Abstract
DNA methylation regulates gene expression and contributes to tumorigenesis in the early stages of cancer. In colorectal cancer (CRC), CpG island methylator phenotype (CIMP) is recognized as a distinct subset that is associated with specific molecular and clinical features. In this study, we investigated the genome-wide DNA methylation patterns among patients with CRC. The methylation data of 1 unmatched normal, 142 adjacent normal, and 294 tumor samples were analyzed. We identified 40,003 differentially methylated positions with 6,933 (79.8%) hypermethylated and 16,145 (51.6%) hypomethylated probes in the genic region. Hypermethylated probes were predominantly found in promoter-like regions, CpG islands, and N shore sites; hypomethylated probes were enriched in open-sea regions. CRC tumors were categorized into three CIMP subgroups, with 90 (30.6%) in the CIMP-high (CIMP-H), 115 (39.1%) in the CIMP-low (CIMP-L), and 89 (30.3%) in the non-CIMP group. The CIMP-H group was associated with microsatellite instability-high tumors, hypermethylation of MLH1, older age, and right-sided tumors. Our results showed that genome-wide methylation analyses classified patients with CRC into three subgroups according to CIMP levels, with clinical and molecular features consistent with previous data.
Keywords: Clinicopathological characteristics, Colorectal cancer, CpG island methylator phenotype, DNA methylation, MutL homolog 1
INTRODUCTION
Colorectal cancer (CRC) is one of the most common cancer types and the leading cause of death worldwide (1). Numerous attempts have been made to better understand the molecular basis of CRC; however, CRC prognosis is currently assessed using TNM staging. The mean 5-year survival rate of patients with localized CRC is 90%, but that of patients with advanced CRC is only approximately 13-17% (2, 3).
CRC tumorigenesis occurs through the accumulation of genetic and epigenetic mutations from adenoma to adenocarcinoma (4). However, not all CRCs arise from conventional genetic mutations in genes such as APC, KRAS, and TP53, suggesting that other mechanisms also contribute to the development of CRC (5). Epigenetic alterations, particularly DNA methylation, are recognized as alternative mechanisms that can lead to inappropriate gene expression and contribute to early-stage tumorigenesis (6, 7). DNA methylation predominantly occurs at CpG sites (present in 70% of human promoters), which generally leads to epigenetic transcriptional silencing (8). In CRC, the CpG island methylator phenotype (CIMP) is a distinct subset associated with BRAF mutations and mismatch repair deficiency due to CIMP-associated methylation of MLH1 (9). Our genome-wide methylation analyses showed that the patients with different CIMP levels had significant correlation with clinical and molecular features consistent with previous data.
RESULTS
Clinicopathological characteristics of the patient cohort
Initially, 299 patients with stage II, III, or IV CRC were included in the study. Patients whose sex and chromosomal data did not match were excluded considering sample contamination (n = 5); thus, 1 unpaired, 142 paired normal, and 294 tumor samples were analyzed.
More than half of the patients (n = 181, 61.6%) were male, with a median age of 62 years (range 20-83). The most common tumor locations were sigmoid colon (n = 113, 38.4%), followed by rectum (n = 58, 19.7%), and ascending colon (n = 57, 19.4%). Approximately half of the patients had stage III disease (n = 165, 56.1%), and a similar number of patients had stage II (n = 65, 22.1%) or IV disease (n = 63, 21.4%). Pathological reports showed that most cases were adenocarcinomas (n = 275, 93.5%), with a few mucinous adenocarcinomas and signet ring carcinomas. Most of the tumors were moderately differentiated (n = 249, 85.9%), 12.4% were poorly differentiated (n = 36), and 1.7% were well differentiated (n = 5). Among patients tested for the presence of KRAS mutations, 40.9% (n = 110/269) had mutations in exons 2, 3, or 4. Only a few patients were tested for BRAF and NRAS; four patients had BRAF mutations, and one patient had an NRAS mutation (Table 1 and Supplementary Fig. 1).
Table 1.
