Web-based predisclosure education for genetic research results is a viable alternative
Abstract
PURPOSE
We developed a web-based education intervention as an alternative to predisclosure education with a genetic counselor (GC) to reduce participant burden and provider costs with return of genetic research results.
METHODS
Women at three sites who participated in 11 gene discovery research studies were contacted to consider receiving cancer genetic research results. Participants could complete predisclosure education through web education or with a GC. Outcomes included uptake of research results, factors associated with uptake, and patient-reported outcomes.
RESULTS
Of 819 participants, 178 actively (21.7%) and 167 passively (20.4%) declined return of results; 474 (57.9%) were enrolled. Most (60.3%) received results although this was lower than the 70% uptake we hypothesized. Passive and active decliners were more likely to be Black, to have less education, and to have not received phone follow-up after the invitation letter. Most participants selected web education (88.5%) as an alternative to speaking with a GC, but some did not complete or receive results. Knowledge increased significantly from baseline to other time points with no significant differences between those who received web versus GC education. There were no significant increases in distress between web and GC education.
CONCLUSION
Interest in web-based predisclosure education for return of genetic research results was high although it did not increase uptake of results. We found no negative patient-reported outcomes with web education, suggesting that it is a viable alternative delivery model for reducing burdens and costs of returning genetic research results. Attention to attrition and lower uptake of results among Black participants and those with less formal education are important areas for future research.
INTRODUCTION
Genetic sequencing studies involving biobanked DNA raise questions about the obligation to share individual research results with participants. 1-7 Arguments supporting return appeal to the principles of beneficence, autonomy, reciprocity, respect for persons, and clinical utility. 4,7-9 Arguments opposing return raise concerns about the distinction between research and clinical care, the actionability of results, the right not to know, and costs. 5,7,9-13 Although debates are ongoing, the consensus favors returning results that could be relevant to participants' health. 2,4,5,14-16
CONTEXT
Key Objective
Is web-based education a viable alternative delivery model for predisclosure education for return of individual genetic research results?
Knowledge Generated
Uptake of web-based predisclosure education for return of individual genetic research results was high among enrolled participants. Among those who completed predisclosure education, most received their research results, which did not differ by web-based education versus education with a genetic counselor. No negative patient-reported outcomes with web education were found.
Relevance (S.B. Wheeler)
As return of genetic test results to patients continues to increase with clinically relevant genetic information becoming more widely available, appropriate and timely education to support interpretation is key. This study offers a potentially impactful and efficient approach to providing that education via web-based platforms.*
*Relevance section written by JCO Associate Editor Stephanie B. Wheeler, PhD, MPH.
Research participants have reported high interest in receiving research results. 16-25 However, lower uptake has been reported, 26 particularly in studies involving biobanks for which return of results was not emphasized during enrollment. 27-34 There are limited patient-reported outcomes (PROs) in the research setting 27-29,35 although some genomic implementation studies have reported no psychological harms with return of genetic findings. 36,37 In RESPECT1, we found favorable cognitive (eg, knowledge) and affective (eg, distress and uncertainty) responses with return of results among patients with breast cancer, but low uptake of results. 38 In addition, we found that one third of participants reported that a self-directed web platform would be an acceptable alternative to speaking with a genetic counselor (GC) and could reduce steps in receiving results. 39
In this multicenter, observational return of results study (RESPECT2), we developed a web-based predisclosure education intervention (web education) as an alternative to speaking with a GC to reduce participant burdens and steps in receiving genetic research results. Our primary aim was to evaluate uptake of genetic research results among research participants who provided a biospecimen for genetic research when using an alternative delivery model incorporating web education. We hypothesized that 70% would receive their results after predisclosure education. Secondary aims included understanding participant factors associated with uptake of results, changes in PROs, and whether outcomes differed by the method of predisclosure education (GC v web education).
METHODS
Participants were English- or Spanish-speaking adult women who had provided a biospecimen for genetic research (11 studies) at the University of Pennsylvania (UPenn), University of Chicago (UChicago), or Columbia University (Columbia) and had not previously had clinical multigene panel testing. Original consents stated that results that could affect participant health would be returned (n = 7) or would not be returned (n = 4). Individuals in the latter group were contacted so as not to assume that they would not want results.
Sequencing included 25 high- and moderate-penetrance genes (APC, BRCA1, BRCA2, CDH1, CDKN2A, PMS2, PTEN, MLH1, MSH2, MUTYH-homozygous, MSH6, STK11, TP53, ATM, BAP1, BARD1, BMPR1A, BRIP1, CHEK2, MRE11A, MUTYH-heterozygous, NBN, PALB2, RAD50, RAD51C, RAD51D) that had potential clinical relevance when the study was conducted. Sequencing was performed in institutional research laboratories. Clinical confirmation testing was recommended to participants for results that could potentially affect medical care (see below).
Institutional Review Board approval was obtained at all sites. Study invitation letters (English and Spanish versions at Columbia) explained that research testing had been completed and that participants could enroll to learn their research results. In eight studies (n = 1,583), the research team could follow up with participants by phone (three to five calls per site standards). In three studies (n = 379), original consents stated that results would not be returned and participants had to call or mail back a response card to be contacted.
Predisclosure Education by Web Intervention or Genetic Provider
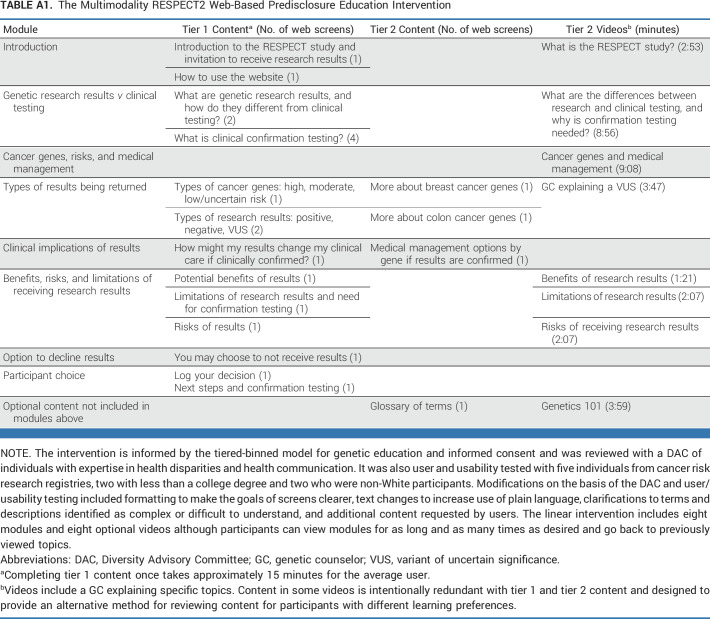
On the basis of participant feedback in RESPECT1, we developed a self-directed web-based alternative for predisclosure education. 39 The intervention was developed to cover the same content as predisclosure genetic counseling. Informed by the tiered-binned model, GCs reached consensus on indispensable tier 1 information that should be presented to all participants and additional or optional tier 2 information that could be provided to support variable information needs. 40 The intervention and genetic counseling checklists included the same tier 1 content. The intervention consisted of seven modules and optional videos and was available in both English and Spanish (Appendix Table A1, online only).
