Abstract
Purpose
More than half of the familial cutaneous melanomas have unknown genetic predisposition. This study aims at characterizing novel melanoma susceptibility gene.
Methods
We performed exome and targeted sequencing on melanoma-prone families without any known melanoma susceptibility genes. We analyzed the expression of candidate gene DENN domain-containing 5A (DENND5A) in melanoma samples in relation to pigmentation and UV signature. Functional studies were carried out using microscopic approaches and zebrafish model.
Results
We identified a novel DENND5A truncating variant that segregates with melanoma within a Swedish family, and two additional rare DENND5A variants, one of which segregates with the disease in an American family. We find that DENND5A is significantly enriched in pigmented melanoma tissue. Our functional studies show that loss of DENND5A function leads to decreases in melanin content in vitro and pigmentation defects in vivo. Mechanistically, harboring the truncating variant or being suppressed, DENND5A loses its interaction with sorting nexin 1 (SNX1) and is unable to transport the SNX1-associated vesicles from melanosomes. Consequently, untethered SNX1-premelanosome protein and redundant tyrosinase are re-directed to lysosomal degradation by default, causing decreases in melanin content.
Conclusion
Our findings provide evidence of a physiological role of DENND5A in the skin context and link its variants to melanoma susceptibility.
Keywords: melanoma, susceptibility gene, pigmentation, DENND5A, SNX1
INTRODUCTION
Approximately 5–10% of melanomas occur in family settings, many of which are attributed to genetic predisposition1. Certain phenotypic traits have been linked to melanoma susceptibility, from fair pigmentation in skin, hair and eyes, poor tanning abilities, to high melanocytic nevus counts or dysplastic nevi2,3. Individuals with such genetic or phenotypic traits are particularly susceptible towards environmental factors such as ultraviolet (UV) exposure4. Known melanoma susceptibility genes are classified into high, medium and low penetrance genes, yet 50% of the familial cases remain unexplained1. Interestingly, many of the medium or low penetrance genes are involved in pigmentation pathways, including the melanocortin 1 receptor (MC1R)5, microphthalmia-associated transcription factor (MITF)6,7, and tyrosinase (TYR)8. Consequently, it has long been established that cutaneous malignant melanoma (CMMs) tumors, tend to have higher burden of pathogenic variants, and are often associated with the characteristic UV signatures - cytidine to thymidine (C>T) substitution at dypyrimidine sites or CC>TT transition9. These pathogenic variant often arise from two major forms of DNA photoproducts, namely cyclobutane pyrimidine dimers (CPDs) and 6–4 pyrimidine-pyrimidone10.
Melanocytes synthesize and store melanin inside a specialized organelle called melanosome11, belonging to a group of lysosome-related organelles (LROs), which are interconnected with the typical endo-lysosomal network. PMEL is the fundamental structural protein that provides the characteristic fibrillar matrix upon which melanin is deposited. More specifically, the PMEL precursor, as a melanosomal cargo, is cleaved into the membrane integrated Mβ fragment and intraluminal Mα fragments and transported to stage I melanosomes (defined by clathrin structure and presence of MLANA)12. Various antibodies targeting different epitopes of PMEL protein have been designed to facilitate the recognition of these isoforms, such as PMEL-C and HMB4513. Previous studies highlighted the significance of mutated PMEL in causing pigmentation defects in zebrafish, mice, dogs and horses11.
In this work, we report a novel germline loss-of-function variant in DENN domain-containing protein 5A (DENND5A) gene and two other candidate gene variants in this gene. We show that DENND5A associates with melanosomes and its dysfunction impedes melanosomal cargo transport and pigmentation pathway. We provide novel mechanistic insights into DENND5A variant-triggered melanoma susceptibility.
MATERIALS AND METHODS
Clinical samples
A four-case melanoma family was identified through a national preventive program for kindreds with familial melanoma and screened for germline variants in known melanoma susceptibility genes (CDKN2A and CDK4) with negative results (Family A in Supplementary Table S1). Within this family, three affected and one unaffected relative were eligible for exome sequencing. We have previously executed a targeted sequencing approach of 59 additional Swedish melanoma families for inherited genetic variants in 120 candidate genes, including DENND5A as described in our previous work14. We have also included an American melanoma family that was involved in exome sequencing15 (Family C in Supplementary Table S1).
Genomic DNA preparation and exome sequencing
Genomic DNA was extracted from peripheral blood for each of the selected samples and paired-end libraries were created according to standard protocols (Illumina Inc., San Diego, CA). Exome enrichment was performed using the TrueSeq Exome Enrichment Kit (Illumina). Enriched libraries were sequenced using HiSeq 2000 (Illumina) generating 2 × 100bp reads. After trimming and quality control the reads were mapped to the hg19 reference genome using Mosaik (v.1.0.1388) (http://bioinformatics.bc.edu/marthlab/Mosaik). More details of the pre-processing part including quality control of reads was described in Höiom et al 16. Variant filtering is described in Supplementary Methods.
Statistical analyses
All statistical analyses were carried out using GraphPad Prism v.7.0 (GraphPad Software, La Jolla, CA, USA). The specific statistical methods used were described in figure legends. Data were presented as mean ± S.E.M. or box plot with line at median.
