ABSTRACT
To mobilize nutrients entrapped into minerals and rocks, heterotrophic bacteria living in nutrient-poor environments have developed different mechanisms based mainly on acidolysis and chelation. However, the genetic bases of these mechanisms remain unidentified. To fill this gap, we considered the model strain Caballeronia mineralivorans PML1(12) known to be effective at weathering. Based on its transcriptomics and proteomics responses in Fe-depleted conditions, we pointed a cluster of genes differentially expressed and putatively involved in the production of siderophores. In this study, we report the characterization of this gene region coding for the production of a non-ribosomal peptide synthetase-independent siderophore (NIS). Targeted mutagenesis associated with functional assays and liquid chromatography coupled to high-resolution tandem mass spectrometry demonstrated the production of a single siderophore, identified as rhizobactin. This siderophore represents the first NIS containing malic acid in its structure. The evidence for the implication of rhizobactin in mineral weathering was demonstrated during a hematite dissolution assay. This study provides the first demonstration of the synthesis of a NIS in the genus Caballeronia and its involvement in mineral weathering. Our conclusions reinforce the idea that strain PML1(12) is particularly well adapted to nutrient-poor environments.
IMPORTANCE
This work deciphers the molecular and genetic bases used by strain PML1(12) of Caballeronia mineralivorans to mobilize iron and weather minerals. Through the combination of bioinformatics, chemical, and phylogenetic analyses, we characterized the siderophore produced by strain PML1(12) and the related genes. This siderophore was identified as rhizobactin and classified as a non-ribosomal peptide synthetase-independent siderophore (NIS). Contrary to the previously identified NIS synthetases that form siderophores containing citric acid, α-ketoglutarate, or succinic acid, our analyses revealed that rhizobactin contains malic acid in its structure, representing, therefore, the first identified NIS with such an acid and probably a new NIS category. Last, this work demonstrates for the first time the effectiveness at weathering minerals of a siderophore of the NIS family. Our findings offer relevant information for different fields of research, such as environmental genomics, microbiology, chemistry, and soil sciences.
KEYWORDS: bacteria, mineral weathering, NRPS-independent siderophore, rhizobactin, malic acid
INTRODUCTION
Iron represents an essential micronutrient required for living organisms. This element is used as a cofactor by many enzymes and is involved in a wide range of physiological processes such as respiration, DNA synthesis, or photosynthesis (1). While iron is the fourth most abundant element in the Earth crust, it is mainly found under insoluble forms (2, 3) in primary (e.g., biotite, granite; Fe3+/Fe2+) and secondary minerals (e.g., hematite, goethite, and magnetite; Fe3+) (4). In addition, iron is usually poorly bioavailable at neutral to basic pH and under oxygenic conditions. To deal with these limiting conditions and their physiological requirements, bacteria have developed a diversity of mechanisms to mobilize iron and other nutrients poorly available or entrapped into minerals and rocks, a global process termed mineral weathering (MWe).
In heterotrophic bacteria, two main MWe mechanisms have been evidenced: acidolysis and chelation (5). The first mechanism is related to the action of the by-products of the primary and secondary metabolisms of bacteria. Indeed, by consuming carbon substrates, bacteria acidify their local environment due to the production and accumulation of organic acids (e.g., citrate, gluconate) and protons. Such acidification allows the dissolution of the minerals and the release of their nutritive content. The effectiveness of this mechanism is clearly associated with the carbon substrates consumed, with glucose usually giving stronger acidification (6, 7). The second mechanism is based on the production of chelating compounds such as organic acids (e.g., oxalate) and siderophores. Bacteria can weather minerals using one or both mechanisms (i.e., acidolysis and chelation) depending on the bacterial strain, the nutrient availability, the type of mineral, and the ionic strength of the local environment (i.e., buffering capacity).
Many microorganisms, such as bacteria and fungi, can produce one or more type(s) of siderophore. These particular metabolites are low-molecular-weight compounds, characterized by a strong affinity for iron (8), although they can also chelate other divalent and trivalent cations such as Al, Mg, or Cr (9, 10). Siderophores are produced when microorganisms experience nutrient limitation and especially iron deficiency (11). In this sense, concentrations up to 2 and 12 nM of siderophores have been measured in the solution of nutrient-poor soils (12). To date, two classes of siderophores have been identified: (i) the non-ribosomal peptide synthetase (NRPS) siderophores (e.g., pyoverdin) and (ii) the NRPS-independent siderophores (NIS) (e.g., aerobactin) (13). The contribution of siderophores to MWe has been known for a long time for different types of molecules (i.e., desferrioxamine) in abiotic experiments (14). However, under such experimental conditions, the use of relatively high concentrations of siderophore did not allow to appreciate how microorganisms produced and regulated this production according to their local environment (i.e., pH, ionic strength, nutrient availability) (9, 15). Despite a constant increase in the number of new siderophores identified, their role in MWe has rarely been assessed in a cellular context (6, 16, 17). Indeed, few studies have considered how the MWe ability of a mutant impaired in siderophore production compares to that of the wild-type (WT) strain and how the nutrient limitations impact the action of the siderophore-mediated weathering.
In this study, we have focused on the model strain PML1(12) from the genus Caballeronia mineralivorans, which has been isolated from a nutrient-poor soil and is known to be effective at MWe and at promoting plant growth (7, 18). However, the molecular basis of its MWe ability is not fully elucidated. The dual transcriptomic and proteomic analyses recently applied on mineral/bacteria interactions highlighted the specific increased expression of a cluster of genes related to iron mobilization and the production of a potential siderophore under Fe-depleted conditions (7). Among these genes, the most highly expressed presented high homology with a putative diaminopimelate decarboxylase, cysteine synthase, and IucA/IucC synthetase in conditions devoid of iron (7). In this context, the objectives of this study were to (i) characterize the cluster of genes conferring the ability to mobilize iron of strain PML1(12); (ii) purify and characterize the siderophore produced by strain PML1(12); (iii) quantify the contribution of siderophore production on mineral weathering; and (iv) study the conservation of the genes involved in siderophore production in the Burkholderia, Caballeronia, Paraburkholderia (BCP) group. To do it, different bioinformatic tools were combined to characterize the cluster of genes related to the production of the NIS produced by strain PML1(12) and to determine the relative conservation of this cluster of genes in the genome of different strains from the BCP group. The use of a mutant impaired in its ability to produce the siderophore allowed for the determination of its role in hematite weathering. Ultra-high-performance liquid chromatography coupled with high-resolution mass spectrometry allowed the identification of the chelating molecule.
RESULTS
A single genomic region is potentially involved in siderophore production
The Chrome Azurol S (CAS) assay performed on strain PML1(12) demonstrated its ability to mobilize iron as stated by the formation of a yellow halo around the colony, suggesting the production of one or several siderophores. An antiSMASH analysis of the PML1(12) genome identified a single siderophore production locus. This region is characterized by 10 genes, among which a single gene encoding a protein of 635 amino acids, we named RhiE, presenting the characteristic signature of the iucA/iucC domain. Such domain is associated with the synthesis of enzymes involved in the production of NIS. The other genes present in the region upstream and downstream of the rhiE gene could be involved in siderophore production (rhiBCD, intermediate production of the siderophore), transport (rhiA, TonB transporter; rhiF, MFS transporter; rhiGHI, ABC systems) and in the regulation of siderophore production (rhiJ). The detail of the functions encoded by each gene of the rhiABCDEFGHIJ cluster is presented in Table 1.
TABLE 1.
Description of the NRPS-independent gene region involved in siderophore production by Caballeronia mineralivorans strain PML1(12)
| Putative function | Accession number (NCBI) | Length | Gene | Annotation (based in BLASTp homology) | Homology |
|---|---|---|---|---|---|
| Transport | KLU26370.1 | 745 | rhiA | TonB-dependent siderophore receptor | 88.19% Paraburkholderia sp. BL23IN1 (WP_120293907.1) |
| Synthesis | KLU26369.1 | 480 | rhiB | Y4yA family PLP-dependent enzyme | 80.62% Paraburkholderia haematera (CAE6738131.1) |
| Synthesis | KLU26368.1 | 346 | rhiC | Cysteine synthase family protein | 93.64% Paraburkholderia sp. BL23IN1 (WP_120293908.1) |
| Synthesis | KLU26367.1 | 388 | rhiD | NAD/NADP octopine/nopaline dehydrogenase family protein | 87.37% Paraburkholderia haematera (WP_236066808.1) |
| Synthesis | KLU26366.1 | 635 | rhiE | iucA/iucC family siderophore biosynthesis protein | 85.83% Paraburkholderia sp. BL23IN1 (WP_120293909.1) |
| Transport | KLU26365.1 | 421 | rhiF | MFS transporter | 85.34% Paraburkholderia haematera (WP_211611243.1) Paraburkholderia sp. BL23IN1 (WP_120293910.1) |
| Transport | KLU26364.1 | 398 | rhiG | ABC transporter substrate-binding protein | 88.69% Paraburkholderia haematera (WP_211611244.1) |
| Transport | KLU26363.1 | 353 | rhiH | Iron ABC transporter permease | 92.63% Paraburkholderia sp. BL23IN1 (WP_120293912.1) |
| Transport | KLU26362.1 | 265 | rhiI | ABC transporter ATP-binding protein | 92.83% Paraburkholderia sp. BL23IN1 (WP_120293913.1) |
| Regulation | KLU26361.1 | 138 | rhiJ | Transcriptional regulator/GNAT family N-acetyltransferase | 87.71% Caballeronia sordidicola (WP_031357714 |
Each gene product of the cluster is presented by its NCBI accession number. The annotation is based on BLASTp homology. NIS synthetases showing the highest homology with RhiE of strain PML1(12) are shown.
