemagglutinating activity that is dependent on sialic acid-containing receptors on erythrocytes. Virology 189:792–794. doi: 10.1016/0042-6822(92)90608-r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schwegmann-Wessels C, Herrler G. 2006. Sialic acids as receptor determinants for coronaviruses. Glycoconj J 23:51–58. doi: 10.1007/s10719-006-5437-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schultze B, Gross HJ, Brossmer R, Herrler G. 1991. The s-protein of bovine coronavirus is a hemagglutinin recognizing 9-O-acetylated sialic-acid as a receptor determinant. J Virol 65:6232–6237. doi: 10.1128/JVI.65.11.6232-6237.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhou HX, Chen YZ, Zhang SY, Niu PH, Qin K, Jia WX, Huang BY, Zhang SY, Lan J, Zhang LQ, Tan WJ, Wang XQ. 2019. Structural definition of a neutralization epitope on the N-terminal domain of MERS-CoV spike glycoprotein. Nat Commun 10:3068. doi: 10.1038/s41467-019-10897-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ströh LJ, Stehle T. 2014. Glycan engagement by viruses: receptor switches and specificity. Annu Rev Virol 1:285–306. doi: 10.1146/annurev-virology-031413-085417 [DOI] [PubMed] [Google Scholar]
- 50. Li W, Hulswit RJG, Widjaja I, Raj VS, McBride R, Peng W, Widagdo W, Tortorici MA, van Dieren B, Lang Y, van Lent JWM, Paulson JC, de Haan CAM, de Groot RJ, van Kuppeveld FJM, Haagmans BL, Bosch B-J. 2017. Identification of sialic acid-binding function for the middle east respiratory syndrome coronavirus spike glycoprotein. Proc Natl Acad Sci U S A 114:E8508–E8517. doi: 10.1073/pnas.1712592114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tortorici MA, Walls AC, Lang Y, Wang C, Li Z, Koerhuis D, Boons G-J, Bosch B-J, Rey FA, de Groot RJ, Veesler D. 2019. Structural basis for human coronavirus attachment to sialic acid receptors. Nat Struct Mol Biol 26:481–489. doi: 10.1038/s41594-019-0233-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Baker AN, Richards S-J, Guy CS, Congdon TR, Hasan M, Zwetsloot AJ, Gallo A, Lewandowski JR, Stansfeld PJ, Straube A, Walker M, Chessa S, Pergolizzi G, Dedola S, Field RA, Gibson MI. 2020. The SARS-CoV-2 spike protein binds sialic acids and enables rapid detection in a lateral flow point of care diagnostic device. ACS Cent. Sci 6:2046–2052. doi: 10.1021/acscentsci.0c00855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Prescher JA, Bertozzi CR. 2006. Chemical technologies for probing glycans. Cell 126:851–854. doi: 10.1016/j.cell.2006.08.017 [DOI] [PubMed] [Google Scholar]
- 54. Yang L, Yu S, Yang Y, Wu H, Zhang X, Lei Y, Lei Z. 2021. Berberine improves liver injury induced glucose and lipid metabolic disorders via alleviating ER stress of hepatocytes and modulating gut microbiota in mice. Bioorg Med Chem 55:116598. doi: 10.1016/j.bmc.2021.116598 [DOI] [PubMed] [Google Scholar]
- 55. Hiss DC, Gabriels GA, Folb PI. 2007. Combination of tunicamycin with anticancer drugs synergistically enhances their toxicity in multidrug-resistant human ovarian cystadenocarcinoma cells. Cancer Cell Int 7:5. doi: 10.1186/1475-2867-7-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li W, van Kuppeveld FJM, He Q, Rottier PJM, Bosch B-J. 2016. Cellular entry of the porcine epidemic diarrhea virus. Virus Res 226:117–127. doi: 10.1016/j.virusres.2016.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchi F. 2010. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol 84:12658–12664. doi: 10.1128/JVI.01542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shirato K, Kawase M, Matsuyama S. 2013. Middle east respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J Virol 87:12552–12561. doi: 10.1128/JVI.01890-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. de Haan CAM, Li Z, te Lintelo E, Bosch BJ, Haijema BJ, Rottier PJM. 2005. Murine coronavirus with an extended host range uses heparan sulfate as an entry receptor. J Virol 79:14451–14456. doi: 10.1128/JVI.79.22.14451-14456.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Belouzard S, Chu VC, Whittaker GR. 2009. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci U S A 106:5871–5876. doi: 10.1073/pnas.0809524106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Licitra BN, Millet JK, Regan AD, Hamilton BS, Rinaldi VD, Duhamel GE, Whittaker GR. 2013. Mutation in spike protein cleavage site and pathogenesis of feline coronavirus. Emerg Infect Dis 19:1066–1073. doi: 10.3201/eid1907.121094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kim J, Yoon J, Park JE. 2022. Furin cleavage is required for swine acute diarrhea syndrome coronavirus spike protein-mediated cell - cell fusion. Emerg Microbes Infect 11:2176–2183. doi: 10.1080/22221751.2022.2114850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Johnson MC, Lyddon TD, Suarez R, Salcedo B, LePique M, Graham M, Ricana C, Robinson C, Ritter DG. 2020. Optimized pseudotyping conditions for the SARS-CoV-2 spike glycoprotein. J Virol 94:e01062-20. doi: 10.1128/JVI.01062-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Regan AD, Shraybman R, Cohen RD, Whittaker GR. 2008. Differential role for low pH and cathepsin-mediated cleavage of the viral spike protein during entry of serotype II feline coronaviruses. Vet Microbiol 132:235–248. doi: 10.1016/j.vetmic.2008.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chen XN, Liang YF, Weng ZJ, Quan WP, Hu C, Peng YZ, Sun YS, Gao Q, Huang Z, Zhang GH, Gong L. 2023. Porcine enteric alphacoronavirus entry through multiple pathways (caveolae, clathrin, and macropinocytosis) requires Rab GTPases for endosomal transport. J Virol 97:e0021023. doi: 10.1128/jvi.00210-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Park J-E, Cruz DJM, Shin H-J. 2014. Clathrin- and serine proteases-dependent uptake of porcine epidemic diarrhea virus into vero cells. Virus Res 191:21–29. doi: 10.1016/j.virusres.2014.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wei X, She G, Wu T, Xue C, Cao Y. 2020. PEDV enters cells through clathrin-, caveolae-, and lipid raft-mediated endocytosis and traffics via the endo-/lysosome pathway. Vet Res 51:10. doi: 10.1186/s13567-020-0739-7 [DOI] [PMC free article] [PubMed] [Google Scholar]