ABSTRACT
Marek’s disease virus (MDV) is a lymphotropic alphaherpesvirus that readily infects chickens and some other poultry, triggering complex disease phenotypes, including paralysis, immunosuppression, and lymphoma leading to death. MDV infection is partially controlled by extensive global vaccination programs but nevertheless, it imposes a significant financial burden on commercial poultry farming. Following infection, the MDV genome integrates into host telomeres, which is associated with the virus entering a latent state. The mechanism of integration remains poorly understood but it is contemporaneous with cellular transformation and lymphoma formation and therefore requires investigation. Here we have developed droplet digital PCR assays to quantify different regions of the MDV genome. We have also used long-range PCR and single telomere amplification to establish the organization and relative orientation of MDV genome regions in integrated MDV (iMDV). These analyses show that following integration, the MDV genome is oriented with the unique short region (US) internal to the unique long region (UL) and that an iMDV-associated telomere forms at the variable repeat array (mTMR) in a terminal a-like sequence. The data also reveal a very wide range of MDV copy numbers in cell lines, including unexpectedly, additional copies of specific MDV genome regions.
IMPORTANCE
Marek’s disease virus (MDV) is a ubiquitous chicken pathogen that inflicts a large economic burden on the poultry industry, despite worldwide vaccination programs. MDV is only partially controlled by available vaccines, and the virus retains the ability to replicate and spread between vaccinated birds. Following an initial infection, MDV enters a latent state and integrates into host telomeres and this may be a prerequisite for malignant transformation, which is usually fatal. To understand the mechanism that underlies the dynamic relationship between integrated-latent and reactivated MDV, we have characterized integrated MDV (iMDV) genomes and their associated telomeres. This revealed a single orientation among iMDV genomes and the loss of some terminal sequences that is consistent with integration by homology-directed recombination and excision via a telomere-loop-mediated process.
KEYWORDS: Marek's disease virus, chicken, telomere, latency, alphaherpesvirus, integration, droplet digital PCR
INTRODUCTION
Marek’s disease virus (MDV) is a widespread, lymphotropic alphaherpesvirus that infects chickens and is a large economic burden to the poultry industry (1, 2). The often complex symptoms of Marek’s disease (MD) include paralysis and immunosuppression (3 – 5). Current vaccines do not provide complete protection; therefore, MDV can replicate and spread between vaccinated birds (2). Prevalent circulating strains of MDV are reported to have become more virulent following widespread vaccination programs against MDV (2, 6). Primary infection by MDV is through inhalation of dander shed by MDV-infected birds (7) and early viral replication occurs predominantly, although not exclusively, in B lymphocytes. Infection strongly regulates gene expression in B cells and decreases the rate of proliferation (8). Linear concatemers of MDV are generated by rolling circle replication and replication peaks 3–7 days post-infection. Unusually, the MDV genome integrates into telomeres of latently infected CD4+ T cells that can undergo cellular transformation prior to lymphoma development. The temporal overlap between integration and transformation has led to the suggestion that integration and latency may be prerequisites for lymphomagenesis (9, 10). Integrated MDV is found in almost all MD lymphomas and where integration is inhibited, lymphoma development was significantly reduced (11). Several viral transcripts and gene products have been implicated in MD pathogenesis and lymphomagenesis. Deletion of meq, the major oncogene of MDV through its interactions with p53 (12), has very little effect on viral replication while completely disrupting tumorigenesis (13 – 15). Similarly, splice variants of phosphoprotein 38 at different stages in the MDV life cycle have been shown to play a role in tumor formation (16). vTR, a viral homolog of telomerase RNA, is also crucial for lymphomagenesis (17, 18).
The MDV genome structure is typical of alphaherpesviruses with a long unique region (UL¸ approximately 115 kb in length) and a short unique region (US, approximately 12 kb), each flanked by inverted repeats (RL, 15 kb and RS, 12 kb) (Fig. 1A). Between the repeats (RL and RS) is an “a-like” region that contains two telomere-like repeat arrays: a shorter array containing exactly six (TTAGGG) repeats (sTMR) and a longer array of variable length (mTMR) that contains (TTAGGG) repeats interspersed with degenerate repeats such as (TGAGGG). The a-like region also contains packaging motifs (PAC1 and PAC2) as well as two copies of DR1, a short sequence downstream of the cleavage site (11, 19 – 21). The mechanism of viral genome integration has not been fully resolved, but the presence of telomere (TTAGGG) repeat arrays in the MDV genome, in particular mTMR, has been shown to be essential for efficient integration, genome maintenance, and reactivation in vivo and in vitro (11, 22 – 25).
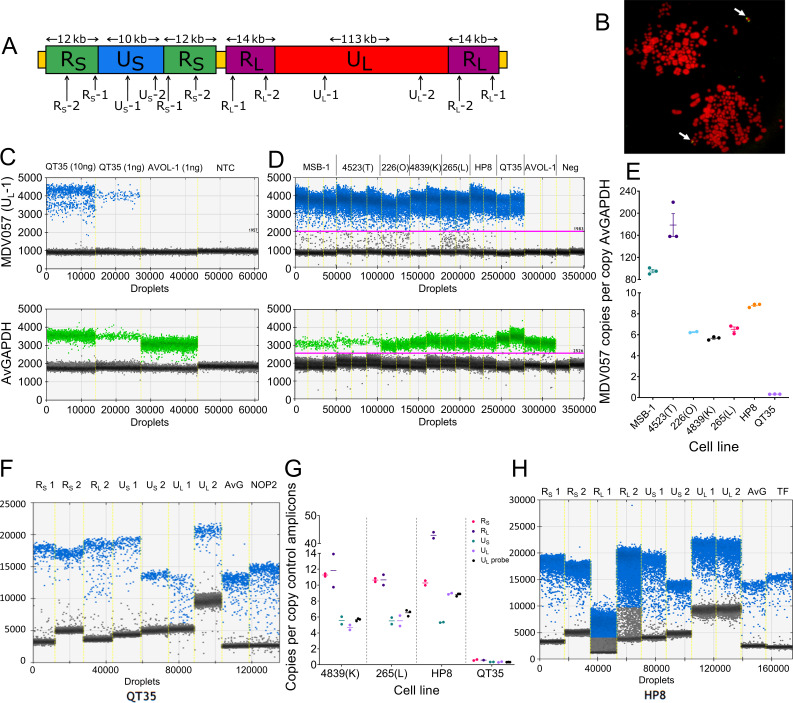
Fig 1.
Assessing MDV copy number and genome organization using droplet digital PCR (ddPCR) assays. (A) Diagram of MDV genome showing the relative positions of amplicons used in ddPCR assays. (B) Fluorescent in situ hybridization image showing that the QT35 cell line contains a single copy of the MDV genome integrated into a terminal location of a micro-chromosome. The arrows indicate the location of the integrated MDV genome in two QT35 metaphase cells. (C) One-dimensional ddPCR plots of duplexed hydrolysis probe assay for MDV057 (upper panel, blue) and AvGAPDH (lower panel, green). Each point of the graph represents an individual droplet generated during the ddPCR setup. The fluorescence intensity within each droplet (MDV057 blue droplets; AvGAPDH green droplets) is shown on the y-axis. Gray droplets show no amplification of either target. Each plot shows four ddPCR reactions with the following DNA input: 10 ng QT35 quail cell line DNA, 1 ng QT35 quail cell line DNA, 1 ng AVOL-1 MDV-negative cell line DNA, and water (no template control, NTC). (D) Replicates of the MDV057-AvGAPDH duplex ddPCR assay for seven chicken cell lines and one quail cell line (QT35). (E) Graph showing MDV057 copy number per cell estimated by ddPCR against AvGAPDH reference amplicon. Error bars are mean ± SEM. (F) One-dimensional ddPCR plots of EvaGreen assays on QT35 DNA. Assays for several amplicons across the MDV genome and the AvGAPDH and NOP2 reference amplicons are shown. (G) Graph showing copy number of each amplicon in four MDV-positive cell lines. The copy number was estimated by ddPCR absolute quantification against two reference amplicons (AvGAPDH and TF for chicken; AvGAPDH and NOP2 for quail). The means shown are the average copy number of both assays in each genome region relative to the average of two control amplicons. Results of UL hydrolysis probe ddPCR are shown (black) for comparison between methods. (H) One-dimensional ddPCR plots of EvaGreen assays using DNA from HP8 indicate a complex organization of the MDV genome in this cell line.