The rhiABCDEFGHIJ region confers the ability to mobilize iron to strain PML1(12)
To investigate the implication of this region in siderophore biosynthesis, a rhiE::Gm knock-out mutant was constructed and tested on solid CAS assay to compare its siderophore production with that of the WT strain (Fig. S1). After 7 days of incubation, the WT strain presented a yellow halo of ca. 1.7 cm, while no halo was visible for the rhiE::Gm mutant strain. The lack of production of chelating molecules by the rhiE::Gm mutant strain was confirmed with the liquid CAS assay, which showed a very slight optical density (OD) measured at 655 nm (OD655nm) decrease from 0.48 ± 0.01 to 0.43 ± 0.01 between the non-inoculated treatment and the rhiE::Gm mutant, when the decrease reached a final value of 0.11 ± 0.01 for the WT strain indicative of a trapping of iron from the CAS-Fe(III) complex.
The rhiABCDEFGHIJ cluster permits the growth of strain PML1(12) in iron-deficient condition
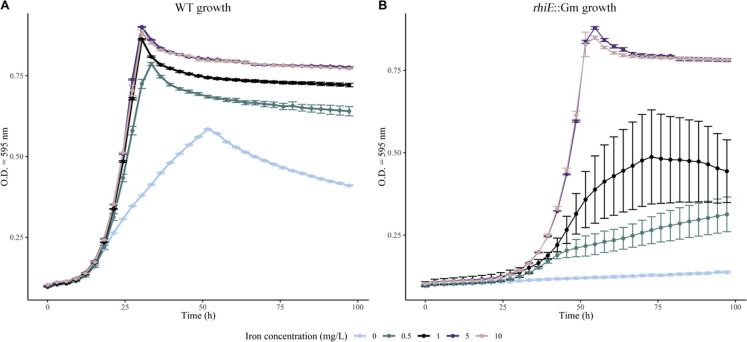
The growth capabilities of the WT and rhiE::Gm strains were measured in a medium devoid of iron (ABm-Fe) amended or not with various concentrations of iron (0, 0.5, 1, 5, and 10 mg L−1 of Fe) (Fig. 1). In absence of iron, the growth of the WT strain was delayed and reached a maximum OD595nm value of 0.58 ± 0.01. The growth rate in absence of iron was significantly lower (P < 0.05) than that observed in the presence of iron at 5 or 10 mg L−1 concentration (OD595nm = 0.88 ± 0.01). The rhiE::Gm mutant strain reached the same OD595nm as the WT strain when growing at the highest iron concentrations (i.e., 5 and 10 mg L−1), but with an important growth delay. The maximal OD595nm was observed at 50 h vs 26 h for the rhiE::Gm and WT strains, respectively. The largest growth differences (i.e., maximal OD595nm and delay) between WT and rhiE::Gm strains were observed when iron concentrations were below 1 mg L−1 in the solution.
Fig 1.
Growth assay. The growth of the WT strain (A) and rhiE::Gm mutant (B) was monitored in presence of different iron concentrations (i.e., 0, 0.5, 1, 5, and 10 mg L−l of Fe) in AB medium depleted of iron. The growth was studied under orbital shaking at 25°C for 90 h. The absorbance was measured at 600 nm every 3 h. Each dot is the mean of independent triplicates.
Inhibition of the siderophore production according to Fe concentration
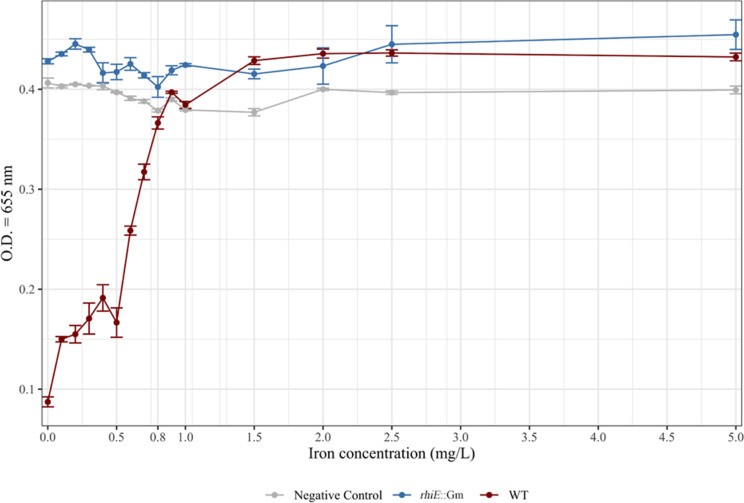
As the production of chelating molecules is usually dependent on Fe concentration in the medium, we assayed the production of siderophores in a range of concentrations of Fe from 0 to 5 mg L−1. After a 3-day incubation period in ABm-Fe amended or not with iron (i.e., FeCl3), the analyses done on the culture supernatant revealed the lack of chelating activity in the non-inoculated controls and in the WT cultures in presence of iron concentrations at 0.8 mg L−1 (Fig. 2), while this activity was detectable at 0.7 mg L−1 and below. Chelating activity reached its maximum in absence of iron in the medium. The same experiment performed with the rhiE::Gm mutant did not reveal any chelating activity whatever the iron concentration tested.
Fig 2.
Impact of increasing concentrations of iron on the chelating activity of the WT and mutant (rhiE::Gm) strains. To determine the concentration of iron inhibiting siderophore production, liquid cultures of WT (red) and rhiE::Gm (blue) strains were performed with different concentrations of iron (i.e., 0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1, 1.5, 2, 2.5, and 5 mg L−1). Pure water (gray) was used as a negative control. After 3 days, the absorbance at 655 nm was measured. The decrease of the measured absorbance means the presence of chelating molecules since the media coloration changes from blue to yellow and the absorbance at 655 nm measures blue color intensity.
Hematite weathering
The ability of the chelating molecule(s) produced by strain PML1(12) to weather minerals was evaluated considering hematite, an iron-rich mineral, as the sole source of iron in our experimental conditions. To limit the impact of acidification on the mineral weathering process, experiments were done in buffered ABm-Fe medium over 7 days of incubation. The pH of the medium was controlled before and after the experiments in presence/absence of bacterial inoculum and hematite and remained stable (pH 6.1). In absence of hematite, the WT strain grew in the buffered ABm-Fe medium to an OD595nm of 0.47 ± 0.01. The presence of hematite in the medium significantly increased growth of the WT strain (OD595nm = 0.82 ± 0.01), while hematite addition had no impact on growth of the rhiE::Gm mutant (OD595nm = 0.45 ± 0.01 in presence vs OD595nm = 0.44 ± 0.02 in absence of the mineral). The CAS assay confirmed that only the WT strain was presenting an active chelating activity in both conditions (e.g., in the presence or in the absence of hematite). Quantification of the iron released in solution from hematite highlighted significantly higher concentrations of iron for the WT strain (Fe = 0.76 ± 0.02 mg L−1; P < 0.05) than for the rhiE::Gm mutant (0.04 ± 0.02 mg L−1) and the non-inoculated control (0.06 ± 0.02 mg L−1) (Fig. 3).
Fig 3.
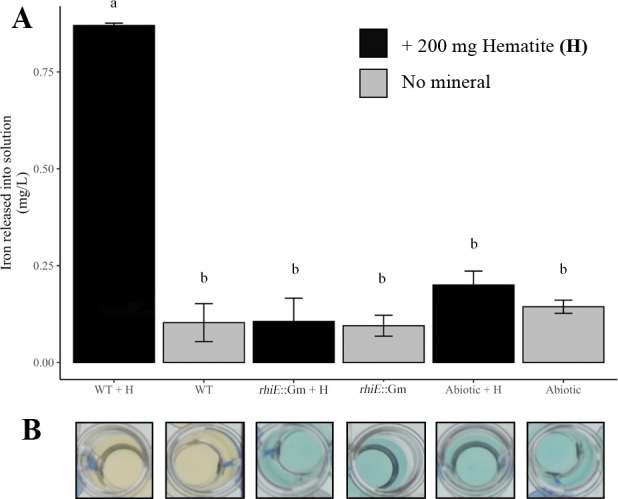
Hematite weathering potential of strain PML1(12) and its rhiE::Gm mutant. The weathering ability was evaluated by the measure of iron released from hematite in ABm medium devoid of iron, after 7 days of incubation at 25°C under agitation (200 rpm). (A) Concentration of iron released in solution. The iron released from hematite was measured by ferrospectral determination (optical density measured at 595nm). Abbreviations were used as follows: WT + H, wild-type strain plus hematite; WT, wild-type strain; rhiE::Gm + H, mutant strain plus hematite; rhiE::Gm, mutant strain; abiotic + h, non-inoculated condition plus hematite; abiotic, non-inoculated condition. Non-inoculated conditions (abiotic control with and without hematite) were used as controls. Samples with the same letter indicate no significant difference (P < 0.05). (B) Siderophore activity. The siderophore activity of the different samples was determined using the CAS method. The yellow color indicates siderophore activity. For each measure, the results are the mean of independent triplicates.
