ABSTRACT
Gut microbiota are fundamentally important for healthy function in animal hosts. Drosophila melanogaster is a powerful system for understanding host-microbiota interactions, with modulation of the microbiota inducing phenotypic changes that are conserved across animal taxa. Qualitative differences in diet, such as preservatives and dietary yeast batch variation, may affect fly health indirectly via microbiota, and may potentially have hitherto uncharacterized effects directly on the fly. These factors are rarely considered, controlled, and are not standardized among laboratories. Here, we show that the microbiota’s impact on fly triacylglyceride (TAG) levels—a commonly-measured metabolic index—depends on both preservatives and yeast, and combinatorial interactions among the three variables. In studies of conventional, axenic, and gnotobiotic flies, we found that microbial impacts were apparent only on specific yeast-by-preservative conditions, with TAG levels determined by a tripartite interaction of the three experimental factors. When comparing axenic and conventional flies, we found that preservatives caused more variance in host TAG than microbiota status, and certain yeast-preservative combinations even reversed effects of microbiota on TAG. Preservatives had major effects in axenic flies, suggesting either direct effects on the fly or indirect effects via media. However, Acetobacter pomorum buffers the fly against this effect, despite the preservatives inhibiting growth, indicating that this bacterium benefits the host in the face of mutual environmental toxicity. Our results suggest that antimicrobial preservatives have major impacts on host TAG, and that microbiota modulates host TAG dependent on the combination of the dietary factors of preservative formula and yeast batch.
IMPORTANCE
Drosophila melanogaster is a premier model for microbiome science, which has greatly enhanced our understanding of the basic biology of host-microbe interactions. However, often overlooked factors such as dietary composition, including yeast batch variability and preservative formula, may confound data interpretation of experiments within the same lab and lead to different findings when comparing between labs. Our study supports this notion; we find that the microbiota does not alter host TAG levels independently. Rather, TAG is modulated by combinatorial effects of microbiota, yeast batch, and preservative formula. Specific preservatives increase TAG even in germ-free flies, showing that a commonplace procedure in fly husbandry alters metabolic physiology. This work serves as a cautionary tale that fly rearing methodology can mask or drive microbiota-dependent metabolic changes and also cause microbiota-independent changes.
KEYWORDS: Acetobacter pomorum, Levilactobacillus brevis, Drosophila melanogaster, diet, microbiota
INTRODUCTION
Fruit flies are a preeminent model for understanding fundamental host-microbiome biology, thanks to experimental tractability, powerful genetic tools, and a simple microbiota dominated by culturable Lactobacillaceae and Acetobacteraceae (1, 2). Flies can be routinely made germ-free (axenic), or selectively reassociated with defined cultures of physiologically- and ecologically-relevant microbiota (gnotobiotic). The fly microbiota is less complex than in vertebrates, yet effects on a plethora of host traits are conserved (3–14), potentially indicating common mechanisms that can be characterized rapidly in the fly.
The microbiota affect fly nutrition, and so variation in microbiota and diet have mutually-interdependent effects (15). Brewer’s yeast is included ubiquitously in fly diets (16). Importantly, yeast is supplied commercially in lots originating from distinct production batches, with potentially variable chemical composition. This potentially introduces nutritional inconsistencies among distinct lots (16), which may modify response to microbiota manipulation.
Fly diets also commonly contain antimicrobial preservatives. Preservative formulae vary both in composition and concentration, and in some microbiota studies they are omitted entirely (10, 13, 14, 17). The commonly used preservative nipagin (methylparaben) affects the density of Acetobacter (18), which may alter growth in fly food, and thereby modify physiological impact. Further, nipagin is dissolved in ethanol, which interacts with variation in the microbiota (19). Acid preservatives are also used, which may modulate fly function through effects on the microbiota (e.g., density, metabolic substrate provision), diet [e.g., pH and nutrient solubility (14, 20–22)] and direct effects on the fly (23).
Here, we test whether the physiological impact of altering the fly microbiota depends on dietary yeast batch and preservatives. We used two lots of one supplier’s yeast, denoted A or B. We either omitted preservatives or added (1) phosphoric acid and propionic acid (15) or (2) nipagin and propionic acid (13). These ingredients were incorporated into an otherwise identical sucrose-yeast-agar (SYA) diet (24). We measured triacylglyceride (TAG) levels, the main storage lipid, which are commonly measured as a metabolic index due to interest in the microbiome’s role in human obesity (25). Within each experiment, we normalized TAG to the mean of axenic flies without preservatives, giving a measure of relative TAG.
