ABSTRACT
Pancreatic ductal adenocarcinoma (PDAC) exhibits elevated levels of autophagy, which promote tumor progression and treatment resistance. ATG4B is an autophagy-related cysteine protease under consideration as a potential therapeutic target, but it is largely unexplored in PDAC. Here, we investigated the clinical and functional relevance of ATG4B expression in PDAC. Using two PDAC patient cohorts, we found that low ATG4B mRNA or protein expression is associated with worse patient survival outcomes, poorly differentiated PDAC tumors and a lack of survival benefit from adjuvant chemotherapy. In PDAC cell lines, ATG4B knockout reduced proliferation, abolished processing of LC3B (also known as MAP1LC3B), and reduced GABARAP and GABARAPL1 levels, but increased ATG4A levels. ATG4B and ATG4A double knockout lines displayed a further reduction in proliferation, characterized by delays in G1-S phase transition and mitosis. Pro-LC3B accumulated aberrantly at the centrosome with a concomitant increase in centrosomal proteins PCM1 and CEP131, which was rescued by exogenous ATG4B. The two-stage cell cycle defects following ATG4B and ATG4A loss have important therapeutic implications for PDAC.
Keywords: Autophagy, ATG4B, ATG4A, Pancreatic cancer, PDAC, CEP131, Centrosome, PCM1, pro-LC3B, GABARAP, Doryphagy
Summary: Low ATG4B expression in pancreatic ductal adenocarcinoma is associated with poor clinical outcomes, and combined loss of ATG4B and ATG4A results in cell cycle defects, with implications for therapeutic strategies.
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is one of the leading causes of cancer-related deaths in North America. The grim survival rate of ∼10–11% for pancreatic cancer is partly due to limited treatment options for patients that are often diagnosed with metastatic disease (American Cancer Society, Cancer Facts & Figures 2022; Canadian Cancer Statistics, 2021). These challenges point out the need for more studies to better understand PDAC tumor biology and identify novel genetic targets.
The intracellular recycling pathway called autophagy has garnered attention as a possible therapeutic avenue for a variety of cancers, including pancreatic cancer (Yun and Lee, 2018). Studies have shown that pharmacological and genetic approaches to inhibit the autophagy pathway significantly reduce tumor growth and increase survival in PDAC mouse models (Yang et al., 2011, 2018). Hydroxychloroquine, which is a general lysosomal inhibitor, has been advocated as an autophagy inhibitor owing to its ability to shut down a late stage in autophagic flux – the fusion of autophagosomes with lysosomes (Mauthe et al., 2018). Although hydroxychloroquine has shown promise as a combination therapy agent in PDAC (Bryant et al., 2019; Kinsey et al., 2019), the non-specific nature of the drug has also been associated with dose-limiting toxicity in both mouse models and patients (Della Porta et al., 2020; Shintani and Klionsky, 2004). These limitations have stimulated efforts to identify alternative and more specific inhibitors of the autophagy pathway, with potential targets including the autophagy-related cysteine protease called ATG4B (Agrotis and Ketteler, 2019).
ATG4B is one of four proteases belonging to the ATG4 family – the others being ATG4A, ATG4C and ATG4D. Together, the ATG4 family members govern the priming and recycling of several substrates, namely the mammalian Atg8 orthologs – the LC3 proteins LC3A (also known as MAP1LC3A), LC3B (also known as MAP1LC3B) and LC3C (also known as MAP1LC3C), as well as GABARAP, GABARAPL1 and GABARAPL2 – which are required for the proper formation of autophagosomes (Weidberg et al., 2010). Although functional redundancy between the ATG4 family members exists, several kinetic studies have confirmed that, compared to the other ATG4 family members, ATG4B is functionally most adept in recognizing and priming all of the substrates mentioned (Agrotis et al., 2019; Kauffman et al., 2018; Li et al., 2011). Additionally, ATG4B expression has been associated with cell survival in a variety of cancers such as breast, colorectal and chronic myeloid leukemia (CML) (Bortnik et al., 2016; Liu et al., 2014; Rothe et al., 2014). However, ATG4B expression in human PDAC tumors has not been investigated extensively, nor has it been evaluated with respect to prognostic or predictive utility.
In pancreatic cancer, the modulation of ATG4B activity, which has been established using a dominant-negative mutant ATG4BC74A, results in complete tumor regression and improved long-term survival in a PDAC mouse model, enhancing the prospect of ATG4B as a potential drug target for pancreatic cancer (Agrotis et al., 2019; Kauffman et al., 2018; Li et al., 2011). Although the ATG4BC74A dominant-negative mutant serves as a useful tool to study the effects of autophagy inhibition, it is not equivalent to ATG4B reduction or loss. The protease activity of the ATG4B dominant-negative mutant is lost due to mutation of the catalytic cysteine residue (C74A), but the mutant protein retains its ability to bind Atg8 orthologs (Fujita et al., 2008; Yang et al., 2018). The formation of stable ATG4BC74A–Atg8 ortholog complexes results in blockade of the ATG7–Atg8 ortholog reaction and the subsequent lipidation of Atg8 orthologs (Fujita et al., 2008). Given that ATG4B preferentially binds to all the substrates compared to the other ATG4 family members, the ATG4B dominant-negative mutant might also restrict the other ATG4 homologs from accessing and processing substrates efficiently, resulting in a drained pool of primed substrates available for autophagic flux. Studies have shown that ATG4B knockout (KO) does not affect all of the substrates in a similar manner due to overlapping functions of the other ATG4 family members (Agrotis et al., 2019). These studies indicate that the effects of the ATG4BC74A dominant-negative mutant are partially distinct from ATG4B inhibition or KO, leaving the consequences of ATG4B loss in PDAC unknown.
While the ATG4–substrate axis has primarily been discussed in the context of autophagy, there is evidence to support autophagy-independent functions in cancer cells. For example, ATG4B-mediated tumor survival in colorectal cancers has been postulated to occur through pathways that affect cell proliferation (Liu et al., 2014). Aside from that, ATG4A expression has been shown to positively associate with the migration and invasion potential of gastric cancer cells both in vitro and in vivo (Yang et al., 2016). The phenotypes, which were found to be independent of autophagy, were shown to be due to an ATG4A–Notch signaling pathway axis, thus revealing a potential autophagy-independent function for ATG4A in gastric cancer (Yang et al., 2016). Additionally, the LC3 and GABARAP subfamily substrates were initially discovered for their functions outside of autophagy, mainly in cellular structural organization, which plays a critical role in many cellular functions including protein trafficking and cell division (Faller et al., 2009; Mansuy et al., 2004; Sagiv et al., 2000; Vernier-Magnin et al., 2005). For example, GABARAP, GABARAPL1 and GABARAPL2 have been shown to interact with several centrosomal proteins such as pericentriolar material 1 (PCM1) and centrosomal protein 131 (CEP131) (Joachim et al., 2015, 2017; Joachim and Tooze, 2017). Hence, the effects of ATG4 proteins go well beyond the canonical autophagy-based interpretations, raising additional questions regarding the potential scope of alterations following ATG4B loss in PDAC.
In this study, we aimed to examine the clinical relevance of ATG4B expression in PDAC patient tumors and the cellular consequences of ATG4B loss in PDAC cells. Using two independent cohorts, low ATG4B expression was shown to be associated with inferior PDAC patient outcomes. Loss of ATG4B, achieved though stable knockdown (KD) or CRISPR–Cas9-mediated KO, resulted in reduced PDAC proliferation, with negligible effects on cell death. ATG4B KO also resulted in increased ATG4A levels, significant reduction in GABARAP and GABARAPL1 levels, and the inhibition of LC3B processing. Double KO of ATG4B and ATG4A further reduced proliferation and GABARAP expression. Defects in proliferation in the PDAC ATG4A and ATG4B double KOs were attributed to delays in two stages of the cell cycle, the G1-S phase transition and mitosis. The mitotic defects were accompanied by alterations in centrosomal proteins that were rescued by ATG4B re-expression. Taken together, these results reveal novel links between ATG4B, ATG4A and cell cycle components in PDAC cell lines, and they indicate that targeting ATG4 family members might have molecular consequences with unexpected therapeutic implications.
RESULTS
Low or undetectable ATG4B expression is associated with worse survival of PDAC patients and poorly differentiated tumors
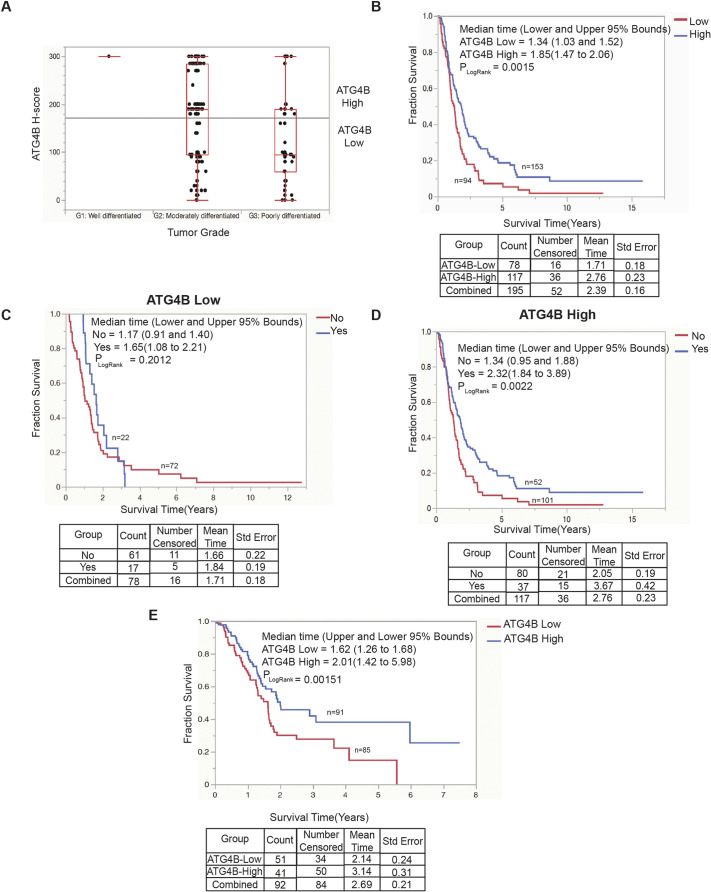
To study ATG4B protein expression in pancreatic tumors, a tissue microarray (TMA) containing 267 patient-derived PDAC specimens was stained and scored for ATG4B protein using a continuous H-score method (see Materials and methods). Twenty cases were excluded due to technical assay failure. Using the continuous H-score data modeled as a continuous variable, ATG4B expression was not found to be significantly associated with age at surgery, sex, invasion status (perineural and lymphovascular), lymph node status or resection status (Table S1). ATG4B expression was associated with histological grade, with grade 2 PDAC specimens having significantly higher ATG4B expression than grade 3 PDAC specimens (P<0.0001; one-way ANOVA) (Fig. 1A; Table S1). A borderline significant association of higher ATG4B expression was found for cases treated with adjuvant chemotherapy (gemcitabine or 5-fluorouracil; P=0.0474; one-way ANOVA). Categorization of cases into ATG4B-low or ATG4B-high expression categories using recursive partitioning yielded an H-score-based threshold of 160 ( ≥160 and <160 for high and low expressers, respectively). Kaplan–Meier analysis of our ATG4B categorized patient cohort (n=247) showed that patients in the ATG4B-low category (n=94; median time=1.34 years) had significantly worse disease-specific survival compared to patients in the ATG4B-high category (n=153; median time=1.85 years) (P=0.0015; log-rank test) (Fig. 1B). To determine whether ATG4B expression was an independent marker of prognosis, multivariable analyses were performed with all clinicopathological parameters. ATG4B was not found to be an independent prognostic marker (risk ratio high=0.84; 95% CI=0.61–1.14; P=0.27; Cox proportional hazards model). Next, ATG4B-low and ATG4B-high staining as potential predictive markers were evaluated for chemotherapy response. For patients with ATG4B-low tumors, there was no significant survival benefit of adjuvant chemotherapy (P=0.20; log-rank test) (Fig. 1C). However, patients with ATG4B-high tumors showed significantly better survival outcomes when treated with adjuvant chemotherapy (P=0.0022; log-rank test) (Fig. 1D), suggesting that ATG4B could have potential predictive utility for adjuvant chemotherapy with the pyrimidine analogs gemcitabine and 5-fluorouracil.
Fig. 1.