Rhizobactin is the siderophore produced by strain PML1(12)
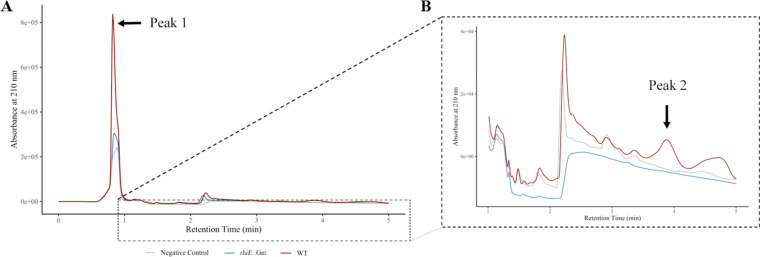
Pre-purified supernatants from 3-day cultures of rhiE::Gm mutant and WT strains were analyzed by high-performance liquid chromatography (HPLC) after purification. For this experiment, the growth was performed in ABs-Fe with succinate as sole carbon source to limit the production of exopolysaccharides, which are not compatible with the purification steps. The comparison of the HPLC chromatograms from the WT and rhiE::Gm strains as well as from the medium alone highlighted one major difference, a peak (peak 1) with a retention time of 0.84 min in the WT supernatant. This peak was 2.8 and 3.8 times more intense for the WT strain than for the rhiE::Gm mutant strain and the non-inoculated medium, respectively (Fig. 4A). To correlate the HPLC chromatograms with the presence of chelating activities, the HPLC fractions were collected and assayed using the CAS assay. The CAS assay was positive for two fractions for the WT samples. The first positive fraction with the CAS assay (OD655nm = 0.06) was collected between 0.77 and 0.97 min corresponding to peak 1 region. The second positive fraction (OD655nm = 0.14) was identified between 3.74 and 3.95 min. Detailed analysis of the HPLC profiles within this retention time highlighted the presence of a small peak (peak 2), only for WT strain (Fig. 4B).
Fig 4.
HPLC analysis and UV detection. The pre-purified supernatant of the WT strain (red), rhiE::Gm mutant (blue) and non-inoculated medium (gray) were monitored at 210 nm after HPLC separation using a Milli-Q water/acetonitrile + 0.2% trifluoroacetic acid gradient for 25 min at 1 mL min−1. To determine the presence of siderophore activity, fractions were collected during the HPLC run and tested using the liquid CAS assay. Positive fractions for siderophore presence are highlighted with black arrows. (A) A major peak (peak 1) at retention time of 0.84 min was identified. (B) A second peak (peak 2) of lower intensity, with a retention time of 3.88 min, was identified after CAS test.
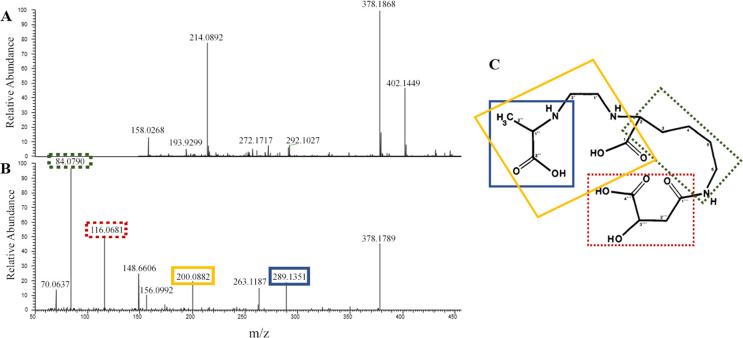
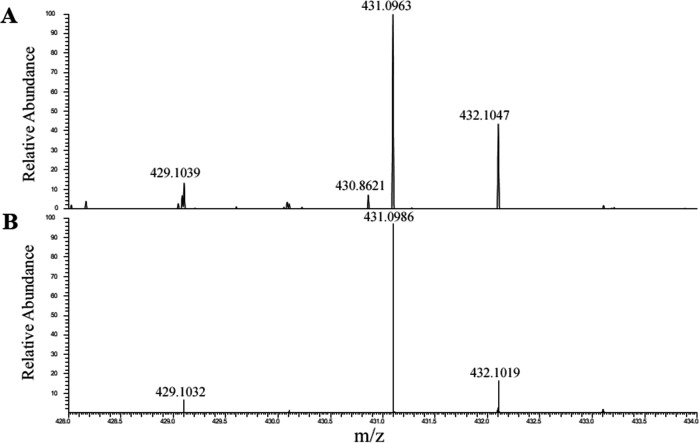
To identify the siderophore produced by strain PML1(12), we performed an HPLC-photodiode array detector (PDA)-high-resolution tandem mass spectrometry (HRMS/MS) analysis of the two fractions mentioned above. High-resolution MS analysis revealed the presence of a single molecule with the same exact mass of 377.1868 Da (monoprotonated ion seen at m/z = 378.1868) in both active fractions and with the theoretical formula C15H25O8N3 (Fig. 5A). Such compound was found only in the WT fractions (and not in the rhiE::Gm strain, nor in the negative control). An additional MS signal with m/z = 431.0982 was also specifically observed in the two active WT fractions and was unambiguously attributed to the ferric complex [(C15H25O8N3-2H) + 56Fe(III)] + (m/ztheo = 377.1788-2 × 1.008 + 55.935 =431.0978, with 56Fe = 55.935 u and 1H = 1.008 u). Indeed, the experimental isotopic pattern was in perfect agreement with the simulated one (Fig. 6), thanks to the clear observation of the contribution of 54Fe isotope at m/z = 429.1029 {[(C15H25O8N3-2H) + 54Fe(III)]+}. The previous mass spectrometric observations allow to conclude with certainty that compound of exact mass M = 377.1788 Da can bind iron (III) efficiently and corresponds to the siderophore produced by strain PML1(12).
Fig 5.
Identification, fragmentation profile, and chemical structure of rhizobactin. (A) Mass spectrum of HPLC positive fractions showing the parent ion of the putative chelating molecule at m/z = 378.1868 (raw formula: C15H25O8N3). (B) Fragmentation pattern of the chelating molecule seen at m/z = 378.1868, revealing a perfect agreement with previous experimental MS2 results done on the rhizobactin identified in Sinorhizobium meliloti DM4 by Smith et al. (19) and in Pseudomonas sp. FEN by Kügler et al. (20). The major product ions are as follows: 289.1351 (in blue, consecutive to alanine loss), 200.0882 (in yellow, formed as a result of loss of ethylenediamine and alanine and an additional carboxylic group), 116.0681 (in red, corresponding to malic acid ion-H2O), and 84.0790 (in green, corresponding to decarboxylated lysine ion). (C) Chemical structure [according to Smith et al. (19)] and fragmentation pattern of the rhizobactin. Same colors as in panel B are used to highlight the correspondence of the chemical moieties of rhizobactin and product ions. Dotted boxes are used to highlight lost ions, while undotted boxes mark the remaining structure of the molecule.
Fig 6.
Isotopic pattern of iron complexes. Comparison between (A) the experimentally observed isotopic pattern of the putative iron(III) siderophore complex ion (m/z = 431.0963) and (B) the theoretical isotopic profile of the complex [(C15H25O8N3-2H)+FeIII]+, C15H25O8N3 being the raw formula associated with the putative siderophore of m/z = 378.1868.
Such exact mass of 377.1868 Da has been previously associated with the siderophore rhizobactin by Smith et al. (19) in a strain of Ensifer meliloti and Kügler et al. (20) in the strain Pseudomonas sp. FEN. In order to elucidate the siderophore highlighted in PML1(12), an MS/MS experiment was performed (Fig. 5B). Noticeably, our analyses identified the same four major product ions characterizing the rhizobactin described by Kügler et al. (20) (Fig. 5C) as follows: (i) m/z = 289.1351 (formed as a result of loss of alanine), (ii) m/z = 200.0882 (formed as a result of loss of ethylenediamine and alanine and an additional carboxylic group), (iii) m/z = 116.0681 (identified as malic acid ion), and (iv) m/z = 84.0790 (identified as decarboxylated lysine ion). Overall, these analyses demonstrate that PML1(12) produces rhizobactin, a siderophore composed of a lysine, an alanine, an ethylenediamine group, and malic acid. This work represents the first demonstration that rhizobactin is an NRPS-independent siderophore. Furthermore, it confirms that rhizobactin is based on malic acid and not on other known NIS substrates (citric acid, α-ketoglutarate, or succinic acid).
Comparative genomics of rhizobactin synthesis
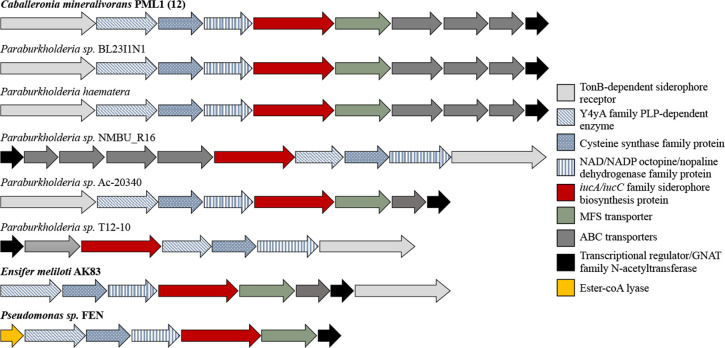
A BlastP search of the nr database using the RhiE protein as template allowed the identification of several homologous proteins presenting a percentage of identity ranging from 64% to 85% and good sequence coverage (95% to 100%). The best hits appeared assigned to the genera Paraburkholderia, Trinickia, Vogesella, Polaromonas, or Pseudomonas. Noticeably, a homolog was found in strain FEN of Pseudomonas sp. (63.5% homolog to RhiE; NCBI accession number WP_191487624.1), a strain described previously for its ability to produce rhizobactin, but in which the synthesis genes had not been identified. Focusing on the Burkholderiaceae, we identified 38 proteins presenting a percentage of identity ranging from 34% to 85% and belonging to the genus Paraburkholderia, Trinickia, Cupriavidus, and Ralstonia. Considering the BCP group, besides the homologs in Caballeronia, we found RhiE homologs only in five strains assigned to Paraburkholderia (P. sp. BL23I1N, P. haematera, P. sp. T12-10, P. sp. NMBU_R16, and P. sp. Ac-20340), organized almost identically as in PML1(12) (i.e., cluster rhiABCDEFJ) (Fig. 7), but with a variable number of genes. Noticeably, the ABC transport systems were not always present.