The genomes of chickens, and other birds, have a large (TTAGGG) repeat component (3–4%) compared to humans (0.3%). This includes interstitial telomere-like repeat arrays and mega-telomeres, up to 2 Mb in length, found at the end of a small number of chromosomes in birds (26, 27). The chicken karyotype is divided into macro-, intermediate-, and micro-chromosomes based on size, and mega-telomeres tend to be found on micro-chromosomes. MDV integrates into many different chromosome ends, often several different chromosome ends in each cell (26, 28, 29), and while there is no strong evidence of integration hot-spots in particular telomeres, lymphomas from the same bird have overlapping integration profiles suggesting clonal expansion (10).
To understand the relationship between MDV telomeric integration and excision, viral latency and reactivation, we have investigated chromosomally integrated MDV (iMDV) and the associated telomere. We used long-range PCR, telomere PCR amplification, and chromatin capture to demonstrate that MDV consistently integrates with the US region closer to the centromere and that the iMDV-associated telomere forms at mTMR in the terminal a-like region. These data support a model of iMDV genome excision by a telomere-loop (t-loop)-driven mechanism.
RESULTS
The MDV genome copy number measured by ddPCR shows extreme variability between cell lines
The copy number of the MDV genome has been shown to vary between tumors and lymphoma-derived cell lines and to change over time in cell culture (26, 28, 29). The approaches used, quantitative PCR, (qPCR) and fluorescent in situ hybridization (FISH), have limitations. To measure the relative MDV copy number per chicken genome, we developed several droplet digital PCR (ddPCR) assays (Fig. 1A) that facilitate sensitive and precise MDV quantification over a wide dynamic range, without the need for comparison to a standard curve (30). QT35, a quail cell line known to harbor a single copy of iMDV per cell (Fig. 1B), was used to establish and assess these assays. First, a duplexed hydrolysis probe assay was used to quantify UL copy number (UL-1 amplicon in ORF MDV057, Fig. 1A) relative to an amplicon in the avian GAPDH gene (amplicon name, AvGAPDH). Figure 1C shows one-dimensional ddPCR plots of UL-1 and AvGAPDH assays as well as the estimated MDV copy number per cell in the QT35 cell line, based on the proportion of positive droplets in each assay. Notably, QT35 has chromosome 1 trisomy (31); consequently, every cell has three copies of AvGAPDH. Therefore, the observed 1:3 ratio of UL-1 to AvGAPDH was expected and consistent with one copy of iMDV per cell (Table 1). Using the same duplexed assay, MDV UL copy number was determined in six MD lymphoma-derived chicken cell lines (226(O), 265(L), 4839(K), 4523(T), MSB-1, and HP8). In four of these cell lines (226(O), 4839(K), 265(L), and HP8), the MDV copy number was measured precisely and ranged from 5 to 10 copies per cell. The other two cell lines (4523(T) and MSB-1) each had a very high copy number and showed greater variation between replicates, particularly the 4523(T) cell line (Fig. 1D and E; Table 1).
TABLE 1.
The copy number of MDV genome regions in one quail and six chicken cell lines
| UL probe (triplicate) a | UL Mean ± SEM f | RS-1 b | RS-2 b | RL-1 b | RL-2 b | US-1 b | US-2 b | UL-1 b | UL-2 b | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MSB-1 | 101.1 | 95.4 | 90.1 | 95.5 ± 3.18 | 190.4 | 193.8 | 118.6 | 152.7 | 80.3 | 106.6 | 95.6 | 116.1 |
| 4523(T) | 158.2 | 158.8 | 220.5 | 179.2 ± 20.67 | 342.7 | 378.3 | 287.3 | 303.9 | 135.4 | 197.8 | 162.0 | 201.2 |
| 226(O) | ND | 6.31 | 6.19 | NA e | 8.26 | 8.64 | 6.56 | 11.42 | 3.31 | 4.90 | 2.81 | 3.80 |
| 4839(K) | 5.50 | 5.72 | 5.80 | 5.67 ± 0.09 | 11.44 | 11.10 | 13.90 | 9.75 | 5.08 | 6.03 | 4.38 | 4.99 |
| 265(L) | 6.10 | 6.64 | 6.83 | 6.52 ± 0.22 | 10.41 | 10.85 | 11.31 | 10.00 | 5.10 | 5.92 | 4.87 | 6.18 |
| HP8 | 8.90 | 8.94 | 8.60 | 8.81 ± 0.11 | 10.00 | 10.56 | 44.05 | 41.88 | 5.35 | 5.29 | 8.84 | 9.00 |
| QT35 | 0.32 | 0.33 | 0.34 | 0.3 ± 0.00 | 0.54 c | 0.67 c | 0.59 c | ND d | 0.34 c | 0.36 c | 0.30 c | 0.39 c |
Copy number was measured with ddPCR using fluorescent hydrolysis probes relative to AvGAPDH amplicon.
Copy number was measured with ddPCR using fluorescent dsDNA intercalating dye relative to the average of AvGAPDH and TF copy number.
Copy number was measured with ddPCR using fluorescent dsDNA intercalating dye relative to the average of the AvGAPDH and NOP2 copy number.
ND: not done.
NA: not applicable.
SEM: standard error of the mean.
To investigate the composition of the MDV genomes within cell lines, absolute quantification ddPCR assays were developed for the UL, US, RL, and RS regions; a reference amplicon in GAPDH (AvGAPDH), common to both chicken and quail; and reference amplicons in Ovotransferrin (TF, Chr 9) and NOP2 nuclear protein (NOP2, Chr 1), specific to chicken and quail, respectively. These ddPCR assays, based on a double-stranded DNA (dsDNA) intercalating dye, were used to measure the copy number of each MDV genome region. For the QT35 cell line, the ratio of UL and US to the avian control amplicons was approximately 1:3 (as expected due to chromosome 1 trisomy) and 2:3 for RS and RL (Fig. 1F; Table 1). Assessment of the MDV genome content in the chicken cell lines with a modest MDV copy number (5–10 copies per cell) was consistent with a 2:1 ratio between MDV repeat and unique regions and comparable with the copy number determined with the hydrolysis probe assay (Fig. 1G; Table 1). The exception to this was an unexpectedly high copy number of RL and UL in the HP8 cell line that revealed a 5:1 ratio of RL:UL and a UL:US ratio of approximately 2:1 (Fig. 1G and H; Table 1).
In summary, the ddPCR assays developed here have demonstrated very high variability of MDV copy number among chicken and quail cell lines, and they extend the range of accurate copy-number estimates without the need for standards. Furthermore, by combining data from multiple ddPCR assays that target different regions in the MDV genome, it is possible to explore MDV genomes that diverge from the expected structure. However, when the MDV copy number per cell is very high, quantification becomes less precise because of the large copy number difference between the MDV target and the reference.
MDV consistently integrates into the host telomere in the same orientation
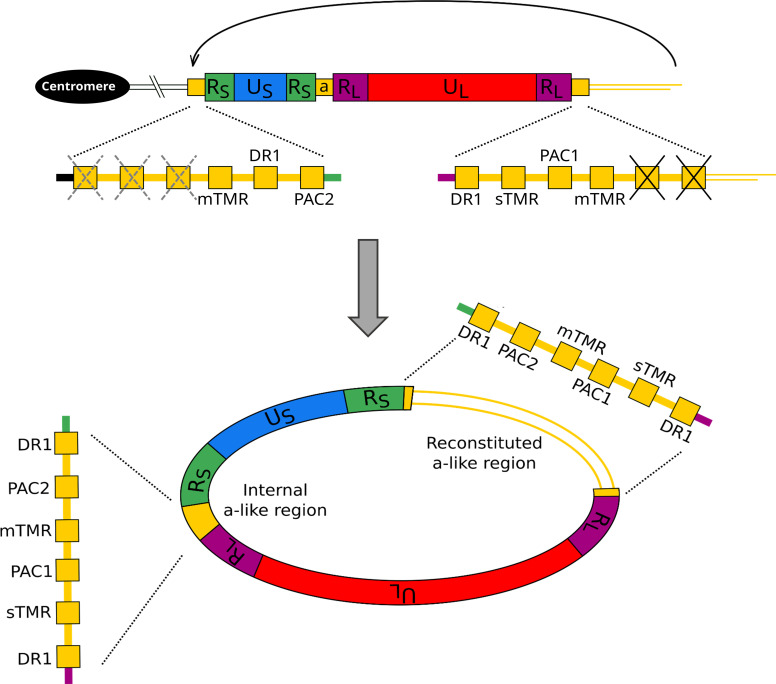
As stated above, the MDV genome contains a-like regions that each contain two telomere-like repeat arrays, the six (TTAGGG) repeat containing sTMR and the variable length degenerate repeat array mTMR. The linear MDV genome has an a-like region at each end (in the same orientation) as well as at least one a-like region between RS and RL (in the opposing orientation, Fig. 2A) (21). When circularized, the MDV genome has at least one a-like region at each RS-RL junction in opposing orientations. If integration of the MDV genome is facilitated by an homology-directed recombination (HDR)-based mechanism, it must integrate with telomere repeats in the same orientation as those in the host, that is 5′-(TTAGGG) n -3′, to produce the essential G-rich overhang at the 3′ ends. Figure 2B shows potential integration points in the MDV linear and circularized genomes. The polarity of the (TTAGGG) n repeats at these potential integration points implies that to ensure homology between the telomere array in MDV and the host telomere, MDV will integrate such that US is closer to the centromere and UL located toward the end of the chromosome, at the telomere (Fig. 2C). If this prediction is correct, following HDR integration the telomere will be present at the a-like region adjacent to RL. Integration of the MDV genome, such that UL is more centromeric, would generate a telomere with a C-rich 3′ overhang and is therefore unlikely as the telomere would not be functional.