Patient characteristics
| Parameter | N (%) |
|---|---|
| Sex | |
| Male | 181 (61.6) |
| Female | 113 (38.4) |
| Age, year | 62.0 (20.0; 83.0)* |
| Location | |
| Cecum | 7 (2.4) |
| Ascending | 57 (19.4) |
| Transverse | 14 (4.8) |
| Descending | 21 (7.1) |
| Rectum | 58 (19.7) |
| Rectosigmoid | 23 (7.8) |
| Sigmoid | 113 (38.4) |
| Synchronous | 1 (0.3) |
| Pathology | |
| Adenocarcinoma | 275 (93.5) |
| Mucinous | 17 (5.8) |
| Mucinous with signet ring carcinoma | 1 (0.3) |
| Signet ring cell carcinoma | 1 (0.3) |
| Differentiation | |
| Well differentiated | 5 (1.7) |
| Moderately differentiated | 249 (85.9) |
| Poorly differentiated | 36 (12.4) |
| N/A | 4 (1.4) |
| MSI status | |
| MSI-H | 16 (5.4) |
| MSI-L | 18 (6.1) |
| MSS | 258 (87.8) |
| N/A | 2 (0.7) |
| KRAS | |
| Mutant | 110 (37.4) |
| Wild type | 159 (54.1) |
| N/A | 25 (8.5) |
| BRAF | |
| Mutant | 4 (1.4) |
| Wild type | 42 (14.3) |
| N/A | 248 (84.4) |
| NRAS | |
| Mutant | 1 (0.3) |
| Wild type | 50 (17.0) |
| N/A | 243 (82.7) |
| Stage (AJCC) | |
| II | 65 (22.1) |
| III | 165 (56.1) |
| IV | 63 (21.4) |
| N/A | 1 (0.3) |
N, number of samples; MSI, microsatellite instability; MSS, microsatellite stability; MSI-H, high microsatellite instability; MSI-L, low microsatellite instability; N/A, not applicable; AJCC, American Joint Committee on Cancer.
*Median age of patients with CRC and overall range in this study.
Preprocessing of the DNA methylation profile of Korean patients with CRC
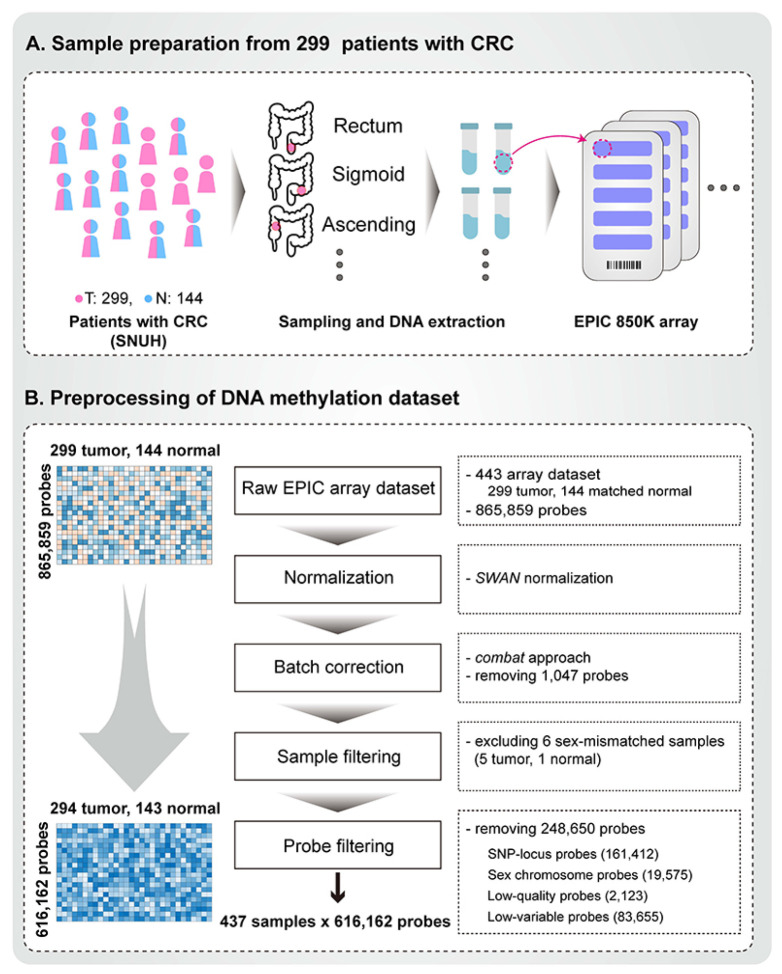
DNA methylation of the CRC samples was performed using a high throughput assay termed EPIC array, which enables examination of over 850,000 methylation sites across the genome. The EPIC array dataset was processed using the previously defined pipeline minfi (10). We initially examined the quality of the EPIC array by inspecting the overall distribution of beta values and control strip plots, including those for bisulfite conversion efficiency, extension quality, and specificity (Supplementary Fig. 2). We then applied subset-quantile within array normalization (SWAN) (11) to correct technical differences between type I and II probes within each array. Next, we addressed known batch effects specific to each EPIC array batch type (with removing 1,047 probes). For downstream studies, we filtered out 2,123 poorly performing probes, 19,575 sex chromosomes, and 161,412 known single nucleotide polymorphism (SNP) locus probes. If the beta range was less than 0.1 in all samples, we excluded these probes (83,655) from further analyses. In total, 616,162 probe methylation profiles of 294 tumor and 143 normal samples (matched 142) were used for downstream analysis (Fig. 1). Using principal component (PC) analysis, we compared the distribution of beta values between the raw and processed probes and found sex- and batch-related biases in the raw beta values (Supplementary Figs. 3A and 4A) prior to preprocessing. Following normalization and filtering procedures, we obtained high-quality data, effectively harmonized the dataset, and eliminated technical noise and sex-based biases. The preprocessing step is illustrated in the PC plots in Supplementary Figs. 3B and 4B.
Fig. 1.
Construction of the colorectal cancer (CRC) methylome profile. (A) Sample collection and preparation: initial processing involved extraction and measurement of 443 raw EPIC array-based methylation profiles. These consisted of 299 tumor samples and 144 matched normal samples from Korean patients with CRC at Seoul National University Hospital. (B) Preprocessing of the methylome profile: the preprocessing procedure adhered to the minfi pipeline protocol. The subset-quantile within array normalization (SWAN) method was used for normalization, and the ComBat method was applied for batch correction. One normal and five tumor samples showing discrepancies between methylation-based sex prediction and clinical information were excluded. After the removal of poorly performing, SNP locus, sex-chromosome, and low variable site probes, the finalized methylation profile comprised 294 tumor and 143 normal samples (142 matched), quantified at 616,162 probe sites.
