Abstract
Aberrant DNA methylation plays a pivotal role in the onset and progression of colorectal cancer (CRC), a disease with high incidence and mortality rates in Korea. Several CRC-associated diagnostic and prognostic methylation markers have been identified; however, due to a lack of comprehensive clinical and methylome data, these markers have not been validated in the Korean population. Therefore, in this study, we aimed to obtain the CRC methylation profile using 172 tumors and 128 adjacent normal colon tissues of Korean patients with CRC. Based on the comparative methylome analysis, we found that hypermethylated positions in the tumor were predominantly concentrated in CpG islands and promoter regions, whereas hypomethylated positions were largely found in the open-sea region, notably distant from the CpG islands. In addition, we stratified patients by applying the CpG island methylator phenotype (CIMP) to the tumor methylome data. This stratification validated previous clinicopathological implications, as tumors with high CIMP signatures were significantly correlated with the proximal colon, higher prevalence of microsatellite instability status, and MLH1 promoter methylation. In conclusion, our extensive methylome analysis and the accompanying dataset offers valuable insights into the utilization of CRC-associated methylation markers in Korean patients, potentially improving CRC diagnosis and prognosis. Furthermore, this study serves as a solid foundation for further investigations into personalized and ethnicity-specific CRC treatments.
Keywords: Cancer characteristics, Colorectal cancer, CpG island methylator phenotype (CIMP), DNA methylation, MutL homolog 1
INTRODUCTION
Colorectal cancer (CRC) is a significant global health concern with incidence and mortality rates of 10.2% and 9.4%, respectively, in 2020 (1, 2). In Korea, CRC is the third most common cancer and poses a significant health challenge (3). CRC develops through a multistep process that involves the accumulation of various genetic and epigenetic alterations, among which aberrant DNA methylation is a key mechanism (4).
DNA methylation primarily occurs at cytosine-phosphate-guanine (CpG) sites and regulates multiple processes, such as gene expression and genome stability. Aberrations in DNA methylation patterns, including hyper- and hypomethylation, are associated with various diseases, including cancer (5). Aberrant DNA methylation has been associated with CRC onset and progression. Hypermethylation of CpG promoters in cancer cells can lead to tumor suppressor gene silencing, inducing mechanisms of carcinogenesis (6, 7). Hypomethylation activates oncogenes and causes genome instability (8). Notably, a subset of CRCs exhibits a distinct feature known as the high-CpG island methylator phenotype (CIMP-H). CIMP-H cancers are characterized by widespread hypermethylation of CpG islands, particularly in the promoter regions, leading to gene inactivation (9, 10); this is linked with distinct clinical and molecular characteristics. These result in a heightened incidence of tumors in the proximal colon, a higher frequency of BRAF gene mutations, and the presence of microsatellite instability (11-13).
Despite efforts made to improve the understanding of the relationship between DNA methylation and CRC, comprehensive clinical and methylome data specific to Korean patients with CRC remains elusive. This information is essential for the identification and validation of CRC-related methylation markers that could enhance disease diagnosis and prognosis. Therefore, this study aimed to construct a comprehensive CRC methylome profile from a large cohort of Korean patients with CRC (n = 172). We determined the methylome distribution and its association with various clinical characteristics. Additionally, we compared our results with those of previous CRC studies to ensure data validity and reliability. Furthermore, we aimed to deepen our understanding of the role of DNA methylation in CRC etiology, prognosis, and treatment of the Korean population.