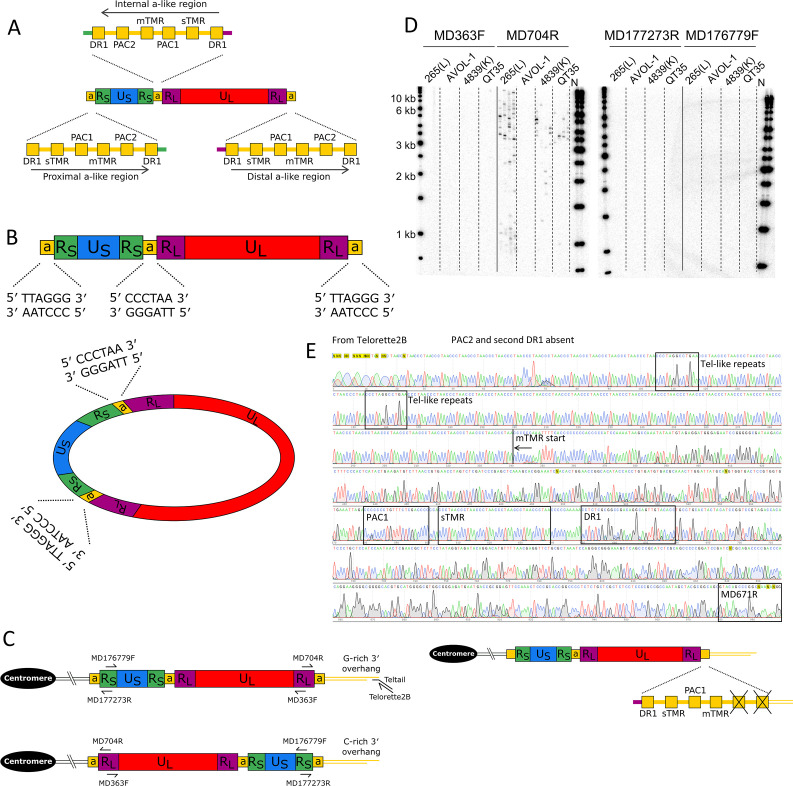
Fig 2.
Exploring integrated MDV genome organization and content. (A) Structures of a-like regions in non-integrated, linear MDV highlighting relative positions of DR1, packaging motifs PAC1 and PAC2, and telomere-repeat containing regions mTMR and sTMR. Arrows show the relative orientation of a-like regions. (B) Diagram showing linear and circularized MDV genomes that each contain two inverted copies of RL and RS. RL and RS are separated by a-like regions that each contain two arrays of telomere-like repeats that could act as a substrate for integration via an HDR-based mechanism. (C) Diagrams showing the predicted organization of the MDV genome following integration via an HDR-based mechanism between the telomere repeat arrays with the same polarity in the host and the viral genome (upper diagram), or via a non-HDR-based mechanism (e.g., non-homologous end joining) between telomere repeat arrays of opposing polarity (lower diagram). The latter is unlikely to lead to a functional telomere as it would produce a C-rich overhang. These diagrams also show the position and orientation of primers used in single telomere amplification (STELA) to determine the location of the integrated MDV-associated telomere. (D) STELA—Southern blots hybridized with a radiolabeled telomere-repeat probe. Shown are STELA reactions for three MDV-positive cell line DNAs (265(L), 4839(K), and QT35) and an MDV-negative cell line (AVOL-1), carried out using four different primers that anneal to the MDV genome. Amplification of the MDV-associated telomere only occurred when using the MD704R primer. (E) Sanger sequence trace obtained from an MDV-associated telomere amplicon, using primer Telorette2B. The sequence shows many copies of (CCCTAA) n telomere repeats with interspersed degenerate telomere-like repeats (black boxes) and the absence of PAC2 and the terminal DR1. The diagram shows the structure of a distal a-like region on an integrated copy of the MDV genome, expanded to show the absence of PAC2 and the second copy of DR1.
Single telomere length analysis (STELA) is a PCR-based approach that can be used to amplify telomeres and measure telomere length (32, 33). Here, STELA has been exploited to amplify MDV-associated telomeres using primers that anneal to either RS or RL. Figure 2D shows that MDV-associated telomeres were amplified from avian cell lines 265(L), 4839(K), and QT35 using the primer MD704R that anneals to RL and is directed toward the a-like region, but not by the MD363F primer that also anneals to RL but oriented in the opposite direction. STELA products were not generated using either MD177273R or MD176779F which anneal to RS. These results demonstrate that, following telomeric integration, the MDV genome is oriented such that US, flanked by the two RS regions, is centromeric to the RL-UL-RL region. Consequently, the MDV-associated telomere is located within the a-like region adjacent to RL.
STELA is typically used to measure the length of individual telomeres at specific chromosome ends and estimate the average length of the specific telomere. However, given the large range in chicken telomere lengths, including extremely long telomeres that are well beyond the limits of PCR amplification, it was considered inappropriate to present estimates of average iMDV-associated telomere lengths. Nevertheless, there is preliminary evidence that the lengths of the iMDV-associated telomeres may vary between cell lines, with the shortest telomeres amplified from cell lines with the highest MDV copy number (4523(T) and MSB-1) (Fig. S1).
In addition to sTMR and mTMR, a-like regions contain packaging signals PAC1 and PAC2, two copies of DR1, and some single-copy sequences (Fig. 2A). To investigate where the iMDV-associated telomere forms within the a-like region, several short STELA products (approximately 1 kb in length) were re-amplified in secondary semi-nested PCRs using Telorette2B and MD671R. Sanger sequences from several such amplicons (example shown in Fig. 2E) reveal that the MDV-associated telomere forms at mTMR, not sTMR and that following integration, the terminal a-like region lacks PAC2 and one copy of DR1. The sequences also showed that these MDV-associated telomeres were composed primarily of perfect (TTAGGG) n repeats with a small number of degenerate, telomere-like repeats.
