Abstract
To evaluate which microorganisms might be responsible for microbial reduction of humic substances in sedimentary environments, humic-reducing bacteria were isolated from a variety of sediment types. These included lake sediments, pristine and contaminated wetland sediments, and marine sediments. In each of the sediment types, all of the humic reducers recovered with acetate as the electron donor and the humic substance analog, 2,6-anthraquinone disulfonate (AQDS), as the electron acceptor were members of the family Geobacteraceae. This was true whether the AQDS-reducing bacteria were enriched prior to isolation on solid media or were recovered from the highest positive dilutions of sediments in liquid media. All of the isolates tested not only conserved energy to support growth from acetate oxidation coupled to AQDS reduction but also could oxidize acetate with highly purified soil humic acids as the sole electron acceptor. All of the isolates tested were also able to grow with Fe(III) serving as the sole electron acceptor. This is consistent with previous studies that have suggested that the capacity for Fe(III) reduction is a common feature of all members of the Geobacteraceae. These studies demonstrate that the potential for microbial humic substance reduction can be found in a wide variety of sediment types and suggest that Geobacteraceae species might be important humic-reducing organisms in sediments.
Recent studies have demonstrated that two Fe(III)-reducing microorganisms, Geobacter metallireducens and Shewanella alga, can substitute humic substances for Fe(III) as the terminal electron acceptor (20). The electron-accepting group(s) on humic substances was not definitively identified. However, it is suspected that quinone moieties might be important electron-accepting components (20). Both G. metallireducens and S. alga conserved energy to support growth by reducing the humic substance analog, 2,6-anthraquinone disulfonate (AQDS), to 2,6-anthrahydroquinone disulfonate (AHQDS). This demonstrated that extracellular quinones can serve as electron acceptors for these organisms.
The process of microbial humic substance reduction is of interest because it may serve as an important mechanism for organic matter oxidation in some environments, especially if Fe(III) is also present (20). This is because reduced humic substances can abiotically transfer the electrons gained from microbial reduction to Fe(III). This regenerates the humic substances in an oxidized form, which may again accept electrons from humic-reducing bacteria. Therefore, in the presence of Fe(III), even low concentrations of humic substances could be a quantitatively significant electron acceptor for organic matter oxidation. Oxidation of the organic matter in this manner may be more rapid than oxidation of organic matter linked directly to Fe(III) because soluble humic substances are more readily accessible for microbial reduction than are insoluble Fe(III) oxides (20). This phenomenon was evident in studies in which the addition of humic substances (26) or AQDS (2) to humic material-poor, Fe(III)-containing sediments from petroleum-contaminated aquifers greatly stimulated the anaerobic degradation of benzene.
Further understanding of the potential importance of microbial humic substance reduction requires knowledge about the distribution and diversity of microorganisms that might be responsible for this reduction in sedimentary environments. In anaerobic environments in which Fe(III) reduction, sulfate reduction, or methanogenesis predominates, acetate is generally the primary electron donor (19). Therefore, acetate-oxidizing humic-reducing bacteria in a variety of sediment types were examined. The results demonstrate that in all the sediments evaluated, the acetate-oxidizing humic-reducing microorganisms recovered were in the family Geobacteraceae.
MATERIALS AND METHODS
Source of sediments.
Grab samples of surficial sediments were collected as previously described (22) from several freshwater and marine habitats. Freshwater sediments were collected from a shallow wetland in Fairfax, Va., that appeared to be contaminated with hydrocarbons, as well as from a depth of 200 ft in Cayuga Lake, Ithaca, N.Y. Marine samples were collected from two previously described sites (5) in San Diego Bay and two sites near the Norfolk Navy Base, Norfolk, Va.
The depth distribution of humic substance-reducing microorganisms was examined in more detail in the previously described (31) freshwater Talladega wetland in Hale County, Ala. Sediments were collected in 5-cm-diameter Plexiglas tubes. The cores were placed in an anaerobic glovebag and fractionated into depth intervals of 0 to 2.5, 5.0 to 7.5, 10 to 15, 15 to 25, and 35 to 55 mm. The comparable depth intervals from six cores were combined.
Culturing techniques.
Standard anaerobic culturing techniques (13, 28) were used throughout. The medium was boiled and then cooled under a stream of N2-CO2 (80:20) to remove dissolved O2 and dispensed into either anaerobic pressure tubes or serum bottles capped with thick butyl rubber stoppers. Unless otherwise stated, all the incubations were under a headspace gas of N2-CO2.