RESULTS
Rare DENND5A germline variants identified in melanoma-prone families
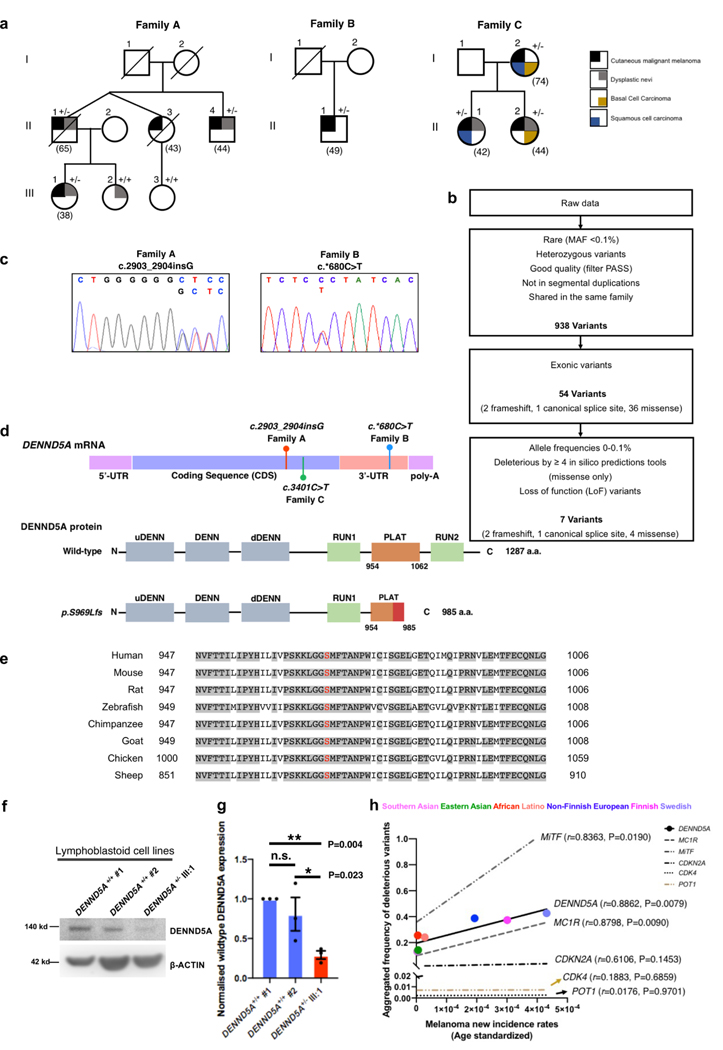
A multi-case cutaneous malignant melanoma (CMM) family (Family A) without carrying any known melanoma-associated pathogenic variants was examined by exome sequencing (Fig. 1a). Three out of four affected family members (II:1, II:4, III:1) and one healthy relative (III:2) were subject to sequencing (Supplementary Table S1). We called more than 288,000 unique single nucleotide variants (SNVs) and small insertion and/or deletion variants (indels). We filtered these variants via the described pipeline (Fig. 1b), keeping only rare heterozygous exonic variants that were predicted to be deleterious by at least four algorithms. Without detecting any shared variants in known melanoma predisposing genes, such as CDKN2A, CDK4, BAP1, POT1 or MITF, we retained 7 candidates, including 4 missense, 1 splice-site and 2 frameshift variants. Upon closer inspection, all four missense variants (C9orf72, PALM2, CLCN1 and LARGE1) were found in the gnomAD database, while the frameshift variant in HYDIN was also detected in the healthy relative in the family. We next performed functional validation on the splice-site variant, yet we could not detect any ambiguous splicing event in the RNA sample of the heterozygote (II:1). These analyses would not necessarily rule out the variants as disease-related in the cases of incomplete penetrance. However, they led us to prioritize the remaining variant, a frameshift variant (1 base pair insertion), that segregated with the disease within this family. The variant was first validated by Sanger sequencing (Fig. 1c), including all individuals analyzed by exome sequencing and one additional healthy family member (III:3). Of note, we subsequently confirmed that the two healthy offsprings (III:2, current age 55; III:3, current age 58), as marked in the pedigree, did not share this variant. This variant, mapped to exon 17 of the DENN domain-containing 5A (DENND5A) gene (Fig. 1d, upper), was predicted to lead to a truncated DENND5A protein (C-del DENND5A), namely p.Ser969fs (Fig. 1d, lower). It is completely novel and absent in population control databases (Table 1) and would cause amino acids changes in a highly conserved region of the functional PLAT domain (Fig. 1e), which could lead to disruption of the PLAT domain and loss of the entire RUN2 domain. In order to examine the presence and stability of the truncated protein, we established lymphoblastoid cell lines from heterozygote III:1 and two unrelated controls (Supplementary Methods), and then probed with a DENND5A antibody recognizing the N-terminus of the protein (156 a.a. - 190 a.a., Fig. 1f). The wild-type DENND5A protein expression around 140 kd is significantly lower in patients harboring the heterozygous variant compared to the unrelated controls (Fig. 1g). There was no detectable protein signal between 100 – 110 kd, where the truncated protein is assumed to be, likely due to the low abundancy of truncated protein and the affinity of the antibody to the truncated form. However, RNA sequencing of the lymph node metastatic melanoma derived from II:1 confirmed the existence of variant transcripts as marked, and also displayed certain degree of nonsense-mediated decay as the variant transcript was less than the wild-type ones (Supplementary Fig. S1a).
Fig. 1. Rare DENND5A variants identified in multi-case melanoma families.
a. Pedigrees of multi-case melanoma families with heterozygous DENND5A variants. +/+ and +/− indicate the tested family members as DENND5A wildtypes and heterozygotes, respectively. b. A flowchart showing the variant filtering of exome sequencing applied to Family A. c. Chromatograms of the validated DENND5A variants found in indicated families. The 1 bp insertion (c.2903_2904insG) found in Family A is shown on the left, the 1 bp substitution (c.*680C>T) found in Family B is shown on the right in blue. d. Schematic diagram shows the positions of identified variants in DENND5A mRNA (upper panel), and the predicted size of truncated protein (p.Ser969fs) derived from the frameshift variant. a.a. = amino acid. e. DENND5A amino acid alignments of a variety of species (Human, Mouse, Rat, Zebrafish, Chimpanzee, Goat, Chicken, Sheep) highlighting the conserved region flanking the frameshift area (shown in red). f. Representative western blotting and quantification (g) of DENND5A protein in EBV-Lymphoblastoid cells established from 2 unrelated controls and DENND5A heterozygote (III:1) from Family A. h. Correlation of cutaneous melanoma incidence rates, and the aggregated deleterious variants frequency in DENND5A, MC1R, MITF, CDKN2A, CDK4 and POT1 across the globe, including Southern Asian, Eastern Asian, African, Latino, Non-Finnish Europeans, Finnish and Swedish people. Two-tailed, paired t test (g), Pearson correlation (h),
Table 1.
DENND5A variants identified in familial melanoma pedigrees
| Family/Pedigree | Family A | Family B | Family C |
|---|---|---|---|
| No. of cases in pedigree | 4 | 3 | 3 |
| No. of heterozygotes/tested cases | 3/3 | 1/1 | 3/3 |
| Genomic changea | g.9167316 _9167317insC | g.9161004C>T | g.9164379G>A |
| Coding variantb | c.2903_2904insG | - | c.3401C>T |
| Amino acid change | Ser969Leufs*17 | - | p.Thr1134Met |
| Affected domain | PLAT, RUN2 | - | RUN2 |
| Variant type | Frameshift, Stop-gain | 3' UTR | Missense |
| Bioinformatic prediction tools | |||
| CADD | 35.00 | 20.90 | 32.00 |
| Mutation Taster | Disease causing | Disease causing | Disease causing |
| FATHMMc | NA | Functional significance | NA |
| SIFT/PolyPhen | NA | NA | Deleterious/Probably damaging |
| MAF in control datasets | |||
| ExAC | Novel | - | - |
| gnomAD NFE | Novel | 0.000032 | 0.000009 |
Reference genome GRCh37/19
The reference transcript, taken from the Ensembl database (release 70), is DENND5A-001 (ENST00000530044)
Non-coding scoring using SNP Nexus, https://www.snp-nexus.org/v4/
To examine the prevalence of the variant, we enrolled DENND5A in a targeted sequencing project where additional 59 melanoma families were examined 14. Among these families, we were able to identify a 3-case melanoma family (Family B, Fig. 1a; For extended pedigree, Supplementary Fig. S1b). In Family B, the only individual (a melanoma patient) that was available for genetic screening, was heterozygous for a 1-bp substitution (c.*680C>T) in the 3’-UTR region of the DENND5A gene, which is predicted to possess mRNA regulatory function (Fig. 1c, Table 1). Through collaboration, we examined rare variants in DENND5A that co-segregate with disease in 98 cutaneous melanoma patients from 27 American melanoma-prone families with at least three melanoma cases/obligate heterozygotes analyzed by exome sequencing. We found a rare DENND5A germline variant that was shared by three melanoma cases in one American family (Family C, Fig. 1a, Supplementary Table S1). The variant was confirmed by Sanger sequencing in the three melanoma patients, while it was absent in four unaffected individuals who were sequenced in this family. Evaluation of exome sequencing data for the co-segregating variants revealed no known cancer susceptibility genes or obvious candidates in this family. The observed p.Thr1134Met variant is a rare missense variant predicted to be deleterious by at least three algorithms (CADD score, MutationTaster and PolyPhen, Table 1). This variant would lead to a change of the first amino acid of the RUN2 domain, slightly downstream of the truncating variant found in Swedish Family A. Of note, several members of family A and C had dysplastic nevi (DN, Fig. 1a). Even though DN did not segregate with the melanomas in these two families, it added to the risk. The age at diagnosis and tumor locations for the melanomas were described in detail in the supplementary data (Supplementary Table S1).