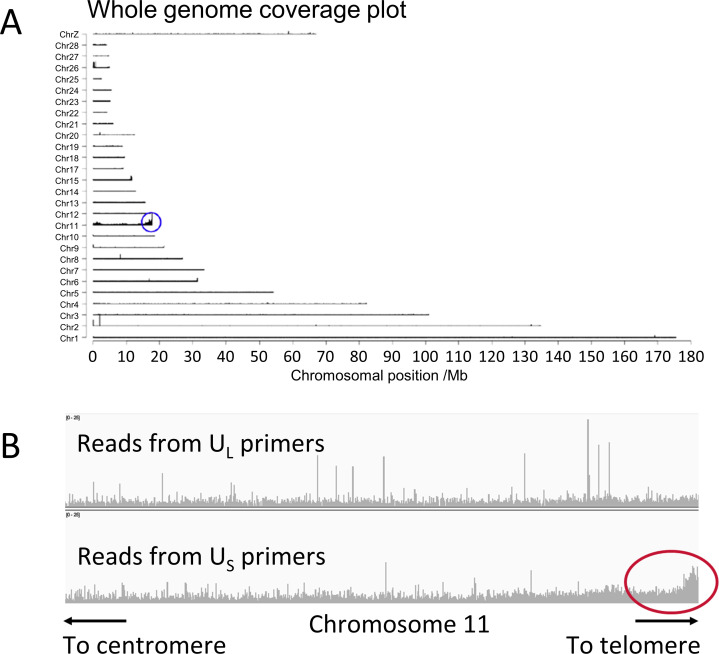
To investigate which chromosome end carries the iMDV in the QT35 cell line, tandem locus amplification (TLA) analysis was conducted (34). TLA analysis is a chromatin-structure capture technology that cross-links DNA in situ, enzymatically digests the genome, and re-ligates DNA fragments that are in close spatial proximity. Subsequent oligonucleotide-directed sequencing of re-ligated DNA reveals sequences that had been close to each other in 3D space by the relative abundance of mapped reads. The whole-genome coverage plot shows where reads from TLA mapped in the Quail genome (Fig. 3A). The depth of coverage was highest on chromosome 11, specifically at the end of the long arm (11q), indicating that the MDV genome is integrated into the 11q telomere in the QT35 cell line. As part of the TLA analysis, the oligonucleotide-directed sequencing was conducted with pairs of primers that anneal to the US or the UL regions far apart in the MDV genome. The increased read depth toward the distal end of the 11q chromosome arm following TLA sequence analysis using US primers (Fig. 3B lower panel) compared with the UL primers (Fig. 3B upper panel), strongly suggests that US is located closer to the chromosome-11-specific sequences in the quail genome than UL. This is consistent with the results obtained from sequencing STELA amplicons, described above (Fig. 2D and E). In summary, our analyses show that following MDV integration into an avian genome, US is located closer to the centromere of the chromosome carrying the integration, than UL. Consequently, one copy of RL occupies a distal location and the MDV-associated telomere is present at mTMR on a terminal, truncated a-like region.
Fig 3.
Tandem locus amplification in the QT35 cell line identifies the MDV integration site. (A) Whole-genome coverage plot obtained from MDV-TLA short read sequence data of QT35. The majority of reads map to chromosome 11 with enrichment at the end of the long arm of chromosome 11, adjacent to the 11q telomere. (B) Read depth across the distal end of chromosome 11 following tandem locus amplification with only the UL primer pair (upper panel) or only US primer pair (lower panel). The increased read-depth toward the telomere following tandem locus amplification with US primer pair is circled in red.
Determining the orientation of US in MDV
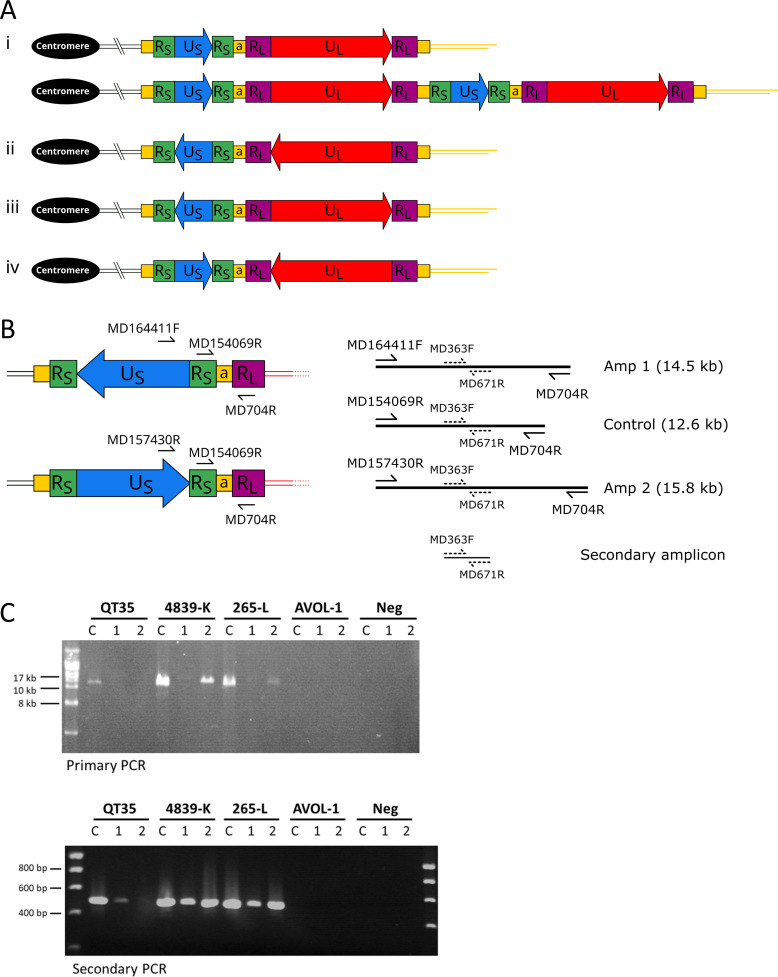
Studies of herpes simplex viruses 1 and 2 (HSV-1 and HSV-2), with similar genome organizations to MDV, have shown that the orientation of US and UL can change with respect to one another. This results in four isomeric forms of the viral genomes thought to be present in equal proportions (35 – 37). The MDV genome is also thought to undergo isomerization facilitated by recombination between the inverted repeats of RL or RS. Isomerization can potentially generate four MDV genome configurations where UL and US are in the same orientation, directed away from each other, or directed toward each other. Furthermore, each isomer could potentially integrate into a telomere as a monomer or concatemer (see example in Fig. 4Ai). To determine the orientation of US in the single copy of MDV in the QT35 cell line, long-range PCR anchored in RL was used to determine which end of US is closest to UL (Fig. 4B). In brief, the MD164411F and MD157430R primers anneal in opposite orientations toward one or the other end of US, and away from the center of US (Fig. 4B). If PCR is carried out with the US MD164411F primer and the MD704R primer in UL, Amplicon 1 will be generated if US is orientated away from UL. By contrast, Amplicon 2 will be generated by amplification with the US MD157430R and UL MD704R primers if US is orientated toward UL. In addition, PCR of a long control amplicon (Rs MD154069R and RL MD704R primers) was included as it should amplify regardless of the orientation of US. To detect the presence of these long amplicons (if sub-visible in the primary PCRs), secondary nested reactions were set up using primers MD363F and MD671R, which are common to all the predicted primary PCR products. Amplicon 1 and the Control amplicon, but not Amplicon 2, were amplified from QT35 DNA (Fig. 3C) thus demonstrating that the orientation of US in the single copy of iMDV is stable and directed toward the interior of the quail chromosome (as shown in Fig. 4A ii or iii). By contrast, Amplicon 1 and Amplicon 2 could both be amplified from 4839(K) and 265(L) DNA, suggesting that both orientations of US are present in different copies of the integrated MDV genome; or that some copies of the viral genome are circularized; or that the MDV genome is integrated as a concatemer; or a combination of these are present in the cell lines.
Fig 4.
Investigating the orientation of US relative to RL in MDV genomes. (A) Diagram showing the four possible relative orientations of US and UL in monomeric iMDV and an example of concatemeric iMDV. The diagrams show US more centromeric than UL (in a telomeric position) with (i) both orientated toward the telomere, (ii) both orientated toward the centromere, (iii) an inverse orientation directed away from each other, or (iv) an inverse orientation directed toward each other. (B) Diagram showing the long-range PCRs used to determine the orientation of US relative to RL. Amplicon 1 (using primers MD164411F and MD704R) will be generated if US is orientated away from RL, Amplicon 2 (primers MD157430R and MD704R) will be generated if US is orientated toward RL. The control amplicon (primers MD154069R and MD704R) for the long-range PCR should generate a product regardless of the US–RL orientation. A secondary amplicon (primers MD363F and MD671R) can be amplified from all three primary amplicons in a fully nested PCR. (C) Images of ethidium bromide-stained agarose gels from the primary PCRs showing detection of the control amplicon (C), Amplicon 1 and Amplicon 2 in MDV-positive QT35, 4839(K) and 265(L), but not in MDV-negative AVOL-1, and water negative controls. Indirect detection of Amplicons 1 and 2 by secondary nested PCR of diluted primary PCR products, using primers MD363F and MD671R.