For all sediments except those from the Talladega wetland, 1-g samples of sediment were inoculated into 9 ml of either APW marine medium (6) or basal freshwater medium (20) with acetate (2 mM) as the electron donor and AQDS (5 mM) as a potential electron acceptor. Isolates were obtained from media solidified with agar. Marine isolates were recovered as individual colonies growing on anaerobic agar plates as previously described (6, 7). Freshwater isolates were obtained by the agar shake tube technique as previously described (7). The isolates were incubated at 30°C in the dark, except for the lake sediments, which were incubated at 25°C.
For the Talladega wetland sediments, aliquots (1 g) from each depth interval were inoculated into triplicate tubes of the acetate-AQDS freshwater medium described above. These initial dilutions also contained sodium pyrophosphate (0.1%) to release cells adsorbed to the sediment particles. Subsequent 10-fold serial dilutions were carried out in triplicate in the same medium without the pyrophosphate. Incubations were at 25°C.
Physiological characterization.
The oxidation of acetate coupled to the reduction of humic acids was determined as previously outlined (20). Briefly, washed suspensions of cells that had been grown in medium with acetate (20 mM) as the electron donor and Fe(III) citrate (50 mM) as the electron acceptor were added to 10 ml of bicarbonate (30 mM) buffer which contained acetate (0.2 mM) amended with [2-14C]acetate (1 μCi, 44.5 mCi/mmol) as the electron donor. The final concentration of cells was ca. 1 mg of protein/ml. Highly purified soil humic acids from the International Humic Substances Society were added (2 mg/ml) as a potential electron acceptor. Acetate oxidation was monitored by measuring the production of 14CO2 over time with a gas proportional counter as previously described (25).
The potential to use electron acceptors other than humic substances was tested by visually monitoring the growth or reduction of the electron acceptor in the isolation medium. Potential electron acceptors were added from sterile anoxic stocks as previously described (7). The potential for growth with electron donors other than acetate was tested in the AQDS isolation medium in which acetate had been replaced with other electron donors. AQDS reduction was monitored visually or by the increase in absorbance at 450 nm.
Phylogenetic analysis of isolates and organisms in MPN enumerations.
Nearly complete 16S rDNA sequences of isolates were amplified with eubacterial primers 8F and 1492R (11, 34) or eubacterial primers 50F (15) and 1391R (16), and sequences were obtained by automated sequencing as previously described (17).
To infer the phylogenetic placement of the acetate-oxidizing, AQDS-reducing microorganisms in the highest positive most-probable-number (MPN) dilutions in the Talladega wetland sediments, cells were collected from a 3-ml aliquot of the dilution by centrifugation. The supernatant was removed, and the pellets were washed with 10 mM Tris buffer (pH 8.0) and resuspended in Tris. The cells were lysed by three rounds of freezing in liquid nitrogen and thawing in a 65°C water bath. The resulting lysate was extracted with phenol, phenol-chloroform-isoamyl alcohol (25:24:1) and chloroform-isoamyl alcohol (24:1) (33). The extracted nucleic acids were amplified by PCR with eubacterial 8F primer (11) with a 40-base GC clamp (29, 30) (5′-CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCGAGAGTTTGATCCTGGCTCAG-3′) and a reverse primer (5′-GTATTACCGCGGCTGCTGG-3′) derived from 519R (16). PCRs were run by touchdown PCR (9) with a hot start to decrease nonspecific PCR products due to the presence of the GC-rich primer (29).
The PCR products were separated by denaturing gradient gel electrophoresis (DGGE) on a denaturing-gradient (50 to 70%) acrylamide gel (7%) as previously described (29). Bands detected by ethidium bromide staining were excised from the gel and reamplified with primers 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) (11) and the reverse primer derived from 519R (16). The PCR products were sequenced as described above.
Preliminary phylogenetic placement of the partial 16S rDNA sequences of isolates and DGGE bands was determined with the Blast program (1). The sequences were manually aligned against 16S rRNA sequences obtained from the Ribosomal Database Project (27). Phylogenetic trees were inferred by using the least-squares algorithm (8) with Jukes-Cantor evolutionary distances (14). GenBank accession numbers are as follows: Desulfobulbus propionicus, M34410; Desulfomonile tiedjei, M26635; Desulfosarcina variabilis, M34407; Desulfovibrio vulgaris, M34399; Desulfovibrio desulfuricans, M37312; Desulfuromonas acetexigens, U23140; Desulfuromonas acetoxidans, M26634; Desulfuromonas palmitatis, U280172; Desulfuromusa bakii, X79412; Desulfuromusa kysingii, X79414; Escherichia coli, J01695; Geobacter chapelleii, U41561; Geobacter hydrogenophilus, U28174; Geobacter metallireducens, L07834; Geobacter sulfurreducens, U13928; Myxococcus xanthus, M34114; Pelobacter acetylenicus, X70955; Pelobacter acidigallici, X77210; Pelobacter carbinolicus, U23141; Pelobacter propionicus, X70954; Pelobacter venetianus, U41562. Aligned sequences were obtained from the Ribosomal Database Project (27).