Differential methylation patterns between normal and tumor samples
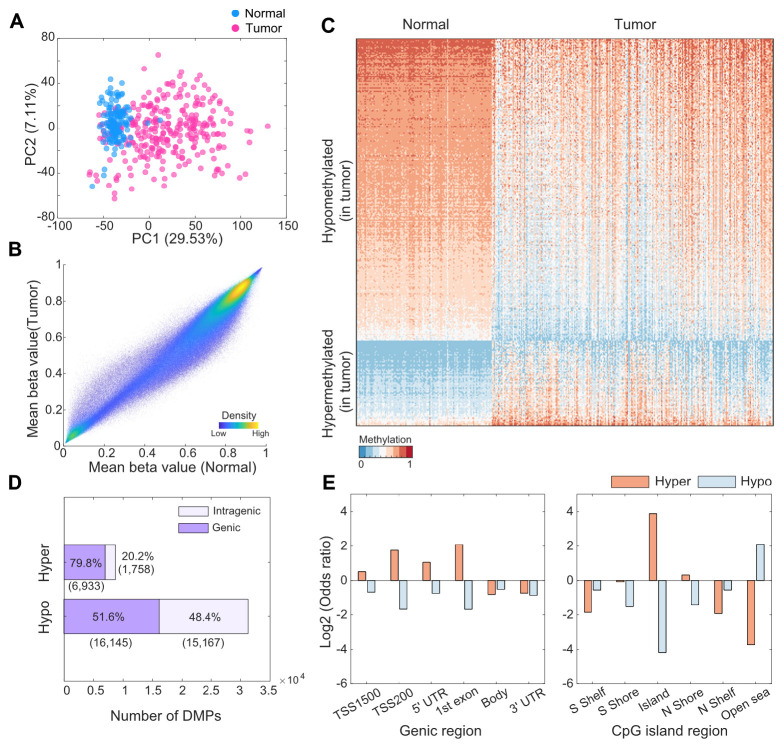
Based on 616,162 processed methylation probes, we observed a clear separation between the tumor and normal samples within the dimensionality reduction plot (Fig. 2A), as indicated by the PCs (explained variance of PC1: 29.53%). The overall methylation levels were slightly higher in the normal samples (0.5848) than in the tumor samples (0.5602; Fig. 2B). Subsequently, we identified 40,003 differentially methylated positions (DMPs) between the tumor and normal samples (Supplementary Tables 1 and 2). There were more hypomethylated (31,312 probes) than hypermethylated (8, 691 probes) sites in the tumor samples (Fig. 2C, D). Among these DMPs, 6,933 (79.8%) hypermethylated probes (i.e., promoter, untranslated region [UTR], and gene body) and 16,145 (51.6%) hypomethylated probes were found in the genic region (Fig. 2D, Supplementary Tables 3 and 4). To investigate the abundance of DMPs in different genic and CpG island regions, we calculated the odds ratio (OR) of DMPs for hyper- and hypomethylated probes in relation to various genomic annotations, such as gene promoter regions, body regions, and islands or shores. In promoter-like regions (TSS1500, TSS200, 5’ UTR, and first exon), the ORs between the number of observed and expected hypermethylated probes were 1.43, 3.38, 2.07, and 4.26, respectively (Fig. 2E and Supplementary Table 3), with significant P-values (< 0.0001). Likewise, hypermethylated probes were highly enriched in the CpG island regions and N shore sites (Fig. 2E and Supplementary Table 5; OR: 14.70 and 1.25, respectively). In contrast, 27,131 hypomethylated probes in tumor samples were predominantly found in open-sea regions (OR: 4.32), which were considerably distant from the CpG island regions (Supplementary Table 6).
Fig. 2.
Differential methylation patterns between tumor and normal tissues. (A) PC plot based on 616,162 preprocessed probes. (B) Scatter plot of mean beta values between tumor and normal samples. (C) Heatmap of the expression of differentially methylated probes. Probes were randomly selected among 5% of total differentially methylated positions (DMPs) in each hypermethylation and hypomethylation. (D) Total number of DMPs between tumor and normal samples. Purple and light purple represent the DMP of the genic (including the promoter region) and intragenic regions, respectively. The numbers within parentheses represent the number of DMPs. (E) Odds ratios of the number of region-dependent DMPs with the total EPIC array positions. Shore represents the region up to 2 kb upstream (N shore) or downstream (S shore) from the start of the CpG island, and shelf represents the region from 2 to 4 kb upstream (N shelf) or downstream (S shelf) from the start of the CpG island.