DISCUSSION
The average copy number of MDV, as measured here by ddPCR, is highly variable across tumor-derived and infected cell lines, consistent with previous studies that used FISH and qPCR (26, 28, 29). ddPCR has advantages over these methods, for example, it is less laborious than FISH, and it can quantify MDV copy per cell without the use of known standards, unlike qPCR. Furthermore, the use of several ddPCR assays across the MDV genome can reveal duplication or deletion events of different genome regions, as shown in the HP8 cell line where more copies of RL and UL are present than expected from the US copy number. These extra copies may have arisen from duplication events facilitated by HDR between the inverted repeats of RL. The low MDV copy number in four chicken cell lines likely represents latent iMDV, whereas the very high copy number in two cell lines (4523(T) and MSB-1) may represent a mixture of iMDV and non-integrated, free circular, or replicating virus (26, 38 – 40). This is consistent with observations that both the 4523(T) and MSB-1 cell lines produce high numbers of infectious virus plaques when co-cultivated on primary chicken embryo fibroblasts in contrast to the HP8 and 226(O) cell lines [(16) and unpublished data]. As the ddPCR assays are reproducible and accurate with low to moderate MDV copy number (particularly when using a hydrolysis probe), they could be used effectively to monitor small changes in copy number to explore viral reactivation in response to drug treatment (41, 42) and to measure the efficacy of vaccines.
Despite recent developments, much of the basic biology of MDV genome integration and release from a telomere remains unknown. Here, we have shown that the MDV genome integrates in such a way that a telomere forms at mTMR in an a-like region. The MDV telomere-associated amplification data presented from six cell lines, together with TLA analysis from the QT35 cell line, show that MDV consistently integrates in one orientation, that is, with RL at the most distal end of the chromosome and with RS in a more centromeric position. Sequencing of re-amplified STELA products showed that the MDV-associated telomere forms at mTMR, the longer variable region of telomere-like repeats within the MDV a-like region.
The potential presence of four different isomers of the MDV genome following MDV infection and rolling circle replication raises the possibility that different isoforms may integrate into distinct telomeres during the transition to latency. We developed long-range PCR assays to distinguish two of the isoforms (Us in different orientations with respect to RL) and identified the isomer present in the single copy of MDV integrated into chromosome 11q in the quail QT35 cell line. Interpretation of similar analysis in two chicken cell lines with modest MDV copy number [265(L) and 4839(K), Table 1] is complicated by the possible presence of circularized or concatemeric forms of the MDV genome, as these can give rise to both amplicons. Therefore, the US orientation analysis (Fig. 4C) does not give definitive proof that these cell lines carry different MDV isomers in individual telomeres. However, the 265(L) cell line, with six MDV copies per cell, is stable and not known to produce a virus. Furthermore, preliminary FISH analysis indicates that the 265(L) cell line carries iMDV in multiple telomeres, as has been shown in many other lymphoma-derived chicken cell lines (26, 29, 38). This supporting evidence strengthens our provisional interpretation that the 265(L) cell line is likely to carry different iMDV isomers in distinct telomeres.
These new observations support a model of MDV genome integration and excision, that is similar but not identical to the model proposed for chromosomally integrated HHV-6 (33, 43). The source of the MDV genome that integrates into a telomere could include a cleaved and packaged MDV genome directly from a viral particle (21); a circularized viral genome that is a substrate for rolling circle replication (44); or a concatemeric form of the MDV genome. Homology-directed recombination between the mTMR in an a-like region and an avian telomere would result in the integration of MDV genome with US positioned centromeric to UL , as observed. We have also shown that the iMDV-associated telomere is present at mTMR in the terminal a-like region that lacks PAC2 and one copy of DR1. In turn, we hypothesize that the internal a-like region is ligated to the avian chromosome at mTMR, and is also truncated, lacking one copy of DR1, sTMR, and PAC1.
Full reactivation of MDV from latency, in a cell that only carries integrated copies of the viral genome (38), requires the release of an iMDV genome (that lacks full-length a-like regions at the ends of the genome) from a telomere and circularization prior to rolling circle replication. Precise excision of the iMDV genome with simultaneous circularization could be achieved via a telomere-loop (t-loop)-mediated mechanism (45, 46). This model is appealing as it also facilitates the reconstitution of a complete a-like region. In this scenario, a t-loop that contains the entire MDV genome would form following strand invasion of the telomeric single-strand overhang into mTMR in the internal a-like region (Fig. 5). Then, as proposed in the chromosomally integrated HHV-6 excision model (33, 43, 47), the base of the t-loop could migrate and be resolved as a double Holliday junction structure to release a “t-circle” containing the entire MDV genome with a reconstituted a-like region. The excised circular MDV genome could then become a template for rolling circle replication. While the telomere-like repeat arrays (sTMR and mTMR) in the centrally located a-like region of an iMDV genome are also possible targets for invasion by the telomeric single-stranded overhang, these repeats are in the opposite orientation (Fig. 2) and in this scenario, excision of a t-circle would lead to an unviable 5′ C-rich overhang. This contrasts with the model of integrated HHV-6 release where partial excision of the integrated viral genome has been shown to occur leaving a 3′ G-rich overhang (48).
Fig 5.
Diagram showing t-loop mediated excision of an iMDV genome from a telomere. The organization of a monomeric iMDV genome is shown. The black crosses in the distal a-like region show the absence of the terminal PAC2 and DR1. The iMDV-associated telomere that starts at the terminal mTMR is shown as parallel yellow lines. The gray dashed crosses on the internal a-like region indicate the hypothetical loss of one copy of DR1, sTMR, and PAC1. The black arrow shows the potential for t-loop formation by the single-strand invasion of the iMDV-associated telomere into the repeats at the junction between iMDV and the host telomere (including mTMR). This would produce a t-loop containing the MDV genome that could be excised as a t-circle. The released, circularized MDV genome would contain an a-like region between both pairs of RS and RL, one reconstituted by a t-loop mediated excision process.
In summary, MDV is an important avian pathogen that has been used to investigate the role of viruses as oncogenic agents (6, 10, 49). As we have shown, MDV genome analyses are also useful to understand mechanisms of telomere integration and excision used by some herpesviruses. In the future, MDV integration and release have the potential to emerge as a model to study telomere dynamics in chicken, and in birds more widely.
MATERIALS AND METHODS
Avian cell lines, culture conditions, and DNA extraction
MDV-positive chicken cell lines 265(L), 4839(K), 4523(T), and 226(O) were established from lymphomas from chickens infected with the RB-1B strain of MDV (50) (16). MDV-positive chicken cell line HP8 (51) was from a GA strain-induced tumor and MSB-1 from a spleen lymphoma induced by BC-1 strain (52). AVOL-1 is an MDV-negative T-cell line (53). The MDV-positive quail cell line QT35 was derived from a methylcholanthrene-induced tumor in Japanese quail (Coturni coturnix japonica) (31).
MSB-1 and AVOL-1 cell lines were cultured in RPMI 1640 + Glutamax I (Gibco-Thermo Fisher Scientific) supplement with 10% heat-inactivated fetal bovine serum (FBS; Sigma-Aldrich), 10% tryptose phosphate broth (Sigma-Aldrich), 1 mM sodium pyruvate (Sigma-Aldrich), and 50 µM 2-mercaptoethanol (Gibco-Thermo Fisher Scientific). The same media supplemented with 15% heat-inactivated chicken serum (Life Science Production) was used to culture the 265(L), 4839(K), 4523(T) and 226(O), and HP8 cell lines. The QT35 cell line was cultured in M199 media with 0.1% glutamine and Earle’s salts (Thermo Fisher Scientific) supplemented with 5% heat- inactivated FBS (Sigma-Aldrich), 10% tryptose phosphate broth (Sigma-Aldrich), and 10% heat-inactivated chicken serum (Life Science Production). All cell lines were cultured at 38.5°C in a 5% CO2, humidified atmosphere.
Harvested cells were pelleted at 1,200 rpm for 8 min, washed twice with phosphate-buffered saline, and snap-frozen on dry ice. High molecular weight DNA was extracted using a standard phenol-chloroform method and precipitated using ethanol. The DNA pellet was washed with 80% ethanol, briefly air-dried and dissolved in sterile pure water.
Fluorescent in situ hybridization
Chromosomal integration of the MDV genome on metaphase spreads from Avian cell lines was detected with full-length MDV genome generated from infectious bacterial artificial chromosome (BAC) clones as hybridization probes using full standard FISH protocols as described previously (54).