Low ATG4B expression is associated with poor PDAC patient outcomes. (A) Graph represents the ATG4B H-score distribution of PDAC specimens classified as either well differentiated (grade 1, G1), moderately differentiated (grade 2, G2) or poorly differentiated (grade 3, G3) tumor grade (P<0.0001, one-way ANOVA). Boxes represent the interquartile range, and the median is marked by a line. Whiskers represent minimum to maximum values. Horizontal line marks the mean of response across all observations. Total n=247. (B) Kaplan–Meier analysis comparing the disease-specific survival outcomes of the British Columbia PDAC patient cohort. Using recursive partitioning, the PDAC specimens were categorized as having either ATG4B-low (red; n=94; H-score<160) or ATG4B-high (blue; n=153; H-score≥160) protein expression (P=0.0015). Total n=247. (C) Disease-specific survival outcomes of PDAC patients with ATG4B-low tumors, stratified by treatment with adjuvant chemotherapy (blue: yes, chemotherapy treatment; red: no chemotherapy treatment). (D) Disease-specific survival outcomes of PDAC patients with ATG4B-high tumors, stratified by treatment with adjuvant chemotherapy (blue: yes, chemotherapy treatment; red: no chemotherapy treatment). (E) Kaplan–Meier analysis comparing the overall survival outcomes of TCGA PDAC patients (data courtesy of Human Protein Atlas, www.proteinatlas.org) with below average ATG4B RNA expression (ATG4B low; n=85) and equal to or above average ATG4B RNA expression (ATG4B high; n=91) (P=0.00151). In B–E, median survival time (years) for each group is shown, and the upper and lower 95% bounds for each category are denoted within brackets, with n shown on the graphs. A log-rank test (Mantel–Cox) was conducted to determine significant differences between survival curves (PLogRank). Details of count, along with mean survival time and standard error (Std error) are shown below the graphs.
To evaluate ATG4B mRNA expression in PDAC, we used pre-mined RNA data from The Cancer Genome Atlas (TCGA), courtesy of Human Protein Atlas (www.proteinatlas.org; Uhlén et al., 2015), to generate Kaplan–Meier curves. The average fragments per kilobase of transcript per million mapped reads (FPKM) of ATG4B RNA expression in 176 PDAC tumors was 9.122. Tumors below the average were classified as ATG4B low, while tumors displaying the average or more were classified as ATG4B high. The overall survival of patients in the ATG4B-low bin (n=85; median time=1.62 years) was found to be significantly lower compared to that of patients in the ATG4B-high bin (n=91; median time=2.01 years) (P=0.00151; log-rank test) (Fig. 1E).
To determine whether ATG4B protein expression in normal pancreas was associated with survival outcomes or other clinicopathological variables in PDAC patients, the levels of ATG4B in matched adjacent normal acinar cells (n=221) were analyzed. ATG4B expression was scored using the continuous H-score outlined for tumor epithelium and analyzed for association with clinicopathological variables. Aside from age at surgery, which showed a positive association with ATG4B H-score in the adjacent stromal component (Spearman's ρ=0.15; P=0.030), all other associations were statistically insignificant (data not shown).
Taken together, these findings indicate that low tumoral expression of ATG4B protein is significantly associated with high tumor grade, and that low expression of either ATG4B mRNA or protein are associated with inferior survival outcomes. ATG4B has potential predictive utility, as patients with high ATG4B tumoral expression appear to have significantly increased benefit from adjuvant chemotherapy.
ATG4B loss reduces proliferation of PDAC cells
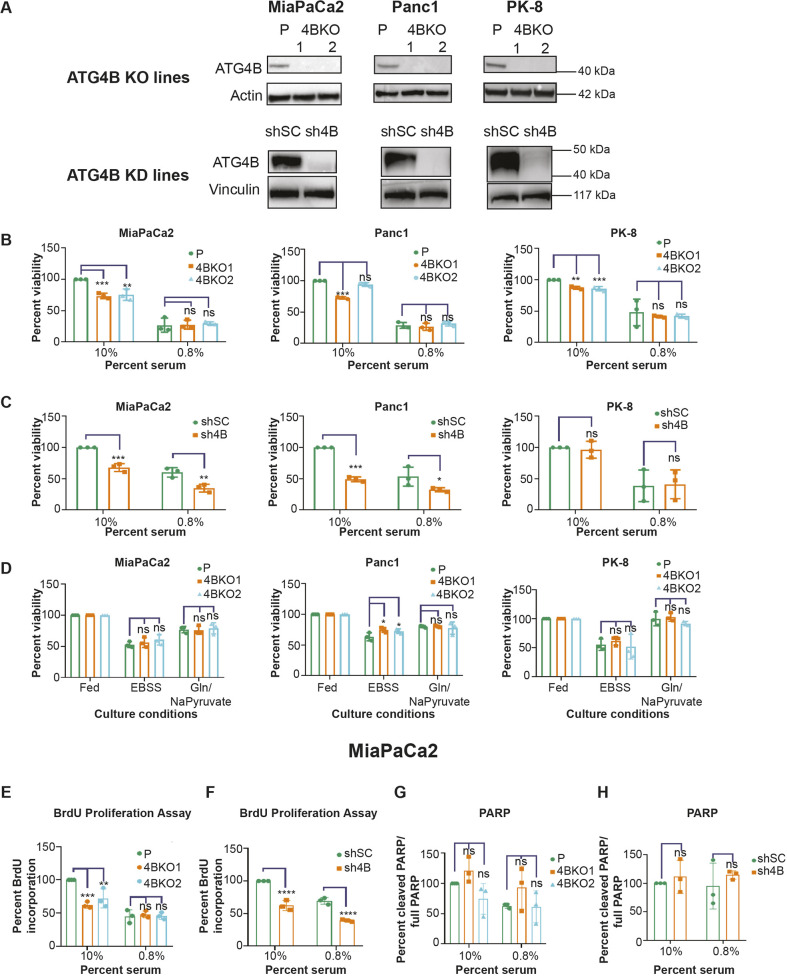
To characterize the phenotypes of low or absent ATG4B expression in pancreatic cancer cells, stable KD (using shRNA) or KO (using CRISPR–Cas9) of ATG4B was established in three PDAC cell lines: MiaPaCa2, Panc1 and PK-8. For each PDAC cell line, we derived a polyclonal population of cells exhibiting stable ATG4B KD (sh4B) or harboring a scramble-shRNA (shSC) control, and two monoclonal cell lines were validated for ATG4B KO (4BKO1 and 4BKO2) (Fig. 2A). A Crystal Violet assay was used to assess cell viability, which was defined as the number of adherent cells at the experiment end point, for each of the ATG4B KD and ATG4B KO lines compared to the respective parental cell line controls. Since autophagy is a stress response mechanism, the effects of ATG4B KD and KO in low serum (0.8% FBS) stress conditions as well as in standard serum (10% FBS) were determined. Five out of six ATG4B KO lines showed significantly reduced viability compared to their respective parental lines under 10% serum conditions (Fig. 2B); the Panc1-derived 4BKO2 line was the outlier that did not show a significant difference compared to its parental line (Fig. 2B). All six ATG4B KO lines displayed a further reduction in cell viability in the 0.8% serum condition, but they were not significantly different compared to their parental lines (Fig. 2B). Whereas the MiaPaCa2 and Panc1 ATG4B KD lines showed significant reduction in cell viability compared to the control lines (shSC) under both 10% and 0.8% serum conditions, the PK-8 ATG4B KD line did not show a significant difference in viability compared to the control under both serum conditions (Fig. 2C).
Fig. 2.
ATG4B loss leads to reduced proliferation of PDAC cells. (A) Representative immunoblots of ATG4B and either actin or vinculin (loading controls) in parental (P), ATG4B KO (4BKO), scramble control shRNA (shSC) and ATG4B KD (sh4B) cell lines in MiaPaCa2, Panc1 and PK-8 backgrounds. Distinct 4BKO clonal lines (4BKO1 and 4BKO2) were tested (n=3 biological replicates). (B,C) Parental and ATG4B KO (B), and stable shSC and sh4B (C) PDAC cell lines (MiaPaCa2, Panc1 and PK-8) were grown under 10% or 0.8% serum conditions, as indicated, for 3 days. Cells were stained with 0.1% Crystal Violet to visualize adherent cells at the experimental end point. Percentage of stained cells, normalized to respective parental lines (B) or shSC (C) in 10% serum conditions, is shown. Mean±s.e.m. of n=3 replicates. (D) Parental and ATG4B KO lines were grown under fed (10% serum) conditions, complete starvation (EBSS) or glutamine and sodium pyruvate deprivation (Gln/NaPyruvate). Each cell line was normalized against their fed control. Percentage stained adherent cells was determined using a Crystal Violet assay. Mean±s.e.m. of n=3 replicates. (E,F) Quantification of BrdU incorporation assay in MiaPaCa2 KO (E) and sh4B (F) cell lines in 10% and 0.8% serum conditions, as indicated. Data are normalized to the parental line (E) and shSC line (F) under 10% serum conditions. Mean±s.e.m. of n=3 replicates. (G,H) Quantification of PARP cleavage assay [(cleaved PARP/full-length PARP)×100] performed in ATG4B KO (G) and sh4B (H) MiaPaCa2 lines in 10% and 0.8% culture conditions, as indicated. Each cell line in G and H is BafA1-treated and was normalized against the shSC (H) or parental (G) line under 10% serum condition. Mean±s.e.m. of n=3 replicates. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001; ns, not significant (two-way ANOVA with Dunnett's multiple comparison tests for comparisons of parental and ATG4B KO lines; two-way ANOVA with Sidak's multiple comparisons test for comparisons of sh4B and shSC lines).
To determine whether the absence of viability differences between the ATG4B KO lines and their parental lines under 0.8% serum was dependent on the type of stress, cell viability was assessed under nutrient deprivation (using EBSS medium) or glutamine and sodium pyruvate-deprived (Gln/NaPyruvate) conditions. Similar to the effects of 0.8% serum, ATG4B KO lines displayed reduced cell viability under both of these conditions, but the reduction in viability was not significantly different compared to the respective parental lines (Fig. 2D). Using a bromodeoxyuridine (BrdU) assay to assess proliferation and a PARP cleavage assay to assess apoptosis, we determined that the reduction in cell viability observed in the ATG4B KD lines (under 10% and 0.8% serum conditions) and ATG4B KO lines (under 10% serum conditions) was due to reduced proliferation and not an increase in cell death (Fig. 2E–H; Fig. S1). Collectively, these findings indicate that ATG4B reduction or complete loss in PDAC cells leads to a modest decrease in viable cells by reducing cell proliferation.
ATG4B loss abolishes LC3B processing and alters levels or lipidation of GABARAP subfamily substrates in PDAC cells
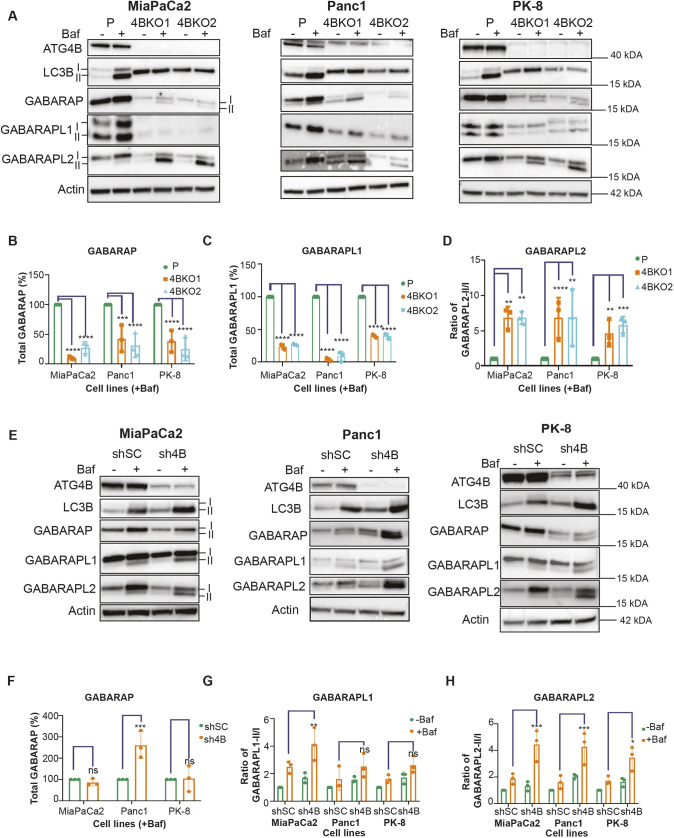
To determine the effects of ATG4B loss on its substrates in PDAC cells, we examined the expression, lipidation and processing of LC3B, GABARAP, GABARAPL1 and GABARAPL2 under standard 10% serum conditions. As expected, LC3B priming was abolished in the ATG4B KO lines of MiaPaCa2, Panc1 and PK-8 (Fig. 3A), as indicated by a single band that has previously been shown to correspond to the pro-form of LC3B (Agrotis et al., 2019; Bosc et al., 2018; Wang et al., 2013). The levels of GABARAP and GABARAPL1 were significantly lower in all the ATG4B KO lines compared to their respective parental lines (Fig. 3A–C), which agrees with previous studies showing that ATG4B is required to stabilize the levels of GABARAP and GABARAPL1 (Skytte Rasmussen et al., 2017). Although protein levels were reduced, we detected an increase in the lipidated form of GABARAP, in the presence of bafilomycin A1 (BafA1), in MiaPaCa2, Panc1 and PK-8 ATG4B KO lines, indicating increased flux (Fig. S2A). GABARAPL1 flux in the KO lines was variable (Fig. S2B). For GABARAPL2, levels of the lipidated form were significantly increased in the KO backgrounds of all three cell lines (Fig. 3D). No consistent changes were observed in protein levels or flux of LC3B, GABARAP and GABARAPL1 across the ATG4B KD lines of MiaPaCa2, Panc1 and PK-8 compared to their respective shSC control lines (Fig. 3E–G; Fig. S2C,D). The exception was with regard to changes in GABARAPL2, where levels of the lipidated form were also significantly increased in all three cell lines in the KD background (Fig. 3H). Although we observed similar substrate alterations in low (0.8%) serum conditions, they were not significantly different compared to the alterations observed in 10% serum conditions (Fig. S2E–M). The consistent increase in levels of lipidated GABARAP and GABARAPL2 across all PDAC ATG4B KO lines suggests that GABARAP and GABARAPL2 play a compensatory role upon ATG4B loss.