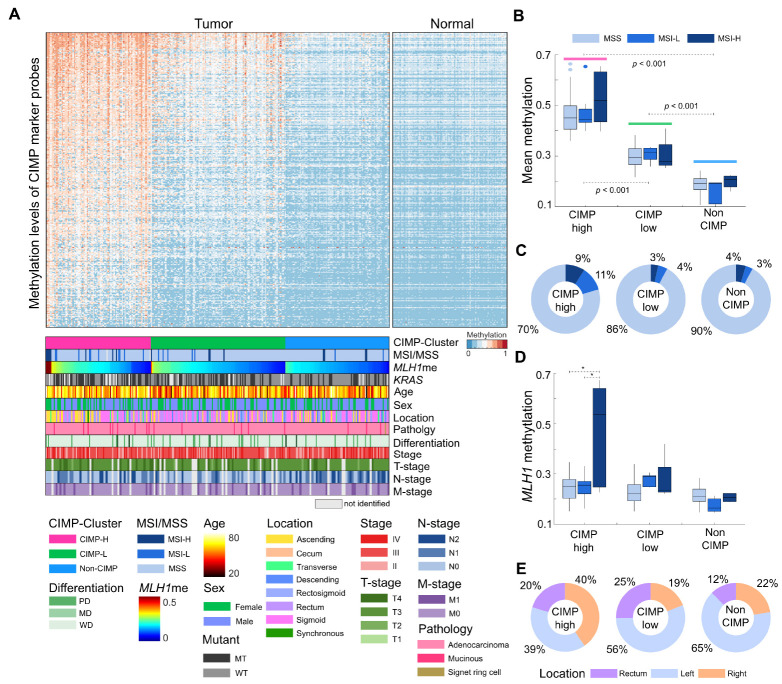
CIMP feature-based clustering for 294 tumor samples
To assess the abundance of CIMP, we used a set of 4,327 CpG island probes from previously defined 258 CIMP gene markers (12). Among the 4,327 probes, we selected 1,930 highly variable sites (standard deviation > 0.15) and clustered CRC samples based on these CIMP marker probes. Consequently, 294 tumor samples were assigned to three clusters: CIMP-high (CIMP-H), CIMP-low (CIMP-L), or non-CIMP, according to the respective mean methylation level of each cluster (Fig. 3A). Based on the established criteria, we identified 90 (30.6%) CIMP-H, 115 (39.1%) CIMP-L, and 89 (30.3%) non-CIMP patients with CRC, and their mean CIMP marker probe methylation levels exhibited significant pairwise differences (Fig. 3B). Next, we compared the CIMP clusters with the clinical characteristics of patients with CRC. In the CIMP-H group, 18 (20%) patients (Fig. 3C) showed high or low microsatellite instability (MSI-H or MSI-L). In contrast, only 7% of the patients in both the CIMP-L and non-CIMP groups presented with MSI-H or MSI-L status. We ascertained that the patients with MSI-H or MSI-L status were significantly enriched in the CIMP-H group (P < 0.05). We next investigated the methylation levels of MLH1, a well-known DNA mismatch repair gene, and found that the overall MLH1 methylation levels were higher in the CIMP-H group (mean methylation: 0.26) than in the CIMP-L (mean methylation: 0.23) and non-CIMP (mean methylation: 0.21) groups. We also observed a significant association between MLH1 methylation and the MSI-H status in the CIMP-H group (Fig. 3D). Notably, in the CIMP-H group, patients with the MSI-H status had a higher mean MLH1 methylation level (0.47) than those with the MSS and MSI-L status (0.24; T-test, P < 0.05). Additionally, the CIMP status correlated with anatomical location and patient age. In the CIMP-H group, 36 (40%) tumor samples were found in the right-sided colon, 35 (39%) in the left colon, and 18 (20%) in the rectum. In contrast, the CIMP-L and non-CIMP groups included 22 (19%) and 20 (2%) right colon tumors, respectively (Fig. 3E). Comparison of age distribution according to the CIMP status revealed that the mean age of CIMP-H patients was 62.9 years, which was slightly higher than that of CIMP-L (58.3 years) and non-CIMP (59.1 years) patients (T-test, P < 0.05). Moreover, in the data under study, a higher frequency of KRAS mutations was observed in patients with CIMP-H compared to those with CIMP-L and non-CIMP. Among CIMP-H patients with confirmed KRAS mutational status, 41 (52.5%) had KRAS mutations. Nevertheless, within the cohort of patients characterized by both CIMP-H and MSI-H, only a single case was observed to harbor the KRAS mutation. Other clinical features, including sex, AJCC stage, differentiation, and T, N, and M stage were not significantly associated with CIMP status (Supplementary Fig. 5). In the survival analysis, the CIMP-H group showed worse prognosis than the CIMP-L and non-CIMP groups (Supplementary Fig. 6).