Droplet digital PCR assays for quantification of regions of the MDV genome
Hydrolysis probe ddPCR reactions (20 µL volume) consisted of 1× ddPCR Supermix for Probes (Bio-Rad Laboratories Ltd), 300 nM of each forward and reverse primer (Sigma-Aldrich), 200 nM of each hydrolysis probe (Eurogentec), 0.25 U XhoI restriction enzyme (New England Biolabs), 0.05× NEB buffer 2.1 (New England Biolabs), and 1 µL of DNA at appropriate concentration. Each 20 µL PCR was partitioned into nanolitre-sized droplets with 70 µL Droplet Generating Oil (Bio-Rad Laboratories Ltd) using QX200 Droplet Generator (Bio-Rad Laboratories Ltd). Thermocycling was carried out in a Veriti 96 cell thermocycler (Applied Biosystems) as follows: 95°C, 10 min; 40 cycles 94°C 30 s, 60°C 1 min; 98°C 10 min. dsDNA intercalating dye (EvaGreen) probe ddPCR reactions (20 µL volume) were set up as above but using 1× ddPCR EvaGreen Supermix (Bio-Rad Laboratories) and with no hydrolysis probe. Thermocycling conditions were as follows: 95°C, 5 min; 45 cycles of 94°C 30 s, 60–62°C 1 min; 4°C 5 min; 90°C 5 min. Droplet fluorescence was measured in one or two channels using QX200 Droplet Reader (Bio-Rad Laboratories Ltd). All data were analyzed using QuantaSoft software (v1.4.0, Bio-Rad Laboratories Ltd), which was also used to generate 1D droplet plots. Primer and probe sequences are shown in Table 2.
TABLE 2.
Primer and probe sequences
| Amplicon name | Primer name | 5–3 sequence | Tm \C |
|---|---|---|---|
| AvGAPDH | AvGAPDH-F1 | AAGCAGGACCCTTTGTTGGA | 60 |
| AvGAPDH-R1 | TCTATCAGCCTCTCCCACCT | ||
| AvGAPDH-Pr | HEX-TGTCTCCCCCACTCCTCCTC-BHQ | ||
| Ovotransferrin (TF) | OvoTF-F | CACTGCCACTGGGCTCTGT | 60 |
| OvoTF-R | GCAATGGCAATAAACCTCCAA | ||
| NOP2 | Q-NOP2-F1 | CTTATGGTTAGAAGCCCGGA | 60 |
| Q-NOP2-R1 | CGAAGAACAGAAGAAGCCAC | ||
| UL-1 | UL-F1 | CATGCAAGTCATTATGCGTGA | 60 |
| UL-R1 | TGTTTCCATTCTGTCTCCAAGA | ||
| UL-probe | FAM-TACGTGTGTGTCAACGACCT-BHQ | ||
| UL-2 | MDVprF4 | TCCATTCATCGGCTGCTTTGTG | 60 |
| MDVprR3 | ATTGTGAAATCCTCCACCGC | ||
| RL-1 | MDV93F | AGACACTTTCCCACTCATAC | 60 |
| Md5-PAC1F | GTCGAGAAACAGGGGGGGTCTA | ||
| RL-2 | MDVprRLR2 | GAGCTGTTGACACCTCTGAT | 62 |
| MDVprRLF3 | GCCAACATTTTCAGACTTTGCC | ||
| US-1 | MDVprUSF2 | CCTGCAAGTCCATCCGTTG | 60 |
| MDVprUSR1 | CGTCAGCGGGTTCTTTAGTA | ||
| US-2 | US-II-F | CCATGAAGGGGCGTGTCAAA | 60 |
| US-II-R | ATTTGCGCCGTAGAGACCAC | ||
| RS-1 | MD154069R | TGCAAGCTCTGAGAACTTCC | 60 |
| MD153927F | AGCTAACCACAGCGTGTCTC | ||
| RS-2 | MDVprRSR1 | TCCGTAAGTGCTTGTCGATG | 60 |
| RS-III-F | GTGACATCCTCCTCTGGCCT | ||
| TLA primers | UL-Fw | TTGCGTACAACCGTGGGATA | |
| UL-Rv | GAGGTGGTTCGGCGCATC | ||
| US-Fw | CCAGTTTCGGTAAGATCAGT | ||
| US-Rv | ACAAGGGTTATGATCGACAC | ||
| US orientation primers | MD164411F | AACGTGCAAGTTCGTCACGA | 57 |
| MD154069R | TGCAAGCTCTGAGAACTTCC | 57 | |
| MD157430R | ATATCCAAGGACAGTGGACA | 57 | |
| MD671R | CCTATTGGCCGGGCTGTAC | 57 a /61 b | |
| STELA primers | MD363F | ACTACATCCGGTTCGTAGAC | 61 |
| MD704R | CTTGTAGCTTCCTCCGCCT | 57 a /61 b | |
| MD176779F | CTGTCATTTCGCAGGGATCT | 61 | |
| MD177273R | GAGTCAGGCATTGCGGAGT | 61 | |
| Telorette2B | TGCTCCGTGCATCTGGCATCTAACCCT | ||
| Teltail | TGCTCCGTGCATCTGGCATC |
Primer annealing temperature in US-RL orientation assays.
Primer annealing temperature in STELA.
Tandem locus amplification analysis of iMDV genome
QT35 cells, 24 h after subculturing, were harvested in fresh media by centrifugation at 1,100 rpm for 8 min with two phosphate-buffered saline washes. Aliquots of 107 cells were resuspended in QT35 culture medium with 10% dimethyl sulfoxide and used for TLA analysis by Cergentis. Sample preparation and data analysis were carried out by Cergentis (34). In brief, genomic DNA was cross-linked in cells resulting in sequences that were in close proximity becoming cross-linked to interacting proteins. The DNA was then digested, ligated to promote circularization, and decross-linked. The ligated DNA fragments and two sets of primers (UL-Fw and UL-Rv, and US-Fw and US-Rv) were used to amplify the DNA. Amplified DNA was then sequenced on an Illumina HiSeq platform and reads were mapped to the quail genome [Corturnix japonica, GCA_001577835.2 (55)] and to the MDV genome (Md5, AF243438) using BWA-SW.
STELA to detect iMDV-associated telomeres
The telomere at the end of the iMDV genome was amplified by STELA using the MD704R primer and Telorette2B/Teltail as described previously (32, 33, 56). 10 µL STELA reactions contained 1× buffer (57), 0.3 µM flanking primers, 0.225 µM Telorette2B, 0.05 µM Teltail, 0.4 U Taq polymerase (Kapa Biosystems Ltd), 0.25 U Pwo (Genaxxon Bioscience), and 1 μL DNA diluted to 10–250 pg. Thermocycling was as follows: 94°C, 1.5 min; 25 cycles 94°C 15 s, 61°C 30 s, 68°C 12 min; 68°C 2 min. STELA products were size separated by agarose gel electrophoresis and amplified telomeres were detected by Southern blot hybridization to a radiolabeled (TTAGGG) n probe. Selected reactions were diluted 1:10 with water and re-amplified in semi-nested secondary PCR with primers Telorette2B and MD671R with the following thermocycling conditions: 94°C, 1.5 min; 25 cycles 94°C 15 s, 61°C 30 s, 68°C 4 min; 68°C 2 min. The reamplified products were sequenced (Source Bioscience) using the Sanger chain termination method.
US orientation determining PCR
Ten microliters of primary PCRs contained 1× buffer (57), 0.3 µM forward and reverse primers, 0.5 U Taq polymerase (Kapa Biosystems Ltd), 0.25 U Pfu polymerase (Thermo Fisher Scientific), and 10 ng DNA. Primary amplicons were amplified using MD704R and MD164411F (Amplicon 1), MD157430R (Amplicon 2), or MD154069R (Control amplicon) with the following conditions: 94°C, 1.5 min; 27 cycles 94°C 15 s, 57°C 30 s, 68°C 14 min; 68°C 2 min. One microliter 1:10 dilutions of primary PCR were used as input for secondary PCR using primer MD363F and MD671R with the following conditions: 94°C, 1.5 min; 25 cycles 94°C 15 s, 61°C 30 s, 68°C 1 min; 68°C 2 min.
ACKNOWLEDGMENTS
M.L.W. was funded by the UK Biotechnology and Biological Sciences Research Council (BBSRC) and Midlands Integrative Biosciences Training Partnership (MIBTP 1645656).
Contributor Information
Nicola J. Royle, Email: njr@leicester.ac.uk.
Anna Ruth Cliffe, University of Virginia, Charlottesville, Virginia, USA .
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.00716-23.