RESULTS
Host TAG is subject to a microbiota*yeast*preservative interaction
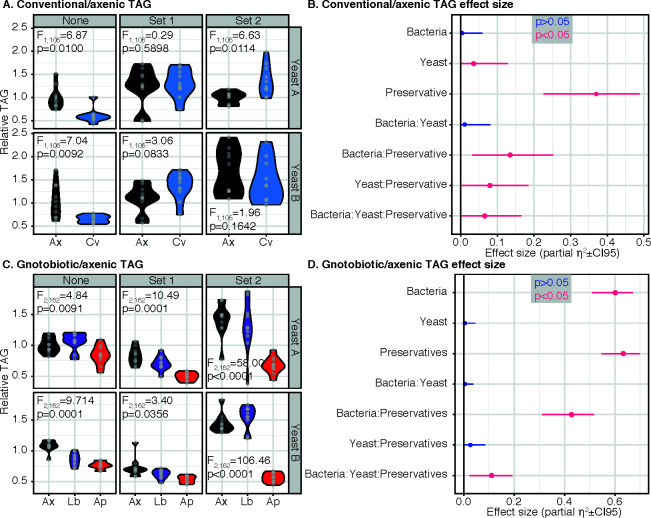
First, we applied a simple microbiome manipulation, comparing relative TAG in conventionally reared and axenic females, 3 days after adult emergence. We analyzed data with ANOVA (Table 1) and post hoc tests with Tukey corrections, implemented in the R “emmeans” package (Table 2). TAG response to bacterial elimination depended on the interaction of yeast batch and preservative formula (ANOVA: bacteria*yeast*preservative F2,106=3.73, P = 0.03; Table 1). This interaction obscured the anticipated main effect of increased TAG in axenics (ANOVA: bacteria F1,106=0.54, P = 0.46, Table 1), suggesting that microbial capacity to modulate TAG depends on a yeast*preservative interaction. To examine specifically how, we stratified our analysis per yeast*preservative combination. Without preservatives, on both yeasts, TAG was elevated in axenics (Table 2). Surprisingly, this response was reversed by a specific yeast*preservative combination, with conventionals showing higher TAG than axenics on yeast A and with preservative formula 2 (Table 2; Fig. 1A). Furthermore, microbial manipulation did not affect TAG in any other condition including preservatives on either yeast (Table 2). Interestingly, preservative formula 2 increased TAG even in axenic flies but only on yeast B (Table 2), suggesting effects via fly or food. Furthermore, the TAG levels were typically more variable when preservatives were present on both yeasts, and this variability was most pronounced on yeast B with preservative set 2 (Fig. 1A).
TABLE 1.
ANOVA (type 3) testing for preservative*bacteria interactions that determine TAG levels in conventionally reared vs axenic flies
| Term | Sum sq | Df | F | Pr(>F) |
|---|---|---|---|---|
| (Intercept) | 155.57 | 1 | 1484.85 | <2.20E-16 |
| Bacteria | 0.06 | 1 | 0.54 | 0.46 |
| Yeast | 0.48 | 1 | 4.57 | 0.034 |
| Preservative | 6.44 | 2 | 30.72 | 2.90E-11 |
| Bacteria:Yeast | 0.13 | 1 | 1.28 | 0.26 |
| Bacteria:Preservative | 1.76 | 2 | 8.40 | 0.0004 |
| Yeast:Preservative | 0.99 | 2 | 4.73 | 0.011 |
| Bacteria:Yeast:Preservative | 0.78 | 2 | 3.73 | 0.027 |
| Residuals | 11.11 | 106 |
TABLE 2.
Effects of microbiota (conventional vs axenic) on TAG levels of flies reared on specific combinations of dietary yeast and preservatives: ANOVA analysis stratified by yeast and preservatives (joint tests)
| Yeast | Preservative | Estimate | SE | df | t ratio | P valuea |
|---|---|---|---|---|---|---|
| A | 1 | 0.231 | 0.428 | 106 | -0.541 | 0.5898 |
| B | 1 | 0.748 | 0.428 | 106 | -1.748 | 0.0833 |
| A | 2 | 1.131 | 0.439 | 106 | -2.575 | 0.0114 |
| B | 2 | 0.599 | 0.428 | 106 | 1.401 | 0.1642 |
| A | None | 1.152 | 0.439 | 106 | 2.622 | 0.0100 |
| B | None | 1.135 | 0.428 | 106 | 2.653 | 0.0092 |
Tukey corrected.
Fig 1.
Metabolic impact of microbiota depends on the combination of yeast batch and preservative formula. (A) and (C) show relative TAG levels in two different experiments, separated by preservative conditions (columns, shown at top), and yeast batch (rows, shown at side). In both experiments, relative TAG was calculated by normalizing TAG density (µg per mg fly wet weight) to the mean of axenic flies without preservatives on yeast A. (B) and (D) show effect size calculations for main effects and interaction terms in the two experiments, color-coded by statistical significance. (A) Comparisons between axenic (Ax) and conventional (Cv) flies show that, on yeasts used in this experiment, relative TAG is reduced only in conventional flies when no preservatives are present. On Yeast A, adding preservative set 2 reversed the sign of the effect of eliminating the microbiota. (B) In the experiment shown in panel A, comparing axenic and conventional flies, preservatives are the biggest source of variance in relative TAG, with both a statistically significant effect (P < 0.05), and the biggest-sized effect. The bacteria-by-preservative interaction is the next biggest-sized effect, suggesting that impacts of eliminating the microbiota are contingent on preservatives. (C) Comparisons of relative TAG between axenic (Ax), Levilactobacillus brevis DmCS003 (Lb), and Acetobacter pomorum DmCS004 (Ap) associated flies show that A. pomorum reduces TAG levels relative to axenic flies in most conditions. Preservative set 2 elevated TAG on both yeasts (noting that it only did so on Yeast B in the first experiment), but A. pomorum abrogated this effect. (D) In the experiment shown in panel C, comparing axenic to monoassociated flies, bacteria and preservatives are equally major contributors to the variance in TAG observed, with their interaction being another significant contributor: again this indicates that the impact of variation in microbiota is contingent on preservatives.
Having identified significant interactions among experimental factors, we asked which of these effects were large and which were small, i.e., what was the relative contribution of each experimental factor and their higher-order interactions to overall variance? We calculated a measure of effect size (partial Eta2) for each experimental variable and their interactions (Fig. 1B). This indicated that preservatives were the biggest source of variance (Fig. 1B). Confidence intervals overlapped for all other significant terms, suggesting equivalent contributions to overall variation. These results indicated that variation in preservatives and their interaction with yeast batch are a hitherto unappreciated factor that affect fly TAG, which can both eclipse and determine effects of microbiota.