Predisclosure, participants were offered access to web education or a GC session (conducted by seven GCs via phone or in person). 38,40 GCs were blinded to participants' research results at predisclosure and completed counseling checklists to ensure fidelity to tier 1 content. 41,42
Disclosure of Genetic Research Results
Participants received results by phone or in person with a GC. The result disclosure visit was separate from the education visit. Fidelity was assessed in 20% of counseling sessions, with a mean fidelity to checklists of 89% (predisclosure sessions) and 88% (disclosure sessions). On the basis of institutional policies, participants at Columbia with a positive result were told that there was a genetic finding but not the specific gene. Participants with a variant of uncertain significance (VUS) or negative result were told that there were no findings that could affect their health. UPenn and UChicago allowed participants with positive or VUS results to learn the specific gene and informed them that they should not change medical care until after confirmation testing.
Confirmation Testing in a Clinical Laboratory Improvement Amendments-Certified Laboratory and Clinical Follow-Up
Participants with a pathogenic or likely pathogenic research result in any gene were recommended to have clinical confirmation testing. We also recommended confirmation for VUS results in high-penetrance genes since a reclassification to pathogenic or likely pathogenic could affect medical care. This approach provided confirmation of the result along with an additional opinion regarding variant calling as clinical laboratories have access to additional data. It also allowed clinical laboratories to be responsible for updates to VUS results. The need for confirmation testing and the importance of not altering medical management until after confirmation was shared in predisclosure education and at disclosure. For discordant results, recommendations for medical management were based on the participant's clinical result and personal and family history, consistent with clinical care. GCs and research staff provided reminders and support to complete confirmation testing. Research funds covered all costs of confirmation testing at UChicago and costs not covered by insurance at UPenn and Columbia. Confirmation testing was completed by mailed saliva kits or phlebotomy during a clinical visit. Confirmation testing results were shared by phone or in person. All participants were recommended to return for follow-up care.
PROs
As previously described, the selection of relevant outcomes after the receipt of genetic research results was informed by our conceptual model, 38,40,41 which is grounded in the Self-Regulation Theory of Health Behavior. 43,44 Our model proposes that uptake of genetic research results and response to (eg, psychosocial adjustment) and use of (eg, performance of health behaviors) genetic information are products of an individual's understanding (eg, knowledge of genetic disease) and perception of disease threat (eg, cancer risk). 41,43 Participants completed surveys at baseline (T0), after predisclosure education (T1), after disclosure counseling (T2), and at 6 months (T3).
Knowledge of genetic disease was evaluated (T0-T3) using an adapted version of the Cancer Genetics Knowledge Scale and ClinSeq knowledge scale 45-47 and included knowledge of inheritance and test interpretation (nine items), benefits (three items) and limitations (six items) of multigene testing, and differences between research and clinical testing (five items; Cronbach's α = .79-.83). 38
Perceived risk of cancer (T0) was measured on a Likert scale in relation to the average woman (much higher, higher, same, lower, much lower) and, in a second item, as a numerical lifetime risk (0%-100%) of getting breast cancer (or breast cancer again).
Psychosocial adjustment included the following: (1) state anxiety (T0-T2), measured with the 20-item State Inventory 48,49 (Cronbach's α = .94-.95); (2) general anxiety and depression (T0-T3), assessed with the Hospital Anxiety and Depression Scale 50 (Cronbach's α = .83-.85 and .81-.83); and (3) cancer-specific distress (T0-T3), evaluated with the 15-item Impact of Events Scale 51 (Cronbach's α = .86-.89).
Satisfaction with genetic services was measured (T2) with a 13-item scale used in related research 38,42,52,53 (Cronbach's α = .81-.83).
Uncertainty was assessed (T0-T3) using a three-item scale adapted from the Multi-Dimensional Impact of Cancer Risk Assessment 54 (Cronbach's α = .84).
Perceived utility was assessed (T0-T3) with a novel scale developed to evaluate patient perceptions of the utility of genetic results, including two 12-item subscales evaluating medical and personal utility, both now and in the future (Cronbach's α = .96-.97). 38,55
Statistical Analyses
We characterized the samples using means, standard deviations, and proportions. We used logistic regressions to examine characteristics associated with responding to enrollment status groups and completion of the intervention. For psychosocial outcomes, we examined linear regressions of change scores between baseline and follow-up times (ie, follow-up minus baseline scores). In regressions, we controlled for variables that were found or hypothesized to be associated with longitudinal follow-up. These included site, Hispanic ethnicity, education, number of children, age, and number of relatives with previously diagnosed cancer. We used multiple imputation methods with 100 multiply imputed data sets to account for missing data in the regression analyses. 56 Analyses were conducted in STATA (StataCorp, College Station, TX) and SAS (SAS Institute, Cary, NC).
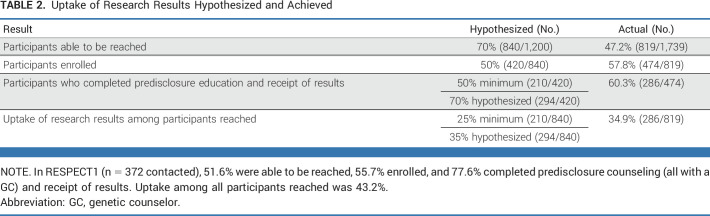
We hypothesized that among 1,200 participants, 70% could be reached, 50% would enroll, and 70% would receive results. With 50% enrollment, we would be able to estimate the true uptake with at least a precision of approximately 4.9%. For secondary analyses, we aimed for a sample size of 420 to have 85% power to detect a standardized odds ratio of 1.5 in logistic regressions. We set the type I error rate and P value to 1% (two-sided) to partially account for multiple hypothesis testing. Additional power calculations are provided in our Protocol (online only).
RESULTS
Participants
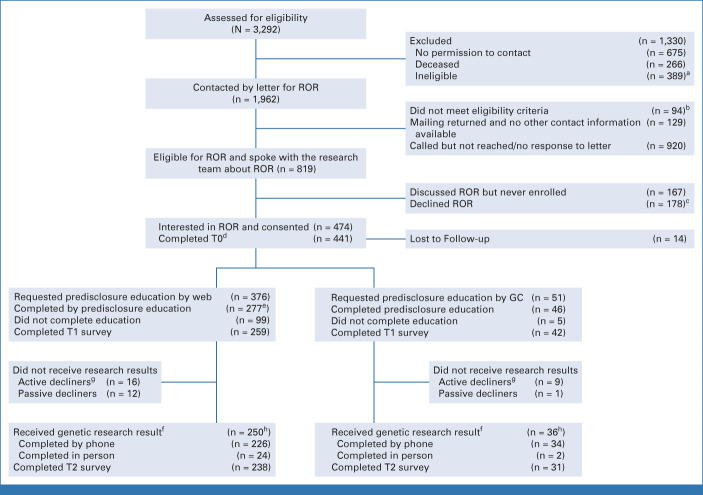
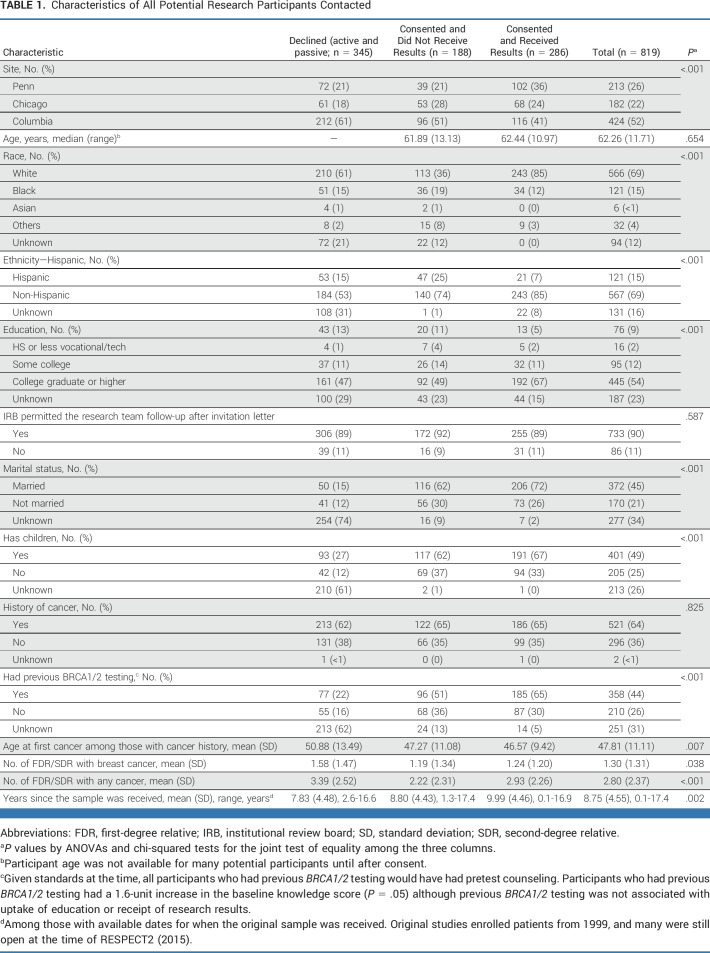
Across three sites, 1,962 potential participants were contacted (April 2015-October 2019; Fig 1). Some letters were returned (6.6%), and 94 participants were found to be deceased or ineligible, leaving 1,739 eligible mailings. We were not able to communicate with 920 research participants to discuss the option to receive research results (ie, nonresponders; 52.9%). Of 819 participants who could be reached, 178 (21.7%) actively declined receipt, 167 (20.4%) passively declined (ie, expressed interest but were lost to follow-up), and 474 (57.8%) enrolled (Table 1).