RESULTS
Clinical characteristics of the study participants
The study consisted of 172 tumor and 128 adjacent normal samples obtained from 172 individuals diagnosed with colorectal adenocarcinoma at the Asan Medical Center (Table 1). In this cohort, 74 were female and 98 were male, with a median age of 59 years (interquartile range: 53-68 years). The cancer was located in the right colon (cecal-splenic flexure of the transverse colon) in 49 patients (28.5%), left colon (splenic flexure of the transverse colon-distal sigmoid colon) in 68 patients (39.5%), and rectum in 55 patients (32.0%). The majority of tumors were T3 (128 patients, 74.4%), N0 (89 patients, 51.6%), M0 (150 patients, 87.2%), fungating in shape (Bormann type I or II, 137 patients, 79.7%), and favorably differentiated (well- or moderately-differentiated, 158 patients, 91.9%). Lymphovascular and perineural invasion were found in 74 patients (43%) and 56 patients (32.6%), respectively. One-hundred and twenty-three patients (71.5%) received chemotherapy after surgery, consisting of single-agent treatment with 5-fluorouracil (5-FU) or capecitabine (n = 39), a combination of 5-FU (or capecitabine) and oxaliplatin (n = 69), or targeted agents with 5-FU and oxaliplatin or irinotecan (n = 15). Twenty-two patients (12.8%) received pre- or post-operative radiotherapy (50.4 Gy in 28 fractions).
Table 1.
Clinical characteristics of the study participants
| Variables | No. of patients (%) or values as median (IQR) (n = 172) |
|---|---|
| Sex, female/male | 74/98 (43.0/57.0) |
| Age at operation, years | 59 (53-68) |
| Preoperative CEA level | 2.5 (1.43-6.28) |
| Tumor location | |
| Right | 49 (28.5) |
| Left | 68 (39.5) |
| Rectum | 55 (32.0) |
| Tumor size, cm | 4.75 (3.5-6.5) |
| Bormann type | |
| I/II | 137 (79.7) |
| III/IV | 35 (20.3) |
| T category | |
| T0 | 1 (0.6) |
| T2 | 14 (8.1) |
| T3 | 128 (74.4) |
| T4 | 29 (16.9) |
| N category | |
| N0 | 89 (51.7) |
| N1 | 48 (27.9) |
| N2 | 35 (20.4) |
| M category | |
| M0 | 150 (87.2) |
| M1 | 22 (12.86) |
| Differentiation | |
| WD/MD | 158 (91.3) |
| PD/Mucinous | 13 (7.6) |
| Unknown | 1 (0.1) |
| Lymphovascular invasion | |
| Negative | 98 (57.0) |
| Positive | 74 (43.0) |
| Perineural invasion | |
| Negative | 116 (67.4) |
| Positive | 56 (32.6) |
| Microsatellite instability | |
| MSS | 142 (82.6) |
| MSI-L | 8 (4.7) |
| MSI-H | 18 (10.5) |
| Unknown | 4 (2.4) |
| Adjuvant chemotherapy | |
| FL or capecitabine | 39 (22.7) |
| FOLFOX or XELOX | 69 (40.1) |
| Targeted agents with FOLFOX or FOLFIRI | 15 (8.7) |
| None | 49 (28.5) |
| Radiotherapy | 22 (12.8) |
IQR, interquartile range; CEA, carcinoembryonic antigen; WD, well-differentiated; MD, moderately-differentiated; PD, poorly-differentiated; MSS, microsatellite instability stable; MSI-L, microsatellite instability low; MSI-H, microsatellite instability high; FL, 5-fluorouracil with leucovorin; FOLFOX, combination of 5-FU and oxaliplatin; FOLFIRI, combination of 5-FU and irinotecan.