STELA of iMDV-associated telomeres in six avian cell lines.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Morrow C, Fehler F. 2004. Marek’s disease: a worldwide problem, p 49–61. In Marek’s disease: An evolving problem. Elsevier. doi: 10.1016/B978-012088379-0/50009-8 [DOI] [Google Scholar]
- 2. Nair V. 2005. Evolution of Marek's disease -- a paradigm for incessant race between the pathogen and the host. Vet J 170:175–183. doi: 10.1016/j.tvjl.2004.05.009 [DOI] [PubMed] [Google Scholar]
- 3. Biggs PM, Nair V. 2012. The long view: 40 years of Marek's disease research and Avian pathology. Avian Pathol 41:3–9. doi: 10.1080/03079457.2011.646238 [DOI] [PubMed] [Google Scholar]
- 4. Calnek BW. 2001. Pathogenesis of Marek’s disease virus infection. Curr Top Microbiol Immunol 255:25–55. doi: 10.1007/978-3-642-56863-3_2 [DOI] [PubMed] [Google Scholar]
- 5. Bertzbach LD, Conradie AM, You Y, Kaufer BB. 2020. Latest insights into Marek's disease virus pathogenesis and tumorigenesis. Cancers (Basel) 12:647. doi: 10.3390/cancers12030647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Osterrieder N, Kamil JP, Schumacher D, Tischer BK, Trapp S. 2006. Marek's disease virus: from miasma to model. Nat Rev Microbiol 4:283–294. doi: 10.1038/nrmicro1382 [DOI] [PubMed] [Google Scholar]
- 7. Ralapanawe S, Walkden-Brown SW, Islam A, Renz KG. 2016. Effects of rispens CVI988 vaccination followed by challenge with Marek’s disease viruses of differing virulence on the replication kinetics and shedding of the vaccine and challenge viruses. Vet Microbiol 183:21–29. doi: 10.1016/j.vetmic.2015.11.025 [DOI] [PubMed] [Google Scholar]
- 8. Trapp-Fragnet L, Schermuly J, Kohn M, Bertzbach LD, Pfaff F, Denesvre C, Kaufer BB, Härtle S. 2021. Marek's disease virus prolongs survival of primary chicken B-cells by inducing a senescence-like phenotype. PLoS Pathog 17:e1010006. doi: 10.1371/journal.ppat.1010006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Robinson CM, Cheng HH, Delany ME. 2014. Temporal kinetics of Marek's disease herpesvirus: integration occurs early after infection in both B and T cells. Cytogenet Genome Res 144:142–154. doi: 10.1159/000368379 [DOI] [PubMed] [Google Scholar]
- 10. Glass MC, Smith JM, Cheng HH, Delany ME. 2021. Marek's disease virus telomeric integration profiles of neoplastic host tissues reveal unbiased chromosomal selection and loss of cellular diversity during tumorigenesis. Genes (Basel) 12:1630. doi: 10.3390/genes12101630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaufer BB, Jarosinski KW, Osterrieder N. 2011. Herpesvirus telomeric repeats facilitate genomic integration into host telomeres and mobilization of viral DNA during reactivation. J Exp Med 208:605–615. doi: 10.1084/jem.20101402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deng X, Li X, Shen Y, Qiu Y, Shi Z, Shao D, Jin Y, Chen H, Ding C, Li L, Chen P, Ma Z. 2010. The Meq oncoprotein of Marek's disease virus interacts with p53 and inhibits its transcriptional and apoptotic activities. Virol J 7:348. doi: 10.1186/1743-422X-7-348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lupiani B, Lee LF, Cui X, Gimeno I, Anderson A, Morgan RW, Silva RF, Witter RL, Kung H-J, Reddy SM. 2004. Marek’s disease virus-encoded Meq gene is involved in transformation of lymphocytes but is dispensable for replication. Proc Natl Acad Sci U S A 101:11815–11820. doi: 10.1073/pnas.0404508101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brown AC, Baigent SJ, Smith LP, Chattoo JP, Petherbridge LJ, Hawes P, Allday MJ, Nair V. 2006. Interaction of MEQ protein and C-terminal-binding protein is critical for induction of lymphomas by Marek's disease virus. Proc Natl Acad Sci U S A 103:1687–1692. doi: 10.1073/pnas.0507595103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tai S-H, Hearn C, Umthong S, Agafitei O, Cheng HH, Dunn JR, Niikura M. 2017. Expression of Marek’s disease virus oncoprotein Meq during infection in the natural host. Virology 503:103–113. doi: 10.1016/j.virol.2017.01.011 [DOI] [PubMed] [Google Scholar]
- 16. Roy P, Moffat K, Nair V, Yao Y. 2021. CRISPR-mediated gene activation (CRISPRa) of pp38/pp24 orchestrates events triggering lytic infection in Marek's disease virus-transformed cell lines. Microorganisms 9:1681. doi: 10.3390/microorganisms9081681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chbab N, Egerer A, Veiga I, Jarosinski KW, Osterrieder N. 2010. Viral control of vTR expression is critical for efficient formation and dissemination of lymphoma induced by Marek's disease virus (MDV). Vet Res 41:56. doi: 10.1051/vetres/2010026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaufer BB, Arndt S, Trapp S, Osterrieder N, Jarosinski KW. 2011. Herpesvirus telomerase RNA (vTR) with a mutated template sequence abrogates herpesvirus-induced lymphomagenesis. PLoS Pathog 7:e1002333. doi: 10.1371/journal.ppat.1002333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tulman ER, Afonso CL, Lu Z, Zsak L, Rock DL, Kutish GF. 2000. The genome of a very virulent Marek’s disease virus. J Virol 74:7980–7988. doi: 10.1128/jvi.74.17.7980-7988.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Spatz SJ, Silva RF. 2007. Sequence determination of variable regions within the genomes of gallid herpesvirus-2 pathotypes. Arch Virol 152:1665–1678. doi: 10.1007/s00705-007-0992-3 [DOI] [PubMed] [Google Scholar]
- 21. Volkening JD, Spatz SJ. 2013. Identification and characterization of the genomic termini and cleavage/packaging signals of gallid herpesvirus type 2. Avian Dis 57:401–408. doi: 10.1637/10410-100312-Reg.1 [DOI] [PubMed] [Google Scholar]
- 22. Jarosinski KW, Tischer BK, Trapp S, Osterrieder N. 2006. Marek's disease virus: lytic replication, oncogenesis and control. Expert Rev Vaccines 5:761–772. doi: 10.1586/14760584.5.6.761 [DOI] [PubMed] [Google Scholar]
- 23. Greco A, Fester N, Engel AT, Kaufer BB. 2014. Role of the short telomeric repeat region in Marek's disease virus replication, genomic integration, and lymphomagenesis. J Virol 88:14138–14147. doi: 10.1128/JVI.02437-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Osterrieder N, Wallaschek N, Kaufer BB. 2014. Herpesvirus genome integration into telomeric repeats of host cell chromosomes. Annu Rev Virol 1:215–235. doi: 10.1146/annurev-virology-031413-085422 [DOI] [PubMed] [Google Scholar]
- 25. You Y, Vychodil T, Aimola G, Previdelli RL, Göbel TW, Bertzbach LD, Kaufer BB. 2021. A cell culture system to investigate Marek’s disease virus integration into host chromosomes. Microorganisms 9:2489. doi: 10.3390/microorganisms9122489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Delecluse HJ, Hammerschmidt W. 1993. Status of Marek’s disease virus in established lymphoma cell lines: herpesvirus integration is common. J Virol 67:82–92. doi: 10.1128/JVI.67.1.82-92.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Delany ME, Krupkin AB, Miller MM. 2000. Organization of telomere sequences in birds: evidence for arrays of extreme length and for in vivo shortening. Cytogenet Cell Genet 90:139–145. doi: 10.1159/000015649 [DOI] [PubMed] [Google Scholar]
- 28. Baigent SJ, Petherbridge LJ, Howes K, Smith LP, Currie RJW, Nair VK. 2005. Absolute quantitation of Marek's disease virus genome copy number in chicken feather and lymphocyte samples using real-time PCR. J Virol Methods 123:53–64. doi: 10.1016/j.jviromet.2004.08.019 [DOI] [PubMed] [Google Scholar]