Fig. 3.
ATG4B KO or KD enhances GABARAPL2 utilization. (A) Representative immunoblots of ATG4B, LC3B, GABARAP, GABARAPL1, GABARAPL2 and actin (loading control) in parental (P) and ATG4B KO (4BKO1 or 4BKO2) MiaPaCa2, Panc1 and PK-8 cell lines. Cells were grown in 10% serum with or without BafA1 (Baf). Lipidated (II) and unlipidated (I) forms are indicated for MiaPaCa2. Blots shown are representative of n=3. (B,C) Quantification of (B) total GABARAP (GABARAP-I and GABARAP-II) or (C) total GABARAPL1 (GABARAPL1-I and GABARAPL1-II) in the presence of Baf from three biological replicates. Each ATG4B KO cell line was normalized to its respective parental line (MiaPaCa2, Panc1 and PK-8). (D) Ratio of lipidated GABARAPL2 (GABARAPL2-II) to unlipidated GABARAPL2 (GABARAPL2-I) in Baf-treated samples in ATG4B KO lines and their respective parental lines. Values were normalized to their respective parental lines (n=3). (E) Representative immunoblots of ATG4B, LC3B, GABARAP, GABARAPL1, GABARAPL2 and actin (loading control) in scramble control (shSC) and ATG4B KD (sh4B) lines in MiaPaCa2, Panc1 and PK-8 backgrounds. Cells were grown in 10% serum with or without BafA1. Lipidated (II) and unlipidated (I) forms are indicated for MiaPaCa2. Blots shown are representative of n=3. (F) Quantitation of total GABARAP (GABARAP-I and GABARAP-II) from the three biological replicates with BafA1. Each cell line was normalized to its respective control (shSC). (G) The ratio of GABARAPL1-II to GABARAPL1-I in sh4B lines and the respective shSC lines with and without BafA1 (n=3). (H) Ratio of lipidated GABARAPL2 (GABARAPL2-II) to unlipidated GABARAPL2 (GABARAPL2-I) with and without BafA1 in the three biological replicates. In G and H, values were normalized to the shSC controls without BafA1 for each cell line. In B–D and F–G, data are presented as mean±s.e.m. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001; ns, not significant (two-way ANOVA with Dunnett's multiple comparisons tests for comparisons of parental and ATG4B KO lines; two-way ANOVA with Sidak's multiple comparisons test for comparisons of sh4B and shSC lines).
ATG4B KO lines have increased ATG4A, which supports PDAC cell viability
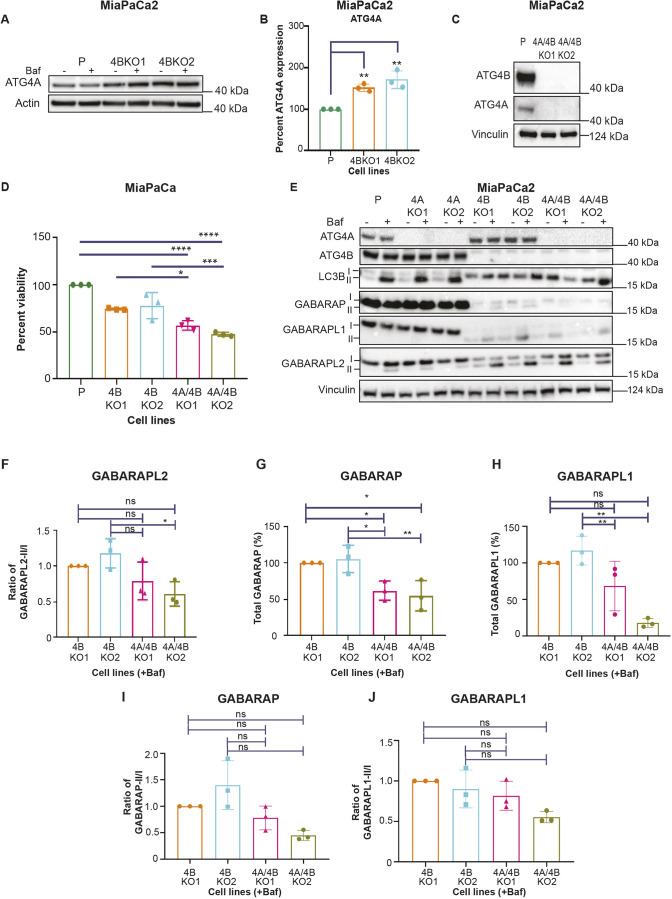
The increased levels of lipidated GABARAP and GABARAPL2 in the ATG4B KO lines prompted us to investigate possible compensation by ATG4A, the ATG4 family member shown to have the highest affinity towards the GABARAP subfamily (Agrotis et al., 2019). We found that the protein level of ATG4A, but not of ATG4C or ATG4D, was significantly increased in the ATG4B KO PDAC cell lines compared to their respective parental lines (Fig. 4A,B; Fig. S3A–E). To study the consequences of ATG4A loss, ATG4A KO lines (4AKO1 and 4AKO2) and ATG4B and ATG4A double KO lines (4A/4BKO1 and 4A/4BKO2, referred to collectively as ATG4A/4B KO) were generated in MiaPaCa2, Panc1 and PK-8 PDAC cells. For MiaPaCa2 and Panc1, two monoclonal ATG4A/4B KO lines each were produced, but only one monoclonal ATG4A/4B KO line was successfully generated for PK-8 (Fig. 4C; Fig. S3F–H).
Fig. 4.
ATG4B KO lines have elevated ATG4A levels, and the combined loss of ATG4B and ATG4A further reduces viability as well as levels of GABARAP and GABARAPL1. (A) Representative immunoblot of ATG4A and actin (loading control) in MiaPaCa2 parental (P) and ATG4B KO lines with (+) and without (−) BafA1 (Baf) treatment (representative of n=3). (B) Bar graph showing quantification of ATG4A levels, from western blots as shown in A (percentage, normalized to the parental line), in samples from BafA1-treated MiaPaCa2 parental and ATG4B KO lines Mean±s.e.m. (n=3). **P<0.01 (ordinary one-way ANOVA with Dunnett's multiple comparisons tests). (C) Immunoblot representation of ATG4B, ATG4A and vinculin in parental and ATG4A/4B KO lines of MiaPaCa2 (representative of n=3). (D) Graphical representation of cell viability (%), determined using the Crystal Violet assay, of MiaPaCa2 parental, ATG4B KO and ATG4A/4B KO lines grown in 10% serum conditions for 72 h. Each sample was normalized to the parental line, but statistical significance was determined by comparing each ATG4A/4B KO line to the ATG4B KO line it was derived from (e.g. 4BKO1 versus 4A/4BKO1) or by comparing parental and ATG4A/4B KO lines. Mean±s.e.m. (n=3). *P<0.05; ***P<0.001; ****P<0.0001 (ordinary one-way ANOVA with Dunnett's multiple comparison tests). (E) Immunoblot of ATG4A, ATG4B, LC3B, GABARAP, GABARAPL1, GABARAPL2 and vinculin (loading control) in MiaPaCa2 parental, ATG4A KO, ATG4B KO and ATG4A/4B KO lines with (+) and without (−) BafA1 treatment (representative of n=3). Lipidated (II) and unlipidated (I) forms are indicated. (F–J) Bar graphs of (F) ratio of lipidated GABARAPL2 (GABARAPL2-II) to unlipidated GABARAPL2 (GABARAPL2-I), (G) total GABARAP (GABARAP-I and GABARAP-II), (H) total GABARAPL1 (GABARAPL1-II and GABARAPL1-I), (I) ratio of lipidated GABARAP (GABARAP-II) to unlipidated GABARAP (GABARAP-I), and (J) ratio of lipidated GABARAPL1 (GABARAPL1-II) to unlipidated GABARAPL1 (GABARAPL1-I), all in MiaPaCa2 ATG4B KO and ATG4A/4B KO lines with BafA1 treatment. Data are normalized to 4BKO1. Mean±s.e.m. (n=3). *P<0.05; **P<0.01; ns, not significant (ordinary one-way ANOVA with Dunnett's multiple comparisons tests).
ATG4A KO alone did not lead to consistent viability changes across all PDAC cell lines (Fig. S4A). However, the combined loss of ATG4B and ATG4A generally resulted in a further reduction in cell viability. We observed that four out of five ATG4A/4B KO lines showed reduced viability when compared to their respective parental line, with three out of those four ATG4A/4B KO lines showing a significant further reduction in cell viability when compared to their respective ATG4B KO line (Fig. 4D; Fig. S3G,H). Loss of ATG4A alone had no detectable effect on the substrates LC3B, GABARAP, GABARAPL1 and GABARAPL2 in MiaPaCa2, Panc1 and PK-8 cells compared to their respective parental lines (Fig. 4E; Fig. S4B–F). The combined loss of ATG4B and ATG4A had no additional effects on flux of GABARAPL2 compared to ATG4B KO alone (Fig. 4E,F). Levels of ATG4C and ATG4D were also not altered in the ATG4A/4B KO line (Fig. S3C–E). GABARAP and GABARAPL1 levels were further reduced in the ATG4A/4B KO background (Fig. 4G,H) but flux was not significantly altered (Fig. 4I,J). The lack of detectable viability effects in ATG4A KO alone, along with a further decrease in viability observed in the ATG4A/4B KO lines, suggests that ATG4A partially compensates for the absence of ATG4B, at least with respect to PDAC cell viability.
The combined loss of ATG4B and ATG4A causes significant cell cycle delay
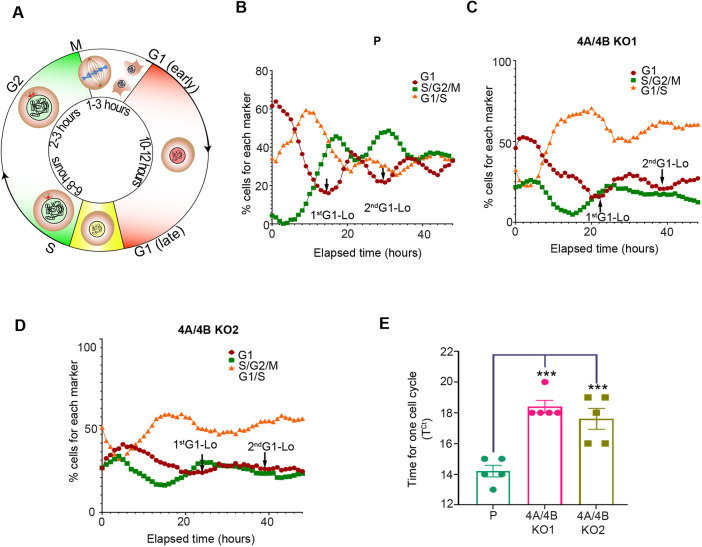
To help determine how the combined loss of ATG4B and ATG4A reduced cell viability, growth kinetics for MiaPaCa2 ATG4A/4B KO lines were analyzed using live-cell and dead-cell sensors NucLight Rapid Red and Cytotox Green, respectively, in an IncuCyte live-cell analysis system. Over the span of 72 h, MiaPaCa2 ATG4A/4B KO lines displayed significantly delayed growth compared to the parental line, but they showed no increase in cell death (Fig. S5A–C). A confluence-based cell proliferation assay showed reduced proliferation of ATG4A/4B KO cells relative to the parent cells (Fig. S5D). To study cell cycle progression, we generated MiaPaCa2 parental and ATG4A/4B KO lines stably expressing a fluorescence ubiquitination-based cell cycle indicator (FUCCI) for real-time visualization of cell cycle phases in the IncuCyte system. This method allowed the detection of cells in G1 phase (red); S phase, G2 phase and G2-M transition (green); and G1-S transition (yellow) of the cell cycle (Fig. 5A). The FUCCI reporter assay showed a delay in the G1-S transition, with an increasing percentage of cells in G1-S in the ATG4A/4B KO lines compared to the parental control (Fig. 5B–D). The time it takes for cells to complete one cell cycle was significantly longer for both ATG4A/4B KO lines compared to the parental line (Fig. 5E). These findings suggest that the combined loss of ATG4B and ATG4A disrupts normal cell cycle progression.