FIG 1.
CONSORT diagram. aTwelve patients already had clinical MGPT, 155 had missing contact information, 103 research samples were not available, 38 were already known to be clinically BRCA+, and 81 were living abroad. bThirty-four patients were found to be deceased, 47 already had clinical MGPT, two were previously disclosed, three had language barriers, two did not have decision making capacity, and six did not have valid consent. cReasons for declining included privacy concerns, concerns about time burdens, not being interested in genetic information, only wanting actionable results, preferring clinical testing, and concerns about uncertainty or distress. dThe time from baseline survey to web education link being sent to participants was a median of 1.0 days and a mean of 4.98 days (SD, 16.06). eTwenty-one were completed with a GC. fFourteen pathogenic or likely pathogenic variants (positive result) were returned, including five high-penetrance genes (BRCA1 [2], BRCA2 [2], MSH6) and nine moderate-penetrance genes (ATM [2], BARD1, CHEK2 [4], NBN, PALB2, RAD51D). There were 23 results with at least one VUS (14 variants in moderate-penetrance genes and nine in high-penetrance genes where confirmation testing was recommended). There were two additional VUS results as a second finding. There were 250 with no findings. One individual received results without completing visit 1. gSixteen withdrew from the study. hTwo hundred twenty-nine of 256 who ultimately completed by web received results, and 56 of 67 who ultimately completed by GC received results. MGPT, multigene panel testing; ROR, return of results; T0, baseline; T1, after predisclosure counseling; T2, after disclosure counseling; VUS, variant of uncertain significance.
TABLE 1.
Characteristics of All Potential Research Participants Contacted
Uptake of Web-Based Predisclosure Education and Genetic Research Results
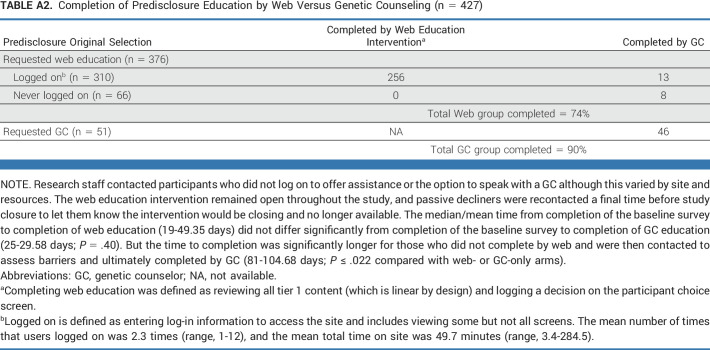
Most participants selected web education (88.5%) as an alternative to speaking with a GC for predisclosure education (Fig 1); 310 logged on, and 82.5% completed web education (Appendix Table A2). Research staff contacted participants who did not log on or complete to offer assistance or the option to speak with a GC. Among participants who selected to speak with a GC, 46 (90.2%) completed.
Among participants who consented to RESPECT2 to learn about receiving research results, 286 (60.3%) received results (Table 2). Most participants (88.2%) who completed predisclosure education received results, which did not differ by method (83.6% with a GC v 89.5% by web education; P = .2). Fourteen (4.9%) participants received a positive result, 23 received a VUS (8.0%), and 250 (87.4%) had no findings (Fig 1).
TABLE 2.
Uptake of Research Results Hypothesized and Achieved
Factors Associated With Uptake of Research Results
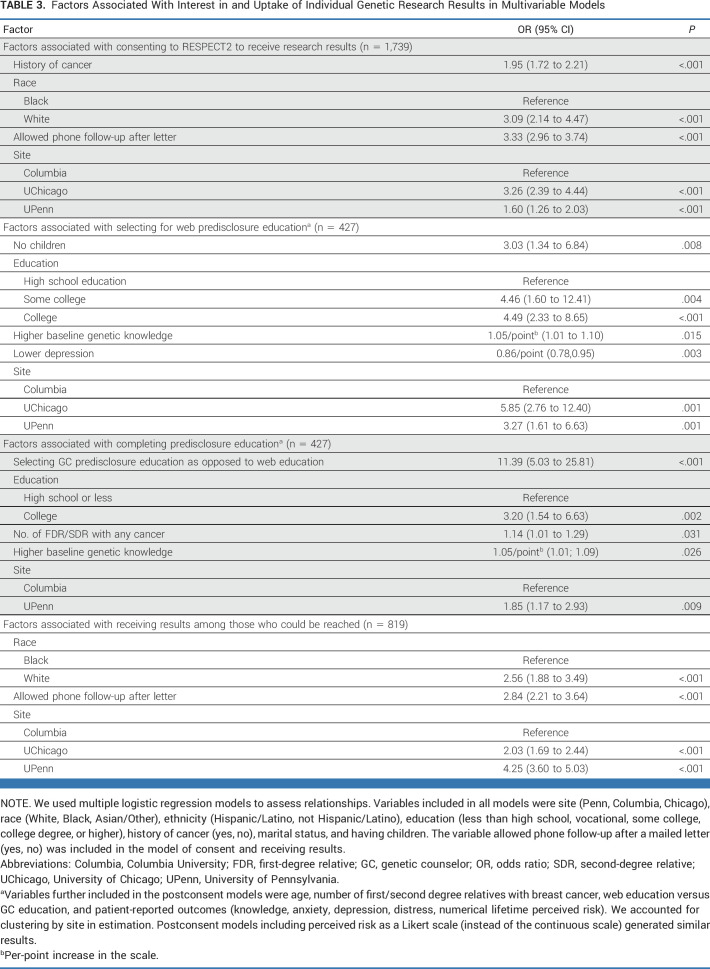
Research participants who could not be reached were more likely to be non-White, have lower education, have no history of cancer, have not allowed phone follow-up, and be at Columbia or UPenn. Research participants who actively or passively declined results were more likely to be Black, have lower education, not allow phone follow-up, and be at Columbia or UChicago (Appendix Table A3).