Methylation patterns in tumors
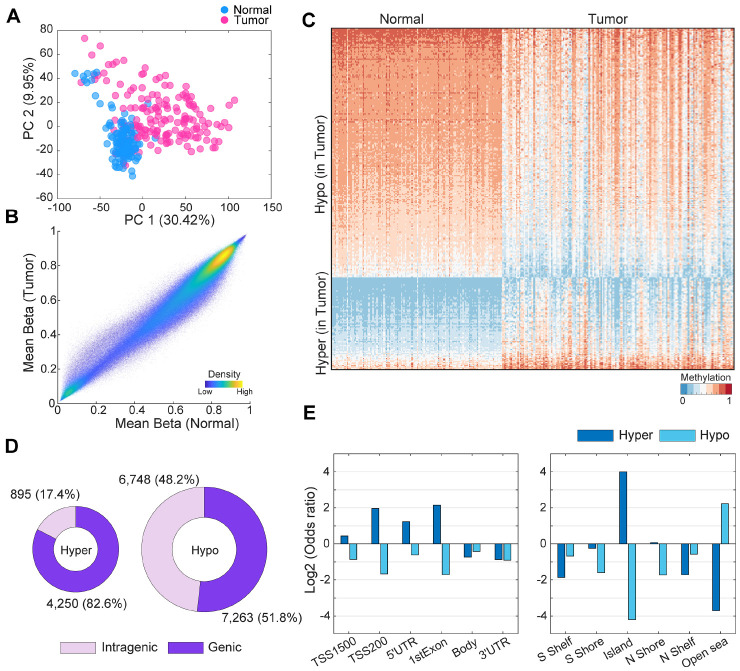
To achieve a robust methylome profile, preprocessing tasks were performed using the well-established and effective minfi pipeline (14) followed by the batch correction with combat (Fig. 1 and Supplementary Figs. 1-3; details in Supplementary Results and Supplementary Methods) (15). From the 610,674 processed methylation probes, we initially observed a distinct separation between tumor and normal samples as seen in the dimensionality reduction plot (Fig. 2A), illustrated by the principal component distribution (explained variance of PC1: 30.42%). Next, we investigated the grand mean methylation levels of the tumor and normal groups for each individual sample. The overall methylation levels were slightly higher in the normal sample group (tumor: 0.5674, normal: 0.5855 [P < 0.0001]; Fig. 2B and Supplementary Fig. 4A). We subsequently identified 19,156 differentially methylated positions (DMPs) between tumor and normal samples (Supplementary Tables 1 and 2). There were a greater number of hypomethylated probes (14,011 probes) than hypermethylated probes (5,145 probes) (Fig. 2C, D). Among the hypermethylated probes, 4,250 (82.6%) were located in the genic region (promoter: 2,956, gene body: 1,645). Among the hypomethylated probes, 7,263 (51.8%) were located in the genic region (promoter: 2,428, gene body: 5,154) (Fig. 2C and Supplementary Fig. 4B). To compare the frequency of detailed regional DMPs, we calculated the odds ratio (OR) for the enrichment of hyper- and hypomethylated probes for various genomic annotations, including gene promoter regions, body regions, and islands or shores. In promoter-like regions, TSS1500, TSS200, 5’ untranslated region (UTR), and first exon, the odds ratios of hypermethylated probes were 1.35, 3.90, 2.36, and 4.43, respectively (Fig. 2E, left; P < 0.0001). Similarly, hypermethylated probes were highly enriched in the CpG island region (Fig. 2E, right), with an odds ratio of 15.98 (P < 0.0001). In contrast, hypomethylated probes in tumor samples were predominantly located in open-sea regions (odds ratio: 4.69) (Fig. 2E, right), which are considerably distant from the CpG island regions. We further validated the methylation patterns through comparison with colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) from The Cancer Genome Atlas (TCGA) (Supplementary Fig. 5) as described in Supplementary Results.
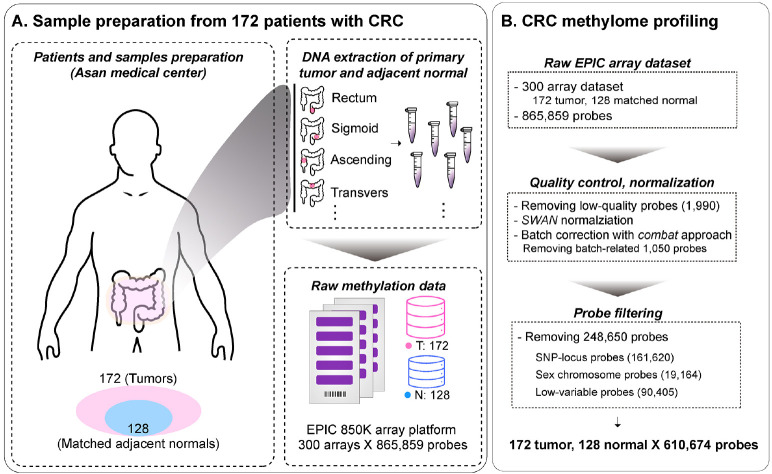
Fig. 1.