Fig. 5.
Loss of ATG4B and ATG4A results in cell cycle delay in PDAC cells. (A) Schematic depiction of the FUCCI indicator at different stages of the cell cycle. Red nuclei indicate cells in G1 phase; green nuclei indicate cells in S phase, G2 phase and G2-M transition; and yellow nuclei indicate cells in G1-S transition. Colorless nuclei indicate cells transitioning between M and G1 phases. (B–D) Red and green fluorescence images of (B) MiaPaCa2 parental (P), (C) 4A/4BKO1 and (D) 4A/4BKO2 cells expressing the FUCCI indicator were acquired on an Incucyte live-cell analysis system every hour for up to 80 h. The percentage of cells with red (G1), green (S/G2/M), or both red and green (overlap, yellow; G1/S) nuclei were calculated as a function of time using the Incucyte basic analyzer software. Representative graphs from five independent replicates are shown. Arrows indicate minima in percentage of cells in G1 across successive cell cycles (1st G1-Lo, 2nd G1-Lo). (E) Time for one cell cycle was calculated for MiaPaCa2 parental, 4A/4BKO1 and 4A/4BKO2 cells. Time between the first and second G1 minimum (G1-Lo, as indicated in B–D) was used to calculate the time taken for one cell cycle (TCt). Mean±s.e.m. of five replicates. ***P<0.001 (ordinary one-way ANOVA with Dunnett's multiple comparisons test).
ATG4A/4B KO-mediated cell cycle delay occurs at two stages and results in increased levels of PCM1 and CEP131
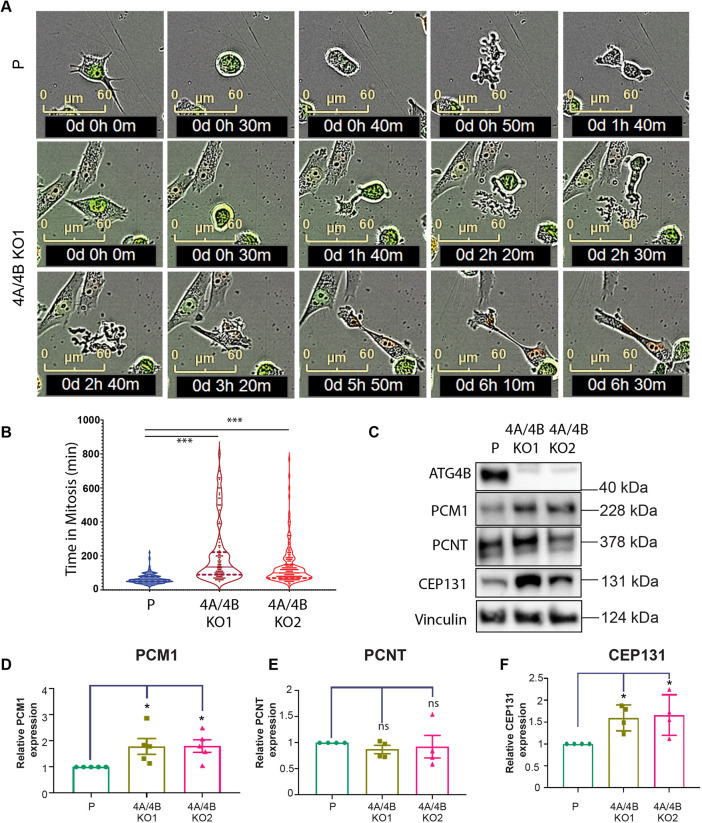
We observed an accumulation of cells in G1-S that was associated with a delay at G1-S transition in the ATG4A/4B KO cells relative to the parental line (Fig. 5B–D). Correspondingly, the percentage of cells in G1 phase or in S, G2 or M (S/G2/M) phases consistently decreased over time in the ATG4A/4B KO lines (Fig. 5B–D). The absence of fluorescence between the M and early G1 phase in the FUCCI fluorescence-based quantification method (Fig. 5A) prevents the detection of alterations in this stage. To address this, the IncuCyte live-imaging system was used to monitor the MiaPaCa2 parental and ATG4A/4B KO lines at 10-min intervals during mitosis (Fig. 6A). A significant delay in mitosis completion was detected in the ATG4A/4B KO lines compared to the parental line (Fig. 6B), particularly during the transition from metaphase to cytokinesis (Fig. 6A; Movies 1 and 2). The selective turnover of centriolar satellite proteins, such as PCM1, via autophagy mediated by GABARAP proteins has previously been shown to be crucial for centrosome integrity and proper chromosome segregation during mitosis (Holdgaard et al., 2019). To study the effects of ATG4B and ATG4A loss on centrosomal proteins, the expression of three centrosomal proteins – PCM1, CEP131 and pericentrin (PCNT) was examined in the MiaPaCa2 parental and ATG4A/4B KO lines. Levels of PCM1 and CEP131, but not of PCNT, were significantly increased in the MiaPaCa2 ATG4A/4B KO lines compared to levels in the parental line (Fig. 6C–F).
Fig. 6.
ATG4A/4B loss leads to mitotic defects, increased time in mitosis and increase in PCM1 and CEP131 levels. (A) Time-lapse imaging of MiaPaCa2 parent (P) and ATG4A/4B KO cells (4A/4BKO1) stably expressing FUCCI. Phase-contrast and fluorescence images were acquired every 10 min using the Incucyte system to determine the time taken for cells to complete mitosis. Examples of a parental MiaPaCa2 cell (40 min in mitosis; rounding at 10 min and division at 50 min, top panel) and an ATG4A/4B KO cell (>2 h in mitosis; rounding at 30 min and division at 2 h 40 m; bottom panels) are shown. See also Movies 1 and 2. (B) Graph represents the quantification of time in mitosis for MiaPaCa2 parent (P; n=229), 4A/4B KO1 (n=132) and 4A/4B KO2 (n=175); total n is a sum of cells from three biological replicates. Violin plots show the distribution of time in mitosis, with the median (line) and the upper and lower quartiles (dashed lines) indicated. (C) Representative western blot of centrosomal proteins PCM1, PCNT and CEP131, along with ATG4B and vinculin as loading control, in MiaPaCa2 parental (P) and ATG4A/4B KO (4A/4BKO1 and 4A/4BKO2). Representative of n=4. (D–F) Expression levels (quantified by western blotting and expressed relative to levels in parental cells) of (D) PCM1, (E) PCNT and (F) CEP131 were quantified in MiaPaCa2 parental and ATG4A/4B KO cell lines (n=4). Mean±s.e.m. *P<0.05; ns, not significant (ordinary one-way ANOVA with Dunnett's multiple comparisons test relative to MiaPaCa2 parental).
Pro-LC3B accumulates in the pericentric region of ATG4A/4B KO cells, and its loss reduces cell viability
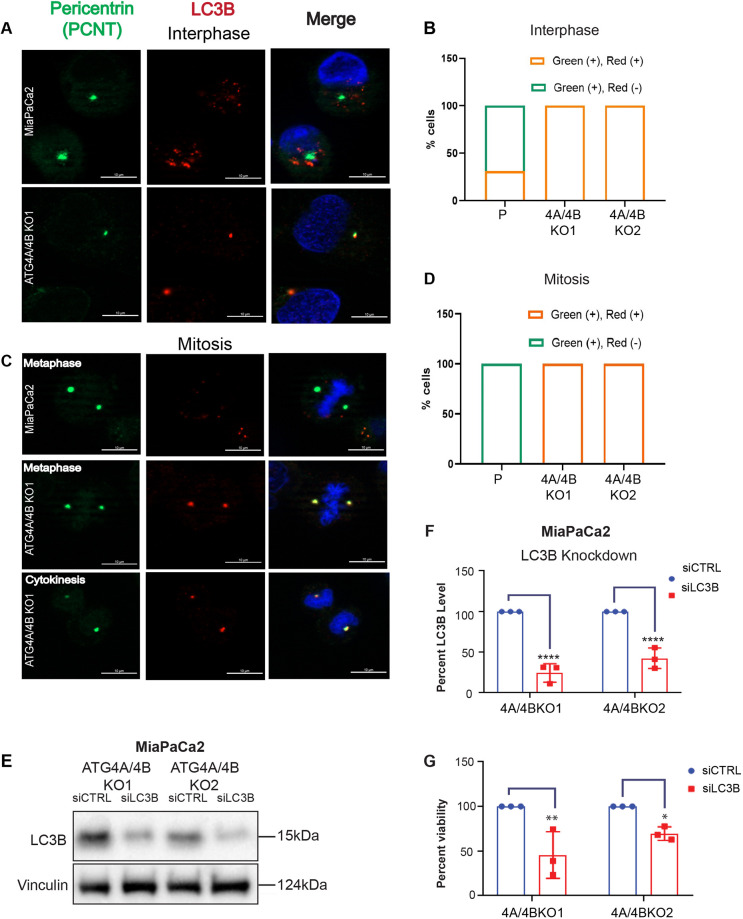
To explore a potential centrosomal relationship in the context of ATG4A/4B KO, the localization of GABARAPL1, GABARAPL2 and LC3B at the centrosomal region was studied. Since PCNT levels were unaltered in the ATG4A/4B KO lines, we used PCNT to label the centrosomal region. MiaPaCa2 parental cells and the ATG4A/4B KO lines showed localization of GABARAPL1 or GABARAPL2 with PCNT (red and green fluorescence, respectively; Fig. S6) at the pericentric region during interphase (>50% of all cells) (Fig. S6B,E) and mitotic phases (100% for all cells) (Fig. S6C,F). In addition, ∼30% of MiaPaCa2 parental cells showed localization of LC3B to the pericentric region (represented by PCNT) during interphase (Fig. 7A,B) but not in the mitotic phase (Fig. 7C,D). Strikingly, in the ATG4A/4B KO cells, pro-LC3B localized to the centrosomal region in 100% of cells both at interphase and mitosis (Fig. 7A–D). These results demonstrate that in the ATG4A/4B KO background, pro-LC3B accumulates aberrantly at centrosomes.
Fig. 7.
Pro-LC3B localizes to centrosomes at both interphase and mitosis and supports viability. (A) Representative images of PCNT (green) and LC3B (red) in MiaPaCa2 parental cells (cleaved LC3B) and 4A/4B KO1 cells (pro-LC3B) at interphase. DNA is stained using DAPI (blue). Scale bars: 10 µm. (B) Quantification of PCNT (green) and LC3B (red) colocalization [green (+), red (+)], as well as PCNT alone [green (+), red (−)], at centrosomes (parental, P, n=74 cells; 4A/4BKO1, n=84 cells; 4A/4BKO2, n=71 cells; sum from at least two biological replicates). (C) Representative images of PCNT (green) and LC3B (red) in MiaPaCa2 parental (cleaved LC3B) and 4A/4B KO1 (pro-LC3B) cells at mitosis. DNA is stained using DAPI (blue). Scale bars:10 µm. (D) Quantification of PCNT (green) and LC3B (red) colocalization [green (+), red (+)], as well as PCNT alone [green (+), red (−)], at centrosomes (parental, P, n=16 cells; 4A/4BKO1, n=19; 4A/4BKO2, n=15; sum from at least two biological replicates). (E) MiaPaCa2 ATG4A/4B KO lines were treated with either siCTRL (scramble) or siLC3B (siRNA targeting LC3B) for 8 days (siRNA replaced every 72 h). Immunoblot of pro-LC3B and vinculin (loading control) in MiaPaCa2 ATG4A/4B KO lines. Representative image of three biological replicates. (F) Quantification of pro-LC3B levels (expressed as a percentage relative to siCTRL) in MiaPaCa2 ATG4A/4B KO lines treated with either siCTRL or siLC3B. Each sample was normalized to the respective siCTRL (n=3). (G) Graph shows the percentage viability of MiaPaCa2 ATG4A/4B KO lines treated with either siCTRL or siLC3B. Cell viability was assessed using Crystal Violet staining at the experimental end point (8 days). Each sample was normalized to the respective siCTRL (n=3). In F and G, data are presented as mean±s.e.m. *P<0.05; **P<0.01; ****P<0.0001 (two-way ANOVA with Sidak's multiple comparisons test).