Enrolled participants who selected web education were more likely to have college education, be at UChicago or UPenn, not have children, have higher baseline knowledge, and have lower depression (Table 3). Completing predisclosure education was associated with selecting a GC, having college education, having higher baseline knowledge, having more relatives with cancer, and being at UPenn (Table 3). Completing predisclosure web education was associated with having higher education, having more relatives with cancer, and being at UPenn. Overall, uptake of research results among those who could be reached was associated with being White, allowing phone follow-up after letter notification, and being at UPenn or Columbia (Table 3).
TABLE 3.
Factors Associated With Interest in and Uptake of Individual Genetic Research Results in Multivariable Models
PROs
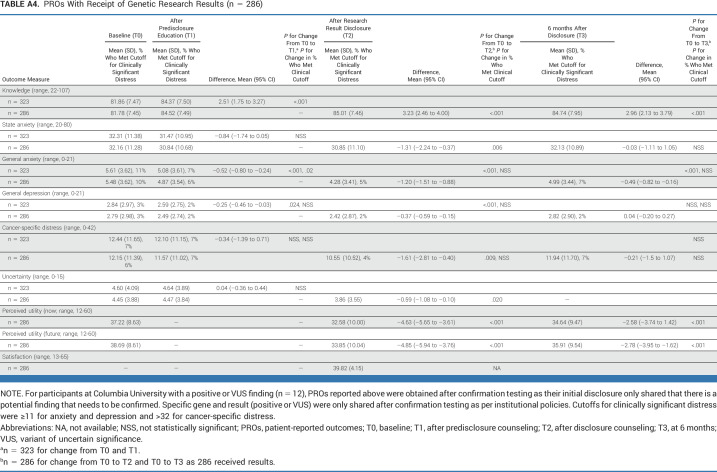
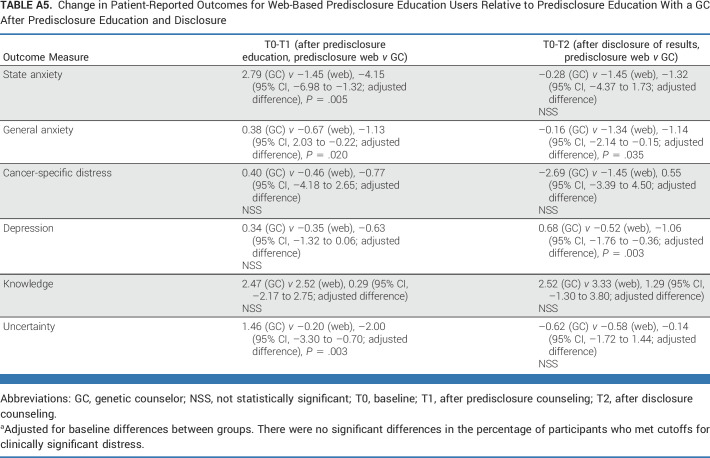
Knowledge increased significantly from baseline to all other time points (Appendix Table A4) and did not differ significantly by web versus GC education (Fig 2A). Similarly, there were no significant increases in distress for those who completed education by web versus GC. There was significantly greater reduction in short-term anxiety and uncertainty (T0-T1) and anxiety and depression (T0-T2) among those who completed by web versus GC although there were no significant differences in the long term (Figs 2A and 2B; Appendix Table A5).
FIG 2.
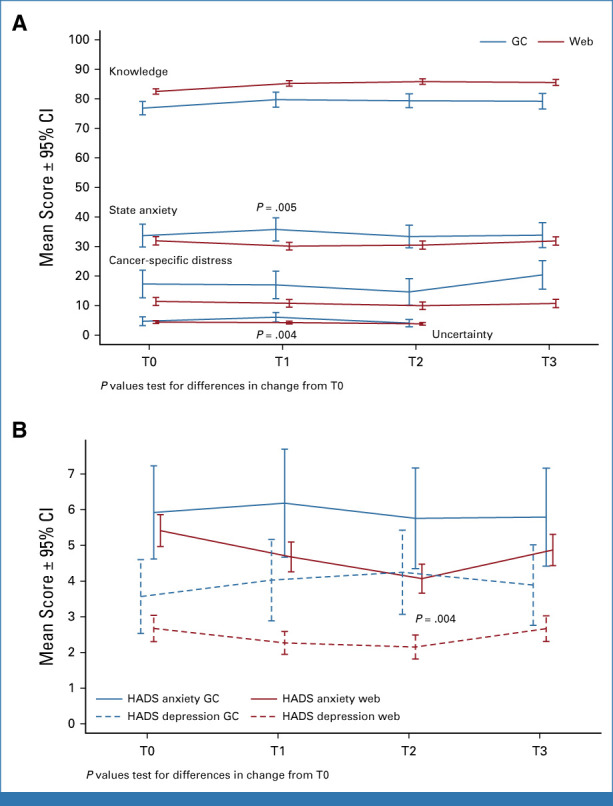
Change in patient-reported outcomes for web-based predisclosure education users relative to predisclosure education with a genetic counselor after predisclosure education and disclosure of results. The x-axis labels denote study wave, with T0 as baseline time point. The y-scale represents the absolute means of the scores. The range differs for each measure, which allowed us to present the results in the same figure to reduce space. The range and means at each time period are reported in Appendix Table A2. There were no significant differences in the percentage of participants who met cutoffs for clinically significant distress between groups. GC, genetic counselor; T0, baseline; T1, after predisclosure counseling; T2, after disclosure counseling; T3, at 6 months.
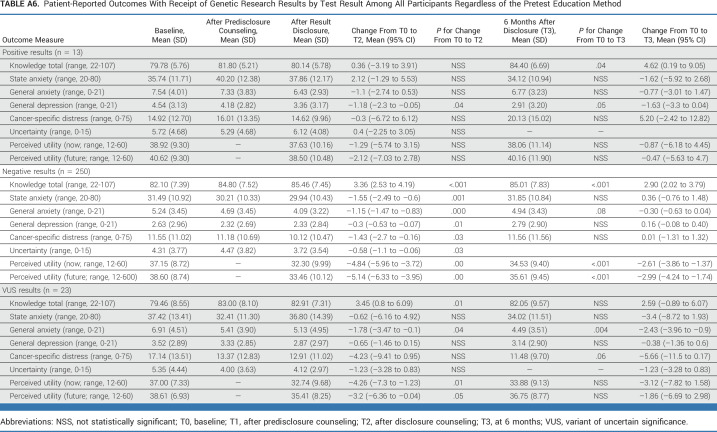
Among participants with a positive result, knowledge increased and depression declined over time (Appendix Table A6). Among those with a VUS or negative result, there was a significant increase in knowledge after the receipt of results. There was also a significant reduction in general anxiety, perceived utility of results (VUS and negative), state anxiety, cancer-specific distress, and depression (negative only) after the receipt of results.
Clinical Confirmation Testing
Most participants (82.6%) underwent recommended clinical confirmation testing (12 of 14 positive results, seven of nine VUS results in high-penetrance genes). Five of 14 (35.7%) participants with a VUS in a low- to moderate-penetrance genes had confirmation testing (testing optional). Among 17 participants for whom testing was submitted to insurance, three (18%) were not covered or had out-of-pocket costs and were covered by research funds.