We next analyzed the significance of harboring DENND5A germline variants in various populations. We pooled the DENND5A germline variants that were predicted by CADD scoring to be deleterious and correlated with melanoma incidence rates from different ethnicity groups, including Southern Asian, Eastern Asian, Latino, and Europeans (Supplementary Methods). As a reference, five other known melanoma susceptibility genes, MC1R, MITF, CDKN2A, CDK4 and POT1 were also analyzed by the same algorithms. Deleterious DENND5A variants were most often seen in the Swedish population (47/100,000 individuals, Fig. 1h). The Aggregated Deleterious variants Frequency (ADF) in DENND5A correlated significantly with melanoma incidence rates worldwide (r=0.8862, P=0.0079). The ADFs in other pigmentation genes also correlated with melanoma incidence rates (MC1R: r=0.8798, P=0.0090. MITF: r=0.8363, P=0.0190). On the other hand, deleterious variants in the non-pigmentation, yet high-risk genes, such as CDKN2A, CDK4 and POT1, did not show significant correlation (CDKN2A: r=0.6106, P=0.1453; CDK4: r=0.1883, P=0.6859; POT1: r=0.0176. P=0.9701).
DENND5A is variably expressed in familial cutaneous melanomas and significantly enriched in pigmented samples
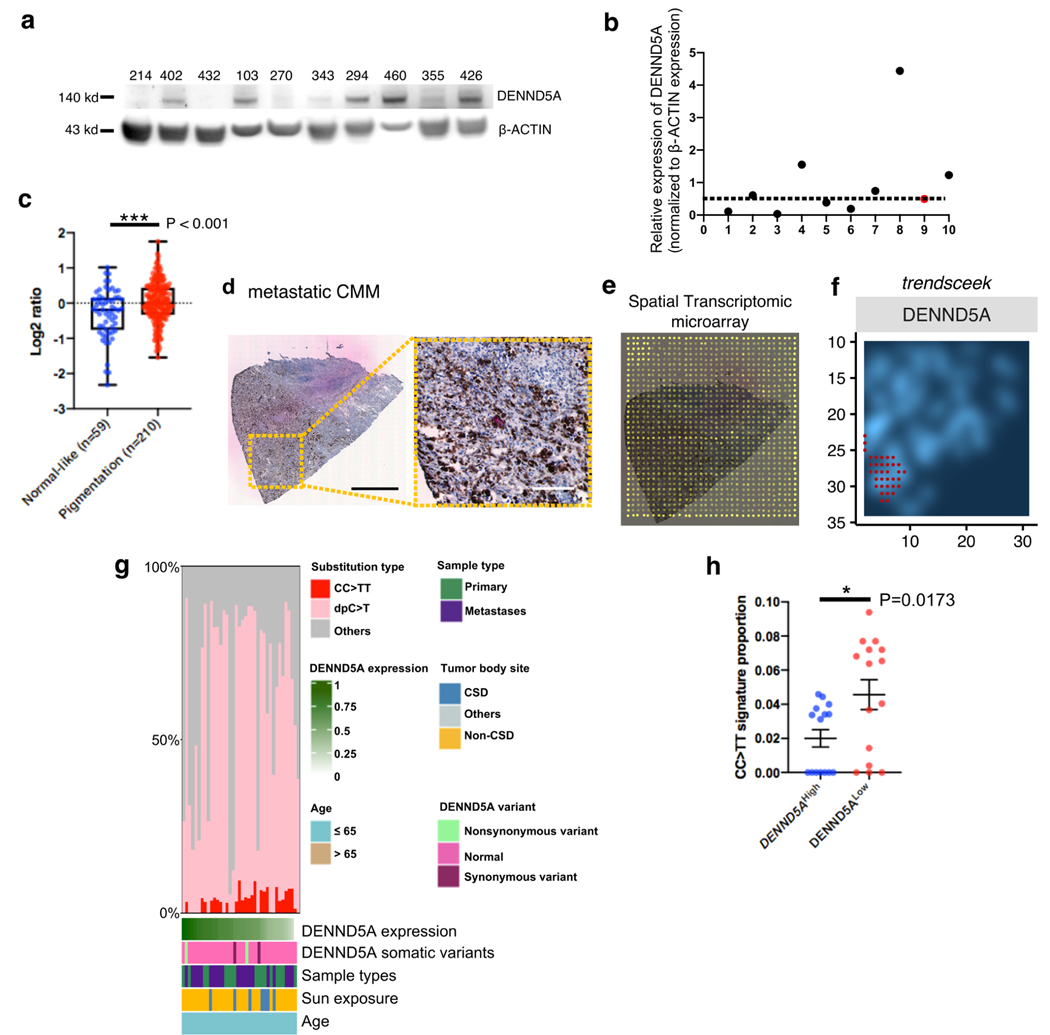
We collected fresh frozen tumors from melanoma-prone families through the Karolinska University Hospital biobank. In total, 10 melanomas from patients belonging to different melanoma-prone families were analyzed for DENND5A protein expression by immunoblotting (Fig. 2a). We noticed considerable variations of DENND5A protein expression among the familial melanoma samples. Among these, the second sample on the right (#355), marked in red (Fig. 2b), derived from the frameshift heterozygote (II:1), potentially harboring only 1 healthy allele. Of note, 3 out of the 10 (30%) melanomas from different patients exhibited substantially suppressed DENND5A protein expression compared to the tumor from the heterozygote (#355, II:1).