Fig 7.
Organization of the cluster of genes involved in rhizobactin biosynthesis in different strains. Each arrow represents one gene, and its size was scaled according to gene length. Gene annotation is shown in the right, and the complete description is provided in Table 1. Light gray, dark gray, and green genes are involved in siderophore transport. In blue, red, and yellow, genes are involved in siderophore biosynthesis. In black, genes involved with siderophore production regulation. In bold, strains for which rhizobactin synthesis was demonstrated.
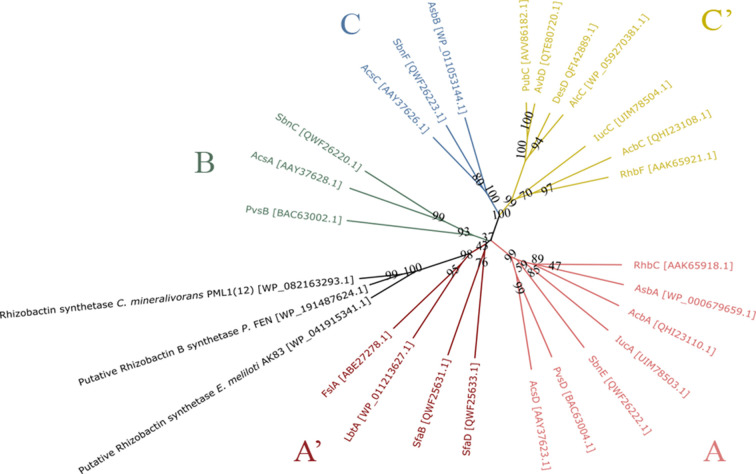
The phylogenetic analysis of NIS synthetases usually clusters them according to their substrate specificity (13). To position the RhiE synthetase of strain PML1(12) in the NIS classification and to determine its relatedness to the known NIS synthetases, we constructed a N-J phylogenetic tree of a representative set of known NIS synthetases (Fig. 8). A total of 24 protein sequences of NIS synthetases which function and substrate are known were considered, as well as RhiE protein sequence of PML1(12) and the two homologous RhiE protein sequences identified in Pseudomonas FEN and Ensifer (Sinorhizobium) meliloti AK83 which are also potentially involved in rhizobactin production. Overall, our results confirm that NIS synthetases are distributed according to their substrate specificity (13, 21–23). The newly characterized RhiE NIS synthetase of strain PML1(12), as well as the two homologs from Pseudomonas and Ensifer, cluster in group A’, forming a well-supported sub-branch within this group. Cluster A’ is the only in the phylogeny which does not seem to follow substrate specificity. Indeed, as demonstrated in the current work, RhiE and homologs synthesize rhizobactin, which is based on the use of malic acid as substrate (see above), while other NIS from this cluster use citric acid to synthesize siderophores such as rhizoferrin. The low support of the tree at the base of this cluster (bootstrap values of 37 and 42) may indicate a problem of phylogenetic resolution using the current set of data.
Fig 8.
Neighbor-joining phylogenetic analysis of NIS synthetases. A total of 24 experimentally validated and two proposed NIS-synthetases were aligned using MUSCLE algorithm, and the phylogenetic tree was constructed using SeaView. The code between brackets corresponds with the NCBI accession number for each protein sequence. NIS synthetases are colored depending on their specificity of substrate and thus their group: A (pink), A’ (red), B (green), C (blue), and C’ (yellow). The enzymes presented in this study (i.e., RhiE and putative rhizobactin synthetases) are shown in black.
The comparison of the complete genomic region of the cluster of genes involved in rhizobactin transport and production (rhiABCDEFGHIJ) (Table 1) found in strain PML1(12) with the genomes of the strains already known to produce rhizobactin (Ensifer meliloti and Pseudomonas sp. FEN) highlighted a partial conservation of this cluster (i.e., rhiBCDEF). The clusters differed, however, in several instances: (i) the presence of a TonB system in the upstream region in strain PML1(12) and at the downstream end in strain AK83 (AEG58212.1) and (ii) the absence of two ABC-transport systems (rhiHI) and the transcriptional regulator (rhiJ) in strains AK83 and FEN (Fig. 7).
DISCUSSION
Rhizobactin identification
Rhizobactin was first described in the plant-associated bacterial strain Ensifer meliloti DM4 (former Sinorhizobium meliloti) (19). This first study provided a detailed chemical composition based on mass spectrometry and NMR analyses, revealing that this carboxylate siderophore was formed by an ethylenediamine group coupled with alanine, lysine, and malic acid. The same siderophore was reported more recently for a bacterial strain isolated from a nutrient-poor environment, the strain Pseudomonas sp. FEN (20). Both studies provided the chemical characterization of this molecule, but neither the genetic characterization nor the classification of rhizobactin. Interestingly, Pseudomonas sp. FEN can produce rhizobactin and rhizobactin B, rhizobactin B being the main siderophore produced. Structurally, these two siderophores differ only by the presence of a methyl group at the C2 position of malic acid, giving a citramalic acid in rhizobactin B instead of a malic acid in rhizobactin. A third rhizobactin type was identified in strain 1021 of Ensifer meliloti (i.e., rhizobactin 1021), but completely different in terms of chemical structure from the one described above. This hydroxamate siderophore of 530 Da is based on a citrate derivative with the carboxyl groups linked to two different side chains (23). In our study, we identified the siderophore produced by strain PML1(12) using a combination of CAS assay, HPLC purification, and MS/MS analyses. This molecule not only binds the different iron isotopes but also presented an exact mass (377.1868 Da) and a fragmentation pattern like that of the rhizobactin identified in Ensifer meliloti strain DM4 and Pseudomonas sp. strain FEN (19, 20). Our rhiE::Gm mutant of strain PML1(12), which is unable to chelate iron, proves the involvement of the rhiE gene cluster in the production of the unique siderophore molecule produced by strain PML1(12). Noticeably, the rhizobactin gene cluster appeared relatively conserved in the two strains already described for the production of rhizobactin, although the strain Pseudomonas sp. FEN presented an additional gene identified as a citrate lyase (WP_191487621.1), which is the first gene of the cluster in this strain, upstream of the rhiE homolog. Citrate lyase may be involved in the process of transformation of malic acid into citramalic acid, explaining why rhizobactin B is the main siderophore produced by strain Pseudomonas sp. FEN, while rhizobactin is produced by Ensifer meliloti AK83 and strain PML1(12).
Rhizobactin as NRPS-independent siderophore
Two groups of bacterial siderophores are currently described according to their biosynthetic pathway. The first pathway uses complex, modular, multidomain enzymes known as NRPS (6, 13, 24). The second relies on NRPS-independent synthetases (13, 25). In comparison to the wealth of information available on the genetics and chemistry of the NRPS siderophores, NIS and their related biosynthetic pathways are still poorly documented (25). This is an important point because siderophores cannot be categorized as NIS or NRPS without a proper genetic characterization. In this sense, rhizobactin was discovered and chemically characterized by Smith et al. (19), as early as 1984, in strain Ensifer meliloti DM4, but could only be classified as an NIS thanks to the current study and its homology at chemical (C15H25O8N3) and genomic level (gene cluster) to that of strain PML1(12). The strain PML1(12) presents a gene cluster of 10 genes (Fig. 7) with the characteristic signature of the NIS synthesis pathway as identified for the first described NIS (i.e., aerobactin) (26, 27). The aerobactin synthetases, IucA and IucC, became, since that, the hallmarks of the presence of NIS in genomes (22), either in a single copy such as for the synthesis of rhizoferrin (e.g., FslA) in Francisella tularensis (28) or rhizobactin in strain PML1(12) (e.g., RhiE, this study) or either in several copies such as for the synthesis of aerobactin (e.g., IucA and IucC) (27). Our findings will enrich the NIS database and improve the siderophore determination using AntiSMASH.
Rhizobactin a new class of NIS?
NIS synthetases traditionally separate in five phylogenetic groups (A, A’, B, C, and C’). This classification which was obtained on experimentally demonstrated NIS synthetases is congruent with their substrate specificity with: (i) type A and A’ specific for citric acid, (ii) type B for α-ketoglutarate intermediates, and (iii) type C and C’ specific for citryl or succinyl-based intermediates (13). The number of experimentally demonstrated NIS synthetases is limited, and consequently, their position in phylogenetic trees may change with the evolution of NIS studies. As an example, the SfaB and SfaD enzymes responsible for the synthesis of staphyloferrin A which presents a citric acid in its structure (29) were initially included in group B, a group of NIS synthetases specific for α-ketoglutarate. With the discovery of other novel citric acid-based and chiral siderophores (e.g.., rhizoferrin, legiobactin), SfaB and SfaD now cluster within a new group, named A’. Our phylogenetic tree agrees with those previously published (13, 21, 22), confirming that the NIS synthetases mostly cluster according to their substrate specificity. In this respect, the rhizobactin synthetase clusters with group A’, which corresponds to citrate-based and chiral siderophores. However, rhizobactin and rhizobactin B synthetases form a well-supported branch within group A’. Malic acid (rhizobactin) and citramalic acid (rhizobactin B) have never been described previously as possible substrates for NIS synthetases. Therefore, the difference of substrate specificity between rhizobactin synthetases (malic for rhizobactin and citramalic acid for rhizobactin B) and synthetases from group A’ (citric acid) and the absence of other already characterized synthetases with malic acid as substrate allow us to propose a new group of NIS called D. The future characterization of new NIS synthetases that use malic/citramalic acid as substrate should make more robust the separation from A’ group, as it happened previously for groups B and A’ (29).