Five of 24 samples (20.8%) sent for confirmation testing were discordant with research testing. These discordant results included a BRCA2 variant, which was classified as pathogenic by the research laboratory and as a VUS by the clinical laboratory (although this variant has been classified as pathogenic in ClinVar), and a common CHEK2 variant, which was not detected by the clinical laboratory because of sample mix-up, or analytic error. Others included MSH6, MUTYH, and PALB2 VUSs, which were not reported by the clinical laboratory. For these, we do not know if discordance was due to a difference in interpretation, sample mix-up or analytic error.
DISCUSSION
To our knowledge, this is the first return of genetic research results study reporting uptake and PROs with a web-based alternative to predisclosure genetic counseling. Although we met our minimum hypothesized uptake of results, as in our previous return of results study, many participants could not be reached. In addition, 88% of participants chose web education although some did not complete it or receive their results. Although uptake of results was not higher among those who requested web education, there was no evidence of misunderstanding or greater distress among those who chose web education as compared with a GC. This suggests that web education is a viable delivery alternative as long as completion rates can be addressed.
Interest in web education was higher than expected on the basis of stakeholder interviews, 39 but was still consistent with other studies of patients with cancer undergoing genetic testing. 57-60 Although most who selected web education completed the intervention, 26% did not and 18% never logged on. This finding is consistent with our stakeholder interviews in which some participants reported that they might be more likely to not complete a web alternative because it is not a scheduled appointment and therefore easier to forget to complete. 39 Participants who never logged on were more likely to have lower education and lower genetic knowledge, raising the concern that eHealth alternatives could increase health disparities. It is possible that staff resources for reminding participants to log on varied and that automated reminders, follow-up, or additional assistance (eg, digital navigators) could be beneficial. In addition, our data suggest that retaining the option to speak with a GC is important for some participants.
Although we expected that web education might decrease barriers and increase uptake of research results, receipt of results among eligible contacted participants was no higher than that in RESPECT1. 38 Other studies have similarly reported difficulty in recontacting research participants, 38,61 with passive or active decline of research results as high as 40%. 27,32-34,38,62 Site differences in reaching research participants and in uptake of research results suggest that the level of engagement with the research cohort may also be important. Lower uptake among non-White participants and those with lower education suggests that disparities continue to exist with respect to interest in receiving genetic information 32,38 and that providing the opportunity to decline genetic research results remains important.
Among those who completed predisclosure education, the majority (88%) chose to receive their research results, which did not differ by web versus GC education. Knowledge did not significantly differ by education method, and there were no increases in negative affective outcomes for web education. In addition, how participants used the intervention (eg, number of times accessed, content accessed) could be informative. More extensive secondary analyses addressing these questions are ongoing.
An additional challenge for research programs that are not sequencing in Clinical Laboratory Improvement Amendments-approved laboratories is the need to confirm research results. Four participants did not complete clinical confirmation testing even with significant support and coverage of costs, highlighting the importance of understanding barriers to confirmation testing. 27,28,38 The need for clinical confirmation was further underscored by the fact that we had at least one pathogenic result that was not confirmed and several results with discordant interpretations. Discordance in our study is likely partially related to the limited data and standards for variant calling at the time the study was conducted. Nevertheless, discordance is not uncommon, even in clinical testing, and provides additional rationale for confirmation testing, including for VUSs. 63
We acknowledge several limitations. This was an observational study, and many research participants could not be reached, creating a potential nonresponse bias. Reaching participants when research results are not immediately available remains a real-world challenge, making understanding barriers to recontact especially important. Because the study was not randomly assigned, there may be unmeasured differences between groups on the basis of participant self-selection. In addition, the study only included women and was focused on those with a personal or family history of breast cancer. As we had few individuals with positive or VUS results, it is important to confirm findings for these subgroups. Although we used standardized counseling checklists, other differences in counseling could affect outcomes. Differences in levels of engagement with the research cohort and site policies regarding the number of follow-up calls may also be relevant. Finally, outcomes for web-based education could be affected by individual use of the intervention.
In conclusion, we found high interest in a web-based alternative for predisclosure education for return of genetic research results although some patients did not complete web education or receive results. Notably, while uptake of results was not higher among those who requested web education, there was no evidence of misunderstanding or greater distress among those who chose web education. Attention to attrition and lower uptake of results among Black participants and those with less formal education are important areas for future research.
ACKNOWLEDGMENT
The authors would like to thank Linda Patrick-Miller, PhD, for her contributions to the study concept and design as well as the implementation of this research.
APPENDIX 1
TABLE A1.
The Multimodality RESPECT2 Web-Based Predisclosure Education Intervention
TABLE A2.
Completion of Predisclosure Education by Web Versus Genetic Counseling (n = 427)
TABLE A3.
Factors Associated With Nonresponse and Declining Research Results
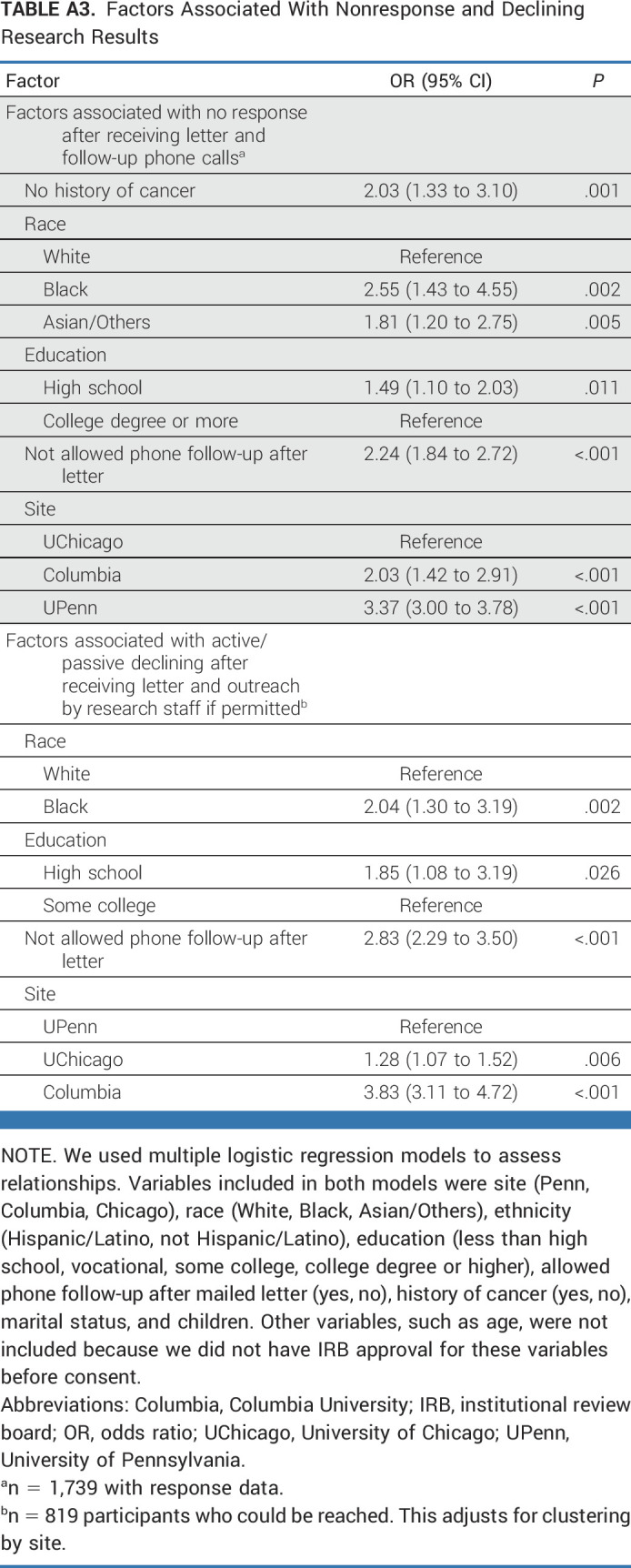
TABLE A4.