Fig. 2. DENND5A is variably expressed in familial melanomas and enriched in pigmented melanoma tissue.
a. Western blotting of DENND5A protein expression in tumors from familial melanomas from a Karolinska University Hospital-based registry since 2000. b. Quantification of relative DENND5A protein expression. Red dot represents DENND5A protein expression in melanoma (#355) derived from heterozygote (II:1). A dotted line is drawn across the DENND5A heterozygote as reference of potential DENND5A suppression. c. DENND5A mRNA expression in the Skin Cutaneous Melanoma (SKCM) cohort of The Cancer Genome Atlas is classified into ‘Normal-like’ and ‘Pigmentation’ subtype by applying ‘Pigmentation gene signature’ and then compared. (d-f). Spatial transcriptomic (ST) analysis on a cutaneous malignant melanoma (CMM). d. (left) Hematoxylin & Eosin stained CMM section and (right) higher magnification showing the pigmentation from the section. Scale bar: 1 cm (left), 0.25 cm (right). e. ST microarray on CMM section with barcoded spots of 100 μm diameter and 200 μm center-to-center distance. f. A density plot of DENND5A expression ran by trendsceek in the same section, with the regions of significantly elevated DENND5A expression marked red. g. A matrix of ultraviolet (UV)-related alterations (CC>TT in red; dpC>T, C>T transition at dipyrimidines, in pink) from all the Cancer Genome Atlas (TCGA) skin cutaneous melanomas (SKCM), arising from both chronic (CSD) and non-chronic (non-CSD) sun-exposed body sites (n=38). Sample types (primary or metastatic), body sites (CSD, or non-CSD) and detected DENND5A somatic variants are color-coded and then sorted after DENND5A mRNA expression. h. Proportion of UV signature (CC>TT transitions) from DENND5AHigh (top 15) and DENND5ALow (lowest 15) mRNA are compared and shown as mean ± S.E.M.; Two-tailed, Unpaired t test (c, h).
We analyzed DENND5A mRNA expression in The Cancer Genome Atlas (TCGA) program with respect to previously described, well-defined melanoma gene signatures17. We showed that DENND5A mRNA expression was significantly higher in ‘Pigmentation’ subtype (n=210) compared to ‘Normal-like’ subtype (n=59) (P < 0.001, Fig. 2c, Supplementary Methods). We also performed correlation analysis on genes that are associated DENND5A mRNA expression in TCGA (Supplementary Fig. S2a), and highlighted genes that were linked to pigmentation defects, such as MYO5A18. Further looking into expression in areas in a single tumor with heterogeneous pigmentation (Fig. 2d), we analyzed spatial transcriptomic data performed on a CMM lymph node metastasis (Fig. 2e, Supplementary Fig. S2b-e, Supplementary Methods). UMAP analysis clearly showed two clusters of expression profiles (Supplementary Fig. S2c), delineating identical pattern of the pigmentation shown in Fig. 2d. DENND5A mRNA expression on each spot was spatially displayed in Supplementary Fig. S2d. Moreover, by utilizing a robust computational method trendsceek (http://github.com/edsgard/trendsceek)19, we found that DENND5A mRNA had a statistically significant spatial pattern and overlapped with the pigmented area of the melanoma tissue (Fig. 2f).
Due to the rarity of germline variants in DENND5A, we sought to explore the relationship between DENND5A mRNA expression and substitution types (such as A>T, A>G, etc.), including the well-established ultraviolet (UV)-related signature (CC>TT substitutions), of sporadic melanomas from The Cancer Genome Atlas (TCGA) skin cutaneous melanoma cohort (SKCM, n=322, Supplementary Fig. S2f)20. Given that genetic load may vary by age of onset and tumor site, we adjusted samples for their age of onset (<=65) and included melanomas arising from both chronic sun-exposed body sites (head and neck) and the non-chronic/intermittently-exposed sites (trunk and proximal extremities, Fig. 2g). After excluding samples without specific body site annotation or arising from sun-shielded body site, we retained 38 melanomas and sorted them after DENND5A mRNA expression. Intriguingly, sporadic melanomas with the lowest DENND5A mRNA expression showed a significantly higher proportion of UV signature (CC>TT transitions, Fig. 2h).
DENND5A and SNX1 are associated with melanosomes
Pigmentation is a unique trait in skin context and implicated in melanoma susceptibility1. To gain a deeper understanding of the role of DENND5A in pigmented CMM samples, we studied the subcellular localization of DENND5A and melanosomal structural proteins ((MLANA and PMEL) by performing immunostaining, immuno-electron microscopy, subcellular fractionation, and immunoprecipitation. The interaction between DENND5A and another critical retrograde transport player sorting nexin 1 (SNX1), was analyzed by immunostaining in the pigmented human melanoma cell line MNT-1. DENND5A as a vesicle trafficking protein originating from the trans-Golgi network (TGN) showed overt perinuclear localization and its positive signals were scattered in cytoplasm (Supplementary Fig. S3a)21. The scattered pattern of SNX1 largely overlapped with a fraction of DENND5A-positive signals. DENND5A staining also co-localized with melanosomal structural proteins MLANA and PMEL (Supplementary Fig. S3b, c). Antibody PMEL-C recognized the PMEL isoforms at TGN (perinuclear pattern), and the membrane of early stage melanosomes (stage I and II, scattered dots, Supplementary Fig. S3c). In addition, SNX1 positive signals significantly overlapped with MLANA and PMEL signals (Supplementary Fig. S3d, e). Differential interference contrast (DIC) microscopy analysis showed that DENND5A and SNX1 trafficked in close vicinity of melanosomes (Supplementary Fig. S3f). Immuno-Electron Microscopy examination on MNT-1 cells captured clusters of DENND5A- and SNX1-colloidal gold particles decorating intermediate to highly pigmented melanosomes (Supplementary Fig. S3g, h, Supplementary Methods). Immunoblotting assays following sucrose gradient ultracentrifugation detected DENND5A and SNX1 in the GM130-negative melanosome-enriched fractions (Supplementary Fig. S3i), concordant with their proximity to melanosomes. GM130 is a specific Golgi marker and here served as a quality control for melanosome purification procedures. Immunoprecipitation showed that SNX1 binds to both DENND5A and melanosome membrane proteins PMEL and MLANA in MNT-1 cells (Supplementary Fig. S3j), suggesting that SNX1 serves as a bridge between DENND5A and early-stage melanosomal cargoes.