Acetobacter pomorum buffers flies against a TAG-promoting effect of preservative set 2
The fly microbiota is dominated by two bacterial genera, with Acetobacter and Lactobacilliaceae exhibiting strain-specific effects on fly physiology (10). Monoassociation with Acetobacter spp., but not Lactobacilliaceae, recapitulates conventional fly TAG levels (10). In conventional flies, the effects of yeast and preservative could potentially be driven by either compositional changes in the microbiota or bacterial physiological changes. We reasoned that compositional changes can be excluded if effects of yeast and preservatives are apparent in gnotobiotic flies monoassociated with a single strain, in which case strain-specific physiological effects might be expected because growth of Acetobacter but not Lactobacilliaceae is impacted by nipagin (18). Could yeast*preservative*microbiota effects on the fly be driven by particular bacterial strains?
We made gnotobiotic flies with A. pomorum (DmCS004) and Levilactobacillus brevis (DmCS003), and axenic controls, and modulated yeast and preservatives, to determine strain*yeast*preservative effects (Fig. 1C) and analyzed TAG levels with ANOVA (Table 3) and post hoc analyses (Tables 4–5). We used the same yeast and preservative set as in the first experiment. We also confirmed that there were no significant differences in standard curves for assays between the two experiments (Supplementary Text, Fig. S2), confirming that our technical detection capacity was the same for the two different experiments. TAG response to bacterial elimination again depended on the interaction of yeast batch and preservative formula (ANOVA: bacteria*yeast*preservative F4,162=4.96, P = 0.0008; Table 3). Across all preservative and yeast conditions, A. pomorum gnotobiotes had lower average TAG than axenics and L. brevis gnotobiotes (Fig. 1C).
TABLE 3.
ANOVA (type 3) testing for preservative*bacteria interactions that determine TAG levels in flies reared either axenically or in association with A. pomorum or L. brevis
| Term | Sum sq | Df | F value | Pr(>F) |
|---|---|---|---|---|
| (Intercept) | 149.958 | 1 | 5370.171 | < 2.2e-16 |
| Bacteria | 6.829 | 2 | 122.2687 | < 2.2e-16 |
| Yeast | 0.02 | 1 | 0.733 | 0.39319 |
| Preservatives | 7.877 | 2 | 141.05 | < 2.2e-16 |
| Bacteria:Yeast | 0.023 | 2 | 0.4116 | 0.66328 |
| Bacteria:Preservatives | 3.393 | 4 | 30.3757 | < 2.2e-16 |
| Yeast:Preservatives | 0.131 | 2 | 2.3448 | 0.09911 |
| Bacteria:Yeast:Preservatives | 0.554 | 4 | 4.9638 | 0.000844 |
| Residuals | 4.524 | 162 |
TABLE 4.
Effects of yeast*preservative interactions on TAG levels under specific microbiota conditions: ANOVA analysis stratified by microbiota status (joint tests)
| Bacteriaa | Term | df1 | df2 | F ratio | P valueb |
|---|---|---|---|---|---|
| Ax | Yeast | 1 | 162 | 0.02 | 0.8871 |
| Ax | Preservatives | 2 | 162 | 78.265 | <0.0001 |
| Ax | Yeast:Preservatives | 2 | 162 | 1.623 | 0.2005 |
| Lb | Yeast | 1 | 162 | 0.235 | 0.6289 |
| Lb | Preservatives | 2 | 162 | 107.785 | <0.0001 |
| Lb | Yeast:Preservatives | 2 | 162 | 9.577 | 0.0001 |
| Ap | Yeast | 1 | 162 | 1.301 | 0.2556 |
| Ap | Preservatives | 2 | 162 | 15.751 | <0.0001 |
| Ap | Yeast:Preservatives | 2 | 162 | 1.072 | 0.3446 |
Ax, axenic; Ap, Acetobacter pomorum DmCS004; Lb, Levilactobacillus brevis DmCS003.
Tukey corrected.
TABLE 5.
Differences in TAG levels of flies reared on different preservatives (none, set 1, set 2) on specific combinations of dietary yeast and microbiota: ANOVA analysis stratified by yeast and microbiota
| Bacteriaa | Yeast batch | Contrast (Preservatives) | Estimate | SE | Df | t ratio | P valueb |
|---|---|---|---|---|---|---|---|
| Ax | A | None vs Set 1 | 0.173 | 0.0747 | 162 | 2.319 | 0.056 |
| Ax | A | None vs Set 2 | 0.417 | 0.0747 | 162 | -5.575 | <0.0001 |
| Ax | A | Set 1 vs Set 2 | 0.59 | 0.0747 | 162 | -7.894 | <0.0001 |
| Lb | A | None vs Set 1 | 0.357 | 0.0747 | 162 | 4.774 | <0.0001 |
| Lb | A | None vs Set 2 | 0.244 | 0.0747 | 162 | -3.27 | 0.0037 |
| Lb | A | Set 1 vs Set 2 | 0.601 | 0.0747 | 162 | -8.043 | <0.0001 |
| Ap | A | None vs Set 1 | 0.35 | 0.0747 | 162 | 4.682 | <0.0001 |
| Ap | A | None vs Set 2 | 0.17 | 0.0747 | 162 | 2.278 | 0.0618 |
| Ap | A | Set 1 vs Set 2 | 0.18 | 0.0747 | 162 | -2.404 | 0.0454 |
| Ax | B | None vs Set 1 | 0.357 | 0.0747 | 162 | 4.782 | <0.0001 |
| Ax | B | None vs Set 2 | 0.367 | 0.0747 | 162 | -4.909 | <0.0001 |
| Ax | B | Set 1 vs Set 2 | 0.724 | 0.0747 | 162 | -9.691 | <0.0001 |
| Lb | B | None vs Set 1 | 0.253 | 0.0747 | 162 | 3.383 | 0.0026 |
| Lb | B | None vs Set 2 | 0.687 | 0.0747 | 162 | -9.188 | <0.0001 |
| Lb | B | Set 1 vs Set 2 | 0.939 | 0.0747 | 162 | -12.571 | <0.0001 |
| Ap | B | None vs Set 1 | 0.236 | 0.0747 | 162 | 3.156 | 0.0054 |
| Ap | B | None vs Set 2 | 0.204 | 0.0747 | 162 | 2.727 | 0.0194 |
| Ap | B | Set 1 vs Set 2 | 0.032 | 0.0747 | 162 | -0.429 | 0.9037 |
Ax, axenic; Ap, Acetobacter pomorum; Lb, Levilactobacillus brevis.