PROs With Receipt of Genetic Research Results (n = 286)
TABLE A5.
Change in Patient-Reported Outcomes for Web-Based Predisclosure Education Users Relative to Predisclosure Education With a GC After Predisclosure Education and Disclosure
TABLE A6.
Patient-Reported Outcomes With Receipt of Genetic Research Results by Test Result Among All Participants Regardless of the Pretest Education Method
Brian L. Egleston
Honoraria: Research Square
Consulting or Advisory Role: Digidence (Inst)
Olufunmilayo Olopade
Employment: CancerIQ
Leadership: CancerIQ
Stock and Other Ownership Interests: CancerIQ, Tempus, 54gene, HealthWell Solutions
Consulting or Advisory Role: Tempus
Research Funding: Roche/Genentech (Inst), Color Genomics (Inst)
Uncompensated Relationships: ACS, Susan G. Komen for the Cure
Open Payments Link: https://openpaymentsdata.cms.gov/physician/1178557
Payal Shah
Stock and Other Ownership Interests: Johnson & Johnson, Novartis, Novo Nordisk, Pfizer, Merck, Amgen
Consulting or Advisory Role: Gilead Sciences, Daiichi Sankyo
Research Funding: AstraZeneca (Inst), Zenith Epigenetics (Inst)
Jane E. Churpek
Honoraria: UpToDate
Patents, Royalties, Other Intellectual Property: My husband has a patent pending (ARCD. P0535US.P2) for risk stratification algorithms for hospitalized patients
Other Relationship: RUNX1 Research Program, MDS Foundation, National Comprehensive Cancer Network
Linda Fleisher
Honoraria: Academy of Oncology Nurse and Patient Navigators
Research Funding: Genentech
Dana Clark
Honoraria: AstraZeneca
Speakers' Bureau: AstraZeneca
Julia Wynn
Employment: BillionToOne
Stock and Other Ownership Interests: BillionToOne
Jessica M. Long
Employment: DePuy Companies
Stock and Other Ownership Interests: DePuy Companies
Jacquelyn Powers
Employment: Carevive Systems
Honoraria: CureConnect, Myriad Genetics
Consulting or Advisory Role: Carevive Systems
Travel, Accommodations, Expenses: Hospital of the University of Pennsylvania
Sarah Nielsen
Employment: Invitae
Stock and Other Ownership Interests: Invitae
Travel, Accommodations, Expenses: Invitae
Susan M. Domchek
Honoraria: AstraZeneca, GlaxoSmithKline
Research Funding: AstraZeneca (Inst), Clovis Oncology (Inst)
Open Payments Link: https://openpaymentsdata.cms.gov/physician/917904
Katherine L. Nathanson
Consulting or Advisory Role: Merck
Angela R. Bradbury
Consulting or Advisory Role: AstraZeneca, Merck
Research Funding: AstraZeneca/Merck (Inst)
No other potential conflicts of interest were reported.
SUPPORT
AUTHOR CONTRIBUTIONS
Conception and design: Brian L. Egleston, Linda Fleisher, Jill Bennett Gaieski, Susan M. Domchek, Katherine L. Nathanson, Angela R. Bradbury
Financial support: Katherine L. Nathanson
Administrative support: Wendy K. Chung, Aileen Espinal, Monica Palese
Provision of study materials or patients: Wendy K. Chung, Olufunmilayo Olopade, Payal Shah, Mary Beth Terry, Katherine L. Nathanson
Collection and assembly of data: Wendy K. Chung, Olufunmilayo Olopade, Kara N. Maxwell, Payal Shah, Jane E. Churpek, Mary Beth Terry, Dominique Fetzer, Jill Bennett Gaieski, Jessica Bulafka, Aileen Espinal, Kelsey Karpink, Sarah Walser, Davone Singleton, Monica Palese, Ilona Siljander, Amanda Brandt, Dana Clark, Carrie Koval, Julia Wynn, Jessica M. Long, Danielle McKenna, Jacquelyn Powers, Sarah Nielsen, Susan M. Domchek, Katherine L. Nathanson, Angela R. Bradbury
Data analysis and interpretation: Madison Kilbride, Brian L. Egleston, Wendy K. Chung, Kara N. Maxwell, Linda Fleisher, Mary Beth Terry, Dominique Fetzer, Jill Bennett Gaieski, Kelsey Karpink, Davone Singleton, Susan M. Domchek, Katherine L. Nathanson, Angela R. Bradbury
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Uptake of Genetic Research Results and Patient-Reported Outcomes With Return of Results Incorporating Web-Based Predisclosure Education
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Brian L. Egleston
Honoraria: Research Square
Consulting or Advisory Role: Digidence (Inst)
Olufunmilayo Olopade
Employment: CancerIQ
Leadership: CancerIQ
Stock and Other Ownership Interests: CancerIQ, Tempus, 54gene, HealthWell Solutions
Consulting or Advisory Role: Tempus
Research Funding: Roche/Genentech (Inst), Color Genomics (Inst)
Uncompensated Relationships: ACS, Susan G. Komen for the Cure
Open Payments Link: https://openpaymentsdata.cms.gov/physician/1178557
Payal Shah
Stock and Other Ownership Interests: Johnson & Johnson, Novartis, Novo Nordisk, Pfizer, Merck, Amgen
Consulting or Advisory Role: Gilead Sciences, Daiichi Sankyo
Research Funding: AstraZeneca (Inst), Zenith Epigenetics (Inst)
Jane E. Churpek
Honoraria: UpToDate
Patents, Royalties, Other Intellectual Property: My husband has a patent pending (ARCD. P0535US.P2) for risk stratification algorithms for hospitalized patients
Other Relationship: RUNX1 Research Program, MDS Foundation, National Comprehensive Cancer Network
Linda Fleisher
Honoraria: Academy of Oncology Nurse and Patient Navigators
Research Funding: Genentech
Dana Clark
Honoraria: AstraZeneca
Speakers' Bureau: AstraZeneca
Julia Wynn
Employment: BillionToOne
Stock and Other Ownership Interests: BillionToOne
Jessica M. Long
Employment: DePuy Companies
Stock and Other Ownership Interests: DePuy Companies
Jacquelyn Powers
Employment: Carevive Systems
Honoraria: CureConnect, Myriad Genetics
Consulting or Advisory Role: Carevive Systems
Travel, Accommodations, Expenses: Hospital of the University of Pennsylvania
Sarah Nielsen
Employment: Invitae
Stock and Other Ownership Interests: Invitae
Travel, Accommodations, Expenses: Invitae
Susan M. Domchek
Honoraria: AstraZeneca, GlaxoSmithKline
Research Funding: AstraZeneca (Inst), Clovis Oncology (Inst)
Open Payments Link: https://openpaymentsdata.cms.gov/physician/917904
Katherine L. Nathanson
Consulting or Advisory Role: Merck
Angela R. Bradbury
Consulting or Advisory Role: AstraZeneca, Merck
Research Funding: AstraZeneca/Merck (Inst)
No other potential conflicts of interest were reported.