Suppressing DENND5A causes a decrease in melanin and dendrite retention
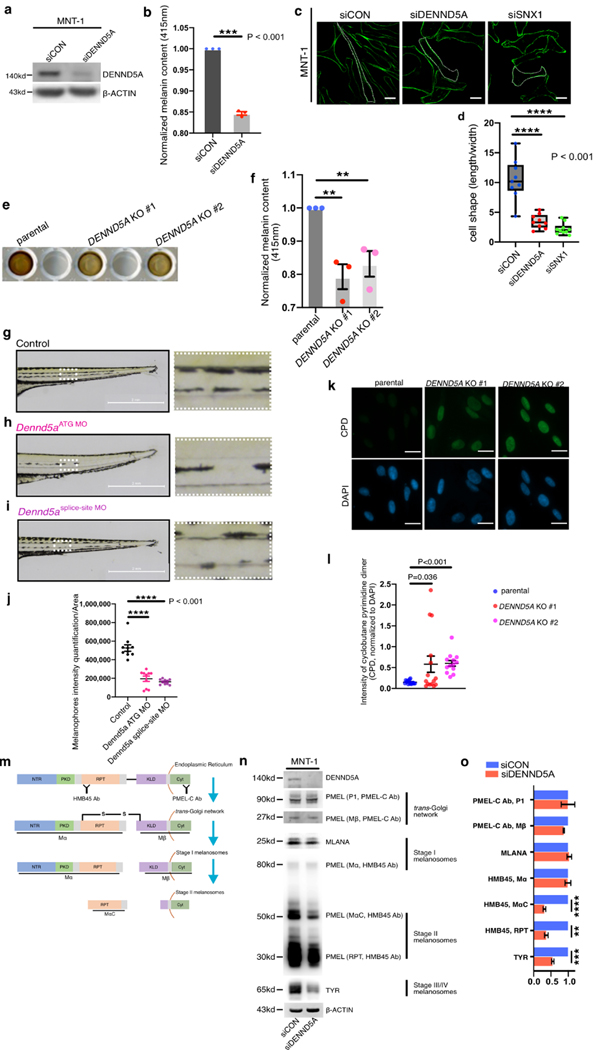
On the basis of these findings, we speculated that DENND5A is involved in melanosomal cargo trafficking. Indeed, down-regulation of DENND5A with small-interfering RNA (siRNA) in MNT-1 markedly decreased the overall melanin content (Fig. 3a, b, Supplementary Methods) and altered cell morphology from an elongated dendritic shape to a rounded, triangular-cuboidal one (Fig. 3c), resembling a well-documented morphological change of retromer malfunction 22,23, as demonstrated by silencing SNX1 in parallel (Fig. 3c, d). By using the CRISPR-Cas9 genome editing technique, we knocked out the DENND5A gene in MNT-1 cells, leading to a dramatic reduction in melanin content and contracted dendrites in DENND5AKO MNT-1 (Fig. 3e, f, Supplementary Fig. S4a-c), consistent with our findings by siRNA silencing. We did not find any significantly altered proliferation, through colony formation assay, in DENND5AKO MNT-1 cells, comparing to the parental MNT-1 cells (Supplementary Fig. S4d, e). By using the IncuCyte live cell imaging system, we also compared the cellular migration of parental and DENND5AKO MNT-1 cells and found similar wound healing capacity (Supplementary Fig. S4f, g).
Fig. 3. DENND5A inhibition leads to melanin reduction, morphological changes and diminished PMEL.
a. Representative western blotting image of DENND5A knockdown efficiency in MNT-1 cells. b. Melanin content of MNT-1 cells before and after knockdown of DENND5A, measured and normalized to corresponding protein yields is shown. c. Phalloidin staining of control (siCON) MNT-1 cells and after silencing DENND5A or SNX1, as indicated. d. Morphological changes are quantified by dividing the length of each cell with its width. e. Melanin samples loaded in a 96-well plate showing the visible difference between MNT-1 parental cells, and DENND5A gene knockout clones (#1, #2) established by CRISPR-Cas9 system. f. Relative melanin content of the parental and DENND5A KO #1 and #2 is quantified and shown. g-i. Brightfield images of (g) wild-type zebrafish (Danio Rerio), (h) Dennd5a-knockdown (Dennd5aATG MO) and (i) Dennd5a-variant mimic (Dennd5asplice-site MO). Higher magnification images of the ventral medial stripes from each group are shown. j. The ventral medial stripes of zebrafish from each group are quantified (10 zebrafish embryos per group) and compared. k. Representative images of the cyclobutane pyrimidine dimer (CPD) staining in parental MNT-1 cells and its DENND5A KO #1 and #2 after UV exposures (30J/m2). l. The intensities of CPD in each area is first quantified and normalized to the intensities of DAPI in the same area and then compared. Scale bar: 20 μm (c), 2 mm (g, h, i). 50 μm (k). m. Schematic chart of the processing of early stage melanosomal cargo PMEL from the ER, trans-Golgi network, to stage I and stage II melanosomes. Antibody PMEL-C recognizes the C-terminus of full-length PMEL precursor and the processed Mβ fragments, both of which are membrane-bound. Antibody HMB45 recognizes the repeated domain (RPT domain) of the full-length PMEL and detects the luminal fragments Mα and MαC, which are crucial for fibrillar matrix formation that is characteristic of Stage II melanosomes. Western blotting (n) and quantification (o) of melanosome markers from different stages of control (siCON) MNT1 and silencing DENND5A (siDENND5A). Quantification is performed from three independent experiments, normalized to controls (b, f, o), and presented as mean ± S.E.M. Two-tailed, Unpaired t test (b, d, f, j, l, o).
Next, we exploited a zebrafish in vivo model to validate our in vitro findings. A specific morpholino, targeting the initiation codon of the DENND5A gene (DENND5AATG-MO) to inactivate its transcription, was delivered to zebrafish embryos. Interestingly, the diminished melanophore formation was noticeable as early as 3 days post-fertilization (dpf). The melanin reduction in ventral medial stripes above cloaca on 5 dpf was shown at higher magnification and quantified (Fig. 3g, h, j). To recapitulate the clinical scenario of the novel frameshift variant in Family A, a splice morpholino targeting the frameshift region of DENND5A was designed and injected (Dennd5a splice-site MO, Fig. 3i). These zebrafish displayed considerably less pigmentation in comparison to the parental zebrafish (Fig. 3j), similar to that observed in DENND5A ATG-MO zebrafish. Interestingly, the DENND5A heterozygote (III:1) had more pale skin, compared to her healthy younger sister (III:2) who did not share the variant (Supplementary Table S1).
Furthermore, we subjected parental and DENND5AKO MNT-1 cells to UV exposure and then compared the induction of potential DNA photodamages through immunostaining of cyclobutane pyrimidine dimers (CPDs). We found significantly elevated levels of intracellular CPD intensities in DENND5AKO MNT-1 cells in comparison to the parental MNT-1 cells (Fig. 3k, l).