Analytical techniques.
HCl-extractable Fe(II) concentrations were determined with ferrozine as previously described (23). Concentrations of AHQDS were determined by measuring the increase in absorbance at 450 nm as described previously (20). Cell growth was determined by direct counts via epifluorescence microscopy (12).
Nucleotide sequence accession numbers.
The partial 16S rDNA sequences of strains Ala-5, FD-1, JW-3, SDB-1, and TC-4 (accession no. AF019928, AF019931, AF019932, AF019933, and AF019935, respectively) and molecular isolates 1, 2, 3, and 4 (accession no. AF019937, AF019938, AF019939, and AF019940, respectively) have been submitted to GenBank.
RESULTS AND DISCUSSION
Enrichment and isolation with AQDS yields humic-reducing Geobacter spp. from freshwater sediments.
No suitable method has yet been devised for directly isolating microorganisms with humic substances as the electron acceptor because of the difficulties in incorporating high concentrations of humic substances in media and in monitoring cell growth in humic substance-containing media. To determine if isolation with the humic substance analog, AQDS, would yield organisms which would also be able to use humic substances as an electron acceptor, enrichments were established with two different freshwater sediments.
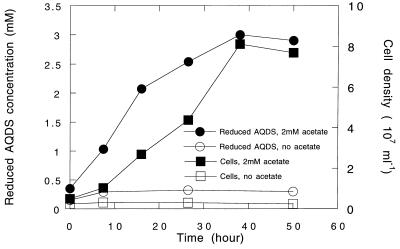
The first of these was from a hydrocarbon-contaminated wetland in Fairfax, Va. Inoculation of sediment into medium with acetate as the sole electron donor and AQDS as the potential electron acceptor resulted in visually apparent AQDS reduction within 5 days. The enrichment was transferred into fresh medium, with a resultant reduction of the AQDS in 2 days. The enrichment was transferred twice more before isolation by the agar shake tube technique (35). When growing in solid acetate-AQDS medium, colonies were typically less than 1 mm in diameter and bright red with an orange halo. Four morphologically identical cultures were isolated. One of these isolates, designated strain JW-3, was selected for further study. Strain JW-3 is a strict anaerobic, nonfermentative, nonflagellated, non-spore-forming, nonmotile, gram-negative rod 1 to 2 μm by 0.5 μm. It grows with acetate as the electron donor and AQDS as the sole electron acceptor (Fig. 1). No growth or AQDS reduction was observed in the absence of acetate. Similarly, no growth was observed with acetate in the absence of a suitable electron acceptor.
FIG. 1.
Growth of strain JW-3 (squares) and AQDS reduction (circles) with acetate (2 mM) as the electron donor. The results of one representative experiment of triplicate determinations are depicted.
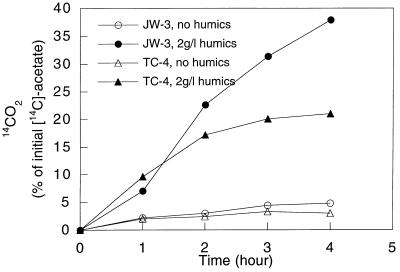
No attempt was made to monitor the growth of strain JW-3 on humic substances because these substances interfere with typical measures of growth such as direct cell counts and protein determinations (20). However, with highly purified soil humic acids from the International Humic Substances Society as the humic substance source, strain JW-3 exhibited humic substance-dependent acetate oxidation (Fig. 2). This result demonstrated that enrichment and isolation of an organism with AQDS as the electron acceptor could yield a humic-reducing microorganism.
FIG. 2.
Humic substance-dependent oxidation of [14C]acetate to 14CO2 by strains JW-3 and TC-4.
To evaluate the AQDS isolation procedure in a different freshwater aquatic sediment, the enrichment and isolation procedure with acetate as the electron donor and AQDS as the electron acceptor was repeated with bottom sediments from Cayuga Lake, Ithaca, N.Y. Four morphologically identical isolates were obtained, one of which, strain TC-4, was selected for further study. Strain TC-4 was morphologically distinct from strain JW-3 in that it was a curved, motile, gram-negative rod 1.5 to 2.5 μm by 0.5 μm. As with strain JW-3, strain TC-4 grew with acetate oxidation coupled to AQDS reduction (4) and oxidized acetate with humic acids as the electron acceptor (Fig. 2).