To further study pro-LC3B in the ATG4A/4B KO lines, an siRNA-mediated KD approach was taken. Multiple siRNA treatments (siLC3B) were required to achieve at least 50% or more KD of pro-LC3B in these cell lines (Fig. 7E,F). Compared to the scramble control (siCTRL), KD of pro-LC3B led to a significant reduction in viability of both MiaPaCa2 ATG4A/4B KO cell lines (Fig. 7G).
ATG4B re-expression rescues Atg8 protein family levels and aberrant accumulation of centrosomal proteins
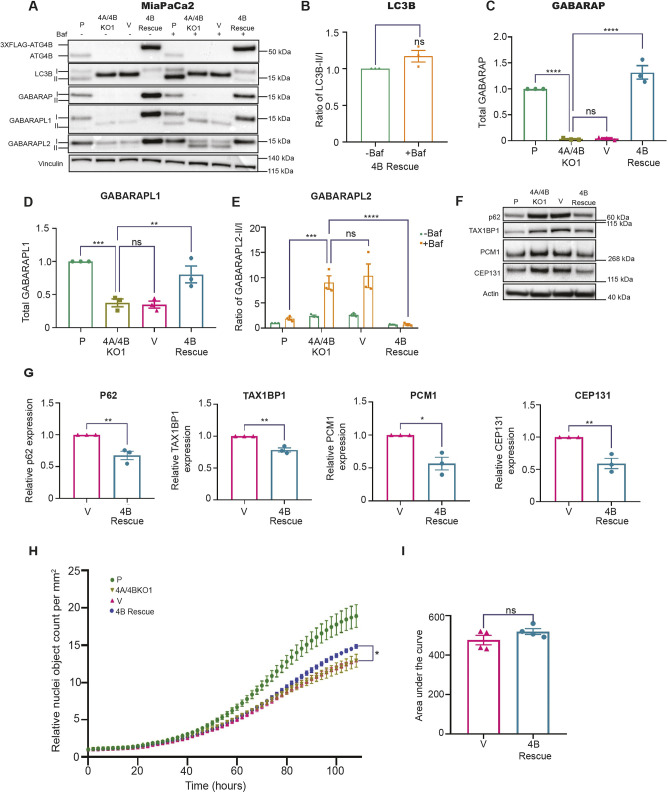
To confirm whether the observed cellular and molecular phenotypes in the ATG4A/4B KO lines were due, in part, to ATG4B loss, we performed ectopic ATG4B rescue experiments. We created stable ATG4A/4B KO lines expressing either N-terminally or C-terminally FLAG-tagged ATG4B (N-3×FLAG–ATG4B or C-3×FLAG–ATG4B, respectively; collectively referred to as 4B rescue), or the corresponding N-3×FLAG or C-3×FLAG vector-only (V) controls (Fig. 8). Using these lines, we measured protein levels of ATG4B, Atg8 family members, p62 (also known as SQSTM1), TAX1BP1, PCM1 and CEP131. First, we examined Atg8 family member expression levels and processing in the absence and presence of BafA1 (Fig. 8A–E). LC3B processing was rescued, as indicated by loss of detectable pro-LC3B levels and detection of the LC3B-I and LC3B-II forms in the ATG4B re-expression line versus the vector-only control (Fig. 8A). GABARAP levels were also significantly restored by ATG4B re-expression (Fig. 8A,C), which is consistent with the stabilization of GABARAP by ATG4B (Skytte Rasmussen et al., 2017). The protein levels of GABARAPL1 and GABARAPL2 were restored by ATG4B re-expression, though the lipidated forms were not detected, even in the presence of BafA1 (Fig. 8A,E). As expected, these findings show that Atg8 family processing and levels can be at least partially attributed to ATG4B.
Fig. 8.
ATG4B re-expression rescues LC3B processing, GABARAP subfamily protein levels, and the accumulation of autophagy receptors and centrosomal proteins. (A) Representative immunoblots of ATG4B, LC3B, GABARAP, GABARAPL1, GABARAPL2 and vinculin (loading control) in MiaPaCa2 parental (P), 4A/4BKO1, 4A/4BKO1 with N-3×FLAG empty vector (V) and 4A/4BKO1 with N-3×FLAG–ATG4B (4B Rescue) cell lines. Cells were grown in 10% serum for 48 h and treated with (+) or without (−) BafA1 (Baf) for 4 h prior to harvesting. Lipidated (II) and unlipidated (I) forms are indicated. Blots shown are representative of n=3. (B) Ratio of lipidated LC3B (LC3B-II) to unlipidated LC3B (LC3B-I) in the absence and presence of BafA1 in the 4B Rescue line, normalized to the no BafA1 condition. Mean±s.e.m., n=3 (ns, not significant; two-tailed unpaired t-test). (C) Quantification of total GABARAP (GABARAP-I and GABARAP-II) in the presence of BafA1 for the indicated cell lines. Values were normalized to parental (P) control. n= 3. (D) Quantification of total GABARAPL1 (GABARAPL1-I and GABARAPL1-II) in the presence of BafA1 for the indicated cell lines. Values were normalized to parental (P) control. n=3. (E) Quantification of the ratio of lipidated GABARAPL2 (GABARAPL2-II) to unlipidated GABARAPL2 (GABARAPL2-I) in the presence and/or absence of BafA1 for the indicated cell lines (n=3). Values were normalized to BafA1-untreated samples. n=3. In C–E, data are presented as mean±s.e.m. **P<0.01; ***P<0.001; ****P<0.0001; ns, not significant (ordinary one-way ANOVA with Dunnett's multiple comparisons test was used in C and D to determine statistical significance between 4A/4BKO1 and P, V and 4B Rescue for GABARAP and GABARAPL1; two-way ANOVA with Tukey's multiple comparisons test was used in E to determine statistical significance between 4A/4BKO1 and P, V and 4B Rescue for GABARAPL2 in BafA1-treated samples). (F) Representative immunoblots of p62, TAX1BP1, PCM1, CEP131 and actin (loading control) in MiaPaCa2 parental (P), 4A/4BKO1, 4A/4BKO1 with N-3×FLAG empty vector (V), and 4A/4BKO1 with N-3×FLAG–ATG4B (4B Rescue) cell lines. Cells were grown in 10% serum for 48 h and harvested without the addition of BafA1. Blots shown are representative of n=3. (G) Quantitation of p62, TAX1BP1, PCM1 and CEP131. Values were normalized to expression levels in the vector-only control (V). Mean±s.e.m., n=3. *P<0.05; **P<0.01 (two-tailed unpaired t-test). (H) Curves representing the total nuclei count of MiaPaCa2 parental (P), 4A/4BKO1, 4A/4BKO1 with C-3×FLAG empty vector (V) and 4A/4BKO1 with C-3×FLAG–ATG4B (4B Rescue) cell lines, generated using the Incucyte system. Cells were grown in 10% serum, stained with NucLight Rapid Red and Cytotox Green dyes, and left to grow for 108 h (4.5 days). Mean±s.e.m., n=4. *P<0.05 (comparison of V versus 4B Rescue at 108 h end point; two-tailed unpaired t-test). (I) Graph showing area under the curve for V and 4B Rescue cell lines in H. Mean±s.e.m, n=4. ns, not significant (two-tailed unpaired t-test).
To determine the effects of ATG4B re-expression on autophagic flux, we next evaluated levels of two autophagy receptors: p62 and TAX1BP1. Relative to levels in the vector-only control, levels of both p62 and TAX1BP1 were significantly reduced by ATG4B re-expression, which indicates an increase in autophagic flux (Fig. 8F,G). The increase in LC3B-II in the presence of BafA1 in the ATG4B re-expression cells (Fig. 8A,B) further supports an increase in autophagic flux. Consistent with the observed increase in autophagic flux, ATG4B re-expression reduced the aberrant accumulation of centrosomal proteins PCM1 and CEP131 (Fig. 8F,G). To determine whether ATG4B re-expression in the 4A/4B KO1 line can at least partly restore cell proliferation, the IncuCyte assay with live-cell and dead-cell sensors was performed. In line with the modest reduction in proliferation upon ATG4B loss alone (Fig. S5), the re-expression of ATG4B showed a reproducible, though modest, increase in proliferation relative to the vector-only control, which reached significance only at the 108 h end point (Fig. 8H,I). Collectively, these findings substantiate a new role for ATG4B in centrosomal protein regulation and also further suggest that ATG4A, in addition to ATG4B, has important roles to play in PDAC cell cycle and proliferation.
DISCUSSION
ATG4B inhibition has previously been shown to result in tumor suppression in pancreatic cancer, making this cysteine protease a potential target for anti-pancreatic cancer therapies (Yang et al., 2018). However, the expression of ATG4B in PDAC tumors and surrounding stroma has not been evaluated in a large cohort, and the consequences of ATG4B loss in PDAC cells have not been fully elucidated. In this study, ATG4B was shown to be variably expressed in PDAC tumors and stroma, and high levels of tumoral ATG4B expression corresponded to a higher patient survival and might have predictive value for adjuvant chemotherapy in PDAC patients. We demonstrated that the loss of ATG4B in PDAC cells leads to decreased levels of proliferation and not an increase in cell death. Double KO of ATG4B and ATG4A led to a further decrease in levels of their substrates – GABARAP and GABARAPL1 – and greater reductions in cell proliferation. Cellular analyses revealed defects at two stages of the cell cycle, the G1-S transition and mitosis. Loss of ATG4B has been reported previously to reduce CCND1 expression and the transition from G1 phase to S phase in colorectal cancer cells (Liu et al., 2014), but a role in mitosis has not been previously described. Our molecular analyses showed that the mitotic alterations were accompanied by an increase in PCM1 and CEP131, as well as the persistent centrosomal accumulation of pro-LC3B. Compared to the vector-only control, ATG4B re-expression in the 4A/4B KO1 line partially rescued the processing of pro-LC3B; the levels of GABARAP, GABARAPL1 and GABARAPL2; and the aberrant accumulation of centrosomal proteins.
We found that ATG4B is widely expressed in PDAC tumors and stroma, and that low levels of ATG4B protein or RNA are associated with worse overall patient outcomes in univariable analyses. These findings differ from the expected outcome based on ATG4B studies in CML and colorectal cancer, where high ATG4B expression is associated with poor treatment response or more aggressive tumors, respectively (Liu et al., 2014; Rothe et al., 2014). However, a previous study has shown that low levels of GABARAP gene expression are associated with inferior survival outcomes in PDAC patients (Manent et al., 2017), and this would be consistent with the findings of our study, and with a previous report showing ATG4B-mediated stabilization of GABARAP (Skytte Rasmussen et al., 2017). In this study, multivariable analyses confirmed that high levels of ATG4B might serve as an independent predictive marker for improved response to adjuvant chemotherapy, and this observation warrants further investigation in additional PDAC cohorts. However, multivariable analyses did not confirm ATG4B as an independent prognostic factor, likely due to its significant association with differentiation status (Table S1). Typically, poorly differentiated tumors tend to be more aggressive and proliferative, whereas well differentiated tumors are relatively slow growing. Since KD or KO of ATG4B in PDAC cell lines resulted in decreased proliferation, one might expect that low levels of ATG4B would be associated with reduced proliferation and a lower tumor grade. Instead, we found that lower levels of ATG4B were associated with poorly differentiated grade 3 tumors, and higher levels of ATG4B were associated with moderately differentiated grade 2 tumors. Although we expect that the role of ATG4B and ATG4A in the cell cycle is likely generalizable to non-PDAC cells, the association between ATG4B and tumor grade might be tissue dependent. Consistent with these possibilities, Liu et al. (2014) have shown there is a positive correlation between ATG4B and the G1-S-transition protein CCND1 in colorectal cancer. The same authors have reported that high ATG4B expression tends to correlate more closely with late-stage colorectal tumors, though sample size in the study was limited (Liu et al., 2014). However, low ATG4B levels are associated with high-grade (grade 3) tumors and the basal subtype of breast cancer (Bortnik et al., 2020). There are a multitude of factors that could explain these differences, including the role of ATG4B in autophagy or the potential compensatory relationship of ATG4A or other ATG4 family members in different tissues and cell types.