Inhibition of DENND5A attenuates processing and secretion of melanosomal cargoes
DENND5A was silenced with small-interfering RNA (siRNA) in MNT-1 cells and probed with antibodies recognizing different stages of melanosomes (Fig. 3m, n). The membrane-integrated P1 and Mβ isoforms of PMEL, recognized by PMEL-C antibody, remained unaltered. Consistently, early stage melanosome structural protein MLANA showed little change after the knockdown. In contrast, MαC and its further processed RPT fragments – isoforms detected by HMB45 antibody - showed significant reduction in siDENND5A-treated MNT-1 cells, indicating lack of fibrillar matrix and insufficient melanin deposition 24. Interestingly, TYR as a stage III/IV melanosomal cargo and rate-limiting enzyme for melanin synthesis 8,11, decreased significantly after knocking down DENND5A (Fig. 3n, o), explaining the reduction of intracellular melanin content. To quantitatively analyze the secreted forms of melanosomes, we collected the conditioned medium of control (siCON-) and siDENND5A-MNT-1 cells (Supplementary Fig. S4h). RAB27A, a well-documented vesicle trafficking protein that regulates melanosome transport to melanocytic dendrites, served as endogenous control for the amounts of secreted vesicles including both melanosomes and possibly exosomes18,25. PMEL (MαC) decreased significantly in the siDENND5A-MNT-1 cells, indicating a reduction in melanosome secretion, whereas the ER-bound P1 isoform of PMEL- as a quality control for isolation - remained undetectable (Supplementary Fig. S4i, j). Moreover, our correlation analysis of DENND5A in TCGA showed that its expression associated with genes that were previously linked to pigmentation defects, such as the core component of retromer (VPS26A, r=0.438) and the metalloproteinase (ADAM17, r=0.341, Supplementary Fig. S2a)12. Further analysis of DENND5A-related genes from spatial transcriptomics also showed that the secretory pathway is significantly associated with DENND5A expression (Supplementary Fig. S2e).
Incomplete function of DENND5A leads to mis-sorting and lysosomal degradation of PMEL and TYR.
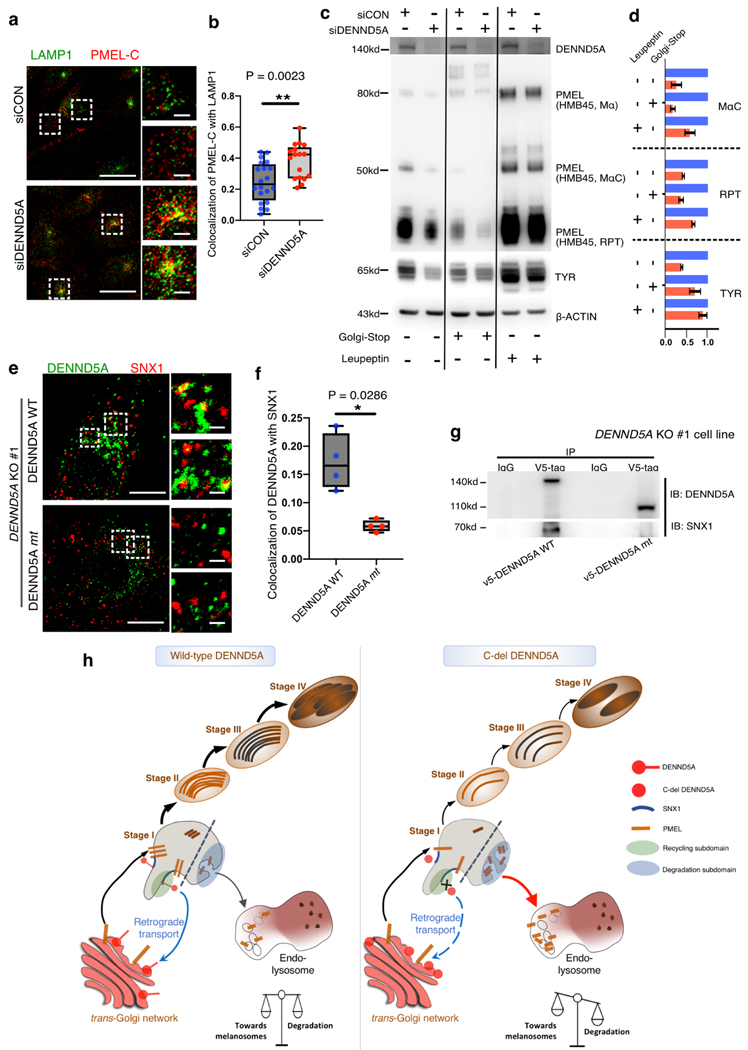
We further investigated the role of DENND5A in the transport and sorting of PMEL. MNT-1 cells (siCON- and siDENND5A) were double immunostained with PMEL-C antibody and lysosomal marker LAMP1. Interestingly, PMEL vesicles were accumulated in LAMP1-positive lysosomes (Fig. 4a, b), suggesting an alternative sorting of melanosomal cargo protein PMEL to lysosome.
Fig. 4. Defective DENND5A triggers lysosomal degradation of melanosomal cargoes and fails to associate with SNX1-decorated vesicles.
a. Representative confocal microscopy images of control MNT-1 (siCON, upper) and after silencing DENND5A (siDENND5A, lower). LAMP1 (green) and PMEL (PMEL-C antibody, red) labeled MNT-1 cells are labeled and shown in merged channels and with higher magnification on the right-hand side. Co-localization is measured using images from two independent experiments and shown in b. Western blotting (c) and quantification (d) of melanosome markers for control (siCON)-MNT1 and siDENND5A-MNT1 after treatment of Golgi-Stop or lysosome protease inhibitor Leupeptin, respectively. e. DENND5A KO #1-MNT-1 cells were transiently introduced with V5-DENND5AWT (upper) and V5-DENND5Amt (lower) respectively. Transfected cells are stained for DENND5A (green) and SNX1 (red). Co-localization co-efficiency is measured using images from two independent experiments and shown in (f). Scale bars: 10 μm (a, e), 1 μm for the magnified panels. Data quantification is presented as mean ± S.E.M. Two-tailed, Unpaired t test (d), Mann-Whitney test (b, f). g. Immunoprecipitation of V5-tag on DENND5A KO #1 cells over-expressing the wild-type (V5-DENND5AWT) and mutated (V5-DENND5Amt) constructs. h. Proposed working model showing (left) the homeostasis of melanosome maturation and lysosomal degradation coordinated by DENND5A-SNX1 interaction. (right) C-del DENND5A loses its interaction with SNX1 and triggers mis-sorting of PMEL from en route to melanosomes to degradation subdomain (stage I melanosome)26, resulting in lysosomal entry and degradation of PMEL.
As the continuous anterograde transport of PMEL from trans-Golgi may mask our observation, we treated siDENND5A-MNT-1 cells with a Golgi-stop reagent. As expected, RPT fragments were further reduced (Fig. 4c, d). Importantly, treatment of siDENND5A-MNT-1 cells with the lysosome protease inhibitor Leupeptin largely restored the expression of melanosomal cargoes PMEL and TYR. These findings collectively demonstrated that DENND5A plays an essential role in maintaining the processing and maturation of melanosomal cargoes, and absence of DENND5A leads to re-direction of melanosomal cargoes to lysosomal degradation.