Further characterization of the humic substance-reducing isolates indicated that in addition to acetate, strain JW-3 used 10 mM formate, 10 mM ethanol, 10 mM pyruvate, 10 mM lactate, 101 kPa of H2, or unidentified electron donors in yeast extract (1 g/liter) for the reduction of AQDS. In addition to AQDS or humic acids, strain JW-3 used 10 mM nitrate, 50 mM Fe(III) citrate, 50 mM Fe(III) oxide, 20 mM Mn(IV), 10 mM elemental sulfur, or 50 mM fumarate as an alternative electron acceptor with acetate as the sole electron donor. Strain JW-3 did not use a variety of other potential electron donors (5 mM propionate, 5 mM butyrate, 10 mM methanol, 0.5 mM phenol, 1 mM palmitate, 0.5 mM benzoate, 1 mM toluene, 10 mM glucose, 1 mM succinate, and 10 mM fumarate) and acceptors (10 mM sulfate, 10 mM malate, and 2 mM selenate). Characterization of TC-4 was not as complete, but it was found that strain TC-4 could grow with Fe(III) chelated with nitrilotriacetate [Fe(III)-NTA] (10 mM) as the sole electron acceptor but not Fe(III) citrate (50 mM).
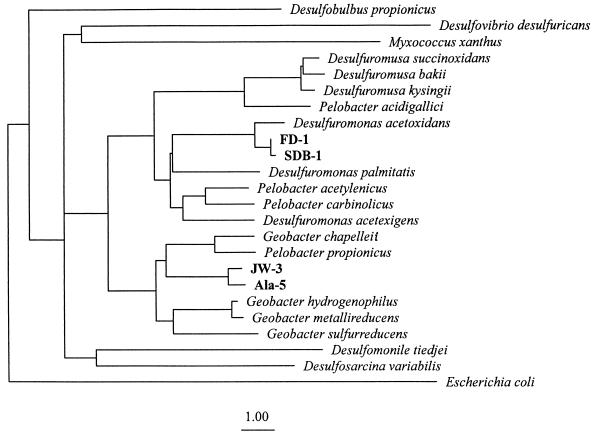
Phylogenetic analysis of the nearly complete 16S rDNA sequence of strain JW-3 placed it within the delta subdivision of the Proteobacteria in the family Geobacteraceae (Fig. 3). The closest previously described relative of strain JW-3 is the dissimilatory Fe(III)-reducer Geobacter chapelleii (93.8% sequence identity; 1,403 nucleotides considered). Detailed inspection of the 16S rDNA sequence of strain JW-3 revealed the presence of nucleotides (positions 122, 239, 286, 453, 454, 681, 690, 822, 859, 878, 888, 1117, 1168, 1254, and 1283; E. coli numbering) characteristic (17) of the genus Geobacter. Phylogenetic analysis of the partial 16S rDNA sequence of strain TC-4 indicated that G. chapelleii was also the closest known relative of TC-4 (97.9% similarity, 434 base positions considered).
FIG. 3.
Phylogenetic tree showing placement of the humic-reducing isolates within the family Geobacteraceae. A total of 812 positions of the 16S rRNA sequences were used to infer the phylogenetic relationships. The sequence of E. coli was included as an outgroup. Bar, one evolutionary distance unit.
Without prior enrichment, the most numerous AQDS reducers recovered from freshwater sediments are also Geobacter spp.
Enrichment prior to isolation may select for microorganisms which grow rapidly in the medium but are not necessarily the most numerous organisms in the environmental sample that can grow in that medium. Therefore, to determine which organisms that could grow in acetate-AQDS medium were the most numerous in the sediments from the Talladega wetland site, the enrichment step was eliminated. The Talladega site was examined because of the importance of Fe(III) reduction in carbon flow in the surficial sediments (31) and because the humic substance-rich waters meant that there was a strong possibility of humic substance-assisted Fe(III) reduction.