Although ATG4B is often acknowledged as the primary cysteine protease in autophagy, studies have shown that the other ATG4 family members display functional redundancy, which might contribute to compensatory mechanisms (Agrotis et al., 2019; Kauffman et al., 2018). Consistent with this, ATG4B loss led to increased expression of ATG4A and increased lipidation of GABARAP and GABARAPL2 in PDAC cell lines. The combined loss of ATG4B and ATG4A still led to an accumulation of lipidated GABARAP and GABARAPL2, indicating a compensatory role for ATG4C and ATG4D in this double KO background, as described previously (Agrotis et al., 2019). Combined loss of ATG4A and ATG4B further reduced cell proliferation in PDAC cell lines. A difference in levels was detected in two substrates – GABARAP and GABARAPL1 – between the ATG4B KO and ATG4A/4B KO lines, suggesting that reduced GABARAP and GABARAPL1 contributes to the enhanced cell proliferation defect in the double KO lines. There are also other substrates, such as LC3A and LC3C, that could potentially contribute to the differences, but these were not evaluated here due to the lack of specific antibodies. In addition to processing substrates for autophagy, ATG4A expression has been implicated in autophagy-independent processes such as epithelial–mesenchymal transition (EMT) and maintenance of cancer stem cell-like properties via the Notch pathway in gastric cancer and breast cancer (Wolf et al., 2013; Yang et al., 2016). Thus, it is possible that ATG4A supports PDAC cell viability in the absence of ATG4B through autophagy-independent mechanisms, further emphasizing the need to study the functional roles of these proteins beyond the scope of autophagy.
The ATG4 protein substrates – LC3 proteins, GABARAP and GABARAPL1 – were initially discovered through their interaction with cellular cytoskeletal proteins such as microtubules and tubulin, which are essential for proper cellular division (Faller et al., 2009; Mann and Hammarback, 1994; Mansuy et al., 2004; Nakaseko and Yanagida, 2001; Wang et al., 1999). Hence, ATG4-mediated alterations to the substrates might affect cell division. Here, we show a novel link between pro-LC3B and centrosomes in PDAC cells. In the absence of LC3B processing, pro-LC3B localized at the centrosome (marked using the centrosomal protein PCNT) regardless of the cell cycle phase, whereas wild-type LC3B localized at the centrosome in ∼30% of parental cells only during interphase. To determine whether LC3B centrosomal localization in mitotic cells is specific to pro-LC3B, it would be beneficial to examine non-cleavable LC3B (i.e. LC3B-G120A; Agrotis et al., 2019) and pre-cleaved LC3B (i.e. LC3B-G120) in parental versus ATG4A/4B KO cells. Since the nature of the pro-LC3B–centrosomal interaction is unknown, it would be important to use untagged LC3B constructs and/or compare the localization of LC3B constructs with tags at either end. LC3 has historically been shown to regulate microtubule dynamicity, favouring growth and stabilization of microtubules. Hence, the localization of pro-LC3B at the centrosome during mitosis could prevent efficient shortening of the microtubules that is required during anaphase (Rusan et al., 2001). Consistent with this, PDAC ATG4A/4B double KO cells exhibited a delay in mitosis specifically between metaphase and cytokinesis, where efficient microtubule regulation is critical. The ATG4 family proteins have not been reported to specifically localize to the centrosome, but this will be an important area for future investigation. GABARAP has been shown to reside at the centrosome and pericentrosomal region, dissociating from the centrosomal region during metaphase (Joachim et al., 2017). Although it is unclear whether GABARAP affects the cell cycle in an autophagy-dependent or -independent manner, GABARAP has been shown to directly interact with the centrosomal proteins PCM1 and CEP131, which regulate doryphagy (Holdgaard et al., 2019; Joachim et al., 2017). Doryphagy, which is a type of selective autophagy, is required for the maintenance of centriolar satellite integrity. Autophagy inhibition by ATG7 KO or ULK1 KO results in accumulation of PCM1 and CEP131, along with the accumulation of abnormal centriolar satellites (Holdgaard et al., 2019). In agreement with this finding, the PDAC ATG4A/4B KO cells, which displayed reduced GABARAP expression, were observed to have significantly higher PCM1 and CEP131 compared to parental cells, suggesting that doryphagy might be reduced in ATG4A/4B KO PDAC cells.
This study provides evidence of a link between two ATG4 family members – ATG4A and ATG4B – and cell cycle regulation in PDAC. The decrease in proliferation observed in ATG4A/4B KO cell lines was accompanied by defects at two stages of the cell cycle, G1-S transition and mitosis. Hence, targeting ATG4B in combination with chemotherapies that rely on rapid cell proliferation, such as gemcitabine, is unlikely to be beneficial. However, ATG4B might still be a promising target when combined with other treatments that are not dependent on enhanced cell proliferation. Taken together, these findings illustrate the broader functions of the ATG4–substrate axis that need to be taken into consideration when targeting the ATG4 family members.
MATERIALS AND METHODS
Cell lines and culture conditions
MiaPaCa2 and Panc1 were obtained from the American Type Culture Collection (ATCC), and PK-8 was purchased from Riken BRC Cell Bank, Japan, for this study. MiaPaCa2 and Panc1 were cultured in Gibco DMEM, and PK-8 was cultured in Gibco RPMI 1640, supplemented with 10% fetal bovine serum (FBS) and 1% Gibco MEM Non-Essential Amino Acids Solution at standard culture conditions (37°C, 5% CO2 in a humidified incubator). All reagents were purchased from Thermo Fisher Scientific. Cell lines were tested periodically for mycoplasma contamination.
Crystal Violet viability assay
Cells were seeded at a concentration of 1.0×105 cells/ml in a 6-well plate. At 24 h following seeding, cells were cultured with either complete medium (fed) or one of the following conditions: DMEM or RPMI 140 supplemented with low FBS serum (0.8%), complete starvation (EBSS; Gibco), or glutamine and sodium pyruvate deprivation (DMEM or RPMI 1640 without glutamine or sodium pyruvate; Gibco). Low serum experiments were left for 3 days, whereas other starvation conditions were terminated after 24 h. Cells were fixed with 4% paraformaldehyde for 15 min and stained with 0.1% Crystal Violet solution. Stained plates were imaged using the ChemiDoc MP System (Bio-Rad). For quantification purposes, retained Crystal Violet staining was solubilized using 10% acetic acid and measured at an absorbance of 590 nm using a plate reader (VersaMax) according to the manufacturer's protocol.
Protein extraction, western blotting and antibodies
Cell pellets were lysed using RIPA lysis buffer (Santa Cruz Biotechnology, SC24948) supplemented with complete protease inhibitor (Roche, 11836153001). Quantification of total protein was determined using the BCA protein assay kit (Thermo Fisher Scientific, 23225), and 20 µg of protein was loaded on 4–12% gradient BOLT Bis-Tris or 3–8% gradient NUPAGE Tris-acetate gels (Invitrogen) for separation and transferred to PVDF membranes (Bio-Rad). Membranes were blocked with 2% milk solution and incubated overnight at 4°C with primary antibodies. Primary antibodies were diluted with Odyssey Blocking Buffer in PBS (LI-COR Inc.).
The following is a list of antibodies and dilutions used for western blot analyses: anti-ATG4B(1:1000; Sigma-Aldrich, A2981), anti-ATG4A (1:1000; Cell Signaling Technology, 7613), anti-ATG4C (1:1000; Novus Biologicals, NBP1-89043), anti-ATG4D (1:1000, Millipore-Sigma, ABC22), anti-LC3B (1:1000; Abcam, ab48394), anti-GABARAP (1:1000; MBL, M135-3), anti-GABARAP-L1 (1:750; Abcam, ab229558; or 1:1000; Cell Signaling Technology, 26632), anti-GABARAP-L2 (1:750; Abcam, ab126607), anti-β-actin (1:10,000; Abcam, ab6276), anti-PARP (1:1000; Cell Signaling Technology, 9542S), anti-p62 (1:1000; Sigma, P0067), anti-PCNT(1:1000; Thermo Fisher Scientific, PA5-37295), anti-PCM1(1:1000; Sigma-Aldrich, SAB1406228), anti-TAX1BP1 (1:1000; Cell Signaling Technology, 5105S), anti-vinculin (1:1000; Abcam, ab129002), anti-CEP131/AZI1 (1:1000; Abcam, ab99379) and anti-FLAG (1:1000; Sigma-Aldrich, F1804).
Membranes incubated with primary antibody were then washed with 1× PBS containing Tween 20 (0.1%) and incubated with the appropriate secondary antibody [goat anti-mouse IgG–horseradish peroxidase (HRP) and goat anti-rabbit IgG–HRP; Santa Cruz Biotechnology] for 1 h. Detection and visualization of protein bands were conducted using the Bio-Rad Clarity Western ECL substrate and a Bio-Rad ChemiDoc MP System. Densitometry was performed using Image Lab software to measure relative levels of protein of interest by normalizing to the loading control (β-actin or vinculin). Images of the full, uncropped blots with molecular mass markers are available in Fig. S7.
Establishment of knockdown and knockout cell lines
To establish KO or KD lines, MiaPaCa2, Panc1 or PK-8 cells were seeded at a density of 8×105 cells in 2 ml medium per well of a 6-well plate and transfected the next day with either 3 µg of pX459 vector (Addgene) carrying either one of the two ATG4B or ATG4A gRNAs (for KO), or 3 µg of pGIPZ shRNA vector (Thermo Fisher Scientific) carrying the shATG4B sequence (for KD) or the control shSC sequence. ATG4B shRNA sequence: 5′-ATCCCTAGATTCTTCTGATGTATAGTGAAGCCACAGATGTATACATCAGAAGAATCTAGGGAC-3′. The scrambled control shRNA sequence, shSC, was 5′-ATCTCGCTTGGGCGAGAGTAAGTAGTGAAGCCACAGATGTACTTACTCTCGCCCAAGCGAGAG-3′. ATG4B and ATG4A gRNAs: ATG4B-KO-1, 5′-TCCTGTCGATGAATGCGTTG-3′; ATG4B-KO-2, 5′-TCCTCAACGCATTCATCGAC-3′; ATG4A-KO-1, 5′-CTGATATAAGTGCTCGTCTA-3′; ATG4A-KO-2, 5′-CATACCAGCTCATCTGTATC-3′.
Transfection was performed using Lipofectamine 3000 (Life Technologies, L3000-008). Cells were incubated with the plasmid for 48 h prior to media replacement and incubation with 10 µg/ml puromycin (plasmid selection agent) for another 48 h. For KO cells, remaining live cells were harvested and seeded at ∼5 cells/well cell density in two 96-well plates. Cells were grown to confluence before being transferred to 24-well plates. Confluent wells were harvested for western blot screening for ATG4B and/or ATG4A. To create the MiaPaCa2 ATG4A/4B double KO lines, confirmed ATG4B KO clones (4BKO1 and 4BKO2) were transfected with either ATG4A-KO-1 or ATG4A-KO-2 gRNAs selected similarly to ATG4B KO clones. For the KD cells, cells were maintained in puromycin until colonies formed and reached confluency. These cells were then flow sorted based on the GFP signal in the vector and maintained in 1 µg/ml puromycin.
ATG4B ectopic expression
pcDNA4/TO plasmids (Invitrogen, V1020-20) modified with N-3×FLAG or C-3×FLAG sequences were used as vector backbones. Human ATG4B ORF was amplified by PCR from an ATG4B-expressing plasmid (GenScript; clone OHu290105) and cloned into the N-3×FLAG pcDNA4/TO vector using BamHI and EcoRI restriction enzymes or the C-3×FLAG pcDNA4/TO vector using BamHI and NotI restriction enzymes. The N-3×FLAG–ATG4B and C-3×FLAG–ATG4B constructs were validated by restriction enzyme digests and gene sequencing. MiaPaCa2 ATG4A/4B KO cells were plated at 3×105 cells/well and transfected with empty vector or plasmid expressing the N- or C-3×FLAG–ATG4B using Lipofectamine 3000 as per the manufacturer's instructions. Cells were subjected to selection using 100 µg/ml Bleocin for up to two weeks. Cells that survived Bleocin treatments were evaluated for expression of ATG4B and FLAG using respective antibodies. Antibiotic selection agent was removed for three passages prior to experiments.