Fig. 3.
CpG island methylator phenotype (CIMP) clustering of 294 tumor samples. (A) Heatmap of 294 CRC samples based on the methylation level of CIMP marker probes. Samples were clustered in each CIMP-based group and sorted according to MLH1 methylation level. (B) Boxplot of the mean methylation levels of CIMP marker genes, according to the CIMP and MSI statuses. (C) Proportions of samples with the MSI and MSS statuses according to the CIMP status. (D) Boxplot of the distribution of MLH1 methylation according to the CIMP and MSI statuses. (E) Distribution of tissue location among the CIMP groups. Tumors at descending, rectosigmoid, and sigmoid colon are considered left-sided, and those at cecum, ascending colon, and transverse colon are considered right-sided. Synchronous tumors occurred as multiple tumors on both left and right sides simultaneously.
We focused on 142 pair-wise matched samples for in-depth analysis. The differences between tumor and adjacent-matched normal tissues demonstrated a strong association with CIMP signals (Supplementary Fig. 7A). We classified the samples into three distinct clusters – C1, C2, and C3 – by pair-differences for the 10k most variable probes. These clusters demonstrated high enrichment of specific CIMP subgroups, with 97.5% for C1, 63.3% for C2, and 49.1% for C3, which corresponds to the CIMP-H, CIMP-L, and non-CIMP categories, respectively (Supplementary Fig. 7B). The methylation differences observed in C1 and C2, which correspond to CIMP-H and CIMP-L, respectively, were characterized by strong hypermethylation in the CpG island region (Supplementary Fig. 7C) and by hypomethylation in the open-sea region (Supplementary Fig. 7D). Interestingly, the C3 cluster, which primarily consisted of non-CIMP and CIMP-L samples, did not show significant differences in methylation in the open-sea region between tumor and normal tissues, in contrast to the methylation patterns of C1 and C2 (Supplementary Fig. 7D). We thus analyzed CpG island and open-sea methylation differences within the CIMP-L samples across C2 and C3. There was no significant difference in CpG island methylation between C2 and C3, whereas open-sea methylation was markedly higher in C2 (Supplementary Fig. 7E).
DISCUSSION
Here, we present genome-wide methylation data from high-throughput microarrays and their clinical implications from a large cohort of patients with CRC. Our analyses showed that hypermethylated and hypomethylated DMPs were located in distinctly different genomic regions and that CIMP-H was associated with MSI-H tumors and MLH1 hypermethylation.
Our genome-wide methylation data of tumors and matched normal tissues identified 40,003 DMPs with 6,933 (79.8%) hypermethylated and 16,145 (51.6%) hypomethylated probes in genic regions. Hypermethylated probes were predominantly found in promoter-like regions, CpG islands, and N shore sites, whereas hypomethylated probes were enriched in open-sea regions. Our findings are consistent with those of previous reports (13, 14) that neoplastic cells often present with methylation at the promoter sites of selected CpG islands, which leads to the silencing of tumor suppressor genes and promotes tumorigenesis (15).
CIMP analyses categorized the tumor samples into three subgroups: CIMP-H, CIMP-L, and non-CIMP. Patients in the CIMP-H group were older and had more frequent right-sided tumors, concordant with the results of a previous study (16). Moreover, CIMP-H tumors were strongly associated with MSI-H and MLH1 hypermethylation, which has also been reported in previous studies (17, 18). CIMP-positive tumors are traditionally categorized into subgroups that exhibit unique molecular characteristics, often marked by prevalent mutations in either KRAS or BRAF genes, as noted in several studies (8, 19-21). Within our study’s patient population, the CIMP-H subgroup demonstrated an elevated frequency of KRAS mutations. However, in the combined subgroup of CIMP-H and MSI-H, we confirmed only one instance of KRAS mutation. This suggests the existence of more intricate sub-molecular classifications within the CIMP-H group (22-24).