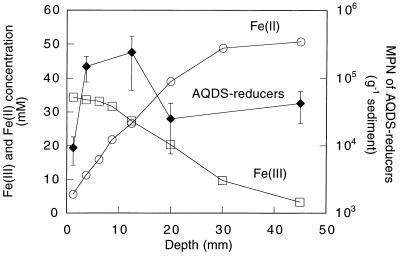
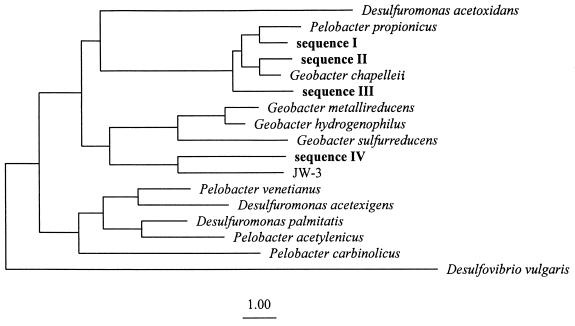
Profiles of Fe(III) and Fe(II) indicated that Fe(III) reduction was an important process at depths between 0 and 5.5 cm (Fig. 4). MPN analysis indicated that the number of acetate-oxidizing AQDS-reducing microorganisms within the Fe(III) reduction zone was 104 to 105 (Fig. 4). PCR-DGGE analysis of 16S rDNA fragments of the highest positive MPN dilutions recovered one to three DGGE bands per depth interval. Phylogenetic analysis indicated that 13 of the 14 16S rDNA fragments resolved by DGGE had sequences that were closely related to those of previously described humic-reducing Geobacter species. These 13 Geobacter bands were composed of four distinct sequences. Three of these were most closely related to G. chapelleii (Fig. 5). They included sequence I, which was recovered from depths of 5 to 7.5, 15 to 25, and 35 to 55 mm (97.6% similarity to G. chapelleii, 333 base positions considered); sequence II, which was recovered from the 10- to 15-mm depth (97.1% similarity to G. chapelleii 306 base positions considered); and sequence III, which was recovered from the 0- to 2.5-mm depth interval (97.0% similarity to G. chapelleii, 334 base positions considered). Sequence IV was most closely related to strain JW-3 (92.9% similarity, 325 base positions considered).
FIG. 4.
Fe(II) and Fe(III) concentration profile and MPN counts of humic reducers with depth at the Talladega Wetland.
FIG. 5.
Phylogenetic placement of Geobacteraceae partial 16S rDNA sequences obtained from acetate-oxidizing, humic-reducing enrichments. A total of 214 positions were considered. The sequence of D. desulfuricans was included as an outgroup. Bar, one evolutionary distance.
The only sequence out of 14 that did not fall within the Geobacteraceae was recovered from the 10- to 15-mm depth interval and was most closely related to Zoogloea ramigera ATCC 25935 (95% similarity, 320 base positions considered) in the beta subclass of the Proteobacteria (32). The DGGE band for this sequence was relatively faint but clearly present. The recovery of an organism closely related to Zoogloea is surprising because Zoogloea species are considered to be obligate aerobes (10) and thus would not be considered capable of anaerobic growth with AQDS as the electron acceptor.
Because of the recovery of a sequence closely related to Zoogloea in the 10- to 15-mm depth interval, it was of interest to determine whether any acetate-oxidizing, AQDS-reducing microorganisms that were not Geobacter species might be isolated from this depth interval. Two isolates obtained from this depth via shake tubes were characterized. These isolates, designated Ala-5 and Ala-6, grew with acetate as the electron donor and AQDS as the electron acceptor. However, 16S rDNA analysis indicated that as with the isolates from other sites, these organisms were members of the genus Geobacter (Fig. 3). Strains Ala-5 and Ala-6 had virtually identical 16S rDNA sequences over unambiguously determined positions (99.9% sequence identity, 1,155 base positions considered) and were most closely related to strain JW-3 (98.1% identity to Ala-5, 1,143 base positions considered). As with strain JW-3, the sequence of strain Ala-5 contained nucleotides (positions 122, 239, 286, 453, 454, 681, 690, 822, 859, 878, 888, 1117, 1168, and 1254; E. coli numbering) characteristic of the Geobacter cluster. These results demonstrate that Geobacter species were the most numerous microorganisms capable of AQDS reduction that could be recovered from the Talladega wetland sediments. However, the results cannot rule out the possibility that the sediments may contain other, as yet undescribed organisms which also oxidize acetate with the reduction of AQDS.
Desulfuromonas spp. are recovered from marine sediments with AQDS.