PDAC patient samples
A retrospective cohort of 267 pancreatic ductal adenocarcinoma patients that was previously described (Riazy et al., 2015) was used for this study. Primary pre-treatment tumor samples associated with defined clinicopathological features and follow-up outcome data were obtained from archives at the Vancouver Coastal Health Region between 1995 and 2014. Duplicate 0.6 mm cores from the epithelial and adjacent normal pancreatic epithelium histological components were used to construct two separate tissue microarrays. All patients in the cohort underwent surgery, and 30% of patients received adjuvant chemotherapy with a nucleoside analog (80% gemcitabine and 20% 5-fluorouracil). Ethical approval for research on this retrospective cohort was obtained from the Research Ethics Board of the University of British Columbia, BC Cancer and Simon Fraser University. A waiver of consent was provided to account for the large proportion of patients that were deceased at the time of the assembly of this cohort. Clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki.
Immunohistochemical staining and scoring
Immunohistochemical staining of ATG4B on 4 µm-thick formalin-fixed paraffin-embedded sections from the pancreatic tissue microarray was performed using the Discovery XT (Ventana Medical Systems, Inc.) automated staining platform following the manufacturer's recommendations. Slides were incubated with the rabbit anti-ATG4B antibody (Sigma-Aldrich, HPA069803) at a dilution of 1:100 for 2 h at room temperature, then washed and incubated with UltraMap anti-rabbit HRP secondary (Ventana Medical Systems, Inc.) for 16 min at room temperature. For all histological components (tumor epithelium and adjacent normal pancreas), staining was quantified by H-score, which represents the product of percent (0–100%) of epithelial cells staining positive and a subjective evaluation of staining intensity (0–3) to yield an H-score range of 0–300. Cases that presented with assay failure due to technical reasons were excluded from the analysis.
Immunofluorescence and imaging
1×105 cells were grown on a coverslip in a 6-well plate. Cells were fixed with ice cold methanol and acetone (1:1) on ice for 5 min, washed with 1×PBS and blocked with 2.5% goat serum (G6767, Sigma-Aldrich) in PBS for 1 h on a rocker, and then stained with appropriate primary antibodies (anti-LC3B: Cell Signaling Technology, 3868S, 1:500; anti-GABARAPL1: Cell Signaling Technology, 26632, 1:500; anti-GABARAPL2: Abcam, 126607, 1:500; anti-PCNT: 1:500, Abcam, 28144, 1:500) in 0.25% goat serum overnight at 4°C. Alexa Fluor 488 (Thermo Fisher Scientific, A11029)- and/or Alexa Fluor 568 (Thermo Fisher Scientific, A11011)-conjugated secondary antibodies (1:2000) were used, followed by mounting with DAPI (D3571, Invitrogen) and SlowFade Diamond Antifade mountant (36967, Invitrogen).
Microscopy images were obtained from a Zeiss Axio Observer (Z1/7) inverted fluorescence microscope equipped with an Apotome.2 and an AxioCam MRm R3 camera (Zeiss). Images were obtained using a 63× objective magnification (with oil immersion for 63×/1.40 NA) using the Zen software (version 2.5, blue edition; Zeiss).
To quantify interphase and mitotic cells, ten images were randomly captured at 63× objective magnification using a Z-stack (ten slices of 15 µM) using DAPI, Alexa Fluor 488 and Alexa Fluor568 channels. Approximately 3–4 slices with cell images were processed with orthogonal projections using default parameters to produce a single image. Cells were manually evaluated to determine whether LC3B, GABARAPL1 or GABARAPL2 staining was overlapping with PCNT. Brightness and contrast were adjusted using Photoshop (CS6; Adobe) and applied to the whole image.
Sequential siRNA transfection for LC3B KD
Cells were plated at 5×104 cells/well in a 12-well plate. The following day, the medium was replaced with DMEM containing 0.1% FBS and cells were transfected with IDT siRNA (LC3B-siRNA1; 288088136) or negative control (NC1; 51011404) at 5 nM with Lipofectamine RNAiMAX transfection reagent (Invitrogen) as per the IDT protocol. The following day, FBS was added to a 10% final concentration. On day 4, all cells were re-plated into respective wells of a 6-well plate. After cells attached (6–8 h), cells were transfected again with 5 nM of respective siRNAs. On day 8, cells were either fixed for Crystal Violet assay or were harvested for western blot analysis.
Bafilomycin A1 treatment
To determine the effects of autophagy inhibition on levels of certain proteins, cells were treated with the autophagy inhibitor bafilomycin A1 (BafA1) 4 h prior to harvest. The concentration of BafA1 used was 80 nM, which was previously determined by establishing a saturation curve of LC3B-II levels. In cases where low-serum starvation was required, the medium was replaced with DMEM or RPM1 with 0.8% FBS along with BafA1 4 h prior to terminating the experiment.
PARP cleavage assay
Cell lines were seeded at 1.0×105 cell density in 6-well plates in 10% or 0.8% serum conditions. Cells were harvested 72 h following cell seeding, and PARP cleavage was determined using western blot analysis.
BrdU proliferation assay
A BrdU Cell Proliferation assay kit (Cell Signaling Technology) was used to study changes in proliferation as per the manufacturer's instructions. Briefly, 5.0×103 cells were plated in 96-well plates under fed or 0.8% serum conditions. At 24 h following cell seeding, BrdU was added to the wells and left to be incorporated into the cells for 24 h. Colorimetric-based measurements were taken after the 24-h incubation with BrdU using a plate reader (Versamax) at an absorbance of 450 nm.
Incucyte cytotoxicity assays
Cells grown in T-75 flasks (Thermo Fisher Scientific) in DMEM plus 10% FBS were subjected to trypsin treatment and plated at 5000 cells/well density in 100 µl volume of DMEM medium supplemented with 10% FBS in a 96-well plate. A 50 µl volume of medium containing IncuCyte Cytotox Green reagent (100 nM; Essen Bioscience, 4633) and IncuCyte NucLight Rapid Red Reagent (1:500 dilution; Essen Bioscience, 4717) was added and incubated at 37°C for ∼2 h. Nuclear (red) and dead (green) cell counts were obtained over time using an IncuCyte ZOOM dual-color live-content imaging system (Model S3, Sartorius). Data were acquired using a 10× objective lens in phase contrast, green fluorescence and red fluorescence channels. Spectral unmixing was set at 8% red signal removed from green signal to better represent the distribution of both green and red channels. Images were acquired from each well at set time intervals. MiaPaCa2 parental and KO cells were analyzed using the following settings to count nuclear red objects (parameter adaptation, threshold adjustment: 1; edge split on; edge sensitivity 50; filter area minimum 20 mm2, maximum 800 mm2; eccentricity maximum 1.0) and nuclear green objects (parameter adaptation, threshold adjustment: 10; edge split on; filter area minimum 5 mm2, maximum 800 mm2; eccentricity maximum 0.9).
Total cell fraction (red positive) at each time point was calculated relative to t=0 (set as 1). The dead-cell fraction was calculated by counting the dead cells (defined as green and red overlap, set at overlap area minimum 50 mm2) and dividing by the total cells (red positive). Total and dead cells over time percent counts were graphed using Prism 8 (GraphPad Software, Inc), and area under the curve (AUC) values were computed.
Time-lapse imaging and cell cycle quantifications
Cell Cycle Green/Red Lentivirus reagent (EF1α-Puro; Essen, 4779; FUCCI) was transduced into MiaPaCa2, ATG4A/4B1 KO and ATG4A/4B2 KO lines derived from MiaPaCa2 cells by the following method. Briefly, a day after seeding cells into a 6-well plate at 50% confluence, the medium was replaced with 4 ml of medium containing 20 µl of the lentivirus reagent and 8 µg/ml polybrene (Sigma-Aldrich, H9268-10G). The plates were centrifuged at 1200 g at 32°C for 1 h and then incubated overnight. The next day, the cells were passaged and selected in 10 µg/ml puromycin for 3 days. Cells were then maintained in 1 µg/ml puromycin, and puromycin was removed for at least two passages before the experiments.
Cells stably expressing the FUCCI construct were plated at 5000 cells per well in a 96-well plate (Corning) and incubated at 37°C. Cells were left to adhere for 2 h prior to images being acquired every hour using a 20× objective for up to 48 h using the following conditions in an IncuCyte (SX3). Image channels: phase-contrast and green fluorescence acquisition time, 350 ms; red fluorescence acquisition time, 450 ms; nine images per scan. Spectral unmixing was set at 8% red signal removed from the green signal to better represent the distribution of both green and red channels.
MiaPaCa2 parental and ATG4A/4B KO cells were analyzed using IncuCyte basic analyzer with the following settings to count nuclear red objects (segmentation, top-hat; radius, 30 µm; threshold adjustment, 0.2; edge split, on; edge sensitivity, −25; filter area minimum, 30 µm2), nuclear green objects (segmentation, top-hat; radius, 20 µm; threshold adjustment 0.5; edge split, on; edge sensitivity, −25; filter area minimum, 30 µm2; eccentricity maximum, 0.97) and phase-contrast objects (segmentation adjustment, 0.5; filter area minimum, 300 µm2). Cells expressing green fluorescence (S, G2 and G2-M) can be separated from the cells expressing red fluorescence (G1), and transition phase (G1-S) can also be identified with high red and green (yellow). Cells in M-G1 cannot be quantified, as these cells do not express fluorescence probes. Phase area confluence (%), green or red object count per mm2, and green and red object count per mm2 values were exported to Excel (Microsoft) for further analyses. To determine pure populations of red or green cells, overlap count was subtracted from each count (e.g. red count−overlap count=pure red population). Total cell count was determined as the sum of the red count and green count, minus the overlap count: [(red+green)−overlap]. Percentage totals of red, green and yellow counts were determined by dividing the pure count by the total cell number multiplied by 100, and were plotted using Prism 8 (GraphPad Software, Inc).
Incucyte time-lapse imaging and quantification of cells in mitosis
Cells stably expressing the FUCCI construct were grown at 5000 cells per well in a 96-well plate (Corning) for 24 h. Images were acquired every 5–10 min using a 20× objective for 24 h using the following conditions in an Incucyte (SX3). Image channels: phase-contrast and green fluorescence acquisition time, 350 ms; red fluorescence acquisition time, 450 ms; two to three images per scan. Spectral unmixing was set at 8% red signal removed from the green signal to better represent the distribution of both green and red channels. Four series of images (size 710×522) from each of two scans per cell line were retained. Time in mitosis from each series was determined manually by quantifying the time from rounding up of cells to dividing into two cells.
Statistics
All cellular data described are from at least three biological replicates unless noted otherwise. When comparing two samples, two-tailed unpaired Student's t-test was used. When comparing multiple samples, ordinary one-way ANOVA or two-way ANOVA was used, with Dunnett's multiple comparisons test or Sidak's multiple comparisons test, as indicated.
H-scores derived from the scoring of immunohistochemical staining were considered as continuous variables in analyses to determine whether significant associations were observed for clinicopathologic covariates. Clinicopathologic covariates modeled as continuous were assessed using univariable linear regression, and those modeled as categorical were assessed with one-way ANOVA. Disease-specific survival time-dependent recursive partitioning was used to categorize ATG4B expression in the tumor epithelium into high and low expressers. The first partition was used as the threshold. Univariable disease-specific survival was modeled with the Kaplan–Meier approach, and differences were assessed with the log-rank statistic. Multivariable disease-specific survival utilized the Cox proportional hazards method. Differences were considered statistically significant at an alpha of <0.05. Immunohistochemical statistical analyses were performed using JMP Pro v16.0 (SAS Institute, Cary, NC, USA).
Supplementary Material
Acknowledgements
The authors thank Gayathri Samarasekera, Chandra Lebovitz, Kevin Yang and Robert Camfield for helpful discussions; Gayathri Samarasekera for assistance with microscopy and comments on the manuscript; and Stephanie McInnis for project management support.
Footnotes
Author contributions
Conceptualization: P.S., S.C., S.E.K., M.A.J., S.M.G.; Methodology: P.S., S.C., S.E.K., J.C., N.E.G.; Formal analysis: P.S., S.C., S.E.K., J.C., N.E.G.; Investigation: P.S., S.C., J.C., N.E.G., M.A.J., C.J.H., T.H., J.X., C.C., D.G.; Resources: S.E.K., C.J.H., J.X., F.D.J., D.J.R., D.F.S.; Writing - original draft: P.S., S.M.G.; Writing - review & editing: P.S., S.C., S.E.K., J.C., N.E.G., M.A.J., C.J.H., T.H., J.X., C.C., D.G., F.D.J., W.W.L., G.B.M., D.J.R., D.F.S., S.M.G.; Visualization: P.S., S.C., S.E.K., J.C.; Supervision: W.W.L., G.B.M., D.J.R., D.F.S., S.M.G.; Funding acquisition: S.M.G.