To recapitulate the novel truncating variant, we investigated the function of C-del DENND5A in a DENND5A KO cell line. DENND5A wild-type (DENND5AWT) and C-del constructs (DENND5Amt) were introduced in DENND5A KO#1 cell line. Notably, C-del DENND5A completely lost its co-localization with SNX1 (Fig. 4e, f). Immunoprecipitation experiments further corroborated this finding and showed that C-del DENND5A lost its interaction with SNX1 (Fig. 4g), indicating that the C-terminus of DENND5A was essential for physical interaction with SNX1. Taken together, our data allowed us to propose a working model (Fig. 4h). Once DENND5A function is compromised, either suppressed or truncated in C-terminus, DENND5A loses its interaction with SNX1 and consequently its interaction with PMEL. Dissociation of DENND5A and SNX1 disrupts PMEL processing, leaving the cargoes accessible for degradation subdomain and default lysosome leakage favored by SNX1-retromer system26.
DISCUSSION
Even though numerous melanoma risk genes have previously been discovered by linkage analyses, massive parallel sequencing and Genome-Wide Association Studies (GWAS), there remain a substantial proportion of melanoma-prone families with unknown genetic background 1. Here, we report rare DENND5A germline variants found in melanoma-prone families. We show that DENND5A is enriched in pigmented melanomas and physically associated with melanosomes. DENND5A protein expression is also frequently suppressed in familial melanoma samples.
DENND5A, also known as the Rab6-interacting protein 1, has previously been shown to be involved in retrograde transport by interacting with Rab6 via its RUN1 domain 21. Interestingly, Patwardhan et al. revealed that Rab6 is exploited in the secretory route to transport Golgi-derived cargoes to lysosome related organelle27.
Overall, loss-of function variants are extremely rare in the general population and DENND5A is considered to be a haploinsufficient gene, defined as being highly intolerant to deleterious variants 28. Previously, germline homozygous variants in DENND5A have been linked to Epileptic Encephalopathy through regulation of the MAPK pathway 29. Recently, two pathogenic homozygous variants in DENND5A were linked to Intellectual Disability (ID)30. We could not find any documentation of neurodevelopmental disorders in our families. This is however understandable, considering those previously reported germline variants were all homozygous, and three out of four (p.Asp173Profs*8, p.Lys850Serfs*11, p.Asp541Gly) occurred much more upstream than the variants we detected in the melanoma families. Our study on DENND5A may expand our knowledge on how disruption of this evolutionarily conserved gene may contribute to other human disorders, such as CMM.
Interestingly, we found that, when DENND5A was suppressed, cells displayed a more oval shape with less properly stretched-out dendrites. The ability to branch out and form dendrites is crucial to melanocytes in vivo, as these increase the contact surface with nearby keratinocytes and therefore facilitate the secretion of mature melanosomes18. The morphological changes might be attributed to the previous finding from C. elegans that DENND5A and SNX1 interacted with component (p150glued) of minus-end directed microtubule motor dynein 31. Suppressing either one of these three led to less longitudinal force and reduced long-range vesicle transport. Notably, our immunoprecipitation experiments suggested that, even though DENND5A and SNX1 interact with each other and also with MLANA (the characteristic, structural protein of early stage melanosome)11, the interaction of DENND5A and membrane-integrated form of PMEL is more likely to be indirect and dependent on SNX1.
By western blotting, we showed that processed forms of PMEL were significantly affected. However, since melanosomal cargo proteins (particularly MLANA) also label the compartments from trans-Golgi network to early endosomes/Stage I melanosomes, our microscopic data could not exclude that impaired DENND5A function may have affected the more upstream sorting of transport intermediates, from early endosomes en route to melanosomes32,33. Although the impact of PMEL dysfunction alone on pigmentation is debated34, reduced MαC and RPT fragments, together with reduction of rate-limiting enzyme TYR are certainly accountable for the visible melanin reduction. At the same time, whether DENND5A plays a more direct role in the secretory pathway of TYR trafficking from endosomes to melanosomes needs further investigation.
We found RPT expression to be restored when using the lysosome protease inhibitor Leupeptin, suggesting that the reduction of PMEL fragments was largely due to lysosomal degradation. Similar mechanisms were also reported when other components of the retromer system were modulated, such as SNX4, SNX27 and SNX1735.
Evidently, while DENND5A germline loss-of-function variants might be rare, suppressed DENND5A protein expression was not uncommon among familial melanoma tumors, which is likely due to additional genomic alterations or epigenetic events. Taken together, we propose a working model highlighting the significance of DENND5A, and more importantly the interaction of DENND5A with SNX1 through its C-terminus. Our model also illustrates how suppressed DENND5A expression could presumably be as defective as having a C-terminal truncated protein.
These individuals presumably tend to have pigmentation deficiency and therefore might be susceptible to UV exposure. The increased intensities of cyclobutane pyrimidine dimers in DENND5AKO-MNT-1 cells after UV exposure, compared to the parental MNT-1 cells, provides further proof of potential photodamages in less pigmented cells. Our discovery of the elevated UV signature in DENND5ALow-sporadic melanomas attributes their melanogenesis to such gene-environment interaction.
Conclusion
We identified a novel loss-of-function germline DENND5A variant and two additional rare variants of this gene in melanoma-prone families. Our study describes the physiological role of DENND5A in the context of the skin. Importantly, we provide mechanistic evidence of DENND5A deregulation contributing to melanoma susceptibility through the pigmentation pathway.
Supplementary Material
Acknowledgements:
We thank patients and their family members for active participation in this work. We thank the Zebrafish Core Facility at Karolinska Institute for the daily maintenance, morpholino injection and sample preparation. We acknowledge the National Genomics Infrastructure (NGI) for next-generation sequencing. The computations and data handling were enabled by resources provided by the Swedish National Infrastructure for Computing (SNIC) at Uppsala Multidisciplinary Center for Advanced Computational Science UPPMAX, in part funded by the Swedish Research Council through grant agreement no. 2018-05973. The authors also acknowledge the research contributions of the Cancer Genomics Research Laboratory for their expertise, execution, and support of this research in the areas of project planning, wet laboratory processing of specimens, and bioinformatics analysis of generated data.
Funding:
This work is supported by grants from the Radiumhemmet’s research funds (2019-194103), the Swedish Cancer Society (CAN 2017-733) and in part with Federal funds from the National Cancer Institute, National Institutes of Health, under NCI Contract No. 75N910D00024. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. M.Y. has received financial support from China Scholarship Council. P.G.E. is financially supported by the Knut and Alice Wallenberg Foundation as part of the National Bioinformatics Infrastructure Sweden at SciLifeLab.
Footnotes
Ethics declarations
Written consents for participation/publication have been received and archived from every individual whose data are included. The study was approved by the ethics committee of Karolinska University Hospital.
Competing Interests
Professor Joakim Lundeberg is scientific consultant for 10X Genomics Inc, holding the IP for the barcoded slides. The rest of the authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data availability
The datasets supporting the conclusions of this article are included within the article and in the supplementary materials. The raw human genetics data are not publicly available due to consent from restrictions. Raw data from RNA sequencing and the significant genetics variant data generated from raw genetics data are available upon request.