Overview of sample preprocessing for CRC methylome profile construction. (A) Preparation of CRC samples for methylome profile. These consisted of 172 tumor samples and 128 adjacent normal samples from patients with CRC at Asan Medical Center. (B) Preprocessing of CRC methylome profile. The minfi pipeline was used for preprocessing, Subset-quantile Within Array Normalization (SWAN) method for normalization, and ComBat approach for batch correction. Poor-performing probes, SNPs, sex chromosomes, and low-variable sites were filtered out. Next, the methylation profiles were finalized. Methylation was quantified at 610,674 probes.
Fig. 2.
Methylation differences between colorectal tumor and normal tissues. (A) Dimension reduction plot of methylation levels of the preprocessed 610,674 probes. (B) Comparisons of mean beta-values between normal and tumor tissue samples. (C) Heatmap of differentially methylated probe expression. Used probes were randomly selected by 5% of total DMPs. (D) Total number of DMPs between tumor and normal tissue samples. Purple and light-purple represents the genic and intragenic regions, respectively. (E) Log2 odds ratios of the number of defined hyper- and hypo-methylated probes according to each genic and CpG Island region.
CIMP tumor sample subgroup
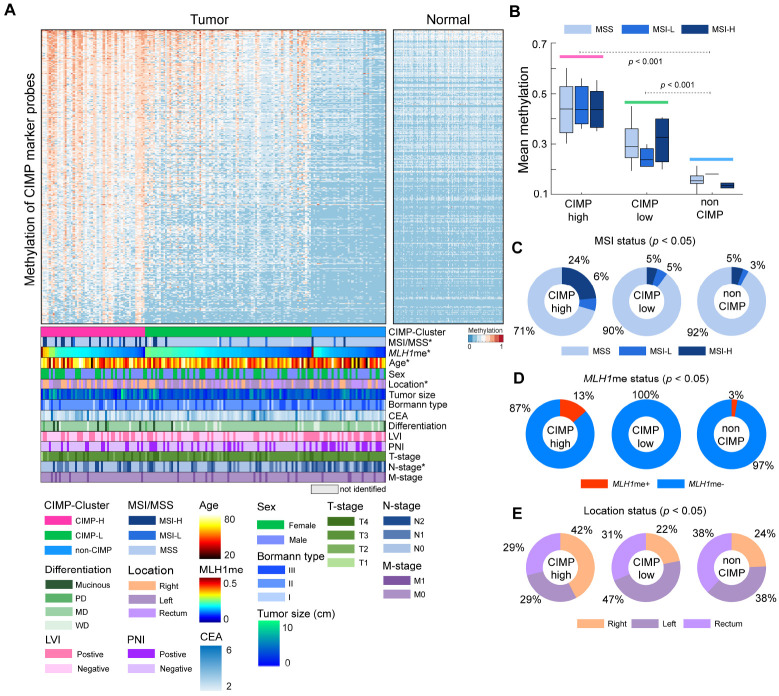
To assess the prevalence of CpG island methylator phenotype (CIMP), we used a CIMP probe set (4,322 probes) based on 258 previously identified CIMP gene markers. From the 4,322 probes, we selected 1,220 highly variable sites (standard deviation > 0.15). Using the K-means algorithm (100 iterations) with the methylation levels of CIMP marker probes, we divided the tumor samples into three clusters and categorized each group as CIMP-high (CIMP-H), CIMP-low (CIMP-L), or non-CIMP, based on the respective mean methylation expression of each cluster (Fig. 3A, B). Based on the predetermined criteria, we identified 52 (30.2%) CIMP-H, 83 (48.2%) CIMP-L, and 37 (21.5%) non-CIMP tumor patients, and their mean CIMP marker probe methylation levels exhibited significant pairwise differences (analysis of variance [ANOVA] with a post-hoc test, P < 0.001) (Fig. 3B). Focusing on microsatellite instability (MSI) status, within the CIMP-H group, 12 (24%) patients displayed high MSI (MSI-H) status (Fig. 3C). In contrast, only 5% of the patients in both the CIMP-L and non-CIMP groups exhibited MSI-H status. This result showed the significant enrichment of MSI-H patients within the CIMP-H group (χ2 test, P < 0.01). Moreover, a significantly higher prevalence of MLH1 silencing was observed in the CIMP-H group as compared to the CIMP-L and non-CIMP groups (χ2 test, P < 0.05) (Fig. 3D). CIMP status was also correlated with the anatomical location of the large intestine and patient age. In the CIMP-H group, 22 tumors (42%) were found in the right colon and 15 tumors each in both the left and rectum were discovered. In the CIMP-L and non-CIMP groups, 18 (22%) and 9 (24%) tumors were found in the right colon, respectively. These findings revealed a higher enrichment of right colon tumor samples in the CIMP-H group (χ2 test, P < 0.01). When comparing age distribution with CIMP status, the mean age of CIMP-H patients was 61.8 years and that of CIMP-L was 60.8 years, slightly higher than that of the non-CIMP (mean age: 55.8 years) group (post-hoc test [CIMP-H vs. non-CIMP], P < 0.05) (Supplementary Fig. 6A). Following this, we explored the correlation between various CRC characteristics and CIMP status. Our results revealed no significant associations between sex, tumor size, preoperative carcinoembryonic antigen (CEA) level, Bormann type, as well as T- and M-stages with CIMP status. Within the non-CIMP groups, a larger proportion exhibited perineural invasion (χ2 test, p = 0.08) and lymphovascular invasion, including instances of N2 stage (χ2 test, P < 0.05) (Supplementary Fig. 6). The differences in both overall and disease-free survival according to CIMP status were not statistically significant. However, we did observe a slight trend toward poorer prognosis in CIMP-H compared to the non-CIMP group (Supplementary Fig. 7).