P value adjustment: Tukey method for comparing a family of 3 estimates.
We again calculated Partial Eta2 (effect size) to indicate impact of experimental variables on overall variation in the experiment, i.e., which effects were significant and large, and which were significant but smaller. Partial Eta2 indicated that the preservative formula and bacterial strain were the leading contributors to TAG variation in this experiment (Fig. 1D). The preservative*bacterial strain interaction had a substantially sized (and statistically significant: P < 2.2e−16, Table 3) effect, suggesting that variation in bacterial strain and preservatives conspired to produce sizeble variation. Altogether, these results indicated that (i) impacts of varying microbiota strains depend on yeast*preservative variation (ii), the lower-order preservative*bacterial strain interactions was a particularly large source of variation, and (iii) the effect of changing preservatives is equivalent to the effect of perturbing the microbiota.
To assess strain-specific impacts of yeast*preservative, we stratified our ANOVA analysis by bacteria (Table 4), revealing yeast*preservative effects in gnotobiotes with L. brevis (F2,162=9.577, P = 0.0001) but not with A. pomorum (F2,162=1.072, P = 0.3446) or in axenic flies (F2,162=1.623, P = 0.2005). Preservative variation had a significant effect across all microbial conditions (Table 4), while yeast had no significant effect in any microbial condition (Table 4).
Why would bacteria*yeast*preservative effects arise? We reasoned it could occur either because (i) a given bacterial strain modulates host TAG only on specific yeast*preservative conditions i.e., indirect effects of preservatives and yeast, or (ii) yeast*preservative conditions affect host TAG, but this effect is buffered by specific bacteria, i.e., direct effects of preservatives and yeast, dependent on microbiota. The finding that yeast*preservative effects were apparent in axenic and L. brevis-associated flies suggested that A. pomorum may indeed buffer an effect of yeast*preservative variation that is apparent in axenic and L. brevis-associated flies. We noted that preservative set 2 appeared to elevate TAG levels in axenic and L. brevis-associated flies but not A. pomorum-associated flies (Fig. 1C: noting that in the first experiment axenic TAG was elevated on Yeast B but not Yeast A), suggesting that A. pomorum may buffer a TAG-promoting effect of these preservatives, in which case TAG should be significantly elevated by these preservatives in axenic or L. brevis-associated flies but not in A. pomorum-associated flies. We tested this prediction using post hoc pairwise tests (Table 5) and found that indeed these preservatives significantly elevated TAG in axenic or L. brevis-associated flies but not in A. pomorum-associated flies; in fact, in the presence of A. pomorum, the effect of these preservatives was reversed, moderately decreasing TAG. This suggested that A. pomorum abrogates a TAG-promoting effect of nipagin and propionic acid contained in preservative set 2.
Elevated TAG in axenic or L. brevis-associated flies suggested that the impact of varying microbial association may be contingent on preservatives and yeast. Specifically, we predicted that the impact of A. pomorum would be greater on preservative set 2 because the starting TAG levels in axenic flies were elevated and these effects are not rescued by L. brevis. We ran F tests for the effect of microbiota status on each yeast*preservative combination (Table 6) and found that indeed F ratios (a measure of effect size) were markedly greater on preservative set 2 (yeast A, F = 58; yeast B, F = 106) than either set 1 or no preservatives (all <10.5). To confirm that this was due to A. pomorum, we ran a series of post hoc tests. We stratified the analysis by yeast and preservatives, and measured pairwise differences in TAG levels among the microbial conditions. As anticipated, t ratios for the difference between A. pomorum and L. brevis conditions, or A. pomorum and axenic conditions, were greater on medium containing preservative set 2 than either set 1 or no preservatives (Table 7). Therefore, we expected that the overall effect of preservative variation would be lesser in the presence of A. pomorum than in the presence of L. brevis or in axenic flies. As expected, when we stratified the analysis by yeast and bacteria, F ratios for effect of preservatives in were substantially reduced by A. pomorum association, relative to axenic flies (~3× lower on yeast A, ~8× lower on yeast B), and relative to L. brevis-associated flies (~3× lower on yeast A, ~14× lower on yeast B) (Table 8). Previous reports suggested that Acetobacter are nipagin-sensitive (18); however, our present results indicated any that A. pomorum nipagin sensitivity did not translate into impaired modulation of host TAG; rather, this strain rescued flies from a TAG-promoting effect of the nipagin-containing preservative set 2.
TABLE 6.
Effects of microbiota (axenic, L. brevis, A. pomorum) on TAG levels of flies reared on specific combinations of dietary yeast and preservatives: ANOVA analysis stratified by yeast and preservatives (joint tests)
| Yeast | Preservatives | df1 | df2 | F ratio | P valuea |
|---|---|---|---|---|---|
| A | None | 2 | 162 | 4.764 | 0.0098 |
| B | None | 2 | 162 | 9.769 | 0.0001 |
| A | Set 1 | 2 | 162 | 10.514 | 0.0001 |
| B | Set 1 | 2 | 162 | 3.643 | 0.0283 |
| A | Set 2 | 2 | 162 | 58.466 | <0.0001 |
| B | Set 2 | 2 | 162 | 106.203 | <0.0001 |
Tukey corrected.