- 29. Robinson CM, Hunt HD, Cheng HH, Delany ME. 2010. Chromosomal integration of an avian oncogenic herpesvirus reveals telomeric preferences and evidence for lymphoma clonality. Herpesviridae 1:5. doi: 10.1186/2042-4280-1-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, Tewari M. 2013. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods 10:1003–1005. doi: 10.1038/nmeth.2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moscovici C, Moscovici MG, Jimenez H, Lai MM, Hayman MJ, Vogt PK. 1977. Continuous tissue culture cell lines derived from chemically induced tumors of Japanese quail. Cell 11:95–103. doi: 10.1016/0092-8674(77)90320-8 [DOI] [PubMed] [Google Scholar]
- 32. Baird DM, Rowson J, Wynford-Thomas D, Kipling D. 2003. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat Genet 33:203–207. doi: 10.1038/ng1084 [DOI] [PubMed] [Google Scholar]
- 33. Huang Y, Hidalgo-Bravo A, Zhang E, Cotton VE, Mendez-Bermudez A, Wig G, Medina-Calzada Z, Neumann R, Jeffreys AJ, Winney B, Wilson JF, Clark DA, Dyer MJ, Royle NJ. 2014. Human telomeres that carry an integrated copy of human herpesvirus 6 are often short and unstable, facilitating release of the viral genome from the chromosome. Nucleic Acids Res 42:315–327. doi: 10.1093/nar/gkt840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Vree PJP, de Wit E, Yilmaz M, van de Heijning M, Klous P, Verstegen M, Wan Y, Teunissen H, Krijger PHL, Geeven G, Eijk PP, Sie D, Ylstra B, Hulsman LOM, van Dooren MF, van Zutven L, van den Ouweland A, Verbeek S, van Dijk KW, Cornelissen M, Das AT, Berkhout B, Sikkema-Raddatz B, van den Berg E, van der Vlies P, Weening D, den Dunnen JT, Matusiak M, Lamkanfi M, Ligtenberg MJL, ter Brugge P, Jonkers J, Foekens JA, Martens JW, van der Luijt R, van Amstel HKP, van Min M, Splinter E, de Laat W. 2014. Targeted sequencing by proximity ligation for comprehensive variant detection and local haplotyping. Nat Biotechnol 32:1019–1025. doi: 10.1038/nbt.2959 [DOI] [PubMed] [Google Scholar]
- 35. Sarisky RT, Weber PC. 1994. Requirement for double-strand breaks but not for specific DNA sequences in herpes simplex virus type 1 genome isomerization events. J Virol 68:34–47. doi: 10.1128/JVI.68.1.34-47.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martin DW, Weber PC. 1996. The a sequence is dispensable for isomerization of the herpes simplex virus type 1 genome. J Virol 70:8801–8812. doi: 10.1128/JVI.70.12.8801-8812.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chang W, Jiao X, Sui H, Goswami S, Sherman BT, Fromont C, Caravaca JM, Tran B, Imamichi T. 2022. Complete genome sequence of herpes simplex virus 2 strain G. Viruses 14:536. doi: 10.3390/v14030536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Delecluse HJ, Schüller S, Hammerschmidt W. 1993. Latent Marek's disease virus can be activated from its chromosomally integrated state in herpesvirus-transformed lymphoma cells. EMBO J 12:3277–3286. doi: 10.1002/j.1460-2075.1993.tb05997.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tanaka A, Silver S, Nonoyama M. 1978. Biochemical evidence of the Nonintegrated status of Marek’s disease virus DNA in virus-transformed lymphoblastoid cells of chicken. Virology 88:19–24. doi: 10.1016/0042-6822(78)90105-8 [DOI] [PubMed] [Google Scholar]
- 40. Kaschka-Dierich C, Nazerian K, Thomssen R. 1979. Intracellular state of Marek’s disease virus DNA in two tumour-derived chicken cell lines. J Gen Virol 44:271–280. doi: 10.1099/0022-1317-44-2-271 [DOI] [PubMed] [Google Scholar]
- 41. Bertzbach LD, Conradie AM, Hahn F, Wild M, Marschall M, Kaufer BB. 2019. Artesunate derivative TF27 inhibits replication and pathogenesis of an oncogenic avian alphaherpesvirus. Antiviral Res 171:104606. doi: 10.1016/j.antiviral.2019.104606 [DOI] [PubMed] [Google Scholar]
- 42. Zhang J, Gu Y, Wu C, Ma H, He J, Bai Y, Li H. 2013. Effects of sodium tanshinone IIA sulfonate against Marek's disease virus in experimentally infected chickens. Int J Biol Macromol 58:258–262. doi: 10.1016/j.ijbiomac.2013.04.029 [DOI] [PubMed] [Google Scholar]
- 43. Wood ML, Royle NJ. 2017. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. Viruses 9:184. doi: 10.3390/v9070184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rziha HJ, Bauer B. 1982. Circular forms of viral DNA in Marek’s disease virus-transformed lymphoblastoid cells. Arch Virol 72:211–216. doi: 10.1007/BF01348966 [DOI] [PubMed] [Google Scholar]
- 45. Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. 1999. Mammalian telomeres end in a large duplex loop. Cell 97:503–514. doi: 10.1016/S0092-8674(00)80760-6 [DOI] [PubMed] [Google Scholar]
- 46. Wang RC, Smogorzewska A, de Lange T. 2004. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119:355–368. doi: 10.1016/j.cell.2004.10.011 [DOI] [PubMed] [Google Scholar]
- 47. Prusty BK, Krohne G, Rudel T. 2013. Reactivation of chromosomally integrated human herpesvirus-6 by telomeric circle formation. PLoS Genet 9:e1004033. doi: 10.1371/journal.pgen.1004033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wood ML, Veal CD, Neumann R, Suárez NM, Nichols J, Parker AJ, Martin D, Romaine SP, Codd V, Samani NJ, Voors AA, Tomaszewski M, Flamand L, Davison AJ, Royle NJ. 2021. Variation in human herpesvirus 6B telomeric integration, excision, and transmission between tissues and individuals. Elife 10:e70452. doi: 10.7554/eLife.70452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McPherson MC, Cheng HH, Delany ME. 2016. Marek’s disease herpesvirus vaccines integrate into chicken host chromosomes yet lack a virus-host phenotype associated with oncogenic transformation. Vaccine 34:5554–5561. doi: 10.1016/j.vaccine.2016.09.051 [DOI] [PubMed] [Google Scholar]
- 50. Zhao Y, Xu H, Yao Y, Smith LP, Kgosana L, Green J, Petherbridge L, Baigent SJ, Nair V, Cullen BR. 2011. Critical role of the virus-encoded microRNA-155 ortholog in the induction of Marek’s disease lymphomas. PLoS Pathog 7:e1001305. doi: 10.1371/journal.ppat.1001305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nazerian K. 1987. An updated list of avian cell lines and transplantable tumours. Avian Pathol 16:527–544. doi: 10.1080/03079458708436402 [DOI] [PubMed] [Google Scholar]
- 52. Akiyama Y, Kato S. 1974. Scanning electron microscopy of lymphoid cell lines and lymphocytes from thymus, bursa and blood of chickens. Biken J 17:193–197. [PubMed] [Google Scholar]
- 53. Yao Y, Zhao Y, Xu H, Smith LP, Lawrie CH, Watson M, Nair V. 2008. MicroRNA profile of Marek's disease virus-transformed T-cell line MSB-1: predominance of virus-encoded microRNAs. J Virol 82:4007–4015. doi: 10.1128/JVI.02659-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kaufer BB. 2013. Detection of integrated herpesvirus genomes by fluorescence in situ hybridization (FISH). Methods Mol Biol 1064:141–152. doi: 10.1007/978-1-62703-601-6_10 [DOI] [PubMed] [Google Scholar]
- 55. Nishibori M, Hayashi T, Tsudzuki M, Yamamoto Y, Yasue H. 2001. Complete sequence of the Japanese quail (Coturnix japonica) mitochondrial genome and its genetic relationship with related species. Anim Genet 32:380–385. doi: 10.1046/j.1365-2052.2001.00795.x [DOI] [PubMed] [Google Scholar]
- 56. Jeyapalan JN, Mendez-Bermudez A, Zaffaroni N, Dubrova YE, Royle NJ. 2008. Evidence for alternative lengthening of telomeres in liposarcomas in the absence of ALT-associated PML bodies. Int J Cancer 122:2414–2421. doi: 10.1002/ijc.23412 [DOI] [PubMed] [Google Scholar]
- 57. Jeffreys AJ, MacLeod A, Tamaki K, Neil DL, Monckton DG. 1991. Minisatellite repeat coding as a digital approach to DNA typing. Nature 354:204–209. doi: 10.1038/354204a0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
STELA of iMDV-associated telomeres in six avian cell lines.