Fig. 3.
CpG Island Methylator Phenotype (CIMP) clustering of 172 colon tumor samples. (A) Heatmap of 172 colon tumor samples based on the methylation expression of CIMP marker probes. Samples were clustered in each CIMP-based group and sorted based on MLH1 methylation. For clinical abbreviations in the figure, MSI and MSS represent microsatellite instability and stability, respectively. LVI, PNI, and CEA represent lymphovascular invasion, perineural invasion, and preoperative carcinoembryonic antigen level, respectively. WD, MD, and PD represent well-differentiated, moderately-differentiated, and poorly-differentiated, respectively. (B) Boxplot represents mean CIMP marker gene methylations based on CIMP and MSI status. (C) Proportion of MSI and MSS status based on CIMP status. (D) Proportion of MLH1 promoter methylation status based on CIMP status. The categorization of samples into MLH1 promoter methylation-positive (MLH1me+) and methylation-negative (MLH1me-) is based on a predetermined methylation level threshold of 0.3. Samples with promoter methylation levels greater than 0.3 were classified as MLH1me+, while those with levels less than 0.3 were classified as MLH1me-. (E) Proportion of tissue location based on CIMP status. In the figure, left colons consist of splenic flexure, descending, sigmoid, and rectosigmoid tumors, and right colons consist of ascending, cecum, hepatic flexure, and transverse tumors. In panel (A), asterisks (*) denote significance (P < 0.05) as determined by the chi-square test, which was used to assess the association between CIMP and clinical characteristics.
DISCUSSION
Here, we provide the DNA methylation profiles of a large cohort of Korean patients with CRC in conjunction with additional cohort collected from another Korean hospital (16), offering valuable insights into the significance of aberrant DNA methylation in CRC. The 172 Korean CRC methylome profiles of matched 128 normal samples confirmed and extended prior findings. First, we compared the overall methylation patterns of tumor and normal samples. We found that hypermethylation in CRC predominantly occurs in the CpG islands and promoter regions. These findings are consistent with those of previous studies showing that DNA hypermethylation in promoter CpG islands leads to tumor suppressor gene silencing, thereby contributing to carcinogenesis (6, 7). Next, we carried out a comparative analysis of the Korean colorectal cancer (CRC) methylome against the methylomes of CRC cases in other ethnic groups observed in previously published studies. Our investigation involved the assessment of overlap with a substantial pool of 450 K probes from TCGA CRC methylome. We observed a significant correlation between the overlapped probes from TCGA and our Korean CRC dataset. This correlation was particularly strong among the DMPs identified in this study. To further validate our findings, we took into account the previously identified diagnostic methylation markers for CRC, which comprised 15 genes (17-23). We found significant differences in the promoter methylation of 13 out of these 15 marker genes between tumor and normal samples in our Korean cohort, suggesting the necessity of identifying more specific Korean methylation markers with consideration of various CRC subtypes.