Role of rhizobactin in mineral weathering
Iron is considered as an essential element, and its deficiency alters the growth of bacteria (6, 30, 31). Strain PML1(12) follows this rule, with a strong delay and decrease in growth in iron deprived conditions compared to non-limiting conditions. To adapt to low-iron conditions, bacteria can produce siderophores, whose production is regulated by the quantity of iron available and stopped at a maximum of 5 mg L−1 of Fe (32). Here, we showed that the production of rhizobactin by strain PML1(12) was detectable until an iron concentration of 0.7 mg L−1. Such iron regulation fits with the observations done by the dual transcriptomic and proteomic analyses on strain PML1(12) in the presence/absence of a mineral carrying iron (i.e., biotite), which evidenced that the genes involved in siderophore production were only up-regulated in Fe-depleted conditions and in the absence of mineral (7). Although minerals represent an important source of iron, the production of siderophores remains strongly determined by the iron bioavailability and the local conditions (pH, ionic strength). Several studies have demonstrated that, in oligotrophic conditions, minerals may be seen as a source of iron if siderophore-producing bacteria are present in the environment (33). The effectiveness of siderophores at solubilizing iron from different minerals is known and demonstrated for a long time in abiotic conditions. As an example, desferrioxamine B was proved to weather Fe from several iron oxides (15). Even more, rhizoferrin and desferrioxamine A and B are not only effective at mobilizing Fe from hydroxide minerals, but also other metals such as chromium (34, 35). However, such abiotic experiments were usually based on high concentrations of siderophores and did not permit to consider all the regulation occurring in bacteria according to nutrient availability, ionic strength of the medium, and mineral type. In contrast, few studies investigated the ability of siderophore-producing bacteria to liberate nutrients trapped in minerals in a cellular context, considering mutants impaired in their ability to produce siderophores. Our study represents the first demonstration of the role of an NIS (i.e., rhizobactin) produced in biotic conditions in the weathering of hematite. Such demonstration completes the list of siderophores already characterized for their role in mineral weathering (6, 16, 17, 33).
Occurrence of the rhizobactin siderophore in the BCP group
Among members of the same genus or species, the type of siderophore produced can strongly differ (i.e., rhizoferrin biosynthesis) (36, 37) or on the contrary be very conserved (i.e., pyoverdine biosynthesis) (38). Members of the Burkholderia sensu lato are well known for the different strategies they use to mobilize iron based on siderophore-independent (Ftr) (39) and siderophore-dependent (e.g., ornibactin) (40, 41) systems, but not yet based on an NIS. Our study represents the first evidence of the production of rhizobactin in this group. Based on this evidence, we evaluated the distribution of a homologue of the Caballeronia mineralivorans strain PML1(12) RhiE NIS synthetase and the conservation of the rhi cluster of genes. Our analysis revealed that homologs of RhiE can be found in many genera among beta-proteobacteria, but the most represented remains the Pseudomonas genus. In this context, Pseudomonas sp. FEN is the only strain characterized for its ability to produce rhizobactin, while many Pseudomonas strains have been described for their ability to produce NRPS siderophores (e.g., pyoverdin) (38). Focusing on the BCP group (including Burkholderia, Paraburkholderia, and Caballeronia), only five Paraburkholderia strains (P. sp. BL23I1N, P. haematera, sp. T12-10, P. sp. NMBU_R16, and P. sp. Ac-20340) presented the rhizobactin cluster, questioning its origin. In terms of ecology, many of the strains we identified to possess a rhizobactin synthesis locus homolog originate from the rhizosphere (e.g., Paraburkholderia sp. T12-10, R16, and Ac-20340 and most of the members of Trinickia, Vogesella, Polaromonas, or Pseudomonas) and nutrient-poor ecosystems suggesting a role in plant nutrition and adaptation to nutrient-poor conditions. The role of siderophore-producing bacteria in plant nutrition and growth promotion has already been demonstrated as in the case of the strains Burkholderia pyrrocinia JK-SH007 (40) or Caballeronia mineralivorans PML1(12) (42, 43).
Conclusion
It is clearer and clearer that the ability of bacteria to weather minerals is finely regulated by local conditions and physiological processes. Nutrient availability, ionic strength of the medium, and the type of mineral and of the carbon substrate metabolized strongly determine the mechanism used by bacteria and the effectiveness at weathering. While the global mechanisms are identified (i.e., acidification, chelation, oxido-reduction), the genetic and geochemical bases remain poorly documented. Demonstrations in a cellular context (i.e., effect of a gene mutation) are essential to cover without a priori the diversity of mechanisms used by phylogenetically different bacteria and to decipher the regulations of these mechanisms occurring according to the mineral type and the local conditions. The combination of molecular and (geo-)chemical techniques used here allowed not only the identification of the siderophore produced by strain Caballeronia mineralivorans PML1(12) as rhizobactin but also its classification as a NIS and its contribution to mineral weathering. Our study provides new information regarding the complexity of mechanisms used by this model strain according to the local conditions and its metabolism. Our results highlight that siderophores play an essential role in mineral weathering under buffered and non-acidifying conditions, while organic acid and proton-based mechanisms represent the main mineral weathering mechanism used by this strain under non-buffered and acidifying conditions. The ability of rhizobactin to weather more complex and less weatherable minerals, with a lower iron content (e.g., biotite) remains to be determined. Together, our results clearly highlight how the strain PML1(12) is well adapted to live in nutrient-poor environments and to develop complex and finely regulated mechanisms to mobilize nutrients from minerals.
MATERIALS AND METHODS
Bacterial strains and growth media
Bacterial strains and plasmid are listed in Table 2. The model Caballeronia mineralivorans strain PML1(12) was isolated from the ectomycorrhizosphere of Scleroderma citrinum associated with oak [for sampling details see Calvaruso et al. (44)].
TABLE 2.
List of bacterial strains, constructions, and primers used in this studya
| Strains, plasmids, or primers | Characteristics | Reference |
|---|---|---|
| Strains | ||
| Caballeronia mineralivorans | ||
| Strain PML1(12) | Wild-type strain | (18) |
| rhiE::Gm [PML1(12)(rhiE::Gm)] | Wild-type strain, with GmR cassette inserted in double crossing over | This study |
| Escherichia coli | ||
| DH5a | supE44, ΔlacU169, (ΦlacZΔM15), recA1, endA1, hsdR17, thi-1, gyrA96, relA1 | Lab collection |
| S17.1λpir | Tpr SmR recA, thi, pro, hsdRM+, RP4::2-Tc::Mu::Km Tn7 λpir | Lab collection |
| Plasmids | ||
| pUC1318 | AmpR, GmR | Lab collection |
| pGEM-T Easy | AmpR | Kit Promega |
| pGEM-rhiE_SmaI | pGEM-T Easy carrying a rhiE gene | This study |
| pGEM-rhiE::Gm | pGEM-rhiE::Gm with insertion in SmaI of the GmR cassette | This study |
| pK19mob | KmR | Lab collection |
| pK19mob-rhiE::Gm | pK19mob containing a EcoRI fragment obtained from pGEM-rhiE::Gm | This study |
| Primers (Tm = 54°C) | ||
| For_rhiE | 5′-ATGAAGACCACTCCTTCGCTATT-3′ | This study |
| Rev_rhiE_SmaI | 5′-GCACCGGTCCCGGGAGCCGCGGCAT-3′ | This study |
| For_rhiE_SmaI | 5′-ATGCCGCGGCTCCCGGGACCGGTGC-3′ | This study |
| Rev_rhiE | 5′-TCAACCCTCGTTGTGCGCCGCCAGC-3′ | This study |
GmR, KmR, SmR, and AmpR indicate gentamicin, kanamycin, spectinomycin, and ampicillin resistance cassettes, respectively.
Strains, with the exception of Escherichia coli, were grown in AB medium (45) supplemented with mannitol (ABm) or succinic acid (ABs) (2 g L−1 final concentration) at 25°C. A version of the AB medium devoid of iron was also used (AB-Fe). E. coli strains were grown in Luria-Bertani at 37°C. When required, gentamycin, kanamycin and ampicillin were added to the media at a final concentration of 20, 50, and 100 µg mL−1, respectively.