To determine whether AQDS-reducing microorganisms could also be recovered from marine environments, marine medium with acetate as the electron donor and AQDS as the electron acceptor was inoculated with sediments from San Diego Bay or the Norfolk Naval Base. Both sites yielded positive enrichments. Two strains from San Diego Bay, SDB-1 and SDB-2, and three strains from Norfolk, FD-1, CD-1, and VES-1, were studied. They all were morphologically identical, strict anaerobic, nonfermentative, nonflagellated, non-spore-forming, nonmotile, gram-negative rods 1 to 2 μm by 0.5 μm. Similar to the freshwater AQDS-reducing strains, colonies on solid AQDS medium were less than 1 mm in diameter and were deep red with an orange halo. All of the marine isolates grew by the oxidation of acetate with AQDS serving as the sole electron acceptor under strictly anaerobic conditions. Evaluation of a variety of potential electron donors other than acetate indicated that all of the marine strains tested could also use ethanol and succinate (Table 1), although AQDS reduction with succinate was much slower than with acetate. In addition to AQDS, strains SDB-2 and CD-1 could couple the oxidation of acetate to the reduction of Fe(III) citrate, elemental sulfur, Mn(IV), fumarate, or malate (Table 2).
TABLE 1.
Compounds used by marine AQDS-reducing isolates as electron donors
| Electron donor (mM) | Utilizationa by strain:
|
|||
|---|---|---|---|---|
| SDB-1 | SDB-2 | FD-1 | CD-1 | |
| H2 (101 kPa) | − | − | − | − |
| Formate (10) | − | − | − | − |
| Acetate (10) | + | + | + | + |
| Propionate (5) | + | + | + | − |
| Butyrate (5) | − | ND | − | ND |
| Methanol (10) | − | − | − | − |
| Ethanol (10) | + | + | + | + |
| Phenol (0.5) | − | − | − | − |
| Lactate (10) | − | − | − | + |
| Palmitate (1) | − | − | − | − |
| Benzoate (0.5) | − | − | − | |
| Toluene (1) | − | − | − | − |
| Glucose (10) | − | − | − | − |
| Yeast extract (1 g/liter) | − | ND | − | + |
| Pyruvate (10) | − | ND | − | ND |
| Succinate (10) | + | + | + | + |
| Fumarate (10) | ND | + | ND | + |
+, utilization of electron donor; −, no utilization; ND, not determined.
TABLE 2.
Compounds used by marine AQDS reducers as alternative electron acceptors
| Electron acceptor (mM) | Utilizationa by strain:
|
|
|---|---|---|
| SDB-2 | CD-1 | |
| Fe(III) citrate (50) | + | + |
| Mn(IV) (20) | + | + |
| Nitrate (10) | − | − |
| Elemental sulfur (10) | + | + |
| Sulfite (10) | − | − |
| Sulfate (10) | − | − |
| Fumarate (50) | + | + |
| Malate (10) | + | + |
| Selenate (2) | − | − |
+, utilization of electron acceptor; −, no utilization.
The potential for the marine AQDS reducers to reduce humic substances could not be evaluated as described above for the freshwater strains. The cells lysed when placed in bicarbonate buffer without marine salts, and the humic substance preparation precipitated in marine salts buffer. However, the fact that these organisms can reduce AQDS demonstrates that they can reduce extracellular quinone-containing compounds, and all organisms tested to date that have the ability to reduce extracellular quinones also have the ability to transfer electrons to humic substances (18, 20).
Analysis of the partial 16S rDNA sequences of the marine isolates indicated that they all were members of the Geobacteraceae. Two strains, SDB-1 and FD-1, were selected for in-depth sequencing and characterization. The 16S rDNA sequences of these strains were nearly identical over unambiguously determined positions (99.4% sequence identity, 1,138 base positions considered). Their closest known relative (Fig. 3) is Desulfuromonas acetoxidans (98.7% similarity, 1,260 base positions considered). The 16S rDNA sequence of strain SDB-1 and FD-1 contained nucleotides (positions 122, 200, 217, 239, 286, 453, 454, 681, 690, 822, 859, 878, 888, 1117, 1122, 1151, 1168, 1254, and 1283; E. coli numbering) and a secondary structure (position 1024; E. coli numbering) characteristic of the Desulfuromonas cluster (17). Analysis of the partial (ca. 350 base positions considered) 16S rDNA sequence of the other marine strains indicated that strains VES-1, CD-1, and SDB-2 were 95.3, 99.3, and 99.1% similar to SDB-1, respectively.
Implications for reduction of humic substances in sediments.
The results demonstrate that microorganisms with the capacity to reduce humic substances live in a diversity of sedimentary environments. Further evidence for this is the finding that acetate-oxidizing AQDS reducers can also be recovered in large numbers from the Fe(III) reduction zone of a petroleum-contaminated aquifer (2).