Recent studies have investigated the role of CIMP in various cancer types including CRC. In CRC, CIMP has been linked with several different molecular alterations, including BRAF mutations and microsatellite instability (24). CRC tumors with CIMP also exhibit distinct clinical features and have been associated with specific locations in the colon (e.g., proximal colon) and with specific histological features. Similarly, in gastric cancer, the CIMP phenotype is associated with MSI-H with MLH1 silencing, and is significantly associated with the presence of Helicobacter pylori and Epstein-Barr virus infection (25, 26). In glioblastoma (GBM), CIMP has been associated with the IDH1 mutation and is observed in a subtype of tumors with generally better prognoses (27, 28). The CIMP-positive GBM is associated with a distinct set of methylated genes, and has a different clinical and molecular profile compared to CIMP-negative GBM. It is important to note that while CIMP status has implications for prognosis and potentially for treatment, further research is required to fully understand the role of CIMP in these different types of cancers. In our Korean CRC cohort, the stratification of patients with CRC based on CIMP status reaffirmed the correlations between CIMP status and clinicopathological characteristics (10, 11, 13). CIMP-high tumors were significantly associated with MSI-H status, right-sided tumors, and older age. Furthermore, we investigated various cancer characteristics according to CIMP status and additionally confirmed that CIMP groups had more MLH1 epigenetically silenced samples, and non-CIMP groups had more perineural invasion (PNI)- and lymphovascular invasion (LVI)-positive samples. CIMP-positive tumors are often associated with methylation of the MLH1 gene promoter, which leads to the silencing of this important DNA mismatch repair (MMR) gene (29, 30). This is a well-established characteristic of a subset of CRCs, leading to MSI-H and a distinct clinical and pathological profile. However, the association between CIMP status and PNI or LVI is not as well-established and may vary across studies (31-34). Given that PNI and LVI are typically associated with more advanced and aggressive disease (35) and may indicate a more complex cancer status, their relationship with CIMP status warrants further investigation including more comprehensive molecular analysis. We noted that our rich CRC methylome profile improves understanding of CIMP-high CRC as a distinct molecular subtype with specific clinical and molecular features. Through developments of efficient and accessible clinical assays such as high-resolution melting curve analysis (36), we believe our discovery has great potential to guide treatment strategies. Overall, this study broadens our current knowledge related to the CIMP-high CRC subtype which allows for improved patient stratification.
Despite these notable findings, this study has several limitations. Although we included a large methylation cohort of Korean patients with CRC, further studies involving other ethnic groups are necessary for the generalization or Korean-specific characterization of the results with multi-omics profiles. Additionally, our CRC methylome focused on investigating the overall Korean CRC landscape from bulk tissue analysis, potentially leading to an oversimplified interpretation of the complex tumor system. In an ideal scenario, single-cell analysis would indeed be a compelling approach, as it could allow us to unravel the complex heterogeneity seen within tumors, delineating specific methylation patterns associated with different cell types. Moreover, treatment strategies vary greatly between patients based on their individual health status, disease stage, and other clinical factors. Examination of post-treatment methylation profiles could be informative, but such analysis would require a comprehensive and longitudinally collected dataset to reflect temporal changes across various treatments.