Preparation of bacterial inoculum of Caballeronia mineralivorans strain PML1(12) and its mutant
For each of the assays presented in this work, the WT strain and the rhiE::Gm mutant strain were recovered from glycerol stock (−80°C) and grown on solid ABm medium (25°C) for 2 days. One isolated colony of each strain was inoculated in 10 mL of liquid ABm medium and incubated 3 days at 25°C under 150 rpm agitation. Liquid cultures were centrifugated at 9,000 × g for 15 min at 4°C, and the pellet was washed three times with sterile Milli-Q water. The pellet was resuspended in sterile Milli-Q water, and the optical density was adjusted to OD595nm= 0.9 ± 0.03 (corresponding to 2.109 cell/mL).
rhiE::Gm mutant construction
Total DNA was extracted from the WT strain PML1(12) using the protocol of Pospiech and Neumann (46). To construct a targeted mutant of the gene homologous to iucA/iucC (rhiE), the total gene was amplified using a combination of primers including For-rhiE and Rev-rhiE for a complete amplification and Rev-rhiE_SmaI and For-rhiE_SmaI to insert a SmaI site (Table 2; Fig. S2 for details). The insertion of a SmaI site in the middle of the rhiE gene was done following three PCR rounds: (i) For-rhiE with Rev-rhiE_SmaI (967 bp), (ii) For-rhiE_SmaI with Rev-rhiE (966 bp), and (iii) For-rhiE and Rev-rhiE (1,908 bp). The PCR products of PCR1 and 2 were purified and used as matrix for PCR 3. After amplification and purification, PCR3 product was ligated into the pGEM-T Easy plasmid (Promega) resulting in the plasmid pGEM-rhiE_SmaI (Table 2). The plasmid pGEM-rhiE_SmaI was then digested with SmaI to introduce the SmaI-digested gentamycin (Gm) resistance cassette from plasmid pUC1318, resulting in the plasmid pGEM-rhiE::Gm. A fragment containing the rhiE::Gm cassette was then obtained by EcoRI restriction and cloned in the plasmid pK19mob, resulting in the pK19mob-rhiE::Gm plasmid. This construction was transferred to E. coli S17.1λpir and then to strain PML1(12) by bi-parental conjugation. ΔrhiE mutants were recovered after 5 days of incubation at 25°C on gentamycin-containing ABm plates. The transconjugants obtained were verified by PCR with For-rhiE and Rev-rhiE primers, to differentiate the single and double cross-over events. Several single and double cross-over mutants were obtained and conserved. The experiments described in our study were done using a double cross-over mutant named rhiE::Gm.
Siderophore activity
Siderophore activity was detected using the liquid and solid versions of the CAS assay according to Schwyn and Neilands (47). For both assays, the color change, sign of the chelating activity of the siderophore for the iron (III) of the CAS-Fe complex, can be quantified by the presence of a halo on the solid CAS assay or through the measurement of the optical density at 655 nm for the liquid assay.
For the solid CAS assay, 5 µL of the bacterial inoculum [both WT and mutant strains; corresponding to 5 × 106 colony-forming units (CFU)] were inoculated in triplicate in agar plates and incubated for 3 days at 25°C. The diameter of colony and the halo surrounding it were measured to determine growth and siderophore(s) production, respectively.
For the liquid CAS assay, 0.5 mL of the bacterial inoculum (both WT and mutant strains; corresponding to 5 × 108 CFU) were inoculated in triplicate in 4.5-mL ABm-Fe. After 3 days of incubation at 25°C under agitation (150 rpm), the cultures were centrifugated at 9,000 × g during 15 min at 4°C to recover culture supernatants. A volume of 100 µL of these supernatants was mixed with 100 µL of CAS-Fe(III) and incubated during 1 h at room temperature in the dark. Then, the OD was measured at 655 nm using a microplate reader (Bio-Rad, model iMark). Decrease of absorbance at this specific wavelength is synonym of siderophore presence [change of media coloration from blue to yellow (6, 47)]. Non-inoculated medium was used as negative control for the liquid assay.
Impact of iron concentration on the growth the WT and rhiE::Gm strains
To determine if the concentration of iron and the mutation of the rhiE gene (rhiE::Gm mutant) affected the growth of strain PML1(12), growth was determined at different iron concentrations (i.e., 0, 0.5, 1, 5, and 10 mg L−l of Fe). A bacterial inoculum of 10 µL (corresponding to 6 × 106 CFU) was added to 190 µL of ABm-Fe and incubated in 96-well microplates for 5 days. This experience was performed in triplicate and conducted at 25°C on orbital shaking in a microplate reader (Tecan infinite M200 pro). The absorbance was measured at 595 nm every 3 h. To evaluate the growth of the strains, the slope of the exponential phase (i.e., growth rate) was calculated for each condition and replicate.
Siderophore production inhibition
Siderophore production is known to be regulated by the availability of nutritive elements in the culture medium, especially iron. To evaluate the concentration of Fe necessary to inhibit the siderophore production, the WT and mutant strains were incubated at different iron concentrations. Non-inoculated conditions were used as negative controls for each concentration. A volume of 10 µL of inoculum was added to 180 µL of ABm-Fe supplemented with 10 µL of different FeCl3 stock solutions ranging from 0 to 5 mg L−1 of Fe to obtain the following final concentrations in the medium: 0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1, 1.5, 2, 2.5, and 5 mg L−1. The 96-well microplates were incubated 3 days at 25°C. Then, microplates were centrifugated at 3,000 × g during 15 min at 4°C, and 100 µL of supernatant was mixed with 100 µL of CAS-Fe(III). After 1-h incubation in the dark, the plates were read at 655 nm using a microplate reader (Bio-Rad, model iMark) as described above.
Hematite weathering assay
Hematite characteristics
Hematite (Fe2O3) is an iron-oxide mineral widely distributed in all soils (48) and characterized by a high iron content. The hematite used in this study comes from a batch extracted in Brazil. The chemical composition was determined on crushed powder by inductively coupled plasma-atomic emission spectrometry (700 Series ICP-AES, Agilent Technologies) as described in Picard et al. (49). This analysis revealed that the hematite used is mainly composed of Fe2O3 (99.3%) and also contains in small amounts: 0.40% TiO2, 0.18% SiO2, 0.16% Al2O3, 0.10% MgO, 0.037% MnO.
Mineral weathering assay
To avoid contamination by residues associated with glass tubes, all glassware was rinsed once with 3.6% HCl and then three times with Milli-Q water. The tubes were filled with 200 mg of hematite and 9 mL of ABm-Fe medium and autoclaved at 121°C. To evaluate the differences in the weathering ability of the rhiE::Gm mutant and the WT strains, 1 mL of each inoculum was added to the tubes (procedure performed in triplicate). Non-inoculated media with and without hematite were used as abiotic and sterility controls for weathering. Samples were incubated for 7 days under agitation at 150 rpm and 25°C.
The cultures were used to determine the following: (i) bacterial growth, (ii) solution pH, (iii) siderophore production, and (iv) iron concentrations. Bacterial growth was estimated directly on the liquid culture by measuring the OD595nm on 200 µL. The other measurements were done after centrifugation at 14,000 × g for 15 min to remove bacterial cells and hematite particles. A volume of 100 µL of supernatant was used to evaluate the level of siderophore(s) production using the liquid CAS assay according to the protocol described above. A volume of 180 µL of supernatant was used to measure the pH by mixing with 20 µL of bromocresol green (1 g L−1) according to Uroz et al. (18). For the measure of the iron concentration, the supernatants were filtered at 0.22 µM (GHP Acrodisc 25 mm syringe filter; PALL) before mixing 180 µL of supernatant with 20 µL of ferrospectral. Both pH and iron determinations are based on optical density measurements at 595 nm using a microplate reader (Bio-Rad, model iMark) and after conversion with the calibration curves done on the bromocresol green and ferrospectral dyes, according to Uroz et al. (18).
Chemical characterization of the siderophore
Siderophore production and purification
Volumes of 50 mL of ABs-Fe were inoculated with each strain (WT and rhiE::Gm) and incubated at 25°C under 150 rpm shaking for 3 days. Succinic acid was used here as sole carbon source to limit the production of exopolysaccharides. A non-inoculated condition was also included as negative control. After the incubation time, cultures were centrifugated at 9,000 × g during 15 min at 4°C. The supernatant was filtered using 0.45 µM filters (Millipore) and treated with a Sep pack Vac 6cc tC18 Cartridge (Waters) to prepurify and concentrate the siderophore(s). The columns were first washed with 10 mL of 100% ethanol and then equilibrated with 10 mL of 0.01% trifluoroacetic acid (TFA). After this equilibration step, a volume of 25 mL of supernatant was added progressively to the column, and finally, the different fractions of the supernatant were eluted by the addition of 5 mL of 10% and then 20% methanol in water. At each step, the eluted volume (ca. 5 mL) was recovered and concentrated in the SpeedVac system. The dried samples were resuspended in 100 µL of Milli-Q water. These pre-purified fractions were used for further characterization using the liquid CAS assay to verify the presence of siderophore.
High-performance liquid chromatography analyses and fractionation
The different pre-purified fractions recovered above (from WT, mutant, and negative control samples) were analyzed by HPLC. A volume of 50 µL of each extract was injected in a Gemini C18 column (150 × 3.0 mm, 5 µM particle size, Phenomenex). Solvent A consisted of Milli-Q water, and solvent B consisted of acetonitrile plus 0.2% of TFA. A linear gradient elution from 2% to 40% of solvent B over 25 min at a flow rate of 1 mL min−1 was used to separate compounds. Absorbance was monitored at 210 nm.
Peaks of potential interest were identified by comparing the chromatogram of the WT sample with that of the rhiE::Gm mutant sample and the negative control. To determine the presence of the siderophore(s) activity, a fraction collection was performed in 96-well microplates using a fraction collector system (FRC-10A fraction collector system, Shimadzu). Fractions of 200 µL were recovered each 0.2 s using the same HPLC conditions as described before. Fraction collection was performed three times independently, and fractions with the same retention times were pooled. The fractions collected were then concentrated using the SpeedVac and resuspended in 100 µL of Milli-Q water. To determine the presence of siderophore(s) activity, a volume of 25 µL of the collected fractions was mixed with 25 µL of liquid CAS, and the absorbance at 655 nm was measured after overnight incubation.