TABLE 7.
Differences in TAG levels among gnotobiotic and axenic flies reared on specific combinations of dietary yeast and preservatives
| Preservatives | Yeast | Contrast (Bacteria) |
Estimate | SE | df | t ratio | P valuea |
|---|---|---|---|---|---|---|---|
| None | A | Ax vs Lb | 0.0617 | 0.0747 | 162 | -0.825 | 0.6881 |
| None | A | Ax vs Ap | 0.1617 | 0.0747 | 162 | 2.164 | 0.0807 |
| None | A | Lb vs Ap | 0.2233 | 0.0747 | 162 | 2.989 | 0.009 |
| None | B | Ax vs Lb | 0.2254 | 0.0747 | 162 | 3.016 | 0.0083 |
| None | B | Ax vs Ap | 0.3218 | 0.0747 | 162 | 4.307 | 0.0001 |
| None | B | Lb vs Ap | 0.0965 | 0.0747 | 162 | 1.291 | 0.4024 |
| Set 1 | A | Ax vs Lb | 0.1218 | 0.0747 | 162 | 1.629 | 0.2362 |
| Set 1 | A | Ax vs Ap | 0.3383 | 0.0747 | 162 | 4.527 | <0.0001 |
| Set 1 | A | Lb vs Ap | 0.2165 | 0.0747 | 162 | 2.897 | 0.0119 |
| Set 1 | B | Ax vs Lb | 0.1208 | 0.0747 | 162 | 1.617 | 0.2414 |
| Set 1 | B | Ax vs Ap | 0.2003 | 0.0747 | 162 | 2.68 | 0.022 |
| Set 1 | B | Lb vs Ap | 0.0795 | 0.0747 | 162 | 1.063 | 0.5383 |
| Set 2 | A | Ax vs Lb | 0.1106 | 0.0747 | 162 | 1.48 | 0.3031 |
| Set 2 | A | Ax vs Ap | 0.7486 | 0.0747 | 162 | 10.017 | <0.0001 |
| Set 2 | A | Lb vs Ap | 0.638 | 0.0747 | 162 | 8.537 | <0.0001 |
| Set 2 | B | Ax vs Lb | 0.0944 | 0.0747 | 162 | -1.263 | 0.4181 |
| Set 2 | B | Ax vs Ap | 0.8925 | 0.0747 | 162 | 11.943 | <0.0001 |
| Set 2 | B | Lb vs Ap | 0.9869 | 0.0747 | 162 | 13.206 | <0.0001 |
P value adjustment: Tukey method for comparing a family of three estimates.
TABLE 8.
Effects of preservatives (none, set 1, set 2) on TAG levels of flies reared on specific combinations of dietary yeast and microbiota: ANOVA analysis stratified by yeast and microbiota (joint tests)
| Bacteria | Yeast | df1 | df2 | F ratio | P valuea |
|---|---|---|---|---|---|
| Ax | A | 2 | 162 | 32.926 | <0.0001 |
| Ax | B | 2 | 162 | 46.962 | <0.0001 |
| Lb | A | 2 | 162 | 32.726 | <0.0001 |
| Lb | B | 2 | 162 | 84.637 | <0.0001 |
| Ap | A | 2 | 162 | 10.965 | <0.0001 |
| Ap | B | 2 | 162 | 5.859 | 0.0035 |
Tukey corrected.
To determine if our strains were indeed differentially sensitive to the two preservative formula, we quantified bacterial colony forming units (CFU) from gnotobiotic adult flies (Fig. S1). One implication of the yeast effects we have documented is that experiments within a given laboratory will be confounded when a given yeast batch is exhausted. In our case, we ran out of yeasts A and B, and could not obtain any more. Therefore, we used three new yeast batches (C–E) to quantify CFU over a wide range of yeast conditions, asking whether CFUs vary by yeast*preservatives and whether these effects are strain-specific. We confirmed that there was indeed a bacteria*yeast*preservative effect (Table 9, GLM with negative binomial distribution, joint tests: F = 11.63, P = 1.18e−07). Next, we applied post hoc tests to assess impacts of preservatives, per yeast, and per bacterium, and determined that both preservative sets reduced A. pomorum CFUs relative to no preservatives but this effect was consistently bigger with nipagin-containing set 2 (Table 10). The t ratios for the preservative set 2 vs no preservative comparison for each yeast batch were lower than those comparing set 1 vs no preservatives per each yeast batch for A. pomorum CFUs. Furthermore, there were consistently significantly more A. pomorum CFUs on preservative set 1 than set 2 (Fig. S1), supplementing previous findings that nipagin limits Acetobacter growth. Taken together, these findings suggest that fly food preservatives can affect fly physiology either directly or via media. However the nature of this effect can depend on batch variation in dietary yeast, and specific bacteria can abrogate these deleterious effects despite themselves enduring negative effects of the preservatives.
TABLE 9.
ANOVA (type 3) testing for preservative*yeast interactions that determine CFUs in gnotobiotic flies colonized with A. pomorum or L. brevis
| Term | Sum sq | Df | F value | Pr(>F) |
|---|---|---|---|---|
| (Intercept) | 2205.28 | 1 | 54503.58 | < 2.2e-16 |
| Bacteria | 18.54 | 1 | 458.1797 | < 2.2e-16 |
| Yeast | 0.98 | 2 | 12.1356 | 2.157e-05 |
| Preservatives | 24.37 | 2 | 301.1093 | < 2.2e-16 |
| Bacteria:Yeast | 0.01 | 2 | 0.1566 | 0.855265 |
| Bacteria:Preservatives | 0.35 | 2 | 4.2667 | 0.016970 |
| Yeast:Preservatives | 0.71 | 4 | 4.3905 | 0.002741 |
| Bacteria:Yeast:Preservatives | 1.88 | 4 | 11.6292 | 1.177e-07 |
| Residuals | 3.64 | 90 |
TABLE 10.