Funding
This work was supported by Pancreas Centre BC [IDEAS grant 2017-2018], the Pancreatic Cancer Action Network [2018 Pancreatic Cancer Action Network Translational Research grant 18-65-GORS] and the BC Cancer Foundation [2022 BC Cancer Research Sustainment Fund, #F22-00948 to S.M.G.]. C.J.H. and J.X. were supported by Canadian Institutes of Health Research (CIHR) Frederick Banting and Charles Best Canada Graduate Scholarship Doctoral Awards, and J.C. was supported by a CIHR Canada Graduate Scholarship – Master's Award. Deposited in PMC for release after 12 months.
Data availability
All relevant data can be found within the article and its supplementary information.
References
- Agrotis, A. and Ketteler, R. (2019). On ATG4B as drug target for treatment of solid tumours—the knowns and the unknowns. Cells 9, 53. 10.3390/cells9010053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrotis, A., Pengo, N., Burden, J. J. and Ketteler, R. (2019). Redundancy of human ATG4 protease isoforms in autophagy and LC3/GABARAP processing revealed in cells. Autophagy 15, 976-997. 10.1080/15548627.2019.1569925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortnik, S., Choutka, C., Horlings, H. M., Leung, S., Baker, J. H. E., Lebovitz, C., Dragowska, W. H., Go, N. E., Bally, M. B., Minchinton, A. I.et al. (2016). Identification of breast cancer cell subtypes sensitive to ATG4B inhibition. Oncotarget 7, 66970-66988. 10.18632/oncotarget.11408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortnik, S., Tessier-Cloutier, B., Leung, S., Xu, J., Asleh, K., Burugu, S., Magrill, J., Greening, K., Derakhshan, F., Yip, S.et al. (2020). Differential expression and prognostic relevance of autophagy-related markers ATG4B, GABARAP, and LC3B in breast cancer. Breast Cancer Res. Treat. 183, 525-547. 10.1007/s10549-020-05795-z [DOI] [PubMed] [Google Scholar]
- Bosc, D., Vezenkov, L., Bortnik, S., An, J., Xu, J., Choutka, C., Hannigan, A. M., Kovacic, S., Loo, S., Clark, P. G. K.et al. (2018). A new quinoline-based chemical probe inhibits the autophagy-related cysteine protease ATG4B. Sci. Rep. 8, 11653. 10.1038/s41598-018-29900-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, K. L., Stalnecker, C. A., Zeitouni, D., Klomp, J. E., Peng, S., Tikunov, A. P., Gunda, V., Pierobon, M., Waters, A. M., George, S. D.et al. (2019). Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat. Med. 25, 628-640. 10.1038/s41591-019-0368-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Porta, A., Bornstein, K., Coye, A., Montrief, T., Long, B. and Parris, M. A. (2020). Acute chloroquine and hydroxychloroquine toxicity: a review for emergency clinicians. Am. J. Emerg. Med. 38, 2209-2217. 10.1016/j.ajem.2020.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faller, E. M., Villeneuve, T. S. and Brown, D. L. (2009). MAP1a associated light chain 3 increases microtubule stability by suppressing microtubule dynamics. Mol. Cell. Neurosci. 41, 85-93. 10.1016/j.mcn.2009.02.001 [DOI] [PubMed] [Google Scholar]
- Fujita, N., Hayashi-Nishino, M., Fukumoto, H., Omori, H., Yamamoto, A., Noda, T. and Yoshimori, T. (2008). An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol. Biol. Cell 19, 4651-4659. 10.1091/mbc.e08-03-0312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdgaard, S. G., Cianfanelli, V., Pupo, E., Lambrughi, M., Lubas, M., Nielsen, J. C., Eibes, S., Maiani, E., Harder, L. M., Wesch, N.et al. (2019). Selective autophagy maintains centrosome integrity and accurate mitosis by turnover of centriolar satellites. Nat. Commun. 10, 4176. 10.1038/s41467-019-12094-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachim, J. and Tooze, S. A. (2017). Centrosome to autophagosome signaling: Specific GABARAP regulation by centriolar satellites. Autophagy 13, 2113-2114. 10.1080/15548627.2017.1385677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachim, J., Jefferies, H. B. J., Razi, M., Frith, D., Snijders, A. P., Chakravarty, P., Judith, D. and Tooze, S. A. (2015). Activation of ULK kinase and autophagy by GABARAP trafficking from the centrosome is regulated by WAC and GM130. Mol. Cell 60, 899-913. 10.1016/j.molcel.2015.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachim, J., Razi, M., Judith, D., Wirth, M., Calamita, E., Encheva, V., Dynlacht, B. D., Snijders, A. P., O'reilly, N., Jefferies, H. B. J.et al. (2017). Centriolar satellites control GABARAP ubiquitination and GABARAP-mediated autophagy. Curr. Biol. 27, 2123-2136.e7. 10.1016/j.cub.2017.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman, K. J., Yu, S., Jin, J., Mugo, B., Nguyen, N., O'Brien, A., Nag, S., Lystad, A. H. and Melia, T. J. (2018). Delipidation of mammalian Atg8-family proteins by each of the four ATG4 proteases. Autophagy 14, 992-1010. 10.1080/15548627.2018.1437341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey, C. G., Camolotto, S. A., Boespflug, A. M., Gullien, K. P., Foth, M., Truong, A., Schuman, S. S., Shea, J. E., Seipp, M. T., Yap, J. T.et al. (2019). Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat. Med. 25, 620-627. 10.1038/s41591-019-0367-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M., Hou, Y., Wang, J., Chen, X., Shao, Z.-M. and Yin, X.-M. (2011). Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J. Biol. Chem. 286, 7327-7338. 10.1074/jbc.M110.199059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P.-F., Leung, C.-M., Chang, Y.-H., Cheng, J.-S., Chen, J.-J., Weng, C.-J., Tsai, K.-W., Hsu, C.-J., Liu, Y.-C., Hsu, P.-C.et al. (2014). ATG4B promotes colorectal cancer growth independent of autophagic flux. Autophagy 10, 1454-1465. 10.4161/auto.29556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manent, J., Banerjee, S., de Matos Simoes, R., Zoranovic, T., Mitsiades, C., Penninger, J. M., Simpson, K. J., Humbert, P. O. and Richardson, H. E. (2017). Autophagy suppresses Ras-driven epithelial tumourigenesis by limiting the accumulation of reactive oxygen species. Oncogene 36, 5576-5592. 10.1038/onc.2017.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann, S. S. and Hammarback, J. A. (1994). Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J. Biol. Chem. 269, 11492-11497. 10.1016/S0021-9258(19)78150-2 [DOI] [PubMed] [Google Scholar]
- Mansuy, V., Boireau, W., Fraichard, A., Schlick, J.-L., Jouvenot, M. and Delage-Mourroux, R. (2004). GEC1, a protein related to GABARAP, interacts with tubulin and GABA(A) receptor. Biochem. Biophys. Res. Commun. 325, 639-648. 10.1016/j.bbrc.2004.10.072 [DOI] [PubMed] [Google Scholar]
- Mauthe, M., Orhon, I., Rocchi, C., Zhou, X., Luhr, M., Hijlkema, K.-J., Coppes, R. P., Engedal, N., Mari, M. and Reggiori, F. (2018). Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 14, 1435-1455. 10.1080/15548627.2018.1474314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko, Y. and Yanagida, M. (2001). Cytoskeleton in the cell cycle. Nature 412, 291-292. 10.1038/35085684 [DOI] [PubMed] [Google Scholar]
- Riazy, M., Kalloger, S. E., Sheffield, B. S., Peixoto, R. D., Li-Chang, H. H., Scudamore, C. H., Renouf, D. J. and Schaeffer, D. F. (2015). Mismatch repair status may predict response to adjuvant chemotherapy in resectable pancreatic ductal adenocarcinoma. Mod. Pathol. 28, 1383-1389. 10.1038/modpathol.2015.89 [DOI] [PubMed] [Google Scholar]
- Rothe, K., Lin, H., Lin, K. B. L., Leung, A., Wang, H. M., Malekesmaeili, M., Brinkman, R. R., Forrest, D. L., Gorski, S. M. and Jiang, X. (2014). The core autophagy protein ATG4B is a potential biomarker and therapeutic target in CML stem/progenitor cells. Blood 123, 3622-3634. 10.1182/blood-2013-07-516807 [DOI] [PubMed] [Google Scholar]
- Rusan, N. M., Fagerstrom, C. J., Yvon, A.-M. C. and Wadsworth, P. (2001). Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-α tubulin. Mol. Biol. Cell 12, 971-980. 10.1091/mbc.12.4.971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagiv, Y., Legesse-Miller, A., Porat, A. and Elazar, Z. (2000). GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J. 19, 1494-1504. 10.1093/emboj/19.7.1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani, T. and Klionsky, D. J. (2004). Autophagy in health and disease: a double-edged sword. Science 306, 990-995. 10.1126/science.1099993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skytte Rasmussen, M., Mouilleron, S., Kumar Shrestha, B., Wirth, M., Lee, R., Bowitz Larsen, K., Abudu Princely, Y., O'Reilly, N., Sjøttem, E., Tooze, S. A.et al. (2017). ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8. Autophagy 13, 834-853. 10.1080/15548627.2017.1287651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén, M., Fagerberg, L., Hallstrom, B. M., Lindskog, C., Oksvold, P., Mardinoglu, A., Sivertsson, A., Kampf, C., Sjöstedt, E., Asplund, A.et al. (2015). Tissue-based map of the human proteome. Science 347, 1260419-1260419. 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Vernier-Magnin, S., Nemos, C., Mansuy, V., Tolle, F., Guichard, L., Delage-Mourroux, R., Jouvenot, M. and Fraichard, A. (2005). Analysis of the guinea-pig estrogen-regulated gec1/GABARAPL1 gene promoter and identification of a functional ERE in the first exon. Biochim. Biophys. Acta Gene Struct. Expr. 1731, 23-31. 10.1016/j.bbaexp.2005.05.002 [DOI] [PubMed] [Google Scholar]
- Wang, H., Bedford, F. K., Brandon, N. J., Moss, S. J. and Olsen, R. W. (1999). GABA(A)-receptor-associated protein links GABA(A) receptors and the cytoskeleton. Nature 397, 69-72. 10.1038/16264 [DOI] [PubMed] [Google Scholar]
- Wang, W., Chen, Z., Billiar, T. R., Stang, M. T. and Gao, W. (2013). The carboxyl-terminal amino acids render pro-human LC3B migration similar to lipidated LC3B in SDS-PAGE. PLoS ONE 8, e74222. 10.1371/journal.pone.0074222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg, H., Shvets, E., Shpilka, T., Shimron, F., Shinder, V. and Elazar, Z. (2010). LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 29, 1792-1802. 10.1038/emboj.2010.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, J., Dewi, D. L., Fredebohm, J., Müller-Decker, K., Flechtenmacher, C., Hoheisel, J. D. and Boettcher, M. (2013). A mammosphere formation RNAi screen reveals that ATG4A promotes a breast cancer stem-like phenotype. Breast Cancer Res. 15, R109. 10.1186/bcr3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S., Wang, X., Contino, G., Liesa, M., Sahin, E., Ying, H., Bause, A., Li, Y., Stommel, J. M., Dell'antonio, G.et al. (2011). Pancreatic cancers require autophagy for tumor growth. Genes Dev. 25, 717-729. 10.1101/gad.2016111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S.-W., Ping, Y.-F., Jiang, Y.-X., Luo, X., Zhang, X., Bian, X.-W., Yu, P.-W., Yang, S.-W., Ping, Y.-F., Jiang, Y.-X.et al. (2016). ATG4A promotes tumor metastasis by inducing the epithelial-mesenchymal transition and stem-like properties in gastric cells. Oncotarget 7, 39279-39292. 10.18632/oncotarget.9827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, A., Herter-Sprie, G., Zhang, H., Lin, E. Y., Biancur, D., Wang, X., Deng, J., Hai, J., Yang, S., Wong, K.-K.et al. (2018). Autophagy sustains pancreatic cancer growth through both cell-autonomous and nonautonomous mechanisms. Cancer Discov. 8, 276-287. 10.1158/2159-8290.CD-17-0952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun, C. W. and Lee, S. H. (2018). The roles of autophagy in cancer. Int. J. Mol. Sci. 19, 3466. 10.3390/ijms19113466 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.