Qualitative HPLC-DAD-MS-MS/MS analysis of siderophores and ferrosiderophores
A Thermo Scientific UHPLC-HRMS/MS system composed of a Vanquish liquid chromatography unit coupled to a PDA and an Orbitrap ID-X Tribrid high-resolution mass spectrometer operating in electrospray ionization mode (ESI) was used to detect and identify the putative siderophores. Ten microliters of each bacterial extract were injected onto a Hichrom Alltima C18 column (150 × 2.1 mm − 5 µM) maintained at 25°C. The flow rate was set at 0.2 mL min−1 and mobile phases consisted of water modified with formic acid (0.1%) for A and acetonitrile modified with formic acid (0.1%) for B. Compounds of interest were eluted using a linear gradient from 5% to 20% of B for 10 min, and then, a 5-min isocratic step was applied at 98% of B to wash the column, before returning to the initial composition of 5% B for 6 min to reach the equilibrium. Mass analysis was carried out in ESI positive ion mode (ESI+), and mass spectrometry conditions were as follows: spray voltage was set at 3.5 kV; source gases were set (in arbitrary units min−1) for sheath gas, auxiliary gas, and sweep gas at 35, 7 ,and 10, respectively; vaporizer and ion transfer tube temperatures were both set at 300°C. Survey scans of precursors were performed from 150 to 2,000 m/z at 60 K resolution (FWHM at 200 m/z) with MS parameters as follows: RF-lens, 35%; maximum injection time, 50 ms; data type, profile; internal mass calibration EASY-IC TM activated; normalized AGC target, 25%. A top speed data-dependent MS2 (0.6 s for the whole cycle time) was carried out using a wide quadrupole isolation (1.5 Th), an HCD fragmentation with a stepped collision energy (20, 35, and 50%) and an Orbitrap measure at 15 K resolution (high-resolution MS/MS analysis). Precursors with an intensity upper than 2.104 were automatically sampled for MS2. Dynamic exclusion was used, and the time of exclusion was set at 2.5 s, with a 10-ppm tolerance around the selected precursor (isotopes excluded). Other MS2 parameters were as follows: data type, profile; normalized AGC target, 20%; AGC target, 10,000.
Mass spectrometer calibration was performed by using the Thermo Scientific Pierce FlexMix calibration solution. MS data acquisition was carried out by using the Xcalibur v. 3.0 software (Thermo Scientific).
Bioinformatic analyses
Genome analysis
Genome analysis was performed using MicroScope MaGe (Magnifying Genomes) (50) and NCBI. The presence of iucA/iucC gene homolog was identified by antiSMASH (51) and confirmed by BLASTP analyses based on sequence homology (52).
Phylogenetic analyses
A total of 27 protein sequences (Table S1) homologous to IucA/IucC, including the homolog of PML1(12) (RhiE), were recovered from NCBI to build a phylogenetic neighbor-joining (N-J) tree using the Seaview platform (version 5.0.4) (53). For this analysis, the protein sequences considered were as follows: (i) sequences which role in siderophore biosynthesis were experimentally demonstrated; (ii) sequences based on their homology with the RhiE protein identified in strain PML1(12). Protein sequences were aligned using MUSCLE algorithm, and then, an N-J tree with 100.000 iterations was constructed.
Statistical analyses
Statistical analyses were performed in R software (Tidyverse packages) (54). Triplicate samples were used in all experiments. Differences between samples means (pH, growth, CAS assay, and released iron) or growth slopes were analyzed by analysis of variance and TukeyHSD tests.
ACKNOWLEDGMENTS
This work was supported by grants from the EC2CO program of the CNRS and the Labex ARBRE from the French National Research Agency (ANR) to S.U. The UMR1136 and UR1138 are supported by the ANR through the Labex Arbre (ANR-11-LABX-0002–01). C.B.N. is supported by a PhD fellowship from the INRAE and Grand Est Region.
The authors thank the MICROSCOPE team (David Roche; Genoscope) for the support provided on the genome management and the SILVATech platform for technical support. The authors also thank the PASM platform for the use of the Orbitrap LC-MS system (Plateau d'Analyse Structurale et Métabolomique - SF4242 EFABA, F-54505 Vandœuvre-lès-Nancy, France).
Contributor Information
Stéphane Uroz, Email: stephane.uroz@inrae.fr.
John R. Spear, Colorado School of Mines, Golden, Colorado, USA
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.00453-23.
Fig. S1 and S2 and Table S1
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Andrews SC, Robinson AK, Rodríguez-Quiñones F. 2003. Bacterial iron homeostasis. FEMS Microbiol Rev 27:215–237. doi: 10.1016/S0168-6445(03)00055-X [DOI] [PubMed] [Google Scholar]
- 2. Desai A, Archana G. 2011. Role of siderophores in crop improvement, p 109–139. In Bacteria in agrobiology: plant nutrient management. doi: 10.1007/978-3-642-21061-7 [DOI] [Google Scholar]
- 3. Radzki W, Gutierrez Mañero FJ, Algar E, Lucas García JA, García-Villaraco A, Ramos Solano B. 2013. Bacterial siderophores efficiently provide iron to iron-starved tomato plants in hydroponics culture. ALJMAO 104:321–330. doi: 10.1007/s10482-013-9954-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Colombo C, Palumbo G, He JZ, Pinton R, Cesco S. 2014. Review on iron availability in soil: interaction of Fe minerals, plants, and microbes. J Soils Sediments 14:538–548. doi: 10.1007/s11368-013-0814-z [DOI] [Google Scholar]
- 5. Uroz S, Picard L, Turpault M-P. 2022. Recent progress in understanding the ecology and molecular genetics of soil mineral weathering bacteria. Trends Microbiol 30:882–897. doi: 10.1016/j.tim.2022.01.019 [DOI] [PubMed] [Google Scholar]
- 6. Picard L, Paris C, Dhalleine T, Morin E, Oger P, Turpault M-P, Uroz S. 2022. The mineral weathering ability of Collimonas pratensis PMB3(1) involves a malleobactin-mediated iron acquisition system. Environ Microbiol 24:784–802. doi: 10.1111/1462-2920.15508 [DOI] [PubMed] [Google Scholar]
- 7. Uroz S, Picard L, Turpault M-P, Auer L, Armengaud J, Oger P. 2020. Dual transcriptomics and proteomics analyses of the early stage of interaction between Caballeronia mineralivorans PML1(12) and mineral. Environ Microbiol 22:3838–3862. doi: 10.1111/1462-2920.15159 [DOI] [PubMed] [Google Scholar]
- 8. Neilands JB. 1995. Siderophores: structure and function of microbial iron transport compounds. J Biol Chem 270:26723–26726. doi: 10.1074/jbc.270.45.26723 [DOI] [PubMed] [Google Scholar]
- 9. Kraemer SM, Duckworth OW, Harrington JM, Schenkeveld WDC. 2015. Metallophores and trace metal biogeochemistry. Aquat Geochem 21:159–195. doi: 10.1007/s10498-014-9246-7 [DOI] [Google Scholar]
- 10. Shiny Matilda C, Mannully ST, Rao VP, Shanthi C. 2021. Chromium binding Bacillus cereus VITSH1 - a promising candidate for heavy metal clean up. Lett Appl Microbiol 72:517–525. doi: 10.1111/lam.13441 [DOI] [PubMed] [Google Scholar]
- 11. Kramer J, Özkaya Ö, Kümmerli R. 2020. Bacterial siderophores in community and host interactions. Nat Rev Microbiol 18:152–163. doi: 10.1038/s41579-019-0284-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Essén SA, Bylund D, Holmström SJM, Moberg M, Lundström US. 2006. Quantification of hydroxamate siderophores in soil solutions of podzolic soil profiles in Sweden. Biometals 19:269–282. doi: 10.1007/s10534-005-8418-8 [DOI] [PubMed] [Google Scholar]
- 13. Carroll CS, Moore MM. 2018. Ironing out siderophore biosynthesis: a review of non-ribosomal peptide synthetase (NRPS)-independent siderophore synthetases. Crit Rev Biochem Mol Biol 53:356–381. doi: 10.1080/10409238.2018.1476449 [DOI] [PubMed] [Google Scholar]
- 14. Torres MA, Dong S, Nealson KH, West AJ. 2019. The kinetics of siderophore‐mediated olivine dissolution. Geobiol 17:401–416. doi: 10.1111/gbi.12332 [DOI] [PubMed] [Google Scholar]
- 15. Cheah SF, Kraemer SM, Cervini-Silva J, Sposito G. 2003. Steady-state dissolution kinetics of goethite in the presence of desferrioxamine B and oxalate ligands: implications for the microbial acquisition of iron. Chem Geol 198:63–75. doi: 10.1016/S0009-2541(02)00421-7 [DOI] [Google Scholar]
- 16. Dehner CA, Awaya JD, Maurice PA, DuBois JL. 2010. Roles of siderophores, oxalate, and ascorbate in mobilization of iron from hematite by the aerobic bacterium Pseudomonas mendocina. Appl Environ Microbiol 76:2041–2048. doi: 10.1128/AEM.02349-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parrello D, Zegeye A, Mustin C, Billard P. 2016. Siderophore-mediated iron dissolution from nontronites is controlled by mineral cristallochemistry. Front Microbiol 7:423. doi: 10.3389/fmicb.2016.00423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Uroz S, Calvaruso C, Turpault MP, Pierrat JC, Mustin C, Frey-Klett P. 2007. Effect of the mycorrhizosphere on the genotypic and metabolic diversity of the bacterial communities involved in mineral weathering in a forest soil. Appl Environ Microbiol 73:3019–3027. doi: 10.1128/AEM.00121-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith MJ, Shoolery JN, Schwyn B, Holden I, Neilands JB. 1985. Rhizobactin, a structurally novel siderophore from Rhizobium meliloti. J Am Chem Soc 107:1739–1743. doi: 10.1021/ja00292a047 [DOI] [Google Scholar]