Differences CFU of flies reared on different preservatives (none, set 1, set 2) on specific combinations of dietary yeast and microbiota: ANOVA analysis stratified by yeast and microbiota
| Bacteriaa | Yeast batch | Contrast (Preservatives) | Estimate | SE | df | t ratio | P valueb |
|---|---|---|---|---|---|---|---|
| Lb | C | Set 1 - Set 2 | 0.267 | 0.116 | 90 | 2.296 | 0.0614 |
| Lb | C | Set 1 - None | 0.951 | 0.116 | 90 | 8.189 | <0.0001 |
| Lb | C | Set 2 - None | 1.218 | 0.116 | 90 | 10.486 | <0.0001 |
| Ap | C | Set 1 - Set 2 | 0.417 | 0.116 | 90 | 3.591 | 0.0015 |
| Ap | C | Set 1 - None | 0.539 | 0.116 | 90 | 4.645 | <0.0001 |
| Ap | C | Set 2 - None | 0.956 | 0.116 | 90 | 8.236 | <0.0001 |
| Lb | D | Set 1 - Set 2 | 0.894 | 0.116 | 90 | 7.698 | <0.0001 |
| Lb | D | Set 1 - None | 0.339 | 0.116 | 90 | 2.92 | 0.0122 |
| Lb | D | Set 2 - None | 1.233 | 0.116 | 90 | 10.618 | <0.0001 |
| Ap | D | Set 1 - Set 2 | 0.315 | 0.116 | 90 | 2.71 | 0.0217 |
| Ap | D | Set 1 - None | 0.534 | 0.116 | 90 | 4.595 | <0.0001 |
| Ap | D | Set 2 - None | 0.848 | 0.116 | 90 | 7.305 | <0.0001 |
| Lb | E | Set 1 - Set 2 | 0.828 | 0.116 | 90 | 7.128 | <0.0001 |
| Lb | E | Set 1 - None | 0.193 | 0.116 | 90 | 1.659 | 0.2265 |
| Lb | E | Set 2 - None | 1.02 | 0.116 | 90 | 8.787 | <0.0001 |
| Ap | E | Set 1 - Set 2 | 0.555 | 0.116 | 90 | 4.775 | <0.0001 |
| Ap | E | Set 1 - None | 1.146 | 0.116 | 90 | 9.867 | <0.0001 |
| Ap | E | Set 2 - None | 1.7 | 0.116 | 90 | 14.642 | <0.0001 |
Lb, L. brevis; Ap, A. pomorum.
Turkey corrected.
DISCUSSION
Our study suggests that microbial regulation of fly TAG is highly dependent not only on media preservatives and constituent yeast batch but also the yeast*preservative interaction. A specific combination of yeast and preservative formula was even sufficient to reverse the effect of microbial elimination in conventionally reared flies, producing a distinct experimental outcome. Preservative formula interfered with microbial effects particularly strongly, with the potential to block microbial regulation of host TAG. The data suggest that these effects are mediated by an impact of nipagin and propionic acid, either directly on the fly or via fly food, which is safeguarded against by A. pomorum (but not L. brevis) despite a cost to the bacteria themselves of compromised growth on the preservatives. These overlooked factors appear to be significant determinants of microbiota-dependent fly phenotypes and bacterial strain colonization densities, as well as major causes of microbiota-independent variation. Factors that we have not measured, such as dietary sugar (15), may further influence these complex interactions.
Our results have implications for future fly research and not only in the microbiota field. Sparse methodological detailing of diet is a persistent problem, e.g., with methods reporting “standard media” when media can in fact vary widely among labs. Preservatives are sometimes not reported, and yeast batch variation receives little attention in the lab or literature. Yet, our results indicate that these variables can determine experimental outcomes, with implications for repeatability. Our results are consistent with the suggestion that variability among labs may result from yeast batch variation (26). We suggest that diet standardization (e.g., chemically defined diet or chemostat-cultured yeast) may mitigate these potential confounding factors. Further studies are required to systematically determine how experimental contexts determine outcomes of manipulating the microbiota.
MATERIALS AND METHODS
Fly rearing and bacterial culturing
All flies were from the Dahomey background, which were originally collected in Dahomey, now Benin. They bore the w1118 mutation and were free of the endosymbiont Wolbachia. All flies (conventional, axenic, and gnotobiotes) were maintained at 25°C on a 12-hour light/dark cycle. SYA fly food was composed of 5% sucrose (Fisher), 10% yeast (MP Biomedicals), and 1.5% agar (Sigma). For the first two experiments, six different SYA diets were used, varying in yeast batch, either lot number S4707 (yeast A) or SR03010 (yeast B). From each batch, preservative-free food was made or food containing preservative set 1 (0.04% phosphoric acid and 0.4% propionic acid) or preservative set 2 (0.3% nipagin and propionic acid). This was repeated for the next set of experiments looking at the bacterial densities in each fly. For these experiments, the following yeast lot numbers were used: S6853 (yeast C), S7760 (yeast D), and U1122284494-1 (yeast E). Levilactobacillus brevis DmCS003 was grown and maintained in YPD medium at 30°C without shaking, while Acetobacter pomorum DmCS004 was grown and maintained in M9 medium with 0.5% DL-lactic acid at 30°C with shaking at 250 rpm.