REFERENCES
- 1. Evans JP, Rothschild BB. Return of results: Not that complicated? Genet Med. 2012;14:358–360. doi: 10.1038/gim.2012.8. [DOI] [PubMed] [Google Scholar]
- 2. Wolf SM. Return of individual research results and incidental findings: Facing the challenges of translational science. Annu Rev Genomics Hum Genet. 2013;14:557–577. doi: 10.1146/annurev-genom-091212-153506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bledsoe MJ, Clayton EW, McGuire AL, et al. Return of research results from genomic biobanks: A call for data. Genet Med. 2013;15:159–160. doi: 10.1038/gim.2012.163. [DOI] [PubMed] [Google Scholar]
- 4. Jarvik GP, Amendola LM, Berg JS, et al. Return of genomic results to research participants: The floor, the ceiling, and the choices in between. Am J Hum Genet. 2014;94:818–826. doi: 10.1016/j.ajhg.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Knoppers BM, Zawati MH, Senecal K. Return of genetic testing results in the era of whole-genome sequencing. Nat Rev Genet. 2015;16:553–559. doi: 10.1038/nrg3960. [DOI] [PubMed] [Google Scholar]
- 6. Bledsoe MJ, Grizzle WE, Clark BJ, et al. Practical implementation issues and challenges for biobanks in the return of individual research results. Genet Med. 2012;14:478–483. doi: 10.1038/gim.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. West KM, Blacksher E, Cavanaugh KL, et al. At the research-clinical interface: Returning individual genetic results to research participants. Clin J Am Soc Nephrol. 2020;15:1181–1189. doi: 10.2215/CJN.09670819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ravitsky V, Wilfond BS. Disclosing individual genetic results to research participants. Am J Bioeth. 2006;6:8–17. doi: 10.1080/15265160600934772. [DOI] [PubMed] [Google Scholar]
- 9.Downey AS, Busta ER, Mancher M, et al., editors. Returning Individual Research Results to Participants: Guidance for a New Research Paradigm. Washington, DC: 2018. [PubMed] [Google Scholar]
- 10. Clayton EW. Sharing individual research results with biospecimen contributors: Counterpoint. Cancer Epidemiol Biomarkers Prev. 2012;21:260–261. doi: 10.1158/1055-9965.EPI-11-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clayton EW, Ross LF. Implications of disclosing individual results of clinical research. JAMA. 2006;295:37. doi: 10.1001/jama.295.1.37-a. [DOI] [PubMed] [Google Scholar]
- 12. Knoppers BM, Joly Y, Simard J, et al. The emergence of an ethical duty to disclose genetic research results: International perspectives. Eur J Hum Genet. 2006;14:1170–1178. doi: 10.1038/sj.ejhg.5201690. [DOI] [PubMed] [Google Scholar]
- 13. Bledsoe MJ, Clayton EW, McGuire AL, et al. Return of research results from genomic biobanks: Cost matters. Genet Med. 2013;15:103–105. doi: 10.1038/gim.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beskow LM, O'Rourke PP. Return of genetic research results to participants and families: IRB perspectives and roles. J Law Med Ethics. 2015;43:502–513. doi: 10.1111/jlme.12292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thorogood A, Dalpe G, Knoppers BM. Return of individual genomic research results: Are laws and policies keeping step? Eur J Hum Genet. 2019;27:535–546. doi: 10.1038/s41431-018-0311-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Edwards KL, Goodman D, Johnson CO, et al. Controversies among cancer registry participants, genomic researchers, and institutional review boards about returning participants' genomic results. Public Health Genomics. 2018;21:18–26. doi: 10.1159/000490235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fernandez CV, Santor D, Weijer C, et al. The return of research results to participants: Pilot questionnaire of adolescents and parents of children with cancer. Pediatr Blood Cancer. 2007;48:441–446. doi: 10.1002/pbc.20766. [DOI] [PubMed] [Google Scholar]
- 18. Facio FM, Eidem H, Fisher T, et al. Intentions to receive individual results from whole-genome sequencing among participants in the ClinSeq study. Eur J Hum Genet. 2013;21:261–265. doi: 10.1038/ejhg.2012.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Halverson CM, Jones SH, Novak L, et al. What results should Be returned from opportunistic screening in translational research? J Pers Med. 2020;10:13. doi: 10.3390/jpm10010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shaibi GQ, Kullo IJ, Singh DP, et al. Developing a process for returning medically actionable genomic variants to Latino patients in a federally qualified health center. Public Health Genomics. 2018;21:77–84. doi: 10.1159/000494488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karlson EW, Boutin NT, Hoffnagle AG, et al. Building the Partners HealthCare Biobank at partners personalized medicine: Informed consent, return of research results, recruitment lessons and operational considerations. J Pers Med. 2016;6:2. doi: 10.3390/jpm6010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hoell C, Wynn J, Rasmussen LV, et al. Participant choices for return of genomic results in the eMERGE Network. Genet Med. 2020;22:1821–1829. doi: 10.1038/s41436-020-0905-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Joffe S, Sellers DE, Ekunwe L, et al. Preferences for return of genetic results among participants in the Jackson Heart Study and Framingham Heart Study. Circ Genom Precis Med. 2019;12:e002632. doi: 10.1161/CIRCGEN.119.002632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carey DJ, Fetterolf SN, Davis FD, et al. The Geisinger MyCode community health initiative: An electronic health record-linked biobank for precision medicine research. Genet Med. 2016;18:906–913. doi: 10.1038/gim.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bollinger JM, Scott J, Dvoskin R, et al. Public preferences regarding the return of individual genetic research results: Findings from a qualitative focus group study. Genet Med. 2012;14:451–457. doi: 10.1038/gim.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wynn J, Martinez J, Bulafka J, et al. Impact of receiving secondary results from genomic research: A 12-month longitudinal study. J Genet Couns. 2018;27:709–722. doi: 10.1007/s10897-017-0172-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blout Zawatsky CL, Shah N, Machini K, et al. Returning actionable genomic results in a research biobank: Analytic validity, clinical implementation, and resource utilization. Am J Hum Genet. 2021;108:2224–2237. doi: 10.1016/j.ajhg.2021.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beil A, Hornsby W, Uhlmann WR, et al. Disclosure of clinically actionable genetic variants to thoracic aortic dissection biobank participants. BMC Med Genomics. 2021;14:66. doi: 10.1186/s12920-021-00902-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leitsalu L, Palover M, Sikka TT, et al. Genotype-first approach to the detection of hereditary breast and ovarian cancer risk, and effects of risk disclosure to biobank participants. Eur J Hum Genet. 2021;29:471–481. doi: 10.1038/s41431-020-00760-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johnson G, Lawrenz F, Thao M. An empirical examination of the management of return of individual research results and incidental findings in genomic biobanks. Genet Med. 2012;14:444–450. doi: 10.1038/gim.2012.20. [DOI] [PubMed] [Google Scholar]
- 31. Keogh LA, Southey MC, Maskiell J, et al. Uptake of offer to receive genetic information about BRCA1 and BRCA2 mutations in an Australian population-based study. Cancer Epidemiol Biomarkers Prev. 2004;13:2258–2263. [PubMed] [Google Scholar]