- 20. Kügler S, Cooper RE, Boessneck J, Küsel K, Wichard T. 2020. Rhizobactin B is the preferred siderophore by a novel Pseudomonas isolate to obtain iron from dissolved organic matter in peatlands. Biometals 33:415–433. doi: 10.1007/s10534-020-00258-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Challis GL. 2005. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochem 6:601–611. doi: 10.1002/cbic.200400283 [DOI] [PubMed] [Google Scholar]
- 22. Oves-Costales D, Kadi N, Challis GL. 2009. The long-overlooked enzymology of a nonribosomal peptide synthetase-independent pathway for virulence-conferring siderophore biosynthesis. Chem Commun 43:6530–6541. doi: 10.1039/b913092f [DOI] [PubMed] [Google Scholar]
- 23. Persmark M, Pittman P, Buyer JS, Schwyn B, Gill PR, Neilands JB. 1993. Isolation and structure of rhizobactin 1021, a siderophore from the alfalfa symbiont Rhizobium meliloti 1021. J Am Chem Soc 115:3950–3956. doi: 10.1021/ja00063a014 [DOI] [Google Scholar]
- 24. Budzikiewicz H. 1997. Siderophores of fluorescent pseudomonads. Z Naturforsch C J Biosci 52:713–720. doi: 10.1515/znc-1997-11-1201 [DOI] [PubMed] [Google Scholar]
- 25. Barry SM, Challis GL. 2009. Recent advances in siderophore biosynthesis. Curr Opin Chem Biol 13:205–215. doi: 10.1016/j.cbpa.2009.03.008 [DOI] [PubMed] [Google Scholar]
- 26. Bindereif A, Neilands JB. 1983. Cloning of the aerobactin mediated iron assimilation system of plasmid ColV. J Bacteriol 153:1111–1113. doi: 10.1128/jb.153.2.1111-1113.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. de Lorenzo V, Bindereif A, Paw BH, Neilands JB. 1986. Aerobactin biosynthesis and transport genes of plasmid ColV-K30 in Escherichia coli K-12. J Bacteriol 165:570–578. doi: 10.1128/jb.165.2.570-578.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ramakrishnan G, Pérez NM, Carroll C, Moore MM, Nakamoto RK, Fox TE. 2019. Citryl ornithine is an intermediate in a three-step biosynthetic pathway for rhizoferrin in Francisella. ACS Chem Biol 14:1760–1766. doi: 10.1021/acschembio.9b00297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cotton JL, Tao J, Balibar CJ. 2009. Identification and characterization of the Staphylococcus aureus gene cluster coding for staphyloferrin A. Biochemistry 48:1025–1035. doi: 10.1021/bi801844c [DOI] [PubMed] [Google Scholar]
- 30. Dehner CA, Barton L, Maurice PA, DuBois JL. 2011. Size-dependent bioavailability of hematite (α-Fe2O3) nanoparticles to a common aerobic bacterium. Environ Sci Technol 45:977–983. doi: 10.1021/es102922j [DOI] [PubMed] [Google Scholar]
- 31. Mridha S, Kümmerli R. 2022. Coordination of siderophore gene expression among clonal cells of the bacterium Pseudomonas aeruginosa. Commun Biol 5:545. doi: 10.1038/s42003-022-03493-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dave BP, Dube HC. 2000. Regulation of siderophore production by iron Fe(III) in certain fungi and fluorescent pseudomonads. Indian J Exp Biol 38:297–299. [PubMed] [Google Scholar]
- 33. Van Den Berghe M, Merino N, Nealson KH, West AJ. 2021. Silicate minerals as a direct source of limiting nutrients: siderophore synthesis and uptake promote ferric iron bioavailability from olivine and microbial growth. Geobiolog 19:618–630. doi: 10.1111/gbi.12457 [DOI] [PubMed] [Google Scholar]
- 34. Akafia MM, Harrington JM, Bargar JR, Duckworth OW. 2014. Metal oxyhydroxide dissolution as promoted by structurally diverse siderophores and oxalate. Geochim Cosmochim Acta 141:258–269. doi: 10.1016/j.gca.2014.06.024 [DOI] [Google Scholar]
- 35. Duckworth OW, Akafia MM, Andrews MY, Bargar JR. 2014. Siderophore-promoted dissolution of chromium from hydroxide minerals. Environ Sci Process Impacts 16:1348–1359. doi: 10.1039/c3em00717k [DOI] [PubMed] [Google Scholar]
- 36. Carroll CS, Grieve CL, Murugathasan I, Bennet AJ, Czekster CM, Liu H, Naismith J, Moore MM. 2017. The rhizoferrin biosynthetic gene in the fungal pathogen Rhizopus delemar is a novel member of the NIS gene family. Int J Biochem Cell Biol 89:136–146. doi: 10.1016/j.biocel.2017.06.005 [DOI] [PubMed] [Google Scholar]
- 37. Sullivan JT, Jeffery EF, Shannon JD, Ramakrishnan G. 2006. Characterization of the siderophore of Francisella tularensis and role of fslA in siderophore production. J Bacteriol 188:3785–3795. doi: 10.1128/JB.00027-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schalk IJ, Rigouin C, Godet J. 2020. An overview of siderophore biosynthesis among fluorescent Pseudomonads and new insights into their complex cellular organization. Environ Microbiol 22:1447–1466. doi: 10.1111/1462-2920.14937 [DOI] [PubMed] [Google Scholar]
- 39. Mathew A, Eberl L, Carlier AL. 2014. A novel siderophore‐independent strategy of iron uptake in the genus Burkholderia. Mol Microbiol 91:805–820. doi: 10.1111/mmi.12499 [DOI] [PubMed] [Google Scholar]
- 40. Min L, Guo L, Ye J. 2019. Mechanism of Burkholderia pyrrocinia JK-SH007 growth-promoting to plant via siderophore-mediation. Journ Nanjing Forest Univ Nat Sci Ed 43:165–172. doi: 10.3969/j.issn.1000-2006.201811058 [DOI] [Google Scholar]
- 41. Alice AF, López CS, Lowe CA, Ledesma MA, Crosa JH. 2006. Genetic and transcriptional analysis of the siderophore malleobactin biosynthesis and transport genes in the human pathogen Burkholderia pseudomallei K96243. J Bacteriol 188:1551–1566. doi: 10.1128/JB.188.4.1551-1566.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Calvaruso C, Turpault M-P, Frey-Klett P. 2006. Root-associated bacteria contribute to mineral weathering and to mineral nutrition in trees: a budgeting analysis. Appl Environ Microbiol 72:1258–1266. doi: 10.1128/AEM.72.2.1258-1266.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Koele N, Turpault MP, Hildebrand EE, Uroz S, Frey-Klett P. 2009. Interactions between mycorrhizal fungi and mycorrhizosphere bacteria during mineral weathering: budget analysis and bacterial quantification. Soil Biol Biochem 41:1935–1942. doi: 10.1016/j.soilbio.2009.06.017 [DOI] [Google Scholar]
- 44. Calvaruso C, Turpault M-P, Leclerc E, Frey-Klett P. 2007. Impact of ectomycorrhizosphere on the functional diversity of soil bacterial and fungal communities from a forest stand in relation to nutrient mobilization processes. Microb Ecol 54:567–577. doi: 10.1007/s00248-007-9260-z [DOI] [PubMed] [Google Scholar]
- 45. Chilton MD, Currier TC, Farrand SK, Bendich AJ, Gordon MP, Nester EW. 1974. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc Natl Acad Sci U S A 71:3672–3676. doi: 10.1073/pnas.71.9.3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pospiech A, Neumann B. 1995. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet 11:217–218. doi: 10.1016/s0168-9525(00)89052-6 [DOI] [PubMed] [Google Scholar]
- 47. Schwyn B, Neilands JB. 1987. Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56. doi: 10.1016/0003-2697(87)90612-9 [DOI] [PubMed] [Google Scholar]
- 48. Dixon JB, Weed SB, Dinauer RC, eds. 1989. Minerals in soil environments. 2nd ed. Soil Science Society of America, Madison, WI, USA. doi: 10.2136/sssabookser1.2ed [DOI] [Google Scholar]
- 49. Picard L, Turpault M-P, Oger PM, Uroz S. 2020. Identification of a novel type of glucose dehydrogenase involved in the mineral weathering ability of Collimonas pratensis strain PMB3(1). FEMS Microbiol Ecol 97:fiaa232. doi: 10.1093/femsec/fiaa232 [DOI] [PubMed] [Google Scholar]
- 50. Vallenet D, Calteau A, Dubois M, Amours P, Bazin A, Beuvin M, Burlot L, Bussell X, Fouteau S, Gautreau G, Lajus A, Langlois J, Planel R, Roche D, Rollin J, Rouy Z, Sabatet V, Médigue C. 2020. MicroScope: an integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res 48:D579–D589. doi: 10.1093/nar/gkz926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP, Medema MH, Weber T. 2021. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res 49:W29–W35. doi: 10.1093/nar/gkab335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 53. Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi: 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- 54. Team RC. 2021. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and S2 and Table S1