CFU counts
Flies were anesthetized 3 days post-eclosion. For each condition, 6 replicates of 8 females were aseptically collected and transferred to a sterile Eppendorfs containing 500 µL 1X PBS. The flies were homogenized using a sterile pestle, and subsequently serially diluted and plated from the 100 to the 103 dilutions. Plates that had 30–300 colonies were counted for CFU determination. The CFUs were then calculated per fly and log10 transformed.
Generation of axenic and gnotobiotic flies
Flies were put in laying cages containing juice agar, transferred to a fresh cage, and allowed to lay eggs for <18 hours. Eggs were collected using PBS and a brush into a sterile chamber with netting. The chamber was incubated in 10% bleach for 3 minutes, followed by 1 minute in sterile dH2O, then 3 minutes in 10% bleach, 1 minute in 100% ethanol, and lastly 1 minute in sterile dH2O. Eggs were collected in sterile 1X PBS and 20 µL was pipetted into sterile T75 flasks with filter caps containing 60 mL of each variation of the SYA diets. Those without bacteria added remained axenic. To generate gnotobiotes, overnight bacterial cultures’ OD600 were measured, normalized to an OD600 = 1, and pelleted. The pellet was washed with sterile 1X PBS, resuspended to an OD600 = 1 in sterile 1X PBS, and then diluted 1:5 to a final concentration of OD600 = 0.2. Two hundred microliters of each bacterium was aseptically added to the surface of the SYA containing the sterilized eggs.
TAG experiments
The eggs were incubated for 10 days, when adult flies emerged. They were then transferred to sterile T75 flasks containing the appropriate diet. After 2 days on the diet (3 days post-eclosion), flies were collected, sorted by sex, and females were collected. Per experimental condition, ten groups of 5 females were weighed and flash frozen in 2 mL screw cap tubes containing 125 µL of TEt Buffer (TE buffer with 0.1% triton X-100). Flies were homogenized for 30 seconds using a Bead Ruptor Elite bead mill homogenizer at speed 6.5, incubated at 72°C for 15 minutes to inactivate endogenous lipases, and spun down for 5 minutes at 4°C at 12,000× g. In a 96-well plate, 3 µL of supernatant or standard was mixed with 300 µL of Infinity Triglycerides Reagent (Thermo Scientific), and plates were covered in foil and incubated at 37°C for 15 minutes. The absorbance at 540 nM was taken using a Thermo Scientific Multiscan FC plate reader. Standard curves were generated using an array of 9 glycerol standards ranging from 1 to 0 µg/µL, and TAG levels were calculated from the best fit line equation. TAG levels were normalized to the weight of the five flies.
Statistical analysis
All data were analyzed in R v4.2.1. Violin plots were produced using ggplot2.
For ANOVA analyses, linear models of the form
were fit using the base function lm, where TAG represented microgram TAG normalized to milligram fly mass, Yeast represented yeast batch, and preservative represented preservative formula. In the first experiment, Bacteria coded whether flies were axenic or conventionally reared. In the second experiment, Bacteria coded whether flies were reared axenically or gnotobiotically with either A. pomorum or L. brevis. All contrasts were set to “contrast sum”. ANOVA tests were applied with car::Anova, test type set to type-3. Post hoc comparisons were applied using emmeans::pairs, specifying comparisons within levels of Yeast and Preservatives.
Effect sizes were calculated using effectsize::eta_squared.
ACKNOWLEDGMENTS
This work was funded by BBSRC (BB/W510658/1), a UKRI Future Leaders Fellowship (MR/S033939/1), and a Lord Kelvin Adam Smith Fellowship from the University of Glasgow.
D.R.S. and A.J.D. designed the research, D.R.S. performed the research, D.R.S. and A.J.D. analyzed the data, D.R.S. drafted the manuscript, and A.J.D. edited the manuscript. We thank Nathan Woodling for helpful comments on the manuscript.
Contributor Information
David R. Sannino, Email: david.sannino@glasgow.ac.uk.
Adam J. Dobson, Email: adam.dobson@glasgow.ac.uk.
Irina S. Druzhinina, Royal Botanic Gardens, Surrey, United Kingdom
DATA AVAILABILITY
R script and data are freely available at https://github.com/dobdobby/preservatives-microbes-yeast
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.00165-23.
Supplementary Text and Supplementary figure legends.
Figure S1 with its legend.