In conclusion, the current study provides a DNA methylation landscape of CRC in the Korean population. Our CRC methylome profile revealed hyper- and hypo-methylation events in specific genomic regions, emphasizing the vital role of aberrant DNA methylation in the development and progression of CRC. Furthermore, by stratifying patients based on their CIMP status, we note the potential of DNA methylation as an epigenetic hallmark for patient classification and personalized treatment strategies. Our findings highlight the need for further investigations into CRC management and pave the way for the development of more effective diagnosis, prognosis, and therapeutic interventions.
MATERIALS AND METHODS
Ethics approval statement
This study was approved by the Asan Medical Center Institutional Review Board (2017-1350) and the Yonsei University Institutional Review Board (approval number: 7001988-201910-BR-727-02), in accordance with the Declaration of Helsinki.
Methylation EPIC array data set and data analysis methods
The detailed materials and methods on EPIC array generation and statistical analyses are provided in Supplementary methods.
Funding Statement
ACKNOWLEDGEMENTS This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science & ICT (grant number: NRF-2017M3A9A7050614). It was additionally supported by a grant from the National Research Foundation of Korea (NRF-2020M3A9I6A01036057).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
DATA AVAILABILITY
Raw IDAT files and processed methylation profiles are available at the Korea BioData Station (KBDS, https://kbds.re.kr/) (37) with the accession ID PRJKA2086325. CRC methylome of TCGA was downloaded from Genomic Data Commons (GDC) data portal (https://portal.gdc.cancer.gov/).
REFERENCES
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Daly MC, Paquette IM. Surveillance, epidemiology, and end results (SEER) and SEER-medicare databases: use in clinical research for improving colorectal cancer outcomes. Clin Colon Rect Surg. 2019;32:61–68. doi: 10.1055/s-0038-1673355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jung KW, Won YJ, Kang MJ, Kong HJ, Im JS, Seo HG. Prediction of cancer incidence and mortality in Korea, 2022. Cancer Res Treat. 2022;54:345–351. doi: 10.4143/crt.2022.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 5.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 6.Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology. 2015;149:1204–1225.e12. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet Spec No 1. 2007;16:R50–R59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- 8.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 9.Toyota M, Ohe-Toyota M, Ahuja N, Issa JP. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci U S A. 2000;97:710–715. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 11.Nazemalhosseini Mojarad E, Kuppen PJ, Aghdaei HA, Zali MR. The CpG island methylator phenotype (CIMP) in colorectal cancer. Gastroenterol Hepatol Bed Bench. 2013;6:120–128. [PMC free article] [PubMed] [Google Scholar]
- 12.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zlobec I, Bihl M, Foerster A, Rufle A, Lugli A. Comprehensive analysis of CpG island methylator phenotype (CIMP)-high, -low, and -negative colorectal cancers based on protein marker expression and molecular features. J Pathol. 2011;225:336–343. doi: 10.1002/path.2879. [DOI] [PubMed] [Google Scholar]
- 14.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 16.Joe S, Kim J, Lee JY, et al. Epigenetic insights into colorectal cancer: comprehensive genome-wide DNA methylation profiling of 294 patients in Korea. BMB Rep. 2023;56:563–568. doi: 10.5483/BMBRep.2023-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lind GE, Danielsen SA, Ahlquist T, et al. Identification of an epigenetic biomarker panel with high sensitivity and specificity for colorectal cancer and adenomas. Mol Cancer. 2011;10:85. doi: 10.1186/1476-4598-10-85.074c02ec5a9444f883efce1c6a2f9981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang E, Park DI, Kim YJ, et al. Detection of colorectal neoplasm using promoter methylation of ITGA4, SFRP2, and p16 in stool samples: a preliminary report in Korean patients. Hepatogastroenterology. 2010;57:720–727. [PubMed] [Google Scholar]