To date, all of the acetate-oxidizing AQDS reducers recovered from sediments are members of the family Geobacteraceae. The freshwater isolates are closely related to previously described Geobacter species, whereas the marine isolates are closely related to previously described Desulfuromonas species. The same pattern was previously observed in studies of freshwater and marine Fe(III)-reducing microorganisms, with Geobacter species predominating in freshwater sediments and Desulfuromonas species predominating in marine environments (6, 7). Geothrix fermentens (17) and Geovibrio ferrireducens (3) are the only two mesophilic organisms outside the Geobacteraceae that are known to oxidize acetate with the reduction of Fe(III), and both of these organisms have only rarely been recovered from sediment samples (21).
Studies with culture conditions other than those used here might have yielded humic substance-reducing bacteria other than members of the Geobacteraceae. For example, the incubation temperatures that were used were selected as the optimum growth temperatures for most mesophilic bacteria, but it is possible that other incubation temperatures would have favored the growth of a greater diversity of microbial types. The marine medium was designed to match the major ion chemistry of San Diego Bay pore water (6) in an attempt to isolate microorganisms adapted for growth in these sediments. The freshwater medium has been successfully used in previous studies to recover a diversity of microorganisms from freshwater environments and is known to support the growth of humic- reducing bacteria (20). Acetate was selected as the electron donor because it is probably the most important electron donor driving anaerobic respiratory processes other than denitrification in sedimentary environments (19, 24). Therefore, microorganisms capable of coupling the oxidation of acetate to the reduction of humic substances are likely to be the most important organisms participating in humic substance reduction. Furthermore, many organisms that have the ability to oxidize acetate with the reduction of humic substances also have the ability to use other electron donors such as H2, fatty acids other than acetate, and aromatic compounds (21). Thus, isolation of acetate-oxidizing humic-reducing microorganisms seemed likely to account for organisms using a variety of other electron donors that might contribute to humic substance reduction. However, humic-reducing bacteria such as Shewanella species that are incapable of oxidizing acetate under anaerobic conditions would not be accounted for under the culture conditions used here.
It would be preferable to attempt to identify the important humic-reducing bacteria in sediments without the need to culture these organisms. However, there is no known method for doing so. For example, not enough is known about the biochemical mechanisms for humic substance reduction to develop molecular probes for humic-reducing enzymes or genes. The capacity for humic substance reduction is found in a number divergent phylogenetic groups, and closely related organisms do not always share the capacity for humic substance reduction (18). Therefore, 16S rRNA-based techniques are not likely to be useful in defining the humic-reducing community. Even though the culturing approaches described here are not ideal, they do provide an initial insight into what organisms might be important in humic substance reduction in sedimentary environments.
In summary, the results suggest that in addition to being the most consistently recovered Fe(III)-reducing microorganisms living in a diversity of sediment types (7), members of the family Geobacteraceae may also be important humic reducers in these environments. The finding that the most important humic reducers might also be the most important Fe(III) reducers leads to the question whether Geobacteraceae and other Fe(III) and humic reducers reduce Fe(III) in sediments and soils primarily via a direct enzymatic reduction or indirectly by the enzymatic reduction of humic substances followed by abiotic reduction of the Fe(III) by the humic substances.
ACKNOWLEDGMENTS
We thank Sue Lootens, Michigan State University Sequencing Facility, for technical assistance.
This research was supported in part with grant N0014-96-1-0382 from the Office of Naval Research.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, R. T. 1997. Unpublished data.
- 3.Caccavo F, Coates J D, Rossello-Mora R A, Ludwig W, Schleifer K H, Lovley D R, McInerney M J. Geovibrio ferrireducens, a phylogenetically distinct dissimilatory Fe(III)-reducing bacterium. Arch Microbiol. 1996;165:370–376. doi: 10.1007/s002030050340. [DOI] [PubMed] [Google Scholar]
- 4.Coates, J. D. Unpublished data.