REFERENCES
- 1.Read J, Wadt KA, Hayward NK. Melanoma genetics. J Med Genet. 2016;53(1):1–14. [DOI] [PubMed] [Google Scholar]
- 2.Titus-Ernstoff L, Perry AE, Spencer SK, Gibson JJ, Cole BF, Ernstoff MS. Pigmentary characteristics and moles in relation to melanoma risk. Int J Cancer. 2005;116(1):144–149. [DOI] [PubMed] [Google Scholar]
- 3.Sulem P, Gudbjartsson DF, Stacey SN, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39(12):1443–1452. [DOI] [PubMed] [Google Scholar]
- 4.Goldstein AM, Tucker MA. Genetic epidemiology of cutaneous melanoma: a global perspective. Arch Dermatol. 2001;137(11):1493–1496. [DOI] [PubMed] [Google Scholar]
- 5.Davie JR, Randerson-Moor J, Kukalizch K, et al. Inherited variants in the MC1R gene and survival from cutaneous melanoma: a BioGenoMEL study. Pigment Cell Melanoma Res. 2012;25(3):384–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertolotto C, Lesueur F, Giuliano S, et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011;480(7375):94–98. [DOI] [PubMed] [Google Scholar]
- 7.Yokoyama S, Woods SL, Boyle GM, et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature. 2011;480(7375):99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudbjartsson DF, Sulem P, Stacey SN, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat Genet. 2008;40(7):886–891. [DOI] [PubMed] [Google Scholar]
- 9.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brenner M, Hearing VJ. The protective role of melanin against UV damage in human skin. Photochem Photobiol. 2008;84(3):539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raposo G, Marks MS. Melanosomes--dark organelles enlighten endosomal membrane transport. Nat Rev Mol Cell Biol. 2007;8(10):786–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bissig C, Rochin L, van Niel G. PMEL Amyloid Fibril Formation: The Bright Steps of Pigmentation. Int J Mol Sci. 2016;17(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leonhardt RM, Vigneron N, Hee JS, Graham M, Cresswell P. Critical residues in the PMEL/Pmel17 N-terminus direct the hierarchical assembly of melanosomal fibrils. Mol Biol Cell. 2013;24(7):964–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuominen R, Engstrom PG, Helgadottir H, et al. The role of germline alterations in the DNA damage response genes BRIP1 and BRCA2 in melanoma susceptibility. Genes Chromosomes Cancer. 2016;55(7):601–611. [DOI] [PubMed] [Google Scholar]
- 15.Goldstein AM, Xiao Y, Sampson J, et al. Rare germline variants in known melanoma susceptibility genes in familial melanoma. Hum Mol Genet. 2017;26(24):4886–4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoiom V, Edsgard D, Helgadottir H, et al. Hereditary uveal melanoma: a report of a germline mutation in BAP1. Genes Chromosomes Cancer. 2013;52(4):378–384. [DOI] [PubMed] [Google Scholar]
- 17.Jonsson G, Busch C, Knappskog S, et al. Gene expression profiling-based identification of molecular subtypes in stage IV melanomas with different clinical outcome. Clin Cancer Res. 2010;16(13):3356–3367. [DOI] [PubMed] [Google Scholar]
- 18.Hume AN, Seabra MC. Melanosomes on the move: a model to understand organelle dynamics. Biochem Soc Trans. 2011;39(5):1191–1196. [DOI] [PubMed] [Google Scholar]
- 19.Edsgard D, Johnsson P, Sandberg R. Identification of spatial expression trends in single-cell gene expression data. Nat Methods. 2018;15(5):339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas N. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161(7):1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miserey-Lenkei S, Waharte F, Boulet A, et al. Rab6-interacting protein 1 links Rab6 and Rab11 function. Traffic. 2007;8(10):1385–1403. [DOI] [PubMed] [Google Scholar]
- 22.Burgo A, Sotirakis E, Simmler MC, et al. Role of Varp, a Rab21 exchange factor and TI-VAMP/VAMP7 partner, in neurite growth. EMBO Rep. 2009;10(10):1117–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohbayashi N, Yatsu A, Tamura K, Fukuda M. The Rab21-GEF activity of Varp, but not its Rab32/38 effector function, is required for dendrite formation in melanocytes. Mol Biol Cell. 2012;23(4):669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watt B, van Niel G, Raposo G, Marks MS. PMEL: a pigment cell-specific model for functional amyloid formation. Pigment Cell Melanoma Res. 2013;26(3):300–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ostrowski M, Carmo NB, Krumeich S, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12(1):19–30; sup pp 11–13. [DOI] [PubMed] [Google Scholar]
- 26.Gershlick DC, Lucas M. Endosomal Trafficking: Retromer and Retriever Are Relatives in Recycling. Curr Biol. 2017;27(22):R1233–R1236. [DOI] [PubMed] [Google Scholar]
- 27.Patwardhan A, Bardin S, Miserey-Lenkei S, et al. Routing of the RAB6 secretory pathway towards the lysosome related organelle of melanocytes. Nat Commun. 2017;8:15835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han C, Alkhater R, Froukh T, et al. Epileptic Encephalopathy Caused by Mutations in the Guanine Nucleotide Exchange Factor DENND5A. Am J Hum Genet. 2016;99(6):1359–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anazi S, Maddirevula S, Faqeih E, et al. Clinical genomics expands the morbid genome of intellectual disability and offers a high diagnostic yield. Mol Psychiatry. 2017;22(4):615–624. [DOI] [PubMed] [Google Scholar]
- 31.Wassmer T, Attar N, Harterink M, et al. The retromer coat complex coordinates endosomal sorting and dynein-mediated transport, with carrier recognition by the trans-Golgi network. Dev Cell. 2009;17(1):110–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Maziere AM, Muehlethaler K, van Donselaar E, et al. The melanocytic protein Melan-A/MART-1 has a subcellular localization distinct from typical melanosomal proteins. Traffic. 2002;3(9):678–693. [DOI] [PubMed] [Google Scholar]
- 33.Raposo G, Tenza D, Murphy DM, Berson JF, Marks MS. Distinct protein sorting and localization to premelanosomes, melanosomes, and lysosomes in pigmented melanocytic cells. J Cell Biol. 2001;152(4):809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hellstrom AR, Watt B, Fard SS, et al. Inactivation of Pmel alters melanosome shape but has only a subtle effect on visible pigmentation. PLoS Genet. 2011;7(9):e1002285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallon M, Cullen PJ. Retromer and sorting nexins in endosomal sorting. Biochem Soc Trans. 2015;43(1):33–47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and in the supplementary materials. The raw human genetics data are not publicly available due to consent from restrictions. Raw data from RNA sequencing and the significant genetics variant data generated from raw genetics data are available upon request.