In-depth analyses of the 142 tumor and normal matched samples suggested that the methylation phenotypes in CRC are not solely governed by CpG island methylation but also by regions beyond CpG islands, such as open-sea regions. Our results imply complexity in CRC methylation phenotypes that extends beyond the conventional CIMP status, underscoring the need for a more nuanced understanding of methylation dynamics of CRC. Our findings also highlight the importance of understanding the global methylation landscape when examining methylation patterns, which potentially offers more comprehensive insights into CRC subtypes and contributes to refined diagnostic and therapeutic strategies. Lastly, we examined the methylation patterns of various MMR-related genes, (including MSH2, MSH3, MSH5, MSH6, MLH3, PMS1, and PMS2) within CIMP-H CRC samples. Interestingly, our results did not reveal significant differences between MSI-H and MSS samples, suggesting that the MSI status in CIMP-H CRC might not be principally driven by the methylation alterations in these MMR genes, excluding MLH1. For a deeper investigation into methylation dynamics, we performed a genome-wide analysis encompassing all 27,365 genes on the EPIC array. We thus identified 207 genes, including MLH1, that exhibited a considerable mean methylation difference (> 0.2, P < 0.05) between MSI-H and MSS samples. Notably, this gene set was enriched for constituents of the WNT signaling pathway, such as DKK1, WNT5A, ROR2, and LEF1 (Supplementary Fig. 8). When analyzing MMR-related genes among CIMP-H samples, we observed that hypomethylation of WNT5A and DKK1, members of the WNT pathway, was associated with the MSI-H subtype of CRC, in accordance with previous studies (25-27). Considering the critical role of WNT signaling in intestinal homeostasis and its deregulation in CRC (28), these epigenetic alterations could potentially contribute to oncogenic processes. Despite the limited number of MSI-H samples, our results uncover potential epigenetic markers for the MSI-H subtype, especially within the CIMP-H group, which broadens our understanding of the intricate interplay between genetic and epigenetic changes in CRC. These findings also provide the basis for future research to validate and further explore epigenetic markers linked to MSI status, which, through the development of efficient diagnostic assay such as high resolution melting curve analysis (29), may enhance the clinical management of CRC. In conclusion, we believe that this report provides a comprehensive analysis of the methylation landscape and its association with clinical characteristics in a CRC cohort, contributing to refined diagnostic and therapeutic strategies. Further investigation of cohort-specific DNA methylation markers and their matched muti-omics will also provide a deeper understanding of the prognosis of CRC based on methylation status.
MATERIALS AND METHODS
Ethics statement
This study was approved by the Institutional Review Boards of the Seoul National University Hospital (IRB No. 2103-121-1206) and Yonsei University Institutional review board (approval number: 7001988-201910-BR-727-02). All methods were performed in accordance with the relevant guidelines and regulations and carried out in accordance with the Declaration of Helsinki.
Methylation EPIC array data set and data analysis methods
The detailed materials and methods on EPIC array generation and statistical analyses are provided in Supplementary Note 1.
Funding Statement
ACKNOWLEDGEMENTS This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science & ICT (grant number: NRF-2017M3A9A7050614 and NRF-2017M3A9A7050610). It was additionally supported by a grant from the National Research Foundation of Korea (NRF-2020M3A9I6A01036057).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
DATA AVAILABILITY
Raw IDAT files and processed methylation profiles are available at the Korea BioData Station (30) (KBDS, https://kbds.re.kr/) under the accession ID PRJKA2086323.