- 32. Fiallos K, Applegate C, Mathews DJ, et al. Choices for return of primary and secondary genomic research results of 790 members of families with Mendelian disease. Eur J Hum Genet. 2017;25:530–537. doi: 10.1038/ejhg.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Keogh LA, Fisher D, Sheinfeld Gorin S, et al. How do researchers manage genetic results in practice? The experience of the multinational Colon Cancer Family Registry. J Community Genet. 2014;5:99–108. doi: 10.1007/s12687-013-0148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Crook A, Plunkett L, Forrest LE, et al. Connecting patients, researchers and clinical genetics services: The experiences of participants in the Australian Ovarian Cancer Study (AOCS) Eur J Hum Genet. 2015;23:152–158. doi: 10.1038/ejhg.2014.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Graves KD, Leventhal KG, Nusbaum R, et al. Behavioral and psychosocial responses to genomic testing for colorectal cancer risk. Genomics. 2013;102:123–130. doi: 10.1016/j.ygeno.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Robinson JO, Wynn J, Biesecker B, et al. Psychological outcomes related to exome and genome sequencing result disclosure: A meta-analysis of seven Clinical Sequencing Exploratory Research (CSER) Consortium studies. Genet Med. 2019;21:2781–2790. doi: 10.1038/s41436-019-0565-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hart MR, Biesecker BB, Blout CL, et al. Secondary findings from clinical genomic sequencing: Prevalence, patient perspectives, family history assessment, and health-care costs from a multisite study. Genet Med. 2019;21:1100–1110. doi: 10.1038/s41436-018-0308-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bradbury AR, Patrick-Miller L, Egleston BL, et al. Returning individual genetic research results to research participants: Uptake and outcomes among patients with breast cancer JCO Precis Oncol 2:1-242018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gaieski JB, Patrick-Miller L, Egleston BL, et al. Research participants' experiences with return of genetic research results and preferences for web-based alternatives. Mol Genet Genomic Med. 2019;7:e898. doi: 10.1002/mgg3.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bradbury AR, Patrick-Miller L, Long J, et al. Development of a tiered and binned genetic counseling model for informed consent in the era of multiplex testing for cancer susceptibility. Genet Med. 2015;17:485–492. doi: 10.1038/gim.2014.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Patrick-Miller LJ, Egleston BL, Fetzer D, et al. Development of a communication protocol for telephone disclosure of genetic test results for cancer predisposition. JMIR Res Protoc. 2014;3:e49. doi: 10.2196/resprot.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bradbury AR, Patrick-Miller LJ, Egleston BL, et al. Randomized noninferiority trial of telephone vs in-person disclosure of Germline cancer genetic test results. J Natl Cancer Inst. 2018;110:985–993. doi: 10.1093/jnci/djy015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shiloh S. Illness representations, self-regulation, and genetic counseling: A theoretical review. J Genet Couns. 2006;15:325–337. doi: 10.1007/s10897-006-9044-5. [DOI] [PubMed] [Google Scholar]
- 44.Leventhal H, Benyamini Y, Brownlee S, et al. Perceptions of health and illness: Current research and applications. In: Petrie KJ, Weinman JA, editors. Illness Representations: Theoretical Foundations. Amsterdam, the Netherlands: Harwood; 1997. pp. 19–46. [Google Scholar]
- 45. Kaphingst KA, McBride CM, Wade C, et al. Patients' understanding of and responses to multiplex genetic susceptibility test results. Genet Med. 2012;14:681–687. doi: 10.1038/gim.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lerman C, Narod S, Schulman K, et al. BRCA1 testing in families with hereditary breast-ovarian cancer. A prospective study of patient decision making and outcomes. JAMA. 1996;275:1885–1892. [PubMed] [Google Scholar]
- 47. Kelly K, Leventhal H, Marvin M, et al. Cancer genetics knowledge and beliefs and receipt of results in Ashkenazi Jewish individuals receiving counseling for BRCA1/2 mutations. Cancer Control. 2004;11:236–244. doi: 10.1177/107327480401100405. [DOI] [PubMed] [Google Scholar]
- 48. Hamilton JG, Lobel M, Moyer A. Emotional distress following genetic testing for hereditary breast and ovarian cancer: A meta-analytic review. Health Psychol. 2009;28:510–518. doi: 10.1037/a0014778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Speilberger CD, Gorsuch RL, Lushene R, et al. Manual for the State-Trait Anxiety Inventory. Palo Alto, CA: Consulting Psychologists Press; 1983. [Google Scholar]
- 50. Zigmond AS, Snaith RP. The hospital anxiety and depression scale. Acta Psychiatr Scand. 1983;67:361–370. doi: 10.1111/j.1600-0447.1983.tb09716.x. [DOI] [PubMed] [Google Scholar]
- 51. Horowitz M, Wilner N, Alvarez W. Impact of Event Scale: A measure of subjective stress. Psychosom Med. 1979;41:209–218. doi: 10.1097/00006842-197905000-00004. [DOI] [PubMed] [Google Scholar]
- 52. Pieterse AH, van Dulmen AM, Beemer FA, et al. Cancer genetic counseling: Communication and counselees' post-visit satisfaction, cognitions, anxiety, and needs fulfillment. J Genet Couns. 2007;16:85–96. doi: 10.1007/s10897-006-9048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Patrick-Miller L, Egleston BL, Daly M, et al. Implementation and outcomes of telephone disclosure of clinical BRCA1/2 test results. Patient Educ Couns. 2013;93:413–419. doi: 10.1016/j.pec.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cella D, Hughes C, Peterman A, et al. A brief assessment of concerns associated with genetic testing for cancer: The Multidimensional Impact of Cancer Risk Assessment (MICRA) questionnaire. Health Psychol. 2002;21:564–572. [PubMed] [Google Scholar]
- 55. Bradbury AR, Egleston BL, Patrick-Miller LJ, et al. Longitudinal outcomes with cancer multigene panel testing in previously tested BRCA1/2 negative patients. Clin Genet. 2020;97:601–609. doi: 10.1111/cge.13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Raghunathan TELJ, Van Hoewyz J, Solenberger P. A multivariate technique for multiply imputing missing values using a sequence of regression models. Surv Methodol. 2001;27:85–95. [Google Scholar]
- 57. Bradbury AR, Lee JW, Gaieski JB, et al. A randomized study of genetic education versus usual care in tumor profiling for advanced cancer in the ECOG-ACRIN Cancer Research Group (EAQ152) Cancer. 2022;128:1381–1391. doi: 10.1002/cncr.34063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Biesecker BB, Lewis KL, Umstead KL, et al. Web platform vs in-person genetic counselor for return of carrier results from exome sequencing: A randomized clinical trial. JAMA Intern Med. 2018;178:338–346. doi: 10.1001/jamainternmed.2017.8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sie AS, van Zelst-Stams WA, Spruijt L, et al. More breast cancer patients prefer BRCA-mutation testing without prior face-to-face genetic counseling. Fam Cancer. 2014;13:143–151. doi: 10.1007/s10689-013-9686-z. [DOI] [PubMed] [Google Scholar]
- 60. Watson CH, Ulm M, Blackburn P, et al. Video-assisted genetic counseling in patients with ovarian, fallopian and peritoneal carcinoma. Gynecol Oncol. 2016;143:109–112. doi: 10.1016/j.ygyno.2016.07.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kochan DC, Winkler E, Lindor N, et al. Challenges in returning results in a genomic medicine implementation study: The Return of Actionable Variants Empirical (RAVE) study. NPJ Genom Med. 2020;5:19. doi: 10.1038/s41525-020-0127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wakefield CE, Thorne H, Kirk J, et al. Improving mutation notification when new genetic information is identified in research: A trial of two strategies in familial breast cancer. Genet Med. 2013;15:187–194. doi: 10.1038/gim.2012.115. [DOI] [PubMed] [Google Scholar]
- 63. Balmana J, Digiovanni L, Gaddam P, et al. Conflicting interpretation of genetic variants and cancer risk by commercial laboratories as assessed by the prospective registry of multiplex testing. J Clin Oncol. 2016;34:4071–4078. doi: 10.1200/JCO.2016.68.4316. [DOI] [PMC free article] [PubMed] [Google Scholar]