- 19.Kim MS, Louwagie J, Carvalho B, et al. Promoter DNA methylation of oncostatin m receptor-beta as a novel diagnostic and therapeutic marker in colon cancer. PLoS One. 2009;4:e6555. doi: 10.1371/journal.pone.0006555.df505dd4c01947cba7d2efc6267b7130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Z, Zhao G, Wang K, et al. Blood leukocytes methylation levels analysis indicate methylated plasma test is a promising tool for colorectal cancer early detection. J Cancer. 2021;12:3678–3685. doi: 10.7150/jca.57114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zou H, Harrington JJ, Shire AM, et al. Highly methylated genes in colorectal neoplasia: implications for screening. Cancer Epidemiol Biomarkers Prev. 2007;16:2686–2696. doi: 10.1158/1055-9965.EPI-07-0518. [DOI] [PubMed] [Google Scholar]
- 22.Xiao W, Zhao H, Dong W, et al. Quantitative detection of methylated NDRG4 gene as a candidate biomarker for diagnosis of colorectal cancer. Oncol Lett. 2015;9:1383–1387. doi: 10.3892/ol.2014.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou H, Allawi H, Cao X, et al. Quantification of methylated markers with a multiplex methylation-specific technology. Clin Chem. 2012;58:375–383. doi: 10.1373/clinchem.2011.171264. [DOI] [PubMed] [Google Scholar]
- 24.Muller D, Gyorffy B. DNA methylation-based diagnostic, prognostic, and predictive biomarkers in colorectal cancer. Biochim Biophys Acta Rev Cancer. 2022;1877:188722. doi: 10.1016/j.bbcan.2022.188722. [DOI] [PubMed] [Google Scholar]
- 25.Matsusaka K, Funata S, Fukayama M, Kaneda A. DNA methylation in gastric cancer, related to Helicobacter pylori and Epstein-Barr virus. World J Gastroenterol. 2014;20:3916–3926. doi: 10.3748/wjg.v20.i14.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Research Network, author. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malta TM, de Souza CF, Sabedot TS, et al. Glioma CpG island methylator phenotype (G-CIMP): biological and clinical implications. Neuro Oncol. 2018;20:608–620. doi: 10.1093/neuonc/nox183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herman JG, Umar A, Polyak K, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim JH, Rhee YY, Bae JM, et al. Subsets of microsatellite-unstable colorectal cancers exhibit discordance between the CpG island methylator phenotype and MLH1 methylation status. Mod Pathol. 2013;26:1013–1022. doi: 10.1038/modpathol.2012.241. [DOI] [PubMed] [Google Scholar]
- 31.Fu T, Liu Y, Li K, et al. Tumors with unmethylated MLH1 and the CpG island methylator phenotype are associated with a poor prognosis in stage II colorectal cancer patients. Oncotarget. 2016;7:86480–86489. doi: 10.18632/oncotarget.13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saadallah-Kallel A, Abdelmaksoud-Dammak R, Triki M, et al. Clinical and prognosis value of the CIMP status combined with MLH1 or p16 INK4a methylation in colorectal cancer. Med Oncol. 2017;34:147. doi: 10.1007/s12032-017-1007-1. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Long Y, Xu Y, et al. Prognostic and predictive value of CpG island methylator phenotype in patients with locally advanced nonmetastatic sporadic colorectal cancer. Gastroenterol Res Pract. 2014;2014:436985. doi: 10.1155/2014/436985.eada09d6c4984be5847a663b4e1ea31d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yagi K, Akagi K, Hayashi H, et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res. 2010;16:21–33. doi: 10.1158/1078-0432.CCR-09-2006. [DOI] [PubMed] [Google Scholar]
- 35.Al-Sukhni E, Attwood K, Gabriel EM, LeVea CM, Kanehira K, Nurkin SJ. Lymphovascular and perineural invasion are associated with poor prognostic features and outcomes in colorectal cancer: a retrospective cohort study. Int J Surg. 2017;37:42–49. doi: 10.1016/j.ijsu.2016.08.528. [DOI] [PubMed] [Google Scholar]
- 36.Kim SC, Kim J, Kim DW, et al. Methylation-sensitive high-resolution melting analysis of the USP44 promoter can detect early-stage hepatocellular carcinoma in blood samples. BMB Rep. 2022;55:553–558. doi: 10.5483/BMBRep.2022.55.11.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee B, Hwang S, Kim PG, et al. Introduction of the Korea BioData Station (K-BDS) for sharing biological data. Genomics Inform. 2023;21:e12. doi: 10.5808/gi.22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.