- 5.Coates J D, Anderson R T, Lovley D R. Anaerobic oxidation of polycyclic aromatic hydrocarbons under sulfate-reducing conditions. Appl Environ Microbiol. 1996;62:1099–1101. doi: 10.1128/aem.62.3.1099-1101.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coates J D, Lonergan D J, Lovley D R. Desulfuromonas palmitatis sp. nov., a long-chain fatty acid oxidizing Fe(III) reducer from marine sediments. Arch Microbiol. 1995;164:406–413. [PubMed] [Google Scholar]
- 7.Coates J D, Phillips E J P, Lonergan D J, Jenter H, Lovley D R. Isolation of Geobacter species from a variety of sedimentary environments. Appl Environ Microbiol. 1996;62:1531–1536. doi: 10.1128/aem.62.5.1531-1536.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Soete G. A least squares algorithm for fitting additive trees to proximity data. Psychometrika. 1983;48:621–626. [Google Scholar]
- 9.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. Touchdown PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dugan P R, Stoner D L, Pickrum H M. The genus Zoogloea. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 3952–3964. [Google Scholar]
- 11.Eden P E, Schmidt T M, Blakemore R P, Pace N R. Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. Int J Syst Bacteriol. 1991;41:324–325. doi: 10.1099/00207713-41-2-324. [DOI] [PubMed] [Google Scholar]
- 12.Hobbie J E, Daley R J, Jasper S. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977;33:1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hungate R E. A roll tube method for cultivation of strict anaerobes. Methods Microbiol. 1969;3B:117–132. [Google Scholar]
- 14.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 15.Lane D J, Field K G, Olsen G J, Pace N R. Reverse transcriptase sequencing of ribosomal RNA for phylogenetic analysis. Methods Enzymol. 1988;167:138–144. doi: 10.1016/0076-6879(88)67015-7. [DOI] [PubMed] [Google Scholar]
- 16.Lane D J, Pace B, Olsen G J, Stahl D, Sogin M L, Pace N R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analysis. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lonergan D J, Jenter H, Coates J D, Schmidt T, Lovley D R. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J Bacteriol. 1996;178:2402–2408. doi: 10.1128/jb.178.8.2402-2408.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lovley, D. R. Unpublished data.
- 19.Lovley D R, Chapelle F H. Deep subsurface microbial processes. Rev Geophys. 1995;33:365–381. [Google Scholar]
- 20.Lovley D R, Coates J D, Blunt-Harris E L, Phillips E J P, Woodward J C. Humic substances as electron acceptors for microbial respiration. Nature. 1996;382:445–448. [Google Scholar]
- 21.Lovley D R, Coates J D, Saffarini D, Lonergan D J. Diversity of dissimilatory Fe(III)-reducing bacteria. In: Winkelman G, Carrano C J, editors. Iron and related transition metals in microbial metabolism. Amsterdam, The Netherlands: Harwood Academic Publishers; 1997. pp. 187–215. [Google Scholar]
- 22.Lovley D R, Phillips E J. Organic matter mineralization with reduction of ferric iron in anaerobic sediments. Appl Environ Microbiol. 1986;51:683–689. doi: 10.1128/aem.51.4.683-689.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lovley D R, Phillips E J P. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl Environ Microbiol. 1988;54:1472–1480. doi: 10.1128/aem.54.6.1472-1480.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lovley D R, Phillips E J P. Requirement for a microbial consortium to completely oxidize glucose in Fe(III)-reducing sediments. Appl Environ Microbiol. 1989;55:3234–3236. doi: 10.1128/aem.55.12.3234-3236.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lovley D R, Woodward J C, Chapelle F H. Stimulated anoxic biodegradation of aromatic hydrocarbons using Fe(III) ligands. Nature. 1994;370:128–131. doi: 10.1038/370128a0. [DOI] [PubMed] [Google Scholar]
- 26.Lovley D R, Woodward J C, Chapelle F H. Rapid anaerobic benzene degradation with a variety of chelated Fe(III) forms. Appl Environ Microbiol. 1996;62:288–291. doi: 10.1128/aem.62.1.288-291.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maidak B, Larsen N, McCaughey M, Overbeek R, Olsen G, Fogel K, Blandy J, Woese C. The ribosomal database project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller T L, Wolin M J. A serum bottle modification of the Hungate technique for cultivating obligate anaerobes. Appl Microbiol. 1974;27:985–987. doi: 10.1128/am.27.5.985-987.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myers R M, Fischer S G, Lerman L S, Maniatis T. Nearly all single base substitutions in DNA fragments joined to a GC-clamp can be detected by denaturing gradient gel electrophoresis. Nucleic Acids Res. 1985;13:3131–3145. doi: 10.1093/nar/13.9.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roden E E, Wetzel R G. Organic carbon oxidation and suppression of methane production by microbial Fe(III) oxide reduction in vegetated and unvegetated freshwater wetland sediments. Limnol Oceanogr. 1996;41:1733–1748. [Google Scholar]
- 32.Rosello-Mora R A, Ludwig W, Schleifer K H. Zoogloea ramigera: a phylogenetically diverse species. FEMS Microbiol Lett. 1993;114:129–134. [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Widdel F, Pfennig N. The genus Desulfuromonas and other gram-negative sulfur-reducing eubacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 3379–3392. [Google Scholar]