REFERENCES
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96:1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 3.Chang GJ, Hu CY, Eng C, Skibber JM, Rodriguez-Bigas MA. Practical application of a calculator for conditional survival in colon cancer. J Clin Oncol. 2009;27:5938–5943. doi: 10.1200/JCO.2009.23.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- 5.Smith G, Carey FA, Beattie J, et al. Mutations in APC, Kirsten-ras, and p53--alternative genetic pathways to colorectal cancer. Proc Natl Acad Sci USA. 2002;99:9433–9438. doi: 10.1073/pnas.122612899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology. 2015;149:1204–1225. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lakshminarasimhan R, Liang G. The role of DNA methylation in cancer. Adv Exp Med Biol. 2016;945:151–172. doi: 10.1007/978-3-319-43624-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 10.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13:R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McInnes T, Zou D, Rao DS, et al. Genome-wide methylation analysis identifies a core set of hypermethylated genes in CIMP-H colorectal cancer. BMC Cancer. 2017;17:228. doi: 10.1186/s12885-017-3226-4.b983699b554c4626aa4eff65786a812f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8:686–700. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu S, Lin S, Ye D, et al. Genome-wide methylation profiling identified novel differentially hypermethylated biomarker MPPED2 in colorectal cancer. Clin Epigenetics. 2019;11:41. doi: 10.1186/s13148-019-0628-y.e1b6e24454be4847ad2dbd248933695a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 16.Jia M, Jansen L, Walter V, et al. No association of CpG island methylator phenotype and colorectal cancer survival: population-based study. Br J Cancer. 2016;115:1359–1366. doi: 10.1038/bjc.2016.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cha Y, Kim KJ, Han SW, et al. Adverse prognostic impact of the CpG island methylator phenotype in metastatic colorectal cancer. Br J Cancer. 2016;115:164–171. doi: 10.1038/bjc.2016.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinoue T, Weisenberger DJ, Lange CP, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22:271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobre M, Salvi A, Pelisenco IA, et al. Crosstalk between DNA methylation and gene mutations in colorectal cancer. Front Oncol. 2021;11:697409. doi: 10.3389/fonc.2021.697409.081157871d384aa89c473e62be9a1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kambara T, Simms LA, Whitehall VL, et al. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137–1144. doi: 10.1136/gut.2003.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toyota M, Ohe-Toyota M, Ahuja N, Issa JP. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci U S A. 2000;97:710–715. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim JH, Rhee YY, Bae JM, et al. Subsets of microsatellite-unstable colorectal cancers exhibit discordance between the CpG island methylator phenotype and MLH1 methylation status. Mod Pathol. 2013;26:1013–1022. doi: 10.1038/modpathol.2012.241. [DOI] [PubMed] [Google Scholar]
- 23.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 24.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rawson J, Mrkonjic M, Daftary D, et al. Promoter methylation of Wnt5a is associated with microsatellite instability and BRAF V600E mutation in two large populations of colorectal cancer patients. Br J Cancer. 2011;104:1906–1912. doi: 10.1038/bjc.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rawson JB, Manno M, Mrkonjic M, et al. Promoter methylation of Wnt antagonists DKK1 and SFRP1 is associated with opposing tumor subtypes in two large populations of colorectal cancer patients. Carcinogenesis. 2011;32:741–747. doi: 10.1093/carcin/bgr020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silva AL, Dawson SN, Arends MJ, et al. Boosting Wnt activity during colorectal cancer progression through selective hypermethylation of Wnt signaling antagonists. BMC Cancer. 2014;14:1–10. doi: 10.1186/1471-2407-14-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wi DH, Cha JH, Jung YS. Mucin in cancer: a stealth cloak for cancer cells. BMB Rep. 2021;54:344–355. doi: 10.5483/BMBRep.2021.54.7.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SC, Kim J, Kim DW, et al. Methylation-sensitive high-resolution melting analysis of the USP44 promoter can detect early-stage hepatocellular carcinoma in blood samples. BMB Rep. 2022;55:553–558. doi: 10.5483/BMBRep.2022.55.11.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee B, Hwang S, Kim PG, et al. Introduction of the Korea BioData Station (K-BDS) for sharing biological data. Genomics Inform. 2023;21:e12. doi: 10.5808/gi.22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.