Figure S2 with its legend.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Douglas AE. 2018. The drosophila model for microbiome research. Lab Anim 47:157–164. doi: 10.1038/s41684-018-0065-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wong CNA, Ng P, Douglas AE. 2011. Low‐diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ Microbiol 13:1889–1900. doi: 10.1111/j.1462-2920.2011.02511.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Broderick NA, Buchon N, Lemaitre B. 2014. Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. mBio 5:e01117-14. doi: 10.1128/mBio.01117-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Broderick NA, Lemaitre B. 2012. Gut-associated microbes of Drosophila melanogaster. Gut Microbes 3:307–321. doi: 10.4161/gmic.19896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bonfini A, Liu X, Buchon N. 2016. From pathogens to microbiota: how Drosophila intestinal stem cells react to gut microbes. Dev Comp Immunol 64:22–38. doi: 10.1016/j.dci.2016.02.008 [DOI] [PubMed] [Google Scholar]
- 6. Consuegra J, Grenier T, Baa-Puyoulet P, Rahioui I, Akherraz H, Gervais H, Parisot N, da Silva P, Charles H, Calevro F, Leulier F. 2020. Drosophila-associated bacteria differentially shape the nutritional requirements of their host during juvenile growth. PLoS Biol 18:e3000681. doi: 10.1371/journal.pbio.3000681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sannino DR, Dobson AJ, Edwards K, Angert ER, Buchon N. 2018. The Drosophila melanogaster gut microbiota provisions thiamine to its host. mBio 9:e00155-18. doi: 10.1128/mBio.00155-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gould AL, Zhang V, Lamberti L, Jones EW, Obadia B, Korasidis N, Gavryushkin A, Carlson JM, Beerenwinkel N, Ludington WB. 2018. Microbiome interactions shape host fitness. Proc Natl Acad Sci U S A 115:E11951–E11960. doi: 10.1073/pnas.1809349115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dobson AJ, Chaston JM, Newell PD, Donahue L, Hermann SL, Sannino DR, Westmiller S, Wong AC-N, Clark AG, Lazzaro BP, Douglas AE. 2015. Host genetic determinants of microbiota-dependent nutrition revealed by genome-wide analysis of Drosophila melanogaster. Nat Commun 6:7296. doi: 10.1038/ncomms8296 [DOI] [PubMed] [Google Scholar]
- 10. Newell PD, Douglas AE. 2014. Interspecies interactions determine the impact of the gut microbiota on nutrient allocation in Drosophila melanogaster. Appl Environ Microbiol 80:788–796. doi: 10.1128/AEM.02742-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dobson AJ, Chaston JM, Douglas AE. 2016. The Drosophila transcriptional network is structured by microbiota. BMC Genomics 17:975. doi: 10.1186/s12864-016-3307-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leitão-Gonçalves R, Carvalho-Santos Z, Francisco AP, Fioreze GT, Anjos M, Baltazar C, Elias AP, Itskov PM, Piper MDW, Ribeiro C. 2017. Commensal bacteria and essential amino acids control food choice behavior and reproduction. PLoS Biol 15:e2000862. doi: 10.1371/journal.pbio.2000862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F. 2011. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab 14:403–414. doi: 10.1016/j.cmet.2011.07.012 [DOI] [PubMed] [Google Scholar]
- 14. Shin SC, Kim S-H, You H, Kim B, Kim AC, Lee K-A, Yoon J-H, Ryu J-H, Lee W-J. 2011. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334:670–674. doi: 10.1126/science.1212782 [DOI] [PubMed] [Google Scholar]
- 15. Wong A-N, Dobson AJ, Douglas AE. 2014. Gut microbiota dictates the metabolic response of Drosophila to diet. J Exp Biol 217:1894–1901. doi: 10.1242/jeb.101725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Le Bourg E. 2022. Drosophila melanogaster flies better know than us the nutrients they need: let them choose. Exp Gerontol 162:111768. doi: 10.1016/j.exger.2022.111768 [DOI] [PubMed] [Google Scholar]
- 17. Sommer AJ, Newell PD. 2019. Metabolic basis for mutualism between gut bacteria and its impact on the Drosophila melanogaster host. Appl Environ Microbiol 85:e01882-18. doi: 10.1128/AEM.01882-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Obadia B, Keebaugh ES, Yamada R, Ludington WB, Ja WW. 2018. Diet influences host–microbiota associations in Drosophila. Proc Natl Acad Sci U.S.A 115. doi: 10.1073/pnas.1804948115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chandler JA, Innocent LV, Martinez DJ, Huang IL, Yang JL, Eisen MB, Ludington WB. 2022. Microbiome-by-ethanol interactions impact Drosophila melanogaster fitness, physiology, and behavior. iScience 25:104000. doi: 10.1016/j.isci.2022.104000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deshpande SA, Yamada R, Mak CM, Hunter B, Soto Obando A, Hoxha S, Ja WW. 2015. Acidic food pH increases palatability and consumption and extends Drosophila lifespan. J Nutr 145:2789–2796. doi: 10.3945/jn.115.222380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Henriques SF, Dhakan DB, Serra L, Francisco AP, Carvalho-Santos Z, Baltazar C, Elias AP, Anjos M, Zhang T, Maddocks ODK, Ribeiro C. 2020. Metabolic cross-feeding in imbalanced diets allows gut microbes to improve reproduction and alter host behaviour. Nat Commun 11:4236. doi: 10.1038/s41467-020-18049-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Consuegra J, Grenier T, Akherraz H, Rahioui I, Gervais H, da Silva P, Leulier F. 2020. Metabolic cooperation among commensal bacteria supports Drosophila juvenile growth under nutritional stress. iScience 23:101232. doi: 10.1016/j.isci.2020.101232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim G, Huang JH, McMullen JG, Newell PD, Douglas AE. 2018. Physiological responses of insects to microbial fermentation products: insights from the interactions between Drosophila and acetic acid. J Insect Physiol 106:13–19. doi: 10.1016/j.jinsphys.2017.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bass TM, Grandison RC, Wong R, Martinez P, Partridge L, Piper MDW. 2007. Optimization of dietary restriction protocols in Drosophila. J Gerontol A Biol Sci Med Sci 62:1071–1081. doi: 10.1093/gerona/62.10.1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gérard P. 2016. Gut microbiota and obesity. Cell Mol Life Sci 73:147–162. doi: 10.1007/s00018-015-2061-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Piper MDW, Blanc E, Leitão-Gonçalves R, Yang M, He X, Linford NJ, Hoddinott MP, Hopfen C, Soultoukis GA, Niemeyer C, Kerr F, Pletcher SD, Ribeiro C, Partridge L. 2014. A holidic medium for Drosophila melanogaster. Nat Methods 11:100–105. doi: 10.1038/nmeth.2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text and Supplementary figure legends.
Figure S1 with its legend.
Figure S2 with its legend.
Data Availability Statement
R script and data are freely available at https://github.com/dobdobby/preservatives-microbes-yeast